Summary
Protein-protein interactions dictate the assembly of the macromolecular complexes essential for functional networks and cellular behavior. Elucidating principles of molecular recognition governing important interfaces such as coiled coils is a challenging goal for structural and systems biology. We report here that two valine-containing mutants of the GCN4 leucine zipper that fold individually as four-stranded coiled coils associate preferentially in mixtures to form an antiparallel, heterotetrameric structure. X-ray crystallographic analysis reveals that the coinciding hydrophobic interfaces of the hetero- and homotetramers differ in detail, explaining their partnering and structural specificity. Equilibrium disulfide exchange and thermal denaturation experiments show that the 50-fold preference for heterospecificity results from a combination of preferential packing and hydrophobicity. The extent of preference is sensitive to the side chains comprising the interface. Thus, heterotypic versus homotypic interaction specificity in coiled coils reflects a delicate balance in complementarity of shape and chemistry of the participating side chains.
Keywords: CHEMBIO, PROTEINS
Introduction
Specific protein-protein interactions underlie fundamental aspects of cellular function and intercellular communication. These interactions mediate the assembly of supramolecular machinery in the essential biological processes of transcription, translation, development, and chemical transformations of small and large molecules. Microarray technology and high-throughput screens have enabled proteomic studies of protein associations on a cellular scale (Uetz et al., 2000). However, despite intensive research efforts, the mechanism by which the interaction properties of proteins are encoded in their sequences has remained elusive. Accurate prediction of protein interaction specificity will require an understanding of fundamental principles governing molecular recognition (Jones and Thornton, 1996) as well as the nature of the interacting surfaces (Clackson and Wells, 1995, Kortemme et al., 2004). However, specificity is determined by the relative thermodynamic stabilities of a target conformation and alternative states. The balance between binding affinity and specificity has become a major thread in tackling the challenging problems of protein interaction prediction and design. For example, both experimental and computational studies have suggested that only a small number of contact residues dominate the free energy contribution of protein-protein interactions (Clackson and Wells, 1995, Ofran and Rost, 2007). Physical models of partnering specificity have shed light on the interaction between pairs of proteins—such as an enzyme and a protein inhibitor, or an antibody and its cognate antigen—to form heterodimeric complexes (Davies and Cohen, 1996, Huber et al., 1974). Much less is known about the assembly of subunits in higher oligomeric proteins because their folding and association are usually tightly coupled.
The α-helical coiled coil offers an attractive simplification of complex oligomeric protein interactions. Restriction of the conformation of the interacting partners to α helices and the symmetry intrinsic to coiled-coil assembly allow for dissection of these interfaces at a high level of detail (Burkhard et al., 2001, Mason and Arndt, 2004, Woolfson, 2005). Coiled coils consist of two or more α helices wrapped around one another with a left-handed superhelical twist. They commonly share a 7 amino acid sequence repeat, labeled a–g (Hodges et al., 1972). Residues at positions a and d comprise the 4-3 hydrophobic repeat characteristic of coiled coils, whereas residues at positions e and g are predominantly charged amino acids that can contribute to intra- or interhelical electrostatic interactions (Mason and Arndt, 2004). At the interface between the helices, the a and d residues make side-by-side contacts in a “knobs-into-holes” pattern to form a hydrophobic core, as first described by Crick (1953). Despite this simple structure and periodicity, coiled coils associate specifically in diverse protein-protein interfaces that regulate transcription, oncogenesis, and membrane fusion (Lupas and Gruber, 2005). How this high degree of partnering selectivity is encoded within the heptad sequence repeat remains an important, unanswered question about the specificity of protein-protein interactions. Specific van der Waals interactions of the core a and d residues, in fact, dictate both helix-orientation preference and oligomerization state (Harbury et al., 1993, Lovejoy et al., 1993). Buried polar side chains at positions a and d can impart a negative design element to disfavor undesired interactions, ensuring specificity toward the target state (Hill et al., 2000, Oakley and Hollenbeck, 2001). Considerable progress has been made in detailed predictions of the interfacial interactions that direct the formation of either homodimers or heterodimers through computational design and experimental characterization (Acharya et al., 2002, Fong et al., 2004, Grigoryan and Keating, 2006, Havranek and Harbury, 2003, Mason et al., 2006, Pokala and Handel, 2005, Summa et al., 2002). Still, predicting the specificity of higher-order coiled-coil protein interactions remains an unsolved challenge, in part due to the paucity of experimental and sequence data on appropriate model systems.
The high incidence of charged residues at the e and g positions in naturally occurring coiled coils was taken to imply a fundamental role for electrostatic interactions in determining the specificity of coiled-coil recognition (Adamson et al., 1993, Mason and Arndt, 2004). For example, the heterodimerization of the Fos-Jun oncoprotein results from the relief of unfavorable interhelical electrostatic interactions between residues of like charge in the homodimers (O'Shea et al., 1989, O'Shea et al., 1992). A de novo heterodimeric coiled coil was successfully designed by minimizing electrostatic repulsion at these sites (O'Shea et al., 1993). Moreover, the presence of apolar amino acids at either the e or g position of the heptad repeat can direct the formation of stable, four-stranded coiled coils (Deng et al., 2006a, Deng et al., 2006b, Fairman et al., 1995, Liu et al., 2006, Solan et al., 2002, Yadav et al., 2006). Structural and energetic analysis of these tetramers reveals a subtle interplay between helix offset and the local packing geometry of the extended core residues (Liu et al., 2007). We have previously characterized the GCN4-pVe and GCN4-pVg peptides that differ from the classical GCN4 leucine zipper, a parallel, two-stranded coiled coil, by valine substitutions at three charged e and g positions, respectively ( Figure 1). GCN4-pVg adopts an antiparallel, tetrameric structure in which the helices are staggered in register by a full turn (Deng et al., 2006a). However, GCN4-pVe forms an unusual parallel tetramer with a 3 residue interhelical offset (Liu et al., 2006). In this study, we demonstrate that these two leucine zipper variants associate preferentially to form a stable, antiparallel, tetraplex coiled coil. We explore the physical basis of coiled-coil association specificity by outlining a model for heterotetramer formation in both structural and energetic terms. We experimentally test this model by characterizing two analogous alanine-containing mutants of the GCN4 leucine zipper that can also form a heterotetramer; however, in this case, heterospecificity is strongly reduced. This work has implications not only for understanding the mechanism of protein interaction specificity, but also for the prediction and design of coiled-coil sequences that preferentially form homo- versus heterotetramers.
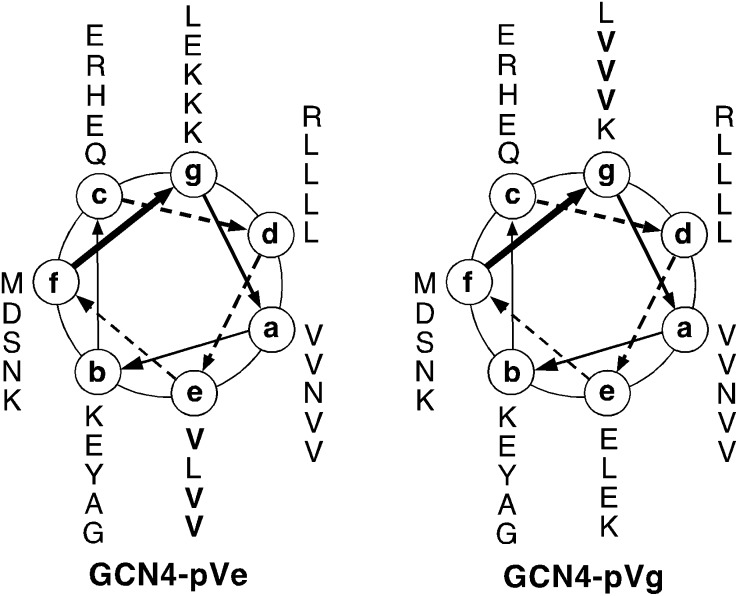
Figure 1.
Axial Helical Projection of the GCN4-pVe and GCN4-pVg Sequences
GCN4-pVe differs from GCN4-pVg only at the three charged e and g positions (bold). The view is from the N termini. Heptad-repeat positions are labeled a–g. The sequence of GCN4-pVe is MK VKQLVDK VEELLSK NYHLVNE VARLVKL VGER; the sequence of GCN4-pVg is MK VKQLEDV VEELLSV NYHLENV VARLKKL VGER.
