Abstract
Our understanding of how the first mammalian cell lineages arise has been shaped largely by studies of the preimplantation mouse embryo. Painstaking work over many decades has begun to reveal how a single totipotent cell is transformed into a multilayered structure representing the foundations of the body plan. Here, we review how the first lineage decision is initiated by epigenetic regulation but consolidated by the integration of morphological features and transcription factor activity. The establishment of pluripotent and multipotent stem cell lines has enabled deeper analysis of molecular and epigenetic regulation of cell fate decisions. The capability to assemble these stem cells into artificial embryos is an exciting new avenue of research that offers a long-awaited window into cell fate specification in the human embryo. Together, these approaches are poised to profoundly increase our understanding of how the first lineage decisions are made during mammalian embryonic development.
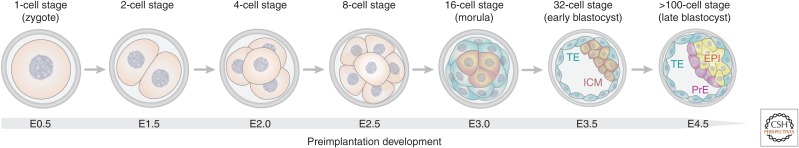
The preimplantation mouse embryo is an exceptional model system for studying the development of the first mammalian cell lineages. Beginning from the single-cell fertilized zygote stage, the mouse embryo can progress through the entire preimplantation stage in vitro without the requirement for maternal input (Whitten and Biggers 1968). Over a period of 4.5 days, the embryo will undergo cleavage divisions without significant growth, activate transcription of the embryonic genome, compact to form a morula, and finally establish the first internal cavity to generate a blastocyst ready for implantation into the uterus (Fig. 1; White et al. 2018). During this time, the cells of the embryo gradually differentiate from the totipotent zygote into three separate lineages. The first two lineages to emerge are the extraembryonic trophectoderm (TE), which will give rise to the placenta and the inner cell mass (ICM). The ICM subsequently differentiates into two lineages, the pluripotent epiblast (EPI), which will give rise to all germ layers of the embryo and the primitive endoderm (PrE), which forms the extraembryonic membranes (Cockburn and Rossant 2010; White et al. 2018). Therefore, at the time of implantation, the blastocyst resembles a hollow multilayered ball consisting of a layer of TE cells that form the outer surface with an inner core of EPI cells at one end, separated from an internal cavity by a layer of PrE cells (Fig. 1). A distinct type of stem cell can be derived from each of these lineages: trophoblast stem cells (TSCs) from the TE (Tanaka et al. 1998), extraembryonic endoderm (XEN) stem cells from the PrE (Kunath et al. 2005), and embryonic stem cells (ESCs) from the EPI or earlier stages of the ICM (Evans and Kaufman 1981; Martin 1981).
Figure 1.
Timeline of mouse preimplantation development. During the first 5 days of preimplantation development, the fertilized zygote undergoes a series of cleavage divisions to form a blastocyst. The first spatial segregation of cells occurs at the 16-cell stage with the formation of a population of inner cells. By the 32-cell stage, the outer cells on the surface of the embryo are committed to becoming trophectoderm (TE) and the inner cells are specified as the inner cell mass (ICM). The first cavity has formed inside the blastocyst. At the late blastocyst stage, the ICM differentiates into primitive endoderm (PrE) and the epiblast (EPI).
The accessibility of the preimplantation mouse embryo combined with its amenability to experimental manipulation has enabled significant progress in understanding how these first cell differentiation events occur. However, although the key molecular differences between the first three lineages have been known for many years (Whitten and Biggers 1968; Palmieri et al. 1994; Russ et al. 2000; Avilion et al. 2003; Mitsui et al. 2003; Niwa et al. 2005; Strumpf et al. 2005; Ralston et al. 2010), how these differences are first established remains the topic of intense investigation. Here, we review our current understanding of how the first cell fate decisions are made during mammalian development from research using preimplantation embryos and stem cell models.
THE FIRST LINEAGE DECISION
The first lineage decision cells must make during mammalian development is the choice between becoming the pluripotent ICM or undergoing the first differentiation event to become TE. Several transcription factors have been identified that are critical for the correct specification of TE and ICM fate. OCT4 is often regarded as a master regulator of a network of pluripotency-associated genes including SOX2 and NANOG; however, OCT4 expression does not become restricted to the ICM until after the start of blastocyst formation (Palmieri et al. 1994; Dietrich and Hiiragi 2007). Loss of OCT4 does not prevent blastocyst formation, but the ICM of these mutant embryos expresses TE markers instead of maintaining pluripotency (Nichols et al. 1998).
The earliest transcription factors to display lineage-specific expression patterns are CDX2 and SOX2 (Strumpf et al. 2005; Wicklow et al. 2014). Expression of SOX2 initiates at the four-cell stage and starts to become restricted to the inner cells at the 16-cell stage, whereas CDX2 is first expressed at the eight-cell stage and is restricted to the outer cells at the late 16-cell stage (White et al. 2016). By the blastocyst stage, SOX2 expression is restricted to those cells of the ICM that will give rise to the EPI, and it is required to maintain these cells in an undifferentiated state (Avilion et al. 2003).
CDX2 expression represses transcription of the ICM-specific genes Oct4 and Nanog in the outer cells and promotes TE maturation (Strumpf et al. 2005; Huang et al. 2017a). Expression of the transcription factor GATA3 is highly correlated with CDX2, and these proteins work both together and independently to activate a TE-specific gene regulatory network (Ralston et al. 2010). Once initial differences in transcription factor expression have been established, they are consolidated by the combined actions of autoregulatory feedback loops that drive Cdx2 and Oct4 expression (Chew et al. 2005), and reciprocal repression of Cdx2 by OCT4, SOX2, and NANOG (Boyer et al. 2005; Loh et al. 2006; Huang et al. 2017a).
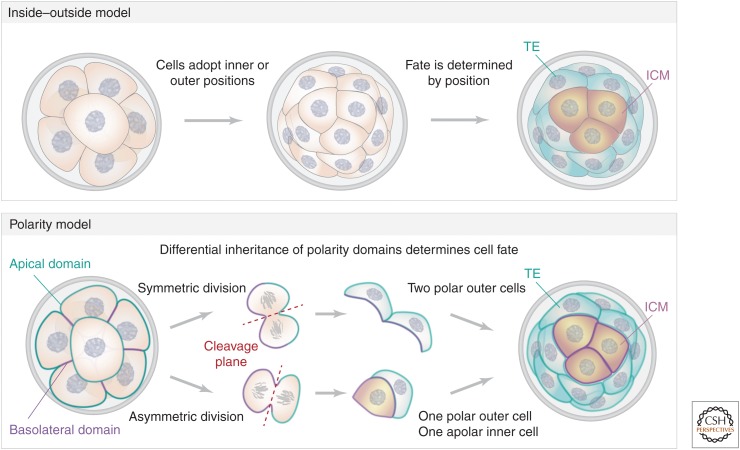
The first lineages begin to spatially segregate at the 16-cell stage when some cells are positioned inside the embryo to give rise to the ICM. Traditionally it was assumed that spatially “asymmetric” cell divisions allocated inner cells by cleaving the parental cell perpendicular to the surface of the embryo and directly pushing one daughter cell inside as a result of the scission (Johnson and Ziomek 1981a; Cockburn and Rossant 2010). However, computational modeling suggests that differentially oriented cell divisions alone are not sufficient to give rise to the observed spatial segregation of inner and outer cells (Krupinski et al. 2011; Holmes et al. 2017), suggesting additional mechanisms are required. Consistent with this, several studies had reported that inner cells could be positioned through cell internalization events (Plusa et al. 2005; Yamanaka et al. 2010; Anani et al. 2014; Watanabe et al. 2014), and the more recent application of quantitative live imaging revealed that most inner cells are allocated through a process of apical constriction (Samarage et al. 2015). Once inside the embryo, the inner cells are completely enclosed by cell–cell contacts and therefore lack apical polarity. The outer cells retain a contactless apical membrane, allowing them to polarize, and they are biased toward the TE lineage. Historically, this seemingly inextricable connection between cell fate, polarity, and position led to proposals that the first lineage decision is directed by cell position: the “inside–outside model” (Tarkowski and Wróblewska 1967), or cell polarity: the “polarity model” (Fig. 2; Johnson and Ziomek 1981a,b). The inside–outside model suggests that cells are exposed to different microenvironments on the basis of their position within the embryo, and this directs their acquisition of TE or ICM identity. Support for this model was provided by the demonstration that repositioning cells within the embryo forces them to adopt the fate appropriate for their new position (Hillman et al. 1972; Kelly 1977; Suwinska et al. 2008). However, the inside–outside model cannot explain the existence of a small population of apolar outer cells that do not become TE, but instead gradually internalize and become ICM (Plusa et al. 2005; Anani et al. 2014).
Figure 2.
The inside–outside and polarity models of the first lineage decision. According to the inside–outside model, cells first adopt positions on the outside or inside of the embryo. The cells are then exposed to different microenvironments on the basis of their position within the embryo, and this directs their differentiation toward either trophectoderm (TE) or inner cell mass (ICM). The polarity model suggests that the differential inheritance of polarity domains during cell division directs cells fate. A symmetric division in which the cleavage plane (red dotted line) aligns with the radial axis of the embryo splits the apical membrane and generates two outer polar cells that are fated to become TE. An asymmetric division in which the plane of cleavage is parallel to the surface of the embryo generates one polar TE cell that inherits the apical membrane and one inner cell that inherits predominantly basolateral membrane and is therefore apolar and fated to become ICM.
