In this study, Hong et al. provide new insights into how NF2 mediates upstream signals to regulate the Hippo pathway. They show that NF2's lipid-binding ability is critical for its function in activating the Hippo pathway in response to osmotic stress in mammalian cells, and identify the PIP5K family as novel regulators upstream of Hippo signaling.
Keywords: Hippo pathway, NF2, phospholipids
Abstract
The Hippo pathway is a master regulator of tissue homeostasis and organ size. NF2 is a well-established tumor suppressor, and loss of NF2 severely compromises Hippo pathway activity. However, the precise mechanism of how NF2 mediates upstream signals to regulate the Hippo pathway is not clear. Here we report that, in mammalian cells, NF2's lipid-binding ability is critical for its function in activating the Hippo pathway in response to osmotic stress. Mechanistically, osmotic stress induces PI(4,5)P2 plasma membrane enrichment by activating the PIP5K family, allowing for NF2 plasma membrane recruitment and subsequent downstream Hippo pathway activation. An NF2 mutant deficient in lipid binding is unable to activate the Hippo pathway in response to osmotic stress, as measured by LATS and YAP phosphorylation. Our findings identify the PIP5K family as novel regulators upstream of Hippo signaling, and uncover the importance of phosphoinositide dynamics, specifically PI(4,5)P2, in Hippo pathway regulation.
The Hippo pathway plays an essential role in maintaining tissue homeostasis and organ size through regulating cell proliferation, migration, and differentiation (Pan 2010; Piccolo et al. 2014; Yu et al. 2015). The mammalian Hippo pathway consists of a kinase cascade that negatively regulates the downstream effectors Yes-associated protein (YAP) and WW domain-containing transcription factor (TAZ). The core components of this kinase cascade include mammalian STE20-like protein kinase (MST1/2), which act in parallel with the mitogen-activated protein kinase kinase kinase kinase (MAP4K) family to phosphorylate and activate the large tumor suppressor (LATS1/2). The activated LATS kinases then phosphorylate YAP and TAZ, resulting in YAP and TAZ binding to 14-3-3 and subsequent sequestration in the cytoplasm, where they are eventually targeted for further phosphorylation, ubiquitination, and degradation. Conversely, when the Hippo pathway kinase cascade is inactive, YAP and TAZ are dephosphorylated and translocate into the nucleus, where they act as transcriptional coactivators to induce expression of downstream target genes CTGF, CYR61, and other prosurvival genes by interacting with transcription factors such as the TEA domain (TEAD) family members (Zhao et al. 2008).
Genetically disrupting components of this Hippo pathway kinase cascade results in uncontrolled tissue overgrowth in both Drosophila and mice (Halder and Johnson 2011). Similarly, dysregulation of the Hippo pathway has also been implicated in several human diseases and cancer (Harvey et al. 2013; Moroishi et al. 2015; Plouffe et al. 2015). For example, elevated YAP protein levels and increased nuclear localization are frequently observed in many types of tumors, suggesting a critical role for YAP in tumor progression (Pan 2010; Plouffe et al. 2015). However, few somatic or germline mutations in any Hippo pathway core components have been identified in human patients (Harvey et al. 2013). As a result, many research efforts have been devoted to identifying novel upstream regulators of the Hippo pathway and to elucidating the Hippo pathway interactome in hopes of better understanding of how YAP and TAZ become dysregulated in disease.
Many Hippo pathway upstream regulators, including neurofibromin 2 (NF2), were shown to localize at the plasma membrane (Yin et al. 2013). The plasma membrane plays an important role in transmitting messages by integrating extracellular environment to induce spatiotemporal organization of signaling proteins (Groves and Kuriyan 2010). How these membrane-associated inputs spatially regulate the Hippo pathway components is unclear. NF2, also known as merlin, is a well-established tumor suppressor and regulator of the Hippo pathway (Hamaratoglu et al. 2006; Zhao et al. 2007; Harvey et al. 2013). In humans, genetic loss-of-function mutations in NF2 have been linked to increased incidence of schwannomas, meningiomas, ependymomas (Xiao et al. 2003), and mesothelioma (Sekido 2011). Mice deficient in NF2 develop cancers such as osteosarcomas, fibrosarcomas, and hepatocellular carcinomas (McClatchey et al. 1998). In the mouse liver, deleting YAP completely blocks tumorigenesis induced by NF2 knockout, suggesting that the Hippo pathway is the major signaling pathway mediating the tumorigenic potential of NF2 inactivation (Zhang et al. 2010). In cell culture, deleting NF2 is sufficient to severely compromise LATS and YAP activity in response to several stimuli known to activate the Hippo pathway (Plouffe et al. 2016). These studies highlight the critical and physiologically relevant connection between NF2 and the Hippo pathway in tumorigenesis. However, the upstream regulators of NF2 and the underlying mechanisms by which NF2 regulates Hippo pathway activity has not yet been fully understood.
Osmotic stress triggers an alteration in membrane curvature and membrane tension (Pedersen et al. 2011). Previously, we observed that osmotic stress induces YAP phosphorylation through both LATS-dependent and LATS-independent mechanisms (Hong et al. 2017). Cells respond to osmotic stress through triggering a series of signal transduction events to maintain cellular homeostasis. These events include the activation of the Hippo pathway, leading to YAP phosphorylation at Ser127 site and thus inhibition, as well as the activation of the Nemo-like kinase (NLK), leading to YAP Ser128 phosphorylation and activation (Hong et al. 2017). The dynamic balance between YAP activation and inhibition is tightly regulated as a cellular adaptive response to stress. The mechanisms by which osmotic stress activates the Hippo pathway remained unknown.
In this study, we focus on the underlining mechanisms of how NF2 relays upstream signals to the Hippo pathway regulation in response to osmotic stress. We discovered that NF2 lipid binding at the plasma membrane is essential for osmotic stress-induced activation of the Hippo pathway. Furthermore, we show that osmotic stress induces ADP-ribosylation factor 6 (ARF6) and type I phosphatidylinositol 4-phosphate 5-kinase (PIP5K) interaction, leading to PIP5K activation and enhanced phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] plasma membrane distribution. Membrane-associated PI(4,5)P2 interacts with NF2 to induce downstream Hippo pathway activation. Our study identifies ARF6 as an osmotic sensor in mammalian cells, and defines a signaling cascade involving ARF6, PIP5K, and PI(4,5)P2 dynamics as upstream regulators of the Hippo pathway.
Results
Osmotic stress activates the Hippo pathway kinase cascade through NF2, but independent of NF2 Ser518 phosphorylation
NF2 is a well-established regulator of the Hippo pathway, both through genetic experiments in Drosophila and mice, which have demonstrated that NF2 acts through the Hippo pathway to control tissue growth (Hamaratoglu et al. 2006; Zhang et al. 2010), as well as in vitro, where deletion of NF2 in human cell lines was sufficient to abolish the Hippo pathway response to serum deprivation, actin disruption, and glucose starvation (Plouffe et al. 2016). However, despite its obvious importance, the precise mechanism of how NF2 is activated under these circumstances is unknown.
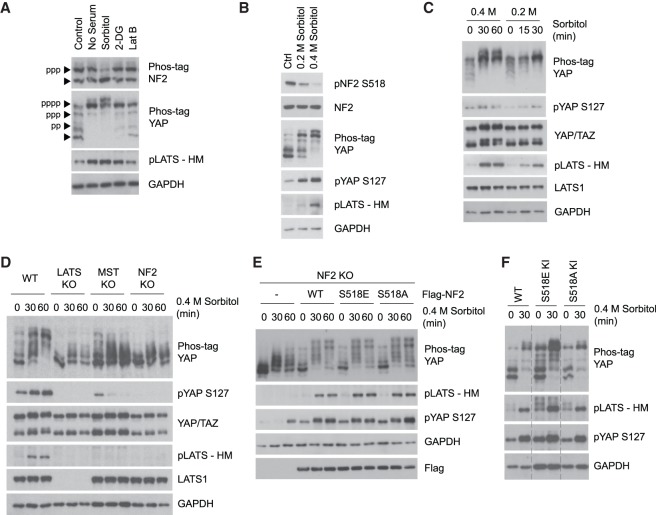
We examined whether NF2 phosphorylation was altered in response to several stimuli known to activate the Hippo pathway. Among the various stimuli tested, including serum deprivation, osmotic stress (sorbitol treatment), energy starvation (2-DG treatment), and actin disruption (Latrunculin B treatment), only osmotic stress induced NF2 dephosphorylation based on the phos-tag gel analysis (Fig. 1A). Hippo pathway activation was assessed by phosphorylation of the LATS hydrophobic motif (HM) and YAP.
Figure 1.
Osmotic stress activates the Hippo pathway through NF2, independent of Ser518 phosphorylation. (A) Osmotic stress specifically induces NF2 dephosphorylation. HEK293A cells were treated with serum starvation, 0.4 M sorbitol, 25 mM 2-DG, or 250 ng/mL Latrunculin B (Lat B) for 30 min. Phos-tag gel was used to assess total phosphorylation of NF2 and YAP based on mobility shift. (B) NF2 is dephosphorylated at Ser518 site in response to increasing osmotic stress. HEK293A cells were treated with either 0.2 or 0.4 M sorbitol for 30 min. Hippo pathway activation is assessed by YAP phosphorylation, based on mobility shift on the phos-tag gel and YAP S127 phospho-specific antibody, and LATS phosphorylation at its hydrophobic motif (HM). NF2 S518 phosphorylation is detected by phospho-specific antibody. (C) Osmotic stress induces LATS and YAP phosphorylation in HEK293A cells. Cells were treated with 0.2 or 0.4 M sorbitol for the indicated times (in minutes). LATS and YAP phosphorylation levels were detected by Western blot. (D) NF2 and MST are important for Hippo pathway activation by osmotic stress. Western blot results for LATS and YAP phosphorylation in wild-type HEK293A, LATS KO, MST KO, and NF2 KO cells treated with 0.4 M sorbitol. (E) NF2 S518 mutants behave similarly as wild type. NF2 WT, S518E, and S518A mutants were transiently expressed in NF2 KO cells, and were then treated with 0.4 M sorbitol. LATS and YAP phosphorylation levels were monitored by Western blot. (F) NF2 S518 phosphorylation status does not determine its function in Hippo pathway regulation. HEK293A WT, S518E knock-in, and S518A knock-in cell lines were treated with 0.4 M sorbitol for 30 min. LATS and YAP phosphorylation levels were monitored by Western blot.