Results and Discussion
Design of Heterotetramerizing Leucine Zipper
Previous studies have shown that the specific topologies of antiparallel, tetrameric coiled coils can be prescribed by the burial of hydrophobic residues at the a, d, and e or a, d, and g positions (Deng et al., 2006a, Hill et al., 2000, Liu et al., 2007, Solan et al., 2002, Yadav et al., 2006). Given the inequivalence of the e and g positions of a 3-3-1 heptad arrangement, there are substantial differences in the interior packing and interhelix register shifts of these tetramers. In the a–d–e pattern, bulky side chains at the a positions form hydrophobic seams linking four antiparallel helices, resulting in the recruitment of interfacial interactions at the d and e positions to complete the tetramer interface (Liu et al., 2007). Likewise, the hydrophobic core of the a–d–g pattern is formed by the interlocking of residues at the central d positions and by lining of the peripherial a and g side chains (Deng et al., 2006a). Because these 3-3-1 sequence motifs form quasi-D2-symmetric homotetramers, we posit that the a–d–e and a–d–g heptad sequences may interact heterotypically rather than homotypically so as to maximize interfacial van der Waals interactions, giving rise to a heterospecific coiled-coil tetramer. To test this hypothesis, we decided to explore the heterotypic interaction between the GCN4-pVe and GCN4-pVg peptides, the dimeric GCN4 leucine zipper variants that differ only at positions e and g (Figure 1).
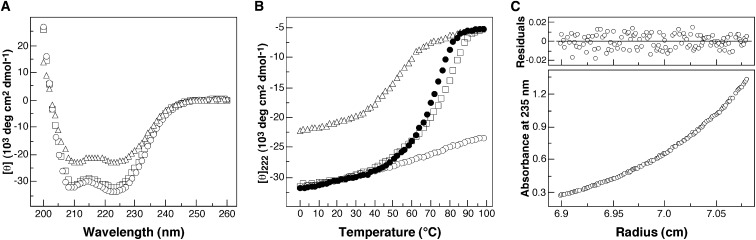
On the basis of circular dichroism (CD) measurements at a 10 μM peptide concentration in TBS buffer (pH 8.0) at 4°C, the individual GCN4-pVe and GCN4-pVg peptides are ∼75% and >90% helical, respectively. In contrast, an equimolar mixture of GCN4-pVe and GCN4-pVg contains >90% helical structure ( Figure 2A), and the ratio of the minima at 208 and 222 nm is typical of spectra observed for coiled coils (Zhou et al., 1992). Under these conditions, the isolated GCN4-pVe and GCN4-pVg peptides exhibit cooperative thermal unfolding transitions with melting temperatures (Tms) of 50°C and 75°C, respectively, whereas the mixture has a thermal stability that exceeds 100°C (Figure 2B). In the presence of the denaturant GuHCl at a 2 M concentration, the mixture melts cooperatively with an apparent Tm of 71°C (Figure 2B), whereas the isolated peptides are essentially unfolded. The significantly higher stability of the mixture relative to the individual samples suggests that a heteromeric, α-helical coiled-coil structure has formed. Sedimentation equilibrium experiments indicate that the GCN4-pVe/GCN4-pVg complex sediments as a discrete dimer of heterodimers: the apparent molecular mass is 16.8 kDa (the expected molecular mass is 15.9 kDa) (Figure 2C). Thus, the GCN4-pVe and GCN4-pVg peptides associate preferentially and fold as an extremely stable, helical heterotetramer.
Figure 2.
The GCN4-pVe and GCN4-pVg Peptides Associate to Form an Extremely Stable, α-Helical Dimer of Heterodimers
(A) CD spectra of GCN4-pVe (open triangles), GCN4-pVg (open squares), and the GCN4-pVe/GCN4-pVg complex (open circles) at 4°C in TBS at a total peptide concentration of 10 μM.
(B) Thermal melts monitored by the CD signal at 222 nm. The filled circles show thermal unfolding data of the GCN4-pVe/GCN4-pVg complex in the presence of 2 M GuHCl.
(C) Equilibrium sedimentation data (27,000 rpm) of a mixture of GCN4-pVe (75 μM) and GCN4-pVg (75 μM) at 20°C in TBS. The data fit closely to a tetrameric complex. The deviation in the data from the linear fit for a tetrameric model is plotted.
Analysis of Heterotetramer Specificity
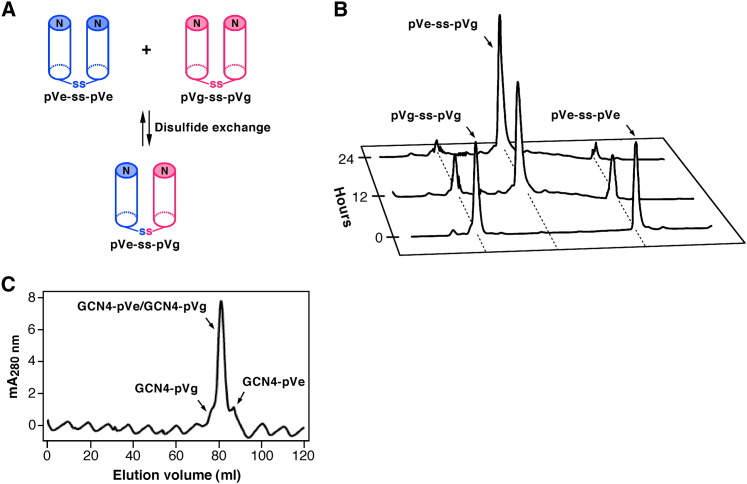
To quantify the specificity of preferential heterotetramer formation, a thiol-disulfide exchange assay was used to directly measure the equilibrium between the disulfide-bonded homodimer and heterodimer forms of the pVe-SH and pVg-SH peptides in which a Gly-Gly-Cys sequence is appended to GCN4-pVe and GCN4-pVg at the C terminus ( Figure 3A). The glycine residues allow for disulfide-bond formation without distortion of the four-helix coiled-coil structure (Harbury et al., 1993). Because CD and sedimentation equilibrium studies demonstrate that the covalently linked heterodimer and homodimers form α-helical dimers, the observed equilibrium reflects the relative stabilities of the disulfide-bonded homotetramer and heterotetramer states of the native peptides. When an equimolar mixture of the covalently linked pVe-SH and pVg-SH homodimers is equilibrated in a redox buffer, a clear preference for heterotetramer formation is evident after the rearrangement is complete (Figure 3B). The degree of specificity can be estimated from Kspec, the equilibrium constant describing the ratio of the heterodimer to the corresponding homodimers. Assuming a two-state model for the dimer-tetramer equilibrium in each case, we can apply the following relationship (Krylov et al., 1994) directly to this system:
| (1) |
where pVe-ss-pVg is the C-terminally disulfide-bonded heterodimer, pVe-ss-pVe and pVg-ss-pVg are the corresponding disulfide-bonded homodimers, and the free energy of specificity for heterotetramer formation (ΔGspec) is equal to −RTlnKspec + RTln2. The factor of two enters into the equilibrium because there are two ways to form the heterodimer and only one way to form the homodimers. Fitting the data to the above equation reveals that ΔGspec = −1.9 kcal/mol, favoring heterotetramer formation. Thus, the GCN4-pVe/GCN4-pVg heterotetramer is preferred over the corresponding homotetramers by ∼50-fold. Size-exclusion chromatography analysis of the refolded sample of the native GCN4-pVe and GCN4-pVg peptides demonstrates that this heterospecificity preference is not affected by the disulfide crosslink (Figure 3C). Evidently, the preferential heterotypic interaction is largely a thermodynamic consequence of the relative stabilities of the hetero- and homotetramers as determined by CD.
Figure 3.
Specificity of the Heterotypic Interaction between the GCN4-pVe and GCN4-pVg Peptides
(A) Preferential formation of a heterodisulfide bond. Assuming that the glycyl linkers allow for random sorting of the C-terminal cysteine residues in a mixture of the pVe-SH and pVg-SH peptides (the variants of GCN4-pVe and GCN4-pVg that have a C-terminal Gly-Gly-Cys sequence), the thermodynamically preferred heterotetramer conformation should favor oxidative heterodisulfide formation.
