Summary
Bacterial vectors, as microscopic living ‘robotic factories’, can be reprogrammed into microscopic living ‘robotic factories’, using a top‐down bioengineering approach to produce and deliver anticancer agents. Most of the current research has focused on bacterial species such as Salmonella typhimurium or Clostridium novyi. However, Escherichia coli Nissle 1917 (EcN) is another promising candidate with probiotic properties. EcN offers increased applicability for cancer treatment with the development of new molecular biology and complete genome sequencing techniques. In this review, we discuss the genetics and physical properties of EcN. We also summarize and analyse recent studies regarding tumour therapy mediated by EcN. Many challenges remain in the development of more promising strategies for combatting cancer with EcN.
Escherichia coli Nissle 1917 (EcN) is a promising candidate with probiotic properties for cancer treatment. This review discuss the genetics and physical properties of EcN, and summarize recent studies regarding tumour therapy mediated by EcN.
![]()
Introduction
Bacteria may be considered programmable ‘robot factories’ that specifically target tumours, and they have unique capabilities that make them well‐suited to be ideal anticancer agents (Forbes, 2010). Recently, the mechanism of action and antitumour effects of bacteria on tumour cells has been studied (Maeda, 2013; Zhang and Forbes, 2015; Zhou et al., 2018). Bacteria exhibit intrinsic antitumour activity, because they express chemotactic receptors, which direct chemotaxis towards molecular signals in the tumour microenvironment. They are also equipped with flagella, which facilitates tissue penetration (Grozdanov et al., 2004; Reister et al., 2014). They can migrate and accumulate far from the vasculature. They may also be engineered to sense and respond to the tumour microenvironment resulting in innate and adaptive antitumour immune responses (Zhou et al., 2018). However, the antitumour effect of bacteria within tumours is generally weak, and different bacteria and treatment strategies have been developed to enhance their antitumour effect (Piñero‐Lambea et al., 2015b). In addition, some bacteria such as Escherichia coli are currently bioengineered using a variety of molecular tools to produce biologically active molecules.
Many studies have focused on the reproductive features of bacteria in combination with their capacity to produce living therapeutics. As a next‐generation therapy, these tiny living factories may decrease production costs, reduce side‐effects, require smaller doses of biological compound and produce more compounds (Pedrolli et al., 2019).
Thus far, bacteria such as Clostridium sp. (Agrawal et al., 2004), Bifidobacterium sp. (Sasaki et al., 2006), Salmonella sp. (Mengesha et al., 2006) and Escherichia sp. (Yu et al., 2004) have been engineered to deliver RNA (Yang et al., 2008), prodrugs (Hedley et al., 2007), cytotoxic agents (Ryan et al., 2009), cytokines (Loeffler et al., 2008), antigens (Nishikawa et al., 2006) and antibodies (Groot et al., 2007). All of these bacteria have been genetically modified to increase the effectiveness of anticancer agents (Fig. 1). A straightforward approach is to engineer bacteria to express proteins such as bacterial toxins to eradicate cancer cells. Bacterial toxins, such as cytolysin A, affect the mammalian cell membranes and induce apoptosis (Jiang et al., 2010). This strategy requires bacterial vectors that specifically target cancer cells or vectors with inducible promoters for better control of gene expression to avoid toxicity to normal tissues (Loessner et al., 2009). Another common strategy is to engineer these bacteria to express prodrug‐converting enzymes. The major advantage of these enzymes is that the resulting cytotoxic products can permeate the cell membrane and diffuse farther inside the solid tumour (Lehouritis et al., 2016).
Figure 1.
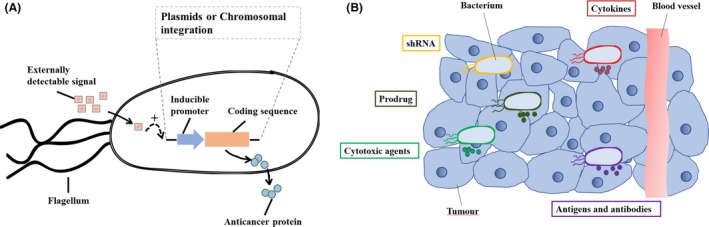
Engineered bacteria for cancer therapy.
