Abstract
Background
Esophageal squamous cell carcinoma (ESCC), a major histological subtype of esophageal cancer, is a common cause of tumor-related deaths in the world. Due to the lack of understanding of the pathogenesis of ESCC, its clinical treatment is still a big challenge. In the present study, we aimed to identify an ESCC-related gene in the GEO dataset, and to explore its function and mechanism in ESCC.
Material/Methods
The GSE dataset (GSE100492) consisting of 10 samples was analyzed using GEO2R for identifying the differentially expressed genes between ESCC and normal samples. Expression levels of mRNA and miRNA in ESCC tissues and cells were detected via quantitative real-time polymerase chain reaction. Protein expression was analyzed by western blot. Cell proliferation viability was determined through MTT and colony formation. Cell distribution and apoptosis was detected by flow cytometry. MiRNA target prediction was analyzed by bioinformatics. The interplay between miR-340-5p and PIK3C3 was validated by dual-luciferase reporter assay.
Results
PIK3C3 was lowly expressed in ESCC tissue and indicated a poor prognosis in patents. Overexpression of PIK3C3 in vitro repressed cell proliferation of KYSE-150 and TE-12 cells. Moreover, PIK3C3 overexpression was demonstrated to enhance the sensitivity of KYSE-150 and TE-12 cells to irradiation. In addition, miR-340-5p was revealed to directly bind and negatively modulate PIK3C3 expression in ESCC. Blockage of miR-340-5p promoted ESCC cell proliferation, while rescue of PIK3C3 reversed this effect. MiR-340-5p was highly expressed in ESCC tissue and it exhibited a negative correlation with PIK3C3 expression.
Conclusions
MiR-340-5p functioned as an oncogene of ESCC by directly binding and repressing the expression of PIK3C3.
MeSH Keywords: Cell Proliferation, Esophageal Neoplasms, MicroRNAs
Background
Esophageal squamous cell carcinoma (ESCC) has become one of the most frequently occurring digestive system tumors in the world [1]. For instance, according to the estimation of the American Cancer Society, approximately 16 940 new ESCC cases (13 360 male and 3580 female) will be diagnosed and 15 690 deaths (12 720 males and 2970 females) will occur every year in the United States [2]. Multiple genetic and environmental factors are thought to contribute to the etiology of ESCC, such as diet, obesity, consumption of tobacco and alcohol, low intake of vegetables, and poor nutrition status [3]. Despite the tremendous progress has been achieved in the research of ESCC during the past several years, its survival prognosis remains poor [4,5]. It is notable that a better understanding of the pathogenesis of ESCC will provide critical information for improving the survival prognosis and life quality of patients.
Phosphoinositide 3-kinases (PI3Ks) can be divided into 3 different classes (I–III) and are a group of enzymes that can phosphorylate the 3-hydroxyl of phosphatidylinositol lipid substrates [6]. PI3Ks has been reported to be correlated with multiple cellular processes, including cell growth, differentiation, apoptosis, and intracellular trafficking, which in turn have been associated with tumor development [7]. PIK3C3, also referred to as vacuolar protein sorting 34, is the only member of the class III of the PI3K family [6]. Previous studies have shown that PIK3C3 might play an important role during the autophagy process of tumor cells, implying the potential utility of PIK3C3 as a therapeutic target for tumor [8,9]. However, up to now, few publications have specifically investigated the role of PIK3C3 in the tumorigenesis of ESCC.
MicroRNAs (miRNAs) are a group of endogenous short non-coding RNAs involved in diverse cellular functions such as cell cycle, survival, differentiation and development [10,11]. By directly binding to their target message RNAs (mRNAs), microRNAs (miRNAs) could specifically modulate the protein expression through suppressing translation or damaging the target mRNAs [12]. Numerous miRNAs were revealed to be dysregulated in various of human tumors, and a part of these dysregulated miRNAs have already been demonstrated to be involved in the pathological conditions in tumor cell [13]. Currently, the implication of miRNAs in ESCC have also been well documented by numerous studies, where more and more ESCC-related miRNAs have been identified, such as miR-27a, miR-33a-5p, miR-134, and miR-4324 [14–17]. Nevertheless, the entire regulatory network of miRNAs in ESCC still remains unknown.
To further investigate the molecular mechanisms of ESCC, we performed differentially expressed genes analysis in the GSE100492 dataset using the GEO2R method. PIK3C3 was identified to be remarkably decreased in ESCC samples compared to adjacent normal tissue. In the functional examinations, we demonstrated that PIK3C3 overexpression not only repressed ESCC cell proliferation, but also enhanced the sensitivity of ESCC cell to irradiation. In the exploration of underlying mechanisms, we found that PIK3C3 could be bind to and be negatively modulated by miR-340-5p, and overexpression of PIK3C3 could reverse the promotive influences of miR-340-5p on ESCC cell proliferation. In conclusion, our findings suggested that PIK3C3 serves as a tumor repressor by interacting with miR-340-5p in ESCC cells in vitro.
Material and Methods
Microarray datasets
The GSE dataset GSE100492 consisting of 10 samples was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and analyzed using GEO2R.
