Abstract
Circulating tumor DNA (ctDNA) describes the fragmented DNA released from tumor cells into the blood. The ctDNA may have the same genetic changes as the primary tumor. Currently, ctDNA has become a popular biomarker for diagnosis, treatment, real-time clinical response monitoring, and prognosis, for solid tumors. Detection of ctDNA is minimally invasive, and repeat sampling can easily be performed. However, due to its low quality and short DNA fragment length, ctDNA detection still faces challenges and requires highly sensitive analytical techniques. Recently, liquid biopsies for the analysis of circulating tumor cells (CTCs) and circulating tumor-derived exosomes have been studied, and nanotechnology techniques have rapidly developed. Compared to traditional analytical methods, these nanotechnology-based platforms have the advantages of sensitivity, multiplex detection, simplicity, miniaturization, and automation, which support their potential use in clinical practice. This review aims to discuss the recent nanotechnological strategies for ctDNA analysis and the design of reliable techniques for ctDNA detection and to identify the potential clinical applications.
MeSH Keywords: Circulating Tumor DNA, Liquid Biopsy, Nanomedicine, Nanotechnology, Nanomaterials
Background
The detection and analysis of tumor-derived biomarkers is a key factor in current patient management and treatment decisions. Recently, blood-based liquid biopsies, which sample circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), circulating tumor-derived exosomes, and tumor-educated platelets (TEPs), have attracted increasing attention due to their non-invasive sampling and ability to serially assess disease progression as an alternative to conventional tissue biopsy or cytology [1,2].
Circulating-free DNA (cfDNA) is fragmented DNA present in the blood, released through cell apoptosis, necrosis, and autophagy [3,4]. The cfDNA from tumor cells, also known as ctDNA, carries tumor-specific genomic alterations, including mutations, methylation, loss of heterozygosity, and microsatellite instabilities [5,6]. Due to their short half-life in the circulation, from between 16 minutes to 2.5 hours [7], ctDNA analysis is applicable to the real-time monitoring of tumor burden. Also, tumor genotyping based on the detection of ctDNA has advantages, particularly in precision medicine, as a safe and minimally invasive alternative to tissue biopsy. For example, there are activating and resistant mutations of the epidermal growth factor receptor (EGFR) gene, which is an important driving gene in non-small cell lung cancer (NSCLC) that responds to EGFR tyrosine kinase inhibitors (EGFR-TKIs) [8]. Specifically, resistant EGFR T790M mutation in NSCLC occurs less commonly in primary tumors compared with activating mutations, such as the exon 19 deletion and EGFR L858R [9]. In 2016, liquid biopsy detection of EGFR mutations was approved for the assessment of treatment response following therapy with EGFR-TKIs in patients with NSCLC [10].
Currently, ctDNA analysis remains challenging. The challenges include large variability in the concentration of plasma cfDNA varying from 0 to more than 1,000 ng/mL in human samples [11], where ctDNA often representing a small fraction of the total cfDNA, which can be as low as 0.01% [12], and the most common ctDNA fragment length is less than 167 bp [13–15]. Therefore, ctDNA detection requires high-sensitive techniques. More importantly, the analytical methods used for the detection of mutant ctDNA must have a high specificity to avoid interference from wild-type genes that are released from normal cells. Therefore, multiplexed detection approaches have significant advantages in a small sample volume and for precision therapy.
Currently, the main techniques for ctDNA analysis are mainly based on sequencing, quantitative real-time polymerase chain reaction (qRT-PCR), or digital PCR (dPCR) platforms. Each one of these techniques has its own advantages and limitations in terms of sensitivity, specificity, and multiplexed detection (Table 1). For example, next-generation sequencing (NGS) can offer high-throughput analysis with modest sensitivity, but is time-consuming [16,17]. The qRT-PCR-based platforms, such as amplification refractory mutation system (ARMS) [18,19] and peptide nucleic acid (PNA) clamping PCR [20,21], have high specificity and wide application due to the rapidity and ease of implementation, but their sensitivity varies greatly in plasma ctDNA analysis. Currently, droplet digital PCR (ddPCR) is an ultra-sensitive and accurate method for the detection of trace amounts of ctDNA and single-molecule analysis [22,23]. However, conventional ddPCR and qRT-PCR are only suitable for a specific type of molecular detection in a single reaction system, which means there is a need to perform multiplex ctDNA analysis by increasing or diluting the required cfDNA sample. Therefore, the key drawbacks of current methods limit their application and partly restrict the application of ctDNA in clinical practice.
Table 1.
Overview of the conventional and nanomaterials-based methods for the detection of circulating tumor DNA (ctDNA).
