In this study, Ritschka et al. investigated whether cellular senescence might play a role in loss of regenerative capacity during liver regeneration. They show that following partial hepatectomy, the senescence-associated genes p21, p16Ink4a, and p19Arf become dynamically expressed in different cell types when regenerative capacity decreases, but without a full senescent response, and that treatment with a senescence-inhibiting drug improves regeneration through targeting aberrantly prolonged p21 expression.
Keywords: senescence, senolytic, liver regeneration, p21, p16Ink4a, ABT-737, hepatocyte, aging
Abstract
Young mammals possess a limited regenerative capacity in some tissues, which is lost upon maturation. We investigated whether cellular senescence might play a role in such loss during liver regeneration. We found that following partial hepatectomy, the senescence-associated genes p21, p16Ink4a, and p19Arf become dynamically expressed in different cell types when regenerative capacity decreases, but without a full senescent response. However, we show that treatment with a senescence-inhibiting drug improves regeneration, by disrupting aberrantly prolonged p21 expression. This work suggests that senescence may initially develop from heterogeneous cellular responses, and that senotherapeutic drugs might be useful in promoting organ regeneration.
Shortly after birth mammals can regenerate some organs, but this capacity declines rapidly, even before reaching adulthood (Han et al. 2008; Porrello et al. 2011; Bassat et al. 2017; Payzin-Dogru and Whited 2018). The mammalian liver is one tissue that retains an increased regenerative capacity for a period of time (Mitchell and Willenbring 2008). Following surgery to remove two-thirds of the liver (2/3 PH [partial hepatectomy]), the consequent tissue damage induces different populations of cells to coordinate regeneration (Su et al. 2002; Michalopoulos 2007; Pedone et al. 2017). However, while this process is effective in young animals, adult mice have a significantly reduced capacity to regenerate. The reasons for such loss of regeneration remain unclear (Mitchell and Willenbring 2008; Enkhbold et al. 2015; Loforese et al. 2017).
Cellular senescence is a form of permanent cell cycle arrest linked with aging and tissue damage (Muñoz-Espín and Serrano 2014; Childs et al. 2015). The aberrant accumulation of senescent cells expressing markers such as p16Ink4a, p19Arf, and p21, as well as secreting inflammatory factors of the senescence-associated secretory phenotype (SASP), contributes to the loss of tissue function seen during aging and in many disease states (Muñoz-Espín and Serrano 2014; Baker et al. 2016; Childs et al. 2017). Recently, senescent cells have been targeted through pharmacological means. Several drugs that alter senescent cell dynamics—collectively called “senotherapeutics,” which includes drugs that kill senescent cells (“senolytics”)—have been used to increase life span, health span, and to improve some disease-associated conditions, including Alzheimer's disease, atherosclerosis, and liver steatosis (Chang et al. 2015; Yosef et al. 2016; Baar et al. 2017; Ogrodnik et al. 2017; Bussian et al. 2018; Musi et al. 2018; Xu et al. 2018). While some of these drugs can target the antiapoptotic machinery in senescent cells, in many cases, their precise cellular or molecular targets in vivo remain unclear.
We used the model of 2/3 PH to assess whether the decrease in regeneration that develops in adult animals is linked to senescence and whether this could be improved with senotherapeutic treatment.
Results and Discussion
p21 and p16Ink4a expression increases in different cell populations as regeneration decreases
To look for a connection between senescence induction and early loss of regenerative capacity, we performed 2/3 PH in young (2- to 3-mo-old) and adult (6- to 8-mo-old) mice and analyzed regeneration in each group up to 10 d after surgery. The livers in young mice regenerated, reaching almost original liver–body weight ratio by 7 d after PH (Fig. 1A). However, the livers of the adult mice exhibited a significant decrease in regeneration (Fig. 1A). Histological assessment in young mice (Fig. 1B) identified regeneration as previously described, without necrosis or inflammation (Mitchell and Willenbring 2008). However, after PH, adult livers showed a severe accumulation of optically empty microvacuolization and macrovacuolization, suggestive of lipids. This was confirmed by Oil Red O (ORO) staining, which demonstrated neutral lipid-accumulation, or lipidosis. This suggests that transient patterns of lipid observed in young animals become misregulated in older animals following PH (Fig. 1B, day 3; Supplemental Fig. S1A; Zou et al. 2012). This is interesting as previous reports demonstrated abnormal lipid accumulation and steatosis in senescent and aged livers (>12 mo with PH, or >18 mo without PH) (Loforese et al. 2017; Ogrodnik et al. 2017).
Figure 1.
