Abstract
An integrated reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) microdevice which consists of microbead-assisted RNA purification and RT-LAMP with real-time monitoring by a miniaturized optical detector was demonstrated. The integrated RT-LAMP microdevice includes four reservoirs for a viral RNA sample (purified influenza A viral RNA or lysates), a washing solution (70% ethanol), an elution solution (RNase-free water), and an RT-LAMP cocktail, and two chambers (a waste chamber and an RT-LAMP reaction chamber). The separate reservoirs for a washing solution, an elution solution, and an RT-LAMP cocktail were designed with capillary valves for stable storage. Three influenza A virus strains (A/H1N1, A/H3N2, and A/H5N1) were used for RNA templates, and RT-LAMP primer sets were designed to detect hemagglutinin (HA) and conserved M gene. Sequential sample flow to the microbeads for RNA purification was achieved by centrifugal force with optimization of capillary valves and a siphon channel. Furthermore, the purified RNA solution was successfully isolated from the waste solution by changing the rotational direction, and combined with the RT-LAMP cocktail in the RT-LAMP reaction chamber for target gene amplification. Total process from the sample injection to the result was completed in 47 min. Influenza A H1N1 virus was confirmed on the integrated RT-LAMP microdevice even with 10 copies of viral RNAs, which revealed 10-fold higher sensitivity than that of a conventional RT-PCR. Subtyping and specificity test of influenza A H1N1 viral lysates were also performed and clinical samples were successfully genotyped to confirm influenza A virus on our proposed integrated microdevice.
Keywords: Centrifugal microdevice, Influenza A virus, Integrated microdevice, Reverse transcriptase loop-mediated ampliciation, Real-time fluorescent detection, Sample pretreatment
Highlights
-
•
An integrated microdevice cosistng of RNA purification and RT-LAMP with real-time monitoring was demonstrated.
-
•
Sequential sample transportation and purified RNA separation were automatically controlled by centrifugal force and optimized microchannels.
-
•
Influenza viruses were isothermally amplified and the products were identified by using a miniaturized fluorescence detector.
-
•
Limit of detection for influenza A H1N1 was 10 copies.
-
•
Clinical samples were successfully analyzed on the microdevice.
1. Introduction
Influenza A virus, one of the zoonotic pathogen, have been rampant periodically throughout the world for centuries. In every year, about 500 million of human infections and 0.25–0.5 million of victims are generated in worldwide from respiratory diseases caused by influenza A viruses (Arias et al., 2009, Medina and Garcia-Sastre, 2011). Pandemic influenza A H1N1 in 2009 generated from swine in Mexico was created by multiple reassortment events (Vijaykrishna et al., 2010). It caused about 18,000 of human victims and was diagnosed in more than 214 nations for about 17 months from April, 2009 to August, 2010 according to the latest update from World Health Organization (WHO) (2009). Thus, early diagnosis of influenza A virus is of paramount importance to prevent human victims as well as economic damages (Lazcka et al., 2007). Among a number of diagnostic tools, the genetic analysis based on reverse transcriptase polymerase chain reaction (RT-PCR) has been considered most sensitive, specific and accurate (Hoffmann et al., 2001). Typically, the PCR method requires three distinct temperature for denature, annealing, and extension for thermal cycling. However, the precise temperature control inevitably increases the cost and bulkiness of the instrumentation, which hinders the on-site genetic analysis capability (Asiello and Baeumner, 2011, Parida et al., 2008). The ramping rate for transitioning between the three temperature zones takes time leading to elongating PCR process. Moreover, the enzyme activity can be reduced as the repeated thermal cycling proceeds continuously. To overcome these limitations of the conventional PCR, isothermal amplification methods such as loop-mediated isothermal amplification (LAMP) (Notomi et al., 2000), nucleic acid sequence-based amplification (NASBA) (Compton, 1991), rolling circle amplification (RCA) (Lizardi et al., 1998), helicase-dependent amplification (HDA) (Vincent et al., 2004), and recombinase polymerase amplification (RPA) (Piepenburg et al., 2006, Rohrman and Richards-Kortum, 2012, Shin et al., 2013) have been recently developed (Morisset et al., 2008). Isothermal amplification employs one mild temperature with specifically designed primer sets to boost up the quantity of the target gene. Among isothermal amplification methods, LAMP uses six primers and a DNA polymerase with strand displacement reaction at 60–65 °C within 1 h (Notomi et al., 2000). Since six primer sets of LAMP recognize eight distinct sequence on the target gene, the specificity of the gene amplification is available. Since a simple water bath or a heating block can substitute a thermal cycler for temperature control, a miniaturized genetic analysis system is more feasible for on-site viral detection (Parida et al., 2008). Regarding the detection of influenza virus, the RNA template is generally targeted and the reverse transcription should be performed as a first step. To this end, reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) was also developed by adding reverse transcriptase enzymes under the identical reaction conditions of LAMP (Mori and Notomi, 2009). Using the RT-LAMP, various epidemics and epizootics such as a severe acute respiratory syndrome (SARS) (Thai et al., 2004), influenza A virus (Ito et al., 2006), and foot and mouth disease virus (FMDV) (Dukes et al., 2006) were identified.
