In this study, Harr et al. use C. elegans to investigate the consequences of a missense mutation (Y45C) in lamin A (encoded by LMNA) found in the human Emery-Dreifuss muscular dystrophy (EDMD) syndrome. Using muscle-specific emerin Dam-ID and other in vivo approaches, the authors report that they were able to counteract the dominant muscle-specific defects provoked by LMNA mutation by the ablation of a lamin-associated H3K9me anchor, suggesting a novel therapeutic pathway for treating EDMD.
Keywords: laminopathy, CEC-4, lamin A, C. elegans, EDMD, nuclear organization, LADs
Abstract
Mutations in the nuclear structural protein lamin A produce rare, tissue-specific diseases called laminopathies. The introduction of a human Emery-Dreifuss muscular dystrophy (EDMD)-inducing mutation into the C. elegans lamin (LMN-Y59C), recapitulates many muscular dystrophy phenotypes, and correlates with hyper-sequestration of a heterochromatic array at the nuclear periphery in muscle cells. Using muscle-specific emerin Dam-ID in worms, we monitored the effects of the mutation on endogenous chromatin. An increased contact with the nuclear periphery along chromosome arms, and an enhanced release of chromosomal centers, coincided with the disease phenotypes of reduced locomotion and compromised sarcomere integrity. The coupling of the LMN-Y59C mutation with the ablation of CEC-4, a chromodomain protein that anchors H3K9-methylated chromatin at the nuclear envelope (NE), suppressed the muscle-associated disease phenotypes. Deletion of cec-4 also rescued LMN-Y59C-linked alterations in chromatin organization and some changes in transcription. Sequences that changed position in the LMN-Y59C mutant, are enriched for E2F (EFL-2)-binding sites, consistent with previous studies suggesting that altered Rb-E2F interaction with lamin A may contribute to muscle dysfunction. In summary, we were able to counteract the dominant muscle-specific defects provoked by LMNA mutation by the ablation of a lamin-associated H3K9me anchor, suggesting a novel therapeutic pathway for EDMD.
Laminopathies represent a large set of rare genetic diseases that lead to differentiated tissue dysfunction, arising from mutations in components of the nuclear lamina, the most frequent being lamin A (LMNA). Point mutations in LMNA, or in other lamin-associated proteins such as emerin (Worman and Bonne 2007), are linked to ∼15 distinct diseases, many of which lead to premature death by cardiac dysfunction, coupled with striated muscular dystrophy, peripheral neuropathy or, more rarely, lipodystrophy (Worman and Bonne 2007). The majority of the ∼450 mapped laminopathy mutations are autosomal dominant missense alleles of LMNA. However, because the mutations occur throughout the gene, without clustering in any one structural domain of the 70-kDa lamin, it has been difficult to link the pathologies to a specific function (Dittmer and Misteli 2011). As a result, there are to date no effective treatments or cures for laminopathic disease.
There are two major hypotheses put forth to explain these diseases, one that implicates structural defects in nuclear organization and a second that implicates lamin A directly in gene regulation (Camozzi et al. 2014; Osmanagic-Myers et al. 2015). These mechanisms are not mutually exclusive, but imply different roles for lamin A. The first argues that lamins, which create a semirigid structure adjacent to the inner nuclear membrane (INM), protect the genome from mechanical stress (Worman and Bonne 2007; Dittmer and Misteli 2011). It is proposed that the compromised scaffold alters chromosomal organization and indirectly compromises tissue function, particularly in muscle, which is subject to stretch and compression. Exactly how mechanical stress changes genome function is unclear. The second hypothesis argues that LMNA mutations themselves provoke changes in gene expression, for instance, by altering lamin A interaction with transcription factors that are needed for maintenance of tissue integrity (Lloyd et al. 2002; Mounkes et al. 2005). Evidence for the former rests on studies showing that lamin A contributes to nuclear shape and the spatial distribution of heterochromatin (Solovei et al. 2013; Cho et al. 2019), while the second is based on the observation that lamins bind a range of transcriptional regulators and/or histone variants (Vlcek and Foisner 2007). These latter interactions involve nucleoplasmic lamin A, as well as the peripheral lamin fibers. Very few experiments address rigorously the relevance of either hypothesis. Indeed, defining the molecular pathways that link lamin A to muscle pathology has been difficult due to the multiplicity of LMNA mutations, the complexity of its interactors, and the fact that mouse models are only moderately successful at recapitulating these diseases (Mounkes et al. 2005; Stewart et al. 2007).
Because laminopathies manifest tissue-specific defects that are often late onset (i.e., manifest in adolescence), the use of model organisms to elucidate disease mechanisms is needed. The nematode Caenorhabditis elegans has proven to be a propitious choice (Wiesel et al. 2008; Mattout et al. 2011; Bank and Gruenbaum 2011a), because worms have only one lamin (LMN-1) and lack adult stem cells in somatic tissues, helping us differentiate reduced muscle function from impaired regeneration. Moreover, we and others have amply demonstrated that condensed, transcriptionally silent heterochromatin is found adjacent to the INM in contact with the nuclear lamina in worms as in other metazoans (Ikegami et al. 2010; Meister et al. 2010b; Towbin et al. 2012). The peripheral tethering of chromatin depends, at least in part, on the interaction of heterochromatin-specific histone modifications with INM components (Harr et al. 2016; Cohen-Fix and Askjaer 2017). In C. elegans embryos this is mediated by CEC-4, a nuclear envelope (NE)-associated protein that has a chromodomain (CD) with 46% identity to the CD of human heterochromatin protein 1α (HP1α) (Gonzalez-Sandoval et al. 2015). CEC-4 specifically tethers nucleosomes bearing histone H3 Lys9 methylation (H3K9me). This mark is essential for peripheral heterochromatin tethering in embryonic nuclei (Towbin et al. 2012; Gonzalez-Sandoval et al. 2015). Intriguingly, after embryonic development a second anchoring pathway is induced, which can be disrupted in L1 stage larvae by ablation of the H3K36me reader, MRG-1, and the release of p300/CBP1 from active chromatin (Cabianca et al. 2019). In vertebrate species, the exact counterparts of these two anchoring pathways remain to be defined, yet two partially redundant pathways that are compromised by ablation of lamin A and lamin B receptor (LBR), respectively, are necessary to maintain heterochromatin at the INM in differentiated cells (Solovei et al. 2013). These also control the expression of genes in early myoblast differentiation (Favreau et al. 2004; Frock et al. 2006; Solovei et al. 2013). An unstructured protein that binds HP1α, called PRR14, may also be involved in both chromatin organization and myoblast differentiation (Poleshko et al. 2013).
We studied the laminopathic mutation (LMNA-Y45C), which results in a degenerative condition in striated muscle called Emery-Dreifuss muscular dystrophy (EDMD). The equivalent mutation was introduced into the worm lamin (LMN-1 Y59C; henceforth, LMN-Y59C), and was shown to compromise striated body muscle integrity in a dominant manner, leading to impaired body movement (Mattout et al. 2011). In both human and worm lamins, the substituted tyrosine is the d residue of one of the most highly conserved heptad repeats within the 1B coiled coil of lamin A/C (Strelkov et al. 2004). The mutant worm protein can still form dimers and filaments, but integrates less stably into the lamin meshwork, and shows more rapid turnover upon FRAP than wild-type LMN-1 (Wiesel et al. 2008). Many of the phenotypes of human EDMD are recapitulated in C. elegans expressing this mutant form in a fusion with GFP (GFP∷LMN Y59C) in the background of wild-type lamin (Mattout et al. 2011). In contrast, strains expressing the equivalent fusion with wild-type lamin (GFP∷LMN-1; henceforth, LMN-WT) were phenotypically indistinguishable from wild-type (N2) C. elegans (Mattout et al. 2011). In both cases the GFP fusion proteins constitute <20% of the total lamin complement throughout embryogenesis and adulthood (Wiesel et al. 2008; Mattout et al. 2011). Coincident with the muscle pathology, the GFP::LMN-Y59C fusion impaired the relocation of an integrated muscle-specific reporter away from the nuclear periphery in muscle cell nuclei (Mattout et al. 2011). This suggests that the mutant lamin hypersequesters or blocks release of this heterochromatic array. However, it was not clear whether this stemmed from a failure to activate the array-borne myo-3 promoter or from an enhanced chromatin–LMN interaction. A programmed cell type-specific release of active tissue-specific promoters from the lamina is documented not only for C. elegans (Meister et al. 2010b; Mattout et al. 2011), but also for tissue-specific genes in a range of mammalian cells (Kosak et al. 2002; Ragoczy et al. 2006; Reddy et al. 2008; Meister et al. 2010b).
Here we investigated the role of chromatin organization in EDMD-like muscle phenotypes using this powerful C. elegans disease model. We examined how the LMN-Y59C mutation modifies genome organization, gene expression, and muscle morphology. To understand whether the observed disease phenotypes are linked to chromatin misorganization, we coupled the gain-of-function Y59C lamin mutation with down-regulation of other INM proteins, most notably with ablation of the heterochromatin anchor, CEC-4. Indeed, the defects of the LMN-Y59C allele can be compensated by deleting cec-4, or ablating the ability of its chromodomain to tether H3K9me-marked chromatin at the INM (Gonzalez-Sandoval et al. 2015). The analysis of lem-2 or emerin null alleles suggests that the effects are specific for cec-4. Thus, we identified compensatory cross-talk between the nuclear lamina and a NE-associated CD protein, showing that the latter is able to counteract the EDMD-like defects in muscle integrity associated with a laminopathy mutation. We suggest that the altered association of chromatin with the NE is causal and not simply a consequence of the disease state.
Results
Mutant lamin alters endogenous genome organization
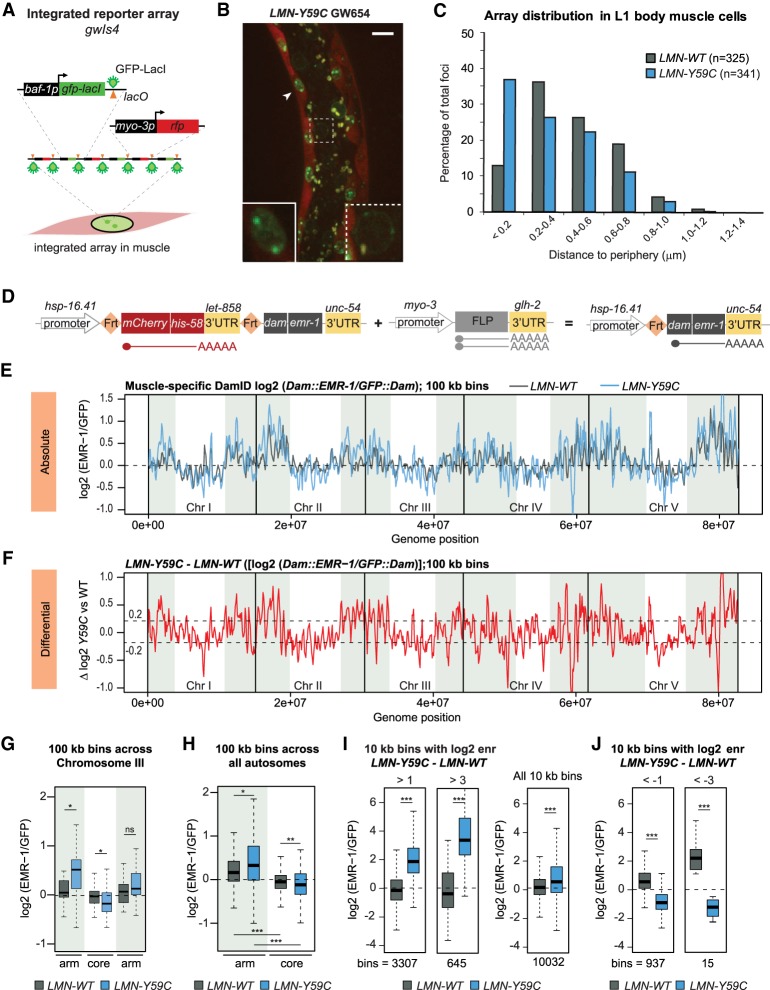
In a wild-type C. elegans background, the expression of an EDMD-linked mutant lamin (GFP::LMN-1 Y59C or LMN-Y59C) altered muscle ultrastructure and impaired body muscle function, coincident with the sequestration of a heterochromatic reporter array bearing a muscle-specific promoter, at the INM in muscle cells (see Fig. 1A–C; Mattout et al. 2011). The mislocalized array consists of ∼280 repeats of an insert that encodes GFP-LacI driven from a baf-1 promoter, and RFP under control of the muscle-specific myo-3 promoter (myo-3p) (Fig. 1A; Towbin et al. 2012). In wild-type muscle cells, with or without LMN-WT expression, the myo-3p-containing array shifts away from the nuclear periphery upon activation of the muscle-specific promoter (Fig. 1C; Meister et al. 2010b). Low-level expression of LMN-WT has no impact on this array behavior nor does it confer any discernible muscle defect (Mattout et al. 2011). In the LMN-Y59C-expressing strain, on the other hand, the array remains at the nuclear periphery in muscle cells despite myo-3p activation; hence, the term “hypersequestration.” We note that the level of GFP-LacI expressed from (and bound to) the reporter is far higher than the GFP signal of the lamin fusion proteins (Fig. 1B).
