Summary
Nematode–bacterial associations are far‐reaching subjects in view of their impact on ecosystems, economies, agriculture and human health. There is still no conclusion regarding which pathogenic bacteria sense nematodes. Here, we found that the pathogenic bacterium Bacillus nematocida B16 was sensitive to C. elegans and could launch smart attacks to kill the nematodes. Further analysis revealed that the spores of B. nematocida B16 are essential virulence factors. Once gaseous molecules (morpholine) produced from C. elegans were sensed, the sporulation of B16 was greatly accelerated. Then, B16 showed maximum attraction to C. elegans during the spore‐forming process but had no attraction until all the spores were formed. The disruption of either the spore formation gene spo0A or the germination gene gerD impaired colonization and attenuated infection in B16. In contrast, complementation with the intact genes restored most of the above‐mentioned deficient phenotypes, which indicated that the spo0A gene was a key factor in the smart attack of B16 on C. elegans. Further, transcriptome, molecular simulations and quantitative PCR analysis showed that morpholine from C. elegans could promote sporulation and initiate infection by increasing the transcription of the spo0A gene by decreasing the transcription of the rapA and spo0E genes. The overexpression of rapA or spo0E decreased the induced sporulation effect, and morpholine directly reduced the level of phosphorylation of purified recombinant RapA and Spo0E compared to that of Spo0A. Collectively, these findings further support a ‘Trojan horse‐like’ infection model. The significance of our paper is that we showed that the soil‐dwelling bacterium B. nematocida B16 has the ability to actively detect, attract and attack their host C. elegans. These studies are the first report on the behaviours, signalling molecules and mechanism of the smart attack of B16 on nematodes and also reveal new insights into microbe–host interactions.
The pathogenic life cycle of the opportunistic bacterial pathogen of nematodes in the soil Bacillus nematocida B16 against C. elegans, which suggested us an active diffusion model of bacteria by actively sensing and responding to the hosts. Our findings in the paper enriched the ‘Trojan horse‐like’ infection mechanism and revealed new insights into microbe‐host interactions.

Introduction
The interactions between pathogenic microorganisms and hosts have always been the focus of biological research. The interaction of bacterial pathogens with their hosts is distinguished from host–microbiota interactions by the resulting host damage (Casadevall and Pirofski, 2000). When a pathogen is established into a new niche that is different from its original environment, the balance is altered, and mutational adaptations occur that change the regulation of virulence and metabolic genes.
Nematodes are ubiquitous organisms that have a significant global impact on ecosystems, economies, agriculture and human health. C. elegans has been an attractive model organism for studying pathogenic bacteria–host interactions since the 1970s (Irazoqui and Ausubel, 2010). The applied importance of nematodes and the experimental tractability of many species have promoted the use of these organisms as models in various research areas, including bacterial pathogen–host interactions (Murfin et al., 2012). Nematode–bacterial associations are far‐reaching research subjects in view of their pervasiveness, experimental tractability or their impact on ecosystems, economies, agriculture and human health. All nematodes interact with bacteria during their evolutionary history and engage in a variety of association types. Interactions between nematodes and bacteria can be positive (mutualistic) (Murfin et al., 2015; Whittaker et al., 2016) or negative (pathogenic/parasitic) (Ganesh and Rai, 2016; Hamid et al., 2017) and may be transient or stably maintained (symbiotic) (Liao et al, 2017). In addition to being a food source, bacteria can be pathogens of nematodes. Many of these are the same or similar to pathogens of humans, which has spurred the use of C. elegans as a model host of human infectious diseases (Pukkila‐Worley and Ausubel, 2012). Cheng et al. (2013) showed that B. xylophilus‐associated bacteria could support nematodes in the degradation of host xenobiotics. Vicente et al. investigated the pinewood nematode‐associated bacteria Serratia spp., which have evolved an elaborate detoxifying system that expresses several antioxidant enzymes to cope with H2O2‐mediated oxidative stress (Vicente et al., 2013).
Bacterial pathogenesis includes the following processes: exposure, attachment, colonization, invasive growth and host immune response/damage (Rahme et al., 2000). In fact, these processes are prerequisites for the recognition between nematodes and bacteria. There are many reports about the recognition mechanism of the nematode C. elegans for bacteria, including differentiating pathogens from food bacteria (Kim, 2015; Nandi et al., 2015; Lee et al., 2017). However, bacteria have also evolved numerous pathways to sense and respond to changing environmental conditions. Huang et al. (2016) reported that the bacterium Comamonas testosteroni CNB‐1 senses and responds to aromatic compounds via binding to the methyl‐accepting chemotaxis protein MCP2901, thereby initiating signal transduction for bacterial chemotaxis. Jia et al. (2016) reported that the gram‐negative bacterium Vibrio brasiliensis senses O2 stress through ligand‐dependent signalling pathways. Willett and Crosson (2017) found atypical modes of bacterial histidine kinase signalling. According to the above reports, it could be inferred that bacteria use various molecular mechanisms to integrate environmental signals to control complex adaptive processes.
The role of C. elegans‐associated bacteria and their interaction with nematodes have recently been under substantial investigation in our research group. In our previous study, we found that the ‘Trojan horse‐like’ infection mechanism of the pathogenic bacterium Bacillus nematocida B16 attacks C. elegans and produces a series of gases (volatile organic compounds, VOCs), such as benzyl benzoate, benzaldehyde, 2‐heptanone and acetophenone, which have functions that attract nematodes (Niu et al., 2010). However, how do the bacteria sense the existence of the worms? Are there also any gas signals produced from nematodes that can be sensed by the B16 bacteria? Which gas signals are the ‘Queen Helene and Prince Paris’ that promote sporulation, trigger the Trojan horse‐like infection and finally result in the destruction of C. elegans as the city of Troy? In this paper, we designed a series of experiments and inadvertently found that bacteria can be induced to sporulate in the presence of worms. The pathogenic bacterium B. nematocida B16 had the ability to sense the presence of nematodes by the signalling molecule morpholine, regulate their metabolism to form spores and finally become ready‐to‐trap nematodes.
Results
Sporulating cells initiate infection by extreme attraction to nematodes
The bacteria B. nematocida B16 and B. subtilis 168, with different incubation times, had varied attraction abilities to C. elegans (Fig. 1A). As a food of C. elegans, B. subtilis 168 shows a low level of attractiveness of nematodes until spores are formed, at which point the bacterium no longer attracts nematodes. However, the attractiveness of B16 was maintained at a similar attractive level as 168 during the initial 36 h and then sharply increased to a maximum level of 10.4 at 48 h. Subsequently, the attraction ability decreased with extended incubation time. Surprisingly, the attractiveness of B16 decreased below zero after 84 h, indicating that the B16 may not be sensed and avoided by worms until after 84 h or a much longer incubation time. Thus, the morphology of the B16 lawns after incubation for 36 h, 48 h and 84 h with an attraction assay was observed (Fig. 1B–D). There were no spores until 36 h (Fig. 1B), while the bacteria began to sporulate after 48 h incubation (Fig. 1C), and almost all the bacteria existed as spores when incubated for 84 h (Fig. 1D). Similar to the attraction experiments, after 48 h incubation, the B16 bacteria began to form spores when the attraction ability to C. elegans also reached a maximum. However, after 36 h, sporulation was not observed; after 84 h, with the approach–avoidance ability, the bacteria were all spores, and almost no vegetative cells were observed.
Figure 1.
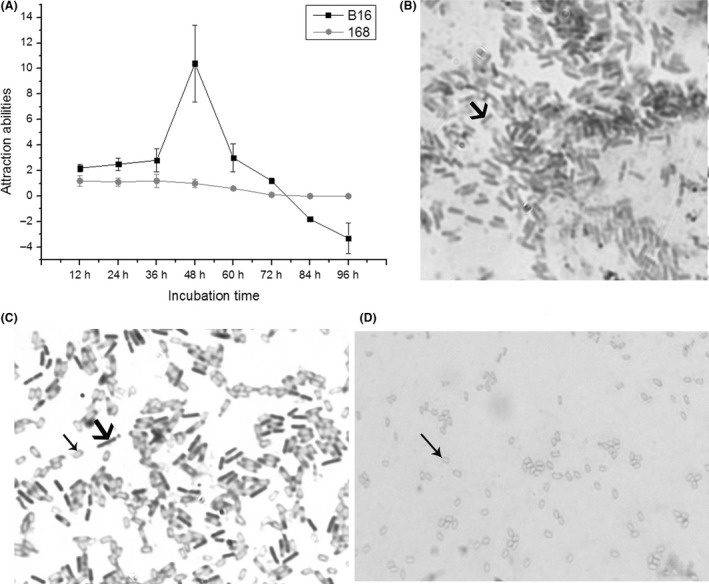
Results of different attraction assays.
A. Results of the attraction assay of the bacteria B16 and 168 to the worm C. elegans with different incubation times.
B–D. Micrographs of sporulation in B16 cells incubated for 36 h (B), 48 h (C) and 84 h (D).
Vegetative cells, sporulating cells and pure spores were selected to determine their attracting capabilities at different states. As shown in Fig. 2, after the nematode suspension was added to the middle of the culture, the worms clearly moved to the fermentation broth of the sporulating cells (Video S1). The majority of the tested worms were near the sporulating bacterial suspension within ten min (Fig. 2B), implying that when the bacteria began to form spores, their ability to attract nematodes was the greatest. These results suggest that B16 exhibits the maximum attraction to the worms once the bacteria begin to sporulate. The changes in the production of attractive VOCs by vegetative cells, sporulating cells and pure spores were further detected. Compared to the vegetative cells, large amounts of VOCs were secreted by the sporulating cells, including 2‐heptanone, 2,5‐dimethyl pyrazine, benzaldehyde, indole, benzyl benzoate and acetophenone, which had been shown to be attractants for C. elegans at low concentrations (Table S3). However, the VOCs from the pure spores were much lower in both quality and quantity than those in the sporulating cells. Our results clearly showed the disappearance of indole (retention time 11.18 min) and a decrease in 2,5‐dimethyl pyrazine (retention time 9.31 min), and both molecules were reported to be attractive. Several new compounds were also detected, such as phenol (retention time 9.09), quinolone (retention time 14.75), butanoic acid 2‐methyl‐ethyl ester (retention time 17.53) and some uncertain compounds, which were possibly derived from the secondary metabolites formed in the spore stage. These results showed that during sporulation, the sporulating cells not only produced the largest amount of attractants but also showed maximum ability to attract nematodes. It was concluded that sporulating cells initiate infection by their extreme attraction of nematodes.
Figure 2.

Comparison of attraction abilities among vegetative cells, sporulating cells and pure spores.