On the other hand, microfluidic technology has demonstrated its high performance in the chemical and biological analysis with low sample consumption, high speed, high integration, automation and portability. Thus, the combination of the microfluidic device with the RT-LAMP content can provide an advanced pathogen analysis methodology (Chang et al., 2013, Hataoka et al., 2004). Fang et al. (2011) reported a PDMS–glass hybrid microdevice for RT-LAMP with a reflected optical sensor to detect multiplex influenza A viruses. Wang et al. (2011) demonstrated an RT-LAMP microdevice which integrates a magnetic bead mediated RNA purification. In this platform, a nervous necrosis virus was analyzed within 60 min and the detection sensitivity was 100-fold higher than a conventional RT-PCR. Full integration of RNA purification by a Flinders Technology Associates (FTA) membrane, an RT-LAMP reaction, and real-time fluorescence detection on a chip was presented to identify HIV particles by Liu et al. (2011). However, the previously reported RT-LAMP microdevices necessitates intermittent manual operation for sample loading, and complex tubing systems for fluidic control, which unavoidably cause the contamination issues during RT-LAMP and reduce the data reproducibility (Chen et al., 2010, Oakley et al., 2009). Thus, an advanced microdevice, which integrates all the necessary steps and is operated automatically without complicated tube networks and human intervention, is desired for more reliable point-of-care pathogen detection.
In this context, the centrifugal microdevice can meet the above requirements. Utilizing the centrifugal force and the microfluidic physics, the loading, splitting, mixing, and separation of the sample solution can be controlled. Simplicity, automation, user-friendliness, low power consumption of the centrifugal microsystem improves the capability of portability, reproducibility, low cost and high-throughput analysis without contamination problems (Cho et al., 2007, Lafleur et al., 2010, Madou et al., 2006, Kazarine et al., 2012). Thus, a variety of biochemical applications on the centrifugal microdevice platform including cell lysis and PCR (Kellogg et al., 2000), ion sensor (Grumann et al., 2005), alcohol assay (Steigert et al., 2006), and immunoassay have been reported (Haeberle et al., 2006). We also developed the centrifugal sample pretreatment microsystem by incorporating silica microbeads on a chip (Jung et al., 2013). As for the pathogen detection applications, Lutz et al. (2011) presented a centrifugal lab-on-a-foil platform for RPA based Staphylococcus aureus identification. Despite of the high sensitivity and rapidity, the DNA extraction step was performed on the off-chip basis. Hoehl et al. (2014) showed a centrifugal LabTube platform to automate DNA purification and LAMP amplification to detect verotoxin-producing Escherichia coli (VTEC). Although all the steps proceeded with reduced human intervention, the used volume of the reagents was equivalent to that of the commercial kit, rendering the rapid and efficient amplification difficult, and the colorimetric detection was limited for quantitative analysis.
In this study, we demonstrated an advanced integrated centrifugal RT-LAMP microdevice. The RNA extraction of the influenza viral lysates, the RT-LAMP reaction, and the real-time laser-induced fluorescence detection were serially operated on a chip by using the sophisticated microchannel structure and the rotation control. Besides the total integration, the intrinsic low dimension of the microreactor enables rapid and efficient isothermal amplification, thereby executing the complete sample-to-result testing in 47 min. Influenza A H1N1 (A/Korea/CJ01/2009 (H1N1)), H3N2 (A/reassortant/NYMC X-175 C (Uruguay/716/2007×Puerto Rico/8/1934) (H3N2)), and H5N1 (A/environment/Korea/W149/2006 (H5N1)) viruses were targeted, and their subtyping on a chip was successfully performed with clinical samples.
2. Experimental
2.1. Design of the integrated RT-LAMP microdevice
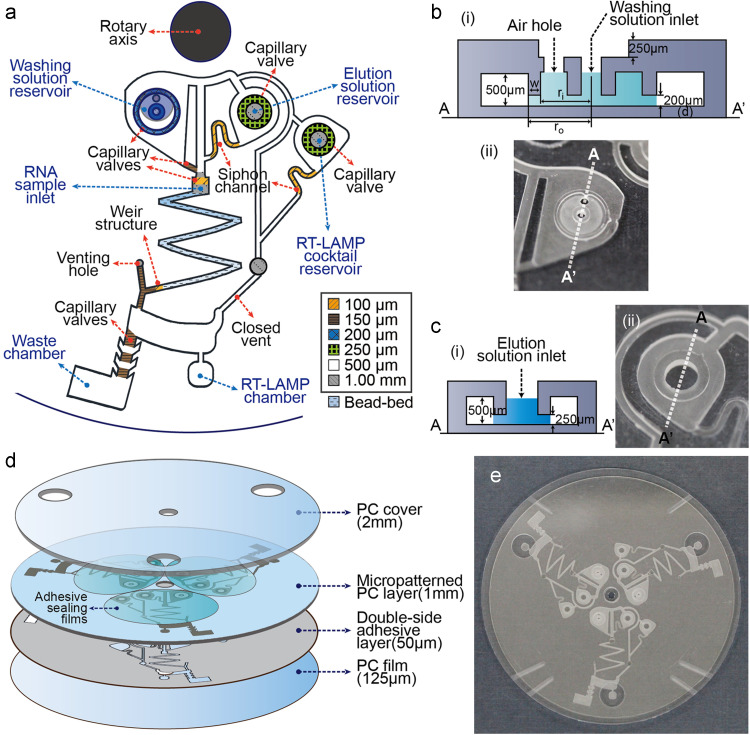
The design of the integrated RT-LAMP microdevice is illustrated in Fig. 1a. Three reservoirs for a RNA sample, a washing solution, and an elution solution were positioned in front of the microbead bed channel which functions as a RNA capture matrix. On the right side, another reservoir for the RT-LAMP cocktail containing an enzyme mix, target-specific primers, and a reaction buffer was designed. The capillary valve (100 μm depth, orange color in Fig. 1a) was patterned in the inner part of the RNA sample inlet to prevent backflow into the washing and elution solution reservoirs. A weir structure (100 μm depth) was fabricated to pack the TEOS-treated microbeads (150–212 μm diameter) according to our previous report (Jung et al., 2013). The washing solution reservoir was connected with the bead-bed microchannel by the capillary valve (580 μm (width)×150 μm (depth), brown color in Fig. 1a). The siphon channel (580 μm (width)×100 μm (depth), orange color on the left in Fig. 1a) bridged between the elution solution reservoir and the bead-bed microchannel. The capillary valve in the elution solution reservoir was employed for storing the liquid safely, while the siphon channel was used prior to the elution solution reservoir in order to sequentially load an RNA sample, a washing solution, and an elution solution. Similarly, the RT-LAMP cocktail reservoir was patterned with a capillary valve and a siphon channel, so that the RT-LAMP cocktail could be moved to the RT-LAMP chamber before the loading of the elution solution (see the details of the flow control in the integrated RT-LAMP microdevice in Supplementary material).
