Abstract
Interferon-induced transmembrane proteins IFITM1 and IFITM3 (IFITM1/3) play a role in both RNA viral restriction and in human cancer progression. Using immunohistochemical staining of FFPE tissue, we identified subgroups of cervical cancer patients where IFITM1/3 protein expression is inversely related to metastasis. Guide RNA-CAS9 methods were used to develop an isogenic IFITM1/IFITM3 double null cervical cancer model in order to define dominant pathways triggered by presence or absence of IFITM1/3 signalling. A pulse SILAC methodology identified IRF1, HLA-B, and ISG15 as the most dominating IFNγ inducible proteins whose synthesis was attenuated in the IFITM1/IFITM3 double-null cells. Conversely, SWATH-IP mass spectrometry of ectopically expressed SBP-tagged IFITM1 identified ISG15 and HLA-B as dominant co-associated proteins. ISG15ylation was attenuated in IFNγ treated IFITM1/IFITM3 double-null cells. Proximity ligation assays indicated that HLA-B can interact with IFITM1/3 proteins in parental SiHa cells. Cell surface expression of HLA-B was attenuated in IFNγ treated IFITM1/IFITM3 double-null cells. SWATH-MS proteomic screens in cells treated with IFITM1-targeted siRNA cells resulted in the attenuation of an interferon regulated protein subpopulation including MHC Class I molecules as well as IFITM3, STAT1, B2M, and ISG15. These data have implications for the function of IFITM1/3 in mediating IFNγ stimulated protein synthesis including ISG15ylation and MHC Class I production in cancer cells. The data together suggest that pro-metastatic growth associated with IFITM1/3 negative cervical cancers relates to attenuated expression of MHC Class I molecules that would support tumor immune escape.
Keywords: Cervical cancer, IFITM1, Interferon, SILAC mass spectrometry, MHC Class I molecule, CAS9 gene editing
Graphical abstract
1. Introduction
Interferons (IFNs) are pleiotropic cytokines produced by the innate immune system as a defensive response [1]. Many biological functions have been described for the interferon pathway. Among them, the best characterized are anti-tumor activity, immunomodulatory effects, anti-pathogen, and anti-viral activity [2]. IFN effects rely on three different types of receptors: Type I (IFNα, IFNβ and IFNω), Type II (IFNγ) and Type III (IFNλ) [3,4]. IFNs increase in response to a broad range of factors such as persistent viral infection or DNA damaging agents which activate the JAK kinase-STAT pathway. Ultimately, this signalling cascade will regulate the transcriptional synthesis of interferon stimulated genes (ISGs) [5,6].
Type I IFNs facilitate anti-proliferative and pro-apoptotic pathways in a wide range of cell types and it has been extensively used as antitumor therapeutic agent. High doses of IFNs are used for cancer therapy and can activate anti-tumor immunity as well as pro-apoptotic and anti-proliferative programs. By contrast, it has been demonstrated that sustained low level of IFN causes a steady-state expression of the interferon resistance DNA damage signature (IRDS) which is comprised of a subset of interferon-stimulated genes [7]. IRDS proteins promote phenotypes that contribute to the tumor development such as resistance to DNA damage, suppression of T cell toxicity, metastasis, and facilitation of epithelial–mesenchymal transition [8].
IFITM1 is a pro-oncogenic receptor which is a component of the IRDS pathway. Basal expression of IFITM proteins is observed in some cells and expression can also be induced by Type I and Type II interferons. IFITM1 is up-regulated during development of radiation resistance, escaping from pro-apoptotic and anti-proliferative effects [9,10]. IFITM1 expression has been extensively reported in many types of cancer; breast, cervix, colon, ovary, brain and oesophagus cancer and its high expression correlates with tumor progression and can lead to a poor outcome [[10], [11], [12], [13], [14], [15], [16]].
IFITM1 was previously known as IFI 17 (interferon induced protein 17), IFI 9–27 (interferon inducible protein 9–27), CD225 (cluster of differentiation) and Leu-13 (leucocyte surface protein). It is a membrane protein that plays a key role in restriction of anti-viral immune response and belongs to the interferon induced transmembrane family (IFITM) [17]. IFITM1 is coded by the IFITM1 gene located on chromosome 11p15.5 and flanked by IFITM2 and IFITM3 genes. The IFITM immunity-related protein family are composed of short amino-terminal and carboxy-terminal domains, two transmembrane domains, and a cytoplasmic domain. IFITM1 is slightly different from IFITM2 and IFITM3, with some studies demonstrating that IFITM1 is uniquely expressed at the cell surface [18,19].
IFITM family members are capable of attenuating the entrance of many human and animal viruses. They suppress the entry of viruses such as influenza A virus (IAV), West Nile virus, dengue virus, SARS coronavirus, filoviruses, VSV, and HCV among others [20]. IFITM family proteins inhibit cytosolic entry of viruses by preventing fusion of viral and host membranes. The protein-protein interaction networks by which IFITM proteins inhibit viral propagation are just emerging. One study using yeast two-hybrid methodology has identified an interaction of IFITM3, IFITM2, and IFITM1 with VAPA that in turn mediates an accumulation of cholesterol in multivesicular structures [21]. This reduces the fusion of intraluminal virion-containing vesicles with endosomal membranes and thereby inhibits virus release into the cytosol. In addition, different family members exhibit specific viral preferences. In the case of IFITM1, it is more active in controlling filoviruses, Influenza A, and SARS [17,20,22]. In this report, we focus on applying methodologies in interactomics, gene editing, SWATH-immunoprecipitation mass spectrometry, and pulse-SILAC mass spectrometry to propose an IFNγ responsive biochemical function for the IFITM1/3 proteins.
2. Experimental procedures
2.1. Cell culture
All chemicals and reagents were obtained from Sigma unless indicated otherwise. All antibodies from Thermofisher unless indicated otherwise. SiHa, IFITM1 null-SiHa, and IFITM1/IFITM3 null-SiHa cell lines were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Labtech) and 1% penicillin/streptomycin (Invitrogen) and incubated at 37 °C with 5% CO2.
2.2. Tissue microarray
Cervical cancer samples were obtained at the Masaryk Memorial Cancer Institute where patients gave written consent for tissue use according to local ethical regulations. Tissues were fixed in 4% formaldehyde for approximately 24 h before processing into paraffin wax and sectioned (4 μm). After sections were cut, the antigens were retrieved, and samples were probed with primary antibodies (including those to p16 protein (data not shown), and IFITM1/3 proteins (using the Mab-MHK); Supplementary Fig. 1). Following this stage, the tissue sections were incubated with secondary antibodies conjugated to streptavidin–HRP. Sections were incubated with DAB (Dako), counterstained with haematoxylin and mounted for visualization, as previously described [23].
Supplementary Fig. 1.
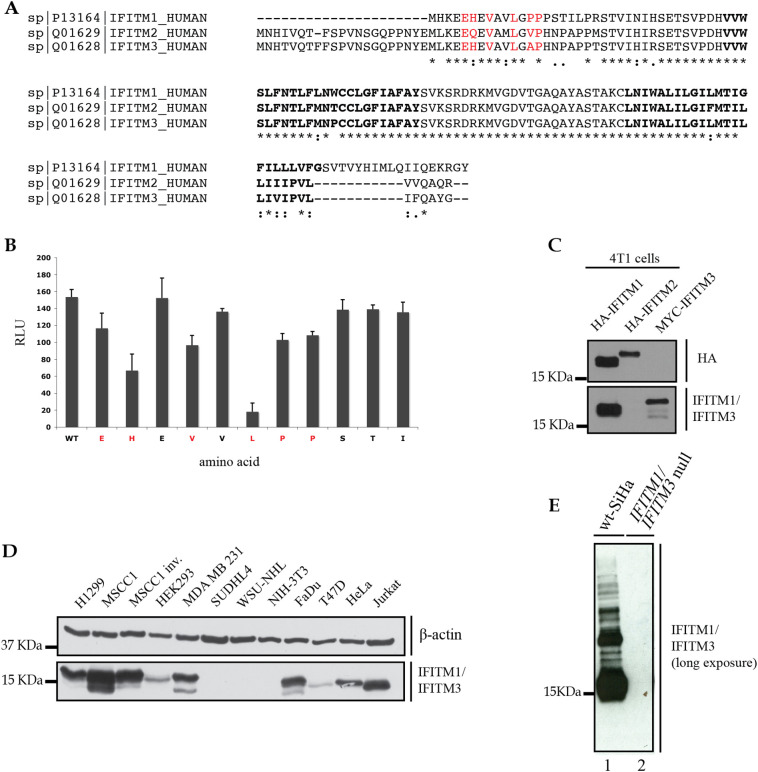
Developing IFITM1/IFITM3 antibody to test IFITM1/IFITM3 protein expression across a set of human cancer cell lines. (A). Homology among the IFITM1 family members, I, 2, and 3. The red amino acids highlight contacts that play a role in Mab-MHK monoclonal antibody binding using alanine substituted peptides, as reviewed in B. (B). An alanine scan ELISA assay using the biotinylated synthetic peptide derived from the N-terminus of IFITM1. The parental 14-mer peptide was obtained from Mimotopes (Australia) and has an N-terminal Biotin followed by the amino acid sequences, SGSGEHEVAVLGPPPSTI. The SGSG sequence form a flexible spacer linking the Biotin to the IFITM1 peptide sequence. Biotinylated peptides containing alanine substitutions at each position (apart from A and G) were assayed in parallel as described previously using streptavidin coated ELISA wells to capture biotinylated peptides [72]. Mab-MHK binding is quantified by RLU (relative light units) using ECL as a function of amino acid substitution. (C). Analysis of Mab-MHK binding to ectopically expressed and tagged IFITM1, 2, and 3 family members. Murine 4T1 cells (IFITM proteins from mouse do not bind to the Mab-MHK antibody) were transfected with the indicated human, tagged expression constructs (HA-IFITM1, HA-IFITM2, and MYC-IFITM3). The cell lysates were immunoblotted with anti-HA tag antibodies (to measure HA-IFITM1 and HA-IFITM2 protein expression) and also blotted with the Mab-MHK that measure expression of HA-IFITM1 and MYC-IFITM3 proteins. The absence of binding of Mab-MHK to IFITM2 protein is rationalized based on the alanine scan peptide ELISA suggesting an important role of the H-Q mutation in reducing antibody binding to IFITM2. (D) Immunoblotting with Mab-MHK in a panel of human cancer cell lines to define those with variable expression of IFITM1 and/or IFITM3. (E). A representative over-exposure of parental SiHa cell lysates (lane 1) and IFITM1/3 double null cells (lane 2), that demonstrate that there are no detectable small molecular mass protein fragments by immunoblotting. This over-exposure is from Fig. 8D (bottom panel that highlights absence of IFITM1/3 expression in the IFITM1/3 double null cells).
2.3. Generation of IFITM1 null and IFITM1/IFITM3 double null cell line using CRISPR/CAS9 technology
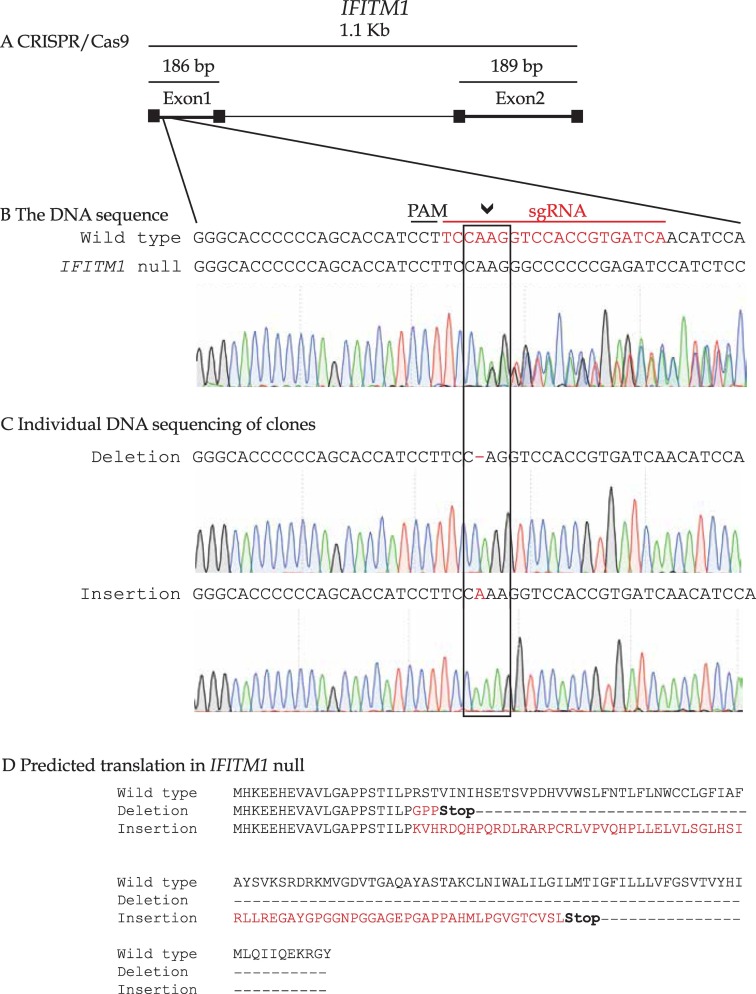
Guide RNA sequences were designed for IFITM1 (5′ TCCAAGGTCCACCGTGATCA 3′) and for IFITM3 (5′ GTCAACAGTGGCCAGCCCCC 3′). The guide RNA was cloned into the lentiCRISPRv2 expression vector [24]. After transfection, cells were selected with puromycin and sorted in a 96-well plate using FACS. After 3 weeks, individual clones were screened for absence of IFITM1 and/or IFITM3 protein expression by Western Blot using the Mab-MHK (Supplementary Fig. 1). Chromosomal DNA from the positive clones were sequenced for a final validation to define the precise gene edit (as in Fig. 2, Fig. 3).
Fig. 2.