(B) HPLC analyses of disulfide rearrangement during the course of the equilibration under redox conditions. Disulfide exchange reactions were initiated from the disulfide-bonded pVe-ss-pVe and pVg-ss-pVg homodimers.
(C) Size-exclusion chromatography profile of the refolded GCN4-pVe/GCN4-pVg sample. An equilmolar mixture of the two peptides was refolded by renaturation from GuHCl and was analyzed by size exclusion on a Superdex 75 column equilibrated with TBS at 4°C. Fractions were analyzed by reverse-phase HPLC. Relative concentrations of the GCN4-pVe/GCN4-pVg complex and the combined GCN4-pVg and GCN4-pVe homotetramers is ∼50:1, as calculated from the peak absorbance at 280 nm.
Structure of the Heterotetramer
To investigate the basis for heterotypic interaction specificity, the X-ray crystal structure of the GCN4-pVe/GCN4-pVg complex was determined at 1.70 Å resolution by molecular replacement ( Table 1). Crystals of the complex contain three tetramers of GCN4-pVe/GCN4-pVg per unit cell. The final 2Fo − Fc map is of excellent quality. The current model includes 387 amino acid residues (21 residues at the chain termini are disordered) and 4 Mg2+ ions (present in the crystallization buffer). The quality of the structure was verified by PROCHECK, with all residues in the most favored α-helical region of the Ramachandran plot. The structural differences between the three copies of the GCN4-pVe/GCN4-pVg heterotetramers are small (e.g., the rmsd between the Cα atom positions of individual pair of heterotetramers is 0.43–0.53 Å). Data collection and refinement statistics are summarized in Table 1.
Table 1.
Summary of Data Collection, Structural Refinement, and Analysis
| GCN4-pVe/GCN4-pVg | GCN4-pAe/GCN4-pAg | |
|---|---|---|
| Resolution (Å) | 45.2−1.70 (1.76−1.70) | 56.2−1.70 (1.74−1.70) |
| Space group | P1 | P4212 |
| Unit cell parameters | a = 48.85 Å, b = 49.11 Å, c = 51.92 Å, α = 115.8°, β = 94.2°, γ = 109.6° | a = b = 79.47 Å, c = 54.95 Å |
| Number of unique reflections | 41,918 (4137) | 19,756 (1281) |
| Multiplicity | 2.0 (2.0) | 9.2 (9.0) |
| Rmerge (%) | 4.2 (41.3) | 5.5 (55.6) |
| I/σI | 12.7 (2.1) | 15.2 (4.9) |
| Completeness (%) | 97.0 (95.8) | 99.7 (99.9) |
| Number of molecules in the asymmetric unit | 12 | 5 |
| Solvent content (%) | 44.7 | 46.0 |
| Number of atoms in refinement | 3163 | 1256 |
| Number of solvent molecules | 405 | 130 |
| Rcryst, Rfree (%) | 17.1, 22.1 | 20.3, 23.0 |
| B protein (Å2) | 30.6 | 37.3 |
| Rmsd from ideal geometry | ||
| Bond lengths (Å) | 0.02 | 0.017 |
| Bond angles (°) | 1.5 | 1.5 |
| B values (Å2) | 3.9 | 3.7 |
Values for the highest-resolution shell are given in parentheses.
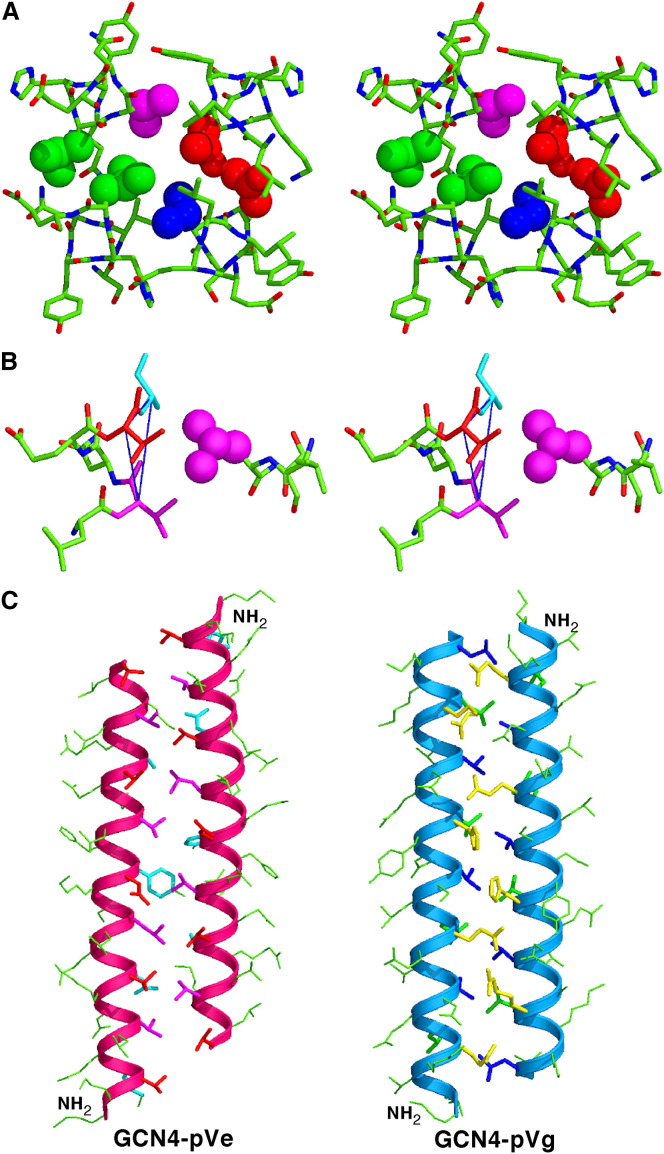
The GCN4-pVe/GCN4-pVg tetramer consists of two interacting, parallel coiled-coil heterodimers that are conjoined in an “up-and-down” conformation through a self-complementary hydrophobic interface ( Figures 4A and 4B). All four helix pairs in the tetramer have crossing angles of near 13°, inducing the four α-helical peptide monomers to wrap tightly around the supercoil axis. This left-handed superhelix creates a cylinder ∼50 Å in length and ∼22–27 Å in diameter. Adjacent parallel hetero-helices have an identical relative orientation and are aligned without lateral displacement. The backbones of neighboring antiparallel homo-helices are shifted from each other by half a turn. The GCN4-pVe and GCN4-pVg chains each can be superimposed on each other with an rmsd for the Cα atoms of 0.23–0.32 Å. An ∼2-fold axis of symmetry is perpendicular to the superhelical axis. The distance between the axes of the parallel hetero-helices is ∼9.4 Å, whereas that between the axes of the neighboring antiparallel homo-helices is ∼9.9 Å.
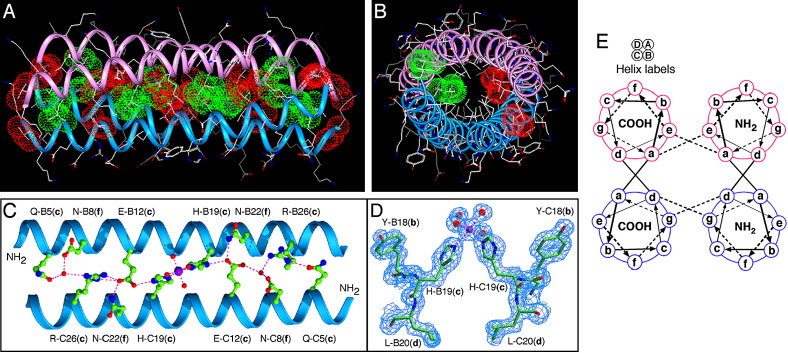
Figure 4.
GCN4-pVe and GCN4-pVg Associate to Form an Antiparallel, Four-Stranded Coiled Coil
(A) Lateral view of the antiparallel heterotetramer (residues 2–31). The Cα backbones of GCN4-pVe (magenta) and GCN4-pVg (blue) are depicted. Red van der Waals surfaces identify residues at the a positions, and green van der Waals surfaces identify residues at the d positions.
