Abstract
The BCL-2 family of proteins control a key checkpoint in apoptosis, that of mitochondrial outer membrane permeabilization or, simply, mitochondrial poration. The family consists of three subgroups: BH3-only initiators that respond to apoptotic stimuli; antiapoptotic guardians that protect against cell death; and the membrane permeabilizing effectors BAX, BAK, and BOK. On activation, effector proteins are converted from inert monomers into membrane permeabilizing oligomers. For many years, this process has been poorly understood at the molecular level, but a number of recent advances have provided important insights. We review the regulation of these effectors, their activation, subsequent conformational changes, and the ensuing oligomerization events that enable mitochondrial poration, which initiates apoptosis through release of key signaling factors such as cytochrome c. We highlight the mysteries that remain in understanding these important proteins in an endeavor to provide a comprehensive picture of where the field currently sits and where it is moving toward.
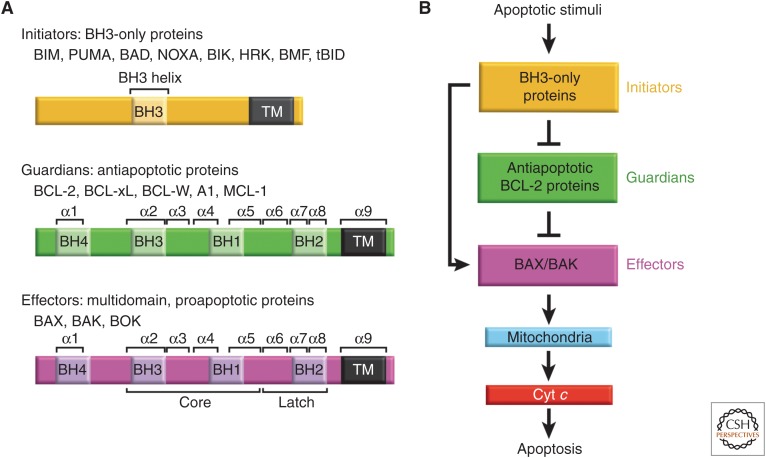
AWAKENING THE BCL-2 EFFECTORS IN APOPTOSIS
Commitment to mitochondrial apoptosis involves activation of effectors, which are uniquely able to directly mediate mitochondrial poration (Fig. 1; Czabotar et al. 2014; Moldoveanu et al. 2014). In most cells, the canonical effectors BAX and BAK are typically expressed in stable dormant conformations in the absence of overt cellular stress (e.g., chemotherapeutic insults, nutrient deprivation, growth factor withdrawal). BAK and BAX have distinct mechanisms of regulation, reflected primarily in their respective steady-state localization at the mitochondrial outer membrane (MOM) and in the cytosol in nonapoptotic live cells. Cellular stress triggers apoptotic stimuli to awaken or activate the effectors, which relocalize and change shape to execute mitochondrial poration thereby initiating apoptosis (Fig. 1). The underlying mechanism of mitochondrial poration by effectors is incompletely defined and debatable as discussed below.
Figure 1.
The BCL-2 family and the intrinsic pathway to apoptosis. (A) Family members of the BCL-2 protein family. The family is made up of three subgroups of proteins related to each other by regions of sequence homology, the so-called Bcl-2 homology (BH) domains. Regions of secondary structure and domains discussed in the text are labeled. (B) BH3-only proteins, which generally only possess the BH3 domain, are up-regulated on apoptotic stimuli to initiate signaling of the pathway. BH3-only proteins interact with both the effectors BAX and BAK, and the antiapoptotic guardians. Guardians can protect against apoptosis by sequestering both the BH3-only proteins, thus inhibiting effector activation, and by neutralizing activated effector proteins directly. If freed, activated effectors oligomerize at the mitochondrial outer membrane leading to permeabilization of this barrier. This enables the release of apoptogenic factors into the cytosol, primarily cytochrome c (cyt c), leading to caspase activation and ensuing apoptosis. Emerging evidence indicates that BOK is a third member of the effector subgroup with alternative mechanisms of regulation (discussed later).
Steady-State BAX on a Leash in the Cytosol
Association of BCL-2 family proteins with mitochondria is largely governed by their carboxy-terminal tails. Tail deletion abolishes mitochondrial localization and severely impacts apoptotic function (Ferrer et al. 2012; Llambi et al. 2016). Although not formally shown, the tails most likely form transmembrane (TM) anchoring helices or may involve association via hydrophobic and hydrophilic interactions with the outer leaflet of the MOM without traversing the bilayer (e.g., as proposed for the BIM carboxy-terminal tail) (Liu et al. 2019). BAX is the only folded BCL-2 family protein that preferentially accommodates its carboxy-terminal TM tail within the canonical hydrophobic groove, protecting the hydrophobic character of this helix and thus enabling the protein to remain soluble in the cytoplasm (Table 1; Fig. 2A; Suzuki et al. 2000; Garner et al. 2016; Robin et al. 2018).
Table 1.
Summary of effector structures
| Protein complex | PDB | References |
|---|---|---|
| Full-length hBAX | 1F16 | Suzuki et al. 2000 |
| ΔNhBAKΔTM | 2IMS | Moldoveanu et al. 2006 |
| BIM SAHB: Full-length hBAX | 2K7W | Gavathiotis et al. 2008 |
| vMIA: Full-length hBAX | 2LR1 | Ma et al. 2012 |
| BID BH3:hBAX | 4BD2 | Czabotar et al. 2013 |
| BAX BH3:hBAX | 4BD6 | Czabotar et al. 2013 |
| Octylmaltoside-induced hBAX dimer (Apo) | 4BD7 | Czabotar et al. 2013 |
| BIM BH3-induced hBAX dimer (Apo) | 4BD8 | Czabotar et al. 2013 |
| hBAX BH3-in-groove dimer (GFP) | 4BDU | Czabotar et al. 2013 |
| BID SAHB:ΔNhBAKΔTM | 2M5B | Moldoveanu et al. 2013 |
| hBAK core/latch dimer | 4U2U | Brouwer et al. 2014 |
| GFP-hBAK(α2–α5) (core domain dimer) | 4U2V | Brouwer et al. 2014 |
| BIM BH3:hBAXΔTM | 4ZIE | Robin et al. 2015 |
| BIM BH3mini:hBAXΔTM | 4ZIF | Robin et al. 2015 |
| BID BH3mini:hBAXΔTM | 4ZIG | Robin et al. 2015 |
| BIM BH3mini:hBAXΔTM | 4ZIH | Robin et al. 2015 |
| BID BH3:hBAXI66AΔTM | 4ZII | Robin et al. 2015 |
| Full-length hBAX P168G | 4S0O | Garner et al. 2016 |
| Full-length hBAX G67R | 4S0P | Garner et al. 2016 |
| BIM-RT:hBAKΔTM core/latch dimer | 5VWV | Brouwer et al. 2017 |
| BIM-RT:hBAKΔTM core/latch dimer | 5VWW | Brouwer et al. 2017 |
| BIM-h3Pc:hBAK monomer | 5VWZ | Brouwer et al. 2017 |
| BIM-h3Pc-RT:hBAK core/latch dimer | 5VWY | Brouwer et al. 2017 |
| chBOK | 5WDD | Ke et al. 2018 |
| hBOK | 6CKV | Zheng et al. 2018 |
| Full-length hBAX P168G | 5W60 | Robin et al. 2018 |
| Full-length hBAX P168G | 5W61 | Robin et al. 2018 |
| Full-length mBAX | 5W62 | Robin et al. 2018 |
| 3C10Ab: full-length hBAX P168G | 5W5X | Robin et al. 2018 |
Figure 2.
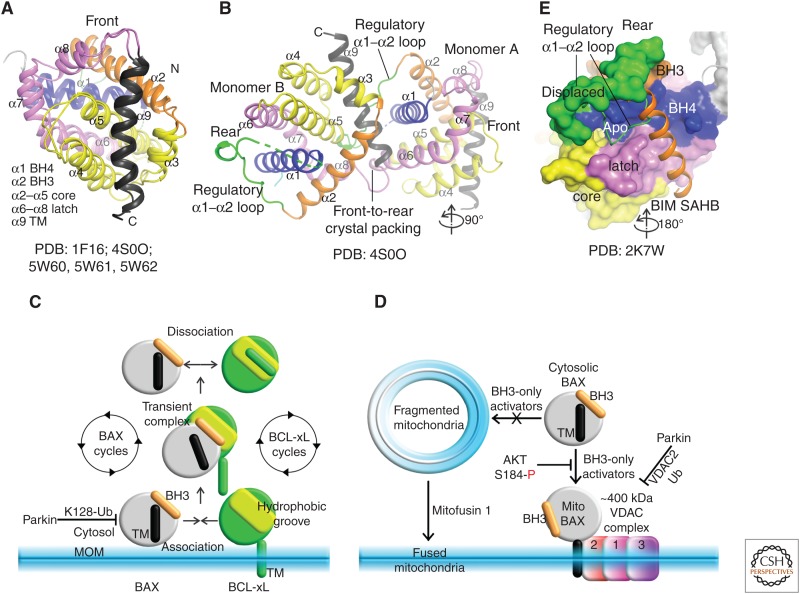
Regulated recruitment of cytosolic BAX to mitochondria. (A) The nuclear magnetic resonance and crystal structures of wild-type (WT) and P168G mutant full-length hBAX (FL BAX) revealed the dormant conformation of cytosolic BAX, with the carboxy-terminal transmembrane (TM) helix α9 bound to and occluding the BAX canonical hydrophobic groove (front view). The same colors have been used in structural figures throughout for different secondary structure elements and regions. PDB identifiers are included (see Table 1 for a summary of effector PDBs). (B) The crystal packing dimer of BAX P168G mutant. The proposed regulatory α1–α2 loop is highly dynamic and found at the rear site. (C) Schematic of retrotranslocation and possible inhibition of mitochondrial BAX by parkin ubiquitylation (Ub). (D) Mitochondrial recruitment is also thought to be regulated by mitochondrial dynamics and by VDAC complex via VDAC2 interactions. Parkin ubiquitylation of VDAC2 blocks BAX recruitment. BAX phosphorylation by AKT at position S184 blocked its mitochondrial association. (E) Chemically stapled BIM, BH3-stabilized α helix of BCL-2 protein (SAHB) has been proposed to displace the regulatory α1–α2 loop at the rear to induce allosteric changes that ultimately destabilized the TM interaction at the front activation groove to drive targeting of BAX to the mitochondria via TM (see D).
