Abstract
Cavally virus (CavV) is a mosquito-borne plus-strand RNA virus in the family Mesoniviridae (order Nidovirales). We present X-ray structures for the CavV 3C-like protease (3CLpro), as a free enzyme and in complex with a peptide aldehyde inhibitor mimicking the P4-to-P1 residues of a natural substrate. The 3CLpro structure (refined to 1.94 Å) shows that the protein forms dimers. The monomers are comprised of N-terminal domains I and II, which adopt a chymotrypsin-like fold, and a C-terminal α-helical domain III. The catalytic Cys-His dyad is assisted by a complex network of interactions involving a water molecule that mediates polar contacts between the catalytic His and a conserved Asp located in the domain II-III junction and is suitably positioned to stabilize the developing positive charge of the catalytic His in the transition state during catalysis. The study also reveals the structural basis for the distinct P2 Asn-specific substrate-binding pocket of mesonivirus 3CLpros.
Keywords: Mesonivirus, Coronavirus, 3C-like protease, Crystal structure, Active site of chymotrypsin-like proteases, Invertebrate RNA virus
Graphical abstract
Highlights
-
•
First structure of a 3CLpro of an invertebrate RNA virus.
-
•
Structural basis of the unique substrate specificity defined by Asn at the P2 position of mesonivirus 3CLpro substrates.
-
•
Emerging role of a conserved Asp residue that assists the Cys-His catalytic dyad in vertebrate and invertebrate 3CLpros.
Abbreviations
- 3Cpro
Picornavirus 3C protease
- 3CLpro
3C-like protease
- CavV
Cavally virus
- EAV
Equine arteritis virus
- HCoV
Human coronavirus
- IBV
Avian infectious bronchitis virus
- MAD
Multi-wavelength anomalous dispersion
- MSA
Multiple sequence alignment
- RMSD
Root-mean-square deviation
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- TGEV
Transmissible gastroenteritis virus
1. Introduction
Proteases with a two-β-barrel fold prototyped by chymotrypsin form one of the largest clans of proteolytic enzymes, called the PA clan, and are encoded by pro- and eukaryotes as well as many viruses (Rawlings et al., 2012). The cellular chymotrypsin-like enzymes invariantly employ a canonical catalytic Ser-His-Asp triad (Hedstrom, 2002; Polgar, 2005) and have evolved, by duplication and diversification, a large spectrum of substrate specificities facilitating their diverse roles in many biological processes. Likewise, RNA viruses with single-stranded RNA genomes of positive polarity (ssRNA + viruses) often employ chymotrypsin-like enzymes to control genome expression and remodel host cell functions by targeting different proteins.
One of the major monophyletic groups of RNA virus chymotrypsin-like enzymes is known as 3C and 3C-like proteases (3C/3CLpro; see below), which evolved by extensive diversification during virus speciation. In contrast to cellular and other viral homologs, these enzymes uniquely diversified the principal catalytic nucleophile residue to employ either the canonical Ser or an unconventional Cys (depending on the virus lineage) (Bazan and Fletterick, 1988; Gorbalenya et al., 1986, 1989a; Malcolm, 1995), while (somewhat counterintuitively) sharing a conserved narrow substrate specificity. In those proteases that employ Cys as the principal nucleophile, the catalytic Asp may be replaced with another residue, either another acidic residue (Glu) (Matthews et al., 1994; Mosimann et al., 1997) or a residue with a side chain that has little or no similarity to that of Asp, further reflecting the pronounced sequence divergence of many ssRNA + virus proteases from their cellular homologs (Gorbalenya and Snijder, 1996). Probably, these replacements have evolved due to the exceptionally large mutation space that RNA viruses of this lineage explored in the course of their evolution (Lauber et al., 2013) and have been fixed to meet the distinct requirements for Ser and Cys residues in mediating the nucleophilic attack during catalysis (McGrath et al., 1989).
3C/3CLpros possess a conserved substrate specificity that is predominantly directed against Gln or Glu in the P1 and a small residue in the P1’ positions of the substrate (Gorbalenya et al., 1989b; Kräusslich and Wimmer, 1988; Ziebuhr et al., 2000) (numbering according to the scheme introduced by Schechter and Berger (1967)). These substrate residues are accommodated in a protease substrate pocket that generally includes a highly conserved His residue (which differs from the catalytic His) and another residue, often Ser or Thr, which together define the S1 subsite homologous to that of cellular chymotrypsin-like proteases (Bazan and Fletterick, 1988; Gorbalenya et al., 1989a). These residues are generally considered valuable targets for the development of broadly acting antiviral drugs (Kuo et al., 2009; Yang et al., 2005; Banerjee et al., 2018; Pillaiyar et al., 2016).