A. The bacterial cell with various inducible systems for payload expression.
B. The bacterial accumulation and replication within solid tumours enabling localized expression of anticancer agents
Bacteria can be genetically engineered in a variety of ways to create a versatile living platform that can deliver a therapeutic payload based on clinical needs. For effective cancer therapy, the initial concern is to select an appropriate bacterial strain (Pedrolli et al., 2019). The facultative anaerobe, Salmonella typhimurium, has been widely studied and engineered to improve its tumour‐targeting ability. It has even been applied in human clinical trials (Toso et al., 2002; Thamm et al., 2005). However, Salmonella, as well as other toxic strains including Clostridium novyi (Dang et al., 2001) and Listeria monocytogenes (Freitag et al., 1993), should be modified to improve its safety profile. Moreover, the effect of Salmonella‐mediated therapy for cancer is smaller than that of virulence‐attenuated Shigella flexneri 21 SC602 and other E. coli strains (Stritzker et al., 2007). Among these bacteria, non‐pathogenic E. coli strains such as K‐12 and Nissle 1917 also exhibit tumour‐targeting activity (Stritzker et al., 2007; Weibel et al., 2008; Piñero‐Lambea et al., 2015a). E. coli Nissle 1917 (EcN) is probably a better choice with its probiotic potential, and it has been widely used and combined into living therapeutics. EcN can be genetically engineered to act as a living therapeutic to treat solid cancers (Singh et al., 2017; Chua et al., 2017). These findings provide a rationale for EcN as a promising probiotic for cancer. This review covers recent methods to engineer EcN for cancer treatment and provides a primary resource for scientists choosing EcN to create new living therapeutics.
Characteristics of E. coli Nissle 1917
Escherichia coli Nissle 1917 (EcN) is a Gram‐negative probiotic, originally isolated by Dr. Alfred Nissle during World War I (Nissle, 1918; Nissle, 1925). EcN is serum‐sensitive and does not produce any enterotoxins or cytotoxins associated with pathogenic E. coli strains (Sonnenborn and Schulze, 2009). It has been licensed as a pharmaceutical for the treatment of diseases such as diarrhoea and colitis ulcerosa (Kruis et al., 2004). Along with the new finding of its biological function, the genetics of EcN has been extensively characterized. It was discovered that EcN harbours two cryptic plasmids, named pMUT1 and pMUT2. Both plasmids have been completely sequenced and shown to be genetically stable and non‐transferable (Blum‐Oehler et al., 2003; Sonnenborn and Schulze, 2009). A plasmid‐free variant of EcN was shown not to be functionally different from the wild‐type EcN strain and may be used as a live vector for recombinant plasmids, based on pMUT1 and pMUT2 (Oswald, 2006).
Analysis of the EcN genome structure has further revealed that the lack of defined virulence factors, such as alpha‐hemolysin, P‐fimbrial adhesins and the semi‐rough lipopolysaccharide phenotype combined with the expression of fitness factors, such as microcins, adhesins and iron uptake systems, may contribute to its probiotic character (Grozdanov et al., 2004). Genomic islands, which synthesize ‘fitness factors’, are located within the EcN chromosome. These islands increase bacterial fitness and are crucial to their ability to colonize a host (Hacker and Carniel, 2001). To further elucidate the molecular basis for EcN’s probiotic nature, the genomic peculiarity of coding sequences and reconstructed metabolic network, inferred from raw genome data, was studied (Sun et al., 2005). In 2014, Reister et al. (2014) reported EcN’s complete and annotated genomic sequence (Genbank accession number: CP007799) and identified the genes and their products essential for its probiotic nature. The size of the EcN genome is 5 441 200 bp, and it contains 5324 predicted genes. Metabolomic studies have revealed that EcN displays substantial metabolic differences as compared with phylogenetically similar pathogenic E. coli strains, thus revealing its potent probiotic characteristics (van der Hooft et al., 2019). Understanding the genetics and characteristics of EcN will lead to engineering methods to effectively manipulate EcN.
The mechanisms of EcN as an antitumour agent
Bacteria use complex mechanisms to target tumours. Different bacterial species share unique intrinsic mechanisms to eliminate cancer. The mechanisms of accumulation within tumours differ and rely on oxygen tolerance. EcN, as a facultative anaerobe, may utilize complex mechanisms to target tumours. The EcN serotype O6:K5:H1 is an excellent example of bacterial genome evolution within the pathogenic E. coli serotype O6 lineage (Behnsen et al., 2013). It is devoid of prominent virulence genes and displays fitness factors that contribute to its colonization efficiency and survival within the host (Sanders, 2003). Moreover, the serum‐sensitive LPS of the EcN membrane ensures the quick elimination of the strain from normal organs, and it is free of immunotoxic side‐effects in patients (Grozdanov et al., 2002). These striking features likely confer protection from clearance by the host immune system.
Another characteristic of EcN is the extracellular K5 capsule, which is important for adhesion and colonization (Burns and Hull, 1998). Bacteria are reported to proliferate preferentially within solid tumours (Pawelek et al., 2003), and this feature probably promotes the targeting of EcN to the tumour, resulting in preferential growth within the tumour microenvironment. It has also been shown that EcN contributes to reduced inflammation by downregulating the expansion of newly recruited T cells into the mucosa (Sturm et al., 2005). This indicates that one mechanism of EcN‐mediated tumour targeting occurs following inflammation.
However, different bacteria in different microenvironments adopt distinct antitumour mechanisms. The specific mechanism may differ depending on which bacterial species is used, tumour type and dynamics of the bacteria‐host interaction (Zhou et al., 2018). The mechanisms of EcN interaction in different environments facilitate accumulation within tumours.
