Abstract
Severe Acute Respiratory Syndrome (SARS), an emerging disease characterized by atypical pneumonia, has recently been attributed to a novel coronavirus. The genome of SARS Coronavirus (SARS-CoV) has recently been sequenced, and a number of genes identified, including that of the nucleocapsid protein (N). It is noted, however, that the N protein of SARS-CoV (SARS-CoV N) shares little homology with nucleocapsid proteins of other members of the coronavirus family [Science 300 (2003) 1399; Science 300 (2003) 1394]. N proteins of other coronavirus have been reported to be involved in forming the viral core and also in the packaging and transcription of the viral RNA. As data generated from some viral systems other than coronaviruses suggested that viral N–N self-interactions may be necessary for subsequent formation of the nucleocapsid and assembly of the viral particles, we decided to investigate SARS-CoV N–N interaction. By using mammalian two-hybrid system and sucrose gradient fractionations, a homotypic interaction of N, but not M, was detected by the two-hybrid analysis. The mammalian two-hybrid assay revealed an approximately 50-fold increase in SEAP activity (measurement of protein–protein interaction) in N–N interaction compared to that observed in either M–M or mock transfection. Furthermore, mutational analyses characterized that a serine/arginine-rich motif (SSRSSSRSRGNSR) between amino acids 184 and 196 is crucial for N protein oligomerization, since deletion of this region completely abolished the N protein self-multimerization. Finally, the full-length nucleocapsid protein expressed and purified from baculovirus system was found to form different levels of higher order structures as detected by Western blot analysis of the fractionated proteins. Collectively, these results may aid us in elucidating the mechanism pertaining to formation of viral nucleocapsid core, and designing molecular approaches to intervene SARS-CoV replication.
Keywords: Severe Acute Respiratory Syndrome, Coronavirus, Nucleocapsid, Protein–protein interactions
The family Coronaviridae is composed of a number of enveloped viruses whose single-stranded positive sense RNA genomes are among the largest of the RNA viruses, ranging from 25 to 30 kb [3], [4]. Coronaviruses typically infect avian or mammalian hosts and are in fact responsible for roughly 30% of mild upper respiratory tract illnesses in humans [4]. Recently, an outbreak of atypical pneumonia in Asia, Canada, and elsewhere, dubbed Severe Acute Respiratory Syndrome (SARS), was attributed to a novel human coronavirus (SARS-CoV) [5], [6]. In contrast to other members of its family, SARS-CoV results in severe morbidity and mortality in up to 10% of infected patients [7]. The genome of SARS-CoV was found to be roughly 29,700 nucleotides long, with a characteristic coronavirus genome organization of 5′-replicase, spike, envelope, membrane, and nucleocapsid-3′ [1], [2]. The nucleocapsid (N) protein of SARS-CoV is 422 amino acids long, sharing only 20–30% homology with the N proteins of other coronaviruses [1], [2]. Previous studies indicate that the N proteins of other coronaviruses are extensively phosphorylated, highly basic, and associated with viral RNA to form a helical ribonucleoprotein (RNP), which comprises the viral core structure [8]. A variety of functional activities have been ascribed to the N proteins of previously known coronaviruses, including participation in transcription of the viral genome, the formation of viral core, and packaging viral RNA [3], [9]. However, no similar functional roles of SARS-CoV have been reported so far in the literature except that SARS-CoV N may selectively activate AP-1 pathway [10]. Furthermore, while it is of note that self-interaction of nucleocapsid proteins (N–N interaction) and N protein interactions with other viral proteins have been reported to be one of the crucial steps in the formation and assembly of viral particles [11], [12], [13], [14], [15], [16], [17], [18]. Such protein–protein interactions, especially N–N self-interaction in coronaviruses, are poorly documented. To better understand SARS-CoV replication, multimerization of the N protein was investigated by both in vitro and in vivo approaches in our current studies. Yeast two-hybrid analysis was first used to identify N protein homotypic interaction, followed by analysis of sequentially truncated proteins to locate the amino acid segments responsible for N–N self-interaction in the mammalian two-hybrid system. We found self-interaction of N, not the viral membrane protein (M), in the aforementioned assays, since a 50-fold increase in the level of SEAP activity (indicator of protein–protein interaction) was observed in N–N interaction compared to M–M interaction and mock transfection. In addition, a serine/arginine-rich region composed of 13 amino acids was found to be crucial for self-interaction of the nucleocapsid protein because deletion of these amino acids completely abolished N multimerization. Finally, multimerization of N was also observed in vitro by using sucrose gradient centrifugation in conjunction with Western blotting detection, which revealed formation of higher order structures of the N proteins. The significance of these findings will be discussed.
