Abstract
Severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) caused a severe outbreak in several regions of the world in 2003. The virus is a novel coronavirus isolated from patients exhibiting atypical pneumonia and may have originated from wild animals such as civet cats in southern China. The genome of SARS-CoV is a positive-sense, single-stranded RNA whose sequence is distantly related to all known coronaviruses that infect humans and animals. Like other known coronaviruses, SARS-CoV is an enveloped virus containing three outer structural proteins, namely the membrane (M), envelope (E), and spike (S) proteins. The nucleocapsid (N) protein together with the viral RNA genome presumably form a helical core located within the viral envelope. The SARS-CoV nucleocapsid (N) protein is a 423 amino-acid, predicted phospho-protein of 46 kDa that shares little homology with other members of the coronavirus family. A short serine-rich stretch, and a putative bipartite nuclear localization signal are unique to it, thus suggesting its involvement in many important functions during the viral life cycle. In this report we have cloned the N gene of the SARS coronavirus, and studied its property of self-association to form dimers. We expressed the N protein as a fusion protein in the yeast two-hybrid system to demonstrate self-association and confirmed dimerization of the N protein from mammalian cell lysates by coimmunoprecipitation. Furthermore, via deletion analysis, we have shown that the C-terminal 209 amino-acid region constitutes the interaction domain responsible for self-association of the N protein to form dimers.
Keywords: SARS coronavirus, Protein–protein interaction, Yeast two-hybrid system, Nucleocapsid protein
Severe acute respiratory syndrome (SARS) is a newly emerging infectious disease. The causative agent of SARS has been identified as a novel coronavirus, namely SARS-associated coronavirus (SARS-CoV) [1], [2], [3]. As of 30 June 2003, 8447 probable SARS cases including 811 deaths have been reported to the World Health Organization (WHO) from 32 countries or regions worldwide.
The SARS viral genome comprises approximately 30,000 nucleotides, which are organized into approximately 13–15 open reading frames (ORFs), taking into consideration only those exceeding 50 amino acids in translational capacity [4], [5]. Sequence comparison with corresponding ORFs of other known coronaviruses reveals a similar pattern of gene organization typical of coronaviruses [6].
The 1259-nucleotide N gene of the SARS virus resides at the 3′ end of the genome. This 46-kDa structural protein has been predicted to interact with the viral RNA, and both constitute the viral nucleocapsid. The N protein of coronaviruses is present in the icosahedral core as well as in the internal helical nucleocapsid [7]. Since no studies have hitherto been reported on the structure and function of the N protein of the SARS coronavirus, we PCR amplified and cloned the N protein of SARS-CoV, and studied its capability to form a dimer by self-association using the yeast two-hybrid system. Here we report that the N protein of the SARS coronavirus is capable of self-association and requires the C-terminal helix-rich 209 amino-acid region to facilitate this protein-protein interaction.
Materials and methods
Growth media, yeast strains, and plasmid constructs. All strains, plasmids, and plasmid constructs used in this study are described in Table 1 . The full-length N gene of the SARS coronavirus (Singapore isolate) was PCR-amplified from a genomic construct of clone NC_004718, and cloned into the pCR-XL-TOPO vector (Invitrogen). The full-length N gene was subjected to DNA sequencing, and the inserts were verified against the corresponding region of the SARS coronavirus complete genome NC_004718. The full-length N gene was excised from the pCR-XL-TOPO-N construct using the restriction enzymes EcoRI and ClaI, and ligated into the pGADT7 vector to generate an N-terminal in-frame fusion with the GAL4 activation domain (AD). To clone N in fusion with the DNA-binding domain of GAL4, pGADT7-N was digested with NcoI and PstI, and the excised fragment was ligated into the pAS2 vector which was linearized with the same pair of restriction enzymes. HA tagged N was cloned into pSGI by digesting pGADT7-N with BglII and ligating into pSGI digested with the same enzyme. Myc-tagged N was produced by ligating the BamHI and ApaI fragment from pCR-XL-TOPO-N into pcDNA 3.1 vector digested with the same enzymes. All DNA manipulations were performed as described by Sambrook et al. [8]. All deletion constructs were generated by subcloning the full-length N gene as summarized in Table 1. All constructs were verified by restriction digestion and sequencing.