Exploration of EcN for tumour‐targeting therapy
Many biological tools such as Red/ET recombination and CRISPR‐Cas9 are currently available for engineering E. coli (Jiang et al., 2013). Based on its ‘tumour‐finding’ nature, E. coli is a programmable delivery vehicle that may be designed to carry multiple genes for therapeutic or diagnostic cancer agents (Pedrolli et al., 2019). EcN is one promising E. coli strain that can be engineered systemically, resulting in exclusive tumour colonization in live mice (Stritzker et al., 2007). Positron emission tomography (PET) and optical imaging have been used to monitor the tumour‐targeting activity of EcN (Brader et al., 2008). Genome sequence analysis has revealed EcN to be a novel bioengineered probiotic with several unique properties such as (i) interaction with the host immune system (Sturm et al., 2005), (ii) antimicrobial activity through secretion of microcins and bacteriocins (Sassone‐Corsi et al., 2016), and (iii) formation of biofilms resulting in the production of defensins (Lasaro et al., 2009). A key step in the engineering of living therapeutics is to choose a suitable chassis, preferably a probiotic with optimal pharmacologic properties (Claesen and Fischbach, 2015). Due to its unique function and high versatility, EcN has provided new opportunities for next‐generation therapeutic and probiotic therapies. Using these modalities, researchers have observed reduced tumour volume, increased survival and eradication of metastatic disease in animal models, while avoiding damage to healthy cells (Table 1).
Table 1.
Current summary of engineered EcN for clinical cancer exploration.
| Effector classes | Effectors or targets | Cancer type | References |
|---|---|---|---|
| Cytotoxic agents | Azurin | Mouse B16 melanoma and human 4T1 breast tumours | Zhang et al. (2012) |
| Colibactin, glidobactin and luminmide | Human U‐2 OS osteosarcoma cells | Li et al. (2019) | |
| p53 | Human hepatoma SMMC‐7721 cells | He et al. (2017) | |
| Synthetic gene circuit | Genomic luxCDABE cassette and lacZ vector to develop a diagnostic platform PROP‐Z | Metastatic murine colorectal cells | Danino et al. (2015) |
| Tumour stroma targeting | Tumstatin | Mouse B16 melanoma cells | He et al. (2017) |
| Human hepatoma SMMC‐7721 cells | He et al. (2019) | ||
| Prodrug‐converting enzymes | Myrosinase | Murine, human and colorectal adenocarcinoma cell lines | Ho et al. (2018) |
| Prodrugs | CB1954, 5‐FC and Fludarabine phosphate | Mouse CT26 colon cells | Lehouritis et al. (2016) |
Expression of therapeutic proteins in EcN
The major strategy of EcN‐mediated tumour therapy is to transform plasmids carrying gene expression cassettes to direct the expression of therapeutic proteins (Table 1). For example, the azurin protein was constitutively expressed to inhibit tumours (Zhang et al., 2012). In this study, BALB/c mice, bearing orthotopic B16 melanoma or 4T1 breast tumours, were administered PBS, EcN and its variants by intravenous (i.v.) injection at a dose of 2 × 107 CFU per mouse. Tumour growth and pulmonary metastasis were efficiently suppressed by azurin release and the resulting inflammatory response without significant toxicity. For the detection of liver metastasis in urine samples, Danino et al. (2015) engineered EcN to carry a genomic luxCDABE cassette containing a high‐expression lacZ vector. This was used to develop a diagnostic platform, PROP‐Z. A murine model of colorectal cancer metastases was used in which spleens from immunocompetent BALB/c mice were surgically injected with metastatic murine colorectal cells (MC26‐LucF). After PROP‐Z was delivered orally, EcN rapidly (within 24 h) translocated across the gastrointestinal tract and specifically colonized within the metastatic tumours present in the liver, but not within healthy organs or fibrotic liver tissue. PROP‐Z expressed high levels of the enzyme lacZ, which cleaves a substrate to produce a small molecule that can be detected in urine. EcN was also selected as a vector to specifically express Tum‐5, which is a suitable tumour‐specific angiogenesis inhibitor. Tum‐5 was expressed in EcN under control by the oxygen‐dependent promoter of the haemoglobin gene (vhb) from Vitreoscilla. The colonization of EcN (Tum‐5) was investigated in C57BL/6 mice bearing B16 melanoma at different time points following i.p. injection of 5×106 CFU/100 µL EcN. Tumour growth and angiogenesis were inhibited by upregulation of Tum‐5 expression (He et al., 2017). In addition, biosynthetic gene clusters encoding cytotoxic compounds such as colibactin, glidobactin and luminmide were introduced into EcN. EcN and its variants were administered by i.v. injection into female NMRI nude mice bearing UT‐SCC‐5 human head and neck squamous tumours at a dose of 1 × 107 CFU per mouse. The colibactin/glidobactins/lumimides‐expressing EcN exhibited significant cytotoxic activity and suppressed tumour growth (Li et al., 2019). The anticancer protein p53 and the anti‐angiogenic factor Tum‐5 were constructed as bifunctional proteins and delivered to solid tumours using EcN (He et al., 2019). In this study, the SMMC‐7721 tumour‐bearing BALB/c nude mice were i.v. injected with EcN and its variants at a dose of 5 × 106 CFU/100 µL. Treatment with the engineered bacteria led to significant inhibitory effects on the growth of orthotopic hepatoma tumours without notable toxicity.
