Summary
Listeria monocytogenes is the causative agent of the foodborne illness listeriosis, which can result in severe symptoms and death in susceptible humans and other animals. L. monocytogenes is ubiquitous in the environment and isolates from food and food processing, and clinical sources have been extensively characterized. However, limited information is available on L. monocytogenes from wildlife, especially from urban or suburban settings. As urban and suburban areas are expanding worldwide, humans are increasingly encroaching into wildlife habitats, enhancing the frequency of human–wildlife contacts and associated pathogen transfer events. We investigated the prevalence and characteristics of L. monocytogenes in 231 wild black bear capture events between 2014 and 2017 in urban and suburban sites in North Carolina, Georgia, Virginia and United States, with samples derived from 183 different bears. Of the 231 captures, 105 (45%) yielded L. monocytogenes either alone or together with other Listeria. Analysis of 501 samples, primarily faeces, rectal and nasal swabs for Listeria spp., yielded 777 isolates, of which 537 (70%) were L. monocytogenes. Most L. monocytogenes isolates exhibited serotypes commonly associated with human disease: serotype 1/2a or 3a (57%), followed by the serotype 4b complex (33%). Interestingly, approximately 50% of the serotype 4b isolates had the IVb‐v1 profile, associated with emerging clones of L. monocytogenes. Thus, black bears may serve as novel vehicles for L. monocytogenes, including potentially emerging clones. Our results have significant public health implications as they suggest that the ursine host may preferentially select for L. monocytogenes of clinically relevant lineages over the diverse listerial populations in the environment. These findings also help to elucidate the ecology of L. monocytogenes and highlight the public health significance of the human–wildlife interface.
Listeria monocytogenes has been extensively studied in food‐processing environments; however, key elements regarding its ecology and reservoirs in the natural environment remain largely uncharacterized. We sampled black bears in the southeastern United States from 2014 to 2017, recovering 777 Listeria spp. isolates, of which most were L. monocytogenes of serotypes commonly encountered in human listeriosis. Our results suggest a potential role for black bears in dissemination and adaptation of L. monocytogenes, with the greatest public health implications in regions of direct overlap between black bears and humans. Urban areas are expanding in the US and other nations, with humans increasingly encroaching into wildlife habitats and enhancing the frequency of human‐wildlife contacts and associated pathogen transfer events. Elucidation of L. monocytogenes harbored in wildlife species will address major gaps in our understanding of the ecology of this pathogen, and will better inform the development of strategies to reduce the risk for listeriosis.

Introduction
Listeria monocytogenes is the causative agent of the severe foodborne disease listeriosis, which can result in stillbirths, meningitis, septicaemia and death in humans and other animals (Painter and Slutsker, 2007; Scallan et al., 2011). Listeria monocytogenes is considered ubiquitous in nature, having been isolated from soil, water and vegetation in diverse geographic regions (Welshimer, 1968; Sauders et al., 2012; Vivant et al., 2013; Cooley et al., 2014; Ferreira et al., 2014). Listeria monocytogenes in food and food processing environments has been extensively studied (Kathariou, 2002; Gandhi and Chikindas, 2007; Carpentier and Cerf, 2011; Ferreira et al., 2014), but the ecology of this pathogen in the natural environment remains poorly understood (Vivant et al., 2013).
Even though there is a notable dearth of information on the prevalence of L. monocytogenes and other Listeria spp. in wildlife (Ivanek et al., 2006; Wesley, 2007; Czuprynski et al., 2010; Chlebicz and Śliżewska, 2018), one of the earliest isolations of L. monocytogenes was from a lethal case of listeriosis in wild gerbils (Pirie, 1927). The gerbil (Meriones unguiculatus) was subsequently shown to be a valuable model for Listeria rhombencephalitis and other listeriosis outcomes (Blanot et al., 1997; Disson et al., 2009). However, while L. monocytogenes is capable of causing severe disease and death among wildlife (Wesley, 2007), infected mammalian species such as red deer (Cervus elaphus) and wild boar (Sus scrofa) may remain asymptomatic (Sasaki et al., 2013; Gnat et al., 2015; Weindl et al., 2016). Importantly, analysis of L. monocytogenes from healthy wild bird populations revealed strain subtypes identical to those detected in foods (Hellström et al., 2008). Although limited, these results suggest a larger, currently uncharacterized ecological role for wildlife as reservoir and vehicle for L. monocytogenes.
American black bears (Ursus americanus) are omnivorous habitat generalists (Karelus et al., 2017; Moeller et al., 2017). In the United States, they use a wide range of habitats and food sources in forested, rural and residential areas. Increasing black bear habitat loss and forest fragmentation due to urban and suburban development have resulted in increased human–bear interactions, as evidenced by observations and property damage (North Carolina Wildlife Resources Commission, 2019). Increased frequency of human–wildlife interactions has major human and animal health implications, as it may lead to greater rates of zoonotic or anthroponotic transmission of pathogens (Patz et al., 2000; Daszak et al., 2001).
While the prevalence of several zoonotic pathogens in black bears has been investigated, the focus has been primarily on viruses and vector‐borne pathogens (Binninger et al., 1980; Ruppanner et al., 1982; Gage et al., 1995; Bronson et al., 2014; Stephenson et al., 2015). A substantial knowledge gap currently exists regarding the incidence of other human pathogens in bear populations, especially in urban and suburban regions of the United States. There is noticeably limited information on the role of black bears as potential reservoirs or vehicles for human foodborne bacterial pathogens such as Salmonella, Campylobacter and Listeria.
In this study, samples obtained in the course of a wildlife mobility and demographic analysis of black bears in the southeastern United States were analysed for L. monocytogenes and other Listeria spp. Samples were obtained through the live capture and monitoring of black bear populations in three separate study sites in North Carolina (NC), Georgia (GA) and Virginia (VA). We determined the prevalence of L. monocytogenes and other Listeria spp. in various types of samples from black bears, including faeces, rectal swabs, nasal swabs and gastrointestinal tracts. To identify important subtypes of Listeria and assess their environmental resilience via previously identified adaptations, we analysed the distribution of different L. monocytogenes serotypes and the incidence of resistance to heavy metals (cadmium and arsenic) and the quaternary ammonium disinfectant benzalkonium chloride (BC). These results will help address a key knowledge gap in the ecology of L. monocytogenes in the natural environment.
Results
Listeria monocytogenes was frequently isolated from black bears in the southeastern United States. We analysed samples from 231 black bear capture events, corresponding to 183 animals. Of the 183 bears, 142 were captured and sampled once, while 38 were sampled multiple times. Most (203/231; 88%) of the capture events involved bears from urban or suburban areas in Asheville, NC (Fig. 1). Thirteen animals were captured in rural areas in GA, and 15 were in temporary captivity at Virginia Tech’s Black Bear Research Center (VT‐BBRC), Blacksburg, VA. Sample types obtained from the live animals included faeces, rectal swabs and nasal swabs.
Figure 1.
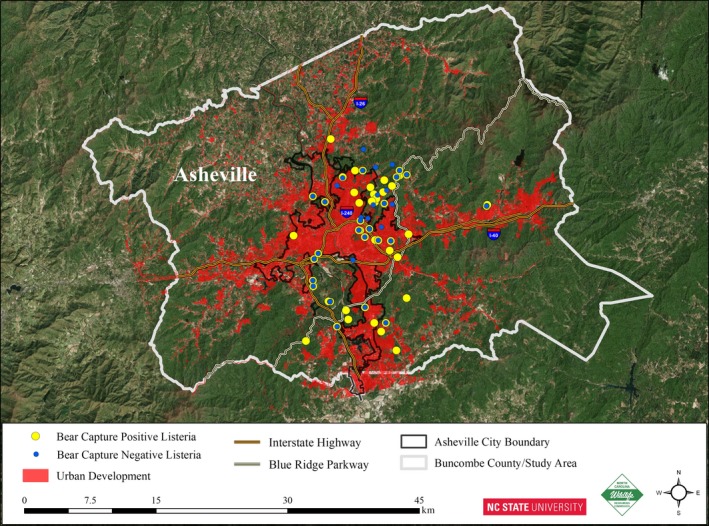
Black bear capture locations in and around Asheville, NC. Yellow and blue symbols represent bears that were positive and negative, respectively, for Listeria. ArcMap 10.5.1 (http://desktop.arcgis.com/en/) was used to generate the map. ArcGIS Desktop: Release 10. Redlands, CA: Environmental Systems Research Institute.
Listeria colonies were identified based on typical morphology on Modified Oxford agar supplemented with Difco™ Modified Oxford Antimicrobic Supplement (MOX) (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Of the 231 black bear capture events, 55% (127/231) yielded Listeria in one or more sample types (Fig. 2). Putative Listeria isolates that failed to yield any amplicons with multiplex PCR serotyping (Doumith et al., 2004) were not detected, and all non‐haemolytic isolates yielded the prs band only. L. monocytogenes was detected in 105 (45%) of the black bear capture events, either alone (74/231; 32%) or, less commonly, together with other Listeria spp. (31/231; 13%) (Fig. 2). Only 22 captures (10%) yielded exclusively Listeria spp. other than L. monocytogenes (Fig. 2). Among the Listeria‐positive black bears, the likelihood of animals being positive for L. monocytogenes was significantly higher than for other Listeria spp. (P < 0.001). The rate of recovery of L. monocytogenes and other Listeria spp. was similar across the three study sites, ranging between 40% and 56% (Fig. 2).
Figure 2.
