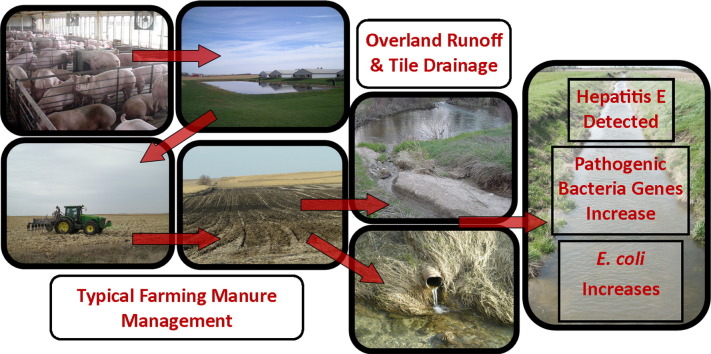
Abstract
Manure application is a source of pathogens to the environment. Through overland runoff and tile drainage, zoonotic pathogens can contaminate surface water and streambed sediment and could affect both wildlife and human health. This study examined the environmental occurrence of gene markers for livestock-related bacterial, protozoan, and viral pathogens and antibiotic resistance in surface waters within the South Fork Iowa River basin before and after periods of swine manure application on agricultural land. Increased concentrations of indicator bacteria after manure application exceeding Iowa's state bacteria water quality standards suggest that swine manure contributes to diminished water quality and may pose a risk to human health. Additionally, the occurrence of HEV and numerous bacterial pathogen genes for Escherichia coli, Enterococcus spp., Salmonella sp., and Staphylococcus aureus in both manure samples and in corresponding surface water following periods of manure application suggests a potential role for swine in the spreading of zoonotic pathogens to the surrounding environment. During this study, several zoonotic pathogens were detected including Shiga-toxin producing E. coli, Campylobacter jejuni, pathogenic enterococci, and S. aureus; all of which can pose mild to serious health risks to swine, humans, and other wildlife. This research provides the foundational understanding required for future assessment of the risk to environmental health from livestock-related zoonotic pathogen exposures in this region. This information could also be important for maintaining swine herd biosecurity and protecting the health of wildlife near swine facilities.
Keywords: Zoonotic pathogens, Hepatitis E virus, Indicator bacteria, Animal agriculture, Water quality, Environmental health
Graphical abstract
Highlights
-
•
Assessment of manure application as a source of pathogens to the environment.
-
•
Overland runoff and tile drainage facilitates pathogen transport to surface waters.
-
•
Detected hepatitis E virus in surface water following manure application.
-
•
Increased pathogen gene detections and indicator bacteria concentrations post-manure.
-
•
Manure application can potentially impair water quality and environmental health.
1. Introduction
Runoff from land application of animal manure is one possible pathway of transport of bacterial, fungal, protozoan, and viral pathogens to surface waters (Khaleel et al., 1980, Vanotti et al., 2007). Animal manure harbors not only animal-specific pathogens (e.g. bovine viral diarrhea virus), but also zoonotic pathogens (e.g. hepatitis E virus, Campylobacter jejuni) capable of infecting humans (Gordoncillo et al., 2013, Haack et al., 2015, United States Environmental Protection Agency, 2013, Ziemer et al., 2010). Many of these pathogens are able to survive, persist, and move through the agricultural landscape. Agricultural runoff whether via overland flow (Davies et al., 2004) or through the outflow of tile-drain systems (Jamieson et al., 2002, Joy et al., 1998, Wilkes et al., 2014) facilitates pathogen transport and can consequently contaminate and significantly impair the water quality of adjacent surface waters (Crane et al., 1983, Meinhardt et al., 1996). Pathogens may not only be transported via stream water, but also subsequently may be adsorbed into streambed sediment and later resuspended when the sediment becomes disturbed with animal movement or increased flow via storm events (Goss and Richards, 2008).
Surface waters and bed sediment can be reservoirs of pathogens that affect both wildlife and human health. In an animal agriculture-dominated region in south Alberta, Canada, researchers found a higher prevalence of zoonotic pathogens in surface waters used for drinking, irrigation, and recreational purposes and also a higher incidence of gastrointestinal illness linked to Escherichia coli O157:H7 and Salmonella than in neighboring non-agricultural regions, suggesting a link between animal agriculture, contaminated surface waters, and human health (Jokinen et al., 2012). Current swine manure management practices utilize injection or tillage to incorporate manure to potentially reduce risks of nutrients or pathogens being directly transported to surface water in runoff after rainfall (Dell et al., 2011, Sterk et al., 2013). Still, this case study along with similar research suggests that both animal and human health are potentially at risk from ongoing manure management practices and subsequent surface water contamination (Gordoncillo et al., 2013, Haack et al., 2016, Haack et al., 2015, United States Environmental Protection Agency, 2013, Ziemer et al., 2010). For example, genes indicative of potentially zoonotic Shiga toxin-producing E. coli, enterohemorrhagic E. coli, and the Enterococcus esp. gene, which is more frequently expressed in isolates from human patients with bacteremia and urinary tract infections (Shankar et al., 1999), were found in surface waters and sediment up to 25 days after a swine manure spill (Haack et al., 2015).
Of additional concern when examining the role of manure in regard to surface water quality and spread of zoonotic pathogens is understanding the potential risk associated with understudied and emerging zoonotic pathogens, such as hepatitis E virus (HEV). Human HEV infections can be serious with a 1% mortality rate and increased mortality rate of 27% among pregnant women (Chau et al., 2006, Kumar et al., 2004). Several HEV outbreaks in developing countries have been linked to waste-contaminated drinking water (Krawczynski, 1993, Satou and Nishiura, 2007) or contaminated food sources (Satou and Nishiura, 2007). Such infections, however, are rare in developed countries (Christou and Kosmidou, 2013, Kase et al., 2009). Among the four HEV genotypes, only genotype 3 has been reported in the United States and has been found in swine and other domestic animals and also in wild rodents, deer, and wild boar (Christou and Kosmidou, 2013, Satou and Nishiura, 2007, Takahashi et al., 2004, Tei et al., 2003, Yugo and Meng, 2013).
Swine are clinically unaffected carriers of HEV and this virus appears to be prevalent among U.S. swine farms and has been detected in swine and in swine manure and waste lagoons (Huang et al., 2002, Kase et al., 2009, Kasorndorkbua et al., 2005). A previous swine study from Iowa documented HEV genotype 3 in approximately 68% of storage pits and 38% of waste lagoons (Kasorndorkbua et al., 2005). This particular genotype is linked to human HEV infections in developed countries (Christou and Kosmidou, 2013, Meng, 2003) and is of particular concern because researchers have found almost genetically indistinguishable swine and human strains of HEV (Kase et al., 2009, Meng et al., 1997). A study examining the prevalence of HEV among midwestern United States swine herds noted that there was sequence homology between swine HEV and clinical isolates from human HEV cases (Meng et al., 1997) suggesting a potential role for swine in human transmission of this virus. In addition, cross-species infection (e.g. swine to human, deer to human, swine to monkey) indicates that animal strains of HEV not only pose a zoonotic risk to humans but also a health risk to other wildlife (Christou and Kosmidou, 2013, Meng et al., 1998, Meng et al., 1997, Yugo and Meng, 2013).
While HEV has been detected in both swine feces and in stored liquid waste that is often applied to agricultural lands (Gentry-Shields et al., 2015, Kase et al., 2009), few studies have been conducted to date to ascertain the environmental occurrence of swine HEV (Gentry-Shields et al., 2015, Kasorndorkbua et al., 2005). Kasorndorkbua et al. (2005) did not detect HEV in 28 surface water samples taken near swine farms during late summer and autumn. The objective of this study was to determine the presence of HEV and other livestock-related bacterial, protozoan, and viral pathogens (Supplemental Tables 1 and 2) in relation to periods of swine manure application in a basin (South Fork Iowa River) with extensive swine production (around 840,000 hogs). This research will provide the foundational understanding required for any future assessment of the risk to environmental health from HEV and other animal agricultural-related pathogen exposures in this region. Such risk could include spread from swine to human, spread from swine to other wildlife (e.g. deer and rodents), or spread among swine farms. This information could be important for maintaining swine herd biosecurity and protecting the health of wildlife near swine facilities.
