Highlights
-
•
RNA-Seq libraries were constructed using three methods.
-
•
RNA virome in ducks was detected.
-
•
RNA virome in mink was detected.
-
•
Various viruses in ducks or minks were possibly first identified.
Abbreviations: AIV, avian influenza virus; IBV, infectious bronchitis virus; NDV, Newcastle disease virus; NGS, next-generation sequencing; PBS, phosphate buffered solution; PCR, polymerase chain reaction; PGM, Personal Genome Machine; RT, reverse transcription; SISPA, sequence independent single primer amplification
Keywords: Detection, Duck, Library, Mink, Next generation sequencing, Virome, Virus
Abstract
Virome (viral megagenomics) detection using next generation sequencing has been widely applied in virology, but its methods remain complicated and need optimization. In this study, we detected the viromes of RNA viruses of one mock sample, one pooled duck feces sample and one pooled mink feces sample on the Personal Genome Machine platform using the sequencing libraries prepared by three methods. The sequencing primers were added through random hybridization and ligation to fragmented viral RNA using a RNA-Seq kit in method 1, through random reverse transcription (RT) and polymerase chain reaction (PCR) in method 2 which was developed in our laboratory, and through hybridization and ligation to fragmented amplicons of random RT-PCR using a single primer in method 3. Although the results of these three samples (nine libraries) all showed that more classified viral families and genera were identified using methods 2 and 3 than using method 1, and more classified viral families and genera were identified using method 2 than using method 3, most of the differences were of no statistical significance. Moreover, 11 mammalian viral genera in minks were possibly identified for the first time through this study.
1. Introduction
Many human and animal infectious diseases, such as influenza, rabies, mumps, acquired immune deficiency syndrome, classical swine fever, foot-and-mouth disease and Newcastle disease, are all caused by RNA viruses (Holmes, 2009, Woolhouse et al., 2016). Moreover, most of emerging infectious diseases, such as Ebola, Nipah virus infection, severe acute respiratory syndrome, Middle East respiratory syndrome, zoonotic H7N9 avian influenza and Zika virus infection, are also caused by RNA viruses (Aziz et al., 2017, Eaton et al., 2006, Gao et al., 2013, Li et al., 2005, Nyakarahuka et al., 2016, Zaki et al., 2012).
Rapid detection of RNA viruses is critical for the diagnosis, treatment, control and prevention of human and animal infectious diseases caused by RNA viruses (Drosten et al., 2002, Torok and Cooke, 2009). Traditionally, rapid detection of RNA viruses has relied on nucleic acid-based techniques like polymerase chain reaction (PCR) using specific primers or probes, and/or serology-based techniques like enzyme linked immunosorbent assay (Aziz et al., 2017). In recent years, the development of next-generation sequencing (NGS) technologies has innovated methods for RNA virus detection (Capobianchi et al., 2013, Shan et al., 2011, Yu et al., 2013, Zhang et al., 2014). NGS can reveal a huge number sequences of nucleic acids (DNA or RNA) within specimens through some random modes, and can detect various kinds of RNA viruses simultaneously thereby. Virome detection has been widely applied to identification of novel RNA viruses and research of RNA viruses (Masson et al., 2014, Paez-Espino et al., 2016, Webster et al., 2015).
Viral RNA in clinical specimens is usually limited in quantity and prone to degradation, and should be transformed into NGS libraries for sequencing. Efficient library preparation is critical for virome detection of RNA viruses and remains challenging. In this study, we detected the viromes of RNA viruses of one mock sample and two pooled authentic samples, using the libraries prepared by the three methods on the Personal Genome Machine (PGM) platform, with the aim to generate data of significance for virome detection of RNA viruses and characterize the viromes of RNA viruses in ducks and minks.
2. Materials and methods
2.1. Ethics statement
This study was conducted according to the animal welfare guidelines of the World Organization for Animal Health and approved by the Animal Welfare Committee of China Animal Health and Epidemiology Center. Fecal samples were collected with permission given by multiple relevant parties, including China Animal Health and Epidemiology Center, the relevant farm owners.