The expression of therapeutic proteins in EcN can successfully regress tumours. However, the accumulation of therapeutic proteins should be synthesized at sufficient concentrations to induce a therapeutic effect, but not high enough to cause systemic toxicity.
Expression of prodrug‐converting enzymes in EcN
Another strategy is to express prodrug‐converting enzymes that can metabolize their corresponding prodrug substrates and convert them into cytotoxic products, thus generating a potent bystander effect (a therapeutic effect on cells that is not influenced by bacteria) (Table 1). Ho et al. (2018) selected alanine‐deficient EcN to co‐express INP‐HlpA (Protein HlpA from Streptococcus gallolyticus with an INP tag) and YebF‐I1 (Myrosinase from Armoracia rusticana with a YebF‐secretion tag) using constitutive promoters. Engineered EcN was orally administered and bound specifically to heparan sulfate proteoglycan on colorectal cancer cells. As a result, secreted myrosinase converted dietary glucosinolate to sulforaphane, an organic molecule with anticancer activity. This combinatorial approach led to an almost complete inhibition of proliferation in murine and human colorectal adenocarcinoma cell lines in vitro. Tumour regression and decreased tumour formation were observed in a murine CRC model fed with the engineered living therapeutic. The efficiency of this strategy relies on the continued high‐level expression of prodrug‐converting enzymes, resulting in the sustained tumour colonization by the bacterial vector. Moreover, EcN has the ability to activate numerous prodrugs and is resistant to prodrug toxicity. Therefore, it was selected to activate multiple prodrugs such as CB1954, 5‐FC and Fludarabine phosphate without genetic modification (Lehouritis et al., 2016). In this study, BALB/c mice bearing subcutaneous CT26 flank tumours were colonized with EcN (5 × 105 CFU/50 µL) by intratumoural injection. CB1954, 5‐FC or a combination of both drugs was also administered i.p. into mice on the same day. The combined use of EcN and prodrugs led to a significant reduction in tumour growth, indicating their potential role in solid tumour treatment.
Engineering of EcN‐derived minicells
In addition to the delivery of EcN payloads, EcN‐derived minicells and bacterial ghosts (BGs) may be modified and filled with tumour‐targeted drugs. MacDiarmid et al. (2007) reported that minicells are nanosized forms of bacteria and contain the same cytoplasmic components as their parental bacteria with the exception of chromosomal DNA. The lack of a genome in the minicells results in a loss of proliferation, but minicells maintain other characteristics inherited from their parental bacteria. Minicells have been used for targeted delivery of siRNA or chemotherapeutic drugs into tumours. These drug‐loaded minicells, which may be modified with antibodies to receptors on cancer cells, can target tumours and release anticancer drugs (MacDiarmid et al., 2009; MacDiarmid et al., 2016). Additionally, Zhang et al. (2018) found that pHLIP‐mosaic minicell treatment resulted in a significant regression of orthotopic breast tumours in a BALB/c mice model along with high biocompatibility and low toxicity. The EcN‐derived minicells were produced by knocking out the minCD gene and enhancing minE expression. A pH (low) insertion peptide (pHLIP) was displayed on the membrane surface through the Lpp‐OmpA’ protein display system to increase targeting efficiency. Then, the EcN‐derived minicells displaying pHLIP were directly extracted from the fermentation broth and loaded with doxorubicin (DOX). EcN was also used as a bacterial carrier to immobilize amphiphilic copolymers through acid‐labile linkers (Xie et al., 2018). The released copolymers were self‐assembled into micelles. These hybrid micelles released both doxorubicin and α‐tocopheryl succinate, which resulted in synergistic antitumour activity in 4T1 tumour‐bearing mice. These studies provide novel strategies for constructing delivery systems by genetically modifying EcN‐derived minicells and utilizing biomaterials that have the ability to penetrate tumours.