Validation of the IFITM1 guide RNA. (A). Gene structure of the IFITM1 gene highlighting its two coding exons and one intron. The genomic region surrounding the gRNA target site (with PAM site and homology region in red) for IFITM1 was amplified and subjected to DNA sequencing. The raw DNA sequence reads in the IFITM1 gene are highlighted in (B). Genomic DNA sequence of the heterozygote IFITM1 indels after cloning the PCR amplified IFITM1 region into a vector and then sequencing individual plasmids for the type of gene edit. One allele of IFITM1 has a one bp insertion and the other a one bp deletion, both resulting in frame-shift stop edits are highlighted in C. Predicted translation of each allele giving rise to premature stop codons are highlighted in D. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
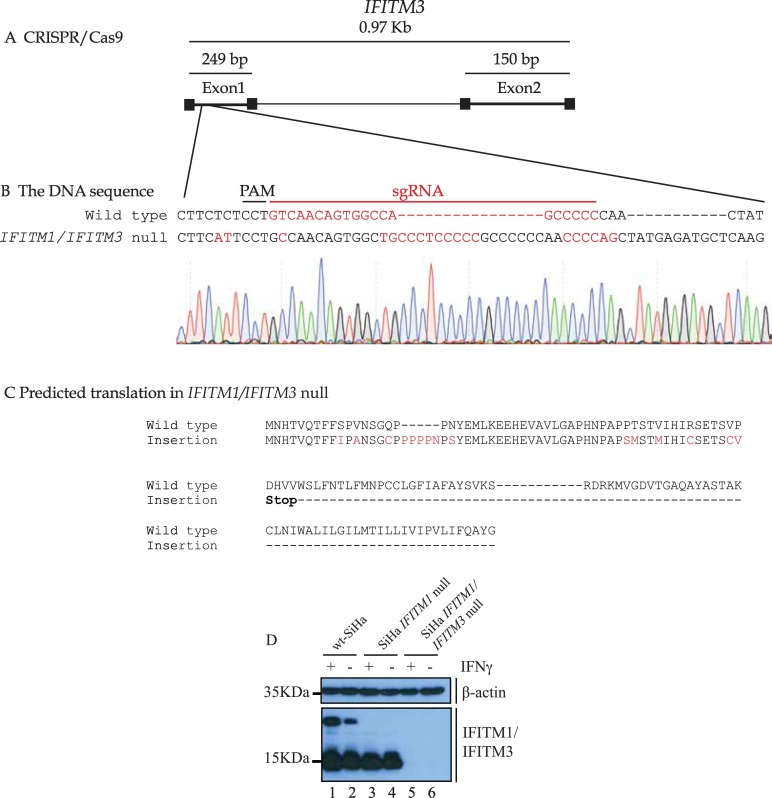
Validation of the IFITM3 guide RNA. (A). Gene structure of the IFITM3 gene highlighting its two coding exons and one intron. The genomic region surrounding the gRNA target site (with PAM site and homology region in red) for IFITM3 was amplified and subjected to DNA sequencing. The raw DNA sequencing reads in the IFITM3 gene are highlighted in (B). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The IFITM3 gene has two insertions that result in a poly-proline frame-shift creating a stop codon (C). (D). Immunoblotting was performed using Mab-MHK with lysates from SiHa cells (lanes 1 and 2); IFITM1 single null cells (lanes 3 and 4); and IFITM1/IFITM3 double null cells (lanes 5 and 6). The cells were either untreated or treated with IFNγ (100 ng/ml) for 24 h and processed for immunoblotting with either a β-actin antibody or Mab-MHK.
2.4. DNA extraction and sequence validation of guide RNA edits
Chromosomal DNA was extracted from frozen cell pellets following the instruction manual (Gentra Puregene Cell kit, Qiagen). Validation of the edited DNA sequence was confirmed by cloning the genomic IFITM1 and IFITM3 PCR products into a holding vector and by amplifying the entire gene and 500 bp surrounding the guide RNA targeting site followed by Sanger sequencing at Source BioScience Service (Scotland).
2.5. Cloning, transfection and affinity purification of SBP-IFITM1 protein
IFITM1 cDNA was cloned by PCR into pEXPR-IBA105 expression vector containing SBP tag at the N-terminus of the coding region (SBP vector (IBA)). SiHa cells were grown in RPMI as a duplicate. For transfection, cells were grown to approximately 80% confluency and transfected using Attractene (Qiagen). Cells were transfected with SBP-empty vector (control cells) and SBP-IFITM1 for 48 h. 48 h after transfection, cells were treated with carrier or with 100 ng/ml IFNγ (Invitrogen) for 24 h in order to “stabilize” potential interferon-activated SBP-IFITM1 interacting proteins. Cells were washed twice in ice cold PBS and scraped into buffer containing 100 mM KCl, 20 mM HEPES pH 7.5, 1 mM EDTA, 1 mM EGTA, 0.5 mM Na3VO4, 10 mM NaF, 10% (v/v) glycerol, protease inhibitor mix, and 0.1% triton X-100, then incubated for 30 min on ice and centrifuged at 13,000 rpm for 15 min at 4 °C. Equal amounts of protein were used for performing the affinity capture. Streptavidin Agarose conjugated beads (Millipore) were pre-washed with in PBS. Then cell lysate was added and incubated for 2 h at RT with gentle rotation. Binding proteins were eluted with a buffer containing 20 mM HEPES pH 8, 2 mM DTT, and 8 M Urea.
2.6. Transient transfection of siRNA
Small interfering RNA directed against the human IFITM1 (Qiagen) and an AllStars negative controls FlexiTube siRNA (Qiagen) were used to transfect SiHa cells for 48 and 72 h. Cells were transfected using HiPerFect (Qiagen) following the manufacturer's instructions.
2.7. SILAC labeling of cells for use in pulse SILAC
For performing “pulse” SILAC, Parental SiHa, IFITM1/IFITM3 double null cells, and IFITM1 single null cells were grown as biological triplicate and incubated with SILAC heavy media for 6 and 24 h with or without 100 ng/ml IFNγ before harvesting [[25], [26], [27]]. Cells were isotopically pulse-labeled using SILAC RPMI-heavy media (Dundee Cell Products, UK); L-[13C614N4] arginine (R6) and L-[13C614N2] lysine (K6). Cells were harvested in a buffer containing 8 M Urea, 0.1 M Tris pH 8.5. Total protein extracts were measured by the Bradford assay [28].
2.8. Antibodies
Proteins were detected using the following primary antibodies: mouse monoclonal anti-bodies generated to a peptide that is identical in IFITM1 and IFITM3 (Mab-MHK) (Moravian Biotechnology, this study is their first description). The antibody we name Mab-MHK can therefore detect co-expression of both IFITM1 and IFITM3 proteins (Supplementary Fig. 1). Other sources include, rabbit monoclonal anti-STAT1 (Cell Signalling), mouse monoclonal anti-IRF1 (BD Transduction Laboratories), rabbit polyclonal anti-HLA-B (Thermo Fisher), rabbit polyclonal antibody anti-ISG15 (Cell Signalling), mouse monoclonal anti-SBP-tag (Sigma), mouse monoclonal anti-β-actin (Sigma), and the mouse monoclonal anti-GAPDH (Abcam).
2.9. Western blotting
Protein from lysed samples was quantified using Protein Assay Dye Reagent (Bio-Rad). Proteins were resolved by SDS-PAGE using 15% gels [29] and transferred onto nitrocellulose membranes (Amersham Protran, GE Healthcare). Immunoblots were processed by enhanced chemiluminescence (ECL).
2.10. Immunofluorescence
Parental SiHa and IFITM1/IFITM3 double null cells were grown over 16 mm diameter glass coverslips. Stimulated cells were treated with 100 ng/ml IFNγ for 24 h. Cells were fixed with 4% (v/v) paraformaldehyde in PBS for 15 min at room temperature and permeabilized using 0.25% triton X-100 in PBS for 10 min. Then, the cells were blocked with 3% BSA in PBS for 30 min. The primary antibody was incubated at a 1:1000 dilution overnight. Alexa Fluor 488 goat anti-rabbit IgG (H + L) (Invitrogen) was incubated as a secondary antibody at 1:2000 dilution for 1 h. The fluorescent signal was detected using a Zeiss Axioplan 2 microscope at 63×. Replicates are described in the Fig. 6 legend. Fluorescence was measured using ImageJ software; cells were selected, and information was extracted on the area, integrated density, and mean gray values by selecting set measurements in the Analyse menu. A region with no fluorescence was selected as background for each image. The following formula was applied for each 50 cells analyzed: CTCF = Integrated Density – (Area of selected cell × Mean fluorescence of background readings); *CTCF = corrected total cell fluorescence.
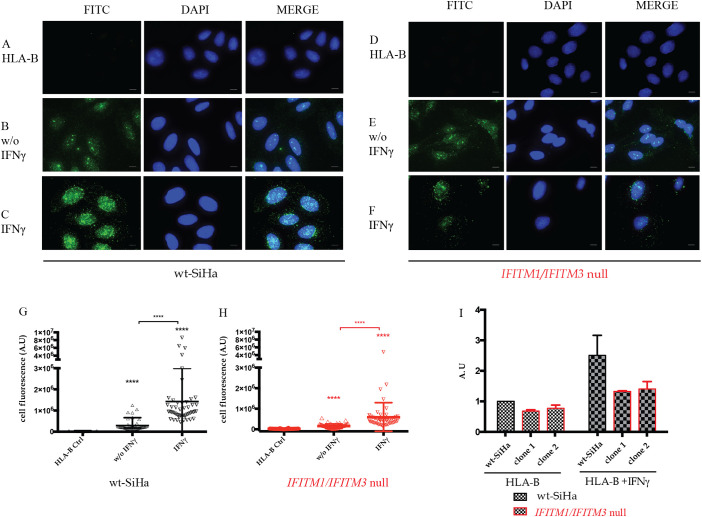
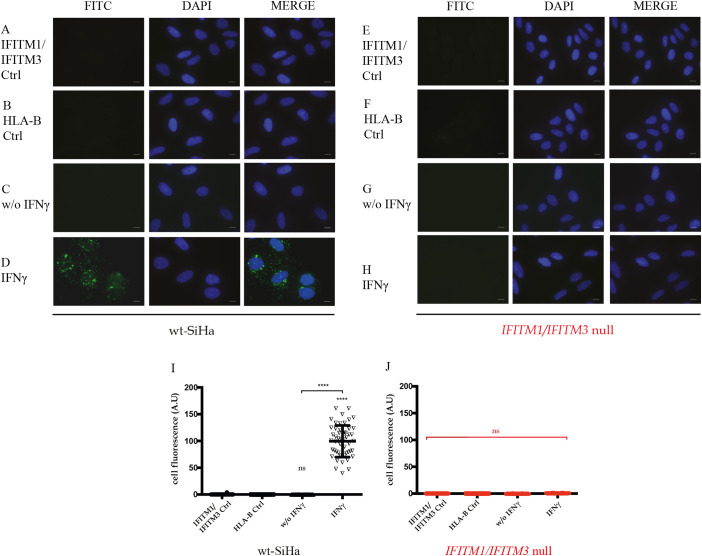
Fig. 6.
Quantitation of HLA-B levels in parental SiHa and IFITM1/IFITM3 double null cells using immunofluorescence. (A–F). The indicated cell panels were fixed without (B and E) or with 24 h of IFNγ treatment (C and F). Samples were fixed, and cells incubated with an HLA-B specific antibody. FITC images identify HLA-B (depicted in green) and DAPI (used for nuclear staining (depicted in blue)). n = 3. Scale bar: 10 μm. (G and H) Quantification of HLA-B total fluorescence per cell in presence or absence of IFNγ stimulation in (G) parental SiHa and (H) IFITM1/IFITM3 double null cells. For quantitation, three independent assays were performed, and each assay had two independent biological replicates. For each assay, fluorescence was measured in at least 50 cells per condition. Fluorescence was measured using ImageJ software. Statistical study was performed with 1-way Anova and Bonferroni correction (p-value < .0001). (I). FACS analysis was performed to measure the relative amount of HLA-B expressed on the surface of non-permeabilized cells treated with carrier or IFNγ for 24 h. The parental SiHa cell was used as a positive control and the two independently isolated IFITM1/IFITM3 double null cells were used as biological replicates. Samples were processed in two independent experiments and HLA-B expression was normalized to 1.0 in the parental SiHa cell line. The data are plotted as relative arbitrary units (AU) as a function of the clone, treated with carrier or with IFNγ. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.11. Proximity ligation assay
Parental SiHa and IFITM1/IFITM3 double null cells were grown and processed as described in the Immunofluorescence method (above). Primary antibodies from different species were incubated with the fixed and permeabilized cells: MHK mouse MAb with rabbit polyclonal anti-HLA-B, at 1:250 dilution overnight. Duolink® assays (Sigma) were carried out following supplier's instructions. The fluorescent signal was detected using a Zeiss Axioplan 2 microscope at 63×. Fluorescence was measured using ImageJ software, as above reviewed for standard immunofluorescence.
2.12. Mass spectrometric experimental screens
2.12.1. Peptide generation using FASP
Cell lysates, immunoprecipitates, or gradient fractions were processed using filter-aided sample preparation protocol (FASP) [30,31]. Urea buffer (8 M Urea in 0.1 M Tris pH 8.5) was added to a 30 kDa spin filter column (MRCPRT010, Microcon). Protein concentration was determined using the RC-DC method (Bio-rad). Normalized sample was added into the spin filter column and was centrifuged at 14000 g for 15 min at 20 °C. Urea buffer was added again with 100 mM Tris (2-carboxyethyl) phosphine hydrochloride (Aldrich) and mixed. The column was left on a thermo-block set at 37 °C shaking at 600 rpm and centrifuged at 12,210 rpm at 20 °C for 15 min. Urea buffer and 300 mM of iodoacetamide (Sigma) were mixed using a thermo-mixer at 600 rpm in the dark for 1 min, then was maintained statically for a further 20 min at room temperature in the dark. The sample was centrifuged at 12,210 rpm at 20 °C for 15 min and the supernatant was discarded. A solution containing 100 mM ammonium bicarbonate were added to the column and then it was centrifuged at 12,210 rpm at 20 °C for 20 min. This step was repeated one more time. The column was placed in a new collecting tube (low binding affinity) and 50 mM ammonium bicarbonate was added along with trypsin diluted in trypsin buffer (Promega) at a 1:100 ratio. The column was incubated at 37 °C overnight. The following day the column was centrifuged at 12,210 rpm at 20 °C for 15 min. Determination of the peptide concentration was performed using the Quantitative Colorimetric Peptide Assay (Pierce, ThermoScientific).
2.13. Desalting peptides using C18 column
Peptides were desalted on micro spin columns C-18 (Harvard Apparatus, USA). C-18 columns were conditioned three times with 100% acetonitrile (AcN) and 0.1% formic acid (FA) and centrifuged at 1200 rpm at room temperature for 2 min. The column was washed with 0.1% FA and centrifuged at 1200 rpm at room temperature for 2 min. The column was hydrated in 0.1% FA for 15 min following centrifugation at 1800 rpm at room temperature for 3 min. The sample was loaded into the column and centrifuged at 2300 rpm for 3 min. After washing the column three times with 0.1% FA, the peptides were eluted in three consecutive centrifugations at 2300 rpm for 3 min using 50%–80% and 100% AcN with 0.1% FA. Subsequently, peptide eluates were evaporated and dissolved in 5% AcN with 0.05% FA.