2.2. Sample collection
Sample 1 was a mock sample mixed from the allantoic fluids of three chicken embryonated eggs (1 mL for each egg) which contained H9N2 subtype avian influenza virus (AIV), a strain of Newcastle disease virus (NDV) and a strain of infectious bronchitis virus (IBV) with unknown viral load, respectively. Sample 2 (approximately 50 mL) was a pool of 300 pieces of fresh duck feces collected from three live poultry markets in July 2015. Sample 3 (approximately 50 mL) was a pool of 1000 pieces of fresh mink feces collected from 13 mink farms in May 2016. The fecal samples were stored in 200 mL phosphate buffered solution (PBS, pH 7.2) containing 10% glycerol at 4 °C.
2.3. Viral RNA extraction
Viral RNA of each sample was extracted within one day after collection. Sample 1 was diluted in 9 mL PBS (pH 7.2) containing 10% glycerol and then filtered through a 0.22-μM filter (Millipore, USA). Samples 2 and 3 were suspended and then clarified by centrifugation at 12,000 rpm for 10 min. The supernatant was filtered through 0.22-μM filters to remove bacteria. The filtered solution was precipitated using 1/10 vol of 50% (w/v) polyethylene glycol 6000 at 4 °C for 2 h. Then the solution was centrifuged at 12,000 rpm at 4 °C for 1 h. The precipitate was suspended in 1 mL PBS solution, and then treated with a mixture of 0.5 μL recombinant DNase I (RNase Free, 5 U/μL) (TaKaRa, Japan) and 0.5 μL Ribonuclease A (RNase A, 10 mg/mL) (TaKaRa, Japan) at 37 °C for 30 min to digest nucleic acids out of cells and viruses. Thereafter, viral nucleic acids were extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions.
2.4. Libraries preparation using method 1
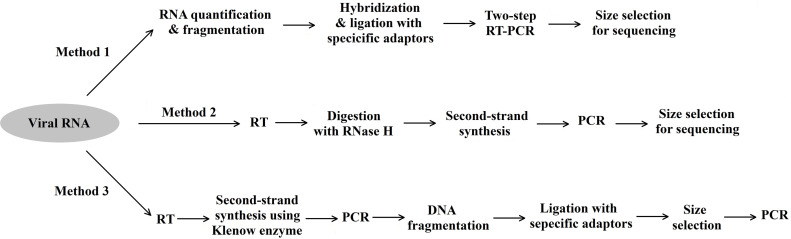
Method 1 for the library preparation was based on the Ion Total RNA-Seq kit v2 (Life Technologies, USA) according to the manufacturer’s instructions. Briefly, the extracted viral RNA was quantified, and then digested using RNase III. The fragmented RNA was ligated with random adaptors, and this was followed by reverse transcription. The cDNA was amplified through PCR, and the amplicons around 450 bp were collected with the E-Gel® SizeSelect™ Agarose Gel (Life Technologies, USA).
2.5. Libraries preparation using method 2
Method 2 for the library preparation was based on a procedure developed by ourselves. The details of the method development will be published elsewhere. It began with random RT reaction: 8 μL viral RNA, 1 μL 100 μM primer A15N6 (Table 1 ), and 2 μL nuclease-free water were mixed and incubated at 72 °C for 5 min. Then the RNA/primer mixture was placed on ice for at least 3 min. Then the mixture was added with 4 μL 5 × first-strand buffer, 1 μL dNTP (100 μM), 2 μL DTT (0.1 M), 1 μL RNaseOUT™ Recombinant Ribonuclease Inhibitor (40 U/μL) and 1 μL SuperScript® III Reverse Transcriptase (200 U/μl) (Invitrogen, USA), and incubated at 25 °C for 15 min and 42 °C for 30 min. The reaction was terminated at 70 °C for 15 min. Then the reaction system was added with 1 μL RNase H (TaKaRa, Japan) and further incubated at 37 °C for 20 min. After purification using DynaMag™-2 Magnet and Agencourt® AMPure® XP Reagent (Beckman Coulter, USA), the purified first-strand cDNA was used for the synthesis of the second-strand cDNA with primer B15N6 (Table 1) at 70 °C for 5 min. Then the mixture was added with 1 μL Klenow fragment (5 U) (NEB, USA), 5 μL 10 × NEBuffer 2, 2 μL dNTP (100 μM) and 1 μL DTT (0.1 M), and then incubated at 37 °C for 30 min. This was followed by PCR amplification using a system containing the double-stranded cDNA template, 1 × Phusion High-Fidelity Buffer, 10 μM primers A30 and B30 (Table 1), 0.5 U Phusion High-Fidelity DNA Polymerase (NEB, USA). The PCR was performed as follows: 2 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 30 s, followed by 14 cycles of 94 °C for 30 s, 65 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 5 min. The library amplicons around 450 bp were collected with the E-Gel® SizeSelect™ Agarose Gel.