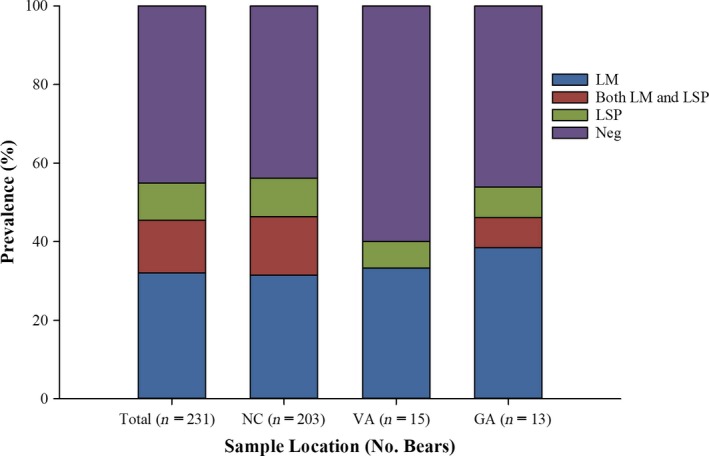
The prevalence of L. monocytogenes and other Listeria spp. in American black bears. Samples were collected in 2014‐2017 from bears in North Carolina (NC), Georgia (GA) and Virginia (VA). Inset: LM, L. monocytogenes; LSP, Listeria spp. other than L. monocytogenes; Neg, Listeria‐negative animals.
Analysis of the data from the NC population that was monitored from 2014 to 2017 failed to reveal noticeable temporal trends in L. monocytogenes recovery. The percentage of black bear capture events yielding L. monocytogenes, alone or in conjunction with other Listeria spp., remained constant throughout all 4 years of the study, varying from 42% to 49% (Fig. 3). The percentage of captures negative for L. monocytogenes but positive for other Listeria spp. also remained stable, varying between ~ 7‐8% in years 1 and 4, and ~10–13% in years 2 and 3 (Fig. 3). Each year, capture events yielding L. monocytogenes were significantly more common than those yielding other Listeria spp. (P < 0.01).
Figure 3.
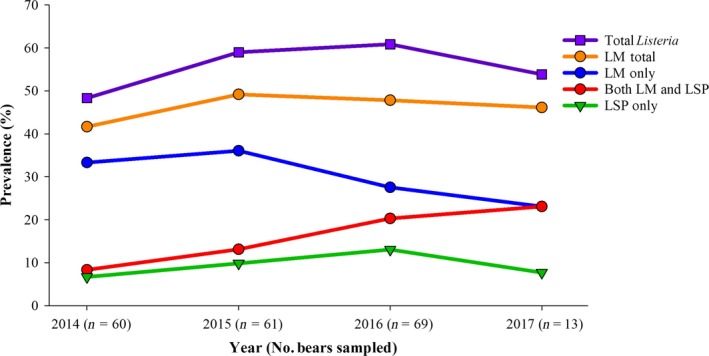
Annual prevalence of L. monocytogenes and other Listeria spp. in American black bears in NC. Inset designations are as in legend to Fig. 2.
L. monocytogenes serotype 1/2a (or 3a) and the 4b complex (4b, 4d, 4e) predominated, with significant representation of the variant IVb‐v1 multiplex PCR profile
A total of 777 isolates were characterized over the course of the study. Employment of the multiplex PCR scheme of Doumith et al. (2004) to determine serotype designations identified several PCR profiles among these isolates (Fig. 4). Based on haemolytic activity and multiplex PCR results, 537/777 (approx. 70%) of the isolates were L. monocytogenes, with the remaining 240 being members of other Listeria spp. (Fig. 5). Serotype 1/2a (or 3a) (hereafter designated ‘1/2a’) was the most prevalent serotype (57%), followed by the serotype 4b complex (4b, 4d or 4e, hereafter designated ‘4b’), which accounted for approximately 33% of the total L. monocytogenes isolates (Fig. 5). Interestingly, almost half of the serotype 4b isolates (82/179; 46%) exhibited the variant IVb‐v1 multiplex PCR profile (Huang et al., 2011; Leclercq et al., 2011; Lee et al., 2012a,b) (Fig. 5). Examination of the locations for capture events yielding L. monocytogenes serotypes 1/2a and 4b in NC showed a relatively even distribution across the capture locations with no obvious geographic bias (Fig. 6A, B). The distribution largely reflected the overall pattern of the capture locations for the Listeria‐positive animals (Fig. 1). Similar findings were obtained with the locations of capture events yielding serotype 4b strains with the typical multiple PCR profile in comparison to those with profile IVb‐v1 (Fig. 6B).
Figure 4.
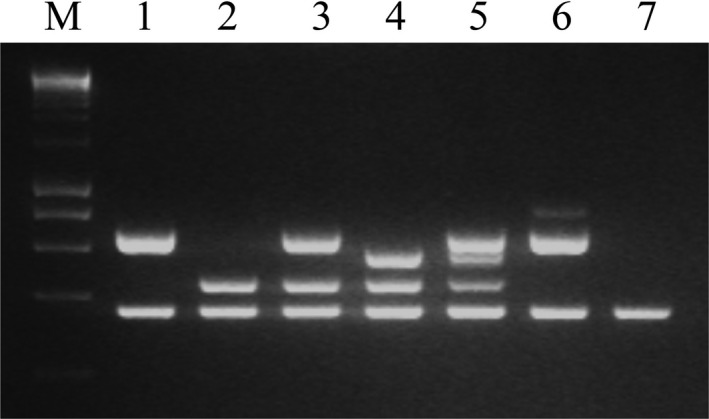
Multiplex PCR‐based serotype designations for L. monocytogenes. The multiplex PCR protocol was based on Doumith et al. (2004). Lanes: 1‐7, black bear‐derived isolates SKB31, SKB781, SKB102, SKB647, SKB341, SKB107 and SKB782 corresponding to serotypes 1/2a, 1/2b, novel multiplex PCR profile with amplicons typical both for serotypes 1/2a and 1/2b (1/2a‐1/2b), 4b, IVb‐v1, 1/2c and lineage III (only prs amplicon), respectively. M, molecular weight marker (HyperLadder 1kb; Bioline, Boston, MA, USA).
Figure 5.
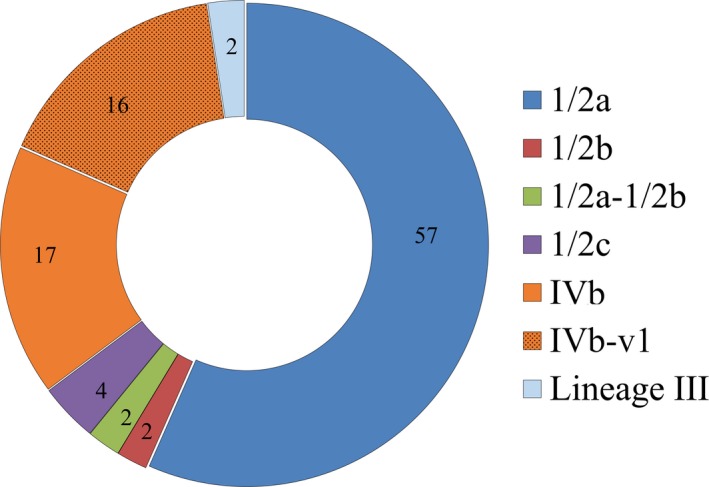
The prevalence of different serotypes and lineage III among L. monocytogenes from American black bears. Numbers indicate the percentage of isolates with a specific multiplex PCR designation. Serotype and putative lineage III designations were determined for 537 isolates by multiplex PCR as described in Experimental Procedures. Bracket indicates total prevalence of serotype 4b (isolates with multiplex PCR profile 4b, indicated as IVb in inset, as well as those with multiplex PCR profile IVb‐v1).
Figure 6.
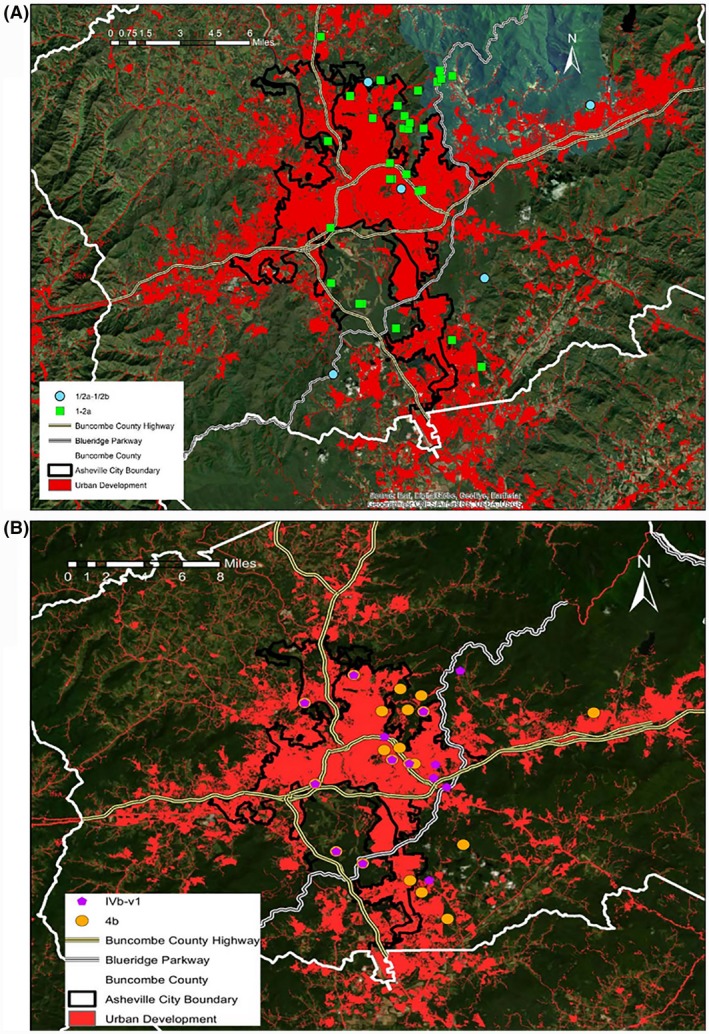
Capture locations of bears yielding isolates with specific multiplex PCR profiles. (A), isolates with profile typical for serotype 1/2a and novel profile with amplicons typical both for serotype 1/2a and serotype 1/2b (1/2a‐1/2b). (B), isolates with profile typical for serotype 4b (designated IVb in the inset) and with variant profile IVb‐v1. Mapping software used was as in legend to Fig. 1.
