Highlights
-
•
Arterivirus RNA synthesis presumably is associated with double-membrane vesicles (DMVs).
-
•
Putative intermediates in DMV formation were detected in infected cells.
-
•
Arterivirus-induced DMVs form a highly interconnected reticulovesicular network (RVN).
-
•
Expression of the nsp2-3 replicase polyprotein fragment induces a comparable RVN.
-
•
Nsp2-7 expression results in smaller DMVs, closer in size to DMVs found in infection.
Keywords: Nidoviruses, Replication complex, Replication structures, Three-dimensional electron microscopy, Correlative light-electron microscopy, Membrane remodelling
Abstract
All eukaryotic positive-stranded RNA (+RNA) viruses appropriate host cell membranes and transform them into replication organelles, specialized micro-environments that are thought to support viral RNA synthesis. Arteriviruses (order Nidovirales) belong to the subset of +RNA viruses that induce double-membrane vesicles (DMVs), similar to the structures induced by e.g. coronaviruses, picornaviruses and hepatitis C virus. In the last years, electron tomography has revealed substantial differences between the structures induced by these different virus groups. Arterivirus-induced DMVs appear to be closed compartments that are continuous with endoplasmic reticulum membranes, thus forming an extensive reticulovesicular network (RVN) of intriguing complexity. This RVN is remarkably similar to that described for the distantly related coronaviruses (also order Nidovirales) and sets them apart from other DMV-inducing viruses analysed to date. We review here the current knowledge and open questions on arterivirus replication organelles and discuss them in the light of the latest studies on other DMV-inducing viruses, particularly coronaviruses. Using the equine arteritis virus (EAV) model system and electron tomography, we present new data regarding the biogenesis of arterivirus-induced DMVs and uncover numerous putative intermediates in DMV formation. We generated cell lines that can be induced to express specific EAV replicase proteins and showed that DMVs induced by the transmembrane proteins nsp2 and nsp3 form an RVN and are comparable in topology and architecture to those formed during viral infection. Co-expression of the third EAV transmembrane protein (nsp5), expressed as part of a self-cleaving polypeptide that mimics viral polyprotein processing in infected cells, led to the formation of DMVs whose size was more homogenous and closer to what is observed upon EAV infection, suggesting a regulatory role for nsp5 in modulating membrane curvature and DMV formation.
1. Membrane modifications to accommodate +RNA virus replication
All positive-stranded RNA (+RNA) viruses of eukaryotes replicate their genome in the cytoplasm of the host cell using a common strategy characterized by the modification of host membranes into organelle-like structures that, for many +RNA viruses, have been directly implicated in viral RNA synthesis (reviewed in Miller and Krijnse-Locker, 2008, Paul and Bartenschlager, 2013, Romero-Brey and Bartenschlager, 2014). Although these virus-induced membrane modifications, often referred to as viral replication organelles or replication structures, have been known for decades, their exact purpose remains enigmatic. Three major advantages of associating viral RNA synthesis with dedicated membranes have been proposed. Firstly, confining viral RNA synthesis in designated compartments could generate optimally suited micro-environments by concentrating the viral proteins and precursors necessary for the process. Furthermore, the anchoring of viral replication complexes to membranes creates a planar geometry for diffusion of metabolites and macromolecules, which can also increase the efficacy of the enzymatic processes. Secondly, compartmentalization could provide a means to spatially separate and coordinate the different stages of the infectious cycle, such as genome translation, replication and packaging. Finally, during viral RNA synthesis several intermediate nucleic acid species such as double-stranded RNA (dsRNA) and 5′ triphosphate-containing RNAs are presumed to be formed, which are potent activators of the innate immune response (Bowie and Unterholzner, 2008, Gurtler and Bowie, 2013). Insulation of these intermediates could therefore prevent or delay detection by the defence systems of the host cell (Neufeldt et al., 2016). Clearly, these proposed functions are not mutually exclusive and, likely, +RNA viruses take advantage of multiple benefits associated with the compartmentalization of their replicative process.
Both viral and cellular factors are thought to be important for the biogenesis of +RNA viral replication organelles, although the specific players and processes involved remain in most cases poorly understood. Multiple +RNA viruses encode non-structural proteins (nsps) that have proven or predicted transmembrane domains and, for some of them, a combination of membrane-associated viral nsps has been shown to be necessary and sufficient for the induction of membrane modifications resembling those found in infected cells (Angelini et al., 2013, Egger et al., 2002, Romero-Brey et al., 2015, Romero-Brey et al., 2012, Salonen et al., 2003, Schwartz et al., 2002, Snijder et al., 2001, Suhy et al., 2000). Besides these viral proteins, a wide variety of host factors have been implicated in the formation and functioning of these structures (reviewed in Belov and van Kuppeveld, 2012, Nagy and Pogany, 2012). Their roles range from the recruitment of viral replication proteins to the induction of membrane modifications. Not surprisingly, the list of identified host factors includes proteins involved in lipid metabolism (e.g. phosphatidylinositol-4-phosphate (PI4P) kinases, fatty acid synthase), specific lipids (e.g. PI4P, sterols), and membrane-shaping proteins (e.g. reticulons and endosomal sorting complexes required for transport (ESCRT) proteins).
A large number of +RNA virus replication organelles have been characterized by electron microscopy (EM) over the last decades. It has become apparent that, despite the large evolutionary distances between these viruses and the different cellular organelles they manipulate, all +RNA viruses seem to induce one of two basic morphotypes of membrane modifications: invaginations or double-membrane vesicles. In the last decade, the study of the architecture of viral replication organelles has been stimulated further by the increasing use of electron tomography (ET). ET enables the three-dimensional (3D) characterization of biological specimens at nanometer resolution by computationally combining images collected at different tilt angles in a transmission electron microscope (Barcena and Koster, 2009). The first +RNA virus replication organelles characterized by ET were those induced by the nodavirus Flock House virus (FHV) (Kopek et al., 2007). FHV induces invaginations in the outer mitochondrial membrane and therefore belong to the first morphotype of +RNA virus replication organelles. Using immuno electron microscopy (IEM) to detect BrUTP incorporated into viral RNA, the interior of these spherules was shown to contain newly-synthesized viral RNA that is thought to be exported to the cytosol through a neck-like channel of ∼10 nm in diameter, which could be clearly visualized in the 3D reconstruction. Since this first study, the membrane modifications induced by, for example, coronaviruses (Knoops et al., 2008, Maier et al., 2013a), arteriviruses (Knoops et al., 2012), flaviviruses (Gillespie et al., 2010, Junjhon et al., 2014, Miorin et al., 2013, Offerdahl et al., 2012, Welsch et al., 2009), hepaciviruses (Romero-Brey and Bartenschlager, 2014), togaviruses (Fontana et al., 2010), picornaviruses (Belov and van Kuppeveld, 2012, Limpens et al., 2011) and tombusviruses (Cao et al., 2015) have been characterized using ET.
The flavivirus dengue virus (DENV) provides another well-characterized example of virus-induced invaginations of cellular membranes (Welsch et al., 2009) that, in this case, occur at the rough endoplasmic reticulum (ER). Despite the different membrane donor organelle, the DENV-induced vesicles share several characteristics with those generated during FHV infection. DENV invaginations are also open to the cytosol through a neck-like connection (∼10 nm in diameter) and contain dsRNA. Apart from these invaginations, DENV also induces the formation of so-called convoluted membranes, which are continuous with the vesicles through ER membranes and are speculated to be a reservoir of proteins and lipids used for DENV replication (Welsch et al., 2009). The formation of ER invaginations that retain a connection to the cytosol has also been shown for other flaviviruses, like tick-borne encephalitis virus (Miorin et al., 2013), West Nile virus (Gillespie et al., 2010), and Langat virus (Offerdahl et al., 2012).
The formation of cytopathic vacuoles (CPVs), which are modified endosomes and lysosomes of about 600–2000 nm that accommodate invaginations on their membranes, is a hallmark for togavirus infection. ET applied to cells infected with rubella virus (RUBV) showed that the CPVs also contained interconnected small vesicles, vacuoles and stacked membranes (Fontana et al., 2010). Furthermore, RUBV recruits rough ER, mitochondria, and Golgi membranes close to the CPVs, presumably to make use of the resources present in these organelles to fuel its genome replication. Semliki Forest virus and Sindbis virus (genus alphavirus) induce early in infection plasma membrane invaginations that appear to be internalized afterwards through the endosomal pathway to produce the CPVs typical of togaviruses (Frolova et al., 2010, Froshauer et al., 1988, Grimley et al., 1968, Kujala et al., 2001).
The morphotype of virus-induced invaginations seems to extend to plant +RNA viruses (reviewed in Laliberte and Sanfacon, 2010; for an ET characterization see Cao et al., 2015). The striking similarities in the morphology of these invaginations across widely different virus groups strongly suggest a highly conserved mechanism behind their formation. In this regard, brome mosaic virus (Bromoviridae), which induces invaginations into the ER, is one of the best studied models for this replication organelle morphotype (Diaz et al., 2012, Schwartz et al., 2002). The insights obtained into the biogenesis and function of BMV-induced invaginations highlighted remarkable parallels with the assembly of retrovirus capsids, which could hint to an ancestral common mechanism to alter cellular membranes that may have been preserved even more broadly among viruses (Ahlquist, 2006).
The second morphotype of replication organelles is strikingly different in architecture to the invagination morphotype and consists of double-membrane vesicles (DMVs). In contrast with the virus-induced invaginations characterized so far, which have the same topology among different virus groups, viral DMVs with different architectures have been found. Thanks to ET, variations in DMV connectivity and topology, not only between different virus groups but sometimes also between subpopulations of DMVs induced by a specific virus, have become apparent (Table 1 ). Moreover, infection with viruses in this group often results in the formation of additional types of membrane modifications, thus creating complex scenarios in which the exact role of each virus-induced structure and their interplay remains elusive (Table 1). An example are the structures induced by the picornaviruses coxsackievirus B3 (CVB3) and poliovirus (PV), two enteroviruses that first generate single-membrane tubular structures that appear to develop into DMVs later in infection, presumably through an enwrapping mechanism (Belov et al., 2012, Limpens et al., 2011).
Table 1.
Arteriviruses and other animal +RNA viruses inducing DMVs.
| Order | Family | Genus | Speciesa | DMVsb |
Additional membrane structuresc | References | ||
|---|---|---|---|---|---|---|---|---|
| Diameter (nm) | Membrane connectionsd | Open or closed to the cytosol | ||||||
| Nidoviruses | Arteriviridae | Arterivirus | EAV | 95e | DMV-ER, DMV-DMV, DMV-PM | Closed | PM | Knoops et al. (2012)h |
| PRRSV | 70–85 | – | – | SMV | Metwally et al. (2010) | |||
| LDV | 100–300 | – | – | Stueckemann et al. (1982) | ||||
| SHFV | 100f | – | – | PM | Wood et al. (1970) | |||
| Coronaviridae | Alphacoronavirus | HCoV-NL63 | 140–210 | – | – | Orenstein et al. (2008) | ||
| HCoV-229E | ∼200g | – | – | Lundin et al. (2014) | ||||
| Betacoronavirus | SARS-CoV | 200–300 | DMV-ER, DMV- DMV, DMV-CM | Closed | CM, VP | Knoops et al. (2008)h | ||
| MHV | 200–350 | – | – | CM | Ulasli et al. (2010) | |||
| MERS-CoV | 150–320 | – | – | CM | de Wilde et al. (2013) | |||
| Gammacoronavirus | IBV | 180e | Isolated DMVs/DMV-ER | Closed | ZM, spherules | Maier et al. (2013b)h | ||
| Picornavirales | Picornaviridae | Enterovirus | CVB3 | 160e | Isolated DMVs | Closed/Open | SMT, MLV | Limpens et al. (2011)h |
| PV | 100–300 | Isolated DMVs | Closed/Open | SMT, MLV | Belov et al. (2012)h | |||
| Unassigned | Flaviviridae | Hepacivirus | HCV | 150e | Isolated DMVs/DMV-ER | Closed/Open | SMV, MLV | Romero-Brey et al. (2012)h |
Equine arteritis virus (EAV), porcine reproductive and respiratory syndrome virus (PRRSV), lactate dehydrogenase elevating virus (LDV), simian haemorrhagic fever virus (SHFV), human coronavirus NL63 (HCoV-NL63), human coronavirus 229e (HCoV-229e), severe acute respiratory syndrome coronavirus (SARS-CoV), mouse hepatitis virus (MHV), Middle East respiratory syndrome coronavirus (MERS-CoV), infectious bronchitis virus (IBV), coxsackievirus B3 (CVB3), poliovirus (PV), hepatitis C virus (HCV).
DMV connections and openings are only specified when 3D studies are available. When different DMV subpopulations have been reported, the distinct features of these subpopulations are separated by a slash.
Paired membranes (PM), single-membrane vesicle (SMV), convoluted membranes (CM), vesicle packets (VP), zippered membranes (ZM), single-membrane tubules (SMT), multilamellar vesicles (MLV).
ER, endoplasmic reticulum.
Average diameter.
Modal diameter.
Not reported, estimate from published images.
3D characterizations by electron tomography.
Notably, whereas the replication organelles for all other members of the family Flaviviridae that have been analysed so far belong to the invagination morphotype, hepatitis C virus (HCV), the prototype of the Hepacivirus genus, predominantly induces DMVs (Ferraris et al., 2013, Romero-Brey et al., 2012). Although single-membrane vesicles could also be detected in HCV-infected cells, the presence of DMVs appeared to correlate with viral RNA replication (Paul et al., 2013, Romero-Brey and Bartenschlager, 2014).These HCV DMVs were described as protrusions from the ER into the cytosol and were often still connected to the ER (Romero-Brey et al., 2012).
Other viruses that, like HCV, seem to use the ER as the membrane donor organelle and also generate DMVs are viruses belonging to the order Nidovirales. This order currently encompasses four virus families, Coronaviridae, Arteriviridae, Roniviridae and Mesoniviridae. Only a limited number of species have been assigned to the latter two families thus far and many aspects of the replication of these viruses remain to be studied in detail (Cowley et al., 2002, Zirkel et al., 2013). In contrast, coronaviruses have been studied extensively because of their role as veterinary pathogens and the large societal impact of the zoonotic disease outbreaks caused by severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). Additionally, several established human coronaviruses are widespread in the population and frequently associated with common colds. Detailed 3D ultrastructural characterizations of virus-induced membranous structures have been published in the past years for some nidoviruses, specifically, for the arterivirus equine arteritis virus (EAV) (Knoops et al., 2012) and the coronaviruses SARS-CoV and infectious bronchitis virus (IBV) (Knoops et al., 2008, Knoops et al., 2010, Maier et al., 2013b). This review focuses on arterivirus replication organelles, for which new data regarding their biogenesis and the viral players involved will be presented. The fundamental analogies and differences between arteriviral and coronaviral replication organelles, as revealed by recent studies, will be analysed in parallel and also in the wider context of DMV-inducing +RNA viruses. Finally, we will discuss some of the open questions and future research directions that, in our view, will be important to decipher the role of these remarkable structures.