It has been reported that cytosolic BAX exists in an equilibrium between monomer and dimer with an estimated KD of ∼30 µm determined by estimating dissociation of BAX dimers into monomers over time from size-exclusion chromatography (Garner et al. 2016). Based on probing of BAX in solution by hydrogen deuterium exchange mass spectrometry and crystal structures, it has been proposed that BAX dimers form by burying the putative regulatory interfaces (canonical groove and rear site) thought to be involved in activation (Fig. 2B; Garner et al. 2016, vide infra). This configuration was interpreted as an autoinhibitory mechanism whereby the dimer is resistant to activation by BH3-only proteins. BAX mutagenesis to break down the autoinhibitory dimer augmented membrane poration and apoptosis (Garner et al. 2016). In contrast, a recent study observed BAX monomers, not dimers in cells, and interpreted the BAX configuration seen in the crystal structure as a crystal-packing artifact (Dengler et al. 2019). Another study also found Bax to be monomeric in cells when solubilized in 1% CHAPS detergent (Dai et al. 2015). Some estimates of BAX cellular levels are lower than the reported dimerization KD (e.g., in overexpression systems at ∼3 µm) (Dussmann et al. 2010), and in several tumor cell lines at ∼20,000–70,000 BAX molecules per cell (∼17–60 nm for average cell volume of ∼2000 µm3) (Dai et al. 2018). How do we reconcile these conflicting reports? One possible explanation is that cytosolic BAX dimers could be stabilized by additional mechanisms, perhaps through posttranslational modifications. Further studies may shed light on this phenomenon.
Dynamics of BAX Association with the Mitochondria
BAX exists in a dynamic equilibrium shuttling between the mitochondria and cytosol, a process termed retrotranslocation (Edlich et al. 2011). In nonapoptotic cells, this equilibrium is weighted such that BAX is primarily cytosolic. Retrotranslocation is thought to occur through interactions between BAX and antiapoptotic BCL-2 proteins such as BCL-xL, BCL-2, and MCL-1, with the TM and hydrophobic grooves of the latter interacting with the BH3 region and TM of the former (Edlich et al. 2011; Todt et al. 2013). This process has not been elucidated structurally, but it appears that antiapoptotic BCL-2 proteins may act as chaperones of BAX, constantly sampling BAX conformations that promote mitochondrial association such as exposure of amino-terminal activation epitopes (6A7 is the antibody that recognizes one such epitope), carboxy-terminal TM association with MOM, and BH3 exposure (Edlich et al. 2011; Todt et al. 2013). Reversal of mitochondrial association by retrotranslocation is thought to occur through transient formation of BAX:antiapoptotic BCL-2 protein complexes that shuttle back into the cytosol before dissociating (Fig. 2C). Remarkably, BAX retrotranslocation appears to occur even if BAX is membrane integral, presumably with the carboxy-terminal TM tail traversing the bilayer (Lauterwasser et al. 2016). It has also been proposed that BAK undergoes a similar shuttling mechanism, although in this case with the equilibrium pushed heavily toward membrane association (Todt et al. 2015). Mitochondrial association also seems to be dependent on mitochondrial morphology, as hyperfragmented mitochondria fail to support BAX association and mitochondrial poration (Renault et al. 2015). This failure has been attributed to the inability of BAX TM tails to interact with fragmented membranes and GTPase mitofusin 1 has been reported to counteract this by mediating mitochondrial fusion (Fig. 2D; Renault et al. 2015).
Several studies have also revealed a role for VDAC2 in the recruitment of BAX to mitochondria (Ma et al. 2014; Lauterwasser et al. 2016; Cakir et al. 2017; Chin et al. 2018; Bernardini et al. 2019). The size of digitonin-solubilized complexes of BAX and VDACs on native PAGE are estimated to be >400 kDa. In biochemical and genetic studies, VDAC2 deletion inhibited the mitochondrial localization and apoptotic function of BAX. Surprisingly, although BAK is also found in a similar high-molecular weight complex that includes VDAC2 (Lazarou et al. 2010; Chin et al. 2018), BAK-mediated apoptosis is not significantly impaired by VDAC2 deletion, but is actually potentiated (Cheng et al. 2003). Structures for these effector:VDAC complexes have not yet been solved, but once these details are understood it is likely that they will provide new strategies for modulating mitochondrial poration and apoptosis.
The Rear of BAX and Mitochondrial Association
The nuclear magnetic resonance (NMR) structure of human full-length BAX predicted that BH3 activators would be unable to displace the TM (helix α9) from the groove in the absence of an energy-driven triggering process to disengage the α9 (Suzuki et al. 2000). In the absence of additional energy, we now know that addition of BH3 activator peptides to full-length BAX induces large aggregates in solution over time (Sung et al. 2015; Garner et al. 2016). These aggregates are readily able to porate membranes (Sung et al. 2015; Garner et al. 2016). We do not know the structure of BAX in this aggregate, but it likely involves a dynamic heterogeneous interaction web mediated by intermolecular associations of displaced hydrophobic TM regions. Similar aggregates are not seen with BAX-ΔTM and BH3 activator peptides, which form 1:1 complexes amenable to structural characterization (Table 1, vide infra). These observations indirectly suggest that BH3 activators are able to displace the TM helix from the groove in solution.
A major point of contention in BAX activation over the past decade has centered on revealing how the TM is displaced from the groove. Allostery has been implicated in this process through ensembles of Bax conformers within the cytosol (Robin et al. 2018; Dengler et al. 2019). To discover allosteric mechanisms for TM displacement from the groove by BH3 domains, chemically stapled BIM BH3 has been deployed in structure–function studies (Gavathiotis et al. 2008, 2010), and chemically stapled BH3 peptides from BID and PUMA were designed to investigate BAX and BAK activation (Edwards et al. 2013; Leshchiner et al. 2013). These low-resolution studies by NMR and photoaffinity labeling mass spectrometry, suggested engagement by BH3 activators to a rear allosteric activation site on BAX (but not BAK) (Fig. 2E). Although the binding of unstapled BH3 activators to BAX is undetectable, stabilization through chemical stapling increased the affinity for the rear allosteric site (KD in µm) (Walensky et al. 2006). Unfortunately, the low-resolution model of the rear activation interface has been refractory to surface probing by site-directed mutagenesis, as none of the substitutions tested have been able to block BAX activation in multiple laboratories (Okamoto et al. 2013; Peng et al. 2013; Garner et al. 2016; Dengler et al. 2019). For instance, mutagenesis to block auto-inhibitory BAX dimers, which should impact BAX activation at the rear site directly, revealed a counterintuitive enhancement in BAX apoptotic activity compared with wild-type (WT) (Garner et al. 2016). It is possible that the models for BH3 activation at the rear site are incomplete, that this interface does not matter significantly for BAX activation, or that the staple itself is facilitating some of these events. From our experience, stapled peptides are most useful when they mimic all of the functional features observed with the unstapled counterparts while still offering enhanced helicity and affinity for the target (Moldoveanu et al. 2013). TM-deleted BAX and full-length BAK show significant interactions with activator BH3 peptides at the canonical groove, which is their main binding site common to both effectors (vide infra) (Edwards et al. 2013; Leshchiner et al. 2013). Future mechanistic high-resolution studies exploring complexes of BAX and BAK with full-length BH3-only proteins, which may show additional regulatory interaction interfaces as recently proposed for full-length BIM and BAX (Chi et al. 2019) and tBID and BAK (Dengler et al. 2019), may resolve these issues.
Getting in (and out of) the Groove—“Hit-and-Run” Effector Unleashing
A better characterized mechanism of BAK and BAX activation involves canonical engagement of their groove by BH3 activators, such as BID, BIM, and PUMA. Because the affinity of BH3 activator peptides for BAK and BAX is relatively low (KD ∼ µm), structural analysis by NMR and X-ray crystallography has lagged behind that of the much tighter complexes occurring between BH3 peptides and antiapoptotic BCL-2 proteins (KD ∼ nm) (Suzuki et al. 2000; Moldoveanu et al. 2006, 2013; Czabotar et al. 2013). The effector activation “hit-and-run” model is based on the weak affinities of these interactions, whereby direct activators bind transiently and are thus not found in stable complexes with effectors, especially in cell-based pull-down or gel-filtration chromatography assays in which protein levels are below the interaction KD (Wei et al. 2000). In retrospect, it is not surprising that effectors were not found bound to direct activators in these early studies.