Table 1.
The details of all the primers used in the paper.
| Primer name | Primer sequence |
|---|---|
| A15N6 | 5′-GTGTCTCCGACTCAGNNNNNN-3′ |
| B15N6 | 5′-TGGGCAGTCGGTGATNNNNNN-3′ |
| A30 | 5′-CCA/TCTCATCCCTGCGTGTCTCCGACTCAG-3′ |
| B30 | 5′-CCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′ |
| SPN8 | 5′-GTTTCCCAGTAGGTCTCNNNNNNNN-3′ |
| SP | 5′-CGCCGTTTCCCAGTAGGTCTC-3′ |
2.6. Libraries preparation using method 3
Method 3 for the library preparation was modified from a procedure reported previously as sequence independent single primer amplification (SISPA) (Allander et al., 2001, Cheng et al., 2010, Djikeng et al., 2008, Palacios et al., 2007) (Fig. 1 ). Briefly, the extracted viral RNA was reversely transcribed using the random primer SPN8 (Table 1), and then the RT system was denaturated at 94 °C for 5 min and on ice for 3 min. The second-strand cDNA synthesis was modified using the denaturated RT system which was added with 1 μL Klenow fragment (5 U) (NEB, USA) and 2.33 μL 10 × NEBuffer, and the synthesis was performed at 37 °C for 60 min and 75 °C for 20 min. This was followed with PCR using the primer SP (Table 1). The PCR amplicons were sheared and then ligated with sequencing adaptors. After that, the ligation products around 450 bp were collected and amplified.
Fig. 1.
Three methods for preparation of NGS libraries.
2.7. Sequencing and analysis
The libraries were sequenced through the Ion Torrent PGM platform (Life Technologies, USA) with the Ion PGM™ Sequencing 400 Kit (Life Technologies, USA) (Merriman et al., 2012). Each library was sequenced separately on an Ion 318™ Chip (Life Technologies, USA). Obtained short reads were deposited in the Sequence Read Archives of NCBI (accession numbers: SRR5078294, SRR5078297, SRR5078298, SRR5078299, SRR5078300, SRR5078301, SRR5078288, SRR4051861 and SRR4051862). The primers SPN8 and SP were trimmed from the reads before assembly for method 3. The reads were assembled using the CLC genomics workbench 8.5.1 (Qiagen, Germany). Assembled contigs shorter than 100 bp were removed, and the remaining contigs were compared to the non-redundant nucleotide databases using the standalone NCBI BLASTn tool (McGinnis and Madden, 2004). The E-value of 10−3 was used as the cutoff value for significant hits. The BLASTn results were parsed using the MetaGenome Analyzer (MEGAN vesion 5.10.5) with the default LCA parameters(Huson et al., 2007), and the taxonomic assignment was based on the first hit. Sequences placed in the roots in Megan diagrams were taken into account. All the contig hits of viruses excluding phages were verified manually through online BLAST at NCBI web station. The sequencing depth (corresponding to the number of hits obtained for a given nucleotide position) and coverage (corresponding to the percentage of the nucleotides in the genomic sequences which have been sequenced) of the three known viruses in sample 1 were analyzed as compared with the genomic sequences of the AIV strain A/chicken/Shaanxi/xa0414/2013 (H9N2) (GenBank accession numbers: KM609865, KM609825, KM609785, KM609585, KM609705, KM609665, KM609625 and KM609745), the NDV strain Du/CH/LGX/280/2013 (GenBank accession number: KM885162), and the IBV strain ck/CH/LHB/130578 (GenBank accession number: KP118890). The reads of sample 1 were mapped to the reference sequences using the software tool BLAT (v 34) (Kent, 2002). The total length of the contigs (the sum of the lengths of all the contigs) plus the sequencing coverage and depth were evaluated using the software tool soap.coverage (2.7.7) (Li et al., 2008).