2. Materials and methods
2.1. Sample sites and sample collection
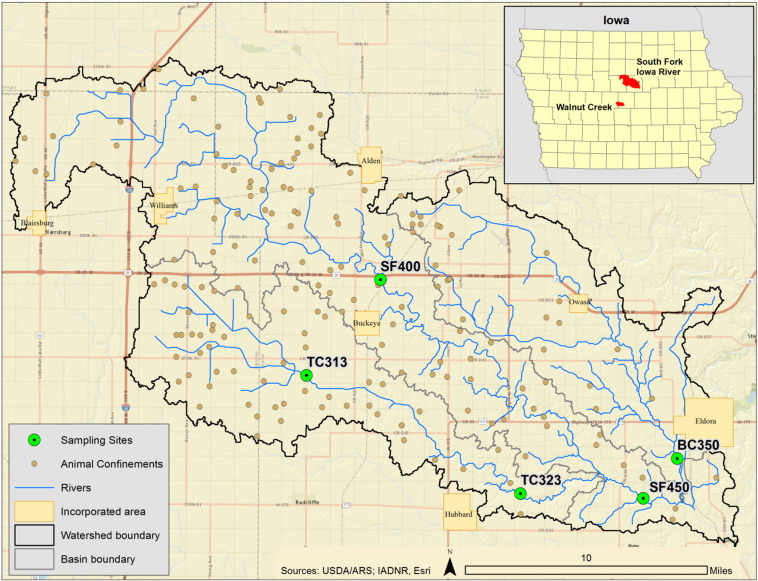
To determine the environmental prevalence of HEV and other microbiological contaminants, 22 water samples from six stream sites in central Iowa were collected (Fig. 1 , Table 1 ). The sampling network included five stream sites located in Beaver Creek, Tipton Creek, and the South Fork Iowa River within the South Fork Iowa River basin and one out-of-basin control site in the Walnut Creek basin where no swine were raised. The South Fork Iowa River basin was selected because: (1) it is an area of intense swine production (roughly 840,000 hogs in the 78,000 ha watershed), (2) about 85% of the basin is in row-crop agriculture and 30 to 60% of the basin receives 93 to 186 m3 ha− 1 of swine manure annually, (3) the basin has an extensive network of subsurface drainage (tiles) to artificially enhance water drainage providing a rapid transport mechanism for contaminants from the land surface to corresponding streams, and (4) a network of streamflow gages were available throughout the basin to provide important ancillary data on streamflow (Tomer et al., 2008). The sampling network in the South Fork Iowa River basin consisted of three tributary and two main-stem sampling sites (Fig. 1). Walnut Creek near Ames, Iowa, was selected as a nearby (within 80 km) out-of-basin control as this basin has similar soils and land use (e.g. tile-drained row crops) but no livestock production and no known swine manure application (Tomer et al., 2003).
Fig. 1.
Diagram of South Fork Iowa River basin sites.
IASF400 (SF400)-South Fork River near Buckeye, Iowa; (TC313)- Tipton Creek near D Avenue in Buckeye, Iowa; (TC323) -Tipton Creek East near Hubbard, Iowa; (SF450)-South Fork Iowa River NE near New Providence, Iowa; (BC350)- Beaver Creek near 250th Street in Eldora, Iowa. The subset shows the location of the South Fork Iowa River basin (top) in reference to the Walnut Creek control (bottom) basin where no swine were raised. Swine confinement operations as of February 2014.
Table 1.
Station IDs, station locations, and environmental parameters at the time of sampling.
| Sampling date | USGS station ID | Station name | Water temperature (°C) | Discharge m3 s− 1 | Dissolved oxygen (mg L− 1) | pH water | Specific conductance (μS cm− 1) |
|---|---|---|---|---|---|---|---|
| 8/09/2011 | 5451110 | SF400 | 27 | 0.096 | 8.8 | 7.9 | 607 |
| 5451140 | TC313 | 26.6 | 0.057 | 13.4 | 8 | 677 | |
| 5451148 | TC323 | 22.4 | 0.184 | 8.5 | 7.9 | 563 | |
| 5451210 | SF450 | 22.4 | 0.623 | 7.9 | 8 | 539 | |
| 5451260 | BC350 | 23.8 | 0.224 | 11.2 | 7.8 | 546 | |
| 8/10/2011 | 5471009 | WC | 15.8 | 0.003 | 8.4 | 7.9 | 801 |
| 11/03/2011 | 5451110 | SF400 | 6.3 | 0.113 | 7.6 | 7.4 | 580 |
| 5451140 | TC313 | 7.9 | 0.014 | 15.1 | 7.8 | 661 | |
| 5451148 | TC323 | 6.2 | 0.108 | 13.1 | 7.8 | 580 | |
| 5451210 | SF450 | 5.6 | 0.190 | 10.6 | 7.3 | 547 | |
| 5451260 | BC350 | 5 | 0.099 | 10.4 | 6.7 | 510 | |
| 3/07/2012 | 5451110 | SF400 | 2.6 | 0.663 | 13 | 7.7 | 582 |
| 5451140 | TC313 | 10.2 | 0.283 | 14 | 7.8 | 615 | |
| 5451148 | TC323 | 7.6 | 0.496 | 13.2 | 7.9 | 648 | |
| 5451210 | SF450 | 6.6 | 1.133 | 11.5 | 7.6 | 581 | |
| 5451260 | BC350 | 7.9 | 0.357 | 12.8 | 7.8 | 587 | |
| 3/08/2012 | 5471009 | WC | 5.3 | 0.011 | 14.8 | 8 | 730 |
| 4/15/2012 | 5451110 | SF400 | 17.2 | 0.906 | 9.9 | 8 | 719 |
| 5451140 | TC313 | 16.6 | 0.736 | 12.6 | 8 | 683 | |
| 5451148 | TC323 | 15.3 | 1.218 | 10.6 | 8 | 609 | |
| 5451210 | SF450 | 13.7 | 2.633 | 8.7 | 7.6 | 532 | |
| 5451260 | BC350 | 13.5 | 1.586 | 9.2 | 7.7 | 604 |
SF400-South Fork River near Buckeye, Iowa; TC313- Tipton Creek near D Avenue in Buckeye, Iowa; TC323-Tipton Creek East in Hubbard, Iowa; SF450-South Fork Iowa River NE near New Providence, Iowa; BC350- Beaver Creek near 250th Street in Eldora, Iowa; and WC- Walnut Creek near 510th Avenue in Ames, Iowa. Water samples were not collected at Walnut Creek for the November 2011 sampling because of a dry streambed or for the April 2012 sampling due to sampling logistics.
Water-quality field measurements were made by use of standard USGS methods (Gibs et al., 2007) (Table 1). Daily mean streamflow was calculated at the sampling sites over the course of the study using methodology described by Tomer et al. (2008). Hydrographs were created for the sampling sites for the date of sampling and seven preceding days (Supplemental Fig. 1).