2. Arteriviruses and the membrane rearrangements they induce
2.1. Arterivirus biology
Although the arterivirus prototype EAV was discovered in 1953 (Bryans et al., 1957), the family Arteriviridae was not established until 1996 (Cavanagh, 1997). The group was long comprised of four members: EAV, porcine reproductive and respiratory syndrome virus (PRRSV), simian haemorrhagic fever (SHFV) and lactase dehydrogenase-elevating virus (LDV). However, over the course of the last five years, several new (tentative) arteriviruses were discovered such as wobbly possum disease virus (WPDV), African pouched rat virus 1 and several new simian arteriviruses that are distantly related to SHFV (Dunowska et al., 2012, Kuhn et al., 2015, Lauck et al., 2013, Snijder et al., 2013). Because of their largest economic impact, EAV and PRRSV have been characterized most extensively and research on these viruses has provided the basis of our current understanding of arterivirus replication.
Arteriviruses have polycistronic +RNA genomes with sizes ranging from 13 to 16 kb (Lauber et al., 2013). The two large 5′-proximal open reading frames (ORF1a and ORF1b), which encompass more than 60% of the genome, encode the replicase or non-structural (poly)proteins (Fig. 1 A). The 3′-proximal part of the genome contains an array of smaller ORFs encoding the structural proteins, which are expressed from a nested set of subgenomic (sg) mRNAs (Fig. 1A) (Snijder et al., 2013). After virus entry, the genome is released into the cytosol and ORF1a can be directly translated into polyprotein 1a (pp1a). Translation can be extended using the downstream ORF1b reading frame when a −1 ribosomal frameshift (RFS) occurs just before the end of ORF1a. This event, which results in the synthesis of pp1ab, is promoted by a specific heptanucleotide shift site and an RNA pseudoknot structure (Brierley et al., 1989, den Boon et al., 1991). The polyproteins pp1a and pp1ab contain the viral nsps, which are released by multiple internal proteases (Li et al., 2014, Ziebuhr et al., 2000). Together they are often referred to as the replicase, as they either are part of or support the functions of the so-called replication and transcription complex (RTC), the aggregate enzymes that replicate the viral genome and produce the sg mRNAs used to express the structural proteins.
Fig. 1.
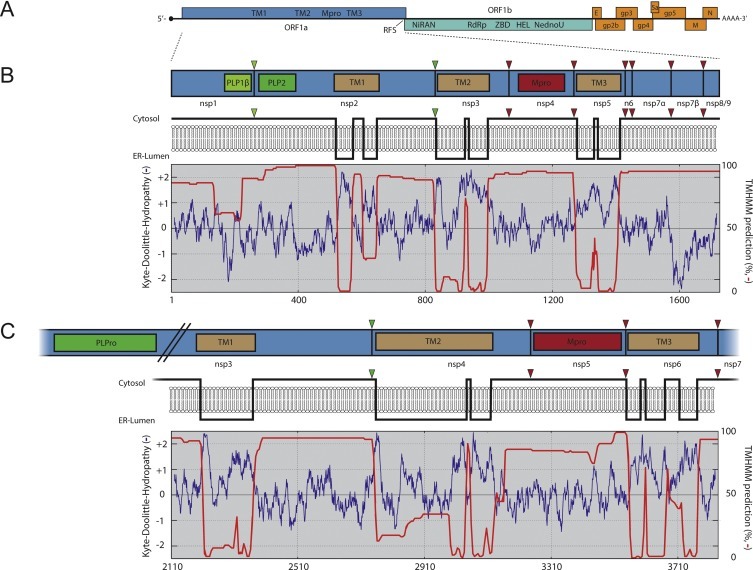
Comparison of transmembrane non-structural proteins of arteri- and coronaviruses. (A) Overview of the EAV genome organization with several conserved domains encoded in ORF1a and ORF1b indicated (see main text for details). The structural protein genes, such as those encoding the viral glycoproteins (GP), envelope (E), membrane (M) and nucleocapsid (N) proteins, are located in the 3′-proximal quarter of the genome (shown in orange). (B) The top panel presents a scaled schematic of EAV pp1a. The three proteases (PLP1β, PLP2 and Mpro) are indicated with coloured boxes and their corresponding cleavage sites in pp1a with triangles. The middle panel shows a model of the membrane topology of EAV ORF1a based on a Kyte & Doolittle hydropathy plot (blue line) and TMHMM predictions (red line; the percentage on the right represents the probability for that part of the protein to be cytosolic), which are both shown in the lower panel. (C) SARS-CoV nsp3-7 region, represented as in B. The region encompassing the TM domains is shown using the same scale as used for EAV pp1a (amino acids 2110–3836 of SARS-CoV pp1a). SARS-CoV nsp3 has been truncated internally to include the protease corresponding to the nsp3/4 cleavage site. Kyte & Doolittle hydropathy plots were generated on www.expasy.org and TMHMM predictions using the Geneious software package both based on database sequences of EAV (NC_002532.2) and SARS-CoV (NC_004718.3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The ORF1b-encoded subunits include the main enzymatic activities required for RNA replication, such as the RNA-dependent RNA polymerase (RdRp) and the helicase (den Boon et al., 1991, Fang and Snijder, 2010, Gorbalenya et al., 2006, Lehmann et al., 2015a, Lehmann et al., 2015b, Snijder et al., 2013). The pp1a subunits seem to support the viral RNA replication more indirectly. Their functions include modulation of host gene expression, proteolytic activity required for polyprotein maturation, and modification of host cell membranes to accommodate viral RNA synthesis.
When considering nidoviruses at large, a conserved array of replicase domains can be defined, consisting of the viral main protease (Mpro) flanked by two transmembrane (TM) domains, the ORF1a/1b ribosomal frameshift site, a nidovirus RdRp-associated nucleotidyltransferase (NiRAN), the RdRp, a zinc-binding domain (ZBD), a superfamily 1 helicase (HEL) and, only in vertebrate nidoviruses, an endoribonuclease (NendoU) (Gorbalenya et al., 2006, Gorbalenya et al., 1989, Lauber et al., 2013, Lehmann et al., 2015a). In all arteri- and coronavirus replicases, a third TM domain (TM1) is present upstream of the hydrophobic regions that flank the main protease (TM2 and TM3; Fig. 1A). Although many other virus groups contain some or most of these elements, the array of (TM1-)TM2-Mpro-TM3-RFS-NiRAN-RdRp-ZBD-HEL is nidovirus specific.
Arteriviruses encode multiple papain-like proteases (PLPs) and a 3C-like or chymotrypsin-like protease that is their Mpro (reviewed in Fang and Snijder, 2010, Snijder et al., 2013, Ziebuhr et al., 2000). The number of active PLP domains varies per virus species (e.g. 2 for EAV, 3 for PRRSV and LDV, and 4 for SHFV) and these proteases all reside in the first 850 residues upstream of TM1, where each of them processes a single downstream cleavage site (Vatter et al., 2014). In the case of EAV, the PLPs located in nsp1 and nsp2 (called PLP1β and PLP2) both swiftly process a single site in the polyprotein (the nsp1/nsp2 and nsp2/nsp3 junctions, respectively; Fig. 1B). Mpro is located in nsp4 and processes the remainder of pp1a/pp1ab (the nsp3-8/nsp3-12 region; Fig. 1B). The C-terminal half of pp1a encompassing nsp3-8 can be processed following two different pathways, each yielding specific end products. The pathway used for nsp3-8 processing depends on the presence or absence of cleaved nsp2 (Wassenaar et al., 1997). When free nsp2 is present, the nsp4/nsp5 junction can be cleaved, which yields the nsp5-7 and nsp5-8 cleavage products that, apart from the slow processing of the nsp7/nsp8 junction, do not seem to undergo further processing. In the absence of nsp2, the nsp4/5 junction remains uncleaved, but instead the nsp5/6, nsp6/7 and possibly the nsp7α/7β sites are processed (Li et al., 2012, Li et al., 2014, van Aken et al., 2006, Wassenaar et al., 1997). In infected cells, the nsp4/5 junction is mostly cleaved, resulting in the majority of nsp5 being present in nsp5-7 and nsp5-8 cleavage products; however, cleavage of sites in nsp5-7, even though occurring in a minority of cases, is critical for virus replication (van Aken et al., 2006).
For the three TM regions in pp1a of arteriviruses, located in nsp2, nsp3 and nsp5, the exact membrane topology has not been established in full experimental detail, but all of them are predicted to span the membrane multiple times (Faaberg and Plagemann, 1996, Pedersen et al., 1999). Furthermore, it is assumed that both termini of all of these nsps must face the cytosol, where the viral proteases that must have access to the cleavage sites are present (Fig. 1B) (Posthuma et al., 2008). Nevertheless, PRRSV TM1 in nsp2 was suggested to span the membrane 5 times after in vitro translation of nsp2 alone in the presence of artificial membranes (Kappes et al., 2015). Such a topology, however, would appear to pose a major problem for polyprotein processing since PLP2 and the nsp2/3 cleavage site would reside on opposite sides of the membrane. The transmembrane nature of EAV TM2 in nsp3 was confirmed by engineering a site for N-linked glycosylation in the first predicted luminal loop, which was indeed glycosylated when pp1a was expressed transiently (Posthuma et al., 2008). This result strongly supports the nsp3 topology model that places the nsp2/3 cleavage site at the cytoplasmic side of the membrane. Translocation of the nsp2C-terminal domain following polyprotein cleavage would be compatible with all these observations; however, evidence supporting this hypothesis remains to be obtained.
When the TM1-TM2-Mpro-TM3 regions of the arterivirus and coronavirus pp1a polyproteins are compared (Fig. 1), their general organization is strikingly similar, despite the 2.5-fold overall pp1a size difference between the two families. In both virus groups, the junction between the nsps containing TM1 and TM2 is cleaved by a PLP located in the TM1-containing nsp (PLP2 in nsp2 for arteriviruses and PLPro in nsp3 for coronaviruses) (Harcourt et al., 2004, Kanjanahaluethai and Baker, 2000, Snijder et al., 1995, Ziebuhr et al., 2001). The other TM-containing nsps (nsp3 and nsp5 for arteriviruses and nsp4 and nsp6 for coronaviruses) are both released from the polyprotein by the Mpro situated between TM2 and TM3 (Ziebuhr et al., 2000). Moreover, the predicted tetraspanning membrane topology of TM2 is very similar, with two ER luminal domains of which the N-terminal one is the largest (Oostra et al., 2007, Posthuma et al., 2008). The fact that the general arrangement of hydrophobic domains in pp1a is conserved between arteriviruses and coronaviruses probably points to a common, and important, function in virus replication.
2.2. Host membrane remodelling in arterivirus infection
The non-structural proteins containing the predicted TM domains (Fig. 1B and C) appear to be the main players in the modification of host cell intracellular membranes (Angelini et al., 2013, Hagemeijer et al., 2014, Pedersen et al., 1999, Snijder et al., 2001) (see also Section 4). These proteins, together with several other nsps like the RdRp-containing nsp9 and the helicase-containing nsp10, were shown by immunofluorescence and IEM to localize to the same membrane structures in the perinuclear region of EAV-infected cells (Pedersen et al., 1999, van der Meer et al., 1998, van Dinten et al., 1996). This suggested that arterivirus RNA synthesis is associated with these intracellular membranes. When a crude membrane fraction was isolated from EAV-infected cells, the viral RTC was retained and RNA synthesis could be reconstituted in vitro, which further supported the concept of membrane-associated viral RNA synthesis (van Hemert et al., 2008a). Interestingly, this in vitro RTC activity was lost after detergent treatment, strongly suggesting that the RTC’s functionality depends on the integrity of the membranes (van Hemert et al., 2008a). Similar data has also been published for coronaviruses (Bost et al., 2000, Brockway et al., 2003, Denison et al., 1999, Harcourt et al., 2004, Prentice et al., 2004b, Shi et al., 1999, Snijder et al., 2006, van der Meer et al., 1999, van Hemert et al., 2008b).
The remarkable ability of arteriviruses to induce the formation of DMVs in the cytoplasm of infected cells was already observed by electron microscopy in the 1970s (Breese and McCollum, 1970, Wood et al., 1970) and has been noticed for all family members analysed by EM to date (Metwally et al., 2010, Pedersen et al., 1999, Pol et al., 1997, Stueckemann et al., 1982, Wada et al., 1995, Weiland et al., 1995). These DMVs, with a diameter of about 100 nm (Table 1), appear in the perinuclear region of the cell and then proliferate in number giving rise to cytoplasmic clusters that often also show an increased presence of free ribosomes (Knoops et al., 2012, Stueckemann et al., 1982, Wood et al., 1970).
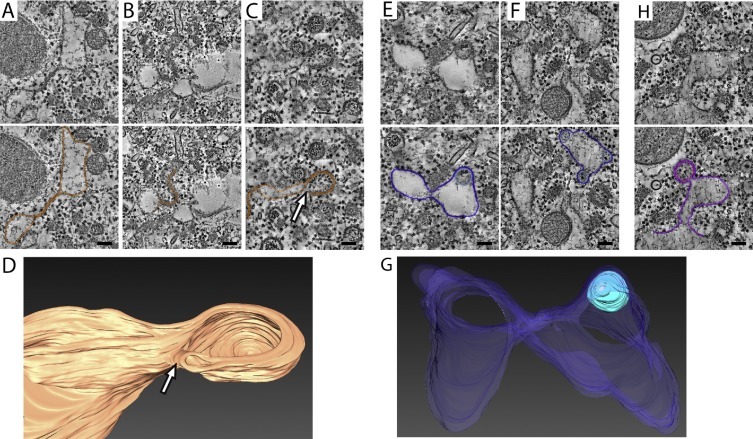
A significant breakthrough in our understanding of the complex architecture and spatial arrangement of arteriviral replication organelles was brought about by applying electron tomography to EAV-infected Vero E6 cells (Knoops et al., 2012) (Fig. 2 A, Supplementary movie S1). More recently these 3D imaging studies were expanded to HuH-7 cells in our laboratory (see Supplementary information) showing that the original observations were not unique for a single cell line (Fig. 2). Occasionally, neck-like connections between the endoplasmic reticulum and the outer membrane of DMVs had been observed in 2D images (Pedersen et al., 1999). However, the 3D analysis showed that these narrow connections are actually rather common. This type of connections (Fig. 2B, white arrows) pointed directly to the ER as the most likely membrane donor organelle, a notion further supported by the fact that ribosomes decorating the outer membrane of the DMVs were regularly observed in the tomograms (Fig. 2C, black arrowheads). Although DMVs are the most prominent and abundant EAV-induced membrane structures, other modified membranes in the form of short stretches of paired membranes or small complex membrane arrangements connecting DMVs to one another or to the ER, were also apparent in the tomograms (Fig. 2D–F). The large majority of the DMVs seemed to be connected to the ER, modified membranes, other DMVs or a combination of the above (Fig. 2B, D–G, white arrows). It should be pointed out that not all of these associations appeared as clear membranous connections but, instead, as tight membrane-membrane interactions that were particularly frequent between DMVs (Fig. 2C and G, black arrows). Only about 10% of the DMVs in the tomograms appeared as separate, free-floating vesicles and this may even be an overestimation since narrow connections in the top or bottom regions of the DMVs cannot be detected due to the intrinsic lower resolution of the tomograms along the Z axis (Barcena and Koster, 2009). Therefore, the general concept that emerges from the 3D analysis is that of a large reticulovesicular network (RVN), consisting of DMVs and modified ER.
