Abstract
The aim of this study was to examine the incidence of Clostridioides (previously Clostridium) difficile and Clostridium perfringens in the feces of diarrheic and non-diarrheic dogs. Also, the presence of other common canine enteropathogens was examined. Toxigenic C. difficile and C. perfringens positive for the NetF-encoding gene (netF) were detected in 11 (11.9%) and seven (7.6%) diarrheic dogs, respectively. Three dogs were diagnosed simultaneously with toxigenic C. difficile and netF-positive C. perfringens. Among other enteropathogens, Giardia sp. was the most common agent detected in dogs positive for toxigenic C. difficile or netF-positive C. perfringens. The results suggest that C. difficile and C. perfringens occur more frequently as a primary cause of diarrhea.
Keywords: Canine diarrhea, Clostridium perfringens, NetF, Clostridioides difficile, Enteric pathogens
There are several reports of enteric disorders caused by Clostridioides (previously Clostridium) difficile in dogs. However, its role in canine diarrhea is still uncertain [1,2]. Similarly, several authors have shown a high prevalence of the enterotoxin (CPE)-encoding gene (cpe) in Clostridium perfringens isolates obtained from diarrheic dogs, which led to the speculation that this toxin was responsible for the pathogenesis of canine C. perfringens-associated diarrhea (CPAD) [3]. Recently, two putative pore-forming toxins (NetE and NetF) were described in strains from cases of fatal canine hemorrhagic gastroenteritis and evidence that NetF was the virulence factor in this syndrome was obtained [4,5].
Although these two agents are commonly described as enteropathogens in dogs [6], their role and the involvement of other enteropathogens remain largely unknown. Thus, the aim of this study was to detect and characterize C. difficile and C. perfringens strains from diarrheic and non-diarrheic dogs. Also, the presence of other common non-clostridial enteropathogens was evaluated to provide a better understanding of the role of these two agents in canine diarrhea.
Stool samples were collected from 154 dogs, of which 92 were diarrheic and 62 were apparently healthy. The samples from diarrheic dogs were obtained directly from the Veterinary Hospital of Universidade Federal de Minas Gerais at the time of the consultation. The clinical history of all animals was recorded. Samples from apparently healthy animals were obtained in city squares in Belo Horizonte (Minas Gerais, Brazil), with prior permission of the owner and only fecal material that did not have contact with the environment was collected. The animals were categorized into four groups based on their age: younger than 6 months (n = 45, 29.2%), from 7 to 12 months (n = 28, 18.1%), from 1 to 5 years (n = 36, 23.3%) and older than 5 years (n = 44, 28.5%). In each age group, at least one apparently healthy dog sample (control) was included for each two diarrheic dog samples.
Isolation of C. difficile was based on previously described protocols [7,8]. All isolates were subjected to a multiplex-PCR for a housekeeping gene (tpi), the toxin A gene (tcdA), the toxin B gene (tcdB) and a binary toxin gene (cdtB) [9] and were PCR ribotyped as previously described [10]. PCR ribotypes for which the reference strains were available are designated by international Cardiff nomenclature, while others are designated by internal nomenclature (BR and number). Stool samples positive for C. difficile isolation were also subjected to toxin A/B detection (C. difficile Tox A/B II - Techlab Inc., USA).
Isolation of C. perfringens was also based on previously described protocols [7,8] and isolates were subjected to a PCR protocol [11] for the detection of genes for typing C. perfringens (alpha, beta, epsilon and iota toxins), and beta-2 toxin (cpb2) and enterotoxin (cpe). PCR protocols described by Refs. [12] and [5] were used for the detection of the NetB-, NetE- and NetF-encoding genes (netB, netE and netF, respectively). Stool samples positive for C. perfringens cpe + strains were also subjected to CPE detection (Ridascreen® Clostridium perfringens Enterotoxin, R-Biopharm, Germany).