2.14. LC-MS/MS analyses
There were three distinct mass spectrometric screens used in the manuscript. The rationale for replicates in each distinct approach is as follows; (i) the SWATH-IP (immunoprecipitation) to identify IFITM1-enriched associated proteins (Fig. 8). The immunoprecipitation and immunoblots of SBP-IFITM1 vs SBP control is representative of experiments performed at least three times. The representative enrichment of IFITM1-associated proteins (in Fig. 8 from Supplementary Table 2) was processed by label free (SWATH) mass-spectrometry in technical triplicates. The measured fold-changes and p-values for quantified proteins are listed in Supplementary Table 2 and Fig. 8; (ii) the siRNA SWATH-MS (Fig. 9A and B). Targeted siRNA to deplete IFITM1 in SiHa cells was performed in at least three separate experiments. A representative depletion of IFITM1 protein (Fig. 9A) was performed in technical triplicates using two different biological states (48 and 72 h). The statistical rationale for performing two biological states (equivalent plating of cell number and differing by time of interferon exposure) rather than two biological replicates at the same time point, was due to the variable induction and suppression of the interferon cascade over this time frame. Thus, any proteins that are observed in two biological states as a function of time are thought to have higher significance than an analysis performed in duplicates at one time point. The samples in the two biological states were processed in technical triplicate for label-free (SWATH) analysis (Fig. 8; Supplementary Table 2); (iii) Pulse SILAC to measure protein synthesis as a function of genotype. Twelve enzymatically digested samples (four samples of parental SiHa, four samples of IFITM1 null, and four samples of IFITM1/IFITM3 double null), each of them as independent biological triplicates, were processed using isotopically labeled amino acids and were separated using LC-MS/MS analysis (Fig. 5; Supplementary Fig. 2; Supplementary Table 1). The statistical rationale for using three biological replicates with one injection per replicate, rather three technical replicates from one biological sample, relates to the dynamics and variability in the interferon dynamics. By using biological triplicates, any common overlaps are deemed to be more significant because of the possible variability in the cell plating and interferon cascade.
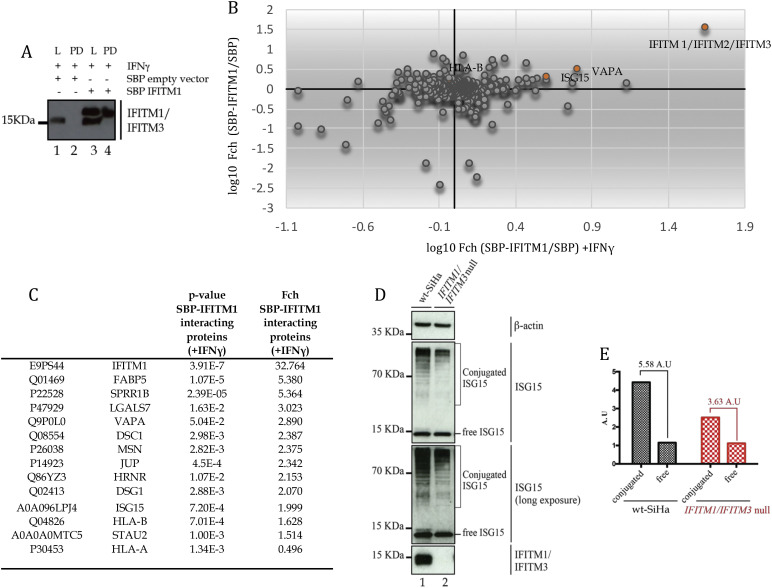
Fig. 8.
Identification of IFITM1 interacting proteins in IFNγ treated cells. (A). SBP vector or SBP-tagged IFITM1 were transfected into parental SiHa cells and treated with IFNγ. Cells were lysed (L) and subjected to immunoprecipitation (PD) and evaluated by immunoblotting with Mab-MHK that can detect untagged or SBP-tagged IFITM1. (B). A 4-quadrant plot showing the relative quantification for binding proteins corresponding to fold change of cells transfected with SBP-IFITM1 versus SBP-empty vector and affinity purified with IFN-γ for 24 h (X-axis) or without IFN-γ stimulation (Y-axis). (C). Table with selected binding proteins detected by mass spectrometry for SBP-IFITM1 enrichment without or with IFN-γ stimulation (from Supplementary Table 3). (D). Cells (parental SiHa; lane 1; or IFITM1/IFITM3 double null (lane 2) were transfected with empty vector (as in 8A) and treated with IFNγ. Samples were processed by immunoblotting with antibodies to ISG15. Free, monomeric ISG15 protein is highlighted, as well as conjugated ISG15. Free ISG15 protein is expressed at similar levels in both cells whilst conjugated ISG15 is attenuated in the IFITM1/IFITM3 double null cell (lane 2 vs 1). This suggests that ISG15ylation protein synthesis is linked to covalent conjugation under these conditions, under conditions where attenuated ISG15 protein synthesis was observed using pulse SILAC (Fig. 5). Antibodies to β-actin and IFITM1/3 proteins were used as loading controls, as indicated. (E). Free and conjugated ISG15 were quantified using ImageJ software. The relative units (in A.U.) define expression as a function of free or conjugated ISG15 in parental and IFITM1/IFITM3 double null cells. The relative change in ISG15 conjugation over free ISG15 was 5.58 A.U. in parental cells. The relative change in ISG15 conjugation over free ISG15 was 3.63 A.U. in IFITM1/3 double null cells.
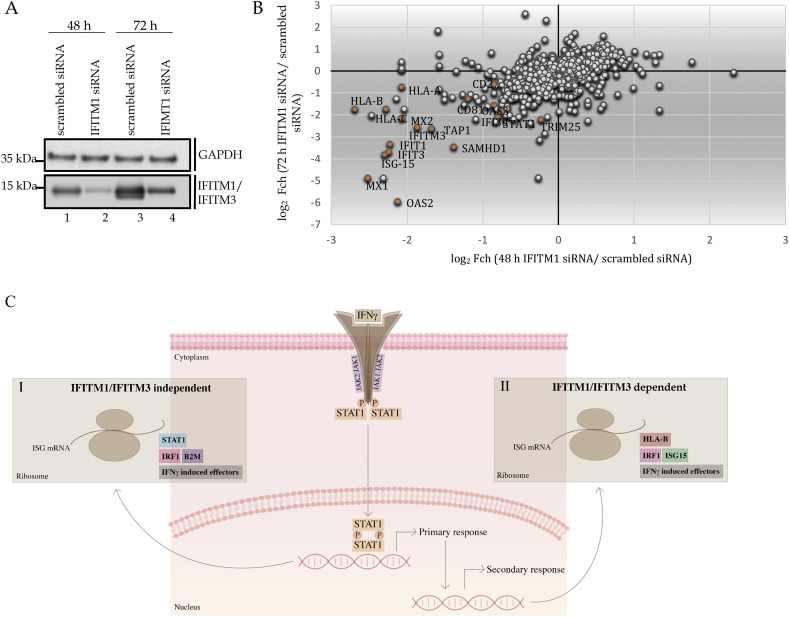
Fig. 9.
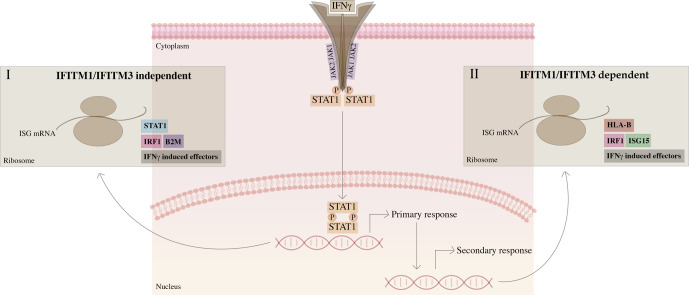
The impact of IFITM1 targeted siRNA on the steady-state proteome. (A). Immunoblotting to demonstrate that IFITM1-targeted siRNA can attenuate IFITM1/3 protein levels. Cells were treated with the indicated control or targeted siRNA for two time points to capture the overlap in the transient dynamics of the interferon signalling cascade. Lysates were immunoblotted with the Mab-MHK antibody (the MHK monoclonal antibody cross reacts to a common N-terminal epitope in IFITM1 and IFITM3, see Supplementary Fig. 1) to quantify IFITM1/3 protein and the loading control (GAPDH), as highlighted. (B). Evaluation of the total steady-state proteome in response to siRNA targeting of IFITM1 in SiHa cells using SWATH-MS (data from Supplementary Table 3). The impact of IFITM1-targeted siRNA treatment for 48 and 72 h. These time points were a point of focus since the siRNA treatment activates the IRF1 transcriptional response over these two-time frames (data not shown). As such, the screen is conducted under experimental conditions in which we consider that the IRF1 response is activated by RNA treatment. The data from these two biological states are plotted as log2 fold change in protein levels (using SWATH-MS) as a function of either the 48 or 72-hour time point. The key proteins whose steady-state levels were suppressed after IFITM1-targeted siRNA treatment are highlighted in red, in the lower left quadrant. (C) The IFITM1 signalling model. Pulse labelling using SILAC methodologies identified STAT1 as a dominant protein synthesized after IFNγ treatment. This forms an internal positive control and is consistent with the classic JAK-STAT response to IFNγ treatment. (I). STAT1 can produce mRNAs that are translated in response to IFNγ treatment including IRF1 and other interferon effectors such as B2M. (II). By contrast to STAT1 and B2M, some of the IFNγ stimulated factors are IFITM1 dependent including MHC Class I molecules and ISG15 (Fig. 5G and L). The siRNA-mediated depletion of IFITM1 represents an orthogonal assay that identified reductions in ISG15 and MHC Class I molecules (Fig. 9A). STAT1 protein reflects a distinct mechanism of control by IFITM1/3. Although pulse SILAC revealed that STAT1 synthesis is IFITM1/3-independent (Fig. 5), the steady state levels of STAT1 protein are reduced after targeted depletion of IFITM1 in SiHa cells (Fig. 9A). These data suggest that turnover of STAT1 protein might be dependent on IFITM1/3, but its synthesis is independent of IFITM1/3. However, these methodologies are complicated to compare directly, since the siRNA methodology uses an intrinsic RNA signal (double stranded RNA) that stimulates IRF1 but without exogenously added IFNγ, whilst the pulse SILAC used IFNγ without RNA ligands. Altogether, these data place IFITM1/3 proteins as a coordinator of the synthesis and/or steady state levels of a sub-set of key players in the IFNγ response. The notable induction of MHC Class I molecules and ISG15 in an IFITM1/3 dependent manner identifies a coordinated signalling pathway with potential clinical relevance (Fig. 1). The recent observation that lowered HLA-A, HLA-B, and HLA-C alleles correlates with poor prognosis and enhanced metastatic growth in cervical cancers [54] is further consistent with the existence of an IFITM1/3:HLA signalling pathway regulating cervical cancer outcomes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
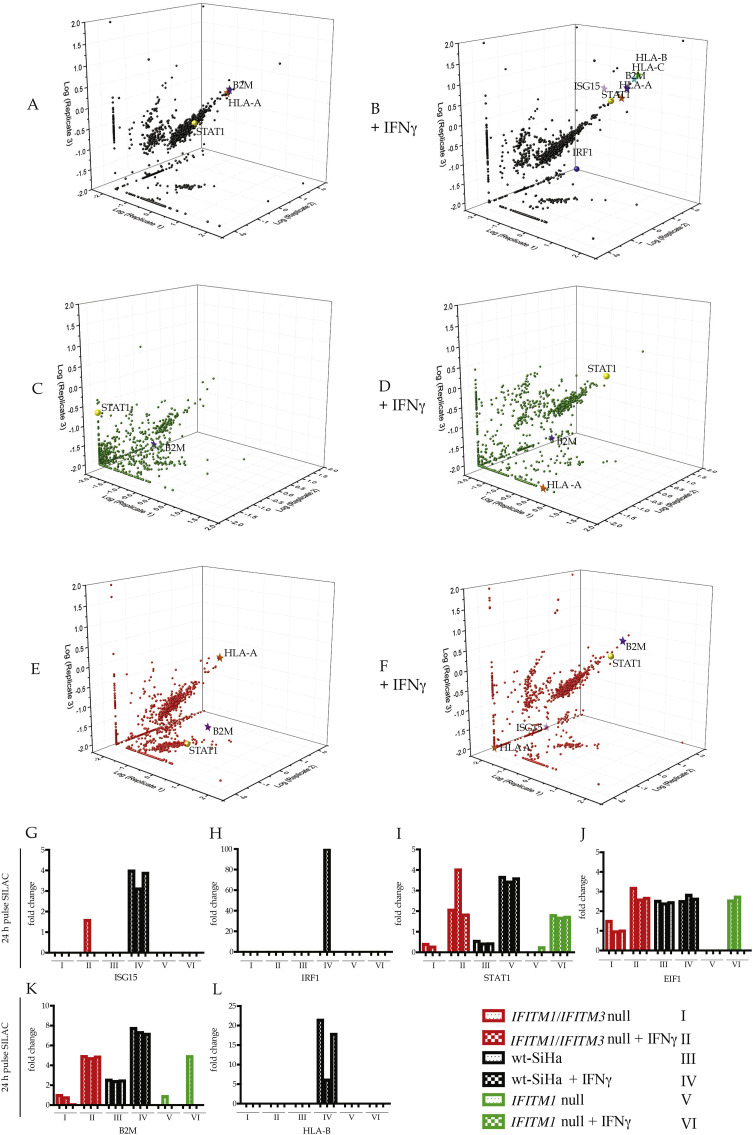
Fig. 5.
Dominant proteins whose synthesis is attenuated after IFN-γ treatment in IFITM1 single null cells, and IFITM1/IFITM3 double null cells. SILAC pulse-labelling for 24 h was performed (A and B) in parental SiHa cells; (C and D) in IFITM1 single null cells; and (E and F) in IFITM1/IFITM3 double null cells (Supplementary Table 1). Cells were incubated with IFN-γ for 24 h in B, D and F. Data were plotted as a function of log10 fold change of heavy/light peptide intensities. Triplicates were represented in the x, y, and z-axis. In samples (G-L), representative peptides used for quantification in biological triplicates are highlighted to demonstrate a protein that is induced independent of IFITM1/IFITM3 (STAT1, I and EIF1, J) and proteins that are IFITM1/3 dependent (ISG15; G, HLA-B, L).
2.15. LC-MS/MS analysis of SILAC labeled samples
SILAC samples were separated using UltiMate 3000 RSLCnano chromatograph (Thermo Fisher Scientific, Massachusetts, USA). Peptides were loaded on a pre-column (μ-precolumn, 30 mm i.d., 5 mm length, C18 PepMap 100, 5 μm particle size, 100 Å pore size) and further separated on an Acclaim PepMap RSLC column (75 mm i.d., length 500 mm, C18, particle size 2 mm, pore size 100 Å) with a 300 nl/min flow rate using a linear gradient: 2% B over 4 min, 2–40% B over 64 min, 40–98% B over 2 min, with A = 0.1% aq. formic acid and B = 80% AcN in 0.08% aq. formic acid. Peptides eluting from the column were introduced into an Orbitrap Elite (Thermo Fisher Scientific, Massachusetts, USA) operating in Top20 data dependent acquisition mode. A survey scan of 400–2000 m/z was performed in the Orbitrap at 120000 resolution with an AGC target of 1 × 106 and 200 ms injection time followed by twenty data-dependent MS2 scans performed in the LTQ linear ion trap with 1 microscan, 10 ms injection time and 10,000 AGC.