Fig. 2.
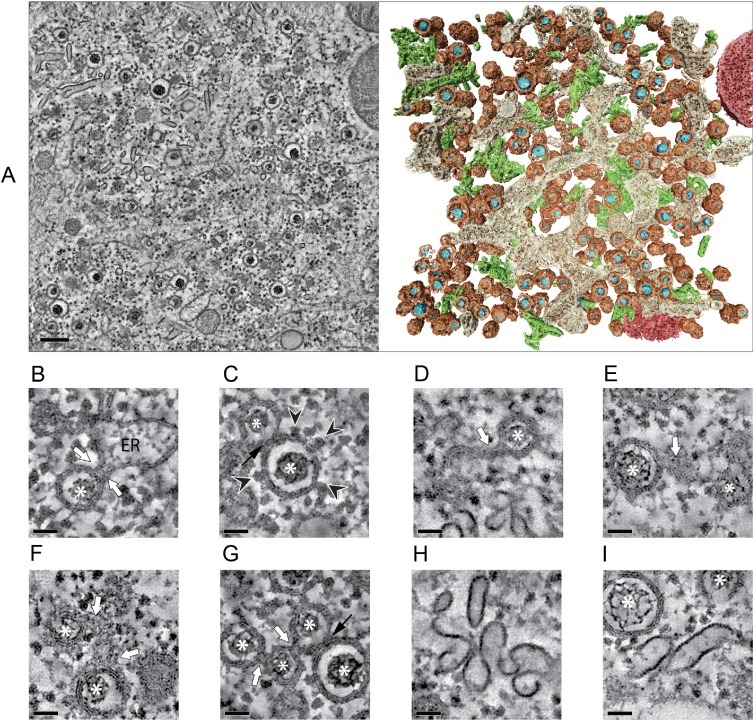
Electron tomography analysis of the EAV-induced RVN. (A) Typical region containing EAV-induced structures in Vero E6 cells at 8 h post-infection (p.i.). Tomographic slice (10 nm thick) extracted from the dual-axis reconstruction of a 200-nm-thick section (left) (see also Supplementary movie S1), and corresponding surface-rendered model (right) displaying DMVs (membranes in brown, electron-dense cores in blue), ER (beige), EAV-induced tubular network (green), and mitochondria (red). (B-I) Close-ups illustrating detailed features of EAV-induced structures. The gallery consists of virtual thin slices extracted from tomograms of infected Vero E6 (B–D,G,H) and HuH-7 cells (E,F,I). In both cell lines, identical features were observed. EAV-induced DMVs often appear connected through their outer membrane (white arrows) to the ER (A), and ribosomes decorate their outer membrane (B, arrowheads), suggesting an ER origin for these membranes. Additional membrane connections of DMVs to paired membranes (D, E) or small complex membranous structures (F), and/or to other DMVs (G) are common. Close membrane-membrane interactions between different DMVs are also frequent (black arrows, C, G). The typical electron-dense cores inside the DMVs are indicated by asterisks. (H, I) Fragments of the tubular network induced by EAV. Note the difference between the single electron-dense layer of the tubular profiles and the trilaminar profile typical of lipid bilayers. This tubular network (in green in the model in 2A) is intertwined with EAV replication structures and contains the nucleocapsid protein (Knoops et al., 2012). Scale bars, 200 nm (A), 50 nm (B–I). For experimental details on EAV-infected HuH-7 samples see Supplementary information. B–D,G: adapted from (Knoops et al., 2012) with permission. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As an additional complexity, a few hours after the appearance of the first DMVs, a rather different EAV-induced structure could be observed between the clusters of DMVs. ET revealed that it consists of a network of branching tubular structures, with an average diameter of approximately 43 nm, (Fig. 2A, right panel; in green; Fig. 2H and I) lacking the typical trilaminar profile of lipid bilayers in EM, and thus most probably representing a proteinaceous structure. Using IEM, these tubules were found to abundantly label for the arteriviral N protein (Knoops et al., 2012). Similar tubules had been detected before in EAV-infected BHK-21 cells, both upon infection (Wada et al., 1995) and upon transfection with EAV RNA replicons (Wieringa et al., 2004). The formation of these tubules seems to depend on expression of the arteriviral N protein as they were absent when the transfected viral RNA lacked the N protein gene. Nevertheless, additional viral components are likely required in the process as N protein expression by itself appeared to be insufficient to generate tubules (Wieringa et al., 2004).
Together with the previously reported observation that similar tubules extended into budding virus particles (Wada et al., 1995), the data suggests that these structures are involved in virus assembly, a process that is still largely uncharacterized for arteriviruses. The fact that this tubular network appears entwined with the RVN could reflect an efficient way of spatially linking the synthesis and packaging of genomic RNA.
Our detailed 3D characterization of the ultrastructure of the EAV-induced RVN revealed additional interesting details with intriguing functional implications. For example, in the DMV interior a previously unreported electron dense core could clearly be distinguished (Fig. 2A, blue; Fig. 2B–I, asterisks). The detection of this core depended on the use of high-pressure freezing (HPF) followed by freeze substitution (FS) for EM sample preparation (Knoops et al., 2012). In chemically fixed samples, the conventional technique used in all previous studies on arterivirus-induced membrane modifications, the DMV interior appeared mainly electron translucent. Using plunge-freezing followed by FS, a granular content became apparent but a dense core was only visible in HPF-FS samples. HPF-FS is considered the gold standard for the ultrastructural preservation of resin-embedded specimens as it has several advantages over alternative methods (McDonald, 2007). In our comparison of different protocols for fixation of EAV-infected cells, it was evident that HPF-FS offered superior preservation, not only of the RVN but also of the cytosolic content and various cellular organelles (Knoops et al., 2012).
The average diameter of the DMV cores (∼50 nm) appears relatively constant during the course of infection and their individual size seems directly proportional to the size of the surrounding DMV. Interestingly, in IEM samples, these cores labelled strongly for dsRNA, a presumed intermediate in viral RNA replication (Fig. 3 A). Consistent with this observation, these structures were shown by 2D and 3D electron spectroscopic imaging (ESI) to be high in phosphorus content (Fig. 3B–D). ESI is an EM technique that takes advantage of the fact that electrons that are inelastically scattered by different elements in the sample lose different amounts of energy. Using this principle, and by collecting images using only electrons that have suffered specific energy losses, images that reveal the location and concentration of a certain element, known as elemental maps, can be obtained (Leapman and Aronova, 2007). In this manner, the average phosphorous content of the cores was determined to be equivalent to about a dozen copies of the EAV RNA genome (Knoops et al., 2012).
Fig. 3.
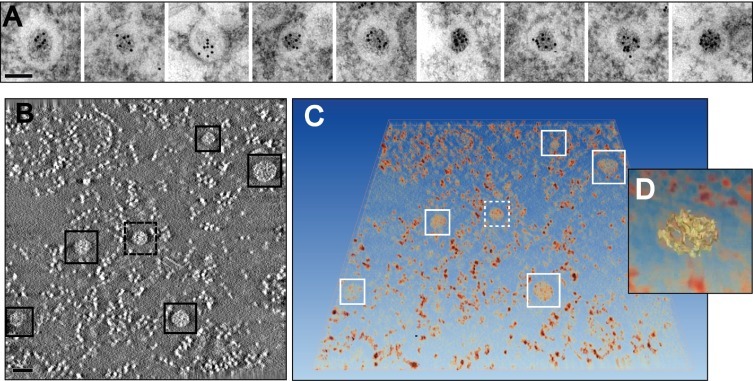
Analysis of the cores in EAV-induced DMVs. (A) Immunogold labelling for dsRNA in EAV-infected Vero E6 cells (7 h p.i.). The gallery of DMVs illustrates the strong labelling for dsRNA of the DMV core. (B–D) Analysis of the phosphorous content in EAV-infected Vero E6 cells (8 h p.i.) by 3D electron spectroscopy imaging (ESI). A tilt series of elemental map images for P was obtained using the jump-ratio approach as described in (Knoops et al., 2012) from a 75-nm thick section of an infected cell. (B) Tomographic slice (∼5 nm thick) extracted from the final ESI reconstruction, showing the high P content (higher intensity) of the DMV cores (boxed). The smaller brighter structures between DMV cores correspond to free ribosomes, also rich in P, which are always particularly abundant in the areas containing DMVs. (C) Direct volume rendering of the ESI tomogram showing the P-content in 3D. (D) Close-up of one of the DMV cores (discontinuous box in B and C), which is highlighted in yellow in isosurface display mode. The resulting isosurface, generated at an intensity threshold level that revealed isolated ribosomes, suggests a thread-like structure of the core RNA content. Scale bars, 100 nm. Adapted from (Knoops et al., 2012) with permission. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In view of the presence of dsRNA inside DMVs, it is tempting to speculate that the DMV interior is the site of viral RNA synthesis as this could provide a propitious environment that would also hide dsRNA replication intermediates from the innate immune sensors of the host. In analogy, for the other morphotype of +RNA virus-induced membrane alterations, the interior of the invaginated spherules seems to be the site of viral RNA synthesis (Friedman and Sreevalsan, 1970, Grimley et al., 1968, Kopek et al., 2007, Kujala et al., 1999, Kujala et al., 2001, Schwartz et al., 2002). However, these invaginations have openings to the cytosol that permit the necessary import of substrates and export of RNA products, whereas in the case of EAV DMVs the inner membrane defines a closed compartment, apparently sealed from the cytosol, thus creating a topological conundrum with respect to RNA synthesis.
The situation is further complicated by the fact that, according to immunolabelling experiments, key replicase subunits, like nsp3 and the RdRp-containing nsp9, localize to DMVs and surrounding RVN membranes but only rarely to the core of DMVs (Knoops et al., 2012). At this point it is important to note that the presence of neither dsRNA nor nsps may be bona fide markers for the localization of the active site of viral RNA synthesis. For example, only a small part of the abundantly expressed nsps is likely to be involved in active RNA-synthesizing complexes, as previously calculated for HCV (Quinkert et al., 2005). Likewise, abundant labelling of DMV cores could reflect accumulation of dsRNA intermediates rather than active RNA synthesis. A less ambiguous alternative to pinpoint viral RNA synthesis is by metabolic labelling of nascent viral RNA followed by EM detection. This latter approach was followed in the past for EAV (Pedersen et al., 1999) by transfecting cells with Br-UTP, after which labelling with a Br-U-specific antibody revealed some signal associated with DMV membranes. However, the size of the DMVs, relatively small when considering the distance that separates the gold markers from the original epitopes (up to ∼30 nm), the limited sample preservation and the qualitative nature of the analysis precluded an accurate localization of the active RTC. Furthermore, signal in the cytosol was also observed, probably reflecting the fact that the labelling times may have been too long to prevent the migration of newly-synthesized RNA.
2.3. Comparison with coronavirus-induced membrane modifications
In the wider context of the order Nidovirales, coronaviruses constitute the only other family for which the putative replication organelles have been analysed in some detail. Coronavirus-induced membrane alterations were first described in the 1960s (David-Ferreira and Manaker, 1965; Svoboda et al., 1962) and have been further investigated in the past decade. In all cases examined so far, also coronaviruses were shown to induce the formation of DMVs. This includes members of three of the four genera of the coronavirus family: alphacoronaviruses (Lundin et al., 2014, Orenstein et al., 2008), betacoronaviruses (de Wilde et al., 2013, Goldsmith et al., 2004, Gosert et al., 2002, Knoops et al., 2008, Snijder et al., 2006, Ulasli et al., 2010), and gammacoronaviruses (Maier et al., 2013b). Considering the pervasiveness of DMVs across the coronavirus and arterivirus families, it seems plausible that this is a shared trait that would extend to other nidoviral families not yet characterized. A striking difference between arterivirus and coronaviruses DMVs concerns their size: coronaviruses induce DMVs that are, on average, around twice the diameter of those in arterivirus-infected cells (Table 1, Fig. 4 A), which corresponds to a ∼8 fold difference in DMV volume.
Fig. 4.
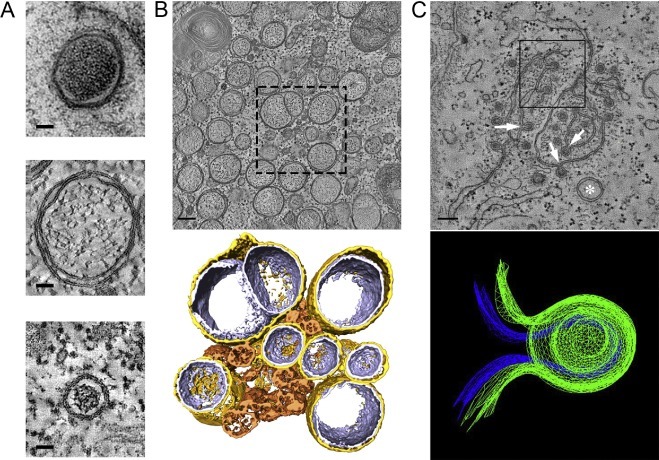
Coronavirus-induced membrane structures. (A) Gallery of nidovirus-induced DMVs represented to scale. The panel shows DMVs in cells infected with (from top to bottom) IBV (CK cells, HPF-FS), SARS-CoV (Vero E6, plunge-freezing and FS) and EAV (HuH-7 cells, HPF-FS). The top image is a 2D projection, while the middle and bottom images are virtual slices (2 nm thick) extracted from tomograms. (B) Membrane modifications induced by SARS-CoV infection in Vero E6 cells (7 h p.i.). The top panel is a 2-nm thick virtual slice from a tomogram. The boxed area represents the region that was used for the 3D surface rendering in the lower panel. In this model, gold represents the outer membrane of the DMV is coloured in gold, the inner membrane in silver, and CM in bronze. (C) Membrane modifications induced by IBV infection in CK cells (16 h p.i., chemical fixation) The upper panel is a virtual slice taken from a tomogram in a region rich in IBV-induced zippered membranes where a DMV (white asterisk) is also present. These zippered membranes often curve to form spherules with a neck that connects their interior to the cytosol (white arrows). The upper spherule in the boxed region was used to create the 3D mesh model presented in the lower panel. Blue represents the inner membrane and green the outer membrane of the spherule. Scale bars, 50 nm (A), 100 nm (B,C). A (middle), B: adapted from (Knoops et al., 2008). A (top), C: adapted from (Maier et al., 2013b). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The betacoronavirus SARS-CoV was the second +RNA virus for which the virus-induced membrane modifications were characterized in 3D by electron tomography (Fig. 4B) (Knoops et al., 2008). This study uncovered DMVs that were connected through their outer membrane with other DMVs and the ER, together forming an RVN of modified ER membranes that is topologically comparable to that described for EAV in the previous section (Knoops et al., 2012). The remarkable similarities between these two distantly-related nidoviruses suggested that the RVN architecture may be a universal feature among nidoviruses. This idea, however, has been challenged by a recent tomographic study of the structures formed upon infection of primary cells with the gammacoronavirus IBV (Fig. 4C) (Maier et al., 2013b). Remarkably, in these tomographic studies, most of the IBV-induced DMVs were found to be isolated vesicles: no DMV-DMV connections were observed and only a small proportion of DMVs appeared to be connected to the ER. Additionally, and in contrast to the case of EAV- and SARS-CoV-induced DMVs, ribosomes were not found to be associated with the outer membrane of IBV-induced DMVs. Whether these features represent specific differences for gammacoronaviruses or also apply to some of the other nidovirus (sub)groups remains to be analysed. In any case, the RVNs generated by SARS-CoV and EAV appear to be a unique example of highly-interconnected membrane modifications among the +RNA viruses that induce DMVs (Table 1). The overall DMV connectivity is decidedly more limited in the case of HCV, for which direct membranous connections between DMVs were not observed (Romero-Brey et al., 2012). Enterovirus-induced modifications appear to be at the opposite end of the spectrum relative to nidoviral RVNs in terms of connectivity, as all of them seem to be isolated compartments with no membranous connections between them or any cellular organelle (Belov et al., 2012, Limpens et al., 2011).