Viral 3C/3CLpros are produced as part of large polyproteins that contain the key replicative domains and several other proteins (for reviews see (Kräusslich and Wimmer, 1988; Ziebuhr et al., 2000; Dougherty and Semler, 1993; Ryan and Flint, 1997)). The individual proteins are released by the 3C/3CLpro in both cis- and trans-cleavage reactions, thereby regulating the formation of viral replication complexes in a timely coordinated manner. In addition, some 3C/3CLpros may mediate the processing of capsid polyproteins to generate the proteins required for virus particle formation as well as cleaving cellular proteins to facilitate virus reproduction. Viruses encoding 3C/3CLpros belong to two large monophyletic orders, the Picornavirales (which infect uni- and multicellular eukaryotes) and the Nidovirales (which infect vertebrates and invertebrates), but also include several other virus families or groups, such as the plant Potyviridae and Sobemovirus, and the vertebrate Caliciviridae and Astroviridae, displaying more distant affinities to one of these two virus orders. Over the past years, our appreciation of the natural 3C/3CLpro diversity and conservation of specific residues has been steadily improved by comparative genomics, while our understanding of the roles and properties of these proteases in viral replication was largely based on just a few viruses of the above taxa, with a strong focus on viruses that (may) infect humans. Specifically, the structural characterization of 3C/3CLpros remained limited to those encoded by several mammalian viruses and one avian virus of the Picornaviridae, Coronaviridae, Arteriviridae, Caliciviridae, and Astroviridae, and plant Potyviridae and Sobemovirus (Matthews et al., 1994; Banerjee et al., 2018; Phan et al., 2002; Nunn et al., 2005; Speroni et al., 2009; Gayathri et al., 2006; Damalanka et al., 2018; Weerawarna et al., 2016; Galasiti Kankanamalage et al., 2017; Takahashi et al., 2013; Kim et al., 2012; Muhaxhiri et al., 2013; Allaire et al., 1994; Anand et al., 2002, 2003; Bergmann et al., 1997; Xue et al., 2008; Yang et al., 2003; Barrette-Ng et al., 2002). All these proteases have substrate specificities that are critically determined by the P1 position.
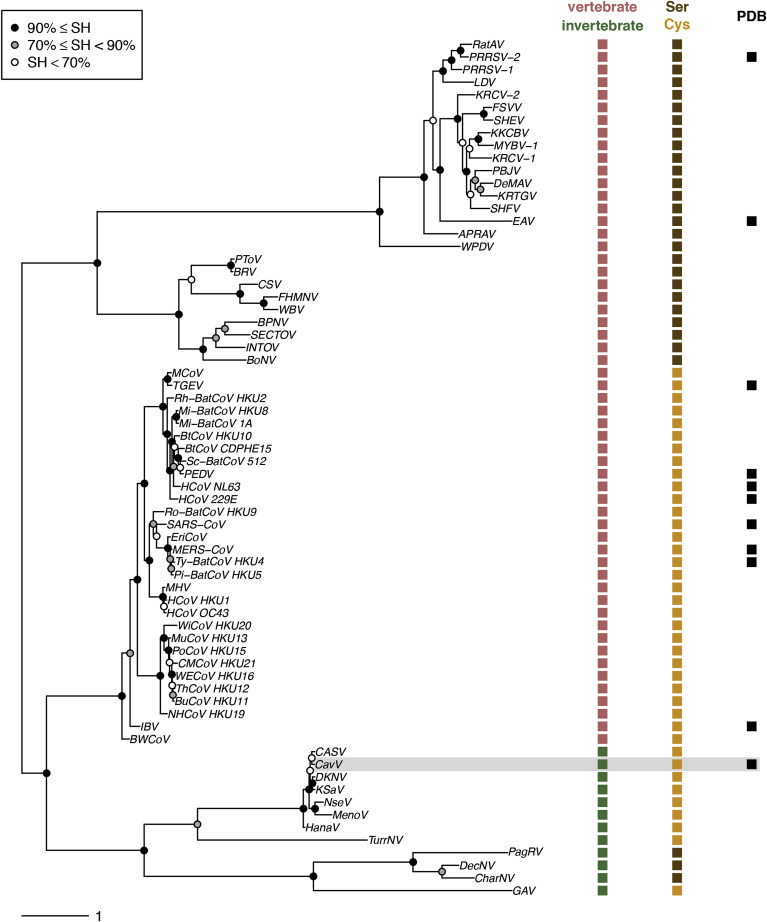
In this study, we sought to address these knowledge gaps by determining the structure of a 3CLpro encoded by Cavally virus, an invertebrate nidovirus. The protease of this virus represents family C107 in the PA(C) subclan of chymotrypsin-like proteases in the MEROPS database (Rawlings et al., 2012) and displays a substrate specificity that is mainly directed toward the P2 position of the substrate (Blanck et al., 2014; Blanck and Ziebuhr, 2016). Cavally virus (CavV) is a member of the genus Alphamesonivirus in the family Mesoniviridae which, together with the families Arteriviridae, Coronaviridae, and Roniviridae as well as five others just approved by ICTV (Siddell et al., 2019), forms the order Nidovirales (Fig. 1 ). A distinct feature of previously characterized proteases from this virus order is the presence of an extra C-terminal domain with regulator function. This domain was shown to adopt different folds in arteri- and coronavirus enzymes (representing protease families S32 and C30, respectively) (Anand et al., 2002; Barrette-Ng et al., 2002) while there is currently no structural information on other nidovirus 3CLpros, such as those of roni-, toro- and bafiniviruses, all of which representing distinct protease families in the MEROPS database (C62, S65, and S75, respectively); toro- and bafiniviruses belong to distinct subfamilies of the newly established family Tobaniviridae.