Table 1.
Yeast strains, plasmids, and recombinant plasmid constructs used in this study
| Strain/plasmid/construct | Genotype/description |
|---|---|
| Strain | |
| AH109 | MATa, ura3-52 leu2-3,112, his3d200, trp1d1, ade2, LYS2::LexA op)-HIS3, ura3::LexA-op)-LacZ, LexA-MS2 coat (TRP1). |
| Constructs | |
| pGADT7 N | pCR-XL-TOPO-N cut withEcoRI and ClaI and cloned into pGADT7 cut with the same. |
| pAS2-1 N | pGADT7 N cut with NcoI and PstI and cloned into pAS2-1 cut with the same. |
| 1-310 N | pGADT7 N cut with SacI, klenowed and vector self-ligated. |
| 1-630 N | pGADT7 N was cut with EcoRI and NheI and fragment cloned into pGADT7 cut with EcoRI and SmaI |
| 1-830 N | pGADT7 N cut with TtHIII, fragment cloned into pGADT7 cut with EcoRI and SmaI |
| 310-1260 N | pGADT7 N cut with SacI and ClaI, fragment cloned into pGADT7 cut with SmaI and ClaI. |
| 630-1260 N | pGADT7 N cut with NheI and ClaI, fragment cloned into pGADT7 cut with SmaI and ClaI |
| pSGI HA N | pGADT7 N cut with BglII and BamHI and cloned into pSGI cut with BglII |
| pCDNA 3.1 myc N | pCR-XL-TOPO-N cut with BamHI and ApaI and cloned into pcDNA3.1 myc cut with the same. |
Yeast two-hybrid techniques. The GAL4-based two-hybrid system, kindly provided by Dr. Stephen Elledge [6], contain pAS2 (DNA binding domain vector) and pACT2 (activation domain vector), together with the yeast reporter strain Saccharomyces cerevisiae AH109 (Table 1) were employed. The host strain containing pAS2-SNF1 and pACT2-SNF4 was used as a positive control [9]. The AH109 host contains integrated copies of both HIS3 and lacZ reporter genes under the control of GAL4 binding sites. The AH109 yeast strain was transformed with the appropriate plasmids using the lithium acetate procedure, and grown on SD plates in the absence of Trp and Leu (SDTrp− and SDLeu−, respectively). Protein interaction was tested on SD plates without Leu, Trp, and His (SDLeu−Trp−His−). After incubation 30 °C for 3 days, individual colonies were streaked out and tested for liquid and filter-lift β-galactosidase activity, by 50 mM oft 3-amino-1,2,3-triazole (3AT) assay, and by the diploid His assay. The filter β-galactosidase assay, a parameter directly reflecting the strength of protein–protein interactions, was performed by streaking doubly transformed yeast colonies onto filter paper and allowing them to grow for 2 days on selection medium. Yeast cells were permeabilized by freezing yeast-impregnated filters in liquid nitrogen and thawing at room temperature. The filter was placed over a second filter that was pre-soaked in 0.1 M phosphate buffer (pH 7.0) containing 300 mg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (CPRG) and 0.27% β-mercaptoethanol. Filters were left for 48 h to develop a blue color which indicated a positive protein-protein interaction. The liquid β-galactosidase activity was determined using the substrate CPRG as described previously [10], [11]. Relative enzymatic activity was determined for five independent transformants. Data for quantitative assays were collected for yeast cell number and are means ± SEM of triplicate assays. Appropriate positive and negative controls, and buffer blanks were used. The AH109 host strain containing pAS2-SNF1 and pACT2-SNF4 was used as a positive control [9].