A common characteristic of coronavirus-infected cells, which is absent in cells infected with arteriviruses, is the formation of relatively disorganized assemblies of tangled, partially double membranes. These structures, now mostly referred to as convoluted membranes (CM), were already described for mouse hepatitis virus (MHV) in 1965 as ‘reticular inclusions’ (David-Ferreira and Manaker, 1965). Since then, they have been further characterized for several betacoronaviruses including MHV (Ulasli et al., 2010), SARS-CoV (Knoops et al., 2008) and the recently emerged Middle East Respiratory Syndrome coronavirus (MERS-CoV) (de Wilde et al., 2013). DMV formation seems to precede CM formation, since the latter has been observed predominantly at later time points after infection (de Wilde et al., 2013, Knoops et al., 2008, Ulasli et al., 2010). CM are found in close proximity to the DMVs, but only their 3D characterization in SARS-CoV-infected cells unambiguously showed the presence of neck-like membranous connections between the CM and surrounding DMVs and ER cisternae (Knoops et al., 2008), which make the CM also an integral part of the coronavirus-induced RVN. Notably, CM have been described so far only for cells infected with betacoronaviruses and could be a feature restricted to this genus. In the few ultrastructural studies of cells infected with alphacoronaviruses, CM were not reported (Lundin et al., 2014, Orenstein et al., 2008); however, these studies focused on virus morphogenesis and DMVs, respectively, and therefore it is unclear whether CM may have been present. For the gammacoronavirus IBV, a thorough search for this structure was reported to be unsuccessful (Maier et al., 2013b).
Interestingly, in cells infected with the gammacoronavirus IBV, a distinct and novel membranous structure was described consisting of double-membrane spherules that were continuous with zippered ER cisternae and had a channel of 4.4 nm in diameter connecting them to the cytosol (Fig. 4C) (Maier et al., 2013b). Although these spherules are delineated by two membranes instead of one, their size (∼60 nm) and overall configuration with an opening to the cytosol was very reminiscent of the single-membrane invaginations induced by a large number of +RNA viruses (see Section 1). Based on this similarity, the authors proposed that viral RNA synthesis would take place inside these spherules. This possibility, which requires further experimental corroboration, is particularly attractive in view of the closed configuration of the DMVs found in coronavirus infection. Both for IBV- and SARS-CoV-infected cells, the tomograms revealed DMVs in which the inner membrane seems to be fully sealed with no apparent openings to the cytosol. This makes it difficult to envision how viral RNA synthesis could take place (at least exclusively) inside these vesicles. This situation is completely analogous to the case of the EAV-induced DMVs discussed in the previous section and the parallels extend to the localization of different viral markers by IEM. Most of the dsRNA signal in SARS-CoV-infected cells was shown to map to the inner compartment of the DMVs, whereas the bulk of the labelling for several replicase proteins was located predominantly in the surrounding CM (Knoops et al., 2008). The CM in MHV-infected cells also abundantly labelled for several nsps (Ulasli et al., 2010). The experiments using metabolic labelling of newly-synthesized viral RNA, which could be crucial to pinpoint the actual site of viral RNA synthesis, are restricted so far to MHV. IEM detection of newly-synthesized RNA using BrUTP as a probe revealed signal associated to the regions of DMV clusters, but the results are difficult to interpret due to the limited sample preservation that resulted in a clear loss of the DMV content and, possibly, also of the CM (Gosert et al., 2002). In a recent study, using incorporation of ethynyl uridine into RNA and its detection through click-chemistry followed by light microscopy (Hagemeijer et al., 2012), a loss of colocalization between the ethynyl-uridine and the dsRNA signal was observed as infection progressed. The ethynyl uridine-containing foci, which early in infection overlapped with the dsRNA signal, became dispersed in the cytoplasm late in MHV infection. This prompted the idea that RNA synthesis could take place, at least during these later stages, in regions of the cell distant from those containing DMVs. The interpretation of these results at the ultrastructural level, however, is not possible in the absence of a parallel EM characterization.
Overall, the question of the actual site of viral RNA synthesis for arteriviruses and coronaviruses appears more enigmatic than ever and it is certainly a key issue to be addressed in the future. New studies that meet the challenge of achieving shorter labelling times without critically reducing signal levels, and that combine improved ultrastructural preservation with quantitative analyses, will be crucial to accurately determine the location of the active RTC. Meanwhile, multiple scenarios remain possible, as originally discussed in (Knoops et al., 2008). If viral RNA synthesis does take place exclusively inside DMVs, a yet unknown mechanism of material exchange with the cytosol should exist, perhaps in the form of a proteinaceous channel spanning the two membranes, similar to the TIM/TOM export complex in mitochondria (Neupert and Herrmann, 2007). The visualisation of such a channel may be possible using advanced techniques like cryo-EM and cryo-ET, which preserve molecular resolution (Barcena and Koster, 2009). On the other hand, if such an exchange mechanism is not present, alternative locations for anchoring the RTC appear necessary. These locations could include the cytosolic face of the DMV outer membrane, but also other membrane structures induced by nidoviruses (e.g. the CM in betacoronaviruses, the open double-membrane spherules of the gammacoronavirus IBV, paired membranes in arteriviruses), and/or even other subcellular membrane structures. In this alternative scenario the DMVs may not even play a critical role in viral RNA synthesis at all, a possibility that may be supported by recent observations with temperature-sensitive MHV mutants. In this study, a direct correlation between the number of DMVs and MHV RNA synthesis could not be established (Al-Mulla et al., 2014), as might have been expected if DMVs would be the sole site of viral RNA synthesis. The fact that DMVs have been detected by EM a few hours later than the onset of viral RNA synthesis could also argue in the same direction; however, this could also simply be due to EM limitations. Incipient modifications, which would likely be rare, would be even less frequent in the cell sections prepared for EM and could easily go undetected.
3. The biogenesis of arterivirus and coronavirus DMVs
A crucial aspect for better understanding virus-induced replication organelles, and possibly for the design of antiviral strategies that target them, is to uncover the steps leading to their formation and, ultimately, to determine the exact nature of the molecular interactions of the viral and host players involved. Compared to DMVs, the +RNA virus-induced invaginations are much better understood in this respect. Mechanistically, their formation entails the induction of negative curvature (i.e. a concave membrane, bending away from the cytosol) on the host membrane-donor organelle to yield a vesicle budding into the luminal space. This negative curvature is compensated by positive curvature (i.e. convex membrane, bent towards the cytosol) at the membranous neck that keeps the vesicle attached to the organelle and connects its interior to the cytosol. Detailed studies on BMV replication organelles have greatly helped to establish a model system for the molecular players and mechanisms involved in moulding the membranes to form this type of invaginations (Diaz et al., 2012, Diaz et al., 2010, Diaz et al., 2015). While the BMV case illustrates the significant progress in understanding the mechanisms involved in producing virus-induced invaginations, our insight into the generation of viral DMVs lags well behind. This is in part due to the more complex architecture of a DMV compared to a single-membrane structure, which implies a more elaborate formation process that would encompass several membrane remodelling steps including fission events. In the context of nidovirus-induced DMV formation, two alternatives have been proposed: enwrapping (also known as protrusion and detachment) and double budding (Pedersen et al., 1999) (Fig. 5 ).
Fig. 5.
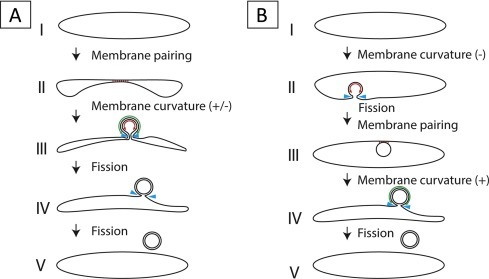
Mechanisms proposed for DMV biogenesis. (A) Different stages in remodelling of ER membranes that would result in DMVs by an enwrapping mechanism. This process would start with the pairing of membranes, which then would start curving until this curvature becomes so pronounced that a double-membrane vase-like compartment is formed. Finally, through a fission event of the inner membrane a closed compartment is formed, while the outer membrane is still continuous with the ER. An additional fission event of the outer membrane would sever the connection with the ER and give rise to a detached DMV. (B) An alternative mechanism that would result in DMV formation is double budding. The first step in this process would be the formation of an invagination in an ER membrane, which then buds into the ER lumen through a fission event. The resulting luminal single-membrane vesicle could subsequently interact with the ER membrane to form stretches of paired membranes. Induction of additional curvature would enable the vesicle to bud out of the ER, thereby acquiring a second membrane. Notice that stages IV and V would be topologically identical in both routes and that similar membrane remodelling events are required for both mechanisms: membrane pairing (represented by red lines), positive and negative membrane curvature (green and red double arrows, respectively) and membrane fission (blue arrowheads). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In the enwrapping model (Fig. 5A) the first crucial step is the pairing of two lipids bilayers in the membrane donor organelle (likely the ER for nidoviruses). These paired membranes would then progressively curve to form a DMV that is still connected to the cytosol in a vase-like configuration. In this step, the outer and inner membranes acquire opposite curvature (positive and negative, respectively). As curvature increases, the opening would get narrower and, eventually, the DMV may be sealed by fission to give rise to an inner compartment containing cytosolic material and an outer lipid bilayer that is still continuous with the ER. The alternative mechanism, the double-budding model (Fig. 5B), would start with the budding of a single-membrane vesicle (SMV) into the lumen of the membrane donor organelle and, therefore, would involve the induction of negative curvature in the ER and fission to release the SMV into the lumen. A second budding event, this time towards the cytosol, would make this vesicle emerge from the ER while acquiring its outer membrane. In this second step, the SMV and the ER would establish membrane-pairing interactions and positive curvature would be induced. This process would result in a DMV outer membrane that is still continuous with the ER, essentially identical to that formed in the enwrapping model. In both models, an additional fission event could lead to the pinching off of a free DMV.
Membrane pairing, fission and induction of both positive and negative membrane curvature are key features of both models, and in fact they only differ in the order in which these events occur. Membrane pairing could be mediated by protein-protein interactions across the lumen. For EAV, this could be potentially achieved by the predicted luminal domains of the transmembrane proteins nsp2 and nsp3, two non-structural proteins that have been shown to engage in a strong interaction (Posthuma et al., 2008, Snijder et al., 1994). Regarding curvature, several processes that can drive bending of cellular membranes have been described (reviewed in Stachowiak et al., 2013). One of them is the generation of asymmetry in the bilayer by differential insertion of proteins or irregularly-shaped lipids, which act as wedges that deform the membrane. Self-oligomerization of transmembrane or soluble scaffold proteins can also result in the induction of membrane curvature. While we currently do not know how arteriviruses and coronaviruses accomplish membrane bending, it seems likely that the transmembrane viral nsps play a prominent role. Other factors could also be involved, for example, a change in lipid composition or homeostasis, as shown for other DMV-inducing viruses. Enteroviruses and HCV alter the lipid composition of the membranes recruited for the formation of replication organelles, which become enriched in phosphatidylinositol-4-phosphate (PIP4) by the specific recruitment of PI4P-kinases (PI4KIIIβ in enteroviruses, PI4KIIIα in HCV), which seems essential for viral replication (Hsu et al., 2010, Reiss et al., 2011). Whether any of the PI4K variants play a role during arterivirus and coronavirus infection would be highly informative and is currently under investigation in our group. A recent siRNA library screen targeting the human kinome in fact identified some proteins involved in the metabolism of complex lipids as factors relevant for SARS-CoV replication. Interestingly, the PI4P-kinases, which appear essential for the replication of other DMV-inducing viruses, did not show an effect on SARS-CoV replication in this study (de Wilde et al., 2015).
3.1. New clues on DMV formation during EAV infection
Clear intermediate structures that could support or argue against one of the two models described above for arterivirus-induced DMV biogenesis have not been reported so far. Historically, the enwrapping model has been favoured due to the detection of short stretches of tightly paired membranes (Pedersen et al., 1999, Weiland et al., 1995), which constitute a specific feature of this mechanism. We now have performed an extensive tomographic analysis which uncovered the 3D structure of some distinct intermediates that further support an enwrapping mechanism for DMV formation. These intermediates include relatively long stretches of paired ER membranes that form sheets in 3D and could represent the first step in DMV formation through enwrapping (Fig. 6 A). In order to transform these sheets into DMVs, curvature must be induced and paired-membrane sheets with different degrees of curvature could indeed be detected in the tomograms (Fig. 6B). A (putative) even more advanced stage, in which the paired curved sheet is already transformed into a nascent DMV that is not yet completely sealed, is shown in Fig. 6C and D.
Fig. 6.