3. Results
3.1. Reads and contigs
As shown in Table 2 , the numbers of reads from the nine sequence libraries varied greatly among the three samples and the three methods for preparing the libraries. The table further showed that the reads using method 1 were shorter than their counterparts using methods 2 and 3 by 20.54%–64.75%. The contigs using method 1 were less than their counterparts using methods 2 and 3 by 47.87%–94.28%, and the total length of the contigs using method 1 was shorter than its counterparts using methods 2 and 3 by 70.43%–99.12%. The contigs using method 1 mapped to viruses including phages were less than their counterparts using methods 2 and 3 by 62.50%–94.77%, and the contigs using method 1 mapped to viruses excluding phages were less than their counterparts using methods 2 and 3 by 46.86%–78.95%.
Table 2.
Analysis of reads and contigs generated in this study.
| Sample 1 |
Sample 2 |
Sample 3 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Method 1 | Method 2 | Method 3 | Method 1 | Method 2 | Method 3 | Method 1 | Method 2 | Method 3 | |
| Number of reads | 5,178,341 | 4,529,496 | 4,859,283 | 1,233,706 | 2,072,054 | 4,014,133 | 924,765 | 4,417,682 | 3,771,214 |
| Average length of reads (bp) | 145.67 | 241.50 | 183.32 | 64.60 | 183.24 | 183.02 | 123.08 | 236.15 | 237.07 |
| Number of contigs | 1884 | 3342 | 6131 | 1197 | 26,361 | 128,514 | 13,994 | 111,203 | 107,111 |
| Total length of contigs (bp) | 262,278 | 886,853 | 1,748,273 | 562,811 | 10,016,341 | 64,285,331 | 2,218,132 | 38,388,478 | 51,957,239 |
| Average length of contigs (bp) | 139.21 | 265.37 | 285.15 | 470.18 | 379.97 | 500.22 | 158.51 | 345.21 | 485.08 |
| Number of contigs mapped to viruses including phages | 51 | 234 | 136 | 131 | 851 | 2503 | 1589 | 6467 | 10,538 |
| The percentage of contigs mapped to viruses including phages | 2.71% | 7.00% | 2.22% | 10.94% | 3.23% | 1.95% | 11.35% | 5.82% | 9.84% |
| Number of contigs mapped to viruses excluding phages | 48 | 228 | 126 | 106 | 457 | 280 | 644 | 1676 | 1212 |
| The percentage of contigs mapped to viruses excluding phages | 2.55% | 6.82% | 2.06% | 8.86% | 1.73% | 0.22% | 4.60% | 1.51% | 1.13% |
| Number of bacteria contigs | 1041 | 1503 | 3910 | 192 | 3204 | 12,627 | 907 | 6423 | 7387 |
| Number of archaea contigs | 0 | 8 | 0 | 9 | 19 | 68 | 0 | 10 | 9 |
| Number of eukaryote contigs | 68 | 90 | 340 | 67 | 933 | 1700 | 1338 | 6533 | 1597 |
| Number of unassigned contigs | 40 | 58 | 92 | 30 | 332 | 902 | 194 | 731 | 940 |
| Number of no-hit contigs | 75 | 1080 | 984 | 690 | 20,576 | 108,003 | 9093 | 85,288 | 80,684 |
| Number of classified viral families identified excluding phages | 2 | 3 | 4 | 8 | 16 | 13 | 13 | 17 | 14 |
| Number of classified viral genera identified excluding phages | 2 | 3 | 4 | 8 | 23 | 19 | 18 | 23 | 19 |
| Accession number (SRR) | 5078294 | 5078297 | 5078298 | 5078299 | 5078300 | 5078301 | 5078288 | 4051861 | 4051862 |
3.2. Viruses excluding phages identified in sample 1
Sample 1 was a mock sample containing three known viruses AIV, NDV and IBV. The AIV and NDV were identified by all the three methods, while the IBV was identified only by methods 2 and 3 (Fig. S1 and Table 3 ). The viral load of IBV in the sample was possible lower than the other two known viruses. The sequencing mean depth of method 1 was only approximately 0.00%–10.19% of its counterparts of methods 2 and 3, and the sequencing coverage of method 1 was only approximately 0.00%–88.51% of its counterparts sequenced by methods 2 and 3, in the sequencing of the three known viruses (Fig. S1 and Table 3).
Table 3.
