In this study, Zhang et al. investigated the mechanism underlying the dysregulation of early neurodevelopment in macrocephaly/autism disorders. They generated Rab39b knockout mice and found that they exhibited cortical neurogenesis impairment, macrocephaly, and hallmark ASD behaviors, in addition to development of mutant human cerebral organoids, which were substantially enlarged due to the overproliferation and impaired differentiation of neural progenitor cells.
Keywords: RAB39b, autism-like behaviors, macrocephaly, neural progenitor cell, PI3K–AKT–mTOR, cerebral organoid, mice
Abstract
Dysregulation of early neurodevelopment is implicated in macrocephaly/autism disorders. However, the mechanism underlying this dysregulation, particularly in human cells, remains poorly understood. Mutations in the small GTPase gene RAB39b are associated with X-linked macrocephaly, autism spectrum disorder (ASD), and intellectual disability. The in vivo roles of RAB39b in the brain remain unknown. We generated Rab39b knockout (KO) mice and found that they exhibited cortical neurogenesis impairment, macrocephaly, and hallmark ASD behaviors, which resembled patient phenotypes. We also produced mutant human cerebral organoids that were substantially enlarged due to the overproliferation and impaired differentiation of neural progenitor cells (NPCs), which resemble neurodevelopmental deficits in KO mice. Mechanistic studies reveal that RAB39b interacts with PI3K components and its deletion promotes PI3K–AKT–mTOR signaling in NPCs of mouse cortex and cerebral organoids. The mTOR activity is robustly enhanced in mutant outer radial glia cells (oRGs), a subtype of NPCs barely detectable in rodents but abundant in human brains. Inhibition of AKT signaling rescued enlarged organoid sizes and NPC overproliferation caused by RAB39b mutations. Therefore, RAB39b mutation promotes PI3K–AKT–mTOR activity and alters cortical neurogenesis, leading to macrocephaly and autistic-like behaviors. Our studies provide new insights into neurodevelopmental dysregulation and common pathways associated with ASD across species.
Autism spectrum disorders (ASDs) are genetically and phenotypically heterogeneous diseases. Several hundreds of autism-associated sequence and structural variants have been identified (Berg and Geschwind 2012; O'Roak et al. 2012). Functional study of rare variants is a valuable approach to understand disease mechanisms and identify convergent molecular pathways in autism (Berg and Geschwind 2012; Willsey et al. 2013). Macrocephaly/autism disorder represents a subset of ASDs coupled with enlarged brain volumes in affected individuals. These brain overgrowth could be due to enlarged brain volume (Courchesne et al. 2003; Shen et al. 2013; Sacco et al. 2015), increased cortical thickness (Zielinski et al. 2014), or increased cortical surface areas, which occur at perinatal or early postnatal stages before the first clinical behavior signs (Ohta et al. 2016; Hazlett et al. 2017). Excess cortical neuron numbers and abnormal cortical patches in young postmortem ASD brains suggest that developmental dysregulation may play a pivotal role in ASD pathogenesis (Courchesne et al. 2011; Stoner et al. 2014). Gene expression alteration in postmortem ASD brains implicates neurodevelopmental genes involved in cell cycle progression, proliferation, and neuronal differentiation (Chow et al. 2012). Characterization of ASD patient iPSCs and brain organoids has revealed altered proliferation, differentiation, and growth of neural progenitor cells (NPCs) (Mariani et al. 2015; Marchetto et al. 2017; Schafer et al. 2019). Functional genomics, genetic, and epidemiological studies have also pointed to early neurodevelopmental deficits in the etiology of neuropsychiatric disorders including ASD and intellectual disability (ID) (Willsey et al. 2013; Casanova and Casanova 2014). Collectively, these observations support the emerging concept that macrocephaly/autism is a progressive disorder of brain development, which is characterized by a cortical surface area hyper-expansion perinatally, brain overgrowth early postnatally, and ultimately the emergence of autistic social deficits (Ernst 2016; Piven et al. 2017; Courchesne et al. 2019).
Prenatal neurodevelopmental abnormalities in ASD are poorly understood due to limited access of human brain tissues (Piven et al. 2017; Courchesne et al. 2019). The majority of current research approaches cannot examine the abnormal neurodevelopmental processes in patients before their clinical phenotypes. Human induced pluripotent stem cells (iPSCs) and iPSC-derived brain organoids provide a platform for experimentally addressing early human neurodevelopmental processes associated with neuropsychiatric disorders in a relevant genetic and cellular context (Lancaster et al. 2013; Wen et al. 2014; Bershteyn et al. 2017; Di Lullo and Kriegstein 2017). Human brain organoids can recapitulate gene expression, neuronal morphology and organization, and functional properties in the developing human brain (Lancaster et al. 2013; Qian et al. 2016). This species-specific ex vivo model is particularly valuable for macrocephaly/autism research because the human cerebral cortex exhibits a dramatic increase in cortical size and complexity during evolution (Rakic 1995). A primate-enriched neurogenic area, named the outer subventricular zone (oSVZ), is located basal to the classic SVZ. The oSVZ neural progenitors, including transient amplifying IPCs and outer radial glia cells (oRGs), are considered to contribute to the majority of upper-layer neurons (Lewitus et al. 2013; Pollen et al. 2015). Although IPCs are conserved between humans and mice, oRGs are abundant in human cerebral cortex but rare in rodent cortex (Wang et al. 2011). Multiple lines of evidences suggest that abundant oRGs and their robust proliferation capacity are critical for evolutionary expansion of cortical size (Reillo et al. 2011; Nonaka-Kinoshita et al. 2013).
Complementary to studies of human brain organoids, investigation of genetically modified mouse models remain an essential strategy to dissect the pathogenesis of ASD, with strengths in circuit and behavioral studies (Nestler and Hyman 2010; Silverman et al. 2010). This approach is enabled by the identification of risk genes linked with ASD. One such gene is RAB39b, which encodes a GTPase belonging to the RAS-like GTPase superfamily. RAB39b is an X-linked gene and mapped to the distal Xq28 locus. Loss of functions of RAB39b mutations lead to macrocephaly, ASD, ID, epilepsy, and early-onset Parkinson disease (Giannandrea et al. 2010; Wilson et al. 2014; Mata et al. 2015; Mignogna et al. 2015; Ciammola et al. 2017; Woodbury-Smith et al. 2017). We previously identified RAB39b as a downstream GTPase associated with a C9orf72/Smcr8-containing complex, which acts as a guanosine diphosphate-guanosine 5′-triphosphate (GDP-GTP) exchange factor (GEF) for RAB39b (Yang et al. 2016). In vitro studies suggest that RAB39b regulates synapse formation and function, α-synuclein levels, and the expression of genes involved in brain development (Giannandrea et al. 2010; Wilson et al. 2014; Mignogna et al. 2015). To investigate the in vivo roles of RAB39b in the brain, we generated Rab39b hemizygous male loss-of-function mice (referred to here as Rab39b−/y due to its X linkage). Rab39b−/y knockout (KO) mice present with macrocephaly and autism-like behaviors. To model human neurodevelopmental dysregulation, we generated RAB39b mutant cerebral organoids, which are substantially enlarged due to the impaired differentiation and overproliferation of NPCs. Mechanistic studies revealed that Rab39b deletion promotes PI3K–AKT–mTOR signaling, inhibition of which rescued enlarged organoid sizes and NPC proliferation caused by RAB39b mutations. Our studies provide new insights into neurodevelopmental dysregulation and common pathways in RAB39b-associated macrocephaly/autism disorders.
Results
Rab39b−/y KO mice exhibit macrocephaly
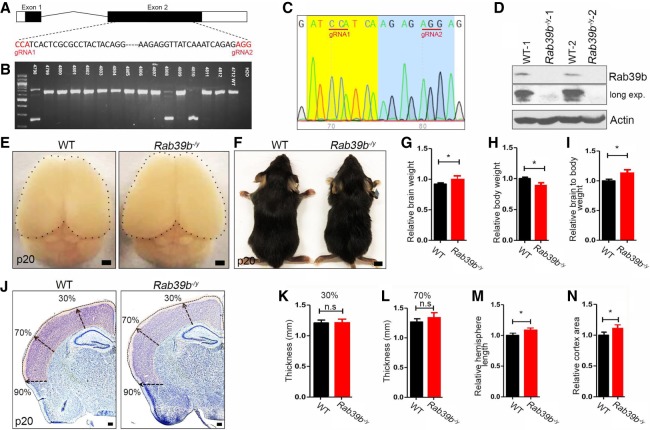
We used two CRISPR–Cas 9 guide RNAs (gRNAs) targeting of C57BL/N6 oocytes to delete exon 2, which is the major Rab39b exon (Fig. 1A). PCR assay identified three independent founder lines (Fig. 1B). Sequence analyses of the lower bands confirmed that all three lines contain the deletion of the major exon 2 of Rab39b gene (Fig. 1C). Mutant carrier lines were crossed with wild-type (WT) C57BL/6N mice. Heterozygous mice were bred more than three generations before subsequent experiments, and multiple litters from these three founder lines were used for the experiments to reduce the potential off-target events.
Figure 1.
Rab39b−/y mouse models exhibit macrocephaly phenotypes. (A) Schematic diagram of mouse Rab39b gene with corresponding guide RNAs 1 and 2 (gRNA1 and gRNA2 in red). (B) PCR genotyping identified three independent founders from CRISPR–Cas9-mediated deletion of Rab39b in mice. (C) Sequencing results confirmed the deletion of the region between gRNA1 and gRNA2. (D) Western blot analyses of Rab39b protein expression in mouse cortex. β-Actin served as the control. (E) Dorsal views of P20 mouse brains. Black dots outline brain surfaces. Scale bars, 1 mm. (F) Representative images of P20 mice. Scale bars, 0.5 cm. (G–I) Quantification of P20 wild type (n = 8) and mutant (n = 6) in brain weights (G), body weights (H), and relative ratios of brain to body weights (I). (J) Representative coronal sections of P20 WT and Rab39b−/y brains with Nissl stain. Arrows represent thickness at 30%, 70%, and 90% distance from the dorsal midline. Scale bars: 300 µm. (K–N) Quantification of WT (n = 4) and mutant (n = 4) cerebral cortex in their thickness at 30% (K) and 70% (L), relative hemisphere length (M), and relative cortex area (N). All data are presented as mean ± SEM from six sections of individual mice. Statistical analyses were performed with nonparametric Mann-Whitney test. (*) P < 0.05; (**) P < 0.01; (n.s) represents no significant difference detected.