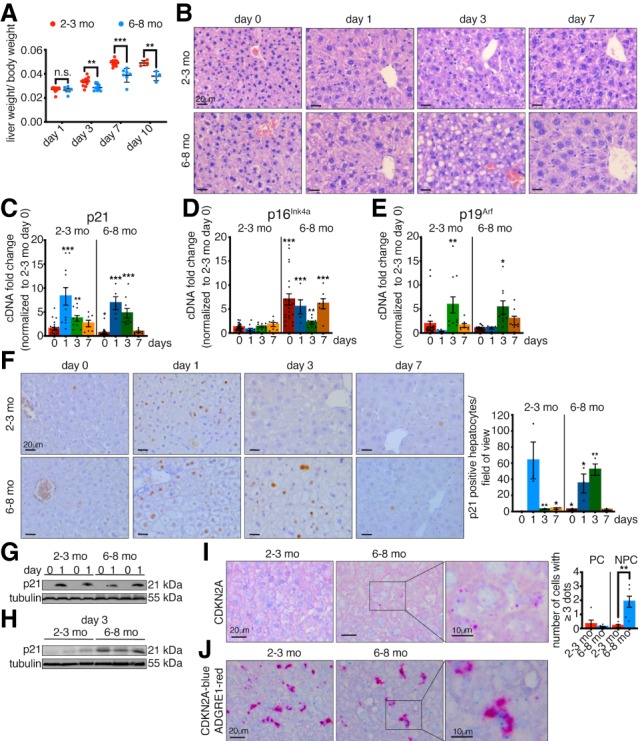
p21 exhibits prolonged expression in adult livers with impaired regeneration. (A) Liver regeneration of young (2- to 3-mo-old) and adult (6- to 8-mo-old) mice was calculated as liver/body weight ratio at different days after PH (n = 4–13). (B) H&E staining of young and adult liver sections before (day 0) and at different time points after PH. All images are representative of at least five biological replicates. Scale bars, 20 µm. (C–E) qPCR analysis for the senescence markers p21 (C), p16Ink4a (D), and p19Arf (E) of young and adult livers at different days after PH normalized to young livers at day 0 (n = 5–27). (F) Immunohistochemistry for p21 in young and adult liver sections at different days after PH and quantification of p21-positive hepatocytes/field of view (n = 3). All images are representative of at least three biological replicates. Scale bars, 20 µm. (G,H) p21 expression in whole liver lysates at 0 d and 1 d (G) and 3 d (H) after PH measured by Western blot. Tubulin was used as a loading control (n = 2–3). (I) RNA in situ hybridization (ISH) staining and quantification for CDKN2A in young and adult liver sections before PH (day 0). Scale bars, 20 µm. Boxed area shows higher magnification of positive staining. Scale bar, 10 µm. All images are representative of at least five biological replicates. Quantification shows number of cells/field of view that have three or more dots/cell. (PC) Parenchymal cells; (NPC) nonparenchymal cells (n = 5–6). (J) RNA co-ISH staining for CDKN2A (blue) and ADGRE1 (red) in young and adult liver sections before PH (day 0). Scale bars, 20 µm. Boxed area shows higher magnification of positive staining. Scale bar, 10 µm. All images are representative of at least three biological replicates. Error bars, mean ± SEM, unpaired two-tailed Student's t-test. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001.
As adult mice exhibit decreased regeneration by 6–8 mo, we assessed for expression of senescence markers p21, p16Ink4a, and p19Arf by qPCR in young and adult livers. In prehepatectomy liver samples (day 0), we found that baseline levels of p21 decreased between 2–3 mo and 6–8 mo of age (Fig. 1C). Conversely however, the senescence marker p16Ink4a significantly increased during this time, while there was no change in p19Arf expression (Fig. 1D,E). Previous reports linked p16Ink4a in the liver with fibrosis (Krizhanovsky et al. 2008). However, no significant change was seen here in adult animals, either before or following PH (Supplemental Fig. S1B,C).
Next, we assessed for dynamic changes of the senescence markers following PH. As previously described for young mice, there was a transient, p53-independent increase in p21 expression that peaked ∼24–48 h after PH and decreased by day 7 (Fig. 1C; Albrecht et al. 1997; Stepniak et al. 2006; Buitrago-Molina et al. 2013). In older animals, this induction of p21 also occurred (Fig. 1C). Surprisingly, similar analysis for p16Ink4a revealed that even though expression was increased in adult prehepatectomy livers, this did not increase further, and even decreased after PH, before returning to the elevated adult prehepatectomy level (Fig. 1D). Finally, p19Arf expression peaked transiently at day 3 after PH in both young and adult animals (Fig. 1E).
We next examined p21-protein expression and localization. In agreement with our qPCR results, there was a clear transient induction of p21 1 d after PH in young animals, predominantly in hepatocytes (Fig. 1F,G; Supplemental Fig. S1D). Surprisingly however, as shown by immunostaining and Western blotting, while a similar induction was seen 24 h after PH in adult livers, p21 expression persisted longer, now also being detectable at day 3 after PH, when p21 expression is lost in the young (Fig. 1F–H; Supplemental Fig. S1D–F).
