Summary Paragraph
Long-lasting, latently-infected, resting CD4+ T cells are the greatest obstacle to cure HIV infection, as they persist despite decades of treatment with ART. Estimates indicate the need for >70 years of continuous, fully suppressive, antiretroviral therapy (ART) to eliminate the HIV reservoir1. Alternatively, induction of HIV from its latent state could accelerate decline of the reservoir, thereby shortening time to eradication. Previous attempts to reactivate latent HIV in preclinical animal models and in clinical trials have measured HIV induction in peripheral blood with minimal focus on tissue reservoirs and had limited effect2-9. Here we show that activation of the non-canonical NF-κB signaling pathway via AZD5582 results in induction of HIV- and SIV-RNA expression in the blood and tissues of ART-suppressed bone marrow/liver/thymus (BLT) humanized mice and rhesus macaques. Analysis of resting CD4+ T cells from tissues after AZD5582 treatment revealed increased SIV-RNA in lymph nodes in macaques and robust induction of HIV in virtually all tissues analyzed in humanized mice including lymph nodes, thymus, bone marrow, liver, and lung. This promising new approach to latency reversal, in combination with appropriate tools for systemic clearance of persistent HIV infection, greatly increases opportunities for HIV eradication.
Latently-infected cells carrying integrated replication-competent provirus that contribute to viral rebound upon ART interruption (termed the “HIV reservoir”) are not detected and eliminated by the immune system or current therapeutics. Therefore, the HIV reservoir has been targeted by approaches to reverse latency and induce viral antigen production (termed “HIV reactivation”)2-9, rendering infected cells susceptible to virus-induced cell death or clearance by the immune system. Previous approaches to HIV reactivation have been modestly effective and have failed to demonstrate reactivation of HIV from resting CD4+ T cells in tissues2-9.
HIV induction in vitro by SMAC mimetics
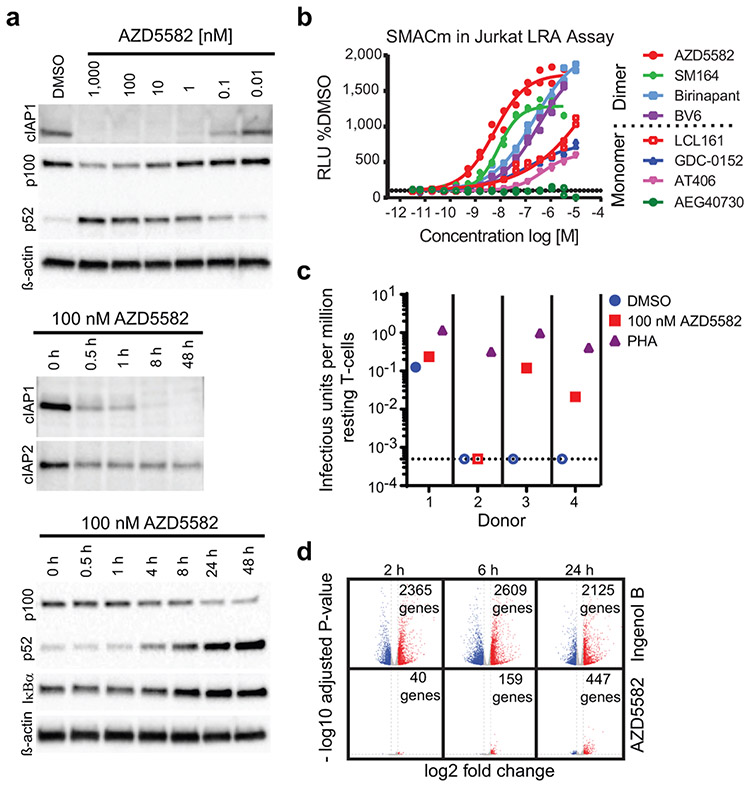
Lack of specificity of molecules that activate the NF-κB pathway as latency reversal agents (LRAs) often results in toxicities that prevent clinical implementation10. We tested the induction of HIV and SIV transcription in latently-infected cells via the non-canonical (nc)NF-κB pathway. This pathway activates of a limited number of cellular genes and a more gradual but persistent activation of NF-κB-driven transcription11. Mimetics of the second mitochondrial-derived activator of caspases (SMAC) activate the ncNF-κB pathway by inhibiting the cellular inhibitor of apoptosis protein 1 (cIAP1) and cIAP2. cIAP1 continually represses the ncNF-κB pathway by constitutively degrading the NF-κB-inducing kinase (NIK) thereby preventing processing of p100 into p5212; this repression can be relieved in CD4+ T cells by in vitro treatment with the SMAC mimetic AZD5582 (Fig. 1a, Extended Data Fig. 1). Compared to other SMAC mimetics, AZD5582 had a superior capacity to reverse HIV latency in vitro (Fig. 1b)13. AZD5582 also induced replication-competent HIV expression from resting CD4+ T cells from ART-suppressed HIV-infected donors (Fig. 1c). AZD5582 induced 5- to 10-fold fewer genes than the protein kinase C agonist Ingenol B (Fig. 1d), a canonical pathway inducer and activator of several transcription factors. By specifically targeting the ncNF-κB signaling pathway, AZD5582 has limited pleotropic impact which may translate into fewer off-target effects14.
Fig. 1. Efficient in vitro AZD5582 target engagement and induction of HIV transcription.
(a) Total CD4+ T cells were treated with a broad range of concentrations (10 pM to 1 μM) of AZD5582 overnight, and cell lysates were analyzed by immunoblot, probing for cIAP1 and p100/p52 as indicated (top panel, representative of 10 experiments). Immunoblot analysis of isolated total CD4+ T cell lysate following treatment with 100 nM AZD5582 examining components of the canonical and ncNF-κB pathway over a 48 h time course post-treatment (middle and bottom panels, representative of 3 and 4 experiments respectively). (b) DMSO-normalized reporter signal induced by a dose titration of a panel of mono- and bivalent SMAC mimetics in a Jurkat luciferase reporter model of HIV-1 latency with 48 h exposure. Symbols represent technical replicates from a single run and are representative of three independent experiments. Lines represent a four-parameter logistic regression model fit. (c) Infectious units per million cells induced by DMSO or 100 nM AZD5582 were determined in a limiting dilution quantitative viral outgrowth assay. Values are infectious units per million resting CD4+ T cells. (d) Volcano plots summarizing average up- and down-regulated genes at 2, 6, and 24 h post-treatment with Ingenol B or AZD5582, with the average log2 fold change on the x-axis and log10 adjusted p-value (two-sided Wald test) on the y-axis as compared to DMSO alone. Dashed lines represent thresholds of log2 fold change of 1 and p-adjusted of 0.05. The data shown represents the average fold change across 4 donors and one experiment. For gel source data, see Supplementary Figure 1.
Latency reversal in BLT humanized mice
BLT mice were infected with HIV-1JR-CSF (Supplementary Table 1) and suppressed with ART15-18 (Fig. 2a,b). Mice were then administered a single intraperitoneal injection of 3 mg/kg AZD5582 or vehicle. No changes in plasma HIV-RNA levels were detected in vehicle control-treated mice at 24 or 48 h nor in AZD5582-treated mice 24 h after AZD5582 administration (Fig. 2c). However, 48 h after AZD5582 treatment increased HIV-RNA was detected in the plasma of 3/6 (50%) and 3/4 (75%) mice in two independent experiments (Fig. 2c). These data demonstrate that a single dose of AZD5582 can induce HIV production, resulting in significant viremia (up to 1,574 HIV-RNA copies/mL plasma) in ART-treated BLT mice (Supplementary Table 2).
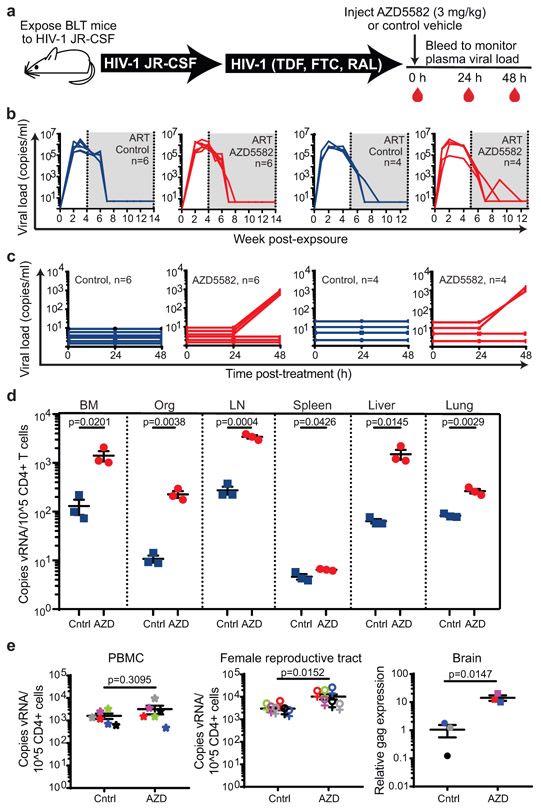
Fig. 2. AZD5582 induces HIV-RNA expression resting CD4+ T cells from tissues of HIV-infected, ART-suppressed BLT mice.
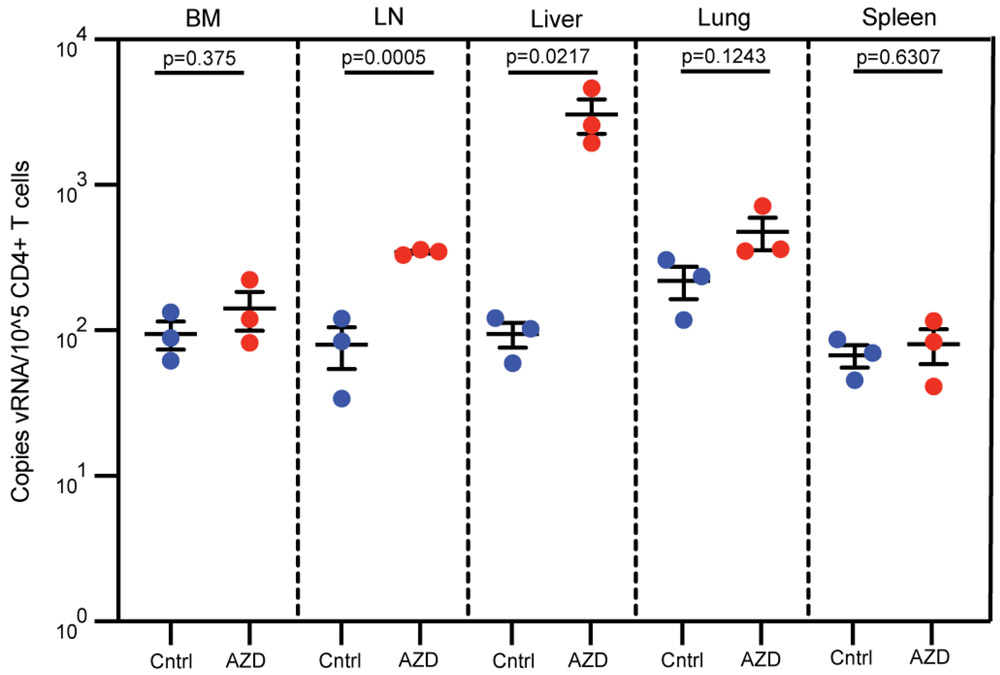
(a) BLT mice were infected with HIV-1JR-CSF. Following 10 weeks of ART treatment, mice were administered vehicle control or AZD5582. (b) HIV-RNA copies/mL plasma in HIV-infected, ART-treated BLT mice prior to treatment with vehicle control (left panels, blue lines) or AZD5582 (right panels, red lines). These are two independent experiments (left panels: n=6 mice/group, right panels: n=4 mice/group). Gray shading: period of ART administration. (c) Plasma HIV-RNA levels in HIV-infected, ART-suppressed mice in panel B treated with vehicle control or AZD5582. (d) HIV-RNA levels in resting CD4+ T cells isolated from the bone marrow (BM), thymic organoid (Org), lymph nodes (LN), spleen, liver and lung of control or AZD5582-treated mice (cells pooled from n=6 mice/group for each tissue) were analyzed in triplicate. Error bars, mean ± s.e.m. Statistical significance was determined with a two-sided student’s T-test. (e) Cell-associated HIV-RNA (vRNA) copies in the blood (PBMCs, n=6), female reproductive tract (FRT, n=6) and brain (n=3). Statistical significance was determined with a two-sided Mann-Whitney test (PBMCs and FRT) or student’s T-test (brain). Colors indicate samples from the same mice. Error bars, mean ± s.e.m.
The hallmark of HIV persistence in humans is the presence of inducible HIV in resting CD4+ T cells. Therefore, resting cells from primary (bone marrow and thymic organoid), secondary (lymph node and spleen) and effector (liver and lung) immune tissues were isolated from HIV-infected, ART-suppressed BLT mice 48 h post administration of vehicle control or AZD558217,19,20. The levels of HIV-RNA in resting CD4+ T cells from AZD5582-treated mice were 11-fold (bone marrow, p=0.0201), 21-fold (thymic organoid, p=0.0038), 12-fold (lymph node, p=0.0004), 1.4-fold (spleen, p=0.0426), 24-fold (liver, p=0.0145), and 3.2-fold (lung, p=0.0029) higher than controls (Fig. 2d). These results were confirmed in a second independent experiment (Extended Data Fig. 2). No notable differences in cell-associated HIV-DNA were noted between mice treated with vehicle control or AZD5582 (Supplementary Table 3). These results demonstrate that AZD5582 induces systemic HIV-RNA production in resting CD4+ T cells, indicative of latency reversal in this critical cellular source of persistent HIV infection. We also isolated cells from peripheral blood, female reproductive tract (FRT) and brain of HIV-infected, ART-suppressed BLT mice 48 h after administration of AZD5582 or vehicle control. As too few CD4+ T cells were available for cell sorting from these compartments, RNA was extracted from total cells and analyzed for the presence of HIV-RNA in each tissue. The levels of HIV-RNA were significantly higher in the FRT (3.4-fold, p=0.0152) and brain (8.7-fold, p=0.0147) of AZD5582-treated mice compared to vehicle controls, but not in blood (p=0.3095) (Fig. 2e). Together, these results show that AZD5582 treatment induces systemic HIV-RNA production throughout BLT mice.