Fig. 1.
(a) Schematic illustration of the integrated RT-LAMP microdevice. (b) (i) Schematic design of the washing solution reservoir and (ii) a digital image of the washing solution chamber from a bottom view. (c) (i) Schematic design of the elution solution reservoir and (ii) a digital images of the elution solution chamber from a bottom view. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
In particular, we carefully designed the washing and elution solution reservoirs to store the solution stably in the designated position. For example, the used washing solution was 70% ethanol which has low contact angle. Due to the high wettability of 70% ethanol, the loaded washing solution tends to uncontrollably move along the channel wall, once loaded. To prevent such a random flow, the washing solution reservoir was patterned as shown in Fig. 1b(i). First, a ring structure (200 μm depth) was fabricated to induce strong capillary force, which can hold the loaded washing solution inside the ring pattern. The capillary force should be strong enough to endure the liquid pressure to inhibit the washing solution leakage. Fig. 1b(ii) shows the bottom view of the washing solution reservoir which displayed the inner and outer ring patterns. The depth of the pattern was roughly calculated in Supplementary material in detail. In order to prevent wetting of a top sealing tape during the injection of a washing solution, a 250 μm deep pattern was fabricated, which ensured the perfect sealing of an adhesive tape to eliminate the contamination and evaporation issues during RT-LAMP process (Focke et al., 2010). Both the elution solution reservoir and the RT-LAMP cocktail reservoir were also designed with a ring pattern with 250 μm depth to firmly store the liquid during the injection step (Fig. 1c(i) and (ii) shows the bottom view of those reservoirs).
The waste chamber and RT-LAMP chamber were designed downstream in the integrated RT-LAMP microdevice. The entrance of the waste chamber contained three capillary valves (dimension: 580 μm (width)×150 μm (depth)×1000 μm (length)) to prevent backflowing of the waste solution into the bead-bed microchannel or the RT-LAMP chamber. On the right side of the waste chamber, the RT-LAMP chamber was patterned with 6 μL volume, and was directly linked with the RT-LAMP cocktail reservoir through a siphon channel. Experimentally, the RT-LAMP cocktail was transported to the RT-LAMP chamber faster than the purified RNA solution.
2.2. Preparation for the integrated RT-LAMP reaction
An RNA sample (influenza A virus lysates or purified viral RNAs), a washing solution (70% ethanol), an elution solution (RNase-free water), and an RT-LAMP reaction cocktail were introduced in the designated reservoirs (Fig. S2a). 3.5 μL of the RNA sample included 0.5 μL influenza A virus sample, 1.25 μL ethanol, and 1.75 μL 6 M Gu-HCl. Upon loading into the sample reservoir, the RNA solution (yellow) was automatically absorbed into the bead-bed microchannel due to the hydrophilicity and capillary forces derived from the packed microbeads, while the capillary valve located on the upper side of the sample inlet prevented the sample solution from back-flowing to the washing or elution solution reservoir. Then, 5 μL of a washing solution (70% ethanol, blue), 2.2 μL of an elution solution (RNase-free water, sky blue), and 4.4 μL of an RT-LAMP reaction cocktail (green) were carefully loaded to each reservoir. Once loaded, the microdevice was positioned on the portable centrifugal system. To block the overflow and evaporation of the solutions, a piece of an adhesive film was used to cover the liquid reservoirs and the venting hole, and finally the PC cover was assembled (Please refer to the section of ‘Fabrication of the integrated RT-LAMP microdevice’ and Fig. S1 in Supplementary material.)
2.3. Operation of the integrated RT-LAMP microdevice
The RNA sample which was absorbed in the bead-bed microchannel started to flow at 5000 revolutions per minute (RPM) for 10 s (counter clockwise). Target RNA and some impurities were captured on the packed microbeads (yellow color in Fig. S2b(i)). Immediately, the washing solution was followed to purify the captured RNAs by removing the remaining salts and proteins adsorbed on the bead-bed (blue color in Fig. S2b(ii)). Centrifugation was conducted further at 5000 RPM for 290 s (counter clockwise) to dry completely any residual ethanol in the microbeads, which might play a role as an inhibitor for gene amplification. At this step, the waste solutions derived from the RNA sample and the washing solution were transported to the waste chamber. Since the flow direction of the waste solution was on the bottom left due to the driving force caused by centrifugal force and Coriolis force, the waste solution moved to the waste chamber, not to the RT-LAMP chamber (Kim et al., 2008, Brenner et al., 2005, Ducree et al., 2007). The sequential strobed images in Fig. S3c were correspondent to the steps in Fig. S2b. Movie 1 in the Supplementary material was supplied to visualize the entire process.
Supplementary material related to this article can be found online at doi:10.1016/j.bios.2014.12.043.
The following is the Supplementary material related to this article Movie S1, Movie S2.
