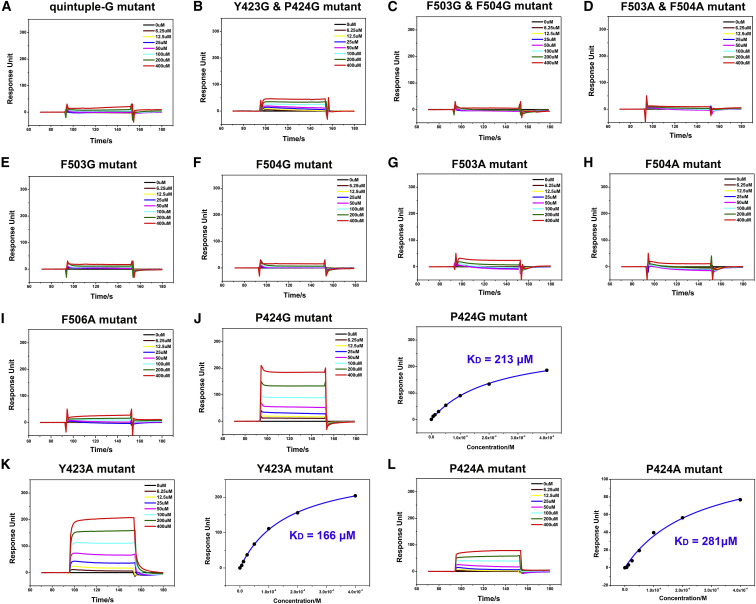
Figure 6.
Alanine/Glycine-Scanning Mutagenesis Experiments on NPC1-C
(A–I) BIAcore diagrams of quintuple-G, Y423G&P424G, F503G&F504G, F503A&F504A, F503G, F504G, F503A, F504A, and Y506A mutants of NPC1-C binding to the GPcl. These nine mutants abolish the binding to the GPcl.
(J) BIAcore diagram and saturation curve of P424G mutant of NPC1-C binding to the GPcl. The P424G mutant can keep binding to the GPcl with an affinity of 213 μM, which is slightly lower than that of wild-type NPC1-C.
(K) BIAcore diagram and saturation curve of Y423A mutant of NPC1-C binding to the GPcl. The Y423A mutant can keep binding to the GPcl with an affinity of 166 μM, which is slightly lower than that of wild-type NPC1-C.
(L) BIAcore diagram and saturation curve of P424A mutant of NPC1-C binding to the GPcl. The P424A mutant can keep binding to the GPcl with an affinity of 281 μM, which is slightly lower than that of wild-type NPC1-C. Response units were plotted against protein concentrations. The KD values were calculated by the BIAcore 3000 analysis software (BIAevaluation Version 4.1).
See also Figures S6 and S7.