Materials and methods
Construction of recombinant vectors. The yeast two-hybrid vectors pGBK/T7 and pGAD/T7, the mammalian two-hybrid vectors pM and pVP16 were obtained from Clontech (Palo Alto, CA). The nucleocapsid gene (GenBank Accession No. AY274119) was amplified by RT-PCR from the SARS-CoV RNA isolated from SARS-infected patients as described [10]. Primers for the amplification are 5′ GTACGAATTCATGTCTGATAATGGACCCCAATC 3′ and 5′ GTACGGATCC-GTGGTCATCATGAGTGTTTATG 3′. The amplified product was then purified with MiniElute PCR Purification kit (Qiagen, Valencia, CA), followed by digestion with EcoRI and BamHI (underlined sequence in primers). The digested DNA fragments were subsequently ligated in-frame into pGBK/T7, pGAD/T7, pM, and pVP16 vectors. As additional controls under the same assay conditions for N, the gene for SARS-CoV membrane (M) was amplified with the following primers: 5′ GTACGAATTCATG-GCAGACAACGGTA 3′ and GTACGGATCC-TTACTGTACTAGCAAAGCA. The PCR product was cloned into the above-noted vectors in EcoRI and BamHI sites. PCR primers for the cloning of the N gene into pAs-Red2 (Clontech, Palo Alto, CA) fluorescent vector (designated as pAsRed-N) are 5′ GTACGAATTCTATGTCTGATAATGGACCCCAATC 3′ and 5′ GTACGGATCCGT-GGTCATCATGAGTGT-TTATG 3′. The expression vector for SARS-CoV M gene is designated as pGBK-M and pGAD-M in this report. To identify putative domains of amino acid sequences required for multimerization, sequential deletion starts from 5′ in the nucleocapsid gene, all sense primers used in the study carry EcoRI restriction site (Table 1 ), and the antisense primer sequence is the same one used for full-length nucleocapsid gene cloning as mentioned above. Primers for these fragments are listed in Table 1. The sequential deletions resulted in an approximately 40 nt truncation (see below for details). In addition to sequential deletion mutants, a serine/arginine-rich motif (amino acid 184–196) was deleted using following primers: 5′ [Phosp]AGAGGCTTGACTGCCG-CCTCTGCTTCC 3′ and 5′ [Phosp]AATTCA-ACTCCTGGCAGCAGTAG 3′. This mutant was designated as ΔpM-N/SR.
Table 1.
Primers for N gene sequential deletion
| ΔN-seq-1 | GTACGAATTCCCCCAAGGTTTACCCAATAATAC |
| ΔN-seq-2 | GTACGAATTCCAAATTGGCTACTACCGAAGAG |
| ΔN-seq-3 | GTACGAATTCGCTAACAAAGAAGGCATCGTATGG |
| ΔN-seq-4 | GTACGAATTCCCAAAAGGCTTCTACGCAGAG |
| ΔN-seq-5 | GTACGAATTCCGAATGGCTAGCGGAGGTGGTG |
| ΔN-seq-6 | GTACGAATTCTCTGCTGCTGAGGCATCTAAAAAG |
| ΔN-seq-7 | GTACGAATTCCAAGGAACTGATTACAAACAT |
| ΔN-seq-8 | GTACGAATTCGACAACGTCATACTGCTGAAC |
Note. Primers used for sequential deletion of the N protein. All primers shown in the table are forward primers. The restriction site EcoRI is underlined. The reverse primer used to generate the deletion products was the one used for the amplification of the full-length N gene shown in Materials and methods.
Yeast transformation and culture. The competent cells of yeast strain AH109 were obtained from Clontech (Palo Alto, CA). Transformations were performed according to the manufacturer’s protocol. Briefly, 500 ng of plasmid DNA was added to 50 ml of competent cells and mixed again with 300 ml lithium acetate for incubation at 30 min at 30 °C. The above mixture was then heat-shocked at 42 °C for 15 min and subsequently spread on drop-out plates in the absence of leucine, tryptophan, adenine, and histidine. Finally, the plates were incubated at 30 °C for 72 h for yeast growth. Positive interactions were determined by the growth of yeast transformants on drop-out plates.