Putative intermediate structures in the formation of DMVs. The greyscale top images are virtual slices (6 nm thick) extracted from tomograms of EAV-infected HuH-7 cells. (A–C) Putative intermediates of an enwrapping process. The membranes of interest are highlighted in orange in the lower images. (A) Paired ER membranes, possibly corresponding to the stage sketched in Fig. 5A as II. This type of paired membranes could progressively curve (B) to form a vase-like double-membrane compartment (C, D) (stage represented in Fig. 5A as III) with an opening to the cytosol (white arrows). (D) 3D-surface rendering of the structure shown in panel C. (E–G) Structures compatible with intermediates in a double-budding mechanism. In the lower panels, the membranes are highlighted in dark blue (ER) and light blue (luminal vesicle). These structures could represent a stage between III and IV in Fig. 5B, in which a luminal vesicle interacts with an ER membrane that curves towards the cytosol, possibly to form the outer membrane of a nascent DMV. (G) 3D surface-rendered model of the structure shown in panel E. (H) Example of a DMV that is connected to the ER though it lacks the electron dense core usually seen in DMVs. The membranes are highlighted with purple (ER membranes) and pink (inner vesicle). Scale bars, 100 nm. See Supplementary information for experimental details. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Given the similarities between arterivirus- and coronavirus-induced DMVs and the presumed common ancestry of their replicase proteins, including the subunits likely to drive the membrane modifications, it is tempting to speculate that the enwrapping mechanism could also be used for the formation of DMVs in coronavirus infection. In this regard, there are some striking similarities between the intermediate paired-membrane structures shown here for EAV and the zippered ER membranes recently documented for the gammacoronavirus IBV (Fig. 4C). These zippered membranes also appear to curve, forming open double-membrane spherules that, nevertheless, were ruled out as DMV precursor by Maier et al. due to their much smaller size (∼60 nm in diameter versus ∼180 nm for IBV DMVs) and their different appearance (Maier et al., 2013b). Interestingly, the tomographic analysis of cells infected with the enteroviruses CVB3 and PV revealed putative intermediate structures supporting an enwrapping mechanism for the biogenesis of the DMVs induced by these viruses (Belov et al., 2012, Limpens et al., 2011). Taken together, these results suggest that enwrapping may well be a general route used for the generation of virus-induced DMVs.
Nevertheless, in our search for intermediates that could shed light on the biogenesis of EAV-induced DMVs, we also detected structures in infected HuH-7 cells that would be compatible with a double-budding mechanism. These consisted of sealed SMVs contained in the ER lumen and closely apposed to the ER membrane to possibly recruit the second membrane through membrane pairing (Fig. 6E–G). We cannot discard, however, an alternative scenario in which these luminal SMVs would result from a distension of the connection between an already formed DMV and the ER. Although the putative double-budding intermediates were considerably rarer in our tomograms than the structures that would fit the enwrapping model, this lower frequency cannot be directly translated into a quantitative assessment about the prevalence of the two mechanisms, as it could simply reflect that double-budding is a faster process that is therefore under-represented in the snapshots that EM provides. Overall, our data suggests that the enwrapping mechanism plays an important role in DMV formation but is also compatible with a model in which both enwrapping and double-budding could coexist. In this regard, it is interesting to emphasize that the fundamental difference between the two proposed mechanisms is simply the order in which essentially the same membrane reshaping events would take place (Fig. 5). It is therefore conceivable that, having the molecular mechanism required for these steps in place, both paths could be used by the virus for the ultimate goal of forming DMVs.
An interesting feature of all putative nascent DMVs observed in the tomograms (Fig. 6C, E and F) is that they lack the electron-dense core that is typical of most DMVs found in EAV infected cells. However, occasionally fully-formed DMVs that do not contain such a core could also be detected. DMVs lacking and containing a core are indistinguishable in size and both types often appear to be connected to the ER (Fig. 6H). In our view, they could well represent early and later stages in the existence of a DMV. In this scenario, the DMV content would be initially identical to the surrounding cytosolic material. Once fully formed and closed, the DMV would accumulate electron-dense material. The exact nature of this material remains unclear although, as described in Section 2.2, one of its components is dsRNA.
3.2. A link with autophagy?
Double-membrane structures are not without precedent in the cell. Organelles delimited by a double membrane in eukaryotic cells include the nucleus, mitochondria and autophagosomes. Although much larger in size (0.5–1.5 μm in diameter), autophagosomes have a similar appearance as virus-induced DMVs. Autophagosome formation (reviewed in Stanley et al., 2014) is a central process in the macroautophagy pathway (hereafter autophagy) (Yang and Klionsky, 2010) and involves the formation of a double-membrane sheet, known as the phagophore, which wraps and sequesters cytoplasmic material, sometimes even whole organelles or bacteria. Recently, it has become clear that the cargo for degradation in autophagosomes is selected specifically by autophagy receptors which bind to the selected cellular components and recruit them to the growing autophagosomal membranes (Stolz et al., 2014). Closure of the phagophore yields the autophagosome, which fuses with late endosomes or lysosomes after maturation to deliver its cargo for degradation and recycling of the engulfed macromolecular components.
Autophagy can be triggered by stress, starvation and infection. While autophagy can act as a cellular defence mechanism for virus clearance in response to infection, several viruses have evolved strategies to avoid or even subvert the autophagy pathway for their own benefit (Chiramel et al., 2013, Kirkegaard et al., 2004, Kudchodkar and Levine, 2009). The parallels between autophagosomes and virus-induced DMVs elicited the hypothesis of a link between autophagy and DMV biogenesis, postulating that DMVs may represent modified autophagosomes.
Infection with arteriviruses and coronaviruses can induce autophagy (Chen et al., 2012, Cottam et al., 2011, Liu et al., 2012, Monastyrska et al., 2013, Prentice et al., 2004a, Sun et al., 2012) but conflicting results on the relationship between autophagy and nidovirus replication have been published, possibly due in part to the differences in the cell lines used. Knock-down of Atg5, an essential protein in the autophagy pathway, was first reported to inhibit MHV replication and preclude the formation of DMVs in embryonic stem cells (Prentice et al., 2004a), but later these effects were not observed in bone marrow-derived macrophages and primary mouse embryonic fibroblasts (MEF) (Zhao et al., 2007), or MEF cells lacking another key autophagy protein, Atg7 (Reggiori et al., 2010). For the avian coronavirus IBV, it was shown that viral replication is not affected by either inhibition or activation of autophagy in monkey kidney cells (Vero) (Cottam et al., 2011, Maier et al., 2013a). Of note, IBV infection induced autophagy in mammalian cells but not in avian cell lines (Maier et al., 2013a), which emphasizes the potential influence of the cell type on the study of the interplay between viral infection and autophagy and, possibly, the consequences of a virus-host mismatch. Likewise, the replication of the arterivirus EAV in Vero cells did not seem to be affected by either Atg7-dependent autophagy or the alternative Atg5/Atg7-independent autophagy pathway (Monastyrska et al., 2013). However, for the arterivirus PRRSV, progeny titers were moderately reduced in another monkey kidney cell line (MARC-145) treated with the autophagy inhibitor 3-methyladenine or when autophagy genes were silenced, whereas activation of autophagy with rapamycin had the opposite effect (Chen et al., 2012, Liu et al., 2012, Sun et al., 2012). Whether these results are dependent on the cell line used or reflect a particularity of PRRSV remains to be investigated. Overall, the data gathered in the last decade seems to strongly suggest that autophagy, or at least the canonical autophagy pathway, is not essential for arterivirus and coronavirus replication and whether it plays a role in DMV biogenesis is therefore unclear. Such a role has not been established for other DMV-inducing viruses either. Whereas in the case of HCV conflicting reports make the influence of autophagy in viral replication rather unclear, for enteroviruses, autophagy appears to have only limited impact on replication. However, autophagy seems to be critically linked to virus assembly, maturation and the non-lytic release of new enterovirus particles (Chen et al., 2015, Jackson et al., 2005, Richards and Jackson, 2012).
Induction of autophagy is often monitored using the autophagy marker LC3 (microtubule-associated protein 1 light chain 3), a key player in autophagosome formation and recruitment of specific cargo for degradation. Upon exposure to autophagy stimuli, the cytosolic form (LC3-I) is converted into a lipidated form (LC3-II) by covalent binding to phosphatidylethanolamine, which is present in the autophagosomal membranes (Yang and Klionsky, 2010). In light microscopy images, this LC3 relocation is reflected in the formation of puncta of fluorescently tagged LC3. Using this approach, the IBV nsp6 replicase protein (but not other IBV nsps) was shown to be capable of inducing autophagosome formation when individually expressed (Cottam et al., 2011, Maier et al., 2013a). This effect was also described for nsp6 of MHV and SARS-CoV, as well as the arteriviral PRRSV nsp5-7 (Cottam et al., 2011). The autophagosomes induced by these proteins appeared to be smaller in size than normal autophagosomes, as judged from light-microscopy images, and therefore closer to viral DMVs (Cottam et al., 2014). However, in contrast with individually expressed nsp6, IBV infection did not induce LC3 relocalization and autophagic signalling in avian cells (Maier et al., 2013a). Therefore the relevance of this nsp6 feature in the context of infection remains unclear,
One of the reasons to relate autophagy to nidovirus replication was the reported colocalization of LC3 with viral replicase proteins or dsRNA in light microscopy images of cells infected with MHV (Prentice et al., 2004a, Reggiori et al., 2010), SARS-CoV (Prentice et al., 2004b, Zhao et al., 2007), PRRSV (Sun et al., 2012) and EAV (Monastyrska et al., 2013), although some other studies with SARS-CoV and IBV did not confirm this finding (Cottam et al., 2011, Snijder et al., 2006). Without accompanying EM data, extrapolating this colocalization, which appears to be partial in confocal images, to the specific ultrastructural location of LC3 is problematic, even more so in the case of coronaviruses considering the variety of membrane structures that they induce during infection (see Section 2.3). Ultrastructural studies using IEM, which are limited so far in the case of EAV to chemically-fixed BHK-21 cells at late time points in infection (Monastyrska et al., 2013), would be essential to establish whether LC3 is associated with virus-induced modifications and, specifically, with DMVs.
Interestingly, data obtained using EAV- and MHV-infected cells indicated that the non-lipidated form of LC3 (LC3-I), which is inactive in autophagosome formation, would be the one that is recruited from the cytosol and that partially colocalizes with viral nsps (Monastyrska et al., 2013, Reggiori et al., 2010). Silencing of LC3 was reported to reduce EAV and MHV replication and this effect could be reversed by transfecting a non-lipidable form of LC3 in MHV-infected cells (Reggiori et al., 2010). This was considered reminiscent of EDEMosomes, a class of short-lived small vesicles that appear to depend on LC3-I recruitment (Cali et al., 2008, Zuber et al., 2007). EDEMosomes have been proposed to serve as a mechanism to tune down the ER-associated protein degradation (ERAD) machinery, which targets misfolded proteins, by transporting some ERAD regulators, like EDEM1 (ER degradation-enhancing α-mannosidase-like protein 1) or OS-9 (osteosarcoma amplified 9), away from the ER for their disposal (Zuber et al., 2007). Exploring these analogies further, EDEM1 and OS-9 were found to partially colocalize with dsRNA in MHV-infected cells, which was interpreted as recruitment of these proteins to DMVs (Reggiori et al., 2010). Similarly, dsRNA and EDEM-1 showed some degree of colocalization in EAV-infected cells (Monastyrska et al., 2013). It was therefore proposed that the ERAD-tuning pathway would supply the membranes for the biogenesis of viral DMVs (Reggiori et al., 2010). The direct conversion of EDEMosomes into DMVs, however, seems difficult to envision, as it would entail the transformation of smooth single-membrane vesicles of about 150 nm in diameter (Zuber et al., 2007) into DMVs that can reach 300 nm in diameter in the case of coronaviruses and whose outer membranes are decorated with ribosomes. The recruitment to virus-induced modifications of some components of the ERAD machinery during arterivirus and coronavirus infection is, nevertheless, plausible, but its functional significance may well be completely unrelated to DMV biogenesis and needs to be further investigated using in-depth EM analysis and molecular biology approaches.
4. Surrogate systems for investigating the biogenesis of arterivirus replication organelles
An important aspect in the biogenesis of virus-induced membrane modifications is the identification of the viral proteins involved in the process and how these proteins interact with host factors to induce the elaborate membrane structures observed during infection. Addressing these questions in the context of infection is complicated by the (potential) number of viral players involved and the fact that targeting the viral proteins with mutagenesis often leads to defects that inhibit or block viral replication. The development of so-called “surrogate systems”, however, does create the possibility to address these questions by essentially uncoupling the expression of the viral proteins under study from their expression from a viable viral genome. In such systems, ectopic expression of a subset of viral proteins is used to mimic the formation of membrane structures that resemble those found during infection, although they obviously lack the viral RNA-synthesizing enzyme complexes associated with these structures in infected cells. For a number of viruses this approach has proven to be productive and, in many cases, instrumental for a better understanding of the biogenesis of virus-induced replication organelles (Angelini et al., 2013, Egger et al., 2002, Romero-Brey et al., 2015, Romero-Brey et al., 2012, Salonen et al., 2003, Schwartz et al., 2002, Snijder et al., 2001, Suhy et al., 2000). These systems can be expanded by using site-directed mutagenesis of the proteins that are expressed in order to investigate the relevance of specific residues, protein domains or interactions.
For EAV, this approach has been instrumental in the identification of the proteins that are likely involved in converting cellular membranes into viral replication organelles. The expression of EAV nsp2-7 in BHK-21 cells was first shown to give rise to membrane rearrangements that were similar to those found in infected cells (Pedersen et al., 1999). The minimal set of viral proteins required for DMV formation was later narrowed down to nsp2 and nsp3 (Snijder et al., 2001) and, since then, expression of these two proteins has been considered a useful surrogate system to mimic and study EAV-induced membrane modifications. Although these studies highlighted the key role of EAV nsp2 and nsp3 in DMV formation, the ultrastructural analysis was limited to 2D electron microscopy of chemically fixed samples. Therefore, the question of whether the detailed architecture of the DMVs induced upon co-expression of nsp2 and nsp3 was comparable to that of the DMVs found in infection was still open. In order to address this question, we now carried out a tomography characterization of the structures induced by different combinations of EAV-encoded non-structural proteins using two protein expression systems in samples prepared by HPF-FS (see Supplementary information for detailed Materials and Methods). The results of these analyses are presented in the next subsections.
4.1. Expression of EAV nsp2-3 using a sindbis virus-based vector
We first analysed the membrane modifications induced by EAV nsp2 and nsp3 using a Sindbis virus-based expression system, essentially identical to that used in previous studies in BHK-21 cells (Posthuma et al., 2008, Snijder et al., 2001). As described in Section 1, alphaviruses like Sindbis virus also alter cellular membranes; however, they modify endosomes, lysosomes and the plasma membrane (Frolova et al., 2010, Froshauer et al., 1988, Spuul et al., 2010) and therefore are thought not to interfere with DMV formation. The system consisted of an RNA replicon (SINrep5) in which a foreign gene (a self-cleaving EAV nsp2-3 polypeptide C-terminally fused to enhanced GFP (eGFP)) replaces the ORF encoding the structural polyprotein of Sindbis virus and is expressed to high levels from a subgenomic mRNA (Bredenbeek et al., 1993). The eGFP tag on nsp3 enabled the application of correlative light and electron microscopy (CLEM), an emerging methodology that allows imaging of the same sample both by light microscopy and high-resolution EM (de Boer et al., 2015). In order to carry out CLEM, we used a freeze-substitution protocol that minimizes staining to preserve the fluorescence throughout EM sample preparation (Kukulski et al., 2011). The eGFP signal facilitated the quick identification of transfected cells and, more specifically, the localization of nsp3-eGFP positive sites in the EM sections (Fig. 7 A–C). In all these areas, some circular double-membrane profiles typical of DMVs were readily observed (Fig. 7C, white asterisks).
