Summary
Severe acute respiratory syndrome (SARS) is caused by a newly emerged coronavirus that infected more than 8000 individuals and resulted in more than 800 fatalities in 2003. Currently, there is no effective treatment for this epidemic. SARS-3CLpro has been shown to be essential for replication and is thus a target for drug discovery. Here, a class of stable benzotriazole esters was reported as mechanism-based inactivators of 3CLpro, and the most potent inactivator exhibited a kinact of 0.0011 s−1 and a Ki of 7.5 nM. Mechanistic investigation with kinetic and mass spectrometry analyses indicates that the active site Cys145 is acylated, and that no irreversible inactivation was observed with the use of the C145A mutant. In addition, a noncovalent, competitive inhibition became apparent by using benzotriazole ester surrogates in which the bridged ester-oxygen group is replaced with carbon.
Keywords: MICROBIO, HUMDISEASE, CHEMBIO
Introduction
Severe acute respiratory syndrome (SARS), a newly emerged infectious disease, first occurred in Guandong (China) in November of 2002 and spread through many countries in 2003; it reemerged in China in December 2003 and in the spring of 2004. This disease is caused by infection with a novel human coronavirus (SARS-CoV) 1, 2, 3, which affected more than 8000 individuals across 32 countries and resulted in more than 800 fatalities in 2003 4, 5, 6. The origin of SARS-CoV is unclear, though studies on the molecular evolution of SARS-CoV indicate that the virus may have emerged from nonhuman species [7]. At present, no efficacious therapy for SARS is available. Therefore, a search for effective antivirals for the SARS-CoV is of current interest.
SARS coronavirus is a positive-strand RNA virus that encodes two polyproteins, pp1a and pp1ab 8, 9, 10, for further proteolytic processing to provide the functional proteins for viral propagation. These processes are mediated primarily by the main protease (Mpro), known as dimeric chymotrypsin-like protease (3CLpro) 11, 12, 13. The active site of 3CLpro contains Cys145 and His41, constituting a catalytic dyad in which the cysteine thiol functions as the nucleophile in the proteolytic process 11, 12, 13. Due to its essential role in viral replication, the protease is an attractive target for the development of therapeutics against SARS.
So far, only a few inhibitors of 3CLpro have been reported, including the HIV protease inhibitor TL-3 [14], zinc-conjugated compounds [15], aryl boronic acids analogs [16], a quinolinecarboxylate derivative [17], a thiophenecarboxylate [18], phthalhydrazide-substituted keto-glutamine analogs [19], and anilides [20]. However, none of these inhibitors exhibits activities in the low nanomolar range, and no preclinical studies were reported.
As a part of our efforts directed toward the development of potent anti-SARS agents, we report here the discovery of a new class of mechanism-based irreversible inactivators with inhibition constants in the nanomolar range, by using the strategy of combinatorial reaction in microtiter plates followed by screening in situ 21, 22, 23. This approach relies on the use of high-yield organic reactions that can be carried out in water or water-miscible, nontoxic solvents on microscales without protecting groups, allowing the product to be assayed directly in situ without isolation and purification. Using this approach, one can quickly modify a lead compound with a small set of building blocks to identify an optimal inhibitor.
Results and Discussion
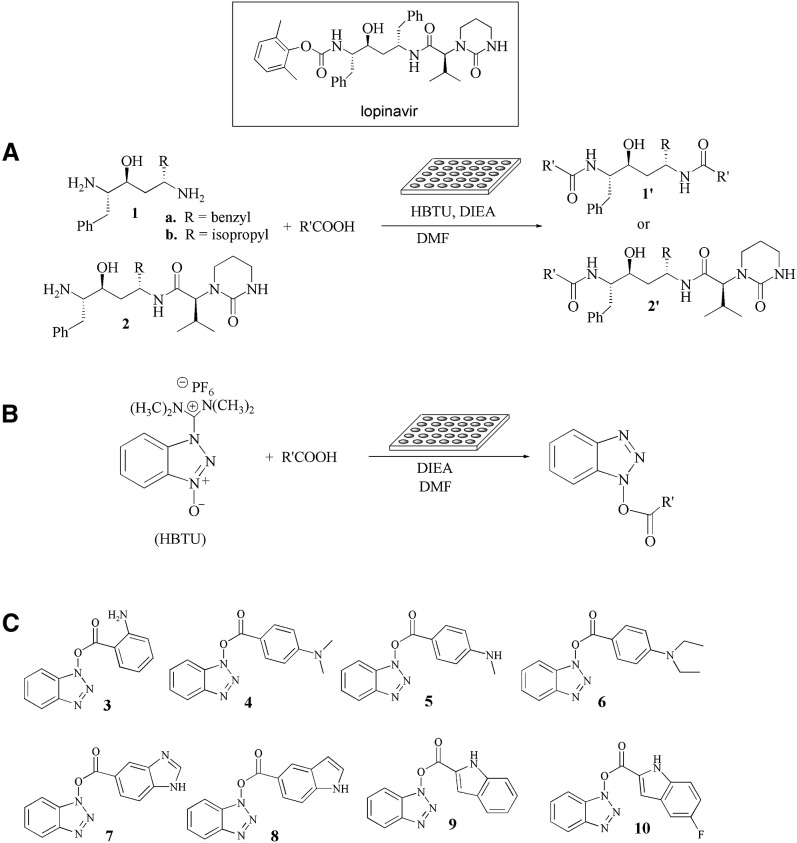
Previously [14], we have reported that the HIV protease inhibitor Lopinavir ( Figure 1) also inhibits 3CLpro with an IC50 of ∼50 μM. In order to find more potent 3CLpro inhibitors, a library of Lopinavir-like compounds was assembled by using either diamine 1 or amine 2 as the core structure [24] for reaction with various acids in microtiter plates followed by screening in situ (Figure 1). In a typical procedure 21, 22, 23, a library of 90 carboxylic acids (see the Supplemental Data available with this article online) (10 μmol each) in a microtiter plate was used to couple with amine 1 or 2 (10 μmol) in the presence of N,N-diisopropylethylamine (22 μmol) and 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, 11 μmol) in DMF (100 μl) for 4 hr. An aliquot of the product, based on a putative 100% conversion of starting materials, was taken and subjected to the 3CLpro inhibition assay in a 20 mM Bis-Tris buffer (pH 7.0) at 25°C. To obtain the IC50 values, the initial velocities of the inhibited reactions with 50 nM of the protease and 6 μM of a fluorogenic substrate were plotted against different inhibitor concentrations. In this study, we noticed that the products derived from 2-aminobenzoic acid (well C9, IC50 = 0.2 μM), 4-(methylamino)benzoic acid (well D5, IC50 = 0.3 μM), 4-(dimethylamino)benzoic acid (well D6, IC50 = 0.5 μM), and 4-(diethylamino)benzoic acid (well D7, IC50 = 0.5 μM) showed the best inhibition and that the degree of inhibition was independent of the amine used.
