Abstract
Severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) is a newly identified member of Family Coronaviridae. Coronavirus envelope spike protein S is a class I viral fusion protein which is characterized by the existence of two heptad repeat regions (HR1 and HR2) (forming a complex called fusion core). Here we report that by using in vitro bio-engineering techniques, SARS-CoV HR1 and HR2 bind to each other and form a typical 6-helix bundle. The HR2, either as a synthetic peptide or as a GST-fusion polypeptide, is a potent inhibitor of virus entry. The results do show that SARS-CoV follows the general fusion mechanism of class I viruses and this lays the ground for identification of virus fusion/entry inhibitors for this devastating emerging virus.
Keywords: SARS-CoV, Coronavirus, Spike(S) protein, Fusion core, Heptad repeat (HR1 and HR2), Inhibition
Severe acute respiratory syndrome coronavirus (SARS-CoV) has been identified as a new distinct pathological entity [1], [2], [3] and the disease infected more than 8000 people and killed 774 worldwide, mostly in Asia, before it was brought under control in July between the winter and spring in 2002–2003 (WHO website: www.who.int). It has been shown that the genomic sequence is unrelated to any other known members of Family Coronaviridae isolated from either humans or animals [4], [5], [6], thereof designated as a new group, Group IV, along with the previous three known groups [4], [5], [6], [7], [8], [9].
Members in Family Coronaviridae are a group of enveloped positive-stranded RNA viruses with largest genome among the RNA viruses and have 3–4 envelope proteins embedded on the surface of the viral envelope lipid membrane [7], [8]. The genomic sequencing reveals that, as with other enveloped RNA viruses, including the coronaviruses [10], [11], [12], SARS-CoV envelope spike (S) protein contains highly conserved heptad repeat regions (HR1 and HR2), which have been shown as important in virus membrane fusion and successfully used as targets for virus entry/fusion inhibitors in a number of viruses [13], [14], [15], [16], [17], [18], including a coronavirus, mouse hepatitis virus (MHV) [12]. The existence of HR regions is also a characteristic of class I viral fusion protein [19]. Currently two classes (classes I and II) of virus fusion proteins have been classified [19].
It is generally believed that the envelope protein undergoes a series of conformational changes during the virus fusion process [13], [20], [21], [22]. The HR1 and HR2 regions are believed to be important modules/domains in this process and show different conformations in different fusion states [13]. Under the current model, there are at least three conformational states of the envelope fusion protein [13]. They are pre-fusion native state, pre-hairpin intermediate state, and post-fusion hairpin state. During these state transitions, the HR1 and HR2 are exposed in an intermediate conformational state but bind to each other to form coiled coil structure in an anti-parallel mode in the post-fusion state. Therefore, the in vitro introduced HR peptides compete with the endogenous HR counterparts in the intermediate state, preventing the transition into the formation of HR1/HR2 coiled coil bundle, the post-fusion state [13]. This coiled coil bundle conformation is believed to be important for bringing two lipid membranes (cellular and viral) into proximity with each other allowing the membrane fusion for virus entry. Membrane fusion is the key step for enveloped virus infection. The HR1/HR2 coiled coil bundle is called the virus fusion core [13]. In this structure, as shown by several crystal structures of fusion cores, including HIV, hRSV, influenza virus A, and Ebola virus [23], [24], [25], [26], [27], [28], [29], three HR1 bind each other to form a trimeric core whereas three HR2 surround this core. As both HR1 and HR2 are structurally α helical in this fusion core, this structure is also called 6-helix coiled coil bundle [13].
To investigate the structural basis of SARS-CoV fusion and entry and identify new fusion inhibitors, we have carried out the analysis of the SARS-CoV HR1 and HR2 fusion core in this study. Both HR1 and HR2 have been expressed in Escherichia coli and the in vitro assembly of the fusion core was analyzed. The GST fusion or chemical synthetic HR2 was tested in virus fusion inhibition. The results do show that SARS-CoV S protein is a typical class I viral fusion protein and HR2 is a potent fusion inhibitor. This is consistent with the recent observation on another coronavirus, MHV [12].