Mammalian cell culture. The African green monkey kidney cell line (Vero cells) was cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). Vero cells were selected because they were susceptible to SARS-CoV infection. All cell cultures were carried out in a humidified 5% CO2 incubator at 37 °C.
Cell transfection. As many as 1 × 105 cells were used for transfection using Effectene transfection reagent according to the manufacturer’s protocol (Qiagen, Valencia, CA). Transfection was conducted as described previously [10]. Briefly, 250 ng plasmid and 2 μl of Enhancer were mixed with 40 μl EC buffer and incubated at room temperature for 5 min. After addition of 3 μl of the Effectene reagent, the transfection mixture was again incubated for 10 min, followed by dropwise addition to the cell culture, which had been washed with serum-free DMEM. The transfection efficiency is routinely monitored by co-transfecting the cells with pEGFP (Clontech, Palo Alto, CA). In addition, the experiments triplicated for each tranfection were repeated at least 3–5 times, with SD (standard deviation) being less than 10% (see figure legends for details).
Western blotting. Protein samples were fractionated on 4–12% Novex NuPAGE SDS–PAGE (Invitrogen, Carlsbad CA) and then transferred to PVDF membrane using semi-dry protein transfer apparatus (Bio-Rad, Hercules, CA). The membrane was blocked for 1 h with 5% skim milk in TBS buffer (20 mM Tris base, 137 mM NaCl, pH 7.6) containing 0.2% Tween 20. Afterwards, the membrane was probed with rabbit anti-His antibodies (Santa Cruz Biotechnologies, Santa Cruz, CA) and subsequently probed with donkey anti-rabbit HRP-conjugated antibodies (AP Biotech, Piscataway, NJ). The results were finally revealed by using Pierce Biotechnology SuperSignal West Femto Maximum Sensitivity Substrate (Rockford, IL).
Mammalian two-hybrid assay. Mammalian two-hybrid assays were performed using Clontech’s (Palo Alto, CA) Mammalian Two-Hybrid Assay Two Kit. The nucleocapsid protein and its mutant were cloned in-frame into pM and pVP16 vectors as described above. The recombinant pM-N constructs were then co-transfected with pVP16-N constructs along with a reporter plasmid, pG5SEAP. The pM vector carries a Gal4 DNA binding domain (BD), while the pVP16 vector carries a herpesvirus VP16 DNA activation domain (AD). In the case of an interaction between the two proteins, the BD and AD domains form a transcriptional activation complex and activate a secreted alkaline phosphatase (SEAP) reporter gene provided on the pG5SEAP. Reporter activity was measured by a chemiluminescent assay (SEAP assay).
SEAP assay. The SEAP assay was used to measure the activity of protein–protein interaction through chemiluminescent readings. Detection of SEAP level in the culture supernatants of the mammalian cell cultures as indicated above would reflect the intensity of protein–protein interaction. The GreatEscAPe SEAP assay was performed according to manufacturer’s protocol (Clontech, Palo Alto, CA).
Baculovirus expression. Detailed expression procedure for baculovirus expression of the N protein has been described. 1 Briefly, we used Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA) for expression of SARS-CoV N. The 6× histidine-tagged N gene was cloned in-frame into pFastBac vector (pFastBac-N) at BamHI and EcoRI sites. The pFastBac-N was subsequently co-transformed with Bacmid DNA into Sf9 cells. A plaque assay was performed to isolate the single clone, which expressed the N protein detectable by Western blot.
Sucrose gradient assay. As many as 2 × 106 of N protein-expressing baculovirus-infected Sf9 cells were lysed by 5 ml of PBS (Invitrogen, Carlsbad, CA) containing 0.2% NP40 (Sigma, St. Louis MO). The lysed cell extract was added to a 10–50% (w/w) fractionated sucrose gradient and subjected to 160,000g force centrifugation for 6 h. The uninfected Sf9 cells were used as the negative control.