Pharmacodynamics and safety in BLT mice
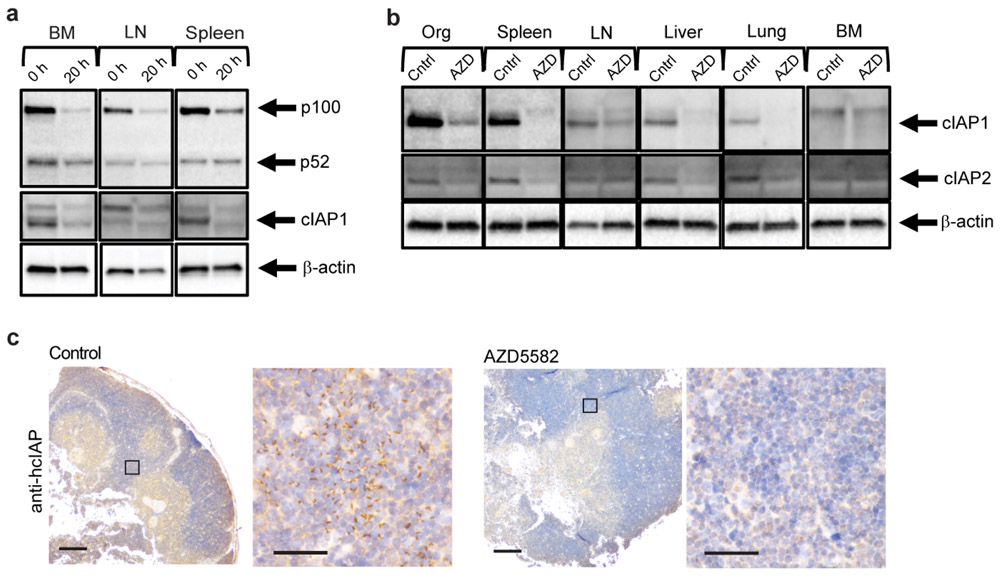
Target engagement after treatment with AZD5582 was confirmed ex vivo by the degradation of cIAP1 (proximal) and p100 (distal) targets of SMAC in the ncNF-κB pathway (Extended Data Fig. 3a). In vivo target engagement was demonstrated in resting CD4+ T cells isolated from thymus, spleen, lymph nodes, liver, lung and bone marrow of BLT mice treated with a single dose of 3 mg/kg AZD5582 or vehicle control (Extended Data Fig. 3b). These results were confirmed by immunohistochemical analysis of the thymic organoid of HIV-infected, ART-suppressed BLT mice demonstrating a dramatic reduction in cIAP1 expression in AZD5582-treated mice (Extended Data Fig. 3c).
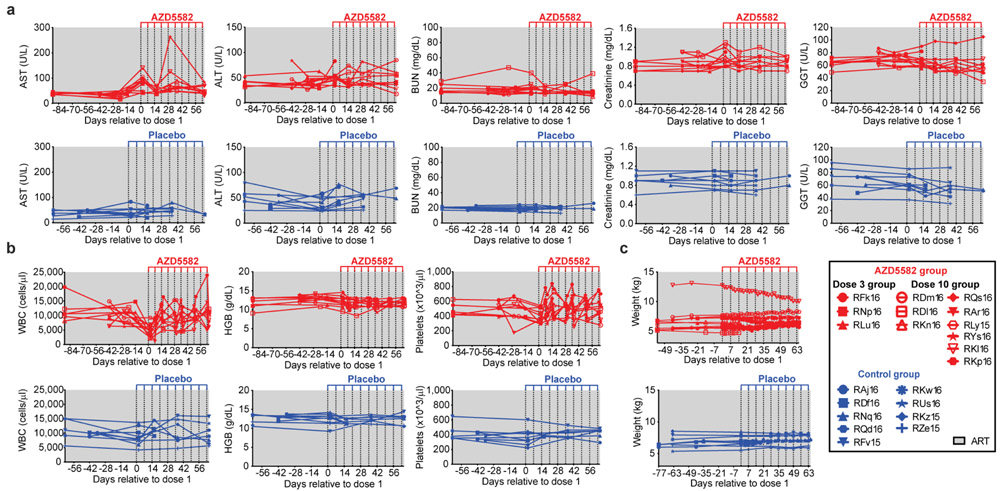
To study off-target or immune-mediated toxicities of AZD5582, we measured serum chemistry, T cell activation, and a panel of plasma cytokines after in vivo treatment of immunocompetent Balb/c mice. AZD5582 administration resulted in mild and transient increases in alanine aminotransferase (ALT) and aspartate aminotransferase (AST) that resolved a few days post-treatment. No other changes in serum chemistries were noted (Supplementary Table 4). In addition, no differences were noted in the levels of activated (CD38+HLA-DR+) CD4+ or CD8+ T cells in BLT mice treated with AZD5582 or vehicle control (Supplementary Table 5) or in plasma levels of 41 human cytokines and chemokines that serve as indicators of systemic activation/inflammation21 (Supplementary Table 6). Together these results demonstrate that AZD5582 does not cause generalized toxicity or activation of the immune system in the BLT model.
Latency reversal in rhesus macaques
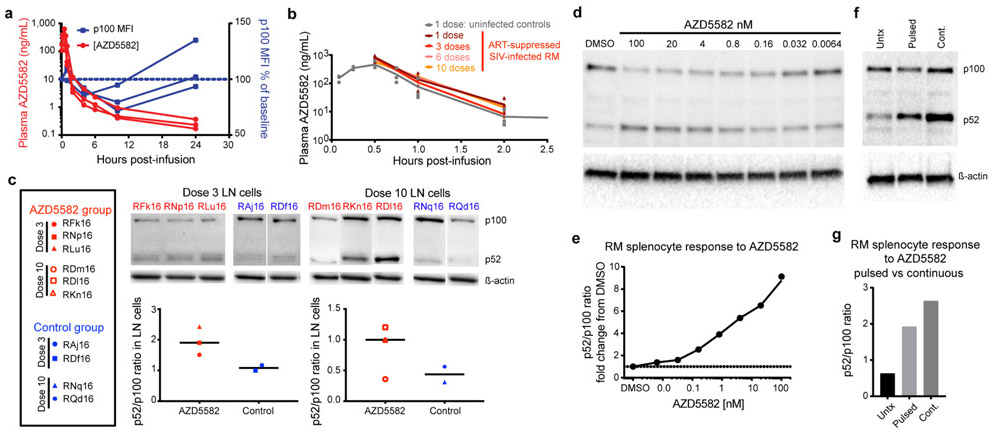
We next evaluated the latency reversal activity of AZD5582 in twenty-one Mamu-B*08 and –B*17 negative rhesus macaques (RMs) infected with SIVmac239 and treated with a potent ART regimen containing tenofovir disoproxil fumarate (TDF), emtricitabine (FTC) and dolutegravir (DTG) initiated eight weeks post infection (Fig. 3a and Supplementary Table 7)49 and 50. Suppression of SIV viremia below 60 copies/mL (standard assay limit of detection) was achieved in all animals in two to 20 weeks and ART was continued for 55 to 67 weeks prior to further treatment (Fig. 3b). Based on pharmacokinetic and pharmacodynamic data in uninfected macaques (Extended Data Fig. 4a) as well as protocols for SMAC mimetics used in oncology, intravenous infusions of 0.1 mg/kg AZD5582 were administered weekly to twelve SIV-infected, ART-suppressed RMs for either three or ten weeks (Fig. 3a). Nine SIV-infected, ART-suppressed RMs served as controls (Fig. 3a). Plasma concentrations of AZD5582 measured after doses one, three, six, and ten indicated drug exposures in SIV-infected, ART-suppressed RMs that were consistent over the dosing period and comparable to those observed in uninfected RMs (Extended Data Fig. 4b).
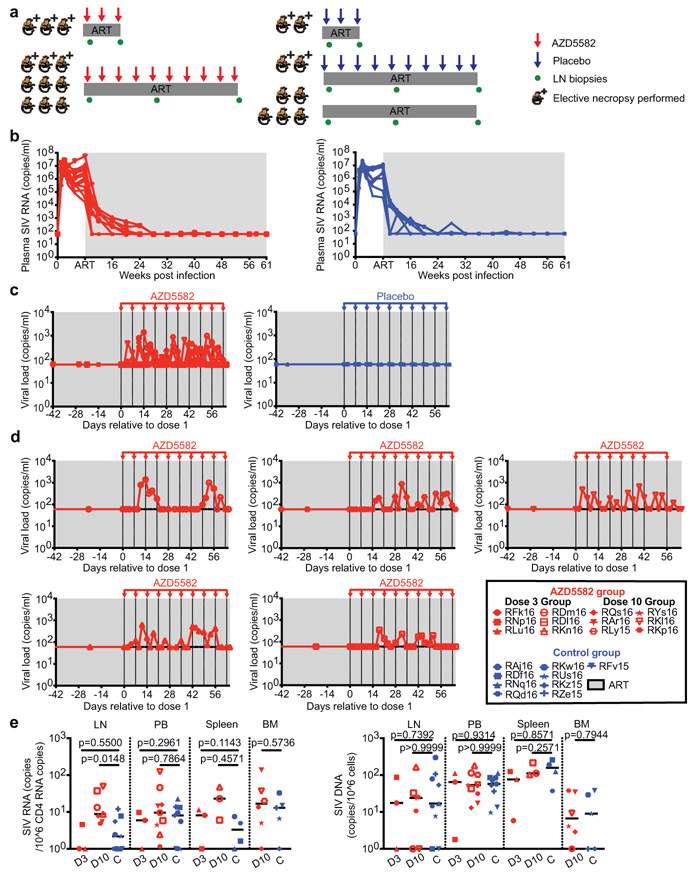
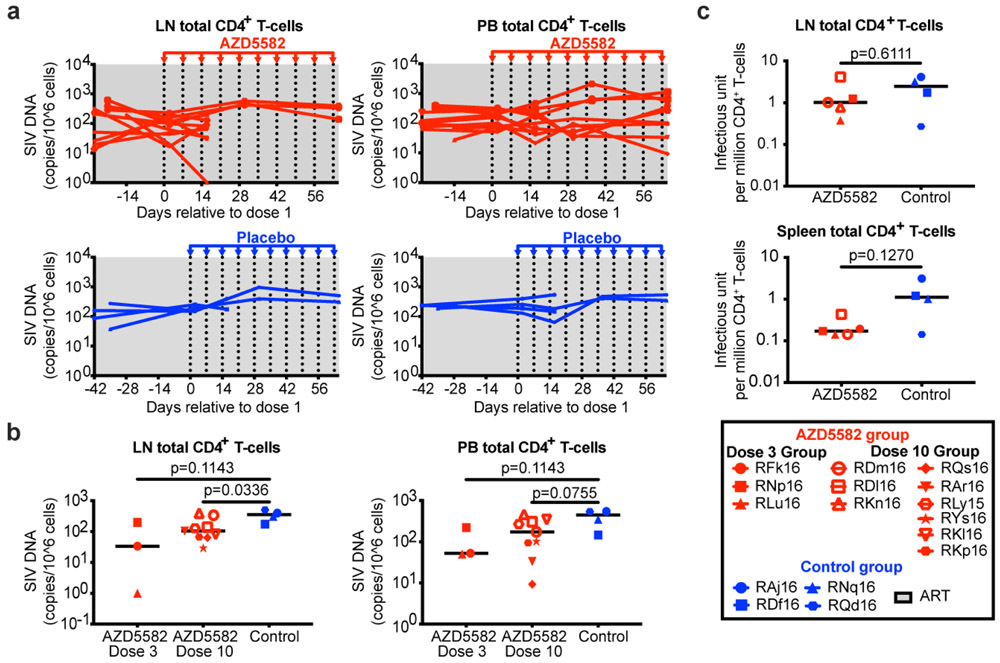
Fig. 3. AZD5582 induces SIV-RNA expression in the plasma and lymph nodes of ART-suppressed SIV-infected RMs.
(a) Experimental design during AZD5582 treatment phase. Three RMs received 3 doses of AZD5582 and were sacrificed 48 h after the last dose. Nine RMs received 10 doses of AZD5582 and 3 were sacrificed 48 h after the last dose. The first dose of AZD5582 was administered after 55 to 67 weeks of ART (3 dose group: 55 weeks; 10 dose group: 55-67 weeks). Four control RMs received a weekly placebo infusion; 2 were sacrificed 48 h after 3 infusions and 2 were sacrificed 48 h after 10 infusions. Five control animals received ART only. (b) Plasma SIV-RNA levels in the 21 SIV-infected RMs prior to treatment with AZD5582 (left, n=12) and equivalent time period for controls (right, n=9). For (b-d): gray shading represents the period of ART administration. (c) Plasma SIV-RNA levels in ART-suppressed SIV-infected RMs during AZD5582 treatment (left, n=12) and the equivalent time period for controls (right, n=9). (d) Individual representation of plasma SIV-RNA levels in the 5 RMs experiencing on-ART viremia during AZD5582 treatment. (e) Cell-associated SIV-RNA (left) and SIV-DNA (right) levels in resting CD4+ T cells isolated from lymph nodes (LN), peripheral blood (PB), spleen and bone marrow (BM) of AZD5582-treated (red) and control (blue) ART-suppressed, SIV-infected RMs. Resting CD4+ T cells were analyzed from AZD5582-treated RMs 48 h after receiving 3 doses (D3, LN, PB and spleen: n=3) or 10 doses (D10, LN: n=7, PB: n=9, spleen: n=3, BM: n=6) of AZD5582. Resting CD4+ T cells were analyzed from control RMs (C, LN and PB: n=9, spleen: n=4, BM: n=5) at equivalent time points. Open symbols indicate AZD5582-treated RMs with on-ART viremia. Statistical significance was determined with a two-sided Mann-Whitney test. Horizontal lines represent the median.