ESCC tumor sample collection and cell culture
Human ESCC tissue samples and matched normal tissue samples were collected from patients who underwent esophagus resection at the First People’s Hospital of Nantong between 2017 and 2019. Tissue samples were kept at −80°C until further study. Written informed consents were provided by all participants. Human ESCC cell lines (KYSE-150, TE-12, EC-109, and EC-9706) and normal human esophageal epithelial cell line (HET-1A) were purchased from the Chinese Academy of Science cell bank (Shanghai, China). All kinds of cell were cultured in RPMI-1640 medium (Invitrogen, USA) supplemented with 10% fetal bovine serum (Gibco, USA) under a humidified atmosphere with 5% CO2.
Quantitative real-time PCR (qRT-PCR)
Total RNAs of treated ESCC tissues and cells were prepared using TRIzol reagent (Invitrogen), and its quality was examined via NanoDro 2000c (Thermo Scientific, USA). Then, 2 μg total RNAs were adopted as template to synthesis cDNA by the use of PrimeScript RT Reagent (Takara, Dalian, China). The levels of PIK3C3 and miR-340-5p was detected through a SYBR Premix Taq (Takara) and miScript SYBR-Green kit (Takara), respectively. PCR reaction was conducted on an ABI 7300 Real-Time system (Ambion, USA) with the following parameters: 95°C for 60 seconds, 35 cycles of 95°C for 20 seconds, 58°C for 35 seconds and 72°C for 40 seconds. Primers are shown in Table 1.
Table 1.
The sequences of primers in this study.
| Gene | Primer | |
|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | |
| GAPDH | TGTTCGTCATGGGTGTGAAC | ATGGCATGGACTGTGGTCAT |
| MiR-340-5p | GCTTATAAAGCAATGAGACTGATT | CTCAACTGGTGTCGTGGA |
| PIK3C3 | TGTGATGTCAATGCCAGTTA | TACTAACAGGTGGAAATGCTC |
| U6 | CTCGCTTCG GCAGCACA | AACGCTTCACGAATTTGCGT |
RNA transfection
The pCMV6/PIK3C3, pCMV6/control, miR-340-5p mimics, miR-340-5p ASO, and negative control (miR-NC) were all designed and provided by GenePharma (Shanghai, China). ESCC cells (2×105 cells/well) were seeded into 96-well plates and cultured at 37°C for at least 8 hours. Next, these cells were transfected with indicated RNAs using the Lipofectamine 3000 (Invitrogen).
Cell proliferation assessment
Cell viability of KYSE-150 and TE-12 cells was estimated by 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and colony formation. For MTT assay, in brief, exponentially growth ESCC cells were harvested and re-seeded into 96-well plates and cultured overnight. MTT solution (20 μL) was added into each well and cultured for another 4 hours. Next, the absorbance of each well was measured through a microplate reader at 490 nm. For colony formation experiment, treated exponentially growth ESCC cells were harvested and re-seeded into 35-mm culture dishes and cultured for 2 weeks. Next, colonies were fixed using 4% paraformaldehyde and stained with Giemsa solution. The colony number was counted manually under a microscope at 10x magnification.
Cell cycle and apoptosis analysis
Cell distribution of ESCC cells was tested via propidium iodide (PI) staining followed by flow cytometry analysis. In brief, after fixed in 75% ethanol for 8 hours, cells were washed using phosphate-buffered saline (PBS) for 3 times (5 minutes per time). Next, cells were stained with PI staining buffer (BD Pharmingen, USA) for cell distribution analysis using a flow cytometry (FACSCalibur, BD Biosciences, USA). For cell apoptosis detection, treated ESCC cells were stained with PI and Annexin V-FITC for 15 minutes at room temperature, then used for cell apoptosis analysis using a flow cytometry.
Western blot analysis and immunohistochemistry (IHC)
Total proteins of treated ESCC cells were extracted using RIPA buffer (Sigma, USA) following the instructions of manufacturers. After qualifying the density of proteins, 50 μg protein samples were loaded and separated by 10% (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) SDS-PAGE. Next, the target proteins were transferred into polyvinylidene difluoride (PVDF) membranes at −4°C followed by 2 hours incubation with 5% non-fat milk and 8 hours incubation with primary antibodies that against PIK3C3 (Rabbit, 1: 3000, ab124905, Abcam, UK). After washed with PBS for 3 times, the membranes were incubated with horse-radish peroxidase (HRP)-conjunct secondary antibodies and the signals were visible using electrochemiluminescence (ECL) reagent (Bio-Rad). Formalin fixed, paraffin embedded tissues were cut into 5-μm thick sections, then blocked with 5% goat serum and incubated for 20 minutes at room temperature before washed. PIK3C3 was detected by rabbit monoclonal primary antibody (EPITOMICS) at a dilution of 1: 400 for 30 minutes. The sections were subsequently incubated with biotinylated, goat anti-rabbit immunoglobulin (Vector Laboratories, Burlingame, CA) and developed with Vectastain Elite BCA kit (Vector Laboratories) as chromogen according to the manufacture’s recommendation.