| Method | Features | Detection limit | Advantages | Limitations |
|---|---|---|---|---|
| NGS | PCR-based sequencing | 0.1–5% [17] | High-throughput analysis, detection of unknown genetic alterations | Time-consuming and expensive detection; low sensitivity |
| ARMS | qRT-PCR based on the 3′-end of the primer targeting the mutant sequence for extension | 0.2% [18] | Ease of use, lower cost | Detection of known mutations, lower sensitivity |
| PNA-PCR | qRT-PCR based on PNA inhibiting the extension of the wild-type sequence | 0.1% [20] | High specificity, ease of use | Detection of known mutations, lower sensitivity |
| ddPCR | DNA templates amplified separately in water-oil droplets and quantified | 0.001% (20/200,000 copies) [22], 0.04% [23] |
High sensitivity, a single molecule analysis, absolute quantitation | Detection of known mutations |
| BEAMing | DNA templates amplified on beads and quantified | 0.01% (1/10,000 copies) [28] | High sensitivity, a single molecule analysis, absolute quantitation | Detection of known mutations, pre-amplification of the target sequence |
| PCR-SERS | PCR products analyzed by SERS detection | 0.1% (10/10,000 copies) [42]; 9.24–59.7 pM [40,41]. |
Multiplexed analysis | Detection of known mutations. |
| PAPL | DNA templates amplified by PNA-aPCR and detected by suspension array | 0.02–0.05% (2/10,000–5/10,000 copies) [47] | Multiplexed analysis, high specificity, high sensitivity | Detection of known mutations, qualitative analysis |
| Fe-Au nanoparticle-coupling strategy | Hybridization-based detection | 0.12% [55] | PCR-independent detection | Lower sensitivity, detection of known mutations |
| Electrochemical DNA biosensor | Detection based on generated electronic signal | 1 aM–1.72 fM (mutations) [60,62]; 2 fM–42 pM (DNA methylation) [59,63,70,71] |
High sensitivity, PCR-independent analysis, detection of methylated DNA without sample pretreatment | Detection of known genetic alterations |
| IC3D ddPCR | ddPCR analysis based on the microfluidic device | 0.00125–0.005% [76] | High sensitivity, a single molecule analysis, miniaturization | Detection of known mutations |
NGS – next-generation sequencing; ARMS – amplification refractory mutation system; qRT-PCR – quantitative real-time polymerase chain reaction; PNA-PCR – peptide nucleic acid clamping polymerase chain reaction; ddPCR – droplet digital polymerase chain reaction; BEAMing – beads, emulsion, amplification, magnetics; SERS – surface-enhanced Raman spectroscopy; PNA – peptide nucleic acid; aPCR – asymmetric polymerase chain reaction; PAPL – peptide nucleic acid clamping asymmetric polymerase chain reaction and liquidchip; IC3D – integrated comprehensive droplet.
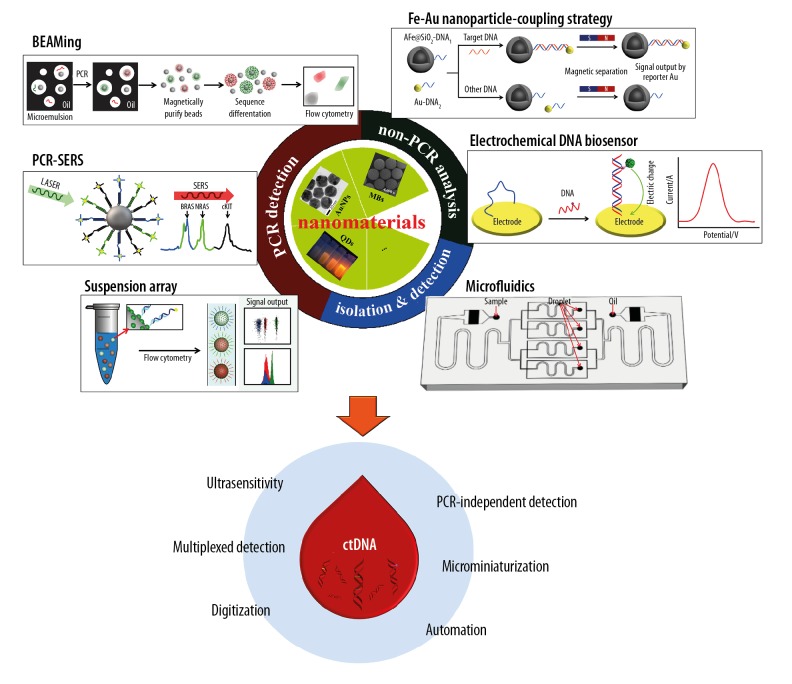
Nanomedicine is a new branch of medicine that has contributed to breakthroughs in both the diagnosis and treatment of human disease. Recently, the rapid development of nanotechnology has led to technological progress in the use of liquid biopsies, particularly in the isolation of CTCs or circulating tumor-derived exosomes [24,25]. Nanometer-sized particles, such as iron oxide nanocrystals, gold (Au) nanoparticles (AuNPs), and quantum dots (QDs) have novel optical, electronic, magnetic, or structural properties that differ from solid compounds [24,26]. Several nanomaterials-based approaches to DNA analysis have been reported, but few have been applied to ctDNA detection. Therefore, this review aims to discuss the recent nanotechnological strategies for ctDNA analysis (Figure 1), the design of reliable techniques for ctDNA detection, and to identify the potential clinical applications.
Figure 1.
Nanomaterials-based strategies for detection and analysis of circulating tumor DNA (ctDNA). This figure shows the beads, emulsion, amplification, magnetics (BEAMing) method, modified from Dressman et al., 2003 [27]; the polymerase chain reaction and surface-enhanced Raman spectroscopy (PCR-SERS) method, modified from Wee et al., 2016 [42]; the suspension array method, modified from Chen et al., 2019 [47]; Fe-Au nanoparticle-coupling strategy, modified from Hu et al., 2018 [55]; the electrochemical DNA biosensor, modified from Campuzano et al., 2019 [57]; and microfluidics method, modified from Ou et al., 2019 [76].