There are many studies suggesting a critical role of serine 518 phosphorylation in NF2 activity regulation (Shaw et al. 2001; Alfthan et al. 2004; Rong et al. 2004; Surace et al. 2004; Okada et al. 2005; Thurneysen et al. 2009; Sher et al. 2012). The Ser518 dephosphorylated NF2 is postulated to be more active in its growth suppressive function. Therefore, we speculated that Ser518 dephosphorylation may play a role in NF2 activation by osmotic stress. We used a phospho-specific antibody to check NF2 Ser518 phosphorylation status and found that hyperosmotic stress led to a dramatic decrease in NF2 Ser518 phosphorylation (Fig. 1B). This NF2 dephosphorylation occurred very rapidly, as it was detected after only 2 min of sorbitol treatment, while YAP was not phosphorylated until 15 min after sorbitol treatment (Supplemental Fig. S1A). These observations prompted us to examine the functional significance of NF2 Ser518 phosphorylation in regulating the Hippo pathway in osmotic stress response.
Hyperosmotic stress, induced by different sorbitol concentrations and assessed at multiple time points, activated the Hippo pathway as indicated by increased LATS and YAP phosphorylation in HEK293A cells and other cell lines (Fig. 1C; Supplemental Fig. S1B,C). To determine whether osmotic stress acts through either the Hippo pathway kinase cascade or NF2, we tested the sorbitol response in LATS, MST, and NF2 knockout (KO) cell lines, which were characterized in our previous studies (Meng et al. 2015; Plouffe et al. 2016). LATS and YAP phosphorylation in the MST and NF2 KO cells were completely abolished in response to low dose sorbitol treatment (Supplemental Fig. S1D) and were strongly compromised in response to high-dose sorbitol treatment (Fig. 1D). Notably, the MST regulatory subunit Salvador 1 (SAV1) was also required for YAP phosphorylation induced by sorbitol, whereas other upstream regulators MAP4K4/6/7 and Angiomotin (AMOT) were not required (Supplemental Fig. S1E). Our result indicates that osmotic stress acts specifically through NF2 and MST-SAV1 complex in mediating LATS and YAP phosphorylation. We further examined the role of NF2 in osmotic stress-induced Hippo pathway activation in two other cell lines MCF7 and HeLa. Similar to HEK293A, osmotic stress activated the Hippo pathway in both MCF7 and HeLa WT cells, while LATS and YAP phosphorylation were completely abolished in NF2 KO MCF7 cells (Supplemental Fig. S1F), and greatly compromised in NF2 KO HeLa cell pools (Supplemental Fig. S1G,H). These results demonstrate that osmotic stress acts through NF2 to regulate the Hippo pathway in a cell-type independent manner.
Next, we tested whether NF2 is generally involved in osmotic stress response or specifically required for Hippo pathway activation. We found that phosphorylation of the p38 MAP kinase, a well-characterized osmotic stress activated kinase, and its substrate MK2, were unaffected by NF2 deletion in HEK293A cells (Supplemental Fig. S1I), indicating a specific role of NF2 in Hippo pathway regulation but not general stress response. Since NF2 Ser518 phosphorylation is known to be inhibitory and is dephosphorylated by osmotic stress, we postulated that osmotic stress may induce NF2 Ser518 dephosphorylation to activate NF2 and the Hippo pathway. To test this hypothesis, we generated constructs containing either wild-type NF2, an NF2 phosphorylation-mimicking S518E mutant, or phosphorylation-deficient S518A mutant. When expressed in the NF2 KO cells, the wild-type NF2 rescued the osmotic stress-induced LATS and YAP phosphorylation (Fig. 1E). Surprisingly, both NF2 S518E and S518A mutant were also able to rescue LATS and YAP phosphorylation in response to sorbitol treatment. This was an unexpected finding as the previous reports showing that Ser518 phosphorylation inhibits NF2 (Shaw et al. 2001; Rong et al. 2004; Surace et al. 2004; Sher et al. 2012).
To avoid potential artifact due to ectopic expression of the NF2 phosphorylation mutants, we performed CRISPR-Cas9 gene editing to knock-in both the NF2 S518E and S518A mutations. HEK293A cells have three alleles for NF2. The NF2 S518E KI cell line is homozygous, with all three alleles containing the S518E mutation (Supplemental Fig. S1J). The S518A KI cell line contains the S518A mutation in two alleles, while the third allele is an out-of-frame deletion (Supplemental Fig. S1K); therefore, the NF2 total protein expression in the S518A KI cells is slightly lower (Supplemental Fig. S1L). Similar to results from the rescue experiments, neither the S518E KI nor S518A KI cells showed observable difference in the phosphorylation of LATS or YAP in response to osmotic stress when compared with the wild-type control cells (Fig. 1F). These results demonstrate that under osmotic stress, Ser518 phosphorylation is unlikely to be involved in mediating NF2 regulation of the Hippo pathway, although Ser518 phosphorylation was reduced. As the Hippo pathway is the major effector of NF2 function, our data question the widely perceived notion that Ser518 phosphorylation plays a critical role in NF2 regulation.
Phospholipid binding is essential for NF2 activity toward the Hippo pathway
NF2 function may be determined by its posttranslational modifications, localization, or interactions with binding partners (Cooper and Giancotti 2014). Based on phosphorylation data from published literatures and high-throughput mass spectrometry cultivated on PhosphoSitePlus, NF2 has several major phosphorylation sites in addition to S518 (Hornbeck et al. 2019). We generated these NF2 phosphorylation mutations; however, an NF2 mutant mimicking phosphorylation of these sites (NF2 6E: S10E, S13E, T230E, S315E, S518E, and S581E) did not preclude LATS and YAP phosphorylation either at the basal or in response to osmotic stress when expressed in the NF2 KO cells (Supplemental Fig. S2). Therefore, NF2 regulation of the Hippo pathway by osmotic stress is most likely independent of NF2 phosphorylation status.
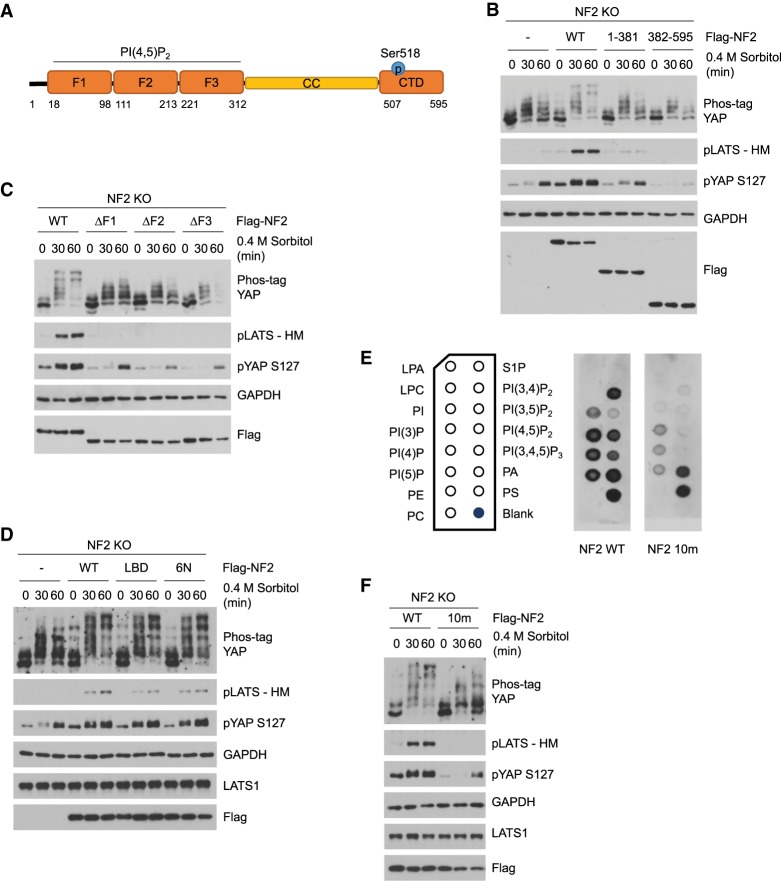
The NF2 protein structure consists of a FERM domain with three subdomains (F1, F2, and F3), followed by a coiled-coil domain (CC) and a C-terminal domain (CTD) (Fig. 2A). It has been shown that the F2 subdomain interacts with the LATS kinases (Li et al. 2015). To determine which domains are essential for NF2 regulation of the Hippo pathway, we generated several truncated NF2 mutants, expressed them in the NF2 KO cells, and monitored LATS and YAP phosphorylation status following sorbitol treatment. Deleting the entire CTD (NF2 1-381) largely blunted the LATS and YAP phosphorylation response, while deleting the N-terminal FERM domain (NF2 382-595) completely blocked any LATS or YAP phosphorylation (Fig. 2B). To determine which FERM subdomains are most critical, we individually deleted each of the F1, F2, and F3 subdomains. Interestingly, we found that all three subdomains were equally important, as deleting any of the subdomains was sufficient to compromise NF2 function as indicated by LATS and YAP phosphorylation (Fig. 2C). That the entire FERM domain is required for NF2 function raises the possibility that NF2 function in relation to the Hippo pathway is primarily determined not by individual posttranslational modifications but rather by the NF2 protein conformation and interactions with its binding partners mediated by the FERM domain.