Dual-luciferase reporter assay
The interplay between miR-340-5p and PIK3C3 was validated by dual-luciferase reporter assay in KYSE-150 and TE-12 cells. The wild type (WT) and mutant (Mut) parts of PIK3C3 3′-UTR containing the miR-340-5p binding sequences were inserted into the pCMV6 (Origene, Rockville, MD, USA) to form pCMV6/PIK3C3-wt and pCMV6/PIK3C3-mut, respectively. pCMV6/PIK3C3-wt or pCMV6/PIK3C3-mut was co-transfected into KYSE-150 and TE-12 cells with miR-340-5p mimics or miR-340-5p ASO. The alterations of luciferase intensity were monitored using a dual-luciferase assay system (Promega) kit.
Statistical analysis
Statistical analysis was conducted on GraphPad Prism (Version 7.0, USA) using one-way ANOVA analysis or Student t-test and data were showed as mean±standard deviation (SD). P<0.05 was considered statistically significant.
Results
PIK3C3 was identified to be downregulated in ESCC
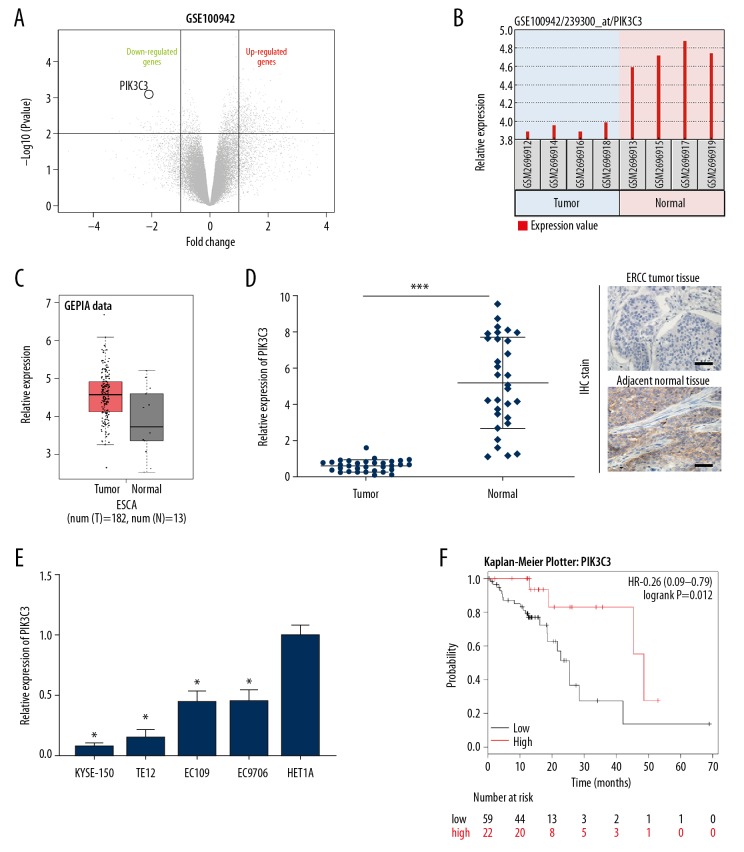
To identify genes that are involved in the tumorigenesis of ESCC, we analyzed the different expressed genes in the GSE100942 dataset. Among these dysregulated genes, we focused our interest on PIK3C3, which was identified to be downregulated in GSE100492 (Figure 1A). We then analyzed the level of PIK3C3 in normal and ESCC samples in GSE100492 by GEO2R, and the results suggested that the PIK3C3 level was significantly lower in the ESCC samples than in the normal tissues samples, the online bioinformatic tool GEPIA also showed the same results (Figure 1B, 1C). To further validate the downregulation of PIK3C3 in ESCC, we detected its expression in 31 pairs of ESCC and normal clinical tissue specimens through qRT-PCR. Compared to the normal group, the level of PIK3C3 was remarkably downregulated in the ESCC group, IHC assay also showed that the expression level of PIK3C3 was downregulated in ESCC tissues compared with matched adjacent normal tissues (*** P<0.001, Figure 1D). Meanwhile, we measured the level of PIK3C3 in 4 ESCC cell lines (KYSE-150, TE-12, EC-109, and EC-9706). qRT-PCR data showed that PIK3C3 was markedly decreased in KYSE-150, TE-12, EC-109, and EC-9706 cell lines compared with HET1A (normal human esophageal epithelial cell line) (* P<0.05, Figure 1E). In addition, we revealed that ESCC patients with low PIK3C3 level exhibited a worse prognosis than those patients with high PIK3C3 level by online tool Kaplan-Meier Plotter (Figure 1F). The correlations between PIK3C3 and clinical characteristics of patients with ESCC were also analyzed (Table 2). These findings indicated that PIK3C3 was decreased in ESCC and associated with a poor prognosis, implying it might be involved in the tumorigenesis of ESCC.
Figure 1.
PIK3C3 was identified to be downregulated in esophageal squamous cell carcinoma (ESCC). (A) Volcano plot showing the dysregulated genes of GSE100942 dataset. (B, C) GEO2R and GEPIA analysis were conducted to examine the expression of PIK3C3 in normal and ESCC samples from GSE100942. (D) PIK3C3 expression was measured by quantitative real-time polymerase chain reaction (qRT-PCR) in 31 pairs of normal and ESCC tissues, *** P<0.001. (E) PIK3C3 expression of 4 ESCC cell lines (KYSE-150, TE-12, EC-109, and EC-9706) were assessed using qRT-PCR, HET1A was used as normal control group, * P<0.05. (F) Survival probability of ESCC patients with low or high PIK3C3 expression.