A. The state when the sample was added at zero min;
B. the movement on the agar plate after 10 min.
Sporulation and the production of attractants are regulated by spo0A
Bioassay and colonization analysis
To verify whether attractants and virulence factors produced in B16 fermentation broth are correlated with the spore‐forming killing of the nematode by B16, vegetative and sporulated cell fermentation supernatants were separately tested. The results showed that 70% of the nematodes could be killed within 24 h by B16 sporulated cell filtrate; after 48 h, all tested nematodes were killed. In contrast, < 15% of the nematodes were killed by B16 vegetative cell filtrate (Table 1). It has been reported that sporulation involves over 500 genes, among which spo0A is one of the master regulators (Galperin et al., 2012). Additionally, it has been suggested that the GerD protein is essential for normal spore germination in B. subtilis, and its absence leads to an obvious decrease in germination (Pelczar et al., 2007; Wang et al., 2011). Therefore, to verify the key functions of spores in infection, spo0A and gerD were knockout, and complemented mutants were separately constructed. The spo0A gene mutant strain B16ks‐9 was defective in sporulation, while the gerD mutant strain B16kg‐8 did not exhibit normal spore germination (Supporting information ‘Spore formation and germination assay of the mutants’). Consistent with our expectations, the mortality of the worms fed on spores of B16g (B16‐GFP) was much higher than that of the nematodes fed on vegetative cells of B16g (Fig. 3A). For experiments with spores of strains B16g and B16cg, over 85% of the tested nematodes were killed after 3 days. However, the majority of worms (over 90%) fed on the vegetative cells of B16g, B16ks and the negative control strain E. coli JM109g were still alive after 5 days. Moreover, approximately 75% of the tested nematodes died over the course of 3 days after feeding on the spores of the complement strain B16cs. To our surprise, the worms that fed on the spores of the gerD mutant strain B16kg had comparatively low mortality (only 40%) after 3 days. Then, we examined the ability of B. nematocida spores to colonize C. elegans by following the kinetics of bacterial accumulation in the nematode intestine over time. The results are shown in Fig. 3B. The population of intestinal B. nematocida, which was infected by the spores of B16g, B16cg and the complementation strain B16cs, reached 104 to 105 bacteria/worm in the first 3 days. The number of bacteria for the worms fed on the spores of strain B16kg did not reach 100 CFU PER worm when infected for 4 days, which was not significantly different from that of the worms fed on the vegetative cells of B16g, B16ks and the negative control strain E. coli 109 g.
Table 1.
Results of the responses of the nematodes towards vegetative cells, sporulating cells and pure spores of B16.
| Different samples | Attraction abilities | Mortalities of nematodes by filtrates (%) (SD) | ||||||
|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 36 h | 48 h | 12 h | 24 h | 36 h | 48 h | |
| Vegetative cells | 1.2 | 1.8 | 1.8 | 1.9 | 5 (0.2) | 10 (1.3) | 12 (1.6) | 14 (1.7) |
| Sporulating cells | 2.2 | 2.8 | 3 | 3.5 | 50 (1.8) | 70 (3.3) | 95 (2.0) | 100 (0) |
| Pure spores | 1.2 | 1.4 | 0 | −1.1 | 20 (1.1) | 30 (1.4) | 55 (1.9) | 70 (2.3) |
| E. coli JM109 | 2.3 | 2.5 | 2.8 | 3.3 | 5 (0.3) | 5 (0.3) | 10 (1.6) | 10 (1.8) |
Figure 3.

Results of mortality and CFU/worm assays by different samples.
A. Killing of the nematode C. elegans by spores or vegetative cells from the wild‐type, knockout and complementary strains.
B. Number of bacteria colonized in the worm C. elegans infected by different samples. Notes: B16g‐s, spores of GFP‐marked B16; B16g‐v, vegetative cells of GFP‐marked B16; B16ks‐v, vegetative cells of spo0A knockout mutant strain; B16kg‐s, spores of gerD knockout mutant strain; B16cs‐s, spores of spo0A complement mutant strain; B16cg‐s, spores of gerD complement mutant strain; 109g, GFP‐marked E. coli JM109.
The worms were visually scored for severity of colonization based on the extent of luminal distention and gfp signal in the intestine (Fig. S5A). As worms have a number of mechanical and chemical mechanisms for restricting bacteria in the gut, individual animals were colonized at different rates (Fig. S5B). During the first 48 h of infection, the spores of the B16kg, B16cs and B16cg mutants colonized C. elegans to a similar degree. After 72 h, the spores of the mutant strain B16kg colonized C. elegans significantly less than those of the wild‐type strain B16g and the mutants B16cs and B16cg (Fig. S5B). In addition, the spores of the wild‐type strain B16g showed notably strong colonization abilities, while the vegetative cells and E. coli strain 109g showed almost no colonization of the worm intestine throughout the whole infection process. Specifically, 90% of animals feeding on the spores of B16g scored in the ‘full’ colonization category, whereas animals constantly exposed to either vegetative cells or E. coli strain 109g only had scores of 10% or less. Furthermore, the full category for the spores of the knockout strain B16kg decreased from approximately 35–20% when we compared the change in the severity of colonization between 48 and 72 h. It was concluded that spores could pass through the digestive system of C. elegans and that the germination of the spore is required for promoting B. nematocida colonization of the C. elegans intestinal lumen.
The involvement of spo0A in the synthesis of attractants
As indicated in Table 1, the results for the tested attraction ability in the gene knockout and complementation mutants were consistent with those for the attraction response assays of the nematode by B16 vegetative and sporulated cell supernatants. Within 8 h, only 8% (40 ± 3.5/500) of C. elegans migrated towards the knockout strain B16ks at the top by arduously climbing the bare wall of the Petri dish. This attraction ability was not significantly different from that in the control JM109 media. When the bacterial lawn consisted of the complementation strain B16cs, 32% (160 ± 8.6/500) of the tested worms moved upwards to the top. Media with strains B16kg and B16cg yielded 30% (150 ± 5.1/500) and 36% (180 ± 7.9/500) worm migration, respectively, which is not significantly different from the worm migration with B16cs media (Fig. 4A). Additionally, the responses of the worm towards B16 vegetative, sporulating and spore cells were individually examined. The attraction abilities of the sporulating cells were 2.2, 2.8 and 3.5 within 12, 24 and 48 h, respectively, whereas the attraction abilities were below 2.0 within 48 h in the vegetative and spore cell groups of B16 (Table 1). These results indicated that the sporulation of strain B16 was closely related to its attraction to C. elegans and that spo0A most likely regulated the attraction process. Moreover, slight aversive behaviour against B16 spores was observed, which might indicate memory behaviour of the worm; the worm may barricade the potential damage caused by bacterial uptake into the intestine of the nematodes.
Figure 4.

A. The results of turnover Petri dish experiments for the mutant strains.
B. The changes in VOCs in the Δspo0A mutant were determined with SPME/GC‐MS. Arrowhead represents the difference in the attractive signal molecules between the wild‐type strain and Δspo0A mutant. Notes: B16ks, spo0A knockout mutant strain; B16kg, gerD knockout mutant strain; B16cs, spo0A complement mutant strain; B16cg, gerD complement mutant strain; JM 109, E. coli JM109.
The changes in the production of attractive volatile organic components (VOCs) in △spo0A mutants were further detected (Niu et al., 2010). The categories and quantities of VOCs in the Δspo0A mutant strain B16ks changed obviously compared with those in the wild‐type B16 strain (Fig. 4B). Our results showed the disappearance of 2‐heptanone (retention time 6.57 min) and benzaldehyde (retention time 9.45 min), and both molecules have been shown to be attractants for C. elegans at low concentrations (Bargmann et al., 1993; Niu et al., 2010). Two new peaks, 3‐methyl butyraldehyde (retention time 3.11 min) and 2‐furanmethanol (retention time 18.13 min), appeared in the B16ks strain, and an uncertain compound (retention time 1.67) decreased. On the other hand, the complement strain B16cs produced the major attractants of 2‐heptanone and benzaldehyde. The results showed that the spo0A gene plays a role in the synthesis of attractants, suggesting that VOCs were prerequisites for successful infection against nematodes.
C. elegans nematodes produce gases that induce the sporulation of B16
The results shown in Fig. 5B, C indicated that the worm C. elegans could effectively induce the formation of B16 spores but not 168 spores in advance. For B16, within 18 h, 20 CFU ml−1 original cell suspensions were observed after incubation at 85°C, while the bacteria without worms did not sporulate at all. Within 34 h, the maximum difference in the spore number was obtained. The number of B16 spores induced by C. elegans was 2.72 × 104 CFU ml−1, while only 84 CFU ml−1 was observed for the bacteria in the negative controls. The largest difference was 324 times that of the negative control. The spore numbers at each testing point were much higher than those in the controls. However, the spore numbers for B. subtilis 168 showed no differences between the samples and the controls.
Figure 5.

Method and results of inducing sporulation experiments.
A. Schematic diagram of the assaying method;
B. the results of the number of bacterial spores under different induction times induced by the nematode C. elegans; C. SEM results of bacterial morphologies.
Further observations of B16 under light microscopy and SEM revealed that, after 18 h, the majority of these bacteria were vegetative cells without C. elegans induction, while spores could be observed at the same time with induction by C. elegans. Most of the B16 spores were observed after incubating with C. elegans induction for 34 h (Fig. 5C). These results suggested that the worms emit some signal molecules that could be sensed by strain B16, which triggered the sporulating process. In total, we identified 25 distinct gases (VOCs) from two samples of the worm C. elegans based on gas chromatography (GC)/MS system data banks (NIST05, NIST98 and Wiley 275, Qual > 85). These gases included ketones, esters, alkanes, phenolic and heterocyclic compounds. Using the C. elegans worms without B16 as the control, of the 25 gases detected in the two samples (Fig. 6), six that were produced by nematodes with B16 were tested individually using 50 ppm commercially available standards. Of these six gases, morpholine displayed the strongest induced abilities, and the inducing abilities reached 49. Two compounds, undecanoic acid propyl ester and hentriacontane, also showed a greater ability to induce sporulation in B16, with inducing abilities of 28 and 25 respectively. 2,6‐Dihydroxyacetophenone revealed almost no attraction activity. To our surprise, 2,6‐Bis (1,1‐dimethylethyl)‐4‐(1‐oxopropyl) phenol not only failed to promote sporulation but also hindered the sporulation ability of B16. The inducing abilities of 2,6‐Bis (1,1‐dimethylethyl)‐4‐(1‐oxopropyl) phenol were only 0.73 (Table 2).
Figure 6.

Gases were determined with GC‐MS by comparing the total ion current traces from different nematode samples.
A. A map of the GC‐MS total ion current traces of the nematode C. elegans in the presence of B16; 25 gases produced by the worm induced B16;
B. a map of the GC‐MS total ion current traces of the nematode C. elegans in the absence of B16; 19 gases were produced by the worm without B16;
C&D indicate the enlarged map of A, and the arrows indicate the 6 candidate gases from C. elegans induced by B16 comparing the total ion current traces from the control B.
Table 2.
Gases potentially emitted by the worm and their ability to induce sporulation.
| Different substances (no.) | RT (min) | % Relative content (SD) | Inducing abilities (SD) |
|---|---|---|---|
| Aziridine | 41.238 | 3.7 (0.4) | 8 (0.5) |
| 2,6‐Dihydroxyacetophenone | 41.968 | 5.8 (0.5) | 1.1 (0.08) |
| 2,6‐Bis(1,1‐dimethylethyl)‐4‐(1‐oxopropyl)phenol | 43.221 | 3.7 (0.4) | 0.73 (0.08) |
| Hentriacontane | 43.702 | 19.3 (0.9) | 25 (1.0) |
| Morpholine | 51.619 | 4.4 (0.4) | 49 (1.9) |
| Undecanoic acid propyl ester | 52.426 | 9.2 (0.7) | 28 (1.1) |
RT, retention time.