Rab39b is an X-linked gene. Male patients carrying RAB39b mutations exhibit robust macrocephaly/autism phenotypes (Giannandrea et al. 2010; Wilson et al. 2014; Woodbury-Smith et al. 2017). Therefore, we first focused on Rab39b−/y KO male mice. Western blot analyses confirmed the absence of Rab39b protein in brain lysates of mutant mice (Fig. 1D). Rab39b KO mice were viable and fertile with indistinguishable morphology from littermate controls. We examined body weights and brain sizes at postnatal day (P) 20, since macrocephaly is a hallmark trait in patients with RAB39b mutations (Giannandrea et al. 2010; Woodbury-Smith et al. 2017). Rab39b deletion in mice resulted in a mild enlargement of the brain (Fig. 1E), which was consistent with increased brain weights in these mice (Fig. 1G, P = 0.0281). Whereas body size appeared normal, Rab39b KO mice had a slightly decreased body weight (Fig. 1H, P = 0.0352), which contributes to increased brain-to-body weight ratios (Fig. 1I, P = 0.0426). A slight increase of brain weight was detected as early as embryonic day (E) 18.5 in mutant mice (Supplemental Fig. S1A–C). We performed Nissl staining (Fig. 1J), and found no significant changes in the cortical thickness (Fig. 1K,L, P > 0.05). Rab39b deletion resulted in a slight but significant increase in hemisphere length and overall cerebral cortex areas (Fig. 1M,N, P = 0.021, P = 0.031). Next, we performed similar studies in homozygous KO female mice. Rab39b deletion led to an increase in brain weight (Supplemental Fig. S1D,F, P = 0.017) and brain-to-body weight ratio (Fig. 1E,H, P = 0.0154), and a decrease in body weight (Fig. 1G, P = 0.0378). Nissle staining revealed an increase in hemisphere length and cortex area without significant alterations in cortical thickness in mutant female brains (Supplemental Fig. S1I–M, P < 0.05). Therefore, Rab39b hemizygous male and homozygous female mice exhibited consistent brain phenotypes. Together, these results suggested that Rab39b−/y KO mice recapitulate macrocephaly phenotypes in patients.
Behavior phenotypes in Rab39b KO male mice
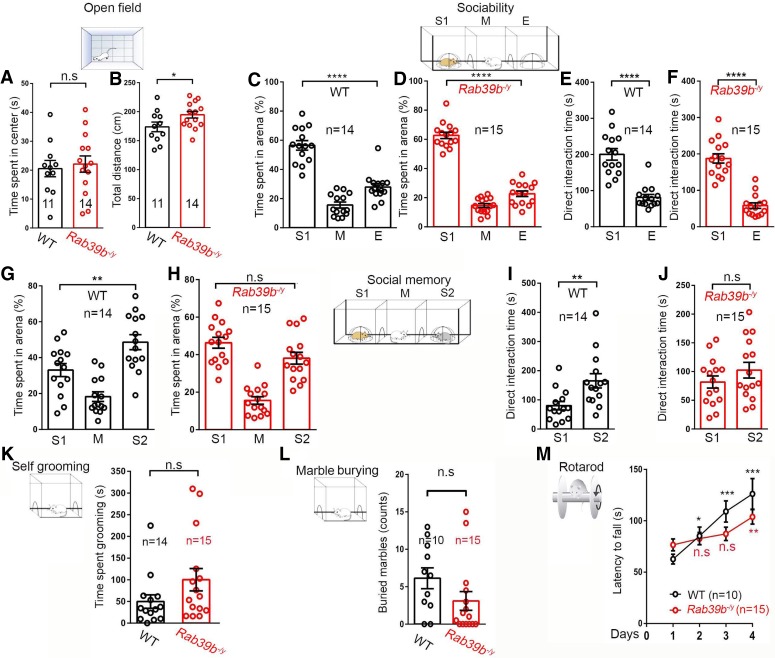
Male patients with RAB39b mutations displayed autism spectrum disorder (ASD) and intellectual disability (ID) (Giannandrea et al. 2010; Woodbury-Smith et al. 2017). Therefore, we conducted a battery of behavioral assays using a cohort of male mice around 2 mo of age. An open field assay was performed to evaluate overall movement and anxiety. Rab39b KO and WT mice spent similar amounts of time in the center of the test device (Fig. 2A, 11 WT, 14 KO, P = 0.0702), and had slightly increased travel distances (Fig. 2B, 11 WT, 14 KO, P = 0.0415), suggestive of no obvious deficits in anxiety and mobility.
Figure 2.
Rab39b−/y mice exhibit deficits in social memory and motor learning. (A,B) Open field test was performed on 2-mo-old male mice to measure time spent in center (A) and the total distance traveled (B). (C–F) Three-chamber task was performed to measure sociability by quantifying the percentage of time the subject mouse spent in the area with a stranger mouse or empty chamber, and quantify the direct interaction time between the subject mouse with the stranger mouse or empty chamber. (G–J) Three-chamber task was performed to measure social memory by quantifying the percentage of time the subject mice spent in the area with the previously stranger (S1) and new stranger (S2) mouse, and quantifying the direct interaction times between the subject mouse, S1 and S2. (K) Quantification of the time spent self-grooming. (L) Quantification of marbles buried in the repetitive behavior test. (M) Accelerating rotarod test was performed on 2-mo-old male mice to examine their latency to fall on four consecutive days. All data are presented as mean ± SEM using numbers (n) of mice as indicated. Statistical analyses were performed with nonparametric Mann-Whitney tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001; (n.s) no significant difference detected.
Behaviors related to ASD were examined using a collection of social and repetitive behavior assays (Crawley 1999; Sukoff Rizzo and Crawley 2017). The percentage of time spent with the novel mouse cage was larger than that with an empty cage, which met the criteria of sociability in this assay for both WT (Fig. 2C, n = 14, P < 0.0001) and KO (Fig. 2D, n = 15, P < 0.0001) male mice. Similarly, Rab39b KO and WT mice had more direct interaction time with the novel mouse (Fig. 2E, n = 14, P < 0.0001) compared with the empty cage (Fig. 2F, n = 15, P < 0.0001). Therefore, sociability appeared normal in the mutant mice. Next, we measured social memory, in which a new stranger mouse (S2) was placed into the empty cage and a previous stranger mouse (S1) became the familiar one. WT mice spent more time with S2 compared with S1 (Fig. 2G, n = 14, P = 0.0058). In contrast, there was no significant difference detected in the percentage of time that mutant mice spent with S1 or S2 (Fig. 2H, n = 15, P = 0.241). WT mice spent more direct interaction time with S2 than S1 (Fig. 2I, n = 14, P = 0.0058), whereas Rab39b KO mice had equal amount of direct interaction time with S1 and S2 (Fig. 2J, n = 15, P > 0.05). Therefore, Rab39b KO mice exhibited social memory deficits.
To assess repetitive behaviors, we used two common behavioral paradigms including self-grooming and marble burying tests. Time spent in grooming was not significantly different between WT (n = 14) and Rab39b KO (n = 15) mice (Fig. 2K, n = 0.1115). Mutant mice buried similar numbers of marbles compared with WT controls (Fig. 2L, 10 WT, 15 KO, P = 0.1208). These results suggest that Rab39b KO mice displayed no substantial repetitive behavior deficits. We next examined their motor coordination and balance using an accelerating (5–40 rpm in 5 min) rotarod test. Mice were given five trials per day, with an intertrial interval of 20 min, for four consecutive days. A decreased trend of latency to fall was detected in mutant mice at days 3 and 4, and there was no significant difference detected between WT and mutants (Fig. 2M, 10 WT, 15 KO), suggesting that mutant mice had normal motor coordination and balance. Because patients carrying RAB39B mutations displayed intellectual disability (Giannandrea et al. 2010; Woodbury-Smith et al. 2017), we examined motor learning, which can be assessed by the number of trials needed to reach the maximal latency to fall on an accelerating rotarod assay (Lalonde et al. 1995). WT mice exhibited an increase in latency to fall over the course of four consecutive days, indicating active motor learning. In contrast, Rab39b KO mice exhibited no significant improvements on days 2 and 3 (Fig. 2M; individual days 2, 3, and 4 were compared with day 1). Together, these results suggest that Rab39b KO mice developed social memory and motor learning deficits.
Rab39b deletion impairs cortical neurogenesis in the developing brain
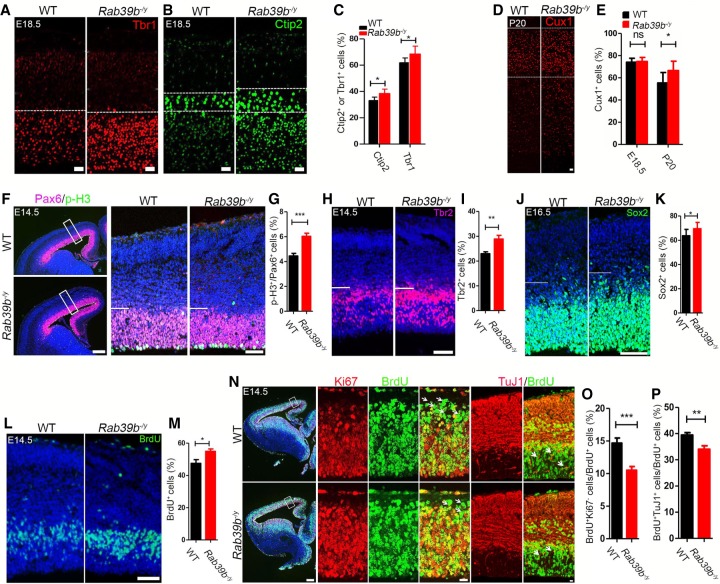
To investigate the causes underlying enlarged brain sizes in Rab39b mutant mice, we examined neurogenesis at different developmental stages. We used Cux1 to label layer II–IV neurons, and Ctip2 and Tbr1 to mark layer V–VI neurons in the cerebral cortex. Wheras no significant differences detected at E14.5 (Supplemental Fig. S2A–D), Tbr1-positive cells were slightly increased in E16.5 mutant cortex (Supplemental Fig. S2E,F, P = 0.0111), and both Tbr1- and Ctip2-positive cells were increased in E18.5 mutant cortex compared with WT controls (Fig. 3A–C, P = 0.021, P = 0.0497). The Cux1-positive cells were significantly increased in P20 but not E18.5 mutant cerebral cortex (Fig. 3D,E, P = 0.0349). The distribution of these layer markers was normal, however, suggesting that the layer organization was not disrupted by the Rab39b deletion. Therefore, we concluded that Rab39b deletion leads to an increase in the numbers of neurons at different layers, which is consistent with the macrocephaly phenotypes.
Figure 3.
Altered cortical neurogenesis in Rab39b−/y mice. (A,B,D) Confocal microscope images of cerebral cortex sections stained with antibodies against Tbr1 (red), Ctip2 (green), and Cux1 (red). Hoechst stains nuclei. Scale bars: 20 µm. (C,E) Quantification of Ctip2-, Tbr1-, and Cux1-positive cells out of total cells from the area surrounded by white dotted lines in A, B, and D. (F,H,J) Representative images of cerebral cortex sections stained with antibodies against Pax6 (purple), p-H3 (green), Tbr2 (red), and Sox2 (green). The right panels are enlargement of boxed areas in left panels. Areas below white lines in the right panels represent ventricular zone (VZ) and subventricular zone (SVZ). Scale bars: left, 100 µm; right, 50 µm. (G,I,K) Quantification of the percentage of p-H3-positive cells out of total Pax6 positive cells (G), Tbr2-positive cells out of total cells in the VZ/SVZ (I), and Sox2-positive cells out of total cells in the VZ/SVZ. (L) Representative images of cerebral cortex sections stained with antibodies against BrdU (green) after 0.5-h pulse. Hoechst stains nuclei. Scale bars, 50 µm. (M) Quantification of the percentage of BrdU-positive cells out of total cells in the VZ/SVZ. (N) Representative images of cerebral cortex sections stained with antibodies against Ki67 (red) plus BrdU (green), or BrdU (green) plus TuJ1 (red). BrdU was administrated at E13.5 followed by a 20-h pulse. Hoechst stains nuclei. The right panels are from boxed areas in left panels. White arrows mark BrdU+Ki67− or BrdU+TuJ1+ cells. Scale bars: left, 100 µm; right, 10 µm. (O,P) Quantification of the percentage of BrdU+Ki67− and BrdU+TuJ1+ cells per total number of BrdU+ cells in the examined areas. Error bars represent SEM of nine sections from three independent experiments (n = 3). Statistical analyses were performed with nonparametric Mann-Whitney tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (ns) no significant difference detected.