Stream water samples were collected in the South Fork Iowa River basin at four time points: before fall manure application (August 9, 2011), after fall manure application (November 3, 2011), before spring manure application (March 7, 2012), and after spring manure application (April 15, 2012). Sampling dates were chosen to capture the first precipitation event causing a rise in the hydrograph after a majority of the manure had been applied to the basin. Water samples were collected from the control site at Walnut Creek on August 10, 2011, and March 8, 2012. Samples were not collected at Walnut Creek for the November 2011 sampling because of a dry streambed or for the April 2012 sampling due to sampling logistics. Water grab samples (n = 22) for enrichment PCR analyses for 19 bacterial antibiotic resistance and pathogen gene markers (bacterial gene markers, BGMs) were collected from the centroid of flow. Water samples (n = 22) for qPCR analyses for HEV and other bacterial, protozoan, and viral targets were collected using a portable pumping system connected to a glass wool filtration with a pre-filter for concentration of bacteria and viruses (Lambertini et al., 2008, Millen et al., 2012). The mean sample volume filtered was 337 L and ranged from 201 to 837 L. A negative control of the glass wool filtration equipment consisted of sterile phosphate buffered saline (1 × PBS) and was performed in the field during every sampling period (four total) and all were negative for all qPCR targets. Bed sediment samples were collected at the same location as the water samples during each sampling period and consisted of the top 2–3 cm of bed material composted from multiple points in the depositional zones (i.e. areas where the bed consisted of finer grained materials). Bed sediment samples were analyzed for indicator bacteria and presence of bacterial pathogen genes. Additionally, hog slurry (manure) samples (n = 2; 1-L in sterile amber glass) were collected from two swine producers in South Fork basin during active 2011 fall manure application. While all sites were visited, fecal samples from wild deer (n = 3) were only found near three sampling sites in March 2012. Deer feces were collected in the riparian zone of the stream adjacent to water sampling locations. Fecal droppings were composited by sampling site in a 1-L sterile amber glass jar and homogenized, and then sub-sampled into three sterile specimen cups. All samples were placed on ice immediately and shipped overnight to Michigan Bacteriological Research Laboratory (MI-BaRL; USGS Michigan Water Science Center, Lansing, MI) for enumeration and enrichment PCR analysis and to the Agricultural Research Service (Marshfield, WI) for qPCR analysis.
2.2. Enumeration of indicator bacteria
Water (50 mL, 10 mL, 3 mL, and 1 mL), sediment and manure (1 and 3 mL of a 1:10, 1:100, and 1:1000 dilution) samples were plated on mFC (fecal coliforms), modified mTEC (E. coli), mEI (enterococci), and Baird Parker (Staphylococcus spp.) for enumeration. Water samples (50 mL) were filtered in the laboratory using sterilized equipment and standard methods for membrane filtration (American Public Health Association, 1998) and subsequent enrichment. Laboratory blanks consisting of sterile PBS (1 ×) were processed with all environmental samples and were absent of contamination.
2.3. Enrichment PCR to detect bacterial gene markers
To determine if selected bacterial gene markers (BGMs) were in each sample, multiple types of growth enrichment were conducted. After indicator bacteria counts were recorded from membrane filtration enumeration, all growth (target and non-target) from the 50-mL filter (or maximum volume) of each enumeration was subsequently used to detect BGMs from target genera which grew upon that media (Supplemental Table 3). Additionally, enrichments were prepared for Campylobacter and Salmonella with a filter through which 50 mL of sample water had been passed, or directly with 1 g sediment or solid waste, or 1 mL liquid waste.
Filters containing colony growth (target and non-target) from mFC (Shigella enrichment), modified mTEC (E. coli enrichment), mEI (enterococci enrichment), and Baird Parker (Staphylococcus enrichment) media were aseptically transferred into sterile PBS, agitated for 15 min, and then centrifuged at 3400 rpm for 20 min at 4 °C to form a pellet. For Bolton broth (Campylobacter enrichment) and Rappaport Vassiliadis R10 broth (Salmonella enrichment), membrane filters were aseptically removed from broth cultures and the remaining enrichment was centrifuged as above to form a pellet. For all samples, the supernatant was decanted and the pellet was resuspended in 1 mL of 20% glycerol/0.5 × PBS. The resulting glycerol stocks of all enrichments were stored at − 70 °C for further analysis.
To determine if BGMs were detected in target and non-target growth from each medium's enrichment (including enumeration media), glycerol stocks were thawed and homogenized and 100 μL (Gram negative cultures) or 400 μL (Gram positive cultures) was used for DNA extraction (Qiagen DNeasy DNA extraction kit; Qiagen, Valencia, CA). Extracted DNA was stored at − 20 °C until PCR analysis was performed.
PCR was used to detect the presence of 19 BGMs from target populations from enrichment media (Supplemental Table 1) using methods referenced in Supplemental Table 1 and elsewhere (Haack et al., 2013). Standard quality control checks of PCR reactions included running a positive and negative PCR control with every 20 reactions. Blanks of DNA extraction methods were also performed for every group of extractions processed. Throughout the study at least one sample that was previously determined to produce a negative PCR result for each virulence gene was spiked with positive-control DNA and repeated to confirm a positive result, and the absence of PCR inhibition (36, 37). All laboratory QC samples produced expected results throughout the study (data not shown).
2.4. Microbiological analyses – qPCR
Using the methods described in Millen et al. (2012), glass wool filters and pre-filters were eluted and the eluent further concentrated by polyethylene glycol. The two final concentrated sample volumes (FCSV) for each pair of glass wool filter and pre-filter composing a sample were combined (mean = 5.9 mL, range 2.4–11.0 mL, n = 22) and stored at − 80 °C until analysis.
An FCSV aliquot of 280 μL was extracted for nucleic acids by first using the freeze-thaw extraction procedure for Cryptosporidium oocyst DNA (Di Giovanni and LeChevallier, 2005) followed by extraction with QIAamp DNA blood mini kit and buffer AVL (Qiagen, Valencia, CA) (Borchardt et al., 2012). The extraction final volume was 150 μL. Every sample was checked for qPCR inhibition and, if necessary, inhibition was mitigated following the methods described by Gibson et al. (2012).
qPCR was conducted for genes specific for the following pathogens: HEV, Influenza A virus, six human viruses (adenovirus, enterovirus, GI norovirus, GII norovirus, hepatitis A virus (HAV), rotavirus), eight bovine viruses (bovine viral diarrhea virus Types 1 and 2 (BVDV1 and BVDV2), group A rotavirus, group C rotavirus, enterovirus, coronavirus, adenovirus, polyomavirus), four bacteria (C. jejuni, Salmonella spp., enterohemorrhagic E. coli, Mycobacterium avium subsp. paratuberculosis, and two protozoa (Giardia lamblia and Cryptosporidium parvum). qPCR standard curve parameters and references for the primers and hydrolysis probes are reported in Supplemental Table 2. qPCR and reverse-transcription (RT) procedures were the same as those described in Borchardt et al. (2012) with one update; the reverse transcriptase was SuperScript® III (Life Technologies, Grand Island, NY). All qPCR assays included no-template controls for extraction, RT, and PCR steps and throughout the study these controls were negative (i.e., no fluorescence). Sources of the positive controls and their enumeration for creating standard curves are described in the Supplemental material.
HEV amplicons from qPCR-positive samples were sequenced to confirm identity. Amplified DNA was visualized by gel electrophoresis and the band size corresponding to HEV was excised from the agarose gel and purified with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Purified DNA was cloned with pGEM-T Easy Vector System (Promega, Madison, WI) and sequencing reactions of the cloned plasmids were conducted in both directions with BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Products were submitted for sequencing to the University of Wisconsin-Madison Biotechnology Center DNA Sequencing Facility (Madison, WI). Consensus sequences were constructed and aligned with Lasergene (DNASTAR, Madison, WI). Sequence identifications were confirmed using BLAST (National Center for Biotechnology Information, Bethesda, MD).
2.5. Statistical analyses
All statistical analyses were conducted in R statistical software using the vegan package (Oksanen et al., 2015, R Core Team, 2015). Mann Whitney U tests were used to determine if there was a statistically significant difference between indicator bacteria (fecal coliforms, E. coli, enterococci, and Staphylococcus spp.) between the Walnut Creek (control site) and the South Fork Iowa River sites. Mann Whitney U tests were also performed to determine the statistical significance between HEV concentration pre- and post- manure application and between main-stem and tributary sites. Paired Wilcoxon signed-rank tests were performed to assess whether there was a significant difference between fecal coliforms, E. coli, enterococci, and Staphylococcus spp. concentrations between pre-manure samplings; between post-manure samplings; and between pre- and post-manure applications. Fisher's exact test was performed to determine whether there was a significant difference in the number (presence/absence) of pathogen gene of BGMs detected pre- and post- manure application. Spearman's rank correlation coefficient (rho) was used to measure the statistical correlation between environmental variables and indicator bacteria concentration, number of BGMs detected, and HEV, C. jejuni, and bovine polyomavirus concentrations. Relations and correlations were considered significant when p < 0.05. Box plots were constructed in R using the reshape package (Wickham, 2007) and the ggplot2 (Wickham, 2009) and cowplot (Wilke, 2015) graphics packages.