Figure 1.
Mutant lamin (LMN-Y59C) leads to alterations in genome organization. (A) Schematic of the array gwIs4 (Meister et al. 2010b). (B) Representative image of C. elegans L1 larvae expressing LMN-Y59C (GW654) with an integrated gwIs4 array. (Arrowhead) Muscle nucleus in inset; (dotted box) intestine nucleus with the gwIs4 array at NE. Low GFP∷LMN-Y59C expression marks the NE. Scale bar, 5 µm. (C) Quantitation of the gwIs4 focus distance from NE in muscle cells of LMN-WT (GW 656) versus Y59C-LMN (GW654). Shortest path from the gwIs4 focus center to NE is scored (n = foci scored). LMN-WT versus LMN-Y59C P < 0.0001, Chi-squared test (peripheral < 0.2 µm vs. central > 0.2 µm). (D) Single-copy transgene insertions for muscle-specific emerin DamID. (Left) Basal hsp-16.41p activity produces very low-level mCherry::his-58 transcript from an FRT-flanked stop cassette. (Right) In muscle cells, FLP recombinase expressed from myo-3p leads to stop cassette excision and low-level Dam::EMR-1 expression. (E) Line plot of log2 Dam∷EMR-1 normalized to GFP∷Dam in LMN-WT (black) and LMN-Y59C (blue) in L3/L4 C. elegans muscle. Signals are averaged over three sliding 100-kb windows. Chromosome arms (see the Materials and Methods) are shaded. The dotted line indicates zero log2 ratio. (F) Line plot of LMN-Y59C–LMN-WT (log2) showing differential EMR-Dam modification in LMN-Y59C versus LMN-WT. Dotted line indicates 0.2 and −0.2 thresholds. (G) Box plots comparing the frequency of contacts of Chr III (log2 [EMR-1/GFP]) in LMN-WT versus LMN-Y59C, averaged over three sliding 100-kb windows. (*) P < 0.01; (**) P < 0.0001; (***) P < 10−10; (ns) nonsignificant using Wilcoxon rank sum tests. (H) As in G, but EMR-Dam contact frequency across all autosomal arms and cores is shown (log2 [EMR-1/GFP]) in LMN-WT and LMN-Y59C, averaged over three sliding 100-kb windows. (I) Box plots comparing the frequency of contacts (log2[EMR-1/GFP]) for 10-kb bins that are enriched (enr) over a threshold of either >1 or >3 for the difference log2(LMN-Y59C)–log2(LMN-WT). (J) As I but comparing the bins for which the values log2(EMR-1/GFP) are below the threshold of −1 or −3 for the difference log2(LMN-Y59C)–log2(LMN-WT).
While the LMN-Y59C mutant gain-of-function sequestration of the myo-3p reporter was intriguing, it was important to determine how LMN-Y59C expression affects the spatial distribution of endogenous chromatin. To test this we performed muscle-specific genome-wide emerin-DamID, a method in which the muscle-specific expression of an INM protein (emerin or EMR-1) fused to the E. coli DNA adenine methyltransferase (Dam) enables mapping of chromatin at the INM of muscle nuclei (Gómez-Saldivar et al. 2016; Muñoz-Jiménez et al. 2017). We expressed Dam∷EMR-1 in a muscle-specific manner thanks to a FLP-FRT system driven by muscle-specific FLP expression (Fig. 1D; Muñoz-Jiménez et al. 2017). This resulted in the preferred methylation of adenines within GATC motifs in chromatin adjacent to the INM in muscle cells only. The use of GmeATC-specific restriction enzymes and ligation-mediated amplification, generated signals that were then normalized to the level of modification upon expression of GFP::Dam alone. The regions enriched for the resulting EMR-DamID signal are equivalent to lamina-associated domains (LADs) mapped by LEM-2 chromatin IP or LMN-1-DamID (González-Aguilera et al. 2014; Gonzalez-Sandoval et al. 2015).
L3/L4 stage larvae expressing LMN-WT or LMN-Y59C and muscle-specific Dam∷EMR-1 were lysed and the meA-bearing DNA was digested, sequenced, and normalized to DNA from similarly staged larvae that express nuclear GFP∷Dam. We plotted the relative enrichment of perinuclear chromatin (log2 [Dam∷EMR-1/GFP∷Dam] referred to as log2 [EMR-1/GFP]) (González-Aguilera et al. 2014). Figure 1E shows normalized enrichments between the LMN-Y59C-expressing strain (blue line) and an isogenic strain expressing the wild-type lamin fusion LMN-WT (black line). Importantly, neither endogenous emerin nor an EMR-1::mCherry fusion showed an altered localization in the LMN-Y59C strain (see Supplemental Fig. S1A; Wiesel et al. 2008). Additionally, endogenous levels of lmn-1, emr-1 and lem-2 are similar in wild-type and the mutant strains used (Supplemental Fig. S1B).
In the LMN-WT muscle cells, the heterochromatin-rich and gene-poor chromosome arms were more closely associated with the INM, while the gene-rich chromosome cores were more central, although the dynamic range of bound and unbound sequences was lower in late larval muscle cells, than in the nuclei of embryos or L1 intestine (cf. Fig. 1E with Gonzalez-Sandoval et al. 2015; Cabianca et al. 2019). This likely reflects active muscle-specific genes found on chromosome arms and technical limitations arising from the nonspherical shape of muscle nuclei. Nonetheless, even the chromosome with the “flattest” pattern, Chr III, showed significantly higher Dam-EMR signal on the chromosome arms than on its core; this trend held up upon averaging across the triplicate DamID assays, for all autosomes (Fig. 1G,H). This is consistent with earlier reports of worm chromosome architecture scored by lamin-DamID and LEM-2 ChIP in whole embryos (Ikegami et al. 2010; Towbin et al. 2012; González-Aguilera et al. 2014; Gonzalez-Sandoval et al. 2015), although the difference between chromosome arms and cores is less pronounced.
In mutant L3/L4 larvae expressing LMN-Y59C we observed subtle but reproducible changes in muscle cell LADs (Fig. 1E,F). When scored by 100-kb bins, 28.5% showed a shift towards the NE (higher [EMR-1/GFP] values), while 20.7% shifted inward (Supplemental Fig. S1C). Intriguingly, 90.2% of the sequences that gained perinuclear signal were found on chromosome arms (distal ∼5 Mb); while 58.4% of the bins shifting inwards map to gene-rich chromosomal cores (Supplemental Fig. S1C,D). The average change across an autosome (Chr III) split into right, left, and core domains, is shown in Figure 1G. In brief, the LMN-Y59C mutation tended to enhance NE-association of sequences on arms and to reduce it for chromosomal cores.
To quantify the changes in peripheral binding sites, we next examined 10-kb domains or “bins,” as they are more likely to reflect the behavior of individual genes and promoters. We grouped all 10-kb bins from both mutant LMN-Y59C and LMN-WT samples that showed a log2 (EMR-1/GFP) ratio > 3 for LMN-Y59C–LMN-WT (645 bins), and asked whether they were already perinuclear in LMN-WT-expressing worms. This was not the case (Fig. 1I), arguing that the LMN-Y59C mutant induces the formation of LADs de novo, rather than simply reinforcing existing contacts. A similar conclusion was drawn for a larger selection of fragments that shift to the periphery with log2 ratios > 1 for LMN-Y59C–LMN-WT (n = 3307). Intriguingly, although the sequences that shifted towards the periphery in the mutant were slightly more enriched in repetitive DNA elements (consistent with their localization along chromosomal arms; Liu et al. 2011), they did not show a striking enrichment for the histone modification H3K9me2 or H3K9me3 based on whole L3 larval ChIP-seq (Supplemental Fig. S1E,F). Finally, we identified sequences that shifted away from the NE in the LMN-Y59C mutant (log2 [EMR-1/GFP] < −1 or −3). For this less-abundant subset, primarily found in chromosomal cores, the LMN-Y59C mutation appears to have the opposite effect; i.e., it counteracted peripheral anchoring (Fig. 1J). Still, none showed significant enrichment for repeats or H3K9me2/3 (Supplemental Fig. S1E,F).
To understand what the newly acquired LADs (log2 [EMR-1/GFP] >1 for LMN-Y59C–LMN-WT) had in common, we screened them for abundant transcription factor motifs. Two of the most highly enriched consensus motifs are recognized by the transcription factor EFL-2, one of two worm E2F homologs, EFL-1 and EFL-2, both of which are bound and inhibited by the retinoblastoma-like pocket protein Rb (LIN-35 in C. elegans) (Supplemental Fig. S2). Intriguingly, the set of 10-kb bins that shifted inward were also enriched for EFL-2 binding sites (Supplemental Fig. S2). Most of EFL-1 is found in complex with DP-1 and the MuvB complex, which recognizes a larger bipartite motif that was not enriched in the LMN-Y59C-sensitive LADs (Supplemental Fig. S2; Narasimhan et al. 2015). It is not known whether EFL-2 is found in multiple complexes, but the fact that its consensus is enriched on sequences that shift position suggests that at least a subset of EFL-2-Rb complexes may be affected by the LMN-Y59C mutation. The presence of binding motifs for an Rb-binding E2F homolog is likely to be relevant, given earlier work showing that mammalian lamin A binds the hypophosphorylated form of Rb (Pekovic et al. 2007). Moreover, the repression of E2F by hypophosphorylated Rb is thought to be needed to arrest myoblast proliferation sufficiently to allow differentiation (Frock et al. 2006; Melcon et al. 2006; Pekovic et al. 2007; Elenbaas et al. 2018). In mammals, the interaction of Rb with lamin A and LAP2α was altered by certain lamin A/C mutants and by the loss of emerin or LAP2α (Markiewicz et al. 2002; Melcon et al. 2006; Pekovic et al. 2007).
Loss of CEC-4, but not LEM-2 or emerin, derepresses a heterochromatic reporter
The nuclear lamina is a collection of lamin- and INM-interacting proteins that form a meshwork of proteins underlying the nucleoplasmic side of the inner nuclear membrane. Some lamin components, such as LAP2α and lamin A in mammals, are also found in the nuclear interior, to varying degrees. C. elegans has most of the major nuclear lamina components found in mammals—i.e., lamin, emerin (EMR-1), SUN domain proteins, and the LEMD2/MAN1 homolog LEM-2 (Fig. 2A)—but it lacks two mammalian components that were previously implicated in muscle differentiation (LAP2α and the lamin B receptor or LBR). On the other hand, worms have a highly specific, NE-associated H3K9 methylation anchor called CEC-4 (Gonzalez-Sandoval et al. 2015).
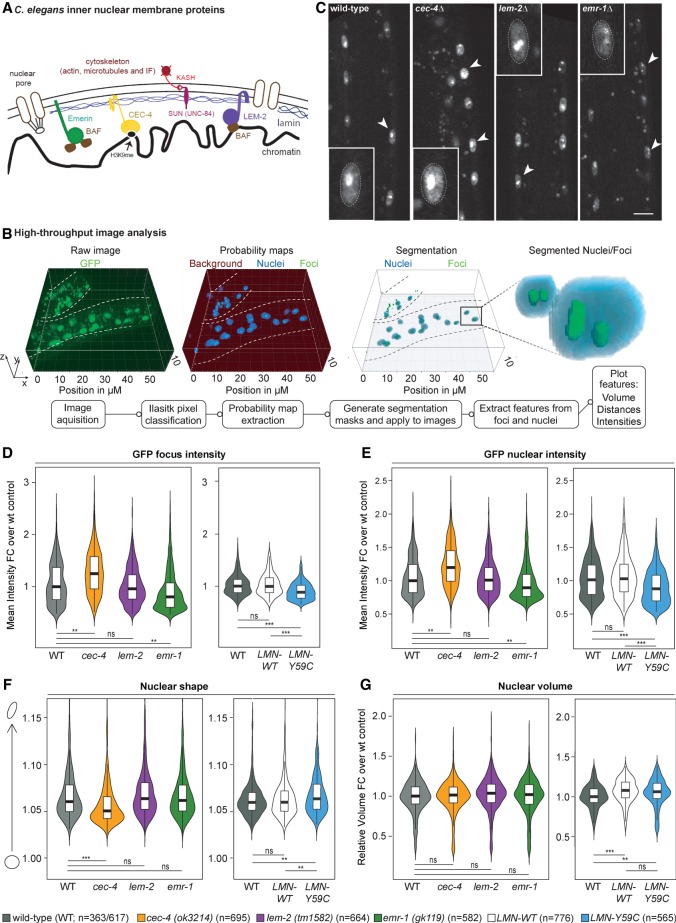
Figure 2.