Serotypes 1/2b and 1/2c were rarely encountered. Isolates of serotype 1/2b accounted for only 11/537 (~2%) of the L. monocytogenes isolates and were obtained from just five capture events in NC, GA and VA, while the 21 serotype 1/2c isolates accounted for ~ 4% of L. monocytogenes and were from nine captures, all in NC (Fig. 5). Lineage III isolates were also uncommon, obtained from only eight captures in NC, GA and VA and accounting for 13 (~2%) of the 537 L. monocytogenes isolates (Fig. 5). Multiplex PCR of these isolates for serotype designations yielded only the prs band (Fig. 4), and all isolates were PCR positive with L. monocytogenes‐specific primers for hly (data not shown).
The multiplex PCR scheme also revealed a novel multiplex PCR profile in 12 isolates, obtained from six captures in NC in 2014 (n = 4) and 2016 (n = 2). This novel profile (designated 1/2a‐1/2b) exhibited a combination of serotype 1/2a and 1/2b amplicons (Fig. 4). All 12 isolates with this profile were PCR positive with L. monocytogenes‐specific primers for hly (data not shown). Capture events yielding isolates with this novel multiplex PCR profile were from several diverse locations (Fig. 6A).
We failed to identify haemolytic isolates that yielded only the prs band but were negative for hly, as would be expected for L. seeligeri, and on blood agar, none of the isolates exhibited the pronounced haemolysis typical for L. ivanovii. As mentioned earlier, we did not identify non‐haemolytic isolates with Listeria‐typical colony morphology on MOX but lacking the prs band, as may be the case for some Listeria spp. that were identified relatively recently (den Bakker et al., 2014) (J. Niedermeyer and S. Kathariou, unpublished).
Temporal stability of dominant serotypes and variant multiplex profile IVb‐v1
The prevalence of the leading serotypes 1/2a and 4b remained high during each year of the study, without significant differences in yearly serotype recovery. Serotype 1/2a was encountered among 49–67% of the L. monocytogenes isolates, while serotype 4b prevalence ranged between 24% and 41% annually (Fig. 7). Recovery of serotype 4b isolates with the IVb‐v1 multiplex PCR profile remained consistent over the study period, ranging between 15% and 17% of the L. monocytogenes isolates annually (Fig. 7). Listeria spp. other than L. monocytogenes accounted for 22–38% of isolates annually (data not shown).
Figure 7.
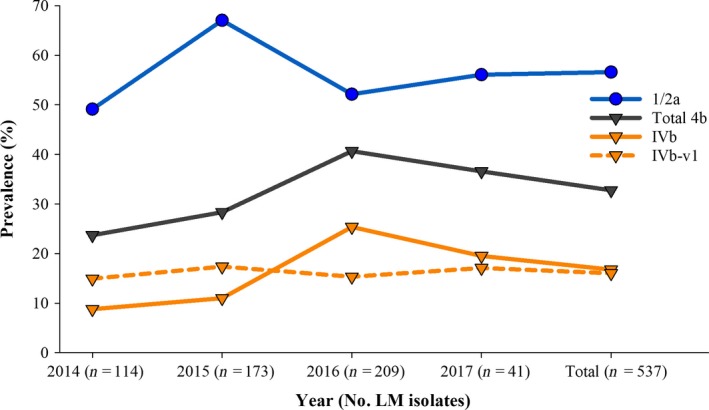
Annual prevalence of the major serotypes of L. monocytogenes recovered from American black bears.
Sample type and enrichment phase impact recovery of listeriae
Listeria was recovered significantly more frequently from primary than secondary enrichments (diff = 0.05, SE = 0.02, t = 2.39, P = 0.04). The listerial community composition (i.e. relative abundance of L. monocytogenes, other Listeria spp. and different serotypes of L. monocytogenes) was ~ 75% identical between primary and secondary enrichment.
Analysis of faeces, rectal swabs and nasal swabs indicated that sample type had the potential to influence Listeria recovery. Recovery of L. monocytogenes was similar between faecal samples and rectal swabs (35% and 33% respectively) and significantly higher than from nasal swabs, of which 19% were positive (rectal swabs: diff = 0.1, SE = 0.3, t = −3.1, P = 0.002, faeces: diff = 0.07, SE = 0.04, t = −2.1, P = 0.04) (Fig. 8). Interestingly, however, the likelihood of encountering L. monocytogenes as opposed to other Listeria spp. was significantly higher in nasal swabs than in faeces (P = 0.0005) or rectal swabs (P = 0.02). Furthermore, the prevalence of samples yielding only Listeria spp. other than L. monocytogenes was higher in faeces and rectal swabs (16% and 8% respectively) than in nasal samples (4%) (Fig. 8).
Figure 8.
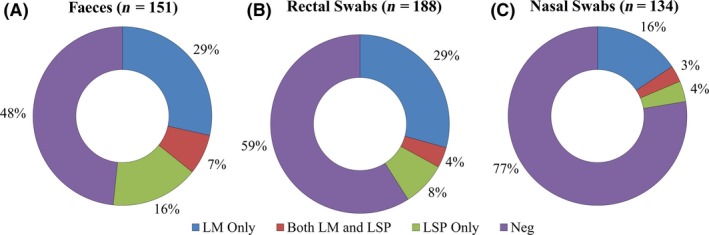
The prevalence of L. monocytogenes and other Listeria spp. in different types of samples from American black bears. Sample types include (A) faeces (n = 151), (B) rectal swabs (n = 188) and (C) nasal swabs (n = 134). Numbers indicate per cent of samples. Inset designations are as in legend to Fig. 2.
The small number of intestinal samples (n = 14; gastrointestinal tracks from vehicle‐killed black bears) and the fact that they had been stored frozen prior to analysis prevented accurate assessments of Listeria prevalence in these samples. The only positive intestinal samples were from the large intestine of two bears and both yielded L. monocytogenes but no other Listeria spp.
Sample type and geographical location may impact recovery of certain serotypes and serotype 4b strains with multiplex PCR profile IVb‐v1
Sample‐dependent differences were noted in the prevalence of certain groups of isolates. Specifically, serotype 1/2a was significantly more common in faeces and rectal swabs than in nasal swabs (faeces and nasal swabs: diff = −13.1, SE = 6.5, Z = −2.0, P = 0.04; rectal and nasal swabs: diff = −16.9, SE = 6.7, Z = 2.5, P = 0.01), with no significant difference in recovery between faeces and rectal swabs (Fig. 9A). Similar trends were noted for serotype 4b isolates with the IVb‐v1 multiplex PCR profile (faeces and nasal swabs: diff = −8.1, SE = 3.9, Z = −2.1, P = 0.04, rectal and nasal swabs: diff = −11.1, SE = 4.3, Z = 2.6, P = 0.01) and for Listeria spp. other than L. monocytogenes (faeces and nasal swabs: diff = −20.6, SE = 5.9, Z = −3.5, P = 0.001, rectal and nasal swabs: diff = −11.9, SE = 5.3, Z = 2.3, P = 0.02) (Fig. 9A).
Figure 9.
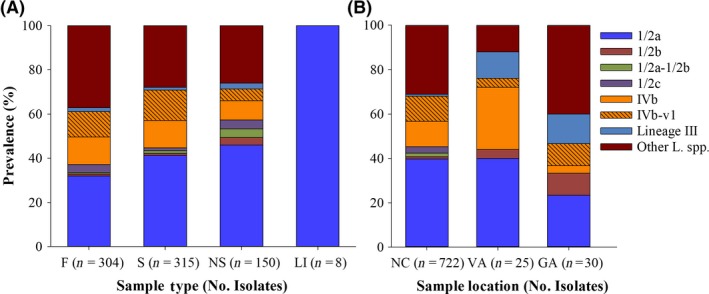
The prevalence of different serotypes and lineage III among L. monocytogenes from American black bears based on (A) sample type and (B) geographical location. Sample types in panel A are faeces (F), rectal swab (S), nasal swab (NS) and large intestine (LI). Geographical locations in panel B are North Carolina (NC), Georgia (GA) and Virginia (VA). Serotype and putative lineage III designations were determined by multiplex PCR as described in Experimental Procedures. Inset designations are as in the legend to Fig. 5. ‘Other L. spp’. refers to species of Listeria other than L. monocytogenes.
Location (NC, GA and VA) did not have significant impacts on the prevalence of serotypes 1/2a and 4b. However, serotype 4b isolates with the IVb‐v1 PCR profile were only recovered in NC and GA, while isolates of serotype 1/2c and those with the novel multiplex PCR profile 1/2a‐1/2b were only recovered from NC (Fig. 9B).
Black bears are transiently colonized by a diverse population of listeriae
While higher resolution techniques will be required to make strain level distinctions between isolates, determination of L. monocytogenes, other Listeria spp. and different L monocytogenes serotypes/multiplex PCR profiles suggested diversity of the Listeria population colonizing bears. As previously stated, 13% of the black bear capture events yielded both L. monocytogenes and other Listeria spp. (Fig. 2). When strains of diverse serotypes were included, 43% (55/127) of the Listeria‐positive captures yielded diverse strains, i.e., L. monocytogenes of different multiplex PCR profiles and Listeria spp. Of the 105 L. monocytogenes‐positive captures, 45 (43%) yielded multiple serotypes of L. monocytogenes. Isolation of L. monocytogenes with different serotypes from the same sample was not uncommon; approximately 20% of L. monocytogenes‐positive samples yielded isolates of multiple serotypes and 11% also yielded other Listeria spp. We based the listerial community composition on Listeria species, and serotype multiplex PCR profile distribution of all recovered isolates, as calculated by the Jaccard similarity index, was 77% similar between rectal swabs and faeces but similarity was lower between nasal and rectal swabs (45%) or faeces (54%).