Table 2.
The association between PIK3C3 expression and clinic pathological parameters in 31 cases of esophageal squamous cell carcinoma patients.
| All cases | PIK3C3 | |||
|---|---|---|---|---|
| High expression | Low expression | P value | ||
| Age (year) | 0.053 | |||
| >60 | 15 | 10 (66.7%) | 5 (33.3%) | |
| ≤60 | 16 | 5 (31.3%) | 11 (68.8%) | |
| Gender | 0.475 | |||
| Male | 14 | 6 (42.9%) | 8 (57.1%) | |
| Female | 17 | 6 (35.3%) | 11 (64.7%) | |
| pN status | 0.012 | |||
| N0 | 24 | 17 (70.8%) | 7 (29.2%) | |
| N1–N2 | 7 | 1 (14.3%) | 6 (85.7%) | |
| pM status | 0.195 | |||
| M0 | 28 | 21 (75.0%) | 7 (25.0%) | |
| M1 | 3 | 1 (33.3%) | 2 (66.7%) | |
| Clinical stage | 0.037 | |||
| I & II | 23 | 16 (69.6%) | 7 (30.4%) | |
| III & IV | 8 | 2 (25.0%) | 6 (75.0%) | |
pN – pathological node; pM – pathological metastasis.
PIK3C3 overexpression suppressed ESCC cell proliferation
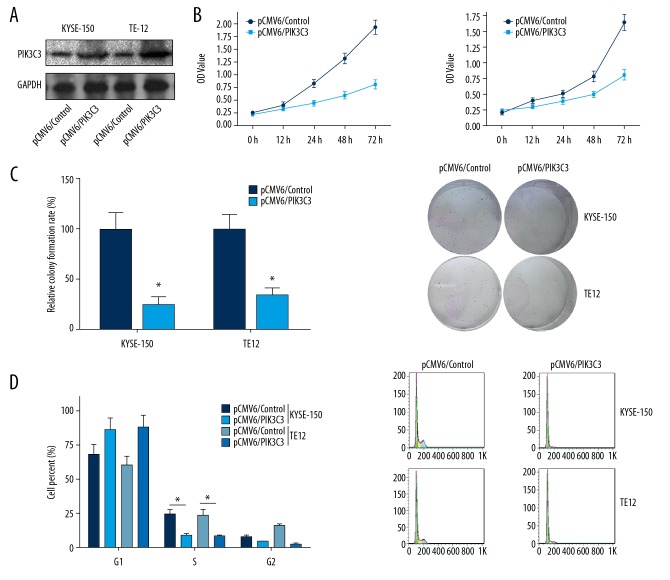
To figure out whether PIK3C3 plays a role during ESCC tumorigenesis, we overexpressed its expression in vitro and then examined its effects on ESCC cell proliferation. We transfected KYSE-150 and TE-12 cells with pCMV6/PIK3C3 followed by examination of overexpression efficiency with western blot. Compared to pCMV6/control transfected groups, PIK3C3 was remarkably upregulated in pCMV6/PIK3C3 transfected groups (Figure 2A). MTT and colony formation assays were adopted to examine the influences of PIK3C3 overexpression on the cell proliferation viability of KYSE-150 and TE-12 cells. MTT data suggested that PIK3C3 overexpression obviously attenuated the cell viability of KYSE-150 and TE-12 cells (* P<0.05, Figure 2B). Colony formation data indicated that PIK3C3 overexpression resulted in a remarkable reduction of colony number in KYSE-150 and TE-12 cells (* P<0.05, Figure 2C). Moreover, flow cytometry was used to analyze the influences of PIK3C3 overexpression on cell cycle of KYSE-150 and TE-12 cells, and results demonstrated that PIK3C3 overexpression remarkably reduced the cell number of S phase in both KYSE-150 and TE-12 cells (* P<0.05, Figure 2D). Taken together, PIK3C3 overexpression was proved to repress ESCC cell proliferation in vitro.
Figure 2.
PIK3C3 overexpression suppressed esophageal squamous cell carcinoma (ESCC) cell proliferation. (A) After 24 hours of transfection with pCMV6/PIK3C3 recombinant plasmid, the KYSE-150 and TE-12 cells subjected to detection of PIK3C3 by western blot. (B) MTT assay was conducted to analyze the cell viability of PIK3C3 overexpressed KYSE-150 and TE-12 cells, * P<0.05. (C) Colony formation experiment was carried out to estimate the influences of PIK3C3 overexpression on ESCC cell proliferation, * P<0.05. (D) Flow cytometry analysis was conducted on PIK3C3 overexpressed KYSE-150 and TE-12 cells to measure the effects of PIK3C3 on ESCC cell cycle, * P<0.05. pCMV6/control transfected cell was used as negative control.