2.16. Database searching and analysis
The data from mass spectrometer were processed either using Proteome Discoverer 1.4 or Proteome Discoverer 2.2 that is employed with imbedded statistical tools (both programs were from Thermo Fisher Scientific, Massachusetts, USA). Proteome Discoverer 1.4 processed the data using Mascot engine with the following search settings: database Swiss-Prot human (April 2016); enzyme trypsin; 2 missed cleavage sites; precursor mass tolerance 10 ppm; fragment mass tolerance 0.6 Da; dynamic modifications: carbamidomethyl [C], oxidation [M], acetyl [protein N-terminus]. The results of the search were further submitted to generate the final report considering 1% FDR on both PSM and peptide group levels. Only unique peptides were used for the protein quantification. SILAC labels of R6 and K6 were chosen for heavy and R0 and K0 for light. The relative quantification value was represented as heavy/light ratio (Supplementary Table 1). In the processing and consensus workflows subsequent nodes were used: the Minora feature detector, the Precursor ion quantifier, and the Feature mapper. The data were processed using Sequest HT engine with the following search settings: database Swiss-Prot 2017-10-25, # sequences 42,252, taxonomy: Homo sapiens (updated February 2018); enzyme trypsin; 2 missed cleavage sites; precursor mass tolerance 10 ppm; fragment mass tolerance 0.6 Da; static modification carbamidomethyl [+57.021 Da, (C)], label 13C(6) [+6.020 Da (K, R)]; dynamic modifications oxidation [peptide terminus, +15.995 Da (M)], Met-loss+Acetyl [protein terminus, −89.030 Da (M)]. The results of the search were further submitted to generate the final report with a 1% FDR using Percolator. For the protein quantification and statistical assessment of the biological triplicates, only unique peptides and razor peptides were used. The relative quantification value is represented as the relative peak area of the peptides with the heavy isotope labels with IFNγ treated cells/IFNγ untreated cells ratios (Supplementary Table 3B).
2.17. SWATH-MS
Label free quantitation was performed using FASP-processed tryptic digests with liquid chromatography coupled to tandem mass spectrometry on an Eksigent Ekspert nanoLC 400 (SCIEX, California, USA) online connected to a TripleTOF 5600+ (SCIEX, Toronto, Canada) mass spectrometer. Cells lysates were processed in technical triplicates. Prior to the separation the peptides were concentrated and desalted on a cartridge trap column (300 μm i.d. × 5 mm) packed with a C18 PepMap100 sorbent with a 5 μm particle size (Thermo Fisher Scientific, Waltham, MA, USA). After a 10 washing using 0.05% trifluoroacetic acid in 5% acetonitrile and 95% water, the peptides were eluted using a gradient of acetonitrile/water (300 nl/min) using a capillary emitter column PicoFrit® nanospray columns 75 μm × 210 mm (New Objective, Massachusetts, USA) self-packed with ProntoSIL 120-3-C18 AQ sorbent with 3 μm particles (Bischoff, Leonberg, Germany). Mobile phase A was composed from 0.1% (v/v) formic acid in water, and mobile phase B was composed of 0.1% (v/v) formic acid in acetonitrile. Gradient elution started at 5% mobile phase B for the first 30 min and then the proportion of mobile phase B increased linearly up to 40%B for the following 120 min. Output from the separation column was directly coupled to an ion source (nano-electrospray). Nitrogen was used as a drying and nebulizing gas. Temperature and flow of the drying gas was set to 150 °C and 12 psi. Voltage at the capillary was 2.65 kV.
Pooled sample for the spectral library was measured in data-dependent positive mode (IDA). The MS precursor mass range was set from m/z 400 up to m/z 1250 and from m/z 200 up to m/z 1600 in MS/MS. Cycle time was 2.3 s and in each cycle 20 most intensive precursor ions were fragmented. Subsequently, their corresponding MS/MS spectra were measured. Precursor exclusion time was set to 12 s. Precursor ions with intensity below 50cps were suspended from the IDA experiment. The extraction of mass spectra from chromatograms, their annotation and deconvolution were performed using Protein Pilot 4.5 (SCIEX, Toronto, Canada). MS and MS/MS data were searched using the Uniprot+swissprot database (02. 2016, 69,987 entries) restricted to Homo sapiens taxonomy. Fixed modification – alkylation on cysteine using iodoacetamide and digestion using trypsin was set for all searches. FDR analysis was performed by searching the shotgun data against the decoy search database. The resulting group file was imported into Peakview 1.2.0.3 (SCIEX, Toronto, Canada), where only proteins with FDR below 1% were imported into the spectral library (465 proteins for SBP-IFITM1 pull down SWATH). In SWATH mode, the mass spectrometer operated in high sensitivity positive mode. Precursor range was set from m/z 400 up to m/z 1200. It was divided into 67 precursor ion windows with the width of 12 Da and 1 Da overlap. Accumulation time was 50 ms per SWATH window and the duty cycle was 3.0 s, which enabled acquisition of at least 10 data points across a chromatographic peak. Product ions were scanned from m/z 400 up to m/z 1600. Data extraction was performed in Peakview 1.2.0.3 (SCIEX, Toronto, Canada) with the spectral library. The retention time window for extraction was manually set to 4 min for SBP-IFITM1 pull down SWATH and for the siRNA SWATH (Fig. 9). Protein quantification was performed using up to 4 peptides and 6 transitions per protein to define the statistically significant proteins. Scope of peptides used for quantification was narrowed to only those with peptide confidence higher than 99% and without any variable modification. Protein summed peak areas were normalized using total area sums option in MarkerView 1.2.1.1 (SCIEX, Toronto, Canada). Samples were compared pairwise using paired t-test. The mass spectrometric data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD007562”. For Reviewers, the details include; Username: reviewer17106@ebi.ac.uk and Password: OfTomSm7.
2.18. Flow cytometry for detection of HLA on the cell surface
The parental SiHa and IFITM1/IFITM3 double null cells were grown in RPMI (supplemented with 10% FBS, pen-strep and pyruvate) to 50% confluence in 6-well plates and treated with 100 ng/ml IFNγ for 24 h. Cells were harvested using Accutase (Sigma-Aldrich, A6964), centrifuged 1000 rpm for 5 min with RPMI and kept on ice in 1% BSA in PBS for 20 min. Primary antibodies (HLA-B: PA5-35345) were diluted in 1% BSA/PBS to 1:100. Cells were centrifuged as before, and cell pellets were resuspended in 100 μl of diluted primary antibodies or in the same amount of 1% BSA/PBS (for control samples) and incubated 45 min a room temperature on a tube rotator. After a triple wash in ice-cold 1% BSA/PBS, cells were incubated with 100 μl of secondary antibody (Abcam, Goat Anti-Rabbit IgG H&L DyLight 488), diluted 1:200, on a tube rotator for 30 min at room temperature. After a triple wash in ice-cold 1% BSA/PBS, cells were resuspended in 500 μl of 1% BSA/PBS and kept on ice before measurement. Samples were measured on FACSVerse (BD Biosciences) and data were analyzed using FACSuite software (BD Biosciences). A negative control without primary antibody was prepared for each sample. HLA expression on the cell surface was counted as FITC median fluorescence intensity (MFI) divided by the MFI of the negative control. Two independent experiments were performed, each with two independently isolated IFITM1/IFITM3 double null cells and two different HLA antibodies (HLA-A (data not shown) and HLA-B (Fig. 6)).
3. Results
3.1. Identifying a clinically relevant model to dissect IFITM1 and IFITM3 (IFITM1/3) signalling
IFITM1 and IFITM3 (IFITM1/3) can function as oncogenic factors in several cancer cells [32,33]. Attenuation of IFITM1 protein expression can inhibit growth, invasion, and/or migration of cancer cells [34]. Patient subgroups with clinically relevant expression data are not well-defined. We developed a panel of monoclonal antibodies to a N-terminal peptide with a high homology between IFITM1 and IFITM3 (Supplementary Fig. 1A, B). This would allow the development of monoclonal antibodies that detect the co-expression of both IFITM1 and IFITM3 proteins. We aimed to use such tools to screen a large panel of human cancers for those that express IFITM1/3 proteins. This would identify clinically relevant models for a focus to dissect IFITM1/3-mediated signalling pathways. The monoclonal antibody chosen (named Mab-MHK; Supplementary Fig. 1B) can bind to IFITM1/3 antigens in a range of human cancer cells (Supplementary Fig. 1C). Some cancer cells exhibit no expression of IFITM1/3 such as the lymphoma cell line WHU-NHL (Supplementary Fig. 1C). Further studies confirmed that Mab-MHK can bind to both IFITM1 and IFITM3 proteins, as defined using single IFITM1 single null and IFITM1/IFITM3 double null cells (see below).
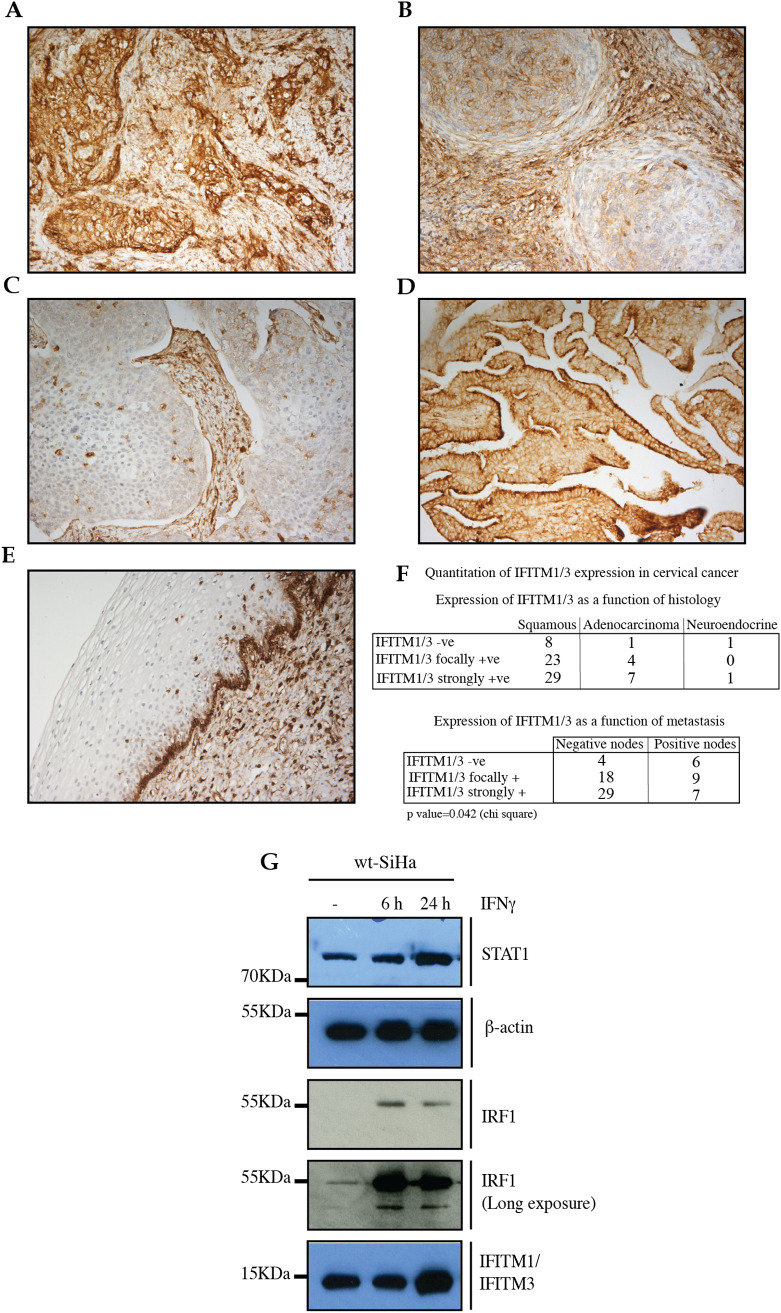
Mab-MHK was used to screen large panels of archival formalin-fixed human cancer tissues to identify potential clinically relevant models. We could detect differential expression of IFITM1/3 in breast cancer, colon cancer, and oesophagus cancer (data not shown). We could also detect differential expression of IFITM1/3 protein in a cervical cancer array (Fig. 1A–F). Squamous cervical cancer samples expressed either high levels of IFITM1/3 (Fig. 1A), lower levels of IFITM1/3 (Fig. 1B), or undetectable levels of the antigens (Fig. 1C). Cervical adenocarcinoma often exhibited high expression (Fig. 1D). Interestingly, some normal squamous cervical epithelium exhibited high expression in the basal ‘stem’ cell or pluripotent layer only (Fig. 1E), but not the differentiating cell layers. We can conclude that 64 out of 74 cervical cancer specimens are positive for IFITM1/3 proteins using the Mab-MHK (Fig. 1F, top panel). What is also interesting is that there is a statistically significant inverse association between IFITM1/3 protein expression and the number of lymph node metastases in patients (Fig. 1F, bottom panel). This will be rationalized in the discussion based on data that emerges below.
Fig. 1.
Immunohistochemical analysis of IFITM1/3 protein expression in cervical cancers using the Mab-MHK. Formalin-fixed, paraffin-embedded cervical carcinoma tissue was processed as stated in the Experimental Procedures using the Mab-MHK that binds to shared epitopes in the N-terminal domains of IFITM1 and IFITM3 (Supplementary Fig. 1A, B). Representative images highlight grades of IFITM1/3 protein expression; (A). high expression in squamous cell carcinoma, (B). medium expression in squamous cell carcinoma, (C). absence of expression in squamous cell carcinoma, (D). high expression in adenocarcinoma, and (E). expression in basal stem cell layer in normal tissue. (F). (Top panel); Quantification of IFITM1/3 protein expression in cervical cancer in relation to histology type and (Bottom panel) IFITM1/3 protein expression in cervical cancer in relation to lymph node metastasis, excluding neuroendocrine. (G). Immunoblotting of protein expression in the parental SiHa cells after IFNγ treatment for the indicated time points. Proteins evaluated include; STAT1; β-actin, IRF1, and IFITM1/3.
3.2. Developing an IFITM1 and IFITM3 double null cell line using a CRISPR-guide RNA methodology
IFITM1 is implicated in IFNγ mediated growth control in some cancer cells with an active p53 pathway [35]. IFITM1 is also implicated in a growth stimulatory role in cervical squamous cancers [36] [37]. The HPV16+ and IFITM1/3 positive cervical cancer cell line SiHa [38] [39] exhibit IFNγ inducible STAT1, IRF1, and IFITM1/3 proteins (Fig. 1G). As such, we focused on using this cervical cancer cell line (SiHa) as a model to identify IFITM1/3 dependent signalling pathways.