To determine where p16Ink4a is expressed, we performed in situ hybridization (ISH) using a probe that recognizes a shared exon in the CDKN2A locus, common to both p16Ink4a and p19Arf. As our qPCR results showed no expression of p19Arf in adult tissue before hepatectomy, the observed signal in the adult livers corresponds to p16Ink4a expression (Fig. 1I; Supplemental Fig. S1G). Similar results were obtained with a lower affinity probe specific for p16Ink4a (data not shown). Surprisingly, however, the staining with the CDKN2A probe clearly showed that p16Ink4a is not in hepatocytes and does not overlap with p21-positive cells (Supplemental Fig. S1H). Next, we performed co-ISH for p16Ink4a with markers of macrophages (Adgre1), Stellate (Desmin), and endothelial (Pecam1) cells. Interestingly, most p16Ink4a expression was in macrophages, in agreement with previous reports (Liu et al. 2019), with some expression in endothelial cells, and little or no expression in stellate cells (Fig. 1J; Supplemental Fig. S1I–K). In addition, we used this same probe to examine the p19Arf expression that was detected in young mice at day 3, when p16Ink4a was not detectable, and found that this was also not localized in the hepatocytes (Supplemental Fig. S1L). Further analysis of senescence and proliferation markers revealed increased expression of proliferation and cell cycle genes in adult livers following PH (Supplemental Fig. S2A–F). However, costaining showed that both the p21- and p16Ink4a-positive cells were negative for proliferation markers (Supplemental Fig. S2G,H). Furthermore, we were unable to reliably detect the senescence marker senescence-associated β-galactosidase (SA- β-gal) (data not shown). Interestingly, by comparison, the level of p16Ink4a expression seen at 6–8 mo was much lower than that typically seen in old mice (Supplemental Fig. S2I,J). This suggests that although there are dynamic changes in senescence-associated genes, this likely does not represent a full senescence-induction.
p21 deficiency partially rescues liver regeneration
To assess whether these genes functionally contributed to the decreased regeneration in adult mice, we performed PH in mice deficient in p21, p19Arf, and p16Ink4a. At 2–3 mo, p21-deficient mice had similar regeneration as their young wild-type controls at day 3, but had impaired regeneration by day 7 (Fig. 2A). However, adult p21-deficient mice showed a significantly increased regenerative capacity at day 3 after PH, which was not maintained to day 7 (Fig. 2A). Histological analysis and ORO staining revealed no major differences in comparison with age-matched controls (Fig. 2B,C; Supplemental Fig. S3A), while these mice exhibited no changes in proliferation or macrophage patterns (Supplemental Fig. S3B–H). Furthermore, qPCR analysis, showed that p21-deficient mice had similar expression patterns of p16Ink4a and p19Arf following PH, as was seen in wild-type mice (Fig. 2D,E). Next, we examined liver regeneration in adult p19Arf-deficient mice, which exhibited similar impaired regeneration as wild-type mice at both days 3 and 7 (Fig. 2A). Interestingly however, adult p16Ink4a-deficient mice seemed to regenerate even more poorly than wild-type mice (Fig. 2F). Together, this suggests that a deficiency of p21 partially improves liver regeneration in adult mice.
Figure 2.
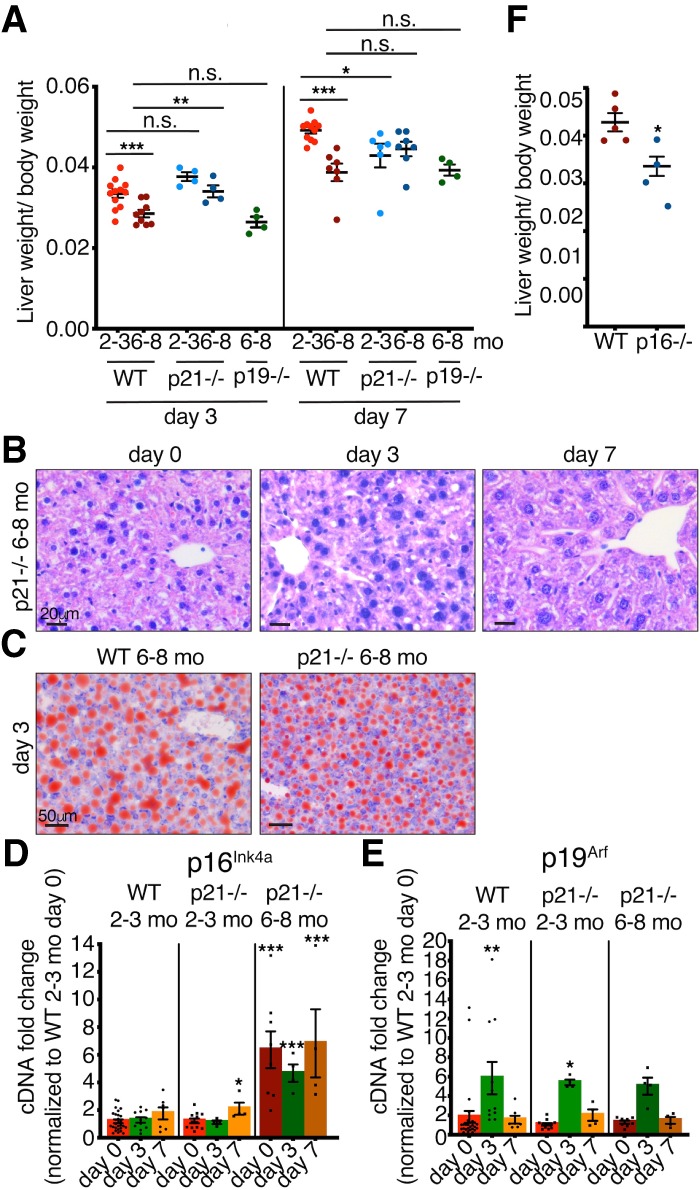