Figure 2.
Phospholipid binding is essential for NF2 activity toward the Hippo pathway. (A) Schematic diagram of the NF2 protein structure. NF2 consists of a FERM domain containing three subdomains (F1, F2, and F3), a coiled-coil (CC) domain, and a C-terminal domain (CTD). NF2 interacting partner and region are listed in the diagram. (B) The FERM domain is essential for NF2 activity. NF2 truncated constructs were transfected into NF2 KO cells. Flag-NF2 1-381 is missing the entire C-terminal domain (CTD), and Flag-NF2 382-595 is missing the entire FERM domain. Cells were treated with 0.4 M sorbitol for the indicated times (in minutes), and LATS and YAP phosphorylation was detected by Western blot. (C) Each of the F1, F2, and F3 subdomains is required for NF2 function. NF2 constructs with deletion of either the F1, F2, or F3 subdomain were transfected into the NF2 KO cells. Cells were treated with 0.4 M sorbitol for the indicated times (in minutes), and LATS and YAP phosphorylation was detected by Western blot. (D) NF2 LBD and 6N mutants show slight decrease in their activities. The NF2 LBD mutant contains the T59V, W60E, R309Q, and R310Q mutations, and the NF2 6N mutant contains the K79N, K80N, K269N, E270N, K278N, and K279N mutations. NF2 wild-type, LBD, and 6N were transiently expressed in NF2 KO cells and treated with 0.4 M sorbitol for the indicated times (in minutes). LATS and YAP phosphorylation levels were detected by Western blot. (E) NF2 10m mutant has defect in binding with phosphoinositides. Flag NF2 WT and 10m proteins were purified and subjected to lipid-binding assay. A strip of phospholipids was used to test for NF2 lipid-binding ability. (F) Lipid binding is essential for NF2 function under osmotic stress. NF2 WT and the 10m mutants were transiently expressed in NF2 KO cells. Cells were treated with 0.4 M sorbitol for the indicated times (in minutes), and LATS and YAP phosphorylation levels were detected by Western blot.
In addition to binding to LATS, the NF2 FERM domain has also been shown to interact with the phospholipid PI(4,5)P2 (Shimizu et al. 2002; Stickney et al. 2004). PI(4,5)P2 binding has been shown to be important for Ezrin function (Nakamura et al. 1999; Barret et al. 2000; Fievet et al. 2004), which is a member of the ezrin–radixin–moesin (ERM) protein family and has a high degree of sequence similarity with NF2 (Rouleau et al. 1993; Trofatter et al. 1993). Two independent sets of mutations in the NF2 FERM domain have been shown to decrease the NF2-binding affinity to PI(4,5)P2 (Mani et al. 2011; Chinthalapudi et al. 2018). In accordance with previous reports and incorporating these mutations, we generated two NF2 mutants: LBD (T59V, W60E, R309Q, and R310Q) and 6N (K79N, K80N, K269N, E270N, K278N, and K279N). We expressed each of these mutants in the NF2 KO cells and evaluated Hippo pathway activity in response to osmotic stress. Each of the mutants were partially defective, as there was a slight decrease in LATS phosphorylation and activity in each (Fig. 2D); however, this reduced LATS activity was still sufficient to phosphorylate YAP.
Since the NF2 LBD and 6N mutants were unable to completely abolish NF2 function on their own, we generated a third mutant, the NF2 10m, which combined the LBD and 6N mutations. We performed a protein–lipid-binding assay to test the binding affinity of the NF2 10m mutant with an array of phospholipids. In brief, NF2 WT and 10m proteins were expressed, purified, and incubated with a membrane strip spotted with different types of phospholipids. The NF2 10m mutant exhibited decreased binding affinity toward all of the phosphatidylinositol groups, especially toward PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3 (Fig. 2E). Finally, to determine the functional activity of the NF2 10m mutant, we transfected both NF2 WT and the NF2 10m mutant into the NF2 KO cells. The NF2 10m mutant completely abolished LATS phosphorylation (Fig. 2F), and similar to NF2 KO cells, YAP phosphorylation remained minimal. These data strongly indicate that NF2 requires its lipid-binding ability to regulate the Hippo pathway in response to sorbitol treatment.
Osmotic stress induces PI(4,5)P2 membrane enrichment and NF2 colocalization
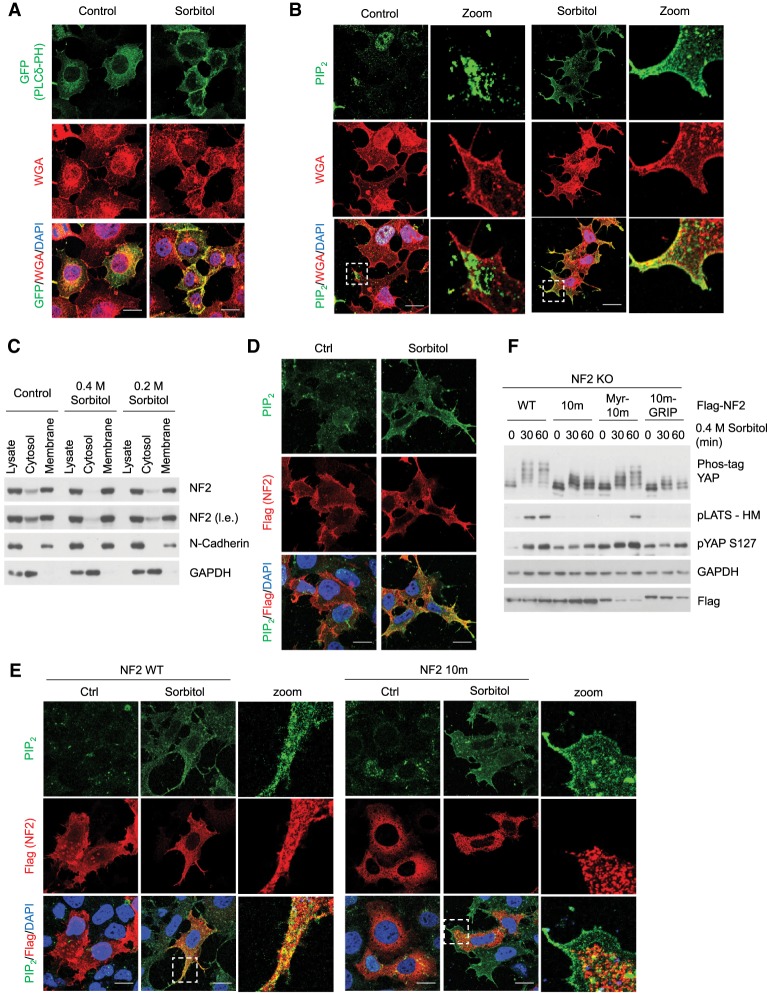
The phospholipids that showed the greatest difference in binding between the NF2 WT and NF2 10m mutant were the phosphoinositides, while other phospholipids such as PS showed less difference (Fig. 2E). Because the spatiotemporal organization of phosphoinositides plays a critical role in their regulation and activity (Di Paolo and De Camilli 2006), we used immunofluorescence staining (IF) to determine whether phosphoinositides are responsive to osmotic stress. PI(4,5)P2 and PI(4)P are the two most abundant phosphoinositides in mammalian cells (Di Paolo and De Camilli 2006). To visualize, GFP PLCδ-PH and GFP P4M-SidM were used as reporters for PI(4,5)P2 and PI(4)P, respectively (Stauffer et al. 1998; Hammond et al. 2014). Distribution of PI(4,5)P2 was strongly enriched in the plasma membrane compartment following sorbitol treatment (Fig. 3A), while the distributions of PI(4)P and the GFP-only control were largely unaffected (Supplemental Fig. S3A). Thus, it seems that hyperosmotic stress selectively induces PI(4,5)P2 membrane localization. A PI(4,5)P2-specific antibody was also tested to further confirm the PI(4,5)P2 enrichment in the plasma membrane compartment after sorbitol treatment (Fig. 3B).
Figure 3.
Osmotic stress induces PI(4,5)P2 plasma membrane enrichment and NF2 colocalization. (A) Osmotic stress induces PI(4,5)P2 membrane localization assayed by PI(4,5)P2 reporter. HEK293A cells were transiently transfected with the PI(4,5)P2 reporter GFP-PLCδ-PH and is shown in green. Plasma membrane marker WGA is shown in red, and the nuclei (DAPI) are shown in blue. Cells were treated with 0.4 M sorbitol for 30 min. Scale bar, 20 µm (for this and other panels in this figure). (B) Osmotic stress induces PI(4,5)P2 membrane localization assayed by PI(4,5)P2 antibody. HEK293A cells were treated with 0.4 M sorbitol for 30 min. PI(4,5)P2 is shown in green, Plasma membrane marker (WGA) is shown in red, and the nuclei (DAPI) are shown in blue. Higher-magnification images of the boxed area are shown at the right. (C) Osmotic stress induces NF2 redistribution to membrane compartment. HEK293A cells were treated with 0.4 or 0.2 M sorbitol for 30 min. Cytosolic and membrane fractions were collected by differential fractionation. GAPDH and N-Cadherin were used as cytosolic and membrane markers, respectively. (l.e.) Long exposure. (D) Osmotic stress induces PI(4,5)P2 and NF2 colocalization. Flag-NF2 was transiently transfected into HEK293A cells for 24 h, and then cells were treated with 0.4 M sorbitol for 30 min. NF2 was stained with an anti-Flag antibody and is shown in red. PI(4,5)P2 is shown in green, and the nuclei (DAPI) are shown in blue. (E) NF2 10m mutant does not colocalize with PI(4,5)P2 upon sorbitol treatment. Flag-NF2 WT and the 10m mutant were transiently transfected into cells for 24 h, and then cells were treated with 0.4 M sorbitol for 30 min. NF2 was stained with an anti-Flag antibody and is shown in red. PI(4,5)P2 is shown in green, and the nuclei (DAPI) are shown in blue. Higher-magnification images of the boxed area are shown at the right. (F) Plasma membrane localization of NF2 10m mutant restores its ability in regulating the Hippo pathway. NF2 WT, 10m, Myr-10m, and 10m-GRIP mutants were transiently expressed in NF2 KO cells. Cells were treated with 0.4 M sorbitol for the indicated times (in minutes), and LATS and YAP phosphorylation were detected by Western blot.