In order to continue to develop a cervical cancer cell model that reflects the clinical data (IFITM1/3 positive or IFITM1/3 negative cancers; Fig. 1F), we set out to develop a double null IFITM1 and IFITM3 cell line model. We first generated an isogenic IFITM1 null cell panel through gene editing to validate the guide RNA. IFITM1 knock-out mice are viable [40] so it was likely we would be able to generate IFITM1 null cells. Guide RNAs targeting exon 1 in the IFITM1 gene (Figure 2A) were cloned into pLentiV2. Cells were transfected and selected for resistance to puromycin to allow stable integration of the IFITM1 targeting guide RNA cassette. Cell clones were chosen for sequencing across the guide RNA targeting site (Fig. 2A) using PCR (Fig. 2B). Both IFITM1 alleles were gene edited in a representative IFITM1-null clone that creates two distinct frameshifts (Fig. 2C and D). We examined four representative IFITM1 null cells in DNA damage response assays and all lines were shown to be either chemosensitive or X-irradiation sensitive (data not shown). Since all single knock-out clones behaved similarly, we chose one representative IFITM1 null cell line for continued study. The IFITM3 gene was next targeted using guide RNA methodologies (Fig. 3A) to to create a IFITM1/IFITM3 double null cell (Fig. 3A–C). IFITM3 null mice, and mice with the entire IFITM chromosomal locus deleted, are also viable [40,41]. This created an isogenic cell model that removed any redundancy of IFITM3 in the IFITM1 interaction landscape, especially as they both are reported to interact with VAPA [21]. Immunoblotting confirmed that IFITM1 and IFITM3 proteins are not detected in the IFITM1/IFITM3 double null cell, respectively (Fig. 3D; lanes 5 and 6). We chose a representative double-null cell line for subsequent studies.
3.3. Pulse SILAC mass spectrometry methodologies identify IFITM1/3 dependent protein synthesis in response to IFNγ signalling
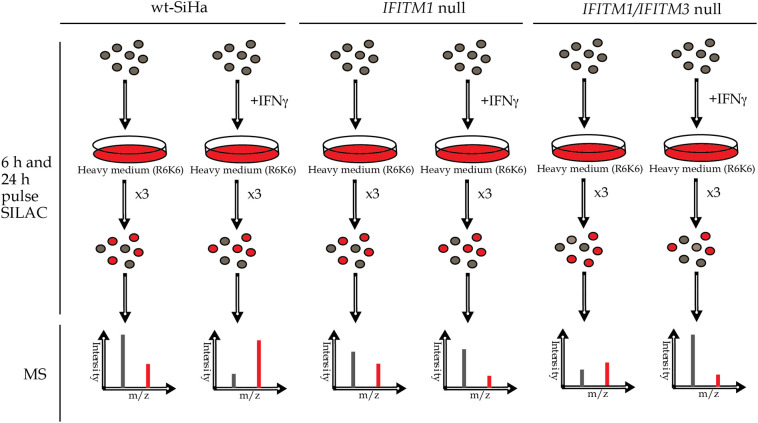
Given that one established effector of IFITM1 is IFNγ [35], we evaluated IFNγ responsive protein synthesis in IFNγ stimulated parental SiHa, IFITM1 single null, and IFITM1/IFITM3 double null cells. The parental SiHa, IFITM1 single null, and IFITM1/IFITM3 double null cells were treated with heavy isotopic amino acid labeling media (the SILAC method) for 6 or 24 h, in the absence or presence of IFNγ (Fig. 4). Cell lysates were then processed by FASP [30,31] and then analyzed for IFITM1/3-dependent protein synthesis using mass spectrometry. The SILAC methodology has been subjected to an analysis of random error associated with the multiple steps in this approach including; cell plating in biological replicates, switch to heavy isotopic media, cell recovery from plastic plates, cell lysis, centrifugation, filter assisted trypsinization, and tryptic peptide recovery and processing. This error can be reduced by employing multiple replicates (n = 3) as highlighted previously [42]. To highlight the importance of biological replicates and the inherent error in this multiple step process, we plot the data not as an average of three replicates, but as individual points from all three replicates (as in Supplementary Fig. 2 and Fig. 5).
Fig. 4.
Methodological approach to identify signalling pathways altered in IFITM1 and IFITM3 double null cells. The indicated cells (parental SiHa, IFITM1 single null, or IFITM1/IFITM3 double null) were pre-treated with carrier or IFNγ for 24 h. The media was replaced with R6K6 isotopically labeled media with carrier or IFNγ for 6 or 24 h. Cells were harvested, lysed, and tryptic peptides processed for analysis by mass spectrometry (MS) as indicated in the Experimental Procedures.
The dominant IFNγ responsive protein to be identified at 6-h post labelling is IRF1 protein (Supplementary Fig. 2B vs 2A) with an attenuation of isotopically labeled IRF1 peptides recovered in the biological triplicates from IFITM1/IFITM3 double null cells (Supplementary Fig. 2H). This suggests that IRF1 is partially dependent upon IFITM1/3 signalling and this was confirmed using IRF1 transcriptional reporter assays (data not shown). This data provides some degree of confidence that the methodology is able to identify a known IFNγ responsive target (IRF1). There are other proteins whose synthesis was detected at 6 hours post-isotopic labelling including STAT1, EIF1, and B2M (Supplementary Fig. 2I–K). EIF1 is not known to be linked to interferon signalling, but it is known to regulate the accuracy of AUG codon selection by the scanning pre-initiation complexes [43]. EIF1 might prove to be involved in regulating interferon dependent anti-viral mRNA selection. Nevertheless, all three proteins are also IFTM1/3-independent (Supplementary Fig. 2I–K). STAT1 and B2M are also both known IFNγ responsive proteins further validating the methodology used to measure quantitative changes in protein synthesis. That all three proteins (STAT1, EIF1, and B2M) exhibit equivalent protein synthesis rates in the parental and double-null cell model indicates that the double null has retained many key regulatory features of the parental cell. This suggests that many IFN regulatory features of the double null cell have been retained despite the selection process creating the cell model.
By 24-h post IFNγ treatment of SiHa cells, HLA family members, B2M, and STAT1 proteins were detected (Fig. 5B vs 5A), again indicating that the methodology can detected known IFNγ inducible proteins. By 24-h post IFNγ treatment, isotopically labeled IRF1 peptides are attenuated (Fig. 5H), which is consistent with the early and transient induction of IRF1 by IFNγ. The synthesis of MHC Class I molecules was IFITM1/3-dependent (Fig. 5B vs 5F; quantified in biological triplicates in Fig. 5L). All three, major HLA alleles exhibited attenuated synthesis in the double null cell, as defined using the tryptic peptide coverage (Supplementary Fig. 3). Isotopically labeled ISG15 tryptic peptides are also not observed in the early interferon response (Supplementary Fig. 2), and the isotopically labeled ISG15 peptide recovery after 24 h is attenuated in the IFITM1/IFITM3 double null cells (quantified in biological triplicates in Fig. 5G) suggesting that ISG15 protein synthesis is largely IFITM1/3 dependent. By contrast, STAT1 protein synthesis at 24 h appears largely IFITM1/3-independent (quantified in biological triplicates in Fig. 5I). Providing another internal control, another well-known inducible IFNγ protein, B2M, exhibits equivalent synthesis in the IFITM1/IFITM3 double null cell (quantified in biological triplicates in Fig. 5K). This indicates that one key regulatory feature of the double null cell, STAT1 production, has remained intact.
3.4. Orthogonal validation of IFITM1/3 dependent induction of HLA-B and ISG15
These data first demonstrate that using the pulse-SILAC methodology, the SiHa cell model reflects the classic IFNγ responsive induction of STAT1, IRF1, B2M, ISG15, and MHC Class I molecules (Fig. 5B vs 5A and Supplementary Fig. 2). Also of note is attenuation of HLA-A, HLA-B, HLA-C, and ISG15 protein synthesis 24 h post-IFNγ treatment in the IFITM1/IFITM3 double null cells compared to parental SiHa (Fig. 5F vs 5B).
In order to determine if IFITM1 deletion alone impacted on this set of gene products, the parental SiHa and IFITM1-null cells were in parallel treated with SILAC heavy-labeling media for 6 or 24 h. As with the double IFITM1/IFITM3 double null cells, IFNγ dependent induction of IRF1 protein synthesis is attenuated 6 hours post labelling in the IFITM1-single null cells (Supplementary Fig. 2D vs 2B). This suggests that IRF1 is dependent upon IFITM1. Elevation of STAT1 protein synthesis are IFITM1-independent based on the equivalent induction of STAT1 in the IFITM1-single null cells (Fig. 5I). HLA-B protein synthesis is attenuated 24 hours post-IFNγ treatment in the IFITM1 single null cells (Fig. 5L). ISG15 synthesis is also attenuated in the IFITM1 single null cell (Fig. 5G). Together, these data suggest that MHC Class I family members and ISG15 require at least IFITM1 for maximal IFNγ stimulated protein synthesis.
The software used to identify HLA orthologues in the pulse-SILAC screen (Fig. 4) can discriminate between HLA-A, HLA-B, and HLA-C based on tryptic peptide sequences (Supplementary Fig. 3). We focus here on HLA-B, which shows accurate identification of HLA-B specific peptides and it is also a member of the IRDS pathway [7]. We thus validated HLA-B protein expression in orthogonal assays to determine whether apparent reductions in HLA-B protein synthesis reduction in the IFITM1/IFITM3 double null cells was reflected in total steady state protein levels and subcellular localization on the plasma membrane.
First, immunofluorescence of HLA-B was defined in parental SiHa and IFITM1/IFITM3 double null cells. Parental SiHa cells revealed significant induction of HLA-B immunoreactivity 24 h after IFNγ treatment (Fig. 6C vs 6B and quantified in 6G). By contrast, basal HLA-B protein expression was attenuated in the IFITM1/IFITM3 double null cells after IFNγ treatment (Fig. 6F vs 6E). Quantitation of the total immunofluorescence in the absence and presence of IFNγ, in the parental SiHa and IFITM1/IFITM3 double null cells, also confirms attenuated HLA-B induction in the null cell panel (Fig. 6H vs 6G). This is consistent with the reduced protein synthesis observed for HLA-B in the pulse SILAC quantitation in the IFITM1/IFITM3 double null cells.
The dominant subcellular localization of HLA-B is thought to reside on the cell surface as an antigen presentation carrier. We therefore evaluated whether HLA-B expression on the plasma membrane was altered in the IFITM1/IFITM3 double null cell using FACS analysis with non-permeabilized cells. Two independent IFITM1/IFITM3 double null cell clones were used as a form of biological replicate in comparison to the parental SiHa cell line. Twenty-four hours post treatment, HLA-B was elevated on the plasma membrane in the parental SiHa cell line (data not shown). Quantitation revealed reduced levels of HLA-B in both independent IFITM1/IFITM3 double null biological replicates in the absence and presence of IFNγ (Fig. 6I). These data indicate that the reduced synthesis of HLA-B in the IFITM1/IFITM3 double null cell (Fig. 5) has an impact on its subcellular localization at the plasma membrane.
We next also examine whether there is any direct protein-protein interaction between IFITM1 and HLA-B since they are both co-synthesized, have transmembrane localizations on the cell surface, and are both IRDS components. In vivo proximity ligation assays are emerging methodologies that have been shown to demonstrate the “association” of two endogenously expressed proteins in fixed cells without the need for harsh lysis [44]. The method can be considered as an in situ mimic of an “immunoprecipitation assay”. Proximity ligation assays can identify a protein-protein interaction/association with a distance of 10–30 nm that is in the upper range of that observed using FRET (5–20 nm) and this methodology can detect authentic endogenous proteins in situ that does not rely on transfected or artificially GFP-tagged protein vectors [45]. We evaluated whether IFITM1/3 and HLA-B co-associate using this methodology using antibodies to HLA-B and Mab-MHK (that can bind to both IFITM1 and IFITM3 proteins; Supplementary Fig. 1). A significant protein-protein interaction was observed in the IFNγ treated cells (representative images; Fig. 7A–D). These foci were absent in the IFITM1/IFITM3 double null cells (Fig. 7E–H). Together, these data validate the pulse-SILAC data that identified HLA-B as a downstream effector of IFITM1/3.
Fig. 7.
Evaluation of the IFITM1/IFITM3:HLA-B protein-protein interaction in situ. Proximity ligation assays were used to study the endogenous interaction between HLA-B and IFITM1/3 proteins in parental SiHa (A–D) and IFITM1/IFITM3 double null cells (E–H). Representative FITC images identify the protein-protein association foci (depicted in green) and DAPI was used for nuclear staining (depicted in blue). (A–B and E–F). Cells were incubated as negative controls using IFITM1/3 or HLA-B antibodies only. (C and G). Cells were incubated with both IFITM1/3 and HLA-B antibodies to define protein-protein foci in non-stimulated cells. (D and H) Cells were incubated with both IFITM1/3 and HLA-B antibodies to define protein-protein foci in IFN-γ stimulated cells. n = 3. Scale bar: 10 μm. (I and J) Quantification of HLA-B and IFITM1/3 total fluorescence per cell in presence or absence of IFNγ stimulation in (I) parental SiHa and (J) IFITM1/IFITM3 double null cells. For quantitation, three independent assays were performed, and each assay had two independent biological replicates. For each assay, fluorescence was measured in at least 50 cells per condition. Fluorescence was measured using ImageJ software. Statistical study was performed with 1-way Anova and Bonferroni correction (p-value < .0001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
ISG15 was not easily visualized using immunofluorescence in situ (data not shown) nor could we identify a protein-protein association between IFITM1/3 proteins and ISG15 using proximity ligations (data not shown). We thus used an independent assay for orthogonal validation of ISG15 induction. We developed a SBP-tagged IFITM1 expression construct in order to ectopically express the protein in the parental SiHa cells and design methodologies for capturing IFITM1 associated proteins. The transfection of the SBP-IFITM1 expression could be detected as migrating at a higher mass (due to the SBP tag) than endogenous IFITM1/3 proteins in the parental SiHa cells (Fig. 8A, lane 3 vs lane 1) and specifically captured after affinity purification following expression in the IFITM1/IFITM3 double null cell (Fig. 8A, lane 4 vs 2).