Adult p21-deficient mice have partially improved regenerative capacity. (A) Liver/body weight ratios of 2- to 3-mo-old and 6- to 8-mo-old WT, p21−/−, and p19−/− mice at 3 and 7 d after PH (n = 4–12). (B) H&E staining of 6- to 8-mo-old p21−/− livers before and at different days after PH. All images are representative of at least four biological replicates. Scale bars, 20 µm. (C) Oil Red O staining of 6- to 8-mo-old WT and p21−/− livers 3 d after PH. All images are representative of at least four biological replicates. Scale bars, 50 µm. (D,E) qPCR analysis for (D) p16Ink4a and (E) p19Arf of 2- to 3-mo-old WT and p21−/− and 6- to 8-mo-old p21−/− livers at different days after PH normalized to 2- to 3-mo-old WT day 0. (n = 4–27). (F) Liver/body weight ratios of 6- to 8-mo-old WT and p16−/− mice 7 d after PH (n = 4–5). Error bars, mean ± SEM, unpaired two-tailed Student's t-test. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001.
ABT-737 treatment improves liver regeneration
Given this associated increase in senescence-associated genes when the adult liver loses its capacity to regenerate, we asked whether treatment with a senotherapeutic drug might affect regeneration. To test this, we devised a strategy to pretreat livers with the senolytic compound ABT-737, or vehicle control, twice over 2 d immediately prior to PH, as such delivery could potentially target both p16Ink4a- and p21-positive cells (Fig. 3A). Treatment of 6–8-month-old mice with vehicle alone showed decreased regenerative function, similar or worse to their untreated controls (Fig. 3B,C). Surprisingly, however, mice that were pretreated with ABT-737 showed significantly increased regenerative capacity by day 7 after hepatectomy (Fig. 3B,C). Supporting this, we found reduced levels of the liver enzymes aspartate transaminase (AST) and alanine transaminase (ALT), which are increased upon PH, tissue damage and aging (Fig. 3D; Enkhbold et al. 2015; Loforese et al. 2017; Raven et al. 2017). Improvements were also evident at the histological level in ABT-737-treated livers, with decreased microvacuolization and macrovacuolization, and decreased ORO staining (Fig. 3E,F; Supplemental Fig. S3A), and a reverse of the increased cell cycle gene expression seen in adult mice (Supplemental Fig. S3I–N). Together, this demonstrates that pretreatment with ABT-737 improves liver regenerative capacity in adult mice.
Figure 3.
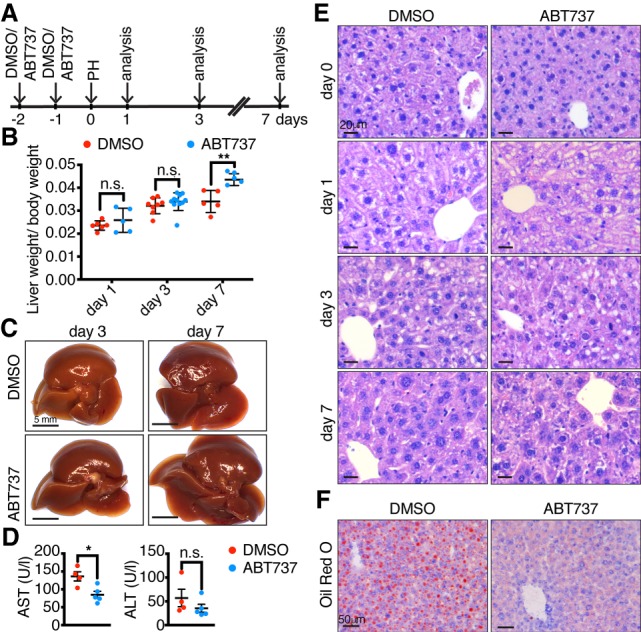
ABT-737 improves regenerative capacity in adult livers. (A) Schematic representation of the experimental protocol. Intraperitoneal injection of DMSO or ABT-737 twice over 2 d was followed 1 d later by PH and mice were analyzed 1, 3, and 7 d after PH. (B) Liver/body weight ratios of 6- to 8-mo-old DMSO- or ABT-737-treated mice at different time points after PH (n = 5–11). (C) Representative macroscopic images of livers from DMSO- or ABT-737-treated mice 3 and 7 d after PH. Scale bars, 5 mm. (D) Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) measured 7 d after PH (n = 4–5). (E) H&E staining of DMSO- and ABT-737-treated livers before and at different days after PH. All images are representative of at least five biological replicates. Scale bars, 20 µm. (F) Oil Red O staining of 6- to 8-mo-old DMSO- or ABT737-treated mice 3 d after PH. All images are representative of at least four biological replicates. Scale bars, 50 µm. Error bars, mean ± SEM, unpaired two-tailed Student's t-test. (*) P ≤ 0.05; (**) P ≤ 0.01.
