Abstract
Viral particles of human severe acute respiratory syndrome coronavirus (SARS CoV) consist of three virion structural proteins, including spike protein, membrane protein, and envelope protein. In this report, virus-like particles were assembled in insect cells by the co-infection with recombinant baculoviruses, which separately express one of these three virion proteins. We found that the membrane and envelope proteins are sufficient for the efficient formation of virus-like particles and could be visualized by electron microscopy. Sucrose gradient purification followed by Western blot analysis and immunogold labeling showed that the spike protein could be incorporated into the virus like particle also. The construction of engineered virus-like particles bearing resemblance to the authentic one is an important step towards the development of an effective vaccine against infection of SARS CoV.
Keywords: Severe acute respiratory syndrome, Coronavirus, Baculovirus, Virus-like particle
A new and deadly disease, named human severe acute respiratory syndrome (SARS), was first recognized in 2002 in the Asian region, and quickly spread to several locations across the world over the following few months. The causative agent for SARS has subsequently been identified as a novel coronavirus [1]. Coronaviruses are among the largest of the enveloped RNA viruses. SARS-associated coronavirus, SARS CoV, has a positive-strand RNA genome of 29 kb and measures about 100 nm in diameter [2], [3]. The viral envelope contains at least three structural membrane proteins, i.e., spike (S), membrane (M), and small membrane or envelope (E) proteins. The S proteins make up the characteristic “club-shaped” or “crown-like” projection of coronaviruses. The S protein is a type I integral membrane glycoprotein, containing a large extracellular domain, a short transmembrane domain, and a small cytoplasmic carboxyl-terminus [4], and is responsible for the entry of the virus into host cells. The M protein is the most abundant virion protein, which is a triple-spanning integral membrane protein with a short ectodomain and a large carboxyl-terminal endodomain [5]. The E protein, the third virion constitution, contains a hydrophobic region flanked by hydrophilic termini [6].
The SARS CoV is phylogenetically distinct from all the three coronavirus groups known today [7]. The receptor binding domain of S of SARS CoV and the receptor of the host cell have recently been identified [8], [9], which is also different from previous studies for other coronaviruses [10], [11], [12]. Thus, although previous studies on coronaviruses showed that possession of the M and E proteins is a minimal requirement for the assembly of viral particles, whereas the S protein is dispensable, but is included when present [13], [14], [15], [16], the demonstration that E and M proteins are sufficient for the assembly of SARS CoV-like particles, and these virus-like particles (VLPs) can further incorporate S proteins are still imperative knowledge and technology for rational strategy (ex. vaccine development) to combat this deadly virus. In this communication, we report the formation and isolation of SARS CoV-like particles through the co-expression of either E and M proteins or E, M, and S proteins using a baculovirus-insect cell system.
Materials and methods
Plasmids and recombinant baculoviruses. The DNA sequences coding the S, E, and M proteins of SARS CoV were obtained from the College of Medicine, National Taiwan University (GenBank Accession No. AY291451). The coding sequences for the three structural proteins were cloned into a modified transfer plasmid derived from pBacPAK8 (Clontech), under the control of a strong viral promoter, polyhedrin promoter. This transfer plasmid also contains a red fluorescent reporter gene, DsRed2 (Clontech), driven by a heat-shock promoter. To generate recombinant baculoviruses using Autographa californica multiple nucleopolyhedrovirus (AcMNPV) viral genome, the individual plasmids containing S, E, and M proteins were co-transfected with linearized viral DNA (BaculoGold, BD Biosciences) by using Lipofectin (Invitrogen) into insect cells, successful recombinants were isolated by the indication of red fluorescence. The recombinant baculoviruses encoding the S, E, and M proteins, assigned as vABhRpS, vABhRpE, and vABhRpM, respectively, were obtained by two- or three-round serial dilutions, and all viral stocks were prepared and manipulated according to the standard protocol described by O’Reilly et al. [17].
Insect cell culture. The Spodoptera frugiperda IPLB-Sf21 (Sf21) insect cell line was cultured as a monolayer in TNM-FH insect medium, containing 8% heat-inactivated fetal bovine serum as described previously [18]. It was used for the propagation and infection of recombinant baculoviruses; titers of viruses were determined by a newly developed quantitative real-time PCR-based method [19].