According to the results above, the compound morpholine is potentially the main inducer. Altogether, these results showed that the gas molecules of C. elegans combined with cell membranes entered cells and induced sporulation.
Transcriptional data reveal candidate genes for inducing sporulation in B16
The differences in the expression of genes related to sporulation were analysed as follows: (i) a total of 27 genes related to sporulation were obviously upregulated, with more than a twofold difference in the B16‐2 versus B16‐5 comparison (Fig. 7), which indicated that in the absence of worm induction, the spore‐forming genes in B16 were increasingly expressed, along with prolonged incubation time. (ii) Nine genes related to sporulation were obviously upregulated by more than a twofold difference in the B16‐2 versus B16C‐2 comparison, which indicated that in the absence of worm induction, the spore‐forming genes in B16 were increasingly expressed, along with prolonged incubation time. Of the nine increasingly expressed genes, the expression of eight genes, including yjcZ, cotH and gerQ, coincided with those in the B16‐2 versus B16‐5 comparison (Table 3). The results showed that the expression of genes involved in sporulation in B16 was advanced in the presence of C. elegans. That is, the spores of bacterial B16 were formed advance. The data were consistent with our observations. (iii) Nine spore‐forming genes were expressed more than twofold in the B16‐5 versus B16C‐5 comparison (Table 4). These results mean that when the incubation time was 34 h, the expression level of B16 spore‐forming genes was higher than that of the control group, which was basically the same as our observations that the higher number of spores was induced by the worms. (iv) Two genes, spo0E and rapA, involved in sporulation regulation were downregulated in the B16‐2 versus B16C‐2 and in B16‐5 versus B16C‐5 comparisons. The expression of the Spo0E phosphatase in induced B16C‐2 was decreased twofold compared with control bacteria B16‐2 for RNA‐seq (Table 3). Another phosphatase, RapA, in induced B16C‐5 was decreased 2.6‐fold compared with the control B16‐5 (Table 4). Spore formation by B. subtilis is triggered via the transfer of phosphoryl groups to Spo0A, and Spo0A ~ P directly promotes sporulation. However, Spo0E inhibited the formation of Spo0A ~ P, which is a negative signal for sporulation.
Figure 7.

Venn diagrams representing the number of upregulated genes expressed between B16‐2 and B16‐5 and between B16‐2 and B16C‐2.
Table 3.
Expression of 9 genes related to sporulation in B16C‐18.
| GeneID | Description | log2 Ratio (B16‐5/B16‐2) | P value (B16‐5/B16‐2) | log2 Ratio (B16C‐2/B16‐2) | P value (B16C‐2/B16‐2) |
|---|---|---|---|---|---|
| Unigene0000105 | Sporulation protein YjcZ | 6.732 | 0 | 1.683 | 0.00063 |
| Unigene0000143 | Spore coat protein CotH | 6.797 | 0 | 1.243 | 0.00112 |
| Unigene0000426 | Small acid‐soluble spore proteins alpha/beta type | 2.737 | 5E‐17 | 1.913 | 5.2E‐05 |
| Unigene0001474 | Small acid‐soluble spore protein | 2.210 | 0 | 1.347 | 0 |
| Unigene0001671 | Spore protein | 1.405 | 1.238 | 1.413 | 1E‐164 |
| Unigene0001803 | Spore coat protein | 3.362 | 0 | 1.046 | 6.7E‐71 |
| Unigene0001844 | Inner spore coat protein | 3.050 | 0 | 1.195 | 5.7E‐84 |
| Unigene0001917 | Spore coat protein GerQ | 1.321 | 0 | 1.006 | 1E‐108 |
| Unigene0000560 | Phosphatase Spo0E | – | – | −1.047 | 3.6E‐07 |
–, represents no expressional difference.
Table 4.
The upregulated expression of genes compared to B16C‐5 and B16‐5.
| GeneID | Description | log2 Ratio(B16C‐5/B16‐5) | P value |
|---|---|---|---|
| Unigene0000001 | Spore coat protein | 1.579021406 | 0 |
| Unigene0000105 | Sporulation protein YjcZ | 1.377068296 | 2.2E‐232 |
| Unigene0000143 | MULTISPECIES: spore coat protein CotH | 1.616124544 | 0 |
| Unigene0000226 | Spore coat protein | 1.405412016 | 0 |
| Unigene0001030 | protein involved in maturation of | ||
| The outermost layer of the spore | 1.001199646 | 1.41E‐25 | |
| Unigene0001460 | Spore coat protein | 1.442324125 | 0 |
| Unigene0001467 | protein involved in maturation of | ||
| The outermost layer of the spore | 1.680168308 | 0 | |
| Unigene0001844 | Inner spore coat protein | 1.212773399 | 0 |
| Unigene0001960 | Spore coat protein D | 1.371361697 | 0 |
| Unigene0001341 | Phosphatase RapA | −1.38507 | 2.60E‐220 |
Morpholine induced the sporulation of B16 through downregulating rapA and spo0E expression
Molecular simulation results
Molecular docking has been widely used in ligand screening and prediction of intermolecular interactions. In this study, the binding free energies between VOCs and protein receptors could be used as a guideline to identify ligand candidates. The K d, which is commonly used to describe the affinity between a ligand and a receptor, for affinity ligands is required to be in the range of 103 to 107 M. Hence, based on the energy criteria and geometry criteria, only the VOCs with a free binding energy less than 4.3 kcal mol−1 (K d < 103 M) were selected as potential ligands. The results of various VOCs binding to different receptors are shown in Table 5. RapA showed the lowest binding free energy with morpholine, which was < 4.3 kcal mol−1, indicating that morpholine had good affinity with RapA. The adsorption behaviour of morpholine and RapA was determined using Gromacs. After 5 ns of simulation (Fig. 8A, B), five morpholine molecules spontaneously bound to RapA. Little fluctuation of the RMSD implied that the system reached equilibrium. The same binding site of morpholine to RapA in molecular docking and simulation showed the consistency of the simulation. The more binding sites of morpholine on RapA in the MD simulation indicated there were multiple adsorption sites in the water environment. The same binding site of morpholine to Spo0E is shown in Fig. 8C, D. After 5 ns of simulation, two morpholine molecules spontaneously bound to the receptor, indicating that the ligand had multiple binding sites in the aquatic environment. According to the literature, the C‐terminal residues of Spo0E are the main functional sites (Grenha et al., 2006). Further geometry criteria analysis showed that morpholine could bind to the catalytic site of Spo0E and interfere with its activity (Fig. 8C). In addition, previous studies have shown that morpholine can penetrate the cell membrane spontaneously. Based on these results, morpholine, RapA and Spo0E were selected for further experiments to determine their interactions.
Table 5.
The free binding energy (E) and inhibition constant of morpholine.
| Morpholine | Rap A | Spo0E | KinA | KinE | Spo0A |
|---|---|---|---|---|---|
| ΔG (kcal mol−1) | −4.37 | −3.27 | −3.33 | −3.82 | −3.44 |
| Inhibition constant | 0.630 × 10−3 | 4.03 × 10−3 | 3.63 × 10−3 | 1.58 × 10−3 | 3.03 × 10−3 |
Figure 8.

Molecular simulations of the morpholine‐protein system.
A. The docking conformation with the lowest free binding energy of morpholine and RapA.
B. Snapshots from a simulation trajectory and RMSD of morpholine and RapA after 5 ns.
C. The docking conformation with the lowest free binding energy of morpholine and Spo0E.
D. Snapshots from a simulation trajectory and RMSD of morpholine and Spo0E after 5 ns. Morpholine is indicated in green, the red circle shows the same binding site in both docking and MD simulations, while the others are labelled in the blue circle.
The transcription of rapA and spo0E decreased in the presence of morpholine
The candidate genes for inducing sporulation in B16 were investigated by transcriptomics. Spore formation by B. subtilis is triggered by the transfer of phosphoryl groups to Spo0A, and Spo0A ~ P directly promotes sporulation. However, Spo0E inhibits the formation of Spo0A ~ P, which is a negative signal for sporulation. Based on these results, three genes, spo0E, rapA and spo0A, were selected for quantitative PCR validation analysis. When the B. nematocida B16 bacteria were exposed to morpholine for 18 h, the formation of opaque colonies, a symptom of spore formation, was faster in B16 than in the control B16 strain without morpholine or in nematodes. Since the phosphorylation of Spo0A is central to the initiation of sporulation, we tested whether morpholine improved the transcription level of the spo0A gene. The RT‐qPCR analysis results in Fig. 9 revealed that the expression level of spo0A was 9.7‐fold higher than that in the control, and this expression was higher than that (6.5‐fold) induced by nematodes. In contrast, the expression levels of the rapA gene decreased to 0.2 and 0.1 after induction by C. elegans and morpholine respectively. Similarly, spo0E decreased to 0.2 and 0.4 after induction by C. elegans and morpholine respectively. These data indicated that morpholine from C. elegans induced the sporulation of B16 through downregulating rapA and spo0E expression.
Figure 9.

qPCR results when the B16 bacteria were separately induced by C. elegans and morpholine.
Morpholine from C . elegans decreased spo0E transcription and expression by simple or facilitated diffusion
Morpholine functions intracellularly
To further investigate whether morpholine works outside or inside of cells, the quantity of morpholine in cells was measured following C. elegans induction. Fig. 10 shows four sample chromatograms from a pooled QC sample analysed by GC‐MS. A total of 1 µl of 100 ppm pure morpholine is shown in Fig. 10A. The insets in Fig. 10B, C revealed the same portion of the two chromatograms from B16 and 168 cells broken by ultrasound. According to the peak area, the amount of morpholine calculated in B16 was approximately one‐tenth of that in the control. Using E. coli OP50 as a negative control, there was no morpholine peak detected by GC‐MS due to its low abundance and overlapping retention time with the three other peaks. The morpholine peak area ratio between B16 and 168 was 8.95, suggesting that morpholine entered B16 cells more than 168 cells. The cellular uptake pathway of small molecules involves the energy‐independent manner of direct translocation and the energy‐dependent manner of endocytosis (Boisguérin et al., 2015). Thus, to further investigate the transport model of morpholine, its concentration was evaluated following incubation at 4°C in medium without sugar. The results based on the experiments from ATP depletion showed that the abundance of morpholine inside the cells was not obviously different from the amount after incubating with sugar at 37°C, indicating that morpholine entered the B16 cells through energy‐independent diffusion.
Figure 10.

The quantity of morpholine in four sample cells was measured following C. elegans induction: A. 1 µl of 100 ppm pure morpholine; B. B16; C. 168; D. E. coli OP50.
To further investigate whether the differences in cell membrane components caused the different morpholine concentrations in the cells of the two strains, the types and quantities of fatty acids of B16 and 168 cells were measured. The results showed that there was no significant difference in the fatty acid composition between the two cell membranes (Table S4, S5). The differences in membrane proteins between B16 and 168 were compared via bioinformatics analysis. In the annotation of the B16 sequenced genome, all membrane proteins have been marked and listed. These membrane protein sequences were extracted and used as query sequences to align with those in 168. The sequences showing no hits were considered completely unmatched and designated as B16‐unique membrane sequences. A total of twelve sequences were obtained, as indicated in Table 6. Taken together, we speculate that morpholine molecules may enter B16 cells by binding these unique membrane proteins.