Fig. 1.
Characteristics of nidovirus 3CLpros in a virus phylogeny and host context. Left, RdRp-based phylogeny of representatives of established nidovirus species. Right, presumed primary host of the respective virus species, 3CLpro principal nucleophile residue, and 3CLpro PDB structure availability. Information on CavV is highlighted using a gray background. Information on nidovirus genomes used for phylogeny reconstruction is provided in the Supplemental Information. Box, SH-aLRT branch support is shown using three ranges of values.
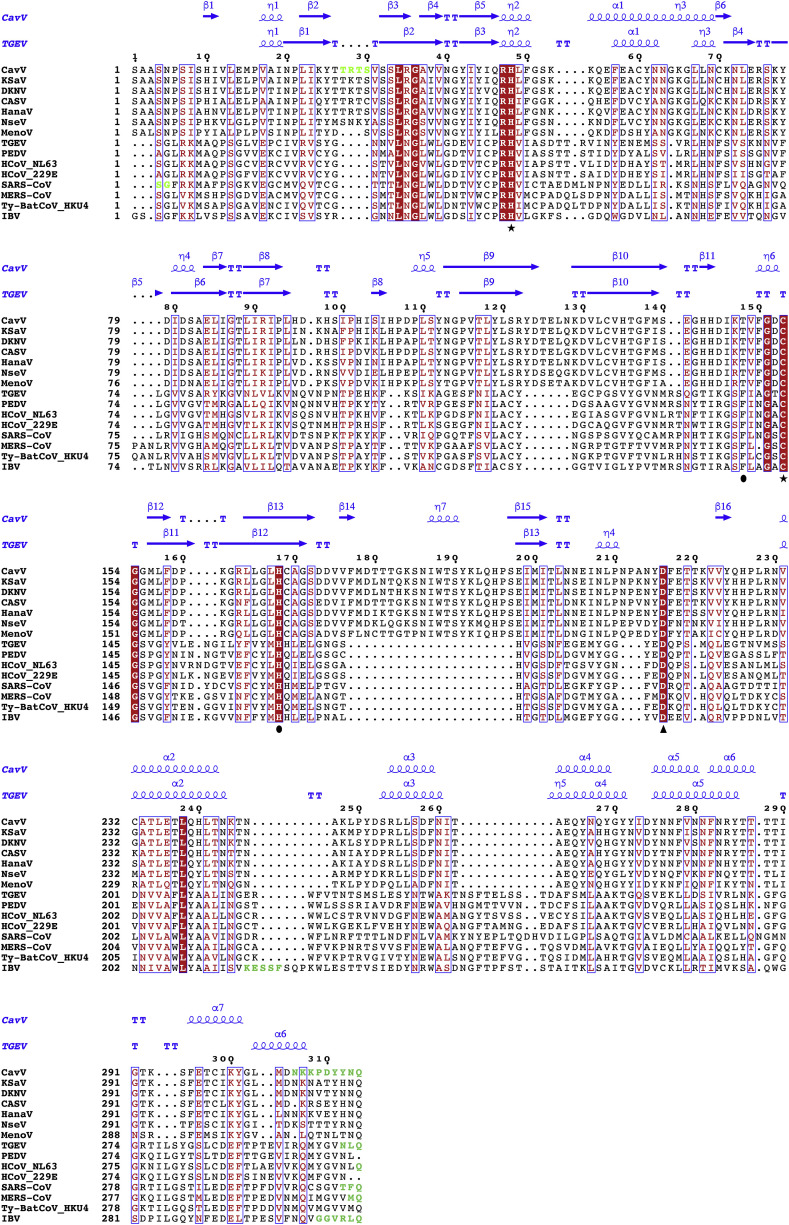
Here, we present the first crystal structures of a mesonivirus 3CLpro, both for the free enzyme and in complex with a covalently bound inhibitor. The protein structure comprises a two-β-barrel fold that is linked to a large C-terminal helical domain of >100 residues. The structure analysis identifies critical residues involved in substrate binding and suggests that mesonivirus 3CLpros employ a catalytic system that depends on (i) Cys153 (principal nucleophile), (ii) His48 (general base), and (iii) a water molecule mediating a polar contact between (iv) Asp216 and the catalytic His48. A strikingly similar arrangement was previously reported for coronavirus 3CLpro structures, which share very low sequence identity (<10%) with 3CLpro orthologs from other nidovirus families, further supporting the universal key roles of these residues in the respective enzymes (Fig. 2 ).
Fig. 2.