Figure 1.
Microtiter Plate-Based Reactions and In Situ Screening
(A) Reactions of 1 or 2 with 90 acids in microtiter plates, followed by in situ screening for inhibitors of SARS-CoV 3CLpro.
(B) Reactions of HBTU and 90 acids in microtiter plates, followed by in situ screening for inhibitors of SARS-CoV 3CLpro.
(C) Molecular structures of inhibitors 3–10 against SARS-CoV 3CLpro.
In order to characterize the inhibitors, we attempted to separately prepare the pure amide derivatives, but we found that the amide formation was very slow, and that the intermediates benzotriazole esters 3−6 were isolated as major products from silica gel columns (the X-ray ORTEP structure of compound 4 is shown in the Supplemental Data). To our surprise, all of the Lopinavir-like compounds showed only modest inhibitory activities toward 3CLpro (IC50 ≥ 10 μM), whereas benzotriazole esters 3−6 showed high inhibition activities. To our knowledge, benzotriazole esters have been used as acylating agents and have never been found to be enzyme inhibitors. We then prepared a series of benzotriazole esters by condensation of HBTU with various carboxylic acids (Figure 1B), and we found that the benzotriazole esters derived from benzoic acid containing electron-withdrawing substituents, e.g., NO2, CN, and CF3, were susceptible to hydrolysis, whereas benzotriazole esters 3–6 and those with electron-donating groups were relatively stable in pH 5.0–8.0 over 24 hr at room temperature. The relative stability of each derivative depends on the pKa of the corresponding benzoic acid and basically follows the Hammett equation 25, 26, 27. These stable benzotriazole esters, esters 3–10 (Figure 1C), were assayed, and their inhibition results against 3CLpro are shown in Table 1.
Table 1.
IC50, Ki, kinact, and CC50 of Benzotriazole Esters
| Compounds | Ki (nM) | kinact (s−1) × 103 | kinact/Ki (M−1s−1) × 10−3 | CC50 (μM) |
|---|---|---|---|---|
| 3 | 19.5 | 1.6 | 82.0 | >100 |
| 4 | 17.4 | 1.3 | 74.7 | >100 |
| 5 | 12.1 | 0.9 | 74.4 | >100 |
| 6 | 11.1 | 0.8 | 72.1 | >100 |
| 7 | 22.9 | 1.1 | 48.0 | >100 |
| 8 | 7.5 | 1.1 | 146.7 | >100 |
| 9 | 12.3 | 0.9 | 73.2 | >100 |
| 10 | 13.8 | 1.2 | 86.9 | >100 |
CC50: The concentration that causes 50% cytotoxicity.
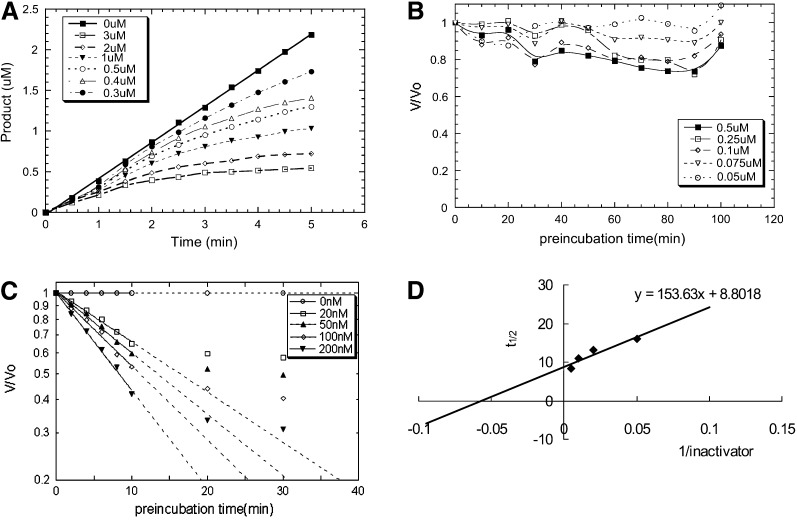
Further study of the inhibition of benzotriazole esters 3–10 showed that there was a time-dependent decrease in enzyme activity as a function of the inhibitor concentration (e.g., the inhibition of compound 4) ( Figure 2A). In the presence of 1 mM DTT (dithiothreitol), the preincubation of enzyme with inhibitor did not affect the enzyme activity, indicating that DTT can protect the enzyme from inactivation (Figure 2B). These experiments indicate an irreversible mode of action and point to the active site Cys being involved in the inactivation process. The kinetic results for k inact and K i determinations of compound 4 are shown in Figures 2C and 2D (the detailed procedures are shown in the Supplemental Data). We used the initial rate of inactivation (0–10 min) showing pseudo first-order kinetics to determine the inhibition constants. The enzyme activity is, however, not reduced to zero over the time period, perhaps because of the decomposition of the thioester intermediate or the existence of two populations of the enzymes. As shown in Table 1, benzotriazole esters 3–10 are strong inhibitors, and, among these, 8 is the most potent, with an inactivation constant of 1.1 × 10−3 s−1 and an inhibition constant of 7.5 nM. In addition, these esters are not toxic to Vero E6 cells, which are often used in the cell-based assay for SARS-CoV [14], at a concentration of 100 μM. Compound 8 represents the most potent mechanism-based 3CLpro inhibitor reported to date.
Figure 2.
Kinetic Studies of Inhibitor 4 and SARS-CoV 3CLpro
(A) The progress curves in the presence of 0.3–3.0 μM inhibitor for reactions initiated by adding enzyme (final concentration of 0.05 μM) into a mixture of substrate (6 μM) and inhibitor 4. Over the entire 5 min time window, the uninhibited enzyme displayed a linear progress curve, whereas the inhibited enzyme with a different concentration of inhibitor showed a time-dependent reduction of activity.