ABT-737 decreases p21 and SASP expression
To explore the mechanisms involved, we examined the expression patterns of the senescence genes following drug treatment. First, examination of the p21 transcript levels revealed a decrease at day 1 after PH in the ABT-737 versus vehicle samples (Fig. 4A). Furthermore, immunostainings for p21 showed a significant reduction in p21 expression, with most drug-treated hepatocytes now negative for p21 (Fig. 4B; Supplemental Fig. S3O). This was most evident by Western blot on whole liver lysates, which demonstrated an overall decrease in p21 expression 1 and 3 d after PH (Fig. 4C,D; Supplemental Fig. S3P,Q). Next, we examined for p16Ink4a expression. Interestingly, while p16Ink4a-positive cells are suggested as a primary target of senolytic treatment in aged tissues, here we found no change in p16Ink4a transcript levels upon ABT-737 treatment (Fig. 4E). In addition, the distribution of p16Ink4a as assessed by ISH was unchanged (Fig. 4F; Supplemental Fig. S3R), suggesting that these cells are not affected by ABT-737 treatment. Similarly, no major changes were detected in p19Arf expression (Fig. 4G). Altogether, this data suggests that ABT-737 mostly affects p21 expression.
Figure 4.
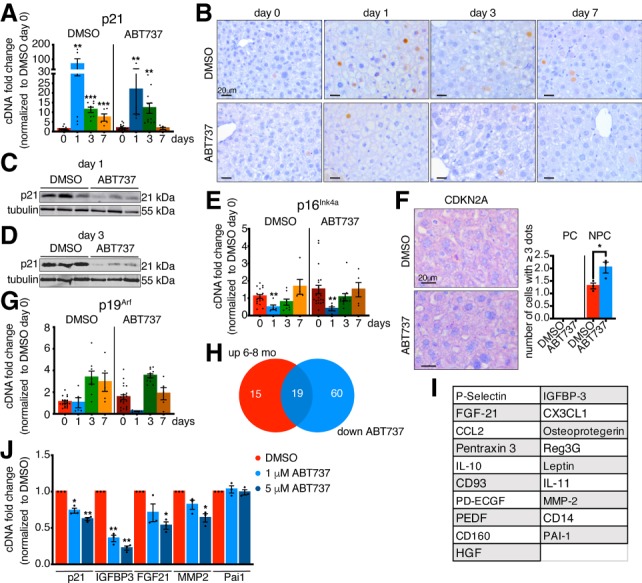
ABT-737 decreases expression of p21 and SASP factors. (A) qPCR analysis for p21 in DMSO- and ABT-737-treated livers at different days after PH normalized to DMSO day 0 (n = 5–21). (B) Immunohistochemistry for p21 in DMSO- and ABT-737-treated livers at different time points after PH. All images are representative of at least four biological replicates. Scale bars, 20 µm. (C,D) p21 expression following treatment with DMSO or ABT-737 in whole liver lysates 1 d (C) and 3 d (D) after PH measured by Western blot. Tubulin was used as a loading control (n = 3). (E) qPCR analysis for p16Ink4a in DMSO- and ABT-737-treated livers at different days after PH normalized to DMSO day 0 (n = 5–21). (F) RNA ISH staining and quantification for CDKN2A in DMSO and ABT737 liver sections before PH (day 0). All images are representative of at least three biological replicates. Scale bars, 20 µm. Quantification shows number of cells/field of view that have three or more dots/cell. (n = 3) (PC) Parenchymal cells; (NPC) nonparenchymal cells. (G) qPCR analysis for p19Arf in DMSO- and ABT-737-treated livers at different days after PH normalized to DMSO day 0 (n = 5–21). (H) Venn diagram showing the overlap in 6- to 8-mo-old liver lysates ≥1.5-fold up-regulated proteins (compared with 2- to 3-mo-old liver lysates) and in ABT737-treated liver lysates ≤0.6-fold down-regulated proteins (compared with DMSO-treated liver lysates) (n = 2). (I) List of 19 overlapping proteins of Venn diagram in H. (J) qPCR analysis for p21 and selected SASP factors in DMSO- or ABT737-treated dermal fibroblasts 4 d after continuous treatment (n = 3). Error bars, mean ± SEM, unpaired two-tailed Student's t-test. (*) P ≤ 0.05; (**) P ≤ 0.01 (***) P ≤ 0.001.