When the rotation of the microdevice stopped, the RNase-free water and the RT-LAMP cocktail were primed to the siphon channel within 30 s (Fig. S2b(iii)). Then, the rotational speed increased up to −5000 RPM in a clockwise direction. The RNase-free water and the RT-LAMP cocktail started to flow (Fig. S2b(iv)). While the RT-LAMP cocktail was directly transported to the RT-LAMP chamber earlier, the RNase-free water passed through the bead-bed microchannel to deliver the purified RNAs into the RT-LAMP chamber afterwards. Since the rotational direction was opposite to the initial step, the driving force was exerted on the bottom right. So, the RNase-free water, which carried the purified RNAs, was moved to the RT-LAMP chamber, not to the waste chamber. We prolonged this step for 90 s to fully recover the residual water on the microbeads as shown in Fig. S2b(iv–vi). Finally, total 6 μL of the RT-LAMP reaction mixture (4 μL of the RT-LAMP cocktail and 2 μL of the purified RNA solution) was collected in the RT-LAMP chamber (blue green color in Fig. S2c), and the RT-LAMP reaction proceeded directly at 64.2 °C for 40 min (See the detailed separation procedure of the purified RNA from the waste in Supplementary material with the sequential strobed images (Fig. S4) and Movie 2.)
2.4. Construction of a temperature controller and a rotational system for an integrated RT-LAMP microdevice
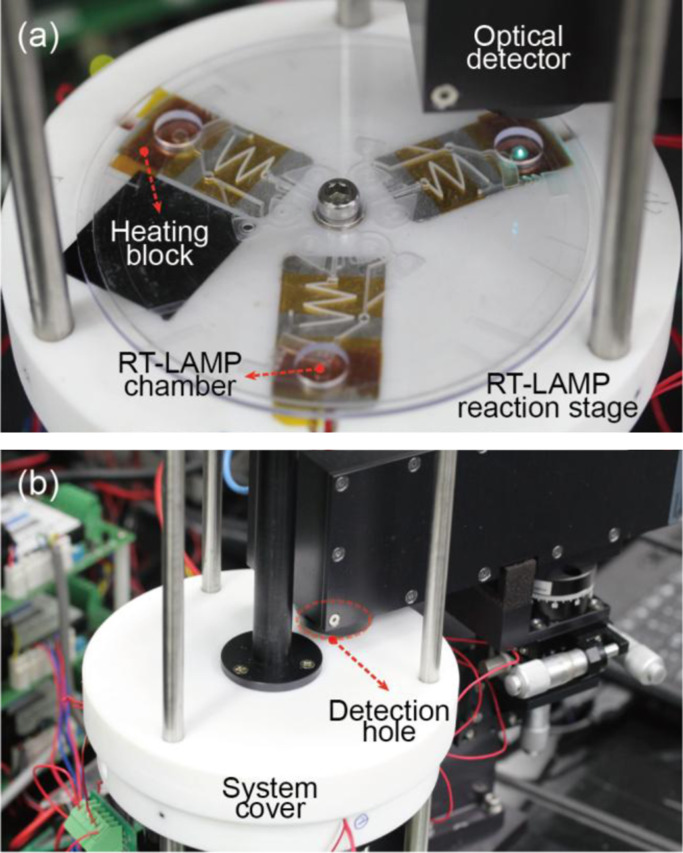
The integrated RT-LAMP microdevice was operated in a custom-made portable rotary genetic analysis microsystem which can perform precise rotation and temperature control (Jung et al., 2012). The centrifugal rotation of the integrated microdevice was driven by a servo motor (AC Servo Motor, HF-KP13, Mitsubishi, Japan) and automatically controlled by an in-house LabVIEW program. The temperature control system was presented in the previous report in details (Jung et al., 2012). Thermal blocks incorporated with a film heater (MINCO™, MN, USA) and a resistive temperature detector (RTD, RdF Corporation, NH, USA) were set up on the rotary stage and used for precise temperature control with a designed electrical circuit and an in-house LabVIEW software. For real-time monitoring of genetic amplification, a miniaturized optical detector was installed to the rotary portable microsystem. The collimated beam from the optical fluorescence detector is focused on the RT-LAMP chamber by emitting 488 nm laser and the fluorescence signal in the range of 500–530 nm was detected by a photomultiplier tube with high sensitivity. As shown in Fig. 2a, the fluorescence emission from the three RT-LAMP chambers were serially measured by the optical detector every 1 min when the chip rotated 120°. A detection hole was fabricated on the system cover made of Teflon for installation of an optical detector. Thus, real-time monitoring of the RT-LAMP amplification could be performed with the system cover tightly sealed as shown in Fig. 2b.
Fig. 2.
A digital image of (a) an RT-LAMP reaction stage including the integrated RT-LAMP microchip, heating blocks, an optical detector, and a rotational axis and (b) the RT-LAMP reaction stage sealed by a system cover equipped with a portable detector.
2.5. Conditions of the real-time RT-LAMP reaction
The RT-LAMP reaction cocktail was prepared containing 10 pmol of each outer primers (F3 and B3), 100 pmol of each inner primers (FIP and BIP), 50 pmol of each loop primers (LF and LB), 12.5 μL of the reaction buffer (2× ThermoPol buffer, 1.6 M betaine, 1.6 mM MgSO4, 2.8 mM dNTPs), 8 U Bst DNA polymerase, 2 U AMV reverse transcriptase, and 1 μL of Loopamp® Fluorescence Reaction Reagent for real-time monitoring of the RT-LAMP amplicons. The RT-LAMP primers were designed as listed in Table S1 (Starick et al., 2000, Sun et al., 2011). 4.4 μL of the RT-LAMP reaction cocktail were loaded to the RT-LAMP cocktail reservoir. In the elution solution reservoir, 2 μL of RNase-free water was introduced, and finally 2 μL of purified RNAs in water were mixed with the 4.4 μL of the RT-LAMP reaction cocktail in the RT-LAMP chamber. The concentration of viral RNAs was controlled from 106 to 10 copies. The RT-LAMP reaction was performed at 64.2 °C for 40 min on the portable genetic analysis microsystem with real-time monitoring. In case of viral lysate samples of influenza A H1N1, A H3N2, and A H5N1, the lysates were directly loaded in the RNA sample inlet. For clinical nasal swab analysis, the clinical samples were first lysed by using a QIAamp Viral RNA Mini kit (Qiagen), and the lysates were injected to the RNA sample inlet.