To determine whether the increased neuronal numbers are derived from the expansion of NPC pool, we used Pax6 and phospho-histone 3 (p-H3) to label apical neural progenitor cells (APCs) and mitotic cells in the ventricular zone and subventricular zone (VZ/SVZ) of embryonic day 14.5 (E14.5) mouse brains (Fig. 3F). Coronal sections positioned similarly along the rostral–caudal axis were analyzed by immunohistochemical (IHC) staining methods. There was no significant difference detected in the numbers of Pax6-positive APCs between WT and mutants (Supplemental Fig. S2G,H). In contrast, the percentage of p-H3- and Pax6-double-positive cells out of the total number of Pax6-positive cells was significantly increased in mutants compared with WT controls (Fig. 3G, P < 0.0001). Next, we examined the intermediate progenitor cells (IPCs) labeled by Tbr2 (Fig. 3H). We used coronal cerebral sections obtained from similar positions on the rostral–caudal axis from E14.5 brains. The numbers of Tbr2-positive IPCs were significantly increased in the VZ/SVZ regions of mutant brains (Fig. 3I, P = 0.0017). Lastly, we examined Sox2 (Fig. 3J), which labels both APCs and IPCs, and found that Sox2-positive NPCs were significantly increased in mutant cortex (Fig. 3K, P = 0.0127). Together, these results suggested that Rab39b deletion results in an increased NPC pool.
Increased p-H3-positive NPCs could be due to an enhancement of cell proliferation or mitotic arrest. To distinguish these two possibilities, we performed BrdU (bromodeoxyuridine/5-bromo-2′-deoxyuridine) labeling studies at E14.5 (Fig. 3L). We conducted IHC staining using coronal sections at matched rostral–caudal axis positions. There was a significant increase in the percentage of BrdU-positive cells out of total cells in the VZ/SVZ regions of E14.5 mutant brains (Fig. 3M, P = 0.0161), suggesting an increased cell proliferation rate in the mutant cerebral cortex. Next, we examined cell cycle exit of NPCs in the developing cerebral cortex. BrdU was administrated at E13.5 pregnant dams followed by IHC staining analyses after a 20-h pulse. Ki67 was used to label all cycling cells in the VZ/SVZ regions, such that BrdU+Ki67− cells represent NPCs exiting the cell cycle (Fig. 3N). There was a significant decrease in the percentage of BrdU+Ki67− cells out of total BrdU-positive cells (Fig. 3O, P = 0.0004), indicating decreased cell cycle exit of NPCs. We examined BrdU+TuJ1+ colabeled cells (Fig. 3N), and found that they were significantly decreased in mutants (Fig. 3P, P = 0.0039), which further supports that mutant NPCs delay their cell cycle exit and neuronal differentiation. Together, these results suggested that Rab39b deletion results in an increased proliferate rate and a decrease in cell cycle exit of NPCs, which in turn lead to the expansion of NPC pool.
RAB39b mutant cerebral organoids model human macroocephaly
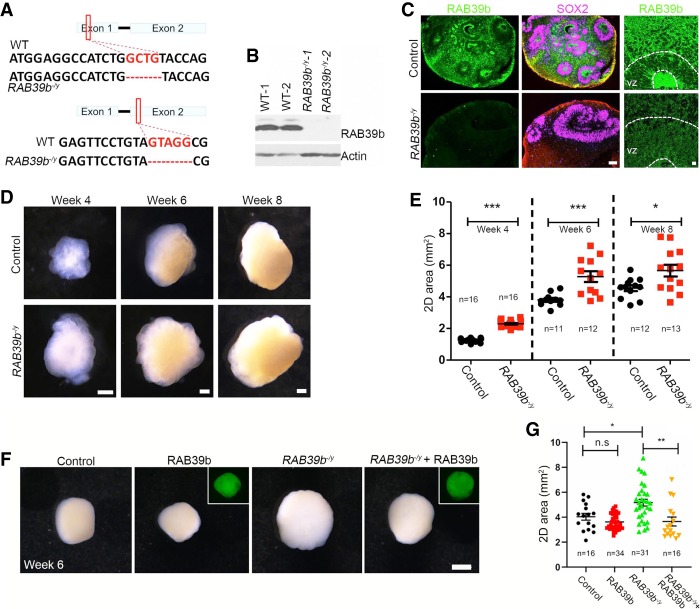
To investigate RAB39b's functions in human brain development, we turned our attention to human pluripotent stem cells (hPSCs) and cerebral organoids (Lancaster et al. 2013; Qian et al. 2016). To delete the human RAB39b gene, we generated mutant PSC lines using the CRISPR/Cas9 approach (Ran et al. 2013). The editing efficiency of gRNA was validated by a T7 endonuclease I assay. We generated two independent hPSC clones, which were derived from induced PSCs or human embryonic stem (hES) cells, including a 4-bp deletion in exon 1 and a 5-bp deletion in exon 2 (Fig. 4A). All of these mutations resulted in a frameshift and led to premature stop codon generation. Western blot confirmed the absence of RAB39b protein in mutant human PSCs (Fig. 4B).
Figure 4.
RAB39b deletion results in enlarged cerebral organoid sizes. (A) CRISPR/Cas9-mediated gene editing of human RAB39b locus in pluripotent stem cells (PSCs), resulting in a 4-bp depletion in exon 1, or 5-bp deletion in exon 2. Each of these deletions resulted in ablation of RAB39b proteins due to premature mature stop codons. (B) Western blot analysis confirmed the absence of RAB39b protein in mutant PSCs. (C) Confocal imaging of cerebral organoid sections stained with antibodies against SOX2 (purple) and RAB39b (green). Scale bars: left, 75 µm; right, 1 µm. (D) Representative images of cerebral organoids at weeks 4, 6, and 8. Scale bars, 0.5 mm. (E) Quantification of 2D surface areas of cerebral organoids. (F) Representative images of week 6 RAB39b mutant organoids with or without rescue by lentivirus expressing RAB39b-GFP. Scale bars, 1 mm. (G) Quantification of 2D surface areas of control and mutant cerebral organoids. Error bars represent SEM of four independent differentiation experiments containing numbers of organoids as indicated. Statistical analyses were performed with nonparametric Mann-Whitney tests. (*) P < 0.05; (***) P < 0.001.
To model RAB39b gene mutation-associated human macrocephaly, we used a cerebral organoid culture system. Dual Smad signaling inhibitors were used in neural induction medium to enhance neuroepithelial expansion (Kadoshima et al. 2013). Embryoid bodies (EBs) were added into droplets of Geltrex to enhance complex tissue formation, followed by the growth in a spinning bioreactor to facilitate oxygen exchange and nutrient absorption (Zhang et al. 2019). To directly compare RAB39b mutant and isogenic controls in organoid formation, equal numbers (∼9000 starting cells) of dissociated single PSCs were used to generate EBs, which resulted in no differences in morphology or surface areas at culture day 12. We analyzed week 4 cerebral organoids and found that RAB39b expression was absent in mutants, and RAB39b was ubiquitously expressed in all cell types in the control organoids (Fig. 4C). Week 4 organoids exhibited transparent and neuroepithelial loop structures that persisted at week 6; the overall organoid sizes gradually increased over time (Fig. 4D). By week 4, RAB39b mutant cerebral organoids were substantially larger in sizes than controls (Fig. 4D; Supplemental Fig. S3A–C). Quantification analyses showed that RAB39b deletion resulted in a significant increase of the organoid surface area at weeks 4, 6, and 8 (Fig. 4E, P < 0.0001, P = 0.0008, P = 0.0154).
To confirm that the enlarged organoid sizes were due to the absence of RAB39b proteins, we performed rescue studies. RAB39b control and mutant human PSCs were infected by lentivirus expressing WT RAB39b-GFP followed by the generation of stable cell lines for the cerebral organoid culture. Western blots confirmed the overexpression of RAB39b protein in the PSC-derived human NPCs (Supplemental Fig. S3D). Analyses of week 6 organoids revealed that enlarged sizes of mutant organoids were rescued by adding back RAB39b proteins (Fig. 4F,G, P = 0.0104, P = 0.0018), whereas RAB39b overexpression in control organoids did not cause a significant size change. These results suggested that the increased size of mutant cerebral organoids was due to loss of function of RAB39b.
Rab39b deletion increases proliferation and expansion of NPCs
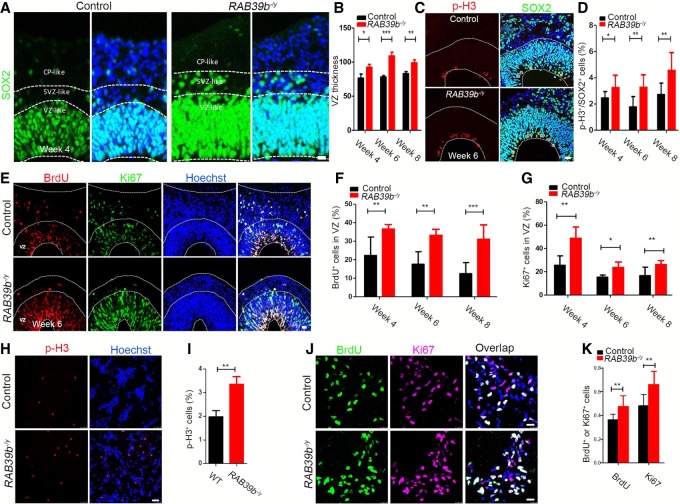
The expansion of the primate cortex in our evolutionary history has been attributed to increased duration and number of NPC divisions (Kornack and Rakic 1998), which may explain the substantial enlargement of mutant human cerebral organoid sizes versus the mild macrocephaly phenotypes in mice with Rab39b mutations. To investigate the mechanisms underlying the enlarged sizes of RAB39b mutant cerebral organoids, we analyzed their neurogenesis. By combing apical neural progenitor markers PAX6/SOX2, IPC marker TBR2, and the deep-layer cortical marker Ctip2, we focused our analyses on VZ-, SVZ-, and CP-like regions. IHC staining was performed using antibodies against SOX2 (Fig. 5A). We quantified the relative thicknesses of SOX2-positive VZ layers according to published methods (Zhang et al. 2019). Statistical analysis showed that mutant organoids had a substantial increase of SOX2-positive VZ thickness at weeks 4, 6, and 8 (Fig. 5B, P = 0.0471, P < 0.0001, P = 0.008), suggesting an increase of the NPC pool in the mutant organoids.
Figure 5.