By contrast, the polarity model proposes that cells that inherit the apical membrane domain during the eight- to 16-cell stage division will become TE. Cells that do not inherit an apical membrane domain will be apolar and form the ICM. Whether a cell inherits the apical membrane was thought to be determined by the plane of cell division (Johnson and Ziomek 1981a). An “asymmetric” division in which the plane of cleavage is parallel to the surface of the embryo would generate one TE cell that inherited the entire apical membrane and was therefore polarized and one ICM cell that only inherited basolateral membrane and was therefore apolar. A symmetric division in which the cleavage plane aligned with the radial axis of the embryo would instead split the apical membrane and generate two polar TE cells (Fig. 2). The recent demonstrations that the apical domain is disassembled just prior to division (Zenker et al. 2018) and that apolar cells can spontaneously polarize many hours after division (Anani et al. 2014; Korotkevich et al. 2017; Zenker et al. 2018) suggest that the polarity model also does not fully explain the segregation of the first two lineages. Although the inside–outside and polarity models have long been presented as distinct ideas, recent work demonstrating how the Hippo signaling pathway directs cell fate in the embryo has largely reconciled these two models.
The Hippo signaling pathway regulates cell fate in response to polarity and cell contact asymmetry (Sasaki 2017; Saini and Yamanaka 2018). Activation of Hippo signaling leads to phosphorylation of the YAP1 transcriptional coactivator by the kinase LATS1/2. The phosphorylation of YAP1 regulates how it shuttles between the nucleus and the cytoplasm (Sasaki 2017). In the outer cells of the embryo, unphosphorylated YAP1 translocates into the nucleus and interacts with the TEAD4 transcription factor to promote expression of TE-specific genes including Cdx2 and Gata3 (Nishioka et al. 2009; Ralston et al. 2010) and repress expression of the pluripotency-associated gene Sox2 (Wicklow et al. 2014; Frum et al. 2018). In the inner cells, phosphorylated YAP1 is excluded from the nucleus, preventing induction of TE-specific gene expression (Nishioka et al. 2009). The phosphorylation status of YAP1 is thought to be regulated by the balance between cell–cell adhesion and cell polarity, which are in turn largely determined by cell position. At the molecular level, these properties are sensed by the cell through the subcellular distribution of angiomotin (AMOT). In the apolar inner cells, AMOT localizes to the adherens junctions enclosing the cell. There, AMOT associates with a complex of NF2, α- and β-catenin, and E-cadherin and is activated through phosphorylation by LATS1/2 kinase (Nishioka et al. 2009; Cockburn et al. 2013; Hirate et al. 2013). Phosphorylation of AMOT inhibits its F-actin binding activity and promotes its interaction with LATS1/2 (Dai et al. 2013; Hirate et al. 2013). Together, phosphorylated AMOT and LATS1/2 enhance the phosphorylation and nuclear exclusion of YAP1, allowing the expression of pluripotency-associated genes.
Conversely, in the polarized outer cells, AMOT is sequestered away from the junctions by the apical domain where it remains unphosphorylated and binds to cortical F-actin (Hirate et al. 2013; Leung and Zernicka-Goetz 2013). RHO kinase also acts to stabilize AMOT binding to F-actin and inhibit the interaction between AMOT and NF2 (Shi et al. 2017). As AMOT is absent from the junctional complexes, Hippo signaling remains inactive and unphosphorylated YAP1 is retained in the nucleus to drive a TE-specific gene expression program (Ralston et al. 2010; Hirate et al. 2013; Sasaki 2017). In this way, the Hippo signaling pathway integrates information about a cell's position and polarity to direct its fate.
However, the Hippo signaling pathway is not the sole regulator of TE-associated gene expression, but instead it acts in parallel with Notch signaling. Activation of the Notch pathway causes translocation of the intracellular domain of the Notch receptor (NICD) into the nucleus where it binds to the transcription factor RBPJ to activate gene expression. Both the YAP1/TEAD and NICD/RBPJ complexes interact with the chromatin modifier SBNO1 to drive the expression of Cdx2 (Rayon et al. 2014; Watanabe et al. 2017). Recent work has demonstrated that Notch signaling is activated heterogeneously between cells as early as the four-cell stage and may account for the onset of Cdx2 expression (Menchero et al. 2019). The differential activation of Hippo signaling following cell polarization and the establishment of inner and outer cell populations may therefore act to reinforce Cdx2 expression in outer cells correctly positioned to become TE while preventing its maintenance in inner cells. Apolar outer cells may resolve their fate by polarizing (Anani et al. 2014; Korotkevich et al. 2017; Zenker et al. 2018) to activate Hippo signaling or internalizing (Plusa et al. 2005; Anani et al. 2014; Samarage et al. 2015) to repress Hippo signaling and become ICM. The factors controlling whether these cells polarize or internalize remain to be determined.
WHEN DOES CELL FATE COMMITMENT OCCUR?
Although the first spatial segregation of cells into inner and outer positions at the 16-cell stage generally designates these cells as future ICM or TE, respectively, their fate is not restricted until subsequent stages of development. Conflicting results were generated by early studies in which inner and outer cells were dissociated from embryos at varying stages of development and then reaggregated to form chimeric embryos (Handyside and Johnson 1978; Hogan and Tilly 1978; Spindle 1978; Rossant and Lis 1979; Tarkowski et al. 2010). However, a consensus appears to be emerging that TE cell fate is largely committed at the 32-cell stage (Suwinska et al. 2008; Posfai et al. 2017), whereas cells of the ICM may be specified at the 32-cell stage but their fate is not committed until the 64-cell stage (Handyside and Johnson 1978; Rossant and Lis 1979; Stephenson et al. 2010; Posfai et al. 2017). These findings are consistent with analysis demonstrating that single cells can be classified as TE or ICM by the 32-cell stage and further categorized into TE, EPI, or PE by the 64-cell stage on the basis of their gene expression (Kurimoto et al. 2006; Guo et al. 2010).
Unlike other organisms such as Caenorhabditis elegans and Arabidopsis, the early mammalian embryo does not follow a stereotyped pattern of cell divisions, meaning that mechanisms are required to correct errors in cell allocation and enable robust development. Furthermore, the mammalian embryo is highly regulative, meaning that it can compensate for the loss or gain of cells (Tarkowski 1959, 1961; Hillman et al. 1972). The gradual nature of cell fate commitment during mammalian preimplantation development has been proposed to create an early time window in which stochastic events can drive correction of cell-specification errors (Holmes et al. 2017; Chen et al. 2018; White et al. 2018). As cell fates gradually become more distinct, the threshold for transitioning between ICM and TE increases and the effects of noise diminish. Intriguingly, cell fate commitment appears to be even more gradual in the human embryo as TE cells are not yet committed even at the late blastocyst stage (De Paepe et al. 2013).
WHEN DO THE FIRST DIFFERENCES ARISE BETWEEN CELLS OF THE EMBRYO?
It has been proposed that differences between cells of the embryo do not arise until cells are spatially segregated at the 16-cell stage (Hiiragi and Solter 2004; Alarcón and Marikawa 2005; Motosugi et al. 2005). Initial studies analyzing the expression of selected candidate genes found no differences between early blastomeres (Dietrich and Hiiragi 2007; Ralston and Rossant 2008; Guo et al. 2010; Wicklow et al. 2014). However, mounting evidence suggests that heterogeneities that bias cells toward TE or ICM may arise far earlier than the 16-cell stage. Several lineage tracing studies revealed that cells of the early embryo may not all contribute equally to all lineages, and some cells display a fate bias as early as the four-cell (Fujimori et al. 2003; Piotrowska-Nitsche et al. 2005; Tabansky et al. 2013) or even two-cell stage (Casser et al. 2017; Boiani et al. 2019), although this may be dependent on genetic background (Alarcón and Marikawa 2005). However, evidence is accumulating for the introduction of molecular heterogeneities between cells of the early embryo from a variety of sources (Fig. 3).
Figure 3.
Molecular heterogeneities in the early preimplantation mouse embryo that influence cell fate. (A, top) Schematic showing how multiplied partitioning errors at each cell division increase the heterogeneity between cells. (Bottom) Graph demonstrating the amplification effect of a small bias in distribution of molecules between cells during the first three cell division cycles. (B) Single-cell RNA-seq and cell-to-cell expression noise in two- to eight-cell mouse and human (C) embryos. Each dot represents a comparison of two cells within the same embryo. (A–C from Shi et al. 2015; adapted, with permission, from Company of Biologists © 2015.) (D) Computational segmentation of a four-cell mouse embryo showing two pairs of sister cells, cell 1 and cell 2 (purple nuclei) and cell 3 and cell 4 (blue nuclei). (E,F) Within each sister pair there is one cell with a larger fraction of Sox2-paGFP engaged in long-lived binding to the DNA (cell 1 and cell 3) and one cell with a smaller fraction (cell 2 and cell 4). Inset in E highlights the differences between the autocorrelation curves in the time window corresponding to DNA binding. (G) Immunostaining of a four-cell stage mouse embryo shows two cells with higher levels of H3R26me2 and two cells with lower levels. (H) Individual data points for the long-lived bound fractions of paGFP-Sox2 (vertical) and levels of H3R26me2 (horizontal) are shown on the same graph. Cells with a larger long-lived bound fraction display higher levels of H3R26me2. (I) CARM1 knockdown decreases inner cell numbers. At the two-cell stage, one cell was microinjected with memb-mCherry RNA and Carm1 siRNAs. Images show computationally segmented 16-cell embryos. Fewer inner cells derive from Carm1 siRNA-injected cells. Scale bar, 10 µm. (D–I from White et al. 2016; adapted, with permission, from Elsevier © 2016.)
Partitioning Errors
The recent application of single-cell RNA sequencing technology to the preimplantation embryo revealed that transcriptional differences between cells of the embryo arise as early as the first cleavage division (Biase et al. 2014; Piras et al. 2014; Shi et al. 2015). At this stage, the zygotic genome has not yet been activated, and development is driven almost exclusively by maternally provided factors deposited in the egg (Zhou and Dean 2015; Gao et al. 2017). During cell division, these factors may be unevenly distributed into each daughter cell in a process known as a partitioning error (Fig. 3A; Huh and Paulsson 2011; Shi et al. 2015). Although some of these errors may be subsequently corrected, genes expressed at low levels are disproportionately affected by partitioning errors (Shi et al. 2015), leading to the establishment of differential gene expression between cells. These early heterogeneities may then be further amplified by transcriptional noise when transcription of the zygotic genome is activated during the two-cell stage (Piras et al. 2014) in mice or the four- to eight-cell stage in humans (Fig. 3B,C; Lee et al. 2014; Shi et al. 2015). Gene expression noise can be introduced intrinsically through transcriptional bursting or extrinsically by the propagation of fluctuations in the expression of one gene leading to fluctuations in the expression of downstream genes. Within the embryo, feedback loops fine-tune initial differences in gene expression to affect the ratio of opposing lineage specifiers and bias cells toward alternate fates (Shi et al. 2015).