PIK3C3 overexpression increased the sensitivity of ESCC cell to irradiation
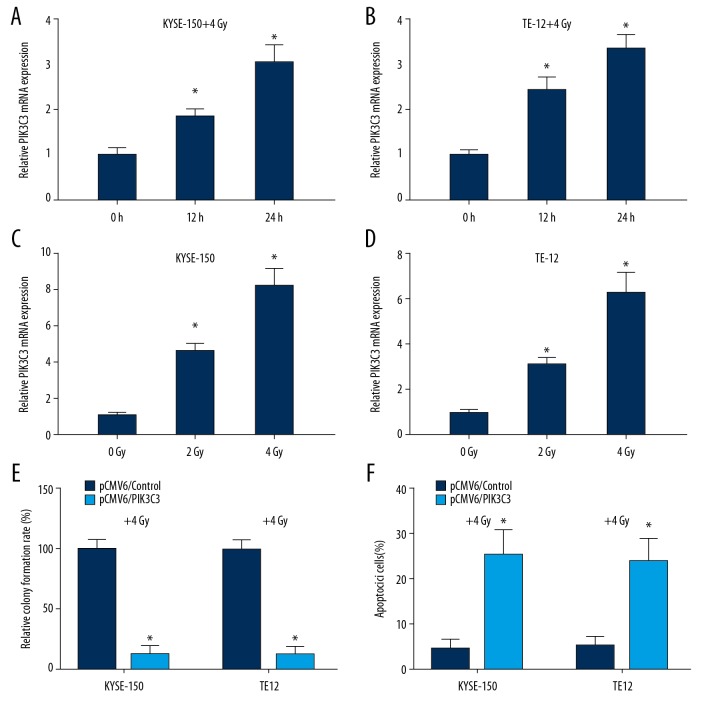
We then examined whether PIK3C3 plays a role in the irradiation treatment of ESCC. By detecting the relative level of PIK3C3 in KYSE-150 and TE-12 cells after 0, 12, and 24 hours of 4 Gy irradiation, we found that relative mRNA expression of PIK3C3 was markedly increased in a time-dependent manner under 4 Gy irradiation in KYSE-150 and TE-12 cells (* P<0.05, Figure 3A, 3B). Moreover, by treating cells with different dose irradiation (0, 2, and 4 Gy), we revealed that the relative mRNA expression of PIK3C3 was increased in a dose-dependent manner in KYSE-150 and TE-12 cells (* v<0.05, Figure 3C, 3D). Therefore, PIK3C3 was demonstrated to be an irradiation-inducible protein in ESCC. We then examine the effects of PIK3C3 overexpression on ESCC cell proliferation and apoptosis under irradiation treatment (4 Gy) via colony formation and flow cytometry analysis. As colony formation data suggested that overexpression of PIK3C3 markedly reduced the number of colonies under 4 Gy irradiation compared to control group (* P<0.05, Figure 3E). Cell apoptosis analysis indicated that PIK3C3 overexpression markedly increased the apoptosis rate of KYSE-150 and TE-12 cells under 4 Gy irradiation treatment (* P<0.05, Figure 3F). Taken together, our findings suggested that PIK3C3 overexpression increased the cellular response to irradiation.
Figure 3.
PIK3C3 overexpression increased the sensitivity of esophageal squamous cell carcinoma (ESCC) cell to irradiation. (A, B) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of PIK3C3 mRNA in KYSE-150 and TE-12 cells treated with irradiation (4 Gy) for 0, 12, and 24 hours, * P<0.05. (C, D) qRT-PCR analysis of PIK3C3 mRNA in KYSE-150 and TE-12 cells treated with different dose of irradiation (0, 2, and 4 Gy), * P<0.05. (E) Colony formation experiment was adopted to analyze the cell proliferation of KYSE-150 and TE-12 cells transfected with pCMV6/PIK3C3 recombinant plasmid under irradiation (4 Gy), * P<0.05. (F) Flow cytometry analysis showing the effects of PIK3C3 overexpression on cell apoptosis of KYSE-150 and TE-12 cells under irradiation (4 Gy), * P<0.05.
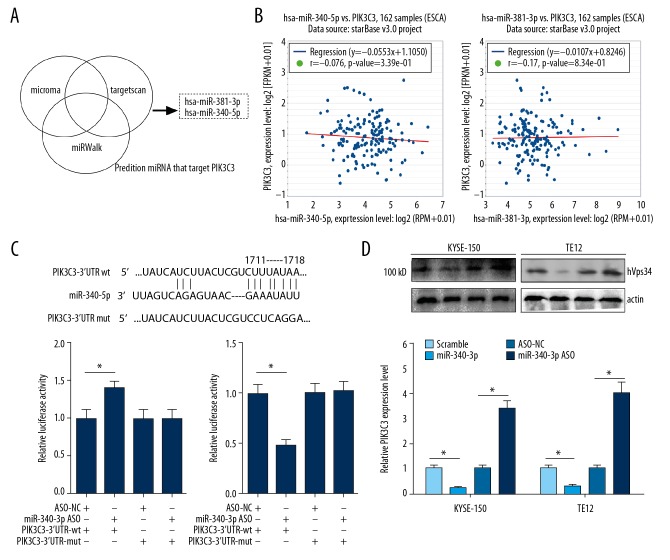
MiR-340-5p targeted and negatively regulated PIK3C3
To understand the underlying molecular mechanisms of PIK3C3 during ESCC, we screened the target miRNAs of PIK3C3 through 3 different software (microRNA, Targetscan and miRWalk). MiR-381-3p and miR-340-5p were identified by all the 3 software (Figure 4A). To evaluate the correlation between miR-381-3p or miR-340-5p and PIK3C3, we conducted Pearson’s correlation analysis in 162 ESCC samples from starBase database. Results suggested that the expression of miR-340-5p or miR-381-3p was negatively or positively correlated with PIK3C3 expression in ESCC samples, and the Pearson’s correlation coefficient was −0.076 and 0.017, respectively (Figure 4B). So, miR-340-5p was selected for further study. Bioinformatics analysis predicted that the 3′-UTR of PIK3C3 possesses miR-340-5p binding sites (Figure 4C). Dual-luciferase experiment was subsequently adopted to validate the interplay between miR-340-5p and PIK3C3. As results indicated that co-transfection of miR-340-5p ASO and PIK3C3-3′-UTR-wt markedly increased the luciferase activity, while co-transfection of miR-340-5p mimics and PIK3C3-3′-UTR-wt markedly attenuated the luciferase intensity (* P<0.05, Figure 4C). The luciferase intensity of cells driven by PIK3C3-3′-UTR-mut was not affected by miR-340-5p mimics and ASO. In addition, we estimated the protein expression level of PIK3C3 in miR-340-5p overexpressed and silenced KYSE-150 and TE-12 cells through western blot. Transfecting KYSE-150 and TE-12 cells with miR-340-5p mimics resulted in a significant downregulation of PIK3C3, while transfection of miR-340-5p ASO exhibited an opposite effect on PIK3C3 expression (* P<0.05, Figure 4D).