Materials and methods
Virus and cell line. SARS-CoV virus isolate WHU was used in this experiment, which was isolated from a SARS patient who died of SARS and the virus genome was confirmed by sequencing (GenBank Accession No. AY394850). The virus causes typical cytopathic effect (CPE) on Vero E6 cell monolayer. Vero E6 cell line was obtained from the American Type Culture Collection (ATCC) (Rockville, MD) and maintained in DMEM (Sigma) containing 2% fetal bovine serum (FBS) (Sigma).
Gene construction. As shown in Fig. 1 , SARS-CoV S protein is a typical type I membrane protein. The HR1 and HR2 regions were predicted using the computer program LearnCoil-VMF (http://nightingale.lcs.mit.edu/cgi-bin/vmf) [30]. The HR1 region covers amino acids 898–1005, whereas the HR2 includes the amino acids 1149–1186 (Fig. 1). The polypeptides were then engineered in vitro and expressed in E. coli. The HR1 was expressed as an N-terminal his-tagged protein in pET30a vector (Novagen) and HR2 (two versions) was expressed as a glutathione-S-transferase (GST) fusion protein. A peptide corresponding to amino acids 1149–1186 (HR2-38) was also synthesized (Sigma-Genosys, UK).
Fig. 1.
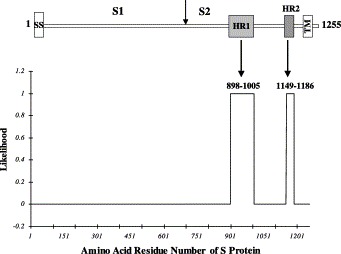
Prediction of the HR regions of SARS-CoV S protein. Schematic diagram of S protein (amino acids 1–1255 for the full-length) is shown in the upper panel. As the basic amino acid cluster required for S1/S2 cleavage is not present it is unlikely that SARS-CoV S protein would be processed further into S1 and S2. However, the positions of the putative S1 and S2 are indicated for the convenience to compare with other coronaviruses. “SS” represents signal sequence and “TM” for transmembrane domain. In the lower panel, likelihood of HR1 and HR2 predicted by LearnCoil-VMF program [30] is shown.
For construct making, His-HR1 construct was cloned into pET30a vector with BamHI and XhoI, thereof resulting in 50 extra amino acids at the N-terminus of the HR1 (same strategy as Wang et al. [31]). GST fusion HR2 constructs (HR2-38 and HR2-44) were cloned into pGEX-6p-1 (Pharmacia) with unique restriction enzyme sites BamHI and NotI (same strategy as Yu et al. [32]). There were two versions of GST-HR2, i.e., GST-HR2-38 and GST-HR2-44. For the latter, there were 6 extra amino acids after the predicted position at the C-terminus from the S gene (amino acids 1149–1192). Cloned constructs were verified by direct DNA sequencing.
Protein expression and purification. All the relevant positive expression vectors (30a-HR1, GST-HR2-38, and GST-HR2-44) were transformed into E. coli strain BL21 (DE3) competent cells and single colony was inoculated into 2 × YT medium containing 100 μg/ml ampicillin at 37 °C for overnight culture. Then the overnight culture was transferred to new 2 × YT medium for large-scale protein production by growing at 37 °C. When the culture density (OD600) reached 0.8, the culture was induced with 0.2 mM IPTG and grown for an additional 5 h at 25 °C before the cells were harvested.
The harvested culture was centrifuged and the bacterial cell pellet was re-suspended in PBS and homogenized by sonication. Triton X-100 was then added to a final concentration of 1%, and the lysate was incubated for 30 min on ice and was subsequently clarified by centrifugation at 12,000g for 15 min at 4 °C. Then the supernatant was loaded onto a glutathione–Sepharose 4B column (Pharmacia) or Ni-chelated Sepharose affinity column (Pharmacia). The protein-loaded column was then washed with 3× column volume of PBS. After that, for GST-fusion protein, the protein was eluted with 10 mM reduced glutathione. For the GST-removed proteins, the GST-3C rhinovirus protease (kindly provided by Drs. J. Heath and K. Hudson) was added into the resin to cleave the GST and the fused target protein and the mixture was incubated with gentle agitation for about 10 h at 4 °C. The target protein was eluted with 10 ml PBS. For the His-tagged HR1, the protein was eluted with 100 mM imidazole.