3. Results
3.1. Detection of bacterial gene markers in fecal sources
Ten of the 19 BGMs (53%) were detected in at least one of the two swine manure samples that were collected from farms within the South Fork Iowa River basin (Supplemental Table 4). In addition to the eaeA (E. coli intimin marker), ddl faecium (E. faecium marker), and ddl faecalis (E. faecalis maker) genes, the Enterococcus esp. (human marker) and hirae (bovine marker) and E. coli STII (swine marker) were detected in both (100%) swine manure samples. All BGMs detected in swine manure samples were subsequently detected in either water and/or sediment samples from subsequent samplings within the South Fork Iowa River basin. Both swine manure samples were positive for HEV RNA with high concentrations of 3.5 × 107 and 2.8 × 106 genomic copies L− 1 (Supplemental Table 5). One of the manure samples was also positive for Influenza A at a concentration of 3.4 × 103 genomic copies L− 1 .
Few BGMs (n = 5) were detected in the three fecal deer pellet samples except for Salmonella invA which was detected in all (100%) of the samples (Supplemental Table 4). All of the BGMs present in deer fecal samples were also detected in at least one of the water and/or sediment samples collected within the South Fork Iowa River basin. The deer fecal samples were also positive for HEV RNA at concentrations of 7.0 × 104 genomic copies g− 1 (Supplemental Table 5). Concentrations of HEV within manure and deer fecal samples were on average 4–5 logs greater than concentrations found in surface waters of the South Fork Iowa River basin.
3.2. Temporal variation in indicator bacteria concentrations
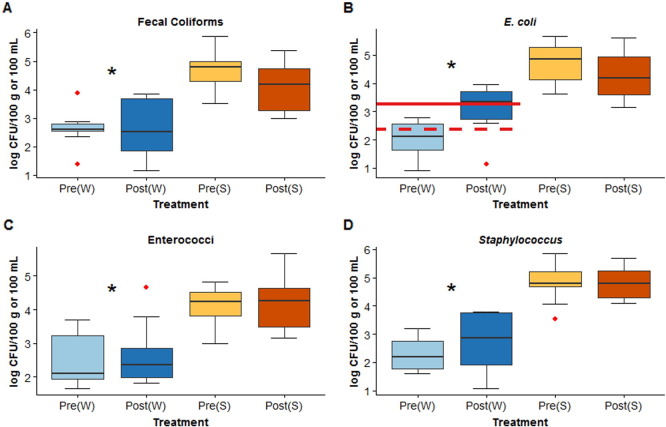
Fecal coliforms, E. coli, enterococci, and Staphylococcus spp. were enumerated in water and sediment samples at all sites pre-and post-manure application (Fig. 2 , Supplemental Table 6). There was no statistically significant difference between the concentrations of these indicator bacteria in water or sediment between both the pre-manure samplings (August 2011 and March 2012) or between both the post-manure samplings (November 2011 and April 2012). Conversely, mean concentrations of fecal coliforms, E. coli, enterococci, and Staphylococcus spp. in all water samples from the South Fork Iowa River basin sites were significantly different between pre- and post- manure application (p = 0.013, p = 0.013, p = 0.009, and p = 0.006 respectively). In water, the largest concentrations of fecal coliforms, E. coli, and Staphylococcus were found in April post-manure application. In sediment, indicator bacteria concentrations were not significantly different between the two pre-manure samplings, between the two post-manure samplings, or between the pre-and post-manure application.
Fig. 2.
Box plot of fecal coliform (A), E. coli (B), enterococci (C), and Staphylococcus (D) concentration in water (log CFU 100 mL− 1) and sediment (log CFU 100 g− 1) from the five sites in the South Fork Iowa River basin. Blue boxes denote pre-manure (August 2011 and March 2012; n = 10) and post manure (November 2011 and April 2012; n = 10) concentrations in water. Brown boxes denote pre-manure (August 2011 and March 2012; n = 10) and post manure (November 2011 and April 2012; n = 10) concentrations in sediment. For E. coli (B), the dashed red line indicates the State of Iowa E. coli water quality standard of 2.35 × 102 CFU 100 mL− 1 in Class A1 (primary contact) waters. The solid red line indicates the E. coli water quality standard of 2.88 × 103 CFU 100 mL− 1 for Class A2 (secondary contact) waters. The lower and upper borders of the box correspond to first and third quartiles (or 25th and 75th percentiles). The whiskers extend to 1.5 × inter-quartile range. Data outside of the whiskers are plotted are outliers and plotted as red points for each box plot. Asterisks indicate statistical significance (p < 0.05; paired Wilcoxon rank-sum test) between the two paired groups (pre-and post-manure).
Considering the State of Iowa's water quality standard for E. coli of 2.35 × 102 CFU 100 mL− 1 for Class A1 (primary contact) waters and 2.88 × 103 CFU 100 mL− 1 for Class A2 (secondary contact) waters (Iowa Department of Natural Resources, 2015a), 80% of South Fork Iowa River basin samples (n = 8) from the August 2011 (pre) and November 2011 (post) and 100% of samples (n = 5) from the April 2012 (post) sampling exceeded the E. coli standard level for Class A1 waters (Fig. 2). Additionally, 80% of the April 2012 (post) water samples (n = 4) exceeded the bacteria standard level for Class A2 waters. Samples (n = 2) collected at the control site at Walnut Creek exceeded the Class A1 E. coli standard level for both the August 2011 and March 2012 (pre-manure application) water samples (Supplemental Table 6). Beaver Creek, Tipton Creek, Walnut Creek, and the South Fork Iowa River all include regions that are classified as Class A1 and Class A2 waters (Iowa Department of Natural Resources, 2015b).
Concentrations of indicator bacteria were similar (μ = 0.54, σ = 0.49 log difference) between the Walnut Creek (control) and South Fork Iowa River basin sites (Supplemental Table 6). The only significant difference between these two basins was observed for enterococci concentrations with the control site having statistically higher enterococci levels (p = 0.04). Indicator bacteria concentrations were only measured at Walnut Creek pre-manure application (August 2011 and March 2012) because of the lack of flow during November 2011 and sampling logistics during April 2012.
3.3. Temporal and spatial variation in pathogen detection
The human viruses enterovirus, GII norovirus, HAV, and rotavirus; the bovine viruses BVDV1, BVDV2, group A rotavirus, group C rotavirus, enterovirus, and coronavirus; M. avium; the protozoan G. lamblia, and the BGM for the enterotoxigenic E. coli ST (human marker) were not detected in any water or sediment samples. Additionally the vanB gene (antibiotic resistance marker) and GI norovirus were both detected at the control site, but not within the South Fork Iowa River basin (Supplemental Tables 4 and 5).