Table 6.
Specific membrane proteins in B. nematocida B16 compared with B. subtilis 168.
| Gene ID | Subcellular localization | Gene length(bp) | Location | NR description |
|---|---|---|---|---|
| gene0271 | Cell membrane | 729 | Chr | Zinc transporter |
| gene0422 | Cell membrane | 675 | Chr | Membrane protein |
| gene0518 | Cell membrane | 1071 | Chr | Urea ABC transporter permease subunit UrtC |
| gene0584 | Cell membrane | 1263 | Chr | PTS ascorbate transporter subunit IIC |
| gene1621 | Cell membrane | 609 | Chr | ABC transporter permease |
| gene2243 | Cell membrane | 465 | Chr | Putative symporter YidK |
| gene2244 | Cell membrane | 180 | Chr | Solute:sodium symporter family transporter |
| gene2444 | Cell membrane | 210 | Chr | Holin |
| gene3189 | Cell membrane | 171 | Chr | Proteolipid membrane potential modulator |
| gene3779 | Cell membrane | 591 | Chr | Membrane protein |
| gene3825 | Cell membrane | 324 | Chr | Branched‐chain amino acid transporter AzlD |
| gene4127 | Cell membrane | 2208 | Chr | Membrane protein |
The induction of sporulation was relieved when the overexpression strains were treated with morpholine
To measure the contribution of the RapA and Spo0E proteins in the induction of sporulation by morpholine molecules, we genetically altered B. nematocida B16 and optimized its growth conditions to overexpress the RapA and Spo0E proteins (Supporting information ‘Overexpression of RapA and Spo0E’). To determine whether the in vivo activities of the overexpressed proteins were increased, immunoblot analysis of whole‐cell lysates of strains expressing the RapA and Spo0E proteins was carried out. The results (shown by a representative gel in Fig. S4D) indicated that the transformant strains ORB16‐1 and OSB16‐1 significantly improved the expression levels of proteins compared with the wild‐type strain, thus showing the successful overexpression of RapA and Spo0E. Additionally, the transcriptional levels of the genes rapA and spo0E were also determined by RT‐qPCR analysis. The results shown in Fig. 11 indicated that the transcriptional levels of rapA and spo0E in the overexpressed strains were 169 and 128 times those of the wild‐type strain at 18 h. After 18 h of morpholine treatment, the transcriptional levels of rapA and spo0E in the mutant strains decreased to three and 22 respectively. The rapA and spo0E transcriptional levels in the wild‐type strain induced by morpholine at the same time were also decreased to 0.233 and 0.267 respectively. With the extension of time from 18 to 34 h, the transcription of rapA in wild‐type B16 decreased from 1 to 0.5 without morpholine treatment and from 169 to 48 in the overexpressed strain. Similarly, the transcription of spo0E in B16 decreased from 1 to 0.6 without morpholine and from 128 to 72 in the overexpressed strain. Under the induction with morpholine, the transcription of rapA in wild‐type B16 decreased from 0.233 to 0.077 and from 3 to 2.3 in the overexpressed strain ORB16. Similarly, the transcription of spo0E in B16 decreased from 0.267 to 0.093 and from 22 to 6.6 in the overexpression strain OSB16. The qPCR analysis results showed that morpholine reduced the transcriptional levels of both rapA and spo0E in the overexpression strains. However, although morpholine decreased the transcription levels of the two genes in the overexpression strains, these levels were still higher than those in the wild‐type strains. Furthermore, the sporulation frequency in the overexpression strain was determined (Table 7). The ability of the RapA and Spo0E proteins to inhibit sporulation was significantly enhanced, which was consistent with previous reports. The sporulation induced by morpholine was significantly suppressed in the overexpression strains ORB16 and OSB16, suggesting the inhibition of sporulation in the overexpression of rapA and spo0E. The data implied that the morpholine targeted the rapA and spo0E genes to improve the sporulation of B16.
Figure 11.

The expression pattern of the rapA and spo0E genes at the level of transcription via qPCR when the wild‐type and overexpressed strains were induced by morpholine.
Table 7.
Sporulation frequencies of strains in the absence and presence of morpholine.
| Strains | 18 h | 34 h | ||||
|---|---|---|---|---|---|---|
| Viable cell count | Spore count | Sporulation (%) | Viable cell count | Spore count | Sporulation (%) | |
| B16 | 5.2 × 108 | 0 | 0 | 2.9 × 1010 | 3.8 × 106 | 0.013 |
| B16 with morpholine | 3.4 × 109 | 7.7 × 104 | 0.002647 | 8.9 × 1010 | 2.2 × 109 | 2.47 |
| ORB16 | 1.9 × 108 | 0 | 0 | 3.6 × 1010 | 0 | 0 |
| ORB16 with morpholine | 5.7 × 108 | 0 | 0 | 4.0 × 1010 | 0 | 0 |
| OSB16 | 2.0 × 108 | 0 | 0 | 2.9 × 1010 | 0 | 0 |
| OSB16 with morpholine | 7.6 × 108 | 0 | 0 | 7.3 × 1010 | 0 | 0 |
Morpholine molecule reduced the phosphatase activity of RapA and Spo0E
We purified the recombinant heterologously expressed RapA and Spo0E proteins to quantitate their activity against purified phosphorylated Spo0A in an in vitro assay. The time‐course for the RapA‐dependent and Spo0E‐dependent dephosphorylation and autodephosphorylation and morpholine treatment of Spo0A ~ P were monitored, and the results are shown in Fig. 12. The data showed that the dephosphorylation of Spo0A ~ P by the RapA and Spo0E proteins was extremely diminished after treatment with morpholine for 30 min compared with that in the no‐morpholine group. The remaining Spo0A ~ P treated with RapA decreased from 90% in the presence of morpholine to 30% in the absence of morpholine at 2 min and from 85% to 25%, respectively, at 4 min. The Spo0E treated with morpholine was approximately 52.6% and 55.6% less active than pure Spo0E at 2 min and 4 min respectively. The remaining Spo0A ~ P treated with morpholine alone presented no significant difference compared to that of the autophosphorylation groups. These results showed that the morpholine directly enters the cell through the membrane, decreasing the transcriptional level of the RapA and Spo0E genes, reducing the RapA and Spo0E activities and inducing the sporulation of B16.
Figure 12.

Dephosphorylation of Spo0A ~ P by Spo0E or Spo0E treated with 100 ppm morpholine for 30 min. Purified Spo0A ~ P (5 M) was incubated in the absence or presence of 2 M Spo0E, and aliquots were withdrawn at 0, 2, 4, 6, 8 and 10 min. All samples were run on a 15% SDS‐PAGE and analysed by ImageQuant software after exposure to a PhosphorImager plate (Amersham Biosciences). The Spo0A ~ P quantity at the zero time point was considered 100%, and the remaining Spo0A ~ P was calculated as a percentage of the initial value.
Discussion
It is a fate for prey to be always preyed on by predators; however, there is no rule that does not have exceptions. Sometimes, the prey becomes the new hunter of the predator. Organisms interact with the environment through different neural functions. The nematode C. elegans, which feeds on various types of bacteria that live in soil and on rotting vegetation by ingesting bacteria in suspension or on detritus (Nicholas, 1975), are bacterivorous. However, some bacterial strains were virulent to C. elegans, including B. nematocida B16, Pseudomonas aeruginosa, Serratia marcescens, B. thuringiensis, B. cereus, B. subtilis, B. pumilus, B. weihenstephanensis, B. mycoides, B. brevis, B. firmus, B. toyonensis, Lysinibacillus sphaericus, Brevibacillus laterosporus and Bacillus sp. (Rae et al., 2010; Meisel and Kim, 2014; Bird et al., 2015; Niu et al., 2015; Niu et al., 2016; Zheng et al., 2016). This coevolution results in a special competition between C. elegans and bacteria. Obviously, many virulence factors and mechanisms of nematode–bacterial interactions are also applicable to human pathogenic bacteria or other hosts. Thus, these phenomena, such as nematode–bacterial interactions, signalling molecules and mechanisms of communication and exchange between the associated partners, have been intensively studied by molecular biology techniques and genome‐scale studies (Mitreva et al., 2011; Tan and Shapira, 2011; Vicente et al., 2013; Koutsovoulos et al., 2014; Bird et al., 2015; Brown et al., 2015; Murfin et al., 2015). Although much has been learned, it is not well understood how pathogenic bacteria sense and process multiple cues of nematode stimuli in the environment.
Great strides in understanding how and why gases are important in mammalian physiology have kick‐started a new area of ‘gasotransmitters’ (King and Weber, 2007; Bowman et al., 2011; Fukuto et al., 2012; Prabhakar and Semenza, 2012; Tinajero‐Trejo et al., 2013). Gaseous signal molecules have been reported to play important roles in many plant responses to biotic and abiotic stresses, mammalian physiology and microbial communications (Hao et al., 2010; Schreiber et al., 2012; Tinajero‐Trejo et al., 2013; Wang et al., 2015). The small gaseous molecules penetrate membranes and play key roles in biology either by direct modification of target proteins or by activation of metalloenzymes (Fukuto et al., 2012; Peng et al., 2017). Additionally, gases were reported as constituting a natural bacterial defence mechanism against antimicrobial compounds (Keren et al., 2013; Liu and Imlay, 2013). Therefore, it is possible that gases are involved in interactions between different microbes or between microbes and their hosts. In our previous study, we found that the pathogen B. nematocida B16 can secrete gases (VOCs) as signalling molecules to attract nematodes and then colonize, proliferate and persist in the intestine of the nematode C. elegans, resulting in the death of the nematodes. The mechanism has been compared to the ‘Trojan horse’ story (Niu et al., 2010). Although the recognition of nematodes by bacteria is a prerequisite for successful infection, both the signalling molecules and mechanism of the bacterial recognition of nematodes have not been reported thus far.