(B) Axial view of the antiparallel heterotetramer. The view is from the N termini of helices A and B, looking down the superhelical axis. The van der Waals surfaces are colored red for Val10(a) of helices A and B and green for Leu27(d) of helices C and D.
(C) The interhelical hydrogen-bonding network formed by a Mg2+ ion, structured waters, and the c and f residues of helices B and C. The Mg2+ ion is represented as a purple ball; water molecules are represented as small, red spheres; and hydrogen bonds are represented by dotted lines.
(D) A portion of the 2Fo − Fc electron density map (contoured at 1.5σ) showing the coordination geometry of the Mg2+ ion (purple ball) connecting the helices B and C. Water molecules are shown as red spheres, and magnesium coordinations are denoted by dotted lines.
(E) Helical wheel representation of the antiparallel heterotetramer. Heptad positions are labeled a–g.
Two parallel heterodimers in the tetramer structure adopt an identical double-stranded coiled-coil conformation. Residues at the a and d positions interact between parallel hetero-helices and stagger axially to form the dimer interface; the heptad repeat is maintained in register through the entire 30 residue region (Lys2–Val31) (Figure 4A). The cross-sectional layers containing valine or asparagine at the a positions alternate with layers containing leucine at the d positions. At the interface between the two heterodimers, nonpolar side chains at the a and e positions of GCN4-pVe and at d and g of GCN4-pVg make unique side-to-side contacts with their corresponding a′–e′ and d′–g′ residues of the neighboring antiparallel helices (where primed letters refer to positions of the neighboring helix), leading to the formation of interlocking hydrophobic seams between supercoiled α-helical ribbons (Figures 4A and 4E). All of the a, d, and e side chains of GCN4-pVe, except Asn17, and all of the a, d, and g side chains of GCN4-pVg, except Val28, assume their well-populated rotamer conformations in α helices (Lovell et al., 2000). Moreover, intrahelical and interhelical salt bridges and charge-stabilized hydrogen bonds coat the surface of the heterospecific tetramer. For example, the network of extended interstrand connections among polar and charged residues at the c and f positions of GCN4-pVg, Mg2+, and ordered water molecules is also involved in close packing and stabilization of interhelical contacts (Figures 4C and 4D). These hydrophilic interactions are different from those in the wild-type GCN4-p1 leucine zipper (O'Shea et al., 1991).
Core Packing Features
The tetramer interface shows combined heterotypic “knobs-into-holes” and homotypic “knobs-into-triangles” packing interactions between triads of the GCN4-pVe and GCN4-pVg peptides ( Figure 5A). The parallel heterodimers adopt classical knobs-into-holes packing characteristics of homodimeric coiled coils; the angles between the Cα–Cα and Cα–Cβ vectors at the a and d layers are 180° and 90°, respectively (Harbury et al., 1998). The interacting surface of the antiparallel homodimers shows a precise interdigitation between side chains at the a, b, and e positions of GCN4-pVe and at c, d, and g of GCN4-pVg. Valine or leucine knobs at the peripheral e positions of GCN4-pVe pack into triangles formed by the a, b, and e residues of the adjacent antiparallel helix (Figure 5B). Similarly, valine or leucine knobs at the peripheral g positions of GCN4-pVg fit into triangles formed by the c, d, and g residues of the adjacent antiparallel helix. The neighboring antiparallel homo-helices are offset by ∼0.25 heptad with respect to each other such that the distance between the a and e′ side chains of GCN4-pVe and between the d and g′ side chains of GCN4-pVg is ∼3.8–4.2 Å (Figure 5C). This helix offset allows for knobs-into-triangles core packing by compensating for the altered Cα–Cβ vectors due to the opposing directions of these interfacial side chains in an antiparallel helical array. Analysis of the heterotetramer by using SOCKET (Walshaw and Woolfson, 2001) at a packing cutoff of 7.0 Å shows that a pair of conjoined parallel heterodimeric coiled coils exhibits pairwise complementary knobs-into-holes interactions, as described above. Using a more liberal cutoff of 8.0 Å, an antiparallel, four-stranded coiled coil was identified with additional peripheral knobs that broaden the helical contacts. In summary, the requirement to satisfy these interleaved triadic packing interactions specifies and stabilizes a heterospecific tetramer structure that has not, to our knowledge, been seen before in naturally occurring coiled coils.
Figure 5.
Extended Knobs-into-Holes Packing in the Antiparallel GCN4-pVe/GCN4-pVg Hetrotetramer
(A) Packing of core residues in the antiparallel heterotetramer. Stereo helix cross-sectional layers centered on positions a, d, and e of GCN4-pVe and on positions a, d, and g of GCN4-pVg are shown in a space-filling representation. The view is from the N termini of helices A and B, looking down the superhelical axis. Residues at positions a (red) and d (green) are packed in register at the interfaces of the parallel A/B and C/D helices with classical coiled-coil interactions. Side chains at positions e of GCN4-pVe (magenta) and g of GCN4-pVg (blue) fit into triangular spaces on the opposite antiparallel helix to form interlocking hydrophobic seams between the antiparallel A/D and B/C helices, respectively.
(B) Stereo view of knobs-into-triangle contacts in the antiparallel heterotetramer. The triangle of the Val21(e) (magenta), Val24(a) (red), and Ala25(b) (cyan) residues on the opposite antiparallel helix, into which the Leu14 side chain nestles, is indicated.
(C) Coiled-coil packing in the antiparallel heterotetramer. The left panel shows the Cα backbones of GCN4-pVe (residues 1–31), with side chains at positions e (magenta) that pack inside triangles of residues at positions a (red), b (cyan), and e of the opposite helix. The right panel shows the Cα backbones of GCN4-pVg (residues 2–31), with side chains at positions g (blue) that pack inside triangles of residues at positions d (green), c (yellow), and g of the opposite helix. Contacting helical turns interdigitate; the vertical offset of heptads is half of a helix turn.
Mechanism of Specificity
What driving force might favor heterotetramer formation by the GCN4-pVe and GCN4-pVg peptides? Varying the salt concentration (between 50 mM and 2 M NaCl) has relatively little effect on the stability of the hetero- and homotetramers in the native peptides, as judged from CD spectra (data not shown). We infer that electrostatic effects do not play a dominant role in discriminating between hetero- and homotetramer structures. Consistent with this argument, the ratio of the disulfide-bonded pVe-ss-pVg heterodimer to the corresponding homodimers measured in the redox equilibrium experiments (Figure 3) is unaffected by increased salt: the preference for heterospecificity remains ∼50:1 in the presence of 2 M NaCl. This is in marked contrast to several studies that implicate stabilizing ionic interactions or avoidance of electrostatic repulsion in determining the partnering specificity of other model dimeric coiled coils (Burkhard et al., 2000, Fairman et al., 1996, Graddis et al., 1993, McClain et al., 2002, Nautiyal et al., 1995, O'Shea et al., 1993); in each of these cases, there is a dramatic salt dependence of heterospecific coiled-coil folding and stability.
One clue to the energetic contribution of interactions to preferential pairing between the GCN4-pVe and GCN4-pVg peptides comes from apparent van der Waals packing differences at the tetramer interfaces of the coiled coils. The supercoil radius, R0; frequency, ω0; and phase angle, ϕ, differ significantly in the hetero- and homotetramer structures. Their superhelical parameters are as follows: R0 = 7.2 Å, ω0 = 129 residues/turn, and ϕ = −20.6° for GCN4-pVe/GCN4-pVg; R0 = 7.4 Å, ω0 = 107 residues/turn, and ϕ = 41.5° for GCN4-pVg; R0 = 6.7 Å, ω0 = 113 residues/turn, and ϕ = 15.9° for GCN4-pVe (the core of the latter structure consists of a pair of conjoined trimers) (Liu et al., 2006). In essence, the backbones of the four associating helices in the heterotetramer flatten out and wrap less tightly around the superhelical axis, allowing the a and d side chains to face inward to create the hydrophobic interface between parallel heterotypic helices. However, the e and g side chains face toward the axis of supercoil rotation and mesh when the two parallel dimers interlock in an antiparallel fashion. As a result, more surface area is buried in the heterotetramer (5950 Å2) than in either the GCN4-pVe (5700 Å2) or GCN4-pVg (5470 Å2) homotetramers. Burial of hydrophobic surface provides one major source of stability in folding (Bryson et al., 1995). If we approximate the free energy change at 25°C for buried nonpolar surface as −15 cal/mole per Å2 (Vallone et al., 1998), then the buried surface free energy change for the heterotetramer relative to the homotetramers would correspond to −5.5 kcal/mole. Correcting for the entropy of mixing between hetero- versus homotypic association would result in a total contribution of nearly 6 kcal/mole, clearly an overestimate. If we assume that polar interactions contribute to the tetramer interface (e.g., Asn17 at the third a position), this value will be reduced, but in any case, the predicted gain in net hydrophobic stabilization energy of the heterotetramer structure can easily account for the extent of heterospecificity that we observe. Thus, the specific interfacial interactions determine the equilibrium transition between hetero- and homotetramers. The mechanism of preferential heterotetramer formation appears to be predominantly hydrophobic in nature.