Engineering of EcN BGs
Bacterial ghosts are considered to be empty and intact non‐living bacterial cell envelopes that may be used as a compound delivery system (Langemann et al., 2010; Kraśko et al., 2017). BGs are devoid of any cytoplasmic content but retain natural outer surface composition. Based on these characteristics, BGs have excellent carrier capacity and immunogenicity, and retain the original targeting functions of parental bacteria (Ganeshpurkar et al., 2014). EcN BGs have been prepared by fusion protein mE‐L‐SNA‐induced lysis and completely retain the intact surface structures required for specific attachment to mammalian cells. EcN BGs were then loaded with the anticancer agent Epothilone B, which induces apoptosis via the mitochondrial pathway in HeLa cells (Zhu et al., 2018). Due to the external immunologic properties of living bacteria, EcN BGs were used as candidate adjuvants. This was done by cell lysate‐based anticancer vaccination of a syngeneic murine lung carcinoma model (Kraśko et al., 2017). These results indicate that EcN BGs are a promising drug delivery carrier for drug candidates in cancer therapy.
Overall, studies of EcN‐mediated tumour therapies have demonstrated that probiotic EcN can be engineered to safely and selectively deliver therapeutic payloads to the tumour microenvironment, and they can be used as an optimal chassis for living cancer therapeutics. The combination of EcN‐derived minicells with ligands against tumour‐associated markers has additional tumour‐targeting effects (Zhang et al., 2018). EcN BGs exhibit excellent immunogenicity and can be used as candidate adjuvants for anticancer vaccination (Kraśko et al., 2017). The choice of a suitable delivery system (EcN, EcN‐derived minicells or BGs shown in Fig. 2) depends on the experimental goals, all of which are aimed at improving the therapeutic index.
Figure 2.
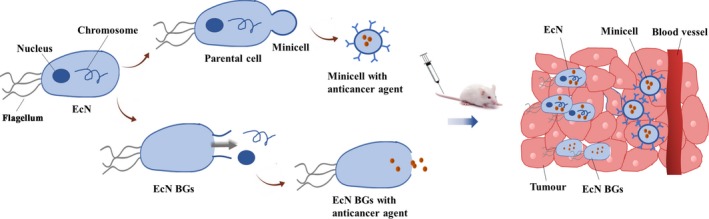
The payload delivery of EcN, EcN‐derived minicells and EcN BGs.
Conclusions
A variety of studies have shown that EcN‐mediated tumour therapies can successfully regress tumours and promote survival in mice. This indicates that EcN is a versatile probiotic that can be adopted for additional clinical applications as living therapeutics. However, numerous challenges remain including genetic instability, targeting efficiency and limited drug production. All of these challenges may be addressed using powerful recombinant DNA and synthetic biology techniques. For example, genetic stability may be improved by incorporating engineered genes into the EcN’s genome and by limiting homologous recombination and horizontal gene transfer with CRISPR‐Cas9 technology. Targeting efficiency may be enhanced by the genetic manipulation of endogenous chemoreceptors. Gene expression is predominantly regulated at the level of transcription. The high‐level constitutive expression of heterologous proteins may lead to a metabolic burden to the bacterial vector, resulting in decreased stability and inefficient colonization. Thus, drug production could be manipulated by optimizing promoter strength, gene copy number, ribosome‐binding sites and bacterial metabolism. Due to its genetic flexibility, EcN may be rationally designed for clinical studies, resulting in a powerful weapon against cancer.
Conflict of interest
None declared.
Acknowledgements
The authors are thankful to laboratory members and to the anonymous reviewers for insightful comments on this manuscript.
Microbial Biotechnology (2020) 13(3), 629–636
Funding information
This work was supported by grants from the National Natural Science Foundation of China 31670047.
Contributor Information
Qingsheng Qi, Email: qiqingsheng@sdu.edu.cn.
Qian Wang, Email: qiqi20011983@sdu.edu.cn.