Nanomaterials-Based Techniques for ctDNA Analysis
Beads, emulsion, amplification, and magnetics (BEAMing)
BEAMing is a digital PCR-based platform that combines magnetic beads-based emulsion PCR and flow-cytometry to identify and quantify mutant DNA [27]. The principal process of the commercial BEAMing (OncoBEAM™) (Sysmex Inostics, San Diego, CA, USA) involves pre-amplification of the genetic regions of interest, the formation of a water/oil emulsion and PCR amplification (emulsion PCR), breaking of the emulsions and hybridization to fluorescent probes, and the flow-cytometry readout [12,27]. A key feature unique to BEAMing is that each created microscopic emulsion droplet contains a single DNA molecule and a primer-coated magnetic bead. Following a conventional PCR amplification, the bead is coated with thousands of target DNA fragments, to provide a digital readout of copy number for further flow-cytometry analysis. BEAMing delivers a high-sensitive detection for rare mutant DNA against a large background of wild-type genes (1/10,000) [28]. Until recently, BEAMing has been used to detect multiple ctDNA mutations, for example, PIK3CA, BRAF, EGFR, KRAS, NRAS, IDH1, and ESR1, which have demonstrated a high concordance between ctDNA in patient blood samples and somatic mutations detected in tumor tissue [29–36]. As a digital PCR-based analysis platform, BEAMing, like ddPCR, is more sensitive than other assays for target sequence detection and is suitable for monitoring disease progression according to the absolute ctDNA quantitation [22,28].
PCR-surface enhanced Raman spectroscopy (SERS)
Raman scattering is a phenomenon that the scattered light has frequency change due to the interaction between oscillation of light and molecular vibration [37,38]. Because a Raman spectrum contains unique molecular fingerprint vibrational information on the sample, Raman spectroscopy contributes to the analysis of the chemical components, the molecular structure, and the interaction between molecules [38]. Although the Raman scattering signal is intrinsically weak, it can be significantly enhanced when the analyte is placed on or near a roughened noble-metal substrate, which is either Au (gold) or Ag (silver), known as SERS [39]. The main SERS mechanisms include electromagnetic and chemical enhancements [38,39]. Electromagnetic enhancement relies on the excitation of the localized surface plasmon resonance (LSPR) of nanostructured or nanoparticle metal surface, and it contributes an average enhancement factor of more than 10,000 [38,39]. In the chemical enhancement, which is now believed to contribute an average enhancement factor of 100, a charge-transfer state is created between the mental and the adsorbate molecules [38,39]. The SERS-based PCR method is suitable for multiplexed ctDNA detection. Li [40,41] and coworkers employed two different SERS-PCR strategies to detect multiple mutant genes. In the study of EGFR mutations, they used multiplexed PCR to amplify target mutant genes with primers labeled with the different fluorescence tags R6G and Cy5, and analyzed the purified PCR products via Ag colloid-based SERS detection [40]. The results showed that the limit of detection (LOD) was 5.97×10−11 M and 9.24×10−12 M for EGFR exon 19 and EGFR exon 21, respectively [40]. A further study used PCR without any modification of primers to amplify BRAF, KRAS, and PIK3CA mutations in colorectal cancer patients, and employed dye-labeled probes to tag mutant sequences for the following SERS detection [41]. The results showed that the LOD of this method was 5.15×10−11 M [41].
In a different approach, in 2016, Wee et al. [42] developed a nanotag-based PCR-SERS assay for multiplexed point mutations detection. In this assay, multiple mutant PCR amplicons were enriched using streptavidin-coated magnetic beads (SMB) in combination with a biotin molecular at the 5′-end of the reverse primer, and traced with specific SERS nanotags complementary to a unique barcode sequence of the forward primer. SERS nanotags are AuNPs modified with DNA probes and Raman reporters such as 4-mercaptobenzoic (MBA), 2,7-mercapto-4-methyl coumarin (MMC), or 4-mercapto-3-nitrobenzoic acid (MNBA). After enrichment with the SMB to remove the excess SERS nanotags, the mutation status was evaluated using Raman spectroscopy, where unique spectral peaks indicate the presence of the mutation of interest. Using this technique, the investigators identified BRAF V600E, c-Kit L576P, and NRAS Q61K mutations in melanoma with a sensitivity of 0.1% (10/10,000) [42].
Recently, in 2019, Lin et al. [43] developed a dual signal amplification SERS method based on the use of metal carbonyls (Re-SCO and Os-SCO) as probes and substrates to detect low concentrations of Epstein-Barr virus (EBV) DNA. In this method, the target sequences captured by biotinylated DNA probes were first combined with the prepared SERS substrate (Au-Os-SCO-Au) via biotin-streptavidin interaction, and chelated by daunorubicin (DNR)-SiO2@Au-Re-SCO. After irradiation with an 808 nm laser, a number of Re-SCO labels were released from mesoporous silica-coated Au nanorods (SiO2@Au) and bound to the surface of the SERS substrate. By measuring the SERS signal intensity of Re-SCO labels at 2113 cm−1, the target DNA was able to be quantified. Their PCR-free SERS results demonstrated that the LOD was 57.74 nM (5.774×10−8 M) [43]. Compared to PCR-free SERS analysis, this method might detect much lower concentrations of DNA if PCR were performed to enrich target sequences.