Fluorescence microscopy. The pAsRed-N vector contains SARS-CoV N gene fused to a red-color fluorescent protein on the amino terminus. The pASRed-N/SR vector is a mutation product of pAsRed-N missing the SR-motif. The pECFP-nuc is a nuclear localization vector (Clontech, Palo Alto, CA) expressing a nuclear localization peptide fused to a cyan-color fluorescent protein. To perform the analysis, the pAsRed-N and pAsRed-N/SR were co-transfected, respectively, with pECFP-nuc into Vero cells. Thirty hours post-transfection, cells were visualized with a Zeiss Axiovert M200 fluorescent microscope (Zeiss, Oberkochen, Germany). The pAsRed-N and pAsRed-N/SR were excited at 545 nm and detected at 620 nm, while the pNuc-cyan was excited at 436 nm and detected at 480 nm.
Results
Yeast two-hybrid analysis shows N protein homotypic interaction
The yeast system was initially chosen because it is one of the most common systems for analysis of protein–protein interactions in vivo. The full-length SARS-CoV N protein was cloned in-frame into both the pGBKT7 (pGBK-N) and pGADT7 (pGAD-N) yeast two-hybrid vectors to investigate N protein self-interaction. Upon co-transformation of both plasmid constructs into Saccharomyces cerevisiae AH109, a phenotype was observed on high stringency nutrition drop-out plates lacking adenine, histidine, leucine, and tryptophan, indicating the presence of N protein homotypic interaction (Fig. 1 ). Clearly, controls with empty plasmids and co-transformation of pGBK-M and pGAD-M, which carry SARS-CoV membrane protein, did not demonstrate cell growth, further suggesting that the presence of SARS-CoV N–N self-interaction was specific in the yeast.
Fig. 1.
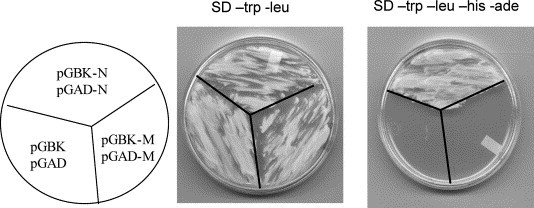
Yeast two-hybrid assay for N protein self-interaction. S. cerevisiae AH109 cells (1.5 ml of overnight culture) were transformed with 0.1 μg pGBK-N and pGAD-N plasmid DNA using the polyethylene glycol/lithium acetate method and plated on SD/LEU/−TRP and SD/−ADE−HIS/−LEU/−TRP drop-out plates and grown at 30 °C for 4 days. pGBK/T7 + pGAD/T7 and pGBK-M + pGAD-M were used as negative controls. Growth of the transformants on the SD/LEU/−TRP plate indicates that both pM- and pVP-related plasmids have been delivered into yeast cells, while the growth on SD/−ADE−HIS/−LEU/−TRP plate reveals activation of reporter genes, and thus interactions of proteins.
Mammalian two-hybrid assay
Mammalian two-hybrid systems are commonly used to confirm protein interactions detected from yeast two-hybrid systems [19], [20], [21]. Since Vero cells are susceptible to SARS-CoV infection, we employed the mammalian two-hybrid system to determine whether the N protein self-interaction also takes place in these cells. To this end, the N gene was cloned in-frame into pM (pM-N) and pVP16 (pVP-N) vectors. In addition, the SARS-CoV M protein was also included in the experiments (pM-M and pVP-M) together with a mock-transfected sample and a sample transfected with pM and pV16 cloning vectors as negative controls. As shown in Fig. 2 , samples co-transfected with pM + pVP16 showed similar level of chemiluminescent activity compared with the mock control, while the pM-N + pVP-N transfected sample showed at least 50 times higher SEAP activity compared with the mock control. Noticeably, no protein–protein interaction was observed with the SARS viral protein (M) under the same experimental conditions. Collectively, these results indicated that SARS-CoV N protein possessed a self-interaction property in vivo.
Fig. 2.
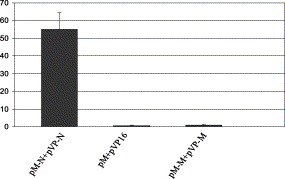
Mammalian two-hybrid analysis on N protein self-interaction. Vero cells were co-transfected with full-length SARS-CoV N protein constructs, pM-N + pVP-N along with pG5SEAP reporter vector, and incubated for 48 h at 37 °C in a CO2 incubator. Co-transfection samples, the cloning vector pM + pVP16, and SARS-CoV M protein constructs pM-M + pVP-M, were designated as negative controls. Error bars represent standard deviation (usually less than 10%).