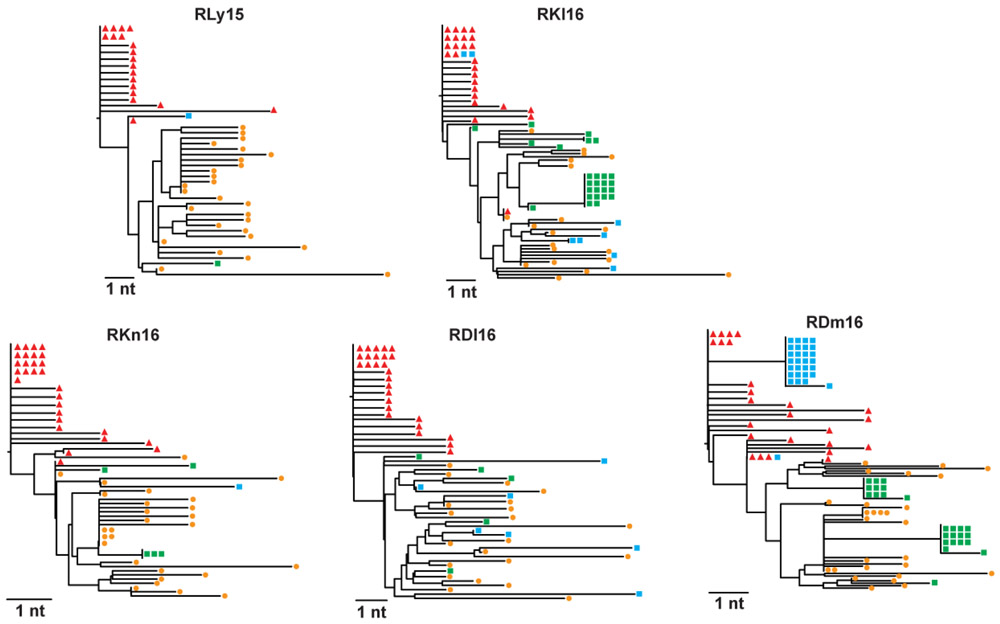
Latency reversal, defined as on-ART viremia increasing from < 60 copies/mL of plasma to > 60 copies/mL of plasma following AZD5582 treatment, was observed as early as 96 h after the first dose and reached levels as high as 1,390 copies/mL in SIV-infected RMs (Fig. 3c,d). On-ART viremia > 60 copies/mL of plasma was observed in 5/12 RMs (42%), corresponding to 5/9 RMs (55%) who received ten doses of AZD5582 (Fig. 3c,d). Multiple instances of sustained viremia > 60 copies/mL between AZD5582 doses were observed. Of 140 viral load measurements performed on the five macaques who exhibited on-ART viremia during AZD5582 treatment, 64 were > 60 copies/mL (46%); in the animal with the greatest frequency of reactivation, this proportion was 15/28 (53%). Longitudinal examination of plasma virus by single genome sequencing analysis (SGA) of the SIVmac239 env gene in all RMs that experienced AZD5582-induced on-ART viremia was performed at four selected time points: two-weeks post infection (near peak viremia), eight-weeks post infection (just prior to ART initiation), and at two time points separated by 26 to 42 days during AZD5582 treatment. Phylogenetic analyses showed several patterns of virus reactivation (Extended Data Fig. 5). In two RMs (RDl16, RKn16), the majority of reactivated virus sequences were phylogenetically closer to sequences at eight-weeks post infection rather than peak viremia and were unique, implying an origin from multiple cells that were seeded at the time of ART initiation22. In two other RMs (RKl16, RDm16), a large fraction of the viruses produced during AZD5582 treatment showed identical sequences, suggesting latency reversal from a single cell or a clonally expanded population of infected cells. These clones clustered with both peak and pre-ART time points and were accompanied by additional unique sequences. In one RM (RLy15) a single virus sequence was amplified at each time point during AZD5582 treatment and these were both phylogenetically similar to sequences found pre-ART. In all, these results indicate that AZD5582 induced virus reactivation from a diverse population of cells, some of which may be clonally expanded23,24.
We quantified cell-associated SIV-RNA and SIV-DNA in resting CD4+ T cells sorted from SIV-infected ART-suppressed RMs treated or not with AZD5582. Cell-associated SIV-RNA levels in resting CD4+ T cells isolated from lymph nodes were significantly higher in animals who received ten doses of AZD5582 compared to controls (p=0.0148) (Fig. 3e). A similar trend was observed in resting CD4+ T cells isolated from the spleens of a subgroup of six animals that were sacrificed. Levels of cell-associated SIV-DNA in resting CD4+ T cells were similar in each compartment across groups (Fig. 3e). To further understand whether latency reversal induced by AZD5582 resulted in a perturbation of the overall level of infected CD4+ T cells, we performed longitudinal measurements of cell-associated SIV-DNA in total (rather than resting) CD4+ T cells isolated from lymph nodes and blood as well as quantitative viral outgrowth assays using CD4+ T cells from lymph nodes and spleen at the end of the treatment period (Extended Data Fig. 6a-c). Despite high-level virus reactivation induced by AZD5582, these experiments did not reveal a consistent reduction in the total or replication-competent SIV reservoir compared to controls.
Pharmacodynamics in rhesus macaques
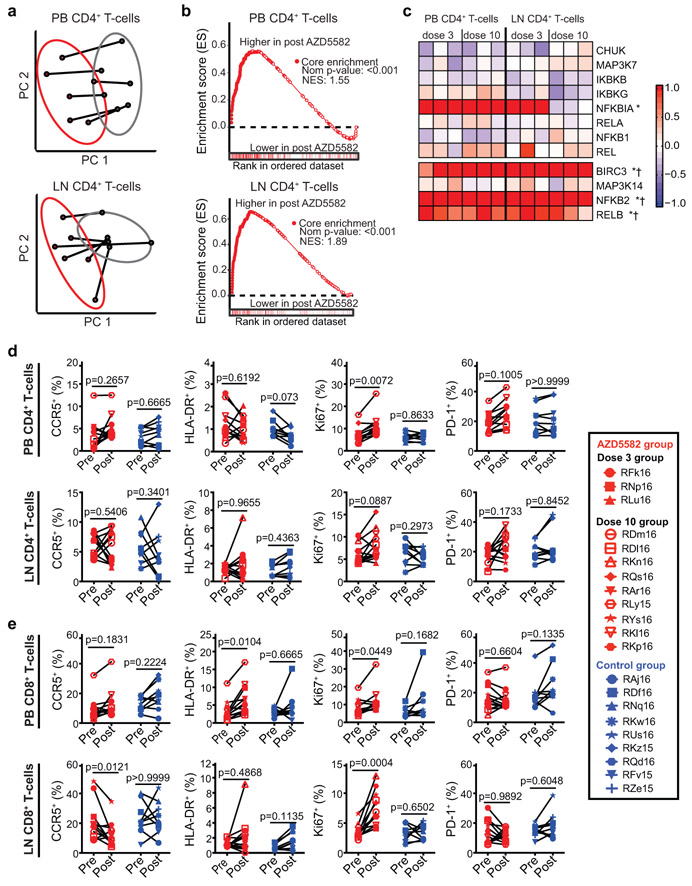
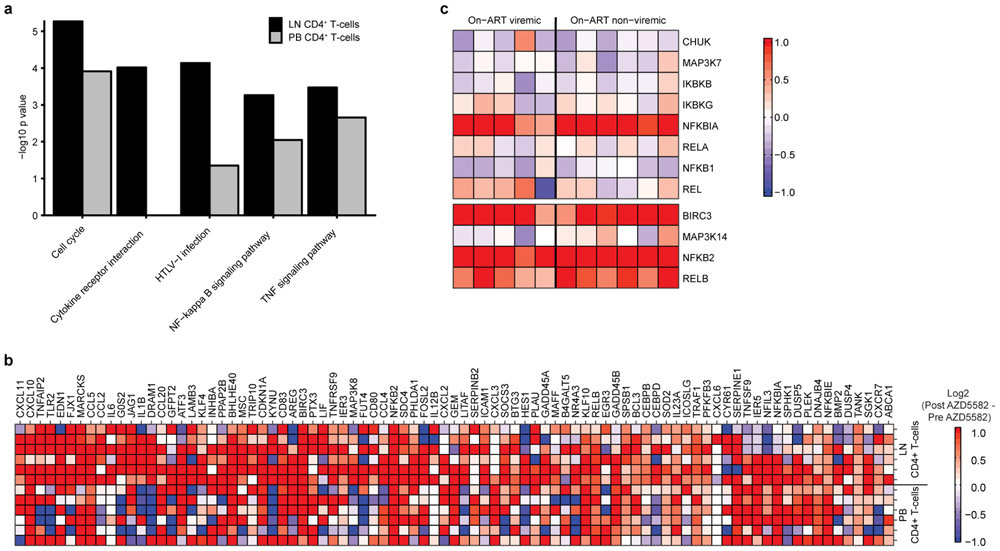
Pharmacologic target engagement of the ncNF-κB pathway was confirmed by Western blot analysis of the degradation of p100 to p52 in lymph node mononuclear cells (LNMCs) after in vivo exposure to AZD5582 (Extended Data Fig. 4c) and in splenocytes treated ex vivo with AZD5582 (Extended Data Fig. 4d-g). Transcriptomic profiling of peripheral blood and lymph node CD4+ T cells isolated from AZD5582-treated RMs showed a distinct effect of AZD5582 on gene expression based on principal component (Fig. 4a) and DAVID pathway analyses (Extended Data Fig. 7a). Enrichment of NF-κB targets after AZD5582 treatment was demonstrated by gene set enrichment analysis (GSEA) (Fig. 4b; heatmap of differentially expressed gene targets of NF-κB is shown in Extended Data Fig. 7b). Hallmark ncNF-κB signaling genes NFKB2 and RELB were significantly upregulated in peripheral blood and lymph node CD4+ T cells following AZD5582 treatment, while cNF-κB signaling molecules were mostly unaffected except for the inhibitor NFKBIA (Fig. 4c). The baculoviral IAP repeat containing 3 (BIRC3) protein, that encodes cIAP2 and regulates both the cNF-κB and ncNF-κB pathways, was also significantly upregulated in peripheral blood and lymph node CD4+ T cells (Fig. 4c). Interestingly, activation of ncNF-κB signaling genes was evident in RMs with and without on-ART viremia > 60 copies/mL (Fig. 4c and Extended Data Fig. 7c) and GSEA showed similar changes in overall gene expression when AZD5582-treated RMs were grouped by presence or absence of on-ART viremia > 60 copies/mL (Supplementary Table 8), suggesting that the lack of virologic response measured by standard viral load assay was not due to compromised target engagement.
Fig. 4. AZD5582 specifically activates the ncNF-κB pathway in SIV-infected, ART-suppressed RMs without generalized T cell activation.
(a-c) Peripheral blood (PB) and lymph node (LN) CD4+ T cell gene expression in SIV-infected, ART-suppressed RMs pre- and post-AZD5582 treatment (n=6 for both LN and PB; for each, n=3 for 3 doses of AZD5582 and n=3 for 10 doses of AZD5582). (a) PCA plots comparing PB (top) and LN (bottom) CD4+ T cell transcriptomes pre- (gray) and post- (red) AZD5582. Ellipses: two standard deviations. (b) GSEA plots of NF-κB-induced genes in PB (top) and LN (bottom) CD4+ T cells. Gene Set: “HALLMARK TNFα SIGNALING VIA NF-κB” (MSigDB). (c) Heatmap of cNF-κB (top) and ncNF-κB (bottom) pathway gene expression. Color scale: log2-fold changes from pre-treatment. Genes differentially expressed post-AZD5582 in PB (asterisks) or LN (crosses) CD4+ T cells are noted. Expression of activation markers in (d) CD4+ and (e) CD8+ T cells in PB (top) and LN (bottom) of SIV-infected, ART-suppressed RMs pre-AZD5582 and 48 h after the final dose (n=12, red) and control RMs (n=9, blue). Statistical significance was determined with a two-sided Mann-Whitney test.
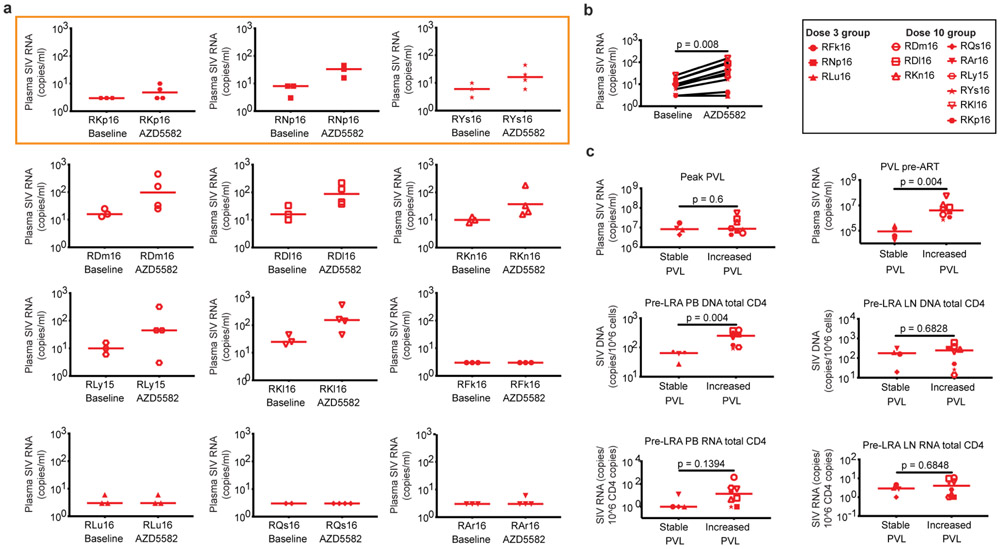
We next used an ultrasensitive assay to measure SIV-RNA in plasma, comparing two to three baseline time points during ART in the four weeks prior to AZD5582 treatment (when plasma SIV-RNA was < 60 copies/mL in all RMs using the standard viral load assay) and three to four time points during AZD5582 treatment (Extended Data Fig. 8a). We found significantly higher plasma SIV-RNA values during the period of AZD5582 treatment compared to pre-treatment (Extended Data Fig. 8b, p=0.008), with 8/12 RMs demonstrating ≥ 2 SIV-RNA measurements above the median of their baseline values. This group of eight RMs with evidence of latency reversal induced by AZD5582 could be segregated from the four RMs with unchanged viral loads based on two parameters: higher levels of plasma SIV-RNA just prior to ART initiation and SIV-DNA in peripheral CD4+ T cells before AZD5582 treatment (p=0.004 for each, Extended Data Fig. 8c).
Safety/immune effects in rhesus macaques
The potential for AZD5582 toxicity in SIV-infected, ART-suppressed RMs was examined and Transient increases in liver enzymes (AST and gamma-glutamyltransferase [GGT]) were observed (Extended Data Fig. 9a). Total white blood cell counts were decreased in all RMs 48 h after the first dose of AZD5582 but returned to normal levels when measured before the second dose and over time (Extended Data Fig. 9b). After the seventh and eighth doses, one animal experienced a reaction characterized by fever, emesis, fatigue, and inappetence, with elevated liver enzyme and creatinine levels and bandemia on laboratory examination. All abnormalities resolved within a two-week period, but this RM was not re-dosed. Importantly, animal weights did not significantly fluctuate over the course of the treatment period (Extended Data Fig. 9c). In summary, 97 doses of AZD5582 were administered to 12 SIV-infected ART-suppressed RMs, of which 95 were well-tolerated and two resulted in a mild adverse reaction.