SWATH-MS analysis of the affinity purified material (SBP-IFITM1 vs SBP-only; material Fig. 8A, lane 4 vs lane 2), from untreated vs interferon treated, identified proteins that were enriched without or with IFN-γ treatment of parental SiHa cells, respectively (Fig. 8B; Supplementary Table 2). The data first highlight the detection of peptides belonging to a homologous sequence for IFITM1/2/3 proteins (Fig. 8B, Fig. 8C). Since IFITM1 was our bait protein we presume that it was the detected isoform, serving as an internal positive control (Fig. 8A). Nevertheless, IFITM1 may also be interacting with IFITM2 or IFITM3. The previously identified IFITM1/2/3 interacting protein VAPA was also detected (Fig. 8B and C), Supplementary Table 2. The proteins with the most significant p-values were enriched in IFN-γ treated cells, relative to non-treated cells (Fig. 8B and C, Supplementary Table 2). These included several proteins involved in cell-cell or cell matrix interactions including cornifin, galectin-7, desmocolin, JUP, hornerin, and desmoglein (Fig. 8C; Supplementary Table 3). This is suggestive of a pathway interaction of IFITM1 with membrane-dependent cell-cell communications. The enrichment of higher confident targets in IFN-γ treated cells also suggests that IFN-γ treatment may be required to fully ‘activate’ the IFITM1 protein interaction landscape. Interestingly, proteins related to IFNγ signalling were also detected; ISG15 and HLA-B (Fig. 8B and C). Immunoblotting also confirmed that ISG15ylation was attenuated in IFITM1/IFITM3 double null cells after IFNγ stimulation (Fig. 8D). By contrast free ISG15 protein remained equivalent in both cells (Fig. 8D), suggesting that the ISG15 synthesis detected using pulse SILAC is directly linked to IFN stimulated protein conjugation. The ratio changes of conjugated to free ISG15 is summarized in Fig. 8E. Together, the orthogonal validation of HLA-B and ISG15 is consistent with the pulse SILAC data; IFITM1/3 proteins are required for maximal induction of HLA-B and ISG15ylation in IFNγ treated cells.
3.5. Orthogonal validation of the IFITM1 signalling protein landscape
We were unable to complement the IFITM1 and IFITM3 gene back into the double-null SiHa cells to stimulate ISG15ylation and HLA-B protein levels in IFN-γ treated cells (data not shown). As such, we took an independent approach to define dominating IFITM1-dependent signalling pathways and whether these overlap with the signalling proteins identified using the pulse-SILAC experimental methods. Although the majority of analyses identified HLA-B and ISG15 protein expression changes using IFITM1/IFITM3 double null cells, we did create an IFITM1 single null cell that revealed similar reductions in ISG15 and HLA-B after IFNγ treatment (Fig. 5D, G, and L). As such, we examined IFITM1 dependence in the steady-state proteome levels using IFITM1 targeted siRNA treatment of parental SiHa cells (Fig. 9A, lanes 4 and 2 vs 3 and 1) to determine if attenuation of IFITM1 by this method gave rise to similar proteome changes. However, there is a caveat to this method. Similar to plasmid transfection [46], double stranded RNA can also induce an IRF-1 dependent transcriptional interferon response [47]. Accordingly, we have also found that siRNA transfection methodologies can induce an IRF1 transcriptional response (data not shown). Nevertheless, siRNA is a powerful tool to determine gene dependencies in cell models. When parental SiHa cells, treated from 48 and 72 h with control siRNA or IFITM1-targeted siRNA, there was striking and selective reduction in interferon-responsive proteins using label free quantitative mass spectrometry at both time points (Fig. 9B and Supplementary Table 3). Some of these proteins, including IFITM1 itself, are also components of the interferon-stimulated DNA damage resistant signature of proteins [9,10]. Interestingly, ISG15 and HLA-B are also lowered after the treatment of SiHa cells with IFITM1 targeted siRNA. However, there is one notable difference in the pulse-SILAC (Fig. 5) vs the siRNA methodology (Fig. 9B); STAT1 and B2M are IFITM1/3-independent as defined using pulse SILAC (Fig. 5); whilst STAT1 and B2M are IFITM1-dependent using the siRNA methodology (Fig. 9B, Supplementary Table 3). This might reflect that fact that the RNA ligand (siRNA) induces additional RNA-activated pathways in the targeted siRNA proteome screen than IFNγ alone.
In conclusion, the pulse SILAC methodology in the IFITM1 single null (Fig. 5) and the siRNA treatment using IFITM1 targeted siRNA in parental SiHa cells (Fig. 9) both gave rise to overlapping proteome changes; mainly ISG15 and HLA-B. These data suggest that IFITM1 might alone function as an effector to these two proteins. However, we cannot rule out a role for IFITM3 in the IFITM1 single null cell, since IFITM3 is also reduced by IFITM1 targeted siRNA treatment of SiHa cells (Fig. 9B). Thus, IFITM1 and IFITM3 might cooperate to mediate these effects.
4. Discussion
The IFITM protein family was identified in whole genome siRNA screens as RNA virus restriction factors [20]. The molecular functions of the IFITM family in the anti-viral response are just beginning to be defined. Yeast-two hybrid, with IFITM3 as a bait, was used as a methodology to discover new protein-protein interactions within the IFITM family that mediate viral restriction [21]; a protein-protein interaction was identified for IFITM3 and the Vesicle-membrane-protein-associated protein A (VAPA). The VAPA interaction occurred with IFITM1, IFITM2, or IFITM3 and these three were not distinguished from each other in this assay. Nevertheless, the study focused on defining how VAPA interaction with IFITM3 results in reduction of VAPA binding to oxysterol-binding protein (OSBP), disrupting cholesterol homeostasis, and preventing viral maturation. As IFITM1 and IFITM2 are also reported to bind to VAPA, it is not yet clear whether they play minor and/or redundant roles in controlling cholesterol-mediated viral maturation [21]. Interestingly, VAPA was also identified in our SWATH-IP using SBP-tagged IFITM1 (Fig. 8), which is consistent with VAPA being a dominant interactor for the IFITM family. In addition to VAPA, palmitoylation of IFITM1/2/3 family members has been reported to be required for some anti-viral functions [48]. Whether the anti-viral associations with VAPA or palmitoylation by the IFITM family impact on cancer growth control is not yet known.
The IFITM family also have reported roles in oncogenic and/or pro-metastatic cancer cell growth. It is not known if the anti-viral and pro-oncogenic functions of the IFITM family overlap. Within the IFITM1 family, IFTM1 is the most well documented to have pro-metastatic roles and it is the only IFITM family member with a partial residence in the plasma membrane [19]. IFITM1 is also the only IFITM family member that is a component of the Interferon-regulated DNA damage resistance (IRDS) pathway [10]. The IRDS contains a subset of approximately 30 interferon responsive genes that are up-regulated late in the viral response, upregulated during development of radiation resistance, and upregulated as a result of chronic exposure to lower levels of type I or type II interferons. IRDS pathway expression is mediated by ‘unphosphorylated’ STAT1 [49] and can be stimulated by IRF9 [50]. Interestingly, two IRDS genes (ISG15 and HLA-B) are induced after IFNγ exposure in a ‘IFITM1-dependent’ manner (Fig. 9C). Our unpublished data also indicate that IFITM1 single null cells are X-ray sensitive or cisplatin-sensitive, consistent with the hypothesis that IFITM1 expression is linked to chemo or irradiation resistance. The mechanisms whereby IFITM1 itself regulates such chemoresistant or pro-metastatic cell signalling events have not been mechanistically defined.
In this report, we begin to set up biochemical assays that could shed light on dominant cancer-associated functions of IFITM1. We first aimed to identify a clinically relevant human cancer model to study IFITM family of proteins. This required us to develop a pan-specific antibody that binds to both IFITM1 and IFITM3 proteins (Supplementary Fig. 1). This resulted in a focus on cervical cancer (Fig. 1) that exhibited high, medium or no expression of IFITM1/3 proteins (Fig. 1F). Of particular interest was the inverse correlation between IFITM1/3 protein expression and lymph node metastasis (Fig. 1F) suggesting that loss of IFITM1/3 protein expression correlates with evasion of the immune system. If IFITM1/3 are ‘pro-oncogenic’, why would cervical cancer panels reveal an inverse correlation between IFITM1/3 expression and cancer-positive lymph nodes in patients?
It is now becoming apparent that there are two distinct modes of metastatic cell growth. The first is the more classically defined metastasis due to enhanced ‘invasion and migration” to secondary tissue niches. The second represents metastasis due to cancer cell escape from immune surveillance [51]. IFITM1/3-positive cancers might indeed be pro-invasive, depending on the microenvironment and tissue type, leading to poor clinical prognosis. The methodologies that highlighted such classic pro-metastatic roles for IFITM1 include over-expression by ectopic transfection of plasmid encoded genes or attenuation of gene expression using targeted siRNA [14]. By such experimental approaches, IFITM1 does indeed promote cancer cell growth and/or ‘cell invasion’ (i.e. a model of metastatic growth) [16]. Consistent with this, using clinical material IFITM1 protein has often been shown in literature to be over-produced in cancers using immunohistochemistry and this often correlates with poor prognosis [12,52].
However, our data reviewed using cervical cancer (Fig. 1) indicate that there can be two distinct states of cervical cancer with respect to IFITM1/3 expression. IFITM1/3 negative cervical cancers might also be more pro-invasive due to immune escape. This is based on the data indicating that the most dominant proteins whose synthesis depends on IFITM1/3 using the pulse SILAC methodology are in fact HLA family members (Fig. 4, Fig. 5). HLA family members are components of the IRDS, they play a role in anti-viral immunity through the presentation of viral peptides through the MHC Class I system, and HLA expression is linked to immune rejection of cancer cells [53]. This suggests that although IFITM1/3 might be pro-oncogenic under some conditions, IFITM1/3-HLA signalling would presumably function as a ‘tumor suppressor’ signal via engagement of CD8+ T-cells. Such IFITM1/3 and HLA positive cancers might not metastasize because they produce neoantigen presenting MHC Class I molecules that keep the primary tumor in a local chronic state of equilibrium with the immune system. IFITM1/3-negative cancers by contrast might be expected to produce lower amounts of MHC Class I molecules following IFNγ stimulation (Fig. 9) resulting in lowered neoantigen expression, immune escape, and metastasis. Such hypotheses are consistent with two clinical observations in cervical cancers. First, an inverse correlation exists between IFITM1/3 expression and lymph-node positive cancers (Fig. 1). Second, a recent study has highlighted that lowered MHC Class I expression also predicts poor prognosis in cervical cancers [54]. This data is also consistent with several studies that highlight elevated rates of metastatic cancer cell growth in vivo are inversely correlated to MHC Class I expression including deletion of the MHC Class I locus [53].
Although expression of IFITM1/3 was detected in cancer cells (Fig. 1), it is interesting that IFITM1/3 protein expression was observed and confined to the basal ‘stem cell’ layer in representative normal tissue controls using immunohistochemistry (Fig. 1 E). Expression was not observed even on one cell layer up from the basal layer. In squamous human skin, this basal layer is reflective of p63 (a squamous stem cell transcription factor) and phosphorylated p53 positive stem cell populations after UV irradiation [55]. Thus, this expression might reflect a role for IFITM1 in squamous stem cell pluripotency. We do not have very many samples reflecting a ‘normal’ human cervical epithelium, but the few cases we have exhibit very strong staining in the basal layer (data not shown). During the course of these studies the Human Protein Atlas has also populated their immunohistochemical library with expression patterns of IFITM1 protein in normal tissues. We can see in this library that IFITM1 protein is also confined to the “basal stem cell” layer in normal squamous oesophagus, cervix, and oral mucosa (Supplementary Fig. 4). Together, these data would suggest that IFITM1 might play a role in squamous stem cell physiology as its expression appears specifically confined to the basal layer and is not expressed in suprabasal normal squamous cells. As the ‘normal’ squamous cervical epithelium we have used is from patients undergoing screening for cervical cancer or dysplasia, we cannot rule out a role for HPV in this expression in normal cervical cells. However, as the oesophagus squamous epithelium also exhibits IFITM1 protein expression in the basal cells of normal squamous epithelium (Protein Atlas, Supplementary Fig. 4), and the normal oesophagus is not noted for HPV infection, we would suggest that the expression of IFITM1 protein in basal cells is not related to HPV status. The signalling pathways that might trigger basal IFITM1 protein expression in squamous stem cell populations are not precisely defined. However, at the mRNA level, differential expression patterns of ifitm1 gene expression were identified in the uteri of mice and there were correlations between the patterns of ifitm1 gene expression and Wnt/β-catenin expression [56]. These data might suggest a role for Wnt signalling as an upstream regulator of IFITM1 in stem cells.
Our focus on cervical cancer as a model to understand the cancer-associated role of IFITM1/3 is interesting considering IFITM1/IFITM3 are themselves RNA viral restriction factors and HPV infection is a risk factor in cervical cancer progression. There is evidence that IFITM1 and IFITM3 might be a positive cofactor for a DNA virus; HPV16 viral propagation [57]. We do not have any data that defines the HPV status of the cervical cancers we have analyzed (Fig. 1), as the main clinically approved assay for diagnostics is p16 positive cancer cells [58]. There is a close relation between persistent viral infection, development of cancer and failure in immune response. As an example, cervical as well as vulvar intraepithelial neoplasia are pre-cancerous conditions characterized by sustained HPV-16 infection. A clinical study shows favorable prognosis related to an increase in IFNγ-producing CD8+ cytotoxic T following to robust cell response induced by vaccination [59]. Vaccination delivers a high dose of specific antigen against HPV-16 oncoproteins E6 and E7 and mediates MHC-binding peptide complex presentation [60]. A similar outcome was also observed after vaccination of a preclinical mouse model of HPV positive cervical cancer [61]. There is a significant correlation between MHC Class I (not found for MHC Class II) expression on malignant cells and T-cell infiltration (TIL) in human ovarian cancer [62], being a positive prognostic factor. Thus, IFITM1 and IFITM3 might have dual roles in stimulating HPV propagation, but also in suppressing cancer escape from immune surveillance.
One of the key approaches we used to evaluate the role of IFITM1 in IFNγ dependent protein production was the utilization of CRISPR-Cas9 guide RNAs to create isogenic knock-out cells. At the outset, we relied less on the use of siRNA to deplete IFITM1 since this would only give a transient reduction in a target protein but also siRNA itself can induce an interferon response (data not shown; [46]). This would have complicated our analysis of the interferon-responsive nature of any IFITM1 protein interactions we visualize and measure. Nevertheless, siRNA was used as a final orthogonal approach to define IFITM1 signalling events (Fig. 9A, B). By contrast, the limitation of using gene knockout tools to reduce expression of a protein requires that loss of the protein is not a lethal event. In the case of IFITM1 and IFITM3 knock-out mice are reported as viable [41], thus it was not unexpected that we were able to generate single IFITM1 null and double IFITM1/IFITM3 double null cell panels (Fig. 2, Fig. 3). However, we were unable to generate single IFITM3 null cells under experiments carried out in parallel to those reported in this manuscript (data not shown).