As indicated above, 38 of the black bears were captured and sampled more than once. Most (33/38, 87%) of these animals were sampled twice, with four sampled three times and one five times (Table 1). Additionally, two of the bears that had been live‐captured were later found to be killed by vehicles one and 4 months after their original capture. Analysis of the samples from the 38 bears sampled on multiple occasions suggested a high degree of transience in the Listeria populations of wild black bears (Table 1). Of the two bears sampled while live and again post‐mortem, one had a Listeria‐positive rectal swab sample during the live capture but the post‐mortem small and large intestine samples were negative. In the 38 black bears that were live‐captured multiple times, results frequently changed from Listeria positive to negative and vice versa between one sampling time and another (Table 1). We identified only one animal (N108) that yielded the same Listeria‐related data upon recapture (Table 1).
Table 1.
Listeria recovery from American black bears that were Listeria positive at multiple sampling times.
| Bear ID | Time between recaptures (Months)a | Initial captureb | Subsequent captures(s)b |
|---|---|---|---|
| N024 | 9 | LM 1/2a | 2nd capture: Listeria spp. |
| N052 | 6 | LM 1/2a and LM 1/2a‐1/2b | 2nd capture: Negative |
| N059 | 6 | LM 1/2c and Listeria spp. | 2nd capture: LM 1/2a and LM IVb |
| N057 | 19 | LM IVb |
2nd capture: Negative 3rd capture: LM 1/2a and LM IVb 4th capture: LM 1/2a 5th capture: Negative |
| N022 | 10 | Negative | 2nd capture: Negative |
| N047 | 8 | Negative | 2nd capture: LM 1/2a |
| N040 | 8 | LM IVb‐v1 | 2nd capture: LM 1/2a, LM IVb and LM 1/2a‐1/2b |
| N043 | 21 | LM 1/2c and LM IVb‐v1 |
2nd capture: LM 1/2a 3rd capture: Negative |
| N048 | 9 | Negative | 2nd capture: Negative |
| N006 | 12 | LM IVb and LM 1/2a |
2nd capture: Negative 3rd capture: Negative |
| N046 | 9 | LM 1/2c | 2nd capture: Negative |
| N063 | 2 | LM 1/2a | 2nd capture: Negative |
| N015 | 13 | Negative | 2nd capture: LM 1/2a |
| N016 | 14 | Listeria spp. | 2nd capture: LM 1/2a |
| N014 | 15 | Negative | 2nd capture: LM 1/2a |
| N060 | 11 | LM IVb‐v1 and Listeria spp. and | 2nd capture: Negative |
| N095 | 7 | LM 1/2a, LM IVb‐v1 and Listeria spp. | 2nd capture: LM 1/2a‐1/2b |
| N056 | 19 | Listeria spp. | 2nd capture: LM 1/2a |
| N075 | 11 | Negative | 2nd capture: Negative |
| N079 | 11 | LM 1/2a, LM Lineage III and Listeria spp. | 2nd capture: LM 1/2a |
| N061 | 17 | LM 1/2a, LM IVb and Listeria spp. |
2nd capture: LM IVb 3rd capture: LM 1/2a, LM IVb‐v1 and Listeria spp. |
| N092 | 10 | Negative |
2nd capture: LM IVb‐v1 3rd capture: Listeria spp. |
| N108 | 2 | LM 1/2a and Listeria spp. | 2nd capture: LM 1/2a and Listeria spp. |
| N051 | 10 | LM IVb and LM IVb‐v1 | 2nd capture: Negative |
| N121 | 2 | Listeria spp. | 2nd capture: LM 1/2a and Listeria spp. |
| N109 | 3 | LM 1/2a and Listeria spp. | 2nd capture: LM 1/2a and LM IVb |
| N087 | 13 | Negative | 2nd capture: LM 1/2a, LM IVb and Listeria spp. |
| N032 | 21 | Negative | 2nd capture: Negative |
| N132 | 2 | Listeria spp. | 2nd capture: LM IVb and Listeria spp. |
| N138 | 2 | Listeria spp. | 2nd capture: LM 1/2a and Listeria spp. |
| N151 | 1 | Negative | 2nd capture: LM 1/2a |
| N037 | 35 | Listeria spp. | 2nd capture: LM 1/2b, LM Lineage III and Listeria spp. |
| BBRC 119 | 5 | LM 1/2a, LM Lineage III | 2nd capture: LM 1/2a, LM IVb |
| BBRC 120 | 5 | Negative | 2nd capture: Negative |
| BBRC 126 | 4 | Negative | 2nd capture: Negative |
| BBRC 127 | 4 | LM IVb and LM Lineage III | 2nd capture: Negative |
| BBRC 128 | 4 | Listeria spp. | 2nd capture: Negative |
| BBRC 129 | 4 | Negative | 2nd capture: Negative |
In the case of animals captured more than twice, time frame spans from first to last capture.
LM, Listeria monocytogenes; Listeria spp., Listeria species other than LM. Designations 1/2a, IVb, 1/2c, IVb‐v1 correspond to multiplex PCR profiles for LM, as in legend to Fig. 5.
Heavy metal and disinfectant resistance were uncommon among listeriae from black bears
Of the 777 isolates, only 24 (3%; three L. monocytogenes and 21 other Listeria spp.) were resistant to cadmium (MIC > 35 µg ml−1). The three L. monocytogenes isolates were all serotype 1/2a from two black bears in NC, 2015; one was from faeces of one bear, while the other two were from the nasal swab of a different bear. The 21 cadmium‐resistant non‐L. monocytogenes Listeria isolates were from six different bears from NC at various times throughout the study (one each in 2014, 2015 and 2017; three in 2016).
PCR of the 24 cadmium‐resistant isolates for the known Listeria cadmium resistance determinants cadA1‐ cadA4 revealed cadA1 (n = 6) and cadA2 (n = 9) in 15 of the 21 non‐L. monocytogenes Listeria isolates. The remaining nine cadmium‐resistant isolates, including the three that were L. monocytogenes, were PCR negative and thus may harbour novel, yet unidentified cadmium resistance determinant(s). Resistance to arsenic or benzalkonium chloride was not encountered among any of the 777 isolates from the bears.
Predictors of L. monocytogenes colonization
We employed models to assess predictors for listerial colonization, including the percentage of agriculture and forest in each occurrence distribution; averages for housing density, temperature and precipitation were used as fixed effects, with year as a random effect. Human housing density (houses per km2) was our measure of urban development and was the only predictor in the best model (Table 2) showing a positive, but not statistically significant relationship with Listeria colonization (ß = 0.33, P = 0.15). This model did not perform significantly better than the null model, suggesting that ‘year’ captures most of the variation and that housing density does not add a significant amount of explanatory power. Occurrence distribution overlap was significantly higher (Wilcoxon χ2 approximation = 81, df = 1, P < 0.0001) between capture events yielding L. monocytogenes than between those that were negative.
Table 2.
Coefficients and associated standard error, critical value and p‐value from the top model based on AICc from a study of L. monocytogenes prevalence in wild American black bears from NC, GA and VA from 2014 to 2017. We fit a generalized linear mixed model to predict the L. monocytogenes colonization status of each bear using the average housing density per km2 within each occurrence distribution with year as a random effect.
| Predictor | β | SE | Z | P‐value |
|---|---|---|---|---|
| (Intercept) | −0.1 | 0.38 | −0.25 | 0.80 |
| Housing density | 0.33 | 0.23 | 1.43 | 0.15 |
Discussion
Our results indicate that L. monocytogenes could be frequently isolated from American black bears in the southeastern United States. We frequently obtained isolates of different serotype‐associated multiplex PCR profiles as well as L. monocytogenes together with other Listeria spp. from the same samples, indicating cohabitation of different Listeria strains and species in the same animal. Analysis of samples from three different states over 4 years revealed that each year nearly half of all black bear capture events yielded L. monocytogenes. This broad temporal and geographic scope suggests that L. monocytogenes was consistently maintained in the black bear population in the study area. Interestingly, occurrence distribution overlap was found to be significantly higher between captures yielding L. monocytogenes than between those that were negative. This suggests potential transfer of L. monocytogenes among individuals who may be more likely to interact, e.g., through shared habitat, behavioural practices or social structure. In an earlier study of Japanese monkeys, potential spread of Listeria spp. among the members of the same troop was also hypothesized (Yoshida et al., 2000).
One of the unexpected findings from our work was the significantly higher prevalence of L. monocytogenes than other Listeria spp. The majority (83%) of the Listeria‐positive capture events yielded L. monocytogenes, either alone or together with other Listeria spp., and bears found to be positive exclusively for Listeria spp. other than L. monocytogenes were uncommonly encountered. This apparent bias for L. monocytogenes over other Listeria spp. was detected in all sample types, but was especially noticeable for nasal swab samples.