RAB39b deletion increases proliferation and expansion of NPCs. (A) Confocal imaging of week 4 organoid sections stained with antibodies against SOX2 (green). Hoechst stains nuclei. Scale bars, 20 µm. White dotted lines mark VZ, SVZ, and cortical plate (CP)-like areas. (B) Quantification of SOX2-positive VZ thickness in organoids. (C) Representative imaging of week 6 organoid sections stained with antibodies against p-H3 (red) and SOX2 (green). Regions below the bottom white dotted lines represent VZ/SVZ-like regions. Hoechst stains nuclei (blue). Scale bars, 20 µm. (D) Quantification of the percentage of p-H3+ cells out of total SOX2+ cells in VZ/SVZ. (E) Representative imaging of week 6 organoid sections stained with antibodies against BrdU (0.5 h; red) and Ki67 (green). Areas below the white dotted lines represent VZ-like regions, respectively. Hoechst stains nuclei (blue). Scale bars, 20 µm. (F,G) Quantification of the percentage of BrdU+ and Ki67+ cells in VZ regions. (H,J) Representative imaging of 2D culture human NPCs stained with antibodies against p-H3 (red), BrdU (green), and Ki67 (purple). Hoechst stains nuclei. Scale bars, 25 µm. (I,K) Quantification of the percentage of p-H3+, BrdU+, or Ki67+ cells out of total cells. Error bars represent SEM of results from four independent sets of cerebral organoids. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, Mann-Whitney tests.
To identify the causes of increased NPC pool in mutant organoids, we examined cell cycle progression. We analyzed p-H3- and SOX2-positive NPCs in the VZ/SVZ regions (Fig. 5C), and found a significant increase in the percentage of p-H3-positive cells out of the total number of SOX2-positive cells in weeks 4, 6, and 8 mutant organoids (Fig. 5D, P = 0.0288, P = 0.0014, P = 0.0011). To determine whether the p-H3 accumulation was due to increased cell proliferation or mitotic arrest, we performed BrdU pulse experiments (Fig. 5E). There was a significant increase in the percentage of BrdU-positive cells in the VZ-like regions of the mutant cerebral organoids in weeks 4, 6, and 8 (Fig. 5F, P = 0.0036, P = 0.0058, P = 0.0001). Ki67 IHC staining revealed that mutant cerebral organoids displayed an increased percentage of cycling cells in the VZ (Fig. 5E–G, P = 0.0024, P = 0.0166, P = 0.0054). To determine the cell-autonomous role of RAB39b in NPC proliferation, we examined 2D-cultured NPCs differentiated from iPSCs. There was a significant increase in the percentage of p-H3-positive cells out of total NPCs (Fig. 5H,I, P = 0.0014). Similar to cerebral organoids, the percentages of S-phase NPCs (BrdU pulse) and total cycling NPCs (labeled by Ki67) were also significantly increased due to RAB39b mutations (Fig. 5J,K, P = 0.0041, P = 0.0016). Together, these results suggest that RAB39b deletion leads to increased NPC proliferation and numbers, resulting in enlarged organoid sizes.
Transiently delayed NPC differentiation and increased neuronal production in mutant organoids
Changes to the G1-S phase transition influence the balance between proliferation and differentiation of NPCs (Lange et al. 2009). The duration and number of NPC divisions are thought to have been enhanced to accommodate the evolutionary expansion of the neocortex in primates compared with rodents (Kornack and Rakic 1998), which is linked with cell differentiation kinetics. Our studies above suggested that our mutant organoids exhibited altered cell cycle kinetics. Therefore, we reasoned that the increased NPC proliferation in mutant organoids could be connected to changes in their differentiation propensity. To test this hypothesis, we examined cell differentiation in mutant organoids. Consistent with published studies (Qian et al. 2016; Li et al. 2017), week 4 organoids exhibited robust NPC differentiation, reflected by the expression of neural differentiation markers MAP2, TuJ1, and DCX, while SOX2-positive NPCs were surrounded by differentiated neurons (Fig. 6A,B). In contrast, RAB39b mutant organoids exhibited minimal staining of MAP2, TuJ1, and DCX (Fig. 6A,B). Quantification confirmed a significant decrease in signal intensities of these neuronal markers (Fig. 6C, P = 0.0349, P = 0.0206, P = 0.0022). These results suggest that RAB39b deletion may cause a transient delay of NPC differentiation in mutant organoids. We reasoned that less differentiated cells could be due to delayed cell cycle exit of NPCs in the mutant organoids. To test this hypothesis, we used a 24-h BrdU pulse to label a subset of NPCs and followed their cycling status by combining the Ki67 and BrdU staining (Fig. 6D). BrdU+Ki67− cells represent the population of NPCs that exist in the cell cycle in VZ/SVZ-like regions. Quantification revealed that the percentage of BrdU+Ki67− cells was significantly reduced in mutant organoids (Fig. 6E, P = 0.0339), suggesting a decrease in cell cycle exit of NPCs. To follow the long-term fates of BrdU-labeled NPCs, we pulsed week 4 organoids with BrdU for 48 h, followed by the IHC staining at week 6 (Fig. 6F). We found a significant increase in BrdU+DCX+ neurons in mutant compared with control organoids (Fig. 6G, P < 0.0001), suggesting that increased NPCs ultimately turned into differentiated neurons despite their transient delays in cell cycle exit.
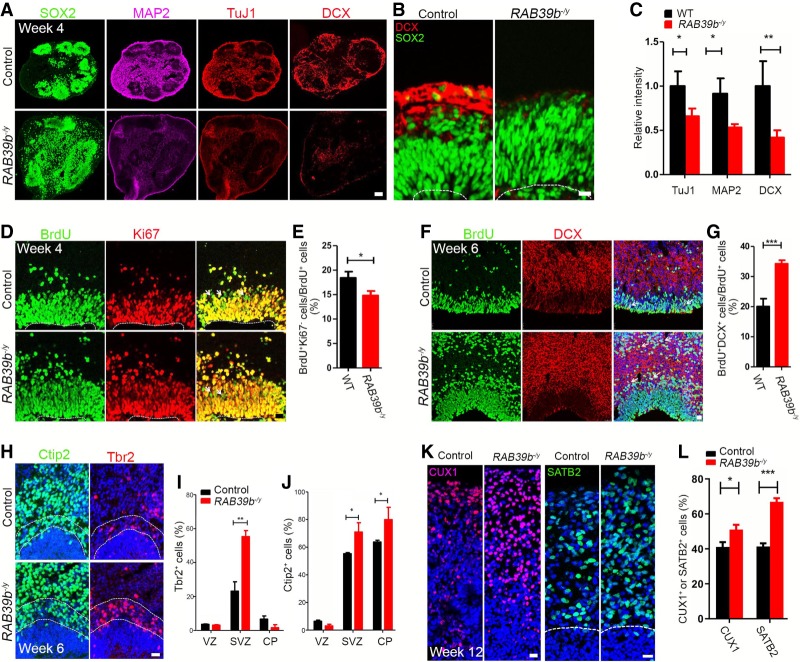
Figure 6.
Delayed differentiation of NPCs and neural overproduction in RAB39b mutant organoids. (A,B) Representative imaging of week 4 organoid sections stained with antibodies against SOX2 (green), MAP2, (purple), TuJ1 and DCX (red). Scale bars: A, 75 µm; B, 10 µm. (C) Quantification of relative intensities of MAP2, TuJ1, and DCX. (D) Representative imaging of week 4 brain organoid sections stained with antibodies against BrdU (24 h; green) and Ki67 (red). Hoechst stains nuclei. White arrowheads mark BrdU+Ki67− cells. Scale bars, 20 µm. (E) Quantification of the percentage of BrdU+Ki67− cells out of total BrdU+ cells. (F) Confocal imaging of week 6 organoid sections stained with antibodies against BrdU (green) and DCX (red). White arrows indicate BrdU+DCX+ cells. Scale bars, 20 µm. Week 4 organoids were administrated with BrdU for 48 h, followed by the experiments at week 6. (G) Quantification of the percentage of BrdU+DCX+ cells out of total BrdU+ cell. (H) Representative imaging of week 6 organoid sections stained with antibodies against Ctip2 (green) and Tbr2 (red). Hoechst stains nuclei. Scale bars: 20 µm. (I,J) Quantification of the percentage of Tbr2+ and Ctip2+ cells out of total cells in the examined areas. (K) Representative imaging of week 12 organoid sections stained with antibodies against CUX1 (red) and SATB2 (green). Hoechst stains nuclei. Scale bars: 20 µm. (L) Quantification of the percentage of CUX1- and SATB2-positive cells out of total cells in the examined areas. Error bars represent SEM of results from four independent sets of cerebral organoids. Statistical analyses were performed with nonparametric Mann-Whitney tests. (*) P < 0.05; (**) P < 0.01.
To further investigate the consequence of the increased neual differentiation, we examined IPCs and neurons in the CP-like regions of week 6 organoids. The CP is adjacent and superior to the TBR2 regions and can be identified by Ctip2 expression. We used Tbr2 to label IPCs, and Ctip2 to mark deep layer neurons in order to define the SVZ and CP-like regions in the organoids. There was a significant increase in the percentage of Tbr2-positive cells in the SVZ-like regions in the mutant organoids (Fig. 6H,I, P = 0.0011). In addition, mutant organoids exhibited an increased percentage of Ctip2-positive deep layer neurons (Fig. 6H,J, P = 0.0366, P = 0.0412). Next, we examined Tbr1-positive deep layer neurons, and upper layer neurons labeled by CUX1 and SATB2 in week 12 organoids (Fig. 6K; Supplemental Fig. S3E). RAB39b deletion resulted in a significant increase in the output of upper and deep layer neurons (Fig. 6L, P = 0.0184, P < 0.0001; Supplemental Fig. S3F, P = 0.0061). Together, these results suggested that RAB39b deletion caused a transient delay of NPC differentiation and an ultimately increased output of neuronal production in the cerebral organoids.
RAB39b regulates PI3K–AKT–mTOR signaling
Beclin 1 and Vps34 are core components of the class III phosphatidylinositol 3-kinase (PI3K) complex, which regulates autophagy and membrane trafficking. RAB39b and its homolog RAB39a have highly sequence similarity, and RAB39a has been shown to interact with class III PI3K component Beclin 1 in cell lines (Seto et al. 2013). To investigate RBA39b's functional mechanisms, we tested whether RAB39b interacts with class III PI3K components. Coimmunoprecipitation (co-IP) studies revealed that RAB39b interacted with neither Beclin 1 nor Vps34 (Supplemental Fig. S4A). Consistently, there were no autophagy defects in RAB39b mutant NPCs, as reflected by normal expression levels of LC3 and p62 that act as makers of autophagosomes and autophagy function, respectively (Supplemental Fig. S4B).
Class I PI3K and its downstream AKT–mTOR signaling acts as a common biological pathway that gives rise to autism-related disorders when disrupted (Huber et al. 2015; Enriquez-Barreto and Morales 2016; Winden et al. 2018). Class I PI3K consists of a 110-kDa (p110α or p110β) catalytic subunit and an 85-kDa (p85α) regulatory subunit. We reasoned that RAB39b might regulate PI3K–AKT–mTOR signaling. To test this hypothesis, we attempted to identify RAB39b's interactions with components in this pathway. Co-IP studies in N2A cell line showed that RAB39b associated with p85α and p110α (Fig. 7A). RAB39b belongs to the RAS-like GTPase superfamily (Mignogna et al. 2015). The constitutively active form of RAB39b (RAB39bQ68L) appears to have a stronger association with p85 compared with the dominant negative one (RAB39bS22N) (Supplemental Fig. S4C). RAB39b failed to interact with the other components of PI3K–AKT–mTOR pathway, including p110γ, Akt1, and Akt3 (Supplemental Fig. S4D). To further validate these interactions, we generated stable iPSC lines with the lentivirus-mediated expression of Flag-tagged WT (RAB39bWT), RAB39bQ68L, and RAB39bS22N. These iPSC lines were differentiated into NPCs followed by co-IP analyses. Again, we found that RAB39b associated with p85α and p110α in NPCs (Supplemental Fig. S4E). Together, these results suggested that RAB39b interacts with PI3K components.