Sucrose gradient fractionation reveals higher order structure of N protein
To further assess the extent of N protein oligomerization, the N gene was cloned into the pBlueBacHis2 vector for recombination into baculovirus and expressed in Sf9 insect cells. Sf9 cell lysate containing recombinant His-tagged N protein was then subjected to sucrose gradient fractionation. The cell lysate was subsequently fractionated on a step-wise sucrose gradient of 10–50% (w/w), with 10%, 20%, 30%, 40%, and 50% fractions. The fractionated proteins were then subjected to Western blotting with anti-His-tag primary antibody. As shown in Fig. 3 , the His-tagged N protein (50 kDa) was found to be sequestered into four of the five sucrose fractions, with a significant amount of N protein being detected in the 30%, 40%, and 50% fraction; only a trace amount of the N protein detected in the 20% fraction; and no detectable amount of the N protein being revealed in the 10% fraction (Fig. 3). Also, no N protein was detected in cells transfected with pBlueBacHis2 alone. The fact that the major amount of the expressed N protein was sequestered in the 30%, 40%, and 50% fractions rather than the 10% and 20% suggests that the majority of the expressed N protein was in the form of higher molecular structures.
Fig. 3.

Western blot analysis of sucrose gradient fractionated His6-tagged N protein. N protein was cloned into the baculovirus pBlueBacHis2 vector and transfected and expressed in Sf21 cells as described in Materials and methods. Roughly 5 × 106 cells were lysed and analyzed by sucrose gradient centrifugation followed by Western blotting detection of the viral proteins. The control sample is a baculovirus construct with pBlueBAcHis2 alone.
Mapping interaction region(s) using mammalian two-hybrid system
To map the regions of interaction within the SARS-CoV N protein, a series of eight nested truncation mutants were generated. The distribution of truncation is shown in Fig. 4A . These mutants’ fragments were cloned into mammalian two-hybrid vector pM. The eight sequential truncation mutants of the SARS-CoV N protein, designated from ΔpM-N1 to ΔpM-N8, were co-transfected with pG5SEAP reporter vector and pVP-N, which contains full-length N gene. Forty-eight hours post-transfection, a chemiluminescence-based assay was conducted to measure the protein–protein interaction activity through the SEAP reporter gene. As shown in Fig. 4B, ΔpM-N1 (aa 43–423), ΔpM-N2 (aa 84–423), ΔpM-N3 (aa 126–423), and ΔpM-N4 (aa 168–423) showed similar levels of chemiluminescence activities compared with full-length N protein. Importantly, it is noted that deletion of 168–208 resulted in a loss of N–N self-interaction, since virtually no chemiluminescence activity was observed in mutants ΔpM-N5, ΔpM-N6, ΔpM-N7, and ΔpM-N8. Because a region composed of 41 amino acids from aa 168 to 208 has been found to be crucial for N multimerization in the above mapping study, we were interested in employing a further investigation on a specific epitope in this region, which is pivotal for the N self-multimerization. It came to our attention from protein sequence analyses that there is a serine/arginine-rich region, which appears to be relatively conserved among most coronaviruses following GenBank searches. We consequently decided to delete this motif and use the mutant ΔpM-N/SR. As shown in Fig. 4C, deletion of the serine/arginine-rich region (aa 184–196) resulted in a complete loss of SEAP activities, suggesting these amino acids are required for N self-interaction. Finally, consistent with results in Fig. 2, no self-interaction was observed in the viral M proteins. Taken together, these results suggest that amino acids 184–196 are indispensable for multimerization of SARS-CoV nucleocapsid protein.
Fig. 4.