A predilection for L. monocytogenes in comparison with other Listeria spp. has rarely been found in other wildlife studies, where in fact the opposite was frequently observed, with L. monocytogenes tending to be noticeably less common than other Listeria spp. (Innoue et al., 1992; Quessy and Messier, 1992; Yoshida et al., 2000; Hellström et al., 2008; Wang et al., 2017; Cao et al., 2019). Only one study of rectal swabs from red fox, beech marten and raccoons in Poland found that, even though overall Listeria prevalence was low (approx. 7.4%), most (64.5%) of the Listeria‐positive samples yielded L. monocytogenes (Nowakiewicz et al., 2016). Even similar prevalence levels between L. monocytogenes and other Listeria spp. are uncommonly reported in wildlife surveys. Fenlon (1985) noted L. monocytogenes, L. innocua and L. seeligeri in 8.4, 4.5 and 3.4%, respectively, of faecal samples from seagulls, primarily from sewage disposal sites, with thus similar prevalence between L. monocytogenes (8.4%) and other Listeria spp. (7.9%). A survey of urban rooks (Corvus frugilegus) in France detected L. monocytogenes, L. innocua and L. seeligeri in 33%, 24% and 8%, respectively, of the samples (33% L. monocytogenes vs. 32% other Listeria spp.) (Bouttefroy et al., 1997). Interestingly, L. seeligeri was not encountered among our bear isolates. Similarly, L. ivanovii accounted for many of the Listeria isolates from rodent surveys in China (Wang et al., 2017; Cao et al., 2019), but was not encountered in our study.
The eco‐physiological determinants underlying the apparent association preference of black bears for L. monocytogenes remain to be elucidated. Surveys of avian Listeria carriage suggested higher prevalence in urban populations, with L. monocytogenes accounting for approximately 36–50% of the Listeria‐positive samples from urban birds (Fenlon, 1985; Bouttefroy et al., 1997; Hellström et al., 2008), while association with rural bird nesting areas was also noted (Quessy and Messier, 1992). In this study, we sampled both suburban and wild black bears and observed significantly higher L. monocytogenes recovery at all locations across the entire study period (Figs 2 and 3). As mentioned above, L. seeligeri was not encountered, even though it was overall the predominant species from both urban and relatively undisturbed natural environments in other studies (Sauders et al., 2012; Linke et al., 2014). It is possible that the foraging behaviour and preferences of black bears preferentially expose them to materials that may serve as reservoirs for L. monocytogenes in the natural environment. It is also conceivable that the nasal and intestinal microbiome of the bears may be selecting for the pathogenic species L. monocytogenes, despite the more diverse Listeria population to which they are exposed.
The prevalence of L. monocytogenes in our study (36%) was noticeably higher than in most other wildlife surveys, where prevalence ranged from 0.3% to 18% (Hayashidani et al., 2002; Lyautey et al., 2007; Wacheck et al., 2010; Nowakiewicz et al., 2016; Weindl et al., 2016; Wang et al., 2017; Cao et al., 2019;). Only two wildlife studies, involving urban birds in Finland and France, respectively, yielded prevalence levels of 33–36% (Bouttefroy et al., 1997; Hellstrom et al., 2008), similar to what we found in the bears. The differences in prevalence between our study and others may reflect different animal species, sample types (e.g., faeces, rectal or nasal swabs), geographical regions and methodologies. Nonetheless, the collective findings support the speculation, first articulated in 1975, that L. monocytogenes may have a cyclic existence between the gut of animals and the natural environment (Weis and Seeliger, 1975).
Employment of both primary and secondary enrichments, multiplex PCR profiling of multiple colonies per positive sample and analysis of multiple sample types from the same animal were important in capturing the diversity of the Listeria population associated with the black bears. Even though Listeria recovery was significantly higher from primary than from secondary enrichments as also reported by others (Weindl et al., 2016), including both enrichments may be useful in capturing diversity. Furthermore, and in agreement with Weindl et al. (2016), examination of different sample types from the same animal proved important in assessments of species (L. monocytogenes vs. other Listeria spp.) and serotype heterogeneity within the Listeria population from the animals.
The preferential recovery of L. monocytogenes from nasal swabs, in comparison with other Listeria spp., was of special interest. It was also noteworthy that even though the overall prevalence of L. monocytogenes was lower in nasal swabs than in faeces or rectal swabs, multiplex PCR profile diversity was higher, with all multiplex PCR profiles encountered among nasal swab‐derived isolates (Fig. 9). Whether nasal passage colonization is mediated by specific genetic adaptations of L. monocytogenes or reflects black bear foraging behaviour that expose nasal passages to materials especially likely to be contaminated by a variety of strains, is worthy of further investigation.
The two serotypes of L. monocytogenes dominant in our study (1/2a and 4b) are among the three (1/2a, 1/2b, and 4b) most commonly implicated in human listeriosis (Swaminathan and Gerner‐Smidt, 2007). Serotypes 1/2a and 4b were also the leading serotypes in other wildlife studies, including surveys of red deer, wild boar and wild birds (Yoshida et al., 2000; Hellström et al., 2008; Weindl et al., 2016). Serotype 1/2b was only encountered in 2% of L. monocytogenes isolates in our study and was also infrequently (5%) recovered by Weindl et al. (2016). Lineage III isolates were also uncommon in our study even though they were reported to show proclivity for non‐human animals (Jeffers et al., 2001; Liu et al., 2006). It was noteworthy that the distribution of serotype 1/2a and 4b isolates in the black bears remained relatively constant over the 4 years of the study. The high representation of serotypes 1/2a and 4b in this and other wildlife hosts is intriguing as it may reflect yet unidentified reservoirs for these serotypes in the natural environment. This high prevalence of serotypes 1/2a and 4b among the bear isolates also has potential public health implications which need to be further assessed, e.g., via high‐resolution genotyping and virulence assessments.
Serotype‐related multiplex PCR profiling of the isolates yielded two surprising findings. First, serotype 4b strains with the variant multiplex profile IVb‐v1 (Leclercq et al., 2011) were repeatedly encountered in our study and accounted for almost half of the serotype 4b isolates. They were recovered from both NC and GA, demonstrating a wide geographic distribution. The IVb‐v1 profile has been detected in several genetically unrelated, emerging clones of serotype 4b, including those implicated in recent outbreaks (Moura et al., 2016; Lee et al., 2018). High‐resolution genotyping is needed to determine the genetic relatedness of the bear‐derived IVb‐v1 strains to those implicated in human listeriosis. The second unexpected finding was the discovery of isolates with a novel multiplex PCR profile that included amplicons typical for both serotypes 1/2a and 1/2b. These isolates were from animals captured at diverse times and locations in NC, and to our knowledge, this represents the first reported encounter of this unique PCR profile.
Resistance to the heavy metals cadmium and arsenic is one of the longest‐known environmental adaptations of L. monocytogenes (Buchanan et al., 1991; Lebrun et al., 1992; McLauchlin et al., 1997), and prevalence of cadmium resistance ranged from 50% to 66% among isolates from food processing facilities (Mullapudi et al., 2008; Ratani et al., 2012; Ferreira et al., 2014; Xu et al., 2014). The determinants that confer cadmium and arsenic resistance in L. monocytogenes are usually harboured on mobile genetic elements such as plasmids, genomic islands or transposons (Lebrun et al., 1994a,b; Kuenne et al., 2010; Dutta et al., 2017; Parsons et al., 2017). Interestingly, none of the 537 L. monocytogenes isolates from the black bears were resistant to arsenic and only three (0.56%) were resistant to cadmium. These low levels of prevalence of resistance may reflect niche‐specific factors that deter acquisition or stable maintenance of mobile genetic elements associated with heavy metal resistance. Cadmium resistance was, to our knowledge, examined in only one other study of Listeria from wildlife, where 28.6% of L. monocytogenes and 7.7% of other Listeria spp. from urban rooks were found to be resistant to cadmium (Bouttefroy et al., 1997). Further studies are needed to determine whether the surprisingly low incidence of resistance in our study is a niche‐specific trait in American black bears.
Our results suggest the potential for black bears to serve as a natural vehicle and perhaps a reservoir for L. monocytogenes and other Listeria spp. Whether the source for the colonization of the black bears originated from the environment (e.g. soil, water or vegetation) or from anthropogenic sources (e.g. bears scavenging around human habitations or agricultural areas) warrants further investigation. Our microbiological results and the occurrence distributions for the black bears which largely overlapped with humans (and accompanying companion animals) in an urban/suburban community suggest that L. monocytogenes may represent an attractive model for the investigation of both anthroponotic and zoonotic transmission of pathogens. Our results also provide novel insights into the complexity of the natural ecology of L. monocytogenes, suggesting a potential role for black bears in the transmission, dissemination and adaptations of this pathogen. Given the increasing human–animal interactions due to urban expansion and the implications for both humans and other animal species to become sick from exposure to L. monocytogenes, further studies of this nature are of clear significance for human and veterinary health.
Experimental procedures
Project locations and study areas
Black bear populations were investigated in three states, (NC, GA and VA) in the southeastern United States from 2014 to 2017. As indicated above, the animals were live‐captured for the purpose of several separate studies focusing on the mobility and demographic patterns of black bear populations in this region. None of the animals were trapped or handled specifically in order to collect samples for isolation of L. monocytogenes or other Listeria spp.
The majority of the samples were obtained through the study in NC, which focused on populations within or adjacent to the city of Asheville, located in Buncombe County in the southern Appalachian mountain range of western NC (Fig. 1). Asheville encompasses 117 km2 with a residential population of approximately 92 000 people and is characterized by heterogeneous topography, temperate climate with high annual precipitation (36.95″ rainfall, 13″ snowfall avg.) and mixed deciduous hardwood and pine‐hardwood forests (Mitchell et al., 2002; Kirk et al., 2012).
The study in GA was conducted within Houston and Twiggs Counties in central GA along the Ocmulgee River drainage (Hooker et al., 2015). This area is located at the boundary of the Piedmont and Coastal Plain regions and is characterized by hardwood forest that dominates the bottomland region and mixed pine‐hardwood forest dominating the uplands. Land west of the study area is dominated by human development including Macon (pop. 91 234), Warner Robins (pop. 72 531) and Bonaire (pop. 13 999; USCB, 2016), whereas land south and east is primarily agricultural. Common crops, several of which are used by bears, are cotton, corn, peaches, peanuts and pecans.