All other BGMs were detected in the main-stem or tributaries of the South Fork Iowa River (Supplemental Table 4). The most frequently detected BGMs in water samples (n = 20) were E. coli eaeA (90%), E. faecium ddl (90%), E. faecalis ddl (65%), and E. coli stx1 (55%) (Table 2 ). It is important to note that the E. coli eaeA BGM (attachment marker) was detected in both (pre) samples and the E. faecium ddl and E. faecalis ddl BGM (species markers) were detected during the March (pre) sampling from the control basin indicating a likely background population of these markers (Supplemental Table 4). The E. coli STII BGM (swine marker) was not detected in any of the water samples, but was found in a single sediment sample from one of the tributaries during post-manure fall (November 2011) application. Additionally, C. jejuni/coli 16SrRNA, E. hirae bov, E. coli STII, E. coli stx2e, and Staphylococcus aureus mecA BGMs were only detected in water and sediment samples from the South Fork Iowa River basin post manure application (Supplemental Table 4). There was an overall significant (p = 0.005) difference in the number of BGMs detected between the pre-manure application (52 BGMs) compared to the post-manure application (90 BGMs) samples. This difference was most notable between the March 2012 pre application and the April 2012 post application samplings (p = 0.01) where a 12% increase (additional 22 BGMs) in the number of BGMs detected was observed. Additionally, there was a significantly (p = 0.03) higher proportion of BGMs detected when water samples exceeded the Class A2 bacteria standard level of 2.88 × 103 E. coli CFU 100 mL− 1, when compared to samples that were below this standard.
Table 2.
Occurrence frequencies (%) of bacterial pathogen genes detected in surface waters by P/A PCR in the South Fork River Basin. Three pathogens genes (vanB, STII, STh) were not detected in any water samples at any of South Fork River Basin sites. The pathogen gene vanB was detected in both water and sediment samples at the WC site during August 2011. The STII gene was detected once in sediment at the TC313 site during November 2011 after fall manure application. Dashes indicate non-detect.
| Sampling date |
C. jejuni or coli |
Enterococcus |
E. coli |
Salmonella |
S. aureus |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S | esp human | vanA | ddl faecalis | ddl faecium | hirae bov | 16S | LTIIa bov | rfb O157 | stx1 | stx2 | stx2e | eaeA | invA | spvC | femA | mecA | |
| 8/09/2011 (pre manure) n = 5 | – | – | – | 20 | 80 | – | 100 | 20 | 20 | 40 | 20 | – | 100 | 20 | 20 | 40 | – |
| 11/03/2011 (post manure) n = 5 | 40 | 20 | – | 60 | 100 | 60 | 100 | 20 | – | – | – | 20 | 80 | 20 | – | – | 20 |
| 3/07/2012 (pre manure) n = 5 | – | – | 20 | 100 | 100 | – | 100 | – | 20 | 80 | – | – | 80 | 40 | – | – | – |
| 4/15/2012 (post manure) n = 5 | 40 | – | 20 | 80 | 80 | 80 | 100 | – | 40 | 100 | 60 | – | 100 | – | – | 60 | 60 |
| All dates (n = 20) | 20 | 5 | 10 | 65 | 90 | 35 | 100 | 10 | 20 | 55 | 20 | 5 | 90 | 20 | 5 | 25 | 20 |
By qPCR, HEV (45%, n = 20), C. jejuni (35%, n = 20), and bovine polyomavirus (25%, n = 20) were the most frequently detected pathogens in the South Fork Iowa River basin (Table 3 , Supplemental Table 5). While C. jejuni and bovine polyomavirus were detected more frequently in the post-manure application samples compared to the pre-manure application samples, this trend was not statistically significant.
Table 3.
Detection frequencies (%) and concentrations (genomic copies L− 1) of the three most commonly detected pathogens by qPCR in the South Fork River Basin. Dashes indicate non-detect.
| Sampling date | HEV |
Campylobacter jejuni |
Bovine polyomavirus |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Frequency | Median | Maximum | Frequency | Median | Maximum | Frequency | Median | Maximum | |
| 8/09/2011 (pre manure n = 5) | 0 | – | – | 20 | – | 2.60 × 101 | 0 | – | – |
| 11/03/2011 (post manure n = 5) | 80 | 9.00 × 100 | 2.99 × 102 | 80 | 2.00 × 100 | 5.20 × 101 | 0 | – | – |
| 3/07/2012 (pre manure n = 5) | 40 | – | 2.50 × 101 | 20 | – | 1.00 × 100 | 20 | – | 3.00 × 100 |
| 4/15/2012 (post manure n = 5) | 60 | 4.20 × 101 | 2.38 × 102 | 20 | – | 1.00 × 101 | 80 | 2.20 × 101 | 1.79 × 102 |
| All dates (n = 20) | 45 | 35 | 25 | ||||||
3.4. Spatial and temporal variation in HEV detection
HEV RNA was detected in 45% of samples (n = 20) collected from the South Fork Iowa River basin (Table 3). This virus was not detected in the two samples collected from the control site at Walnut Creek (Supplemental Table 5). Within the South Fork Iowa River basin, 25% of the main-stem river (SF450 and SF400) samples and 58% of tributary (BC350, TC313, and TC323) were positive for HEV RNA. Prior to manure application (August 2011 and March 2012), HEV detection rates were similar between the main-stem (25%) and tributary (17%) samples. After manure application (November 2011 and April 2012), HEV detections were significantly different (p = 0.01) between main-stem (25%) and tributary (100%) samples. The occurrence and concentration of HEV in surface waters increased significantly (p = 0.04) following manure application with HEV being detected in 20% of samples prior to manure application and in 80% of samples after manure application (Table 3).
For all environmental samples collected, an attempt was made to further identify the positive HEV detections by sequencing. Five of the nine HEV-positive surface water samples could be sequenced; amplicon quantity was insufficient for successful cloning and sequencing of the other four surface water samples. There was 100% sequence similarity of the HEV RNA recovered from all deer fecal, all manure, and the five surface water samples. However, the amplicon sequences were only 70 base pairs long and within a highly-conserved region of the HEV genome (Jothikumar et al., 2006) so it was not possible to discern whether the HEV genotypes in the three sample sources (i.e. deer, swine, and stream) were identical. There was no attempt to culture HEV-positive samples to determine infectivity.
3.5. Relation between indicator bacteria and BGMs, HEV, and bovine polyomavirus
The concentrations of fecal coliforms, E. coli, and Staphylococcus were all significantly correlated with each other (Table 4 ). There were significant (rho > 0.68) correlations between the concentration of the indicator bacteria (fecal coliforms, E. coli, and Staphylococcus) and the number of BGMs detected in the surface waters of the South Fork Iowa River basin (Table 4). As with the number of BGMs detected, there was a significant correlation (rho > 0.62) between the concentration of indicator bacteria (fecal coliforms, E. coli and Staphylococcus) and the concentration of bovine polyomavirus. Additionally, there was a significant relation (rho = 0.69) between the number of BGMs detected and the concentration of bovine polyomavirus. There were no significant correlations between HEV and the concentration of any of the indicator bacteria, number of BGMs detected, or the concentration of bovine polyomavirus (Table 4).
Table 4.
Spearman rank correlation coefficient (rho) between indicator bacteria concentration (fecal coliforms, E. coli, enterococci, and Staphylococcus spp.), number of bacterial gene markers (BGMs), hepatitis E virus (HEV) concentration, Campylobacter jejuni concentration, and bovine polyomavirus concentration in the South Fork Iowa River basin. Bold values indicate a statistically significant relation where p < 0.05.
| Target | n | E. coli | Enterococci | Staphylococcus | BGMs | HEV | C. jejuni | Bovine polyomavirus |
|---|---|---|---|---|---|---|---|---|
| Fecal coliforms | 20 | 0.98 | 0.42 | 0.68 | 0.69 | 0.23 | 0.47 | 0.62 |
| E. coli | 20 | 0.41 | 0.71 | 0.68 | 0.16 | 0.35 | 0.68 | |
| Enterococci | 20 | 0.73 | 0.41 | 0.43 | 0.27 | 0.35 | ||
| Staphylococcus | 20 | 0.71 | 0.30 | 0.19 | 0.75 | |||
| BGMs | 20 | 0.36 | 0.37 | 0.69 | ||||
| HEV | 20 | 0.52 | 0.25 | |||||
| C. jejuni | 20 | − 0.09 |
3.6. Environmental influence on indicator bacteria, BGMs, HEV, and bovine polyomavirus
Stream flows during the August (fall pre-manure) and November 2011 (fall post-manure) samplings were much lower than flows during the March (spring pre-manure) and April 2012 (spring post-manure) samplings (Supplemental Fig. 1). The August 2011 sample was collected during a period of stable to decreasing flow conditions with total precipitation of 23 mm in the 7 days preceding sampling and 21 mm on the day of sampling. The November 2011 sample was collected during a period of minor increasing flow conditions and directly following the first rain event (14 mm rainfall) after a substantial manure application. However, there was only a total of 1 mm of rain over the preceding and two weeks and the rain event would likely not have produced much overland flow. The increase in streamflow is likely due to rain falling directly into the steam. While there could have been a contribution of flow from tiles in the basin, no tiles near the sampling were observed to be flowing at the time.