INM proteins EMR-1, LEM-2, and CEC-4 have distinct effects on array expression and nuclear shape. (A) Sketch of the C. elegans INM components (Bank and Gruenbaum 2011a; Cohen-Fix and Askjaer 2017). (B) Summary of automated image segmentation and analysis used in D–G. (C) Examples of confocal fluorescence imaging of C. elegans L1 larvae [wild type; gwIs4 (GW76), cec-4Δ; gwIs4 (GW833), lem-2Δ; gwIs4 (GW201), and emr-1Δ; gwIs4 (GW164)]. Arrowheads indicate typical muscle nuclei, some being enlarged to show intensity and position of the gwIs4 array. Scale bar, 5 µm. (D–G) Violin plots resulting from the automated analysis monitoring GFP-LacI foci intensity (D), total nuclear GFP-LacI intensity (E), muscle nuclear shape (F), or volume (G). Strains used are indicated below the box plots; the mutants are null alleles, except for strains expressing LMN-WT (GW656) or LMN-Y59C (GW654) from integrated arrays. These are included and compared with GW76, which carries gwIs4 only. cec-4 (ok3214) is referred to as cec-4Δ in the text. (**) P < 0.0001; (***) P < 10−8; (ns) nonsignificant, versus wild type, Tukey post-hoc tests.
Given that mutations in either emerin or lamin A can generate EDMD in humans, we asked whether the loss of other INM components of C. elegans mimic the nuclear changes observed with the LMN-Y59C allele in muscle cell nuclei. Using a standardized, high-throughput image processing pipeline that segments and analyzes confocal 3D fluorescence imaging data in an automated fashion (Fig. 2B), we examined the effects of EMR-1, LEM-2, or CEC-4 ablation, compared with the low-level expression of LMN-WT or LMN-Y59C, by monitoring muscle cell nuclei in the appropriate L1 larvae. We scored nuclear size, shape, and the expression of GFP-LacI from the gwIs4 reporter (Fig. 2C–G). Under none of the conditions tested [the presence of mutant lamin (LMN-Y59C) or null alleles cec-4 (ok3214), lem-2 (tm1592), or emr-1 (gk119)] was nuclear volume significantly altered (Fig. 2G). On the other hand, cec-4 (ok3124) (hereafter cec-4Δ) led to an increase in the intensity of GFP-LacI at the array focus and throughout the nucleus (Fig. 2D,E), indicative of array derepression. There was no significant change in GFP fluorescence seen in lem-2 (tm1592) worms, while the ablation of emerin [emr-1 (gk119)] had a repressive effect, even stronger than LMN-Y59C. We could exclude that the phenotypes arise from altered expression of the genes encoding these INM proteins (Supplemental Fig. S1B). In addition to GFP up-regulation, cec-4Δ altered the typical oblong shape of muscle nuclei, making them more spherical, while lem-2 (tm1592), emr-1 (gk119), and the expression of the mutant LMN-Y59C protein did not (Fig. 2F). LMN-WT expression in all cases resembled wild-type (N2) worms. In summary, whereas emerin ablation yielded muscle nuclear defects reminiscent of LMN-Y59C, cec-4Δ had the opposite effect.
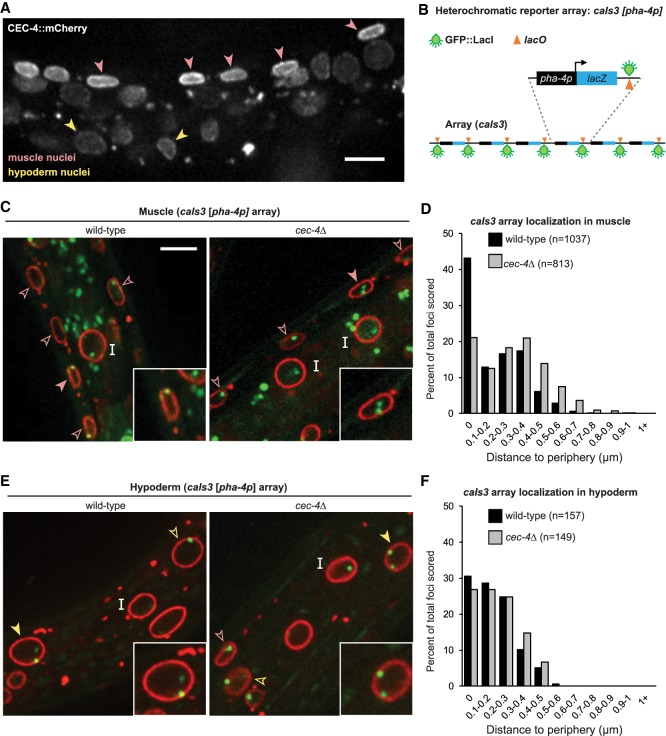
CEC-4's role in heterochromatin localization was first identified and characterized in C. elegans embryos, although the protein is present throughout development (Gonzalez-Sandoval et al. 2015). Indeed, in L1 larvae CEC-4 is more abundant in muscle cells than in other tissues (Fig. 3A; Gonzalez-Sandoval et al. 2015). Given its abundance and impact on muscle nuclear morphology (Fig. 2F), we examined whether CEC-4 plays a role in sequestering heterochromatic arrays at the nuclear periphery in muscle. To do this, we monitored the position of the heterochromatic array caIs3, which contains the promoter pha-4p, which is silent in both muscle and hypoderm (Fig. 3B). We scored the position of the GFP-LacI-tagged cals3 reporter in the nuclei of differentiated tissues of both wild-type and cec-4Δ larvae, relative to emerin-mCherry, a marker for the INM. In muscle cells, the cec-4Δ mutation compromised the peripheral enrichment of the array (80% of 813 foci in wild type, vs. 57% of 1037 in cec-4Δ) (Fig. 3C,D), demonstrating a role for CEC-4 in heterochromatin tethering beyond embryogenesis. This is consistent with effects monitored for cec-4Δ in the peripheral anchoring of endogenous sequences in L1 intestinal cells, although silent arrays in these cells remained peripheral (Cabianca et al. 2019). The silent caIs3 array in hypodermal nuclei behaved similarly: The arrays remained at the NE of cec-4Δ larvae (Fig. 3E,F; Supplemental Fig. S3A,B). In conclusion, CEC-4 has exceptionally high levels in differentiated muscle cell nuclei and is active in heterochromatin retention at the NE in muscle, where it also contributes to array repression and the maintenance of nuclear shape.
Figure 3.
CEC-4 contributes to heterochromatin perinuclear sequestration in muscle. (A) Fluorescence microscopy of a C. elegans L1 larvae expressing CEC-4∷mCherry capturing both muscle (pink arrow) and hypoderm (yellow arrow) nuclei. Average mCherry signal intensity = 2.1 arbitrary units (a.u.) for muscle, 1.5 a.u. for intestine, and 1.9 a.u. for hypoderm (Gonzalez-Sandoval et al. 2015). The larvae is twisted such that muscle nuclei appear closer together than usual. Scale bar, 5 µm. (B) Sketch of the caIs3 reporter integrated in ∼200 copies. This array expresses LacZ from the gut-specific pha-4 promoter and contains one lacO consensus per repeat. A second integrated array (gwIs39) expresses GFP∷LacI from baf-1p and lacks lacO. (C,E) Fluorescence confocal images of C. elegans L1 larvae expressing the pha-4 reporter (cals3; green) in either wild-type (GW1394) or cec-4 (ok3124; cec-4Δ; GW1395) worms expressing EMR-1∷mCherry (red). Arrowheads indicate muscle cells (C) and hypoderm cells (E) and “I” indicates intestine cell nuclei. Filled arrowheads mark nuclei in insets. Scale bar, 5 µm. (D) Quantitation of subnuclear positioning of the cals3 array in muscle cells of wild-type or cec-4Δ larvae from at least two biological replicates. Wild type versus cec-4Δ P < 0.0001, χ2 (peripheral < 0.2 µm vs. central > 0.2 µm). (F) As in D but for hypoderm cells. P-value wild type versus cec-4Δ is not significant.
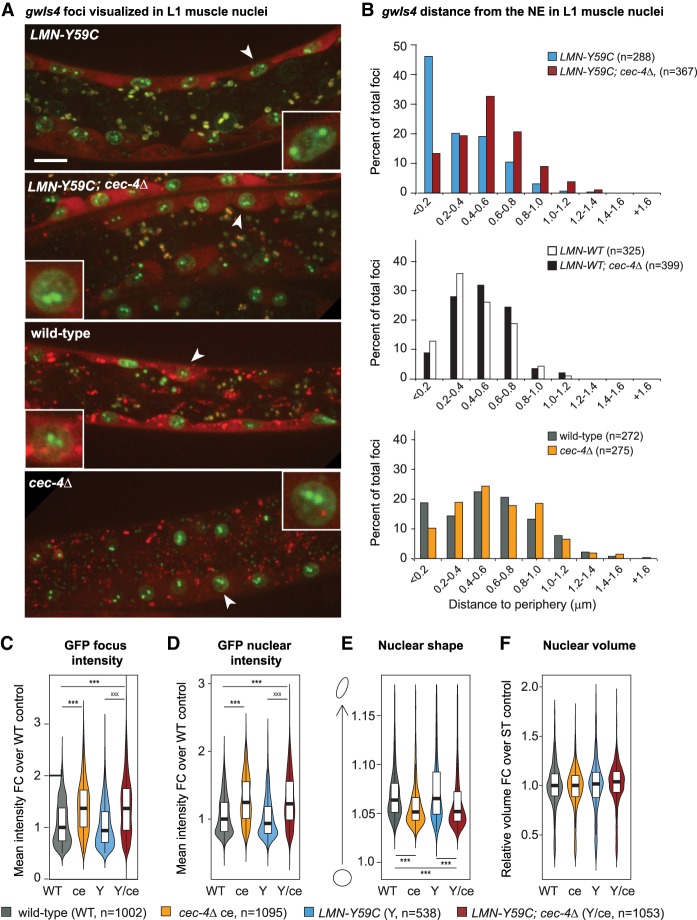
Given that the effects of cec-4Δ on the reporter in muscle were often the opposite of those scored in LMN-Y59C expressing worms, we examined the effect of combining the two mutations in one strain (Fig. 4). Remarkably, in the double mutant, the LMN-Y59C-mediated gwIs4 array sequestration at the NE was reversed: The deletion of cec-4 restored an internal localization for the active myo-3p-containing array, as was seen in N2 and/or LMN-WT muscle nuclei (Fig. 4A,B). In LMN-WT or N2 backgrounds, since in both cases the gwIs4 array was already internal, cec-4Δ had very minor effects on array localization (Fig. 4B). On the other hand, cec-4Δ released the heterochromatic from the NE in early embryos expressing LMN-Y59C, as it did in wild-type embryonic nuclei (Supplemental Fig. S3C,D). We subjected the double mutant to the high-throughput analysis of GFP intensity, nuclear volume, and nuclear shape, as carried out in Figure 2, and found that the coupling of cec-4Δ with LMN-Y59C enhanced the GFP-LacI signal on the array and throughout the nucleus (Fig. 4C,D). Moreover, muscle nuclear shape became more spherical, although nuclear volume was unchanged (Fig. 4E,F). Again, we could rule out indirect changes due to lamin misregulation in the single or double mutant (Supplemental Fig. S1A,B). In summary, cec-4Δ suppressed or counteracted the hypersequestration of a muscle-specific promoter-carrying array (myo-3p on gwIs4) in muscle nuclei, provoked by the gain-of-function LMN-Y59C mutation.
Figure 4.