The spores of many bacteria (such as B. anthracis) are considered potent bioterrorism agents because the spores could contribute to the ability to adhere to surfaces and show extreme resistance to all types of environmental stresses (Roffey et al., 2002; Joshi et al., 2012); (Setlow, 2006; Setlow, 2007). When such spores are placed under appropriate environmental conditions, they can also germinate and grow rapidly (Paidhungat et al., 2002; Moir, 2006). Sporulation is a last‐resort adaptive process that is tightly regulated by complex cell–cell signalling, which requires several hours to complete (Gonzalez‐Pastor et al., 2003). Therefore, sporulation is often a temporal, spatial and dynamic decision‐making process (Errington, 2003). Indole and indole derivatives induced sporulation in Stigmatella aurantiaca (Gerth et al., 1993), while another report showed that indole inhibited spore maturation of Paenibacillus alvei (Yong‐Guy et al., 2011). Thus, different bacterial species have developed unique regulatory systems to form endospores. Sporulation‐related genes in B. nematocida B16 were considered an extraordinarily important part of the colonization process, which was determined by constructing a random mutant library of B. nematocida B16‐GFP and functional screening (Niu et al., 2012). Usually, bacterial sporulation is often triggered by starvation. Here, we interestingly found that the nematode C. elegans could induce the sporulation of B. nematocida B16 but was not able to induce the sporulation of B. subtilis 168. The gases secreted from C. elegans are signalling molecules that induce sporulation. To find the gaseous components, we performed a gas composition analysis using SPME‐GC‐MS. We confirmed that six molecules of C. elegans induce sporulation in pathogenic bacteria, among which, morpholine produced from C. elegans showed the strongest sporulation‐inducing capabilities. Further results of molecular simulations indicated that morpholine may have affinity for RapA and interfere with the bioactivity of Spo0E. The pathogenic bacterium B. nematocida B16 had the ability to sense the presence of nematodes. However, B. subtilis 168 lacks the membrane protein receptors of gas signal molecules, which leads to failure to recognize the host and exhibit the corresponding response. Most dramatically, both the gene expression and sporulation of B16 in advance were regulated by the gases of nematodes, finally resulting in the trap of nematodes. Furthermore, transcriptome data revealed nine candidate genes related to sporulation that were upregulated > 2‐fold with statistical significance between the bacteria induced by the worm and the controls. The expression of both the RapA and Spo0E genes decreased to 0.2, while the expression of Spo0A increased 9.7 times compared to the control bacteria, as confirmed by real‐time PCR. Moreover, cell with a disruption in the Spo0A gene lost the ability to form spores. The Spo0A mutant also caused impaired attraction, colonization and attenuation. In contrast, complementation with the intact genes restored most of the above‐mentioned deficient phenotypes. The translocation of VOCs was an ATP‐free process in B. nematocida B16. In contrast to B16, morpholine or other VOCs could not penetrate the cell membrane freely in B. subtilis 168, which further prevented bacteria 168 from detecting the signal molecules and responding to the worms.
Recent molecular genetics and behavioural studies have revealed the mechanisms of the genetic model organism C. elegans in sensing and responding to pathogenic bacteria at the molecular or cellular level; these studies have included a C. elegans strain that senses different bacterial food sources and how C. elegans learns to avoid pathogenic bacteria (Zhang, 2008). As shown in Fig. 13, we also reported the pathogenic life cycle of the particular bacterium B. nematocida B16 against C. elegans. Once morpholine (nematode secretion) was detected, the pathogen B16 formed spores as concurrently secreted attractants to lure C. elegans. Once the worms arrived, the spores of B16 were preyed on and consumed. The spore coat protein provided protection in vivo against predation by C. elegans (Laaberki and Dworkin, 2008). Then, the bacteria avoided their hosts by producing repellents. Unlike vegetative cells, which are usually destroyed by nematodes, the B16 spores escaped and germinated in the intestinal tract of nematodes where they proliferated and subsequently killed the worms. Finally, the worms were degraded, and the bacteria propagated again. Furthermore, if the nutrients were exhausted, then the spores were formed again, resulting in a diffusion and migration process (Fig. 13). Apparently, B16 sensed, attracted, infected and destroyed the nematodes, completing its own reproduction. Here, the prey becomes the new hunter of the predator. In addition, our findings also suggested that the B16 bacteria could actively but not randomly diffuse. This purposeful diffusion may lead to a more general understanding of bacterial transmission in nature. These findings further enriched the Trojan horse‐like infection mechanism and tell a story similar to how the Trojan horse‐like story starts. Morpholine was similar to Prince Paris while attractants secreted by B16 were Helen the Beauty. Like the story, the VOCs trigger the entire life cycle, resulting in the ruin of C. elegans, similar to the destiny of Troy. This study is the first report on the new function of the VOC signals produced from worms for inducing the sporulation of B. nematocida. The signalling molecules and mechanism of the pathogenic life cycle were completely investigated and analysed by multiple methods. The interactions of cocompetition and coevolution between bacteria and host, prey and predator were clarified. This manuscript provides novel insights that may help improve the understanding of host–microbe interactions, which will deepen our understanding of how nematode–bacterium interactions evolve and how these interactions impact our environment. It will be helpful to construct new ecological environments and new biological drugs.
Figure 13.

A. B. subtilis 168 are ground and digested by C. elegans and become nematode food;
B. schematic diagram of the pathogenic life cycle of the particular bacterium B. nematocida B16 against C. elegans;
C. molecular mechanism of the sensing and response to nematodes in B. nematocida B16: morpholine from C. elegans penetrates the B16 membrane, inducing the sporulation of B16 by inhibiting the activity of RapA and Spo0E.
Experimental procedures
The effect of spores on the ability to attract C . elegans
The relationship between spores and the ability to attract C. elegans was analysed as follows: 10 µl of the wild‐type strain B16 grown overnight in LB medium was spread onto LB agar plates prepared in Petri dish lids. The bacterial lawns were individually incubated for 12, 24, 36, 48, 60, 72, 84 and 96 h at 37°C. The bacteria at different incubation times were used to quantitatively assay the attraction capabilities based on previously described methods (Ishikawa et al., 1986). Furthermore, Petri dish lids containing LB agar medium were separately inoculated with the knockout and complementation strains and incubated for 2 days at 37°C. The inoculated lid was inverted on top of another lid containing a 1‐day‐old adult hermaphrodite nematode suspension, sealed with air‐permeable adhesive tape, maintained at 26°C and 80% humidity and incubated in a dark chamber to assess the attraction capabilities (Niu et al., 2010).
To investigate the attraction abilities in different bacterial states, bacteria were first incubated in LB medium for different times, and vegetative cells, sporulating cells and pure spores were obtained by microscopic observation. The culture media in different states were diluted to the same concentration, followed by attraction tests: 300 µl of three kinds of bacterial suspensions was first dropped onto sterile filter paper to allow gradual vapourization of the sample during the test. Then, the filter paper with different bacterial samples was separately placed on one side of T‐glass tubing containing 1.5% agar (sample side), and the same quantity of LB medium alone was placed on the other side of the agar (control side). The nematode suspension (approximately 2000) was dropped into the middle of the two sides. Then, the worms, which had moved away from the middle to either side of the T‐tubing, were collected and counted individually from each side. The attracting activity was determined with the equation (A‐B)×10/B, where A and B represent the number of worms on the sample and control side respectively.
Spore preparation and germination
The spores were prepared as described by Kenny and Couch, with some modifications (Kenney and Couch, 1981). The bacterial strains were grown in tryptone soy broth (TSB) at 30°C overnight. The cell suspension was spread onto spore preparation agar (peptone, 3.3 g l−1; beef extract powder, 1.0 g l−1; NaCl, 5.0 g l−1; K2HPO4, 2.0 g l−1; KCl, 1.0 g l−1; MgSO4. 7H2O, 0.25 g l−1; MnSO4, 0.01 g l−1; lactose, 5 g l−1; agar 15 g l−1) using a sterile cotton swab and incubated at 28°C for 4–8 days. To collect the spores, 5 ml of sterile distilled water was added to the plate, and the spores were suspended in water using a micropipette. The spore suspension was then incubated at 85°C for 15 min to kill the vegetative cells. The spore concentration in the suspension was determined by diluting the suspension with 1 × PBS (phosphate buffer solution) and spreading the dilution onto triplicate plates of tryptone soy agar (TSA) to count the CFU. The spores were stored at −20°C and tested for germination between 2 and 6 weeks after preparation.
The method to indirectly assay spore germination was carried out by measuring the optical density of the broth medium at 600 nm (OD600). Since the optical density of liquid media inoculated with spores of Bacillus decreases rapidly as spores germinate (Powell, 1950; Powell, 1951), a qualitative criterion method was used to measure the decrease in OD600 of media inoculated with the spore suspension after incubation.
Sporulation frequencies were determined by the heat‐resistance assay (Harwood and Cutting, 1990).
Bacterial colonization assay
The final concentration of the spore suspension was adjusted with sterile water to approximately 1.0 × 1010 CFU ml−1 for the infection and colonization experiments. The same quantities of vegetative cells and spores of the wild‐type and recombinant strains were fed to 1‐day‐old adult hermaphrodite worms.
First, the kinetics of the colonization of C. elegans by constant exposure to bacteria was determined by fluorescence microscopy at 200× magnification using the method described by Alegado and Tan (2008). At each time point, three sets of 10 infected worms were observed to score the colonization situations. Worms with fluorescent bacteria in the entire lumen were scored as ‘full’; worms without any green fluorescence signal in the lumen were scored as ‘undetectable’; and worms harbouring bacteria between these two extremes were scored as ‘partial’.
The mortality bioassay analysis was conducted following established methods (Aballay et al., 2000; Garigan et al., 2002). As described above, the wild‐type strain B16 was first incubated in LB medium for different times, and different bacterial states, including vegetative cells, sporulating cells and pure spores, were confirmed by microscopic observation. Then, approximately fifty worms were separately placed on bacterial plates with different forms, and the plates were incubated at 25°C for 4 h. The worms were finally removed from the plates, washed twice in M9 buffer and transferred to NG agar plates for 3–4 days. The mortality of the worms was counted every 12 h with a light microscope, and death was determined when a worm no longer responded to touch (Kenyon et al., 1993). In addition, killing assays of fermentation supernatants against nematodes were performed as follows: 150 μl of fermentation supernatants was placed in a 1.5 ml Eppendorf tube, and approximately 200 nematode suspensions were transferred to each tube. After incubating the tubes at 25°C, the number of dead nematodes was determined every 12 h to count mortalities as described above.
Finally, the number of colonizing bacteria within the C. elegans digestive tract was measured using a previously described method (Garsin et al., 2001; Alegado and Tan, 2008). Three sets of 10 infected worms were picked at each time point and surface sterilized by placing worms on an agar plate containing 100 μg ml−1 gentamicin. The worms were then washed with M9 buffer containing 100 μg ml−1 gentamicin and 25 mM levamisole for paralysis. Pharyngeal pumping and expulsion were inhibited to prevent the entry of gentamicin into the intestinal lumen and the release of luminal bacteria. Then, the animals were washed twice more with M9 buffer alone and homogenized with M9 containing 1% Triton X‐100 to recover bacteria within the worm intestine (Portal‐Celhay et al., 2012). The washed nematodes were then mechanically disrupted using a grinding rod. After appropriate dilution, the lysates were plated onto selective media. Each data point represents the mean CFU from triplicate samples, and the error bars represent the standard deviation.
Assessment of induced sporulation
The flow diagram of this experiment is indicated in Fig. 5A. First, the original bacteria were streaked onto LB agar plates and incubated overnight (18 h) at 37°C. Then, a colony of a bacterial strain was inoculated into 4.5 ml of liquid LB medium in a 16 × 125 mm test tube that lacked visible scratches. After mixing the contents of the tube thoroughly, the culture was incubated at 37°C with vigorous aeration. The incubation was not stopped until the OD650 was between 0.4 and 0.6. Next, 0.05 ml of this culture was transferred to 15 ml of LB agar medium prepared in a 5 × 5 cm Petri dish lid based on a previously described attraction assessment method (Niu et al., 2010). The inoculated lid was inverted on top of another lid with water agar medium containing a drop of nematode suspension in the centre (∼500 nematodes). The two lids were sealed with air‐permeable adhesive tape and incubated in a dark chamber at 26°C. Within 8–24 h, the number of spores of bacterial lawns on the upper lid was assessed using the following methods: 10 ml of sterile distilled water was added to the plate, and the spores were suspended in water using a micropipette. The spore suspension was then incubated at 85°C for 15 min to kill the vegetative cells. The spore concentration in the suspension was determined by diluting the suspension with 1 × PBS and spreading the dilution onto triplicate plates of tryptone soy agar (TSA); the CFU was counted after incubating overnight at 37°C. The same diameter of blank water agar medium without worms in the lower lid was used as a negative control. The activity of the induced bacterial spores was determined with the following equation: inducing activity = A/B, where A is the number of spores on the sample present with nematodes and B is the number on the control side. The experiments were performed with five parallels and repeated three times.