Coimmunoprecipitation assay and Western blotting. COS-1 cells were transfected with the pSGI-N or pCDNA3.1-N plasmid or both using the lipofectine reagent. At 48 h post-transfection, cells were washed once in PBS and then lysed in lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerol-PO4, 1 mM Na3SO4), with protease inhibitor cocktail. Equal amounts of protein were incubated with respective antibodies overnight at 4 °C. To the samples was added 100 μl of 10% suspension of protein A–Sepharose. The mixture was allowed to shake for 1 h at 4 °C following which the beads were washed four times in lysis buffer, and protein was eluted in 2× SDS dye by boiling the sample for 5 min. Samples were resolved by 10% SDS–PAGE and transferred to a nitrocellulose membrane. The membrane was blocked using 0.5% non-fat dried milk in PBST for 1 h, and incubated overnight with primary antibody. The blot was then washed thrice in PBST, incubated with anti-mouse IgG HRPO for 1 h and washed thrice and bands were detected using the ECL detection method as recommended by the manufacturer (Cell Signalling Technology).
Results and discussion
The full-length N protein can form a dimer
The nucleocapsid (N) protein of the SARS coronavirus (Singapore isolate) was cloned into the yeast two-hybrid vectors (Table 1) resulting in an N-terminal in-frame fusion of the GAL4-DNA binding domain and the GAL4 activation domain to the N protein. In order to confirm the reading frame and expression of the N protein, it was expressed in vitro using the rabbit reticulocyte-based, coupled transcription–translation system (Promega) and immunoprecipitated using anti-HA antibody (data not shown). S. cerevisiae AH109 (MATa trp1-901 his3 leu2-3, 112 ura3-52 ade2 gal4 gal80URA3::GAL-lacZ LYS2::GAL-HIS3) cells were transformed with single plasmids, or cotransformed with the GAL4 BD- and AD-vectors containing SARS-CoV N. The AH109 host strain containing pAS2-SNF1 and pACT2-SNF4 was used as positive controls [9]. AH109 contains integrated copies of both HIS3 and lacZ reporter genes under the control of GAL4-binding sites. The results of the two-hybrid assay are shown in Fig. 1 . Single transformants used in this assay were AH109 yeast cells containing BD-N, and cells containing AD-N. Yeast host cells containing only BD- and only AD-vectors, were also used as negative controls (Fig. 1). All these transformants grew on the YPD non-selective media. The untransformed host cells were also plated as negative controls. Single transformants containing the BD-vector alone or as a fusion with N showed growth on the synthetic dextrose Trp− plate (SDTrp−). Correspondingly, single transformants containing the AD-vector alone or as a fusion with N showed growth on the synthetic dextrose Leu− plate (SDLeu−). The cotransformants were similarly plated on YPD and synthetic dextrose medium lacking Trp or Leu or both, and the resultant colonies were subsequently plated on His− medium (SDTrp−Leu−His−) to test for His prototrophy. Growth of the cotransformants, containing the BD-N and AD-N constructs, in both SDTrp− and SDLeu− plates simply showed that both plasmids were present in the transformed cells. Growth of these clones on the SDTrp−Leu−His− media showed that the transcription of the HIS3 gene was switched on by the reconstitution of the GAL4 transactivator due to a specific N–N interaction. Colonies were transferred onto nitrocellulose filters and a β-galactosidase filter assay was performed [12], [13], [14]. The cotransformants containing both the BD-N and AD-N constructs together with all positive and negative controls were tested by this assay. Results obtained from the β-galactosidase filter assay were in agreement with the results obtained from the SDHis− growth experiments.
Fig. 1.

Yeast two-hybrid results showing full-length homotypic interactions of the N protein of SARS-CoV. YPD yeast peptone dextrose media (no selection), SDTrp−, SDLeu−, SDTrp−Leu−, and SDHis− are restrictive growth media lacking Trp, Leu, Trp Leu, and Trp Leu His, respectively. β-Galactosidase filter assay results are also shown.
The liquid β-galactosidase assay was conducted and activity was determined with the substrate chlorophenol red-β-d-galactopyranoside (CPRG) as described previously [15], [16]. The host strain AH109 alone, together with single transformants containing the AD-N and BD-N and cotransformants containing AD-/BD- without a fusion protein, BD-/AD- and single transformants with either BD-N or AD-N were tested. Negative controls (host untransformed cells) showed virtually no liquid β-galactosidase activity. BD-1111/AD-11112 was the positive control whereas the clones containing BD-N/AD-N were the test samples in this experiment (Fig. 2 ). Relative enzymatic activity was determined in five independent transformants from each group. Our results from this assay indicate a strong protein–protein interaction between AD-N and BD-N proteins.