The VA study included 15 animals that were kept in temporary captivity (6–8 months) at VT‐BBRC in Blacksburg, VA (Mesa‐Cruz, 2018) and then released into the wild. These black bears were captured from different counties in the Appalachian Mountains of western VA by the VA Department of Game and Inland Fisheries (VDGIF).
Sample collection
All animal capture and handling protocols were approved by the Institutional Animal Care and Use Committee at North Carolina State University (14‐019‐O), the University of Georgia (A2011 10‐004‐A1) and Virginia Tech (12‐112 and 15‐162). Bears were live‐captured from April 2014 to August 2017 in NC and June to July 2014 in GA. In VA, the bears were sampled from October 2014 to March 2015 and from October 2015 to February 2016. Live‐captured bears were chemically immobilized with a combination of 5 ml of tiletamine‐zolazepam (100 mg ml−1) with 4.0 ml of ketamine hydrochloride and 1.0 ml of xylazine hydrochloride. Bears captured in Georgia were immobilized with Telazol® (equal parts tiletamine and zolazepam; Fort Dodge Animal Health, Fort Dodge, IA, USA) or large animal xylazine (100 mg ml−1) combined with Telazol® delivered intramuscular (IM) via blowdart. Each bear in the NC and GA study was given uniquely numbered ear tags, matching lip tattoo, passive integrated transponder (PIT) tags and GPS radio collars (Vectronic, Berlin, Germany or Lotek Wireless Inc., Newmarket, ON, Canada) to track movement. Weight, sex, reproductive status and body measurements were recorded for each bear. Temperature, pulse and blood oxygen levels were monitored throughout immobilizations to ensure the health/stability of the bears. Anaesthesia was reversed using a reversal agent such as yohimbine hydrochloride (0.15–0.3 mg kg−1, I.M. or I.V.). Anaesthesia of bears immobilized with Telazol in GA was reversed approximately 45 min after initial immobilization using latipamazole hydrochloride (5 mg ml−1; Antisedan®, Orion Pharma, Orion Corporation, Espoo, Finland) and diazepam (5 mg ml−1) delivered IM via hand injection.
When available, fresh faecal samples were collected in Ziploc® bags and shipped overnight on ice to North Carolina State University (NCSU). Rectal and nasal swabs were collected, while the animals were anesthetized, using sterile techniques with pre‐sterilized swabs inside collection tubes (Becton, Dickinson, and Company) and similarly shipped to NCSU.
A small number (n = 14) of bears killed by vehicular collisions in Buncombe County (NC) were also sampled. GI tract samples from these bears were kept frozen at −20°C and shipped on ice in batches to the NCSU laboratory.
Microbiological sample processing and enrichment
Samples were enriched for Listeria using the ISO Method as described (Azizoglu et al., 2014) with minor alterations. Specifically, faecal samples were diluted 1:10 (1.25 g 11.25 ml−1) in Half Fraser Broth supplemented with Half Fraser Selective Supplement (Oxoid, Hampshire, UK) and incubated at 30°C for 24‐48 h for primary enrichment. Swab samples were similarly enriched in 11.25 ml of the primary enrichment broth. The primary enrichment (0.1 ml) was then transferred into 10 ml of Full Fraser Broth supplemented with Fraser selective supplement (Oxoid) and incubated at 37°C for 48 h for secondary enrichment. Putative positive primary or secondary enrichments, evidenced by colour change in the medium, were streaked (20 µl) on MOX and incubated at 37°C for 48 h. Up to six putative Listeria colonies from the MOX plates were streaked on tryptic soy agar (TSA) with 5% sheep blood (Remel, San Diego, CA, USA) and incubated at 37°C for 36‐48 h to purify and assess haemolytic activity. Pure cultures were preserved at −80°C in brain heart infusion (BHI) with 20% glycerol (Fisher Scientific, Fairlawn, NJ, USA). Cultures were routinely grown on Tryptic Soy Broth (Becton, Dickinson and Company) supplemented with 0.7% yeast extract (Fisher Scientific) and 1.2% agarose (Becton, Dickinson and Company; TSAYE).
Determination of serotype designations and susceptibility to heavy metals and benzalkonium chloride
Listeria spp. verification and serotype designations of 1/2a (or 3a), 1/2b (or 3b), 1/2c (or 3c) and 4b (or 4d, 4e) were determined via multiplex PCR (Doumith et al., 2004), as was the serotype 4b variant profile, IVb‐v1 (Huang et al., 2011; Leclercq et al., 2011; Lee et al., 2012a,b). Haemolytic isolates yielding only the prs PCR product were further tested with PCR using L. monocytogenes‐specific hly primers for Listeriolysin O, to determine whether they were L. monocytogenes lineage III isolates serotype 4a or 4c or L. seeligeri (Eifert et al., 2005). Susceptibility to cadmium and arsenic was determined on Iso‐Sensitest™ Agar (Oxoid) with 35 and 70 µg ml−1 cadmium chloride or 500 µg ml−1 sodium arsenite, respectively, while BC susceptibility was determined on TSAYE with 10 µg ml−1 BC; positive and negative control strains were used for each agent in each test for quality assurance, as described (Lee et al., 2013). Isolates exhibiting resistance to cadmium were tested by PCR for known resistance determinants using previously described primers (Lee et al., 2013; Parsons et al., 2017).
Assessing predictors of Listeria colonization
Black bear capture and trap locations in and around Asheville, NC, are shown in Fig. 1. We analysed movement data from GPS‐collared bears in this area, as it accounted for the preponderance of the samples, and we lacked movement data for the GA and VA collection sites. We restricted movement data to within 2 weeks after the sample collection date and used the ctmm package (Calabrese et al., 2016; Fleming and Calabrese, 2016) in r (v. 3.3.1) (R Foundation for Statistical Computing, 2016) to construct 95% occurrence distributions (OD). Thus, we estimated the area that each animal actually used during each 2‐week period as opposed to constructing a home range. We used the raster package (Hijmans et al., 2016) in R to determine the percentage of agriculture and forest (reclassified from GAP land cover to include all forest types) (U.S. Geological Survey, 2011) in each OD and the average housing density (houses per km2, SILVIS) (Hammer et al., 2004). We obtained daily temperature and precipitation data from Weather Underground using the rwunderground package in r (Shum, 2018). Initial assessments using a chi‐squared test for independence in r indicated that sex (χ2 = 437.6, df = 1, P < 0.001) and age (χ2 = 286.2, df = 1, P < 0.001) of each bear were not strong predictors of L. monocytogenes colonization and these were omitted from our models.
We ran a generalized linear mixed model to predict the L. monocytogenes colonization status of each bear using the percentage of agriculture and forest in each OD, the average housing density, average temperature and average precipitation as fixed effects, with year as a random effect. Correlation between covariates was assessed using a Pearson correlation matrix in r to verify that all covariates were correlated < 50% and to also verify that where > 50% those predictors did not appear in the same model. We ran a suite of seven models and chose the best model for the data based on Akaike’s information criterion (AIC) (Burnham et al., 2002). All continuous predictors were centred and scaled prior to running the model.
Because contact with other L. monocytogenes‐positive bears could lead to pathogen spread, either through direct contact or via the faecal contamination of shared spaces, we assessed whether L. monocytogenes‐colonized black bears shared space with other non‐colonized black bears. We used the adehabitatHR package in r (Calenge, 2006) to compute indices of home range overlap from locations as described in Fieberg and Kochanny (2005), specifically estimating the proportion of the home range of one animal covered by the home range of another, separately for each year. We separated the black bears into those that yielded at least one L. monocytogenes‐positive sample and those from which all tested samples were negative for L. monocytogenes, then determined whether the average home range overlap between the groups was significantly different using a Wilcoxon rank‐sum test in r.
Statistical analysis of black bear sampling data
Differences in recovery based on sample type were assessed using a Wilcoxon rank‐sum test in r. Similarity in Listeria community composition (L. monocytogenes, other Listeria spp. and serotype) was assessed via a Jaccard similarity index where 1 is no similarity and 0 is identical, using the vegan (Oksanen et al., 2017) package in r. Tests for overall positive or negative status for sample type or enrichment type were performed using a logistic regression with bear ID as a random effect and sample type or enrichment as fixed effect (α = 0.05). Differences in yearly recovery were assessed using a chi‐squared test. Differences in recovery of L. monocytogenes vs. other Listeria spp. were assessed using a Pearson’s chi‐squared test in SPSS (IBM, Armonk NY, USA). Differences in mean yearly prevalence of L. monocytogenes vs. other Listeria spp. were assessed using an independent samples t test in SPSS (IBM).
Conflict of interest
None declared.
Acknowledgements
This work is supported by the AFRI‐ELI under award # 2017‐67012‐26001 from the USDA National Institute of Food and Agriculture. Any opinions, findings, conclusions or recommendations expressed are those of the authors and do not necessarily reflect the view of the USDA. Funding for the Georgia portion of this project was graciously provided by the Georgia Department of Transportation, the Georgia Department of Natural Resources–Wildlife Resources Division, and the Warnell School of Forestry and Natural Resources at the University of Georgia. The field portion of this project was funded by the Pittman Robertson Federal Aid to Wildlife Restoration Grant and was a joint research project between the North Carolina Wildlife Resources Commission (NCWRC) and the Fisheries, Wildlife, and Conservation Biology (FWCB) Program at North Carolina State University (NCSU). We thank the homeowners who granted us permission and access to their properties. We thank numerous other staff from the NCWRC and the FWCB program at NCSU for their ongoing assistance and support. The Virginia Department of Game and Inland Fisheries, the Alcinda Acorn Foundation, and the Faile Foundation provided funds to temporarily house bears at the VT‐BBRC.