The GCN4-pAe/GCN4-pAg Heterotetramer
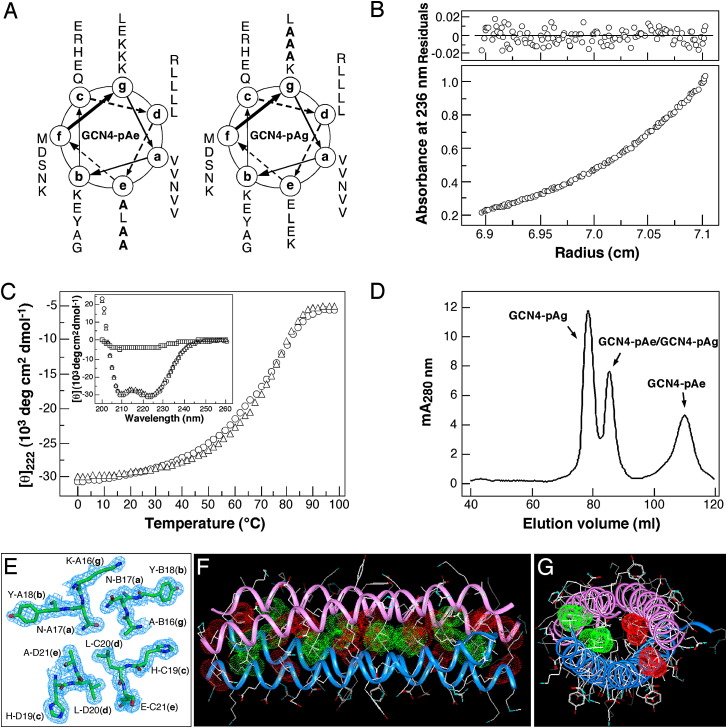
The above-described studies indicate that interfacial van der Waals interactions provided by valine residues that form knobs at the flanking e and g positions of GCN4-pVe and GCN4-pVg mediate preferential heterotetramer formation. To probe this partnering specificity further, we characterized the interaction properties of the GCN4-pAe and GCN4-pAg peptides containing alanine substitutions at three corresponding e and g positions ( Figure 6A). Alanine was selected for its minimal apolar side chain and relatively low hydrophobicity. Our interpretation of the structural basis of heterotetramer formation in the case of valine would predict significant weakening of the contribution from packing and hydrophobicity at these sites in the Val → Ala mutants. Our previous studies show that GCN4-pAe forms a relatively unstable dimer in aqueous solution (Deng et al., 2007), whereas GCN4-pAg folds into a stable, antiparallel, four-stranded coiled coil (Deng et al., 2006a). To explore the heterotypic interaction between GCN4-pAe and GCN4-pAg, a mixture of the two peptides was refolded by renaturation from GuHCl solution and fractionated by gel-filtration chromatography. The peptide mixture forms a clean tetramer, as determined by sedimentation equilibrium experiments (Figure 6B). CD measurements at a 10 μM protein concentration in TBS show that the peptide mixture is >90% helical at 4°C and undergoes a cooperative unfolding transition with a Tm of 70°C, compared with a Tm of 72°C for GCN4-pAg at the same peptide concentration (Figure 6C). Under the same conditions, GCN4-pAe is predominantly unfolded (Figure 6C). Serendipitously, reverse-phase HPLC analysis of the peptide mixture reveals a 1:5 ratio of GCN4-pAe and GCN4-pAg. Size-exclusion chromatography of the refolded peptide sample under equilibrium conditions shows a 1:2 ratio of the GCN4-pAe/GCN4-pAg complex and the GCN4-pAg tetramer; their retention volumes are distinct from the GCN4-pAe dimer (Figure 6D). Taken together, these data demonstrate that the hetero- and homotetrameric species coexist in solution and that the GCN4-pAg tetramer is preferred over the heterotetramer by 2-fold.
Figure 6.
Heterotetramer Formation by the GCN4-pAe and GCN4-pAg Peptides
(A) Coiled-coil helical wheel representation of the GCN4-pAe and GCN4-pAg sequences. They differ from the dimeric GCN4 leucine zipper by alanine substitutions (bold) at three e and three g positions, respectively. The sequence of GCN4-pAe is MK VKQLADK VEELLSK NYHLANE VARLAKL VGER; the sequence of GCN4-pAg is MK VKQLEDA VEELLSA NYHLENA VARLKKL VGER.
(B) Equilibrium sedimentation data (27,000 rpm) of the GCN4-pAe/GCN4-pAg mixture (150 μM) at 20°C in TBS. The data fit closely to a tetrameric complex. The deviation in the data from the linear fit for a tetrameric model is plotted.
(C) Thermal melts of the GCN4-pAe/GCN4-pAg mixture (circles) and GCN4-pAg (triangles) monitored by the CD signal at 222 nm at a total protein concentration of 10 μM. The insert shows the CD spectra of the GCN4-pAe/GCN4-pAg mixture (circles), GCN4-pAg (triangles), and GCN4-pAe (squares) at 4°C.
(D) Size-exclusion chromatography profile of the refolded GCN4-pAe/GCN4-pAg sample. An equilmolar mixture of the two peptides was refolded by renaturation from GuHCl solution and was analyzed by size exclusion on a Superdex 75 column equilibrated with TBS at 4°C. Fractions were analyzed by reverse-phase HLPC and equilibrium sedimentation. Relative concentrations of the GCN4-pVe/GCN4-pVg complex and the GCN4-pVg tetramer were calculated from the peak absorbance at 280 nm.
(E) Crystal structure of the GCN4-pAe/GCN4-pAg complex. The 1.70 Å 2Fo − Fc electron density map at 1.5σ contour shows a cross-section of the antiparallel heterotetramer.
(F) Lateral view of the antiparallel heterotetramer. The Cα backbones of GCN4-pAe (magenta) and GCN4-pAg (blue) are depicted. Red van der Waals surfaces identify residues at the a positions, and green van der Waals surfaces identify residues at the d positions.
(G) Axial view of the antiparallel heterotetramer. The view is from the N termini of helices A and B, looking down the superhelical axis. The van der Waals surfaces are colored red for Val10(a) of helices A and B and green for Leu27(d) of helices C and D.
We determined the crystal structure of the GCN4-pAe/GCN4-pAg mixture at 1.70 Å resolution (Figure 6E; Table 1). The unit cell contains two distinct tetrameric species, consistent with the solution data. The heterotetramer is made up from chains A/D (GCN4-pAe) and B/C (GCN4-pAg), arranged with an approximate 2-fold symmetry perpendicular to the superhelical axis (Figure 6F), whereas the homotetramer contains chain E (GCN4-pAg) and its symmetry-related set generated by crystallographic 222 axes. The GCN4-pAg tetramer adopts the same antiparallel, four-stranded coiled-coil conformation as in the previously determined crystal structure (rmsd for equivalent Cα positions of 0.30 Å) (Deng et al., 2006a). In the case of the GCN4-pAe/GCN4-pAg heterotetramer, the superhelix again forms an overall rod-shaped structure ∼50 Å in length with a diameter of ∼24 Å (Figures 6F and 6G). The crossing angles between pairs of adjacent helices is 30° and 11° for the parallel heterodimers and the antiparallel homodimers, respectively. Significantly, the combined knobs-into-holes and knobs-into-triangles packing interactions in the GCN4-pAe/GCN4-pAg structure are virtually identical to those of the GCN4-pVe/GCN4-pVg heterotetramer (Figure 5). We conclude that mixed heptad sequence repeats with triadic a–d–e and a–d–g hydrophobic residue patterns can favor a heterospecific, antiparallel, four-helix coiled-coil structure.