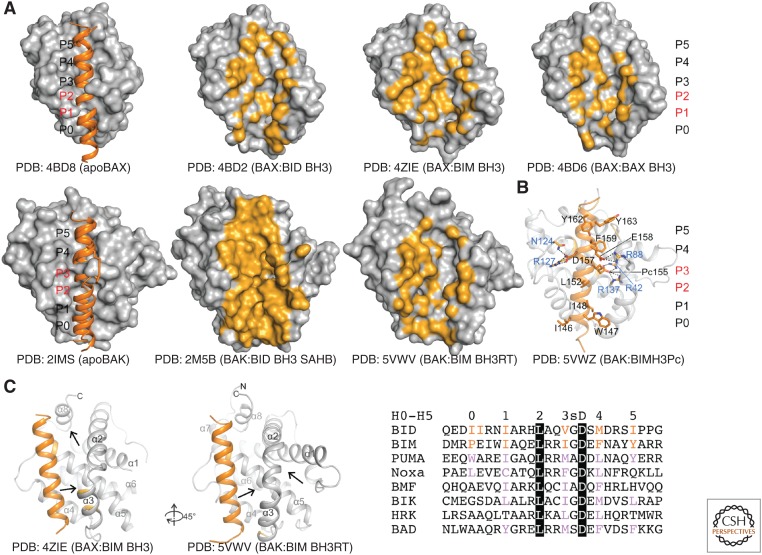
To this end, three breakthroughs toward stabilization of effector:activator complexes have been made to achieve their high-resolution structures: (1) domain-swapped dimers of BAX-ΔTM and BAK-ΔTM, which are more stable than monomers and crystallize more readily in complex with activators (Czabotar et al. 2013; Robin et al. 2015; Brouwer et al. 2017); (2) chemically stabilized activator peptides with enhanced helicity and affinity for effectors (Moldoveanu et al. 2013); and (3) mutant activator peptides with enhanced solubility and affinity (Robin et al. 2015; Brouwer et al. 2017). These efforts have generated 11 high-resolution structures of effector:activator complexes (Table 1). These structures have captured BAK and BAX in several groove-destabilized conformations, whose intramolecular contacts throughout the groove are rearranged or disrupted relative to apo structures (Fig. 3A). Similar to their complexes with antiapoptotic BCL-2 proteins, these structures have revealed a six-turn activator helix bound via six to eight hydrophobic residues to hydrophobic pockets in effector grooves, stabilized by a conserved arginine-aspartate salt bridge (Fig. 3B). Extensive activator mutagenesis corroborated these structures and has been adopted to turn BH3-only sensitizers such as BAD into direct effector activators (Czabotar et al. 2013; Moldoveanu et al. 2013). Biochemical studies have suggested that most BH3 peptides are able to activate BAX and BAK (Kuwana et al. 2002, 2005; Kim et al. 2009; Du et al. 2011; Chen et al. 2015; Hockings et al. 2015), and sequence alignments in light of the structural studies corroborate that most BH3 peptides have some conserved features allowing them to partly engage canonical grooves (Fig. 3C). How full-length BH3-only proteins contribute to effector activation remains a pressing question. A recent biochemical study suggests that there are contributions from the carboxy-terminal mitochondrial targeting sequence of BIM in BAX activation (Chi et al. 2019).
Figure 3.
Structural basis of BAX and BAK direct activation through the canonical BH3 binding groove and selective human BAK inhibition. (A) BH3 peptide complexes with BAX (top) and BAK (bottom), aligned on apoBAK, reveal the overall similar arrangement of BH3 helices at the activation grooves. Six to eight hydrophobic residues of the BH3 peptides engage six hydrophobic pockets (numbered P0–P5). BAX and BAK grooves are occluded in apo forms at P1–P2, and P2–P3, respectively. Chemical stapling induced a slight shift of the P0–P3 portion of the BID BH3 helix at the activation groove of BAK. BH3 binding involves opening of the occluded grooves in BAX and BAK and formation of deep pockets not engaged by peptide residues (van der Walls contacts 4 Å from the nearest peptide atom) (orange). (B) Structure-guided design of BIM BH3-based molecular glue that locks BAK in an inactive conformation similar to apoBAK by introducing stabilizing salt bridges that block helix α1 release. (C) BAX and BAK engage BH3 peptides similarly at the canonical groove yet they show different peptide-induced changes, which manifest by formation of engorged cavities at the regions indicated by the arrows. Destabilized regions in BAX are at the groove region near helix α3 and α8, and in BAK are on either side of the carboxyl terminus of helix α2 and the amino terminus of helix α3. Rotation is relative to the front view. BH3 peptide alignments indicate the position of the hydrophobic residues (H0–H5) that make contact with hydrophobic pockets (P0–P5) in BAX and BAK. H0–H5 residues in BID and BIM (red) were deduced from structures of respective peptides in complex with BAX and BAK. Purple residues are hydrophobic residues found at the putative positions in other BH3 peptides expected to more weakly activate effectors compared with BH3 peptides from BID and BIM. A conserved BH3 aspartic acid (black highlight) forms a stabilizing salt bridge with a conserved arginine in the groove of BAX and BAK (B). A small residue (glycine or alanine, labeled s), allows tight contact between the BH3 helix and the groove next to the conserved salt bridge.
Gluing Inhibitors at the Groove of BAK
Two studies revealed how the canonical activation groove can also act as a site of inhibition for BAK. The original study showed biochemically that when the BID BH3 carboxyl terminus was cross-linked to the carboxyl terminus of BAK-ΔTM via engineered cysteines, the corresponding complex was intrinsically inactive, yet it was activatable through competition with excess-free BID BH3 peptide (Moldoveanu et al. 2013). More recently, structure-guided engineering of BIM-like BH3 peptides selective for human BAK provided a strategy for BAK inhibition. This study rationally introduced nonnatural acidic amino acid residues at the BIM H3 position to interact with two buried basic residues deep in the P3 pocket of BAK (Brouwer et al. 2017). Although the KD of these inhibitory complexes range from µm to nm, the peptides show molecular glue characteristics through stabilizing contacts that keep BAK inactive by preventing release of helix α1 (Fig. 3B). This study also shows that replacement of H0 amino acids from BIM-like to BID-like residues confers a significant boost in affinity (KD ∼1 µm to ∼20 nm) indicating major contributions to stabilizing interactions at this position (Fig. 3B,C). Based on the original study, modifications in BH3 peptides to enhance affinity for effectors may be applicable more broadly to effector inhibition (Moldoveanu et al. 2013), and similar design strategies may be adopted in groove-targeted effector inhibitory small-molecule chemical probes (Moldoveanu et al. 2013; Brouwer et al. 2017).
HOW ACTIVE EFFECTORS EXECUTE MITOCHONDRIAL PORATION
From Monomers to Dimers, the Particle on Which the Oligomer Builds
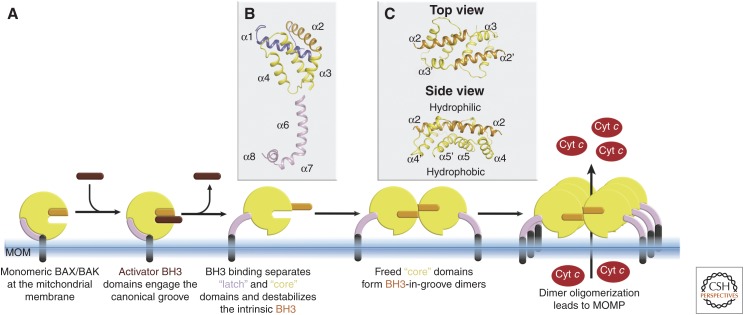
Once activated and at the MOM, Bax and Bak go through a series of conformational changes that involve the release of their α1 helix (Hsu and Youle 1998; Llambi et al. 2011), partial exposure of their BH3 domain (Dewson et al. 2008, 2012; Llambi et al. 2011) and disengagement of core (α2–α5) from latch (α6–α8) regions (Fig. 4A,B; Czabotar et al. 2013; Brouwer et al. 2014). The order of these events remains unclear, and some or all may occur simultaneously. Antiapoptotic guardians, if not already complexed with BH3-only relatives, can neutralize the activated forms of BAX and BAK at this step by binding to their exposed BH3 domains to prevent homodimerization (Oltvai et al. 1993; Wang et al. 1998; Fletcher et al. 2008; Llambi et al. 2011). Increasing concentrations of competing BH3-only proteins, decreasing concentrations of prosurvival proteins (e.g., by degradation) (Willis et al. 2005; Czabotar et al. 2007), or increasing concentrations of activated BAX and/or BAK lead to the formation of homodimers, or possibly in some settings BAX/BAK heterodimers (Dewson et al. 2012).
Figure 4.
Structural transitions during BAX and BAK activation. (A) Activation of monomeric BAX and BAK leads to a series of structural transitions eventuating in dimerization, oligomerization, and ultimately pore formation. Of this sequence, high-resolution structural details are available for monomeric forms of the proteins (Fig. 2A), BH3 activation at the canonical groove (Fig. 3), core from latch detachment (B), and core domain dimers as in C. (B) One of the structurally characterized conformation changes is disengagement of the core domain from the latch region. (C) Once released, core domains dimerize through a symmetric BH3-in-groove protein–protein interface. The dimer produced has a hydrophilic surface dominated by α2–α3 and a hydrophobic surface lined by α4–α5. The hydrophobic surface is thought to engage the outer mitochondrial membrane as a step toward permeabilization. Structures shown throughout are for BAX, but similar transitions have been shown for BAK as indicated in the text.
Homodimer formation is a key step in the process to oligomerization and pore formation. Early cross-linking studies showed that these formed through insertion of the BH3 of one monomer into the groove of a neighboring molecule (Dewson et al. 2008, 2012; Bleicken et al. 2010; Zhang et al. 2010). Subsequent crystallographic studies revealed the structure of these symmetric BH3-in-groove dimers (Czabotar et al. 2013; Brouwer et al. 2014), later also confirmed through biophysical proximity measurements (Bleicken et al. 2014; Mandal et al. 2016). A consequence of this transition from monomer to dimer is the conversion from a globular entity with a largely hydrophilic surface to a platform-like structure, which is amphipathic in nature (Fig. 4C). It has been proposed that the hydrophobic surface of this platform is responsible for interacting with lipids of the outer mitochondrial membrane surface as a step toward permeabilization (Czabotar et al. 2013; Brouwer et al. 2014) and/or to line the pore of the lumen subsequently formed (Bleicken et al. 2014; Mandal et al. 2016).