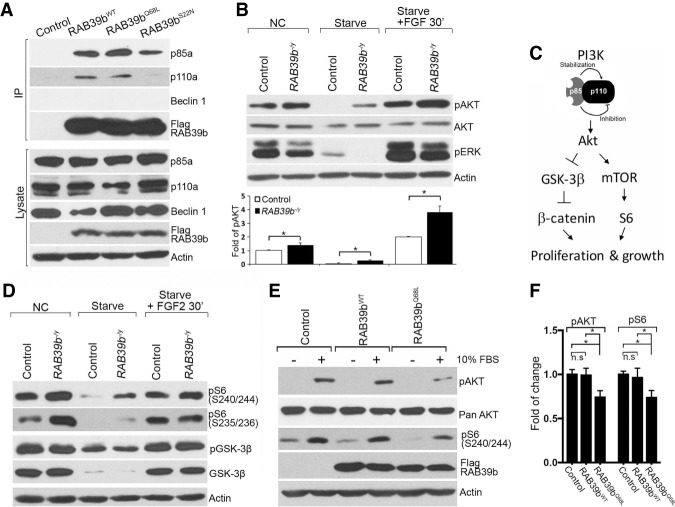
Figure 7.
RAB39b regulates PI3K–AKT–mTOR signaling. (A) Coimmunoprecipitation (co-IP) analysis revealed that WT (Rab39bWT), constitutively active (Rab39bQ68L), and dominant-negative (Rab39bS22N) Rab39b interact with p85α and p110α in N2A cells. (B) Western blot analysis showed that RAB39b deletion increases the pAKT signaling activity in mutant human NPCs in normal culture (NC), under starvation conditions, and in response to FGF stimulation conditions. (C) Diagram of the PI3K-AKT downstream GSK-3β and mTOR signaling pathways. (D) RAB39b deletion results in the up-regulation of mTOR signaling, reflected by increased pS6 (S240/244) and pS6 (S235/236) expression levels in human NPCs in normal culture (NC), under starvation conditions and in response to FGF stimulation conditions. (E) The constitutively active RAB39bQ68L decreases pAKT and pS6 (S240/244) expression levels in human NPCs. Actin serves as the loading controls. (F) Quantification of pAKT and pS6 (S240/244) expression levels in experiment E. Error bars represent SEM of results from three independent experiments. Statistical analyses were performed with nonparametric Mann-Whitney tests. (*) P < 0.05; (n.s) no significant difference detected.
AKT activation occurs downstream from PI3K signaling (Fruman et al. 2017). To determine whether RAB39b mutations disrupt PI3K signaling, we examined AKT signaling in NPCs using antibodies against phosphorylated AKT (pAKT at Ser473), a marker of AKT activation (Manning and Cantley 2007). Western blot analyses showed a significant increase in the expression level of pAKT in mutant NPCs in normal culture, under starvation conditions, and in response to FGF stimulation conditions (Fig. 7B). The expression levels of AKT and pERK remained unchanged in all three conditions, suggesting the specificity of pAKT up-regulation (Fig. 7B). AKT signaling regulates cellular responses through multiple downstream effectors, including activation of mTOR pathway and inhibition of GSK-3β signaling (Fig. 7C; Manning and Cantley 2007; Chen et al. 2015). We used phosphorylation of S6 (pS6 at Ser240/244 or Ser235/236) and GSK-3β (pGSK-3β at Ser9) to monitor the activation of the AKT–mTOR and AKT–GSK-3β axes, respectively. There was a significant increase in the expression levels of pS6 in the mutant NPCs in normal culture, under starvation conditions, and in response to FGF2 stimulation, whereas the phosphorylated and total GSK-3β levels appeared unchanged (Fig. 7D).
To investigate whether RAB39b is sufficient to inhibit PI3K–AKT–mTOR activity, we turned our attention to RAB39bWT and RAB39bQ68L (RAB39b-overexpressing) human NPCs. Whereas no substantial changes were detected in RAB39bWT expression cells, constitutively active RAB39bQ68L significantly decreased the expression of pAKT and pS6 (Ser240/244) (Fig. 7E,F, P < 0.05). Next, we examined RAB39b loss-of-function NPCs with dominant-negative RAB39bS22N expression, and detected an up-regulation of pAKT and pS6 (Ser240/244) (Supplemental Fig. S4F,G). These results suggest that activation or inhibition of RAB39b GTPase activity is sufficient to decrease or increase AKT–mTOR signaling, respectively. Together, these results support the notion that RAB39b deletion promotes PI3K–AKT–mTOR signaling activity, which may enhance the NPC proliferation and lead to macrocephaly.
AKT–mTOR activation mediates the enhanced NPC proliferation and organoid sizes
To validate the in vitro finding of AKT–mTOR signaling up-regulation in vivo, we examined E14.5 cerebral cortex using pS6 as the readout. As reported in previous studies (Poduri et al. 2012), the expression of p-S6 was enriched in mitotic NPCs, which are labeled by phospho-Vimentin (p-VIM) staining (Fig. 8A). Therefore, we quantified p-S6-positive mitotic NPCs and found that the percentage of p-S6-positive mitotic NPCs out of total mitotic NPCs was significantly increased in mutant mouse cerebral cortex (Fig. 8B, P = 0.0007). Next, we performed similar studies in the cerebral organoids. As reported in published studies (Li et al. 2017), p-S6 expression was enriched in the VZ/SVZ-like regions in the cerebral organoids (Fig. 8C). The percentage of p-S6-positive mitotic NPCs (labeled by p-VIM) was significantly increased in mutant organoids (Fig. 8D, P = 0.0061). Together, these results suggest that RAB39b deletion leads to up-regulation of PI3K–AKT–mTOR signaling activity in mice and in cerebral organoids.
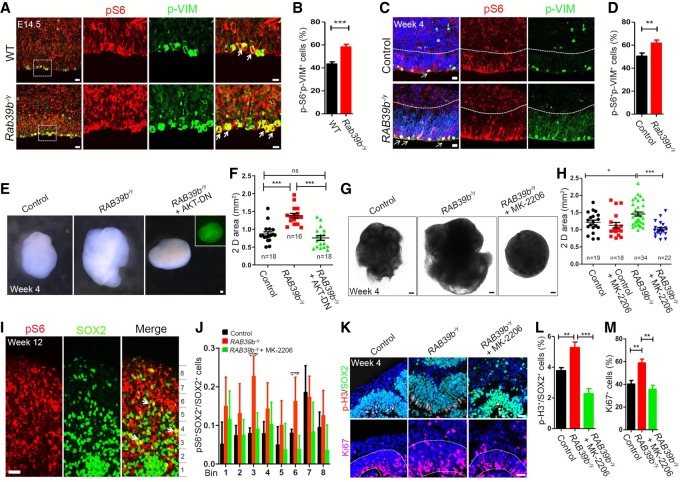
Figure 8.
AKT–mTOR signaling up-regulation mediates NPC overproliferation and organoid enlargement. (A,C) Representative imaging of E14.5 cerebral cortex (A) or week 4 organoid (C) sections stained with antibodies against p-S6 (red) and p-VIM (green). Hoechst stains nuclei (blue). The right panels are enlargements of boxed areas in leftmost panels. White arrowheads mark NPCs with the colocalization of p-S6 and p-VIM. Scale bars: left panels, 10 µm; other panels, 20 µm. (B,D) Quantification of the percentage of p-S6;p-VIM double-positive cells out of total p-VIM cells in the examined areas. (E) Representative week 4 organoids from iPSCs of control and mutants with or without infection of lentivirus expressing dominant-negative AKT (GFP). Scale bars, 0.1 mm. (F) Quantification of 2D surface areas of cerebral organoids. (G) Representative imaging of cerebral organoids with or without AKT inhibitor MK-2206. Scale bars, 0.2 mm. (H) Quantification of 2D surface areas of cerebral organoids. (I) Representative imaging of week 12 cerebral organoid sections stained with antibodies against pS6 (red) and SOX2 (green). White arrows represent pS6+SOX2+ cells. Scale bars, 20 µm. (J) Quantification of the number of pS6+SOX2+ cells out of total SOX2+ cells in different bins as indicated in experiment I. (K) Representative imaging of week 4 organoid sections stained with antibodies against SOX2 (green), p-H3 (red), and Ki67 (red). Scale bars, 20 µm. (L) Quantification of the percentage of p-H3+ cells out of total SOX2+ cells. (M) Quantification of the percentage of Ki67+ cells out of total cells in areas surrounded by white dotted lines in K. Error bars represent SEM of results from independent mice (n = 3) or organoids (n = 4). Statistical analyses were performed with nonparametric Mann-Whitney test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (ns) no significant difference detected.
Outer radial glia cells (oRGs) are essential for the evolutionary expansion of the human brains (Noctor et al. 2007). Multiple regulators of PI3K–AKT–mTOR pathway are specifically enriched in the oRGs, including the p-S6 (a canonical readout of mTOR activation), in comparison with other NPCs such as APCs and INPs (Nowakowski et al. 2017). Human radial glial cells exhibited elevated activation of mTOR signaling compared with those of other primates and rodents (Pollen et al. 2019). We hypothesized that oRGs may be particularly vulnerable to the RAB39b deletion-mediated activation of mTOR signaling. To test this hypothesis, we examined mTOR effector p-S6 expression in SOX2-labeled radial glia cells in the outer SVZ as in published studies (Pollen et al. 2019). We first confirmed that the majority of SOX2-positive cells were colabeled by oRG marker PTPRZ1 (Supplemental Fig. S5A). We quantified pS6 levels in equal bins across VZ/SVZ and outer SVZ regions (Supplemental Fig. S5B–D). Results showed that RAB39b deletion significantly increased the percentage of pS6-positive oRG cells in cerebral organoid (Fig. 8J).
To investigate whether AKT–mTOR activation is responsible for the enlarged organoid sizes, we performed rescue studies by inactivating AKT signaling. Human iPSCs were transduced with lentivirus expressing dominant-negative AKT followed by the generation of cerebral organoids (Fig. 8E). Quantificaton results showed that the inhibition of AKT activity significantly reduced mutant organoid sizes (Fig. 8F). Next, we used pharmacological approaches by treating day 12 cultured cells with MK-2206 (100 nM), a highly potent and specific inhibitor of AKT activation (Hirai et al. 2010; Ji et al. 2017). Whereas a high concentration of inhibitors was detrimental to cellular growth, lower concentrations of MK-2206 allowed the generation of human cerebral organoids with morphology and sizes comparable with those of DMSO-treated controls (Fig. 8G,H). Quantification results showed that MK-2206 treatment restored the mutant organoid surface areas to control levels (Fig. 8G,H); the mTOR activity, monitored by p-S6, was also rescued by MK-2206 (Fig. 8I,J; Supplemental Fig. S5B–D). Next, we examined MK-2206's rescuing effects on cellular behaviors. MK-2206 treatment reduced NPC proliferation, reflected by the decrease of p-H3-positive NPCs labeled by SOX2 in mutant cerebral organoids (Fig. 8K,L). Ki67 staining results showed that MK-2206 restored the cycling cells (labeled by Ki67) to control levels (Fig. 8K,M), which may contribute to the rescued cerebral organoid sizes. Together, these results suggested that RAB39b deletion led to the upregulation of PI3K–AKT–mTOR signaling activity, which was responsible for the overproliferation of NPCs and ultimate overgrowth of mutant cerebral organoids.