Isolation of coronavirus-like particles. Insect cells were co-infected with vABhRpE and vABhRpM at a multiplicity of infection (moi) of 1:5. At 4 days post-infection (dpi), cells were collected by a cell scraper (Costar) and then re-suspended in Tris-buffered saline (TBS), containing a cocktail of protease inhibitor (1:1000 dilution, SET III, Calbiochem), and lysed by sonication. The post-nuclear supernatant (PNS) was obtained by centrifugation at 1000 rpm for 10 min, and was then placed on a 30% (w/w) sucrose cushion for centrifugation at 34,000 rpm for 20 h. Pellets were washed twice with TBS and re-suspended in the same buffer, and then subjected to a 20–60% (w/w) sucrose gradient at 34,000 rpm for 60 h. Thirty fractions, monitored at a wavelength of 280 nm, were extracted from the centrifuged sucrose gradient using a density gradient fractionation system (ISCO). The fraction containing VLPs was subsequently examined by Western blot analysis and by electron microscope as described below.
Western blot analysis. Each fraction of sucrose gradient was analyzed by an 8% and 14% of SDS–PAGE, respectively; the separated proteins were transferred onto a PVDF membrane (Immobilin-P, Millipore). The membrane was blocked with 5% non-fat milk for 1 h and then probed (at a dilution of 1:5000) by antiserum at 4 °C overnight. The antiserum used to probe S protein was developed by immunizing rabbits with the amino acid fragment 510–1195 of spike protein (a gift from Dr. Pei-Jer Chen, Graduate School of Clinical Medicine, College of Medicine, National Taiwan University), whereas those used to probe E protein were raised by immunizing rabbits with synthetic oligopeptides (LVKPTVYVYSRVKNL, C-terminal of E protein, Genesis Biotech). After three washes with 0.1% Tween 20 containing phosphate-buffered saline (TPBS), a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG was added at a dilution ratio of 1:2500, incubating at room temperature for 1 h. The blot was then washed with TPBS four times, followed by visualization using chemiluminescent reagent (Western Lightning, Perkin–Elmer) and developed on an X-ray film (BioMax, Kodak).
Characterization of VLPs by electron microscope. Ultra-structure of cells co-infected with recombinant baculoviruses was collected, fixed, and then visualized by electron microscopy as described in our previous studies [20]. To visualize purified VLPs using transmission electron microscopy with negative staining, an aliquot of 10 μl of the VLP preparation was loaded onto a carbon-coated grid, letting standstill for 5 min. Grids were then stained with 2% of phosphotungstic acid (PTA) for 1 min; after excess PTA was drained and the grid was examined directly under electron microscope (EM). For immunogold labeling, VLPs were loaded onto a collodion-coated EM grid for 5 min. After the removal of excess of sample solution by gently blotting with a filter paper at the edge of the grid, an antibody specific against S protein (developed by immunizing rabbits with synthetic peptides corresponding to amino acids 19–35 of S protein; IMG-541, Imgenex) was added onto the grid and incubated for 1 h at room temperature. Grids then underwent 10 s wash six times in Sorensen’s phosphate buffer at room temperature and were incubated with 12 nm gold conjugated anti-rabbit IgG for 1 h. After six 10 s washes in Sorensen’s phosphate buffer, the samples were stained with 2% PTA for 1 min, then drained, and examined under the EM.
Results and discussion
Formation of VLPs by co-infection with recombinant baculoviruses expressing the E and M proteins in insect cells
To express E, M, and S proteins of SARS CoV in baculovirus-insect system, we first cloned the individual sequences into three different plasmids, under control of a strong viral promoter, polyhedrin promoter (see Materials and methods). Recombinant baculoviruses encoding E, M, and S protein genes were used to infect Sf21 insect cell, and the expression of each protein was checked by Western blot at 4 dpi (data not shown).