Building the Larger Oligomer and Forming a Pore
The precise nature by which BAX and BAK dimers form into larger oligomers and then permeabilize the outer mitochondrial membrane remains one of the principal questions within the field. From some studies, a daisy chain model of Bax and Bak oligomerization has been proposed (Bogner et al. 2010; Pang et al. 2012), with the front of one Bax monomer interacting with the rear of a neighbor. However, this model is inconsistent with the now well-characterized symmetric BH3-in-groove dimers, as a daisy chain interface is asymmetric, and thus is unlikely to represent the reality of the larger oligomer. Other studies showed that residues in the α6 of Bax and Bak could be cross-linked in a fashion consistent with these helices running parallel to each other in a larger complex (Dewson et al. 2009, 2012). Further cross-linking studies showed that linkage between dimers was not restricted to only this region, and that in fact a large number of residues across a range of different regions of the proteins could indeed be cross-linked (Zhang et al. 2010, 2016; Aluvila et al. 2014; Iyer et al. 2015; Uren et al. 2017). In all cases, these interdimer cross-links do not saturate, unlike the previously observed intra-dimer cross-links, suggesting that these contacts do not necessarily represent defined protein–protein interfaces, as observed and structurally characterized for the BH3-in-groove dimer.
The mechanism by which larger BAX and BAK oligomers build, how they form pores, and the nature of the pores themselves has also been a matter of much conjecture. Atomic force microscopy, superresolution microscopy, and electron tomography show that BAX oligomers can form a variety of sizes and shapes including clusters, rings, lines, and arcs (Grosse et al. 2016; Salvador-Gallego et al. 2016; Ader et al. 2019). Recently, it was discovered that pores of extremely large apertures can be induced by BAX (McArthur et al. 2018; Riley et al. 2018). These “macropores” enable the release of mtDNA into the cytoplasm resulting in activation of the cGAS/STING pathway and leading to type I interferon production (Rongvaux et al. 2014; White et al. 2014). It is still unclear how BAX and BAK initiate these heterogeneic membrane ruptures, but once this event has occurred it is likely that oligomers line the pore in some manner to stabilize the lumen. Protein-lined membrane pores can be either proteinaceous, in which the lumen is entirely lined by protein, or toroidal, where the lumen is lined by both protein and lipid headgroups; mounting evidence supports the latter for BAX and BAK (Terrones et al. 2004; Qian et al. 2008; Aluvila et al. 2014; Bleicken et al. 2014; Salvador-Gallego et al. 2016; Li et al. 2017). Spectroscopic experiments have been used to understand the alignment of dimers on the pore lumen and within the oligomer, leading to two distinct models. One involves dimers straddling the membrane bilayer such that the α9 TM helices run antiparallel to each other, the so-called clamp model (Bleicken et al. 2014). A second model involves dimers concentrated on the upper edge of the lumen leaving α9 TM helices to enter the membrane from the cytosolic side of the membrane and thus run parallel to each other (Mandal et al. 2016). It is currently unclear which of the two models represents the situation in cells, with further work required to understand this important aspect of membrane permeabilization. For further details and discussion of these opposing models, we refer the reader to a recent comprehensive review by Bleicken et al. (2018).
TOWARD CHEMICAL PROBING CANONICAL EFFECTORS
BAX and BAK are potential therapeutic targets that may be activated or inhibited to initiate or block apoptosis in disease. Understanding their molecular mechanisms of action importantly informs the development of small-molecule chemical probes to selectively target their activity. Accordingly, the effector field is now following in the footsteps of groups that developed antagonist of antiapoptotic BCL-2 proteins as exquisite chemical probes and therapeutics (Oltersdorf et al. 2005; Tse et al. 2008; Lessene et al. 2013; Souers et al. 2013; Huang et al. 2019).
Several small molecules have been discovered in academic laboratories through screening efforts against BAX and BAK. It is early for many of these compounds, particularly compared with those targeting antiapoptotic proteins, and future applicability requires these to be carefully tested by multiple investigators for potency, selectivity, and a proven mechanism of action (Arrowsmith et al. 2015). Nonetheless, these published compounds potentially inform on the dynamics, accessibility, and drugability of effector binding sites. Compound binding at these sites may induce, block, or cooperate with putative mechanisms of effector activation, inhibition, functional homodimerization, and higher order oligomerization. We briefly summarize features of small-molecule effector modulators below (Fig. 5).
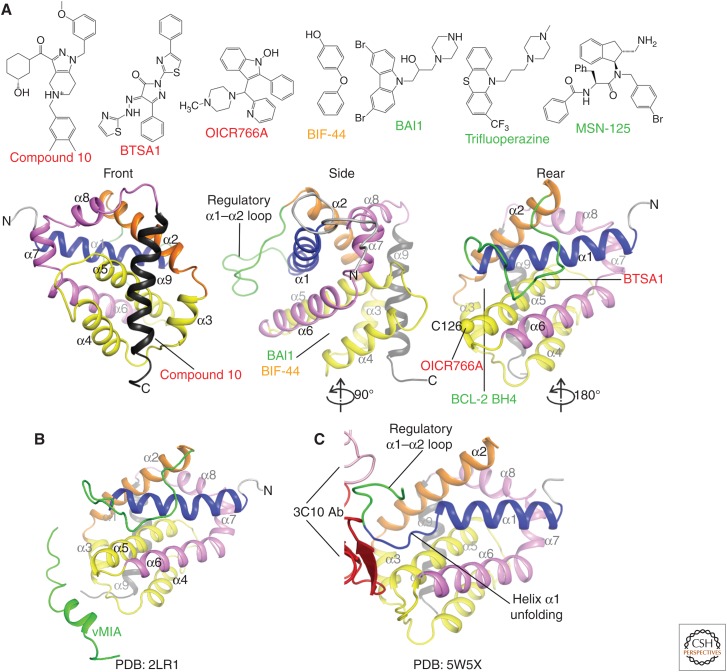
Figure 5.
Effector activation and inhibition with molecules other than BH3 peptides. (A) Proposed small-molecule activators (red), inhibitors (green), and allosteric modulators (orange) of BAX and BAK membrane permeabilization, and their possible interaction sites. The postulated inhibitory site of interaction for BCL-2 BH4 SAHB is also indicated. (B) The human cytomegalovirus protein viral mitochondrial-localized inhibitor of apoptosis (vMIA) inhibits BAX through a short helical peptide. (C) Antibody-mediated activation of BAX involves binding of an epitope of the regulatory α1–α2 loop, which unfolds the carboxyl terminus of helix α1. The heavy and light chains of the 3C10 Ab are colored red and pink, respectively.
BAX Activators Aimed at Its Front and Rear Sites
Compound 106 (ZINC 14750348) was identified in virtual screens for binders of the front, canonical BAX activation site and induced BAX-dependent but not BAK-dependent cytotoxicity in bak−/−bax−/− mouse embryonic fibroblasts (MEFs) expressing human BAX or human BAK (Fig. 5A; Zhao et al. 2014). Compound 106 destabilized BAX in thermal shift assays and induced BAX-dependent cyt c release in vitro and human tumor apoptosis in tissue culture and in vivo. Others have discovered additional BAX-selective activator compounds with in vitro and in vivo activity in virtual screens as supposed modulators of the front site interaction with the TM tail (Xin et al. 2014), although their direct interaction with, and functional activation of, BAX was not rigorously shown. Two of these compounds contain alkene groups that could form Michael addition adducts with many intracellular nucleophiles and therefore are not likely selective for BAX.
Compounds designed as rear site activators of BAX, including BAM7 (Gavathiotis et al. 2012) and its second iteration BTSA1 (Reyna et al. 2017), were identified through in silico and subsequent analog searches (Fig. 5A). These compounds displace BIM SAHB:BAX complexes with IC50 of 3 µm and 250 nm in fluorescence polarization assays, respectively. Evidence that they behave as bona fide BAX activators includes their induction of 6A7 antibody epitope exposure, oligomerization in solution over time similar to that induced by BH3 activators, membrane translocation, membrane permeabilization, BAX-selectivity over BAK in cells, and cell-based and in vivo activity against acute myeloid leukemia tumors (Reyna et al. 2017). On the other hand, their binding to BAX by 2D NMR is less compelling with very small chemical shift perturbations (<0.015 ppm) for most of the resonances deemed significant and overlapping the rear activation site (Gavathiotis et al. 2012; Reyna et al. 2017). Might these compounds engage the canonical activation site in BAX? We do not currently know, but this question should be included in experimental design whenever BAX modulators are pursued, as we expect that canonical groove engagement and destabilization to be essential in BAX activation. Interestingly, BTSA1 contains an imine group known to be prone to hydrolysis that may release the hydrazino-thiazole moiety (Gaulton et al. 2017). A caveat of effector activation in cells is that many chemicals may trigger effectors indirectly by up-regulating and activating the upstream BH3-only proteins. Such possibilities could be explored through profiling of BCL-2 proteins ± drug to ensure direct rather than indirect effector activation.
BAX Inhibitors Glue the Bottom of the Groove
BAX inhibitory compounds (BAIs) were discovered early (Bombrun et al. 2003), and were recently reported to inhibit activated truncated BID (tBID)-induced BAX-mediated liposome permeabilization with IC50 ∼4 µm (Garner et al. 2019). BAIs bind to a pocket at the bottom of the canonical groove between helices α3, α4, α5, and α6 with KD ∼15 µm, established by microscale thermophoresis, although they induce small chemical shift perturbations (<0.015 ppm) in BAX detectable by 2D NMR (Fig. 5A; Garner et al. 2019). These inhibitors are thought to block the conformational changes associated with BAX activation, thus preventing its mitochondrial translocation and oligomerization, and selectively inhibit BAX- but not BAK-dependent apoptosis with an IC50 of ∼2 µm (Garner et al. 2019). Interestingly, trifluoroperazine (TFP) is a distant structural analog of BAIs, discovered as an inhibitor of BAK-mediated liposome permeabilization with an IC50 ∼10 µm (Song et al. 2014). TFP binds BAK-ΔTM with KD ∼70 µm in isothermal titration calorimetry assays, and blocks BAK oligomerization in liposomes, but its mode of binding to BAK has not been established (Song et al. 2014).