Epigenetic Modifications and Transcription Factor Binding
Some of the first differences demonstrated between cells of the early embryo occur at the level of epigenetic modifications. At the four-cell stage, an asymmetry between cells in the levels of dimethylation of histone H3 at arginines 17 and 26 (H3R17 and H3R26) and in the expression of the chromatin modifiers CARM1 (Torres-Padilla et al. 2007) and PRDM14 (Burton et al. 2013) becomes apparent. H3 methylation levels are proposed to influence cell fate determination, as cells displaying higher levels of methylation at H3R17 and H3R26 at the four-cell stage tend to contribute more progeny to the ICM of the embryo (Torres-Padilla et al. 2007; Burton et al. 2013). CARM1 overexpression leads to increased H3R26 methylation and is associated with positioning of progeny within the ICM and elevated expression of ICM fate markers NANOG and SOX2 (Torres-Padilla et al. 2007).
Recently, it has become possible to probe transcription factor dynamics in vivo using photoactivatable fluorescence correlation spectroscopy (paFCS) techniques (Plachta et al. 2011; Kaur et al. 2013; Zhao et al. 2017). The development of this technology has provided insights into how differences in epigenetic modifications may contribute to lineage divergence. Application of paFCS within the embryo revealed that higher levels of CARM1-mediated histone H3 methylation promotes DNA binding of the pluripotency-associated transcription factor SOX2, likely by increasing the accessibility of SOX2 binding sites (Fig. 3D–I; White et al. 2016). This increased SOX2–DNA binding is proposed to up-regulate SOX2-dependent gene expression linked to pluripotency and bias cells toward an embryonic fate (Goolam et al. 2016; White et al. 2016). How the variability in histone H3 methylation is first established at the four-cell stage is not entirely clear, but it may depend partly on the heterogenous expression of a long noncoding RNA (lncRNA) called LincGET (Wang et al. 2018). During the two- to four-cell stage, LincGET is transiently and asymmetrically expressed. It can physically interact with CARM1, promoting its nuclear localization and increasing histone methylation and chromatin accessibility. Like CARM1, the overexpression of LincGET in one cell at the two-cell stage biases the injected cells toward an ICM fate. Knocking down LincGET reduces the contribution of the injected cells to the ICM, although the total number of cells in the ICM remains unchanged and embryos can still develop to the blastocyst stage (Wang et al. 2018). An additional lncRNA called Neat1 is also required for CARM1-dependent histone methylation and promotion of pluripotent cell fate (Hupalowska et al. 2018). Neat1 acts as a scaffold for nuclear foci called paraspeckles, which recruit CARM1 and are heterogenous between cells of the four-cell stage embryo in a manner that correlates with H3R26 methylation levels. Disrupting paraspeckles leads to reduced levels of histone methylation, increased expression of Cdx2, a failure to correctly specify the first cell lineages, and developmental arrest (Hupalowska et al. 2018). Although LincGET and Neat1 appear to play pivotal roles in the first cell lineage decision, the factors underlying the initial differences in these lncRNAs between cells of the embryo remain to be determined.
Therefore, the evidence to date suggests that the initial heterogeneities in histone modifications may originate from an interplay between the first differences in gene expression (Biase et al. 2014; Piras et al. 2014; Shi et al. 2015), variations in cell cleavage orientations (Zernicka-Goetz et al. 2009), the asymmetric distribution of lncRNAs such as LincGET (Wang et al. 2018) and Neat 1 (Hupalowska et al. 2018), and potentially other unidentified factors.
REGULATORS OF PLURIPOTENCY IN THE PREIMPLANTATION EMBRYO
For the past two decades it has been known that the preimplantation stage of mouse development is characterized by significant epigenetic changes (Thompson et al. 1995; Stein and Schultz 2000; Ma et al. 2001). Parental epigenetic marks inherited by the zygote from the terminally differentiated gametes must be extensively remodeled to establish a zygotic epigenome required for subsequent embryo development (Huang et al. 2015; Dahl et al. 2016; Eid et al. 2016). Recent technological advances are now revealing how changes in chromatin accessibility and three-dimensional (3D) organization, histone modifications, DNA methylation, and expression of retrotransposons are dynamically regulated in the early embryo (Xu and Xie 2018).
Global Chromatin Remodeling
At the most global level, changes in chromatin structure and nuclear architecture control the accessibility of key regulatory DNA sequences to transcription factors and RNA polymerase to regulate gene expression (Tsompana and Buck 2014). The fundamental unit of chromatin is the nucleosome, which is formed by wrapping DNA around an octamer of histone proteins. Posttranslational modifications of the histones and incorporation of histone variants modulate the degree of chromatin compaction or relaxation and recruit regulatory proteins (Schneider and Grosschedl 2007; Bannister and Kouzarides 2011). Chromatin structure is very relaxed after fertilization in both the mouse and human zygote (Wu et al. 2018) and then slowly compacts throughout preimplantation development (Du et al. 2017). The relaxed chromatin state is supported by the high mobility of histone proteins, which gradually decreases throughout preimplantation development, in line with the idea that increasing condensation of chromatin is associated with differentiation during development (Boskovic et al. 2014; Ooga et al. 2016).
The most obvious features of such chromatin reorganization in the early embryo can be observed directly using microscopy to visualize stained DNA (Borsos and Torres-Padilla 2016). In zygotes and early two-cell-stage embryos, most of the chromatin is uncompacted with only a small amount of dense heterochromatin surrounding the nuclear envelope or nucleoli (Ahmed et al. 2010). Concurrent with activation of transcription of the zygotic genome at the late two-cell stage, the perinucleolar chromatin begins to cluster into patches that dissociate from the nucleoli during the four-cell stage and are dispersed throughout the nucleus from the eight-cell stage onward (Martin et al. 2006a; Aguirre-Lavin et al. 2012). A similar global reorganization of the chromatin also occurs in embryos generated by parthenogenesis and somatic cell nuclear transfer (SCNT), suggesting that it may be associated with reprogramming (Martin et al. 2006b; Merico et al. 2007). Although the purpose of the early perinucleolar organization of heterochromatin and the molecular mechanisms underlying its redistribution are not fully understood, disrupting this subnuclear localization leads to serious defects in embryo development (Jachowicz et al. 2013).
Histone Modifications
Following fertilization, the maternal and paternal genomes undergo extensive epigenetic remodeling, and an asymmetry in histone modifications is established (Beaujean 2014). The paternal genome becomes rapidly hyperacetylated on lysines 5, 8, 12, and 16 of histone H4 and lysines 9, 14, 18, and 27 of histone H3. Acetylation of lysine neutralizes its positive charge and is thought to promote an open chromatin structure and gene transcription (Clayton et al. 2006). Consistent with this, there is increased transcriptional activation of the paternal genome at the one-cell stage (Aoki et al. 1997). By contrast, the maternal genome is characterized by increased methylation on lysines 4, 9, 27, 36, and 64 of histone H3 and lysine 20 of histone H4. Histone methylation does not alter the binding of DNA to histones but can recruit regulatory proteins that act in association with chromatin remodeling complexes (Bannister and Kouzarides 2011; Musselman et al. 2012). This asymmetry between the parental epigenomes can be detected until the four-cell stage in the mouse embryo and has been proposed to be necessary for early development, yet this remains to be demonstrated (Beaujean 2014; Burton and Torres-Padilla 2014).
Although the initial asymmetries between the parental genomes are gradually equalized for most histone modifications following the first cell cleavages, by the blastocyst stage a new epigenetic asymmetry develops between the lineages (Burton and Torres-Padilla 2010). The cells of the ICM have globally higher levels of methylation on both the DNA and histone H3, as well as reduced phosphorylation of histone H2A and H4 compared to the TE cells (Santos et al. 2002; Erhardt et al. 2003; Sarmento et al. 2004). In addition, activating histone modifications are enriched on the Oct4 and Nanog promoters and a repressive histone mark labels the Cdx2 promoter in ICM cells compared with TE cells, establishing a correlation between histone methylation and gene expression in the blastocyst (O'Neill et al. 2006). Studies demonstrating that disrupting the methyltransferases responsible for the DNA and histone methylations in mouse embryos results in more severe defects in the embryonic tissues than the TE provide further evidence of the functional importance of these epigenetic asymmetries between the lineages (Li et al. 1992; Okano et al. 1999; Tachibana et al. 2002; Erhardt et al. 2003).
DNA Methylation
Beyond methylation of histones, genomic DNA can also be methylated by a covalent modification of cytosine (5mC) that occurs mainly on the CpG dinucleotide and is often associated with transcriptional repression (Bird 1986). 5mC is established and maintained by the DNA methyltransferases DNMT3A/B/L and DNMT1, respectively, and removed either passively by dilution during genome replication or actively by Ten-eleven translocation (TET) proteins (Bhutani et al. 2011; Jurkowska et al. 2011). Inhibiting the activity of DNMT3A/B or TET3 in oocytes leads to disruption of DNA methylation and embryonic defects, confirming the importance of regulating DNA methylation during early development (Okano et al. 1999; Gu et al. 2011).
In human embryos, a wave of global DNA demethylation occurs within 10–12 h after fertilization and acts predominantly on the paternal genome (Zhu et al. 2018). An additional two waves of DNA demethylation occur during the late zygote to two-cell stage and the eight-cell to morula stage. Within this context of global demethylation, there are also two waves of focused methylation occurring during the early male pronuclear to mid-pronuclear stage and from the four-cell to eight-cell stage. This de novo methylation occurs largely within retrotransposons such as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and long terminal repeats (LTRs) (Zhu et al. 2018).