Structure-based MSA of mesonivirus and coronavirus 3CLpros. Strictly conserved residues are indicated using red background color, partially conserved residues are indicated in red. Secondary structure elements are shown for CavV (PDB ID: 5LAC, chain B) and TGEV (PDB ID: 1LVO, chain A) in blue above the alignment. Residues that were omitted from the respective structure models are indicated in green. Residue numbers given above the alignment refer to the CavV 3CLpro sequence corresponding to the CavV 3CLpro structure (PDB ID: 5LAC, chain B). Crucial CavV 3CLpro residues are indicated below the alignment: catalytic residues – by asterisks; key residues in the S1 subsite of the substrate-binding pocket – by filled circles; the Asp residue that interacts with a conserved water molecule in the active site – by a black triangle. PDB IDs used to produce the structure-based MSA are given in the Supplemental Information.
2. Results
2.1. Overall structure of CavV 3CLpro
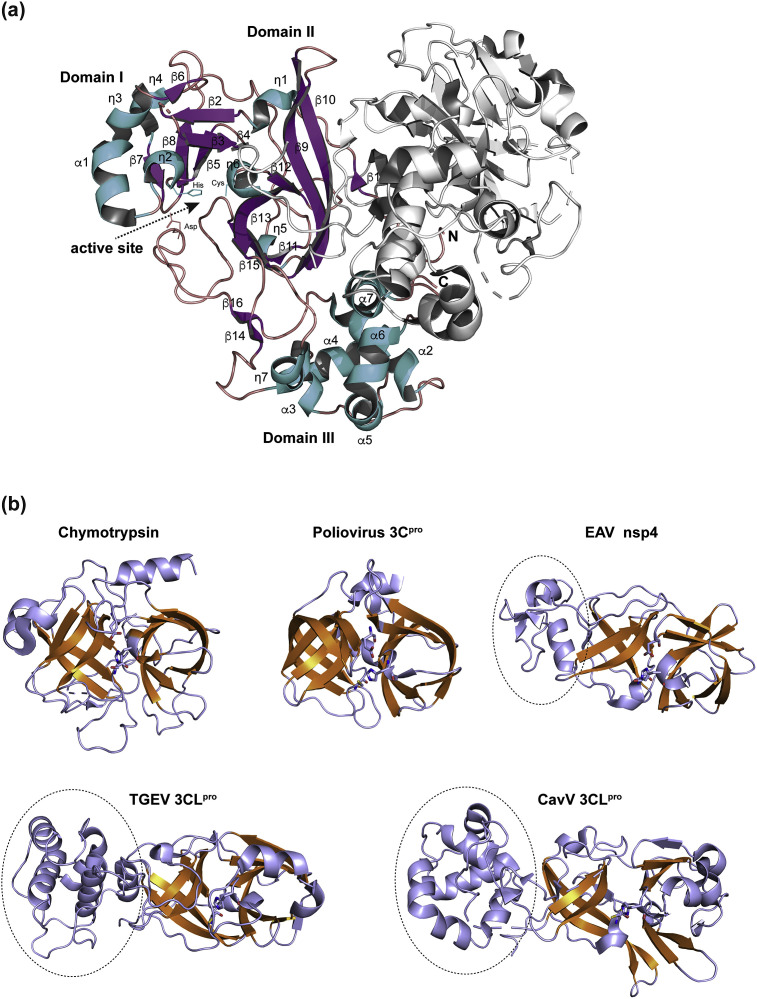
The CavV 3CLpro crystal structure determined at 1.94 Å resolution shows two 3CLpro molecules in the asymmetric unit (Fig. 3 A). The two molecules, named A (residues 1–314) and B (residues 1–305), form a dimer with an overall contact surface of 1728 Å2, involving 4 salt bridges and 24 hydrogen bonds as calculated by PDBePISA (Krissinel and Henrick, 2007). Molecules A and B are quite similar, with a root-mean-square deviation (RMSD) of 0.59 Å (as determined for all equivalent Cα atoms).
Fig. 3.
(a) Schematic view of the secondary structure elements of the CavV 3CLpro (PDB ID: 5LAC). Chain B is shown with helices colored in blue, β-strands colored in purple, and loops colored in red; chain A is shown in white. This figure and the following figures were generated using PyMol. (b) Structural homology of nidovirus 3CLpros with chymotrypsin and picornavirus 3C proteases. The double β-barrel that characterizes members of the PA superfamily of proteases is highlighted in orange. Shown are the structures of equine arteritis virus (EAV) nsp4 (PDB ID: 1MBM), transmissible gastroenteritis virus (TGEV) 3CLpro (PDB ID: 1LVO), CavV 3CLpro (PDB ID: 5LAC), chymotrypsin (PDB ID: 5J4Q), and poliovirus 3Cpro (PDB ID: 1L1N). Dotted lines indicate the extra C-terminal helical domains conserved in main proteases of the Nidovirales, represented in this figure by EAV (family Arteriviridae), TGEV (family Coronaviridae), and CavV (family Mesoniviridae). Catalytic residues are shown as sticks.
Each molecule is comprised of three domains, called I, II, and III, respectively. Domains I and II have a chymotrypsin-like, two-β-barrel fold that is formed by seven and six β-strands, respectively (β2-β8 and β9-β13 plus β15). The principal catalytic residues, Cys153 and His48 (Blanck et al., 2014), are part of strand β9 and 310 helix ƞ6, respectively, and are positioned in the center of the active-site cleft located between domains I and II. Domain II is connected to the C-terminal domain III, the latter being formed by seven helices (ƞ7 and α2 to α7) (Fig. 3A). Due to partially poor electron density, residues 51–55, 191–194, and 214–220 of chain A as well as residues 27–30 and 306–314 of chain B were omitted from the model. The C-terminal part of chain A wraps around chain B and thereby comes in close contact to the active site of chain B.