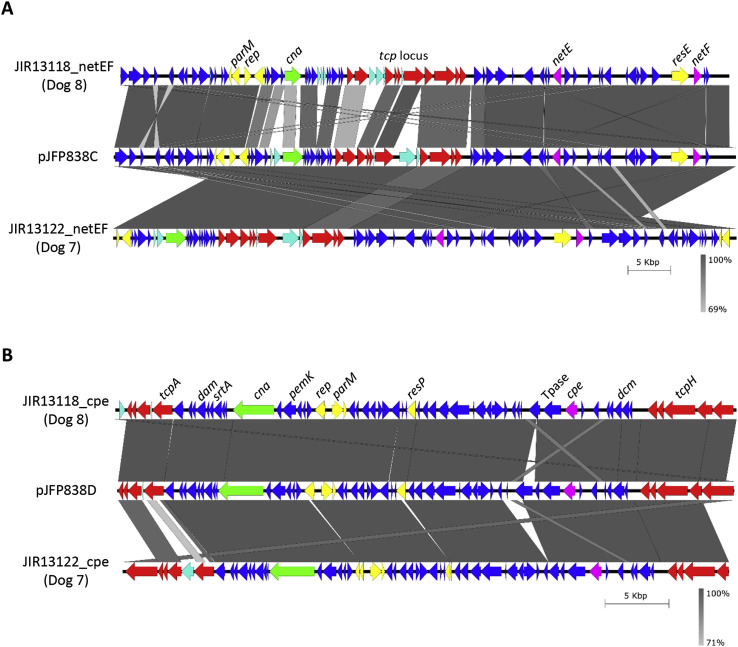
Two C. perfringens netF + isolates (from diarrheic dogs 7 and 8) were selected for whole genome sequencing. Both developed bloody diarrhea without an identifiable predisposing factor and recovered after regular treatment. Library preparations (Nextera kit, Illumina, USA) from genomic DNA [13], were sequenced using a Miseq (Illumina, USA). Reads were trimmed for quality (Nesoni, Paul Harrison (http://www.vicbioinformatics.com/software.nesoni.shtml),), assembled using A5-miseq [14] and auto-annotated using Prokka [15]. Pseudomolecules were generated by scaffolding against the plasmid sequences pJFP838C and pJFP838D [4] for the netE-, netF- and cpe-encoding plasmids, respectively, and manually inspected using Artemis [16]. Plasmid comparisons were generated using the Blastn tool of EasyFig [17]. Plasmid sequences were lodged with NCBI under the following accession numbers (MG456813, MG456814, MG456815 and MG456816).
The presence of parvovirus (CPV), rotavirus, coronavirus and Giardia spp. was evaluated by commercial lateral flow tests (Ecodiagnostica, Brazil). For the isolation of Salmonella spp., stool samples were inoculated into tetrathionate broth followed by plating on Hektoen enteric agar (Oxoid, USA). Sulfide-producing colonies were subjected to a previously described PCR assay to detect the Sulmonella ompC gene [18]. For Escherichia coli detection, stool samples were plated on MacConkey agar (Difco, USA) and characteristic lactose-fermenting colonies were analyzed by PCR to detect several genes associated with diarrheagenic E. coli [19].
In this survey, diarrheic dogs were more than five times more likely to be positive for C. difficile than apparently healthy animals (Table 1 ). Toxins A and B were detected in eight fecal samples (8.7% of the diarrheic animals) that were PCR positive for toxigenic C. difficile, confirming the diagnosis of C. difficile infection (CDI) in these animals. Although antibiotic therapy is a known risk factor for CDI in humans and dogs [2,20,21], only one animal that was positive for toxigenic C. difficile had been treated with antibiotics (Trimethoprim/Sulfamethoxazole) before the onset of diarrhea. The isolation rate of toxigenic C. difficile was slightly higher in adult dogs (older than 1 year) than in young dogs (p = .043).
Table 1.
Frequency of enteric pathogens and detection of selected virulence factors and virulence genes in diarrheic (n = 92) and apparently healthy (n = 62) dogs.