It is reported that phosphatidic acid (PA) inhibits the Hippo pathway by directly binding to NF2 (Han et al. 2018). We performed liposome-binding assay that mimics the lipid-bilayer membrane environment in the cell (Putta et al. 2016) to test for NF2-binding ability with PA. We found that NF2 association with liposome was enhanced by the presence of PI(4,5)P2 but not PA (Supplemental Fig. S3B), indicating that NF2 has low binding affinity toward PA. We further used IF to determine PA localization using a PA-specific reporter RFP-PASS (Zhang et al. 2014). No significance difference in PA levels or distribution was observed upon sorbitol treatment (Supplemental Fig. S3C). These data suggest that PA is not regulated by osmotic stress and unlikely to contribute to Hippo pathway activation induced by osmotic stress.
Regulation of membrane dynamic through reversible recruitment of cytosolic proteins to specific membranes is crucial for cellular functions (Schink et al. 2016). We speculate that PI(4,5)P2 enrichment at the membrane compartment in response to sorbitol treatment may recruit NF2 to the plasma membrane, where NF2 stimulates the Hippo pathway. To test this hypothesis, we performed subcellular fractionation to separate cytosol and membrane compartments and assayed NF2 localization by Western blot. We found that NF2 cytosolic portion decreased under both 0.4 M and 0.2 M sorbitol treatment (Fig. 3C), suggesting that osmotic stress induces NF2 membrane translocation. We then transfected and stained for Flag-NF2 and found that osmotic stress not only led to increased PI(4,5)P2 membrane localization, but also increased PI(4,5)P2 colocalization with NF2 (Fig. 3D). Consistent with the protein–lipid-binding assay in which the NF2 10m mutant showed greatly reduced binding to PI(4,5)P2, we found that the Flag-NF2 10m mutant failed to colocalize with PI(4,5)P2 or localize to the plasma membrane following sorbitol treatment (Fig. 3E). To further test whether the NF2 plasma membrane localization recruited by PI(4,5)P2 plays a role in Hippo pathway activation, we targeted the lipid-binding-deficient NF2 10m mutant to the plasma membrane by inserting an N-terminal myristoylation tag (Myr-10m), or to the Golgi compartment with a C-terminal GRIP domain (10m-GRIP). Our data show that only NF2 Myr-10m regained its activity toward Hippo pathway activation upon sorbitol treatment (Fig. 3F; Supplemental Fig. S3D). This result adds strong support to our model that NF2 plasma membrane recruitment by PI(4,5)P2 contributes to Hippo pathway activation. On the other hand, both the NF2 6N and LBD mutants managed to partially localize to the plasma membrane, albeit to a lesser degree than the NF2 WT (Supplemental Fig. S3E); this may account for why the NF2 6N and LBD mutants retain some activity to induce LATS and YAP phosphorylation (Fig. 2D). Note that plasma membrane targeted NF2 is not constitutively active as overexpression of Myr-NF2 did not show higher basal LATS and YAP phosphorylation than NF2 WT, and it was able to restore osmotic stress response in NF2 KO cells (Supplemental Fig. S3F). We speculate that in addition to plasma membrane distribution, NF2 needs to be localized to specific plasma membrane subdomain to activate the Hippo pathway.
PIP5K is an upstream regulator of the Hippo pathway in response to osmotic stress
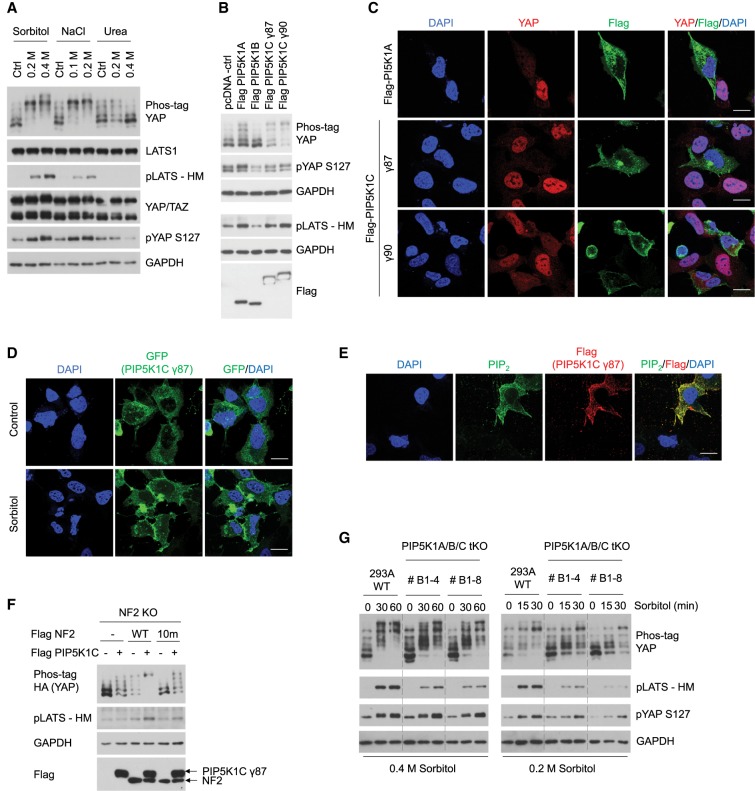
Hyperosmotic stress can be induced by a variety of methods, such as using inorganic salts or organic osmolytes. However, these methods can trigger distinct downstream mechanisms. It has been reported that NaCl-induced, but not urea-induced, osmotic stress activates PIP5Ks, which are the primary kinases that synthesize PI(4,5)P2 by phosphorylating PI(4)P (Yamamoto et al. 2006). If osmotic stress indeed relies specifically on PI(4,5)P2 to activate the Hippo pathway, then we might expect that urea treatment would not activate the Hippo pathway. This prediction is supported by our observation that urea failed to increase LATS or YAP phosphorylation (Fig. 4A). By treating the cells with p38 inhibitor SB203580 together with JNK inhibitor SP600125, we ruled out the possibility that p38 and JNK, another MAP kinase that is known to be activated by osmotic stress, are responsible for Hippo pathway activation by osmotic stress (Supplemental Fig. S4A). Together, our results suggest a key role of PIP5Ks in osmotic stress-induced Hippo pathway activation.
Figure 4.
PIP5Ks act as upstream regulators of the Hippo pathway in response to osmotic stress. (A) Hyperosmotic stress induced by urea does not activate the Hippo pathway. HEK293A cells were treated with different dosage of sorbitol, NaCl, or urea for 30 min. LATS and YAP phosphorylation was detected by Western blot. (B) PIP5K1A and PIP5K1C activate the Hippo pathway. PIP5K1A, PIP5K1B, PIP5K1C γ87, and PIP5K1C γ90 were overexpressed in HEK293A cells. Expression levels of PIP5Ks were detected using an anti-Flag antibody. LATS and YAP phosphorylation levels were detected by Western blot. (C) PIP5K1A and PIP5K1C induce YAP cytoplasmic localization. Flag-PIP5K1A, Flag-PIP5K1C γ87, and Flag-PIP5K1C γ90 were transiently transfected into HEK293A cells. PIP5Ks were stained using an anti-Flag antibody and are shown in green, YAP is shown in red, and nuclei (DAPI) are shown in blue. Scale bar, 20 µm (for this and other panels in this figure). (D) Osmotic stress induces PIP5K1C γ87 plasma membrane localization. GFP-PIP5K1C γ87 was transiently transfected into the cells and is shown in green. Cells were treated with 0.4 M sorbitol for 30 min. Nuclei (DAPI) are shown in blue. (E) PIP5K expression induces PI(4,5)P2 levels and plasma membrane localization. Flag-PIP5K1C γ87 was transiently transfected into the cells. PIP5K1C γ87 was stained using an anti-Flag antibody and is shown in red, PI(4,5)P2 is shown in green, and the nuclei (DAPI) are shown in blue. (F) NF2 lipid binding is required for PIP5K1C to activate the Hippo pathway. NF2 WT, NF2 10m mutant, and PIP5K1C γ87 were transiently transfected into NF2 KO cells in different combinations. LATS and YAP phosphorylation was determined by Western blot. (G) PIP5K1A/B/C tKO shows impaired Hippo pathway activation in response to osmotic stress. PIP5Ks KO cells with deletion of PIP5K1A, PIP5K1B, and PIP5K1C were generated by CRISPR-Cas9 gene editing. LATS and YAP phosphorylation in wild-type, PIP5Ks KO clone #B1-4, and clone #B1-8 were detected by Western blot following treatment with 0.4 M or 0.2 M of sorbitol at indicated times (in minutes).