Retrotransposon Activation
In both mouse and human embryos, there is significant stage-specific activation of retrotransposon transcription during preimplantation development that is proposed to contribute to zygotic genome activation and cellular plasticity (De Iaco et al. 2017; Jachowicz et al. 2017; Rodriguez-Terrones and Torres-Padilla 2018). In the mouse oocyte, LTRs are the most highly expressed class of retrotransposon; however, by the blastocyst stage, LINE-1 becomes the most abundant retrotransposon transcript (Fadloun et al. 2013). In addition to DNA methylation, retrotransposon expression during preimplantation development seems to be modulated by varying levels of histone modifications, but the molecular mechanisms regulating this remain poorly understood (Fadloun et al. 2013).
Within the family of LTRs, endogenous retroviruses (ERVs) are the most abundant element, comprising ∼8% of the human genome (Lander et al. 2001). LTRs are able to act as powerful promoters that drive the transcription of nearby genes in the oocytes and embryos of both mice and humans (Peaston et al. 2004). This developmentally regulated LTR expression may trigger sequential reprogramming of the embryonic genome and is required for the maternal-to-zygotic transition in mouse embryos (Huang et al. 2017b). ERVs can also encode lncRNAs such as LincGET, which is essential for embryo development (Wang et al. 2016) and interacts with CARM1 to increase chromatin accessibility and promote ICM cell fate (Wang et al. 2018).
Temporal regulation of LINE-1 transcription is required for mouse preimplantation development as either premature silencing of LINE-1 in the zygote or preventing LINE-1 silencing after the two-cell stage leads to embryo arrest (Jachowicz et al. 2017). The transcriptional activation of LINE-1 promotes global remodeling of chromatin accessibility, suggesting that retrotransposons contribute to shaping chromatin architecture during early development.
Together, these studies are building a clearer picture of the underlying mechanisms that drive the progression from totipotency to pluripotency and then on to the establishment of the first cell lineages in the mammalian embryo. The earliest event in this cascade is the reorganization of chromatin architecture by epigenetic modifications and activation of retrotransposon transcription to establish a permissive chromatin environment for cell fate choices. Against a backdrop of ongoing chromatin remodeling, differences in transcription factor binding and gene expression gradually begin to emerge between cells. The first lineage decision appears to be initiated by epigenetic regulation, but the cell fate choice is only consolidated by transcription factor expression later in development.
MOUSE PLURIPOTENT STEM CELLS
In 1981, pluripotent ESCs were first established as nontransformed cell lines from the ICM of mouse blastocysts (Evans and Kaufman 1981; Martin 1981). These cells were both self-renewing and pluripotent as they could form teratomas when implanted into an immunocompromised mouse and chimeric animals when injected into a preimplantation mouse embryo (Beddington and Robertson 1989). Although ESCs retain the capacity to differentiate into all three germ layers of the embryo, they typically do not contribute to the extraembryonic membranes or placenta. Furthermore, ESCs exhibit significant differences in epigenetic regulation, gene expression, and cell signaling when compared to their in vivo counterparts within the ICM (Rugg-Gunn et al. 2010; Tang et al. 2010; Munoz-Descalzo et al. 2015).
Pluripotent cells can be isolated until the gastrulation stage of the mouse embryo; however, there is a temporal transition in their pluripotency state (Fig. 4). ESCs isolated from the ICM of the early blastocyst are considered to be in a state of “naive” pluripotency that retains the potential to differentiate into the three germ layers both in vitro and in vivo (Nichols and Smith 2009). These cells can be derived from the preimplantation EPI from ∼E3.75 to E4.75 (Boroviak et al. 2014) and maintained in culture through the addition of leukemia inhibitory factor (LIF) and small molecule inhibitors of MEK and GSK3 kinases (2i/LIF conditions) (Ying et al. 2008; Leitch et al. 2013).
Figure 4.
Derivation of embryonic stem cells (ESCs) from the mouse preimplantation embryo. Expanded potential stem cells (EPSCs) can be derived from the mouse embryo at the two- to eight-cell stages and give rise to both embryonic and extraembryonic lineages. Trophoblast stem cells (TSCs) and extraembryonic endoderm (XEN) stem cells can be isolated from the trophectoderm (TE) and primitive endoderm (PrE), respectively, and give rise to only extraembryonic lineages. ESCs are derived from the epiblast (EPI) of the preimplantation embryo and can give rise to all embryonic tissues. ESCs are thought to be in a naive state of pluripotency. Stem cells derived from the postimplantation EPI are referred to as EPI-derived stem cells (EpiScs). These cells are considered to be in a primed state of pluripotency that is fated, but not committed, toward specific embryonic lineages.
By contrast, ESCs isolated from the EPI after embryo implantation are considered to be in a “primed” state of pluripotency that is fated, but not committed, toward embryonic lineages and differentiates into all three germ layers in vitro but not when injected into the blastocyst (Huang et al. 2012). These primed ESCs are also referred to as EPI-derived stem cells (EpiSCs), and they can be derived from the mouse embryo from E5.5 to E7.5 (Brons et al. 2007; Tesar et al. 2007). The primed pluripotent state is favored by culturing ESCs in FGF/ACTIVIN (Tesar et al. 2007). Although EpiSCs were initially thought to correspond to postimplantation EPI at E5.0–E6.0, their transcriptional profile is most similar to gastrulating embryos at E7.25–E8.0 (Kojima et al. 2014). EpiSCs exhibit higher expression of genes related to adhesion, FGF/mitogen-activated protein kinase (MAPK), TGF-β, and WNT signaling and different methylation status of specific promoters compared to the postimplantation EPI, likely as a result of in vitro culture conditions (Kojima et al. 2014; Veillard et al. 2014).
Both ESCs and EpiSCs utilize the Oct4/Sox2/Nanog transcription factor circuit, but they can be distinguished by differences in their growth factor requirements, morphology, metabolism, epigenetic modifications, chromatin accessibility, transcriptomes, and activation status of the X chromosome (Tesar et al. 2007; Nichols and Smith 2009; Weinberger et al. 2016; Schlesinger and Meshorer 2019). Naive ESCs grow as dome-shaped colonies, express the “naive” transcription factors KLF4, KLF2, ESRRB, TFCP2L1, TBX3, and GBX2, utilize the Oct4 distal enhancer, have low levels of DNA methylation, and exhibit two active X chromosomes in female cells. EpiSCs grow as a monolayer, repress the expression of naive transcription factors, utilize the Oct4 proximal enhancer, have high levels of DNA methylation, and exhibit X-chromosome inactivation in female cells. Many of these parameters, however, are influenced by the culture conditions and do not necessarily reflect the in vivo pluripotent state (Rugg-Gunn et al. 2010; Tang et al. 2010; Veillard et al. 2014; Morgani et al. 2017; Schlesinger and Meshorer 2019).
ESCs can be differentiated into EpiSCs in vitro, but the heterogenous differentiation and high levels of cell death accompanying this process suggest it is not a direct conversion (Guo et al. 2009). In line with the temporal separation of the naive and primed pluripotent states within the mouse embryo, an intermediate “formative” state has been proposed to exist between ESCs and EpiSCs (Kalkan and Smith 2014; Smith 2017). This transitional period may be required for cells to lose expression of naive pluripotency markers and undergo epigenetic remodeling to gain competency to respond to lineage-specification cues. During the in vitro differentiation of mouse ESCs, a short-lived population of cells was identified that had gained competence for differentiation into primordial germ cells (Hayashi et al. 2011). These cells were referred to as EPI-like cells (EpiLCs) and were subsequently shown to display epigenetic profiles that are intermediate between ESCs and EpiSCs (Kurimoto et al. 2015; Shirane et al. 2016; Kalkan et al. 2017). Whether these EpiLCs can be reliably maintained as a pure population in culture remains to be determined.
Optimizing the culture conditions and inhibiting the TGF-β pathway enabled the efficient isolation of ESCs from individual cells of embryos at the two- to eight-cell stage; however, these cells were demonstrated to contribute only to the embryonic germ layers in vivo (Hassani et al. 2014). More recently, it was shown that by inhibiting MAPKs, SRC and Hippo signaling pathways, and poly-ADP-ribosylation regulators, ESCs with an expanded developmental potential (EPSCs) could be generated (Yang et al. 2017). EPSCs have similar transcriptomes and DNA methylation characteristics to four- to eight-cell-stage embryos and can give rise to both embryonic and extraembryonic lineages in vitro and in vivo. However, despite these molecular similarities, EPSCs still do not perfectly recapitulate the transcriptome and epigenome of the early embryo, again probably because of the influence of in vitro culture conditions.
Multipotent stem cell lines have also been established from mouse extraembryonic tissues. TSCs were derived from the TE (Tanaka et al. 1998), and XEN cells were derived from the PrE (Kunath et al. 2005). These stem cell lines are able to be maintained indefinitely in vitro and contribute to the expected extraembryonic lineages in vivo after transfer into mouse blastocysts. TSC and XEN cells are separated from ESCs and EpiSCs by robust DNA methylation and stable repression of regulators of embryonic development. It has been proposed that these epigenetic modifications may act as a barrier that bifurcates the embryonic and extraembryonic lineages (Senner et al. 2012).