The March 2012 sample was collected during a period of generally decreasing flow conditions. Streamflow does appear a bit variable on the site hydrographs (Supplemental Fig. 1) and is due to early spring snowmelt conditions with 24 mm precipitation in the 7 days preceding sampling . High streamflow was recorded during the April 2012 sampling following a storm event (35 mm rainfall) with flow rate doubling from that of the previous day. Rainfall in March and April generally coincides during periods of high soil moisture content, and therefore, are likely to generate more runoff and drainage than rainfall occurring in August or November. This increased rainfall during April sampling coupled with high streamflow documents a “run-off event” resulting in subsequent increased BGM occurrence in both the water and sediment and increased concentrations of HEV and bovine polyomavirus. Despite seasonal differences in streamflow there were few statistically significant correlations (p < 0.05) between the measured environmental parameters (daily mean streamflow, water temperature, discharge, dissolved oxygen, pH, and specific conductance) and indicator bacteria, bacterial gene markers, HEV and Campylobacter jejuni (Table 5 ). However, there were significant correlations between the detected concentrations of bovine polyomavirus and daily mean streamflow (rho = 0.65) and discharge (rho = 0.71).
Table 5.
Spearman rank correlation coefficient (rho) between indicator bacteria (fecal coliforms, E. coli, enterococci, and Staphylococcus spp.), array of bacterial gene markers (BGMs), hepatitis E virus (HEV), and Campylobacter jejuni, bovine polyomavirus with measured environmental parameters for South Fork Iowa River basin sites.
Bold values indicate a statistically significant relation where p < 0.05.
| Target | n | Daily mean flow | Water temperature | Discharge | Dissolved oxygen | pH | Specific conductance |
|---|---|---|---|---|---|---|---|
| Fecal coliforms | 20 | 0.14 | 0.11 | 0.09 | − 0.39 | − 0.18 | − 0.23 |
| E. coli | 20 | 0.16 | 0.18 | 0.14 | − 0.47 | − 0.10 | − 0.16 |
| Enterococci | 20 | 0.06 | − 0.55 | 0.14 | 0.02 | − 0.50 | − 0.16 |
| Staphylococcus | 20 | 0.27 | − 0.21 | 0.37 | − 0.19 | − 0.13 | 0.07 |
| Array of BGMs | 20 | 0.31 | − 0.21 | 0.35 | 0.23 | − 0.08 | 0.21 |
| HEV | 20 | 0.16 | − 0.36 | 0.09 | 0.38 | − 0.19 | 0.11 |
| C. jejuni | 20 | − 0.12 | − 0.29 | − 0.19 | 0.21 | − 0.59 | − 0.52 |
| Bovine polyomavirus | 20 | 0.65 | 0.25 | 0.71 | − 0.20 | 0.26 | 0.37 |
4. Discussion
The occurrence of numerous BGMs and significantly-increased frequency and concentration of HEV in stream water after swine-manure application and runoff generating rain indicates a link between swine manure management and zoonotic pathogens to the environment. In two previous studies, HEV in surface waters adjacent to animal-based agriculture was detected once at a single site (Gentry-Shields et al., 2015, Kasorndorkbua et al., 2005). However, HEV was detected more frequently in the current study suggesting a higher incidence of the virus in swine farms in this region. There are several other factors that may have contributed to the increased detection of HEV in the South Fork Iowa River basin including 1) differences in manure management, 2) the intensity of hog production with 30 to 60% of the basin receiving manure, and 3) that the basin is extensively tile drained facilitating rapid transport of these pathogens to streams. HEV was not detected in the Walnut Creek control basin also suggesting that swine are a source of this virus to the South Fork Iowa River watershed. Additionally, deer fecal samples were positive for HEV RNA suggesting that deer may also be a potential source of HEV to the environment.
The E. coli STII (swine marker) BGM and HEV were detected in swine farm manure, but were not always detected in adjacent surface waters. Although Haack et al. (2015) found that the STII BGM increased in concentration and persisted in surface water following a swine manure spill, this BGM was also found upstream of the manure spill suggesting that STII may be endemic in this stream dominated by a multitude of swine sources. The current study supports this finding by detecting the STII BGM once in the sediment post-manure application, but not in a similar broad distribution that was found after a swine manure spill (Haack et al., 2015).
The E. hirae (bovine) BGM was detected in both swine manure samples and frequently in sediment and water samples post manure applications. Cattle are present in this watershed particularly at TC313 and TC323 where in some cases the cattle have direct access to the stream. However, this particular marker has been detected previously in stream water following a swine manure spill suggesting that this marker is not solely specific to cattle manure (Haack et al., 2015, Oksanen et al., 2015). Furthermore, the E. coli LTIIa (bovine) BGM which is another bovine indicator gene was not detected in the swine manure samples. Likewise, the esp. (human-infection associated Enterococcus) BGM which was detected in a single post-manure water sample and in both swine manure samples has also been previously detected in swine and other manure (Haack et al., 2015, Willems et al., 2001).
Elevated concentrations of indicator bacteria after manure application exceeding the Iowa's state bacteria water quality standard indicate that swine manure contributes to diminished water quality and that risk to human health may be increased from waters exceeding this standard. These E. coli and enterococci concentrations are consistent with previous reports on this watershed (Tomer et al., 2008) where investigators found the highest concentrations of E. coli and enterococci in summer and lower concentrations in winter. Other researchers have documented that manure application can result in dramatic increases in water quality impairments (Dean and Foran, 1992). However, concentrations of culturable E. coli and enterococci in the same range in the Walnut Creek control site (where no animal agriculture existed) and the study sites and detections of Enterococcus-specific BGMs in Walnut Creek suggest additional sources are present (i.e. humans and wildlife).
Indicator bacteria and selected BGMs were detected in the control Walnut Creek basin where there was no known swine or livestock manure applications taking place. However, there are wildlife in this basin that may be a vector of indicator bacteria and BGMs. Previous studies found that background levels of fecal coliforms could be attributed to wildlife and changing environmental conditions (Farnleitner et al., 2010, Gessel et al., 2004, Somarelli et al., 2007). Additionally, there was a single detection of GI norovirus within this basin that potentially suggests septic influence. Septic systems may also have influenced concentrations of indicator bacteria (Sterk et al., 2013).
Although there were no significant correlations between daily streamflow or discharge and HEV and BGMs, increased detection and concentration of HEV and the number of BGMs detected post-manure application suggests that runoff and tile input are important transport mechanisms of both HEV and BGMs to corresponding surface waters. This finding is consistent with the hydrology of the South Fork Iowa River which is dominated by subsurface drainage that artificially enhances the transport of water to corresponding streams. Approximately 90% of baseflow in this basin is from subsurface drainage (Green et al., 2006). Overland flow after storm events is also a potential transport pathway and both surface and subsurface flow contribute to the transport of E. coli and Enterococcus (Tomer et al., 2010). Thus, it is likely that both subsurface drainage and overland flow contributed to the transport of HEV and other manure-related constituents to adjacent surface water.