Figure 4.
MiR-340-5p targeted and negatively regulated PIK3C3. (A) Target miRNAs of PIK3C3 were predicted by microRNA (miRNA), miRWalk and Targetscan, and the Venn diagram showing the intersection. (B) The correlation between miR-340-5p or miR-381-3p and PIK3C3 in 162 esophageal squamous cell carcinoma (ESCC) samples. Figures were downloaded from starBase database. (C) Upper panel, the wild type and mutant sequence of predicted binding sites of miR-340-5p in the 3′-UTR of PIK3C3. Lower panel, the interplay between miR-340-5p and PIK3C3 was verified by luciferase reporter assay. (D) Protein expression of PIK3C3 was detected by western blot in miR-340-5p overexpressed and blocked KYSE-150 and TE-12 cells.
PIK3C3 reversed the promotive effects of miR-340-5p on ESCC cell proliferation
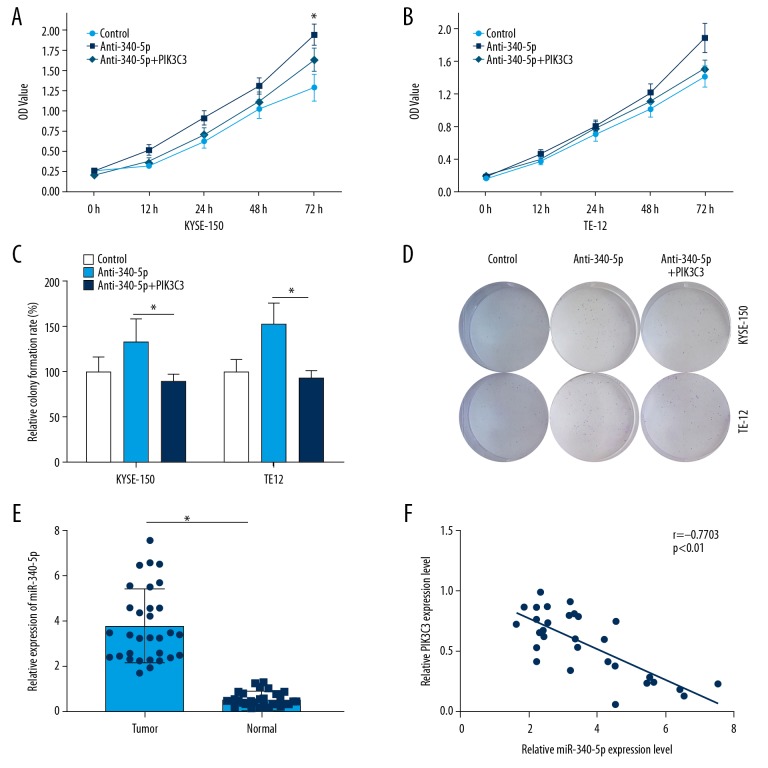
Next, we examined the function of miR-340-5p in ESCC cell proliferation by silencing its expression in vitro. By using MTT assay, we found that cell viability of KYSE-150 and TE-12 cells treated with anti-miR-340-5p were markedly increased compared to control group (* P<0.05, Figure 5A, 5B). Moreover, co-transfection of anti-miR-340-5p and PIK3C3 reversed the upregulation of cell viability of KYSE-150 and TE-12 cells induced by miR-340-5p knockdown (Figure 5A, 5B). In the colony formation experiment, we demonstrated that miR-340-5p knockdown could remarkably increase the colony number in KYSE-150 and TE-12 cells, however, co-transfection of anti-miR-340-5p and PIK3C3 blocked this phenomenon (* P<0.05, Figure 5C, 5D). qRT-PCR analysis showed that miR-340-5p was markedly upregulated in the ESCC tissue samples compared to the normal counterparts (* P<0.05, Figure 5E). The correlations between miR-340-5p and clinical characteristics of patients with ESCC were also analyzed (Table 3). In addition, a negative correlation was observed between the level of miR-340-5p and PIK3C3 in ESCC samples (Figure 5F). These results indicated that PIK3C3 could reverse the promotive effects of miR-340-5p on ESCC cell proliferation.