HR1–HR2 complex assembly and gel-filtration analysis. Equal molar purified His (30a)-HR1 and GST-removed HR2 were mixed at room temperature for 1 h. The mixture was then loaded onto a Superdex G200 column (Pharmacia). One single peak (60–70 kDa) was detected and the eluted fractions were analyzed on Tris–Tricine SDS–PAGE.
GST pull-down experiment. Purified GST-HR2-38 was mixed with equal molar 30a-HR1 on ice for 1 h and then loaded onto a glutathione–Sepharose 4B column. The column was washed with 10 column volumes of PBS and then eluted with 10 mM reduced glutathione. The existence of both GST-HR2-38 and 30a-HR1 in the elution was tested on SDS–PAGE.
Circular dichroism spectroscopy. Circular dichroism (CD) spectra were measured on a Jasco J-715 spectrophotometer in PBS. Wavelength spectra were recorded at 25 °C using a 0.1 cm pathlength cuvette. For the thermodynamic stability, the HR1–HR2 complex protein was measured at 222 nm by monitoring the CD signal in the temperature ranging from 25 to 95 °C with a scan rate of 1 °C per minute.
Fusion inhibition assay. For the inhibition experiments, Vero E6 cell cultures growing in 96-well microplates were infected with 100× TCID50/well (TCID50 was virus titers causing 50% of CPE on Vero E6 cell monolayer and the TCID50 used in this experiment was 1.263 × 106/ml). The serial 10-fold dilutions of polypeptides (8 repeats for each dilution) were added at the same time as virus adsorption for 1 h at 37 °C. Then the mixtures of virus and the polypeptides were replaced by DMEM containing 2% FBS and continued to cultivate for 96 h until calculating the well numbers of CPE and inhibition of CPE. The IC50 was calculated according to Reed–Muench method [33].
Results and discussion
HR1 binds to HR2 to form a complex
All three polypeptides, 30a-HR1, GST-HR2-38, and GST-HR2-44, were expressed in E. coli as soluble proteins in this study. As our previous work has shown that the extra amino acids in the 30a expressed protein in hRSV had no effect on the binding to HR2 [31] we carried out all the binding experiments with this 30a-HR1 preparation. GST-removed HR2-38 and HR2-44 were both soluble proteins in PBS. When equal molars of 30a-HR1 and HR2-38 were mixed a unique complex peak could be detected by gel filtration and the peak molecular weight was estimated approximately as 60 kDa (Fig. 2A ). Both 30a-HR1and HR2 were detected in the peak on SDS–PAGE (Fig. 2A). This matches the calculation of 3× 30a-HR1 and 3× HR2-38, implying the formation of 6-helix bundle complex. We also tested the GST pull-down experiments and showed the tight binding of 30a-HR1 to GST-HR2-38 (data not shown).
Fig. 2.