Using these isogenic cell panels, we were able to determine whether there were defects in IFNγ dependent protein synthesis using pulse SILAC methodologies. We chose to use a pulse-SILAC approach to identify the isotopically labeled tryptic peptides with the most significant fold change after IFNγ treatment and which are altered in the double IFITM1/IFITM3 or IFITM1 single null cells. The most significantly suppressed proteins in the double IFITM1/IFITM3 or IFITM1 single null cells after IFNγ treatment were MHC Class I orthologues encoded by the HLA-A, HLA-B, and HLA-C genes (Fig. 5). As rationalized above, the data suggested an inverse correlation between IFITM1/3 protein and HLA expression with metastatic growth in cervical cancer.
The interferon-responsive protein ISG15 was also attenuated in the double IFITM1/IFITM3 null cells or single IFITM1 single null cells after IFNγ treatment (Fig. 5G). The fact that ISG15 and HLA-B are enriched in the SBP-IFITM1 protein affinity purification after IFNγ treatment (Fig. 8) suggests a co-operative activity exists between the two proteins. Indeed, as HLA-B can interact in situ with IFITM1/3 after IFNγ treatment (Fig. 7), and as ISG15 is a high-confident IFITM1-associated protein using SWATH-IP mass spectrometry (Fig. 8), these data suggest that the two proteins are directly involved in the IFITM1/3 dependent IFNγ response. ISG15 is also a component of an interferon and immune responsive gene cluster that are suppressed by stem cell pluripotent gene product expression [63], suggesting that, like HLA suppression, ISG15 suppression might be co-incident with immune escape. In the case of IFITM1/3 signalling, we do not see defects in ‘free’ monomeric ISG15 in the double IFITM1/IFITM3 null cells, but reductions in the conjugation of higher molecular mass ISG15ylated adducts (Fig. 8D). These data suggest that conjugation of proteins to ISG15 during interferon stimulation might play a coordinated role in the IFITM1/3 dependent immune-tumor cell interactions. Overproduction of ISG15 has been reported previously to stabilize IFITM3 [64], consistent with our data that ISG15 is detected in the SBP-IFITM1 complex (Fig. 8). Ubiquitination of IFITM3 might counteract the stimulatory effect of ISG15 on the anti-viral functions of the protein [65]. How ubiquitination and ISG15ylation regulate IFITM1 and/or IFITM3 in a coordinated fashion is not defined.
These data together provide a novel biochemical pathway relevant for cancer associated functions of IFITM1/3 that correlates with the interferon-responsive nature of IFITM1/3 signalling; they can mediate IFNγ dependent protein production of MHC Class I proteins and ISG15 (Fig. 5), whilst the maintenance of STAT1 protein in response to IFNγ involves by a different signalling mechanism that is IFITM1/3-independent (Fig. 9B). Both antigen presentation and ISG15ylation signalling events are important for anti-viral signalling as well as immune regulation of cancer cells at the immune-cancer synapse [[66], [67], [68], [69], [70], [71]]. Further research will shed light on how reductions in HLA and ISG15ylation can impact on both oncogenic signalling and/or anti-viral activity in response to IFITM1/3 expression.
The following are the supplementary data related to this article.
Dominant proteins whose synthesis is attenuated after IFN-γ treatment in IFITM1 single null cells, and IFITM1/IFITM3 double null cells. SILAC pulse-labelling was performed for 6 h (A and B) in parental SiHa cells; (C and D) in IFITM1 single null cells; and (E and F) in IFITM1/IFITM3 double null cells (Supplementary Table 1). Cells were incubated with IFN-γ for 24 h in B, D and F. Data were plotted as a function of log10 fold change of heavy/light peptide intensities. Triplicates were represented in the x, y, and z- axis. In samples (G-L), representative peptides used for quantification in biological triplicates are highlighted to demonstrate a protein that is induced independent of IFITM1/3 (STAT1, I and EIF1, J) and proteins that are partially IFITM1/3 dependent (IRF; H).
Tryptic peptides identified for HLA-A, HLA-B and HLA-C. Underlined are the peptides identified in HLA-A, HLA-B and HLA-C for parental SiHa, parental SiHa interferon-γ stimulated, IFITM1/IFITM3 double null, IFITM1/IFITM3 double null interferon-γ stimulated, IFITM1 single null, IFITM1 single null interferon-γ stimulated cells. Additionally, highlighted (*) are the unique peptides characteristic for each MHC Class I orthologue.
Examples of IFITM1 protein expression in normal squamous epithelium from the Human Protein Atlas. (A) oesophagus; (B) cervix; and (C) oral mucosa. The brown staining in each panel highlights the predominant IFITM1 protein expression in the basal squamous epithelium cell layer, which is similar to the typical expression pattern we observed in the basal squamous epithelium of the cervix (Fig. 1E). The data is suggestive of IFITM1 stem cell expression pattern in these tissues. The web link to each tissue from the Human Protein Atlas is imbedded in the figure.
Relative quantification values (heavy vs light ratios) in parental SiHa, ifitm1 single null, ifitm1-ifitm3 double null cells untreated or IFN-γ stimulated for 6 and 24 h and pulse labeled in heavy-SILAC media for 6 and 24 h. All samples were processed as biological triplicates. Comparisons (heavy/light) were performed from pulse-labeled newly synthesized protein (heavy) vs total protein amount in the cell (light) before treatment. Each excel spread sheet tab exported from Proteome Discoverer 1.4 shows one condition, from left to right; parental SiHa (6 h); parental SiHa (6 h with IFNγ); IFITM1/IFITM3 null (6 h); IFITM1/IFITM3 -null (6 h with IFNγ; IFITM1 null (6 h); IFITM1-null (6 h with IFNγ; parental SiHa (24 h); parental SiHa (24 h with IFNγ; IFITM1/IFITM3 null (24 h); IFITM1/IFITM3 -null (24 h with IFNγ IFITM1 null (24 h); IFITM1-null (24 h with IFNγ). The data in the multiconsensus report are represented, as in line 1; Accession number, Description (gene name), ΣCoverage (the percent peptide coverage of an identified protein), Σ# Proteins (number of proteins identified in the protein group; introduced is the master protein that is identified by a set of peptides that are not included in any other protein group), Σ# Unique Peptides (number of peptides that are only contained in protein group), Σ# Peptides (number of distinct peptides in protein group), Σ# PSMs (peptide spectrum matches, the total number of identified peptides for the protein),. The line continues with values characterized quantification for each biological replicate (A, B, and C): Area (peak area for any quantified peptide), Heavy/Light (the heavy to light ratio of peak areas), Heavy/Light Count (the number of peptide ratios that were used to calculate a particular protein ratio), Heavy/Light Variability [%] (the variability of the peptide ratios that were used to calculate a particular protein ratio),; then for each replicate were calculated: Score (XCorr score was calculated by Sequest HT search engine for peptide matches); Coverage, # Peptides, and # PSM Three last columns characterize identified protein by its # AAs (the number of amino acids in the protein sequence), MW [kDa] (molecular weight), and calc. pI (calculated value of its isoelectric point). The data in this table was the source for the data in Fig. 5 and Supplementary Fig. 2.
Identified IFITM1 interacting proteins performed in parental SiHa cells by label-free SWATH analysis. The data are summarized as peak name, group (gene name), p-value, fold change (SBP-IFITM1/SBP control), and log10 (fold change). Cells were ectopically transfected under two biological states; (i) with SBP-IFITM1 or SBP-empty vectors for 24 h (columns A to E) with carrier treatment for 24 h or (ii) with SBP-IFITM1 or SBP-empty vectors for 24 h including IFN-γ stimulation for the final 24 h (columns G to K). Samples were lysed and processed for mass spectrometric analysis (SWATH). These two conditions (i and ii) were used to cover the variability inherent in transfection of plasmid DNA into cells whose mRNA expression can induce a general interferon response. Accordingly, the p-values of the IFITM1 associated proteins obtained from biological state ii with IFNγ were of higher significance than those from biological state i (without IFNγ stimulation). The data in this table were used to derive Fig. 8B and C.
Identification of IFITM1 dependent signalling proteins depleting IFITM1 protein using siRNA in parental SiHa cells. Parental SiHa cells transfected with IFITM1-targeted-siRNA or scrambled siRNA for 48 (row 1, columns G to K) and 72 h (row 1, columns A to E). The data acquired from these two biological states were processed in technical triplicates and after SWATH analysis that data are summarized in row 2 as: peak name, group (gene name), p-value, fold change (IFITM1 siRNA/con siRNA), and log10 fold change. The data in this table was used to derive the data in Fig. 9B.
Acknowledgements
This work was supported by the Medical Research Scotland (MGH); Ministry of Education Youth and Sports (MEYS) NPS-I – LO1413 and LM2015089, with Czech Science Foundation (GACR) 18-23773Y (MN, JF) and MH CZ - DRO (MMCI, 00209805), the BBSRC (BB/C511599/1; United Kingdom); The International Centre for Cancer Vaccine Science project is carried out within the International Research Agendas programme of the Foundation for Polish Science cofinanced by the European Union under the European Regional Development Fund.
References
- 1.Takaoka A., Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8(6):907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 2.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 3.Bekisz J., Schmeisser H., Hernandez J., Goldman N.D., Zoon K.C. Human interferons alpha, beta and omega. Growth Factors. 2004;22(4):243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 4.Borden E.C., Sen G.C., Uze G., Silverman R.H., Ransohoff R.M., Foster G.R., Stark G.R. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell J.E., Jr., Kerr I.M., Stark G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 6.Williams B.R. Transcriptional regulation of interferon-stimulated genes. Eur. J. Biochem. / FEBS. 1991;200(1):1–11. doi: 10.1111/j.1432-1033.1991.tb21041.x. [DOI] [PubMed] [Google Scholar]
- 7.Cheon H., Borden E.C., Stark G.R. Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol. 2014;41(2):156–173. doi: 10.1053/j.seminoncol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace T.A., Martin D.N., Ambs S. Interactions among genes, tumor biology and the environment in cancer health disparities: examining the evidence on a national and global scale. Carcinogenesis. 2011;32(8):1107–1121. doi: 10.1093/carcin/bgr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khodarev N.N., Roizman B., Weichselbaum R.R. Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin. Cancer Res. 2012;18(11):3015–3021. doi: 10.1158/1078-0432.CCR-11-3225. an official journal of the American Association for Cancer Research. [DOI] [PubMed] [Google Scholar]
- 10.Weichselbaum R.R., Ishwaran H., Yoon T., Nuyten D.S., Baker S.W., Khodarev N., Su A.W., Shaikh A.Y., Roach P., Kreike B., Roizman B., Bergh J., Pawitan Y., van de Vijver M.J., Minn A.J. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T.H., Schreiber K., Arina A., Khodarev N.N., Efimova E.V., Rowley D.A., Weichselbaum R.R., Schreiber H. Progression of cancer from indolent to aggressive despite antigen retention and increased expression of interferon-gamma inducible genes. Cancer Immun. 2011;11:2. [PMC free article] [PubMed] [Google Scholar]
- 12.Borg D., Hedner C., Gaber A., Nodin B., Fristedt R., Jirstrom K., Eberhard J., Johnsson A. Expression of IFITM1 as a prognostic biomarker in resected gastric and esophageal adenocarcinoma. Biomark Res. 2016;4:10. doi: 10.1186/s40364-016-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gyorffy B., Dietel M., Fekete T., Lage H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int. J. Gynecol. Cancer. 2008;18(6):1215–1233. doi: 10.1111/j.1525-1438.2007.01169.x. [DOI] [PubMed] [Google Scholar]
- 14.Sari I.N., Yang Y.G., Phi L.T., Kim H., Baek M.J., Jeong D., Kwon H.Y. Interferon-induced transmembrane protein 1 (IFITM1) is required for the progression of colorectal cancer. Oncotarget. 2016;7(52):86039–86050. doi: 10.18632/oncotarget.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogony J., Choi H.J., Lui A., Cristofanilli M., Lewis-Wambi J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res.: BCR. 2016;18(1):25. doi: 10.1186/s13058-016-0683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu F., Xie D., Ng S.S., Lum C.T., Cai M.Y., Cheung W.K., Kung H.F., Lin G., Wang X., Lin M.C. IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer Lett. 2015;368(1):135–143. doi: 10.1016/j.canlet.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 17.Bailey C.C., Zhong G., Huang I.C., Farzan M. IFITM-family proteins: the cell's first line of antiviral Defense. Annu. Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia R., Ding S., Pan Q., Liu S.L., Qiao W., Liang C. The C-terminal sequence of IFITM1 regulates its anti-HIV-1 activity. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weston S., Czieso S., White I.J., Smith S.E., Kellam P., Marsh M. A membrane topology model for human interferon inducible transmembrane protein 1. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H., Shi M., Huang I.C., Farzan M., Jung J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13(4):452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1) doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nenutil R., Smardova J., Pavlova S., Hanzelkova Z., Muller P., Fabian P., Hrstka R., Janotova P., Radina M., Lane D.P., Coates P.J., Vojtesek B. Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J. Pathol. 2005;207(3):251–259. doi: 10.1002/path.1838. [DOI] [PubMed] [Google Scholar]
- 24.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doherty M.K., Beynon R.J. Protein turnover on the scale of the proteome. Expert Rev. Proteomics. 2006;3(1):97–110. doi: 10.1586/14789450.3.1.97. [DOI] [PubMed] [Google Scholar]
- 26.Doherty M.K., Hammond D.E., Clague M.J., Gaskell S.J., Beynon R.J. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 2009;8(1):104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Liang Y., Liu L., Shi J., Zhu H.J. Targeted absolute quantitative proteomics with SILAC internal standards and unlabeled full-length protein calibrators (TAQSI) Rapid Commun. Mass Spectrom. 2016;30(5):553–561. doi: 10.1002/rcm.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruger N.J. The Bradford method for protein quantitation. Methods Mol. Biol. 1994;32:9–15. doi: 10.1385/0-89603-268-X:9. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Manza L.L., Stamer S.L., Ham A.J., Codreanu S.G., Liebler D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5(7):1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 31.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 32.Li D., Peng Z., Tang H., Wei P., Kong X., Yan D., Huang F., Li Q., Le X., Li Q., Xie K. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin. Cancer Res. 2011;17(11):3558–3568. doi: 10.1158/1078-0432.CCR-10-2729. an official journal of the American Association for Cancer Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seyfried N.T., Huysentruyt L.C., Atwood J.A., 3rd, Xia Q., Seyfried T.N., Orlando R. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: a membrane proteomics approach. Cancer Lett. 2008;263(2):243–252. doi: 10.1016/j.canlet.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu F., Ng S.S., Chow B.K., Sze J., Lu G., Poon W.S., Kung H.F., Lin M.C. Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J. Neuro-Oncol. 2011;103(2):187–195. doi: 10.1007/s11060-010-0377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang G., Xu Y., Chen X., Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26(4):594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 36.Pan Z., Chen S., Pan X., Wang Z., Han H., Zheng W., Wang X., Li F., Qu S., Shao R. Differential gene expression identified in Uigur women cervical squamous cell carcinoma by suppression subtractive hybridization. Neoplasma. 2010;57(2):123–128. doi: 10.4149/neo_2010_02_123. [DOI] [PubMed] [Google Scholar]