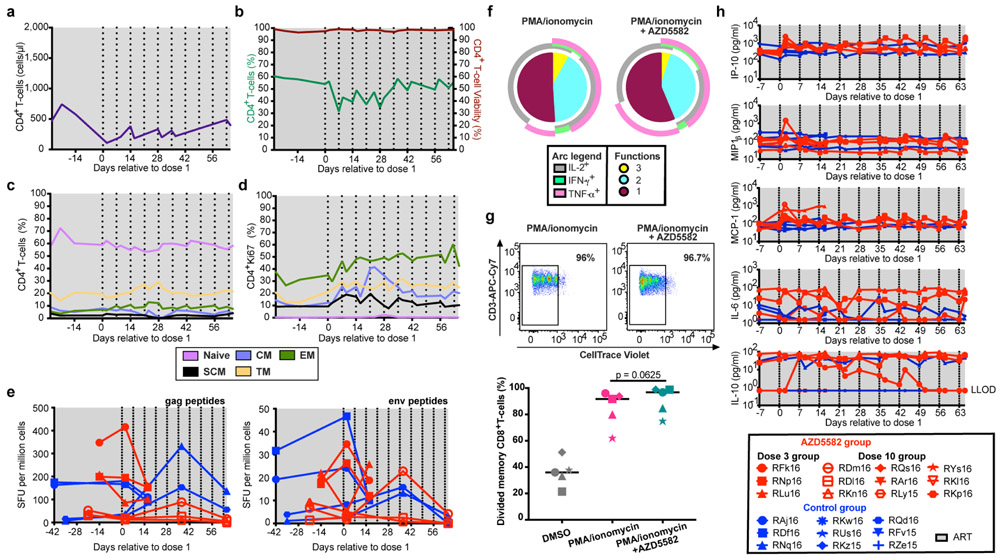
Markers of T cell activation (both CD4+ and CD8+) were assessed in AZD5582-treated RMs and controls, with expression of CCR5, HLA-DR, and PD-1 on CD4+ T cells from blood and lymph nodes being similar before and after treatment with AZD5582 (Fig. 4d,e). Levels of intracellular Ki-67 were increased in CD4+ T cells in the blood and CD8+ T cells in blood and lymph nodes after AZD5582 treatment (Fig. 4d,e). CD8+ T cell expression of HLA-DR was higher in blood following exposure to AZD5582 (Fig. 4e). These results indicate that AZD5582 does not induce global CD4+ T cell activation in RMs but the combined pharmacologic and virologic effects may have a stimulatory effect on CD8+ T cells. Longitudinal analyses of CD4+ T cell counts and frequencies, CD4+ T cell viability, CD4+ T cell subset frequencies and their Ki-67 expression are shown in Extended Data Fig. 10a-d. The increased Ki-67 observed within CD4+ T cells following AZD5582 treatment may sound a note of caution as the proliferation of latently-infected memory CD4+ T cells is hypothesized to contribute to the maintenance of the viral reservoir over time24,25; however, we did not find an increase in infected cells after AZD5582 treatment (Fig. 3e and Extended Data Fig. 6).
We next sought to determine if AZD5582 would impair SIV-specific T cell responses in SIV-infected, ART-suppressed RMs, a potential adverse outcome of treatment that has been suggested for other LRAs26. However, the frequency of SIV Gag- or Env-specific CD8+ T cells measured by IFNγ ELISPOT did not decline following AZD5582 treatment (Extended Data Fig. 10e), and CD8+ T cell polyfunctionality and proliferative responses were largely unaffected by AZD5582 treatment ex vivo, with the exception of IL-2+IFNγ+TNFα+ triple positive cells being somewhat reduced (p=0.0476, Extended Data Fig. 10f,g). Further, longitudinal assessment of plasma levels of inflammatory cytokines and chemokines did not reveal profound changes induced by AZD5582 (Extended Data Fig. 10h). Overall, our work in the RM model indicates that AZD5582 treatment is safe in a majority of animals and can induce appreciable increases in plasma SIV-RNA and SIV expression in lymph node resting CD4+ T cells during ART.
Discussion
Eradication of HIV infection after prolonged viral suppression is the focus of intense research and latency reversal has been a cornerstone of this effort. LRAs have been widely recognized as critical (yet before now mostly theoretical) tools to induce HIV expression in resting human CD4+ T cells. Future clinical applications of HIV cure strategies must be relevant to the majority of people living with HIV for whom the approaches taken with the Berlin and London patients27,28 (whose non-HIV life-threatening hematologic malignancies warranted aggressive, toxic therapies) pose an unacceptable level of risk. Therefore, LRAs must be identified that are highly effective but have minimal side effects. Here, we used two different but highly complementary animal models48 to show that treatment with AZD5582 had minimal and transient side effects but resulted in significant increases in HIV/SIV-RNA levels both in plasma and in resting CD4+ T cells isolated from all tissues analyzed from BLT mice and from the lymph nodes of macaques. Our results represent the first in vivo evidence of systemic HIV/SIV latency reversal from resting CD4+ T cells in humans or any animal model. The concordance between the results obtained in two fundamentally different animal models highlight the robust and reproducible nature of the effect of AZD5582 on HIV and SIV reservoirs. The fact that there is little toxicity associated with the use of AZD5582 strongly suggests that activators of the ncNF-κB pathway may be well suited for HIV eradication approaches in humans.
Methods
Experimental design
The purpose of this study was to determine the efficacy of the SMAC-mimetic AZD5582 as an HIV/SIV latency reversing agent in vivo. To this end, two animal models of HIV-1 infection were utilized: the HIV/BLT humanized mouse model and the SIV/rhesus macaque (RM; Macaca mulatta) nonhuman primate (NHP) model. In each model, animals were infected with HIV-1 or SIV and viremia was durably suppressed by ART. AZD5582 was administered to infected, ART-suppressed animals that were then assayed for changes associated with reactivation of the viral reservoir. Mice were maintained under specific pathogen-free conditions by the Division of Comparative Medicine at the University of North Carolina, Chapel Hill. Mouse experiments were conducted in accordance with NIH guidelines for the housing and care of laboratory animals and in accordance with protocols reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of North Carolina, Chapel Hill. Healthy RMs for pharmacokinetic studies were housed at GlaxoSmithKline and all procedures were conducted in accordance with the GlaxoSmithKline Policy on the Care, Welfare, and Treatment of Laboratory Animals and were reviewed by the IACUC at GlaxoSmithKline. RMs infected with SIV were housed at the Yerkes National Primate Research Center (Atlanta, GA) and treated in accordance with Emory University and Yerkes National Primate Research Center IACUC regulations (PROTO201800308). Animal care facilities are accredited by the U.S. Department of Agriculture (USDA) and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International.
Preparation of Jurkat HIV-luciferase cell clones
Cell clones Jurkat-C16 and Jurkat-I15 were prepared by infecting Jurkat cells (Jurkat Clone E6–1 cells, American Type Culture Collection TIB-152, authenticated by morphological identification and virus susceptibility profiles, tested for mycoplasma by the supplier) with a full length, infectious HIV-1NL4–3-based virus engineered to express a luciferase reporter in place of the HIV-1 nef gene (NLCH-Luci). The Jurkat-N6 cell clone was made using the same virus as described above with an additional mouse heat stable antigen (mHSA) reporter located just downstream of the luciferase open reading frame and separated by a T2A element (NLCH-Luci-HSA). NLCH, kindly provided by R. Swanstrom, is the parent molecular infectious clone used to make the Jurkat clones and is a modification of HIV-1NL4–3 (GenBank U26942) where flanking sequences were removed. All viruses were derived by transfection of human embryonic kidney 293 cells (HEK 293T, European Collection of Authenticated Cell Cultures, authenticated by morphological identification, tested for mycoplasma by the supplier) with 1μg of the HIV-1NL4–3 derived infectious molecular plasmid DNAs using the FuGENE HD Transfection reagent (Promega) per the manufacturer’s recommendations. Supernatants were collected 48 h post transfection, passed through a 0.2 micron filter, and used to infect wild-type Jurkat cells. Following infection, cells expressing high levels of HIV-encoded mHSA were removed using biotin-labeled rat anti-mouse CD24 antibody (clone M1/69, BD Biosciences) that was adsorbed to streptavidin-labeled magnetic Dynabeads M-280 (Life Technologies) per the manufacturer’s recommendations. Negatively selected, HIV-infected Jurkat cells were then limit-diluted at 0.5 cells/well in 96-well plates, and individual cell clones were expanded for two to four weeks in culture in the presence of 500 nM efavirenz (EFV). Clones were profiled for baseline reporter level and responsiveness to benchmark LRAs, with C16, I15, and N6 representing the most quiescent but inducible clones obtained.
Cell culture and Jurkat HIV-luciferase assay
Jurkat HIV-luciferase clones were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco by Life Technologies) containing 10% (vol/vol) fetal bovine serum (SAFC/Sigma-Aldrich) and 25 units/mL penicillin, 25 units/mL streptomycin (Gibco by Life Technologies), and were split 1:4 every three to four days to maintain a cell density of ~0.3 to 1 million cells/mL. The Jurkat clones were maintained with the addition of 500 nM EFV in the medium. Three Jurkat cell clones (C16, I15, and N6), each harboring one or two integrated HIV proviruses expressing the luciferase reporter gene, were added at equal amounts for a total of 5,000 cells per well to 384-well plates containing compound titrations. Dose-response testing was performed on compounds dissolved in dimethyl sulfoxide (DMSO; Fisher Scientific) dispensed in duplicate serial three-fold, 14-point titrations using a D300e Digital Droplet Dispenser (Hewlett-Packard) to give final assay concentrations of 10 μM to 2.1 pM in 50 μL of medium at 0.5% DMSO (vol/vol) final concentration. Cells and compound were incubated at 37°C for 48 h, unless otherwise indicated, followed by the addition of 20 μL of Steady-Glo® Luciferase (Promega). Luminescence resulting from the induction of the virally expressed luciferase was measured using an EnVision™ 2102 Multilabel Plate Reader (Perkin Elmer). Dose-response relationships were analyzed with GraphPad Prism (version 6) using a four-parameter logistic regression model to calculate the concentration of compound that gives half-maximal response (EC50) and the maximal percent activation compared to the vehicle control.
Immunoblot analyses
For the immunoblot assays, 10 μg of cell lysate was loaded per well into 4–20% Tris-Glycine SDS-PAGE gels. Protein from the SDS-PAGE gels were transferred to Turbo Midi PVDF Transfer Packs (BioRad) using the “Mixed MW” protocol for one Midi Format Gel (constant 2.5A up to 25V, for 7 min) of the Trans-Blot® Turbo™ Transfer System (BioRad) with premade Trans-Blot per the manufacturer’s instructions. After transfer, PVDF membranes were blocked in 5% bovine serum albumin (BSA) in 1x Tris-buffered saline (TBS) (BioRad) with 0.1% TWEEN® 20 for 1 h at room temperature with gentle rocking. Primary antibodies were added and incubated overnight at 4°C (anti-cIAP1, 1:1,000 (Abcam); anti-p100/p52, 1:1,000 (Cell Signaling Technology); anti-IκBα, 1:1,000 (Cell Signaling Technology); anti-cIAP2, 1:1,000 (Abcam); anti-actin-HRP conjugate, 1:30,000 (Abcam). Following primary staining, the membrane was washed three times with 1x TBS+0.1% TWEEN® 20, 10 min each wash. After washing, the membrane was incubated in 5% BSA in 1x TBS+0.1% TWEEN® 20 with the appropriate secondary antibody for 2 h at room temperature. Following secondary stain, the membrane was washed twice for 10 min with 1x TBS+0.1% TWEEN® 20 followed by a 10 min wash with 1xTBS. The membrane was then patted dry with filter paper and an image was captured of the undeveloped membrane on the ChemiDoc™ MP Imaging System using Image Lab software (version 6.0.1, BioRad). Sufficient ECL reagent (GE Healthcare) was used to cover the membrane and a series of images were taken with increasing exposure times until the luminescence from the developed membrane saturated the image. The developed membrane was then washed three times with 1x TBS for 5 min to remove the residual ECL reagent and then stored at 4°C in sufficient 1x TBS to submerse the entire membrane. Densitometry of images of the developed membrane were then carried out using the Image Lab software (version 6.0.1, BioRad). Some membranes were stripped for 1 min with One Minute® Plus Western Blot Stripping Buffer (GM Biosciences) and then washed three times for 10 min with 1x TBS. The stripped membranes were then blocked in 5% BSA in 1x TBS+0.1% TWEEN® 20 for an hour and re-probed overnight with a new primary antibody. To normalize samples for loading a 1:20,000 dilution of beta-actin (Abcam) was run on the stripped membranes.
Target gene quantitative RT-PCR
Two million normal donor CD4+ T cells were treated with a range of concentrations of AZD5582. Total RNA was isolated using the RNEasy® Mini kit (Qiagen) per the manufacturer’s instructions. The following TaqMan primer probe sets were sourced from Applied Biosystems: Hs00985031_g1 (BIRC3), Hs00174517_m1 (NFKB2), and Hs02800695 (HPRT1). TaqMan-based real-time PCR (Fast Virus 1-Step Master Mix, Applied Biosystems) was used to amplify host genes of interest and acquire signal on a QuantStudio 3 Real-Time PCR thermocycler (ThermoFisher). Gene expression was normalized to HPRT1 and comparative threshold cycle (CT) method (ΔΔCT) was used for relative quantification of gene expression. The relative quantification was analyzed by QuantStudio™ 3 Real-Time PCR System software (version 1.4.3, ThermoFisher).
HIV quantitative viral outgrowth
All human peripheral blood mononuclear cell (PBMC) samples were obtained under a specimen procurement protocol reviewed and approved by the University of North Carolina Biomedical Institutional Review Board and the McGill University Health Centre Ethical Review Board. Informed consent was obtained from all participants. Human PBMCs for quantitative viral outgrowth were obtained via continuous flow leukapheresis. Resting CD4+ T cells were isolated and virus outgrowth assays performed as previously described29,30 with some modifications. Briefly, 20–50 x 106 highly purified resting CD4+ T cells were stimulated with PHA, IL-2 (60 U/mL) and irradiated PBMCs from a seronegative donor, or with either 100 nM AZD5582, 335 nM vorinostat, or 0.003% DMSO (vehicle control) in limiting dilution for 24 h. Cultures were washed to remove drugs and CCR5 high, CD8 depleted, PHA stimulated PBMC from an uninfected donor were added twice to amplify virus outgrowth. Culture supernatants were assayed for HIV p24 expression by ELISA on day 15 and confirmed on day 19. A maximum likelihood method was used to estimate the frequency of inducible virus and is reported as infectious units per million (IUPM)31.