In addition to isolating stem cells from the embryo, they can also be produced by SCNT, or by forcing the expression of specific transcription factors in somatic cells to produce induced pluripotent stem cells (iPSCs). In SCNT, the nucleus of a relatively differentiated cell is transferred into an enucleated oocyte, allowing it to be reprogrammed to totipotency by factors within the oocyte cytoplasm. The donor cell's chromatin undergoes global chromatin remodeling with the replacement of somatic histones by maternally stored histones and reprogramming of histone modifications and DNA methylation to resemble the zygotic epigenome (Nashun et al. 2011; Liu et al. 2016; Djekidel et al. 2018; Matoba et al. 2018). SCNT is able to produce entire cloned animals from a single adult cell, demonstrating that under the right conditions differentiated cells retain the potential for totipotency (Matoba and Zhang 2018). This technique, however, remains extremely inefficient, indicating there are barriers to reprogramming. Some such barriers have already been identified, including aberrant X-chromosome inactivation (Inoue et al. 2010), persistence of repressive histone methylation (Matoba et al. 2014), and loss of gene imprinting (Okae et al. 2014). Creating strategies to overcome these epigenetic barriers produces stem cells that more closely resemble the zygote and improves the development of cloned embryos (Matoba et al. 2011, 2014; Liu et al. 2016).
iPSCs were first created through in vitro reprogramming of fibroblast cells through the ectopic expression of the OCT4, SOX2, MYC, and KLF4 transcription factors (Takahashi and Yamanaka 2006). Subsequently, many transcription factors and small molecules have been shown to facilitate reprogramming or even replace the original four “Yamanaka factors,” suggesting that pluripotency can be attained through multiple distinct routes (Buganim et al. 2014; Theunissen and Jaenisch 2014). During their formation, iPSCs do not undergo a complete global demethylation process, and they may retain more of the epigenetic signature of their somatic parental cell than ESCs or nuclear-transfer embryonic stem cells (NT-ESCs) generated through SCNT (Kim et al. 2010; Lister et al. 2011; Ohi et al. 2011), although some of these differences may resolve over time in culture (Chin et al. 2010).
In addition to reprogramming cells into an ESC-like pluripotent state, forced expression of transcription factors can also direct induction of multipotent stem cells resembling XEN cells and TSCs (Benchetrit et al. 2015; Kubaczka et al. 2015; Parenti et al. 2016). Somewhat unexpectedly, forced expression of the Yamanaka factors in fibroblasts was shown to direct cellular reprogramming along two distinct pathways. Although some cells are induced to become pluripotent (iPSCs), others are reprogrammed into multipotent endodermal stem cells (iXEN) (Parenti et al. 2016). These iXEN cells have not been derived from iPSCs but represent an alternative route of reprogramming that may be favored by increased expression of GATA6. A different complement of transcription factors is required to reprogram cells into induced trophoblast stem cells (iTSCs). The minimal transcription factor set comprises GATA3, EOMES, and TFAP2C but the speed or efficiency of reprogramming can be increased through the inclusion of ETS2 (Kubaczka et al. 2015) or MYC, respectively, (Benchetrit et al. 2015). Most recently, it was demonstrated that expressing the transcription factor combination GATA3, EOMES, TFAP2C, MYC, and ESRRB can reprogram fibroblasts to produce iPSCs, iXENs, and iTSCs concomitantly; however, only the iPSCs and iTSCs were functionally demonstrated to contribute to the expected lineages in vivo (Benchetrit et al. 2019). Previous work has demonstrated that iXEN cells can contribute to the expected extraembryonic lineages in chimeric blastocysts (Parenti et al. 2016), and together with iPSCs and iTSCs, they provide the possibility of generating synthetic embryos created entirely from reprogrammed somatic cells.
STEM CELL–DERIVED ARTIFICIAL EMBRYOS
The ability of ESCs to recapitulate embryogenesis has been explored since shortly after they were first discovered (Fig. 5). Initially, mouse ESCs were aggregated into free-floating embryoid bodies (EBs) (Evans and Kaufman 1981; Martin 1981) that can establish self-organizing morphogen gradients (ten Berge et al. 2008). Although these EBs develop anteroposterior polarity and form a primitive streak-like structure, they form an internal cavity through an apoptotic process that does not occur during in vivo development (Bedzhov and Zernicka-Goetz 2014). Culturing individual ESCs in a 3D matrix in vitro instead enabled the reproduction of in vivo lumen formation though a sequence of cell polarization, rosette formation, and vesicular exocytosis (Bedzhov and Zernicka-Goetz 2014). Despite the remarkable ability of ESCs to self-organize into structures that mimic certain features of embryogenesis, the developmental potential of these structures is limited by the absence of extraembryonic tissues. Overlaying EBs with TSCs allows the creation of blastoid structures that resemble the blastocyst and can even initiate implantation when transferred to pseudopregnant mice (Rivron et al. 2018).
Figure 5.
Stem cells models of the mouse embryo. Schematic shows comparison between E5.5 postimplantation mouse embryo and stem cell–derived models. Suspension culture of embryonic stem cells (ESCs) and trophoblast stem cells (TSCs) generates a blastoid that is morphologically similar to a blastocyst. Combining ESCs and TSCs within a matrix allows formation of an ETS embryo with a cylindrical structure. The ETS embryo contains embryonic and extraembryonic compartments but lacks primitive endoderm (PrE). Combining ESCs, TSCs, and XEN cells within a matrix allows formation of an ETX-embryo that mostly closely replicates the tissue architecture of the in vivo embryo. (XEN) extraembryonic endoderm, (EPI) epiblast.
Early postimplantation embryonic development can be modeled by allowing ESCs and TSCs to combine within a 3D matrix to produce embryo-like structures called ETS embryos (Harrison et al. 2017). These structures mimic the formation of the proamniotic cavity and the asymmetric induction of mesoderm and primordial germ cell markers but do not reproduce key features of gastrulation. Incorporating XEN cells with the ESCs and TSCs circumvents the requirement to provide exogenous extracellular matrix components and generates embryo-like structures called ETX embryos. These embryo-like formations proceed through the early stages of gastrulation and can initiate implantation in utero (Sozen et al. 2018; Zhang et al. 2019). Improvements in culture conditions are likely to result in embryoid structures that reconstruct developmental processes even more faithfully in vitro. Finally, applying these approaches to combinations of iPSCs, iTSCs, and iXEN cells may enable the creation of artificial embryo-like structures created entirely from somatic cells. This would be of particular interest for research into early human development in which the use of embryonic cells is undesirable for ethical reasons.
HUMAN PLURIPOTENT STEM CELLS
Pluripotent stem cell lines have been isolated from human blastocysts (Thomson et al. 1998), induced from somatic cells, human-induced pluripotent stem cells (hiPSCs) (Takahashi et al. 2007), or created by SCNT using fetal (Tachibana et al. 2013) or adult somatic cells (Chung et al. 2014; Yamada et al. 2014). However, these lines differ significantly from mouse ESCs in terms of morphology, differentiation, and molecular characteristics (Rossant and Tam 2017). Furthermore, although they have been shown to differentiate into derivatives of the three germ layers in vitro and form teratomas when injected into immunocompromised mice (Thomson et al. 1998; Takahashi et al. 2007), it is not practical to test their contribution to human chimeric embryos, the gold standard for demonstrating functional pluripotency. Interspecies chimeras have been made by injecting human embryonic stem cells (hESCs) into mouse blastocysts but it is unclear whether the relatively low efficiency reflected a species barrier or limited in vivo pluripotency of the hESCs (James et al. 2006). The first hESC lines isolated are generally considered to be in a primed state of pluripotency, analogous to mouse EpiSCs (Brons et al. 2007; Tesar et al. 2007) and transcriptionally most similar to postimplantation EPI (Nakamura et al. 2016). hESCs and mouse EpiSCs share similar morphology, a dependency on ACTIVIN/NODAL signaling to maintain pluripotency (Xu et al. 2008), and X-chromosome inactivation (Silva et al. 2008) and are more similar to each other in Oct4 promoter occupancy than they are to mouse ESCs (Tesar et al. 2007). However, the interpretation that hESCs are equivalent to mouse EpiSCs has been questioned because hESCs exhibit a range of molecular characteristics that may reflect a continuous spectrum of pluripotent states (Davidson et al. 2015), and significant differences between mouse and human preimplantation development mean these comparisons may be of limited value. Nevertheless, many attempts have been made to capture hESCs in an earlier naive state of pluripotency by converting primed hESCs or hiPSCs, direct conversion of somatic cells to pluripotency through transcription factor expression, or deriving new hESC lines from human embryos at earlier developmental stages (for review, see Ware 2017). Comparison of the transcriptomes of these cells lines with single cells of the human preimplantation embryo revealed that the hESCs that most resemble the in vivo naive pluripotent state are those that were converted through transcription factor expression or chemical treatment (Takashima et al. 2014; Guo et al. 2017), derived from earlier embryonic stages (Guo et al. 2016), or maintained in a chemical cocktail containing five kinase inhibitors (Theunissen et al. 2014; Stirparo et al. 2018).
In addition to hESCs, human trophoblast stem cells (hTSCs) have also been isolated from human placenta and blastocysts, albeit with significantly different requirements for culture conditions than mouse TSCs (Okae et al. 2018). This may reflect the marked differences in the transcription factor network and signaling pathways underlying trophoblast formation in mouse and humans. To date, human XEN cell lines have not been successfully established; however, once this is achieved, the tools will be in place to substantially expand our understanding of human embryonic development by reconstructing the earliest stages using stem cells in vitro.
CONCLUDING REMARKS
The preimplantation mouse embryo has long been used as a model system to study the earliest developmental processes in mammals. For seven decades, these embryos have been observed, pulled apart, and reassembled with ever-evolving technologies. In recent years, the establishment of pluripotent stem cell lines has provided an entirely new window into the events driving differentiation of the first cell lineages. The possibility to create large amounts of starting materials, combined with advances in “omics” and single-cell approaches, affords the opportunity to analyze cellular, molecular, and epigenetic features at vastly greater depth. Perhaps most exciting is the capacity to assemble artificial embryos entirely from stem cells. Not only can these synthetic embryos replicate key features of development, they are scalable and highly accessible for manipulation and enable the study of emergent properties within the developing system. Further advances in in vitro culture conditions will enable more accurate recapitulation of developmental processes. Although currently only in their infancy, these stem cell models of embryonic development are likely to have a profound impact on our understanding of how the first cell lineages are established in the human embryo.
ACKNOWLEDGMENTS
This work was supported by grants from A*STAR, the European Molecular Biology Organization, and the Howard Hughes Medical Institute-Wellcome Trust. We apologize to those colleagues whose work we are unable to review here because of space limitations.