Fig. 7.
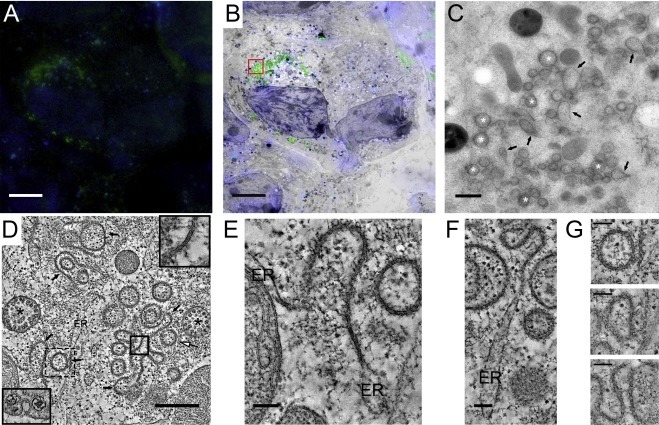
Characterization of membrane structures induced by EAV nsp2-3 expression from a Sindbis virus-based replicon. (A–C) Correlative light-electron microscopy (CLEM) analysis of transfected cells expressing an eGFP-tagged self-cleaving nsp2-3 polyprotein. (A) Fluorescence microscopy image of a 200 nm section on an EM grid showing nuclear DNA staining and a clear cytosolic eGFP signal. (B) Merging of the fluorescence image and the corresponding electron microscope images. (C) Higher magnification image of the boxed area in panel B, showing the structures underlying the eGFP signal. Several circular profiles typical of DMVs are clearly visible (white asterisks), as well as other more irregular double-membrane structures (black arrows). (D–G) 3D architecture of nsp2-3 induced membrane rearrangements. (D) Tomographic slice (2 nm thick) of EAV nsp2-3-induced structures (see also Supplementary movie S2). These consist of the characteristic double-membrane vesicles (white arrow), as well as double-membrane sheets (black arrows) that are particularly abundant in this region. The membrane structures induced by Sindbis non-structural proteins are also visible (black asterisks), and consist of invaginations in modified endosomes that have a neck-like channel connecting their interior with the cytosol, (lower left inset). The inset at the upper right corner is a close-up of the boxed area in (D) (continuous line) and highlights the two tightly-apposed membranes that form these structures. (E,F) Gallery of tomographic slices (6 nm thick) highlighting the continuity of the paired-membrane sheets with ER cisternae. (G) Tomographic slices through the structure highlighted in (D) with a dashed box (relative z height of the slices, from top to bottom: 0, 35 and 60 nm). Notice how at one level (top image) the structure appears indistinguishable from a closed DMV, while the successive tomographic slices reveal that it is actually open to the cytosol and more akin to a highly curved fragment of the paired membrane sheet network to which it is connected. Scale bars, 10 μm (A and B), 500 nm (C and D); 100 nm (E–G). See Supplementary information for experimental details.
Intriguingly, numerous irregular structures that were also delineated by two tightly apposed membranes were readily apparent in the regions containing membrane modifications (Fig. 7C, black arrows). Electron tomography revealed that this type of irregular structures consist of strikingly long stretches of paired membrane sheets (Fig. 7D, black arrows) which appear to derive from the ER, with which they are continuous (Fig. 7E and F). Although the majority of the circular double-membrane profiles observed in 2D projection images were shown in the tomograms to be closed DMVs (Fig. 7D, white arrow) and thus analogous to those found in EAV infection, many apparent DMVs turned out to be patches of these membrane sheets that locally curved creating a secluded internal space reminiscent of a DMV but still continuous with the cytosol (Fig. 7G). In fact, these DMV-like structures, which could be easily mistaken for bona fide DMVs in 2D projection images, appeared to be part of a large network of double-membrane sheets weaving through the cytoplasm in the perinuclear area (Supplementary movie 2).
This profusion of membrane sheets is a phenotype that had been described before in the SINrep5nsp2-3 expression system but only when a mutation creating an N-linked glycosylation site was introduced in the first luminal loop of nsp3 (T873N) (Posthuma et al., 2008) and not in cells expressing wild-type EAV nsp2-3 (Posthuma et al., 2008, Snijder et al., 2001). The reasons for this discrepancy are unclear and, although it may be partially attributed to the lack of 3D data in previous studies, it could also be related to differences in the experimental set-up. In any case, these differences were not related to the cell line used, nor were they due to the eGFP tag on nsp3 since similar double-membrane sheet networks were detected using BHK-21 and Vero E6 cells and upon transfection with the same construct lacking the eGFP fluorescent tag (data not shown).
4.2. Cell lines for the inducible expression of EAV nsps2-3 and nsp2-7
In order to avoid possible undesired side-effects derived from the expression of Sindbis virus non-structural proteins and to obtain a surrogate system that is more convenient, reproducible and suitable for a wider range of experimental setups, we created stable cell lines which can be induced to express the self-cleaving EAV nsp2-3 polyprotein from a cytomegalovirus (CMV) promoter.
Clusters of DMVs were readily observed 24 h after induction of expression in HuH-7 cells (Fig. 8 A, white asterisks). Both the inner and outer membranes of these DMVs were decorated with ribosomes (Fig. 8B and C, black arrows), which, as in the infected cell, hints at the ER as the membrane donor for DMV formation. In fact, some of these DMVs showed a direct connection with ER cisternae, with which they were continuous through their outer membrane (Fig. 8D). As in the case of DMVs found in infected cells, neck-like membrane connections between DMVs were detected in the tomograms (Fig. 8E, black arrowheads). Finally, the similarities with DMVs observed in EAV infection extended to the closed configuration of these compartments, as no openings could be detected in the DMVs induced by nsp2-3 expression in this system.
Fig. 8.
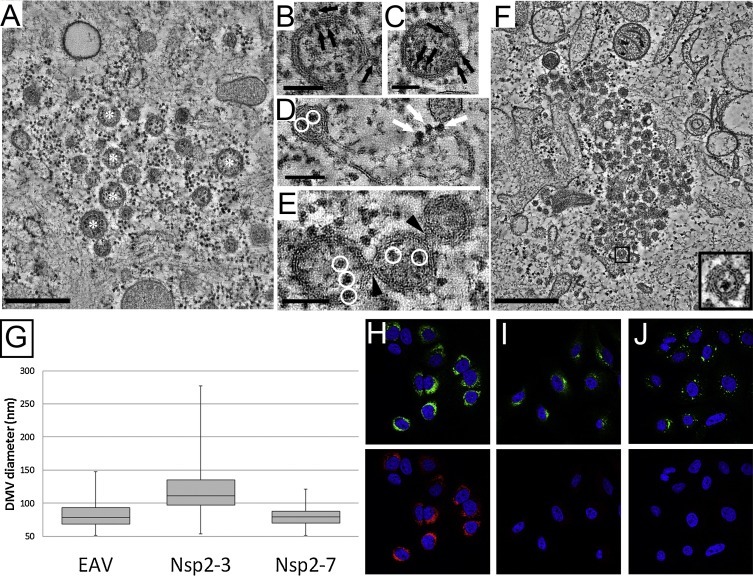
Characterization of membrane structures induced after EAV nsp2-3 and nsp2-7 expression using inducible HuH-7 cell lines. (A) Tomogram slice (2 nm thick) showing an overview of the structures induced upon nsp2-3 expression. The structures consist of DMVs, some of which are indicated by white asterisks. Note that the membrane sheet network observed in the SINrep5 expression system (Fig. 7) is absent. (B–E) Close-up tomographic slices (6 nm thick) highlighting several features of the DMVs induced by nsp2-3 expression. (B, C) Ribosomes decorate both the inside and the outside of the DMVs (black arrows). (D) An example of membrane continuity between rough ER and DMV, similar to what is found for DMVs induced in EAV infection (Fig. 2B). The ribosomes decorating the surface of the ER membrane are indicated by white arrows, while the ribosomes in the interior of the vesicle are encircled in white. Another feature, highlighted in (E), is the propensity for the DMV to form interconnections similar to those found in EAV-induced DMVs (black arrowheads). (F) Virtual slice (2 nm thick) showing an overview of the typical structures seen upon induction of nsp2-7 expression, which consist of clusters of DMVs. The inset shows a close-up of the DMV in the boxed area, which appears to contain a ribosome. (G) Boxplot illustrating the diameter distribution for DMVs induced by EAV infection, nsp2-3 expression and nsp2-7 expression in HuH-7 cells, as measured from the tomographic reconstructions from samples prepared in parallel in the same experiment. The middle of the boxplot represents the median value for each population, the bottom and top of the boxplot represent the first and third quartile respectively. The whiskers extend to the minimum and maximum measured values. (H–J) Immunofluorescence labelling of viral markers in EAV-infected HuH-7 cells (H) and in the inducible HuH-7 cell lines expressing nsp2-3 (I) or nsp2-7 (J). A perinuclear nsp2 labelling (top, green) was apparent in the three experimental conditions, while dsRNA signal (bottom, red) was only detected in EAV-infected cells. Scale bars, 500 nm (A, I); 100 nm (B–E). See Supplementary information for experimental details. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite these general similarities, some anticipated differences with the structures found in infected cells were apparent. No tubular structures (Section 3, Fig. 2H and I) were detected, which is in agreement with the notion that their formation depends on N protein expression (Knoops et al., 2012, Wieringa et al., 2004). Also, the DMVs formed upon expression of nsp2-3 lack the electron dense core that is typical of the DMVs in EAV-infected cells. The absence of this structure is consistent with the interpretation that the DMV cores that appear in infected cells, which have a high phosphorus content and label for dsRNA (Fig. 3), contain viral RNA. The content of the nsp2-3-induced DMVs appears quite similar to the surrounding cytosolic material, although slightly more electron dense and, like the cytosol, it contains densities that may be free ribosomes (Fig. 8D and E white circles). Taken together, our results show that the expression of nsp2-3 suffices to reproduce the double-membrane architecture, topology and connectivity of the DMVs found in infected cells, and therefore constitutes a useful system to study these features of the EAV-induced RVN. This also implies that the membrane-remodelling capacities necessary to generate the RVN reside in these two arteriviral nsps, likely with the assistance of recruited host factors.
The results with this nsp2-3 expression system raise the question of the role in DMV formation, if any, of the third (presumed) transmembrane subunit of the arterivirus replicase: nsp5. To investigate this, using the same approach as above, we created a stable inducible HuH-7 cell line expressing nsp2-7. Since most nsp5 molecules in infected cells are present in the form of nsp5-7 or nsp5-8 processing intermediates (Li et al., 2014, Wassenaar et al., 1997) (Section 2.1), expression of a self-cleaving nsp2-7 was chosen to mimic the course of events in infected cells most closely (Supplementary Fig. 1). As in the case of nsp2-3, induction of nsp2-7 expression gave rise to the appearance of DMV clusters in close proximity to the ER (Fig. 8F). Notably, these DMVs observed in HuH-7 cells at 24 h after induction were strikingly smaller than those observed upon expression of nsp2 and nsp3 only. A detailed DMV size comparison was carried out using the 3D-reconstructions (Fig. 8G, Table 2 ). This method provides a much more reliable estimate of vesicle sizes than 2D measurements because, inevitably, not every DMV is fully contained in a cell section. Only a 3D analysis allows exclusion of DMVs whose equator is not present in the cell section. In this manner, the average diameter of the nsp2-7-induced DMVs was determined to be 79 nm (±13 nm, n = 145) versus 119 nm (±32 nm, n = 145) for nsp2-3-induced DMVs. The average size difference between the two populations, which translates into a decrease in DMV volume of about 70%, was found to be statistically significant using an independent two-sample t-test (Table 2). Remarkably, expression of nsp2-7 seems to bring the average diameter of the DMVs closer to the average diameter of DMVs formed in EAV infection (83 ±21 nm, n = 145). Additionally, the population of nsp2-7-induced DMVs appeared to be significantly more homogenous in size than the DMVs produced upon expression of just nsp2 and nsp3, where particularly large DMVs (up to ∼270 nm in diameter) were detected (Fig. 8G). The DMVs in the nsp2-7-expressing cell line also lacked the electron dense core typical of DMVs formed upon EAV infection, although their content appeared slightly more electron dense than the surrounding cytosolic material. The dark content often seemed to correlate with the presence of densities that resembled ribosomes (Fig. 8F, inset). In any case, immunofluorescence microscopy confirmed that this darker interior is not due to the presence of dsRNA. As expected for systems lacking viral RNA replication, no dsRNA signal could be detected in cells expressing either nsp2-3 or nsp2-7, in contrast with the abundant dsRNA labelling found in EAV-infected HuH-7 cells (Fig. 8H–J).
Table 2.
Diameters of DMVs in EAV infection, nsp2-3 expression and nsp2-7 expression.
| n | Median | Average | Standard deviation | p-valuea | p-valueb | p-valuec | |
|---|---|---|---|---|---|---|---|
| EAV infection | 145 | 78 | 83 | 21 | – | <0.001 | <0.05 |
| Nsp2-3 | 145 | 111 | 119 | 32 | <0.001 | – | <0.001 |
| Nsp2-7 | 145 | 80 | 79 | 13 | <0.05 | <0.001 | – |
Comparison with diameters of DMVs formed upon EAV infection.
Comparison with diameters of DMVs formed upon nsp2-3 expression.
Comparison with diameters of DMVs formed upon nsp2-7 expression.
Overall, our results show that, while nsp2 and nsp3 are capable of inducing all the membrane-remodelling steps required to generate the virus-induced RVN, the presence of additional nsps can modulate the process and produce DMVs more similar in size to those found in EAV-infected cells. The significantly smaller DMVs found upon nsp2-7 expression relative to those induced by nsp2-3 suggest a role for the extra proteins in enhancing the curvature of the membranes used for DMV formation, while the narrower size distribution suggests a tighter regulation of the process. Although several additional nsps and intermediates are expressed in the nsp2-7 system, it seems likely that, nsp5, given its presumed transmembrane nature, is the primary cause of the observed differences.
4.3. Comparison with the role of coronavirus nsps in membrane remodelling
In terms of host cell membrane remodelling, the coronaviral non-structural proteins 3, 4 and 6 that contain three conserved transmembrane domains (TM1, TM2 and TM3) are considered to be the functional equivalents of arterivirus nsp2, nsp3 and nsp5, respectively (Gorbalenya et al., 2006), see also Section 2.1 and Fig. 1.