References
- Agrawal, N. , Bettegowda, C. , Cheong, I. , Geschwind, J.F. , Drake, C.G. , Hipkiss, E.L. , et al (2004) Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proc Natl Acad Sci USA 101: 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen, J. , Deriu, E. , Sassone‐Corsi, M. , and Raffatellu, M. (2013) Probiotics: properties, examples, and specific applications. Cold Spring Harb Perspect Med 3: a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum‐Oehler, G. , Oswald, S. , Eiteljörge, K. , Sonnenborn, U. , Schulze, J. , Kruis, W. , et al (2003) Development of strain‐specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res Microbiol 154: 59–66. [DOI] [PubMed] [Google Scholar]
- Brader, P. , Stritzker, J. , Riedl, C.C. , Zanzonico, P. , Cai, S. , Burnazi, E.M. , et al (2008) Escherichia coli Nissle 1917 facilitates tumor detection by positron emission tomography and optical imaging. Clin Cancer Res 14: 2295–2302. [DOI] [PubMed] [Google Scholar]
- Burns, S.M. , and Hull, S.I. (1998) Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun 66: 4244–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, K.J. , Kwok, W.C. , Aggarwal, N. , Sun, T. , and Chang, M.W. (2017) Designer probiotics for the prevention and treatment of human diseases. Curr Opin Chem Biol 40: 8–16. [DOI] [PubMed] [Google Scholar]
- Claesen, J. , and Fischbach, M.A. (2015) Synthetic microbes as drug delivery systems. ACS Synth Biol 4: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, L.H. , Bettegowda, C. , Huso, D.L. , Kinzler, K.W. , and Vogelstein, B. (2001) Combination bacteriolytic therapy for the treatment of experimental tumors. Proc Natl Acad Sci USA 98: 15155–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino, T. , Prindle, A. , Kwong, G.A. , Skalak, M. , Li, H. , Allen, K. , et al (2015) Programmable probiotics for detection of cancer in urine. Sci Transl Med 7: 289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, N.S. (2010) Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer 10: 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag, N.E. , Rong, L. , and Portnoy, D.A. (1993) Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell‐to‐cell spread. Infect Immun 61: 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshpurkar, A. , Ganeshpurkar, A. , Pandey, V. , Agnihotri, A. , Bansal, D. , and Dubey, N. (2014) Harnessing the potential of bacterial ghost for the effective delivery of drugs and biotherapeutics. Int J Pharma Investig 4: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot, A.J. , Mengesha, A. , van der Wall, E. , van Diest, P.J. , Theys, J. , and Vooijs, M. (2007) Functional antibodies produced by oncolytic clostridia. Biochem Biophys Res Commun 364: 985–989. [DOI] [PubMed] [Google Scholar]
- Grozdanov, L. , Zahringer, U. , Blum‐Oehler, G. , Brade, L. , Henne, A. , Knirel, Y.A. , et al (2002) A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J Bacteriol 184: 5912–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov, L. , Raasch, C. , Schulze, J. , Sonnenborn, U. , Gottschalk, G. , Hacker, J. , et al (2004) Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186: 5432–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, J. , and Carniel, E. (2001) Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep 2: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Yang, H. , Liu, F. , Chen, Y. , Tang, S. , Ji, W. , et al (2017) Escherichia coli Nissle 1917 engineered to express Tum‐5 can restrain murine melanoma growth. Oncotarget 8: 85772–85782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L. , Yang, H. , Tang, J. , Liu, Z. , Chen, Y. , Lu, B. , et al (2019) as a targeted vehicle for delivery of p53 and Tum‐5 to solid tumors for cancer therapy. J Biol Eng 13: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley, D. , Ogilvie, L. , and Springer, C. (2007) Carboxypeptidase‐G2‐based gene‐directed enzyme prodrug therapy: a new weapon in the GDEPT armoury. Nature Rev Cancer 7: 870–879. [DOI] [PubMed] [Google Scholar]
- Ho, C.L. , Tan, H.Q. , Chua, K.J. , Kang, A. , Lim, K.H. , Ling, K.L. , et al (2018) Engineered commensal microbes for diet‐mediated colorectal‐cancer chemoprevention. Nat Biomed Eng 2: 27–37. [DOI] [PubMed] [Google Scholar]
- van der Hooft, J.J.J. , Goldstone, R.J. , Harris, S. , Burgess, K.E.V. , and Smith, D.G.E. (2019) Substantial extracellular metabolic differences found between phylogenetically closely related probiotic and pathogenic strains of Escherichia coli . Front Microbiol 10: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S.N. , Phan, T.X. , Nam, T.K. , Nguyen, V.H. , Kim, H.S. , Bom, H.S. , et al (2010) Inhibition of tumor growth and metastasis by a combination of Escherichia coli‐mediated cytolytic therapy and radiotherapy. Mol Ther 18: 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Bikard, D. , Cox, D. , Zhang, F. , and Marraffini, L.A. (2013) RNA‐guided editing of bacterial genomes using CRISPR‐Cas systems. Nat Biotechnol 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraśko, J.A. , Zilionyte, K. , Darinskas, A. , Strioga, M. , Rjabceva, S. , Zalutsky, I. , et al (2017) Bacterial ghosts as adjuvants in syngeneic tumour cell lysate‐based anticancer vaccination in a murine lung carcinoma model. Oncol Rep 37: 171–178. [DOI] [PubMed] [Google Scholar]