Moreover, the morphology of B16 bacteria at different times was observed by SEM examination. The bacteria were fixed in 4% glutaraldehyde for 2 h and then in 2% osmic acid for 40–60 min. After dehydration in a series of ethanol solutions, the samples were dried, sputter‐coated with gold and examined under SEM (Philips XL30) in the environmental mode operating at 10–20 kV.
Transcriptome analysis of B. nematocida B16 in response to C. elegans
RNA extraction, RNA‐seq library construction and high‐throughput sequencing
C. elegans induced sporulation in B. nematocida B16 at 18 h (B16‐2), and the number of spores reached the maximum difference when induced at 34 h (B16‐5). To address the differential gene expression, we separately selected the bacterial samples induced by C. elegans at 18 h (B16C‐2) and 34 h (B16C‐5). B16 bacteria without C. elegans induction were used as controls. Total RNA was extracted using TRIzol, and RNA quality was measured using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano Kit (Agilent Technologies, Beijing, China). Equal amounts of RNA from each sample were used to construct the cDNA library with the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (cat: E7420) according to the manufacturer’s instructions. mRNA was enriched by removing rRNA with the Ribo‐Zero™ Magnetic Kit (Epicentre). Then, the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse‐transcribed into cDNA with random primers. Second‐strand cDNA was synthesized by DNA polymerase I, RNase H, dNTPs and buffer. Then, the cDNA fragments were purified with a QiaQuick PCR extraction kit and end‐repaired; a poly(A) was added and the fragments were ligated to Illumina sequencing adapters. The ligation products were size‐selected by agarose gel electrophoresis, PCR amplified and sequenced using the Illumina HiSeqTM 4000 System with the 2 × 150 bp paired‐end read module by Gene Denovo Biotechnology Co. (Guangzhou, China). Transcriptome assembly, transcriptome characterization and differential gene expression analysis were carried out according to standard procedures.
Differential gene expression analysis
For each sample, the reads were mapped to the assembled transcriptome using Bowtie2 (version: 2.2.6) to obtain overall transcript expression values (Tan and Shapira, 2011), and the transcript expression was calculated and normalized to RPKM (reads per kilobase million) (Zhang, 2008). Differential transcript expression was performed by comparing each sample to the other sample. The calculations were performed with the edgeR package (version: 3.10.2) with default parameters (Niu et al, 2012). Transcripts with a fold change ≥ 2 and a false discovery rate (FDR) < 0.05 were considered significant DEGs (differentially expressed genes). The lists of differentially expressed genes for each sample were analysed for enrichment of Gene Ontology categories using BLAST2GO (version: 2.3.5), and the terms were deemed significant when FDR < 0.05. Based on the KEGG database annotation of unigenes, the differentially expressed genes for each comparison were analysed for pathway enrichment, and pathways with FDR < 0.05 were considered significant enrichment pathways.
Morpholine import assayed by GC
To test whether morpholine molecules can penetrate the cell membrane properly, solid‐phase microextraction (SPME) in combination with GC‐MS was used to collect and assay the quantity of morpholine molecules in B16 cells. Briefly, freshwater agar and LB agar plates were prepared in Petri dish lids. Lids containing LB agar medium were inoculated with a bacterial lawn (5 × 5 cm) and incubated for 2 days at 37°C. The inoculated lid was inverted on top of another lid without medium but containing a drop of 100 ppm morpholine suspension in the centre. The two lids were sealed with air‐permeable adhesive tape; after 8 h of acting on bacteria, the morpholine molecule was removed, and the bacteria were collected. The morpholine molecule was collected and identified by comparing the mass with the equivalent standard morpholine product. The bacterium B. subtilis 168 without nematocidal activities was used as a comparison. The nematode food E. coli OP50 was used as a negative control.
Molecular simulations
Based on the results of transcriptome analysis, proteins with significant differences in the expression levels of genes concerned with sporulation were selected for docking simulations. The protein structures for KinA, KinE, RapA, Spo0E and Spo0A (2VLG, 3D36, 4I9E, 2C0S and 1LQ1) were downloaded from the Protein Data Bank (http://www.rcsb.org/). The file for the VOC morpholine was generated by Visualizer Studio 3.1 molecular docking to investigate the interactions between the VOCs and proteins by using AutoDock 4.2 (Autodock Molecular Graphics Laboratory, the Scripps Research Institute, La Jolla, CA, USA) (Morris et al., 2009). Parameters, including the Lamarckian genetic algorithm (LGA) with a population size of 100 individuals, a maximum of 2 500 000 energy evaluations and a maximum of 27 000 generations, were employed during molecular docking, and all other parameters were set as default.
The MD simulations were carried out using the gromacs 4.5.4 package (Van Der Spoel et al., 2005). Molecular topologies of morpholine for gromacs (Gromacs Science For Life Laboratory, Stockholm University and KTH, Stockholm, Sweden) were generated using the prodrg2 online server (van Aalten et al., 1996). The simulations for the system of morpholine‐protein were performed for 5 ns using the Gromos96 G53A1 force field in combination with the extended simple point charge (SPCE) water model. System stability was verified by the root‐mean‐square deviation (RMSD). The simulation results were visualized using visual molecular dynamics (VMD) and DSV. All simulations were performed on the Lenovo Think Station T910 (Lenovo, Hong kong, China).
Plasmid construction, subcloning, heterologous expression and overexpression of RapA and Spo0E
The 1.1 kb gene rapA from B. nematocida was amplified by PCR with a SalI‐linked sense primer, 5‐ACGCGTCGACATGGAGCAATTAATTCCGTC‐3, and an XhoI‐linked antisense primer, 5‐CCGCTCGAGAATTTCATATAAACACTCTC‐3. A second gene, spo0E of 256 bp, was amplified by the following primers with BamHI and HindIII sites respectively: 5‐CGCGGATCCATGGGCGGTTCTTCTGAAC‐3 and 5‐CCCAAGCTTTTTATTTGCATCATATGCTG‐3. The PCR products were purified from agarose gels and ligated into the pMD19‐T vector (Promega) to generate the pMD19‐rapA or pMD19‐spo0E plasmids respectively. After digestion with SalI and XhoI or BamHI and HindIII, the rapA or spo0E fragments were inserted into the bacterial expression vector pET‐32a. Transformed cells were then grown at 37°C in Luria–Bertani (LB) medium supplemented with kanamycin (50 µg/ml) to a cell density of A660 = 0.4–0.6. The protein expression in E. coli BL21 was induced by 1.0 mM IPTG (Sigma), and incubation was continued for 12 h at 25°C with shaking at 260 rpm. The expressed proteins were obtained from inclusion bodies and purified using a protocol for 6 × His‐tagged protein purification by Ni‐NTA affinity chromatography according to a pET system manual. The correct conformations of the purified proteins were regenerated according to the protocol of Yokoyama et al. (2002) and dissolved in renaturation buffer at 4°C. The renaturation buffer was readjusted to pH 7.0 using Tris–HCl buffer, and the dephosphorylation activities of the purified proteins were determined. The purified recombinant proteins were sent to the Laboratory for Monoclonal Antibodies at Yunnan University to produce the mouse polyclonal antibody. The construction process of overexpression was very similar to that of heterologous expression (Supporting information).
Dephosphorylation assay
To test the dephosphorylation activities of the proteins RapA and Spo0E, the spo0A gene was also heterologously expressed by using the same method. Primers OA5NcoI (CCACCATGGGGGAGGAAGAAACGTGGAG) and OA3XhoI (CTCCTCGAGACTGCTGGCATTTCCGCTGACC) were used to amplify the spo0A gene, which was cloned into the pET32a expression vector. The Spo0A protein and its phosphorylated form (Spo0A ~ P) were also purified as previously described (21). Dephosphorylation assays using purified Spo0A ~ P were carried out as previously described (Stephenson and Perego, 2002). First, Spo0A was phosphorylated in a reaction mixture with ATP for 45 min at room temperature (Grimshaw et al., 1998). Then, the dephosphorylation activities of RapA were assayed by adding RapA or RapA treated with morpholine to the reactions. Meanwhile, the treatment of Spo0A ~ P with morpholine served as a negative control. The reactions were stopped at different times by adding SDS loading buffer and immediately frozen in a dry ice–ethanol bath. The remaining Spo0A ~ P activities were analysed by 15% SDS‐PAGE, followed by PhosphorImager quantitation by ImageQuant software (General Electric Company, Boston, MA, USA). The dephosphorylation activities of Spo0E were measured using the same method.
Conflict of interest
None declared.
Supporting information
Fig. S1. Process of gene knockout. (A) Use of the integration vector pCP115 to construct a knockout mutation in a hypothetical open reading frame orfA. (B) PCR analysis of spo0A mutant. M, Molecular marker; lane 1, B16g; lane 2, the mutant transformants B16ks‐3; lane 3, B16ks‐9; lane 4, B16ks‐10. (C) Agarose gel electrophoresis analysis of PCR products for analysis of gerD mutant. M, Molecular marker; lane 1, B. nematocida strain B16g; lane 2‐5: the mutant transformants B16kg‐4; B16kg‐7; B16kg‐8; B16kg‐10.
Fig. S2. Process of gene complementation. (A) Physical map of the vector pHY300‐plk used to construct complementary plasmids; (B) Target gene containing promoter and the ORF inserted into the vector pHY300‐plk; (C) Validation of the plasmid construction and complementary transformants. M, Molecular marker; lane1, pHYspo0A digestion product; lane 2, pHYgerD digestion product; lane 3, PCR product of the transformant B16cs‐2; lane 4, PCR product of the transformant B16cg‐4.
Fig. S3. Sporulation analysis on different strains of B. nematocida by SEM examination. (A) Wild type strain B16g cultured for 48h; (B) spo0A knockout strain B16ks‐9 cultured for 5 day; (C) gerD knockout strain B16kg‐8 cultured for 48h; (D) Complementary strain B16cs‐2 cultured for 48h.
Fig. S4. Construction and verification of overexpression mutants for RapA and Spo0E. (A) Working diagram of how an ectopic integration vector inserts a hypothetical open reading frame, orfA, into a target locus on the chromosome. (B) Physical mapping of the overexpression vector pAX01 in B. nematocida. The cassette was integrated by double recombination between the plasmid and chromosomal lacA sequence. (C) Transformants validated by PCR: lane M represents DNA marker (DL5000), lanes 1 and 5 represent PCR amplification from the wild‐type strain B16 as the template (negative control), and lanes 2‐4 and 5‐9 represent PCR results from the genomic DNA templates of three transformants (OSB16‐3, OSB16‐2 and OSB16‐1) overexpressing Spo0E and four transformants overexpressing RapA (ORB16‐4, ORB16‐3, ORB16‐2 and ORB16‐1), respectively. D. Western blot analysis of B. nematocida strains expressing RapA and Spo0E wild type and mutant proteins. Total cell lysates of strains expressing the wild type or mutant rapA and spo0E genes were run on 15% SDS‐PAGE, and the RapA and Spo0E proteins were detected with anti‐RapA and anti‐Spo0E polyclonal antibodies. Lane 1, wild‐type RapA and Spo0E; lanes 2‐3 represent ORB16‐1, OSB16‐1 and the negative control strain CB16.