Microbial Biotechnology (2020) 13(3), 706–721
Funding Information
This work is supported by the AFRI‐ELI under award # 2017‐67012‐26001 from the USDA National Institute of Food and Agriculture. Funding for the Georgia portion of this project was graciously provided by the Georgia Department of Transportation, the Georgia Department of Natural Resources–Wildlife Resources Division, and the Warnell School of Forestry and Natural Resources at the University of Georgia. The Virginia Department of Game and Inland Fisheries, the Alcinda Acorn Foundation, and the Faile Foundation provided funds to temporarily house bears at the VT‐BBRC.
References
- Azizoglu, R.O. , Gorski, L. , and Kathariou, S. (2014) Isolation of Listeria monocytogenes from food and water: official and experimental protocols. Curr Protoc Microbiol 33: 9B–9B5. [DOI] [PubMed] [Google Scholar]
- den Bakker, H.C. , Warchocki, S. , Wright, E.M. , Allred, A.F. , Ahlstrom, C. , Manuel, C.S. , et al (2014) Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int J Syst Evol Microbiol 64: 1882–1889. [DOI] [PubMed] [Google Scholar]
- Binninger, C.E. , Beecham, J.J. , Thomas, L.A. , and Winward, L.D. (1980) A serologic survey for selected infectious diseases of black bears in Idaho. J Wildl Dis 16: 423–430. [DOI] [PubMed] [Google Scholar]
- Blanot, S. , Joly, M.M. , Vilde, F. , Jaubert, F. , Clement, O. , Frija, G. , and Berche, P. (1997) A gerbil model for rhombencephalitis due to Listeria monocytogenes . Microb Pathog 23: 39–48. [DOI] [PubMed] [Google Scholar]
- Bouttefroy, A. , Lemaître, J.P. , and Rousset, A. (1997) Prevalence of Listeria sp. in droppings from urban rooks (Corvus frugilegus). J Appl Microbiol 82: 641–647. [DOI] [PubMed] [Google Scholar]
- Bronson, E. , Spiker, H. , and Driscoll, C.P. (2014) Serosurvey for selected pathogens in free‐ranging American black bears (Ursus americanus) in Maryland, USA. J Wildl Dis 50: 829–36. [DOI] [PubMed] [Google Scholar]
- Buchanan, R.L. , Klawitter, L.A. , Bhaduri, S. , and Stahl, H.G. (1991) Arsenite resistance in Listeria monocytogenes . Food Microbiol 8: 161–166. [Google Scholar]
- Burnham, K.P. , Anderson, D.R. , and Burnham, K.P. (2002) Model selection and multimodel inference: a practical information‐theoretic approach. J Wildlife Manag 67: 655. [Google Scholar]
- Calabrese, J.M. , Fleming, C.H. , and Gurarie, E. (2016) ctmm: an r package for analyzing animal relocation data as a continuous‐time stochastic process. Methods Ecol Evol 7: 1124–1132. [Google Scholar]
- Calenge, C.C. (2006) The package “adehabitat” for the R software: a tool for the analysis of space and habitat use by animals. Ecol Modell 197: 516–519. [Google Scholar]
- Cao, X. , Wang, Yan , Wang, Yi , Li, Hui , Luo, L. , Wang, P. , et al (2019) Prevalence and characteristics of Listeria ivanovii strains in wild rodents in China. Vector‐Borne Zoonotic Dis 19: 8–15. [DOI] [PubMed] [Google Scholar]
- Carpentier, B. , and Cerf, O. (2011) Review ‐ Persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145: 1–8. [DOI] [PubMed] [Google Scholar]
- Chlebicz, A. , and Śliżewska, K. (2018) Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as zoonotic foodborne diseases: A Review. Int J Environ Res Public Health 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M.B. , Quiñones, B. , Oryang, D. , Mandrell, R.E. , and Gorski, L. (2014) Prevalence of shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front Cell Infect Microbiol 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuprynski, C.J. , Kathariou, S. , and Poulsen, K. (2010) Listeria In Pathogenesis of Bacterial Infections in Animals. Oxford, UK: Wiley‐Blackwell, pp. 167–187. [Google Scholar]
- Daszak, P. , Cunningham, A.A. , and Hyatt, A.D. (2001) Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop 78: 103–116. [DOI] [PubMed] [Google Scholar]
- Disson, O. , Nikitas, G. , Grayo, S. , Dussurget, O. , Cossart, P. , and Lecuit, M. (2009) Modeling human listeriosis in natural and genetically engineered animals. Nat Protoc 4: 799–810. [DOI] [PubMed] [Google Scholar]
- Doumith, M. , Buchrieser, C. , Glaser, P. , Jacquet, C. , and Martin, P. (2004) Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42: 3819–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, V. , Lee, S. , Ward, T.J. , Orwig, N. , Altermann, E. , Jima, D.D. , et al (2017) Genome sequences of Listeria monocytogenes strains with resistance to arsenic. Genome Announc 5: e00327–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifert, J.D. , Curtis, P.A. , Bazaco, M.C. , Meinersmann, R.J. , Berrang, M.E. ,Kernodle, et al (2005) Molecular characterization of Listeria monocytogenes of the serotype 4b complex (4b, 4d, 4e) from two turkey processing plants. Foodborne Pathog Dis 2: 192–200. [DOI] [PubMed] [Google Scholar]
- Fenlon, D.R. (1985) Wild birds and silage as reservoirs of Listeria in the agricultural environment. J Appl Bacteriol 59: 537–543. [DOI] [PubMed] [Google Scholar]
- Ferreira, V. , Wiedmann, M. , Teixeira, P. , and Stasiewicz, M.J. (2014) Listeria monocytogenes persistence in food‐associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77: 150–70. [DOI] [PubMed] [Google Scholar]
- Fieberg, J. , and Kochanny, C.O. (2005) Quantifying home range overlap: The importance of the utilization distribution. J Wildl Manage 69: 1346–1359. [Google Scholar]
- Fleming, C.H. , and Calabrese, J.M. (2016) ctmm: Continuous‐Time Movement Modeling. R Packag version 033 https://https//CRANR-project.org/package=ctmm . [Google Scholar]
- Gage, K.L. , Ostfeld, R.S. , and Olson, J.G. (1995) Nonviral vector‐borne zoonoses associated with mammals in the United States. J Mammal 76: 695. [Google Scholar]
- Gandhi, M. , and Chikindas, M.L. (2007) Listeria: a foodborne pathogen that knows how to survive. Int J Food Microbiol 113: 1–15. [DOI] [PubMed] [Google Scholar]
- Gnat, S. , Trościańczyk, A. , Nowakiewicz, A. , Majer‐Dziedzic, B. , Ziółkowska, G. , Dziedzic, R. , et al (2015) Experimental studies of microbial populations and incidence of zoonotic pathogens in the faeces of red deer (Cervus elaphus). Lett Appl Microbiol 61: 446–452. [DOI] [PubMed] [Google Scholar]
- Hammer, R.B. , Stewart, S.I. , Winkler, R.L. , Radeloff, V.C. , and Voss, P.R. (2004) Characterizing dynamic spatial and temporal residential density patterns from 1940–1990 across the North Central United States. Landsc Urban Plann 69: 183–199. [Google Scholar]
- Hayashidani, H. , Kanzaki, N. , Kaneko, Y. , Okatani, A.T. , Taniguchi, T. , Kaneko, K. , and Ogawa, M. (2002) Occurrence of Yersiniosis and Listeriosis in wild boars in Japan. J Wildl Dis 38: 202–205. [DOI] [PubMed] [Google Scholar]
- Hellström, S. , Kiviniemi, K. , Autio, T. , and Korkeala, H. (2008) Listeria monocytogenes is common in wild birds in Helsinki region and genotypes are frequently similar with those found along the food chain. J Appl Microbiol 104: 883–888. [DOI] [PubMed] [Google Scholar]
- Hijmans, R.J. , van Etten, J. , Mattiuzzi, M. , Sumner, M. , Greenberg, J.A. , Lamigueiro, O.P. , et al (2016) Raster: Geographic data analysis and modeling. R Packag version. [Google Scholar]
- Hooker, M.J. , Laufenberg, J.S. , Ashley, A.K. , Sylvest, J.T. , and Chamberlain, M.J. (2015) Abundance and density estimation of the American black bear population in central Georgia. Ursus 26: 107–115. [Google Scholar]
- Huang, B. , Fang, N. , Dimovski, K. , Wang, X. , Hogg, G. , and Bates, J. (2011) Observation of a new pattern in serogroup‐related PCR typing of Listeria monocytogenes 4b isolates. J Clin Microbiol 49: 426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanek, R. , Gröhn, Y.T. , and Wiedmann, M. (2006) Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog Dis 3: 319–36. [DOI] [PubMed] [Google Scholar]
- Jeffers, G.T. , Bruce, J.L. , McDonough, P.L. , Scarlett, J. , Boor, K.J. , and Wiedmann, M. (2001) Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147: 1095–1104. [DOI] [PubMed] [Google Scholar]
- Karelus, D.L. , McCown, J.W. , Scheick, B.K. , Van De Kerk, M. , Bolker, B.M. , and Oli, M.K. (2017) Effects of environmental factors and landscape features on movement patterns of Florida black bears. J Mammal 98: 1463–1478. [Google Scholar]
- Kathariou, S. (2002) Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Prot 65: 1811–1829. [DOI] [PubMed] [Google Scholar]
- Kirk, R.W. , Bolstad, P.V. , and Manson, S.M. (2012) Spatio‐temporal trend analysis of long‐term development patterns (1900–2030) in a Southern Appalachian County. Landsc Urban Plan 104: 47–58. [Google Scholar]
- Kuenne, C. , Voget, S. , Pischimarov, J. , Oehm, S. , Goesmann, A. , Daniel, R. , et al (2010) Comparative analysis of plasmids in the genus Listeria . PLoS ONE 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun, M. , Loulergue, J. , Chaslus‐Dancla, E. , and Audurier, A. (1992) Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl Environ Microbiol 58: 3183–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun, M. , Audurier, A. , and Cossart, P. (1994a) Plasmid‐borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917 . J Bacteriol 176: 3049–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun, M. , Audurier, A. , and Cossart, P. (1994b) Plasmid‐borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J Bacteriol 176: 3040–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq, A. , Chenal‐Francisque, V. , Dieye, H. , Cantinelli, T. , Drali, R. , Brisse, S. , and Lecuit, M. (2011) Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb‐v1. Int J Food Microbiol 147: 74–77. [DOI] [PubMed] [Google Scholar]
- Lee, S. , Ward, T.J. , Graves, L.M. , Wolf, L.A. , Sperry, K. , Siletzky, R.M. , and Kathariou, S. (2012a) Atypical Listeria monocytogenes serotype 4b strains harboring a lineage II‐specific gene cassette. Appl Environ Microbiol 78: 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Ward, T.J. , Siletzky, R.M. , and Kathariou, S. (2012b) Two novel type II restriction‐modification systems occupying genomically equivalent locations on the chromosomes of Listeria monocytogenes strains. Appl Environ Microbiol 78: 2623–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Rakic‐Martinez, M. , Graves, L.M. , Ward, T.J. , Siletzky, R.M. , and Kathariou, S. (2013) Genetic determinants for cadmium and arsenic resistance among Listeria monocytogenes serotype 4B isolates from sporadic human listeriosis patients. Appl Environ Microbiol 79: 2471–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Chen, Y. , Gorski, L. , Ward, T.J. , Osborne, J. , and Kathariou, S. (2018) Listeria monocytogenes source distribution analysis indicates regional heterogeneity and ecological niche preference among serotype 4b clones. MBio: 9: pii: e00396‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke, K. , Rückerl, I. , Brugger, K. , Karpiskova, R. , Walland, J. , Muri‐Klinger, S. , et al (2014) Reservoirs of Listeria species in three environmental ecosystems. Appl Environ Microbiol 80: 5583–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Lawrence, M.L. , Wiedmann, M. , Gorski, L. , Mandrell, R.E. , Ainsworth, A.J. , and Austin, F.W. (2006) Listeria monocytogenes subgroups IIIA, IIIB, and IIIC delineate genetically distinct populations with varied pathogenic potential. J Clin Microbiol 44: 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyautey, E. , Hartmann, A. , Pagotto, F. , Tyler, K. , Lapen, D.R. , Wilkes, G. , et al (2007) Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can J Microbiol 53: 1158–1167. [DOI] [PubMed] [Google Scholar]
- McLauchlin, J. , Hampton, M.D. , Shah, S. , Threlfall, E.J. , Wieneke, A. , and Curtis, G.D. (1997) Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J Appl Microbiol 83: 381–388. [DOI] [PubMed] [Google Scholar]
- Mesa‐Cruz, J. B. (2018) Assessment of physiological challenges in overwintering American black bears (Ursus americanus): active gestation, neonatal growth, and skeletal muscle conservation.