Other Means to Effector Structure–Function Modulation
Several modulators of BAK and BAX were discovered based on screening using liposome permeabilization assays. OICR766A activated BAX with an EC50 of ∼100 nm (liposomes) and ∼900 nm (mitochondria), was shown to induce oligomerization and membrane localization of BAX, and showed a strict requirement for residue C126 for activity (Fig. 5A; Brahmbhatt et al. 2016). OICR766A reportedly induced apoptosis in a BAX- or BAK-dependent manner and was inactive in bak−/−bax−/− baby mouse kidney (BMK) cells, suggesting that it directly or indirectly activated BAX/BAK. MSN-125 and its analogs partially inhibited BAX-mediated liposome permeabilization with an IC50 of ∼4 µm, BAX- or BAK-mediated mitochondrial poration with an IC50 of ∼10 µm and ∼20 µm, respectively, and inhibited BAK- and BAX-dependent apoptosis (Niu et al. 2017). Interestingly, MSN-125 appeared to act by preventing higher order oligomerization beyond dimerization as examined in cross-linking studies (Fig. 5A).
Allosteric modulation of effector activity has been reported with: (1) a helical peptide from human cytomegalovirus viral mitochondria-localized inhibitor of apoptosis (vMIA), which binds a surface pocket at the Bax α3–α4 and α5–α6 hairpins, presumably acting by allosterically blocking conformational changes, or activation at the canonical groove (Fig. 5B; Ma et al. 2012); (2) antibodies that bind epitopes in the α1–α2 loop, inducing protein unfolding to directly activate BAK and BAX (Fig. 5C; Iyer et al. 2016; Robin et al. 2018); (3) a stapled BCL-2 BH4 helix peptide that binds a site at the bottom of the canonical groove composed of the α1, α1–α2 loop, and α5–α6 hairpin presumably blocking BAX activation (Fig. 5A; Barclay et al. 2015); and (4) a BIF-44 fragment binding a surface pocket at the center of α5–α6 helices, which synergizes with peptide-based BH3 activators (Fig. 5A; Pritz et al. 2017). These studies have revealed multiple modalities for modulating BAK and BAX conformations, potentially impacting their apoptotic function either independently or through synergism with BH3 activators. Ultimately, chemical probe effector modulators need to evolve via structure-based design to simplify and guide a structure–activity relationship (SAR). Moreover, effector binding site mutagenesis must be thoroughly performed to test the role of surface pocket engagement in effector modulation. Some of the sites may be undruggable because of their small size and/or peripheral involvement in effector modulation, factors that would limit the development of potent (KD ∼ nm) and selective (e.g., ∼30-fold differences for BAX vs. BAK) chemical probes.
POSTTRANSLATIONAL EFFECTOR MODIFICATIONS DOWN-REGULATE APOPTOSIS
Posttranslational modifications have been implicated in BAK and BAX activity, although this area of the field has not been exhaustively probed. It has been reported that phosphorylation of BAX S184 in the carboxy-terminal TM tail prevents BAX targeting to mitochondria in tumor cell lines (Gardai et al. 2004; Xin and Deng 2005; Wang et al. 2010; Quast et al. 2013). Through exposure of the canonical groove, this phosphorylation event could render cytosolic BAX a better receptor for BH3-only activators. Such unproductive BH3-only protein:BAX interactions would down-regulate apoptosis induced by BH3 mimetics that disrupt or prevent formation of complexes between pro- and antiapoptotic BCL-2 proteins (Fig. 2D; Kale 2018).
Ubiquitylation of BAK, BAX, and BOK (vide infra) has been reported to down-regulate effector-mediated apoptosis (Cakir et al. 2017; Bernardini et al. 2019). Monoubiquitylation of BAK's single lysine, K113, in the canonical groove by the E3 ligase parkin during mitophagy has been shown to down-regulate the ability of BAK to permeabilize membranes in vitro and in cells by interfering with activation, dimerization, and oligomerization (Bernardini et al. 2019). In contrast, VDAC2 ubiquitination by parkin inhibited BAX mitochondrial localization, possibly through steric hindrance of the interaction between BAX and ubiquitylated VDAC2 and/or through reduced BAX recruitment because of reduced VDAC2 levels (Bernardini et al. 2019), although this has not been formally tested. BAX regulation by ubiquitylation is more complex and context-dependent and may involve direct (Cakir et al. 2017) and indirect targeting of the BAX apoptotic axis (Fig. 2C,D; Bernardini et al. 2019). BAX K128 has been implicated in ubiquitylation and degradation by ectopic parkin expression, which may protect against BAX-mediated apoptosis. K128R BAX was resistant to parkin-mediated degradation suggesting that pharmacologic targeting of parkin may modulate BAX-dependent apoptosis (Cakir et al. 2017). In contrast, VDAC2 ubiquitination by parkin inhibited BAX mitochondrial localization by presumed steric hindrance of the interaction between BAX and ubiquitylated VDAC2 (Bernardini et al. 2019). Future high-resolution studies may shed light on the molecular mechanisms of inhibition of effectors by posttranslational modifications.
BOK—THE LONE WOLF EFFECTOR
BOK is a BCL-2 family member that most resembles BAX and BAK at the sequence level, but for many years its role in apoptosis was unclear largely because its upstream triggers were unknown (Ke et al. 2012; Carpio et al. 2015; Fernández-Marrero et al. 2016). Early studies indicated that it was predominately located at the endoplasmic reticulum (ER) in complex with IP3 receptors rather than at the mitochondria (Echeverry et al. 2013; Schulman et al. 2013, 2016). Additionally, a role for BOK in mitochondrial fusion has also been suggested (Schulman et al. 2019). However, recently, an effector role for BOK has emerged from genetic, biochemical, and structural studies (Fig. 6). Genetically, BOK has been implicated as an effector in studies that combined deletion of BAK, BAX, and BOK in hematopoiesis and during embryonic development (Ke et al. 2015, 2018). Combined deletion of BAK and BOK or BAX and BOK only revealed a possible role for BOK in ovaries (Ke et al. 2013). In lethally irradiated mice transplanted with fetal liver cells from bak−/−bax−/− double knockout (DKO) and bak−/−bax−/−bok−/− triple knockout (TKO) mice, TKO reconstituted mice had more peripheral blood lymphocytes, and lymphoid infiltration, supporting BOK's functional redundancy with BAK and BAX (Ke et al. 2015). The most compelling genetic demonstration for a role of BOK as an effector was the comparative investigation of DKO and TKO mice (Ke et al. 2018). These suggested that intrinsic apoptosis defects are greatly exacerbated in TKOs, which ultimately contributed to widespread embryonic and perinatal lethality. Surprisingly, a small fraction of TKO mice lived normally to adulthood, showing no apparent phenotypes, suggesting that intrinsic apoptosis is dispensable for normal organ development and homeostasis. Other forms of cell death may be redundant with intrinsic apoptosis, although neither extrinsic apoptosis, necroptosis, pyroptosis, or autophagy were up-regulated in the TKO mice during development.
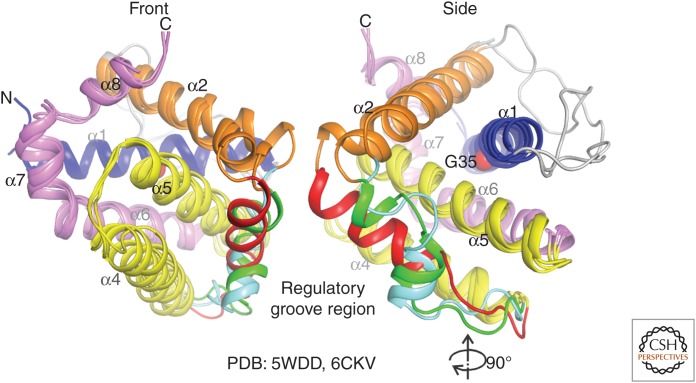
Figure 6.
Dormant BOK is intrinsically unstable. Overlay of chicken and human BOK structures reveals three possible arrangements of the regulatory groove region between helices α3–α4. Additionally, human BOK has instability at the α1 helix, through G35, whose mutation to alanine significantly inhibits membrane association and permeabilization, and blocks BAK- and BAX-independent apoptosis.
Biochemical demonstration for the effector role of BOK involved discovery of BOK's degradation machinery, the ER-associated degradation (ERAD) gp78 E3 ligase proteasome system (Llambi et al. 2016). Accordingly, proteasome inhibition stabilized and up-regulated BOK protein (Llambi et al. 2016; Zheng et al. 2018), and this could also be achieved through a combination of certain ER stress drugs (tunicamycin + PERK inhibitors), which dismantle the gp78 E3 ligase complex, inhibition of VCP AAA-ATPase, which blocks substrate shuttling from the E3 ligase to the proteasome, or simply through knockdown of the gp78 subunits or VCP (Llambi et al. 2016). Unlike BAX and BAK, BOK interactions with BCL-2 family proteins appear to be weak and functionally fruitless. Mitochondrial poration and apoptosis induction by BOK does not appear to be enhanced by direct activators such as activated BID, or inhibited by antiapoptotic BCL-2 proteins (Llambi et al. 2016; Zheng et al. 2018). However, it should be noted that some studies report that activated BID or BID BH3 can promote BOK activity in liposome settings (Fernández-Marrero et al. 2017; Zheng et al. 2018). Also, it has been reported that antiapoptotic BCL-2 proteins may modulate BOK activity through their TM region (Stehle et al. 2018), more so than through their canonical grooves, which have weak affinity for BOK BH3 (KD ∼ µm) (Llambi et al. 2016).