Suspension array (liquidchip)
Suspension array, or liquidchip, is a high-throughput, large-scale, and multiplexed screening platform utilized to evaluate biomolecules. This technical platform uses encoded microspheres and permits the parallel testing of multiple gene variants, as each type of microsphere has a single identification based on variations in the optical properties [44]. The first commercial suspension array (Luminex xMAP Technology) incorporates 5.6 μm polystyrene microspheres loaded with two organic dyes with different wavelengths to create a library of barcodes using flow cytometry [45]. Using these encoded microspheres, Li et al. [46] developed a SurPlex®-xTAG70plex-EGFR liquidchip platform to detect 70 alleles of EGFR, KRAS, BRAF, and PIK3CA mutations from formalin-fixed and paraffin-embedded tissue sections. The method involves multiplex PCR, allele-specific primer extension, hybridization, and flow-cytometry. However, this high-throughput analysis method only detects 1,000 copies of mutant DNA against a background of 20,000–100,000 copies of wild-type DNA (sensitivity, 1–5%). Compared with other high-throughput DNA analysis platforms such as sequencing and microarray, suspension assay is more rapid and less costly cost. However, the lower sensitivity restricts its clinical application.
Suspension array for nucleic acid analysis is a hybridization-based platform. A useful measure to increase its sensitivity is to enhance the special enrichment of target sequences. From this perspective, our group developed PNA-aPCR-liquidchip (PAPL) as a unique method for multiple mutant ctDNA detection consisting of multiplex PNA clamping asymmetric PCR (PNA-aPCR) amplification and liquidchip test [47]. PNA is a DNA mimic wherein the deoxyribose-phosphate backbone is replaced by an oligoamide formed of N-(2-aminoethyl)-glycine units [48]. The complementary PNA/DNA heteroduplex has higher thermal stability than the corresponding DNA/DNA duplex, and even a base-pair mismatch could result in the unstable binding between PNA and DNA [49,50]. Consequently, there is a unique amplification of the target Cy5-labeled mutant sequences in PNA-aPCR process due to the PNAs as well as special primers blocking their wild-type sequences. Without purification or enzyme digestion, the whole multiplex PCR products directly hybridize with oligonucleotide-coupled dual-encoded magnetic beads (DEMBs). The formed Cy5-DNA/DEMBs complexes are detected by flow cytometer to perform multiplexed analysis according to high fluorescent signals on beads. Different from the commercial Luminex xTAG platform, DEMBs developed in our lab consist of QDs-encoded host particle and fluorescein-5-isothiocyanate (FITC)-labelled guest nanoparticles [51,52]. QDs, also known as nanoscale semiconductor crystals, are generally composed of elements from groups II–VI and III–V in the periodic table, including Ag, Cd, Hg, Ln, P, Pb, Se, Te, and Zn [53]. Compared with conventional organic dyes, QDs have narrow and symmetrical emission spectra, broad excitation wavelength, emission wavelengths that can be tuned, and high brightness [44,54]. These characteristics make QDs ideal candidates for the creation of a diverse barcode array. When QDs and organic dyes are used together, the potential fluorescence interference can be identified using a barcode library. In addition to two separate fluorescent building blocks, DEMBs are assembled by simple and rapid magnetic separation, rather than centrifugation, due to their excellent magnetic host particles. We applied the PAPL assay to detect three high-frequent EGFR mutations, exon 19 deletion, L858R, and T790M, in patients with NSCLC. We identified that 2–5 copies of mutant EGFR were distinguished from 10,000 copies of wild-type EGFR, with a sensitivity of between 0.02–0.05%. Also, there was a high concordance rate of 88–95% between the EGFR mutational state in plasma cfDNA by PAPL and that by ddPCR. These findings show that the PAPL assay can be an effective analytical technique for multiplexed detection of mutant ctDNA.
Fe-Au nanoparticle-coupling strategy
Currently, almost all DNA analysis methods involve PCR amplification to enrich target DNA molecules to achieve highly sensitive detection. PCR is the most commonly used technique for DNA analysis, which has undergone three main technological advances that include qualitative analysis, relative quantitation (qRT-PCR), and absolute quantitation (dPCR), respectively. However, these methods have limitations, such as contamination of the PCR sample and non-specific amplification. With the rapid development in molecular technology, there are now many PCR-independent approaches to DNA analysis. For example, in 2018, Hu et al. [55] used AFe@SiO2 and AuNPs to develop a Fe-Au nanoparticle-coupling strategy to identify the KRAS mutations in serum samples from lung cancer patients. In this technique, target single-stranded DNA (ssDNA) hybridizes to capture both DNA-coupling AFe@SiO2 (AFe@SiO2-DNA1) and AuNPs (Au-DNA2) [55]. AFe@SiO2 magnetic nanoparticles are used to separate and enrich the formed AFe@SiO2-DNA1/target DNA/Au-DNA2 complex from the solution [55]. Since AuNPs serve as reporters, the quantity in the separated products enables the quantification of the target DNA. However, although the Fe-Au nanoparticle-coupling strategy is a PCR-independent and hybridization-based platform, it exhibited high specificity and sensitivity for the mutant KRAS detection at a low concentration (0.1 pg/mL) [55]. To enhance the detection of trace mutant ctDNA in serum samples, PNA-functionalized magnetic nanoparticles can be used to remove wild-type sequences and complementary strands of the target KRAS mutant sequences before the nanoparticle-coupling step [55]. The results showed that Fe-Au nanoparticle-coupling technology detected KRAS mutations down to 0.12%, with the findings in agreement with those obtained with the ddPCR assay [55].