(B) The same experiments as performed in (A), but with 1 mM DTT in the preincubation mixture.
(C) Preincubation time dependence of the fractional velocity of the protease-catalyzed reaction in the presence of 0.02–0.2 μM time-dependent inhibitor 4.
(D) Kitz and Wilson replot of the half-life (t1/2) of enzyme inactivation as a function of the reciprocal of the slow inactivator concentration. The kinact is 0.0013 s−1 and Ki is 17.4 nM for the time-dependent inactivator 4 based on the kinetic data.
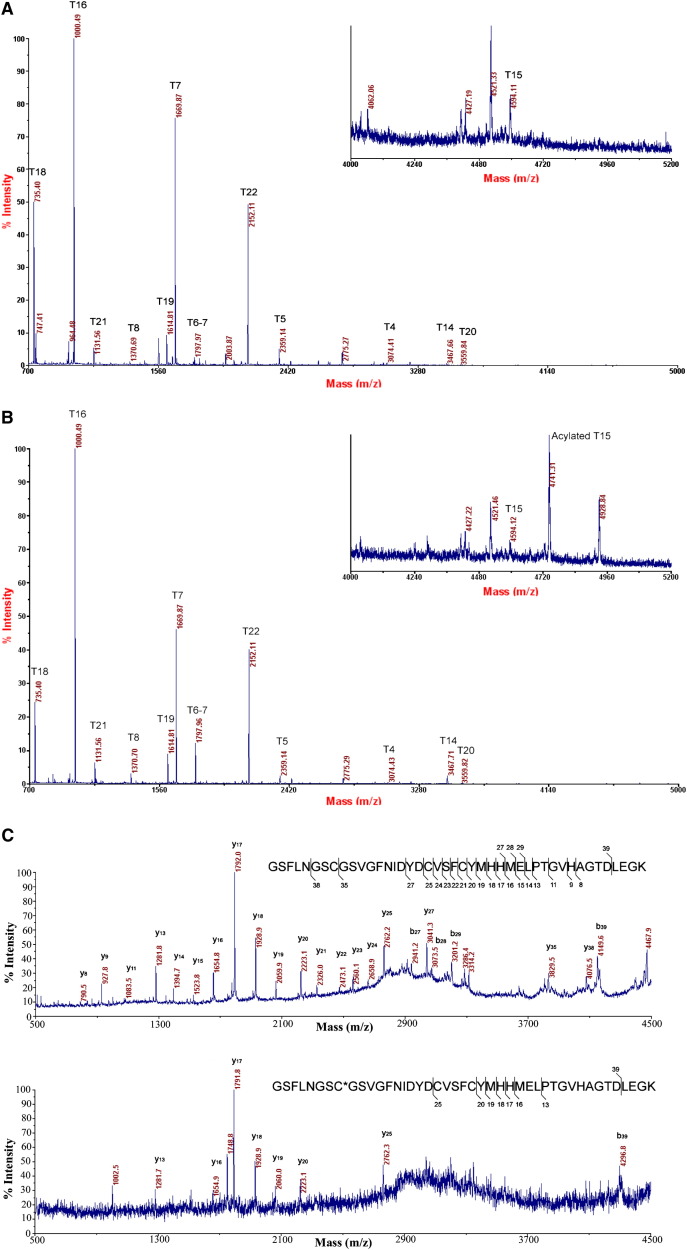
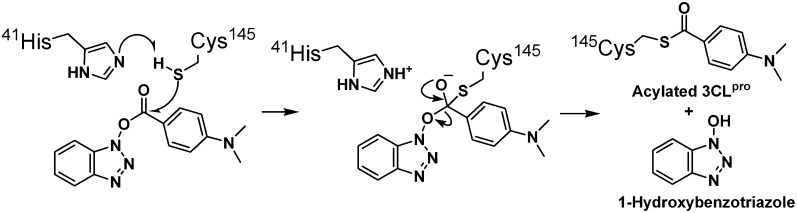
We also investigated the nature of the inhibited enzyme by mass spectrometry. Electrospray ionization mass spectra of wild-type 3CLpro and 3CLpro treated with 4-(dimethylamino)benzoyl ester 4 (2 hr of incubation and 18 hr of dialysis) and their deconvoluted mass spectra were determined (see the Supplemental Data). The mass difference of 148.6 Da between the peaks of 33847.35 ([M + H]+) and 33995.93 ([M + H]+) implies the acylation of 3CLpro with a 4-(dimethylamino)benzoyl moiety (mass 147.12). To further investigate the acylation site, MALDI-TOF mass spectrometric analysis of the trypsin digest of 3CLpro and 3CLpro treated with 4-(dimethylamino)benzoyl ester 4 was performed. From the MALDI spectra of the tryptic 3CLpro and the tryptic acylated 3CLpro of G138-K180 peptide fragments, a mass shift of 147 Da between T15 (4594.11 Da) and acylated T15 (4741.31 Da) indicates that this peptide fragment contains an acylated residue ( Figures 3A and 3B). In order to determine the acylation site on the acylated T15 peptide, we performed a sequence analysis by MALDI MS/MS for peptides T15 and acylated T15 as depicted in Figure 3C. There is no mass difference among y series fragment ions up to y27, but a mass shift of 147.2 Da (4149.6 Da versus 4296.8 Da) on b39 clearly shows that Cys145 is the only acylation site. This is consistent with the observation of no mass shift of mutant C145A 3CLpro treated with 4-(dimethylamino)benzoyl ester 4. All of these results support the mechanism of irreversible inhibition of 3CLpro by 4-(dimethylamino)benzoyl ester 4 via acylation of Cys145 ( Figure 4).
Figure 3.
MALDI Spectrum of Inhibitor 4 and SARS 3CLpro
(A) MALDI spectrum of tryptic 3CLpro.
(B) MALDI spectrum of tryptic acylated 3CLpro.
(C) MALDI MS/MS spectra of T15 and acylated T15 peptides (GSFLNGSC∗GSVGFNIDYDCVSFCYMHHMELPTGVHAGTDLEGK) showed a mass shift of 147.2 Da (4149.6 Da versus 4296.8 Da) on b39, indicating that Cys145 (C∗) is the acylation site.