Fig. 2.
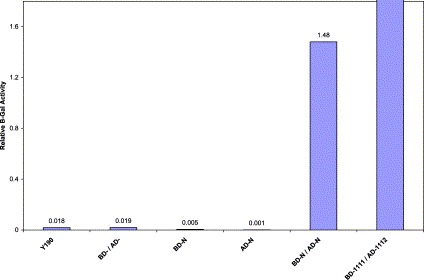
Liquid β-galactosidase assay results. Single and cotransformants were analyzed for a liquid β-galactosidase assay and were compared with others. Values are given in arbitrary units. The numbers above each bar represent the mean of five independent transformants.
To further confirm these interactions in a mammalian cell environment, we performed a coimmunoprecipitation assay using N protein fused with two different tags. COS-1 cells were transfected with amino-terminal HA-tagged N or carboxy-terminal myc-tagged N or both. Lysates were subsequently immunoprecipitated using respective antibodies and immunoblotted using HA antibody. Lane 2 in Fig. 3 shows a cotransfected sample immunoprecipitated using anti-myc antibody. Lane 1 shows the HA-tagged N-transfected sample immunoprecipitated using anti-HA antibody which served as a positive control. Lane 3 shows myc-tagged N-transfected sample immunoprecipitated with anti-HA antibody which served as a negative control. Lane 4 shows myc-tagged N immunoprecipitated using myc antibody. These results clearly demonstrate that the N protein is capable of homodimerization in a mammalian cellular environment. In order to form the capsid of SARS-CoV, the nucleocapsid protein has to oligomerize. Hence our data provide a functional verification to earlier predictions regarding the N protein of the SARS virus. Further studies on the molecular dissection of N were carried out to map the region of N responsible for this interaction.
Fig. 3.

COS1 cells were transfected with HA-N,MYC-Nor both, immunoprecipitated using respective antibodies, resolved by 10% SDS–PAGE and Western blotted using anti-HA antibody as described in Materials and methods. Lane 1 shows HA-N immunoprecipitated with the same antibody, lanes 2 and 3 show cotransfected cells immunoprecipitated using anti-myc or anti-HA antibody, respectively, lane 4 shows myc-N immunoprecipitated using anti-myc antibody.
A 210 amino-acid carboxy-terminal region is responsible for N dimerization
Various deletions were designed to characterize the homodimerization domain of the N protein. Thus, plasmids pGADT7 N 1-310, pGADT7 N 1-630, pGADT7 N 1-830, pGADT7 N 310-1260, and pGADT7 N 630-1260 were constructed as described in Table 1. We investigated the strength of all these interactions for the full-length and truncated N proteins by measuring the HIS3 reporter gene in the presence of 0, 5, 10, 20, 25, and 50 mM of 3-aminotriazole (AT). These results indicated the strength of the protein–protein interaction as a function of His prototrophy. Cells containing both fusions of full-length N proteins showed growth on SD His− 50 mM AT plates, thereby confirming strong homodimerization.
Our two-hybrid experiments each consisted of cotransformation of one of the pGADT7 N deletions with the full-length fusion protein pGADT7 N. The results of these experiments are shown in Fig. 4 . The AD 1-830, AD 1-630, and AD 1-310 constructs cotransformed with the full-length N construct (BD-N) lacked reporter gene activation. In contrast, the AD-N 310-1260 and AD-N 630-1260 constructs when tested with full-length BD-N, revealed reporter gene activity. The AD-N 310-1260 and AD-N 630-1260 when tested with BD-N (full-length) showed reporter gene activity. However, the AD 1-830, AD 1-630, and AD 1-310, when cotransformed with their corresponding full-length N construct (BD-N), showed no reporter gene activation. However, the strength of interaction between 630-1260 and BD-N was found to be lower than that of full-length N–N or AD N 310-1260 and full length N interaction. Hence, we mapped the interaction domain for the N protein dimerization to reside in C terminal one-third amino acids. Upon investigation of the predicted secondary structure model for N, we found this region to be helix-rich and contained the putative nuclear localization signal.