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Aguirre-Lavin T, Adenot P, Bonnet-Garnier A, Lehmann G, Fleurot R, Boulesteix C, Debey P, Beaujean N. 2012. 3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development. BMC Dev Biol 12: 30 10.1186/1471-213X-12-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE 5: e10531 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón VB, Marikawa Y. 2005. Unbiased contribution of the first two blastomeres to mouse blastocyst development. Mol Reprod Dev 72: 354–361. 10.1002/mrd.20353 [DOI] [PubMed] [Google Scholar]
- Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y. 2014. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 141: 2813–2824. 10.1242/dev.107276 [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. 1997. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181: 296–307. 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140. 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujean N. 2014. Histone post-translational modifications in preimplantation mouse embryos and their role in nuclear architecture. Mol Reprod Dev 81: 100–112. 10.1002/mrd.22268 [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. 1989. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105: 733–737. [DOI] [PubMed] [Google Scholar]
- Bedzhov I, Zernicka-Goetz M. 2014. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 156: 1032–1044. 10.1016/j.cell.2014.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchetrit H, Herman S, van Wietmarschen N, Wu T, Makedonski K, Maoz N, Yom Tov N, Stave D, Lasry R, Zayat V, et al. 2015. Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell 17: 543–556. 10.1016/j.stem.2015.08.006 [DOI] [PubMed] [Google Scholar]
- Benchetrit H, Jaber M, Zayat V, Sebban S, Pushett A, Makedonski K, Zakheim Z, Radwan A, Maoz N, Lasry R, et al. 2019. Direct induction of the three pre-implantation blastocyst cell types from fibroblasts. Cell Stem Cell 24: 983–994.e7. 10.1016/j.stem.2019.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. 2011. DNA demethylation dynamics. Cell 146: 866–872. 10.1016/j.cell.2011.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biase FH, Cao X, Zhong S. 2014. Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res 24: 1787–1796. 10.1101/gr.177725.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. 1986. CpG-rich islands and the function of DNA methylation. Nature 321: 209–213. 10.1038/321209a0 [DOI] [PubMed] [Google Scholar]
- Boiani M, Casser E, Fuellen G, Christians ES. 2019. Totipotency continuity from zygote to early blastomeres—a model under revision. Reproduction 10.1530/REP-19-0462 [DOI] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J. 2014. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 16: 516–528. 10.1038/ncb2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsos M, Torres-Padilla ME. 2016. Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev 30: 611–621. 10.1101/gad.273805.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EV, Meshorer E, Torres-Padilla ME. 2014. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev 28: 1042–1047. 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al. 2014. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell 15: 295–309. 10.1016/j.stem.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME. 2010. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct Genomics 9: 444–454. 10.1093/bfgp/elq027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME. 2014. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 15: 723–734. 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- Burton A, Muller J, Tu S, Padilla-Longoria P, Guccione E, Torres-Padilla ME. 2013. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep 5: 687–701. 10.1016/j.celrep.2013.09.044 [DOI] [PubMed] [Google Scholar]
- Casser E, Israel S, Witten A, Schulte K, Schlatt S, Nordhoff V, Boiani M. 2017. Totipotency segregates between the sister blastomeres of two-cell stage mouse embryos. Sci Rep 7: 8299 10.1038/s41598-017-08266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Shi J, Tao Y, Zernicka-Goetz M. 2018. Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat Commun 9: 1819 10.1038/s41467-018-04155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al. 2005. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25: 6031–6046. 10.1128/MCB.25.14.6031-6046.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE. 2010. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell 7: 263–269. 10.1016/j.stem.2010.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Eum JH, Lee JE, Shim SH, Sepilian V, Hong SW, Lee Y, Treff NR, Choi YH, Kimbrel EA, et al. 2014. Human somatic cell nuclear transfer using adult cells. Cell Stem Cell 14: 777–780. 10.1016/j.stem.2014.03.015 [DOI] [PubMed] [Google Scholar]
- Clayton AL, Hazzalin CA, Mahadevan LC. 2006. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 23: 289–296. 10.1016/j.molcel.2006.06.017 [DOI] [PubMed] [Google Scholar]
- Cockburn K, Rossant J. 2010. Making the blastocyst: lessons from the mouse. J Clin Invest 120: 995–1003. 10.1172/JCI41229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn K, Biechele S, Garner J, Rossant J. 2013. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr Biol 23: 1195–1201. 10.1016/j.cub.2013.05.044 [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. 2016. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537: 548–552. 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL, et al. 2013. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem 288: 34041–34051. 10.1074/jbc.M113.518019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson KC, Mason EA, Pera MF. 2015. The pluripotent state in mouse and human. Development 142: 3090–3099. 10.1242/dev.116061 [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49: 941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe C, Cauffman G, Verloes A, Sterckx J, Devroey P, Tournaye H, Liebaers I, Van de Velde H. 2013. Human trophectoderm cells are not yet committed. Hum Reprod 28: 740–749. 10.1093/humrep/des432 [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. 2007. Stochastic patterning in the mouse pre-implantation embryo. Development 134: 4219–4231. 10.1242/dev.003798 [DOI] [PubMed] [Google Scholar]
- Djekidel MN, Inoue A, Matoba S, Suzuki T, Zhang C, Lu F, Jiang L, Zhang Y. 2018. Reprogramming of chromatin accessibility in somatic cell nuclear transfer is DNA replication independent. Cell Rep 23: 1939–1947. 10.1016/j.celrep.2018.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547: 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Eid A, Rodriguez-Terrones D, Burton A, Torres-Padilla ME. 2016. SUV4-20 activity in the preimplantation mouse embryo controls timely replication. Genes Dev 30: 2513–2526. 10.1101/gad.288969.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130: 4235–4248. 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20: 332–338. 10.1038/nsmb.2495 [DOI] [PubMed] [Google Scholar]
- Frum T, Murphy TM, Ralston A. 2018. HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo. eLife 7: e42298 10.7554/eLife.42298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. 2003. Analysis of cell lineage in two- and four-cell mouse embryos. Development 130: 5113–5122. 10.1242/dev.00725 [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu X, Tang B, Li C, Kou Z, Li L, Liu W, Wu Y, Kou X, Li J, et al. 2017. Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep 21: 3957–3969. 10.1016/j.celrep.2017.11.111 [DOI] [PubMed] [Google Scholar]
- Goolam M, Scialdone A, Graham SJ, Macaulay IC, Jedrusik A, Hupalowska A, Voet T, Marioni JC, Zernicka-Goetz M. 2016. Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165: 61–74. 10.1016/j.cell.2016.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610. 10.1038/nature10443 [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063–1069. 10.1242/dev.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18: 675–685. 10.1016/j.devcel.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A, Nichols J. 2016. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Rep 6: 437–446. 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, von Meyenn F, Rostovskaya M, Clarke J, Dietmann S, Baker D, Sahakyan A, Myers S, Bertone P, Reik W, et al. 2017. Epigenetic resetting of human pluripotency. Development 144: 2748–2763. 10.1242/dev.146811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handyside AH, Johnson MH. 1978. Temporal and spatial pat terns of the synthesis of tissue-specific polypeptides in the preimplantation mouse embryo. J Embryol Exp Morphol 44: 191–199. [PubMed] [Google Scholar]
- Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka-Goetz M. 2017. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356: eaal1810 10.1126/science.aal1810 [DOI] [PubMed] [Google Scholar]
- Hassani SN, Pakzad M, Asgari B, Taei A, Baharvand H. 2014. Suppression of transforming growth factor β signaling promotes ground state pluripotency from single blastomeres. Hum Reprod 29: 1739–1748. 10.1093/humrep/deu134 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Hiiragi T, Solter D. 2004. First cleavage plane of the mouse egg is not predetermined but defined by the topology of the two apposing pronuclei. Nature 430: 360–364. 10.1038/nature02595 [DOI] [PubMed] [Google Scholar]
- Hillman N, Sherman MI, Graham C. 1972. The effect of spatial arrangement on cell determination during mouse development. J Embryol Exp Morphol 28: 263–278. [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al. 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol 23: 1181–1194. 10.1016/j.cub.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Tilly R. 1978. In vitro development of inner cell masses isolated immunosurgically from mouse blastocysts. I: Inner cell masses from 3.5-day p.c. blastocysts incubated for 24 h before immunosurgery. J Embryol Exp Morphol 45: 93–105. [PubMed] [Google Scholar]
- Holmes WR, Reyes de Mochel NS, Wang Q, Du H, Peng T, Chiang M, Cinquin O, Cho K, Nie Q. 2017. Gene expression noise enhances robust organization of the early mammalian blastocyst. PLoS Comput Biol 13: e1005320 10.1371/journal.pcbi.1005320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Osorno R, Tsakiridis A, Wilson V. 2012. In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep 2: 1571–1578. 10.1016/j.celrep.2012.10.022 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang H, Wang X, Dobbs KB, Yao J, Qin G, Whitworth K, Walters EM, Prather RS, Zhao J. 2015. Impairment of preimplantation porcine embryo development by histone demethylase KDM5B knockdown through disturbance of bivalent H3K4me3-H3K27me3 modifications. Biol Reprod 92: 72 10.1095/biolreprod.114.122762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Guo G, Yuan P, Ralston A, Sun L, Huss M, Mistri T, Pinello L, Ng HH, Yuan G, et al. 2017a. The role of Cdx2 as a lineage specific transcriptional repressor for pluripotent network during the first developmental cell lineage segregation. Sci Rep 7: 17156 10.1038/s41598-017-16009-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kim JK, Do DV, Lee C, Penfold CA, Zylicz JJ, Marioni JC, Hackett JA, Surani MA. 2017b. Stella modulates transcriptional and endogenous retrovirus programs during maternal-to-zygotic transition. eLife 6: e22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Paulsson J. 2011. Random partitioning of molecules at cell division. Proc Natl Acad Sci 108: 15004–15009. 10.1073/pnas.1013171108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalowska A, Jedrusik A, Zhu M, Bedford MT, Glover DM, Zernicka-Goetz M. 2018. CARM1 and paraspeckles regulate pre-implantation mouse embryo development. Cell 175: 1902–1916.e13. 10.1016/j.cell.2018.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, et al. 2010. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330: 496–499. 10.1126/science.1194174 [DOI] [PubMed] [Google Scholar]