Figure 4.
Proposed Mechanism for Inhibition of SARS-CoV 3CLpro by Acylation with Benzotriazole Esters
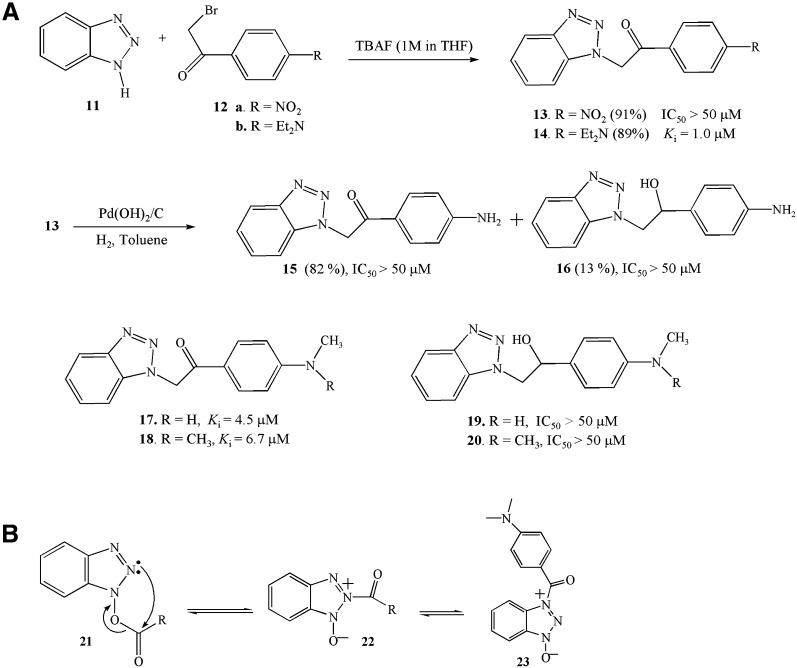
In order to develop stable, noncovalent inhibitors based on the benzotriazole esters discovered in this study, we synthesized compounds 13 and 14 by using TBAF-assisted N-alkylation of 1H-benzotriazole 11 28, 29. In addition, compound 13 was hydrogenated with Pd(OH)2/C as a catalyst at room temperature to obtain compounds 15 (82%) and 16 (13%). Compounds 15 and 16 were methylated or dimethylated to compounds 17–20 by using TBAF as a reagent ( Figure 5A). Compounds 13–20 have all the features of benzotriazole ester inhibitors, except that the ester oxygen is replaced by a carbon. Inhibition analysis shows that compounds 13–20 are noncovalent competitive inhibitors, albeit relatively weak ones compared to the corresponding esters; of these, compound 14 is most potent, with a K i of 1.0 μM. Reduction of the carbonyl group, however, results in a significant loss of activity (Figure 5A). It is noted that the benzotriazole compounds contain three equilibrium structures in solution (Figure 5B) 25, 26, and that compounds 13–20 may mimic the ester forms instead of the three equilibrium forms found in solution.
Figure 5.
Synthesis of Noncovalent Inhibitors
(A) Synthesis of compounds 13–20 and their IC50s or Kis for SARS-CoV 3CLpro.
(B) The equilibrium structures of the benzotriazole compounds in solution.
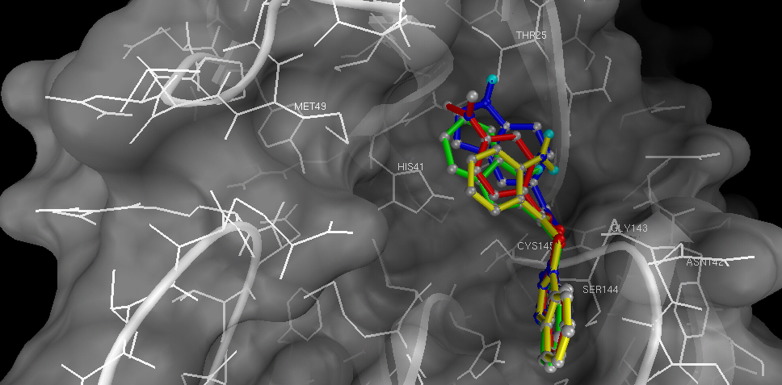
To gain further insight into the mode of inhibition, a docking experiment based on computer modeling (Autodock version 3.0.5) [30] for the binding of compounds 3, 4, 8, and 9 with 3CLpro (1uk4) [12] was carried out, and the result indicated that the benzotriazole moiety was disposed in the pocket formed by Cys145, Ser144, and Gly143 in the active site ( Figure 6). The γ-S atom of Cys145 was close enough, within 3.5 Å in a rigid model, to the carbonyl group of the benzotriazole ester to render a nucleophilic attack. The aminophenyl group (for 3 and 4) and the indole moiety (for 8 and 9) were in the region surrounded by Thr25, Thr26, His41, Thr45, Ala46, and Met49. The NH group of the indole moiety of 8 was hydrogen bonded with the side chain OH of Thr25. In comparison, the indole group of 9 lacks such hydrogen bonding and thus shows a weaker affinity toward 3CLpro than does isomer 8. Compounds 13–20 do not fit into the pocket well enough to interact with the residues mentioned above, and the calculated minimal energies of binding are ∼0.5–1.0 kcal/mol higher than those of the corresponding esters, consistent with the inhibition assay result (see the Supplemental Data).
Figure 6.
A Modeling Complex of SARS 3CLpro and Benzotriazole Esters
Binding modes of compounds 3 (yellow), 4 (red), 8 (blue), and 9 (green) in the active site of SARS-CoV 3CLpro (PDB luk4). Models were generated by Autodock and displayed by MGLTOOLS (MGL, Scripps).
Significance
Severe acute respiratory syndrome (SARS) is a newly emerged disease caused by a novel human coronavirus. Currently, no effective antiviral agents exist against this deadly epidemic. The main protease of SARS-CoV, 3CLpro, is an attractive target for drug discovery due to its essential role in viral replication. We have discovered several stable benzotriazole esters as a new class of irreversible enzyme inhibitors, and, to our knowledge, these compounds are the most potent mechanism-based 3CLpro inhibitors known to date. The mode of action has been studied and has been shown to proceed through acylation of the active site Cys145 assisted by the catalytic dyad.