Increased storm event flows coupled with increased flow from seasonal snowmelt prior to the March sampling (pre manure), may have resulted in the resuspension of bacteria containing BGMs in the sediment and increased detection of BGMs during this pre-manure sampling. Several BGMs including E. faecalis ddl, E. faecium ddl, E. hirae bov, E. coli stx1, Salmonella invA, and S. aureus femA were routinely detected in the sediment pre- and post-manure application suggesting that the sediment may be a sink and reservoir for BGMs as has been documented previously (Haack et al., 2015). High stream flows and storm events after the April 2012 manure application flushed freshly applied manure-derived constituents into the environment resulting in this study's highest pathogen concentration and number of BGMs detected.
Indicator bacteria concentrations for fecal coliforms, E. coli, and Staphylococcus and the number of BGMs detected were significantly correlated suggesting that these constituents have similar transport pathways and transport rates in the environment. Although bovine polyomavirus concentration was related to increased indicator bacteria concentrations and BGM detections, HEV concentration was not correlated to the detection or concentrations of these constituents. There is little documentation of virus transport in tile drainage, but Wilkes et al. (2014) reported the detection of coliphage in water from tile-drained watersheds. Previous studies have shown that various viruses, including hepatitis A, can be transported through soil to groundwater (Borchardt et al., 2012, De Serres et al., 1999) and from surface to groundwater (Bradford et al., 2013).
5. Conclusion
This study detected several antibiotic resistance and pathogen gene markers from bacteria (Table 2, Supplemental Table 4) that can pose mild to serious health risks to swine, humans, and wildlife were detected including Shiga-toxin producing E. coli, C. jejuni, enterococci, and S. aureus (based on detection of mecA and femA genes). The spread of HEV and other zoonotic pathogens in the environment is of concern to both animal and human health. Manure management is linked to pathogen load and indirectly influences pathogen survival and transport through the environment (Goss and Richards, 2008). Application of fresh manure containing bacterial and viral pathogens to fertilize agricultural land can potentially impair groundwater, surface water quality and consequently influence environmental and human health. Further research is needed to: 1) better understand the persistence and survival of manure-related zoonotic pathogens in water and sediment, 2) assess the risk to animals and humans from manure-related zoonotic pathogens, and 3) ascertain manure management techniques that can reduce zoonotic pathogen load to the environment.
Acknowledgments
This study was funded by the Toxic Substances Hydrology Program, U.S. Geological Survey. We thank Kevin Cole (USDA); Jessica Garrett and Shannon Meppelink (USGS Iowa Water Science Center; Iowa City, IA); Heather Johnson (USGS Michigan-Ohio Water Science Center; Lansing, MI); and Hana Millen, Jordan Gonnering, and Jackson Borchardt (Marshfield) for sample collection and processing assistance. We thank Donna Francy (USGS Michigan-Ohio Water Science Center; Columbus, OH) and Dale Griffin (USGS; St. Petersburg, FL) for their technical comments. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Editor: D. Barcelo
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.scitotenv.2016.05.123.
Appendix A. Supplementary data
Supplementary material.
References
- American Public Health Association . 21st ed. American Public Health Association; Washington D.C.: 1998. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- Borchardt M.A., Spencer S.K., Kieke B.A., Jr., Lambertini E., Loge F.J. Viruses in nondisinfected drinking water from municipal wells and community incidence of acute gastrointestinal illness. Environ. Health Perspect. 2012;120:1272. doi: 10.1289/ehp.1104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford S.A., Morales V.L., Zhang W., Harvey R.W., Packman A.I., Mohanram A. Transport and fate of microbial pathogens in agricultural settings. Crit. Rev. Environ. Sci. Technol. 2013;43:775–893. [Google Scholar]
- Chau T.N., Lai S.T., Tse C., Ng T.K., Leung V.K.S., Lim W. Epidemiology and clinical features of sporadic hepatitis E as compared with hepatitis A. Am. J. Gastroenterol. 2006;101:292–296. doi: 10.1111/j.1572-0241.2006.00416.x. [DOI] [PubMed] [Google Scholar]
- Christou L., Kosmidou M. Hepatitis E virus in the Western world—a pork-related zoonosis. Clin. Microbiol. Infect. 2013;19:600–604. doi: 10.1111/1469-0691.12214. [DOI] [PubMed] [Google Scholar]
- Crane S.R., Moore J.A., Grismer M.E., Miner J.R. Bacterial pollution from agricultural sources: a review. Trans. ASAE. 1983;26:858–866. [Google Scholar]
- Davies C.M., Ferguson C.M., Kaucner C., Krogh M., Altavilla N., Deere D.A. Dispersion and transport of Cryptosporidium oocysts from fecal pats under simulated rainfall events. Appl. Environ. Microbiol. 2004;70:1151–1159. doi: 10.1128/AEM.70.2.1151-1159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Serres G., Cromeans T.L., Levesque B., Brassard N., Barthe C., Dionne M. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J Infect Dis. 1999;179:37–43. doi: 10.1086/314565. [DOI] [PubMed] [Google Scholar]
- Dean D.M., Foran M.E. The effect of farm liquid waste application on tile drainage. J. Soil Water Conserv. 1992;47:368–369. [Google Scholar]
- Dell C.J., Meisinger J.J., Beegle D.B. Subsurface application of manures slurries for conservation tillage and pasture soils and their impact on the nitrogen balance. J. Environ. Qual. 2011;40:352–361. doi: 10.2134/jeq2010.0069. [DOI] [PubMed] [Google Scholar]
- Di Giovanni G.D., LeChevallier M.W. Quantitative-PCR assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 2005;71:1495–1500. doi: 10.1128/AEM.71.3.1495-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnleitner A.H., Ryzinska-Paier G., Reischer G.H., Burtscher M.M., Knetsch S., Kirschner A.K. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 2010;109:1599–1608. doi: 10.1111/j.1365-2672.2010.04788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry-Shields J., Myers K., Pisanic N., Heaney C., Stewart J. Hepatitis E virus and coliphages in waters proximal to swine concentrated animal feeding operations. Sci. Total Environ. 2015;505:487–493. doi: 10.1016/j.scitotenv.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessel P.D., Hansen N.C., Goyal S.M., Johnston L.J., Webb J. Persistence of zoonotic pathogens in surface soil treated with different rates of liquid pig manure. Appl. Soil Ecol. 2004;25:237–243. [Google Scholar]
- Gibs J., Wilde F.D., Heckathorn H. Use of multiparameter instruments for routine field measurements. In: Wilde F.D., editor. Field Measurements: US Geological Survey Techniques of Water-Resources Investigations. Vol. 9. 2007. [Google Scholar]
- Gibson K.E., Schwab K.J., Spencer S.K., Borchardt M.A. Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 2012;46:4281–4291. doi: 10.1016/j.watres.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Gordoncillo M.J.N., Donabedian S., Bartlett P.C., Perri M., Zervos M., Kirkwood R. Isolation and molecular characterization of vancomycin-resistant Enterococcus faecium from swine in Michigan, USA. Zoonoses Public Health. 2013;60:319–326. doi: 10.1111/zph.12008. [DOI] [PubMed] [Google Scholar]
- Goss M., Richards C. Development of a risk-based index for source water protection planning, which supports the reduction of pathogens from agricultural activity entering water resources. J. Environ. Manag. 2008;87:623–632. doi: 10.1016/j.jenvman.2006.12.048. [DOI] [PubMed] [Google Scholar]
- Green C.H., Tomer M.D., Di Luzio M., Arnold J.G. Hydrologic evaluation of the soil and water assessment tool for a large tile-drained watershed in Iowa. Trans. ASABE. 2006;49:413–422. [Google Scholar]
- Haack S.K., Fogarty L.R., Stelzer E.A., Fuller L.M., Brennan A.K., Isaacs N.M. Geographic setting influences Great Lakes beach microbiological water quality. Environ. Sci. Technol. 2013;47:12054–12063. doi: 10.1021/es402299a. [DOI] [PubMed] [Google Scholar]