Altered array positioning in the LMN-Y59C mutant is reversed by cec-4Δ. (A) Confocal fluorescence microscopy of C. elegans muscle nuclei bearing the muscle-specific gwIs4 array and expressing LMN-Y59C in wild-type or cec-4Δ backgrounds, as indicated [LMN-Y59C (GW654), LMN-Y59C; cec-4Δ (GW888), wild type (GW76) and cec-4Δ (GW833)]. Image of LMN-Y59C worm is the same as in Figure 1. Arrowheads indicate muscle nuclei enlarged to show array position. Scale bar, 5 µm. (B) Quantitation of subnuclear position of gwIs4 in muscle cells of indicated genotypes from at least two biological replicates. Distances measured are shortest distance from focus to the NE. (n) Number of nuclei scored. LMN-Y59C versus LMN-Y59C; cec-4Δ, P < 0.0001; LMN-WT versus LMN-WT; cec-4Δ, P > 0.01; WT versus cec-4Δ, P = 0.004, by χ2 test (peripheral < 0.2 µm vs. central > 0.2 µm). (C–F) Violin plots for gwIs4 focus GFP-LacI intensity. (WT) Wild type; (ce) cec-4Δ; (Y) LMN-Y59C; and (Y/ce) LMN-Y59C; cec-4Δ. (C) GFP focus intensity. (D) Muscle nuclear GFP intensity. (E) Muscle nuclear shape/roundness. (F) Muscle nuclear volume. (n) Number of nuclei counted for each strain. P-values were calculated and are indicated as in Figure 2, D–G
Loss of cec-4 specifically reverses the Y59C mutant defects in mobility and muscle structure
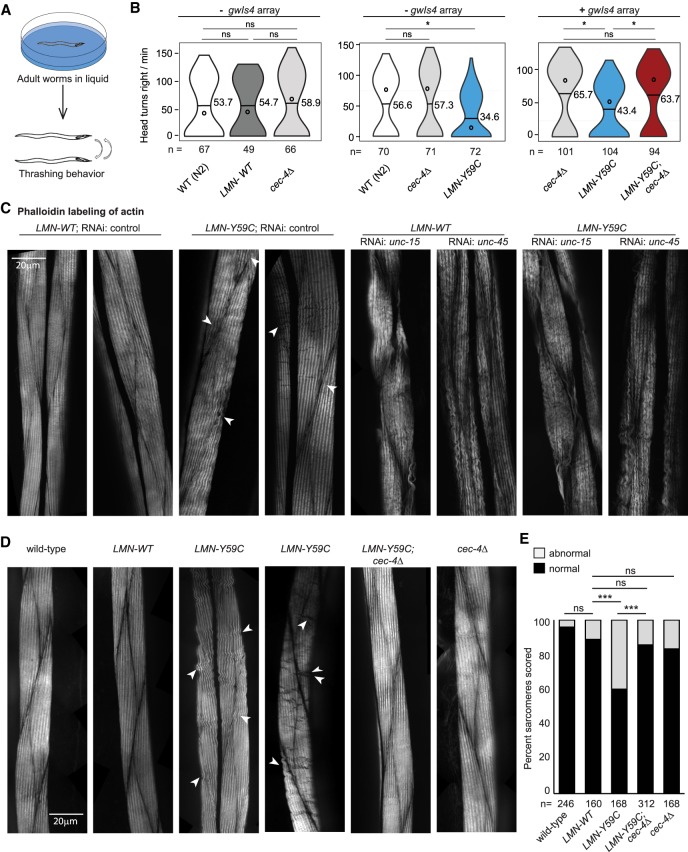
Given its suppression of the LMN-Y59C-induced array phenotype, we next asked whether cec-4Δ suppresses defective muscle function as well. To this end, we examined head thrashing and muscle structure in C. elegans, in both single- and double-mutant backgrounds. Previous work showed that the ectopic expression of LMN-Y59C altered actin filament organization and compromised mobility, which is monitored by the frequency of rapid head movements to the right when adult worms are placed in liquid (Fig. 5A; Mattout et al. 2011). We tested L4 larvae from wild-type, cec-4Δ, LMN-WT, LMN-Y59C, and LMN-Y59C;cec-4Δ double-mutant strains, scoring head bend frequency during 30 sec. Populations from at least six independent assays were quantified, and to minimize variability, only three genotypes were measured in parallel. In addition, strains with and without the gwIs4 array were monitored. We found no significant difference between wild-type, LMN-WT, and cec-4Δ worms (mean head bend frequency ranged from 53.7 to 58.9 in different assays, [P > 0.5]); however, we scored a significant reduction of head bend frequency in the mutant LMN-Y59C (34.6; P < 0.01 as compared with wild type) (Fig. 5B). Importantly, combining cec-4Δ with LMN-Y59C restored rates of head-thrashing equivalent to either wild type or the cec-4Δ single mutant (Fig. 5B).
Figure 5.
Motility and actin filament organization disrupted by LMN-Y59C can be rescued by cec-4Δ. (A) Sketch of the head thrashing (swimming) assay using L4 larval stage worms (see Materials and Methods). (B) Left, individual C. elegans L4 larva of wild-type (N2), LMN-WT (GW656), and cec-4Δ (GW828) genotypes were transferred to liquid, and head bends at the right were counted for 30 sec. (Horizontal bar) Mean; (open circle) median. Data are from at least three independent assays. Grouped genotypes were quantified in parallel. Middle graph as left, with strains wild type (N2), cec-4Δ (GW828), and LMN-Y59C (GW653). None of these strains carry gwIs4. (Right) Swimming assays on strains cec-4Δ (GW833), LMN-Y59C (GW654), and LMN-Y59C; cec-4Δ (GW888), which carry the gwIs4 array. (*) P < 0.01; (ns) nonsignificant by Student's t-test. (C) Representative images of phalloidin–rhodamine staining in L4/young adult C. elegans, illustrating normal and abnormal phenotypes. RNAi for control, unc-15, or unc-45 was done in either a LMN-WT (GW656) or LMN-Y59C (GW653) background. Arrowheads indicate mild abnormal phenotypes in mutant lamin strains. Scale bar, 20 µm. (D) Representative images of phalloidin-rhodamine staining in L4/young adult C. elegans from wild type (N2), cec-4Δ (GW828), LMN-WT (GW655), LMN-Y59C (GW653), and LMN-Y59C; cec-4Δ (GW1468). Arrowheads and bar are as above. (E) Quantitation of actin organization in muscle cells of indicated strains from three or more biological replicas. (***) P < 0.0001; (ns) nonsignificant (P > 0.01), by χ2 tests.
Restoration of muscle function by cec-4Δ correlates with restored sarcomere structure
To link the defects in body muscle function in the single and double mutants (LMN-Y59C vs. LMN-Y59C; cec-4Δ) to muscle anatomy, we used fluorescence imaging to monitor both actin and myosin organization in the sarcomeres of young adult C. elegans. Sarcomeres consist of a bundle of thick filaments (myosin) and thin filaments (actin) that are interdigitated. Using young adults, we could easily score changes in actin organization of muscle cells by tracking high-resolution images of phalloidin–rhodamine-stained striated muscle (Fig. 5C–E). We scored actin organization in each sarcomere as normal or abnormal, with the latter presenting disordered filaments (waves, gaps, and fusions in the actin filaments, Fig. 5C). To convince ourselves that these reflect sarcomere pathologies and not artefacts, we compared phalloidin staining of wild-type and LMN-WT or LMN-Y59C-expressing worms with the same structures stained in N2 worms treated with RNAi against unc-15 or unc-45 (Waterston et al. 1977; Venolia and Waterston 1990). These genes are involved in the enervation of muscle and lead to worms that cannot move (Waterston et al. 1977; Venolia and Waterston 1990). Whereas the phalloidin staining of wild type and LMN-WT (with and without control RNAi) revealed uniform and smooth actin filaments, the treatment with unc-15 or unc-45 RNAi showed extensive waves, ripples, breaks, and irregularities in the actin-myosin filaments (Fig. 5C). The LMN-Y59C worms (with and without control RNAi) showed a much less pronounced, but reproducible, presence of similar fractures, waves, and irregularities within sarcomeres and at sarcomere junctions (Fig. 5C,D). The abnormalities were subtle but consistently observed in LMN-Y59C, unlike LMN-WT and N2 worms. We quantified the frequency of aberrant sarcomeres, comparing wild-type, cec-4Δ, LMN-Y59C, and the LMN-Y59C; cec-4Δ double-mutant strains. The percentage of sarcomeres containing abnormal actin organization increased robustly only in the LMN-Y59C strain (P < 0.001; Fig. 5E), and by combining cec-4Δ with LMN-Y59C we found the higher frequency of waves, ripples, and breaks (Fig. 5E), as well as the defects scored at sarcomere junctions (Supplemental Fig. S4A,B, arrowheads), to be suppressed. To ensure that this phenomenon was not influenced by the fixation required for phalloidin staining, we monitored sarcomere structure by a GFP-tagged muscle-specific myosin expressed in the same mutants. Live imaging of young adult C. elegans expressing GFP::MYO-3 showed abnormal myosin organization, particularly at sarcomere junctions, and uniquely in muscle of the LMN-Y59C worms (Supplemental Fig. S4C,D). Once again, the defect was suppressed by the deletion of cec-4.
As shown above, the LMN-Y59C and cec-4 mutant effects are most pronounced in muscle and may be cell type-specific (cf. Fig. 3). To see whether the removal of another INM protein would confer or suppress these EDMD-like phenotypes, we examined the deletion of lem-2 (human LEMD2 or MAN-1) in more detail, scoring for both functional and structural defects in muscle. Unlike cec-4, we found that lem-2 deficiency on its own reduced head-thrashing rates, very much like the Y59C-LMN mutant (mean head bends to the right: lem-2Δ = 37.5 vs. LMN-Y59C = 34.7) (Supplemental Fig. S4E; see also Barkan et al. 2012). Interestingly, the combination of lem-2 with LMN-Y59C showed epistasis, rather than suppression of the muscle defects (mean head bends 40.9, P > 0.5) (Supplemental Fig. S4E). Moreover, a detailed analysis of sarcomere and junction integrity by phalloidin staining showed no significant variance from wild type for either phenotype in lem-2 deficient worms (Supplemental Fig. S4F). We conclude that the LMN-Y59C-induced defect in sarcomere structure that can be reversed by cec-4Δ is not mimicked by lem-2Δ.
To see whether the cec-4Δ suppression stems from the loss of binding to the H3K9 methylation mark, we made use of a strain expressing a mutant form of CEC-4 in which the two crucial tyrosine residues of its chromodomain (Y87/Y111) have been replaced by alanines. These substitutions impair binding to the methylated histone H3K9 peptide in vitro, and are sufficient to release heterochromatin from the INM in embryos (Supplemental Fig. S5A; Gonzalez-Sandoval et al. 2015). The LMN-Y59C mutant in a cec-4-null background was complemented either with the Y87A/Y111A mutant form (cec-4(cd2YA)::mCherry) or with wild-type CEC-4 (cec-4::mCherry). The expression of the CD-deficient cec-4(cd2YA) resulted in a partial suppression of the LMN-Y59C phenotypes in the head-thrashing assay (Supplemental Fig. S5B), while the WT control (cec-4::mCherry) did not (Supplemental Fig. S5B). Suppression was also scored for the LMN-Y59C-induced sequestration of the gwIs4 array in muscle nuclei (Supplemental Fig. S5C,D, green bars). We conclude that loss of the CEC-4-H3K9me binding helps suppress the LMN-Y59C phenotypes, although another presently unknown function of CEC-4 may also contribute.