Electrochemical DNA biosensors
The electrochemical DNA biosensor is an analytical device that converts a target DNA response into an electronic signal [56]. Based on the linear target DNA concentrations, electrochemical biosensors not only assess the LOD, but also quantify target DNA molecules. The electrochemical DNA sensing platform consists mainly of capture probes immobilized on a surface of an electrode to capture target sequences and signal probes using electrochemical tags for signal generation. The high specificity of electrochemical biosensors relies on a special combination between capture probes and target DNA molecules. Currently, the reported capture probes in electrochemical biosensors involve ssDNA, DNA tetrahedron, and PNA [57]. The PNA probe has a higher specificity due to its increased thermal stability compared with that of the DNA probe [58–61]. The high sensitivity of electrochemical biosensors is due to signal amplification. One of the popular amplification strategies is the introduction of nanoparticles, such as AuNPs, carbon nanomaterials, and MoS2 nanosheets [58,62–64]. Functional nanomaterials can not only produce a synergic effect in catalytic activity, conductivity, and biocompatibility to accelerate the signal transduction as electrode materials, but also amplify biorecognition events with specifically designed signal tags, leading to high sensitivity detection [65]. Until recently, electrochemical biosensors could detect DNA mutations at 1×10−18 M with a combination of nanoparticles and biological amplification, such as the hybridization chain reaction (HCR), and the dual enzyme (ribonuclease H II and terminal deoxynucleotidyl transferase) assay [60,66].
DNA methylation is an important epigenetic modification that governs gene expression and usually occurs in the cytosine within the CpG dinucleotide to form 5-methycytosine (5-mC) [67]. Appropriate DNA methylation is essential for cell development and function. Therefore, any abnormalities in this process may lead to disease, including cancer [67,68]. Conventional methods for DNA methylation analysis include qRT-PCR, sequencing, and microarray, which require DNA sample pretreatment using bisulfite, methylation-sensitive restriction enzymes, or affinity purification [69]. Without sample pretreatment, electrochemical biosensors use a monoclonal antibody to 5-mC as the biorecognition element to detect methylated DNA. In 2018, Povedano et al. [63] presented two electrochemical biosensing platforms, the DNA sensor and the immunosensor, for the detection of methylated DNA using functionalized magnetic beads (MBs), the antibody to 5-mC as affinity bioreceptor, and amperometric detection of screen-printed carbon electrodes (SPCE) using the hydrogen peroxide/hydroquinone (H2O2/HQ) system. The DNA sensor is formed from DNA probes on the MB surface to capture the special methylated sequences, and the methylation in the captured target DNA is recognized by the specific antibody to 5-mC tagged with a secondary antibody, the horseradish peroxidase (HRP)-conjugated anti-mouse IgG [63]. The LOD of the DNA sensor was reported to be 26 pM (2.6×10−11 M) for the O-6-methylguanine-DNA methyltransferase (MGMT) gene and 42 pM (4.2×10−11 M) for Ras association domain family 1 isoform A (RASSFIA), respectively [63]. Different from the DNA sensor, the immunosensor uses two different antibodies that include the antibody to 5-mC immobilized on the MB surface to capture any methylated ssDNA sequence, and a second antibody conjugated with horseradish peroxidase (HRP) anti-ssDNA or HRP anti-FITC Fab, used as the detector [63,70]. Also, the immunosensor assay can detect global methylation using the HRP-anti-ssDNA as the detection antibody and identifies the locus-specific methylation with the HRP-anti-FITC Fab antibody. In 2019, Povedano et al. reported that methylated MGMT analysis indicated that the LOD of their immunosensor was 1.2 pM (1.2×10−12 M) [70]. Daneshpour et al. [71] described a previously developed nanobiosensor to identify target DNA methylation using the antibody to 5-mC immobilized on a SPCE surface and DNA probe-labeled Fe3O4/N-trimethyl/gold (Fe3O4/TMC/Au) nanocomposites as the gene-trapping and signal amplification unit. Based on the linear range of RASSF1A concentrations (1×10−14 to 5×10−9 M), the estimated LOD of this DNA nanobiosensor was reported to be 2×10−15 M [71]. Cai et al. developed another assay in 2018 [59], which uses gold PNA probe nanoparticles (PNA-AuNPs) and a lead phosphate apoferritin (LPA)-based dual biomarker detection platform to identify tumor-specific mutations and methylation of PIK3CA. The PNA probe and the antibody to 5-mC are used to recognize the different parts of ctDNA [59]. However, the AuNPs and the LPA are introduced for dual signal amplification in a biosensor, and the DNA biosensor yielded a linear current response to ctDNA concentrations (5×10−14 to 1×10−11 M) with a LOD of 1×10−14 M [59]. Therefore, electrochemical biosensing is an interesting approach to mutations and methylated ctDNA detection due to its speed, simplicity, low sample requirement, sensitivity, miniaturization, low cost, and PCR-independent detection method.
Microfluidics
Microfluidics is a technique for handling small volumes of liquids in picoliters to nanoliters [72]. The overall reaction within the microfluidic system involves increased heat transfer, higher surface-to-volume ratio, and controllable mixing efficiency and diffusion rates, resulting in higher energy transfer to reactive species [73]. Microfluidic devices are miniaturized devices that integrate multiple processes. There have been some recently published studies that have reported cfDNA purification on microfluidic devices through MBs-mediated enrichment. For example, in 2018, Kim et al. [74] developed a centrifugal lab-on-a-disc system equipped with electromagnetically-actuated and reversible diaphragm values to isolate cfDNA from whole blood (3 mL) in a fully automated manner with the entire process of cfDNA purification including plasma separation, protein lysis, cfDNA binding, multiple washing steps, and elution of the cfDNA in less than 30 minutes [74]. Also, in 2019, Gwak et al. [75] developed a microfluidic chip with a gradient magnetic-activated cfDNA sorter (the vortex-GMACS) to spontaneously isolate cfDNA using silica magnetic particles in a method that took 19 minutes without any limitation of the sample volume.