The N-terminal domains I and II of CavV 3CLpro resemble those of related enzymes from the Coronaviridae and Arteriviridae and the 3C/3CLpro enzymes from other RNA + viruses that lack an extra C-terminal domain (Fig. 3B) (Matthews et al., 1994; Mosimann et al., 1997; Phan et al., 2002; Speroni et al., 2009; Muhaxhiri et al., 2013; Allaire et al., 1994; Anand et al., 2002, 2003; Bergmann et al., 1997; Xue et al., 2008; Yang et al., 2003; Barrette-Ng et al., 2002; Zhao et al., 2008).
2.2. CavV 3CLpro active site
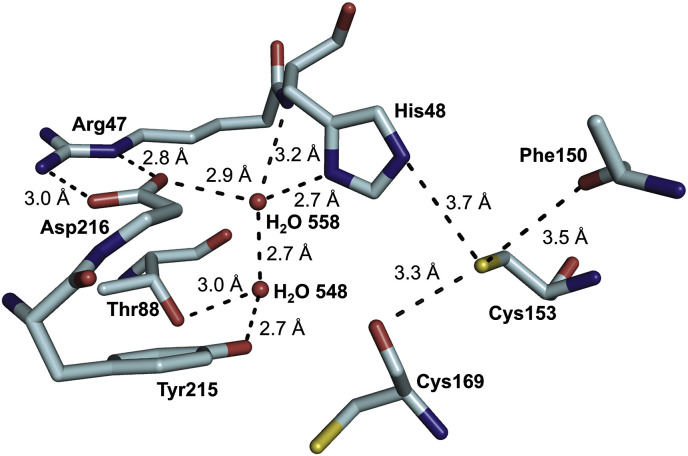
The Sγ atom of the catalytic Cys153 in chain B is in H-bond distance to the carbonyl oxygen of Phe150 (3.5 Å), the carbonyl oxygen of Cys169 (3.3 Å), and the Nε of the catalytic His48 (3.7 Å), respectively. The Nδ of catalytic His48 makes also contact to the carboxylate side chain of Asp216 through water molecule 558 with distances of 2.7 Å and 2.9 Å, respectively, forming an angle of 118.9° (chain B). This water molecule establishes another H-bond to water molecule 548 (2.7 Å), which is also hydrogen-bonded to Oγ of Thr88 (3.0 Å) and the side chain oxygen of Tyr215 (2.7 Å) (Fig. 4 ). Furthermore, Asp216 forms a salt bridge to Arg47 with H-bond distances of 2.8 and 3.0 Å, respectively. In monomer A, the residues of the active site exhibit a higher flexibility compared to the residues of chain B, which is indicated by (i) a slightly more disordered electron density for His48, (ii) an alternative side chain conformation for Cys153 (56% conformer A, 44% conformer B), and (iii) the lack of visible Asp216.
Fig. 4.
Representation of an extensive H-bond network involving a conserved water molecule and the catalytic Cys153 and His48 in the active site of the CavV 3CLpro structure (PDB ID: 5LAC; chain B). Only amino acids involved in hydrogen bonding are shown as sticks and color-coded by atom type. Dashed lines indicate key hydrogen bonds. For clarity, the backbone carbonyl group of Asp216 and the backbone amide of Arg47 were omitted from this representation.