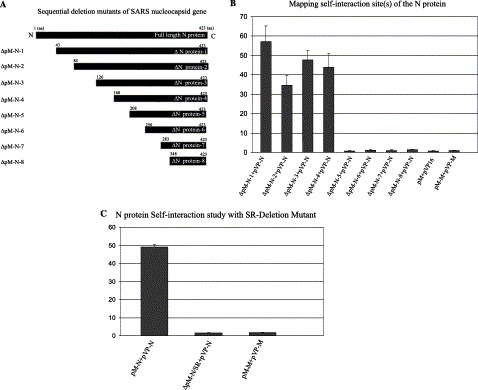
(A) Schematic description of N gene sequential deletion. N gene was truncated from 5′, primer sequences are indicated in Table 1. (B) Mammalian two-hybrid analysis of truncation mutant constructs of N protein in comparison with full-length N protein. pM-N mutants were co-transfected with pVP16-N and the reporter vector pG5SEAP into Vero cells. Chemiluminescent activity of secreted alkaline phosphatase present in cell culture medium was assayed as described in Materials and methods. Data represent an increase in chemiluminescence as compared to co-transfected vector controls. Error bars represent standard deviation (usually less than 10%). (C) Mammalian two-hybrid analysis of the requirement of the serine/arginine-rich region for the N protein self-interaction. The co-transfected samples were full-length N protein constructs pM-N and pVP-N, serine/arginine-rich region deletion mutant ΔpM-N/SR and pVP-N, and SARS-CoV M protein pM-M and pVP-M. The reporter vector pG5SEAP was also co-transfected in each group. Chemiluminescent activity of secreted alkaline phosphatase in cell culture medium was analyzed as described in Materials and methods. Error bars represent standard deviation (usually less than 10%).
Phenotypic changes of SR-motif deletion mutant
To characterize the impact of the alteration of N protein multimerization towards the virus assembly, we studied the sub-cellular localization of the N protein and its SR-deletion mutant. Sub-cellular localization of the viral protein is one of the commonly used approaches to characterize the involvement of the protein in the virus assembly [22], [23], [24], [25]. The N protein and its SR-deletion mutant were cloned into a red-color fluorescent vector pAs-Red2; the two vectors were, respectively, co-transformed with a nuclear localization vector pECFP-nuc. As indicated in Fig. 5A , the wild-type N protein was localized around the nucleus, forming an eclipse-like protein localization, while the SR-deletion mutant (Fig. 5B) demonstrated a completely different distribution pattern with scattered localization surrounding the nucleus. This result implicated that the SR region in the SARS-CoV N protein was not only responsible for the multimerization of the protein, but also pertinent to the sub-cellular localization of the protein, which is important for the virus assembly.
Fig. 5.
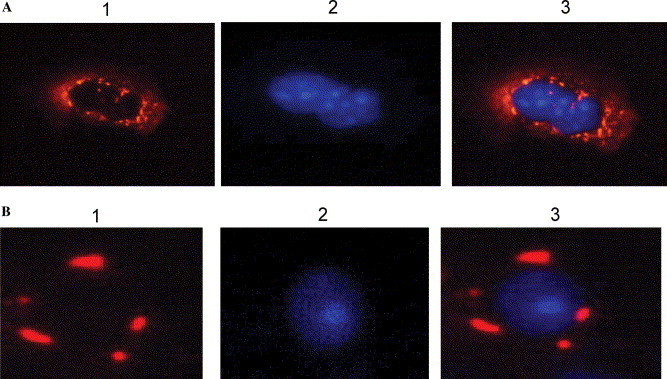
Co-localization analysis of SARS-CoV N protein. Constructs of pAsRed-N and ΔpAsRed-N/SR were co-transfected with the nuclear localization vector pECFP-nuc, respectively. Thirty hours post-transfection, cells were visualized with a fluorescent microscope. From A1 to A3 are the N protein, pECFP-nuc, and the super imposed image of A1 and A2. From B1 to B3 are the SR-deletion mutant, pECFP-nuc, and the superimposed image of B1 and B2.