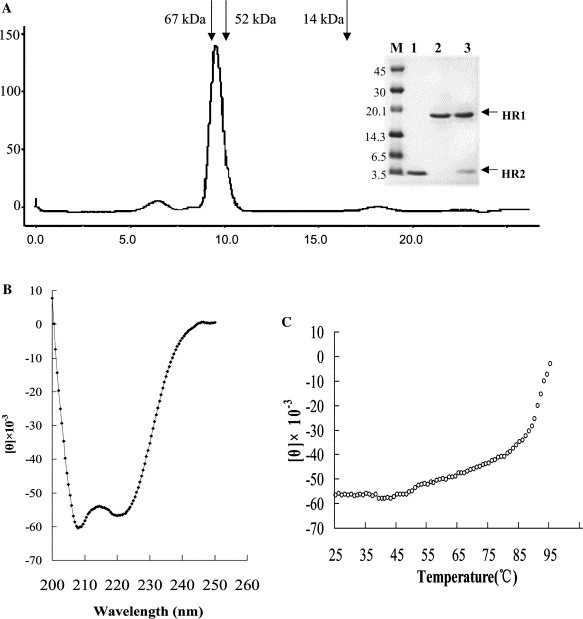
Characterization of the HR1–HR2 6-helix bundle. (A) The 30a-HR1 bound to GST-removed HR2. On gel-filtration, the equal-molar mixture of HR1 and HR2 forms 6-helix bundle complex. The molecular weight of the complex on gel filtration is approximately 67 kDa and the peak shows the existence of both HR1 and HR2 in similar stoichiometry (inset picture). Inset picture is a Tris–Tricine SDS–PAGE gel. Lane M, protein standard marker in kDa; lane 1, purified GST-removed HR2; lane 2, purified his-HR1; and lane 3, peak from gel-filtration, clearly showing the co-existence of the HR1 and HR2 bands from a single peak. (B) Assembled HR1/HR2 forms typical α-helix coiled coil as shown by circular dichroism (CD) spectroscopy (double minima at 208 and 222 nm). (C) Thermodynamic stability of the complex. The melting temperature is over 85 °C, typical stable coiled coil bundle.
HR1 and HR2 complex is a stable α-helix coiled coil
CD spectroscopic profile of the HR1–HR2 complex shows a typical α-helix structure, with double minima at 208 and 222 nm (Fig. 2B). From previous work on other viral fusion core, this structure is coiled coil which is characterized by its extreme stability. Thermodynamic measurements of this HR1–HR2 complex showed it remains structured up to 85 °C (Fig. 2C), indicating its extraordinary stability.
HR2 inhibits SARS-CoV fusion
In the presence of HR2 polypeptides (GST-fusion forms or synthetic peptide), we observed an analogous, strong inhibitory effect of SARS-CoV HR2 which blocked virus entry (Fig. 3 ). The 50% effective doses (IC50) for inhibition of the CPE in cultured cells were 0.5–5 nM for the synthetic HR2 peptide and 66.2–500 nM for the GST fusion HR2 forms (Fig. 3). Despite our extensive experiments, using both the synthetic and bio-engineered HR1s, we did not see any virus-infection inhibition effect from HR1 (data not shown). This might reflect the difference between HR1 and HR2 in the virus entry inhibition as observed in some paramyxoviruses [31], [32], [34], [35], [36]. The mechanism underlying this difference needs to be further investigated in the future. It is noteworthy that, in a recent study on MHV, only HR2 inhibition has been observed [12].
Fig. 3.
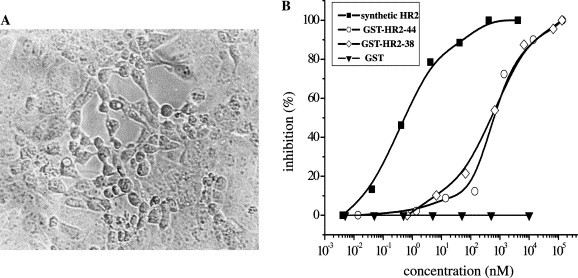
Peptide fusion–inhibition curve. (A) Typical CPE of SARS-CoV (Strain WHU) formed on Vero E6 cells. (B) CPE-inhibition curves of HR2 peptides. The IC50 of the different polypeptides were, respectively: synthetic HR2-38 (amino acids 1149–1186), 0.5–5 nM; GST-HR2-38 (amino acids 1149–1186), 66.2 nM; GST-HR2-44 (amino acids 1149–1192, 6-extra amino acids from the program prediction at the C-terminus), 500 nM; and GST protein itself was used as control and no inhibitions were observed.
Implications for coronavirus fusion and fusion inhibitor design
Coronavirus entry into the cells starts with viral and cellular membrane fusion which is mediated by the binding of the viral S protein with the cellular receptor/s [7]. Receptor-induced conformational changes in the S protein are crucial in this step [20], [21], [22]. In this study, we have shown that the putative HR1 and HR2 of SARS-CoV bind to each other and form a 6-α-helix bundle (trimer of HR1/HR2 dimer), a characteristics of class I viral fusion proteins. The importance of the HR regions in MHV fusion has been demonstrated with site-directed mutagenesis study by Luo and Weiss [10], [11]. As the receptor for SARS-CoV has been identified recently [37], receptor-induced conformational changes of S protein should be pursued in the near future, esp. the conformations of HR1 and HR2 fusion core in different stages during the fusion process.