Protein Interaction Specificity
How proteins discriminate their natural binding partners from a plethora of possible competitors with similar sequences and structures remains an outstanding general issue in the field of protein-protein interactions. Recent advances in array technology and high-throughput screens have resulted in a large body of protein-protein interaction data at the cell level that needs to be analyzed and interpreted (Newman and Keating, 2003, Uetz et al., 2000, Tarassov et al., 2008). Existing knowledge of the determination of dimeric coiled coils provides a high-resolution structural framework with which to approach one important class of protein interaction domains (Acharya et al., 2002, Fong et al., 2004, Grigoryan and Keating, 2006, Havranek and Harbury, 2003, Mason et al., 2006, Pokala and Handel, 2005, Summa et al., 2002). Despite the seeming simplicity of coiled-coil structure, folding can potentially generate a multiplicity of alternative conformations that are close in free energy. This situation leads to the potential coexistence of mixed species in solution; in fact, several reported examples of conformational transitions in coiled coils involve heterogeneous mixtures as one reaction component (Gonzalez et al., 1996, Lovejoy et al., 1993). Current design algorithms have difficulty discriminating among such degenerate states and ensuring that a chosen sequence will specifically assume a target fold (Havranek and Harbury, 2003). In contrast to the seemingly facile repacking of the core in globular proteins (Behe et al., 1991), compatibility of the buried side chains with the restricted packing spaces within coiled coils confers selectivity for stoichiometry, helix orientation, and stagger, and now hetero- versus homotetrameric association. Our results reinforce the conclusion that finely tuned interfacial interactions can contribute to functionally specific protein-protein recognition (Clackson and Wells, 1995, Ofran and Rost, 2007). Thus, principles governing oligomeric contacts within the extended hydrophobic cores should assist in both the prediction of protein interaction partners and the development of an improved physical model for the free energy of protein-protein interactions.
The present case highlights the need to extend the current rules of coiled-coil domain recognition to include nonpolar side chains at the e and g positions, as discussed extensively by Alber, Woolfson, and coworkers (Woolfson and Alber, 1995, Walshaw and Woolfson, 2003). Our previous studies suggest that the structural selectivity of conserved, charged residues at the e and g positions of the GCN4 leucine zipper results not only from favorable interhelical electrostatic interactions, but also from necessary destabilization (negative design) of alternate highly stable coiled-coil conformations (Deng et al., 2006b). The GCN4-pVe/GCN4-pVg peptides may represent a minimalist system for modeling heterospecificity through interactions of the core side chains in a direct and “positive” manner. Deletion of the valine side chain as in the GCN4-pAe/GCN4-pAg complex causes the equilibrium to favor mixed hetero- and homotetrameric states. Antiparallel, four-stranded coiled coils are involved in homo- and hetero-protein complex assembly and targeting throughout nature (Fairman et al., 1995, Deng et al., 2006b), including the well-known lac repressor (Fairman et al., 1995); thus, principles underlying heterospecificity learned from our model peptide system can be used to modulate biologically important conformational switches in general. For example, the intermediate filament proteins desmin and vimentin individually form homotetramers, but yield a distinctly different heterotetrameric complex when allowed to coassemble (Wickert et al., 2005). Our work here provides a model system with which to investigate the molecular basis for this protein interaction specificity. An additional important example of heterospecific coiled-coil tetramer structure comes from the SARS coronavirus S2 envelope protein (Deng et al., 2006b). This antiparallel, four-helix rod is implicated in driving a conformational change in S2 during fusion activation and may therefore be of fundamental importance in membrane apposition and fusion (Deng et al., 2006b). As well, cellular membrane fusion processes mediated by SNARE proteins rely on four-helix bundle formation to achieve apposition of the vesicle and target membranes (Sutton et al., 1998). Finally, the GCN4-pVe/GCN4-pVg peptides described here can be used to mediate hetero- and homotetramer states, providing oligomerization scaffolds for exploring biological roles of these protein complexes.
Significance
The coiled coil is a widespread structural motif for mediating specific protein-protein interactions in molecular recognition and for the assembly of multiprotein complexes. One important aspect of coiled-coil interaction specificity centers on the determination of homo- versus heteromeric association. Despite intensive analysis, however, the rules and mechanisms that govern the pairing and structural specificity of coiled coils are still incompletely understood. Recent experiments show that the presence of nonpolar amino acids at either of the normally charged e and g positions of the dimeric GCN4 leucine zipper can direct the formation of stable, four-stranded coiled coils. Here, we demonstrate that the GCN4-pVe and GCN4-pVg peptides that differ only at the e and g positions preferentially form (by 50-fold) a heterotetramer over homotetramers. In contrast to the dominant role of electrostatics found in earlier studies, the basis for this preference appears to reside in a global restructuring of the interface, leading to more favorable hydrophobic and van der Waals interactions in the heterotetramer. The novel packing arrangement displayed by the GCN4-pVe/GCN4-pVg structure is also found in that of the GCN4-pAe and GCN4-pAg peptides with alanine substitutions at the corresponding e and g positions. In the latter case, the heterotetramer and the GCN4-pAg homotetramer are nearly equivalent in stability, whereas GCN4-pAe is incompletely folded. Its instability likely provides a thermodynamic driving force for heterotetramer formation. Evidently, additional dimensions of regulation and diversity of coiled-coil interactions can be generated by local interactions among side chains at each of the core a, d, e, and g positions of the heptad repeat. These new heterospecific tetramers allow us to begin detailed exploration of structural principles underlying the folding and association of higher-order heterotypic coiled coils. A comprehensive understanding of such principles can be used to predict coiled-coil partnering specificity, to design new protein-protein interfaces with specific biological functions, and to develop inhibitors of such assemblies.
Experimental Procedures
Cloning and Expression
The sequence of pVe-SH is MK VKQLVDK VEELLSK NYHLVNE VARLVKL VGER-GGC; the sequence of pVg-SH is MK VKQLEDV VEELLSV NYHLENV VARLKKL VGER-GGC. “SH” denotes a (Gly-Gly-Cys) linker. The pVe-SH and pVg-SH constructs were derived from plasmids pGCN4-Ve (Liu et al., 2006) and pGCN4-Vg (Deng et al., 2006a), respectively. Mutations were introduced by using the method of Kunkel et al. (1987) and were verified by DNA sequencing. All peptides were expressed in E. coli BL21(DE3)/pLysS cells, purified to homogeneity by reverse-phase HPLC on a Vydac preparative C18 column, and lyophilized. Peptide identities were confirmed by electrospray mass spectrometry. Protein concentrations were determined by using the method of Edelhoch (1967).
CD Measurements
CD studies were performed in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl (TBS) on an Aviv 62A/DS CD spectrometer at 10 μM total peptide concentration. Thermal stability were assessed by monitoring [θ]222 as a function of temperature, with the addition of 2 M GuHCl to facilitate unfolding. For studies of the ionic strength dependence, melting curves were recorded in 50 mM Tris-HCl (pH 8.0) in the presence of the indicated concentration of NaCl. Melting profiles are reversible; >90% of the CD signal was regained upon cooling. Tm values were estimated by evaluating the maximum of the first derivative of [θ]222 versus temperature data (Cantor and Schimmel, 1980).
Analytical Ultracentrifugation
Sedimentation equilibrium measurements were carried out on a Beckman XL-A analytical ultracentrifuge at 20°C. Protein samples were dialyzed overnight against TBS at room temperature and were loaded at initial concentrations of 15, 50, and 150 μM into cells placed in an An-60 Ti rotor. Data were acquired at two wavelengths at rotor speeds of 24,000 and 27,000 rpm and were processed globally for the best fit to a single-species model of absorbance versus radial distance by using the Origin software provided by the manufacturer. The residuals were analyzed to reveal the presence of species other than the tetramer; in no case were systematic deviations of the residuals observed. Protein partial specific volumes and solvent densities were calculated as described by Laue et al. (1992).