There are three major isoforms of PIP5Ks: PIP5K1A, PIP5K1B, and PIP5K1C (Ishihara et al. 1996, 1998; Loijens and Anderson 1996). To determine which members of the PIP5K family could regulate the Hippo pathway, Flag-tagged PIP5Ks were transiently transfected into HEK293A cells. Ectopic expression of PIP5K1A, PIP5K1C γ87, or PIP5K1C γ90, but not PIP5K1B, increased LATS and YAP phosphorylation (Fig. 4B) and YAP cytoplasmic translocation (Fig. 4C). The reduced YAP staining in the PIP5K transfected cells is consistent with our previous study that phosphorylation can induce YAP ubiquitination and degradation (Zhao et al. 2010). Next, we examined the effect of osmotic stress on PIP5K subcellular localization, as PIP5Ks are activated upon membrane association to produce PI(4,5)P2 (van den Bout and Divecha 2009; Liu et al. 2016). We found that the GFP-tagged PIP5K1C γ87 was enriched in the plasma membrane compartment, especially at the cell–cell junctions, following sorbitol treatment (Fig. 4D). As expected, overexpression of PIP5K1C γ87 resulted in increased PI(4,5)P2 levels (Fig. 4E). These data support the notion that PIP5K is activated and increases plasma membrane PI(4,5)P2 levels in response to hyperosmotic stress.
In order to further support that PIP5K activity is upstream of PI(4,5)P2 and NF2, we tested whether the lipid-binding ability of NF2 is required for PIP5Ks to activate the Hippo pathway. Indeed, overexpressing PIP5K1A or PIP5K1C γ87 dramatically increased LATS and YAP phosphorylation in NF2 KO cells that were rescued with wild-type NF2, while NF2 10m mutant rescued cells only exhibited a blunted response (Fig. 4F; Supplemental Fig. S4B). Together, these data support the model that PIP5Ks act through PI(4,5)P2 and NF2 to regulate the Hippo pathway.
Additionally, we generated PIP5K1A/B/C tKO cell lines using CRISPR–Cas9 (Supplemental Fig. S4C [clone #B1-4], Supplemental Fig. S4D [clone #B1-8]). Both of these cell lines exhibited a marked decrease in LATS phosphorylation in response to sorbitol treatment compared with the wild-type cells (Fig. 4G), supporting an important role for PIP5K in mediating the osmotic stress response to activate the Hippo pathway. Notably, the effect on YAP phosphorylation in these KO cell lines was relatively modest (Fig. 4G). A possible explanation is that a low level of LATS kinase activity can induce substantial YAP phosphorylation. In addition, although PI(4,5)P2 is primarily synthesized by the PIP5K family, there are other pathways that can lead to increased PI(4,5)P2 levels, such as phosphatidylinositol 5-phosphate 4-kinase (PIP4K) phosphorylating PI(5)P, or phosphatase and tensin homolog (PTEN) dephosphorylating PI(3,4,5)P3 (van den Bout and Divecha 2009). These compensatory pathways may contribute to PI(4,5)P2 production in PIP5K KO cells.
ARF6 acts in coordination with PIP5K to stimulate the Hippo pathway
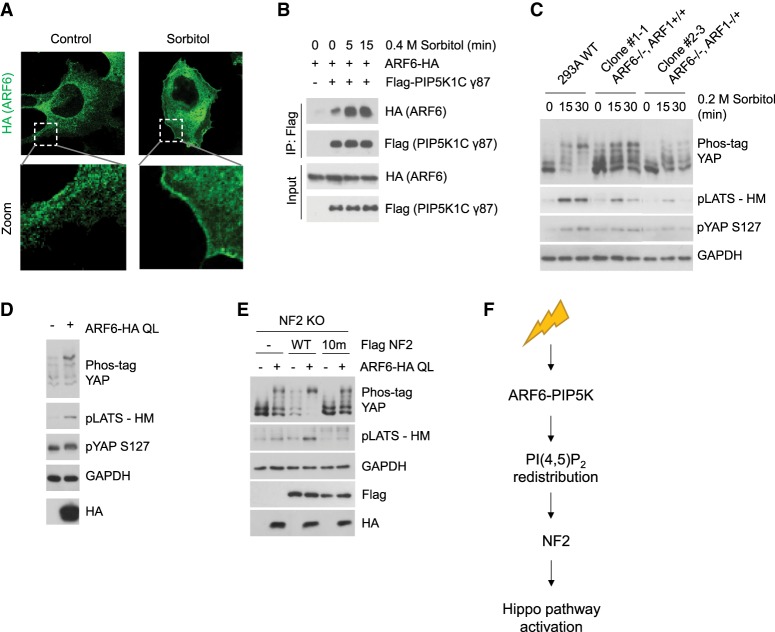
The small GTPase ARF6 has been reported to regulate PI(4,5)P2 synthesis through direct activation of PIP5K (Honda et al. 1999; Jones et al. 2000; Funakoshi et al. 2011). Interestingly, ARF6 also responds to membrane curvature and is involved in cytoskeleton rearrangements (Boshans et al. 2000; Lundmark et al. 2008). Since cell volume change and cytoskeleton remodeling are key hallmarks of the osmotic stress response, we speculated that ARF6 may mediate the osmotic stress response to regulate PIP5K, hence the Hippo pathway. Similar to PIP5K, we found that ARF6 translocated to the plasma membrane following sorbitol treatment (Fig. 5A). Increased ARF6 and PIP5K interaction has also been reported to induce PIP5K activation (Aikawa and Martin 2003). By coimmunoprecipitation, we found that osmotic stress rapidly induced ARF6 and PIP5K association (Fig. 5B), suggesting a possible mechanism that osmotic stress promotes ARF6 interaction and thus activation of PIP5K.
Figure 5.
ARF6 acts in coordination with PIP5K to activate the Hippo pathway. (A) Osmotic stress induces ARF6 plasma membrane localization. ARF6-HA was transiently transfected into HEK293A cells. Cells were treated with 0.4 M sorbitol for 30 min. ARF6 was stained with an anti-HA antibody and is shown in green. Higher-magnification images of the boxed area are shown at the bottom. Scale bar, 20 µm. (B) Osmotic stress induces ARF6 and PIP5K1C interaction. HEK293A cells were transiently transfected with HA-tagged ARF6 and Flag-tagged PIP5K1C γ87, and were treated with 0.4 M sorbitol at different time points (in minutes). PIP5K1C γ87 was immunoprecipitated with the Flag antibody, and ARF6 and PIP5K1C were detected using the anti-HA and anti-Flag antibodies, respectively, by Western blot. (C) ARF6 deletion compromises Hippo pathway activation under sorbitol treatment. ARF1/6 KO clone #1-1 cells have complete loss of ARF6, and clone #2-3 has complete loss of ARF6 and partial deletion of ARF1. LATS and YAP phosphorylation in wild-type HEK293A, ARF1/6 KO clone #1-1, and clone #2-3 was detected by Western blot following treatment with 0.2 M sorbitol for the indicated times (in minutes). (D) ARF6 induces LATS and YAP phosphorylation. Constitutively active ARF6 (ARF6 QL) was overexpressed in HEK293A cells. LATS and YAP phosphorylation was detected by Western blot. (E) NF2 lipid binding is important for Hippo pathway activation by ARF6. NF2 WT, NF2 10m mutant, and ARF6 QL were transiently transfected into NF2 KO cells in different combinations. LATS and YAP phosphorylation was detected by Western blot. (F) A proposed model linking osmotic stress to the Hippo pathway.
To determine whether ARF6 is involved in Hippo pathway regulation, we used CRISPR-Cas9 gene editing to delete ARF6 and ARF1, which is the closest homolog of ARF6 and also known to activate PIP5K (Jones et al. 2000; Skippen et al. 2002). Complete ARF1/6 double KO cells could not be maintained (Supplemental Fig. S5A,B). However, we were able to obtain a complete ARF6 KO clone (clone #1-1: ARF6−/−, ARF1+/+) (Supplemental Fig. S5C) and a complete ARF6 KO clone with a heterozygous KO of ARF1 (clone #2-3: ARF6−/−, ARF1−/+) (Supplemental Fig. S5D). Both cell lines, especially clone #2-3, showed compromised Hippo pathway activation in response to low-dose sorbitol treatment (Fig. 5C), while only the ARF6−/−, ARF1−/+ cell line (clone #2-3) showed diminished Hippo pathway activation in response to the high-dose sorbitol treatment (Supplemental Fig. S5E). These results suggest that both ARF1 and ARF6 play a role in Hippo pathway regulation by osmotic stress, and that there exists some functional redundancy between ARF1 and ARF6. Finally, to further support the role of ARF6, we overexpressed the ARF6 constitutive-active Q67L mutant. ARF6 Q67L induced both LATS and YAP phosphorylation (Fig. 5D). Collectively, our data indicate that ARF6 serves as an upstream regulator and activator of the Hippo pathway.
Next, we tested the function of NF2 for Hippo pathway activation by ARF6. ARF6 overexpression induced a weak phosphorylation of LATS and YAP in NF2 KO cells. This increased LATS and YAP phosphorylation by ARF6 can be potentiated by re-expression of wild-type NF2, but not the NF2 10m mutant (Fig. 5E). These data suggest that NF2 lipid binding is important for the Hippo pathway activation by ARF6, which is consistent with our model that ARF6 activates the Hippo pathway through regulating PI(4,5)P2. Notably, overexpression of ARF6 was still able to induce some YAP phosphorylation in NF2 KO cells (Fig. 5E), indicating that ARF6 may have additional mechanisms to induce YAP phosphorylation. In summary, we propose that ARF6 and PIP5K act together to generate plasma membrane domains enriched in PI(4,5)P2, which then recruits NF2 to activate the Hippo pathway under hyperosmotic stress conditions (Fig. 5F).