Figure 5.
PIK3C3 reversed the promotive effects of miR-340-5p on esophageal squamous cell carcinoma (ESCC) cell proliferation. (A, B) MTT assay was adopted to analyze the cell viability in control, anti-340-5p and anti-340-5p+PIK3C3 treated KYSE-150 and TE-12 cells, * P<0.05. (C, D) Colony formation assay was utilized to estimate the cell proliferation in control, anti-340-5p, and anti-340-5p+PIK3C3 transfected KYSE-150 and TE-12 cells, * P<0.05. (E) Relative expression of miR-340-5p was estimated by qRT-PCR in ESCC and normal tissue samples, * P<0.05. (F) Correlation between miR-340-5p and PIK3C3 in ESCC samples was assessed.
Table 3.
The association between miR-340-5p expression and clinic pathological parameters in 31 cases of esophageal squamous cell carcinoma patients.
| All cases | MiR-340-5p | |||
|---|---|---|---|---|
| High expression | Low expression | P value | ||
| Age (year) | 0.422 | |||
| >60 | 15 | 6 (40.0%) | 9 (60.0%) | |
| ≤60 | 16 | 8 (50.0%) | 8 (50.0%) | |
| Gender | 0.276 | |||
| Male | 14 | 9 (64.3%) | 5 (35.7%) | |
| Female | 17 | 8 (47.1%) | 9 (52.9%) | |
| pN status | 0.058 | |||
| N0 | 24 | 7 (29.2%) | 17 (70.8%) | |
| N1–N2 | 7 | 5 (71.4%) | 2 (28.6%) | |
| pM status | 0.027 | |||
| M0 | 28 | 7 (25.0%) | 21 (75.0%) | |
| M1 | 3 | 3 (100.0%) | 0 (0.0%) | |
| Clinical stage | 0.037 | |||
| I & II | 23 | 7 (30.4%) | 16 (69.6%) | |
| III & IV | 8 | 6 (75.0%) | 2 (25.0%) | |
pN – pathological node; pM – pathological metastasis.
Discussion
PIK3C3 is known as an autophagy-related protein, inhibition of PIK3C3 was reported to prevent autophagy and synergize with MTOR inhibition in cancer cells [18]. Several PI3K blockers, including 3-methyladenine, LY294002 and wortmannin, have been developed as autophagy repressor, however, they exhibit extremely low selectivity in PIK3C3 [19,20]. Miller et al. (2010) identified the structure of PIK3C3, not only offering a novel insight into the catalytic mechanisms of PIK3C3, but also providing physical support to the development of effective and selective PIK3C3 repressors [21]. Currently, SAR405 is considered to be one of the most effective PIK3C3 repressor with a binding equilibrium constant of 1.5 nmol/L [22]. Research on SAR405 indicates that the catalytic activities of PIK3C3 are indispensable for keeping the size of late endosomes/lysosomes and properties of lysosomes during vesicle trafficking. Moreover, repressing PIK3C3 via SAR405 could suppress the process of autophagy caused by nutrient starvation [18]. All these findings suggest that PIK3C3 might be involved in the tumorigenesis of various human tumors such as bladder cancer, leukemia, and so on [9,23]. Nevertheless, there is almost no publication addressing the role of PIK3C3 during the tumorigenesis of ESCC. In a pilot case-control research of ESCC patients and matched controls, a total of 38 single nucleotide polymorphisms correlated with ESCC were found in 33 genes, which including PIK3C3 [24]. In the present study, PIK3C3 was proven to be downregulated in ESCC. In order to confirm its role in ESCC, we overexpressed its expression in vitro followed by the examination of cell proliferation. Results indicated that PIK3C3 severs as a tumor repressor of ESCC, which, up to the best of our knowledge, is the first publication reporting the function of PIK3C3 in ESCC.
To further identify the mechanism of PIK3C3 in ESCC, we predicted its target miRNAs through bioinformatics methods, and miR-340-5p was screened and proven to directly bind to the 3′-UTR area of PIK3C3. MiR-340-5p was previously demonstrated to affect the progression of various human cancers, such as glioblastoma, non-small cell lung cancer, and osteosarcoma, through multiple distinct mechanisms [25–27]. Recently, it was predicted to be a potential prognostic indicator of colorectal cancer [28]. The implication of miR-340-5p in ESCC has not been reported so far. In this study, miR-340-5p was found to negatively regulated the expression of PIK3C3, and overexpression of PIK3C3 was shown to abolish the promotive influences of miR-340-5p on ESCC cell proliferation in vitro.
Conclusions
In summary, we provided in vitro evidences of miR-340-5p/PIK3C3 axis in regulating ESCC cell proliferation, implying that miR-340-5p/PIK3C3 axis might be a potential therapeutic target of ESCC. However, in vivo studies should be performed to further confirm the biological functions of miR-340-5p/PIK3C3 axis.
Footnotes
Conflict of interest
None.