Altered genome organization in the LMN-Y59C mutant is rescued by loss of CEC-4
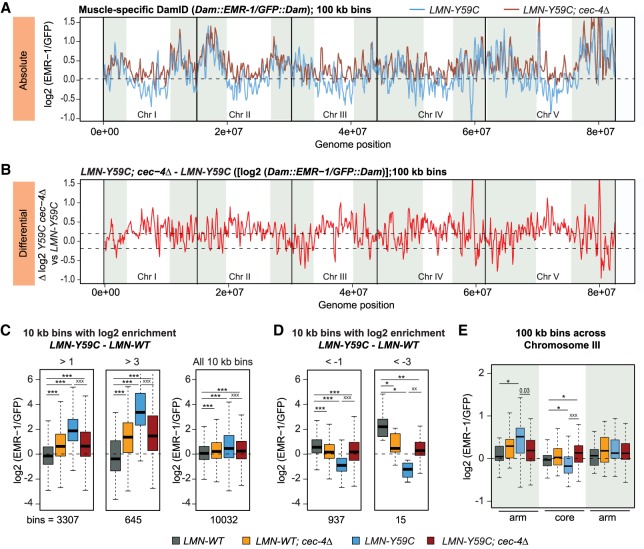
Given the ability of cec-4Δ to suppress the muscle defects of the LMN-Y59C mutant, we asked whether cec-4Δ acts by rescuing or suppressing the misorganization of genomic sequences that we scored by muscle-specific EMR-DamID in the LMN-Y59C mutant (Fig. 1). We performed the same EMR-DamID in worms expressing either LMN-Y59C or LMN-WT coupled with cec-4Δ. We confirmed that neither the expression nor the localization of emerin or lamin itself was altered by cec-4Δ (Fig. 3C,E; Supplemental Fig. S1A,B). Still, the combination of LMN-Y59C and cec-4Δ elevated the EMR-Dam signals in chromosomal cores, and did not only reduce LAD signals, as one might have expected from previous work in embryos (Fig. 6A,B; Gonzalez-Sandoval et al. 2015). Indeed, as many 100-kb bins shifted outwards as inwards upon cec-4 ablation in the LMN-Y59C background (Supplemental Fig. S6A). Using a low-level cutoff (log2 ratio < −0.2), 92.6% of the bins shifting inward in cec-4Δ were found on chromosome arms, while most 100-kb bins shifting toward the periphery map to chromosomal cores (Supplemental Fig. S6B–D). A similar trend was observed for cec-4Δ in the LMN-WT background (Supplemental Fig. S6B–D). At higher resolution (i.e., 10-kb bins), the cec-4Δ effect was even more pronounced. We focused on the 10-kb bins that were identified as having a strong shift either toward or away from the INM in the LMN-Y59C background compared with LMN-WT (log2 [EMR-1/GFP] ratio >1 or >3, or else <−1 or <−3 for LMN-Y59C–LMN-WT). For these LMN-Y59C-sensitive sequences, we observed a striking reversal of the LMN-Y59C-induced change upon cec-4 ablation (Fig. 6C,D). The bins showing enhanced EMR-contact in LMN-Y59C (blue) were systematically shifted inwards in the double mutant (dark red), restoring the position that was scored in cec-4Δ alone (Fig. 6C). The converse was also true (bins that shifted inward in the LMN-Y59C mutant, were restored to the INM by cec-4 ablation; Fig. 6D). This cec-4Δ rescue of aberrant chromatin architecture induced by LMN-Y59C was not dependent on the size of the sequence window used; the effect was also clear in 100-kb bins along the left (pairing) arm of chromosome III (Fig. 6E).
Figure 6.
LMN-Y59C-induced EMR-DamID phenotypes in muscle are suppressed by cec-4Δ. (A) Line plot of log2 of EMR-1-Dam normalized to GFP-Dam from LMN-Y59C (blue) and LMN-Y59C; cec-4Δ (dark red) C. elegans L3/L4 muscle cells. Signal is averaged over three sliding 100-kb windows. Chromosome arms are shaded (see the Materials and Methods); dotted line indicates zero log2 ratio. (B) Line plot of EMR-1/GFP (log2) of differences between LMN-Y59C; cec-4Δ versus LMN-Y59C. The dotted line indicates 0.2 and −0.2 log2 ratio thresholds. (C) Box plots comparing the frequency of contacts (log2 [EMR-1/GFP]) for 10-kb bins that are enriched for log2(LMN-Y59C)–log2(LMN-WT) over a threshold of either >1 or >3 in LMN-WT, LMN-WT; cec-4Δ, LMN-Y59C, and LMN-WT; cec-4Δ strains. (D) Same as in C but comparing the frequency of contacts (log2 [EMR-1/GFP]) of 10-kb bins depleted for log2(LMN-Y59C)–log2(LMN-WT) below a threshold of either <−1 or <−3 in LMN-WT and LMN-Y59C strains. (E) Box plot comparing the frequency of EMR-Dam contacts along chromosome III normalized to GFP-Dam in the indicated strains. Arm versus core designation and averaging as in A. Probability values from Wilcoxon rank sum tests are indicated: LMN-WT (Dam∷EMR-1/GFP∷Dam; GW1480/GW1481), LMN-WT; cec-4Δ (Dam∷EMR-1/GFP∷Dam; GW1482/GW1483), LMN-Y59C (Dam∷EMR-1/GFP∷Dam; GW1508/GW1485), and LMN-Y59C; cec-4Δ (Dam∷EMR-1/GFP∷Dam; GW1486/GW1487). (*) P < 0.01; (**) P < 0.0001; (***) P < 10−10, when compared with LMN-WT. Significance level compared with LMN-Y59C mutants (x), are also given.
Given the partial phenotype suppression by the CEC-4 Y87A/Y111A chromodomain mutant and the fact that chromosomal arms shifted more than the chromosomal cores (Supplemental Fig. S6A,B), we asked whether cec-4Δ preferentially acted on repetitive sequences or domains enriched for H3K9me2 or H3K9me3. We were able to score a marginal enrichment for repeat elements, but not for H3K9 methylation (Supplemental Fig. S6E,F). This may reflect limitations in sensitivity, because H3K9me was mapped in whole larvae (on mixed tissues), in which muscle is a minor fraction, while the EMR-DamID changes are muscle-specific. Nonetheless, it suggests that genes and promoters, rather than repeat elements per se, respond to LMN-Y59C and cec-4Δ.
Importantly, the effect of cec-4 ablation was more pronounced in the LMN-Y59C mutant than in the LMN-WT background: While 12.3% of sequence bins were released from the periphery in the LMN-WT; cec-4Δ double mutant, twice as many shifted in the LMN-Y59C; cec-4Δ strain (24.8%) (Supplemental Fig. S6A,B). Indeed, most chromatin showed no change in position in the LMN-WT background upon cec-4 deletion, and surprisingly, if position changed, more shifted toward than away from the INM. The opposite was true in the LMN-Y59C strain. This suggests that the aberrant positioning of endogenous chromatin triggered by LMN-Y59C might actually be mediated by CEC-4, given that cec-4Δ is able to reverse the effects.
Once again, we examined whether transcription factors were enriched on the promoters of genes that shifted position in the LMN-Y59C; cec-4Δ double mutant. We plotted the relative log2 (EMR-1/GFP) ratio of genes bearing E2F consensus motifs (either EFL-2 or EFL-1 and EFL-2), in LMN-Y59C strains with or without cec-4. E2F motif-containing genes that were peripheral in LMN-Y59C shifted to a more central position in cec-4Δ, and vice versa (Supplemental Fig. S7A). If we performed the analysis the other way around, and searched for transcription factor motif enrichment in the cec-4 sensitive sequences, we again identified the consensus for EFL-2, but not for EFL-1, as being strongly enriched (Supplemental Fig. S7B). This may indicate a special role for EFL-2 in nuclear organization in muscle. Of note, 44% of all muscle-expressed genes contain E2F motifs in their promoters (Supplemental Fig. S7C).
In summary, we found that EMR-DamID signals that change in the LMN-Y59C mutant are efficiently reverted upon cec-4 ablation (Fig. 6). We conclude that cec-4Δ compensates for gain-of-function effects of LMN-Y59C with respect to global genome architecture. The genes and promoters most significantly affected are enriched for the motif recognized by EFL-2, an Rb-binding E2F homolog, or else for a nuclear receptor transcription factor, NHR-66 (Supplemental Fig. S7B).
Altered transcription in a lamin mutant can be reversed by cec-4 deletion
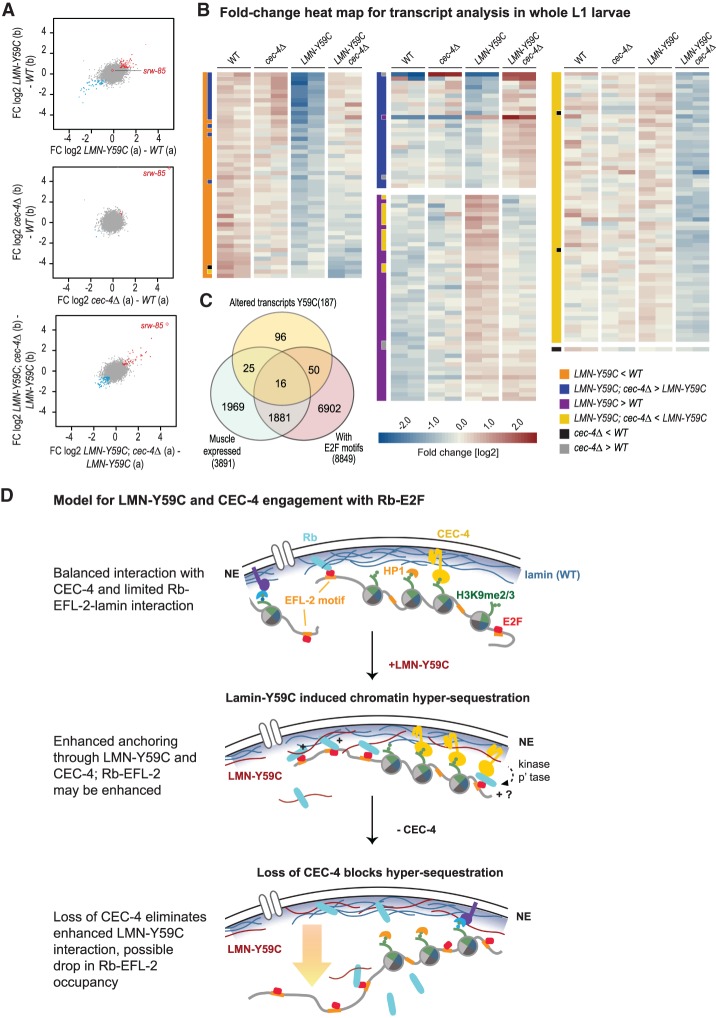
We next examined transcriptional changes in the LMN-Y59C worms and asked whether they were rescued by cec-4 deletion. We performed RNA-seq on whole worm L1 larvae in wild-type, cec-4Δ, LMN-Y59C and the LMN-Y59C; cec-4Δ double-mutant strains (Fig. 7), determining significant transcript level changes by edgeR with an FDR < 0.01 and FC > 1.5 (Materials and Methods). The transcriptional changes in LMN-Y59C and in the double mutant, while not dramatic, were reproducible between replicate sample pairs (Fig. 7A), allowing us to identify 187 genes with >1.5-fold change in transcript levels in each replica of the pairwise comparisons indicated in the heat map legend (Fig. 7B). There were not many transcript changes meeting this stringent criteria, 49 genes were up-regulated and 48 genes down-regulated >1.5-fold in the comparison of LMN-Y59C versus WT L1 larvae (Supplemental Tables S1–S4). Importantly, and consistent with a previous report on cec-4Δ embryos (Gonzalez-Sandoval et al. 2015), we found that the cec-4 ablation alone provoked very little change in steady state transcript abundance over WT L1 larvae (Fig. 7A), with the striking exception of a gene encoding a transmembrane G-coupled receptor, swr-85 (see also Gonzalez-Sandoval et al. 2015). Nonetheless, a large fraction of the transcripts whose levels were altered in the LMN-Y59C mutant now showed sensitivity to the deletion of cec-4, with mRNA levels shifting in the opposite direction from that scored in the LMN-Y59C mutant. That is, ∼30% of the genes shown in Figure 7B as transcripts that changed significantly upon LMN-Y59C expression showed compensatory effects in the LMN-Y59C; cec-4Δ double mutant. In some cases the LMN-Y59C led to down-regulation, and cec-4Δ raised expression levels; in others, LMN-Y59C induced expression that cec-4Δ then reversed.
Figure 7.