- Haack S.K., Duris J.W., Kolpin D.W., Fogarty L.R., Johnson H.E., Gibson K.E. Genes indicative of zoonotic and swine pathogens are persistent in stream water and sediment following a swine manure spill. Appl. Environ. Microbiol. 2015;81:3430–3441. doi: 10.1128/AEM.04195-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack S.K., Duris J.W., Kolpin D.W., Focazio M.J., Meyer M.T., Johnson H.E. Contamination with bacterial zoonotic pathogen genes in US streams influenced by varying types of animal agriculture. Sci. Total Environ. 2016;563:340–350. doi: 10.1016/j.scitotenv.2016.04.087. [DOI] [PubMed] [Google Scholar]
- Huang F.F., Haqshenas G., Guenette D.K., Halbur P.G., Schommer S.K., Pierson F.W. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J. Clin. Microbiol. 2002;40:1326–1332. doi: 10.1128/JCM.40.4.1326-1332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iowa Department of Natural Resources . Environmental Protection Commission; 2015. Iowa Administrative Code. 61. [Google Scholar]
- Iowa Department of Natural Resources . Environmental Services Division; 2015. Surface Water Classification. [Google Scholar]
- Jamieson R.C., Gordon R.J., Sharples K.E., Stratton G.W., Madani A. Movement and persistence of fecal bacteria in agricultural soils and subsurface drainage water: a review. Can. Biosyst. Eng. 2002;44:1–9. [Google Scholar]
- Jokinen C.C., Edge T.A., Koning W., Laing C.R., Lapen D.R., Miller J. Spatial and temporal drivers of zoonotic pathogen contamination of an agricultural watershed. J. Environ. Qual. 2012;41:242–252. doi: 10.2134/jeq2011.0203. [DOI] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T.L., Robertson B.H., Meng X.J., Hill V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods. 2006;131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Joy D.M., Lee H., Reaume C.M., Whiteley H.R., Zelin S. Microbial contamination of subsurface tile drainage water from field applications of liquid manure. Can. Agric. Eng. 1998;40:153–160. [Google Scholar]
- Kase J., Correa M., Sobsey M. Detection and molecular characterization of swine hepatitis E virus in North Carolina swine herds and their faecal wastes. J. Water Health. 2009;7:344–357. doi: 10.2166/wh.2009.137. [DOI] [PubMed] [Google Scholar]
- Kasorndorkbua C., Opriessnig T., Huang F.F., Guenette D.K., Thomas P.J., Meng X.-J. Infectious swine hepatitis E virus is present in pig manure storage facilities on United States farms, but evidence of water contamination is lacking. Appl. Environ. Microbiol. 2005;71:7831–7837. doi: 10.1128/AEM.71.12.7831-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleel R., Reddy K.R., Overcash M.R. Transport of potential pollutants in runoff water from land areas receiving animal wastes: a review. Water Res. 1980;14:421–436. [Google Scholar]
- Krawczynski K. Hepatitis E. Hepatology. 1993;17:932–941. [PubMed] [Google Scholar]
- Kumar A., Beniwal M., Kar P., Sharma J.B., Murthy N.S. Hepatitis E in pregnancy. Int. J. Gynaecol. Obstet. 2004;85:240–244. doi: 10.1016/j.ijgo.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Lambertini E., Spencer S.K., Bertz P.D., Loge F.J., Kieke B.A., Borchardt M.A. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 2008;74:2990–2996. doi: 10.1128/AEM.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt P.L., Casemore D.P., Miller K.B. Epidemiologic aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiol. Rev. 1996;18:118–136. doi: 10.1093/oxfordjournals.epirev.a017920. [DOI] [PubMed] [Google Scholar]
- Meng X. Xeno-transplantation. Springer; 2003. Swine hepatitis E virus: cross-species infection and risk in xenotransplantation; pp. 185–216. [DOI] [PubMed] [Google Scholar]
- Meng X.-J., Purcell R.H., Halbur P.G., Lehman J.R., Webb D.M., Tsareva T.S. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. U. S. A. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.-J., Halbur P.G., Shapiro M.S., Govindarajan S., Bruna J.D., Mushahwar I.K. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 1998;72:9714–9721. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen H.T., Gonnering J.C., Berg R.K., Spencer S.K., Jokela W.E., Pearce J.M. Glass wool filters for concentrating waterborne viruses and agricultural zoonotic pathogens. J. Vis. Exp. 2012 doi: 10.3791/3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O'Hara R.B. 2015. Vegan: Community Ecology Package. [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Satou K., Nishiura H. Transmission dynamics of hepatitis E among swine: potential impact upon human infection. BMC Vet. Res. 2007;3:9. doi: 10.1186/1746-6148-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar V., Baghdayan A.S., Huycke M.M., Lindahl G., Gilmore M.S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarelli J.A., Makarewicz J.C., Sia R., Simon R. Wildlife identified as major source of Escherichia coli in agriculturally dominated watersheds by BOX A1R-derived genetic fingerprints. J. Environ. Manag. 2007;82:60–65. doi: 10.1016/j.jenvman.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Sterk A., Schijven J., de Nijs T., de Roda Husman A.M. Direct and indirect effects of climate change on the risk of infection by water-transmitted pathogens. Environ. Sci. Technol. 2013;47:12648–12660. doi: 10.1021/es403549s. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Kitajima N., Abe N., Mishiro S. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology. 2004;330:501–505. doi: 10.1016/j.virol.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Tei S., Kitajima N., Takahashi K., Mishiro S. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362:371–373. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- Tomer M.D., Meek D.W., Jaynes D.B., Hatfield J.L. Evaluation of nitrate nitrogen fluxes from a tile-drained watershed in central Iowa. J. Environ. Qual. 2003;32:642–653. doi: 10.2134/jeq2003.6420. [DOI] [PubMed] [Google Scholar]
- Tomer M.D., Moorman T.B., Rossi C.G. Assessment of the Iowa River's South Fork watershed: part 1. Water quality. J. Soil Water Conserv. 2008;63:360–370. [Google Scholar]
- Tomer M.D., Wilson C.G., Moorman T.B., Cole K.J., Heer D., Isenhart T.M. Source-pathway separation of multiple contaminants during a rainfall-runoff event in an artificially drained agricultural watershed. J. Environ. Qual. 2010;39:882–895. doi: 10.2134/jeq2009.0289. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency . US Environmental Protection Agency Office of Water; Washington, DC: 2013. Literature Review of Contaminants in Livestock and Poultry Manure and Implications for Water Quality. [Google Scholar]
- Vanotti M.B., Szogi A.A., Hunt P.G., Millner P.D., Humenik F.J. Development of environmentally superior treatment system to replace anaerobic swine lagoons in the USA. Bioresour. Technol. 2007;98:3184–3194. doi: 10.1016/j.biortech.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Wickham H. Reshaping data with the reshape package. J. Stat. Softw. 2007;21:1–20. [Google Scholar]
- Wickham H. New York; 2009. ggplot2: Elegant graphics for data analysis: Springer. [Google Scholar]
- Wilke C.O. 2015. Cowplot: Streamlined Plot Theme and Plot Annotations for ggplot2. [Google Scholar]
- Wilkes G., Brassard J., Edge T.A., Gannon V., Gottschall N., Jokinen C.C. Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired-agricultural watersheds under controlled and conventional tile drainage management. Appl. Environ. Microbiol. 2014;80:3708–3720. doi: 10.1128/AEM.00254-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems R.J., Homan W., Top J., van Santen-Verheuvel M., Tribe D., Manzioros X. Variant esp gene as a marker of a distinct genetic lineage of vancomycin resistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357:853–855. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- Yugo D.M., Meng X.-J. Hepatitis E virus: foodborne, waterborne and zoonotic transmission. Int. J. Environ. Res. Public Health. 2013;10:4507–4533. doi: 10.3390/ijerph10104507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemer C.J., Bonner J.M., Cole D., Vinje J., Constantini V., Goyal S. Fate and transport of zoonotic, bacterial, viral, and parasitic pathogens during swine manure treatment, storage, and land application. J. Anim. Sci. 2010;88:E84–E94. doi: 10.2527/jas.2009-2331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.