Discussion
Through creating the first genetic mouse and organoid models for RAB39b-associated macrocephaly/autism, here we discovered RAB39b-dependent dysregulation of cortical neurogenesis due to aberrant activation of PI3K–AKT–mTOR signaling, which contributes to macrocephaly and autistic-like behaviors. Our studies provide new insights into the mechanisms of neurodevelopmental dysregulation and common pathways associated with macrocephaly/autism disorders across species.
Rab39b KO mouse models behavioral deficits in macrocephaly/autism disorders
Consistent with the predicted effects of loss of function of RAB39b mutations in human genetic data, our Rab39b null mouse model exhibited deficits in social memory and motor learning, which partially recapitulated patient ASD/ID phenotypes (Giannandrea et al. 2010; Woodbury-Smith et al. 2017). Our Rab39b KO mice displayed similar autism-like behaviors to those seen in the mutant mouse model of Chd8 (Platt et al. 2017), mutations of which are strongly associated with ASD. Despite its enrichment in neurons, Rab39b appears to be ubiquitously expressed in the different regions of the brain. These observations raise the question of how mutation of a ubiquitously expressed gene leads to selective behavior deficits. A potential possibility is that certain cell types and brain regions are particularly vulnerable to the functional loss of Rab39b. Future studies should identify which cell type(s) or brain region(s) are particularly sensitive to the loss of function of Rab39b, resulting in specific behavior deficits.
Early neurodevelopmental dysregulation in macrocephaly/autism disorders
Our studies provided new insights into neurodevelopmental dysregulation associated with macrocephaly/autism, which supports the emerging notion that ASD is a progressive disorder of brain development. ASD represents a spectrum of genetically and phenotypically heterogenenous diseases. It is unclear which neurodevelopmental processes are disrupted by individual mutations, and how these disruptions ultimately contribute to ASD pathophysiology. We found that RAB39b mutations result in overproliferation and differentiation deficits of NPCs in both organoids and mice, which is consistent with what has been observed in patient iPSCs and animal models carrying different ASD mutant genes (Mariani et al. 2015; Gompers et al. 2017; Marchetto et al. 2017). Excess NPC proliferation is expected to generate a larger than normal number of neurons. Indeed, Rab39b KO mice exhibited an increase in different layer neurons as well as macrocephaly reflected by an increase in hemisphere length and cortical surface area, which is consistent with reported brain overgrowth in macrocephaly/autism patients (Hazlett et al. 2011; Zielinski et al. 2014; Ohta et al. 2016). The proliferation of Pax6-positive NPCs is enhanced, which is coupled with increased Tbr2-positive IPCs resulting in the expansion of progenitor pools in mutant mice. We detected an increase in Tbr1/Ctip2-positive neurons at E18.5 but not E14.5 mutant mouse brains. The Cux1-positive neurons were increased at P20 but not E18.5 mutant mice. These results suggested that deep layer neurons could be still generated at later time points, and the increase of upper layer neurons was manifested at later developmental stages in Rab39b mutant mice, which is consistent with the transient delay of cell cycle exit of mutant NPCs. Future studies should determine how Rab39b deletion changes NPC differentiation dynamics in the developing brain.
RAB39b mutation-associated macrocephaly appears more severe in mutant organoids than that in KO mouse models, which highlights the strength of cerebral organoids in modeling early human neurodevelopment. Why might RAB39b deletion generate more severe developmental phenotypes in human than mouse models? One reason might be that the oRG neural progenitors are abundant in the human cerebral cortex but not in that of mice; they are highly proliferative, resulting in their progenitor contribution to the majority of upper layer neurogenesis (Lewitus et al. 2013; Pollen et al. 2015). The expansion of oRGs in human organoids due to RAB39b mutation may lead to the promotion of neurogenesis output specifically in the human cerebral cortex in comparison with that of mice. This is supported by a more pronounced increase in NPC pool and upper layer neurons in mutant organoids than that in mutant mice, which could explain the dramatic size disparity between mutant and control organoids relative to the mild macrocephaly seen in mice upon RAB39b deletion. One caveat for this comparison is that developmental timing is not comparable between mutant organoids and mutant mice. It will be informative to perform side by side comparisons between mouse and human cerebral organoids without or without Rab39b deletion.
RAB39b regulates the PI3K–AKT–mTOR pathway
ASD exhibits striking heterogeneity in genetics and clinical presentations. Studying rare mutations has proven to be valuable in identifying common pathways implicated in autism (Berg and Geschwind 2012; Willsey et al. 2013). Our work has connected RAB39b mutation-associated macrocephaly/autism with PI3K–AKT–mTOR signaling, a convergent pathway implicated in ASD pathology across divergent etiologies (Huber et al. 2015; Magdalon et al. 2017). RAB39b interacts with PI3K components p85α and p110α, and its deletion promotes PI3K–AKT–mTOR signaling activity in human NPCs, cerebral organoids, and mouse cortex. Inhibition of AKT is sufficient to rescue the NPC proliferation and organoid size deficits caused by RAB39b mutations, suggesting that activation of the PI3K–AKT–mTOR pathway is responsible for the overproduction of NPCs and macrocephaly in RAB39b-associated ASD. Furthermore, our RAB39b mutant organoid studies revealed human-specific regulation of PI3K–AKT–mTOR signaling, which has a high basal level activity in oRGs. The human oRG cells have enriched expression of several regulators of mTOR signaling pathway (Nowakowski et al. 2017); human-specific activators of mTOR pathway exist in oRGs (Pollen et al. 2019), suggesting that the activity of mTOR signaling has evolved recently in human oRGs. This raises the possibility that oRG cells could be especially vulnerable to mutations affecting mTOR pathway (Nowakowski et al. 2017; Pollen et al. 2019). Indeed, increased PI3K–AKT–mTOR signaling was detected in SOX2-positive oRGs and may promote self-renewal/proliferation of oRGs leading to robust organoid size enlargement in RAB39b mutant organoids. Recent studies reported that dysregulation of β-catenin/Brn2 transcriptional cascade leads to increased NPC proliferation and transient embryonic brain overgrowth contributing to social deficits in adults (Belinson et al. 2016; Marchetto et al. 2017). Therefore, it is possible that embryonic overgrowth due to Rab39b mutations may similarly play pathological roles in postnatal cognitive behavioral abnormalities. Taken together, these studies suggest that different genetic mutations act on common pathways, converging on the disruption of NPC proliferation and differentiation processes and contributing to autistic behaviors in adults.
Materials and methods
All animal procedures were approved by the University of Southern California Animal Care and Use Committee and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Rab39b knockout mouse generation
Two gRNAs (CCTGTAGTAGGCGCGAGTGA and AAGAGGTTATCAAATCAGAG) were designed to target exon 2 of mouse Rab39b. The designed guide RNA and Cas9 mRNA were injected into C57BL/6N zygotes. Fifteen pups were obtained from the zygotes injection and were genotyped. Three of them (one male and two females) showed large deletion-carrying mutations. Sanger sequencing showed the mutants were harboring 397-bp deletions in Rab39b exon 2. This sequence maps uniquely to the targeting site via BLAST, reducing the likelihood of off-target mutations. The PCR primers used for genotyping were Rab39b primer-F (GGACTGTCAGGAATCAGGAACACTAG) and Rab39b primer-R (GCCTAGGAAGAAGGCTCATTTATTATCC).
Cell culture
A human iPSC line was characterized and obtained from NINDS Human Cell and Data Repository (NHCDR). H9 human ES cells were ordered from the Wisconsin International Stem Cell (WISC) bank. ES cells or iPSCs (referred to here collectively as human pluripotent stem cells [hPSCs]) were cultured in mTeSR medium (Stem cell technologies), and were passaged 5 d onto a new plate coated with Geltrex (Thermo Fisher Scientific). The hPSCs were detached from the plate by incubation with ReLeSR (Stem cell technologies) for 1 min. The cells were then dissociated into small cell aggregates by manual pipetting. The medium was changed every day. N2A and 293T cells were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM (Life technolgies) supplemented with 10% FBS.
Generation of RAB39b KO hPSC lines
For genome targeting using Crispr–Cas9 appraoches, gRNAs were designed using the online software GPP sgRNA Designer (Broad Institute) and cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 vector. Exon1-gRNA (CCATCTGGCTGTACCAGTTC) and Exon 2-gRNA (CCATCACTCGCGCCTACTAC) were chosen for the generation of RAB39b KO hPSCs. PX459 V2.0 was a gift from Dr. Feng Zhang's laboratory (Addgene plasmid #62988). RAB39b KO hPSCs were generated using a previously published method (Ran et al. 2013). Briefly, hPSCs were treated with ROCK inhibitor Y27632 (10 µM; Selleckchem) for 24 h prior to electroporation. The cells were dissociated into single cells using Accutase. Three micrograms of plasmids was electroporated into 8.0 × 105 cells using Amaxa Nucleofector II eletroporator (Lonza) with human stem cell nucleofector kit 1 (Lonza), then immediately plated on Geltrax-coated plates and cultured in mTeSR medium containing Y27632 (50 µM) for the first 24 h. Puromycin (0.5 µg/mL) was added to the medium for 2-d selection. Subsequently, hPSCs were maintained in medium without puromycin until colonies emerged. Individual colonies were picked up and expanded. PCR products were amplified and subjected to Sanger sequencing to identify mutant clones. The primers were Rab39b-Forward (GTCTACGCGGGGGATTACAG) and Rab39b-Reverse (TGTTATTGACCGGCCTTCCC).
Human NPC induction and culture
Human NPC induction from hPSCs was performed as previously described with minor modifications (Shi et al. 2012). Briefly, hPSCs were cultured until 80% confluence, and then were passaged into cell aggregates to form embryoid bodies (EBs). These EBs underwent suspended culture for 2 wk in neural induction medium, which consisted of N2B27 medium supplemented with dual Smad inhibitors SB431542 (10 µM) and LDN-193189 (0.1 µM; Selleckchem). N2B27 medium was 50% DMEM/F12, 50% Neurobasal medium, 0.5% N2 supplement, 1% B27 supplement, 1% glutamax, 1% penicillin–streptomycin, and 1% NEAA (Thermo Fisher Scientific).Then EBs were attached to the Geltrex-coated plates and cultured in neural induction medium containing 20 ng/mL bFGF until the neural rosettes emerged. Neural rosettes were manually picked up and dissociated into individual cells by Accutase, which were then plated on Geltrex-coated plates. The NPCs were maintained in N2B27 medium with 20 ng/mL bFGF and 20 ng/mL EGF, and the medium was changed every 3 d.