To study if VLPs are able to form in insect cells, we first used vABhRpE and vABhRpM to co-infect Sf21 cells at different moi (i.e., E:M=1:10–10:1), and examined these co-infected cells by electron microscopy (data not shown). We found the formation of VLPs was satisfactory at moi of 1:5. Fig. 1A shows a typical electron micrograph of a cell co-infected with recombinant baculoviruses expressing both E and M proteins, in which the boxed region was further magnified in Fig. 1B. The VLPs were observed in a large lightly stained vesicle (solid arrow). A higher magnification showed that several heavily stained smaller vesicles with VLPs were also found (open arrows) near the lightly stained vesicle. These lightly and darkly stained structures are similar to the electron microscopic studies of a specimen from SARS patient [21]. The distribution of VLPs in the baculovirus-insect cell system was also consistent with a previous study demonstrating the assembly of hepatitis C virus-like particles in insect cells [22]. In Figs. 1C and D, the cytoplasm of a cell is covered with VLPs, some of which are arbitrarily indicated by arrowheads. This result suggested that VLP-containing vesicles (open arrow in (B)) were disrupted and VLPs were released all over in the cytoplasm in some cells.
Fig. 1.
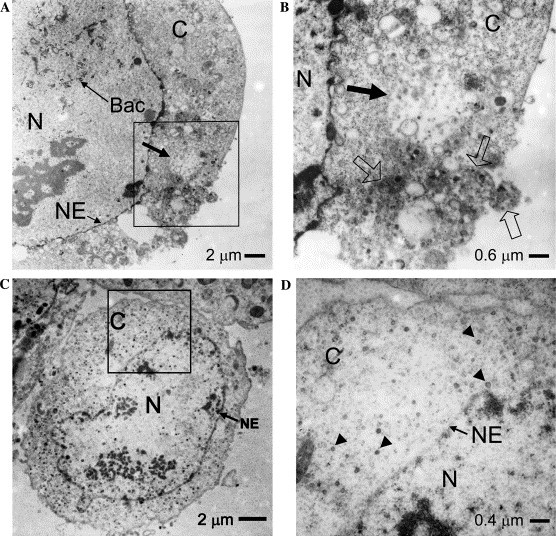
Electron microscopic analysis of VLPs in insect cells. (A) The formation of VLPs in the cytoplasm of an insect cell upon co-infection with E- and M-expressing recombinant baculoviruses. Bundles of baculoviruses were found in the nucleus of the cell (indicated by Bac). C, cytoplasm; N, nucleus; and NE, nuclear envelope. Bar=2 μm. (B) The boxed region of (A) was further magnified to show the formation of VLPs in a vesicle (solid arrow). Smaller dense vesicles filled with VLPs were also found in this region (open arrow). Bar=0.6 μm. (C) Extensive cytoplasmic spreading of VLPs in a cell. Bar=2 μm. (D) Higher magnification of the boxed region in (C). Some of VLPs were arbitrarily indicated by arrowheads. Bar=0.4 μm.
Previously, it was shown that E protein is not essential for the replication of murine coronavirus. However, a virus with E protein mutation gave rise to smaller plaques with at least 3 orders of magnitude lower titer than those typically observed for the wild type virus [6]. In our study, baculovirus which only expresses M protein was used to infect insect cells. Electron microscopic analysis showed that VLPs were not found in the infected insect cells (data not shown). This result suggests that E protein is required for the efficient formation of VLP of SARS CoV, although, we cannot rule out the possibility that VLP may still be formed in the cells with significantly reduced numbers and thus difficult to detect.
The S protein was located at the periphery of VLPs
The S protein is the major antigen determinant site and the protein responsible for the recognition by host cells. In order to determine if S protein could be recruited into VLPs for further usage as vaccine or delivery tool to target cells of the SARS CoV, insect cells were co-infected with vABhRpS, vABhRpE, and vABhRpM at an moi of 1:0.5:5, respectively. After harvesting the infected cells, cell lysate was purified through a sucrose gradient centrifugation to isolate VLPs. Aliquots of sucrose gradients (450 μl each) were fractionated, and the chromatograph was monitored at 280 nm (Fig. 2A ). The absorbance reached a peak at tube 17, by which the density was calculated as 1.2 mg/ml, corresponding to 43% (w/w) of sucrose.
Fig. 2.