Structural studies of apoBOK-ΔTM suggest that dormant BOK is poised for spontaneous conformational changes, captured by NMR spectroscopy and X-ray crystallography, providing a potential explanation for BOK poration of the mitochondrial membrane in the absence of direct activation (Fig. 6; Ke et al. 2018; Zheng et al. 2018). Like BAX and BAK, BOK can form large pores that are likely toroidal in nature (Fernández-Marrero et al. 2017). Our understanding of BOK function and activity lags far behind that of BAX and BAK. There is much that remains to be discovered, for example, what are the contributions of BOK to apoptosis in cells in which BAX and/or BAK are present, and does BOK permeabilize membranes in a similar fashion to BAX and BAK. This is likely an area in which significant discoveries will be made in the field in coming years.
FUTURE DIRECTIONS
Mitochondrial poration is moving into a new structure–function phase with the pursuit of high-resolution structural details for effector conformations and complexes at membranes. Targets include structures for effectors alone and posttranslationally modified effector: activator complexes, effector:antiapoptotic complexes, and effector:VDAC complexes. Innovative multipronged high-resolution approaches and careful functional probing will be necessary to achieve these aims, and these efforts will fuel the discovery of new chemical probes for apoptosis research. We anticipate a rich decade ahead in effector research as we delve deeper into mechanisms underlying the key event of MOM permeabilization.
Footnotes
Editors: Kim Newton, James M. Murphy, and Edward A. Miao
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ader NR, Hoffmann PC, Ganeva I, Borgeaud AC, Wang C, Youle RJ, Kukulski W. 2019. Molecular and topological reorganizations in mitochondrial architecture interplay during Bax-mediated steps of apoptosis. eLife 8: e40712 10.7554/eLife.40712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluvila S, Mandal T, Hustedt E, Fajer P, Choe JY, Oh KJ. 2014. Organization of the mitochondrial apoptotic BAK pore: oligomerization of the BAK homodimers. J Biol Chem 289: 2537–2551. 10.1074/jbc.M113.526806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME, et al. 2015. The promise and peril of chemical probes. Nat Chem Biol 11: 536–541. 10.1038/nchembio.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay LA, Wales TE, Garner TP, Wachter F, Lee S, Guerra RM, Stewart ML, Braun CR, Bird GH, Gavathiotis E, et al. 2015. Inhibition of Pro-apoptotic BAX by a noncanonical interaction mechanism. Mol Cell 57: 873–886. 10.1016/j.molcel.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini JP, Brouwer JM, Tan IK, Sandow JJ, Huang S, Stafford CA, Bankovacki A, Riffkin CD, Wardak AZ, Czabotar PE, et al. 2019. Parkin inhibits BAK and BAX apoptotic function by distinct mechanisms during mitophagy. EMBO J 38: e99916 10.15252/embj.201899916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Classen M, Padmavathi PV, Ishikawa T, Zeth K, Steinhoff HJ, Bordignon E. 2010. Molecular details of Bax activation, oligomerization, and membrane insertion. J Biol Chem 285: 6636–6647. 10.1074/jbc.M109.081539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, García-Sáez AJ, Bordignon E. 2014. Structural model of active Bax at the membrane. Mol Cell 56: 496–505. 10.1016/j.molcel.2014.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Assafa TE, Stegmueller C, Wittig A, Garcia-Saez AJ, Bordignon E. 2018. Topology of active, membrane-embedded Bax in the context of a toroidal pore. Cell Death Differ 25: 1717–1731. 10.1038/s41418-018-0184-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner C, Leber B, Andrews DW. 2010. Apoptosis: embedded in membranes. Curr Opin Cell Biol 22: 845–851. 10.1016/j.ceb.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Bombrun A, Gerber P, Casi G, Terradillos O, Antonsson B, Halazy S. 2003. 3,6-Dibromocarbazole piperazine derivatives of 2-propanol as first inhibitors of cytochrome c release via Bax channel modulation. J Med Chem 46: 4365–4368. 10.1021/jm034107j [DOI] [PubMed] [Google Scholar]
- Brahmbhatt H, Uehling D, Al-Awar R, Leber B, Andrews D. 2016. Small molecules reveal an alternative mechanism of Bax activation. Biochem J 473: 1073–1083. 10.1042/BCJ20160118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer JM, Westphal D, Dewson G, Robin AY, Uren RT, Bartolo R, Thompson GV, Colman PM, Kluck RM, Czabotar PE. 2014. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol Cell 55: 938–946. 10.1016/j.molcel.2014.07.016 [DOI] [PubMed] [Google Scholar]
- Brouwer JM, Lan P, Cowan AD, Bernardini JP, Birkinshaw RW, van Delft MF, Sleebs BE, Robin AY, Wardak A, Tan IK, et al. 2017. Conversion of Bim-BH3 from activator to inhibitor of Bak through structure-based design. Mol Cell 68: 659–672.e9. 10.1016/j.molcel.2017.11.001 [DOI] [PubMed] [Google Scholar]
- Cakir Z, Funk K, Lauterwasser J, Todt F, Zerbes RM, Oelgeklaus A, Tanaka A, van der Laan M, Edlich F. 2017. Parkin promotes proteasomal degradation of misregulated BAX. J Cell Sci 130: 2903–2913. 10.1242/jcs.200162 [DOI] [PubMed] [Google Scholar]
- Carpio MA, Michaud M, Zhou W, Fisher JK, Walensky LD, Katz SG. 2015. BCL-2 family member BOK promotes apoptosis in response to endoplasmic reticulum stress. Proc Natl Acad Sci 112: 7201–7206. 10.1073/pnas.1421063112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Kanai M, Inoue-Yamauchi A, Tu HC, Huang Y, Ren D, Kim H, Takeda S, Reyna DE, Chan PM, et al. 2015. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat Cell Biol 17: 1270–1281. 10.1038/ncb3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. 2003. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301: 513–517. 10.1126/science.1083995 [DOI] [PubMed] [Google Scholar]
- Chi X, Pemberton J, Nguyen D, Osterlund EJ, Liu Q, Brahmbhatt H, Zhang Z, Lin J, Leber B, Andrews D. 2019. The carboxyl-terminal sequence of Bim enables Bax activation and killing of unprimed cells. bioRxiv 10.1101/554907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin HS, Li MX, Tan IKL, Ninnis RL, Reljic B, Scicluna K, Dagley LF, Sandow JJ, Kelly GL, Samson AL, et al. 2018. VDAC2 enables BAX to mediate apoptosis and limit tumor development. Nat Commun 9: 4976 10.1038/s41467-018-07309-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC, Fairlie WD, Hinds MG, Colman PM. 2007. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci 104: 6217–6222. 10.1073/pnas.0701297104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, et al. 2013. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152: 519–531. 10.1016/j.cell.2012.12.031 [DOI] [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. 2014. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15: 49–63. 10.1038/nrm3722 [DOI] [PubMed] [Google Scholar]
- Dai H, Ding H, Meng XW, Peterson KL, Schneider PA, Karp JE, Kaufmann SH. 2015. Constitutive BAK activation as a determinant of drug sensitivity in malignant lymphohematopoietic cells. Genes Dev 29: 2140–2152. 10.1101/gad.267997.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Ding H, Peterson KL, Meng XW, Schneider PA, Knorr KLB, Kaufmann SH. 2018. Measurement of BH3-only protein tolerance. Cell Death Differ 25: 282–293. 10.1038/cdd.2017.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler MA, Robin AY, Gibson L, Li MX, Sandow JJ, Iyer S, Webb AI, Westphal D, Dewson G, Adams JM. 2019. BAX activation: Mutations near its proposed non-canonical BH3 binding site reveal allosteric changes controlling mitochondrial association. Cell Rep 27: 359–373.e6. 10.1016/j.celrep.2019.03.040 [DOI] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Sim HW, Puthalakath H, Adams JM, Colman PM, Kluck RM. 2008. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell 30: 369–380. 10.1016/j.molcel.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM. 2009. Bak activation for apoptosis involves oligomerization of dimers via their α6 helices. Mol Cell 36: 696–703. 10.1016/j.molcel.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Dewson G, Ma S, Frederick P, Hockings C, Tan I, Kratina T, Kluck RM. 2012. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ 19: 661–670. 10.1038/cdd.2011.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wolf J, Schafer B, Moldoveanu T, Chipuk JE, Kuwana T. 2011. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem 286: 491–501. 10.1074/jbc.M110.167148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussmann H, Rehm M, Concannon CG, Anguissola S, Würstle M, Kacmar S, Völler P, Huber HJ, Prehn JH. 2010. Single-cell quantification of Bax activation and mathematical modelling suggest pore formation on minimal mitochondrial Bax accumulation. Cell Death Differ 17: 278–290. 10.1038/cdd.2009.123 [DOI] [PubMed] [Google Scholar]
- Echeverry N, Bachmann D, Ke F, Strasser A, Simon HU, Kaufmann T. 2013. Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ 20: 785–799. 10.1038/cdd.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzner A, Tjandra N, Youle RJ. 2011. Bcl-xL retrotranslocates Bax from the mitochondria into the cytosol. Cell 145: 104–116. 10.1016/j.cell.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AL, Gavathiotis E, LaBelle JL, Braun CR, Opoku-Nsiah KA, Bird GH, Walensky LD. 2013. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol 20: 888–902. 10.1016/j.chembiol.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marrero Y, Ke F, Echeverry N, Bouillet P, Bachmann D, Strasser A, Kaufmann T. 2016. Is BOK required for apoptosis induced by endoplasmic reticulum stress? Proc Natl Acad Sci 113: E492–E493. 10.1073/pnas.1516347113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marrero Y, Bleicken S, Das KK, Bachmann D, Kaufmann T, Garcia-Saez AJ. 2017. The membrane activity of BOK involves formation of large, stable toroidal pores and is promoted by cBID. FEBS J 284: 711–724. 10.1111/febs.14008 [DOI] [PubMed] [Google Scholar]