Last, we wanted to know whether this ARF6–PIP5K–PI(4,5)P2–NF2 signaling axis is specific to osmotic stress. Two other stimuli that are known to activate the Hippo pathway, serum starvation, and energy stress (2-DG treatment), were treated to wild-type HEK293A, NF2 KO, PIP5K1A/B/C tKO, and ARF1/6 KO (clone #2-3) cells, and Hippo pathway activity was assayed by Western blot. Our results show that both stimuli require NF2 and PIP5Ks to stimulate the Hippo pathway, as NF2 KO cells completely abolished LATS and YAP phosphorylation, and PIP5K tKO cells exhibited a reduced LATS phosphorylation (Supplemental Fig. S5F,G). Similar to osmotic stress, energy stress acts through ARF1 and ARF6 to activate the Hippo pathway, indicated by the decreased LATS phosphorylation in ARF1/6 KO (clone #2-3) cells. In contrast, serum starvation activates the Hippo pathway independent of ARF1 and ARF6. From these observations, we conclude that NF2 and PIP5K are more broadly involved in Hippo pathway regulation, whereas ARF6 is required for Hippo pathway activation induced by a subset of stimuli, such as energy stress and osmotic stress.
Discussion
The Hippo pathway plays an essential role in maintaining tissue homeostasis by balancing cell proliferation and differentiation. Exploring the upstream regulatory network of the Hippo pathway is critical for understanding its role in cellular signaling and how it becomes dysregulated in disease. NF2 is a well-established tumor suppressor and key component upstream of the Hippo pathway, and mutations in NF2 have been frequently observed in human cancer. Previous studies have demonstrated that NF2 is necessary for Hippo pathway activation, and that loss of NF2 significantly disrupts LATS and YAP phosphorylation. However, despite extensive research on NF2 regulation, the precise mechanism of how NF2 activates the Hippo pathway remain poorly understood. This report reveals the critical function and mechanism of NF2 in mediating osmotic stress signal to Hippo pathway activation.
It is widely reported that NF2 phosphorylation at Ser518 determines its activity and tumor suppressor function; however, few of these studies have examined NF2 activity in the context of Hippo pathway regulation. To investigate whether NF2 S518 phosphorylation is critical in regulating the Hippo pathway, we generated NF2 phospho-mimetic (S518E) and NF2 phospho-deficient (S518A) knock-in cell lines. Ultimately, neither of these NF2 knock-in cell lines showed any difference in Hippo pathway activity in response to various conditions, including serum starvation, cell detachment (data not shown), or osmotic stress. These observations suggest that NF2 S518 dephosphorylation alone does not regulate NF2 activity toward the Hippo pathway. Activation of the Hippo pathway represents a major mechanism for the tumor suppressor function of NF2. Our data raise concerns about the essentiality of S518 phosphorylation status in determining NF2 function in the context of cell growth regulation.
Through domain mapping, we identified that the NF2 FERM domain is essential for NF2 regulation of the Hippo pathway, as osmotic stress fails to induce LATS or YAP phosphorylation if NF2 is missing any of its three FERM subdomains. Also, consistent with previous reports that NF2 is localized to lipid rafts by binding to phospholipids (Stickney et al. 2004; Mani et al. 2011), we found that NF2 recruitment to the plasma membrane through lipid binding is required for Hippo pathway activation, and that an NF2 mutant deficient in phosphoinositide binding fails to activate the Hippo pathway in response to osmotic stress. Introduction of 10 point mutations to NF2 might alter its conformation. However, our data suggest that NF2 10m mutant is unlikely to be drastically misfolded, as it can still interact with Kibra (data not shown), and targeting this mutant to the plasma membrane by inserting a myristoylation tag (Myr-10m) can rescue its activity in Hippo pathway activation (Fig. 3D; Supplemental Fig. S3D). Nonetheless, we could not rule out the possibility of a partial conformational alteration. Although NF2 has been previously shown to interact with PI(4,5)P2 and that this interaction may modulate NF2 function (Mani et al. 2011; Chinthalapudi et al. 2018), to our knowledge, no physiological signals have been identified that relay NF2 and PI(4,5)P2 interaction to Hippo pathway activation. Our study emphasizes the spatial regulation of NF2 in coordination with phospholipid dynamics as a key mechanism in Hippo pathway regulation. We posit that NF2 plasma membrane association through binding to PI(4,5)P2 potentiates NF2 in Hippo pathway activation.
PI(4,5)P2 levels are dynamically maintained through regulated synthesis and degradation. PI(4,5)P2 is primarily synthesized by PIP5K phosphorylation of PI(4)P. In this report, we present evidence that the PIP5K family, along with its activator ARF6, also serve as upstream regulators of the Hippo pathway. We propose a model that osmotic stress induces a series of signaling cascades, including ARF6 and PIP5K activation, which in turn enhances plasma membrane localization of PI(4,5)P2 and increases PI(4,5)P2 interaction with NF2. ARF6 may function as a sensor by detecting alteration in membrane curvature that is caused by osmotic stress to initiate the osmosignaling and Hippo pathway activation (Lundmark et al. 2008). NF2 binding to PI(4,5)P2 then induces downstream Hippo pathway activation. This report uncovers a novel pathway in which osmotic stress activates the Hippo pathway through phospholipids, which highlights the complexity of Hippo pathway regulation and opens new doors for future inquiries.
Although we identified PI(4,5)P2 as an important contributor to NF2 activity and signaling, clearly there are other phospholipids capable of interacting with NF2. It will be interesting to explore how other phospholipids may regulate the Hippo pathway in other contexts. We found that NF2 also interacts with phosphatidylserine (PS), which is a major structural phospholipid component for membrane bilayer. We speculate that the interaction with PS may provide a nonselective and low-affinity binding for NF2 association with membrane bilayer, while the interaction with PI(4,5)P2 provides specificity for NF2 to be recruited to PI(4,5)P2-enriched subdomains on the plasma membrane, where NF2 recruits and activates Hippo pathway components such as LATS (Yin et al. 2013).
In addition to NF2, we also observed a critical role of MST in Hippo pathway activation by osmotic stress. Loss of MST1 and MST2 abolished osmotic stress-induced Hippo pathway activation even in the presence of MAP4Ks. This is a rather interesting observation because for many other stimuli, including serum deprivation, glucose starvation, or actin disruption, MST1/2 and MAP4Ks show functional redundancy as deletion of both MST1/2 and MAP4K4/6/7 is required to completely abolish Hippo pathway activation (Meng et al. 2015). Therefore, osmotic stress may serve as a useful tool for future mechanistic and regulatory studies of the MST kinases.
Materials and methods
Cell culture and transfection
Most cell lines (HEK293A, including KI and KO cells, MEF, HaCaT, HeLa, and MCF7) were cultured in Dulbecco's modified Eagle's medium DMEM (Gibco 11965-092) with 10% FBS (Gibco A31606-02) and 100 U/mL penicillin–streptomycin (Gibco 15140-122). Cells were incubated in a humidified incubator at 37°C with 5% CO2. MCF10A cells were cultured in DMEM-F12 (Gibco11330-032) supplemented with 5% horse serum (Gibco 26050088), 20 ng/mL EGF (Peprotech AF-100-15), 0.5 μg/mL hydrocortisone (Sigma H4001), 10 μg/mL insulin (Sigma I1882), 100 ng/mL cholera toxin (Sigma C8052), and 100 U/mL penicillin–streptomycin. MCF10A cells were also maintained at 37°C with 5% CO2.
Transfection
Cells were transfected with plasmid DNA using PolyJet reagent (SignaGen Laboratories 50-478-8) according to the manufacturer's instructions. Cells were harvested 24 h after transfection for protein analysis.
Antibodies and chemicals
The following antibodies were purchased from Cell Signaling and used at the indicated dilution for Western blot analysis: Merlin (NF2) (1:1000; 6995), phospho-Merlin Ser518 (1:1000; 9163), YAP (1:1000; 14074), pYAP-Ser127 (1:1000; 4911), LATS1 (1:1000; 3477), phospho-LATS HM (1:1000; 8654), phospho-MK2 Thr334 (1:1000; 3007), phospho-p38 Thr180/Tyr182 (1:1000; 4511), and HA HRP-conjugated (1:10,000; 2999). GAPDH (1:2000) and YAP/TAZ (1:500) were obtained from Santa Cruz Biotechnology (sc-25778 and sc-101199, respectively). Vinculin (1:5000) and Flag HRP conjugated (1:10,000) were from Sigma (V9131 and A8592, respectively. N-Cadherin (1:1000) was from BD Biosciences (610920).
The following antibodies were used for immunofluorescent microscopy experiments at the indicated dilutions: GFP (1:200) was from Abcam (ab6673), PI(4,5)P2 (1:200) was from Echelon Biosciences (Z-G045), Flag (1:500) and HA (1:500) were from Cell Signaling (14793 and 3724, respectively), and YAP (1:200) was from Santa Cruz Biotechnology (sc-101199).
Secondary antibodies Alexa fluor 488 and 555 and phalloidin were from Invitrogen and used in 1:1000 dilution.
Chemicals sorbitol was from Fisher Scientific (S459-500), latrunculin B (Lat B) was from Abcam (ab144291), 2-deoxy-D-glucose (2-DG) was from Sigma (D8375), and p38 inhibitor SB203580 (1202) and JNK inhibitor SP600125 (1496) were from Tocris.