- Jachowicz JW, Santenard A, Bender A, Muller J, Torres-Padilla ME. 2013. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev 27: 2427–2432. 10.1101/gad.224550.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla ME. 2017. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet 49: 1502–1510. 10.1038/ng.3945 [DOI] [PubMed] [Google Scholar]
- James D, Noggle SA, Swigut T, Brivanlou AH. 2006. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol 295: 90–102. 10.1016/j.ydbio.2006.03.026 [DOI] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA. 1981a. The foundation of two distinct cell lineages within the mouse morula. Cell 24: 71–80. 10.1016/0092-8674(81)90502-X [DOI] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA. 1981b. Induction of polarity in mouse 8-cell blastomeres: specificity, geometry, and stability. J Cell Biol 91: 303–308. 10.1083/jcb.91.1.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowska RZ, Jurkowski TP, Jeltsch A. 2011. Structure and function of mammalian DNA methyltransferases. Chembiochem 12: 206–222. 10.1002/cbic.201000195 [DOI] [PubMed] [Google Scholar]
- Kalkan T, Smith A. 2014. Mapping the route from naive pluripotency to lineage specification. Philos Trans R Soc Lond B Biol Sci 369: 20130540 10.1098/rstb.2013.0540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T, Olova N, Roode M, Mulas C, Lee HJ, Nett I, Marks H, Walker R, Stunnenberg HG, Lilley KS, et al. 2017. Tracking the embryonic stem cell transition from ground state pluripotency. Development 144: 1221–1234. 10.1242/dev.142711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Costa MW, Nefzger CM, Silva J, Fierro-González JC, Polo JM, Bell TD, Plachta N. 2013. Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy. Nat Commun 4: 1637 10.1038/ncomms2657 [DOI] [PubMed] [Google Scholar]
- Kelly SJ. 1977. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J Exp Zool 200: 365–376. 10.1002/jez.1402000307 [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467: 285–290. 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DA, Jones V, Hor A, de Alencastro G, Logan GJ, et al. 2014. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 14: 107–120. 10.1016/j.stem.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F, Hiiragi T. 2017. The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell 40: 235–247.e7. 10.1016/j.devcel.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinski P, Chickarmane V, Peterson C. 2011. Simulating the mammalian blastocyst—molecular and mechanical interactions pattern the embryo. PLoS Comput Biol 7: e1001128 10.1371/journal.pcbi.1001128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubaczka C, Senner CE, Cierlitza M, Araúzo-Bravo MJ, Kuckenberg P, Peitz M, Hemberger M, Schorle H. 2015. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell 17: 557–568. 10.1016/j.stem.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, Yamanaka Y, Heard E, Gardner RL, Avner P, Rossant J. 2005. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132: 1649–1661. 10.1242/dev.01715 [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M. 2006. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res 34: e42 10.1093/nar/gkl050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Hayashi K, Ohta H, Kiyonari H, Mitani T, Moritoki Y, Kohri K, Kimura H, Yamamoto T, et al. 2015. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell 16: 517–532. 10.1016/j.stem.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20: 311–316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M. 2013. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Commun 4: 2251 10.1038/ncomms3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73. 10.1038/nature09798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu X, Wang C, Gao Y, Gao R, Kou X, Zhao Y, Li J, Wu Y, Xiu W, et al. 2016. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov 2: 16010 10.1038/celldisc.2016.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440. 10.1038/ng1760 [DOI] [PubMed] [Google Scholar]
- Ma J, Svoboda P, Schultz RM, Stein P. 2001. Regulation of zygotic gene activation in the preimplantation mouse embryo: global activation and repression of gene expression. Biol Reprod 64: 1713–1721. 10.1095/biolreprod64.6.1713 [DOI] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Beaujean N, Brochard V, Audouard C, Zink D, Debey P. 2006a. Genome restructuring in mouse embryos during reprogramming and early development. Dev Biol 292: 317–332. 10.1016/j.ydbio.2006.01.009 [DOI] [PubMed] [Google Scholar]
- Martin C, Brochard V, Migné C, Zink D, Debey P, Beaujean N. 2006b. Architectural reorganization of the nuclei upon transfer into oocytes accompanies genome reprogramming. Mol Reprod Dev 73: 1102–1111. 10.1002/mrd.20506 [DOI] [PubMed] [Google Scholar]
- Matoba S, Zhang Y. 2018. Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell 23: 471–485. 10.1016/j.stem.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, et al. 2011. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci 108: 20621–20626. 10.1073/pnas.1112664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. 2014. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159: 884–895. 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, et al. 2018. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23: 343–354 e345. 10.1016/j.stem.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menchero S, Rollan I, Lopez-Izquierdo A, Andreu MJ, de Aja J S, Kang M, Adan J, Benedito R, Rayon T, Hadjantonakis AK, et al. 2019. Transitions in cell potency during early mouse development are driven by Notch. eLife 8: e42930 10.7554/eLife.42930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico V, Barbieri J, Zuccotti M, Joffe B, Cremer T, Redi CA, Solovei I, Garagna S. 2007. Epigenomic differentiation in mouse preimplantation nuclei of biparental, parthenote and cloned embryos. Chromosome Res 15: 341–360. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642. 10.1016/S0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- Morgani S, Nichols J, Hadjantonakis AK. 2017. The many faces of pluripotency: In vitro adaptations of a continuum of in vivo states. BMC Dev Biol 17: 7 10.1186/s12861-017-0150-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motosugi N, Bauer T, Polanski Z, Solter D, Hiiragi T. 2005. Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev 19: 1081–1092. 10.1101/gad.1304805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Descalzo S, Hadjantonakis AK, Arias AM. 2015. Wnt/ss-catenin signalling and the dynamics of fate decisions in early mouse embryos and embryonic stem (ES) cells. Semin Cell Dev Biol 47-48: 101–109. 10.1016/j.semcdb.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19: 1218–1227. 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M. 2016. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537: 57–62. 10.1038/nature19096 [DOI] [PubMed] [Google Scholar]
- Nashun B, Akiyama T, Suzuki MG, Aoki F. 2011. Dramatic replacement of histone variants during genome remodeling in nuclear-transferred embryos. Epigenetics 6: 1489–1497. 10.4161/epi.6.12.18206 [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. 2009. Naive and primed pluripotent states. Cell Stem Cell 4: 487–492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391. 10.1016/S0092-8674(00)81769-9 [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. 2009. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16: 398–410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123: 917–929. 10.1016/j.cell.2005.08.040 [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. 2011. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13: 541–549. 10.1038/ncb2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H, Matoba S, Nagashima T, Mizutani E, Inoue K, Ogonuki N, Chiba H, Funayama R, Tanaka S, Yaegashi N, et al. 2014. RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum Mol Genet 23: 992–1001. 10.1093/hmg/ddt495 [DOI] [PubMed] [Google Scholar]
- Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T. 2018. Derivation of human trophoblast stem cells. Cell Stem Cell 22: 50–63.e56. 10.1016/j.stem.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- O'Neill LP, VerMilyea MD, Turner BM. 2006. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet 38: 835–841. 10.1038/ng1820 [DOI] [PubMed] [Google Scholar]
- Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. 2016. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics 11: 85–94. 10.1080/15592294.2015.1136774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol 166: 259–267. 10.1006/dbio.1994.1312 [DOI] [PubMed] [Google Scholar]
- Parenti A, Halbisen MA, Wang K, Latham K, Ralston A. 2016. OSKM induce extraembryonic endoderm stem cells in parallel to induced pluripotent stem cells. Stem Cell Rep 6: 447–455. 10.1016/j.stemcr.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606. 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. 2005. Four-cell stage mouse blastomeres have different developmental properties. Development 132: 479–490. 10.1242/dev.01602 [DOI] [PubMed] [Google Scholar]
- Piras V, Tomita M, Selvarajoo K. 2014. Transcriptome-wide variability in single embryonic development cells. Sci Rep 4: 7137 10.1038/srep07137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. 2011. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol 13: 117–123. 10.1038/ncb2154 [DOI] [PubMed] [Google Scholar]
- Plusa B, Frankenberg S, Chalmers A, Hadjantonakis AK, Moore CA, Papalopulu N, Papaioannou VE, Glover DM, Zernicka-Goetz M. 2005. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci 118: 505–515. 10.1242/jcs.01666 [DOI] [PubMed] [Google Scholar]
- Posfai E, Petropoulos S, de Barros FRO, Schell JP, Jurisica I, Sandberg R, Lanner F, Rossant J. 2017. Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. eLife 6: e22906 10.7554/eLife.22906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Rossant J. 2008. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol 313: 614–629. 10.1016/j.ydbio.2007.10.054 [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. 2010. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137: 395–403. 10.1242/dev.038828 [DOI] [PubMed] [Google Scholar]
- Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Canon S, Sasaki H, Hadjantonakis AK, de la Pompa JL, et al. 2014. Notch and Hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 30: 410–422. 10.1016/j.devcel.2014.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset JC, Korving J, Vivie J, Truckenmuller RK, van Oudenaarden A, van Blitterswijk CA, Geijsen N. 2018. Blastocyst-like structures generated solely from stem cells. Nature 557: 106–111. 10.1038/s41586-018-0051-0 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Torres-Padilla ME. 2018. Nimble and ready to mingle: transposon outbursts of early development. Trends Genet 34: 806–820. 10.1016/j.tig.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Rossant J, Lis WT. 1979. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev Biol 70: 255–261. 10.1016/0012-1606(79)90022-8 [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PPL. 2017. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell 20: 18–28. 10.1016/j.stem.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. 2010. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci 107: 10783–10790. 10.1073/pnas.0914507107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. 2000. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404: 95–99. 10.1038/35003601 [DOI] [PubMed] [Google Scholar]