Sequencing depth and coverage of three known viruses in sample 1 detected using three methods.
| AIV |
NDV |
IBV |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Method 1 | Method 2 | Method 3 | Method 1 | Method 2 | Method 3 | Method 1 | Method 2 | Method 3 | |
| Mean depth | 32.85 | 322.41 | 1,613.21 | 0.25 | 10.84 | 95.94 | 0.00 | 0.01 | 0.21 |
| Coverage (%) | 88.50 | 100.00 | 99.99 | 9.47 | 93.48 | 99.75 | 0.00 | 1.22 | 17.12 |
3.3. Viruses excluding phages identified in sample 2
Sample 2 was a pool of duck feces collected from a live bird market. As shown in Table 2, from this sample, the classified families of viruses excluding phages identified through method 1 were approximately 50.00% − 61.54% of those identified through method 2 or 3, and the classified genera of viruses excluding phages identified through method 1 were approximately 34.78%–42.11% of those identified through method 2 or 3.
Taken the results of the three methods together, 17 classified families and 27 classified genera of viruses, as well as some unclassified viruses, were detected in sample 2 (Table S1). As shown in Fig. S2, 41.18% of the above 17 classified families and 25.93% of the above 27 classified genera of viruses were identified by all the three methods, and some other viral families and genera were identified only by one or two of the three methods.
Among the 27 viral genera, Picobirnavirus, Rotavirus, Orthoreovirus, Avulavirus, Influenzavirus A, Avastrovirus, Gammacoronavirus, Sapelovirus, Megrivirus, Tremovirus, Gallivirus, Bocaparvovirus, Dependoparvovirus, Aviadenovirus and Circovirus were assumed to be duck viruses, because similar duck or avian viruses had been identified previously (Table S1). Regarding some other viral genera (Aparavirus, Cripavirus, Iflavirus, Potyvirus, Carmovirus, Tobamovirus, Ambidensovirus and Iteradensovirus), they were assumed to be from other sources (e.g., duck feed or environment), as similar viruses had been identified from plants, insects or protozoa previously (Table S1).
3.4. Viruses excluding phages identified in sample 3
Sample 3 was a pool of mink feces collected from 13 mink farms. As shown in Table 2, from this sample, the classified families and genera of viruses excluding phages of identified through method 1 were only one less than those identified through method 3, and four or five less than those identified through method 2.
Taken the results of the three methods together, 20 classified families and 29 classified genera of viruses, as well as some unclassified viruses, were detected in sample 3 (Table S2). As shown in Fig. S3, 45.45% of the above 20 classified families and 44.83% of the above 29 classified genera of viruses were identified by all the three methods, and some other viral families and genera were identified only by one or two of the three methods.
Among the 29 viral genera, Mamastrovirus, Norovirus, Sapovirus, Vesivirus, Hepevirus, Alphacoronavirus, Cardiovirus, Kobuvirus, Picobirnavirus, Rotavirus, Orthoreovirus, Gyrovirus, Ambidensovirus, Amdoparvovirus and Bocaparvovirus, were assumed to mammalian viruses, because similar mammalian viruses (e.g. Aleutian mink disease virus within Amdoparvovirus) had been identified previously (Table S2). Through investigation of the mink farmers, we knew that the mink in the farms had not been fed with any mammalian meat. Therefore, we assumed that all mammalian viruses identified in the mink feces sample were possibly mink viruses. Of these mammalian viral genera, Mamastrovirus, Norovirus, Sapovirus, Vesivirus, Cardiovirus, Picobirnavirus, Rotavirus, Orthoreovirus, Gyrovirus, Ambidensovirus and Bocaparvovirus from mink had not been reported previously. Some other viral genera, like Betanodavirus, Cripavirus, Chlorovirus, Trichovirus, Marafivirus, Tobamovirus, Giardiavirus and Pelamoviroid, were assumed to be from other sources (e.g., mink feed or environment), as similar viruses from plants, fishes, insects or unicellular eukaryotic organisms had been identified previously (Table S2). Some avian viruses in the genera of Influenzavirus A, Avastrovirus, Gammacoronavirus and Avibirnavirus which were identified in sample 3 might be from mink feed which contained fresh meat, giblets and eggs of chickens and ducks. It should be mentioned that avian influenza virus can replicate in mink.
4. Discussion
Various NGS platforms, such as Illumina HiSeq, MiSeq and NovaSeq, Ion Torrent PGM, Proton and S5, BGI BGISeq-500, have been commercially available (Quail et al., 2012). Among these platforms, Ion Torrent PGM is competitive for detection of viruses and bacteria with respect to instrumental price, sequencing cost and simplicity of operation, although its sequencing throughput is lower than MiSeq and Proton.