Disulfide Exchange Experiments
The HPLC-purified pVe-SH and pVg-SH peptides were air oxidized in 50 mM Tris-HCl (pH 8.6) plus 6 M GuHCl. The disulfide-bonded pVe-ss-pVe and pVg-ss-pVg homodimers were repurified by HPLC as described above. An equimolar mixture of pVe-ss-pVe (50 μM) and pVg-ss-pVg (50 μM) was incubated in redox buffer consisting of 500 μM reduced glutathione, 125 μM oxidized glutathione, 50 mM Tris-HCl (pH 8.6) in the presence of the indicated concentration of NaCl at room temperature under an argon atmosphere. Redox exchange reactions were equilibrated for 12–24 hr and were quenched by the addition of concentrated acetic acid to a final concentration of 10% by volume (pH < 2). The reaction products were analyzed by reverse-phase HPLC on a C18 analytical column. Relative concentrations of the pVe-ss-pVg heterodimer and the pVe-ss-pVe and pVg-ss-pVg homodimers were determined by integration of the corresponding peaks (absorbance at 229 nm was monitored). Results from disulfide exchange reactions with the reduced and oxidized pVe-SH and pVg-SH peptides as the starting reactants agreed to within 0.1 kcal/mol, indicating that equilibrium had been reached (O'Shea et al., 1989).
Crystallization and Structure Determination
The GCN4-pVe/GCN4-pVg complex was refolded by renaturation from GuHCl and was purified by size exclusion on a Superdex 200 column. Purified complex was crystallized from 0.1 M Tris-HCl (pH 8.5), 0.2 M MgCl2, 30% PEG 4000 by using the hanging-drop method. Crystals were cryoprotected in 0.1 M Tris-HCl (pH 8.5), 0.2 M MgCl2, 32.5% PEG 4000, 15% glycerol and were subsequently frozen in liquid nitrogen. X-ray diffraction data were collected on X4C at the National Synchrotron Light Source (NSLS). Intensities were integrated and scaled by using DENZO and SCALEPACK (Otwinowski and Minor, 1997). Initial phases were determined by molecular replacement with Phaser (Storoni et al., 2004) by using the structure of the GCN4-pVg tetramer (PDB entry 2B22) as a search model. Density interpretation and manual model building were performed with O (Jones et al., 1991). Crystallographic refinement was done by using Refmac (Murshudov et al., 1997), resulting in an Rfree of 28.7% and an Rwork of 23.6% between 45.2 and 1.70 Å resolution. At this stage, solvent molecules were incorporated into the model. Four electron density peaks in the difference Fourier maps could not be adequately accounted for by water molecules. On the basis of the size of each peak and the components of the crystallization conditions, these electron densities were modeled as Mg2+ ions. The individual B factor refinements of the modeled magnesium ions converged at 13.7, 21.9, 22.6, and 33.2 Å2 for the B/C, F/G, J/K, and H helices, respectively. The first three Mg2+ cations are hexahedrally coordinated by the two Nɛ2 atoms of His19 of GCN4-pVg and four water molecules, whereas the coordination of the last metal ion involves five water molecules and the carbonyl oxygen of the Asp8 side chain of GCN4-pVe. The Mg-N and Mg-O distances are in the range of 2.0–2.4 Å. The final model includes 12 monomers in the asymmetric unit. Refinement was concluded with an overall anisotropic thermal factor correction by using TLS groups for each monomer (Schomaker and Trueblood, 1998). All protein residues occupy the most favored regions of the Ramachandran plot.
An equimolar mixture of the GCN4-pAe and GCN4-pAg peptides was dissolved in TBS plus 6 M GuHCl and was refolded by dilution into TBS. The peptide sample was purified by size exclusion on a Superdex 200 column equilibrated with TBS, exchanged into 10 mM Tris-HCl (pH 8.0), and concentrated to 10 mg/ml by ultrafiltration. Crystals were grown from 0.1 M Tris-HCl (pH 9.8), 50 mM MgCl2, 13% PEG 4000. The crystals were cryoprotected in 0.1 M Tris-HCl (pH 9.8), 40 mM MgCl2, 21% PEG 4000, 25% glycerol and frozen in liquid nitrogen. X-ray diffraction data were collected on X4A at NSLS and were indexed and scaled by using DENZO and SCALEPACK, respectively. Initial phases were determined by molecular replacement with Phaser by using the structure of the GCN4-pAe monomer (PDB entry 2NRN) as a search model. Five monomers corresponding to the two GCN4-pAe and three GCN4-pAg molecules were oriented and placed in the asymmetric unit with a Z score of 10.9 and a final refined log-likelihood gain of 482. This model and the data set for the peptide sample were directly fed to Arp/Warp (Lamzin and Wilson, 1993), which allowed for ∼90% of the final model to be automatically traced. Iterative rounds of model building with O and refinement with Refmac, as well as the addition of ordered solvent, clarified the trace, except the helix-terminal regions, which are not visible in the electron density maps and therefore must be disordered. An overall anisotropic thermal factor correlation was applied by using TLS groups for each monomer. All protein residues are in the most favored regions of the Ramachandran plot.
Structural Analysis
Coiled-coil parameters were calculated by using TWISTER (Strelkov and Burkhard, 2002). Knobs-into-holes packing interactions were analyzed by using SOCKET (Walshaw and Woolfson, 2001). The rmsds were calculated with LSQKAB in CCP4i program suite (Potterton et al., 2003). Buried surface areas were calculated from the difference of the accessible side chain surface areas of the tetramer structure and of the individual helical monomers by using CNS 1.0 (Brunger et al., 1998). Figures were generated by using MOLSCRIPT (Kraulis, 1991), Raster 3D (Merritt and Bacon, 1997), and SETOR (Evans, 1993).
Acknowledgments
We thank John Schwanof at the National Synchrotron Light Source for support at beamline X4A and a referee for her especially perceptive comments on the manuscript. This work was supported by National Institutes of Health grant AI042382.
Published: September 19, 2008
Accession Numbers
The coordinates and structure factors have been deposited in the PDB with entries 3CK4 and 3CRP.