Fig. S5. Kinetics of colonization of C.elegans by constant exposure to B.nematocida. (A) Colonization categories: A. the ‘full’ scored worms; B. the ‘partial’ scored worms; C. the ‘undetectable’ worms. (B) Colonization of the worms by different bacterial samples.
Table S1. Strains and plasmids.
Table S2. Gene‐specific primers used in this study.
Table S3. The kinds and amounts of compounds produced from bacteria B16 in different states.
Table S4. The types and quantities of fatty acids of B. nematocida B16 cells.
Table S5. The types and quantities of fatty acids of 168 cells.
Vidoe S1. In the video, the Eppendorf tubes left, right and below were put into B. nematocida B16 suspension of vegetative cells, pure spores and sporulating cells, respectively. One minute after the nematodes were added to the central of the plate, we observed that most of the nematodes moved toward the below tube containing B16 of sporulating cells.
Acknowledgements
We wish to thank Prof. Daniel R. Zeigler for providing the plasmid pCP115, and Dr. Qinggang Guo Dr. Qinggang in Hebei Academy of Agricultural and Forestry Sciences for providing the plasmid pHY300PLK.
This work was supported by the projects from the National Natural Science Foundation Programme of the People’s Republic of China (31570120, 31100104), by Innovation Scientists and Technicians Troop Construction Projects (Sustainable Utilization of Energy Microbial Resources) of Henan Province and by the programme for Science & Technology Innovation Talents in Universities of Henan Province, HASTIT (17HASTIT041).
Microbial Biotechnology (2020) 13(3), 683–705
Funding information
This work was supported by the projects from the National Natural Science Foundation Programme of the People’s Republic of China (31570120, 31100104), by Innovation Scientists and Technicians Troop Construction Projects (Sustainable Utilization of Energy Microbial Resources) of Henan Province and by the programme for Science & Technology Innovation Talents in Universities of Henan Province, HASTIT (17HASTIT041).
References
- Aballay, A. , Yorgey, P. , and Ausubel, F.M. (2000) Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans . Curr Biol 10: 1539–1542. [DOI] [PubMed] [Google Scholar]
- Alegado, R.A. , and Tan, M.W. (2008) Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol 10: 1259–1273. [DOI] [PubMed] [Google Scholar]
- Bargmann, C.I. , Hartwieg, E. , and Horvitz, H.R. (1993) Odorant‐selective genes and neurons mediate olfaction in C. elegans . Cell 74: 515–527. [DOI] [PubMed] [Google Scholar]
- Bird, D.M. , Jones, J.T. , Opperman, C.H. , Kikuchi, T. , and Danchin, E.G. (2015) Signatures of adaptation to plant parasitism in nematode genomes. Parasitology 142(Suppl 1): S71–8S84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisguérin, P. , Deshayes, S. , Gait, M.J. , O'Donovan, L. , Godfrey, C. , Betts, C.A. , et al (2015) Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv Drug Deliv Rev 87: 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, L.A. , McLean, S. , Poole, R.K. , and Fukuto, J.M. (2011) The diversity of microbial responses to nitric oxide and agents of nitrosative stress close cousins but not identical twins. Adv Microb Physiol 59: 135–219. [DOI] [PubMed] [Google Scholar]
- Brown, A.M. , Howe, D.K. , Wasala, S.K. , Peetz, A.B. , Zasada, I.A. , and Denver, D.R. (2015) Comparative genomics of a plant‐parasitic nematode endosymbiont suggest a role in nutritional symbiosis. Genome Biol Evol 7: 2727–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall, A. , and Pirofski, L.A. (2000) Host‐pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infec Immun 68: 6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.Y. , Tian, X.L. , Wang, Y.S. , Lin, R.M. , Mao, Z.C. , Chen, N. , et al (2013) Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci Rep ‐UK 3: 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington, J. (2003) Regulation of endospore formation in Bacillus subtilis . Nat Rev Microbiol 1: 117–126. [DOI] [PubMed] [Google Scholar]
- Fukuto, J.M. , Carrington, S.J. , Tantillo, D.J. , Harrison, J.G. , Ignarro, L.J. , Freeman, B.A. , et al (2012) Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Cheml Res Toxicol 25: 769–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, M.Y. , Mekhedov, S. , Puigbo, P. , Smirnov, S. , Wolf, Y.I. , and Rigden, D.J. (2012) Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation‐specific genes. Environ Microbiol 14: 2870–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh, P.S. , and Rai, R.V. (2016) Inhibition of quorum‐sensing‐controlled virulence factors of Pseudomonas aeruginosa by Murraya koenigii essential oil: a study in a Caenorhabditis elegans infectious model. J Med Microbiol 65: 1528–1535. [DOI] [PubMed] [Google Scholar]
- Garigan, D. , Hsu, A.L. , Fraser, A.G. , Kamath, R.S. , Ahringer, J. , and Kenyon, C. (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat‐shock factor and bacterial proliferation. Genetics 161: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin, D.A. , Sifri, C.D. , Mylonakis, E. , Qin, X. , Singh, K.V. , Murray, B.E. , et al (2001) A simple model host for identifying Gram‐positive virulence factors. Proc Natl Acad Sci USA 98: 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerth, K. , Metzger, R. , and Reichenbach, H. (1993) Induction of myxospores in Stigmatella aurantiaca (Myxobacteria): inducers and inhibitors of myxospore formation, and mutants with a changed sporulation behavior. J Gen Microbiol 139: 865–871. [Google Scholar]
- Gonzalez‐Pastor, J.E. , Hobbs, E.C. , and Losick, R. (2003) Cannibalism by sporulating bacteria. Science 301: 510–513. [DOI] [PubMed] [Google Scholar]
- Grenha, R. , Rzechorzek, N. J. , Brannigan, J.A. , de Jong, R.N. , Ab, E. , Diercks, T. , et al (2006) Structural characterization of Spo0E‐like protein‐aspartic acid phosphatases that regulate sporulation in bacilli. J Biol Chem 281: 37993–38003. [DOI] [PubMed] [Google Scholar]
- Grimshaw, C.E. , Huang, S. , Hanstein, C.G. , Strauch, M.A. , Burbulys, D. , Wang, L. , et al (1998) Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis . Biochemistry 37: 1365–1375. [DOI] [PubMed] [Google Scholar]
- Hamid, M.I. , Hussain, M. , Wu, Y. , Zhang, X. , Xiang, M. , and Liu, X. (2017) Successive soybean‐monoculture cropping assembles rhizosphere microbial communities for the soil suppression of soybean cyst nematode. FEMS Microbiol Ecol 93: fiw222. [DOI] [PubMed] [Google Scholar]
- Hao, F. , Zhao, S. , Dong, H. , Zhang, H. , Sun, L. , and Miao, C. (2010) Nia1 and Nia2 are involved in exogenous salicylic acid‐induced nitric oxide generation and stomatal closure in Arabidopsis . J Integr Plant Biol 52: 298–307. [DOI] [PubMed] [Google Scholar]
- Harwood, C.R. , and Cutting, S.M. (1990) Molecular Biological Methods for Bacillus. Sussex, UK: Wiley. [Google Scholar]
- Huang, Z. , Ni, B. , Jiang, C.Y. , Wu, Y.F. , He, Y.Z. , Parales, R.E. , and Liu, S.J. (2016) Direct sensing and signal transduction during bacterial chemotaxis toward aromatic compounds in Comamonas testosteroni . Mol Microbiol 101: 224–237. [DOI] [PubMed] [Google Scholar]
- Irazoqui, J.E. , and Ausubel, F.M. (2010) 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Caenorhabditis elegans as a model to study tissues involved in host immunity and microbial pathogenesis. Clin Exp Immunol 160: 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, M. , Shuto, Y. , and Watanable, H. (1986) β‐Myrcene, a potent attractant component of pine wood for the pine woodnematode, Bursaphelenchus xylophilus . Agric Biol Chern 50: 1863–1866. [Google Scholar]
- Jia, X. , Wang, J.B. , Rivera, S. , Duong, D. , and Weinert, E.E. (2016) An O2‐sensing stressosome from a Gram‐negative bacterium. Nat Commun 7: 12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, L.T. , Phillips, D.S. , Williams, C.F. , Alyousef, A. , and Baillie, L. (2012) Contribution of spores to the ability of Clostridium difficile to adhere to surfaces. Appl Environ Microb 78: 7671–7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney, D.S. , and Couch, T.L. (1981) Mass production of biological agents for plant disease, weed and insect control In Biological Control in Crop Production BARC Symposium No. 5. Papavizas G. C. (ed.). Totowa, NJ: Allenheld and Osmum, pp. 143–150. [Google Scholar]
- Kenyon, C. , Chang, J. , Gensch, E. , Rudner, A. , and Tabtiang, R. (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Keren, I. , Wu, Y. , Inocencio, J. , Mulcahy, L.R. , and Lewis, K. (2013) Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339: 1213–1216. [DOI] [PubMed] [Google Scholar]
- Kim, D. H. (2015) Signal transduction: a different kind of Toll is in the BAG. Curr Biol 25: R767–769. [DOI] [PubMed] [Google Scholar]
- King, G.M. , and Weber, C.F. (2007) Distribution, diversity and ecology of aerobic CO‐oxidizing bacteria. Nat Rev Microbiol 5: 107–118. [DOI] [PubMed] [Google Scholar]
- Koutsovoulos, G. , Makepeace, B. , Tanya, V.N. , and Blaxter, M. (2014) Palaeosymbiosis revealed by genomic fossils of Wolbachia in a strongyloidean nematode. PLoS Genet 10: e1004397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laaberki, M.H. , and Dworkin, J. (2008) Role of spore coat proteins in the resistance of Bacillus subtilis spores to Caenorhabditis elegans predation. J Bacteriol 190: 6197–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Kim, Y.G. , Kim, M. , Kim, E. , Choi, H. , Kim, Y. , et al (2017) Indole‐associated predator‐prey interactions between the nematode Caenorhabditis elegans and bacteria. Environ Microbiol 19: 1776–1790. [DOI] [PubMed] [Google Scholar]
- Liao, C. , Gao, A. , Li, B. , Wang, M. , and Shan, L. (2017) Two symbiotic bacteria of the entomopathogenic nematode Heterorhabditis spp. against Galleria mellonella . Toxicon 127: 85–89. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , and Imlay, J.A. (2013) Cell death from antibiotics without the involvement of reactive oxygen species. Science 339: 1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel, J.D. , and Kim, D.H. (2014) Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans . Trends Immunol 35: 465–470. [DOI] [PubMed] [Google Scholar]
- Mitreva, M. , Jasmer, D.P. , Zarlenga, D.S. , Wang, Z. , Abubucker, S. , Martin, J. , et al (2011) The draft genome of the parasitic nematode Trichinella spiralis . Nat Genet 43: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir, A. (2006) How do spores germinate? J Appl Microbiol 101: 526–530. [DOI] [PubMed] [Google Scholar]