3. Results and discussion
3.1. Fluorescence detection on the integrated RT-LAMP microdevice
The prepared RT-LAMP reaction mixture consisting of the purified RNA and the RT-LAMP cocktail was isothermally amplified at 64.2 °C for 40 min on a chip. The top thick PC cover (Fig. S1a) not only helps to prevent the chip from being bent, but also makes the RT-LAMP chambers contacted to the heating blocks for efficient heat transfer. To estimate the temperature of the RT-LAMP chamber, Fourier's law was utilized assuming that there is no heat loss at steady state. The middle of the RT-LAMP chamber was calculated as 64.2 °C, which was suitable for the RT-LAMP reaction (Figs. S5 and S6). The calculation process was shown in Supplementary material in detail.
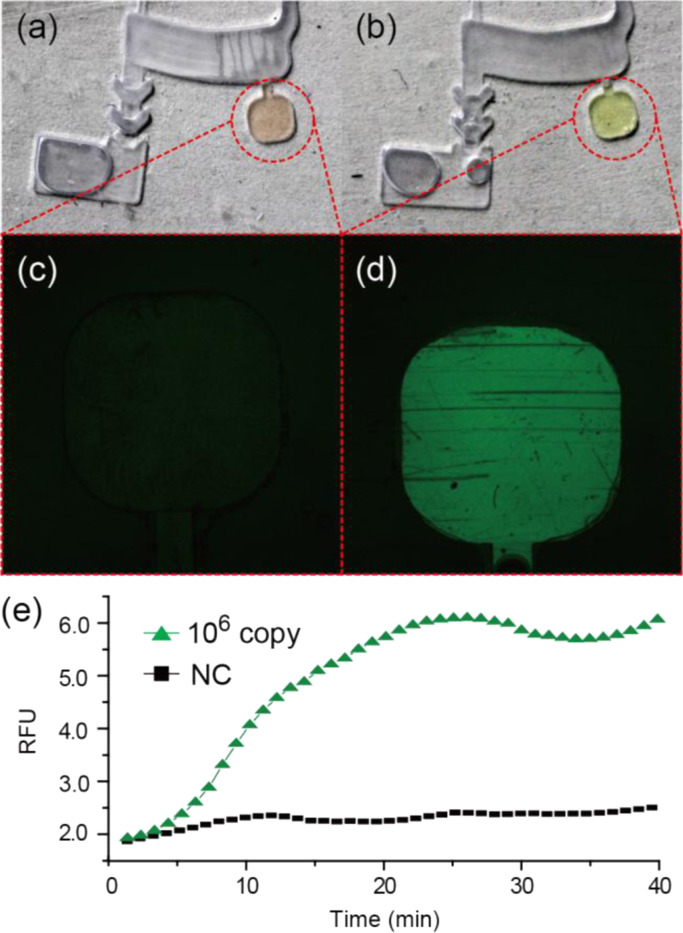
Digital and fluorescence images of the RT-LAMP chamber before and after the RT-LAMP reaction are shown in Fig. 3a–d. The initial state of the RT-LAMP chamber has orange color due to the quenched calcein metal indicator which was combined with manganous ions (Mn2+) (Fig. 3a). At this stage, the chamber did not emit any fluorescence signal (λ ex=488 nm/λ em=515 nm) (Fig. 3c). As the RT-LAMP reaction was carried out for 40 min, the color of the reaction mixture was changed to yellow. The reason for the color change is that the generated pyrophosphate ions () during the RT-LAMP deprived of the combined Mn2+ ions, and the calcein was complexed with magnesium ion (Mg2+) in the reaction mixture, which produced strong fluorescence (Fig. 3b and d) (Tomita et al., 2008).
Fig. 3.
Digital images of the RT-LAMP chamber (a) before (Orange color) and (b) after the RT-LAMP reaction (Yellow color). Confocal images of the RT-LAMP chamber (c) before and (d) after the RT-LAMP reaction. (e) Real-time RT-LAMP profiles obtained from the RT-LAMP chamber (RFU: relative fluorescence units, NC: negative control). (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
A fluorescence signal coming from the RT-LAMP chamber was in situ monitored every 1 min by the miniaturized optical detector during isothermal amplification. Fig. 3e shows the isothermal amplification profile of the conserved M gene of influenza A H1N1 with 106 copies of RNAs. Compared with the negative control which omitted the input of the RNA template in the RT-LAMP cocktail, the intensity of the M gene amplicon gradually increased, meaning that the viral RNA was successfully purified and the target gene was isothermally amplified.