Fig. 4.
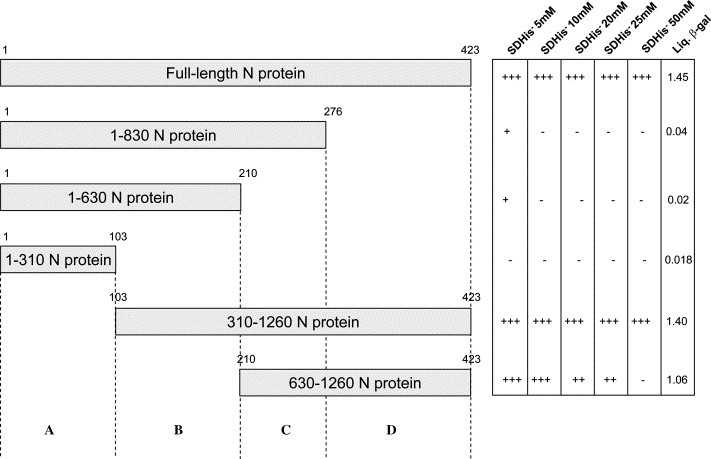
Deletion analysis for isolation of the interaction domain of the N protein of SARS-CoV. Using the yeast two-hybrid approach, each of the shown deletions was tested along with a full-length N protein and scored for marker gene activity. Histidine prototrophy was studied at 5, 10, 20, 25, and 50 mM AT concentrations on SDHis− media. Liquid β-galactosidase assay was conducted on the cotransformants and mean of five samples is displayed for each deletion of the N protein. The deletions divide the N protein into four regions A, B, C, and D. Numbers above each deletion bar depict amino-acids.
Self-association of the nucleocapsid protein has been observed in many viruses. This process leads to the formation of a viral capsid which protects the genome from extracellular agents. In this report, we have used two independent assays to establish a direct interaction between the nucleocapsid molecules. This dimerization process is likely to lead to multimerization of the N protein and to subsequent capsid assembly. While the functional relevance of this interaction in an in vivo context is unknown, our report nonetheless clearly proves a direct physical interaction between the SARS N protein molecules. Moreover, the strong capability to self-associate was demonstrated by the N–N cotransformed yeast cells which exhibited the ability to grow at 50 mM concentration of 3AT in the yeast two-hybrid assay. However, the exact mechanism of interaction is not known. In a wide range of viruses disulfide bond mediated self-association of nucleocapsid has been observed [17], [18], [19], [20], but the N protein of SARS is unable to do so since it does not have any cysteine as observed from its amino acid composition. Observations have been gathered from N proteins of other RNA virus genomes (e.g., equine arthritis virus and simian hemorrhagic fever virus). The disulfide bond in the nucleocapsid protein has been proposed to enhance the stability of virions: e.g., hepatitis B virus [18] and human papilloma virus [19]. Interestingly, N proteins of other corona viruses contain cysteine residues, e.g., MHV N has 2 cysteine residues. Although it is not known whether cysteine plays a role in self-association of nucleocapsid in other corona viruses, the complete absence of cysteine residues in the N protein of SARS implies that it is unique among other corona viruses. It is quite possible that the SARS virus may have adopted an alternative conformational strategy to circumvent the problem of missing cysteine. In an in silico model for secondary predictions, both N proteins of MHV and SARS show a hydrophilic, helix rich C-terminal. This distinct secondary structure may be responsible for tight intramolecular self-association; however, the exact mechanism and forces driving the described self-association of the nucleocapsid proteins need further investigation. Further, since the N protein of SARS has been predicted to play other regulatory functions [21] besides capsid assembly, dimerization could be an activating switch for regulation of protein function. Moreover, in MHV, the RNA binding domain has been found to reside in similar locations between amino acids 163–229 [20], which includes the C-terminal helix-rich region. We have identified the dimerization domain of the N protein of SARS to be localized to the C-terminal helix rich region thus suggesting a similar strategy by the SARS virus for nucleocapsid formation.