| Enteropathogens | Dogs (n = 154) |
p value | ||
|---|---|---|---|---|
| Diarrheic (n = 96) (%) |
Non diarrheic (n = 62) (%) |
|||
| Clostridioides difficile | A+B+CDT- | 11 (11.9)* | 0 (0) | 0.032 |
| A−B- CDT- | 8 (8.7) | 3 (4.8) | 0.526 | |
| A/B toxinsb | 8/11 (72.3) | – | – | |
| Clostridium perfringens | Type A | 46 (50) | 22 (34.5) | 0.099 |
| cpe+ | 10 (10.8)* | 0 (0) | 0.006 | |
| cpe+netF+ | 7 (7.6)* | 0 (0) | 0.042 | |
| CPEa | 5/10 (50) | – | – | |
| Escherichia coli | Enteropathogenic (EPEC) | 11 (11.9) | 11 (17.7) | 0.352 |
| Shiga Toxin-Producing (STEC) | 1 (0.9) | 2 (3.2) | 0.565 | |
| Enterotoxigenic (ETEC) | 3 (3.2) | 1 (1.6) | 0.648 | |
| Atypical | 1 (0.9) | 0 (0) | 1.000 | |
| Salmonella sp. | 0 (0) | 0 (0) | – | |
| Parvovirus (CPV) | 10 (10.8)* | 0 (0) | 0.006 | |
| Canine Coronavirus | 2 (2.1) | 2 (3.2) | 1.000 | |
| Rotavirus | 1 (0.9) | 1 (1.6) | 1.000 | |
| Giardia sp. | 10 (10.8) | 2 (3.2) | 0.124 | |
*Chi-square test or Fisher's exact test were used to evaluate possible association between enteropathogens in diarrheic and healthy dogs or age groups. P values of <0.05 were considered significant (in bold).
Enterotoxin (CPE) detection by commercial EIA on unthawed aliquots of stool samples positive for C. perfringens cpe+isolation.
A/B toxin detection by commercial EIA on unthawed aliquots of stool samples positive for toxigenic C. difficile isolation.
Among toxigenic C. difficile isolates, ribotypes 014/020 and 106 were the most common. Ribotype 014/020 seems to be the most frequent in several canine studies and is commonly implicated in human CDI worldwide [32,22,23]. In Brazil, ribotype 106 is common in humans and has also been described in some animals, including dogs [32]. Other authors have suggested pets as a potential source of community acquired CDI in humans due to high genetic similarity between dog and human disease isolates [2,24].
The isolation of cpe-positive C. perfringens was associated with diarrhea to at least some extent (Table 1), corroborating previous studies [7,8,25]. In addition, C. perfringens strains positive for NetE- and NetF-encoding genes were detected in seven (7.6%) samples from diarrheic dogs (p = .042). The netE and netF genes were identified only in C. perfringens cpe + strains and were exclusively isolated from adult dogs with bloody diarrhea, corroborating the results of [5]. Genomic sequencing of two netE + netF + cpe + strains from different diarrheic dogs revealed that plasmids in both of these Brazilian strains had significant identity (Fig. 1 ) to plasmids previously identified from dogs in Canada [5]. These results are consistent with recent findings that showed that the netEF plasmids, and to a lesser extent the cpe plasmids, are highly conserved in canine and equine isolates of C. perfringens from diarrheic animals [4] and support the hypothesis that C. perfringens netF + strains are enteropathogenic in adult dogs.
Fig. 1.
Comparison of the plasmids from Dog 7 and 8 with plasmids pJFP838C and pJFP838D: A) Blastn analysis using EasyFig to align pJFP838C against the pseudomolecules encoding the netEF genes from the genome sequences obtained from dog 7 and 8 isolates. B) An EasyFig alignment of pJFP838D compared to pseudomolecules encoding the cpe gene from genome data derived from dog 7 and 8. Legend: grey shaded regions indicate nucleotide identity. ORFs are represented as arrows and are coloured as follows: magenta – toxin genes (as indicated), red – conjugation locus, yellow – plasmid replication and maintenance, green - putative collagen adhesion, dark blue – conserved ORF, light blue – poorly or non-conserved ORFs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
All dogs positive for C. perfringens netF + were apparently healthy before the onset of diarrhea. Additional stool samples were collected from four dogs, five to eight months after the diarrheal episode. In two instances, dogs housed together with the previously affected animals were also sampled. The netF + gene was not detected in any of these healthy dogs. These results are consistent with the finding that netF- + C. perfringens strains have only been isolated from dogs during diarrheal episodes [5].
In the present study, toxigenic C. difficile and netF-positive C. perfringens were detected together in three animals (Table 2 ). These are the first confirmed coinfections of these strains in dogs and two of these cases have been described in more details in a case report [26]. Among other enteropathogens, Giardia sp. was the most common agent detected in dogs positive for toxigenic C. difficile or netF-positive C. perfringens (Table 2), with dogs positive for Giardia sp. more likely to be positive for C. perfringens (p = .0057), similar to a previous report [27]. Veterinary practitioners commonly recognize giardiasis as a cause of mild and uncomplicated diarrhea in dogs, but the role of this parasite as a predisposing factor for acute diarrhea associated with other enteropathogens is not known.