3.2. Sensitivity of the integrated RT-LAMP assay
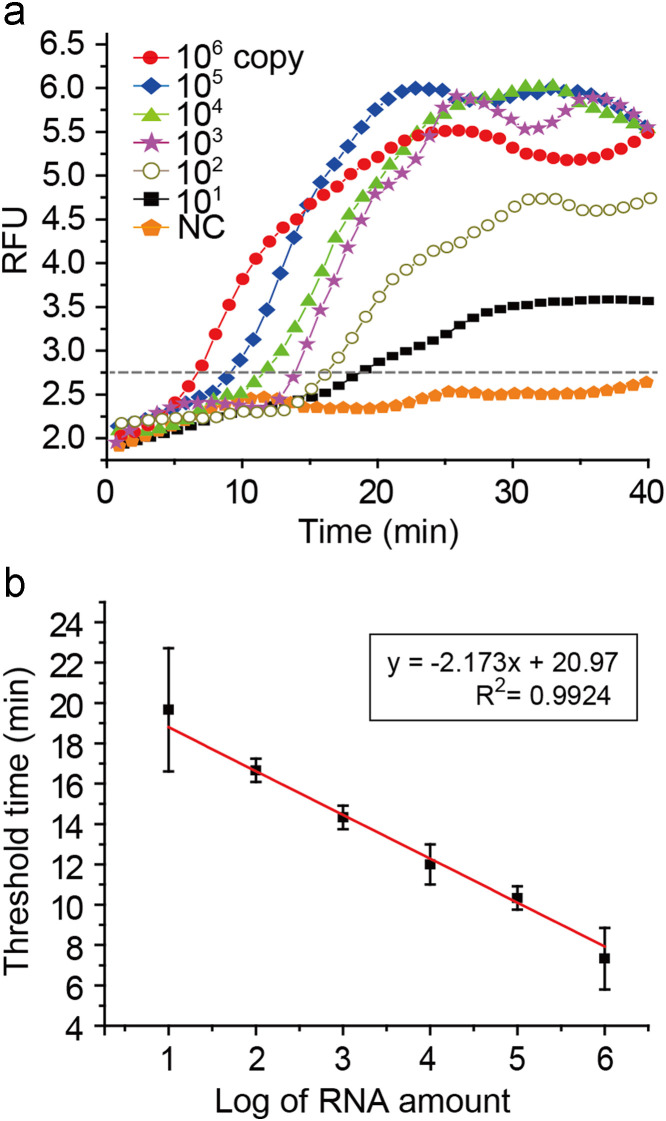
Considering that the low number of the viral particles can cause significant damages, sensitive detection of influenza virus is of importance. To demonstrate the capability of our methodology for ultrasensitive detection, we performed a limit of detection (LOD) test on the integrated RT-LAMP microdevice by using serially diluted influenza A H1N1 viral RNA templates from 106 to 10 copies, and conserved M gene was targeted. The real-time RT-LAMP profiles were depicted as shown in Fig. 4a. The fluorescence signals gradually increased even with 10 copies of influenza A H1N1 viral RNAs. The reaction time for transcending the threshold value (a dashed line in Fig. 4a) was reduced as the number of RNA templates increased. The positive fluorescence signals were observed within 20 min, and the signal intensities became plateau after 30 min in all cases. For quantitative analysis, a standard curve was established by plotting the threshold time versus the logarithm of RNA copy number with triplicate experiments (R 2=0.9924) (Fig. 4b). Total processing time was 47 min including 7 min for the RNA purification and 40 min for the RT-LAMP reaction. However, since the saturation of fluorescence signal in the real-time RT-LAMP reaction was achieved within 30 min (Fig. 4a), the total time can be reduced to 37 min.
Fig. 4.
(a) Real-time RT-LAMP profiles using purified RNAs ranging from 106 to 10 copies. (b) A standard curve by plotting the threshold time versus the logarithm of RNA copy number (RFU: relative fluorescence units, NC: negative control).
The detection sensitivity of the integrated RT-LAMP assay was compared to that of the conventional real-time RT-PCR method, and the real-time RT-PCR profiles are presented in Fig. S7. The used reaction volume was 20 μL (Jung et al., 2013), and the purified RNA by a RNA extraction kit was employed for the conventional real-time RT-PCR. The influenza A virus could be identified with 102 copies in the real-time RT-PCR method. Thus, the detection sensitivity of our integrated centrifugal microsystem was enhanced by 10-fold compared with that of the conventional real-time RT-PCR. Whereas the conventional PCR requires time-consuming thermal cycling steps, the isothermal amplification needs only single reaction temperature. In addition, rapid amplification can be realized in the microfluidic-based platform owing to the smaller thermal inertia, low thermal mass, and high surface to volume ratio of the reaction chamber. Thus, our microfluidic based isothermal amplification system produced higher sensitivity than that of the conventional real-time RT-PCR (Chang et al., 2013).
3.3. Subtyping of influenza A virus
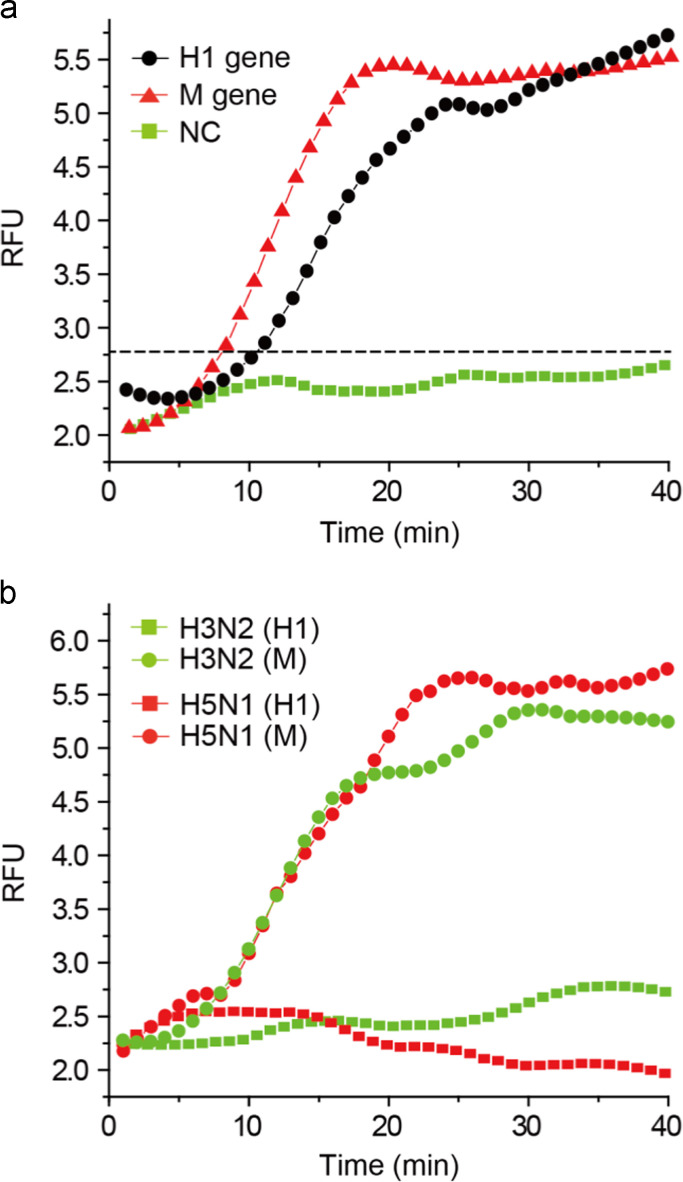
Rapid subtyping of influenza A virus is critical for early medical pretreatment and prognosis. To this end, the subtyping test of an influenza A H1N1 virus was investigated on the integrated RT-LAMP microsystem. H1 gene and M gene were targeted to identify influenza A H1N1 virus. The RT-LAMP cocktails which contained H1 gene specific primer sets and M gene specific primer sets were loaded in the RT-LAMP cocktail reservoirs separately, and the RNA purification and the RT-LAMP reaction were conducted simultaneously. In this case, we used viral lysates directly as a sample solution instead of purified RNAs. As shown in Fig. 5a, both the H1 and M gene were successfully