RNA-Seq of human cells
Total CD4+ T cells were isolated from the peripheral blood mononuclear cells (PBMCs) of four ART-treated, aviremic patients by negative selection (EasySep Human CD4+ T cell Enrichment Kit, StemCell) according to the manufacturer’s instructions. Dead cells and other debris was removed using a Dead Cell Removal Kit (Miltenyi Biotec) according to the manufacturer’s instructions. Cells from each patient were treated with 0.05% DMSO, 100 nM AZD5582, or 25 nM Ingenol B and harvested at 2 h, 6 h, and 24 h after exposure. RNA was isolated from the harvested cells using AllPrep DNA/RNA Mini Kit (Qiagen). Two hundred ng of RNA from each sample was checked for quality using an Agilent Bioanalyzer, with RNA integrity number (RIN) scores typically >9.0 suggesting high quality RNA. These total RNA samples were then processed into stranded, mRNA libraries using the KAPA library preparation kit (KAPA BioSystems, F. Hoffmann-La Roche Ltd). Final libraries were checked by Qubit (Thermo Fisher) for concentration, and with a BioAnalyzer HS-DNA chip (Agilent) for fragment size distribution (mean size 359 bp). Samples were then sequenced using an Illumina Hiseq 4000 sequencer using a paired-end 50 bp x 50 bp run. Samples were successfully demultiplexed and then quality assurance and quality controlled (QAQC’ed) using FASTQC (version 0.11.1) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Raw reads were mapped to the human genome and transcriptome (GRCh38.p7) using STAR32 and Salmon (version 0.7.2)33. Data was normalized and interrogated for changes in gene expression using DESeq234 package in R. P-values were adjusted for multiple testing using a false discovery rate using the Benjamini-Hochberg method35. Data was analyzed both jointly and within each treatment compared to the vehicle control. Differential expression of outliers was assessed and found insignificant in overall effect. Thresholds applied to call a significant response were average log2 fold change >1 and p-adjusted <0.05. Graphs and summary tables were built in R using ggplot. Gene set enrichment was performed using GSEA (version 2.2.3) and GO analysis (GO PANTHER version 11.1)36. Results shown are the average responses of the four donors tested.
Construction and maintenance of BLT mice
BLT mice were prepared as previously reported18,37,38. Briefly, a 1–2 mm piece of human liver tissue was sandwiched between two pieces of autologous thymus tissue (Advanced Bioscience Resources) under the kidney capsule of sub-lethally irradiated (200 cGy) 12–15-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG; The Jackson Laboratory) mice. Following implantation, mice were transplanted intravenously with CD34+ hematopoietic stem and progenitor cells isolated from autologous human liver tissue. Human immune cell reconstitution was monitored in the peripheral blood of BLT mice by flow cytometry every three to four weeks21. For the study that examined the impact of AZD5582 administration on T cell activation, the mean weight of the mice used was 26.88 grams and they were approximately one year old at the initiation of the study. For the study that examined the impact of AZD5582 administration on plasma and tissue viremia during ART suppression, the mean weight of the mice used was 23.14 grams and the mice were approximately seven months old at the initiation of the study. Mice were randomized for assignment to either experimental or control groups (random.org).
HIV infection of BLT mice
Stocks of HIV-1JR-CSF were prepared as follows. The proviral clone was transfected into human embryonic kidney (HEK)293T cells using Lipofectamine™ 2000 (Invitrogen) following manufacturer’s protocols. Viral supernatant was collected 48 h after transfection. Viral supernatant was tittered by infecting TZM-bl cells (NIH AIDS Reagent Program, authenticated by morphological identification and virus susceptibility profiles, tested for mycoplasma by the supplier) by at multiple dilutions. Virus containing medium was removed the next day and replaced with fresh Dulbecco’s Modified Eagle Medium (DMEM)(ThermoFisher) plus 10% fetal bovine serum (FBS) and the incubation continued for 24 h. The cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and blue cells were counted directly to determine infectious particles per mL. Each titer of these viral stocks was performed in triplicate and at least two different titer determinations were performed for each virus stock. Exposure of BLT mice to HIV-1JR-CSF was conducted via tail vein injection with 3x104 tissue culture infectious units (TCIU) of virus. Plasma viral load in peripheral blood of infected mice was monitored longitudinally by quantitative real-time PCR using TaqMan® RNA to-CT™ 1-step kit (Applied Biosystems). The sequences of the forward and reverse primers and the TaqMan™ probe for PCR amplification and detection of HIV gag RNA were: 5′-CATGTTTTCAGCATTATCAGAAGGA-3′, 5′-TGCTTGATGTCCCCCCACT-3′, and 5′-FAM CCACCCCACAAGATTTAAACACCAT- GCTAA-Q-3′, respectively. For viral load analysis, 40 μl of plasma was collected and analyzed with a sensitivity of 350 copies/mL. All samples were run and analyzed on an ABI 7500 Fast Real Time PCR System (Applied Biosystems).
SIV infection of NHPs
Twenty-one male and female Indian RMs, three to six years of age, with exclusion of Mamu B*08 and B*17 positive animals, were enrolled in this study (Supplementary Table 7). RMs were infected intravenously with 3x103 TCID50 of SIVmac239. SIVmac239 stock was titrated in vitro for viral infectivity by standard endpoint titration on CEMx174 cells. The 50% tissue culture infectious dose (TCID50) was calculated by the method of Reed and Meunch. Standard SIVmac239 plasma viral load quantification was performed regularly throughout the study and three times per week during the AZD5582 treatment period in the Translational Virology Core Laboratory of the Emory Center for AIDS Research using a standard quantitative real-time PCR (qPCR) assay (limit of detection: 60 copies/mL plasma) as described previously39. Ultrasensitive SIVmac239 plasma viral load quantification (limit of detection: 3 copies/mL plasma) was performed for two to three time points before AZD5582 treatment and three to four time points during AZD5582 treatment as described previously40,41.
ART and AZD5582 treatment of BLT mice and NHPs
ART was administered to BLT mice via 1/2″ pellets of irradiated Teklad chow consisting of emtricitabine (1,500 mg/kg), tenofovir disoproxil fumarate (1,560 mg/kg), and raltegravir (600 mg/kg) (Research Diets). Both the AZD5582 and the vehicle control (10% sterile captisol dissolved in sterile distilled water) were prepared fresh for each administration. AZD5582–2HCl was obtained from ChemieTek and dissolved at 5 mg/mL in sterile distilled water (Gibco Life Technologies) containing 10% Captisol β-Cyclodextrine Sulfobutyl Ethers Sodium Salts (Cydex Pharmaceuticals). All injections performed were intraperitoneal at a dose of 3 mg/kg.
RMs were treated with a potent 3-drug ART regimen initiated 56 days post-infection consisting of two reverse transcriptase inhibitors, tenofovir disoproxil fumarate (5.1 mg/mL) and emtricitabine (40 mg/mL) plus the integrase inhibitor dolutegravir (2.5 mg/mL). ART was administered once daily at 1 mL/kg body weight via the subcutaneous route. Twelve RMs were treated with AZD5582 and nine RMs served as controls. AZD5582 was infused weekly intravenously at 0.1 mg/kg. Three RMs received three doses of AZD5582 and were sacrificed 48 h after the last dose. Nine RMs received 10 doses of AZD5582 and three animals were sacrificed 48 h after the last dose. Among the control RMs, four animals received a weekly placebo infusion, two animals were sacrificed 48 h after three infusions and two animals were sacrificed 48 h after ten infusions. The remaining five control animals received ART only.
Resting CD4+ T cell enrichment
Human resting CD4+ T cells were enriched from total cells isolated from BLT mouse tissues as follows. Each tissue from each mouse was processed individually and then tissues were pooled for immunomagnetic sorting. Each pooled tissue was first enriched for human cells with the EasySep™ Mouse/Human Chimera Kit (Stem Cell Technologies) and then for resting CD4+ T cells with a human custom selection kit which included the following antibodies: (CD8, CD14, CD16, CD19, CD20, CD36, CD56, CD123, Glycophorin A, CD66b, CD25, and HLA-DR; Stem Cell Technologies). Flow cytometry was performed prior to and following the enrichment to confirm efficacy of the sort and purity of the sorted samples.
RM resting CD4+ T cells were isolated from the peripheral blood, bone marrow, lymph node and spleen. Prior to sorting CD4+ T cells were enriched with the use of magnetic beads and column purification (Miltenyi Biotec). Enriched CD4+ T cells were then stained with previously determined volumes of the following fluorescently conjugated antibodies: CD3-AF700 (clone SP34–2), CD8-APC-Cy7 (clone SK1), CD69-PE-CF594 (clone FN50), HLA-DR- PerCP-Cy5.5 (clone G46–6), from BD Bioscience; and CD4-BrilliantViolet650 (clone OKT4), CD25-PE-Cy7 (clone BC96) from Biolegend. Resting CD4+ T cells were defined as CD3+CD4+CD8-CD69-CD25-HLA-DR-. Sorting was performed on a FACSAria LSR II (BD Biosciences) equipped with FACSDiva™ software.
Plasma cytokine and chemokine analysis
Human plasma cytokine and chemokine analysis in BLT mice was performed by the University of North Carolina, Chapel Hill Center for AIDS Research (CFAR) Virology Core Laboratory. Plasma was tested undiluted in singular wells using a Milliplex® MAP kit (Millipore # HCYTMAG-60K-PX41) on a Luminex MAGPIX™ instrument. The following markers were tested: EGF, Eotaxin, FGF-2, Flt-3L, Fractalkine, G-CSF, GM-CSF, GRO, IFNα2, IFN-γ, IL-1α, IL-1β, IL-1RA, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17A, IP-10, MCP-1, MCP-3, MDC, MIP-1α, MIP-1β, PDGF-AA, PDGF-AB/BB, RANTES, sCD40L, TGFα, TNFα, TNFβ, VEGF.
Rhesus Macaque plasma levels of proinflammatory cytokines and chemokines were evaluated using the Nonhuman Primate MSD V-Plex assay systems developed by MesoScale Discovery. The two validated kits used were the V-PLEX Plus Proinflammatory Panel 1 NHP Kit (K15056D) that evaluates IFN-γ, IL-10, IL-1b, IL-2, IL-6, IL-8 as well as a custom Chemokine NHP Kit that evaluates IP-10, MCP-1, and MIP-1β (K15055G). The manufacturer provided protocol was followed with a few modifications. Since the plasma samples were infectious, Triton™ X-100 at a final concentration of 1% was added not only to the subject samples but to assay diluents provided with the MSD kit prior to use. Calibration standards were reconstituted according to the protocol provided with the assay diluent that had the Triton™ X-100 added to it so that all standards and samples had the same components present. Plasma (60 μl) was diluted two-fold with assay diluent and then 50 μl of diluted sample was added to the assay plate along with 50 μl of the calibrator according manufacturer’s protocol. The plate was then covered and incubated on a shaker for two h. After the two h incubation, plates were washed at least 6x with the supplied kit wash buffer and then 25 μl of detection antibodies were added according to kit protocol, and the plate was covered and incubated on a shaker for two h at room temperature. After the detection-incubation was over, plates were then washed 6x and 150 μl of 2x Read buffer was added to each well and plates were read on the Sector s600 MSD plate reader. Data analysis was performed using MSD Discovery Workbench analysis software.
Cell-associated HIV-RNA quantification in BLT mice
For tissue RNA analysis, RNA was extracted using QIAamp® viral RNA columns (Qiagen) according to the manufacturer’s protocol including an optional treatment with RNase-free DNase and analyzed using one-step reverse transcriptase real-time PCR [ABI custom TaqMan™ Assays-by-Design]42. Known quantities of HIV gag RNA standards were run in parallel, creating a standard curve for HIV gag and sample RNA was quantified by extrapolation from the standard curve. All samples were run and analyzed on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems™). Due to the relatively low number of human cells found in the brain, HIV-RNA levels were quantified for only three vehicle control and three AZD5582-treated animals.
Immunohistochemical analysis in BLT mice
Tissues for immunohistochemical analysis were harvested from BLT mice and fixed in 10% formalin for 16 to 24 h at 4°C. Samples were then embedded in paraffin, cut into 5-μm sections, and mounted onto poly-L-lysine–coated glass slides. Following paraffin removal, antigen retrieval (DIVA Decloaker, Biocare Medical), and blocking of nonspecific Ig-binding sites (Background Sniper, Biocare Medical), tissue sections were stained with anti-cIAP1 antibody (R&D Systems) overnight at 4°C. To detect cIAP1, sections were probed with a goat-on-rodent HRP-polymer (Biocare Medical) and developed with diaminobenzidine (ImmPact™ DAB Peroxidase Substrate, Vector Laboratories). As an isotype control, tissue sections were stained with polyclonal goat IgG (R&D Systems) negative control antibodies. Tissue sections were imaged with a Nikon Eclipse Ci microscope using Nikon Elements BR software (version 4.30.01) and a Nikon Digital Sight DS-Fi2 camera.
Serum chemistry analysis in mice
Twenty-four hours following AZD5582 administration, serum chemistry analysis in female 20 week old BALB/c mice (BALB/cJ, The Jackson Laboratory) was performed by the University of North Carolina, Chapel Hill Animal Histopathology Core Laboratory of the Lineberger Comprehensive Cancer Center. All clinical chemistry was performed on the Alpha Wasserman Vet Axcel® analyzer using Alfa Wasserman reagents. Methodology performed for each analyte as follows: Alkaline phosphatase: Tietz optimized Bowers and McComb assay; aspartate aminotransferase: Henry modification of Karmen’s assay; alanine aminotransferase: Henry modification of Wroblewski and LaDue assay; creatinine: Jaffe reaction; blood urea nitrogen: enzymatic assay; albumin- Doumas and Briggs modification of the bromcresol green (BCG) dye method; amylase: based on use of chromogenic 2-chloro-p-nitrophenol linked with maltotriose; calcium- Calcium-Arsenazo assay phosphorus- based on method of Daly and Ertingshausen with modifications by Armador and Urban; total bilirubin: Walters and Gerarde modification of the dimethyl sulfoxide method; total protein: modification of Weichselbaum’s biuret reagent.
Immunophenotyping of BLT mice by flow cytometry
Immunophenotyping was performed on peripheral blood samples collected longitudinally and at harvest, on blood and mononuclear cells isolated from the tissues of BLT mice. All flow cytometry data were collected on either a BD LSR Fortessa™ or BD FACSCanto™ instruments using BD FACSDiva™ software (version 6.1.3) and data were analyzed with FlowJo™ software (version 10.4.2). Antibodies used for longitudinal monitoring of human cells in peripheral blood include anti-CD45 APC (clone HI30; BD Biosciences), anti-CD3 FITC (clone HIT3a; BD Biosciences), anti-CD4 PE (clone RPA-T4; BD Biosciences), and anti-CD8 PerCP (clone SK1; BD Biosciences). Flow cytometric gating for expression of lineage specific antigens on human leukocytes was performed as follows: (step 1) forward and side scatter properties were utilized to set a live cell gate; (step 2) live cells were then analyzed for expression of the human pan-leukocyte marker CD45; (step 3) human leukocytes were then analyzed for human CD3+ T cells and (step 4) T cells were analyzed for human CD4 and CD8 expression (Supplementary Figure 2a).
At harvest, peripheral blood and cells isolated from each individual tissue were stained with antibodies to detect human CD45, CD3, CD4, CD8, CD38 (anti-CD38 APC, clone HB7; BD Biosciences) and HLA-DR (anti-HLA-DR PE, clone TU36; BD Biosciences) to assess T cell activation. The following gating strategy was employed: (step 1) forward and side scatter properties were utilized to set a live cell gate; (step 2) live cells were then analyzed for expression of the human pan-leukocyte marker CD45; (step 3) human leukocytes were then analyzed for human CD3+ T cells; (step 4) T cells were analyzed for human CD4 and CD8 expression; (step 5) either CD4+ or CD8+ were analyzed for the expression of CD38 and HLA-DR (Supplementary Figure 2b).