- Mitchell, M.S. , Zimmerman, J.W. , and Powell, R.A. (2002) Test of a habitat suitability index for black bears in the southern Appalachians. Wildl Soc Bull 30: 794–808. [Google Scholar]
- Moeller, K.T. , Moeller, A.K. , Moyano, F. , and Lundgren, E.J. (2017) Observation of an American black bear eating odonates in Yosemite National Park. West North Am Nat 77: 99–101. [Google Scholar]
- Moura, A. , Criscuolo, A. , Pouseele, H. , Maury, M.M. , Leclercq, A. , Tarr, C. , et al (2016) Whole genome‐based population biology and epidemiological surveillance of Listeria monocytogenes . Nat Microbiol 2: 16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullapudi, S. , Siletzky, R.M. , and Kathariou, S. (2008) Heavy‐metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey‐processing plants. Appl Environ Microbiol 74: 1464–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North Carolina Wildlife Resources Commission (2019) Bears in Residential Areas. URL https://www.ncwildlife.org/Learning/Species/Mammals/Black-Bear/Bears-in-Residential-Areas. [Google Scholar]
- Nowakiewicz, A. , Zięba, P. , Ziółkowska, G. , Gnat, S. , Muszyńska, M. , Tomczuk, K. , et al (2016) Free‐Living species of carnivorous mammals in Poland: red fox, beech marten, and raccoon as a potential reservoir of Salmonella, Yersinia, Listeria spp. and coagulase‐positive Staphylococcus . PLoS ONE 11: e0155533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, F. J. , Blanchet, F.G. , Friendly, M. , Kindt, R. , Legendre, P. , Mcglinn, D. , et al (2017) vegan: Community Ecology Package. R package version 2.4‐4. [Google Scholar]
- Painter, J. , and Slutsker, L. (2007) Listeriosis in humans In Listeria, Listeriosis and Food Safety. Boca Raton, FL, USA: CRC Press, pp. 85–109. [Google Scholar]
- Parsons, C. , Lee, S. , Jayeola, V. , and Kathariou, S. (2017) Novel cadmium resistance determinant in Listeria monocytogenes . Appl Environ Microbiol 83: e02580–16. 10.1128/AEM.02580-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patz, J.A. , Graczyk, T.K. , Geller, N. , and Vittor, A.Y. (2000) Effects of environmental change on emerging parasitic diseases. Int J Parasitol 30: 1395–1405. [DOI] [PubMed] [Google Scholar]
- Pirie, J.H. (1927) A new disease of veld rodents. “Tiger river disease”. Publ S Afr Inst Med Res 3: 163–187. [Google Scholar]
- Quessy, S. , and Messier, S. (1992) Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring‐billed gulls (Larus delawarensis). J Wildl Dis 28: 526–531. [DOI] [PubMed] [Google Scholar]
- R Foundation for Statistical Computing (2016) R: A Language and Environment for Statistical Computing. Vienna: Austria: R A Lang Environ Stat Comput 331. [Google Scholar]
- Ratani, S.S. , Siletzky, R.M. , Dutta, V. , Yildirim, S. , Osborne, J.A. , Lin, et al (2012) Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl Environ Microbiol 78: 6938–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppanner, R. , Jessup, D.A. , Ohishi, I. , Behymer, D.E. , and Franti, C.E. (1982) Serologic survey for certain zoonotic diseases in black bears in California. J Am Vet Med Assoc 181: 1288–91. [PubMed] [Google Scholar]
- Sasaki, Y. , Goshima, T. , Mori, T. , Murakami, M. , Haruna, M. , Ito, K. , and Yamada, Y. (2013) Prevalence and antimicrobial susceptibility of foodborne bacteria in wild boars (Sus scrofa) and wild deer (Cervus nippon) in Japan. Foodborne Pathog Dis 10: 985–991. [DOI] [PubMed] [Google Scholar]
- Sauders, B.D. , Overdevest, J. , Fortes, E. , Windham, K. , Schukken, Y. , Lembo, A. , and Wiedmann, M. (2012) Diversity of Listeria species in urban and natural environments. Appl Environ Microbiol 78: 4420–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan, E. , Hoekstra, R.M. , Angulo, F.J. , Tauxe, R.V. , Widdowson, M.A. , Roy, S.L. , et al (2011) Foodborne illness acquired in the United States‐Major pathogens. Emerg Infect Dis 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum, A. (2018) R Interface to Weather Underground API. [Google Scholar]
- Stephenson, N. , Higley, J.M. , Sajecki, J.L. , Chomel, B.B. , Brown, R.N. , and Foley, J.E. (2015) Demographic characteristics and infectious diseases of a population of American black bears in Humboldt county, California. Vector‐Borne Zoonotic Dis 15: 116–123. [DOI] [PubMed] [Google Scholar]
- Swaminathan, B. , and Gerner‐Smidt, P. (2007) The epidemiology of human listeriosis. Microbes Infect 9: 1236–1243. [DOI] [PubMed] [Google Scholar]
- U.S. Geological Survey (2011) National Land Cover, Version 2. Gap Anal Progr. [Google Scholar]
- USCB (2016) US Census Bureau 2010 Census. [Google Scholar]
- Vivant, A.‐L. , Garmyn, D. , and Piveteau, P. (2013) Listeria monocytogenes, a down‐to‐earth pathogen. Front Cell Infect Microbiol 3: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacheck, S. , Fredriksson‐Ahomaa, M. , König, M. , Stolle, A. , and Stephan, R. (2010) Wild Boars as an important reservoir for foodborne pathogens. Foodborne Pathog Dis 7: 307–312. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Lu, L. , Lan, R. , Salazar, J.K. , Liu, J. , Xu, J. , and Ye, C. (2017) Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg Microbes Infect 6: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindl, L. , Frank, E. , Ullrich, U. , Heurich, M. , Kleta, S. , Ellerbroek, L. , and Gareis, M. (2016) Listeria monocytogenes in different specimens from healthy red deer and wild boars. Foodborne Pathog Dis 13: 391–397. [DOI] [PubMed] [Google Scholar]
- Weis, J. , and Seeliger, H.P. (1975) Incidence of Listeria monocytogenes in nature. Appl Microbiol 30: 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshimer, H.J. (1968) Isolation of Listeria monocytogenes from vegetation. J Bacteriol 95: 300–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, I. (2007) Listeriosis in animals In Listeria, Listeriosis, and Food Safety. Elliot T., and Ryser E. H. M. (eds). Boca Raton: CRC Press, p. c2007. [Google Scholar]
- Xu, D. , Li, Y. , Zahid, M.S.H. , Yamasaki, S. , Shi, L. , Li, J. , and Yan, H. (2014) Benzalkonium chloride and heavy‐metal tolerance in Listeria monocytogenes from retail foods. Int J Food Microbiol 190: 24–30. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Sugimoto, T. , Sato, M. , and Hirai, K. (2000) Incidence of Listeria monocytogenes in wild animals in Japan. J Vet Med Sci 62: 673–675. [DOI] [PubMed] [Google Scholar]