amplified, resulting in the improved fluorescence signal along with the reaction time. The threshold time gap (8 min for M gene and 11 min for H1 gene) may be due to the difference of the primer binding affinity on the target template and the amplification efficiency during the RT-LAMP reaction. Since our integrated microdevice could perform three RT-LAMP reactions at one time (Fig. 2a), the subtyping of influenza A virus was possible. Following the previous subtyping of influenza A virus, we tested the specificity of H1 targeting primer sets by applying them for amplifying H3 and H5 genes. Influenza A H3N2 and H5N1 viral lysates were used as RNA templates, and the fluorescence signal was real-time monitored on a chip (Fig. 5b). In both samples, there were no amplicons produced, showing the fluorescence intensities at the background level even after 40 min. These results indicate that the H1 targeting primer sets were specific to the H1 gene (Fig. 5a). On the other hand, when the M gene targeting primer sets were used, the M gene was amplified in both influenza A H3N2 and H5N1 viruses as the reaction time went by. Thus, the designed M gene primers were successfully targeting the conserved region of M gene, and universal to identify the influenza A virus. The RT-LAMP microdevice allows us to pathotype and subtype the influenza A virus at one time.
Fig. 5.
The real-time RT-LAMP profiles for (a) subtyping of influenza A H1N1 virus and (b) specificity (RFU: relative fluorescence units, NC: negative control).
3.4. Clinical sample analysis on the integrated RT-LAMP microsystem
Three clinical samples with nasal swabs were analyzed in the integrated RT-LAMP microsystem. We analyzed the samples using the conventional real-time RT-PCR beforehand to confirm that they were infected by influenza A virus by detecting the conserved M gene (Fig. S8a). The profiles of real-time RT-LAMP on a chip were depicted in Fig. S8b, showing that the M gene was amplified in all the samples. These results were perfectly matched with those of the real-time RT-PCR data, confirming that three clinical samples were infected by influenza A virus.
4. Conclusion
In summary, we have demonstrated an integrated rotary genetic analysis microsystem, which consisted of an integrated RT-LAMP microdevice, a rotational platform, and a miniaturized optical detector. Through the sophisticated microfluidic design, and the rotation scheme, we could serially perform the RNA extraction for 7 min and the real-time RT-LAMP reaction for 40 min in a simplified polycarbonate microdevice. Using influenza A H1N1, H3N2 and H5N1 viral samples, the target gene amplification (H1 and M genes) was in situ detected with 10 copies of LOD, which sensitivity was 10-fold higher than that of a conventional RT-PCR. The on-site pathotyping and subtyping of the influenza A virus on a chip could be realized with high speed, automation, and sensitivity, providing an advanced genetic analysis platform in the clinical and biomedical diagnostic fields.
Acknowledgment
This work was supported by Center for BioNano funded by the Ministry of Science, ICT and Future Planning (MSIP) of Korea as Global Frontier Project (H-GUARD_2013M3A6B2078964), and by the Engineering Research Center of Excellence Program of Korea Ministry of Science, ICT & Future Planning (MSIP)/National Research Foundation of Korea (NRF) (Grant NRF_2014R1A5A1009799).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bios.2014.12.043.