Experimental Procedures
Materials and Methods
SARS-CoV 3CL protease was prepared according to the previously described procedure [31]. Reactions requiring dry conditions were carried out under an inert atmosphere by using standard techniques. All of the reagents and solvents were reagent grade and were used without further purification unless otherwise specified. THF was distilled from sodium benzophenone ketyl under N2. HRMS values were obtained by using the EI as the ionization source.
General Procedure for the Preparation of Benzotriazole Esters
To a solution of acid compound (1.0 equiv.), 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, 1.1 equiv) in DMF (1.0 ml), as well as N,N-diisopropylethylamine (DIEA, 1.1 equiv.), was added. After the solution was stirred at 25°C for 3.0 hr, the reaction mixture was added to 1 M NaHCO3 (10 ml) and extracted with ethyl acetate (EA) (15 ml × 3), and the organic layer was collected and concentrated under reduced pressure. The residue was purified by use of column chromatography on silica gel to provide the desired 1-hydroxybenzotriazole esters.
2-Aminobenzoic Acid Benzotriazol-1-yl Ester, 3
The standard procedure was followed by use of 2-aminobenzoic acid (C9, 34.8 mg, 0.2537 mmol, 1.0 equiv.), HBTU (105.9 mg, 0.2791 mmol, 1.1 equiv.), and DIEA (49 μl, 0.2791 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (5% MeOH in chloroform as eluant). Benzotriazole ester 3 (57.4 mg, 0.2258 mmol) was obtained in 89% yield as a yellow solid: TLC Rf = 0.81 (5% MeOH in CHCl3 as eluant); 1H NMR (CDCl3, 400 MHz) δ = 5.31 (brs, 2 H, NH2), 6.73–6.78 (m, 2 H, 2 × ArH), 7.41 (t, 2 H, J = 6.0 Hz, 2 × ArH), 7.46 (d, 1 H, J = 6.6 Hz, ArH), 7.52 (t, 1 H, J = 6.0 Hz, ArH), 8.06 (d, 2 H, J = 6.7 Hz, ArH), 8.14 (d, 1 H, J = 6.7 Hz, ArH); 13C NMR (CDCl3, 100 MHz) δ = 104.26, 108.45, 116.82, 116.98, 120.42, 124.78, 128.66, 129.00, 130.84, 136.71, 143.50, 152.23, 163.65; IR (KBr) 3456 (m, NH), 3322 (m, NH), 1736 (s, C=O), 1632 (s), 1566 (m), 1485 (s), 1372 (m), 1228 (s), 1153 (s), 977 (s), 740 (s) cm−1; HRMS [M + 1] calcd for C13H11N4O2: 255.0875, found 255.0882.
4-Dimethylamino-Benzoic Acid Benzotriazol-1-yl Ester, 4
The standard procedure was followed by use of (dimethylamino)benzoic acid (D6, 120.3 mg, 0.7283 mmol, 1.0 equiv.), HBTU (303.8 mg, 0.8011 mmol, 1.1 equiv.), and DIEA (140 μl, 0.8011 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 4 (191.2 mg, 0.6773 mmol) was obtained in 93% yield as a light-yellow solid: TLC Rf = 0.50 (CHCl3 as eluant); 1H NMR (CDCl3, 400 MHz) δ = 3.13 (s, 6 H, 2 × CH3), 6.74 (d, J = 8.9 Hz, 2 H, 2 × ArH), 7.42 (t, J = 6.7 Hz, 1 H, ArH), 7.47–7.55 (m, 2 H, 2 × ArH), 8.07–8.12 (m, 3 H, 3 × ArH); 13C NMR (CDCl3, 100 MHz) δ = 40.01, 108.64, 110.02, 111.06, 120.35, 124.54, 128.36, 129.11, 132.73, 143.57, 154.38, 163.51; IR (KBr) 2914 (w), 1768 (s, C=O), 1609 (s), 1537 (s), 1441 (s), 1386 (s), 1263 (s), 1183 (s), 957 (s), 770 (s) cm−1; HRMS [M + 1] calcd for C15H15N4O2: 283.1195, found 283.1199.
4-Methylamino-Benzoic Acid Benzotriazol-1-yl Ester, 5
The standard procedure was followed by use of (4-methylamino)benzoic acid (D5, 53.5 mg, 0.3539 mmol, 1.0 equiv.), HBTU (147.7 mg, 0.3894 mmol, 1.1 equiv.), and DIEA (68 μl, 0.3894 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 5 (88.3 mg, 0.3291 mmol) was obtained in 93% yield as a white solid: TLC Rf = 0.28 (CHCl3 as eluant); 1H NMR (CDCl3, 400 MHz) δ = 2.92 (d, 3 H, J = 1.5 Hz, CH3N), 6.63 (d, 2 H, J = 8.7 Hz, 2 × ArH), 7.41 (t, 1 H, J = 7.6 Hz, ArH), 7.46–7.53 (m, 2 H, 2 × ArH), 8.04–8.07 (m, 3 H, 3 × ArH); 13C NMR (CDCl3, 100 MHz) δ = 29.91, 108.61, 111.05, 111.52, 120.29, 124.62, 128.43, 129.05, 133.00, 143.52, 154.70, 162.70; IR (KBr) 3346 (m, NH), 3010 (m), 2907 (w), 1764 (s, C=O), 1602 (s), 1545 (s), 1497 (s), 1446 (s), 1362 (s), 1238 (s), 1068 (s), 957 (s), 791 (s) cm−1; HRMS [M + 1] calcd for C14H13N4O2: 269.1039, found 269.1032.