Due to high cost for comparing these three methods for library preparation, only three samples (one mock sample and two authentic samples) were detected using the PGM platform in this study. The results of these three samples (nine libraries) all showed that more classified viral families and genera were identified using methods 2 and 3 than using method 1, and largely more classified viral families and genera were identified using methods 2 than using method 3. However, all of the differences were of no statistical significance by the Chi-square test (P > 0.05), except that significantly more viral families and genera were identified by method 2 than by method 1 for samples 2 and 3 (P < 0.05).
Methods 1 and 3 require specific commercial kits for fragmentation of nucleic acids and ligation of sequencing adaptors. Therefore, they are more costly than method 2 which does not require any commercial kits. Moreover, method 1 is of great technical difficulty as it requires RNA fragmentation whose quality is difficult to control. Theoretically, methods 2 and 3 can be started from viral RNA of a concentration below RNA quantification limits, while method 1 should be started from viral RNA above the RNA quantification limits. Since most clinical samples collected from a single person or animal contains viral RNA below the RNA quantification limit, method 1 is not suitable for most clinical samples, although it may be suitable for other applications, e.g., transcriptome detection. We selected the three samples containing much amount of viral RNA in this study in order to obtain NGS data using all the three methods. In the future, it is of significance to compare methods 2 and 3 in detecting viromes of RNA viruses in some clinical samples containing limited viral RNA.
Although all the three methods were designed for detection of viromes of RNA viruses, some DNA viruses were also identified using the three methods. This may be attributed to that the sequencing primers could be added to viral DNA at one or more steps for preparing the libraries (e.g. the step of RT or PCR amplification of method 2). Nevertheless, most (96.65%) hits of viruses excluding phages detected in this study were from RNA viruses (Tables S1 and S2).
Detection of viromes of duck guts has been reported recently and 18 classified viral families were identified (Fawaz et al., 2016). Seven of these 18 families (Circoviridae, Dicistroviridae, Iflaviridae, Parvoviridae, Picornaviridae, Phycodnaviridae, and Virgaviridae) were also identified in the duck feces sample through this study. Hits of Mimiviridae and Retroviridae were also identified in the duck feces sample through this study, but they were assumed to be unreliable through our online BLAST analysis. The remaining nine families including Baculoviridae, Herpesviridae, Iridoviridae, Marseilleviridae, Nodaviridae, Papillomaviridae, Partitiviridae, Poxviridae and Totiviridae were not identified through this study, while ten families including Paramyxoviridae, Orthomyxoviridae, Astroviridae, Coronaviridae, Potyviridae, Tombusviridae, Picobirnaviridae, Reoviridae, Adenoviridae and Avsunviroidae identified in this study were not identified in the recent report. The differences can be attributed to that the samples were collected at different sites, regions and time and detected using different methods. It is of value to conduct further studies to make clear whether some viruses identified in the duck feces belong to new viral species.
Detection of viromes of minks has not been reported previously, and a total of 11 mammalian viral genera in minks might be reported for the first time through this study. It is of value to conduct further studies to make clear clinical significance and taxonomic status of these viruses.
Ducks and minks are economically important for their feather, fur, egg and/or meat. Detection of viromes of ducks and minks increases our understanding of the viral diversity in the animals, and provides novel clues for further studies regarding diagnosis of infectious diseases, identification of novel viruses and research of host-virus relationships.
Compliance with ethical standards
Conflict of Interest: Kai-Cheng Wang declares that she has no conflict of interest. Run Wu declares that he has no conflict of interest. Yuan Qiu declares that he has no conflict of interest. Ji-Ming Chen declares that he has no conflict of interest. Tong Wang declares that he has no conflict of interest. Qing-Ye Zhuang declares that she has no conflict of interest. Guang-Yu Hou declares that he has no conflict of interest.