Generation of cerebral organoids
Cerebral organoids were generated with published protocols with minor modifications (Lancaster et al. 2013; Zhang et al. 2019). Two mutant PSC lines were used, including human iPSC line CR0000010 with the 4-bp deletion in exon 1 and human H9 ES cell line with the 5-bp deletion in exon 2. The cerebral organoids were cultured in four different spinning culture bottles (n = 4). Individual culture bottle contained 15–25 cerebral organoids, of which three to four organoids were embedded for sectioning. A total of eight sections for each organoid and two to three intact cortical structures from each section was used for the analyses. The hPSC colonies were dissociated into single cells using Accutase. On day 1, a total of 9000 cells was plated into each well of an ultralow-attachment 96-well plate (Thermo) for single EB formation. The EB formation medium consisted of DMEM/F12, 20% knockout serum replacement, 1% GlutaMAX, 1%NEAA, 50 µM Y27632, and 4 ng/mL bFGF (Perpotech). On day 4, the EBs were cultured in EB formation medium without Y27632 and bFGF. Starting on day 7, the EBs were cultured in neural induction medium for 5 d. The neural induction medium consisted of DMEM/F12, 0.5% N2 supplement, 1% GlutaMAX, 1% nonessential amino acids, 1% penicillin–streptomycin, and 10 µg/mL heparin with dual Smad inhibitors A83-01(1 µM) and LDN-193189 (0.1 µM). On day 12, the EBs were embedded into Geltrex droplets (Thermo) and cultured for 4 d in medium containing 50% DMEM/F12, 50% neurobasal medium, 0.5% N2 supplement, 1% B27 supplement without vitamin A, 1% GlutaMAX, 1% NEAA, 1% penicillin–streptomycin, and 2.5 ng/mL human insulin (Sigma). On day 16, the organoids were transferred to a spinner flask rotating continuously at 60 rpm and cultured in N2B27 medium with 2.5 ng/mL human insulin. The medium was changed every week. For long-term culture (more than 2 mo) of cerebral organoids, we followed a published protocol (Quadrato et al. 2017). Briefly, 5000 cells were used to generate EBs. After neural induction and Matrigel embedding, the cerebral organoids were transferred to the spinner flask and cultured in N2B27 medium containing 10 ng/mL BDNF. Medium was changed every week.
Immunostaining and immunohistochemical staining
For human NPC immunostaining, cells were fixed in 4% PFA for 15 min at room temperature, washed twice with PBS, and then incubated with primary antibodies in blocking buffer (2% goat serum, 1% BSA, 0.1% TritonX-100 in PBS) overnight at 4°C before secondary antibody incubation. Brain organoids were fixed in 4% PFA for 30 min at room temperature. Brain organoids were washed three times with PBS, incubated in 30% sucrose solution overnight at 4°C, and then embedded in O.C.T. solution followed by dry ice freezing. The frozen organoids were sectioned into 10-µm-thick slices for immunostaining. We measured SOX2+ ventricular zone (VZ) thickness according to published methods (Qian et al. 2016) in which measurements were taken at 45° angles to obtain the mean value. Histological processing and immunohistochemical labeling of cryosections were performed using cerebral cortex sections from different stages of embryos as described previously (Yang et al. 2016). The primary antibodies are listed in Supplemental Table 1. The secondary antibodies used were Alexa 488, Alexa 555, and Alexa 647 conjugated to specific IgG types (Invitrogen Molecular Probes). All of the experiments were repeated at least three times, and representative images are shown in the individual figures.
BrdU labeling and analysis
E14.5 pregnant mice were injected intraperitoneally with BrdU (Sigma) at 25 mg/kg body weight. The animals were sacrificed at 0.5 h after the injection. The embryo brains were dissected out and fixed in 4% PFA for 1.5 h on ice. Subsequently, the brains were stored in 30% sucrose overnight and embedded in the O.C.T. solution. For in vitro labeling, the brain organoids were incubated in the medium containing Brdu (100 µM) for 2 h, then washed with PBS and fixed in 4% PFA for 30 min at room temperature. The samples were stained with Brdu after sectioning. The Brdu+ cells were counted in the VZ/SVZ-like region.
Cell cycle exit
Pregnant mice were intraperitoneally injected at E13.5 with 50 mg/kg body weight Brdu. After 20 h, mice were anesthetized and embryo brains fixed with 4% PFA for 1.5 h on ice. The brain organoids and NPCs were treated with Brdu (100 µM) for 24 or 48 h and fixed with 4% PFA for 30 min at room temperature. After sectioning, the samples were costained with BrdU and Ki67. The cell cycle exit was determined by the ratio of Brdu+Ki67− cells to total BrdU+ cells in VZ/SVZ-like region.
RAB39b overexpression vector construction
The vectors pcDNA3.1-RAB39bWT, pcDNA3.1-RAB39bQ68L, and pcDNA3.1-RAB39bS22N were generated using a site mutagenesis kit. The Flag tag was introduced after initiation codon ATG of the RAB39b by PCR. The Flag-tagged RAB39bWT, pcDNA3.1-RAB39bQ68L, or pcDNA3.1-RAB39bS22N fragement amplified by PCR were digested with BamHI and AgeI, then inserted into lentivirus vector. The lentivirus vector backbone was derived from LentiCRISPRv2GFP vector, and C-terminal of RAB39b, is linked to the GFP through P2A in the vector.The LentiCRISPRv2GFP was a gift from David Feldser (Addgene plasmid # 82416).
Lentivirus production and transduction
To produce lentivirus, a total of 15 µg of Flag-tagged RAB39bWT, RAB39bQ68L, or RAB39bS22N lentiviral vectors were transfected into 293T cells together with 6 µg VSVG and 9 µg pspax2 using using CaPO4 precipitation. The viruses in the supernatant were collected at 24 and 48 h after transfection, filtered through a 0.45-µm filter, and concentrated via ultracentrifugation. To transduce hPSCs with lentivirus, hPSCs were cultured in mTeSR medium containing lentivirus, Y27632 (10 µM) and polybrene (8 µg/mL). After 12 h of infection, medium was changed. The hPSCs were dissociated into singles cells and continue to cultured. GFP positive clones were picked up and expanded.
RAB39b KO rescue experiment
For RAB39b overexpression rescue experiments, the RAB39b KO cell lines were overexpressed with Flag-tagged Rab39bWT GFP by lentivirus infection, followed by the generation of cerebral organoids. For the inhibitor rescued experiment, the PI3K–AKT inhibitor MK-2206 (100 nM; Selleck) was added into the medium starting at day 1 of cerebral organoid generation. The control group was treated with an equal amount of DMSO. For nonpharmacological manipulation of the PI3K-AKT singaling, the lentiviral expression plasmid pHRIG-AktDN, which contains dominant negative Akt1, was used for lentivirus production. The iPS cell lines were infected by lentivirus followed by the generation of cerebral organoids. pHRIG-AktDN was a gift from Heng Zhao (Addgene plasmid # 53597)
Western blot analysis
Isolated mouse cortex and human iPS-derived NPCs were lysed in buffers containing 50 mM Tris HCl (pH 8), 140 nM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, and 0.25% Triton with protease inhibitor cocktail (Thermo). After sonication, samples were spun down, and the supernatants were used for a BCA Bradford assay (Bio-Rad) with the Spectramax iD3 plate reader to assess protein concentration using a standard curve. Thirty micrograms of protein was used on a 8%–12% SDS-PAGE using the Western blotting system (Bio-Rad).
Immunoprecipitation
Flag-tagged RAB39bWT, RAB39bQ68L and RAB39bS22N plasmids were transfected into N2A cells using JetPRIME (Poly plus transfection). After 48 h, the transfected N2A cells were lysed in RIPA buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris at pH 8.0, one tablet protease inhibitor [Thermo] per 10 mL). Cell debris was pelleted at 15,000 rpm for 15 min at 4°C. For immunoprecipitation, 1–2 mg of supernatants from the centrifugation of the lysate was incubated with anti-Flag M2 affinity resin (Sigma) overnight at 4°C, followed by washing the immunoprecipitates three times with lysis buffer. The immunoprecipitates were eluted in SDS-PAGE loading buffer by boiling for 10 min before Western blot analysis.
Behavioral testing
An open field test was performed using an overhead SMART video tracking system (Panlab), which measures distance traveled, time spent in each zone, and speed. The apparatus consisted of a gray open top plastic box (w × d × h: 45 × 45 × 40 cm) divided into four equal arenas. After a 2-h acclimatization to the behavioral testing room, each animal was placed in the center of the open field and left undisturbed for 30 min. The apparatus was wiped between trials with a 70% ethanol solution.
For the rotarod test, an accelerating rotarod (Panlab) was used to analyze motor coordination and balance. Mice were trained three times on the rotarod at 4 rpm for a maximum of 180 sec per day before testing. During test conditions, we measured the latency of the mouse falling from the rotating beam while ramping up the rotation speed, starting at 4 rpm and accelerating to 40 rpm over each 5-min trial period. Mice were given three trials per day, with an intertrial interval of 20 min. The average time before falling in the three trials was used to evaluate latency.
For the three-chamber test, a rectangular Plexiglas box (w × d × h: 52 × 25 × 23 cm) was divided into three chambers. In the habituation trial (5 min), empty wire cages (h × d: 12 × 8 cm) that could provide contact with another mouse were present in the left and right chambers visible from the middle chamber. In the sociability trial (10 min), a novel mouse from same strain that had never been observed before by the subject mouse was placed in the wire cage in the left chamber, whereas no mouse was placed in the wire cage in the right chamber. For the social memory test, a variant of the social novelty trial (10 min) was conducted wherein another novel mouse was placed in the wire cage of the right chamber. The test mouse was placed in the central chamber and allowed to explore all three chambers for each trail. The time spent in each chamber was recorded. At the end of each test, all the equipment was cleaned with paper towels and 70% ethanol.
For the marble burying test, 20 glass marbles (diameter: 15 mm) were arranged in a symmetrical 4 × 5-cm grid on top of 2- to 3-cm-deep bedding in a clean standard mouse cage (w × d × h: 27 × 16.5 × 12.5 cm). Each mouse was placed in the center of the cage for a 30-min exploration period and at the end of this period each marble was scored as buried if two-thirds of its surface area was covered by bedding.
For evaluation of self-grooming behaviors, each mouse was placed individually into a standard mouse cage. A front-mounted camera was placed 1 m from the cages to record the 20-min sessions. The first 10-min period was habituation and was not scored. Each subject was scored for cumulative time spent grooming all body regions during the second 10 min of the session.
Supplementary Material
Acknowledgments
We thank Chen laboratory colleagues for stimulating discussions. We are grateful for Bridget Samuels's critical reading of the manuscript. The Chen laboratory is supported by funds from the Associate Dean of Research Fund from the Center for Craniofacial Molecular Biology, Herman Ostrow School of Dentistry at the University of Southern California, and grants R01NS097231 (J.C.) and R01NS096176 (J.C.) from the National Institute of Health.
Author contributions: W.Z., L.M., M.Y., Q.S., J.-F.C. conceived and performed all experiments. J.X., Z.L., Z.Z., R.C., and Y.C. helped with the manuscript writing. J.-F.C. designed and interpreted the experiments and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.332494.119.
Freely available online through the Genes & Development Open Access option.