To determine the success of the sort and purity of sorted human resting CD4+ T cells, the following antibodies were used to presort and post sort samples: anti-CD3 BV421 (clone UCHT1; BD Biosciences), anti-HLA-DR PerCP (clone L243; BD Biosciences), anti-CD4 BV605 (clone RPA-T4; BD Biosciences), anti-CD8 APC-Cy7 (clone SK1; BD Biosciences), anti-CD25 APC (clone 2A3; BD Biosciences), and anti-CD45 V500 (clone H130; BD Biosciences). Antibodies used as isotype controls: anti-mouse IgG1κ APC (clone MOPC-21; BD Biosciences), anti-mouse IgG2ακ PerCP (clone X39; BD Biosciences), anti-mouse IgG1κ PE (clone MOPC-21; BD Biosciences), anti-mouse IgG1κ PE-Cy7 (clone MOPC-21; BD Biosciences), and anti-mouse IgG2ακ FITC (clone G155–178; BD Biosciences). Flow cytometric gating was performed as follows: (Step 1) live cells were gated based on forward scatter and side scatter; (step 2) human hematopoietic cells were gated based on expression of human CD45; (step 3) human T cells were gated based on expression of CD3; (step 4) T cell subsets were gated based on the expression of human CD8 and CD4; (step 5) expression of activation markers (CD25 and HLA-DR) were analyzed on the surface of CD4+ T cells (Supplementary Figure 2c).
NHP sample collection and processing
EDTA-anticoagulated blood samples were collected regularly and used for a complete blood count, routine chemical analysis and immunostaining, with plasma separated by centrifugation within 1 h of phlebotomy. At the end of the studies, tissue samples were collected including lymph nodes (21 RMs), spleen (10 RMs), and bone marrow (11 RMs). After two washes in RPMI and removal of connective and fat tissues, lymph nodes were grinded using a 70-μm cell strainer. PBMCs and bone marrow mononuclear cells were prepared by density gradient centrifugation. CD4+ T cells were negatively selected from fresh or frozen cell suspension using magnetically labeled microbeads and subsequent column purification according to the manufacturer’s protocol (Miltenyi Biotec).
AZD5582 pharmacokinetics in NHPs
Three healthy male rhesus macaques (Macaca mulatta) of Indian origin, age six to seven years, 10.5–13.0 kg, were utilized for the study. Fasted RM received 0.1 mg/kg, dose volume 0.25 mL/kg at 0.40 mg/mL of AZD5582 in 10% Captisol + ≤ 5% DMSO, filtered during administration (0.22 μM PES in-line filter, Millex), via a 30 min saphenous vein infusion (infusion pump, Harvard Apparatus). Blood from femoral venipuncture or saphenous catheters was collected to obtain samples for plasma PK (K2 EDTA microtainers spun at 13k RPM for 5 min to obtain plasma), flow cytometry (Cytochex tubes [Streck]) and PBMC isolation (CPT blood collection tubes, (BD Biosciences) processed according to the manufacturers’ instructions and a dry pellet of cells snap frozen on dry ice and stored at −80°C at various times after administration. AZD5582 was extracted from macaque plasma samples with an isotopically labeled internal standard (rilpivirine-d6) using protein precipitation. The compound was then eluted from a Waters Atlantis T3 (50×2.1 mm, 3 μm particle size) analytical column under reverse-phase conditions and detected on an AB Sciex API-5000™ triple quadrupole mass spectrometer under TurboIonspray mode. Standards were prepared in singlet and quality controls (QC) in duplicate, and a calibration curve was generated using a weighted 1/(x2) linear regression of concentration (x) vs analyte:internal standard peak area ratio (y). Concentrations of QCs and study samples were calculated from this calibration curve using Sciex Analyst® Software (version 1.6.2). The acceptance criterion of the assay was ± 25% of nominal concentration for standards and QCs, and the quantifiable range was 2–2,000 ng/mL.
Ex vivo analysis of AZD5582 activity in NHP cells
Cryopreserved RM splenocytes initially processed as above and frozen in FBS+10% DMSO were thawed and exposed to a range of concentrations of AZD5582 for either 48 h continuously or for 1 h followed by drug removed through washing and additional culture for 47 h. In preparation for Western blot analysis, cells were lysed on ice with intermittent vortex mixing using NP40 Cell Lysis Buffer (Invitrogen, Part# FNN0021) supplemented with properly diluted 10x Protease Inhibitor Cocktail (Sigma, Part# P-2714), 1 mM PMSF (Sigma, Part# 93482) and 1 mM DTT (Sigma, Part# 43816) to create a final concentration of 1 mM each in the complete lysis buffer. The lysed cells were then centrifuged at 13,000 RPM for 10 min at 4°C and then supernatants (soluble fraction lysates) removed and stored at −80° or utilized immediately for protein concentration and Western blot analysis.
Flow cytometry assay for p100 protein
Whole blood collected in Cyto-Chex® blood collection tubes (Myriad RBM) was added to ACK lysis buffer to remove red blood cells, then washed with PBS and resuspended in a staining cocktail including anti-CD3 BV421 (clone SP34-2; BD Biosciences), anti-CD16 BV605 (clone SG8; BD Biosciences), anti-CD4 BV711 (clone L200; BD Biosciences), anti-CD14 BV786 (clone M5E2; BD Biosciences), anti-CD123 PerCP-Cy5.5 (clone 7G3; BD Biosciences), anti-CD20 PE-CF594 (clone 2H7; BD Biosciences), anti-CD8 PE-Cy7 (clone SK1; BD Biosciences), anti-CD11c Alexa700 (clone 3.9; Ebioscience), and anti-HLA-DR APC-Cy7 (clone L243; BD Biosciences). Following surface staining, cells were washed twice with phosphate-buffered saline (PBS), then permeabilized using the Cytofix/Cytoperm kit (BD Biosciences, kit used as directed), and stained intracellularly with anti-p100 (clone EPR18756; Abcam). Cells were washed twice and stained with a secondary chicken anti-rabbit Alexa Fluor 488 antibody (Invitrogen). Samples were acquired using a 4-laser Fortessa™ flow cytometer (BD Biosciences) and analyzed with FlowJo™ software (version 9.7.6, Treestar).
SIV env sequencing and analysis
To generate env cDNA, reverse transcription of viral RNA was performed using SuperScript III reverse transcriptase according to the manufacturer’s directions (Invitrogen) and the gene specific primer SIVEnvR1 5’-TGT AAT AAA TCC CTT CCA GTC CCC CC-3’). Single genome amplification of SIV env was performed by serially diluting this cDNA among independent PCR reactions to identify a dilution where amplification occurred in <30% of the total number of reactions. PCR amplification was performed with 1X PCR buffer, 2 mM MgSO4, 0.2 mM of each deoxynucleoside triphosphate, 0.2 μM of each primer, and 0.025 U/μl Platinum Taq High Fidelity polymerase (Invitrogen) in a 20-μl reaction. First round PCR was performed with primer SIVEnvF1 5’-CCT CCC CCT CCA GGA CTA GC-3’ and antisense primer SIVEnvR1 under the following conditions: 1 cycle of 94°C for 2 min, 35 cycles at 94°C for 15 s, 55°C for 30 s, and 68°C for 5 min, followed by a final extension of 68°C for 10 min. Next, 1 μl from the first-round PCR product was added to a second-round PCR reaction that included the sense primer SIVEnvF2 5’- TAT AAT AGA CAT GGA GAC ACC CTT GAG GGA GC-3’ and antisense primer SIVEnvR2 5’-ATG AGA CAT RTC TAT TGC CAA TTT GTA-3’ performed under the same conditions used for first-round PCR, but with a total of 45 cycles. Correct sized amplicons were identified by agarose gel electrophoresis and directly sequenced with second round PCR primers and nine SIV specific primers using BigDye™ Terminator technology (Applied Biosystems). To confirm PCR amplification from a single template, chromatograms were manually examined for multiple peaks, indicative of the presence of amplicons resulting from PCR-generated recombination events, Taq polymerase errors or multiple variant templates. Alignment and phylogenetic trees were implemented in Geneious (Biomatters) using the Muscle algorithm and the Neighbor-Joining method with the Tamura-Nei genetic distance model respectively.
Cell-associated SIV-RNA and -DNA quantification in NHPs
Cell-associated SIV-RNA and -DNA were measured simultaneously in resting CD4+ T cells isolated from peripheral blood, spleen, bone marrow and lymph nodes (52,279 to 500,000 cells) or in total CD4+ T cells isolated from peripheral blood or lymph nodes (100,000 to 500,000 cells), lysed in Buffer RLT+ (Qiagen), and stored at −80°C. Nucleic acids were extracted using the AllPrep DNA/RNA mini kit (Qiagen) according to the manufacturer’s recommendations with an on-column DNase digestion step. Cell-associated DNA quantification of SIVmac gag DNA was performed on the extracted cell-associated DNA by quantitative PCR using a 5′ nuclease (TaqMan™) assay with SIV gag primers and normalized to the RM albumin gene, as previously described43. For cell-associated RNA quantification, RNA was reverse transcribed using the ThermoScientific’s High Capacity cDNA Reverse Transcription kit and random hexamers. SIV gag and the RM CD4 gene were quantified by qPCR of the resultant cDNA using ThermoScientific’s Taqman™ Universal Mastermix II. The CD4 primer and probe sequences were Rh-CD4-F: 5’-ACATCGTGGTGCTAGCTTTCCAGA-3’, Rh-CD4-R: 5’-AAGTGTAAAGGCGAGTGGGAAGGA-3’, and Rh-CD4-Probe: 5’-AGGCCTCCAGCACAGTCTATAAGAAAGAGG-3’. The means of two replicate wells were used in all analyses. Samples with undetectable SIV-DNA or -RNA were assigned a level of 1 copy per million cells or million CD4 RNA copies, respectively, for graphical purposes.
SIV quantitative viral outgrowth assay
Replication-competent SIV reservoirs were measured by the Viral Reservoir Core Laboratory of the Emory Center for AIDS Research. Latently-infected cells were quantified by using a limiting dilution culture assay in which CD4+ T cells, enriched from lymph node or spleen cells using magnetic beads and column purification (Miltenyi Biotec) which were cocultured with CEMx174 cells in five-fold serial dilutions ranging from as many as 5x106 cells per well to as few as 4x105 cells per well. The cells were cultured in RPMI containing 10% fetal bovine serum and 100 U/mL IL-2 (Sigma). The ratio of target cells added was 4:1 for the two highest dilutions. A constant number of 1x106 CEMx174 cells was added to all other wells. The cultures were split every seven days, and fresh medium was added. After 21 days, growth of virus was detected by qRT-PCR. SIV-RNA was isolated from 400 μL of culture supernatant using the Zymo Viral RNA isolation Kit (Zymo Research). DNase treatment was performed using RQ1 RNase-free DNase kit (Promega). A one-step qRT-PCR targeting SIVgag was performed using an Applied Biosystems 7500 Real Time PCR System (Applied Biosystems) and the Taqman™ Fast Virus 1-step Master Mix (Thermo Scientific) for qRT-PCR with the following primers and probe: SIVgagFwd: 5'-GCAGAGGAGGAAATTACCCAGTAC-3'; SIVgagRev: 5'-CAATTTTACCCAGGCATTTAATGTT-3'; SIVgag-probe: 5’−6FAM-TGTCCACCTGCCATTAAGCCCGA-3IBFQ-3’. The frequencies of infected cells were determined by the maximum-likelihood method44 and were expressed as infectious units per million (IUPM) CD4+ T cells.
RNA-Seq of NHP cells
RNA-Seq analysis was conducted at the Yerkes Nonhuman Primate Genomics Core Laboratory (http://www.yerkes.emory.edu/nhp_genomics_core/). RNA was purified from 50,000 peripheral blood or lymph node derived CD4+ T cells, which were purified by flow cytometry and lysed in in 350 μl of RLT buffer at −80°C, using Qiagen Micro RNEasy columns, and RNA quality was assessed using an Agilent Bioanalyzer. Two nanograms of total RNA were used as input for mRNA amplification using 5′ template switch PCR with the Clontech SMART-Seq® v4 Ultra® Low Input RNA kit according to manufacturer’s instructions. Amplified mRNA was fragmented and appended with dual-indexed bar codes using Illumina NexteraXT DNA library preparation kits. Libraries were validated by capillary electrophoresis on an Agilent 4200 TapeStation, pooled, and sequenced on an Illumina HiSeq 3000 using 100-bp SR at an average depth of 25 M reads.
RNA-Seq statistical analyses
RNA-Seq data were mapped to the MacaM (version 7.8) assembly of the Indian RM genome45 (available at https://www.unmc.edu/rhesusgenechip/index.htm) and alignment was performed with STAR (version 2.5.2) using the annotation as a splice junction reference. Transcripts were annotated using the MacaM (version 7.8.2) annotation. Transcript abundance was estimated within STAR using the htseq-count algorithm and differential expression analyses were performed using DESeq2. For significance testing by DESeq2, the samples from three- and ten-dose RMs were grouped together, separately for peripheral blood and lymph node, and compared to their pre-treatment samples. Genes determined to be differentially expressed by DESeq2 (padj < 0.05 and fold-change > ± 1.5) were tested for enrichment of molecular pathways using the DAVID database46 (https://david.ncifcrf.gov/) and by using GSEA with the desktop module (https://www.broadinstitute.org/gsea/). GeneSets for GSEA analysis were selected from the MSigDB database (http://software.broadinstitute.org/gsea/msigdb/index.jsp) and from the “blood transcriptome modules” described by Li et al47. Heatmaps, DAVID enrichment bar charts and GSEA enrichment plots were generated with the R (version 3.5.0) package ggplot2. Principal component analysis was performed using Partek Genomics Suite software (version 6.6) using a covariance matrix.
Immunophenotyping of NHPs by flow cytometry
Multicolor flow cytometric analysis was performed on whole blood and lymph node mononuclear cells using predetermined optimal concentrations of the following fluorescently conjugated monoclonal antibodies: CD3-APC-Cy7 (clone SP34–2), Ki-67-AF700 (clone B56), HLA-DR-PerCP-Cy5.5 (clone G46–6), CCR5-APC (clone 3A9, CCR7-FITC (clone A20 ), CD45RA-PE-Cy7 (clone 5H9), and CD62L-PE (clone SK11) from BD Biosciences; CD8-BV711 (clone RPA-T8), CD4-BV650 (clone OKT4), CD95-BV605 (clone DX2) and PD-1-BV421 (clone EH12.2H7) from Biolegend and CD28-PE-Cy5.5 (clone CD28.2) from Beckman Coulter. Flow cytometric acquisition and analysis of samples was performed on at least 100,000 events on an LSRII flow cytometer driven by the FACSDiva™ software package (BD Biosciences). Analyses of the acquired data were performed using FlowJo™ software (Tree Star, version 10.0.4).