Ethical approval: All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Acknowledgement
This work was supported with the Innovation Fund of CAHEC (Number: 2015IF-0004FF).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2017.05.003.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. U. S. A. 2001;98(20):11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz H., Zia A., Anwer A., Aziz M., Fatima S., Faheem M. Zika virus: global health challenge, threat and current situation. J. Med. Virol. 2017;89(6):943–951. doi: 10.1002/jmv.24731. [DOI] [PubMed] [Google Scholar]
- Capobianchi M.R., Giombini E., Rozera G. Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 2013;19(1):15–22. doi: 10.1111/1469-0691.12056. [DOI] [PubMed] [Google Scholar]
- Cheng W.X., Li J.S., Huang C.P., Yao D.P., Liu N., Cui S.X., Jin Y., Duan Z.J. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One. 2010;5(10):e13583. doi: 10.1371/journal.pone.0013583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Halpin R., Kuzmickas R., Depasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., Spiro D.J. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gottig S., Schilling S., Asper M., Panning M., Schmitz H., Gunther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40(7):2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton B.T., Broder C.C., Middleton D., Wang L.F. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 2006;4(1):23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawaz M., Vijayakumar P., Mishra A., Gandhale P.N., Dutta R., Kamble N.M., Sudhakar S.B., Roychoudhary P., Kumar H., Kulkarni D.D., Raut A.A. Duck gut viral metagenome analysis captures snapshot of viral diversity. Gut Pathog. 2016;8:30. doi: 10.1186/s13099-016-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K., Xu X., Lu H., Zhu W., Gao Z., Xiang N., Shen Y., He Z., Gu Y., Zhang Z., Yang Y., Zhao X., Zhou L., Li X., Zou S., Zhang Y., Li X., Yang L., Guo J., Dong J., Li Q., Dong L., Zhu Y., Bai T., Wang S., Hao P., Yang W., Zhang Y., Han J., Yu H., Li D., Gao G.F., Wu G., Wang Y., Yuan Z., Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013;368(20):1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Holmes E.C. Oxford University Press; 2009. The Evolution and Emergence of RNA Viruses. [Google Scholar]
- Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Masson P., Hulo C., de Castro E., Foulger R., Poux S., Bridge A., Lomax J., Bougueleret L., Xenarios I., Le Mercier P. An integrated ontology resource to explore and study host-virus relationships. PLoS One. 2014;9(9):e108075. doi: 10.1371/journal.pone.0108075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–25. doi: 10.1093/nar/gkh435. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriman B., Ion Torrent R., Team D., Rothberg J.M. Progress in ion torrent semiconductor chip based sequencing. Electrophoresis. 2012;33(23):3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- Nyakarahuka L., Kankya C., Krontveit R., Mayer B., Mwiine F.N., Lutwama J., Skjerve E. How severe and prevalent are Ebola and Marburg viruses? A systematic review and meta-analysis of the case fatality rates and seroprevalence. BMC Infect. Dis. 2016;16(1):708. doi: 10.1186/s12879-016-2045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering earth's virome. Nature. 2016;536(7617):425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Palacios G., Quan P.L., Jabado O.J., Conlan S., Hirschberg D.L., Liu Y., Zhai J., Renwick N., Hui J., Hegyi H., Grolla A., Strong J.E., Towner J.S., Geisbert T.W., Jahrling P.B., Buchen-Osmond C., Ellerbrok H., Sanchez-Seco M.P., Lussier Y., Formenty P., Nichol M.S., Feldmann H., Briese T., Lipkin W.I. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 2007;13(1):73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R., Bertoni A., Swerdlow H.P., Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Li L., Simmonds P., Wang C., Moeser A., Delwart E. The fecal virome of pigs on a high-density farm. J. Virol. 2011;85(22):11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok E., M.E, Cooke F. Oxford University Press; 2009. Oxford Handbook of Infectious Diseases and Microbiology. [Google Scholar]
- Webster C.L., Waldron F.M., Robertson S., Crowson D., Ferrari G., Quintana J.F., Brouqui J.M., Bayne E.H., Longdon B., Buck A.H., Lazzaro B.P., Akorli J., Haddrill P.R., Obbard D.J. The discovery, distribution, and evolution of viruses associated with drosophila melanogaster. PLoS Biol. 2015;13(7):e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M.E., Brierley L., McCaffery C., Lycett S. Assessing the epidemic potential of RNA and DNA viruses. Emerg. Infect. Dis. 2016;22(12):2037–2044. doi: 10.3201/eid2212.160123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.M., Li X.Y., Ao Y.Y., Li L.L., Liu N., Li J.S., Duan Z.J. Identification of a novel picornavirus in healthy piglets and seroepidemiological evidence of its presence in humans. PLoS One. 2013;8(8):e70137. doi: 10.1371/journal.pone.0070137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang W., Li L., Deng X., Kapusinszky B., Delwart E. What is for dinner Viral metagenomics of US store bought beef, pork, and chicken. Virology. 2014;468–470:303–310. doi: 10.1016/j.virol.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.