- Morris, G.M. , Huey, R. , Lindstrom, W. , Sanner, M.F. , Belew, R.K. , Goodsell, D.S. , and Olson, A.J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfin, K.E. , Dillman, A.R. , Foster, J.M. , Bulgheresi, S. , Slatko, B.E. , Sternberg, P.W. , and Goodrich‐Blair, H. (2012) Nematode‐bacterium symbioses–cooperation and conflict revealed in the "omics" age. Biol Bull 223: 85–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfin, K.E. , Lee, M.M. , Klassen, J.L. , McDonald, B.R. , Larget, B. , Forst, S. , et al (2015) Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. MBio 6: e00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, M. , Selin, C. , Brassinga, A.K. , Belmonte, M.F. , Fernando, W.G. , Loewen, P.C. , and de Kievit, T.R. (2015) Pyrrolnitrin and hydrogen cyanide production by Pseudomonas chlororaphis strain PA23 exhibits nematicidal and repellent activity against Caenorhabditis elegans . PLoS ONE 10: e0123184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, W. (1975) The Biology of Free‐living Nematodes. Oxford, UK: Clarendon Press. [Google Scholar]
- Niu, Q. , Huang, X. , Zhang, L. , Xu, J. , Yang, D. , and Zhang, K.‐Q. (2010) A Trojan horse mechanism of bacterial pathogenesis against nematodes. Proc Natl Acad Sci USA 107: 16631–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Q. , Huang, X. , Hui, F. , Huang, S. , Ke, T. , Zhang, K.‐Q. , et al (2012) Colonization of Caenorhabditis elegans by Bacillus nematocida B16, a bacterial opportunistic pathogen. J Mol Microb Biotech 22: 258–267. [DOI] [PubMed] [Google Scholar]
- Niu, Q. , Zheng, H. , Zhang, L. , Qin, F. , Facemire, L. , Zheng, Q. , et al (2015) Knockout of the adp gene related with colonization in Bacillus nematocida B16 using customized transcription activator‐like effectors nucleases. Microb Biotechnol 8: 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Q. , Zhang, L. , Zhang, K. , Huang, X. , Hui, F. , Kan, Y. , et al (2016) Changes in intestinal microflora of Caenorhabditis elegans following Bacillus nematocida B16 infection. Sci Rep‐UK 6: 20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat, M. , Setlow, B. , Daniels, W.B. , Hoover, D. , Papafragkou, E. , and Setlow, P. (2002) Mechanisms of induction of germination of Bacillus subtilis spores by high pressure. Appl Environ Microb 68: 3172–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelczar, P.L. , Igarashi, T. , Setlow, B. , and Setlow, P. (2007) Role of GerD in germination of Bacillus subtilis spores. J Bacteriol 189: 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y.J. , Zhang, X. , Gridina, A. , Chupikova, I. , McCormick, D.L. , Thomas, R.T. , et al (2017) Complementary roles of gasotransmitters CO and H2S in sleep apnea. Proc Natl Acad Sci USA 114: 1413–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal‐Celhay, C. , Bradley, E.R. , and Blaser, M.J. (2012) Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans . BMC Microbiol 12: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J.F. (1950) Factors affecting the germination of thick suspensions of Bacillus subtilis spores in L‐alanine solution. Microbiology 4: 330–338. [DOI] [PubMed] [Google Scholar]
- Powell, J. F. (1951) The sporulation and germination of a strain of Bacillus megatherium . Microbiology 5: 993–1000. [DOI] [PubMed] [Google Scholar]
- Prabhakar, N.R. , and Semenza, G.L. (2012) Gaseous messengers in oxygen sensing. J Mol Med 90: 265–272. [DOI] [PubMed] [Google Scholar]
- Pukkila‐Worley, R. , and Ausubel, F.M. (2012) Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol 24: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae, R. , Iatsenko, I. , Witte, H. , and Sommer, R.J. (2010) A subset of naturally isolated Bacillus strains show extreme virulence to the free‐living nematodes Caenorhabditis elegans and Pristionchus pacificus . Environment Microbiol 12: 3007–3021. [DOI] [PubMed] [Google Scholar]
- Rahme, L.G. , Ausubel, F.M. , Cao, H. , Drenkard, E. , Goumnerov, B.C. , Lau, G.W. , et al (2000) Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA 97: 8815–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffey, R. , Lantorp, K. , Tegnell, A. , and Elgh, F. (2002) Biological weapons and bioterrorism preparedness: importance of public‐health awareness and international cooperation. Clin Microbiol Infec 8: 522–528. [DOI] [PubMed] [Google Scholar]
- Schreiber, F. , Wunderlin, P. , Udert, K.M. , and Wells, G.F. (2012) Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front Microbiol 3: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow, P. (2006) Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101: 514–525. [DOI] [PubMed] [Google Scholar]
- Setlow, P. (2007) I will survive: DNA protection in bacterial spores. Trends Microbiol 15: 172–180. [DOI] [PubMed] [Google Scholar]
- Stephenson, S.J. , and Perego, M. (2002) Interaction surface of the Spo0A response regulator with the Spo0E phosphatase. Mol Microbiol 44: 1455–1467. [DOI] [PubMed] [Google Scholar]
- Tan, M.W. , and Shapira, M. (2011) Genetic and molecular analysis of nematode‐microbe interactions. Cellular Microbiol 13: 497–507. [DOI] [PubMed] [Google Scholar]
- Tinajero‐Trejo, M. , Jesse, H.E. , and Poole, R.K. (2013) Gasotransmitters, poisons, and antimicrobials: it's a gas, gas, gas! F1000prime Rep 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aalten, D.M. , Bywater, R. , Findlay, J.B. , Hendlich, M. , Hooft, R.W. , and Vriend, G. (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aid Mol Des 10: 255–262. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel, D. , Lindahl, E. , Hess, B. , Groenhof, G. , Mark, A.E. , and Berendsen, H.J.C. (2005) GROMACS: fast, flexible, and free. J Comput Chem 26: 1701–1718. [DOI] [PubMed] [Google Scholar]
- Vicente, C.S. , Ikuyo, Y. , Mota, M. , and Hasegawa, K. (2013) Pinewood nematode‐associated bacteria contribute to oxidative stress resistance of Bursaphelenchus xylophilus . BMC Microbiol 13: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Yi, X. , Li, Y.Q. , and Setlow, P. (2011) Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins, which are important in spore germination. J Bacteriol 193: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Du, Y. , Hou, Y.J. , Zhao, Y. , Hsu, C.C. , Yuan, F. , et al (2015) Nitric oxide negatively regulates abscisic acid signaling in guard cells by S‐nitrosylation of OST1. Proc Natl Acad Sci USA 112: 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, J.H. , Robertson, A.P. , Kimber, M.J. , Day, T.A. , and Carlson, S.A. (2016) Intestinal Enterobacteriaceae that protect nematodes from the effects of Benzimidazoles. J Bacteriol Parasitol 7: 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett, J.W. , and Crosson, S. (2017) Atypical modes of bacterial histidine kinase signaling. Mol Microbiol 103: 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong‐Guy, K. , Jin‐Hyung, L. , Moo Hwan, C. , and Jintae, L. (2011) Indole and 3‐indolylacetonitrile inhibit spore maturation in Paenibacillus alvei . BMC Microbiol 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. (2008) Neuronal mechanisms of Caenorhabditis elegans and pathogenic bacteria interactions. Curr Opin Microbiol 11: 257–261. [DOI] [PubMed] [Google Scholar]
- Zheng, Z. , Zheng, J. , Zhang, Z. , Peng, D. , and Sun, M. (2016) Nematicidal spore‐forming Bacilli share similar virulence factors and mechanisms. Sci Rep‐UK 6: 31341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Process of gene knockout. (A) Use of the integration vector pCP115 to construct a knockout mutation in a hypothetical open reading frame orfA. (B) PCR analysis of spo0A mutant. M, Molecular marker; lane 1, B16g; lane 2, the mutant transformants B16ks‐3; lane 3, B16ks‐9; lane 4, B16ks‐10. (C) Agarose gel electrophoresis analysis of PCR products for analysis of gerD mutant. M, Molecular marker; lane 1, B. nematocida strain B16g; lane 2‐5: the mutant transformants B16kg‐4; B16kg‐7; B16kg‐8; B16kg‐10.
Fig. S2. Process of gene complementation. (A) Physical map of the vector pHY300‐plk used to construct complementary plasmids; (B) Target gene containing promoter and the ORF inserted into the vector pHY300‐plk; (C) Validation of the plasmid construction and complementary transformants. M, Molecular marker; lane1, pHYspo0A digestion product; lane 2, pHYgerD digestion product; lane 3, PCR product of the transformant B16cs‐2; lane 4, PCR product of the transformant B16cg‐4.
Fig. S3. Sporulation analysis on different strains of B. nematocida by SEM examination. (A) Wild type strain B16g cultured for 48h; (B) spo0A knockout strain B16ks‐9 cultured for 5 day; (C) gerD knockout strain B16kg‐8 cultured for 48h; (D) Complementary strain B16cs‐2 cultured for 48h.
Fig. S4. Construction and verification of overexpression mutants for RapA and Spo0E. (A) Working diagram of how an ectopic integration vector inserts a hypothetical open reading frame, orfA, into a target locus on the chromosome. (B) Physical mapping of the overexpression vector pAX01 in B. nematocida. The cassette was integrated by double recombination between the plasmid and chromosomal lacA sequence. (C) Transformants validated by PCR: lane M represents DNA marker (DL5000), lanes 1 and 5 represent PCR amplification from the wild‐type strain B16 as the template (negative control), and lanes 2‐4 and 5‐9 represent PCR results from the genomic DNA templates of three transformants (OSB16‐3, OSB16‐2 and OSB16‐1) overexpressing Spo0E and four transformants overexpressing RapA (ORB16‐4, ORB16‐3, ORB16‐2 and ORB16‐1), respectively. D. Western blot analysis of B. nematocida strains expressing RapA and Spo0E wild type and mutant proteins. Total cell lysates of strains expressing the wild type or mutant rapA and spo0E genes were run on 15% SDS‐PAGE, and the RapA and Spo0E proteins were detected with anti‐RapA and anti‐Spo0E polyclonal antibodies. Lane 1, wild‐type RapA and Spo0E; lanes 2‐3 represent ORB16‐1, OSB16‐1 and the negative control strain CB16.
Fig. S5. Kinetics of colonization of C.elegans by constant exposure to B.nematocida. (A) Colonization categories: A. the ‘full’ scored worms; B. the ‘partial’ scored worms; C. the ‘undetectable’ worms. (B) Colonization of the worms by different bacterial samples.
Table S1. Strains and plasmids.
Table S2. Gene‐specific primers used in this study.
Table S3. The kinds and amounts of compounds produced from bacteria B16 in different states.
Table S4. The types and quantities of fatty acids of B. nematocida B16 cells.
Table S5. The types and quantities of fatty acids of 168 cells.
Vidoe S1. In the video, the Eppendorf tubes left, right and below were put into B. nematocida B16 suspension of vegetative cells, pure spores and sporulating cells, respectively. One minute after the nematodes were added to the central of the plate, we observed that most of the nematodes moved toward the below tube containing B16 of sporulating cells.