Appendix A. Supplementary material
Supplementary material
References
- Arias C.F., Escalera-Zamudio M., Rio M.D., Cobian-Guemes A.G., Isa P., Lopez S. Arch. Med. Res. 2009;40:643–654. doi: 10.1016/j.arcmed.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Asiello P.J., Baeumner A.J. Lab Chip. 2011;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- Brenner T., Glatzel T., Zengerle R., Ducree J. Lab Chip. 2005;5:146–150. doi: 10.1039/b406699e. [DOI] [PubMed] [Google Scholar]
- Chang C.-M., Chang W.-H., Wang C.-H., Wang J.-H., Mai J.D., Lee G.-B. Lab Chip. 2013;13:1225–1242. doi: 10.1039/c3lc41097h. [DOI] [PubMed] [Google Scholar]
- Chen G.D., Alberts C.J., Rodriguez W., Toner M. Anal. Chem. 2010;82:723–728. doi: 10.1021/ac9024522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.K., Lee J.-G., Park J.-M., Lee B.-S., Lee Y., Ko C. Lab Chip. 2007;7:565–573. doi: 10.1039/b616115d. [DOI] [PubMed] [Google Scholar]
- Compton J. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Ducree J., Haeberle S., Lutz S., Pausch S., Stetten F. v., Zengerle R. J. Micromech. Microeng. 2007;17:S103–S115. [Google Scholar]
- Dukes J.P., King D.P., Alexandersen S. Arch. Virol. 2006;151:1093–1106. doi: 10.1007/s00705-005-0708-5. [DOI] [PubMed] [Google Scholar]
- Fang X., Chen H., Yu S., Jiang X., Kong J. Anal. Chem. 2011;83:690–695. doi: 10.1021/ac102858j. [DOI] [PubMed] [Google Scholar]
- Focke M., Stumpf F., Faltin B., Reith P., Bamarni D., Wadle S., Muller C., Reinecke H., Schrenzel J., Francois P., Mark D., Roth G., Zengerle R., Stetten F.v. Lab Chip. 2010;10:2519–2526. doi: 10.1039/c004954a. [DOI] [PubMed] [Google Scholar]
- Grumann M., Geipel A., Riegger L., Zengerle R., Ducree J. Lab Chip. 2005;5:560–565. doi: 10.1039/b418253g. [DOI] [PubMed] [Google Scholar]
- Haeberle S., Brenner T., Zengerle R., Ducree J. Lab Chip. 2006;6:776–781. doi: 10.1039/b604145k. [DOI] [PubMed] [Google Scholar]
- Hataoka Y., Zhang L.H., Mori Y., Tomita N., Notomi T., Baba Y. Anal. Chem. 2004;76:3689–3693. doi: 10.1021/ac035032u. [DOI] [PubMed] [Google Scholar]
- Hoehl M.M., Weibert M., Dannenberg A., Nesch T., Paust N., Stetten F. v., Zengerle R., Slocum A.H., Steigert J. Biomed. Microdevices. 2014;16:375–385. doi: 10.1007/s10544-014-9841-9. [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Ito M., Watanabe M., Nakagawa N., Ihara T., Okuno Y. J. Virol. Methods. 2006;135:272–275. doi: 10.1016/j.jviromet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Choi S.J., Park B.H., Choi Y.K., Seo T.S. Lab Chip. 2012;12:1598–1600. doi: 10.1039/c2lc21269b. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Park B.H., Choi Y.K., Seo T.S. Lab Chip. 2013;13:3383–3388. doi: 10.1039/c3lc50266j. [DOI] [PubMed] [Google Scholar]
- Kazarine A., Kong M.C.R., Templeton E.J., Salin E.D. Anal. Chem. 2012;84:6939–6943. doi: 10.1021/ac301421k. [DOI] [PubMed] [Google Scholar]
- Kellogg G.J., Arnold T.E., Carvalho B.L., Duffy D.C., Sheppard N.F. Proc. MicroTAS. 2000:239–242. [Google Scholar]
- Kim J., Kido H., Rangel R.H., Madou M. Sens. Actuators B. 2008;128:613–621. [Google Scholar]
- Lafleur J.P., Rackov A.A., McAuley S., Salin E.D. Talanta. 2010;81:722–726. doi: 10.1016/j.talanta.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Lazcka O., Campo F.J.D., Munoza F.X. Biosens. Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- Liu C., Geva E., Mauk M., Qiu X., Abrams W.R., Malamud D., Curtis K., Owen S.M., Bau H.H. Analyst. 2011;136:2069–2076. doi: 10.1039/c1an00007a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi P.M., Huang X., Zhu T., Bray-Ward P., Thomas D.C., Ward D.C. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Muller C., Mark D., Roth G., Munday P., Armes N., Piepenburg O., Zengerle R., Stetten F. v. Lab Chip. 2011;11:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- Madou M., Zoval J., Jia G., Kido H., Kim J., Kim N. Annu. Rev. Biomed. Eng. 2006;8:601–628. doi: 10.1146/annurev.bioeng.8.061505.095758. [DOI] [PubMed] [Google Scholar]
- Medina R.A., Garcia-Sastre A. Nat. Rev. Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Notomi T. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisset D., Stebih D., Cankar K., Zel J., Gruden K. Eur. Food Res. Technol. 2008;227:1287–1297. [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley J.A., Shaw K.J., Docker P.T., Dyer C.E., Greenman J., Greenway G.M., Haswell S.J. Lab Chip. 2009;9:1596–1600. doi: 10.1039/b820553a. [DOI] [PubMed] [Google Scholar]
- Parida M., Sannarangaiah S., Dash P.K., Rao P.V.L., Morita K. Rev. Med. Virol. 2008;(18):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrman B.A., Richards-Kortum R.R. Lab Chip. 2012;12:3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Perera A.P., Kim K.W., Park M.K. Lab Chip. 2013;13:2106–2114. doi: 10.1039/c3lc50129a. [DOI] [PubMed] [Google Scholar]
- Starick E., Romer-Oberdorfer A., Werner O. J. Vet. Med. Ser. B. 2000;47:295–301. doi: 10.1046/j.1439-0450.2000.00386.x. [DOI] [PubMed] [Google Scholar]
- Steigert J., Grumann M., Brenner T., Riegger L., Harter J., Zengerle R., Ducree J. Lab Chip. 2006;6:1040–1044. doi: 10.1039/b607051p. [DOI] [PubMed] [Google Scholar]
- Sun Y., Dhumpa R., Bang D.D., Hogberg J., Handberg K., Wolff A. Lab Chip. 2011;11:1457–1463. doi: 10.1039/c0lc00528b. [DOI] [PubMed] [Google Scholar]
- Thai H.T.C., Le M.Q., Vuong C.D., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna D., Poon L.L.M., Zhu H.C., Ma S.K., Li O.T.W., Cheung C.L., Smith G.J.D., Peiris J.S.M., Guan Y. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Xu Y., Kong H. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-H., Lien K.-Y., Wang T.-Y., Chen T.-Y., Lee G.-B. Biosens. Bioelectron. 2011;26:2045–2052. doi: 10.1016/j.bios.2010.08.083. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO), 2009. Pandemic (H1N1) 2009 – Update, 112 Weekly Update, 〈http://www.who.int/csr/don/2010_08_06/en/〉 (accessed August 2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material