Senescent cells are associated with an increased inflammatory environment and secretory capacity, known as the SASP. By qPCR we could not detect increased SASP factors such as IL6, Ccl2, IL1, or Pai1 prior to PH in adult tissue (Supplemental Fig. S4A). Our data pointed to an aberrant expression of p21 3 d after PH in adult tissue. Based on this, we analyzed the expression of 111 proteins, including many SASP factors, by cytokine array. Interestingly, we found that many of these showed increased expression in day 3 adult livers compared with their young counterparts (Fig. 4H; Supplemental Fig. S4B). We then asked whether ABT-737-treated livers with improved regeneration exhibit similar inflammatory profiles. Interestingly, cytokine array analysis demonstrated a large reduction in the expression of many of the same SASP-associated factors, including Pai1, Ccl2, IGFBP-3, and MMP2 (Fig. 4H,I; Supplemental Fig. S4C). This suggests that ABT-737 decreases both p21 and SASP-associated proteins. Such drugs are suggested to induce apoptosis in fully senescent cells. However, in treated adult livers with improved regeneration, we were unable to detect increased apoptosis, nor changes in macrophage number (Supplemental Fig. S5A,B). This suggests that ABT-737 may also interfere with p21 and SASP expression. To investigate this possibility, we treated proliferating p21-positive dermal fibroblasts with ABT-737 and by qPCR, found decreased expression of p21 and some of the same SASP-associated factors including IGFBP3, FGF21, and MMP2, without noticeable cell death (Fig. 4J; Supplemental Fig. S5C). This suggests that this drug can decrease p21 and SASP-levels.
Altogether, this study shows that p21 and SASP-like proteins become aberrantly expressed following PH in adult mice, which, interestingly, appears independent of the other senescence-associated genes p16Ink4a and p19Arf. Importantly, we show that treatment with ABT-737 can improve regenerative capacity, introducing the concept that such drugs could be explored further in regeneration strategies.
In the liver, models of severe damage such as extended hepatectomy (Lehmann et al. 2012), acetaminophen treatment (Bird et al. 2018), Mdm2 deletion (Lu et al. 2015), β1-integrin loss (Raven et al. 2017), or p21 overexpression (Raven et al. 2017), each induce a pronounced p21 expression in hepatocytes, which results in decreased regeneration, senescence, and senescence spreading. It will be interesting to determine whether senotherapeutics can also improve such cases of severe damage. Indeed, senolytics were shown to function in the liver (Ovadya et al. 2018) and in ameliorating age-related hepatic steatosis (Ogrodnik et al. 2017). Also, although future studies testing different drugs with different stages of delivery (before and after injury) are needed, based on these findings it will be interesting to explore whether senotherapeutics can improve regeneration in human liver and other tissues.
Our study suggests that p21-positive cells may be a primary target of senotherapeutics. Interestingly, we found that ABT-737 can decrease levels of p21 and SASP genes, suggesting that this is partly how it might enable apoptosis in senescent cells and ultimately act as a senolytic. Unexpectedly, we see no effect of ABT-737 treatment on the increased expression of p16Ink4a that is present in adult mice. However, as this appears to be primarily in macrophages at this stage and at lower levels than are detectable in advanced age, it is likely that these cells are not senescent. However, as we see prolonged p21 expression (a key mediator of senescence induction) in hepatocytes after injury, this possibly gives clues as to how full senescence arrest might develop. Perhaps with advanced age or chronic damage, p21 and p16Ink4a become expressed at higher levels, or in the same cell types (Wang et al. 2014), and what we witnessed here was an early stage in a cumulative and progressive decline that becomes more complex over time.
Materials and methods
Animal use
Wild-type C57Bl6/J and FVB, p21−/−, p19−/−, and p16−/− mice were used. Breeding and maintenance of mice were performed in the accredited Institut de Génétique et de Biologie Moléculaire et Cellulaire/Mouse Clinical Institute animal house, in compliance with French and European Union regulations on the use of laboratory animals for research, under the supervision of Dr. Bill Keyes, who holds animal experimentation authorizations from the French Ministry of Agriculture and Fisheries. All animal experiments were approved by the ethical committee Com'Eth (Comité d'Ethique pour l'Expérimentation Animale, Strasbourg, France).
Partial hepatectomy (PH)
The mice used in this study were 8- to 14-wk-old (young) and 24- to 32 wk-old (adult) male and female mice. Two-thirds partial hepatectomy was performed under isoflurane anesthesia and according to standard procedures, as described in supplemental material (Mitchell and Willenbring 2008).
Elimination of senescent cells in vivo
For elimination of senescent cells, 6- to 8-mo-old mice were injected once per day intraperitoneally (i.p.) on two consecutive days immediately before PH with DMSO (2%) or ABT-737 (25 mg/kg body weight; AdooQ Bioscience). DMSO and ABT-737 were prepared in the following working solution: 30% propylene glycol, 5% Tween 80, 5% dextrose in water (pH 4–5), essentially as previously reported (Ovadya et al. 2018).
Histology and immunohistochemistry
Fixed liver tissues were washed in PBS and then processed for paraffin embedding and hematoxylin and eosin (H&E) staining. Immunohistochemistry was performed using standard procedures, as described in supplemental material.