Discussion
Self-interactions of viral nucleocapsid protein (N–N interaction) or multimerization, have been well documented and suggested to be critical to the formation of viral nucleocapsid core, an important composition in the viral particle assembly and maturation in a variety of viral systems [11], [12], [13], [14], [15], [16], [17], [18]. The multimerization of nucleocapsid proteins of coronaviruses is possibly a functional form of the protein, which may be involved in the virion assembly by stabilizing the helical structure of the N protein [26]. A study on another coronavirus, murine hepatitis virus (MHV), revealed that a multimerized form of the nucleocapsid protein was observed from Western blot analysis [26]. Although N–N in MHV has been identified in two independent groups, functional domains for MHV N–N interactions remain to be defined [9]. Two lines of evidence prompted us to conduct the current studies on SARS-CoV N protein: (1) the scarcity of published data on the N–N interaction in other coronaviruses hinders the elucidation of the mechanism involving formation of coronavirus nucleocapsid core; (2) little homology between SARS-CoV N and N proteins of other coronaviruses makes it difficult to link the data obtained from those coronaviruses to the understanding of structures and functions of SARS-CoV N protein, and may even intricate the rationalization of anti-SARS design. To facilitate investigations on the functional roles of SARS-CoV replication we set out to determine whether there is an existence of SARS-CoV N–N self-interaction, by using both yeast and mammalian two-hybrid systems, in addition to the mapping of the amino acid sequences responsible for protein multimerization.
We present here the first evidence of multimerization of the N protein of SARS-CoV. Our data showed that there was a 50-fold increase in chemiluminescence activity with co-transfection of pM-N + pVP-N compared to negative controls during mammalian two-hybrid analysis, implicating a strong homotypic interaction of the N protein (Fig. 4). Notably, no such interaction was observed for another viral protein, M, and consistent results were obtained in both yeast and mammalian two-hybrid systems, ruling out the possibility that identification of the N self-interaction was due to experimental deviation. Additional analysis with sucrose gradient revealed that N protein forms a range of higher order structures, with a majority of N proteins present in multimeric form, confirming that N multimerization does exist both in vitro and in vivo.
The identification of the amino acid region from 168 to 208 for protein multimerization during the initial screening process using sequential deletion mutants largely facilitated the subsequent localization of a much shorter stretch of 13 amino acids rich in serine and arginine (SSRSSSRSRGNSR). It is clear in our studies that deletion of these amino acids completely abolished N protein multimerization (Fig. 4C). Sub-cellular localization of the viral protein is widely used to characterize the involvement of the protein in the virus assembly [22], [23], [24], [25]. In addition, we also found that deletion of the SR-motif in the N protein resulted in a dramatic change of the sub-cellular localization of the N protein compared to the wild type, further implicating that this motif could be important for viral replication (Fig. 5). Our search for similar protein domains in public protein database (Entrez and ExPASy) yielded little information pertaining to known functional activities associated with similar aa sequences. In MHV (Accession No. NC-001846), a similar motif in the N gene shares only 46% homology with what we have found to be critical for N–N interaction in SARS-CoV. However, it is unknown whether those amino acids are involved in MHV N–N interaction. Clearly, further studies on these residues with respect to the exact mechanism involved in N self-interaction are necessary, and are ongoing in our laboratories.
The functional roles of SARS-CoV N in viral replication and disease development remain largely unknown. We recently reported that the N protein can selectively activate AP-1 pathway [10], suggesting that the virus has encoded a strategy to regulate cellular signaling processes. A variety of functional activities have been ascribed to the nucleocapsid protein of other known coronaviruses, including virion assembly, and RNA-dependent RNA transcription and translation [3], [27], [28]. Noticeably, N protein self-association may be important for initiating RNP (ribonucleoprotein) formation leading to encapsidation [26], [27], [29]. As SARS-CoV has a remote phylogenetic relationship with other known coronaviruses, vigorous studies on the genetics and functions of SARS-CoV structural proteins are necessary to fully elucidate the mechanisms involving SARS-CoV replication. In this report, we presented the data that SARS-CoV N protein has intrinsic properties of self-interaction, and that a region rich in serine and arginine residues has been found to be critical for the N protein multimerization.