References
- Acharya A., Ruvinov S.B., Gal J., Moll J.R., Vinson C. A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry. 2002;41:14122–14131. doi: 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- Adamson J.G., Zhou N.E., Hodges R.S. Structure, function and application of the coiled-coil protein folding motif. Curr. Opin. Biotechnol. 1993;4:428–437. doi: 10.1016/0958-1669(93)90008-k. [DOI] [PubMed] [Google Scholar]
- Behe M.J., Lattman E.E., Rose G.D. The protein-folding problem: the native fold determines packing, but does packing determine the native fold? Proc. Natl. Acad. Sci. USA. 1991;88:4195–4199. doi: 10.1073/pnas.88.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bryson J.W., Betz S.F., Lu H.S., Suich D.J., Zhou H.X., O'Neil K.T., DeGrado W.F. Protein design: a hierarchic approach. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Meier M., Lustig A. Design of a minimal protein oligomerization domain by a structural approach. Protein Sci. 2000;9:2294–2301. doi: 10.1110/ps.9.12.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld J., Strelkov S.V. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- Cantor C., Schimmel P. W.H. Freeman and Co.; New York: 1980. Biophysical Chemistry, Volume 3. [Google Scholar]
- Clackson T., Wells J.A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Crick F.H.C. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- Davies D.R., Cohen G.H. Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Liu J., Zheng Q., Eliezer D., Kallenbach N.R., Lu M. Antiparallel four-stranded coiled coil specified by a 3–3-1 hydrophobic heptad repeat. Structure. 2006;14:247–255. doi: 10.1016/j.str.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Liu J., Zheng Q., Yong W., Lu M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure. 2006;14:889–899. doi: 10.1016/j.str.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Zheng Q., Liu J., Cheng C.S., Kallenbach N.R., Lu M. Self-assembly of coiled-coil tetramers in the 1.40 Å structure of a leucine-zipper mutant. Protein Sci. 2007;16:323–328. doi: 10.1110/ps.062590807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Evans S.V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Fairman R., Chao H.G., Mueller L., Lavoie T.B., Shen L., Novotny J., Matsueda G.R. Characterization of a new four-chain coiled-coil: influence of chain length on stability. Protein Sci. 1995;4:1457–1469. doi: 10.1002/pro.5560040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman R., Chao H.G., Lavoie T.B., Villafranca J.J., Matsueda G.R., Novotny J. Design of heterotetrameric coiled coils: evidence for increased stabilization by Glu(−)-Lys(+) ion pair interactions. Biochemistry. 1996;35:2824–2829. doi: 10.1021/bi952784c. [DOI] [PubMed] [Google Scholar]
- Fong J.H., Keating A.E., Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez L., Jr., Brown R.A., Richardson D., Alber T. Crystal structures of a single coiled-coil peptide in two oligomeric states reveal the basis for structural polymorphism. Nat. Struct. Biol. 1996;3:1002–1009. doi: 10.1038/nsb1296-1002. [DOI] [PubMed] [Google Scholar]
- Graddis T.J., Myszka D.G., Chaiken I.M. Controlled formation of model homo- and heterodimer coiled coil polypeptides. Biochemistry. 1993;32:12664–12671. doi: 10.1021/bi00210a015. [DOI] [PubMed] [Google Scholar]
- Grigoryan G., Keating A.E. Structure-based prediction of bZIP partnering specificity. J. Mol. Biol. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Zhang T., Kim P.S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Plecs J.J., Tidor B., Alber T., Kim P.S. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- Havranek J.J., Harbury P.B. Automated design of specificity in molecular recognition. Nat. Struct. Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- Hill R.B., Raleigh D.P., Lombardi A., DeGrado W.F. De novo design of helical bundles as models for understanding protein folding and function. Acc. Chem. Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges R.S., Sodek J., Smillie L.B., Jurasek L. Tropomyosin: amino acid sequence and coiled-coil structure. Cold Spring Harb. Symp. Quant. Biol. 1972;37:299–310. [Google Scholar]
- Huber R., Kukla D., Bode W., Schwager P., Bartels K., Deisenhofer J., Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 Å resolution. J. Mol. Biol. 1974;89:73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- Jones S., Thornton J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kortemme T., Joachimiak L.A., Bullock A.N., Schuler A.D., Stoddard B.L., Baker D. Computational redesign of protein-protein interaction specificity. Nat. Struct. Mol. Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- Krylov D., Mikhailenko I., Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A., Roberts J.D., Zakour R.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lamzin V.S., Wilson K.S. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- Laue T.M., Shah B.D., Ridgeway T.M., Pelletier S.L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S.E., Rowe A.J., Horton J.C., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- Liu J., Deng Y., Zheng Q., Cheng C.S., Kallenbach N.R., Lu M. A parallel coiled-coil tetramer with offset helices. Biochemistry. 2006;45:15224–15231. doi: 10.1021/bi061914m. [DOI] [PubMed] [Google Scholar]
- Liu J., Zheng Q., Deng Y., Li Q., Kallenbach N.R., Lu M. Conformational specificity of the lac repressor coiled-coil tetramerization domain. Biochemistry. 2007;46:14951–14959. doi: 10.1021/bi701930d. [DOI] [PubMed] [Google Scholar]
- Lovejoy B., Choe S., Cascio D., McRorie D.K., DeGrado W.F., Eisenberg D. Crystal structure of a synthetic triple-stranded α-helical bundle. Science. 1993;259:1288–1293. doi: 10.1126/science.8446897. [DOI] [PubMed] [Google Scholar]
- Lovell S.C., Word J.M., Richardson J.S., Richardson D.C. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- Lupas A.N., Gruber M. The structure of α-helical coiled coils. Adv. Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Mason J.M., Arndt K.M. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- Mason J.M., Schmitz M.A., Muller K.M., Arndt K.M. Semirational design of Jun-Fos coiled coils with increased affinity: universal implications for leucine zipper prediction and design. Proc. Natl. Acad. Sci. USA. 2006;103:8989–8994. doi: 10.1073/pnas.0509880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain D.L., Gurnon D.G., Oakley M.G. Importance of potential interhelical salt-bridges involving interior residues for coiled-coil stability and quaternary structure. J. Mol. Biol. 2002;324:257–270. doi: 10.1016/s0022-2836(02)01072-0. [DOI] [PubMed] [Google Scholar]
- Merritt E.A., Bacon D.J. Raster 3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likehood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nautiyal S., Woolfson D.N., King D.S., Alber T. A designed heterotrimeric coiled coil. Biochemistry. 1995;34:11645–11651. doi: 10.1021/bi00037a001. [DOI] [PubMed] [Google Scholar]
- Newman J.R., Keating A.E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- O'Shea E.K., Rutkowski R., Stafford W.F., 3rd, Kim P.S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245:646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- O'Shea E.K., Klemm J.D., Kim P.S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- O'Shea E.K., Rutkowski R., Kim P.S. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- O'Shea E.K., Lumb K.J., Kim P.S. Peptide ‘Velcro’: design of a heterodimeric coiled coil. Curr. Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- Oakley M.G., Hollenbeck J.J. The design of antiparallel coiled coils. Curr. Opin. Struct. Biol. 2001;11:450–457. doi: 10.1016/s0959-440x(00)00232-3. [DOI] [PubMed] [Google Scholar]
- Ofran Y., Rost B. ISIS: interaction sites identified from sequence. Bioinformatics. 2007;23:e13–e16. doi: 10.1093/bioinformatics/btl303. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pokala N., Handel T.M. Energy functions for protein design: adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J. Mol. Biol. 2005;347:203–227. doi: 10.1016/j.jmb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Potterton E., Briggs P., Turkenburg M., Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Schomaker V., Trueblood K.N. Correlation of internal torsional motion with overall molecular motion in crystals. Acta Crystallogr. B. 1998;54:507–514. [Google Scholar]
- Solan A., Ratia K., Fairman R. Exploring the role of alanine in the structure of the Lac repressor tetramerization domain, a ferritin-like Alacoil. J. Mol. Biol. 2002;317:601–612. doi: 10.1006/jmbi.2002.5427. [DOI] [PubMed] [Google Scholar]
- Storoni L.C., McCoy A.J., Read R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Strelkov S.V., Burkhard P. Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- Summa C.M., Rosenblatt M.M., Hong J.K., Lear J.D., DeGrado W.F. Computational de novo design, and characterization of an A(2)B(2) diiron protein. J. Mol. Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Tarassov K., Messier V., Landry C.R., Radinovic S., Molina M.M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S.W. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vallone B., Miele A.E., Vecchini P., Chiancone E., Brunori M. Free energy of burying hydrophobic residues in the interface between protein subunits. Proc. Natl. Acad. Sci. USA. 1998;95:6103–6107. doi: 10.1073/pnas.95.11.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshaw J., Woolfson D.N. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 2001;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- Walshaw J., Woolfson D.N. Extended knobs-into-holes packing in classical and complex coiled-coil assemblies. J. Struct. Biol. 2003;144:349–361. doi: 10.1016/j.jsb.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Wickert U., Mucke N., Wedig T., Muller S.A., Aebi U., Herrmann H. Characterization of the in vitro co-assembly process of the intermediate filament proteins vimentin and desmin: mixed polymers at all stages of assembly. Eur. J. Cell Biol. 2005;84:379–391. doi: 10.1016/j.ejcb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Woolfson D.N. The design of coiled-coil structures and assemblies. Adv. Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- Woolfson D.N., Alber T. Predicting oligomerization states of coiled coils. Protein Sci. 1995;4:1596–1607. doi: 10.1002/pro.5560040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M.K., Leman L.J., Price D.J., Brooks C.L., III, Stout C.D., Ghadiri M.R. Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino Acid substitution. Biochemistry. 2006;45:4463–4473. doi: 10.1021/bi060092q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N.E., Zhu B.Y., Kay C.M., Hodges R.S. The two-stranded α-helical coiled-coil is an ideal model for studying protein stability and subunit interactions. Biopolymers. 1992;32:419–426. doi: 10.1002/bip.360320419. [DOI] [PubMed] [Google Scholar]