- Kruis, W. , Fric, P. , Pokrotnieks, J. , Lukás, M. , Fixa, B. , Kascák, M. , et al (2004) Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemann, T. , Koller, V. J. , Muhammad, A. , Kudela, P. , Mayr, U. B. , and Lubitz, W. (2010) The Bacterial Ghost platform system: production and applications. Bioeng Bugs 1: 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasaro, M.A. , Salinger, N. , Zhang, J. , Wang, Y. , Zhong, Z. , Goulian, M. , et al (2009) F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol 75: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehouritis, P. , Stanton, M. , McCarthy, F.O. , Jeavons, M. , and Tangney, M. (2016) Activation of multiple chemotherapeutic prodrugs by the natural enzymolome of tumour‐localised probiotic bacteria. J Control Release 222: 9–17. [DOI] [PubMed] [Google Scholar]
- Li, R. , Helbig, L. , Fu, J. , Bian, X. , Herrmann, J. , Baumann, M. , et al (2019) Expressing cytotoxic compounds in Escherichia coli Nissle 1917 for tumor‐targeting therapy. Res Microbiol 170: 74–79. [DOI] [PubMed] [Google Scholar]
- Loeffler, M. , Le'Negrate, G. , Krajewska, M. , and Reed, J.C. (2008) IL‐18‐producing Salmonella inhibit tumor growth. Cancer Gene Ther 15: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner, H. , Leschner, S. , Endmann, A. , Westphal, K. , Wolf, K. , Kochruebe, K. , et al (2009) Drug‐inducible remote control of gene expression by probiotic Escherichia coli Nissle 1917 in intestine, tumor and gall bladder of mice. Microbes Infect 11: 1097–1105. [DOI] [PubMed] [Google Scholar]
- MacDiarmid, J.A. , Mugridge, N.B. , Weiss, J.C. , Phillips, L. , Burn, A.L. , Paulin, R.P. , et al (2007) Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell 11: 431–445. [DOI] [PubMed] [Google Scholar]
- MacDiarmid, J.A. , Amaro‐Mugridge, N.B. , Madrid‐Weiss, J. , Sedliarou, I. , Wetzel, S. , Kochar, K. , et al (2009) Sequential treatment of drug‐resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat Biotechnol 27: 643–651. [DOI] [PubMed] [Google Scholar]
- MacDiarmid, J.A. , Langova, V. , Bailey, D. , Pattison, S.T. , Pattison, S.L. , Christensen, N. , et al (2016) Targeted doxorubicin delivery to brain tumors via minicells: proof of principle using dogs with spontaneously occurring tumors as a model. PLoS ONE 11: e0151832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, H. (2013) The link between infection and cancer: tumor vasculature, free radicals, and drug delivery to tumors via the EPR effect. Cancer Sci 104: 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengesha, A. , Dubois, L. , Lambin, P. , Landuyt, W. , Chiu, R.K. , Wouters, B.G. , et al (2006) Development of a flexible and potent hypoxia inducible promoter for tumor‐targeted gene expression in attenuated Salmonella . Cancer Biol Ther 5: 1120–1128. [DOI] [PubMed] [Google Scholar]
- Nishikawa, H. , Sato, E. , Briones, G. , Chen, L. M. , Matsuo, M. , Nagata, Y. , et al (2006) In vivo antigen delivery by a Salmonella typhimurium type III secretion system for therapeutic cancer vaccines. J Clin Invest 116: 1946–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissle, A. (1918) Die antagonistische Behandlung chronischer Darmstörungen mit Colibakterien. Med Klin 2: 29–30. [Google Scholar]
- Nissle, A. (1925) Weiteres über Grundlagen und Praxis der Mutaflorbehandlung. DMW‐Deutsche Medizinische Wochenschrift 51: 1809–1813. [Google Scholar]
- Oswald, S. (2006) Molekularbiologische Untersuchungen des probiotischen Escherichia coli Stammes DSM 6601 und Entwicklung der stammeigenen Plasmide als Klonierungsvektoren. Dissertation, Julius‐Maximilians‐Universität Würzburg.
- Pawelek, J.M. , Low, K.B. , and Bermudes, D. (2003) Bacteria as tumour‐targeting vectors. Lancet Oncol 4: 548–556. [DOI] [PubMed] [Google Scholar]
- Pedrolli, D.B. , Ribeiro, N.V. , Squizato, P.N. , de Jesus, V.N. , Cozetto, D.A. , Team AQA Unesp at iGEM 2017 (2019) Engineering microbial living therapeutics: the synthetic biology toolbox. Trends Biotechnol 37: 100–115. [DOI] [PubMed] [Google Scholar]
- Piñero‐Lambea, C. , Bodelón, G. , Fernández‐Periáñez, R. , Cuesta, A.M. , Álvarez‐Vallina, L. , and Fernández, L.Á. (2015a) Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth Biol. 4: 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero‐Lambea, C. , Ruano‐Gallego, D. , and Fernández, L.Á. (2015b) Engineered bacteria as therapeutic agents. Curr Opin Biotechnol 35: 94–102. [DOI] [PubMed] [Google Scholar]