RNA in situ hybridization
In situ RNA hybridization was performed using RNAscope probes (Advanced Cell Diagnostics) for CDKN2A (411011), PPIB-positive control (313911) and bacterial Dapb-negative control (310043) and Adgre1 (460651-C2), Pecam1 (316721-C2), Desmin (407921-C2), Ki67 (416771-C2), CDKN1A (408551-C2), PPIB-C1/POLR2A-C2-positive control (321651), and bacterial Dapb-negative control (320751), as per the manufacturer's instructions. RNAscope 2.5 HD reagent kit-RED (322360) and RNAscope 2.5 HD duplex kit (322430) was used for chromogenic labeling and hematoxylin for counterstaining. Images were obtained on a Leica DM 1000 LED microscope.
Cytokine array
Cytokine levels from tissue lysates were analyzed using the mouse XL cytokine array kit (R&D Systems), following the manufacturer's instructions, as described in the Supplemental Material.
RT-qPCR and analysis
Frozen tissue was homogenized in TRI reagent (MRC), purified using Direct-zol RNA miniprep kit (Zymo Research), and reverse-transcribed using qScript cDNA Supermix (VWR International SAS). Values were normalized toward Actinb and Gapdh quantification. Real-time qPCR was performed using gene-specific primers (Supplemental Table 1) and a LightCycler 480 (Roche).
Tissue culture
Primary mouse dermal fibroblasts were isolated from 1- to 2-day-old mice. Cells were treated with 0.05% DMSO, 1 µM ABT737 and 5 µM ABT737 (both with 0.05% DMSO). Medium with treatment agents was changed daily. After 4 d, RNA was isolated using NucleoSpin RNA plus (Machery-Nagel).
Statistical analysis
Statistical analysis was performed using the Prism 8 software (GraphPad Software, Inc.). Results are presented as mean ± S.E.M. Statistical significance was determined by the two-tailed unpaired Student's t-test (P ≤ 0.05 [*]; P ≤ 0.01; [**], and P ≤ 0.001 [***]).
Supplementary Material
Acknowledgments
We thank Muriel Rhinn for help with procedures. We thank the mouse facility at the Mouse Clinical Institute (ICS), the Histology Facilities at the Centre for Genomic Regulation and the Institut de Génétique et de Biologie Moléculaire et Cellulaire/ICS, and the phenotyping platform at the ICS for excellent technical support. Work in the Keyes lab was funded in part by grants from the Spanish Ministry for Economy and Competitiveness (SAF2013-49082-P), La Fondation Recherche Medicale (FRM) (AJE20160635985), Fondation ARC (PJA20181208104), IDEX Attractivité-University of Strasbourg (IDEX2017), and La Fondation Schlumberger pour l'Education et la Recherche (FSER) (FSER 19-Year 2018), and ANR (ANR-19-CE13-0023-03). Work was also supported by grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d'Avenir (ANR-10-IDEX-0002-02). We acknowledge the support of the Spanish Ministry of Science and Innovation to the EMBL partnership, the Centro de Excelencia Severo Ochoa and the CERCA Programme/Generalitat de Catalunya.
Author contributions: B.R., M.P.C., and W.M.K. conceived and designed the study. B.R. performed most of the experiments. T.K.-M., A.M., J.-L.P., D.S.G., M.D., U.D.V., and E.P. performed additional experiments and techniques. H.J. performed histopathological analysis. B.R. and W.M.K. wrote the manuscript. M.P.C. and W.M.K. supervised the project and secured funding. All authors approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.332643.119.
References
- Albrecht JH, Meyer AH, Hu MY. 1997. Regulation of cyclin-dependent kinase inhibitor p21WAF1/Cip1/Sdi1 gene expression in hepatic regeneration. Hepatology 25: 557–563. 10.1002/hep.510250311 [DOI] [PubMed] [Google Scholar]
- Baar MP, Brandt RMC, Putavet DA, Klein JDD, Derks KWJ, Bourgeois BRM, Stryeck S, Rijksen Y, van Willigenburg H, Feijtel DA, et al. 2017. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169: 132–147.e16. 10.1016/j.cell.2017.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, et al. 2016. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530: 184–189. 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, et al. 2017. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547: 179–184. 10.1038/nature22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TG, Müller M, Boulter L, Vincent DF, Ridgway RA, Lopez-Guadamillas E, Lu WY, Jamieson T, Govaere O, Campbell AD, et al. 2018. TGFβ inhibition restores a regenerative response in acute liver injury by suppressing paracrine senescence. Sci Transl Med 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago-Molina LE, Marhenke S, Longerich T, Sharma AD, Boukouris AE, Geffers R, Guigas B, Manns MP, Vogel A. 2013. The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology 58: 1143–1152. 10.1002/hep.26412 [DOI] [PubMed] [Google Scholar]
- Bussian TJ, Aziz A, Meyer CF, Swenson BL, van Deursen JM, Baker DJ. 2018. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 562: 578–582. 10.1038/s41586-018-0543-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Wang Y, Shao L, Laberge R, Demaria M, Campisi J, Janakiraman K, Sharpless NE, Ding S, Feng W, et al. 2015. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med 22: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs BG, Durik M, Baker DJ, Van Deursen JM. 2015. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med 21: 1424–1435. 10.1038/nm.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs BG, Gluscevic M, Baker DJ, Laberge RM, Marquess D, Dananberg J, Van Deursen JM. 2017. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov 16: 718–735. 10.1038/nrd.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkhbold C, Morine Y, Utsunomiya T, Imura S, Ikemoto T, Arakawa Y, Saito Y, Yamada S, Ishikawa D, Shimada M. 2015. Dysfunction of liver regeneration in aged liver after partial hepatectomy. J Gastroenterol Hepatol 30: 1217–1224. 10.1111/jgh.12930 [DOI] [PubMed] [Google Scholar]