A recent study addressed the question of the membrane-remodelling abilities of the SARS-CoV proteins nsp3, nsp4 and nsp6, using (co-)transfection of plasmids encoding separate nsps to express them either individually or in various combinations, followed by an ultrastructural analysis of the cells by EM (Angelini et al., 2013). In this experimental set-up, co-expression of these three nsps appeared to be required for the formation not only of DMVs but also of structures resembling the CM that are typical of SARS-CoV infection (Fig. 4B). This outcome differs from the results obtained with arteriviruses, where expression of just nsp2 and nsp3 was sufficient, as described above, for the formation of closed DMVs in the expression systems tested. The corresponding combination of coronavirus nsps, namely SARS-CoV nsp3 and nsp4, gave rise to membrane alterations termed maze-like bodies, which consisted of clusters of curved parallel-running profiles of ER-derived paired membranes (Angelini et al., 2013). Interestingly, these were interspersed with circular profiles delineated by a double membrane. The circular profiles had an average diameter similar to the average spacing of the curved parallel double-membrane profiles (∼80 nm), and they were interpreted, respectively, as cross or longitudinal sections of double-membrane walled tubules. However, in the absence of 3D-EM data, the authors recognized that is was not possible to firmly establish this point. In our opinion, the circular double-membrane profiles may alternatively represent DMVs interspersed with regions of paired membranes sheets, similar to what is observed upon expression of EAV nsp2-3 in the Sindbis virus-based expression system (Section 4.1, Fig. 7). This is a particularly interesting possibility as it would imply that, both for arteriviruses and coronaviruses, the proteins containing TM1 and TM2 would be sufficient to induce DMV formation. In this alternative scenario, the nsp comprising TM3 may merely have a modulating role in DMV formation and, in coronaviruses, could be important for the generation of CM.
In any case, the presence of maze-like bodies reflects that expression of coronavirus nsp3 in combination with nsp4 is able to induce membrane-pairing, which is likely an essential step in DMV formation (Section 3). The membrane pairing observed in the maze-like bodies appeared to strictly depend on the co-expression of nsp3 and nsp4, suggesting that it could be the result of heterotypic interactions between these proteins (Angelini et al., 2013). This type of interaction between nsp3 and nsp4 has been biochemically detected by mammalian two-hybrid assay (Pan et al., 2008) and co-immunoprecipitation (Hagemeijer et al., 2011), and may resemble the nsp2-nsp3 interaction documented for arteriviruses (Posthuma et al., 2008, Snijder et al., 1994).
Additional experiments provided some interesting hints regarding the specific domains that could be important for the nsp3-nsp4 interaction possibly leading to membrane-pairing. The co-expression of MHV nsp4 and a C-terminal fragment of nsp3 that contained the two predicted transmembrane domains changed the localization of these proteins from the ER to discrete perinuclear foci (Hagemeijer et al., 2011). At the ultrastructural level, the two nsps mapped to regions of highly curved, ER-derived membranes that were vaguely reminiscent of the maze-like bodies described by Angelini et al. (2013) (Hagemeijer et al., 2014). Using truncated mutants, the critical regions for the relocation of these proteins to foci could be narrowed down to the luminal loop of nsp3 and the first luminal loop of nsp4 (Hagemeijer et al., 2014) (Fig. 1). This large N-terminal nsp4 loop contains ten conserved cysteines, which could be important in inter- or intra-molecular interactions. Mutations in each of these cysteines severely impaired the relocation of nsp4 and the nsp3C-terminal fragment, suggesting an important direct or indirect role of these cysteines in the nsp3-nsp4 interaction (Hagemeijer et al., 2014). Like the coronavirus nsp4, the corresponding arterivirus protein (nsp3) contains a cluster of four conserved cysteine residues in its first luminal loop, and each of them appeared to be critical for DMV formation and virus replication (Posthuma et al., 2008). The latter study also showed, by mutating a catalytic residue in PLP2 (H332Y), that the cleavage between nsp2 and nsp3, although essential for virus replication, was dispensable for DMV formation in an expression system. When expressed from the SINrep5 vector in BHK-21 cells, an nsp2-3 construct containing an N glycosylation site in the cysteine-containing loop induced the abundant formation of curved paired membranes, which was interpreted as partial inhibition of DMV formation (Posthuma et al., 2008). These modifications, however, closely resemble the results now obtained using wild-type nsp2-3 expressed from a Sindbis virus-based vector (Section 4.1) and therefore perhaps reflect peculiarities of this expression system rather than an effect of the mutation. This latter possibility seems to be supported by preliminary results for this mutant when expressed in an inducible HuH-7 cell line, in which long curved paired membranes could not be detected whereas regular DMV clusters were present (data not shown).
One major difference between studies on coronaviruses and arteriviruses (Pedersen et al., 1999, Posthuma et al., 2008, Snijder et al., 2001), Sections 4.1 and 4.2 is that coronavirus nsps were expressed independently from separate plasmids, whereas arterivirus proteins were expressed as (more natural) self-cleaving polyproteins. Obviously, the former poorly mimics the situation in infected cells and could influence the course of events during which translation, polyprotein cleavage, nsp translocation, and membrane remodelling occur, likely in a defined order. Also processing intermediates could play an important role in membrane rearrangement, for example, by keeping nsps in close proximity to each other to favour key interactions in the membrane remodelling process. Additionally, it should be noted that proteins made from polyproteins are produced at equimolar ratios, something that is virtually impossible to control when co-transfecting multiple plasmids. Unfortunately, the exceptionally large size of coronavirus nsp3 (∼2000 amino acids long) has so far complicated the production of the nsps from a self-cleaving polyprotein in an expression system. Future studies that overcome this technical difficulty may be pivotal to definitively establish the similarities and differences between coronaviruses and arteriviruses regarding the membrane-remodelling abilities of their nsps.
5. Conclusions and future perspectives
The replication organelles induced by +RNA viruses are examples of the remarkable level of sophistication in host membrane manipulation that viruses can achieve. Their complex architecture, revealed by electron tomography, poses additional challenges, for example concerning DMV structure-function interpretation. In this respect, arteriviruses and coronaviruses represent perhaps the most intriguing case (Table 1). The RVNs that EAV and SARS-CoV induce are intricate labyrinths of modified membranes that have no parallel in the +RNA virus world thus far. Additionally, and in contrast with other +RNA viruses that induce mixed populations of open and closed DMVs, the closed configuration of nidovirus-induced DMVs defies the straightforward concept of viral RNA synthesis occurring inside these compartments, and puts at the forefront the open question of the location of the viral enzyme complexes actively engaged in replication/transcription. Until this question is unambiguously answered experimentally, multiple possibilities remain open, from a scenario in which RNA synthesis would take place solely inside DMVs while a still undetected macromolecular channel would accomplish the necessary import/export processes, to a situation in which the active nidoviral RTC would be located in alternative sites and the DMVs would not even play any direct role in viral RNA synthesis. Along these lines, the possibility of DMVs simply being a by-product of the high expression levels of the viral non-structural proteins cannot be completely discarded. However, even if the DMVs are not critically involved in viral RNA synthesis, less passive roles seem also plausible. These RNA-containing structures could perhaps be used to shield an excess of viral RNA from the cellular innate immune sensors and/or be the result of the subversion of cellular pathways for the benefit of virus replication. Clearly, additional experimental data are required to provide key information on the precise role of nidovirus-induced DMVs, which may well extend to other +RNA viruses.
The biogenesis of the DMVs formed upon nidovirus infection is another aspect that we are just beginning to address. The results presented here for EAV document clear putative intermediates in DMV formation by enwrapping of paired ER membranes as well as structures also compatible with a double-budding mechanism. This suggests that, maybe more than a strict sequence of events, the virus could “direct” basic membrane remodelling events (e.g. membrane pairing and induction of membrane curvature) that could be used in different routes ultimately leading to the same (or similar) structures. Viral proteins are likely to drive RVN biogenesis with the aid of host factors that, in the case of nidoviruses, remain to be identified. It seems clear now that both in arteri- and coronaviruses the TM1- and TM2-containing nsps are sufficient to produce membrane pairing (Angelini et al., 2013, Snijder et al., 2001) and, in arteriviruses, also suffice for the formation of DMVs that have a comparable architecture to those found in infection. Our results suggest that the TM3-containing nsp, which has been proposed to be essential for DMV formation in coronaviruses (Angelini et al., 2013), would merely have a curvature modulating role in DMV biogenesis in the case of arteriviruses. Similar electron tomography studies with coronaviral nsp expression systems may help to firmly establish the similarities and differences between arterivirus- and coronavirus-induced DMV formation. This kind of 3D-ultrastructural studies, possibly complemented with similar analyses in vitro using membrane model systems (Wang et al., 2013), could be a significant step towards the mechanistic dissection of the role of the nsps and their different domains in RVN biogenesis.
The stable inducible cell lines ectopically expressing EAV nsps that we present in this study, besides closely mimicking the morphology of DMVs in EAV-infected cells, open multiple possibilities for future research. These ‘surrogate systems’, inducibly expressing self-cleaving nsp constructs, could be employed, for example, to investigate the interplay between the virus-induced membrane structures and different host factors and pathways (e.g. innate immunity, autophagy), to study how viral proteins affect cellular lipid homeostasis, as well as in the search for new antiviral drugs that specifically target viral replication organelles.
Acknowledgements
We are grateful to Kèvin Knoops and Helena Maier for kindly providing high-resolution originals for some of the figures in this article. We thank Ronald Limpens for general technical assistance and discussions. This work was supported in part by the Netherlands Organization for Scientific Research through a TOP grant from the Council for Chemical Sciences (NWO-CW grant 700.57.301 to EJS and AJK) and a MEERVOUD grant (NWO-ALW grant 836.10.003 to MB) from the Council for Earth and Life Sciences.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2016.04.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
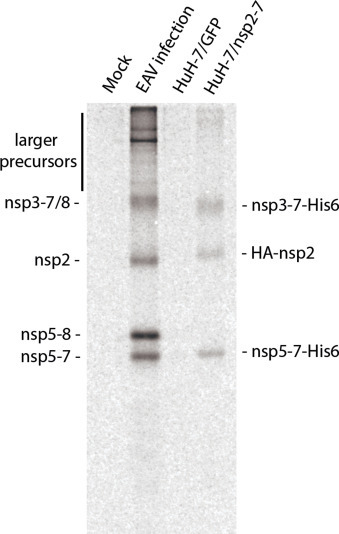
References
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nature reviews. Microbiology. 2006;4(5):371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla H.M., Turrell L., Smith N.M., Payne L., Baliji S., Zust R., Thiel V., Baker S.C., Siddell S.G., Neuman B.W. Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. mBio. 2014;5(2):e01107–e01113. doi: 10.1128/mBio.01107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(4) doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena M., Koster A.J. Electron tomography in life science. Semin. Cell Dev. Biol. 2009;20(8):920–930. doi: 10.1016/j.semcdb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., van Kuppeveld F.J. (+)RNA viruses rewire cellular pathways to build replication organelles. Curr. Opin. Virol. 2012;2(6):740–747. doi: 10.1016/j.coviro.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Nair V., Hansen B.T., Hoyt F.H., Fischer E.R., Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 2012;86(1):302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost A.G., Carnahan R.H., Lu X.T., Denison M.R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 2000;74(7):3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nature reviews. Immunology. 2008;8(12):911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Frolov I., Rice C.M., Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 1993;67(11):6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese S.S.J., McCollum W.H. Electron microscopy characterization of equine arteritis virus. In: Bryans J.T., Gerber H., editors. Proceedings of the 2nd International Conference on Equine Infectious Diseases; S. Karger Basel; 1970. pp. 133–139. [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77(19):10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryans J.T., Crowe M.E., Doll E.R., McCollum W.H. Isolation of a filterable agent causing arteritis of horses and abortion by mares; its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47(1):3–41. [PubMed] [Google Scholar]
- Cali T., Galli C., Olivari S., Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 2008;371(3):405–410. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- Cao X., Jin X., Zhang X., Li Y., Wang C., Wang X., Hong J., Wang X., Li D., Zhang Y. Morphogenesis of endoplasmic reticulum membrane-Invaginated vesicles during beet black scorch virus infection: role of auxiliary replication protein and new implications of three-dimensional architecture. J. Virol. 2015;89(12):6184–6195. doi: 10.1128/JVI.00401-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142(3):629–633. [PubMed] [Google Scholar]
- Chen Q., Fang L., Wang D., Wang S., Li P., Li M., Luo R., Chen H., Xiao S. Induction of autophagy enhances porcine reproductive and respiratory syndrome virus replication. Virus Res. 2012;163(2):650–655. doi: 10.1016/j.virusres.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Du W., Hagemeijer M.C., Takvorian P.M., Pau C., Cali A., Brantner C.A., Stempinski E.S., Connelly P.S., Ma H.C., Jiang P., Wimmer E., Altan-Bonnet G., Altan-Bonnet N. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160(4):619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiramel A.I., Brady N.R., Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2(1):83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7(11):1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 2002;83(Pt 4):927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- David-Ferreira J.F., Manaker R.A. An electron microscope study of the development of a mouse hepatitis virus in tissue culture cells. J. Cell Biol. 1965;24:57–78. doi: 10.1083/jcb.24.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Spaan W.J., van der Meer Y., Gibson C.A., Sims A.C., Prentice E., Lu X.T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 1999;73(8):6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P., Hoogenboom J.P., Giepmans B.N. Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods. 2015;12(6):503–513. doi: 10.1038/nmeth.3400. [DOI] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65(6):2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W., Posthuma C.C., van der Meer Y., Barcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94(Pt 8):1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Wannee K.F., Scholte F.E., Goeman J.J., Ten Dijke P., Snijder E.J., Kikkert M., van Hemert M.J. A kinome-wide small interfering RNA screen identifies proviral and antiviral host factors in severe acute respiratory syndrome Coronavirus replication, including double-Stranded RNA-Activated protein kinase and early secretory pathway proteins. J. Virol. 2015;89(16):8318–8333. doi: 10.1128/JVI.01029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Wang X., Ahlquist P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. U. S. A. 2010;107(37):16291–16296. doi: 10.1073/pnas.1011105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Gallei A., Ahlquist P. Bromovirus RNA replication compartment formation requires concerted action of 1a’s self-interacting RNA capping and helicase domains. J. Virol. 2012;86(2):821–834. doi: 10.1128/JVI.05684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Zhang J., Ollwerther A., Wang X., Ahlquist P. Correction: host ESCRT proteins are required for bromovirus RNA replication compartment assembly and function. PLoS Pathog. 2015;11(4):e1004845. doi: 10.1371/journal.ppat.1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunowska M., Biggs P.J., Zheng T., Perrott M.R. Identification of a novel nidovirus associated with a neurological disease of the Australian brushtail possum (Trichosurus vulpecula) Vet. Microbiol. 2012;156(3–4):418–424. doi: 10.1016/j.vetmic.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D., Wolk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002;76(12):5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faaberg K.S., Plagemann P.G. Membrane association of the C-terminal half of the open reading frame 1a protein of lactate dehydrogenase-elevating virus. Arch. Virol. 1996;141(7):1337–1348. doi: 10.1007/BF01718835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Snijder E.J. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154(1-2):61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris P., Beaumont E., Uzbekov R., Brand D., Gaillard J., Blanchard E., Roingeard P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell. Mol. Life Sci. 2013;70(7):1297–1306. doi: 10.1007/s00018-012-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J., Lopez-Iglesias C., Tzeng W.P., Frey T.K., Fernandez J.J., Risco C. Three-dimensional structure of Rubella virus factories. Virology. 2010;405(2):579–591. doi: 10.1016/j.virol.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.M., Sreevalsan T. Membrane binding of input arbovirus ribonucleic acid: effect of interferon or cycloheximide. J. Virol. 1970;6(2):169–175. doi: 10.1128/jvi.6.2.169-175.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E.I., Gorchakov R., Pereboeva L., Atasheva S., Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010;84(22):11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froshauer S., Kartenbeck J., Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988;107(6 Pt 1):2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie L.K., Hoenen A., Morgan G., Mackenzie J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010;84(20):10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucl. Acids Res. 1989;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimley P.M., Berezesky I.K., Friedman R.M. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 1968;2(11):1326–1338. doi: 10.1128/jvi.2.11.1326-1338.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21(8):413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Ulasli M., Vonk A.M., Reggiori F., Rottier P.J., de Haan C.A. Mobility and interactions of coronavirus nonstructural protein 4. J. Virol. 2011;85(9):4572–4577. doi: 10.1128/JVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J., de Haan C.A. Visualizing coronavirus RNA synthesis in time by using click chemistry. J. Virol. 2012;86(10):5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J., Reggiori F., de Haan C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu N.Y., Ilnytska O., Belov G., Santiana M., Chen Y.H., Takvorian P.M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C.E., Ehrenfeld E., van Kuppeveld F.J., Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141(5):799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Jr., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junjhon J., Pennington J.G., Edwards T.J., Perera R., Lanman J., Kuhn R.J. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 2014;88(9):4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Baker S.C. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 2000;74(17):7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes M.A., Miller C.L., Faaberg K.S. Porcine reproductive and respiratory syndrome virus nonstructural protein 2 (nsp2) topology and selective isoform integration in artificial membranes. Virology. 2015;481:51–62. doi: 10.1016/j.virol.2015.01.028. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Taylor M.P., Jackson W.T. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nature reviews. Microbiology. 2004;2(4):301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Swett-Tapia C., van den Worm S.H., Te Velthuis A.J., Koster A.J., Mommaas A.M., Snijder E.J., Kikkert M. Integrity of the early secretory pathway promotes, but is not required for, severe acute respiratory syndrome coronavirus RNA synthesis and virus-induced remodeling of endoplasmic reticulum membranes. J. Virol. 2010;84(2):833–846. doi: 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Barcena M., Limpens R.W., Koster A.J., Mommaas A.M., Snijder E.J. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012;86(5):2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5(9):e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar S.B., Levine B. Viruses and autophagy. Rev. Med. Virol. 2009;19(6):359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Lauck M., Bailey A.L., Shchetinin A.M., Vishnevskaya T.V., Bao Y., Ng T.F., LeBreton M., Schneider B.S., Gillis A., Tamoufe U., Diffo J.L., Takuo J.M., Kondov N.O., Coffey L.L., Wolfe N.D., Delwart E., Clawson A.N., Postnikova E., Bollinger L., Lackemeyer M.G., Radoshitzky S.R., Palacios G., Wada J., Shevtsova Z.V., Jahrling P.B., Lapin B.A., Deriabin P.G., Dunowska M., Alkhovsky S.V., Rogers J., Friedrich T.C., O'Connor D.H., Goldberg T.L. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2015 doi: 10.1007/s00705-015-2672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala P., Ahola T., Ehsani N., Auvinen P., Vihinen H., Kaariainen L. Intracellular distribution of rubella virus nonstructural protein P150. J. Virol. 1999;73(9):7805–7811. doi: 10.1128/jvi.73.9.7805-7811.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala P., Ikaheimonen A., Ehsani N., Vihinen H., Auvinen P., Kaariainen L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75(8):3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W., Schorb M., Welsch S., Picco A., Kaksonen M., Briggs J.A. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 2011;192(1):111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberte J.F., Sanfacon H. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 2010;48:69–91. doi: 10.1146/annurev-phyto-073009-114239. [DOI] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., Parquet Mdel C., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9(7):e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauck M., Sibley S.D., Hyeroba D., Tumukunde A., Weny G., Chapman C.A., Ting N., Switzer W.M., Kuhn J.H., Friedrich T.C., O'Connor D.H., Goldberg T.L. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J. Virol. 2013;87(1):688–691. doi: 10.1128/JVI.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leapman R.D., Aronova M.A. Localizing specific elements bound to macromolecules by EFTEM. Methods Cell Biol. 2007;79:593–613. doi: 10.1016/S0091-679X(06)79024-0. [DOI] [PubMed] [Google Scholar]
- Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucl. Acids Res. 2015;43(17):8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K.C., Snijder E.J., Posthuma C.C., Gorbalenya A.E. What we know but do not understand about nidovirus helicases. Virus Res. 2015;202:12–32. doi: 10.1016/j.virusres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tas A., Snijder E.J., Fang Y. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J. Gen. Virol. 2012;93(Pt 4):829–839. doi: 10.1099/vir.0.039289-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Tas A., Sun Z., Snijder E.J., Fang Y. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res. 2014 doi: 10.1016/j.virusres.2014.12.027. [DOI] [PubMed] [Google Scholar]
- Limpens R.W., van der Schaar H.M., Kumar D., Koster A.J., Snijder E.J., van Kuppeveld F.J., Barcena M. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio. 2011;2(5) doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Qin Y., Zhou L., Kou Q., Guo X., Ge X., Yang H., Hu H. Autophagy sustains the replication of porcine reproductive and respiratory virus in host cells. Virology. 2012;429(2):136–147. doi: 10.1016/j.virol.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergstrom T., Kann N., Adamiak B., Hannoun C., Kindler E., Jonsdottir H.R., Muth D., Kint J., Forlenza M., Muller M.A., Drosten C., Thiel V., Trybala E. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10(5):e1004166. doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Cottam E.M., Stevenson-Leggett P., Wilkinson J.A., Harte C.J., Wileman T., Britton P. Visualizing the autophagy pathway in avian cells and its application to studying infectious bronchitis virus. Autophagy. 2013;9(4):496–509. doi: 10.4161/auto.23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4(5):e00801–e00813. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. Cryopreparation methods for electron microscopy of selected model systems. Methods Cell Biol. 2007;79:23–56. doi: 10.1016/S0091-679X(06)79002-1. [DOI] [PubMed] [Google Scholar]
- Metwally S., Mohamed F., Faaberg K., Burrage T., Prarat M., Moran K., Bracht A., Mayr G., Berninger M., Koster L., To T.L., Nguyen V.L., Reising M., Landgraf J., Cox L., Lubroth J., Carrillo C. Pathogenicity and molecular characterization of emerging porcine reproductive and respiratory syndrome virus in Vietnam in 2007. Transbound. Emerg. Dis. 2010;57(5):315–329. doi: 10.1111/j.1865-1682.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nature reviews. Microbiology. 2008;6(5):363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Romero-Brey I., Maiuri P., Hoppe S., Krijnse-Locker J., Bartenschlager R., Marcello A. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J. Virol. 2013;87(11):6469–6481. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., Ulasli M., Rottier P.J., Guan J.L., Reggiori F., de Haan C.A. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9(2):164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nature reviews. Microbiology. 2012;10(2):137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt C.J., Joyce M.A., Van Buuren N., Levin A., Kirkegaard K., Gale M., Jr., Tyrrell D.L., Wozniak R.W. The hepatitis C virus-Induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12(2):e1005428. doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Offerdahl D.K., Dorward D.W., Hansen B.T., Bloom M.E. A three-dimensional comparison of tick-borne flavivirus infection in mammalian and tick cell lines. PLoS One. 2012;7(10):e47912. doi: 10.1371/journal.pone.0047912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81(22):12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein J.M., Banach B., Baker S.C. Morphogenesis of Coronavirus HCoV-NL63 in cell culture: a transmission electron microscopic study. Open Infect. Dis. J. 2008;2:52–58. doi: 10.2174/1874279300802010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3(10):e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013;2(2):32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Hoppe S., Saher G., Krijnse-Locker J., Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 2013;87(19):10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K.W., van der Meer Y., Roos N., Snijder E.J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 1999;73(3):2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol J.M., Wagenaar F., Reus J.E. Comparative morphogenesis of three PRRS virus strains. Vet. Microbiol. 1997;55(1–4):203–208. doi: 10.1016/s0378-1135(96)01329-6. [DOI] [PubMed] [Google Scholar]
- Posthuma C.C., Pedersen K.W., Lu Z., Joosten R.G., Roos N., Zevenhoven-Dobbe J.C., Snijder E.J. Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J. Virol. 2008;82(9):4480–4491. doi: 10.1128/JVI.02756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279(11):10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinkert D., Bartenschlager R., Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005;79(21):13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Cali T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7(6):500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.S., Longerich T., Diehl S., Ramirez F., Balla T., Rohr K., Kaul A., Buhler S., Pepperkok R., Lengauer T., Albrecht M., Eils R., Schirmacher P., Lohmann V., Bartenschlager R. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9(1):32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A.L., Jackson W.T. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 2012;8(11):e1003046. doi: 10.1371/journal.ppat.1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6(7):2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Merz A., Chiramel A., Lee J.Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S., Walther P., Antony C., Krijnse-Locker J., Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8(12):e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Berger C., Kallis S., Kolovou A., Paul D., Lohmann V., Bartenschlager R. NS5A domain 1 and polyprotein cleavage kinetics are critical for induction of double-Membrane vesicles associated with hepatitis C virus replication. mBio. 2015;6(4):e00759. doi: 10.1128/mBio.00759-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Vasiljeva L., Merits A., Magden J., Jokitalo E., Kaariainen L. Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment. J. Virol. 2003;77(3):1691–1702. doi: 10.1128/JVI.77.3.1691-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;9(3):505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- Shi S.T., Schiller J.J., Kanjanahaluethai A., Baker S.C., Oh J.W., Lai M.M. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J. Virol. 1999;73(7):5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., Spaan W.J. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 1994;68(9):5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., Spaan W.J., Gorbalenya A.E. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 1995;270(28):16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Roos N., Pedersen K.W. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 2001;82(Pt 5):985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94(Pt 10):2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- Spuul P., Balistreri G., Kaariainen L., Ahola T. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 2010;84(15):7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak J.C., Brodsky F.M., Miller E.A. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat. Cell Biol. 2013;15(9):1019–1027. doi: 10.1038/ncb2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley R.E., Ragusa M.J., Hurley J.H. The beginning of the end: how scaffolds nucleate autophagosome biogenesis. Trends Cell Biol. 2014;24(1):73–81. doi: 10.1016/j.tcb.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Ernst A., Dikic I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- Stueckemann J.A., Ritzi D.M., Holth M., Smith M.S., Swart W.J., Cafruny W.A., Plagemann G.W. Replication of lactate dehydrogenase-elevating virus in macrophages. 1. Evidence for cytocidal replication. J. Gen. Virol. 1982;59(Pt 2):245–262. doi: 10.1099/0022-1317-59-2-245. [DOI] [PubMed] [Google Scholar]
- Suhy D.A., Giddings T.H., Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74(19):8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.X., Huang L., Wang R., Yu Y.L., Li C., Li P.P., Hu X.C., Hao H.P., Ishag H.A., Mao X. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy. 2012;8(10):1434–1447. doi: 10.4161/auto.21159. [DOI] [PubMed] [Google Scholar]
- Svoboda D., Nielson A., Werber A., Higginson J. An electron microscopic study of viral hepatitis in mice. Am. J. Pathol. 1962;41:205–224. [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12(6):844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aken D., Zevenhoven-Dobbe J., Gorbalenya A.E., Snijder E.J. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 2006;87(Pt 12):3473–3482. doi: 10.1099/vir.0.82269-0. [DOI] [PubMed] [Google Scholar]
- van Dinten L.C., Wassenaar A.L., Gorbalenya A.E., Spaan W.J., Snijder E.J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 1996;70(10):6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., de Wilde A.H., Gorbalenya A.E., Snijder E.J. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J. Biol. Chem. 2008;283(24):16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4(5):e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer Y., van Tol H., Locker J.K., Snijder E.J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 1998;72(8):6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J., Locker J.K. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 1999;73(9):7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatter H.A., Di H., Donaldson E.F., Radu G.U., Maines T.R., Brinton M.A. Functional analyses of the three simian hemorrhagic fever virus nonstructural protein 1 papain-like proteases. J. Virol. 2014;88(16):9129–9140. doi: 10.1128/JVI.01020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R., Fukunaga Y., Kondo T., Kanemaru T. Ultrastructure and immuno-cytochemistry of BHK-21 cells infected with a modified Bucyrus strain of equine arteritis virus. Arch. Virol. 1995;140(7):1173–1180. doi: 10.1007/BF01322744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ptacek J.B., Kirkegaard K., Bullitt E. Double-membraned liposomes sculpted by poliovirus 3AB protein. J. Biol. Chem. 2013;288(38):27287–27298. doi: 10.1074/jbc.M113.498899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar A.L., Spaan W.J., Gorbalenya A.E., Snijder E.J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 1997;71(12):9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland F.H., Granzow M., Wieczorek-Krohmer M., Weiland E. Electron microscopic studies on the morphogeneis of PRRSV in infected cells—comparative studies. Schwyzer M., Ackermann M., Bertoni G., Kocherhands R., McCullough K., Engels M., Wittek R., Zanoni R., editors. Immunobiology of Viral Infections. Proceedings of the 3rd Congress of the European Society of Veterinary Virology. 1995:499–502. [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa R., de Vries A.A., van der Meulen J., Godeke G.J., Onderwater J.J., van Tol H., Koerten H.K., Mommaas A.M., Snijder E.J., Rottier P.J. Structural protein requirements in equine arteritis virus assembly. J. Virol. 2004;78(23):13019–13027. doi: 10.1128/JVI.78.23.13019-13027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood O., Tauraso N., Liebhaber H. Electron microscopic study of tissue cultures infected with simian haemorrhagic fever virus. J. Gen. Virol. 1970;7(2):129–136. doi: 10.1099/0022-1317-7-2-129. [DOI] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W.t. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3(6):581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(Pt 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276(35):33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Roth H., Kurth A., Drosten C., Ziebuhr J., Junglen S. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 2013;87(11):6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C., Cormier J.H., Guhl B., Santimaria R., Hebert D.N., Roth J. EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites. Proc. Natl. Acad. Sci. U. S. A. 2007;104(11):4407–4412. doi: 10.1073/pnas.0700154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