In addition to ctDNA isolation, microfluidic devices demonstrate enormous potential for ctDNA detection. Also, in 2019, Ou et al. [76] developed an ultrasensitive IC3D droplet digital detection PCR (IC3D ddPCR) system that combines droplet microfluidics, fluorescence multiplex PCR amplification, and a high-throughput 3D particle counting system on a microfluidic chip to detect ctDNA. The results from this recent methodological study showed that the IC3D ddPCR system could identify KRAS G12D mutant alleles against a background of wild-type genomes at the high sensitivity of 0.00125–0.005% [76]. Compared to the commercial ddPCR kit (Bio-Rad, Hercules, CA, USA), this ddPCR platform accommodates a larger sample volume (20 μg/mL) and greater numbers of partitions (18 million reactions per mL) [76]. Microfluidic devices have attracted increasing attention in liquid biopsies due to their automation, microminiaturization, portability, high sensitivity, low-sample requirement, and speed of use.
Conclusions
Currently, ctDNA is an attractive substitute for conventional early tumor tissue molecular diagnosis, the evaluation of treatment efficacy, monitoring of tumor dynamics, monitoring of tumor progression, and prediction of cancer recurrence. There are different diagnostic methods to detect tumor-specific DNA in carcinoma patients. However, only a few approaches are suitable for ctDNA detection. Due to its short fragment length and low quantity, ctDNA analysis requires more sensitive techniques to ensure its reliability compared to methods used in tissue biopsies. It is worth noting that the existing techniques for ctDNA analysis focus mainly on the identification of mutations in advanced cancer treatment, but rarely on early diagnosis at a low concentration of ctDNA. Other genetic alterations, for example, methylation, loss of heterozygosity, and microsatellite instability are not currently analyzed. Therefore, ctDNA analysis is facing significant technological challenges, which may ultimately determine the potential clinical application. The nanotechnological platforms for ctDNA analysis discussed in this review have highlighted their advantages in terms of analytical sensitivity, multiplexed detection, simplicity, and cost benefits (Table 1). Generally, nanomaterials are used to enhance traditional analytical methods and develop new detection platforms. However, current nanomaterial-based ctDNA analysis is in its early stages and faces technical challenges in terms of stability, sensitivity, and specificity. In addition to BEAMing, PCR-SERS, the suspension assay, the Fe-Au nanoparticle-coupling strategy, electrochemical DNA biosensors, and microfluidics remain in use in academic research studies and have yet to be evaluated with a large number of clinical specimens. Therefore, there is great potential for nanotechnology-based approaches to ctDNA analysis, and it is only a matter of time before they are approved for clinical use. With the rapid development of this technology, we anticipate that there will be some novel platforms for ctDNA analysis with unique characteristics, including the non-PCR amplification, multiplexed detection, simplicity, rapidity, micro miniaturization, automation, and digital detection to meet future clinical demands.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by the Shanghai Jiao Tong University Fund (No. YG2015ZD11), and the Innovation Group Project of Shanghai Municipal Health Commission (No. 2019CXJQ03)
References
- 1.Vaidyanathan R, Soon RH, Zhang P, et al. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2018;19:11–34. doi: 10.1039/c8lc00684a. [DOI] [PubMed] [Google Scholar]
- 2.Poulet G, Massias J, Taly V. Liquid biopsy: General concepts. Acta Cytol. 2019;63:449–55. doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 3.Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–42. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 4.Thierry AR, Messaoudi SEI, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–76. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 6.Crowley E, Nicolantonio FD, Loupakis F, et al. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–84. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 7.To EW, Chan KC, Leung SF, et al. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res. 2003;9:3254–59. [PubMed] [Google Scholar]
- 8.Lee DH. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): The road to a success, paved with failures. Pharmacol Ther. 2017;174:1–21. doi: 10.1016/j.pharmthera.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Shea M, Costa DB, Rangachari D. Management of advanced non-small cell lung cancers with known mutations or rearrangements: Latest evidence and treatment approaches. Ther Adv Respir Dis. 2016;10:113–29. doi: 10.1177/1753465815617871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5:46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer – a survey. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102:16368–73. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang P, Lo YMD. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. 2016;32:360–71. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto Y, Uemura M, Fujita M, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019;110:617–28. doi: 10.1111/cas.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: Focus on non-small cell lung cancer patients. Transl Lung Cancer Res. 2016;5:505–10. doi: 10.21037/tlcr.2016.10.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vollbrecht C, Lehmann A, Lenze D, et al. Validation and comparison of two NGS assays for the detection of EGFR T790M resistance mutation in liquid biopsies of NSCLC patients. Oncotarget. 