Recently, Rottier and co-workers [12] reported the formation of HR1/HR2 6-helix bundle complex of MHV S protein. This and our present result on SARS virus are the first evidence for a 6-helix bundle formation of HR1 and HR2 and the HR2 fusion inhibitor derived from Family Coronaviridae, implying that coronavirus adopts a similar fusion or entry mechanism to other RNA viruses [13]. Taking the fact into account that SARS is an acute disease, this kind of peptide inhibitors might act better than that in chronic infection HIV because the therapeutic peptide can be used through intranasal route. The conformational changes of the S protein are very likely to be crucial for coronavirus entry [20], [21], [22] and so this provides a most plausible target for drug design. The strong inhibition of the peptide in the GST (25 kDa) fusion form indicates that there is significant space for HR2 to interact with HR1 in the intermediate state of the S protein during the process of virus fusion. This has also been observed in our previous work on Newcastle disease virus and hRSV, members of Family Paramyxoviridae [31], [32]. Our results do provide a drug lead for SARS-CoV and its potential clinical application should be rigorously pursued in the near future. A clear crystal structure of this fusion core of SARS-CoV would also make a great deal of contribution to our understanding of coronavirus fusion and fusion inhibitor discovery.
Acknowledgements
This work was supported by National Frontier Research Program (Project 973) of the Ministry of Science and Technology of China (Grant No. 2003CB514116), The Key Project of the Knowledge Innovation Program of Chinese Academy of Sciences (CAS), and The Special SARS Research Fund of Wuhan University. We thank Prof. Rodney Phillips, Dr. Bent Jakobsen, and Dr. Catherine Zhang for comments.
References
- 1.Ksiazek T.G, Erdman D, Goldsmith C.S, Zaki S.R, Peret T, Emery S, Tong S, Urbani C, Comer J.A, Lim W, Rollin P.E, Dowell S.F, Ling A.E, Humphrey C.D, Shieh W.J, Guarner J, Paddock C.D, Rota P, Fields B, DeRisi J, Yang J.Y, Cox N, Hughes J.M, LeDuc J.W, Bellini W.J, Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Peiris J.S, Lai S.T, Poon L.L, Guan Y, Yam L.Y, Lim W, Nicholls J, Yee W.K, Yan W.W, Cheung M.T, Cheng V.C, Chan K.H, Tsang D.N, Yung R.W, Ng T.K, Yuen K.Y. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt H.R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R.A, Berger A, Burguiere A.M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J.C, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk H.D, Osterhaus A.D, Schmitz H, Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Ruan Y.J, Wei C.L, Ee A.L, Vega V.B, Thoreau H, Su S.T, Chia J.M, Ng P, Chiu K.P, Lim L, Zhang T, Peng C.K, Lin E.O, Lee N.M, Yee S.L, Ng L.F, Chee R.E, Stanton L.W, Long P.M, Liu E.T. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M.M.C. Lai, K.V. Holmes, in: D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, Straus, (Eds.), Coronaviridae: The Viruses and Their Replication, fourth ed., Fields Virology, Lippincott Williams & Wilkins Publishers, Philadelphia, PA, 2001, pp. 1163-1185
- 8.L. Enjuanes, D. Brian, D. Cavanagh, K.V. Holmes, M.M.C. Lai, H. Laude, P. Masters, P.J.M. Rottier, S.G. Siddell, W.J.M. Spaan, F. Taguchi, P. Talbot, in: M.H.V. van Regenmortel, C.M. Fauquet, D.H.L. Bishop, E.B. Carstens, M.K. Estes, S.M. Lemon, J. Maniloff, M.A. Mayo, D.J. McGeoch, C.R. Pringle, R.B. Wickner, (Eds),Virus Taxonomy: Classification and Nomenclature of Viruses, 7th Report, seventh ed., Academic Press, San Diego, 2000, pp. 835–849