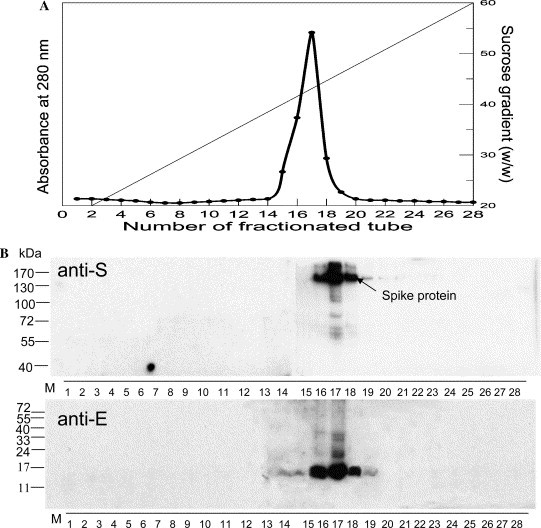
Isolation and identification of VLPs. The VLPs were isolated from cells co-expressing S, E, and M proteins. After sucrose gradient centrifugation, 28 fractions were collected and their chromatograph was shown in (A). Fractionation was monitored with a wavelength of 280 nm; the dotted line represents weight percentage of sucrose. (B) Equal amounts of gradient fractions were separately resolved in 8% (anti-S) and 14% (anti-E) of polyacrylamide gels, followed by Western analyses. Numbers on the left sides of the sub-panels are the molecular weights of protein markers (M). Western analysis confirmed the presence of S and E proteins mainly in fractions 16–18.
The contents of centrifuged fractions were analyzed by 8% and 14% SDS–PAGE, respectively, and then analyzed by Western blot. The presence of S and E proteins was evident in fractions 16–18 (Fig. 2B). These results implicated that VLPs contained S protein. To verify the incorporation of S protein onto VLPs, fractionated tube 17 was further analyzed by electron microscopy. The isolated VLPs were observed by negative staining in electron microscope (Fig. 3 ), whereas the location of S protein was depicted by immunogold labeling using the S protein-specific antibody. As shown in Fig. 3, the VLP, ca. 110 nm in diameter, was specifically labeled with gold particles. Our Western and electron microscopic analyses accordingly demonstrated that the S protein can be incorporated into the VLP and is located to the periphery of VLP.
Fig. 3.
Ultrastructural characterization of VLPs. (A) VLPs purified by sucrose gradient centrifugation were analyzed by negative staining. (B) Immunogold labeling of purified VLP. A baculovirus particle (Bac) was shown to serve as a negative control. VLP, virus-like particle.
By Western blot analysis, we were able to detect a notable band corresponding to a molecular weight of about 160 kDa, which is larger than the predicted molecular weight of S protein amino acid sequence (ca. 139 kDa) by ∼20 kDa. The difference could be attributed to the extent of glycosylation since there are about 15 potential N-glycosylated asparagine residues in S protein (data not shown). Taking advantage of post-translational modifications, baculovirus-insect cell system can be used to generate functional spike proteins, supposedly in an oligomeric form on the surface of the VLPs, to raise neutralizing antibodies in the human body to combat SARS CoV infection.
To obtain protective immunity at maximum efficiency, the ideal immunogens used to develop vaccines should elicit immunity, resemble the pathogenic viruses, and be non-pathogenic. VLPs closely fit an ideal immunogen. One promising application for the production of non-pathogenic VLPs is to act as most effective immunogens to induce protective immunity and to develop immunogen-based assays [23], [24], [25]. Another possible application is to define the possible packaging signal of SARS CoV. The packaging signal of coronavirus RNA was previously identified by using defective interfering viral particles [26], since the SARS-CoV is a highly infectious virus, it would be much more feasible to identify this signal by using VLPs. Moreover, S protein-containing VLPs should be able to specifically target to the host cell of the SARS CoV and serve as a safe and efficient tool to deliver potential therapeutic agents (e.g., siRNA) for effective treatment of SARS in the future.
Acknowledgements
We thank Dr. Pei-Jer Chen for providing genes and Dr. S.H. Yeh for providing antiserum against S protein, which are necessary for these experiments. We also thank Drs. Michael M. Lai and Pei-Jer Chen for their comments. This work was supported by grants from the National Science Council (NSC92-2751-B-001-001-Y) and Academia Sinica (IMB 3/15 and AS93-AB-IMB-01).