RNA-seq shows compensatory effects of cec-4Δ on LMN-Y59C-induced transcriptional changes. (A) Scatter plots visualizing the replicate concordance of log2 fold changes (FC) in differential transcript abundances contrasting the two replicas (shown in A,B). (Top) LMN-Y59C (GW653) compared with wild type (WT, N2). (Middle) cec-4Δ (GW828) compared with WT. (Bottom) LMN-Y59C; cec-4Δ (GW1468) compared with LMN-Y59C (GW653). (Blue) Differential genes (P < 0.01) down in both replica pairs [<−log2(1.5×)]; (red) differential genes (P < 0.01) up in both [> log2(1.5×)]. (Red open diamond) srw-85, a gene up-regulated in cec-4Δ embryos (Gonzalez-Sandoval et al. 2015). (B) Heat map visualizing relative transcript abundances in L1 whole larval RNA in two replicates of each strain (see A). Each row represents a gene identified as differential by edgeR (FDR < 0.01 and FC > 1.5 over the indicated comparison based on average values). The two-column bar at the left of each gene set indicates the comparisons used to define the given gene set. Each subset is sorted by the FC between “LMN-Y59C” over “LMN-Y59C; cec4Δ” (last four columns). The color scale in the heat map indicates the FC deviation from the gene-wise average transcript abundance over all samples. (C) Venn diagram of altered transcripts in mutant LMN-Y59C L1 larvae, muscle-expressed genes (Blazie et al. 2015), and genes with putative E2F sites (EFL-1 and EFL-2 pooled). (D) A model for LMN-Y59C and CEC-4 interaction with possible role for Rb-EFL-2. The top sketch indicates the INM components and chromatin components relevant for sequence positioning in differentiated muscle of C. elegans. In purple is a hypothesized unknown anchor. (Middle and bottom) The gain-of-function LMN-Y59C mutation confers enhanced Rb-EFL-2 and/or enhanced CEC-4 binding, and the loss of CEC-4 counteracts this. (p'tase) Phosphatase.
We examined the prominent LMN-Y59C-sensitive genes for functional pathways or relevant GO terms; 22% (41/187) of the LMN-Y59C-sensitive genes are muscle expressed, as defined by Blazie et al. (2015). This is not a strong enrichment, but given that RNA-seq was carried out in whole larvae, where muscle cells comprise only 10% of the somatic tissues, changes in low to moderately expressed muscle genes may fall below our limit of detection. There was no significant enrichment for ELF-2 motifs among the sequences or genes that change level in LMN-Y59C expressing larvae, although 27% of all ELF-2 motif-containing promoters are found upstream of muscle-expressed genes. If we broadened the range to include both EFL-1 and EFL-2 motifs, we found that 35% of the LMN-Y59C misregulated genes and 49% of muscle-expressed genes bear such motifs within the genomic range of −1.5 kb upstream of to +0.5 kb downstream from the annotated transcription start sites (Fig. 7C). Thus, even without a statistically compelling link between the LMN-Y59C induced changes in position and changes in expression, both assays identify EFL-2—or potentially an indirect effect on a complex containing EFL-1—as relevant downstream factors. Importantly, both E2F homologs bind Rb, a repressor that was shown to bind mammalian Lamin A.
Discussion
Lamin A is a highly conserved coiled-coil intermediate filament protein of the nuclear lamina meshwork found adjacent to the nuclear envelope. Lamins are present in all eukaryotes that have an open mitosis, for the presence of at least one lamin family member is necessary to ensure proper nuclear reassembly after telophase (Nigg 1992). DamID data implicate lamins in the distribution of genomic domains bearing specific heterochromatic marks, most notably histone H3K9me2/3 (Kind et al. 2013; Solovei et al. 2013), even though lamin itself does not have a domain that can selectively bind silent chromatin. Indeed, the molecular recognition of chromatin by the nuclear lamina is still poorly defined in vertebrates (Bank and Gruenbaum 2011b; Dittmer and Misteli 2011; Camozzi et al. 2014).
Compromised integrity of the nuclear lamina leads to tissue-specific pathologies in a range of human diseases called laminopathies, some with severe pathologies, such as EDMD (Worman and Bonne 2007). We previously showed that the expression of a mutant form of C. elegans lamin (LMN-Y59C) generates a number of EDMD phenotypes in worms, mimicking the human disease-linked gain-of-function mutation, LMNA-Y45C. Not only did LMN-Y59C-expressing worms show perturbed body muscle function and compromised muscle ultrastructure, but the mutant lamin interfered with the relocation of an activated muscle-specific promoter away from the INM to the nuclear interior in adult muscle cells (Mattout et al. 2011). In most laminopathy studies, links that are proposed to exist between chromatin misorganization and the physiological dysfunction of muscle have remained on the level of correlation (Bank and Gruenbaum 2011b; Camozzi et al. 2014; Perovanovic et al. 2016), as no clear mechanisms were identified that linked chromatin misorganization to the disease.
Here we showed by means of muscle-specific emerin-DamID that the worm mutation equivalent to the human LMNA Y45C mutation (LMN-Y59C) changes INM-chromatin interactions and genome architecture in C. elegans muscle nuclei. The LMN-Y59C effect is dominant, given that the mutant contributes at most 20% of the endogenous wild-type lamin complement, yet the phenotypes are reproducibly manifest. Whereas the vast majority (>90%) of the genome does not appear to shift position upon LMN-Y59C expression, we found that ∼4% of sequences genome-wide shift to the nuclear periphery, while ∼4% become more central. Most of the sequences that become peripheral are enriched on the heterochromatin-rich autosomal arms (Supplemental Fig. S6A,B), yet there is no statistical enrichment of H3K9 methylation on these sequences (Supplemental Fig. S1E,F). Therefore, we were prompted to look for transcription factor enrichment on the mislocalized chromatin. This revealed a selective enrichment for the binding consensus of the E2F homologue, EFL-2 (Supplemental Fig. S2A).
The E2F family of transcription factors is regulated by the formation of an inhibitory complex with Rb, a key tumor suppressor in mammals. As an activator, E2F triggers key cell cycle events, including apoptosis, and controls differentiation in certain tissues. This was particularly well-characterized for the C. elegans DREAM complex, which contains a E2F-DP heterodimer (EFL-1 and DPL-1), a retinoblastoma-like pocket protein (LIN-35), and a five-subunit MuvB complex (LIN-9, LIN-37, LIN-52, LIN-53, and a DNA-binding subunit, LIN-54) (Ceol and Horvitz 2001; Petrella et al. 2011; Kudron et al. 2013; Latorre et al. 2015; Goetsch et al. 2017). E2F-binding sites are found throughout the C. elegans genome, and the binding of the worm E2F homologs, EFL-1 or EFL-2, can lead to both up-regulation and down-regulation of gene expression, although the loss of E2F or Rb alone does not necessarily alter gene expression (Petrella et al. 2011; Latorre et al. 2015; Goetsch et al. 2017). In LMN-Y59C-affected sequences, we found a specific enrichment for a smaller motif, that of EFL-2, which lacks the additional LIN-54 recognition motif. Thus, we propose a role of EFL-2, and possibly the EFL-2/Rb complex, in muscle nuclear organization that is distinct from the activities of the DREAM complex (Ceol and Horvitz 2001). In emerin-deficient mice the misregulation of E2F by Rb has been implicated in muscle-related pathologies (Melcon et al. 2006), and lamin A mutants were shown to impair muscle differentiation in vitro in an Rb-dependent manner (Favreau et al. 2004; Markiewicz et al. 2005). While suggestive, these earlier studies did not explore links to chromatin organization, even though Rb had been reported to bind the nuclear lamina.
We show here that the changes in the spatial organization of the genome likely cause the muscle laminopathy linked to LMN-Y59C (Y45C in human LMNA), because we are able to counteract the LMN-Y59C-induced phenotypes by deleting the C. elegans heterochromatin binding protein, CEC-4. CEC-4 is a H3K9methyl-binding chromodomain protein that is associated with the nuclear lamina (Gonzalez-Sandoval et al. 2015). It also has a large disordered C-terminal domain of unknown function that is necessary for proper INM localization. Ablation of cec-4 releases both heterochromatic arrays and endogenous heterochromatin from the nuclear periphery in embryos (Gonzalez-Sandoval et al. 2015) and triggers reporter array expression in muscle cells (Fig. 2). CEC-4 contributes actively to the positioning of endogenous sequences in muscle cells (Fig. 6), and its ablation largely reverses the changes in chromatin distribution brought about by laminopathy mutant LMN-Y59C. Intriguingly, the binding motif of the E2F-homolog EFL-2, is enriched in the sequences that shift position in the LMN-Y59C mutant. With respect to gene expression, changes provoked by the LMN-Y59C mutation are, in some cases, reversed by cec-4Δ, arguing that CEC-4 is a downstream effector of the pathology-provoking lamin variant (Fig. 7A; Supplemental Fig. S7A). Whether CEC-4 interacts with, or modulates EFL-2 function remains to be explored.
There are at least two hypotheses that could explain these observations. There may be a gain (or, respectively, loss) of H3K9 methylation in the LMN-Y59C mutant that leads to chromatin relocation by altering interaction with CEC-4. Alternatively, CEC-4 may have an alternative function in the LMN-Y59C-expressing strain that interferes with the effect of the lamin mutation on gene or locus positioning (Fig. 7D). The fact that chromatin domains shift both toward and away from the INM in the LMN-Y59C mutant, and that cec-4 deletion compensates for both, argues that CEC-4 may have more than one mode of action. Nonetheless, there are roughly threefold as many highly significant shifts (difference log2 > 3) toward the nuclear periphery in the LMN-Y59C mutant as away from it (log2 < −3). Given that the loss of cec-4 suppresses not only aspects of chromatin positioning, but other muscle-relevant LMN-Y59C-induced defects, the double mutant provides a strong argument that the muscle pathology correlated with LMN-Y59C stems from, and not only correlates with, altered patterns of chromatin organization.
We performed whole worm RNA-sequencing on L1 larvae to monitor transcriptional changes in LMN-Y59C and their suppression by cec-4Δ. Around 100 genes showed significant up-regulation or down-regulation upon expression of the LMN-Y59C mutant, and by combining cec-4Δ with LMN-Y59C, we could compensate or reverse a large fraction of these (Fig. 7; see Supplemental Table S1–S4). Consistent with earlier work in embryos (Gonzalez-Sandoval et al. 2015), we found few transcriptional differences provoked by cec-4Δ alone (Fig. 7). Nonetheless, it is unclear which of the affected genes may have relevance for the observed muscle pathology. Roughly 22% of the transcripts altered are muscle-expressed (Fig. 7C), and 17 out of the 41 genes that are misregulated by LMN-Y59C and counteracted by cec-4Δ, harbor E2F sites. While striking, the percentage of muscle-expressed genes bearing E2F sites is even higher (48%). Once again, in the affected sequences, the motif for the less common E2F homolog, EFL-2, was far more enriched than that of the more common E2F homolog, EFL-1 (Supplemental Fig. S7C). Thus, EFL-2 may have a unique role in muscle homeostasis (Narasimhan et al. 2015).
We propose that at least part of the LMN-Y59C-sensitive genes depend on E2F and Rb for subnuclear positioning, and possibly also for transcriptional regulation, although the two do not strictly correlate. Unfortunately, in worms, Rb and EFL-1 deletions have many other pleiotropic effects on development, including sterility and embryonic lethality, rendering their roles difficult to test (Petrella et al. 2011; Kudron et al. 2013; Goetsch et al. 2017). EFL-2 is less well studied, but efl-2 RNAi did not mimic loss of efl-1 (Ceol and Horvitz 2001). Whereas the CEC-4 CDY2A mutant, which ablates CEC-4 binding to H3K9me (Gonzalez-Sandoval et al. 2015), partially suppresses array mislocalization and head-thrashing defects, we do not rule out that CEC-4 might interact with other factors through its unstructured C-terminal domain. These factors may be histone acetyltransferases or deacetylases, kinases or phosphatases, or transcription factors themselves. Preliminary pull-down assays with a tagged version on CEC-4 identified only the expected INM/lamina components as strong interactors. Nonetheless, other interactions are likely to occur, and may alter chromatin-lamina interactions.
We searched the literature for the mammalian equivalent of CEC-4, especially with respect to chromatin interaction with the nuclear lamina. Based on functional and structural homology, the mammalian proline-rich 14 protein (PRR14) has been proposed to represent a CEC-4 functional homolog in that it is associated with lamin A/C and tethers heterochromatin to the INM during interphase and mitotic exit (Poleshko et al. 2013). The ablation of PRR14 impeded muscle differentiation, while its overexpression enhanced muscle differentiation in vitro (Yang and Yuan 2015), apparently by enhancing the activity of MyoD. Unlike CEC-4, PRR14 can bind HP1α, and thus the differentiation effects were thought to be due to a loss of heterochromatin stabilization. No genetic interaction with lamin A/C nor synergy with laminopathic mutations has been reported for prr14 knockdown, thus its relationship to our study remains unclear. Unlike PRR14, CEC-4 appears to actually compete with HP-1 for binding H3K9 methylated chromatin (Gonzalez-Sandoval et al. 2015).