- Ferrer PE, Frederick P, Gulbis JM, Dewson G, Kluck RM. 2012. Translocation of a Bak C-terminus mutant from cytosol to mitochondria to mediate cytochrome C release: Implications for Bak and Bax apoptotic function. PLoS ONE 7: e31510 10.1371/journal.pone.0031510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JI, Meusburger S, Hawkins CJ, Riglar DT, Lee EF, Fairlie WD, Huang DC, Adams JM. 2008. Apoptosis is triggered when prosurvival Bcl-2 proteins cannot restrain Bax. Proc Natl Acad Sci 105: 18081–18087. 10.1073/pnas.0808691105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardai SJ, Hildeman DA, Frankel SK, Whitlock BB, Frasch SC, Borregaard N, Marrack P, Bratton DL, Henson PM. 2004. Phosphorylation of Bax Ser184 by Akt regulates its activity and apoptosis in neutrophils. J Biol Chem 279: 21085–21095. 10.1074/jbc.M400063200 [DOI] [PubMed] [Google Scholar]
- Garner TP, Reyna DE, Priyadarshi A, Chen HC, Li S, Wu Y, Ganesan YT, Malashkevich VN, Cheng EH, Gavathiotis E. 2016. An autoinhibited dimeric form of BAX regulates the BAX activation pathway. Mol Cell 63: 485–497. 10.1016/j.molcel.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner TP, Amgalan D, Reyna DE, Li S, Kitsis RN, Gavathiotis E. 2019. Small-molecule allosteric inhibitors of BAX. Nat Chem Biol 15: 322–330. 10.1038/s41589-018-0223-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, et al. 2017. The ChEMBL database in 2017. Nucleic Acids Res 45: D945–D954. 10.1093/nar/gkw1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, et al. 2008. BAX activation is initiated at a novel interaction site. Nature 455: 1076–1081. 10.1038/nature07396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. 2010. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell 40: 481–492. 10.1016/j.molcel.2010.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES, Walensky LD. 2012. Direct and selective small-molecule activation of proapoptotic BAX. Nat Chem Biol 8: 639–645. 10.1038/nchembio.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse L, Wurm CA, Bruser C, Neumann D, Jans DC, Jakobs S. 2016. Bax assembles into large ring-like structures remodeling the mitochondrial outer membrane in apoptosis. EMBO J 35: 402–413. 10.15252/embj.201592789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockings C, Anwari K, Ninnis RL, Brouwer J, O'Hely M, Evangelista M, Hinds MG, Czabotar PE, Lee EF, Fairlie WD, et al. 2015. Bid chimeras indicate that most BH3-only proteins can directly activate Bak and Bax, and show no preference for Bak versus Bax. Cell Death Dis 6: e1735 10.1038/cddis.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YT, Youle RJ. 1998. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J Biol Chem 273: 10777–10783. 10.1074/jbc.273.17.10777 [DOI] [PubMed] [Google Scholar]
- *.Huang DCS, Gong J-N, Fairbrother WJ. 2019. BH3 mimetic drugs to treat cancers. Cold Spring Harb Perpect Biol 10.1101/cshperspect.a036327 [DOI] [Google Scholar]
- Iyer S, Bell F, Westphal D, Anwari K, Gulbis J, Smith BJ, Dewson G, Kluck RM. 2015. Bak apoptotic pores involve a flexible C-terminal region and juxtaposition of the C-terminal transmembrane domains. Cell Death Differ 22: 1665–1675. 10.1038/cdd.2015.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Anwari K, Alsop AE, Yuen WS, Huang DC, Carroll J, Smith NA, Smith BJ, Dewson G, Kluck RM. 2016. Identification of an activation site in Bak and mitochondrial Bax triggered by antibodies. Nat Commun 7: 11734 10.1038/ncomms11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale J, Kutuk O, Brito GC, Andrews TS, Leber B, Letai A, Andrews DW. 2018. Phosphorylation switches Bax from promoting to inhibiting apoptosis thereby increasing drug resistance. EMBO Rep 19 10.15252/embr.201745235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke F, Voss A, Kerr JB, O'Reilly LA, Tai L, Echeverry N, Bouillet P, Strasser A, Kaufmann T. 2012. BCL-2 family member BOK is widely expressed but its loss has only minimal impact in mice. Cell Death Differ 19: 915–925. 10.1038/cdd.2011.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke F, Bouillet P, Kaufmann T, Strasser A, Kerr J, Voss AK. 2013. Consequences of the combined loss of BOK and BAK or BOK and BAX. Cell Death Dis 4: e650 10.1038/cddis.2013.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke F, Grabow S, Kelly GL, Lin A, O'Reilly LA, Strasser A. 2015. Impact of the combined loss of BOK, BAX and BAK on the hematopoietic system is slightly more severe than compound loss of BAX and BAK. Cell Death Dis 6: e1938 10.1038/cddis.2015.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke FFS, Vanyai HK, Cowan AD, Delbridge ARD, Whitehead L, Grabow S, Czabotar PE, Voss AK, Strasser A. 2018. Embryogenesis and adult life in the absence of intrinsic apoptosis effectors BAX, BAK, and BOK. Cell 173: 1217–1230.17. 10.1016/j.cell.2018.04.036 [DOI] [PubMed] [Google Scholar]
- Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. 2009. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell 36: 487–499. 10.1016/j.molcel.2009.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111: 331–342. 10.1016/S0092-8674(02)01036-X [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. 2005. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 17: 525–535. 10.1016/j.molcel.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Lauterwasser J, Todt F, Zerbes RM, Nguyen TN, Craigen W, Lazarou M, van der Laan M, Edlich F. 2016. The porin VDAC2 is the mitochondrial platform for Bax retrotranslocation. Sci Rep 6: 32994 10.1038/srep32994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Stojanovski D, Frazier AE, Kotevski A, Dewson G, Craigen WJ, Kluck RM, Vaux DL, Ryan MT. 2010. Inhibition of Bak activation by VDAC2 is dependent on the Bak transmembrane anchor. J Biol Chem 285: 36876–36883. 10.1074/jbc.M110.159301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner ES, Braun CR, Bird GH, Walensky LD. 2013. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci 110: E986–E995. 10.1073/pnas.1214313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessene G, Czabotar PE, Sleebs BE, Zobel K, Lowes KN, Adams JM, Baell JB, Colman PM, Deshayes K, Fairbrother WJ, et al. 2013. Structure-guided design of a selective BCL-XL inhibitor. Nat Chem Biol 9: 390–397. 10.1038/nchembio.1246 [DOI] [PubMed] [Google Scholar]
- Li MX, Tan IKL, Ma SB, Hockings C, Kratina T, Dengler MA, Alsop AE, Kluck RM, Dewson G. 2017. BAK α6 permits activation by BH3-only proteins and homooligomerization via the canonical hydrophobic groove. Proc Natl Acad Sci 114: 7629–7634. 10.1073/pnas.1702453114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Oesterlund EJ, Chi X, Pogmore J, Leber B, Andrews DW. 2019. Bim escapes displacement by BH3-mimetic anti-cancer drugs by double-bolt locking both Bcl-XL and Bcl-2. eLife 8: e37689 10.7554/eLife.37689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR. 2011. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 44: 517–531. 10.1016/j.molcel.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, Parsons MJ, Zheng JH, Brown SA, Pelletier S, et al. 2016. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell 165: 421–433. 10.1016/j.cell.2016.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Edlich F, Bermejo GA, Norris KL, Youle RJ, Tjandra N. 2012. Structural mechanism of Bax inhibition by cytomegalovirus protein vMIA. Proc Natl Acad Sci 109: 20901–20906. 10.1073/pnas.1217094110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SB, Nguyen TN, Tan I, Ninnis R, Iyer S, Stroud DA, Menard M, Kluck RM, Ryan MT, Dewson G. 2014. Bax targets mitochondria by distinct mechanisms before or during apoptotic cell death: a requirement for VDAC2 or Bak for efficient Bax apoptotic function. Cell Death Differ 21: 1925–1935. 10.1038/cdd.2014.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal T, Shin S, Aluvila S, Chen HC, Grieve C, Choe JY, Cheng EH, Hustedt EJ, Oh KJ. 2016. Assembly of Bak homodimers into higher order homooligomers in the mitochondrial apoptotic pore. Sci Rep 6: 30763 10.1038/srep30763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, Geoghegan ND, Chappaz S, Davidson S, San Chin H, et al. 2018. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359: eaao6047 10.1126/science.aao6047 [DOI] [PubMed] [Google Scholar]
- Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K. 2006. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell 24: 677–688. 10.1016/j.molcel.2006.10.014 [DOI] [PubMed] [Google Scholar]