CRISPR knockout
CRISPR genomic editing technology was used for generating knockout (KO) cell lines. The gene-specific guide sequences were designed at https://benchling.com. Single-guide RNA (sgRNA) sequences were cloned into expression vector pSpCas9(BB)-2A-Puro (PX459) (Addgene 48139) or lentiCRISPR v2 (Addgene 52961). The constructed plasmids were transfected into HEK293A and HeLa cells, and transduced into MCF7 cells. Twenty-four hours after transfection or transduction, cells were enriched by 1 μg/mL puromycin (Gibco, #A11138-03) selection for 2–3 d and single-cell-sorted by FACS into 96-well plate format. Single clones were expanded for HEK293A and MCF7 cells, and genomic DNA were extracted using Purelink genomic DNA minikit (Invitrogen K182002). Genomic DNA were PCR amplified and sent out for sequencing. sgRNA sequences for each KO cell line are as follows: PIP5K1A (5′-TCCCTTACCATGTAGTATCC-3′), PIP5K1B (5′-TGCATAAGAACATCTCGTTC-3′), PIP5K1C (5′-CGGATGCGTCCACACCTCGA-3′), ARF1 for clone #1-3 (5′-CTTAAGCTTGTAGAGGATCG-3′), ARF1 for clone #2-3 (5′-AGAACATCAGCTTCACTGTG-3′), ARF6 for clone #1-1 and #1-3 (5′-GTGTAGTAATGCCGCCAGAG-3′), and ARF6 for clone #2-3 (5′-GAAACCCACAGTGGGAATGG-3′).
CRISPR knock-in
CRISPR genomic editing technology was used for generating knock-in (KI) cell lines. Gene-specific sgRNA and homologous recombination (HR) templates were designed using Benchling at https://benchling.com. Single-guide RNA (sgRNA) sequence was cloned into expression vector pSpCas9(BB)-2A-Puro (PX459). sgRNA sequence for both NF2 S518E and S518A KI was 5′- TGACATGAAGCGGCTTTCCA-3′. The template sequence for S518E KI was 5′-AACCCAATTCCAGCACCGTTGCCTCCTGACATACCAAGCTTCAACCTCATTGGTGACAGCCTGTCTTTCGACTTCAAAGATACAGACATGAAGAGATTGGAAATGGAGATAGAGAAAGAAAAGTATGTAGCCCCCTGTGCCCTGCTGTGGGCAGCTGTGAACTAGACTGAGTGATTGGGGCCTTGGGAAGCTGGGGCAGA-3′. The template sequence for S518A KI was 5′-AACCCAATTCCAGCACCGTTGCCTCCTGACATACCAAGCTTCAACCTCATTGGTGACAGCCTGTCTTTCGACTTCAAAGATACAGATATGAAGAGGTTGGCTATGGAGATAGAGAAAGAAAAGTATGTAG CCCCCTGTGCCCTGCTGTGGGCAGCTGTGAACTAGACTGAGTGATTGGGGCCTTGGGAAGCTGGGGCAGA-3′.
HEK293A cells were transfected with sgRNA and HR template, selected with puromycin for 2–3 d, and single-cell-sorted by FACS into 96-well plate format. Single clones were expanded and genomic DNA were extracted using Purelink Genomic DNA Mini Kit. Genomic DNA were PCR amplified and sent out for sequencing.
Lipid-binding assay
PIP strips membrane (Echelon Biosciences P-6001) was blocked in blocking solution (3% BSA in TBST) for 1 h at room temperature. Protein (0.5 µg/mL) in blocking solution was incubated with the membrane for 2 h at room temperature with gentle mixing. Membrane was then washed with TBST three times and subjected to primary antibody incubation in blocking solution with gentle mixing. Membrane was washed again as above and lipid-associated proteins were detected by ECL according to the manufacturer's instructions.
Immunofluorescence staining and confocal microscopy
HEK293A cells were plated on fibronectin-coated (Sigma F1141) coverslips 1 d prior to experimentation. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences 15710) for 15 min and permeabilized with 0.1% saponin (Sigma 47036) for 5 min. Note that in Figure 3, A and B, 2 μg/mL wheat germ agglutinin (WGA) (Biotium 29076) was added before permeablization for 10 min, and in Figure 4C, samples were permeabilized with 0.1% Triton X-100 for visualizing YAP localization. After blocking in 3% BSA in PBS for 30 min, cells were incubated with primary antibodies diluted in 3% BSA overnight at 4°C. After three washes with PBS, cells were incubated with Alexa fluor secondary antibodies at 1:1000 dilution (Invitrogen) for 1 h in the dark at room temperature. Actin is stained with phalloidin conjugated with fluorescent dye (Invitrogen). Phalloidin was added at 1:1000 dilution together with the secondary antibodies. Coverslips were mounted with ProLong Gold antifade mountant with DAPI (Invitrogen P36931). Images were captured with Nikon Eclipse Ti confocal microscope using a 60× oil-immersion objective. Each image is a single Z section at the same cellular level. Fields with moderate cell confluence were randomly selected for imaging. Figures were made and analyzed with ImageJ software.
Liposome-binding assay and protein purification
TRX-tagged Merlin (human, full-length) was expressed in Escherichia coli with induction of expression by 0.1 mM IPTG overnight at 16°C. Proteins were purified using Ni2+-nitrilotriacetic acid agarose (Ni-NTA) column followed by size-exclusion chromatography (Superdex 200 column from GE Healthcare) in a sample buffer containing 50 mM Tris-HCl (pH 7.8), 250 mM NaCl, 1 mM DTT, and 1 mM EDTA.
Defined liposomes were reconstituted from synthetic PC (Avanti Polar Lipid 850375) and PS (Avanti Polar Lipid 840029) with or without certain concentrations of PI(4,5)P2 (Avanti Polar Lipid 850155) or PA (Avanti Polar Lipid 840875). Lipids dissolved in chloroform were mixed in a glass tube at an appropriate ratio defined in each experiment, and the solvent was evaporated under a stream of N2 gas at 4°C. The sample buffer was added to bring the final lipid concentration to 2.5 mg/mL. The lipid solution was rigorously vortexed for 5 min, and then the mixture was hydrated by 10 cycles of freeze and thaw with liquid N2. Ten micromolar defined liposomes and protein samples were mixed and incubated for 1 h at 4°C followed by centrifugation at 100,000g for 30 min at 4°C in a Beckman TLA100.1 rotor. The supernatant fractions were removed for determination of proteins not bound to liposomes. The pellet fractions were washed twice with the same buffer and brought up to the same volume as the supernatant. The proteins in the supernatant and pellet were analyzed by SDS-PAGE with Coomassie blue staining.
Subcellular fractionation
To separate cytosolic and membrane-associated proteins, cells were subjected to S100/P100 fractionation. Cells were treated with 0.4 or 0.2 M sorbitol for 30 min, then washed with PBS and incubated in S100/P100 buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 2.5 mM EDTA, 1 mM EGTA, 1 mM PMSF, 1 mM DTT) supplemented with protease inhibitor (Roche 11873580001) for 20 min at 4°C. After homogenization using tissue grinder homogenizer (Kontes), nuclei were removed by centrifugation at 1000g for 2 min at 4°C. The supernatant was then centrifuged at 100,000g for 60 min at 4°C in a Beckman TLA100.3 rotor. Equal amounts of supernatant (cytoplasmic fraction) and pellet (membrane fraction) were analyzed by Western blot.
Coimmunoprecipitation
Cells were lysed with buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 10 mM KCl, 1% Triton X-100) supplemented with 0.25% sodium deoxycholate, protease inhibitor (Roche 11873580001), phosphatase inhibitor (Thermo Scientific 88667), and 1 mM PMSF (Sigma P7626) for 10 min on ice and centrifuged at 12,000g for 15 min at 4°C. The supernatants were incubated with the Flag antibody (Sigma F1804) overnight at 4°C and protein A/G magnetic beads (Thermo Scientific 88802) were added for 1 h. Proteins were washed with lysis buffer without sodium deoxycholate three times and were eluted with SDS-PAGE sample buffer. Samples were followed by Western blot analysis.
Competing interest statement
K.L.G. is a cofounder of and has equity interest in Vivace Therapeutics. The terms of this arrangement have been reviewed and approved by the University of California at San Diego in accordance with its conflict of interest policies.
Supplementary Material
Acknowledgment
We thank Dr. Guangwei Du (University of Texas Health Science Center at Houston) for providing PA reporter RFP-PASS. We also thank all the members in the Guan laboratory for intellectual inputs and providing reagents, and the Sanford Consortium Stem Cell Core Facility for technical support. A.W.H. is supported in part by the T32 GM007752 training grant. M.Z. is supported by grants from Research Grants Council of Hong Kong (AoE-M09-12 and C6004-17G). K.-L.G. is supported by grants from National Institute of Health (CA196878, GM51586, DEO15964, and P30CA023100).
Author contributions: A.W.H. carried out the experiments and wrote the manuscript. Z.M. contributed to intellectual inputs. S.W.P. proofread the manuscript. Z.L. and M.Z. carried out the experiments and provided reagents. K.-L.G. supervised the project and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.333435.119.