- 9.Holmes K.V. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 10.Luo Z, Weiss S.R. Virology. 1998;244:483–494. doi: 10.1006/viro.1998.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z.L, Weiss S.R. Adv. Exp. Med. Biol. 1998;440:17–23. doi: 10.1007/978-1-4615-5331-1_3. [DOI] [PubMed] [Google Scholar]
- 12.Bosch B.J, van der Zee R, de Haan C.A, Rottier P.J. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert D.M, Kim P.S. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 14.Lamb R.A, Joshi S.B, Dutch R.E. Mol. Membr. Biol. 1999;16:11–19. doi: 10.1080/096876899294715. [DOI] [PubMed] [Google Scholar]
- 15.Bentz J. Biophys. J. 2000;78:886–900. doi: 10.1016/S0006-3495(00)76646-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skehel J.J, Wiley D.C. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 17.Skehel J.J, Wiley D.C. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 18.Weissenhorn W, Dessen A, Calder L.J, Harrison S.C, Skehel J.J, Wiley D.C. Mol. Membr. Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 19.Lescar J, Roussel A, Wien M.W, Navaza J, Fuller S.D, Wengler G, Rey F.A. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 20.Sturman L.S, Ricard C.S, Holmes K.V. J. Virol. 1990;64:3042–3050. doi: 10.1128/jvi.64.6.3042-3050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weismiller D.G, Sturman L.S, Buchmeier M.J, Fleming J.O, Holmes K.V. J. Virol. 1990;64:3051–3055. doi: 10.1128/jvi.64.6.3051-3055.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelus B.D, Schickli J.H, Blau D.M, Weiss S.R, Holmes K.V. J. Virol. 2003;77:830–840. doi: 10.1128/JVI.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bullough P.A, Hughson F.M, Skehel J.J, Wiley D.C. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 24.Chan D.C, Fass D, Berger J.M, Kim P.S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 25.Tan K, Liu J, Wang J, Shen S, Lu M. Proc. Natl. Acad. Sci. USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X, Singh M, Malashkevich V.N, Kim P.S. Proc. Natl. Acad. Sci. USA. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissenhorn W, Carfi A, Lee K.H, Skehel J.J, Wiley D.C. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 28.Malashkevich V.N, Schneider B.J, McNally M.L, Milhollen M.A, Pang J.X, Kim P.S. Proc. Natl. Acad. Sci. USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weissenhorn W, Dessen A, Harrison S.C, Skehel J.J, Wiley D.C. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 30.Singh M, Berger B, Kim P.S. J. Mol. Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang E, Sun X, Qian Y, Zhao L, Tien P, Gao G.F. Biochem. Biophys. Res. Commun. 2003;302:469–475. doi: 10.1016/s0006-291x(03)00197-9. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, Wang E, Liu Y, Cao D, Jin N, Zhang C.W, Bartlam M, Rao Z, Tien P, Gao G.F. J. Gen. Virol. 2002;83:623–629. doi: 10.1099/0022-1317-83-3-623. [DOI] [PubMed] [Google Scholar]
- 33.Welkos S, O’Brien A. Methods Enzymol. 1994;235:29–39. doi: 10.1016/0076-6879(94)35128-7. [DOI] [PubMed] [Google Scholar]
- 34.Young J.K, Hicks R.P, Wright G.E, Morrison T.G. Virology. 1997;238:291–304. doi: 10.1006/viro.1997.8834. [DOI] [PubMed] [Google Scholar]
- 35.Rapaport D, Ovadia M, Shai Y. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wild T.F, Buckland R. J. Gen. Virol. 1997;78:107–111. doi: 10.1099/0022-1317-78-1-107. [DOI] [PubMed] [Google Scholar]
- 37.Li W, Moore M.J, Vasilieva N, Sui J, Wong S.K, Berne M.A, Somasundaran M, Sullivan J.L, Luzuriaga K, Greenough T.C, Choe H, Farzan M. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]