- Reister, M. , Hoffmeier, K. , Krezdorn, N. , Rotter, B. , Liang, C. , Rund, S. , et al (2014) Complete genome sequence of the gram‐negative probiotic Escherichia coli strain Nissle 1917. J Biotechnol 187: 106–107. [DOI] [PubMed] [Google Scholar]
- Ryan, R.M. , Green, J. , Williams, P.J. , Tazzyman, S. , Hunt, S. , Harmey, J.H. , et al (2009) Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther 16: 329–339. [DOI] [PubMed] [Google Scholar]
- Sanders, M.E. (2003) Probiotics: Considerations for human health. Nutr Rev 61: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T. , Fujimori, M. , Hamaji, Y. , Hama, Y. , Ito, K. , Amano, J. , et al (2006) Genetically engineered Bififidobacterium longum for tumor‐targeting enzyme‐prodrug therapy of autochthonous mammary tumors in rats. Cancer Sci 97: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone‐Corsi, M. , Nuccio, S.P. , Liu, H. , Hernandez, D. , Vu, C.T. , Takahashi, A.A. , et al (2016) Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540: 280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, B. , Mal, G. , and Marotta, F. (2017) Designer probiotics: paving the way to living therapeutics. Trends Biotechnol 35: 679–682. [DOI] [PubMed] [Google Scholar]
- Sonnenborn, U. , and Schulze, J. (2009) The non‐pathogenic Escherichia coli strain Nissle 1917‐features of a versatile probiotic. Microb Ecol Health Dis 21: 122–158. [Google Scholar]
- Stritzker, J. , Weibel, S. , Hill, P.J. , Oelschlaeger, T.A. , Goebel, W. , and Szalay, A.A. (2007) Tumor‐specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol 297: 151–162. [DOI] [PubMed] [Google Scholar]
- Sturm, A. , Rilling, K. , Baumgart, D.C. , Gargas, K. , Abou‐Ghazale, T. , Raupach, B. , et al (2005) Escherichia coli Nissle 1917 distinctively modulates T‐cell cycling and expansion via Toll‐like receptor 2 signaling. Infect Immun 73: 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Gunzer, F. , Westendorf, A.M. , Buer, J. , Scharfe, M. , Jarek, M. , et al (2005) Genomic peculiarity of coding sequences and metabolic potential of probiotic Escherichia coli strain Nissle 1917 inferred from raw genome data. J Biotechnol 117: 147. [DOI] [PubMed] [Google Scholar]
- Thamm, D.H. , Kurzman, I. D. , King, I. , Li, Z. , Sznol, M. , Dubielzig, R.R. , et al (2005) Systemic administration of an attenuated, tumor‐targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin Cancer Res 11: 4827–4834. [DOI] [PubMed] [Google Scholar]
- Toso, J.F. , Gill, V.J. , Hwu, P. , Marincola, F.M. , Restifo, N.P. , Schwartzentruber, D.J. , et al (2002) Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol 20: 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel, S. , Stritzker, J. , Eck, M. , Goebel, W. , and Szalay, A.A. (2008) Colonization of experimental murine breast tumours by Escherichia coli K‐12 significantly alters the tumour microenvironment. Cell Microbiol 10: 1235–1248. [DOI] [PubMed] [Google Scholar]
- Xie, S. , Chen, M. , Song, X. , Zhang, Z. , Zhang, Z. , Chen, Z. , et al (2018) Bacterial microbots for acid‐labile release of hybrid micelles to promote the synergistic antitumor efficacy. Acta Biomater 78: 198–210. [DOI] [PubMed] [Google Scholar]
- Yang, N. , Zhu, X. , Chen, L. , Li, S. , and Ren, D. (2008) Oral administration of attenuated S. typhimurium carrying shRNA‐expressing vectors as a cancer therapeutic. Cancer Biol Ther 7: 145–151. [DOI] [PubMed] [Google Scholar]
- Yu, Y.A. , Shabahang, S. , Timiryasova, T.M. , Zhang, Q. , Beltz, R. , Gentschev, I. , et al (2004) Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light‐emitting proteins. Nature Biotech 22: 313–320. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , and Forbes, N.S. (2015) Trg‐defificient Salmonella colonize quiescent tumor regions by exclusively penetrating or proliferating. J Control Release 199: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, Y. , Xia, L. , Zhang, X. , Ding, X. , Yan, F. , et al (2012) Escherichia coli Nissle 1917 targets and restrains mouse B16 melanoma and 4 T1 breast tumors through expression of azurin protein. Appl Environ Microbiol 78: 7603–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Ji, W. , He, L. , Chen, Y. , Ding, X. , Sun, Y. , et al (2018) E. coli Nissle 1917‐derived minicells for targeted delivery of chemotherapeutic drug to hypoxic regions for cancer therapy. Theranostics 8: 1690–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S. , Gravekamp, C. , Bermudes, D. , and Liu, K. (2018) Tumour‐targeting bacteria engineered to fight cancer. Nat Rev Cancer 18: 727–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Hao, L. , Liu, X. , Borrás‐Hidalgo, O. , and Zhang, Y. (2018) Enhanced anti‐proliferative efficacy of epothilone B loaded with Escherichia coli Nissle 1917 bacterial ghosts on the HeLa cells by mitochondrial pathway of apoptosis. Drug Dev Ind Pharm 44: 1328–1335. [DOI] [PubMed] [Google Scholar]