Source of support: This research was financially supported by the Project of Jiangsu Province “Six Talents Summit” (No.2014-wsn-075)
References
- 1.Lin J. Esophageal squamous cell carcinoma and adenocarcinoma: At the Gates of Mordor. J Thorac Cardiovasc Surg. 2017;154(4):1444–45. doi: 10.1016/j.jtcvs.2017.07.048. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Prabhu A, Obi KO, Rubenstein JH. The synergistic effects of alcohol and tobacco consumption on the risk of esophageal squamous cell carcinoma: A meta-analysis. Am J Gastroenterol. 2014;109(6):822–27. doi: 10.1038/ajg.2014.71. [DOI] [PubMed] [Google Scholar]
- 4.Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis. 2001;22(11):1737–46. doi: 10.1093/carcin/22.11.1737. [DOI] [PubMed] [Google Scholar]
- 5.Baba Y, Yoshida N, Shigaki H, et al. Prognostic impact of postoperative complications in 502 patients with surgically resected esophageal squamous cell carcinoma: A retrospective single-institution study. Ann Surg. 2016;264(2):305–11. doi: 10.1097/SLA.0000000000001510. [DOI] [PubMed] [Google Scholar]
- 6.Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. 2014;127(Pt 5):923–28. doi: 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11(5):329–41. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 8.Kim TM, Baek JH, Kim JH, et al. Development of in vitro PIK3C3/VPS34 complex protein assay for autophagy-specific inhibitor screening. Anal Biochem. 2015;480:21–27. doi: 10.1016/j.ab.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Chen CH, Changou CA, Hsieh TH, et al. Dual inhibition of PIK3C3 and FGFR as a new therapeutic approach to treat bladder cancer. Clin Cancer Res. 2018;24(5):1176–89. doi: 10.1158/1078-0432.CCR-17-2066. [DOI] [PubMed] [Google Scholar]
- 10.Razmara E, Bitaraf A, Yousefi H, et al. Non-coding RNAs in cartilage development: An updated review. Int J Mol Sci. 2019;20(18) doi: 10.3390/ijms20184475. pii: E4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calderon-Dominguez M, Belmonte T, Quezada-Feijoo M, et al. Emerging role of microRNAs in dilated cardiomyopathy: Evidence regarding etiology. Transl Res. 2019;215:86–101. doi: 10.1016/j.trsl.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 13.Lin YH. MicroRNA networks modulate oxidative stress in cancer. Int J Mol Sci. 2019;20(18) doi: 10.3390/ijms20184497. pii: E4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, An D, Liu X, et al. MicroRNA-27a downregulates the expression of Hsp90 and enhances the radiosensitivity in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:5967–77. doi: 10.2147/OTT.S197456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Wang L, Yang J, et al. MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1. Eur J Pharmacol. 2019;861:172590. doi: 10.1016/j.ejphar.2019.172590. [DOI] [PubMed] [Google Scholar]
- 16.Wang WW, Zhao ZH, Wang L, et al. MicroRNA-134 prevents the progression of esophageal squamous cell carcinoma via the PLXNA1-mediated MAPK signalling pathway. EBioMedicine. 2019;46:66–78. doi: 10.1016/j.ebiom.2019.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Zhou J, Zhu J, Jiang G, et al. Downregulation of microRNA-4324 promotes the EMT of esophageal squamous-cell carcinoma cells via upregulating FAK. Onco Targets Ther. 2019;12:4595–604. doi: 10.2147/OTT.S198333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasquier B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy. 2015;11(4):725–26. doi: 10.1080/15548627.2015.1033601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YT, Tan HL, Shui G, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285(14):10850–61. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yap TA, Bjerke L, Clarke PA, Workman P. Drugging PI3K in cancer: Refining targets and therapeutic strategies. Curr Opin Pharmacol. 2015;23:98–107. doi: 10.1016/j.coph.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller S, Tavshanjian B, Oleksy A, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327(5973):1638–42. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10(12):1013–19. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 23.Kristensen L, Kristensen T, Abildgaard N, et al. High expression of PI3K core complex genes is associated with poor prognosis in chronic lymphocytic leukemia. Leuk Res. 2015;39(6):555–60. doi: 10.1016/j.leukres.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Ng D, Hu N, Hu Y, et al. Replication of a genome-wide case-control study of esophageal squamous cell carcinoma. Int J Cancer. 2008;123(7):1610–15. doi: 10.1002/ijc.23682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Choi JY, Seok HJ, et al. MiR-340-5p Suppresses aggressiveness in glioblastoma multiforme by targeting Bcl-w and Sox2. Mol Ther Nucleic Acids. 2019;17:245–55. doi: 10.1016/j.omtn.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G, Zhang Y. MicroRNA-340-5p suppresses non-small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell Mol Biol Lett. 2019;24:34. doi: 10.1186/s11658-019-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rongxin S, Pengfei L, Li S, et al. MicroRNA-340-5p suppresses osteosarcoma development by down-regulating the Wnt/beta-catenin signaling pathway via targeting the STAT3 gene. Eur Rev Med Pharmacol Sci. 2019;23(3):982–91. doi: 10.26355/eurrev_201902_16985. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Men WL, Yan KM, et al. MiR-340-5p is a potential prognostic indicator of colorectal cancer and modulates ANXA3. Eur Rev Med Pharmacol Sci. 2018;22(15):4837–45. doi: 10.26355/eurrev_201808_15619. [DOI] [PubMed] [Google Scholar]