2018;9:18529–39. doi: 10.18632/oncotarget.24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Xu H, Su S, et al. Clinical validation of a highly sensitive assay to detect EGFR mutations in plasma cell-free DNA from patients with advanced lung adenocarcinoma. PLoS One. 2017;12:e0183331. doi: 10.1371/journal.pone.0183331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng WN, Gu WQ, Zhao N, et al. Comparison of the SuperARMS and droplet digital PCR for detecting EGFR mutation in ctDNA from NSCLC patients. Transl Oncol. 2018;11:542–45. doi: 10.1016/j.tranon.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guha M, Castellanos-Rizaldos E, Makrigiorgos GM. DISSECT method using PNA-LNA clamp improves detection of EGFR T790m mutation. PLoS One. 2013;8:e67782. doi: 10.1371/journal.pone.0067782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han JY, Choi JJ, Kim JY, et al. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer. 2016;16:627. doi: 10.1186/s12885-016-2678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe M, Kawaguchi T, Isa S, et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21:3552–60. doi: 10.1158/1078-0432.CCR-14-2151. [DOI] [PubMed] [Google Scholar]
- 23.Zhu G, Ye X, Dong Z, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015;17:265–72. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Bhana S, Wang Y, Huang X. Nanotechnology for enrichment and detection of circulating tumor cells. Nanomedicine. 2015;10:1973–90. doi: 10.2217/nnm.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma A, Khatun Z, Shiras A. Tumor exosomes: Cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine. 2016;11:421–37. doi: 10.2217/nnm.15.210. [DOI] [PubMed] [Google Scholar]
- 26.Raju GSR, Dariya B, Mungamuri SK, et al. Nanomaterials multifunctional behavior for enlightened cancer therapeutics. Semin Cancer Biol. :2019. doi: 10.1016/j.semcancer.2019.08.013. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Dressman D, Yan H, Traverso G, et al. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100:8817–22. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17:7808–15. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 29.Higgins MJ, Jelovac D, Barnathan E, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–69. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santiago-Walker A, Gagnon R, Mazumdar J, et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res. 2016;22:567–74. doi: 10.1158/1078-0432.CCR-15-0321. [DOI] [PubMed] [Google Scholar]
- 31.Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686) Clin Cancer Res. 2016;22:2386–95. doi: 10.1158/1078-0432.CCR-15-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3375–82. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grasselli J, Elez E, Caratu G, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28:1294–301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325–32. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen WW, Balaj L, Liau LM, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tycova A, Prikryl J, Foret F. Recent strategies toward microfluidic-based surface-enhanced Raman spectroscopy. Electrophoresis. 2017;38:1977–87. doi: 10.1002/elps.201700046. [DOI] [PubMed] [Google Scholar]
- 38.Zong C, Xu M, Xu LJ, et al. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem Rev. 2018;118:4946–80. doi: 10.1021/acs.chemrev.7b00668. [DOI] [PubMed] [Google Scholar]
- 39.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Surface-enhanced Raman spectroscopy. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:601–26. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Yang T, Li CS, et al. Detection of EGFR mutation in plasma using multiplex allele-specific PCR (MAS-PCR) and surface enhanced Raman spectroscopy. Sci Rep. 2017;7:4771. doi: 10.1038/s41598-017-05050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Yang T, Li CS, et al. Surface enhanced Raman spectroscopy (SERS) for the multiplex detection of Braf, Kras, and Pik3ca mutations in plasma of colorectal cancer patients. Theranostics. 2018;8:1678–89. doi: 10.7150/thno.22502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wee EJ, Wang Y, Tsao SC, Trau M. Simple, sensitive and accurate multiplex detection of clinically important melanoma DNA mutations in circulating tumour DNA with SERS nanotags. Theranostics. 2016;6:1506–13. doi: 10.7150/thno.15871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin D, Gong T, Qiu S, et al. A dual signal amplification nanosensor based on SERS technology for detection of tumor-related DNA. Chem Commun. 2019;55:1548–51. doi: 10.1039/c8cc07461e. [DOI] [PubMed] [Google Scholar]
- 44.Leng Y, Sun K, Chen X, Li W. Suspension arrays based on nanoparticle-encoded microspheres for high-throughput multiplexed detection. Chem Soc Rev. 2015;44:5552–95. doi: 10.1039/c4cs00382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li G, Luo X, He J, et al. A novel liquidchip platform for simultaneous detection of 70 alleles of DNA somatic mutations on EGFR, KRAS, BRAF and PIK3CA from formalin-fixed and paraffin-embedded slides containing tumor tissue. Clin Chem Lab Med. 2011;49:191–95. doi: 10.1515/CCLM.2011.040. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Zhang DS, Wang L, et al. Multiplexed detection of mutant circulating tumor DNA using peptide nucleic acid clamping asymmetric polymerase chain reaction and liquidchip. J Biomed Nanotechnol. 2019;15:1578–88. doi: 10.1166/jbn.2019.2794. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen PE, Egholm M. An introduction to peptide nucleic acid. Curr Issues Mol Biol. 1999;1:89–104. [PubMed] [Google Scholar]
- 49.Hyrup B, Nielsen PE. Peptide nucleic acids (PNA): Synthesis, properties and potential applications. Bioorg Med Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 50.Kyger EM, Krevolin MD, Powell MJ. Detection of the hereditary hemochromatosis gene mutation by real-time fluorescence polymerase chain reaction and peptide nucleic acid clamping. Anal Biochem. 1998;260:142–48. doi: 10.1006/abio.1998.2687. [DOI] [PubMed] [Google Scholar]