- Saini D, Yamanaka Y. 2018. Cell polarity-dependent regulation of cell allocation and the first lineage specification in the preimplantation mouse embryo. Curr Topics Dev Biol 128: 11–35. 10.1016/bs.ctdb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Samarage CR, White MD, Alvarez YD, Fierro-Gonzalez JC, Henon Y, Jesudason EC, Bissiere S, Fouras A, Plachta N. 2015. Cortical tension allocates the first inner cells of the mammalian embryo. Dev Cell 34: 435–447. 10.1016/j.devcel.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241: 172–182. 10.1006/dbio.2001.0501 [DOI] [PubMed] [Google Scholar]
- Sarmento OF, Digilio LC, Wang Y, Perlin J, Herr JC, Allis CD, Coonrod SA. 2004. Dynamic alterations of specific histone modifications during early murine development. J Cell Sci 117: 4449–4459. 10.1242/jcs.01328 [DOI] [PubMed] [Google Scholar]
- Sasaki H. 2017. Roles and regulations of Hippo signaling during preimplantation mouse development. Dev Growth Differ 59: 12–20. 10.1111/dgd.12335 [DOI] [PubMed] [Google Scholar]
- Schlesinger S, Meshorer E. 2019. Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev Cell 48: 135–150. 10.1016/j.devcel.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. 2007. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21: 3027–3043. 10.1101/gad.1604607 [DOI] [PubMed] [Google Scholar]
- Senner CE, Krueger F, Oxley D, Andrews S, Hemberger M. 2012. DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells 30: 2732–2745. 10.1002/stem.1249 [DOI] [PubMed] [Google Scholar]
- Shi J, Chen Q, Li X, Zheng X, Zhang Y, Qiao J, Tang F, Tao Y, Zhou Q, Duan E. 2015. Dynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seq. Development 142: 3468–3477. 10.1242/dev.123950 [DOI] [PubMed] [Google Scholar]
- Shi X, Yin Z, Ling B, Wang L, Liu C, Ruan X, Zhang W, Chen L. 2017. Rho differentially regulates the Hippo pathway by modulating the interaction between Amot and Nf2 in the blastocyst. Development 144: 3957–3967. 10.1242/dev.157917 [DOI] [PubMed] [Google Scholar]
- Shirane K, Kurimoto K, Yabuta Y, Yamaji M, Satoh J, Ito S, Watanabe A, Hayashi K, Saitou M, Sasaki H. 2016. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev Cell 39: 87–103. 10.1016/j.devcel.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. 2008. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci 105: 4820–4825. 10.1073/pnas.0712136105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. 2017. Formative pluripotency: the executive phase in a developmental continuum. Development 144: 365–373. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B, Amadei G, Cox A, Wang R, Na E, Czukiewska S, Chappell L, Voet T, Michel G, Jing N, et al. 2018. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat Cell Biol 20: 979–989. 10.1038/s41556-018-0147-7 [DOI] [PubMed] [Google Scholar]
- Spindle AI. 1978. Trophoblast regeneration by inner cell masses isolated from cultured mouse embryos. J Exp Zool 203: 483–489. 10.1002/jez.1402030315 [DOI] [PubMed] [Google Scholar]
- Stein P, Schultz RM. 2000. Initiation of a chromatin-based transcriptionally repressive state in the preimplantation mouse embryo: lack of a primary role for expression of somatic histone H1. Mol Reprod Dev 55: 241–248. [DOI] [PubMed] [Google Scholar]
- Stephenson RO, Yamanaka Y, Rossant J. 2010. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development 137: 3383–3391. 10.1242/dev.050195 [DOI] [PubMed] [Google Scholar]
- Stirparo GG, Boroviak T, Guo G, Nichols J, Smith A, Bertone P. 2018. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development 145: dev158501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132: 2093–2102. 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. 2008. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol 322: 133–144. 10.1016/j.ydbio.2008.07.019 [DOI] [PubMed] [Google Scholar]
- Tabansky I, Lenarcic A, Draft RW, Loulier K, Keskin DB, Rosains J, Rivera-Feliciano J, Lichtman JW, Livet J, Stern JN, et al. 2013. Developmental bias in cleavage-stage mouse blastomeres. Curr Biol 23: 21–31. 10.1016/j.cub.2012.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791. 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. 2013. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153: 1228–1238. 10.1016/j.cell.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158: 1254–1269. 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282: 2072–2075. 10.1126/science.282.5396.2072 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. 2010. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6: 468–478. 10.1016/j.stem.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski AK. 1959. Experiments on the development of isolated blastomers of mouse eggs. Nature 184: 1286–1287. 10.1038/1841286a0 [DOI] [PubMed] [Google Scholar]
- Tarkowski AK. 1961. Mouse chimaeras developed from fused eggs. Nature 190: 857–860. 10.1038/190857a0 [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Wróblewska J. 1967. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. Development 18: 155–180. [PubMed] [Google Scholar]
- Tarkowski AK, Suwinska A, Czolowska R, Ozdzenski W. 2010. Individual blastomeres of 16- and 32-cell mouse embryos are able to develop into foetuses and mice. Dev Biol 348: 190–198. 10.1016/j.ydbio.2010.09.022 [DOI] [PubMed] [Google Scholar]
- ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. 2008. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3: 508–518. 10.1016/j.stem.2008.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Jaenisch R. 2014. Molecular control of induced pluripotency. Cell Stem Cell 14: 720–734. 10.1016/j.stem.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15: 524–526. 10.1016/j.stem.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EM, Legouy E, Christians E, Renard JP. 1995. Progressive maturation of chromatin structure regulates HSP70.1 gene expression in the preimplantation mouse embryo. Development 121: 3425–3437. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445: 214–218. 10.1038/nature05458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsompana M, Buck MJ. 2014. Chromatin accessibility: a window into the genome. Epigenetics Chromatin 7: 33 10.1186/1756-8935-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillard AC, Marks H, Bernardo AS, Jouneau L, Laloe D, Boulanger L, Kaan A, Brochard V, Tosolini M, Pedersen R, et al. 2014. Stable methylation at promoters distinguishes epiblast stem cells from embryonic stem cells and the in vivo epiblasts. Stem Cells Dev 23: 2014–2029. 10.1089/scd.2013.0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li X, Wang L, Li J, Zhao Y, Bou G, Li Y, Jiao G, Shen X, Wei R, et al. 2016. A novel long intergenic noncoding RNA indispensable for the cleavage of mouse two-cell embryos. EMBO Rep 17: 1452–1470. 10.15252/embr.201642051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang L, Feng G, Wang Y, Li Y, Li X, Liu C, Jiao G, Huang C, Shi J, et al. 2018. Asymmetric expression of LincGET biases cell fate in two-cell mouse embryos. Cell 175: 1887–1901 e1818. 10.1016/j.cell.2018.11.039 [DOI] [PubMed] [Google Scholar]
- Ware CB. 2017. Concise review: lessons from naive human pluripotent cells. Stem Cells 35: 35–41. 10.1002/stem.2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Biggins JS, Tannan NB, Srinivas S. 2014. Limited predictive value of blastomere angle of division in trophectoderm and inner cell mass specification. Development 141: 2279–2288. 10.1242/dev.103267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Miyasaka KY, Kubo A, Kida YS, Nakagawa O, Hirate Y, Sasaki H, Ogura T. 2017. Notch and Hippo signaling converge on Strawberry Notch 1 (Sbno1) to synergistically activate Cdx2 during specification of the trophectoderm. Sci Rep 7: 46135 10.1038/srep46135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. 2016. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 17: 155–169. 10.1038/nrm.2015.28 [DOI] [PubMed] [Google Scholar]
- White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V, Plachta N. 2016. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165: 75–87. 10.1016/j.cell.2016.02.032 [DOI] [PubMed] [Google Scholar]
- White MD, Zenker J, Bissiere S, Plachta N. 2018. Instructions for assembling the early mammalian embryo. Dev Cell 45: 667–679. 10.1016/j.devcel.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Whitten WK, Biggers JD. 1968. Complete development in vitro of the pre-implantation stages of the mouse in a simple chemically defined medium. J Reprod Fertil 17: 399–401. 10.1530/jrf.0.0170399 [DOI] [PubMed] [Google Scholar]
- Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, Ralston A. 2014. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 10: e1004618 10.1371/journal.pgen.1004618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, Huang B, Wang X, Li T, Shi S, et al. 2018. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557: 256–260. 10.1038/s41586-018-0080-8 [DOI] [PubMed] [Google Scholar]
- Xu Q, Xie W. 2018. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol 28: 237–253. 10.1016/j.tcb.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. 2008. NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3: 196–206. 10.1016/j.stem.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, et al. 2014. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature 510: 533–536. 10.1038/nature13287 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. 2010. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137: 715–724. 10.1242/dev.043471 [DOI] [PubMed] [Google Scholar]
- Yang J, Ryan DJ, Wang W, Tsang JC, Lan G, Masaki H, Gao X, Antunes L, Yu Y, Zhu Z, et al. 2017. Establishment of mouse expanded potential stem cells. Nature 550: 393–397. 10.1038/nature24052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker J, White MD, Gasnier M, Alvarez YD, Lim HYG, Bissiere S, Biro M, Plachta N. 2018. Expanding actin rings zipper the mouse embryo for blastocyst formation. Cell 173: 776–791 e717. 10.1016/j.cell.2018.02.035 [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW. 2009. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 10: 467–477. 10.1038/nrg2564 [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen T, Chen N, Gao D, Shi B, Kong S, West RC, Yuan Y, Zhi M, Wei Q, et al. 2019. Implantation initiation of self-assembled embryo-like structures generated using three types of mouse blastocyst-derived stem cells. Nat Commun 10: 496 10.1038/s41467-019-08378-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZW, White MD, Alvarez YD, Zenker J, Bissiere S, Plachta N. 2017. Quantifying transcription factor-DNA binding in single cells in vivo with photoactivatable fluorescence correlation spectroscopy. Nat Protoc 12: 1458–1471. 10.1038/nprot.2017.051 [DOI] [PubMed] [Google Scholar]
- Zhou LQ, Dean J. 2015. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol 25: 82–91. 10.1016/j.tcb.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Guo H, Ren Y, Hou Y, Dong J, Li R, Lian Y, Fan X, Hu B, Gao Y, et al. 2018. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat Genet 50: 12–19. 10.1038/s41588-017-0007-6 [DOI] [PubMed] [Google Scholar]