Table 2.
Details of all dogs positive for Clostridium perfringens cpe+ or toxigenic Clostridium difficile.
| ID | Age (months) | Feacal characteristic |
Clostridium difficile |
Clostridium perfringens |
Other enteropathogens | Outcome | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A/B toxin | Isolation | Ribotypea | cpe | netE/F | netG | CPE | |||||
| 1 | 48 | Mushy | – | – | – | + | – | – | + | – | Recovered. |
| 2 | 3 | Bloody | – | – | – | + | – | – | + | Coronavirus | Died |
| 3 | 125 | Bloody | – | – | + | – | – | – | – | Recovered. | |
| 4 | 61 | Bloody | – | – | – | + | + | – | – | Giardia sp. | Recovered. |
| 5 | 18 | Bloody | – | – | – | + | + | + | + | – | Recovered. |
| 6 | 133 | Bloody | – | – | – | + | + | – | + | – | Recovered. |
| 7 | 18 | Bloody | – | – | – | + | + | – | – | – | Recovered |
| 8 | 19 | Bloody | + | A+B+CDT- | 014/020 | + | + | – | – | – | Recovered. |
| 9 | 145 | Bloody | + | A+B+CDT- | 014/020 | + | + | + | – | – | Died. |
| 10 | 36 | Bloody | + | A+B+CDT- | 106 | + | + | + | + | EPEC (eae+) | Died. |
| 11 | 19 | Bloody | + | A + B + CDT- | 014/020 | – | – | – | – | – | Recovered |
| 12 | 181 | Bloody | + | A + B + CDT- | BR1 | – | – | – | – | CPV-2b and Giardia sp. | Recovered |
| 13 | 6 | Bloody | + | A + B + CDT- | BR1 | – | – | – | – | – | Died |
| 14 | 168 | Mushy | + | A + B + CDT- | 014/020 | – | – | – | – | – | Recovered |
| 15 | 48 | Bloody | + | A + B + CDT- | 106 | – | – | – | – | – | Died |
| 16 | 121 | Bloody | – | A + B + CDT- | BR2 | – | – | – | – | – | Died |
| 17 | 5 | Bloody | – | A + B + CDT- | BR3 | – | – | – | – | – | Died |
| 18 | 23 | Mushy | – | A + B + CDT- | BR4 | – | – | - | - | Giardia sp. | Recovered. |
| 19 | 120 | Mushy | – | A + B + CDT- | 602 | – | – | – | – | – | Recovered. |
Legend: EPEC – Enteropathogenic E. coli; CPV-2b – Canine parvovirus type 2b; CPE – C. perfringens enterotoxin; A – Toxin A encoding gene (tcdA); B – Toxin B encoding gene (tcdB); CDT – binary toxin gene (cdtB).
PCR ribotypes for which the reference strains were available are designated by international Cardiff nomenclature, while others are designated by internal nomenclature (BRA and number).
Canine coronavirus, CPV and enteropathogenic E. coli were also detected in association with toxigenic C. difficile or netF-positive C. perfringens (Table 2). Despite previous reports that suggested that CPV could be an important predisposing factor for CPAD in dogs [28,29], enterotoxigenic C. perfringens was not recovered from any CPV-positive animals. However, the only adult dog positive for CPV in the present study (Dog 12) was also positive for CDI and Giardia sp (Table 2). Sequencing of the VP2-encoding gene from this sample revealed the involvement of CPV-2b [30]. These data suggest that, in addition to CPV-2c, CPV-2b can also cause infection in repeatedly vaccinated dogs and highlight the need for broad studies focusing on a possible synergism of C. difficile infection and canine CPV in adult dogs [31].
In summary, the present study reinforces the association of netF-positive C. perfringens and toxigenic C. dificile as enteropathogens in dogs, with these isolates commonly associated with bloody diarrhea in adult individuals. A possible coinfection of these two agents and also with other enteropathogens is feasible based on these data, but further studies are required to assess the significance of this association.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by funds from CAPES, CNPq, FAPEMIG and PRPQ/UFMG.