- 51.Zhang DS-z, Jiang Y, Yang H, et al. Dual-encoded microbeads through a host-guest structure: Enormous, flexible, and accurate barcodes for multiplexed assays. Advanced Functional Materials. 2016;26:6146–57. [Google Scholar]
- 52.Lu S, Zhang DS, Wei D, et al. Three-dimensional barcodes with ultrahigh encoding capacities: A flexible, accurate, and reproducible encoding strategy for suspension arrays. Chem Mater. 2017;29:10398–408. [Google Scholar]
- 53.Shekhar N, Bhanoji Rao ME. Quantum dot: novel carrier for drug delivery. International Journal of Research in Pharmaceutical and Biomedical Sciences. 2011;2:448–56. [Google Scholar]
- 54.Matea CT, Mocan T, Tabaran F, et al. Quantum dots in imaging, drug delivery and sensor applications. Int J Nanomedicine. 2017;12:5421–31. doi: 10.2147/IJN.S138624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu P, Zhang S, Wu T, et al. Fe-Au nanoparticle-coupling for ultrasensitive detections of circulating tumor DNA. Adv Mater. 2018;30:e1801690. doi: 10.1002/adma.201801690. [DOI] [PubMed] [Google Scholar]
- 56.Bellassai N, Spoto G. Biosensors for liquid biopsy: Circulating nucleic acids to diagnose and treat cancer. Anal Bioanal Chem. 2016;408:7255–64. doi: 10.1007/s00216-016-9806-3. [DOI] [PubMed] [Google Scholar]
- 57.Campuzano S, Serafin V, Gamella M, et al. Opportunities, challenges, and prospects in electrochemical biosensing of circulating tumor DNA and its specific features. Sensors (Basel) 2019;19:E3762. doi: 10.3390/s19173762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das J, Ivanov I, Sargent EH, Kelley SO. DNA clutch probes for circulating tumor DNA analysis. J Am Chem Soc. 2016;138:11009–16. doi: 10.1021/jacs.6b05679. [DOI] [PubMed] [Google Scholar]
- 59.Cai C, Guo Z, Cao Y, et al. A dual biomarker detection platform for quantitating circulating tumor DNA (ctDNA) Nanotheranostics. 2018;2:12–20. doi: 10.7150/ntno.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang H, Gao Y, Wang S, et al. In situ hybridization chain reaction mediated ultrasensitive enzyme-free and conjugation-free electrochemcial genosensor for BRCA-1 gene in complex matrices. Biosens Bioelectron. 2016;80:450–55. doi: 10.1016/j.bios.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, Chen F, Zhang D, et al. Single copy-sensitive electrochemical assay for circulating methylated DNA in clinical samples with ultrahigh specificity based on a sequential discrimination-amplification strategy. Chem Sci. 2017;8:4764–70. doi: 10.1039/c7sc01035d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Fan X, Xu S, et al. Low fouling label-free DNA sensor based on polyethylene glycols decorated with gold nanoparticles for the detection of breast cancer biomarkers. Biosens Bioelectron. 2015;71:51–56. doi: 10.1016/j.bios.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 63.Povedano E, Vargas E, Montiel VR, et al. Electrochemical affinity biosensors for fast detection of gene-specific methylations with no need for bisulfite and amplification treatments. Sci Rep. 2018;8:6418. doi: 10.1038/s41598-018-24902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Dai Z, Liu X, Yang J. High-performance electrochemical sensing of circulating tumor DNA in peripheral blood based on poly-xanthurenic acid functionalized MoS2 nanosheets. Biosens Bioelectron. 2018;105:116–20. doi: 10.1016/j.bios.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 65.Zhu C, Yang G, Li H, et al. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Analyt Chem. 2014;87:230–49. doi: 10.1021/ac5039863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang HF, Ma RN, Sun F, et al. A versatile label-free electrochemical biosensor for circulating tumor DNA based on dual enzyme assisted multiple amplification strategy. Biosens Bioelectron. 2018;122:224–30. doi: 10.1016/j.bios.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 67.Kulis M, Esteller M. DNA methylation and cancer. Ady Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 68.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Syedmoradi L, Esmaeili F, Norton ML. Towards DNA methylation detection using biosensors. Analyst. 2016;141:5922–43. doi: 10.1039/c6an01649a. [DOI] [PubMed] [Google Scholar]
- 70.Povedano E, Montiel VR, Valverde A, et al. Versatile electroanalytical bioplatforms for simultaneous determination of cancer-related DNA 5-methyl- and 5-hydroxymethyl-cytosines at global and gene-specific levels in human serum and tissues. ACS Sens. 2019;4:227–34. doi: 10.1021/acssensors.8b01339. [DOI] [PubMed] [Google Scholar]
- 71.Daneshpour M, Moradi LS, Izadi P, et al. Femtomolar level detection of RASSF1A tumor suppressor gene methylation by electrochemical nano-genosensor based on Fe3O4/TMC/Au nanocomposite and PT-modified electrode. Biosens Bioelectron. 2016;77:1095–103. doi: 10.1016/j.bios.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 72.Rackus DG, Riedel-Kruse IH, Pamme N. “Learning on a chip:” Microfluidics for formal and informal science education. Biomicrofluidics. 2019;13:041501. doi: 10.1063/1.5096030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knapp KA, Nickels ML, Manning HC. The current role of microfluidics in radiofluorination chemistry. Mol Imaging Biol. :2019. doi: 10.1007/s11307-019-01414-6. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 74.Kim CJ, Park J, Sunkara V, et al. Fully automated, on-site isolation of cfDNA from whole blood for cancer therapy monitoring. Lab Chip. 2018;18:1320–29. doi: 10.1039/c8lc00165k. [DOI] [PubMed] [Google Scholar]
- 75.Gwak HKJ, Cha S, Cheon YP, et al. On-chip isolation and enrichment of circulating cell-free DNA using microfluidic device. Biomicrofluidics. 2019;13:024113. doi: 10.1063/1.5100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ou CY, Vu T, Grunwald JT, et al. An ultrasensitive test for profiling circulating tumor DNA using integrated comprehensive droplet digital detection. Lab Chip. 2019;19:993–1005. doi: 10.1039/c8lc01399c. [DOI] [PMC free article] [PubMed] [Google Scholar]