4-Diethylamino-Benzoic Acid Benzotriazol-1-yl Ester, 6
The standard procedure was followed by use of (diethylamino)benzoic acid (D7, 48.4 mg, 0.2505 mmol, 1.0 equiv.), HBTU (104.5 mg, 0.2755 mmol, 1.1 equiv.), and DIEA (48 μl, 0.2755 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 6 (71.5 mg, 0.2304 mmol) was obtained in 92% yield as a light-yellow solid: TLC Rf = 0.50 (CHCl3 as eluant); 1H NMR (CDCl3, 400 MHz) δ = 1.17–1.25 (m, 6 H, 2 × CH3), 3.38–3.49 (m, 4 H, 2 × CH2), 6.70 (d, 2 H, J = 8.8 Hz, 2 × ArH), 7.40 (t, J = 7.4 Hz, 1 H, ArH), 7.42–7.53 (m, 2 H, 2 × ArH), 6.70 (d, 1 H, J = 8.8 Hz, ArH), 8.06 (d, J = 8.8 Hz, 2 H, 2 × ArH); 13C NMR (CDCl3, 100 MHz) δ = 12.39, 44.66, 108.63, 110.64, 120.29, 124.53, 128.33, 128.89, 132.23, 133.01, 143.53, 152.61, 162.69; IR (KBr) 2980 (m), 1774 (s, C=O), 1602 (s), 1529 (s), 1445 (s), 1380 (s), 1260 (s), 1155 (s), 963 (s), 766 (s) cm−1; HRMS [M + 1] calcd for C17H19N4O2: 311.1508, found 311.1503.
1H-Benzoimidazole-5-Carboxylic Acid Benzotriazol-1-yl Ester, 7
The standard procedure was followed by use of 5-benzimidazolecarboxylic acid (G5, 39.1 mg, 0.2411 mmol, 1.0 equiv.), HBTU (100.6 mg, 0.2652 mmol, 1.1 equiv.), and DIEA (46 μl, 0.2652 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 7 (48.5 mg, 0.1737 mmol) was obtained in 72% yield as a light-brown solid: TLC Rf = 0.21 (10% MeOH in CHCl3 as eluant); 1H NMR (d4-methanol + CDCl3, 400 MHz) δ = 7.47–7.53 (m, 2 H, 2 × ArH), 7.79–7.84 (m, 2 H, 2 × ArH), 8.04 (d, 1 H, J = 8.5 Hz, ArH), 8.17 (dd, 1 H, J = 8.5, 1.6 Hz, ArH), 8.49 (s, 1 H, N=CH), 8.61 (d, 1 H, J = 1.6 Hz, ArH); 13C NMR (d4-methanol + CDCl3, 100 MHz) δ = 108.53, 110.19, 115.19, 117.00, 118.35, 119.38, 124.66, 125.11, 125.96, 126.82, 128.88, 143.11, 144.83, 163.12; IR (KBr) 3099 (m, NH), 2901 (w), 1778 (s, C=O), 1620 (s), 1576 (m), 1421 (s), 1362 (s), 1281 (s), 1155 (s), 993 (s), 739 (s) cm−1; HRMS [M + 1] calcd for C14H10N5O2: 280.0834, found 280.0834.
1H-Indole-5-Carboxylic Acid Benzotriazol-1-yl Ester, 8
The standard procedure was followed by use of indole-5-carboxylic acid (70.6 mg, 0.4381 mmol, 1.0 equiv.), HBTU (182.8 mg, 0.4819 mmol, 1.1 equiv.), and DIEA (84 μl, 0.4819 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 8 (107.3 mg, 0.3855 mmol) was obtained in 88% yield as a brown solid: 1H NMR (d6-acetone, 400 MHz) δ = 6.78 (d, 1 H, J = 3.08 Hz,), 7.52 (t, 1 H, J = 7.1 Hz, ArH), 7.59 (t, 1 H, J = 2.3 Hz, ArH), 7.65 (t, 1 H, J = 8.4 Hz, ArH), 7.68 (d, 1 H, J = 8.6 Hz, =CH), 7.79 (d, 1 H, J = 8.4 Hz, ArH), 8.01 (d, 1 H, J = 8.4 Hz, ArH), 8.10 (d, 1 H, J = 8.6 Hz, =CH), 8.68 (s, 1 H, ArH); 13C NMR (d6-acetone, 100 MHz) δ = 104.06, 109.58, 112.82, 115.65, 120.64, 123.78, 125.56, 125.79, 128.53, 128.90, 129.50, 129.78, 141.00, 144.27, 164.87; IR (KBr) 3213 (m, NH), 2907 (w), 1767 (s, C=O), 1614 (s), 1582 (m), 1446 (s), 1359 (s), 1266 (s), 1157 (s), 990 (s), 789 (s) cm−1; HRMS [M + 1] calcd for C15H11N4O2: 279.0882, found 279.0878.
1H-Indole-2-Carboxylic Acid Benzotriazol-1-yl Ester, 9
The standard procedure was followed by use of indole-2-carboxylic acid (F1, 74.9 mg, 0.4648 mmol, 1.0 equiv.), HBTU (193.9 mg, 0.5113 mmol, 1.1 equiv.), and DIEA (89 μl, 0.5113 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 9 (115.1 mg, 0.4136 mmol) was obtained in 89% yield as a light-yellow solid: TLC Rf = 0.82 (5% MeOH in CHCl3 as eluant); 1H NMR (CDCl3, 400 MHz) δ = 7.24 (t, 1 H, J = 8.0 Hz, ArH), 7.41–7.46 (m, 2 H, 2 × ArH), 7.49–7.58 (m, 3 H, 3 × ArH), 7.69 (s, 1 H, =CH), 7.78 (d, 1 H, J = 8.2 Hz, ArH), 8.10 (1 H, J = 8.4 Hz, ArH), 9.97 (br, 1 H, NH); 13C NMR (CDCl3, 100 MHz) δ = 38.58, 108.37, 112.48, 113.28, 120.45, 120.69, 121.69, 123.11, 124.90, 127.06, 127.29, 128.83, 138.60, 143.42, 157.99; IR (KBr) 3282 (m, NH), 1773 (s, C=O), 1670 (s), 1517 (s), 1459 (s), 1389 (s), 1340 (s), 1161 (s), 1052 (s), 778 (s) cm−1; HRMS [M + 1] calcd for C15H11N4O2: 279.0882, found 279.0878.