2.3. Structure of a CavV 3CLpro/inhibitor complex
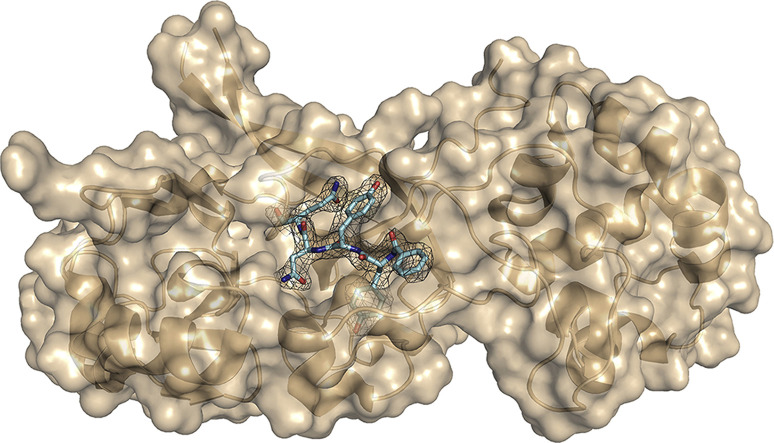
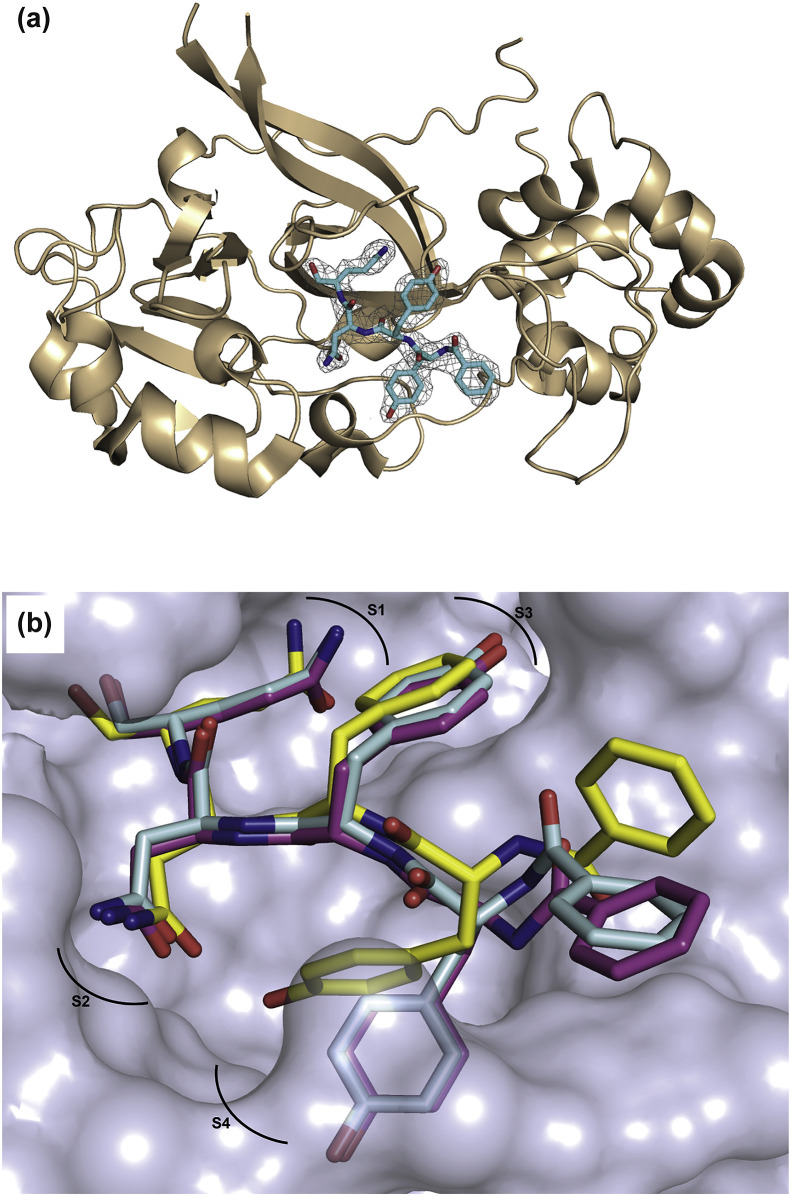
Overall structure. The structure of CavV 3CLpro in complex with Bz-YYNQ-H, a peptide aldehyde inhibitor representing the P1–P4 residues of the C-terminal CavV 3CLpro autoprocessing site, contains four rather than two molecules in the asymmetric unit. The molecules form a tetramer (a dimer of dimers) with RMSDs of 0.53 Å (for dimer A-B) and 0.65 Å (for dimer C-D), respectively. RMSD values of all four monomers are low and range between 0.33 and 0.56 Å, suggesting that the four monomers adopt the same fold. The total contact areas between monomers A and B and between C and D were calculated to be 1770 Å2 and 1640 Å2, respectively. Dimer A/B is stabilized by four salt bridges and 29 hydrogen bonds whereas, in the contact surface of C/D, three salt bridges and 24 hydrogen bonds are present as determined by PDBePISA (Krissinel and Henrick, 2007). With only 494 Å2, the contact area between the two dimers is significantly smaller and involves three salt bridges and eight hydrogen bonds. In both monomers A and C, Arg28 was found to reach into the substrate-binding pocket of another molecule (C and A, respectively), resulting in different interactions between enzyme and ligand in those cases (see below). (In monomer C, the side chain of Arg28 was omitted due to missing electron density, but it would interact with molecule A if present.) As expected, the ligand Bz-YYNQ-H has reacted with the sulfhydryl moiety of the catalytic Cys153 and the resulting thiohemiacetal is clearly detectable in the active site with well-defined density in the 2mFo-DFc map for monomers A, B, and C (Fig. 5 A). Also for monomer D, the Fo-Fc map indicates that the ligand is bound but, in this case, the electron density is relatively poor. We therefore omitted the ligand from the final model. The binding mode of the ligand to the three subsites S1, S2, and S3 is nearly identical for monomers A, B, and C, while slight variations exist among the three monomers with respect to the S4 subsite (Fig. 5B).
Fig. 5.
(a) Crystal structure of Bz-YYNQ-H (cyan, color-coded by atom type) in complex with CavV 3CLpro (cartoon, wheat). The electron-density map (2mFo-DFc) for the ligand is displayed at a σ level of 1.0 as gray mesh. (b) Superposition of the peptide aldehyde bound to chain A (blue), chain B (yellow), and chain C (pink), respectively, each represented as sticks and color-coded by atom type. The substrate-binding pocket of chain C (light blue) is shown in surface representation. Subpockets S1 to S4 are indicated.