Acknowledgements
This work was supported jointly by internal funds from the International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, and a grant from the Department of Biotechnology to S.K.L. The visiting resource persons program sponsored by ASM-UNESCO and support from Microbiology Department, National University of Singapore are gratefully acknowledged. M.S. is a senior research fellow of the CSIR. Technical support from Yeo Wee Ming is gratefully acknowledged.
Footnotes
Abbreviations: AD, activation domain; BD, binding domain; 3-AT, 3-aminotriazole; N, nucleocapsid.
References
- 1.Buttner J. Evaluation of the diagnostic value of laboratory investigations. J. Clin. Chem. Clin Biochem. 1977;15:1–12. [PubMed] [Google Scholar]
- 2.A.T. Fleischauer, CDC SARS Investigative Team, Outbreak of severe acute respiratory syndrome—worldwide, Morb. Mortal Wkly. Rep. 52 (2003) 226–228 [PubMed]
- 3.Cumulative number of SARS probable cases in Taiwan, SARS Online Information Centre, Center for Disease Control, Taiwan [Accessed July 2, 2003]. Available from: URL: http:/www.cdc.gov.tw/sarsen
- 4.Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Lai M.M.C, Cavanagh D. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risco C, Anton I.M, Enjuanes L, Carrascosa J.L. J. Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E.F, Maniatis T. second ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 9.Harper J.W, Adami G.R, Wei N, Keyomarsi K, Elledge S.J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 10.Rose M.D, Winston F, Hieter P. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1990. Methods in Yeast Genetics. [Google Scholar]
- 11.Bai C, Elledge S.J. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S, Korkaya H, Zafrullah M, Jameel S, Lal S.K. The phosphorylated form of ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 2002;277:22759–22767. doi: 10.1074/jbc.M200185200. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava R, Lal S.K. A liquid synchronized growth culture assay for the identification of true positive and negative yeast three-hybrid transformants. Lett. Appl. Microbiol. 2002;34:300–303. doi: 10.1046/j.1472-765x.2002.01085.x. [DOI] [PubMed] [Google Scholar]
- 14.Tyagi S, Salier J.P, Lal S.K. The liver-specific human α1-microglobulin/bikunin precursor (AMBP) is capable of self-association. Arch. Biochem. Biophys. 2002;399:66–72. doi: 10.1006/abbi.2001.2745. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Jameel S, Lal S.K. The full-length and N-terminal deletion of ORF2 protein of hepatitis E Virus can dimerize. Biochem. Biophys. Res. Commun. 2001;286:214–221. doi: 10.1006/bbrc.2001.5256. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S, Jameel S, Lal S.K. Self-association and mapping of the interaction domain of the hepatitis E virus ORF3 protein. J. Virol. 2001;75:2493–2498. doi: 10.1128/JVI.75.5.2493-2498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron M.D, Forsell K. Oligomerization of structural proteins of rubella virus. Virology. 1991;185:811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 18.Jeng K.S, Hu C.P, Chang C.M. Differential formation of disulfide linkages in the core antigen of extracellular and intracellular hepatitis B virus core particles. J. Virol. 1991;65:3924–3927. doi: 10.1128/jvi.65.7.3924-3927.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Beard P.A, Estes P.A, Lyon M.K, Garcia R.L. Intercapsomeric disulfide bonds in papilloma virus assembly and disassembly. J. Virol. 1998;72:2160–2167. doi: 10.1128/jvi.72.3.2160-2167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He R, Leeson A, Andonov A, Li Y, Bastien N, Cao J, Osiowy C, Dobie F, Cutts T, Ballantine M, Li X. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wooton S.K, Yoo D. Homooligomerization of the Porcine reproductive and respiratory syndrome virus nucleocapsid protein and the role of disulfide linkages. J. Virol. 2003;77:4546–4557. doi: 10.1128/JVI.77.8.4546-4557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