Handling Editor: TG Nagaraja
References
- 1.Marks S.L., Rankin S.C., Byrne B.A., Weese J.S. Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J. Vet. Intern. Med. 2011;25:1195–1208. doi: 10.1111/j.1939-1676.2011.00821.x. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez C., Van Broeck J., Taminiau B., Delmée M., Daube G. Clostridium difficile infection: early history, diagnosis and molecular strain typing methods. Microb. Pathog. 2016;97:59–78. doi: 10.1016/j.micpath.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Silva R.O., Lobato F.C. Clostridium perfringens: a review of enteric diseases in dogs, cats and wild animals. Anaerobe. 2015;33:14–17. doi: 10.1016/j.anaerobe.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Gohari M.I., Kropinski A.M., Weese S.J., Whitehead A.E., Parreira V.R., Boerlin P., Prescott J.F. NetF-producing Clostridium perfringens: clonality and plasmid pathogenicity loci analysis. Infect. Genet. Evol. 2017 Apr;49:32–38. doi: 10.1016/j.meegid.2016.12.028. [DOI] [PubMed] [Google Scholar]
- 5.Gohari I.M., Parreira V.R., Nowell V.J., Nicholson V.M., Oliphant K., Prescott J.F. A novel pore-forming toxin in type a Clostridium perfringens is associated with both fatal canine hemorrhagic gastroenteritis and fatal foal necrotizing enterocolitis. PLos One. 2015;10 doi: 10.1371/journal.pone.0122684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weese J.S. Bacterial enteritis in dogs and cats: diagnosis, therapy, and zoonotic potential. Vet Clin North Am Small Anim Pract. 2011;41:287–309. doi: 10.1016/j.cvsm.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Silva R.O., Ribeiro M.G., Palhares M.S., Borges A.S., Maranhão R.P., Silva M.X., Lucas T.M., Olivo G., Lobato F.C. Detection of A/B toxin and isolation of Clostridium difficile and Clostridium perfringens from foals. Equine Vet. J. 2013;45:671–675. doi: 10.1111/evj.12046. [DOI] [PubMed] [Google Scholar]
- 8.Silva R.O., Santos R.L., Pires P.S., Pereira L.C., Pereira S.T., Duarte M.C., de Assis R.A., Lobato F.C. Detection of toxins A/B and isolation of Clostridium difficile and Clostridium perfringens from dogs in Minas Gerais, Brazil. Braz. J. Microbiol. 2014;44:133–137. doi: 10.1590/S1517-83822013005000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva R.O.S., Salvarani F.M., Cruz Júnior E.C.C., Pires P.S., Santos R.L.R., Assis R.A., Guedes R.M.C., Lobato F.C.F. Detection of enterotoxin A and cytotoxin B, and isolation of Clostridium difficile in piglets in Minas Gerais, Brazil. Ciência Rural. 2011;41:1130–1135. [Google Scholar]
- 10.Janezic S., Rupnik M. Molecular typing methods for Clostridium difficile: pulsed-field gel electrophoresis and PCR ribotyping. In: Mullany P., Roberts A., editors. Vol. 646. Humana Press; 2010. pp. 55–66. (Clostridium difficile, Methods and Protocols; Springer Protocols – Methods in Molecular Biology (Walker JM, Series Ed.)). [DOI] [PubMed] [Google Scholar]
- 11.Vieira A.A.S., Guedes R.M.C., Salvarani F.M., Silva R.O.S., Assis R.A., Lobato F.C.F. Genotipagem de Clostridium perfringens isolados de leitões diarréicos. Arq Instit Biol. 2008;75:513–516. [Google Scholar]
- 12.Keyburn A.L., Boyce J.D., Vaz P., Bannam T.L., Ford M.E., Parker D., Di Rubbo A., Rood J.I., Moore R.J. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor J.R., Lyras D., Farrow K.A., Adams V., Powell D.R., Hinds J., Cheung J.K., Rood J.I. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 2006;61:1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- 14.Coil D., Jospin G., Darling A.E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 15.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2004;30:2068–2069. [Google Scholar]