- Moldoveanu T, Grace CR, Llambi F, Nourse A, Fitzgerald P, Gehring K, Kriwacki RW, Green DR. 2013. BID-induced structural changes in BAK promote apoptosis. Nat Struct Mol Biol 20: 589–597. 10.1038/nsmb.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T, Follis AV, Kriwacki RW, Green DR. 2014. Many players in BCL-2 family affairs. Trends Biochem Sci 39: 101–111. 10.1016/j.tibs.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Brahmbhatt H, Mergenthaler P, Zhang Z, Sang J, Daude M, Ehlert FGR, Diederich WE, Wong E, Zhu W, et al. 2017. A small-molecule inhibitor of Bax and Bak oligomerization prevents genotoxic cell death and promotes neuroprotection. Cell Chem Biol 24: 493–506.e5. 10.1016/j.chembiol.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Zobel K, Fedorova A, Quan C, Yang H, Fairbrother WJ, Huang DC, Smith BJ, Deshayes K, Czabotar PE. 2013. Stabilizing the pro-apoptotic BimBH3 helix (BimSAHB) does not necessarily enhance affinity or biological activity. ACS Chem Biol 8: 297–302. 10.1021/cb3005403 [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. 2005. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435: 677–681. 10.1038/nature03579 [DOI] [PubMed] [Google Scholar]
- Oltvai ZN, Milliman CL, Korsmeyer SJ. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programed cell death. Cell 74: 609–619. 10.1016/0092-8674(93)90509-O [DOI] [PubMed] [Google Scholar]
- Pang YP, Dai H, Smith A, Meng XW, Schneider PA, Kaufmann SH. 2012. Bak conformational changes induced by ligand binding: Insight into BH3 domain binding and Bak homo-oligomerization. Sci Rep 2: 257 10.1038/srep00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Tong JS, Li H, Yue B, Zou F, Yu J, Zhang L. 2013. Targeting Bax interaction sites reveals that only homo-oligomerization sites are essential for its activation. Cell Death Differ 20: 744–754. 10.1038/cdd.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritz JR, Wachter F, Lee S, Luccarelli J, Wales TE, Cohen DT, Coote P, Heffron GJ, Engen JR, Massefski W, et al. 2017. Allosteric sensitization of proapoptotic BAX. Nat Chem Biol 13: 961–967. 10.1038/nchembio.2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Wang W, Yang L, Huang HW. 2008. Structure of transmembrane pore induced by Bax-derived peptide: evidence for lipidic pores. Proc Natl Acad Sci 105: 17379–17383. 10.1073/pnas.0807764105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast SA, Berger A, Eberle J. 2013. ROS-dependent phosphorylation of Bax by wortmannin sensitizes melanoma cells for TRAIL-induced apoptosis. Cell Death Dis 4: e839 10.1038/cddis.2013.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault TT, Floros KV, Elkholi R, Corrigan KA, Kushnareva Y, Wieder SY, Lindtner C, Serasinghe MN, Asciolla JJ, Buettner C, et al. 2015. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell 57: 69–82. 10.1016/j.molcel.2014.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna DE, Garner TP, Lopez A, Kopp F, Choudhary GS, Sridharan A, Narayanagari SR, Mitchell K, Dong B, Bartholdy BA, et al. 2017. Direct activation of BAX by BTSA1 overcomes apoptosis resistance in acute myeloid leukemia. Cancer Cell 32: 490–505.e10. 10.1016/j.ccell.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JS, Quarato G, Cloix C, Lopez J, O'Prey J, Pearson M, Chapman J, Sesaki H, Carlin LM, Passos JF, et al. 2018. Mitochondrial inner membrane permeabilisation enables mtDNA release during apoptosis. EMBO J 37: e99238 10.15252/embj.201899238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin AY, Krishna Kumar K, Westphal D, Wardak AZ, Thompson GV, Dewson G, Colman PM, Czabotar PE. 2015. Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction. Cell Death Dis 6: e1809 10.1038/cddis.2015.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin AY, Iyer S, Birkinshaw RW, Sandow J, Wardak A, Luo CS, Shi M, Webb AI, Czabotar PE, Kluck RM, et al. 2018. Ensemble properties of Bax determine its function. Structure 26: 1346–1359.e5. 10.1016/j.str.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, et al. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159: 1563–1577. 10.1016/j.cell.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Gallego R, Mund M, Cosentino K, Schneider J, Unsay J, Schraermeyer U, Engelhardt J, Ries J, Garcia-Saez AJ. 2016. Bax assembly into rings and arcs in apoptotic mitochondria is linked to membrane pores. EMBO J 35: 389–401. 10.15252/embj.201593384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JJ, Wright FA, Kaufmann T, Wojcikiewicz RJ. 2013. The Bcl-2 protein family member Bok binds to the coupling domain of inositol 1,4,5-trisphosphate receptors and protects them from proteolytic cleavage. J Biol Chem 288: 25340–25349. 10.1074/jbc.M113.496570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JJ, Wright FA, Han X, Zluhan EJ, Szczesniak LM, Wojcikiewicz RJ. 2016. The stability and expression level of Bok are governed by binding to inositol 1,4,5-trisphosphate receptors. J Biol Chem 291: 11820–11828. 10.1074/jbc.M115.711242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman JJ, Szczesniak LM, Bunker EN, Nelson HA, Roe MW, Wagner LE II, Yule DI, Wojcikiewicz RJH. 2019. Bok regulates mitochondrial fusion and morphology. Cell Death Differ. 10.1038/s41418-019-0327-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SS, Lee WK, Aluvila S, Oh KJ, Yu YG. 2014. Identification of inhibitors against BAK pore formation using an improved in vitro assay system. Bull Korean Chem Soc 35: 419–424. 10.5012/bkcs.2014.35.2.419 [DOI] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. 2013. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19: 202–208. 10.1038/nm.3048 [DOI] [PubMed] [Google Scholar]
- Stehle D, Grimm M, Einsele-Scholz S, Ladwig F, Johänning J, Fischer G, Gillissen B, Schulze-Osthoff K, Essmann F. 2018. Contribution of BH3-domain and transmembrane-domain to the activity and interaction of the pore-forming Bcl-2 proteins Bok, Bak, and Bax. Sci Rep 8: 12434 10.1038/s41598-018-30603-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung TC, Li CY, Lai YC, Hung CL, Shih O, Yeh YQ, Jeng US, Chiang YW. 2015. Solution structure of apoptotic BAX oligomer: oligomerization likely precedes membrane insertion. Structure 23: 1878–1888. 10.1016/j.str.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. 2000. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell 103: 645–654. 10.1016/S0092-8674(00)00167-7 [DOI] [PubMed] [Google Scholar]
- Terrones O, Antonsson B, Yamaguchi H, Wang HG, Liu J, Lee RM, Herrmann A, Basañez G. 2004. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J Biol Chem 279: 30081–30091. 10.1074/jbc.M313420200 [DOI] [PubMed] [Google Scholar]
- Todt F, Cakir Z, Reichenbach F, Youle RJ, Edlich F. 2013. The C-terminal helix of Bcl-xL mediates Bax retrotranslocation from the mitochondria. Cell Death Differ 20: 333–342. 10.1038/cdd.2012.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todt F, Cakir Z, Reichenbach F, Emschermann F, Lauterwasser J, Kaiser A, Ichim G, Tait SW, Frank S, Langer HF, et al. 2015. Differential retrotranslocation of mitochondrial Bax and Bak. EMBO J 34: 67–80. 10.15252/embj.201488806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. 2008. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68: 3421–3428. 10.1158/0008-5472.CAN-07-5836 [DOI] [PubMed] [Google Scholar]
- Uren RT, O'Hely M, Iyer S, Bartolo R, Shi MX, Brouwer JM, Alsop AE, Dewson G, Kluck RM. 2017. Disordered clusters of Bak dimers rupture mitochondria during apoptosis. eLife 6: e19944 10.7554/eLife.19944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. 2006. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell 24: 199–210. 10.1016/j.molcel.2006.08.020 [DOI] [PubMed] [Google Scholar]
- Wang K, Gross A, Waksman G, Korsmeyer SJ. 1998. Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing. Mol Cell Biol 18: 6083–6089. 10.1128/MCB.18.10.6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun SY, Khuri F, Curran WJ, Deng X. 2010. Mono- or double-site phosphorylation distinctly regulates the proapoptotic function of Bax. PLoS ONE 5: e13393 10.1371/journal.pone.0013393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev 14: 2060–2071. [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, et al. 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159: 1549–1562. 10.1016/j.cell.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. 2005. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev 19: 1294–1305. 10.1101/gad.1304105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Deng X. 2005. Nicotine inactivation of the proapoptotic function of Bax through phosphorylation. J Biol Chem 280: 10781–10789. 10.1074/jbc.M500084200 [DOI] [PubMed] [Google Scholar]
- Xin M, Li R, Xie M, Park D, Owonikoko TK, Sica GL, Corsino PE, Zhou J, Ding C, White MA, et al. 2014. Small-molecule Bax agonists for cancer therapy. Nat Commun 5: 4935 10.1038/ncomms5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu W, Lapolla SM, Miao Y, Shao Y, Falcone M, Boreham D, McFarlane N, Ding J, Johnson AE, et al. 2010. Bax forms an oligomer via separate, yet interdependent, surfaces. J Biol Chem 285: 17614–17627. 10.1074/jbc.M110.113456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Subramaniam S, Kale J, Liao C, Huang B, Brahmbhatt H, Condon SG, Lapolla SM, Hays FA, Ding J, et al. 2016. BH3-in-groove dimerization initiates and helix 9 dimerization expands Bax pore assembly in membranes. EMBO J 35: 208–236. 10.15252/embj.201591552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Zhu Y, Eno CO, Liu Y, Deleeuw L, Burlison JA, Chaires JB, Trent JO, Li C. 2014. Activation of the proapoptotic Bcl-2 protein Bax by a small molecule induces tumor cell apoptosis. Mol Cell Biol 34: 1198–1207. 10.1128/MCB.00996-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JH, Grace CR, Guibao CD, McNamara DE, Llambi F, Wang YM, Chen T, Moldoveanu T. 2018. Intrinsic instability of BOK enables membrane permeabilization in apoptosis. Cell Rep 23: 2083–2094.e6. 10.1016/j.celrep.2018.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]