References
- Belinson H, Nakatani J, Babineau BA, Birnbaum RY, Ellegood J, Bershteyn M, McEvilly RJ, Long JM, Willert K, Klein OD, et al. 2016. Prenatal β-catenin/Brn2/Tbr2 transcriptional cascade regulates adult social and stereotypic behaviors. Mol Psychiatry 21: 1417–1433. 10.1038/mp.2015.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Geschwind DH. 2012. Autism genetics: searching for specificity and convergence. Genome Biol 13: 247 10.1186/gb-2012-13-7-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR. 2017. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20: 435–449.e4. 10.1016/j.stem.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova EL, Casanova MF. 2014. Genetics studies indicate that neural induction and early neuronal maturation are disturbed in autism. Front Cell Neurosci 8: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Huang W-C, Séjourné J, Clipperton-Allen AE, Page DT. 2015. Mutations alter brain growth trajectory and allocation of cell types through elevated β-catenin signaling. J Neurosci 35: 10252–10267. 10.1523/JNEUROSCI.5272-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow ML, Pramparo T, Winn ME, Barnes CC, Li H-R, Weiss L, Fan J-B, Murray S, April C, Belinson H, et al. 2012. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet 8: e1002592 10.1371/journal.pgen.1002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciammola A, Carrera P, Di Fonzo A, Sassone J, Villa R, Poletti B, Ferrari M, Girotti F, Monfrini E, Buongarzone G, et al. 2017. X-linked Parkinsonism with intellectual disability caused by novel mutations and somatic mosaicism in RAB39B gene. Parkinsonism Relat Disord 44: 142–146. 10.1016/j.parkreldis.2017.08.021 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N. 2003. Evidence of brain overgrowth in the first year of life in autism. JAMA 290: 337–344. 10.1001/jama.290.3.337 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, Barnes CC, Pierce K. 2011. Neuron number and size in prefrontal cortex of children with autism. JAMA 306: 2001–2010. 10.1001/jama.2011.1638 [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pramparo T, Gazestani VH, Lombardo MV, Pierce K, Lewis NE. 2019. The ASD living biology: from cell proliferation to clinical phenotype. Mol Psychiatry 24: 88–107. 10.1038/s41380-018-0056-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. 1999. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res 835: 18–26. 10.1016/S0006-8993(98)01258-X [DOI] [PubMed] [Google Scholar]
- Di Lullo E, Kriegstein AR. 2017. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18: 573–584. 10.1038/nrn.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Barreto L, Morales M. 2016. The PI3K signaling pathway as a pharmacological target in Autism related disorders and Schizophrenia. Mol Cell Ther 4: 2 10.1186/s40591-016-0047-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C. 2016. Proliferation and differentiation deficits are a major convergence point for neurodevelopmental disorders. Trends Neurosci 39: 290–299. 10.1016/j.tins.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. 2017. The PI3K pathway in human disease. Cell 170: 605–635. 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannandrea M, Bianchi V, Mignogna ML, Sirri A, Carrabino S, D'Elia E, Vecellio M, Russo S, Cogliati F, Larizza L, et al. 2010. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet 86: 185–195. 10.1016/j.ajhg.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompers AL, Su-Feher L, Ellegood J, Copping NA, Riyadh MA, Stradleigh TW, Pride MC, Schaffler MD, Wade AA, Catta-Preta R, et al. 2017. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci 20: 1062–1073. 10.1038/nn.4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet C, Piven J. 2011. Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry 68: 467–476. 10.1001/archgenpsychiatry.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, Elison JT, Swanson MR, Zhu H, Botteron KN, et al. 2017. Early brain development in infants at high risk for autism spectrum disorder. Nature 542: 348–351. 10.1038/nature21369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Pan B-S, et al. 2010. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther 9: 1956–1967. 10.1158/1535-7163.MCT-09-1012 [DOI] [PubMed] [Google Scholar]
- Huber KM, Klann E, Costa-Mattioli M, Zukin RS. 2015. Dysregulation of mammalian target of rapamycin signaling in mouse models of autism. J Neurosci 35: 13836–13842. 10.1523/JNEUROSCI.2656-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Lin W, Wang L, Ni Z, Jin F, Zha X, Fei G. 2017. Combined targeting of mTOR and Akt using rapamycin and MK-2206 in the treatment of tuberous sclerosis complex. J Cancer 8: 555–562. 10.7150/jca.17205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. 2013. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci 110: 20284–20289. 10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. 1998. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci 95: 1242–1246. 10.1073/pnas.95.3.1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde R, Bensoula AN, Filali M. 1995. Rotorod sensorimotor learning in cerebellar mutant mice. Neurosci Res 22: 423–426. 10.1016/0168-0102(95)00916-H [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. 2009. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5: 320–331. 10.1016/j.stem.2009.05.026 [DOI] [PubMed] [Google Scholar]
- Lewitus E, Kelava I, Huttner WB. 2013. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front Hum Neurosci 7: 424 10.3389/fnhum.2013.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, Gehrke L, Knoblich JA, Jaenisch R. 2017. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 20: 385–396.e3. 10.1016/j.stem.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalon J, Sánchez-Sánchez SM, Griesi-Oliveira K, Sertié AL. 2017. Dysfunctional mTORC1 signaling: a convergent mechanism between syndromic and nonsyndromic forms of autism spectrum disorder? Int J Mol Sci 18: 659 10.3390/ijms18030659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. 2007. AKT/PKB signaling: navigating downstream. Cell 129: 1261–1274. 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria K, Beltrao-Braga P, Trujillo CA, Mendes APD, Padmanabhan K, et al. 2017. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry 22: 820–835. 10.1038/mp.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al. 2015. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162: 375–390. 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata IF, Jang Y, Kim C-H, Hanna DS, Dorschner MO, Samii A, Agarwal P, Roberts JW, Klepitskaya O, Shprecher DR, et al. 2015. The RAB39B p.G192R mutation causes X-linked dominant Parkinson's disease. Mol Neurodegener 10: 50 10.1186/s13024-015-0045-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignogna ML, Giannandrea M, Gurgone A, Fanelli F, Raimondi F, Mapelli L, Bassani S, Fang H, Van Anken E, Alessio M, et al. 2015. The intellectual disability protein RAB39B selectively regulates GluA2 trafficking to determine synaptic AMPAR composition. Nat Commun 6: 6504 10.1038/ncomms7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. 2010. Animal models of neuropsychiatric disorders. Nat Neurosci 13: 1161–1169. 10.1038/nn.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeño V, Kriegstein AR. 2007. Neural stem and progenitor cells in cortical development. Novartis Found Symp 288: 59–73. discussion 73–8–96–8. [PubMed] [Google Scholar]
- Nonaka-Kinoshita M, Reillo I, Artegiani B, Martínez-Martínez MÁ, Nelson M, Borrell V, Calegari F. 2013. Regulation of cerebral cortex size and folding by expansion of basal progenitors. EMBO J 32: 1817–1828. 10.1038/emboj.2013.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. 2017. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358: 1318–1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Nordahl CW, Iosif A-M, Lee A, Rogers S, Amaral DG. 2016. Increased surface area, but not cortical thickness, in a subset of young boys with autism spectrum disorder. Autism Res 9: 232–248. 10.1002/aur.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. 2012. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622. 10.1126/science.1227764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piven J, Elison JT, Zylka MJ. 2017. Toward a conceptual framework for early brain and behavior development in autism. Mol Psychiatry 22: 1385–1394. 10.1038/mp.2017.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Zhou Y, Slaymaker IM, Shetty AS, Weisbach NR, Kim J-A, Sharma J, Desai M, Sood S, Kempton HR, et al. 2017. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep 19: 335–350. 10.1016/j.celrep.2017.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. 2012. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 74: 41–48. 10.1016/j.neuron.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. 2015. Molecular identity of human outer radial glia during cortical development. Cell 163: 55–67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, et al. 2019. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176: 743–756.e17. 10.1016/j.cell.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165: 1238–1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, et al. 2017. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545: 48–53. 10.1038/nature22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 1995. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci 18: 383–388. 10.1016/0166-2236(95)93934-P [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR–Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V. 2011. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex 21: 1674–1694. 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- Sacco R, Gabriele S, Persico AM. 2015. Head circumference and brain size in autism spectrum disorder: a systematic review and meta-analysis. Psychiatry Res 234: 239–251. 10.1016/j.pscychresns.2015.08.016 [DOI] [PubMed] [Google Scholar]
- Schafer ST, Paquola ACM, Stern S, Gosselin D, Ku M, Pena M, Kuret TJM, Liyanage M, Mansour AA, Jaeger BN, et al. 2019. Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat Neurosci 22: 243–255. 10.1038/s41593-018-0295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Sugaya K, Tsujimura K, Nagata T, Horii T, Koide Y. 2013. Rab39a interacts with phosphatidylinositol 3-kinase and negatively regulates autophagy induced by lipopolysaccharide stimulation in macrophages. PLoS One 8: e83324 10.1371/journal.pone.0083324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MD, Nordahl CW, Young GS, Wootton-Gorges SL, Lee A, Liston SE, Harrington KR, Ozonoff S, Amaral DG. 2013. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain 136: 2825–2835. 10.1093/brain/awt166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Livesey FJ. 2012. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 7: 1836–1846. 10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. 2010. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci 11: 490–502. 10.1038/nrn2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, Courchesne E. 2014. Patches of disorganization in the neocortex of children with autism. N Engl J Med 370: 1209–1219. 10.1056/NEJMoa1307491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukoff Rizzo SJ, Crawley JN. 2017. Behavioral phenotyping assays for genetic mouse models of neurodevelopmental, neurodegenerative, and psychiatric disorders. Annu Rev Anim Biosci 5: 371–389. 10.1146/annurev-animal-022516-022754 [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai J-W, LaMonica B, Kriegstein AR. 2011. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci 14: 555–561. 10.1038/nn.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim N-S, Yoon K-J, Shin J, Zhang C, et al. 2014. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515: 414–418. 10.1038/nature13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. 2013. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155: 997–1007. 10.1016/j.cell.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GR, Sim JCH, McLean C, Giannandrea M, Galea CA, Riseley JR, Stephenson SEM, Fitzpatrick E, Haas SA, Pope K, et al. 2014. Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with α-synuclein pathology. Am J Hum Genet 95: 729–735. 10.1016/j.ajhg.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden KD, Ebrahimi-Fakhari D, Sahin M. 2018. Abnormal mTOR activation in autism. Annu Rev Neurosci 41: 1–23. 10.1146/annurev-neuro-080317-061747 [DOI] [PubMed] [Google Scholar]
- Woodbury-Smith M, Deneault E, Yuen RKC, Walker S, Zarrei M, Pellecchia G, Howe JL, Hoang N, Uddin M, Marshall CR, et al. 2017. Mutations in RAB39B in individuals with intellectual disability, autism spectrum disorder, and macrocephaly. Mol Autism 8: 59 10.1186/s13229-017-0175-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, Chen J-F. 2016. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv 2: e1601167 10.1126/sciadv.1601167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yang S-L, Yang M, Herrlinger S, Shao Q, Collar JL, Fierro E, Shi Y, Liu A, Lu H, et al. 2019. Modeling microcephaly with cerebral organoids reveals a WDR62–CEP170–KIF2A pathway promoting cilium disassembly in neural progenitors. Nat Commun 10: 2612 10.1038/s41467-019-10497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski BA, Prigge MBD, Nielsen JA, Froehlich AL, Abildskov TJ, Anderson JS, Fletcher PT, Zygmunt KM, Travers BG, Lange N, et al. 2014. Longitudinal changes in cortical thickness in autism and typical development. Brain 137: 1799–1812. 10.1093/brain/awu083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.