1H-Benzoimidazole-5-Carboxylic Acid Benzotriazol-1-yl Ester, 10
The standard procedure was followed by use of 5-fluoroindole-2-carboxylic acid (F4, 26.1 mg, 0.1457 mmol, 1.0 equiv.), HBTU (60.8 mg, 0.1603 mmol, 1.1 equiv.), and DIEA (28 μl, 0.1603 mmol, 1.1 equiv.). After the reaction mixture was worked up, the residue was purified by use of column chromatography (chloroform as eluant). Benzotriazole ester 10 (39.3 mg, 0.1327 mmol) was obtained in 91% yield as a light-yellow solid: TLC Rf = 0.83 (5% MeOH in CHCl3 as eluant); 1H NMR (d6-DMSO and CDCl3, 400 MHz) δ = 7.15 (dd, 1 H, J = 7.4, 2.4 Hz, ArH), 7.41–7.52 (m, 3 H, 3 × ArH), 7.58–7.65 (m, 2 H, ArH + =CH), 7.71 (d, 1 H, J = 8.4 Hz, ArH), 8.07 (1 H, J = 8.4 Hz, ArH); 13C NMR (d6-DMSO and CDCl3, 100 MHz) δ = 106.15 (d, J = 24.0 Hz), 108.60, 111.92 (d, J = 6.0 Hz), 114.14 (d, J = 10.0 Hz), 115.61 (d, J = 27.0 Hz), 119.66, 121.80, 124.79, 126.35 (d, J = 11.0 Hz), 128.26, 128.77, 135.58, 142.65, 156.32 (d, J = 235.0 Hz), 157.34; IR (KBr) 3231 (m, NH), 2981 (w), 1782 (s, C=O), 1681 (s), 1521 (m), 1434 (s), 1342 (s), 1222 (s), 1186 (s), 921 (s), 757 (s) cm−1; HRMS [M + 1] calcd for C15H10FN4O2: 297.0779, found 297.0788.
General Procedure for the TBAF-Assisted Benzotriazole N-Alkylation, 14
Benzotriazole (100 mg, 0.84 mmol) and α-bromo-4-(diethylamino)acetophenone (272 mg, 1.00 mmol, 1.2 equiv.) were placed in a 5 ml flask with a stirring bar, followed by the addition of TBAF (1.00 ml, 0.84 mmol, 1.2 equiv., 1 M in THF) at room temperature. After being stirred for 2 hr at room temperature, the reaction was directly loaded into the column, and the product was eluted with a solution of 6:1 hexane:ethyl acetate to yield 217.4 mg (84%) of product as a pale-yellow solid. 1H NMR (500 MHz, CDCl3) δ 8.05 (d, J = 8.5 Hz, 1 H), 7.96 (d, J = 9.2 Hz, 2 H), 7.50–7.30 (m, 3 H), 6.68 (d, J = 9.2 Hz, 2 H), 5.98 (s, 2 H), 3.42 (q, J = 7.0 Hz, 4 H), 1.19 (t, J = 7.0 Hz, 6 H); 13C NMR (125 MHz, CDCl3) δ 187.92, 152.46, 146.42, 134.38, 131.37, 128.00, 124.21, 121.41, 120.25, 110.91, 110.46, 53.76, 45.06, 12.87; ESI-MS calculated for C18H20N4O 308.16, found 308.12.
Inhibition Assay against the SARS-CoV 3CL Protease
A fluorometric assay [31] was utilized to determine the inhibition constants of the prepared samples. Briefly, a fluorogenic peptide, Dabcyl-KTSAVLQSGFRKME-Edans, was used as the substrate [31], and the enhanced fluorescence due to cleavage of this substrate catalyzed by the protease was monitored at 538 nm with excitation at 355 nm. The IC50 value of individual inhibitors was measured in a reaction mixture containing 50 nM SARS 3CL protease and 6 μM fluorogenic substrate in 20 mM Bis-Tris (pH 7.0). The enzyme stock solution was kept in 12 mM Tris-HCl (pH 7.5) containing 120 mM NaCl, 0.1 mM EDTA, plus 7.5 mM β-ME before being added to the assay solution. The Ki measurements (for the noncovalent inhibitors case) were performed at two fixed inhibitor concentrations and various substrate concentrations. In the mechanism-based inactivator cases, we used t1/2 versus 1/[inactivator] for Ki measurements (for more detailed procedures for the behavior of inhibitors, please see the Supplemental Data).
Expression and Purification of SARS-CoV 3CLpro
Wild-type and the C145A mutant of the SARS protease were cloned in pET 28 with N-terminal Trx, His tag, and FXa site. The tags were removed by FXa protease after the proteins were purified with NiNTA column chromatography. For more detailed experimental procedures, please see [32].
Mass Spectrometric Analysis
The ESI-MS experiments were conducted on a Bruker Daltonics BioTOF III high-resolution mass spectrometer, equipped with a home-built nanoESI source. Mass resolution was better than 20,000 on one-pass mode with a mass range of ∼100–3,000. The samples were diluted to 1.0 μM with bidistilled aqueous solution containing 40% methanol and 0.1% formic acid (v/v/v) and were infused with a syringe pump at a flow rate of 300 nl/min. The actual amount of samples consumed was less than 300 fmole.
The MALDI-MS measurements were performed on a MALDI-TOF/TOF mass spectrometer (Applied Biosystems 4700 Proteomics Analyzer). Tryptic digest solution was mixed 1:1 with matrix solution (CHCA [α-cyano-4-hydroxycinnamic acid] 10 mg/ml in solvents 49.9/50/0.1 H2O/CH3CN/TFA), and 1.0 μl was then spotted in each well. Each MALDI spectrum was accumulated from up to 4,000 laser shots from a random sampling of 40 positions per well.
Computer Modeling of SARS-CoV 3CL Protease Inhibition
Docking was performed by using Autodock, version 3.05 [30]. Precomputed energy grid maps with grid point spacing of 0.375 Å and 50 × 50 × 50 grid points centered at the active site were used (autogrid tool in Autodock, version 3.05). During a docking experiment, each compound was kept flexible (except their rings and amide bonds), and the built-in LGA method was adopted. In each compound structure, 1.5 × 106 energy was evaluated, and 40 poses were selected from 2.7 × 105 generations per run.
The crystal structure of SARS-CoV 3CL protease in complex with a substrate-analog inhibitor (coded 1uk4) was obtained from The Protein Data Bank (PDB; http://www.rcsb.org/pdb/) (for more detailed procedures, please see the Supplemental Data).
Acknowledgments
This work is supported by the National Science Council, Taiwan and Genomics Research Center, Academia Sinica. We also thank Ms. Hsien-Hua Hsu for her help with the cytotoxicity assay.