Binding mode. In monomers A, B, and C, the ligand is covalently bound to Cys153 with C–S bond lengths of 1.9 Å (chain A), 1.8 Å (chain B), and 1.8 Å (chain C), respectively. The oxyanion hole that stabilizes the noncovalent Michaelis complex (Otto and Schirmeister, 1997) and the transition state includes the amide nitrogens of Gly151 (2.9 Å to the oxygen of the thiohemiacetal) and Cys153 (3.1 Å to the oxygen of the thiohemiacetal) in molecule A. Similar distances are observed in molecule C for the oxygen atom of the thiohemiacetal (2.8 Å to Gly151 and 2.9 Å to Cys153), while the distances in molecule B are much longer (4.4 Å to Gly 151 and 4.5 Å to Cys153) due to R-configuration. The ligand is suitably positioned to facilitate the nucleophilic attack of the sulfhydryl moiety at the prochiral carbonyl carbon, resulting in the formation of the (S)-configured thiohemiacetal in molecules A and C. Overall, the binding pockets of the monomers have a similar shape, except for two flexible regions, Tyr215-Val222 and a small region around the oxyanion hole (Phe150-Cys153).
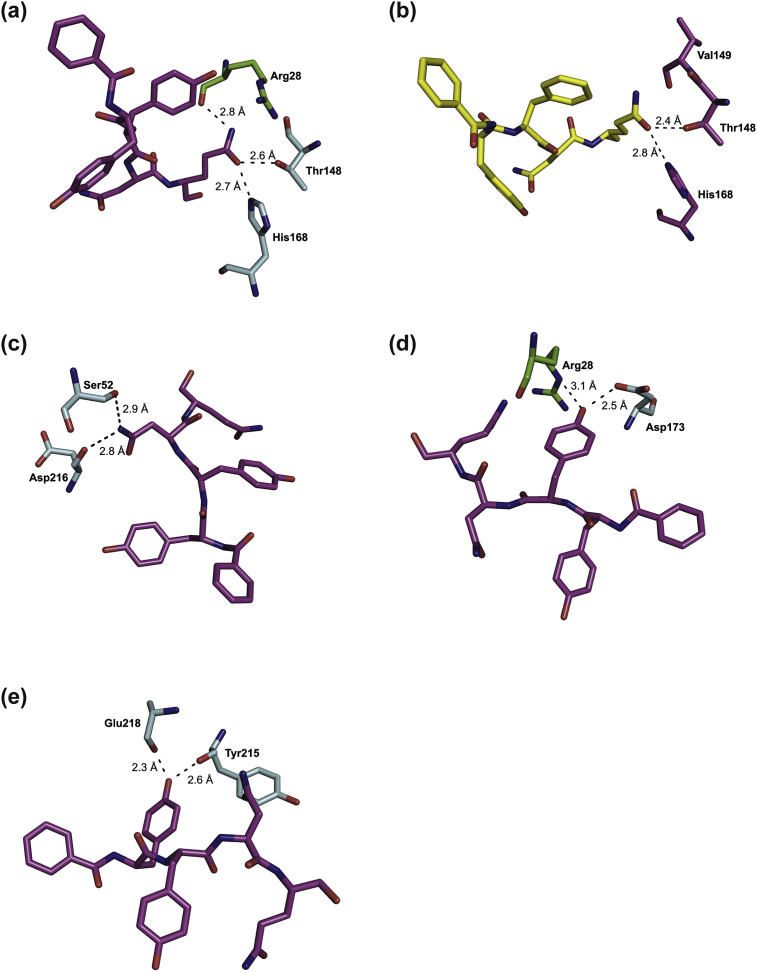
S1 subpocket. In monomer A, the S1 subpocket is formed by His168 and Thr148, plus Arg28 of chain C, with the main chain carbonyl oxygen of Arg forming a hydrogen bond to the Nε of the P1 Gln of the ligand with a distance of 2.8 Å. As the side chain of Arg28(C) was not clearly defined in the electron density, it was not considered further.
In contrast, the side chain of Arg28 of chain A is well defined in the density of the S1 site of monomer C (Fig. 6 A), which is organized in a manner similar to that of monomer A. The side chain closes the S1 site and the main chain carbonyl oxygen of Arg28 forms a hydrogen bond to the Nε of the P1 Gln (2.8 Å). The Oε of the P1 Gln establishes H-bonds to the Nε of His168 (C) at a distance of 2.7 Å and to the Oγ of Thr148 (C) at a distance of 2.6 Å.
Fig. 6.
Key interactions of the ligand in the S1 (a), S2 (c), S3 (d), and S4 (e) subpockets, respectively, of monomer C and in the S1 subpocket of monomer B (b) of the CavV 3CLpro (PDB ID: 5LAK). Atoms of the ligand (pink) and protease (chain C in light blue and chain A in green) are represented as sticks and color-coded by atom types. In panel (b), the ligand is shown in yellow and residues of chain B are shown in pink. Only residues involved in hydrogen bonding are shown.