ELISPOT for SIV-specific IFNγ production in NHPs
IFNγ production was evaluated following stimulation of PBMC with SIVmac239 Gag and SIVmac239 Env peptide pools. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: SIVmac239 Env Peptide Pool from NIAID, DAIDS as well as SIVmac239 Gag Peptide Pool. IFNγ expression we used the Monkey Elispot kit from MABTECH. The manufacturer’s instructions were followed as provided with the exception that Concanavalin A (final concentration of 2.5 μg/mL) was used as the positive control agent. ELISPOT plates were blocked for 30 min with 100 μl of RPMI 1640 supplemented with 10%FBS. PBMCs (4x105 per well) were incubated with 1μg/mL of DMSO, SIVmac239 Gag Peptide Pool, or SIVmac239 Env Peptide Pool for 18 h prior to running the assay. For each plate Concavalin A was used for the positive control wells. Samples were run in duplicate.
Intracellular cytokine staining
PBMCs from five ART-suppressed SIV-infected RMs were thawed. Pretreatment with AZD5582 was performed for 1 h at 100 nm. After two washes, PMA and ionomycin were added for 1 h at 500 ng/mL and 10 μg/mL, respectively. DMSO treatment controls were prepared in parallel. After 1 h, brefeldin-A (BFA) and Golgi stop solution were added following manufacturer’s recommendations (BD GolgiStop™ Protein Transport Inhibitor, BD Biosciences). Cells were incubated at 37°C, 5% CO2 in R10 for 6–8 h before staining with the following antibodies: CD3-APC-Cy7 (clone SP34–2), CD8-BV711 (clone RPA-T8), CD4-BV650 (clone OKT4), from Biolegend, IFN-γ-PE (clone B7), TNF-α-AF700 (clone Mab11) and IL-2-BV605 (clone MQ1–17H12) from BD Biosciences. Flow cytometric acquisition and analysis of samples was performed on at least 100,000 events on an LSRII flow cytometer driven by the FACSDiva software package (BD Biosciences). Analyses of the acquired data were performed using FlowJo™ (TreeStar, version 10.0.4) and Simplified Presentation of Incredibly Complex Evaluations (SPICE, version 6.0) software.
Proliferation assay
PBMCs from five ART-suppressed SIV-infected RMs were thawed and labeled with CellTrace™ Violet Proliferation Kit according to the manufacturer instruction (Molecular Probes). Cells were plated in R10 in 96-well round bottom plate at 4 million cells per mL. Pretreatment with AZD5582 was performed for 1 h at 100 nm. After two washes, phorbol 12-myristate 13-acetate (PMA) and ionomycin were added for 1h at 500 ng/mL and 10 μg/mL, respectively. DMSO treatment controls were prepared in parallel. Cells were incubated at 37°C, 5% CO2 in R10 supplemented with 20 IU/mL IL-2. After 5 days, cells were stained with CD3-APC-Cy7 (clone SP34–2), CD8-BV711 (clone RPA-T8), CD4-BV650 (clone OKT4), from Biolegend and analyzed by flow cytometry on an LSRII instrument. Data were analyzed using FlowJo™ software (TreeStar, version 10.0.4).
Statistical analysis
Statistical analyses were performed using GraphPad Prism Software (version 7). A p value of 0.05 or less was considered statistically significant. At least three samples were used for each group, the minimum to achieve statistical significance. No statistical methods were used to predetermine sample size. Investigators were not blinded to group allocations or when assessing outcomes. In some instances, cells were pooled from individual humanized mice for each tissue and experimental group for the isolation of resting CD4+ T cells (Fig. 2d and Extended Data Fig. 2). To test the statistical significance of the differences we observed in PCR data in BLT mouse brains in Fig. 2e and grouped tissue PCR data in Fig. 2d, unpaired, two-sided T tests were employed. To test the statistical significance of the differences, we observed in PCR data in PBMC and FRT in Fig. 2e, T cell activation markers in Supplementary Table 5 and plasma cytokines/chemokines in Supplementary Table 6, we employed an unpaired, two-sided Mann-Whitney U comparison. To assess the statistical significance of the differences observed over time in serum enzymes in Supplementary Table 4, we employed the paired, nonparametric Wilcoxon matched-pairs signed rank test. To test the statistical significance of the differences in activation marker levels expressed on T cells in RMs in Fig. 4d,e, as well as the ultrasensitive plasma viral load results in Extended Data Fig. 8b and the ex vivo proliferation assay in Extended Data Fig. 10g, we used a Wilcoxon matched-pairs signed rank test. To assess the statistical significance of the differences observed between SIV-DNA and -RNA levels in resting or total CD4+ T cells in RMs in Fig. 3e and Extended Data Fig. 6b, respectively, as well as for the quantitative viral outgrowth results in Extended Data Fig. 6c, we employed an unpaired, two-sided Mann-Whitney U comparison. An unpaired, two-sided Mann-Whitney U test was also used to compare RMs with stable vs. increased viremia in Extended Data Fig. 8c.
Extended Data
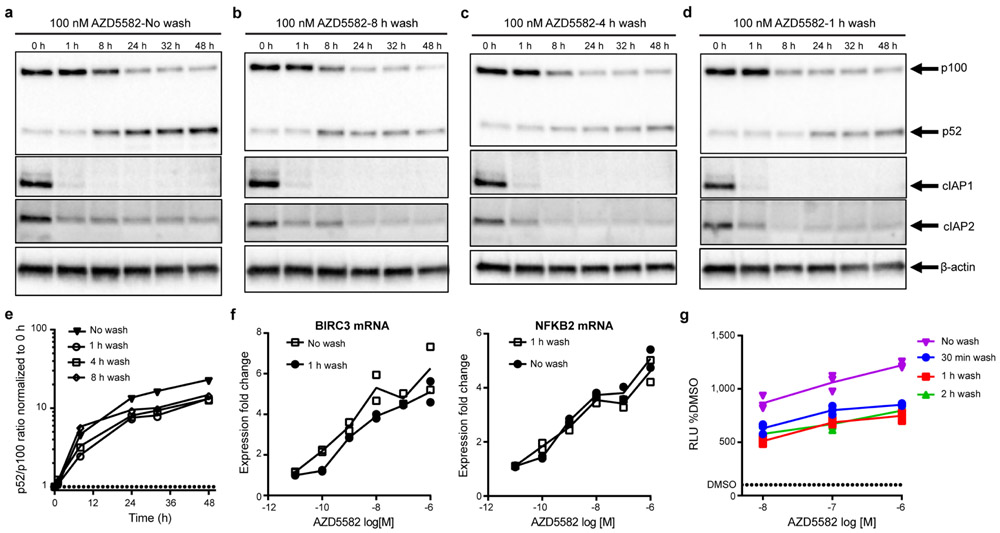
Extended Data Fig. 1. Short duration exposure to AZD5582 activates the ncNF-κB pathway.
(a-d) Isolated total human CD4+ T cells treated with 100 nM of AZD5582 and then either not washed (a) or washed three times with PBS at (b) 8 h, (c) 4 h, or (d) 1 h post-treatment. Whole cell lysates were then analyzed by immunoblot for components of the ncNF-κB pathway. (e) Densitometry analysis of the ratio of p52 to p100 from the pulse-wash assay immunoblots. Points represent values for the densitometric ratio from one western blot, representative of several independent experiments. (a-e) Entire set representative of one experiment with 5 replicates of the 1 h wash condition conducted. (f) Fold induction of ncNF-κB target gene expression in isolated CD4+ T cells from an uninfected donor treated with the indicated concentration of AZD5582, and either washed after 1 h or not washed, then cultured for 24 h as measured by quantitative RT-PCR. Points represent two technical replicates and lines represent the average. The data presented are representative of three independent experiments. (g) DMSO-normalized induction of luciferase activity from the Jurkat reporter model after exposure to AZD5582 (10, 100, or 1000 nM) for either 30 min (blue), 1 h (red), 2 h (green), or continued exposure (purple). Points represent three replicates in one assay run, representative of two independent experiments. Lines represent the average of the three replicates. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 2. AZD5582 induces HIV-RNA expression in resting CD4+ T cells from tissues of HIV-infected, ART-suppressed BLT mice.
HIV-RNA levels in resting CD4+ T cells isolated from the bone marrow (BM), lymph nodes (LN), liver (Liv), lung (lng), and spleen (spl) of control (Cntrl, blue circles) or AZD5582-treated (red circles) mice (cells pooled from n=6 mice/group for each tissue) were analyzed in triplicate. Statistical significance was determined with a two-sided student’s T test. The mean fold increase in viral RNA levels in resting CD4+ T cells from tissues between the experiment shown in Figure 2 of the manuscript was 12.1 (±3.7) and for Extended Data Figure 2 it was 8.4 (±6.0). These values were not statistically different (p=0.4286, two-sided Mann-Whitney test). Error bars, mean ± s.e.m.
Extended Data Fig. 3. AZD5582 ex vivo target engagement.
(a) Western blot analysis of p100, p52 and cIAP1 protein levels in cells isolated from the bone marrow (BM), lymph node (LN), and spleen of BLT mice prior to and 20 h post ex vivo treatment with AZD5582. Loading control: β-actin. Representative of two experiments. (b) cIAP expression in resting CD4+ T cells isolated from the thymus, spleen, lymph nodes (LN), liver, lung and bone marrow (BM). Loading control: β-actin. Representative of two experiments. (c) cIAP expression in the thymic organoid of HIV-infected, ART-suppressed BLT mice 48 h post administration of vehicle control or AZD5582 (three control and one AZD5582-treated analyzed). Positive cells: brown. 4x and 40x magnifications, scale bars: 100 μm (4x) and 50 μm (40x). Boxes on 4x images indicate regions corresponding to 40x magnification images. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 4. Pharmacokinetic and pharmacodynamic assessment of AZD5582 in RMs.
(a) AZD5582 (0.1 mg/kg) was administered to healthy RMs (n=3) by intravenous infusion. Plasma concentrations of AZD5582 (left Y axis) are shown at the indicated time points. Flow cytometry was used to measure intracellular p100 levels, shown as geometric mean fluorescence intensity (gMFI) in CD4+ T cells and plotted as percentage of baseline p100 gMFI (right Y axis). (b) Plasma concentrations of AZD5582 after 1 (dark red), 3 (red), 6 (pink) or 10 (orange) doses in 6 SIV-infected, ART-treated RMs and after 1 dose in 3 uninfected control RMs (gray). Individual values are shown with symbols. (c) Western blot analyses of inactive p100 and active p52 forms of NF-κB2 in lymph node mononuclear cells collected 48 h after the third or tenth dose of AZD5582 in SIV-infected, ART-suppressed RMs (red; n=3 for both 3 dose and 10 dose groups) or at equivalent time points for placebo controls (blue; n=2 for both 3 dose and 10 dose groups). Immunoblots are shown in the top panels and densitometry analyses of the p52:p100 ratios are shown in the bottom panels. Line represents the median. (d) Cryopreserved control RM splenocytes were treated with the indicated concentration of AZD5582 for 48 h, then p100/p52 levels analyzed by Western blotting to measure engagement of the ncNF-κB pathway. For (d and e): the experiments were performed in duplicate. (e) DMSO-normalized densitometric p52:p100 ratio versus the AZD5582 concentration. (f) Cryopreserved RM splenocytes were exposed to DMSO alone (Untx), 100 nM AZD5582 washed off after 1 h and cultured for 47 h (Pulsed), or continuous 100 nM AZD5582 for 48 h (Cont.) then studied by Western blot for p100 and p52 levels. (g) Densitometric p52:p100 ratio. For (f and g): data represents a single experiment. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 5. Phylogenetic trees based on full envelope sequencing.
Plasma virus was sequenced from four time points per animal (n=5): near peak viremia (2 wks post infection; red), immediately prior to ART (8 wks post infection; orange), and two time points of on-ART viremia during AZD5582 treatment (green, blue). All sequences per animal were phylogenetically analyzed and the resulting phylogenetic trees are shown for each animal. The horizontal bar indicates the genetic distance, nt: nucleotide.
Extended Data Fig. 6. SIV-DNA levels in total CD4+ T cells and replication-competent reservoir size in ART-suppressed SIV-infected RMs.
(a) Longitudinal assessment of cell-associated SIV-DNA levels in total CD4+ T cells isolated from peripheral blood (PB) and lymph nodes (LN) of AZD5582-treated (n=12, red) and control (n=4, blue,) ART-suppressed SIV-infected RMs. Gray shading represents the period of ART administration. (b) Comparison of cell-associated SIV-DNA levels in total CD4+ T cells isolated from the LN and PB of AZD5582-treated and control ART-suppressed, SIV-infected RMs. Total CD4+ T cells were analyzed from AZD5582-treated RMs 48 h after receiving 3 doses (LN and PB: n=3) or 10 doses (LN and PB: n=9) of AZD5582. Total CD4+ T cells were analyzed from placebo control RMs (LN and PB: n=4) at equivalent time points. Open symbols indicate AZD5582-treated RMs with on-ART viremia above 60 copies/mL of plasma. Statistical significance was determined with a two-sided Mann-Whitney test. (c) Quantitative viral outgrowth assays (QVOA) were performed from AZD5582-treated RMs 48 h after receiving three doses (LN: n=2, spleen: n=3) or 10 doses (LN: n=3, spleen: n=2) of AZD5582. QVOAs were performed from control RMs (LN and spleen: n=4) at equivalent time points. Open symbols indicate AZD5582-treated RMs with on-ART viremia. Statistical significance was determined with a two-sided Mann-Whitney U test. Horizontal lines represent the median (b,c).
Extended Data Fig. 7. AZD5582-induced gene pathways and genes in SIV-infected, ART-suppressed RMs.
(a) DAVID analysis showing pathways significantly enriched in genes differentially expressed after AZD5582 treatment relative to baseline in CD4+ T cells isolated from LN (black bars) and peripheral blood (gray bars). N=6 for both LN and PB; for each, n=3 for 3 doses of AZD5582 and n=3 for 10 doses of AZD5582. (b) Leading edge genes from the “HALLMARK TNFα SIGNALING VIA NF-κB” pathway from MSigDB. Genes were identified in the leading edge of LN CD4+ T cell samples pre-treatment vs. AZD5582-treated samples (shown in Fig. 4b). The contrast depicted is the fold-change of each gene for each animal’s post-treatment sample relative to its pre-treatment values, with LN CD4+ T cells above and PB CD4+ T cells below. N=6 for both LN and PB; for each, n=3 for 3 doses of AZD5582 and n=3 for 10 doses of AZD5582. (c) Heatmap of cNF-κB (top) and ncNF-κB (bottom) pathway gene expression in RMs with (left, n=5) or without (right, n=6) on-ART viremia > 60 copies/mL plasma. One RM without on-ART viremia was excluded from this analysis for technical issues (higher than expected unmapped and multi-mapped reads, and lower than expected unique identified reads compared to the means). Color scale: log2-fold changes from pre-treatment.