Published online: March 24, 2006
Footnotes
Supplemental data including 90 different acids, the X-ray data for compound 4, synthesis and characterization of compounds 15–20, a supplemental enzyme kinetic study, and the detailed molecular docking data are available at http://www.chembiol.com/cgi/content/full/13/3/261/DC1/.
Contributor Information
Po-Huang Liang, Email: phliang@gate.sinica.edu.tw.
Chi-Huey Wong, Email: wong@scripps.edu.
Supplemental Data
References
- 1.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F. A major outbreak of severe acute respiration syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 5.Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M.N. Canadian severe acute respiratory syndrome study, identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 6.Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 7.He J.-F., Peng G.-W., Min J., Yu D.-W., Liang W.-J., Zhang S.-Y., Xu R.-H., Zheng H.-Y., Wu X.-W., Xu J. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 8.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 9.Marra M.A., Jones S.J.M., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S.N., Khattra J., Asano J.K., Barber S.A., Chan S.Y. The genome sequence of the SARS-associate coronovirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 10.Ruan Y., Wei C.L., Lin A.E., Vega V.B., Thoreau H., Se-Thoe S.Y., Chia J.M., Ng P., Chiu K.P., Lim L. Comparative full-length genome sequence analysis of 14 SARS coronovirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main protease (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 12.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H. The crystal structure of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou K., Wei D., Zhong W. Binding mechanism of coronavirus main protease with ligands and its implication to drug design against SARS. Biochem. Biophys. Res. Commun. 2003;308:148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C.-Y., Jan J.-T., Ma H.-H., Kuo C.-J., Juan H.-F., Cheng Y.-S.E., Hsu H.-H., Huang H.-C., Wu D., Brik A. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu J.T.-A., Kuo C.-J., Hsieh H.-P., Wang Y.-C., Huang K.-K., Lina C.P.-C., Huang P.-F., Chen X., Liang P.-H. Evaluation of metal-conjugated compounds as inhibitors of 3CL protease of SARS-CoV. FEBS Lett. 2004;574:116–120. doi: 10.1016/j.febslet.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 17.Kao R.Y., Tsui W.H.W., Lee T.S.W., Tanner J.A., Watt R.M., Huang J.-D., Hu L., Chen G., Chen Z., Zhang L. Identification of novel small-molecular inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain R.P., Pettersson H.I., Zhang J., Aull K.D., Fortin P.D., Huitema C., Eltis L.D., Parrish J.C., James M.N.G., Wishart D.S. Synthesis and evaluation of keto-glutamine analogues as potent inhibitors of severe acute respiratory syndrome 3CLpro. J. Med. Chem. 2004;47:6113–6116. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 20.Shie J.-J., Fang J.-M., Kuo C.-J., Kuo T.-H., Liang P.-H., Huang H.-J., Yang W.-B., Lin C.-H., Chen J.-L., Wu Y.-T. Discovery of potent anilide inhibitors against the severe acute respiratory syndrome 3CL protease. J. Med. Chem. 2005;48:4469–4473. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 21.Brik A., Lin Y.-C., Elder J., Wong C.-H. A quick diversity-oriented amide-forming reaction to optimize p-subsite residues of HIV protease inhibitors. Chem. Biol. 2002;9:891–896. doi: 10.1016/s1074-5521(02)00184-9. [DOI] [PubMed] [Google Scholar]
- 22.Wu C.-Y., Chang C.-F., Chen J.S.-Y., Wong C.-H., Lin C.-H. Rapid diversity-oriented synthesis in microtiter plates for in situ screening: discovery of potent and selective α-fucosidase inhibitors. Angew. Chem. Int. Ed. 2003;42:4661–4664. doi: 10.1002/anie.200351823. [DOI] [PubMed] [Google Scholar]
- 23.Chang C.-F., Ho C.-W., Wu C.-Y., Chao T.-A., Wong C.-H., Lin C.-H. Discovery of picomolar slow tight-binding inhibitors of α-fucosidase. Chem. Biol. 2004;11:1301–1306. doi: 10.1016/j.chembiol.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Stoner E.J., Cooper A.J., Dickman D.A., Kolaczkowski L., Lallaman J.E., Liu J.-H., Oliver-Shaffer P.A., Patel K.M., Paterson J.B., Jr., Plata D.J. Synthesis of HIV protease inhibitor ABT-378 (Lopinavir) Org. Proc. Res. Dev. 2000;4:264–269. [Google Scholar]
- 25.McCarthy D.G., Hegarty A.F., Hathaway B.J. N-hydroxy-compounds as acyl transfer agents. Part 1. kinetics and mechanism of nucleophilic displacements on 1-hydroxybenzotriazole esters and crystal and molecular structure of 1-benzoyloxybenzotriazole. J. Chem. Soc. Perkin II. 1977;2:224–231. [Google Scholar]
- 26.Barlos K., Papaioannou D., Voliotis S. Crystal structure of 3-(Nα-tritylmethionyl)-benzotriazole 1-oxide, a synthon in peptide synthesis. J. Org. Chem. 1985;50:696–697. [Google Scholar]
- 27.Hansh C., Leo A., Taft R.W. A survey of Hammett substituent constants and resonance and field parameters. Chem. Rev. 1991;91:165–195. [Google Scholar]
- 28.Brik A., Wu C.-Y., Best M.D., Wong C.-H. Tetrabutylammonium fluoride-assisted rapid N9-alkylation on purine ring: application to combinatorial reactions in microtiter plates for the discovery of potent sulfotransferase inhibitors in situ. Bioorg. Med. Chem. 2005;13:4622–4626. doi: 10.1016/j.bmc.2005.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C.-Y., Brik A., Wang S.-K., Chen Y.-S., Wong C.-H. Tetrabutylammonium fluoride-mediated rapid alkylation reaction in microtiter plates for discovery of enzyme inhibitors in situ. Chembiochem. 2005;6:2176–2180. doi: 10.1002/cbic.200500295. [DOI] [PubMed] [Google Scholar]
- 30.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olsen A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 31.Kuo C.-J., Chi Y.-H., Hsu T.-A., Liang P.-H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004;318:862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu M.-F., Kuo C.-J., Chang K.-T., Chang H.-C., Chou C.-C., Ko T.-P., Shr H.-L., Chang G.-G., Wang A.H.-J., Liang P.-H. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280:31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.