References
- 1.Kuiken T, Fouchier R.A, Schutten M, Rimmelzwaan G.F, van Amerongen G, van Riel D, Laman J.D, de Jong T, van Doornum G, Lim W, Ling A.E, Chan P.K, Tam J.S, Zambon M.C, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra J.C, Stohr K, Peiris J.S, Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 3.Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4.Godeke G.J, de Haan C.A, Rossen J.W, Vennema H, Rottier P.J. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 2000;74:1566–1571. doi: 10.1128/jvi.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan C.A, Kuo L, Masters P.S, Vennema H, Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo L, Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eickmann M, Becker S, Klenk H.D, Doerr H.W, Stadler K, Censini S, Guidotti S, Masignani V, Scarselli M, Mora M, Donati C, Han J.H, Song H.C, Abrignani S, Covacci A, Rappuoli R. Phylogeny of the SARS coronavirus. Science. 2003;302:1504–1505. doi: 10.1126/science.302.5650.1504b. [DOI] [PubMed] [Google Scholar]
- 8.Wong S.K, Li W, Moore M.J, Choe H, Farzan M. A 193-amino-acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2003 doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Moore M.J, Vasilieva N, Sui J, Wong S.K, Berne M.A, Somasundaran M, Sullivan J.L, Luzuriaga K, Greenough T.C, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonavia A, Zelus B.D, Wentworth D.E, Talbot P.J, Holmes K.V. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan K, Zelus B.D, Meijers R, Liu J.H, Bergelson J.M, Duke N, Zhang R, Joachimiak A, Holmes K.V, Wang J.H. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 2002;21:2076–2086. doi: 10.1093/emboj/21.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultze B, Herrler G. Recognition of cellular receptors by bovine coronavirus. Arch. Virol. Suppl. 1994;9:451–459. doi: 10.1007/978-3-7091-9326-6_44. [DOI] [PubMed] [Google Scholar]
- 13.Vennema H, Godeke G.J, Rossen J.W, Voorhout W.F, Horzinek M.C, Opstelten D.J, Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bos E.C, Luytjes W, van der Meulen H.V, Koerten H.K, Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corse E, Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Reilly D.R, Miller L.K, Luckow V.A. Oxford University Press; New York: 1994. Baculovirus Expression Vectors: A Laboratory Manual. [Google Scholar]
- 18.Lee J.C, Chen H.H, Chao Y.C. Persistent baculovirus infection results from deletion of the apoptotic suppressor gene p35. J. Virol. 1998;72:9157–9165. doi: 10.1128/jvi.72.11.9157-9165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo H.R, Chao Y.C. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 2004;20:354–360. doi: 10.1021/bp034132i. [DOI] [PubMed] [Google Scholar]
- 20.Lin J.L, Lee J.C, Chen S.S, Wood H.A, Li M.L, Li C.F, Chao Y.C. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J. Virol. 1999;73:128–139. doi: 10.1128/jvi.73.1.128-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ksiazek T.G, Erdman D, Goldsmith C.S, Zaki S.R, Peret T, Emery S, Tong S, Urbani C, Comer J.A, Lim W, Rollin P.E, Dowell S.F, Ling A.E, Humphrey C.D, Shieh W.J, Guarner J, Paddock C.D, Rota P, Fields B, DeRisi J, Yang J.Y, Cox N, Hughes J.M, LeDuc J.W, Bellini W.J, Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 22.Baumert T.F, Ito S, Wong D.T, Liang T.J. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touze A, Enogat N, Buisson Y, Coursaget P. Baculovirus expression of chimeric hepatitis B virus core particles with hepatitis E virus epitopes and their use in a hepatitis E immunoassay. J. Clin. Microbiol. 1999;37:438–441. doi: 10.1128/jcm.37.2.438-441.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang J, Wunschmann S, George S.L, Klinzman D, Schmidt W.N, LaBrecque D.R, Stapleton J.T. Recombinant hepatitis C virus-like particles expressed by baculovirus: utility in cell-binding and antibody detection assays. J. Med. Virol. 2002;68:537–543. doi: 10.1002/jmv.10237. [DOI] [PubMed] [Google Scholar]
- 25.Noad R, Roy P. Virus-like particles as immunogens. Trends Microbiol. 2003;11:438–444. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 26.Kim K.H, Narayanan K, Makino S. Characterization of coronavirus DI RNA packaging. Adv. Exp. Med. Biol. 1998;440:347–353. doi: 10.1007/978-1-4615-5331-1_45. [DOI] [PubMed] [Google Scholar]