Extended Data Fig. 8. Relationship between virological status pre-intervention and virological response to AZD5582 treatment in SIV-infected, ART-suppressed RMs.
(a) Individual representations of plasma SIV-RNA levels measured by ultrasensitive assay (limit of detection 3 copies/mL of plasma) before and during AZD5582 treatment in SIV-infected, ART-suppressed RMs (n=12). “Baseline” shows two or three plasma viral loads before AZD5582 treatment and “AZD5582” shows three or four plasma viral loads during AZD5582 treatment. Open symbols indicate the five AZD5582-treated RMs with on-ART viremia > 60 copies of SIV-RNA/mL of plasma using the standard viral load assay (confirmed with ultrasensitive assay). The orange box highlights an additional three RMs who did not experience on-ART viremia > 60 copies/mL of plasma but showed ≥ 2 SIV-RNA measurements above baseline values using the ultrasensitive quantification. (b) Comparison of the medians of baseline and during AZD5582 treatment plasma SIV-RNA levels measured by ultrasensitive assay for ART-suppressed SIV-infected RMs (n=12). Statistical significance was determined with a Wilcoxon matched-pairs signed rank test. (c) Comparison of the levels of plasma SIV-RNA at peak, plasma SIV-RNA before ART initiation, SIV-DNA in PB CD4+ T cells before AZD5582 treatment (pre-LRA), SIV-DNA in LN CD4+ T cells pre-LRA, SIV-RNA in PB CD4+ T cells pre-LRA, and SIV-RNA in LN CD4+ T cells pre-LRA in RMs who showed increased plasma viral loads (PVL) by standard and/or ultrasensitive assay during AZD5582 treatment (increased PVL, n=8) vs. RMs who did not demonstrate increased viral loads (stable PVL, n=4). Statistical significance was determined with a two-sided Mann-Whitney test. Horizontal lines represent the median (a,c).
Extended Data Fig. 9. AZD5582 can be safely administered in SIV-infected, ART-suppressed RMs.
Longitudinal assessment of (a) serum chemistries, (b) complete blood counts, and (c) weight in SIV-infected, ART-treated RMs dosed with AZD5582 (red, n=12) compared to controls (blue, n=9). Gray shading represents the period of ART administration. AST: aspartate aminotransferase, ALT: alanine aminotransferase, BUN: blood urea nitrogen, GGT: gamma-glutamyltransferase, WBC: white blood cell, HGB: hemoglobin.
Extended Data Fig. 10. Immunologic impact of AZD5582 in SIV-infected, ART-suppressed RMs.
Longitudinal assessment of CD4+ T cell (a) count, (b) frequency and viability in SIV-infected, ART-treated RMs during the period of AZD5582 treatment (n=12). Lines represent median values. CD4+ T cell naïve and memory subset frequencies (c) and their expression of Ki-67 (d) in SIV-infected, ART-treated RMs during the period of AZD5582 treatment (n=12). Lines represent median values. (e) SIVgag-specific (left) and SIVenv-specific (right) CD8+IFNγ+ T cell responses in SIV-infected, ART-suppressed, AZD5582-treated (red, n=6) and control (blue, n=4) RMs. SFU: spot forming units. (f) Pie charts depicting the ability of memory CD8+ T cells isolated from SIV-infected, ART-suppressed control RMs (n=5) to produce IFNγ, IL-2 and/or TNFα in response to PMA/ionomycin stimulation in the absence (left) or presence (right) of AZD5582 pre-treatment. (g) Memory CD8+ T cell proliferative response to PMA/ionomycin stimulation with or without AZD5582 pre-treatment. Top panels show representative flow cytometry dot plots gated on memory CD8+ T cells. Bottom panel shows the comparison of divided cells 5 d post stimulation in each group (n=5 for each). Statistical significance was determined with a Wilcoxon matched-pairs signed rank test. Horizontal lines represent the median. (h) Longitudinal assessment of plasma levels of IP-10, MIP-1β, MCP-1, IL-6 and IL-10 by multiplex assay in SIV-infected, ART-suppressed RMs treated with AZD5582 (red, n=6) or control RMs (blue, n=4). An additional four analytes (IFNγ, IL-8, IL-1β, and IL-2) were undetectable in all animals. (a-e, h): gray shading represents the period of ART administration. Dashed lines represent AZD5582 or placebo infusions. LLOD: lower limit of detection.
Supplementary Material
Acknowledgments:
We thank Garcia and Chahroudi laboratory members, the Animal Histopathology & Laboratory Medicine Core at the University of North Carolina-Chapel Hill (UNC-CH), supported in part by an NCI Center Core Support Grant (5P30CA016086-41) to the UNC Lineberger Comprehensive Cancer Center and technicians from the Department of Comparative Medicine at UNC-CH. We also thank the HIV/STD Laboratory Core and the Clinical Pharmacology and Analytical Chemistry Core of the UNC Center for AIDS Research (CFAR) (P30 AI050410). We thank D. Hazuda, B. Howell and S. Barrett (Merck & Co.) for assistance with antiretroviral-containing chow. We thank Yerkes Animal and Research Resources. We acknowledge the Children’s Healthcare of Atlanta and Emory University Pediatric Flow Cytometry Core, Emory CFAR Translational Virology and Reservoir Cores, and the Quantitative Molecular Diagnostics Core of the AIDS and Cancer Virus Program, Frederick National Laboratory, as well as GSK for TDF, FTC, and DTG. Funding: This work was supported by the National Institutes of Allergy and Infectious Diseases (NIAID)(AI123010, AI096113, AI111899, AI117851, and P30 AI050410) and Mental Health (NIMH) (MH108179). This work was supported by the Emory Consortium for Innovative AIDS Research in Nonhuman Primates (UM1 AI124436), amfAR (109353-59-RGRL), the Yerkes National Primate Research Center (P51 OD011132), and the Translational Virology and Reservoir Cores of the Center for AIDS Research at Emory University (P30 AI050409). Research was also supported by Qura Therapeutics and by CARE, a Martin Delaney Collaboratory (1UM1AI126619-01) of the NIAID, NINDS, NIDA, and NIMH. By the Natural Science Foundation of Guangdong Province, China (2016A030310108), UNC-South China STD Research Training Center (1D43TW009532) and Chinese National Key Technologies R&D Program for the 13th Five-year Plan (2017ZX10202101003). Federal funds were used for this research from the National Cancer Institute, NIH (Contracts HHSN261200800001E and 75N91019F00129). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Competing Interests: The authors have no competing interests.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Source data for Figs 1-4, Extended Data Figs. 1-2, 4, 6-10, and Supplementary Tables 4-6 are provided with the manuscript. Gene expression data are available at the Gene Expression Omnibus (GEO) repository (accession numbers GSE141546 and GSE142774). Any other data are available from corresponding authors on reasonable request.
Main References
- 1.Finzi D et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5, 512–517, doi: 10.1038/8394 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Archin NM et al. Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J Clin Invest 127, 3126–3135, doi: 10.1172/JCI92684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archin NM et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485, doi: 10.1038/nature11286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott JH et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10, e1004473, doi: 10.1371/journal.ppat.1004473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutierrez C et al. Bryostatin-1 for latent virus reactivation in HIV-infected patients on antiretroviral therapy. AIDS 30, 1385–1392, doi: 10.1097/QAD.0000000000001064 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Kulkosky J et al. Intensification and stimulation therapy for human immunodeficiency virus type 1 reservoirs in infected persons receiving virally suppressive highly active antiretroviral therapy. J Infect Dis 186, 1403–1411, doi: 10.1086/344357 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Prins JM et al. Immuno-activation with anti-CD3 and recombinant human IL-2 in HIV-1-infected patients on potent antiretroviral therapy. AIDS 13, 2405–2410 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen TA et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1, e13–21, doi: 10.1016/S2352-3018(14)70014-1 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Sogaard OS et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS Pathog 11, e1005142, doi: 10.1371/journal.ppat.1005142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke R, Conway JM, Margolis DM & Perelson AS Determinants of the efficacy of HIV latency-reversing agents and implications for drug and treatment design. JCI Insight 3, doi: 10.1172/jci.insight.123052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun SC The noncanonical NF-kappaB pathway. Immunol Rev 246, 125–140, doi: 10.1111/j.1600-065X.2011.01088.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulda S Molecular pathways: targeting death receptors and smac mimetics. Clin Cancer Res 20, 3915–3920, doi: 10.1158/1078-0432.CCR-13-2376 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Pache L et al. BIRC2/cIAP1 Is a Negative Regulator of HIV-1 Transcription and Can Be Targeted by Smac Mimetics to Promote Reversal of Viral Latency. Cell Host Microbe 18, 345–353, doi: 10.1016/j.chom.2015.08.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennessy EJ et al. Discovery of a novel class of dimeric Smac mimetics as potent IAP antagonists resulting in a clinical candidate for the treatment of cancer (AZD5582). J Med Chem 56, 9897–9919, doi: 10.1021/jm401075x (2013). [DOI] [PubMed] [Google Scholar]
- 15.Honeycutt JB et al. T cells establish and maintain CNS viral infection in HIV-infected humanized mice. J Clin Invest 128, 2862–2876, doi: 10.1172/JCI98968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessing CF et al. In Vivo Suppression of HIV Rebound by Didehydro-Cortistatin A, a “Block-and-Lock” Strategy for HIV-1 Treatment. Cell Rep 21, 600–611, doi: 10.1016/j.celrep.2017.09.080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai P et al. In vivo analysis of the effect of panobinostat on cell-associated HIV RNA and DNA levels and latent HIV infection. Retrovirology 13, 36, doi: 10.1186/s12977-016-0268-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melkus MW et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med 12, 1316–1322, doi: 10.1038/nm1431 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Choudhary SK et al. Latent HIV-1 infection of resting CD4(+) T cells in the humanized Rag2(−)/(−) gammac(−)/(−) mouse. J Virol 86, 114–120, doi: 10.1128/JVI.05590-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denton PW et al. Generation of HIV Latency in BLT Humanized Mice. J Virol 86, 630–634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahl A et al. Precision mouse models with expanded tropism for human pathogens. Nat Biotechnol 37, 1163–1173, doi: 10.1038/s41587-019-0225-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahams MR et al. The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci Transl Med 11, doi: 10.1126/scitranslmed.aaw5589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferris AL, Wells DW, Guo S, Del Prete GQ, Swanstrom AE, Coffin JM, Wu X, Lifson JD, Hughes SH. Clonal expansion of SIV-infected cells in macaques on antiretroviral therapy is similar to that of HIV-infected cells in humans. PLoS Pathog. 2019. July 10;15(7):e1007869. doi: 10.1371/journal.ppat.1007869. eCollection 2019 Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson EM & Maldarelli F The role of integration and clonal expansion in HIV infection: live long and prosper. Retrovirology 15, 71, doi: 10.1186/s12977-018-0448-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo HH & Lichterfeld M Recent progress in understanding HIV reservoirs. Curr Opin HIV AIDS 13, 137–142, doi: 10.1097/COH.0000000000000441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clutton GT & Jones RB Diverse Impacts of HIV Latency-Reversing Agents on CD8+ T-Cell Function: Implications for HIV Cure. Front Immunol 9, 1452, doi: 10.3389/fimmu.2018.01452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta RK et al. HIV-1 remission following CCR5Delta32/Delta32 haematopoietic stem-cell transplantation. Nature 568, 244–248, doi: 10.1038/s41586-019-1027-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutter G et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360, 692–698, doi: 10.1056/NEJMoa0802905 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Archin NM et al. Expression of Latent HIV Induced by the Potent HDAC Inhibitor Suberoylanilide Hydroxamic Acid. AIDS Research and Human Retroviruses 25, 207–212, doi: 10.1089/aid.2008.0191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keedy KS et al. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol 83, 4749–4756, doi: 10.1128/JVI.02585-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trumble IM et al. SLDAssay: A software package and web tool for analyzing limiting dilution assays. J Immunol Methods 450, 10–16, doi: 10.1016/j.jim.2017.07.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419, doi: 10.1038/nmeth.4197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y & Hochberg Y Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 57, 289–300 (1995). [Google Scholar]
- 36.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denton PW et al. Antiretroviral pre-exposure prophylaxis prevents vaginal transmission of HIV-1 in humanized BLT mice. PLoS Med 5, e16, doi: 10.1371/journal.pmed.0050016 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denton PW et al. One Percent Tenofovir Applied Topically to Humanized BLT Mice and Used According to the CAPRISA 004 Experimental Design Demonstrates Partial Protection from Vaginal HIV Infection, Validating the BLT Model for Evaluation of New Microbicide Candidates. J Virol 85, 7582–7593, doi:JVI.00537–11 [pii] 10.1128/JVI.00537-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palesch D et al. Short-Term Pegylated Interferon alpha2a Treatment Does Not Significantly Reduce the Viral Reservoir of Simian Immunodeficiency Virus-Infected, Antiretroviral Therapy-Treated Rhesus Macaques. J Virol 92, doi: 10.1128/JVI.00279-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen SG et al. Addendum: Immune clearance of highly pathogenic SIV infection. Nature 547, 123–124, doi: 10.1038/nature22984 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Li H et al. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc Natl Acad Sci U S A 113, E3413–3422, doi: 10.1073/pnas.1606636113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krisko JF, Martinez-Torres F, Foster JL & Garcia JV HIV restriction by APOBEC3 in humanized mice. PLoS Pathog 9, e1003242, doi: 10.1371/journal.ppat.1003242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cartwright EK et al. CD8(+) Lymphocytes Are Required for Maintaining Viral Suppression in SIV-Infected Macaques Treated with Short-Term Antiretroviral Therapy. Immunity 45, 656–668, doi: 10.1016/j.immuni.2016.08.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenbloom DIS, Hill AL, Laskey SB & Siliciano RF Re-evaluating evolution in the HIV reservoir. Nature 551, E6–E9, doi: 10.1038/nature24634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimin AV et al. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 9, 20, doi: 10.1186/1745-6150-9-20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Li J et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood 124, 3636–3645, doi: 10.1182/blood-2014-07-588806 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nixon CC, Mavigner M, Silvestri G & Garcia JV In Vivo Models of Human Immunodeficiency Virus Persistence and Cure Strategies. J Infect Dis 215, S142–S151, doi: 10.1093/infdis/jiw637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mavigner M et al. Simian Immunodeficiency Virus Persistence in Cellular and Anatomic Reservoirs in Antiretroviral Therapy-Suppressed Infant Rhesus Macaques. J Virol 92, doi: 10.1128/JVI.00562-18 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mavigner M et al. Pharmacological modulation of the Wnt/B-catenin pathway inhibits proliferation and promotes differentiation of long-lived memory CD4+ T cells in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 94, doi: 10.1128/JVI.01094-19 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.