With respect to the LMNA-Y45/LMN-Y59C allele, we note that this mutation is in one of the most highly conserved α-helical domains of lamin A/C, and that a cytosine replaces a key hydrophobic contact in the heptad repeat of the 1B coiled-coil domain. The LMN-Y59C lamin forms filaments in vitro, and when expressed in worms, it forms a perinuclear ring together with wild-type lamin. However, consistent with the prediction that the dimers might be less stable, FRAP data confirms a more rapid turnover of LMN-Y59C within the lamina (Wiesel et al. 2008; Mattout et al. 2011). Lamin A, like LMN-1 in worms, functions both at the INM and in the nucleoplasm, and through its affinity for Rb, it may activate or repress genes regulated by E2F. The 1B coiled-coil domain itself is unlikely to alter Rb interaction, as Rb binding was mapped to aa 247–355 in lamin A (Ozaki et al. 1994). However, a more rapid turnover of the LMN-Y59C dimer could change the distribution of Rb-bound promoters, releasing some sites, while allowing others to be sequestered. Those sequences showing NE-sequestration, like the heterochromatic array, are released upon cec-4 ablation. For this reason we also entertain the model that CEC-4 may promote Rb-EFL-2 stability (Fig. 7D). A failure to release and activate tissue-specific promoters in a muscle-specific manner may also be relevant for human patients carrying the LMN-Y45C mutation (Melcon et al. 2006; Camozzi et al. 2014).
In summary, here we provided novel evidence for a genetic suppressor of an EDMD-like disease provoked by a dominant missense mutation in lamin A, and its suppression by loss of a CD protein with specificity for histone H3K9me, i.e., CEC-4. The simplest interpretation is that this deletion compensates for aberrant nuclear positioning of crucial sequences at the INM. We confirmed this for muscle using muscle-specific DamID. This mechanism of suppression is consistent with major hypotheses concerning the effects of lamin mutations on tissue integrity (Worman and Bonne 2007; Bank and Gruenbaum 2011a; Camozzi et al. 2014) and argues that specific lamin A mutations have both gain-of-function activity (generating promiscuous LADs) and dominant-negative activity (an inability to initiate and spread LADs), which has consequences during the differentiation of cell fate (Perovanovic et al. 2016). The Perovanovic et al. (2016) study in mammalian cells also used DamID to map LADs in laminopathy mutants, and proposed that the diseases caused by certain lamin mutations resulted from epigenetic changes. They documented changes in histone H3 methylation and DNA methylation at a large number of sites. Our contribution goes one step further, in that we show that one can reverse the changes in genome organization (i.e., LADs) by the deletion of a specific NE protein that reads H3K9 methylation. With this, we have substantiated the hypothesis that chromatin domain organization is a crucial player in laminopathic disease. Our study suggests that antagonizing such phenomena may open therapeutic options for treating laminopathies.
Materials and methods
Strains and crosses
C. elegans strains (Supplemental Table S5) were maintained and manipulated as previously described (Brenner 1974). Strains were maintained at 22.5 °C for all studies unless otherwise noted. Worms were grown on NGM plates and fed OP50 bacteria except for emerin (EMR-1)-DamID experiments.
Head thrashing assay
To score C. elegans motility, L4 staged worms were gently transferred by pick to 30 µL of nuclease-free water (Qiagen) in an 18-well uncoated slide (Ibidi 81821) with two worms/well. Fifteen minutes after the last worm was transfered, the number of head movements to the right per 30 sec was counted under a bright light microscope. Room temperature was monitored closely and maintained within 21°C–23°C. Worms for each experiment were processed and analyzed in parallel as small temperature changes alter locomotion, by a single-blind observer. No more than three strains were analyzed in parallel to ensure consistency. Worm health was verified by touch response to the pick; unreactive worms were excluded.
RNAi
RNAi experiments were performed by placing L1 larvae on feeding plates as described previously (Timmons et al. 2001). The progeny of the RNAi fed C. elegans were scored for phenotypes. Sequence specificity of RNAi clones used were sequenced for confirmation. As a mock RNAi control, the L4440 vector (Fire vector library) was modified by removing an EcoRV fragment containing 25 bp identical to GFP-LacI.
Staining
For phalloidin staining, C. elegans were synchronized by bleaching adults and seeding L1 larvae after hatching overnight. L1 larvae were collected at the young adult stage (before eggs start to form). For all centrifugation steps, a swinging-bucket rotor was used at 800g and at room temperature. Worms were washed twice in M9 and transferred to a low-bind 1.5-mL tube. Following fixation with 4% paraformaldehyde (EMS Cat#15710) in 0.1 M Na2HPO4 for 15 min with rotation, worms were pelleted, and then permeabilized with 500 µL of cold acetone (100% at −20 °C) for 5 min at −20°C. Worms were pelleted and washed by rotation in PBS containing 0.5% Triton X-100 and 30 mM glycine (PBS-TG) for 10 min. Staining was with 500 µL of tetramethylrhodamine-phalloidin (1:250; Molecular Probes R415) in PBS-TG for 30 min, rotating at room temperature in the dark. Following centrifugation and three washes with PBS-TG at room temperature, they were left to settle on poly-L-lysine-coated slides for 5 min without drying. Excess liquid was removed and mounting medium (ProLong Gold antifade mountant, Thermo Scientific P36930) was added. The monitoring of defective sarcomeres was performed by a single-blind observer, with partial double-blind checking by an independent assistant.
Microscopy
Microscopy was carried out on spinning-disk multipoint confocal microscopes (AxioImager M1 [Carl Zeiss] + Yokogawa CSU-22 scan head, Plan-Neofluar 100×/1.45 NA oil objective, EM-CCD camera [Cascade II, Photometrics], and MetaMorph 7.7.2 software or Olympus IX81 + Yokogawa CSU-X1 scan head, PlanApo 100×/1.45 TIRFM objective, 2× back-illuminated EM-CCD EvolveDelta (Photometrics), and VisiView software GFP) and mCherry fluorophores were excited using a Toptica iBeamSmart 491-nm and 561-nm lasers. For live microscopy, plates were washed twice with M9 to remove adults and larvae and leave only eggs on the bacteria layer. L1 larvae hatched during 2–3 h and were collected by M9 wash. For live imaging, animals were mounted on slides coated with 2% agarose pads, supplemented with 0.1% NaN3 and 1 mM levamisole. For early adult live microscopy, worms were synchronized by bleaching gravid adults, and seeding L1 larvae after hatching overnight. Once worms reached early adult stage (before formation of eggs) they were washed twice in M9 and mounted on coverslips in the presence of levamisole for imaging.
Image analysis
Single- or multiplane (maximum intensity Z projections) images and analysis were generated using Fiji/ImageJ software 6. Quantitation of gwIs4 and cals3 reporter distribution on focal stacks of images was done with plug-in PointPicker (http://bigwww.epfl.ch/thevenaz/pointpicker) as described previously (Meister et al. 2010a). For high-throughput image analysis, 3D stacks were processed and analyzed as described previously (Gonzalez-Sandoval et al. 2015; Hauer et al. 2017). For statistical analysis and data representation, only nuclei with two foci were taken into account. Nuclei touching the image boarders at the X-, Y-, or Z-axis were excluded from the analysis. Table operations, data filtering (two foci per nucleus), and normalization were performed with KNIME (Dietz and Berthold 2016). The control (WT) condition was set to 1 and data derived from mutants or other experimental conditions are shown relative to the control. Distributions were plotted with R as violin plot graphs.
Muscle-specific emerin (EMR-1) DamID
DamID was performed and analyzed largely as described in Gómez-Saldivar et al. (2016) and Cabianca et al. (2019). In this study, three biological replicas were prepared and all strains were grown in parallel in each replica and for all strains the analysis was performed in biological triplicates. C. elegans strains were synchronized by seeding four 10-cm NGM plates with E. coli GM119 Dam bacteria with ∼4000 L1s per plate. Animals were incubated for 26 h at 22.5°C until L3/L4 stage, before collection for DamID procedure. The triplicates were prepared and sequenced separately. Individual samples within each replica were barcoded, pooled, and then sequenced on a HiSeq 2500 (Illumina) sequencer with 50-cycle single-end reads. Most analyses were performed in R Studio (version 1.0.153) using the damid.seq.R pipeline (Sharma et al. 2016; available at http://www.github.com/damidseq/RDamIDSeq). This pipeline identifies and maps specifically those NGS reads that contain the DamID adapters (adapt.seq = “CGCGGCCGAG”) and corresponds to genomic fragments flanked by GATC sites (restr.seq = “GATC”). Mapping was performed to the C. elegans genome release WBCel235 (species = “BSgenome.Celegans.UCSC.ce11”). The pipeline provides absolute and relative read counts both per GATC fragment and per user-defined bin size. The number of reads obtained for each replicate in each strain ranged from 35,000 to 32.3 million (specifically, one was ∼35,000 and all others were >200,000), but all were well above the level required for robust results (10,000 reads) for 10-kb DamID resolution (Sharma et al. 2016). The pipeline computed the log2 ratio between relative Dam::EMR-1 and GFP::Dam reads within each replica and the Spearman correlation coefficients calculated pairwise between replicas ranged from 0.56 to 0.99 (median = 0.93). Worm chromosomes have no repeat-rich centromere, and repeat elements are enriched on chromosome arms. It is therefore standard to compare chromosome arms versus cores for LAD behavior (Ikegami et al. 2010). Following standard practice, we used the following border coordinates: ChrI 3745632 and 10809938, ChrII 4708341 and 11877168, ChrIII 3508994 and 9947268, ChrIV 7317812 and 12176625, ChrV 8125434 and 13849337, and ChrX 41919362. Pairwise Wilcoxon rank test values were calculated in R.
RNA-seq samples preparation
For RNA-seq, synchronized L1 larvae were obtained by bleaching gravid adults and the eggs recovered were left to hatch 16 h at room temperature in M9. The L1 larvae were refed for 2.5 h, washed three times in M9, resuspended in 100 µL of M9 and 400 µL of Trizol (Ambion), and snap-frozen in N2(liq) for RNA extraction. Extraction of RNA used four freeze–thaw cycles from N2(liq) to a 42°C heat bath, followed by the addition of 200 µL of Trizol to each sample. Vigorous vortexing at room temperature (five cycles at 30-sec vortex, 30 sec on ice), was followed by 5 min at room temperature. RNA extraction was by 140 µL of chloroform (15 sec of shaking and 2 min at room temperature); after centrifugation at 4°C, the aqueous phases were transferred to fresh tubes. An equal volume of 70% EtOH was added slowly and the homogeneous mixture was transferred to a Qiagen RNeasy spin column (RNeasy kit, Qiagen 74104), including a subsequent 30-min DNase treatment. Stranded library preparation was performed with a TruSeq Illumina mRNA-seq library preparation kit. The quality of the resulting libraries was assessed with an Agilent Bioanalyzer and concentrations were measured with a Qubit fluorometer prior to pooling. Fifty-nucleotide single-end sequencing was done on an Illumina HiSeq 2500.
RNA-seq analysis
For each genotype the RNA-seq data from two biological replicate samples were mapped to the C. elegans genome (ce10) using the R package QuasR v1.22.0. QuasR includes the short read aligner bowtie to consider only uniquely mapping reads. Count tables of reads mapping within annotated exons in WormBase (WS220) were normalize by division by the total number of reads in each library and multiplied by the average library size. Transformation into log2 space was performed after the addition of a pseudocount of eight in order to minimize large changes in abundance fold change (FC) caused by low count numbers. The edgeR package v3.24 was applied to select genes with differential transcript abundances between the indicated genotypes (contrasts) based on false discovery rates (FDR < 0.01) and fold changes (FC ≥1.5), independently, in each of two replicate sample pairs. For each differentially expressed transcript (duplicate calls in different contrasts excluded) the average abundance over all samples was computed, and the FC deviations were represented in heat maps.
Transcription factor motif analysis
A compendium of transcription factor weight matrices (Narasimhan et al. 2015) was downloaded (http://hugheslab.ccbr.utoronto.ca/supplementary-data/CeMotifs) to examine the set in the file “TF_Information.txt.” These were used to scan the C. elegans ce10 genome using the matchPWM function in the Bioconductor package Biostrings. Only hits with a minimum score of 10 were considered unless the maximum obtainable score by the weight matrix was <10. In that case, the maximum obtainable score was required. The resulting binding sites were then intersected with gene promoter annotation (WS220, 1500 bp 5′, and 500 bp 3′ of transcription start site) to determine the number of sites for each transcription factor and gene.
Data availability
RNA sequencing and EMR-1-DamID data are available through Gene Expression Omnibus (GEO), SuperSeries GSE136577 with SubSeries GSE136576 and GSE135850.