- 16.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;10:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwang J., Littledike E.T., Keen J.E. Use of the polymerase chain reaction for Salmonella detection. Lett. Appl. Microbiol. 1996;22:46–51. doi: 10.1111/j.1472-765x.1996.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 19.Puño-Sarmiento J., Medeiros L., Chiconi C., Martins F., Pelayo J., Rocha S., Blanco J., Blanco M., Zanutto M., Kobayashi R., Nakazato G. Detection of diarrheagenic Escherichia coli strains isolated from dogs and cats in Brazil. Vet. Microbiol. 2013;166:676–680. doi: 10.1016/j.vetmic.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Duijvestijn M., Mughini-Gras L., Schuurman N., Schijf W., Wagenaar J.A., Egberink H. Enteropathogen infections in canine puppies: (Co-)occurrence, clinical relevance and risk factors. Vet. Microbiol. 2016;195:115–122. doi: 10.1016/j.vetmic.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hensgens M.P., Keessen E.C., Squire M.M., Riley T.V., Koene M.G., de Boer E., Lipman L.J., Kuijper E.J. European society of clinical microbiology and infectious diseases study group for Clostridium difficile (ESGCD). Clostridium difficile infection in the community: a zoonotic disease? Clin. Microbiol. Infect. 2012;18:635–645. doi: 10.1111/j.1469-0691.2012.03853.x. [DOI] [PubMed] [Google Scholar]
- 22.Janezic S., Ocepek M., Zidaric V., Rupnik M. Clostridium difficile genotypes other than ribotype 078 that are prevalent among human, animal and environmental isolates. BMC Microbiol. 2012;27(12):48. doi: 10.1186/1471-2180-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janezic S., Zidaric V., Pardon B., Indra A., Kokotovic B., Blanco J.L., Seyboldt C., Diaz C.R., Poxton I.R., Perreten V., Drigo I., Jiraskova A., Ocepek M., Weese J.S., Songer J.G., Wilcox M.H., Rupnik M. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol. 2014;14:173. doi: 10.1186/1471-2180-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone N.E., Sidak-Loftis L.C., Sahl J.W., Vazquez A.J., Wiggins K.B., Gillece J.D., Hicks N.D., Schupp J.M., Busch J.D., Keim P., Wagner D.M. More than 50% of Clostridium difficile isolates from pet dogs in flagstaff, USA, carry toxigenic genotypes. PLos One. 2016;11 doi: 10.1371/journal.pone.0164504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamoto Y., Dhanani N., Markel M.E., Steiner J.M., Suchodolski J.S. Prevalence of Clostridium perfringens, Clostridium perfringens enterotoxin and dysbiosis in fecal samples of dogs with diarrhea. Vet. Microbiol. 2014;174:463–473. doi: 10.1016/j.vetmic.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Diniz A.N., Silva R.O., Oliveira Junior C.A., Pierezan F., Lobato F.C. Clostridium perfringens type A netF and netE positive and Clostridium difficile co-infection in two adult dogs. Anaerobe. 2016;38:94–96. doi: 10.1016/j.anaerobe.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupont S., Butaye P., Claerebout E., Theuns S., Duchateau L., Van de Maele I., Daminet S. Enteropathogens in pups from pet shops and breeding facilities. J. Small Anim. Pract. 2013;54:475–480. doi: 10.1111/jsap.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva R.O.S., Dorella F.A., Figueiredo H.C.P., Costa É.A., Pelicia V., Ribeiro B.L.D., Ribeiro M.G., Paes A.C., Megid J., Lobato F.C.F. Clostridium perfringens and C. difficile in parvovirus-positive dogs. Anaerobe. 2017;48:66–69. doi: 10.1016/j.anaerobe.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turk J., Fales W., Miller M., Pace L., Fischer J., Johnson G., Kreeger J., Turnquist S., Pittman L., Rottinghaus A. Enteric Clostridium perfringens infection associated with parvoviral enteritis in dogs: 74 cases (1987-1990) J. Am. Vet. Med. Assoc. 1992;200:991–994. [PubMed] [Google Scholar]
- 30.Buonavoglia C., Martella V., Pratelli A., Tempesta M., Cavalli A., Buonavoglia D., Bozzo G., Elia G., Decaro N., Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J. Gen. Virol. 2001;82(2001):3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 31.Decaro N., Buonavoglia C. Canine parvovirus–a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet. Microbiol. 2012 Feb 24;155(1):1–12. doi: 10.1016/j.vetmic.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva R.O., Rupnik M., Diniz A.N., Vilela E.G., Lobato F.C. Clostridium difficile ribotypes in humans and animals in Brazil. Mem Inst Oswaldo Cruz. 2015;110:1062–1065. doi: 10.1590/0074-02760150294. [DOI] [PMC free article] [PubMed] [Google Scholar]