- 37.Zheng W., Zhao Z., Yi X., Zuo Q., Li H., Guo X., Li D., He H., Pan Z., Fan P., Li F., Liao Y., Shao R. Down-regulation of IFITM1 and its growth inhibitory role in cervical squamous cell carcinoma. Cancer Cell Int. 2017;17:88. doi: 10.1186/s12935-017-0456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker C.C., Phelps W.C., Lindgren V., Braun M.J., Gonda M.A., Howley P.M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 1987;61(4):962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phelps W.C., Howley P.M. Transcriptional trans-activation by the human papillomavirus type 16 E2 gene product. J. Virol. 1987;61(5):1630–1638. doi: 10.1128/jvi.61.5.1630-1638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey C.C., Huang I.C., Kam C., Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8(9) doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Everitt A.R., Clare S., McDonald J.U., Kane L., Harcourt K., Ahras M., Lall A., Hale C., Rodgers A., Young D.B., Haque A., Billker O., Tregoning J.S., Dougan G., Kellam P. Defining the range of pathogens susceptible to Ifitm3 restriction using a knockout mouse model. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G., Fenyo D., Neubert T.A. Evaluation of the variation in sample preparation for comparative proteomics using stable isotope labeling by amino acids in cell culture. J. Proteome Res. 2009;8(3):1285–1292. doi: 10.1021/pr8006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thakur A., Hinnebusch A.G. eIF1 loop 2 interactions with met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex. Proc. Natl. Acad. Sci. U. S. A. 2018;115(18):E4159–E4168. doi: 10.1073/pnas.1800938115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weibrecht I., Leuchowius K.J., Clausson C.M., Conze T., Jarvius M., Howell W.M., Kamali-Moghaddam M., Soderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics. 2010;7(3):401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 45.Raykova D., Koos B., Asplund A., Gelleri M., Ivarsson Y., Danielson U.H., Soderberg O. Let there be light! Proteomes. 2016;4(4) doi: 10.3390/proteomes4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huerfano S., Ryabchenko B., Forstova J. Nucleofection of expression vectors induces a robust interferon response and inhibition of cell proliferation. DNA Cell Biol. 2013;32(8):467–479. doi: 10.1089/dna.2012.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X.Q., Li X.N., Liang J.J., Cai X.B., Tao Q., Li Y.X., Qin Q., Xu S.P., Luo T.R. IRF1 up-regulates isg15 gene expression in dsRNA stimulation or CSFV infection by targeting nucleotides −487 to −325 in the 5′ flanking region. Mol. Immunol. 2018;94:153–165. doi: 10.1016/j.molimm.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 48.Narayana S.K., Helbig K.J., McCartney E.M., Eyre N.S., Bull R.A., Eltahla A., Lloyd A.R., Beard M.R. The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 2015;290(43):25946–25959. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheon H., Stark G.R. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U. S. A. 2009;106(23):9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nan J., Wang Y., Yang J., Stark G.R. IRF9 and unphosphorylated STAT2 cooperate with NF-kappaB to drive IL6 expression. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15):3906–3911. doi: 10.1073/pnas.1714102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Lee J., Goh S.H., Song N., Hwang J.A., Nam S., Choi I.J., Shin A., Kim I.H., Ju M.H., Jeong J.S., Lee Y.S. Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am. J. Pathol. 2012;181(1):43–52. doi: 10.1016/j.ajpath.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 53.McGranahan N., Rosenthal R., Hiley C.T., Rowan A.J., Watkins T.B.K., Wilson G.A., Birkbak N.J., Veeriah S., Van Loo P., Herrero J., Swanton C., Consortium T.R. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171(6) doi: 10.1016/j.cell.2017.10.001. (1259–1271 e11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferns D.M., Heeren A.M., Samuels S., Bleeker M.C.G., de Gruijl T.D., Kenter G.G., Jordanova E.S. Classical and non-classical HLA class I aberrations in primary cervical squamous- and adenocarcinomas and paired lymph node metastases. J. Immunother. Cancer. 2016;4:78. doi: 10.1186/s40425-016-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finlan L.E., Nenutil R., Ibbotson S.H., Vojtesek B., Hupp T.R. CK2-site phosphorylation of p53 is induced in DeltaNp63 expressing basal stem cells in UVB irradiated human skin. Cell Cycle. 2006;5(21):2489–2494. doi: 10.4161/cc.5.21.3393. [DOI] [PubMed] [Google Scholar]
- 56.Park H.J., Kuk I.S., Kim J.H., Kim J.H., Song S.J., Choi B.C., Kim B., Kim N.H., Song H. Characterisation of mouse interferon-induced transmembrane protein-1 gene expression in the mouse uterus during the oestrous cycle and pregnancy. Reprod. Fertil. Dev. 2011;23(6):798–808. doi: 10.1071/RD10086. [DOI] [PubMed] [Google Scholar]
- 57.Warren C.J., Griffin L.M., Little A.S., Huang I.C., Farzan M., Pyeon D. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0096579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H.R., Li Y.C., Guo H.Q., Yu L.L., Wu Z., Yin J., Liao G.D., Qu Y.M., Jiang Y., Wang D., Chen W. A cocktail of p16(INK4a) and Ki-67, p16(INK4a) and minichromosome maintenance protein 2 as triage tests for human papillomavirus primary cervical cancer screening. Oncotarget. 2017;8(48):83890–83899. doi: 10.18632/oncotarget.19870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenter G.G., Welters M.J., Valentijn A.R., Lowik M.J., Berends-van der Meer D.M., Vloon A.P., Essahsah F., Fathers L.M., Offringa R., Drijfhout J.W., Wafelman A.R., Oostendorp J., Fleuren G.J., van der Burg S.H., Melief C.J. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 2009;361(19):1838–1847. doi: 10.1056/NEJMoa0810097. [DOI] [PubMed] [Google Scholar]
- 60.Melief C.J., van der Burg S.H. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat. Rev. Cancer. 2008;8(5):351–360. doi: 10.1038/nrc2373. [DOI] [PubMed] [Google Scholar]
- 61.Zwaveling S., Ferreira Mota S.C., Nouta J., Johnson M., Lipford G.B., Offringa R., van der Burg S.H., Melief C.J. Established human papillomavirus type 16-expressing tumors are effectively eradicated following vaccination with long peptides. J. Immunol. 2002;169(1):350–358. doi: 10.4049/jimmunol.169.1.350. [DOI] [PubMed] [Google Scholar]
- 62.Kooi S., Zhang H.Z., Patenia R., Edwards C.L., Platsoucas C.D., Freedman R.S. HLA class I expression on human ovarian carcinoma cells correlates with T-cell infiltration in vivo and T-cell expansion in vitro in low concentrations of recombinant interleukin-2. Cell. Immunol. 1996;174(2):116–128. doi: 10.1006/cimm.1996.0301. [DOI] [PubMed] [Google Scholar]
- 63.Farshchian M., Matin M.M., Armant O., Geerts D., Dastpak M., Nakhaei-Rad S., Tajeran M., Jebelli A., Shahriyari M., Bahrami M., Fallah A., Yaghoobi V., Mirahmadi M., Abbaszadegan M.R., Bahrami A.R. Suppression of dsRNA response genes and innate immunity following Oct4, Stella, and Nanos2 overexpression in mouse embryonic fibroblasts. Cytokine. 2018;106:1–11. doi: 10.1016/j.cyto.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 64.Chesarino N.M., McMichael T.M., Yount J.S. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 2015;11(8) doi: 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yount J.S., Karssemeijer R.A., Hang H.C. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012;287(23):19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassani-Sternberg M., Braunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., Martignoni M.E., Werner A., Hein R., H Busch D., Peschel C., Rad R., Cox J., Mann M., Krackhardt A.M. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 2016;7:13404. doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burks J., Reed R.E., Desai S.D. Free ISG15 triggers an antitumor immune response against breast cancer: a new perspective. Oncotarget. 2015;6(9):7221–7231. doi: 10.18632/oncotarget.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dos Santos P.F., Mansur D.S. Beyond ISGlylation: functions of free intracellular and extracellular ISG15. J. Interf. Cytokine Res. 2017;37(6):246–253. doi: 10.1089/jir.2016.0103. [DOI] [PubMed] [Google Scholar]
- 69.Okumura A., Lu G., Pitha-Rowe I., Pitha P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U. S. A. 2006;103(5):1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Montfoort N., van der Aa E., Woltman A.M. Understanding MHC class I presentation of viral antigens by human dendritic cells as a basis for rational design of therapeutic vaccines. Front. Immunol. 2014;5:182. doi: 10.3389/fimmu.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wahl A., Schafer F., Bardet W., Buchli R., Air G.M., Hildebrand W.H. HLA class I molecules consistently present internal influenza epitopes. Proc. Natl. Acad. Sci. U. S. A. 2009;106(2):540–545. doi: 10.1073/pnas.0811271106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Worrall E.G., Wawrzynow B., Worrall L., Walkinshaw M., Ball K.L., Hupp T.R. Regulation of the E3 ubiquitin ligase activity of MDM2 by an N-terminal pseudo-substrate motif. J. Chem. Biol. 2009;2(3):113–129. doi: 10.1007/s12154-009-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dominant proteins whose synthesis is attenuated after IFN-γ treatment in IFITM1 single null cells, and IFITM1/IFITM3 double null cells. SILAC pulse-labelling was performed for 6 h (A and B) in parental SiHa cells; (C and D) in IFITM1 single null cells; and (E and F) in IFITM1/IFITM3 double null cells (Supplementary Table 1). Cells were incubated with IFN-γ for 24 h in B, D and F. Data were plotted as a function of log10 fold change of heavy/light peptide intensities. Triplicates were represented in the x, y, and z- axis. In samples (G-L), representative peptides used for quantification in biological triplicates are highlighted to demonstrate a protein that is induced independent of IFITM1/3 (STAT1, I and EIF1, J) and proteins that are partially IFITM1/3 dependent (IRF; H).
Tryptic peptides identified for HLA-A, HLA-B and HLA-C. Underlined are the peptides identified in HLA-A, HLA-B and HLA-C for parental SiHa, parental SiHa interferon-γ stimulated, IFITM1/IFITM3 double null, IFITM1/IFITM3 double null interferon-γ stimulated, IFITM1 single null, IFITM1 single null interferon-γ stimulated cells. Additionally, highlighted (*) are the unique peptides characteristic for each MHC Class I orthologue.
Examples of IFITM1 protein expression in normal squamous epithelium from the Human Protein Atlas. (A) oesophagus; (B) cervix; and (C) oral mucosa. The brown staining in each panel highlights the predominant IFITM1 protein expression in the basal squamous epithelium cell layer, which is similar to the typical expression pattern we observed in the basal squamous epithelium of the cervix (Fig. 1E). The data is suggestive of IFITM1 stem cell expression pattern in these tissues. The web link to each tissue from the Human Protein Atlas is imbedded in the figure.
Relative quantification values (heavy vs light ratios) in parental SiHa, ifitm1 single null, ifitm1-ifitm3 double null cells untreated or IFN-γ stimulated for 6 and 24 h and pulse labeled in heavy-SILAC media for 6 and 24 h. All samples were processed as biological triplicates. Comparisons (heavy/light) were performed from pulse-labeled newly synthesized protein (heavy) vs total protein amount in the cell (light) before treatment. Each excel spread sheet tab exported from Proteome Discoverer 1.4 shows one condition, from left to right; parental SiHa (6 h); parental SiHa (6 h with IFNγ); IFITM1/IFITM3 null (6 h); IFITM1/IFITM3 -null (6 h with IFNγ; IFITM1 null (6 h); IFITM1-null (6 h with IFNγ; parental SiHa (24 h); parental SiHa (24 h with IFNγ; IFITM1/IFITM3 null (24 h); IFITM1/IFITM3 -null (24 h with IFNγ IFITM1 null (24 h); IFITM1-null (24 h with IFNγ). The data in the multiconsensus report are represented, as in line 1; Accession number, Description (gene name), ΣCoverage (the percent peptide coverage of an identified protein), Σ# Proteins (number of proteins identified in the protein group; introduced is the master protein that is identified by a set of peptides that are not included in any other protein group), Σ# Unique Peptides (number of peptides that are only contained in protein group), Σ# Peptides (number of distinct peptides in protein group), Σ# PSMs (peptide spectrum matches, the total number of identified peptides for the protein),. The line continues with values characterized quantification for each biological replicate (A, B, and C): Area (peak area for any quantified peptide), Heavy/Light (the heavy to light ratio of peak areas), Heavy/Light Count (the number of peptide ratios that were used to calculate a particular protein ratio), Heavy/Light Variability [%] (the variability of the peptide ratios that were used to calculate a particular protein ratio),; then for each replicate were calculated: Score (XCorr score was calculated by Sequest HT search engine for peptide matches); Coverage, # Peptides, and # PSM Three last columns characterize identified protein by its # AAs (the number of amino acids in the protein sequence), MW [kDa] (molecular weight), and calc. pI (calculated value of its isoelectric point). The data in this table was the source for the data in Fig. 5 and Supplementary Fig. 2.
Identified IFITM1 interacting proteins performed in parental SiHa cells by label-free SWATH analysis. The data are summarized as peak name, group (gene name), p-value, fold change (SBP-IFITM1/SBP control), and log10 (fold change). Cells were ectopically transfected under two biological states; (i) with SBP-IFITM1 or SBP-empty vectors for 24 h (columns A to E) with carrier treatment for 24 h or (ii) with SBP-IFITM1 or SBP-empty vectors for 24 h including IFN-γ stimulation for the final 24 h (columns G to K). Samples were lysed and processed for mass spectrometric analysis (SWATH). These two conditions (i and ii) were used to cover the variability inherent in transfection of plasmid DNA into cells whose mRNA expression can induce a general interferon response. Accordingly, the p-values of the IFITM1 associated proteins obtained from biological state ii with IFNγ were of higher significance than those from biological state i (without IFNγ stimulation). The data in this table were used to derive Fig. 8B and C.
Identification of IFITM1 dependent signalling proteins depleting IFITM1 protein using siRNA in parental SiHa cells. Parental SiHa cells transfected with IFITM1-targeted-siRNA or scrambled siRNA for 48 (row 1, columns G to K) and 72 h (row 1, columns A to E). The data acquired from these two biological states were processed in technical triplicates and after SWATH analysis that data are summarized in row 2 as: peak name, group (gene name), p-value, fold change (IFITM1 siRNA/con siRNA), and log10 fold change. The data in this table was used to derive the data in Fig. 9B.