Supplementary Material
Acknowledgments
We dedicate this paper to our dear colleague and coauthor, Michael Hauer, who passed away during revisions. We thank the Caenorhabditis Genetics Center (CGC) funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440) for strains. We are grateful to P. Sartre for technical help and to Y. Gruenbaum, I. Katic, and members of the Friedrich Miescher Institute Genomics and Microscopy Facilities for advice and support. J.C.H. was supported by an FP7 Marie Curie Action Intra-European Fellowship and by a grant from the Foundation Suisse de Recherche sur les Maladies Musculaires. A.M. was funded by the same and by a Marie Heim-Vögtlin grant from the Swiss National Science Foundation. C.M.-J., R.R.-B., and P.A. thank the Spanish State Research Agency and the European Regional Development Fund (BFU2016-79313-P and MDM-2016-0687) for support. This project received funding from the European Research Council under the European Union's Horizon 2020 Research and Innovation program (Epiherigans grant no. 743312 to S.M.G.).
J.C.H. and S.M.G. wrote the manuscript. J.C.H. and A.M. designed and performed experiments and interpreted results. C.M.-J. and R.R.-B. performed DamID experiments and analysis. V.K. contributed to microscopy and motility experiments and invaluable technical support. A.G.-S. created strains for CEC-4(CD) rescue experiments. J.P. and C.D.S. performed bioinformatic analyses and interpreted results. M.H.H. did the high-throughput image analysis. P.A. analyzed DamID data, interpreted results, and provided funding. S.M.G. interpreted results, revised the paper, and provided funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.332213.119.
Freely available online through the Genes & Development Open Access option.
References
- Bank EM, Gruenbaum Y. 2011a. Caenorhabditis elegans as a model system for studying the nuclear lamina and laminopathic diseases. Nucleus 2: 350–357. 10.4161/nucl.2.5.17838 [DOI] [PubMed] [Google Scholar]
- Bank EM, Gruenbaum Y. 2011b. The nuclear lamina and heterochromatin: a complex relationship. Biochem Soc Trans 39: 1705–1709. 10.1042/BST20110603 [DOI] [PubMed] [Google Scholar]
- Barkan R, Zahand AJ, Sharabi K, Lamm AT, Feinstein N, Haithcock E, Wilson KL, Liu J, Gruenbaum Y. 2012. Ce-emerin and LEM-2: essential roles in Caenorhabditis elegans development, muscle function, and mitosis. Mol Biol Cell 23: 543–552. 10.1091/mbc.e11-06-0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie SM, Babb C, Wilky H, Rawls A, Park JG, Mangone M. 2015. Comparative RNA-seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol 13: 4 10.1186/s12915-015-0116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Muñoz-Jiménez C, Kalck V, Gaidatzis D, Padeken J, Seeber A, Askjaer P, Gasser SM. 2019. Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature 569: 734–739. 10.1038/s41586-019-1243-y [DOI] [PubMed] [Google Scholar]
- Camozzi D, Capanni C, Cenni V, Mattioli E, Columbaro M, Squarzoni S, Lattanzi G. 2014. Diverse lamin-dependent mechanisms interact to control chromatin dynamics. Nucleus 5: 427–440. 10.4161/nucl.36289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. 2001. Dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol Cell 7: 461–473. 10.1016/S1097-2765(01)00194-0 [DOI] [PubMed] [Google Scholar]
- Cho S, Vashisth M, Abbas A, Majkut S, Vogel K, Xia Y, Ivanovska IL, Irianto J, Tewari M, Zhu K, et al. 2019. Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle Arrest. Dev cell 49: 920–935.e5. 10.1016/j.devcel.2019.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Askjaer P. 2017. Cell biology of the Caenorhabditis elegans nucleus. Genetics 205: 25–59. 10.1534/genetics.116.197160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz C, Berthold MR. 2016. KNIME for open-source bioimage analysis: a tutorial. Adv Anat Embryol Cell Biolpp 219: 179–197. 10.1007/978-3-319-28549-8_7 [DOI] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. 2011. The lamin protein family. Genome Biol 12: 222 10.1186/gb-2011-12-5-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenbaas JS, Bragazzi Cunha J, Azuero-Dajud R, Nelson B, Oral EA, Williams JA, Stewart CL, Omary MB. 2018. Lamin A/C maintains exocrine pancreas homeostasis by regulating stability of RB and activity of E2F. Gastroenterology 154: 1625–1629.e8. 10.1053/j.gastro.2018.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau C, Higuet D, Courvalin JC, Buendia B. 2004. Expression of a mutant lamin A that causes Emery-Dreifuss muscular dystrophy inhibits in vitro differentiation of C2C12 myoblasts. Mol Cell Biol 24: 1481–1492. 10.1128/MCB.24.4.1481-1492.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Kudlow BA, Evans AM, Jameson SA, Hauschka SD, Kennedy BK. 2006. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev 20: 486–500. 10.1101/gad.1364906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch PD, Garrigues JM, Strome S. 2017. Loss of the Caenorhabditis elegans pocket protein LIN-35 reveals MuvB's innate function as the repressor of DREAM target genes. PLoS Genet 13: e1007088 10.1371/journal.pgen.1007088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Saldivar G, Meister P, Askjaer P. 2016. DamID analysis of nuclear organization in Caenorhabditis elegans. Methods Mol Biol 1411: 341–358. 10.1007/978-1-4939-3530-7_22 [DOI] [PubMed] [Google Scholar]
- González-Aguilera C, Ikegami K, Ayuso C, de Luis A, Íñiguez M, Cabello J, Lieb JD, Askjaer P. 2014. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol 15: R21 10.1186/gb-2014-15-2-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A, Towbin Benjamin D, Kalck V, Cabianca Daphne S, Gaidatzis D, Hauer MH, Geng L, Wang L, Yang T, Wang X, et al. 2015. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163: 1333–1347. 10.1016/j.cell.2015.10.066 [DOI] [PubMed] [Google Scholar]
- Harr JC, Gonzalez-Sandoval A, Gasser SM. 2016. Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man. EMBO Rep 17: 139–155. 10.15252/embr.201541809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer MH, Seeber A, Singh V, Thierry R, Sack R, Amitai A, Kryzhanovska M, Eglinger J, Holcman D, Owen-Hughes T, et al. 2017. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nature Struct Mol Biol 24: 99–107. 10.1038/nsmb.3347 [DOI] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120 10.1186/gb-2010-11-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome–nuclear lamina interactions. Cell 153: 178–192. 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162. 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- Kudron M, Niu W, Lu Z, Wang G, Gerstein M, Snyder M, Reinke V. 2013. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol 14: R5 10.1186/gb-2013-14-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre I, Chesney MA, Garrigues JM, Stempor P, Appert A, Francesconi M, Strome S, Ahringer J. 2015. The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev 29: 495–500. 10.1101/gad.255810.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21: 227–236. 10.1101/gr.115519.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DJ, Trembath RC, Shackleton S. 2002. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Human Mol Gen 11: 769–777. 10.1093/hmg/11.7.769 [DOI] [PubMed] [Google Scholar]
- Markiewicz E, Dechat T, Foisner R, Quinlan RA, Hutchison CJ. 2002. Lamin A/C binding protein LAP2α is required for nuclear anchorage of retinoblastoma protein. Mol Biol Cell 13: 4401–4413. 10.1091/mbc.e02-07-0450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E, Ledran M, Hutchison CJ. 2005. Remodelling of the nuclear lamina and nucleoskeleton is required for skeletal muscle differentiation in vitro. J Cell Sci 118: 409–420. 10.1242/jcs.01630 [DOI] [PubMed] [Google Scholar]
- Mattout A, Pike Brietta L, Towbin Benjamin D, Bank Erin M, Gonzalez-Sandoval A, Stadler Michael B, Meister P, Gruenbaum Y, Gasser Susan M. 2011. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol 21: 1603–1614. 10.1016/j.cub.2011.08.030 [DOI] [PubMed] [Google Scholar]
- Meister P, Gehlen LR, Varela E, Kalck V, Gasser SM. 2010a. Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol 470: 535–567. 10.1016/S0076-6879(10)70021-5 [DOI] [PubMed] [Google Scholar]
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. 2010b. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev 24: 766–782. 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcon G, Kozlov S, Cutler DA, Sullivan T, Hernandez L, Zhao P, Mitchell S, Nader G, Bakay M, Rottman JN, et al. 2006. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum Mol Genet 15: 637–651. 10.1093/hmg/ddi479 [DOI] [PubMed] [Google Scholar]
- Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. 2005. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Human Mol Genet 14: 2167–2180. 10.1093/hmg/ddi221 [DOI] [PubMed] [Google Scholar]
- Muñoz-Jiménez C, Ayuso C, Dobrzynska A, Torres-Mendéz A, Ruiz PDLC, Askjaer P. 2017. An efficient FLP-based toolkit for spatiotemporal control of gene expression in Caenorhabditis elegans. Genetics 206: 1763–1778. 10.1534/genetics.117.201012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Lambert SA, Yang AW, Riddell J, Mnaimneh S, Zheng H, Albu M, Najafabadi HS, Reece-Hoyes JS, Fuxman Bass JI, et al. 2015. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. Elife 4: e06967 10.7554/eLife.06967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. 1992. Assembly and cell cycle dynamics of the nuclear lamina. Semin Cell Biol 3: 245–253. 10.1016/1043-4682(92)90026-R [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Dechat T, Foisner R. 2015. Lamins at the crossroads of mechanosignaling. Genes Dev 29: 225–237. 10.1101/gad.255968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T, Saijo M, Murakami K, Enomoto H, Taya Y, Sakiyama S. 1994. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene 9: 2649–2653. [PubMed] [Google Scholar]
- Pekovic V, Harborth J, Broers JLV, Ramaekers FCS, van Engelen B, Lammens M, von Zglinicki T, Foisner R, Hutchison C, Markiewicz E. 2007. Nucleoplasmic LAP2α–lamin A complexes are required to maintain a proliferative state in human fibroblasts. J Cell Biol 176: 163–172. 10.1083/jcb.200606139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perovanovic J, Dell'Orso S, Gnochi VF, Jaiswal JK, Sartorelli V, Vigouroux C, Mamchaoui K, Mouly V, Bonne G, Hoffman EP. 2016. Laminopathies disrupt epigenomic developmental programs and cell fate. Sci Transl Med 8: 335ra58 10.1126/scitranslmed.aad4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S. 2011. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138: 1069–1079. 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, Katz RA. 2013. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep 5: 292–301. 10.1016/j.celrep.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. 2006. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20: 1447–1457. 10.1101/gad.1419506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247. 10.1038/nature06727 [DOI] [PubMed] [Google Scholar]
- Sharma R, Ritler D, Meister P. 2016. Tools for DNA adenine methyltransferase identification analysis of nuclear organization during C. elegans development. Genesis 54: 151–159. 10.1002/dvg.22925 [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kozlov S, Fong LG, Young SG. 2007. Mouse models of the laminopathies. Exp Cell Res 313: 2144–2156. 10.1016/j.yexcr.2007.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. 2004. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol 343: 1067–1080. 10.1016/j.jmb.2004.08.093 [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Venolia L, Waterston RH. 1990. The unc-45 gene of Caenorhabditis elegans is an essential muscle-affecting gene with maternal expression. Genetics 126: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlcek S, Foisner R. 2007. A-type lamin networks in light of laminopathic diseases. Biochim Biophys Acta 1773: 661–674. 10.1016/j.bbamcr.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Waterston RH, Fishpool RM, Brenner S. 1977. Mutants affecting paramyosin in Caenorhabditis elegans. J Mol Biol 117: 679–697. 10.1016/0022-2836(77)90064-X [DOI] [PubMed] [Google Scholar]
- Wiesel N, Mattout A, Melcer S, Melamed-Book N, Herrmann H, Medalia O, Aebi U, Gruenbaum Y. 2008. Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc Natl Acad Sci 105: 180–185. 10.1073/pnas.0708974105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. 2007. ‘Laminopathies’: a wide spectrum of human diseases. Exp Cell Res 313: 2121–2133. 10.1016/j.yexcr.2007.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Yuan Z-M. 2015. A novel role of PRR14 in the regulation of skeletal myogenesis. Cell Death Dis 6: e1734 10.1038/cddis.2015.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing and EMR-1-DamID data are available through Gene Expression Omnibus (GEO), SuperSeries GSE136577 with SubSeries GSE136576 and GSE135850.