Footnotes
Y. Li et al., manuscript in preparation.
References
- 1.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Kattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 2.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with Severe Acute Respiratory Syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 3.Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes K.V. Chapter 36: Coronaviruses. In: Knipe D.M., Howley P.M., editors. vol. 1. Williams & Wilkins; Baltimore, MD: 2001. pp. 1187–1203. (Fields Virology). [Google Scholar]
- 5.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguiere A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra S., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.-W. Identification of a novel coronavirus in patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Tsang K.W., Mok T.Y., Wong P.C., Ooi G.C. Severe acute respiratory syndrome (SARS) in Hong Kong. Respirology. 2003;8(3) doi: 10.1046/j.1440-1843.2003.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. A major outbreak of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 8.Macneughton M.R., Davies H.A. Ribonucleoprotein-like structures from coronavirus particles. J. Gen. Virol. 1978;39(3):545–549. doi: 10.1099/0022-1317-39-3-545. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Zhang X. The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo. Virology. 1999;265(1):96–109. doi: 10.1006/viro.1999.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., Li X. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311(4):870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfadhli A., Steel E., Finlay L., Bachinger H.P., Barklis E. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 2002;277(30):27103–27108. doi: 10.1074/jbc.M203395200. [DOI] [PubMed] [Google Scholar]
- 12.Bisho D.H.L. Biology and molecular biology of bunyaviruses in the family Bunyaviridae. In: Elliott R.M., editor. The Bunyaviridae. Plenum Press; New York, NY: 1996. pp. 1–15. [Google Scholar]
- 13.Kaukinen P., Koistinen V., Vapalahti O., Vaheri A., Plyusnin A. Interaction between molecules of hantavirus nucleocapsid protein. J. Gen. Virol. 2001;82(Part 8):1845–1853. doi: 10.1099/0022-1317-82-8-1845. [DOI] [PubMed] [Google Scholar]
- 14.Luban J., Alin K.B., Bossolt K.L., Humaran T., Goff S.P. Genetic assay for multimerization of retroviral gag polyproteins. J. Virol. 1992;66(8):5157–5160. doi: 10.1128/jvi.66.8.5157-5160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto M., Hwang S.B., Jeng K.S., Zhu N., Lai M.M. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology. 1996;218(1):43–51. doi: 10.1006/viro.1996.0164. [DOI] [PubMed] [Google Scholar]
- 16.Myers T.M., Pieters A., Moyer S.A. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP–NP binding domain. Virology. 1997;229(2):322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- 17.Uhrig J.F., Soellick T.R., Minke C.J., Philipp C., Kellmann J.W., Schreier P.H. Homotypic interaction and multimerization of nucleocapsid protein of tomato spotted wilt tospovirus: identification and characterization of two interacting domains. Proc. Natl. Acad Sci. USA. 1999;96(1):55–60. doi: 10.1073/pnas.96.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimatsu K., Lee B.-H., Araki K., Morimatsu M., Ogino M., Ebihara H., Arikawa J. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J. Virol. 2003;77(2):943–952. doi: 10.1128/JVI.77.2.943-952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshima T., Yura T., Yanagi H. The trimerization domain of human heat shock factor 2 is able to interact with nucleoporin p62. Biochem. Biophys. Res. Commun. 1997;240(1):228–233. doi: 10.1006/bbrc.1997.7662. [DOI] [PubMed] [Google Scholar]
- 20.Ema M., Hirota K., Mimura J., Abe H., Yodoi J., Sogawa K., Poellinger L., Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18(7):1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto H., Rahman M., Takatera H., Kang H.Y., Yeh S., Chang H.C., Nishimura K., Fujimoto N., Chang C. A dominant-negative mutant of androgen receptor coregulator ARA54 inhibits androgen receptor-mediated prostate cancer growth. J. Biol. Chem. 2002;277(7):4609–4617. doi: 10.1074/jbc.M108312200. [DOI] [PubMed] [Google Scholar]
- 22.Mahalingam S., Van Tine B., Santiago M.L., Gao F., Shaw G.M., Hahn B.H. Functional analysis of the simian immunodeficiency virus Vpx protein: identification of packaging determinants and a novel nuclear targeting domain. J. Virol. 2001;75(1):362–374. doi: 10.1128/JVI.75.1.362-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75(19):9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77(19):10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee P., Hruby D.E. Trans processing of vaccinia virus core proteins. J. Virol. 1993;67(7):4252–4263. doi: 10.1128/jvi.67.7.4252-4263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins S.G., Frana M.F., McGowan J.J., Boyle J.F., Holmes K.V. RNA-binding proteins of coronavirus MHV: detection of monomeric and multimeric N protein with an RNA overlay-protein blot assay. Virology. 1986;150(2):402–410. doi: 10.1016/0042-6822(86)90305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 28.Escors D., Izeta A., Capiscol C., Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J Virol. 2003;77(14):7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risco C., Anton I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 1996;70(7):4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