- Han M, Yang X, Lee J, Allan CH, Muneoka K. 2008. Development and regeneration of the neonatal digit tip in mice. Dev Biol 315: 125–135. 10.1016/j.ydbio.2007.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. 2008. Senescence of activated stellate cells limits liver fibrosis. Cell 134: 657–667. 10.1016/j.cell.2008.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K, Tschuor C, Rickenbacher A, Jang JH, Oberkofler CE, Tschopp O, Schultze SM, Raptis DA, Weber A, Graf R, et al. 2012. Liver failure after extended hepatectomy in mice is mediated by a p21-dependent barrier to liver regeneration. Gastroenterology 143: 1609–1619.e4. 10.1053/j.gastro.2012.08.043 [DOI] [PubMed] [Google Scholar]
- Liu JY, Souroullas GP, Diekman BO, Krishnamurthy J, Hall BM, Sorrentino JA, Parker JS, Sessions GA, Gudkov AV, Sharpless NE. 2019. Cells exhibiting strong p16INK4a promoter activation in vivo display features of senescence. Proc Natl Acad Sci 116: 2603–2611. 10.1073/pnas.1818313116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loforese G, Malinka T, Keogh A, Baier F, Simillion C, Montani M, Halazonetis TD, Candinas D, Stroka D. 2017. Impaired liver regeneration in aged mice can be rescued by silencing Hippo core kinases MST1 and MST2. EMBO Mol Med 9: 46–60. 10.15252/emmm.201506089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W-Y, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, et al. 2015. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 17: 971–83. 10.1038/ncb3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. 2007. Liver regeneration. J Cell Physiol 213: 286–300. 10.1002/jcp.21172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Willenbring H. 2008. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 3: 1167–1170. 10.1038/nprot.2008.80 [DOI] [PubMed] [Google Scholar]
- Muñoz-Espín D, Serrano M. 2014. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 15: 482–496. 10.1038/nrm3823 [DOI] [PubMed] [Google Scholar]
- Musi N, Valentine JM, Sickora KR, Baeuerle E, Thompson CS, Shen Q, Orr ME. 2018. Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell 17: e12840 10.1111/acel.12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrodnik M, Miwa S, Tchkonia T, Tiniakos D, Wilson CL, Lahat A, Day CP, Burt A, Palmer A, Anstee QM, et al. 2017. Cellular senescence drives age-dependent hepatic steatosis. Nat Commun 8: 15691 10.1038/ncomms15691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadya Y, Landsberger T, Leins H, Vadai E, Gal H, Biran A, Yosef R, Sagiv A, Agrawal A, Shapira A, et al. 2018. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat Commun 9: 5435 10.1038/s41467-018-07825-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payzin-Dogru D, Whited JL. 2018. An integrative framework for salamander and mouse limb regeneration. Int J Dev Biol 62: 393–402. 10.1387/ijdb.180002jw [DOI] [PubMed] [Google Scholar]
- Pedone E, Olteanu VA, Marucci L, Muñoz-Martin MI, Youssef SA, de Bruin A, Cosma MP. 2017. Modeling dynamics and function of bone marrow cells in mouse liver regeneration. Cell Rep 18: 107–121. 10.1016/j.celrep.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. 2011. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O'Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, et al. 2017. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547: 350–354. 10.1038/nature23015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepniak E, Ricci R, Eferl R, Sumara G, Sumara I, Rath M, Hui L, Wagner EF. 2006. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev 20: 2306–2314. 10.1101/gad.390506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG. 2002. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci 99: 11181–11186. 10.1073/pnas.122359899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MJ, Chen F, Li JX, Liu CC, Zhang HB, Xia Y, Yu B, You P, Xiang D, Lu L, et al. 2014. Reversal of hepatocyte senescence after continuous in vivo cell proliferation. Hepatology 60: 349–361. 10.1002/hep.27094 [DOI] [PubMed] [Google Scholar]
- Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, et al. 2018. Senolytics improve physical function and increase lifespan in old age. Nat Med 24: 1246–1256. 10.1038/s41591-018-0092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef R, Pilpel N, Tokarsky-amiel R, Biran A, Ovadya Y, Cohen S, Vadai E, Dassa L, Shahar E, Condiotti R, et al. 2016. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat Commun 7: 1–11. 10.1038/ncomms11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Bao Q, Kumar S, Hu M, Wang GY, Dai G. 2012. Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration. PLoS One 7: e30675 10.1371/journal.pone.0030675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