References
- Aikawa Y, Martin TF. 2003. ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. J Cell Biol 162: 647–659. 10.1083/jcb.200212142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfthan K, Heiska L, Grönholm M, Renkema GH, Carpén O. 2004. Cyclic AMP-dependent protein kinase phosphorylates merlin at serine 518 independently of p21-activated kinase and promotes merlin-ezrin heterodimerization. J Biol Chem 279: 18559–18566. 10.1074/jbc.M313916200 [DOI] [PubMed] [Google Scholar]
- Barret C, Roy C, Montcourrier P, Mangeat P, Niggli V. 2000. Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP2) binding site in the NH2-terminal domain of ezrin correlates with its altered cellular distribution. J Cell Biol 151: 1067–1080. 10.1083/jcb.151.5.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshans RL, Szanto S, van Aelst L, D'Souza-Schorey C. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol Cell Biol 20: 3685–3694. 10.1128/MCB.20.10.3685-3694.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinthalapudi K, Mandati V, Zheng J, Sharff AJ, Bricogne G, Griffin PR, Kissil J, Izard T. 2018. Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2. Nat Commun 9: 1338 10.1038/s41467-018-03648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J, Giancotti FG. 2014. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett 588: 2743–2752. 10.1016/j.febslet.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M. 2004. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol 164: 653–659. 10.1083/jcb.200307032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y, Hasegawa H, Kanaho Y. 2011. Regulation of PIP5K activity by Arf6 and its physiological significance. J Cell Physiol 226: 888–895. 10.1002/jcp.22482 [DOI] [PubMed] [Google Scholar]
- Groves JT, Kuriyan J. 2010. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol 17: 659–665. 10.1038/nsmb.1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. 2011. Hippo signaling: growth control and beyond. Development 138: 9–22. 10.1242/dev.045500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol 8: 27–36. 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol 205: 113–126. 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Qi R, Zhou JJ, Ta AP, Yang B, Nakaoka HJ, Seo G, Guan KL, Luo R, Wang W. 2018. Regulation of the Hippo pathway by phosphatidic acid-mediated lipid-protein interaction. Mol Cell 72: 328–340.e8. 10.1016/j.molcel.2018.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nature reviews Cancer 13: 246–257. 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, et al. 1999. Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99: 521–532. 10.1016/S0092-8674(00)81540-8 [DOI] [PubMed] [Google Scholar]
- Hong AW, Meng Z, Yuan HX, Plouffe SW, Moon S, Kim W, Jho EH, Guan KL. 2017. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep 18: 72–86. 10.15252/embr.201642681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Latham V, Murray B, Nandhikonda V, Nord A, Skrzypek E, Wheeler T, Zhang B, Gnad F. 2019. 15 years of PhosphoSitePlus®: integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res 47: D433–D441. 10.1093/nar/gky1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Shibasaki Y, Kizuki N, Katagiri H, Yazaki Y, Asano T, Oka Y. 1996. Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J Biol Chem 271: 23611–23614. 10.1074/jbc.271.39.23611 [DOI] [PubMed] [Google Scholar]
- Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. 1998. Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J Biol Chem 273: 8741–8748. 10.1074/jbc.273.15.8741 [DOI] [PubMed] [Google Scholar]
- Jones DH, Morris JB, Morgan CP, Kondo H, Irvine RF, Cockcroft S. 2000. Type I phosphatidylinositol 4-phosphate 5-kinase directly interacts with ADP-ribosylation factor 1 and is responsible for phosphatidylinositol 4,5-bisphosphate synthesis in the golgi compartment. J Biol Chem 275: 13962–13966. 10.1074/jbc.C901019199 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou H, Li F, Chan SW, Lin Z, Wei Z, Yang Z, Guo F, Lim CJ, Xing W, et al. 2015. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res 25: 801–817. 10.1038/cr.2015.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Sui D, Wu D, Hu J. 2016. The activation loop of PIP5K functions as a membrane sensor essential for lipid substrate processing. Sci Adv 2: e1600925 10.1126/sciadv.1600925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loijens JC, Anderson RA. 1996. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J Biol Chem 271: 32937–32943. 10.1074/jbc.271.51.32937 [DOI] [PubMed] [Google Scholar]
- Lundmark R, Doherty GJ, Vallis Y, Peter BJ, McMahon HT. 2008. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J 414: 189–194. 10.1042/BJ20081237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani T, Hennigan RF, Foster LA, Conrady DG, Herr AB, Ip W. 2011. FERM domain phosphoinositide binding targets merlin to the membrane and is essential for its growth-suppressive function. Mol Cell Biol 31: 1983–1996. 10.1128/MCB.00609-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Saotome I, Mercer K, Crowley D, Gusella JF, Bronson RT, Jacks T. 1998. Mice heterozygous for a mutation at the Nf2 tumor suppressor locus develop a range of highly metastatic tumors. Genes Dev 12: 1121–1133. 10.1101/gad.12.8.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, et al. 2015. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 6: 8357 10.1038/ncomms9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Hansen CG, Guan KL. 2015. The emerging roles of YAP and TAZ in cancer. Nature reviews Cancer 15: 73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Huang L, Pestonjamasp K, Luna EJ, Furthmayr H. 1999. Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol Biol Cell 10: 2669–2685. 10.1091/mbc.10.8.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Lopez-Lago M, Giancotti FG. 2005. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J Cell Biol 171: 361–371. 10.1083/jcb.200503165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen SF, Kapus A, Hoffmann EK. 2011. Osmosensory mechanisms in cellular and systemic volume regulation. J Am Soc Nephrol 22: 1587–1597. 10.1681/ASN.2010121284 [DOI] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94: 1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Plouffe SW, Hong AW, Guan KL. 2015. Disease implications of the Hippo/YAP pathway. Trends Mol Med 21: 212–222. 10.1016/j.molmed.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe SW, Meng Z, Lin KC, Lin B, Hong AW, Chun JV, Guan KL. 2016. Characterization of Hippo pathway components by gene inactivation. Mol Cell 64: 993–1008. 10.1016/j.molcel.2016.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putta P, Rankenberg J, Korver RA, van Wijk R, Munnik T, Testerink C, Kooijman EE. 2016. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim Biophys Acta 1858: 2709–2716. 10.1016/j.bbamem.2016.07.014 [DOI] [PubMed] [Google Scholar]
- Rong R, Surace EI, Haipek CA, Gutmann DH, Ye K. 2004. Serine 518 phosphorylation modulates merlin intramolecular association and binding to critical effectors important for NF2 growth suppression. Oncogene 23: 8447–8454. 10.1038/sj.onc.1207794 [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. 1993. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 363: 515–521. 10.1038/363515a0 [DOI] [PubMed] [Google Scholar]
- Schink KO, Tan KW, Stenmark H. 2016. Phosphoinositides in control of membrane dynamics. Annu Rev Cell Dev Biol 32: 143–171. 10.1146/annurev-cellbio-111315-125349 [DOI] [PubMed] [Google Scholar]
- Sekido Y. 2011. Inactivation of Merlin in malignant mesothelioma cells and the Hippo signaling cascade dysregulation. Pathol Int 61: 331–344. 10.1111/j.1440-1827.2011.02666.x [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Paez JG, Curto M, Yaktine A, Pruitt WM, Saotome I, O'Bryan JP, Gupta V, Ratner N, Der CJ, et al. 2001. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev Cell 1: 63–72. 10.1016/S1534-5807(01)00009-0 [DOI] [PubMed] [Google Scholar]
- Sher I, Hanemann CO, Karplus PA, Bretscher A. 2012. The tumor suppressor merlin controls growth in its open state, and phosphorylation converts it to a less-active more-closed state. Dev Cell 22: 703–705. 10.1016/j.devcel.2012.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Seto A, Maita N, Hamada K, Tsukita S, Tsukita S, Hakoshima T. 2002. Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. J Biol Chem 277: 10332–10336. 10.1074/jbc.M109979200 [DOI] [PubMed] [Google Scholar]
- Skippen A, Jones DH, Morgan CP, Li M, Cockcroft S. 2002. Mechanism of ADP ribosylation factor-stimulated phosphatidylinositol 4,5-bisphosphate synthesis in HL60 cells. J Biol Chem 277: 5823–5831. 10.1074/jbc.M110274200 [DOI] [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol 8: 343–346. 10.1016/S0960-9822(98)70135-6 [DOI] [PubMed] [Google Scholar]
- Stickney JT, Bacon WC, Rojas M, Ratner N, Ip W. 2004. Activation of the tumor suppressor merlin modulates its interaction with lipid rafts. Cancer Res 64: 2717–2724. 10.1158/0008-5472.CAN-03-3798 [DOI] [PubMed] [Google Scholar]
- Surace EI, Haipek CA, Gutmann DH. 2004. Effect of merlin phosphorylation on neurofibromatosis 2 (NF2) gene function. Oncogene 23: 580–587. 10.1038/sj.onc.1207142 [DOI] [PubMed] [Google Scholar]
- Thurneysen C, Opitz I, Kurtz S, Weder W, Stahel RA, Felley-Bosco E. 2009. Functional inactivation of NF2/merlin in human mesothelioma. Lung Cancer 64: 140–147. 10.1016/j.lungcan.2008.08.014 [DOI] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. 1993. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 75: 826 10.1016/0092-8674(93)90501-G [DOI] [PubMed] [Google Scholar]
- van den Bout I, Divecha N. 2009. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci 122: 3837–3850. 10.1242/jcs.056127 [DOI] [PubMed] [Google Scholar]
- Xiao GH, Chernoff J, Testa JR. 2003. NF2: the wizardry of merlin. Genes Chromosomes Cancer 38: 389–399. 10.1002/gcc.10282 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Chen MZ, Wang YJ, Sun HQ, Wei Y, Martinez M, Yin HL. 2006. Hypertonic stress increases phosphatidylinositol 4,5-bisphosphate levels by activating PIP5KIβ. J Biol Chem 281: 32630–32638. 10.1074/jbc.M605928200 [DOI] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154: 1342–1355. 10.1016/j.cell.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19: 27–38. 10.1016/j.devcel.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang Z, Lu M, Yonekubo Y, Liang X, Zhang Y, Wu P, Zhou Y, Grinstein S, Hancock JF, et al. 2014. Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol Cell Biol 34: 84–95. 10.1128/MCB.00987-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP. Genes Dev 24: 72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.