In monomer B, the ligand is bound slightly differently (Fig. 6B). The S1 subpocket is formed by His168 (B), Thr148 (B), and Val149 (B). The polar interactions between ligand and S1 subsite are very similar to those observed for monomers A and C. The binding is stabilized by H-bonds between the Oε of the P1 Gln to both the Oγ of Thr148 (B; 2.4 Å) and the Nε of His168 (B; 2.8 Å). We consider it likely that the observed interactions of Arg28 (of chain C and A, respectively) with the S1 subpockets of another monomer (A and C, respectively) result from the crystal packing and thus represent crystallographic artifacts that are unlikely to occur in solution.
S2 subpocket. The P2 Asn residue in monomer C of the Bz-YYNQ-H ligand points toward the S2 subpocket (Fig. 6C) formed by Asp216 and Ser52. Polar contacts are established between the Nδ of the ligand P2 Asn and the carbonyl oxygen of Asp216 (chain A: 2.8 Å; chain B: 2.9 Å; chain C: 2.9 Å). In monomers A and B, a second hydrogen bond is formed with the Oγ of Ser52 (chain A: 2.9 Å; chain B: 3.4 Å). Because of insufficient electron density, the latter interaction could not be confirmed for monomer C.
S3 subpocket. The S3 pocket (Fig. 6D) is formed by Asp173, with a hydrogen bond between Oδ of Asp173 and the oxygen of the P3 tyrosine (monomer A: 2.7 Å; monomer C: 2.5 Å) stabilizing the binding to the ligand. In monomer C, an additional hydrogen bond (3.1 Å) to Nε of Arg28 (A) is formed. A similar binding mode was observed for the P3 tyrosine in molecule B, albeit the side chain OH was not included in the model.
S4 subpocket. The S4 pocket (Fig. 6E) is formed by Leu209, Tyr 215, and Val222 and flanked by a loop consisting of residues Asp216 to Asn221. The hydroxyl functionality of the ligand establishes hydrogen bonds to the carbonyl oxygen of Tyr215 (monomer A: 2.7 Å; monomer C: 2.6 Å) and to the carbonyl oxygen of Glu218 (chain A: 3.3 Å; chain C: 2.3 Å). The observed binding mode of the ligand differs in monomer B, where the residues of the loop region adopt a different conformation, resulting in a smaller S4 pocket, which, in turn, leads to a relocation of the P4 tyrosine. For the ligand in monomer B, an intramolecular hydrogen bond to the P2 Asn residue is observed (2.5 Å).
For the N-terminal benzoyl group of the ligand, different conformations were observed for each monomer, resulting in different interactions. While the ligand does not establish any H-bonds in monomer A, H-bonds are formed to the amide nitrogen of Val222 (3.6 Å) in monomer B and to the carbonyl oxygen atom of Thr220 (2.8 Å) in monomer C.
3. Discussion
In this study, we present the first crystal structure of a 3CLpro from a mosquito-borne nidovirus representing the family Mesoniviridae in the Nidovirales, a profoundly divergent order of RNA + viruses. The structure supports and extends previous studies suggesting that nidovirus 3C-like main proteases share a three-domain domain organization involving a chymotrypsin-like fold (domains I and II) and a C-terminal domain III (Anand et al., 2002; Barrette-Ng et al., 2002; Blanck et al., 2014; Blanck and Ziebuhr, 2016; Lauber et al., 2012; Nga et al., 2011; Zirkel et al., 2011, 2013). Domain III is absent (or much smaller) in the related 3C/3CLpros of other RNA + viruses, while the N-terminal chymotrypsin-like fold (Birktoft and Blow, 1972; Matthews et al., 1967) is conserved in all these enzymes (Matthews et al., 1994; Mosimann et al., 1997; Muhaxhiri et al., 2013; Allaire et al., 1994). Despite their conserved structural organization, nidovirus 3CLpros are remarkably diverse with respect to their catalytic residues, again illustrating the profoundly divergent evolution of the various subfamilies of the order Nidovirales. Thus for example, arterivirus, torovirus and bafinivirus 3CLpros employ a canonical Ser-His-Asp triad (Barrette-Ng et al., 2002; Smits et al., 2006; Ulferts et al., 2011), while coronavirus, mesonivirus, and ronivirus 3CLpros use a Cys-His catalatic dyad that, as shown previously for coronavirus (Anand et al., 2002) and, in this study, for mesonivirus 3CLpro, is assisted by a water molecule interacting with an Asp residue located in a noncanonical position in the primary structure, C-proximal of the catalytic Cys (see below and Fig. 2).
Database searches for related structures using the DALI server (Holm and Rosenström, 2010) revealed the 3C-like cysteine proteases of the subfamily Coronavirinae as the closest structural homologs of the CavV 3CLpro (PDB ID: 5LAC, chain A). Using the HCoV-HKU1 3CLpro structure (PDB ID: 3D23, chain A) (Zhao et al., 2008), an RMSD (Cα) of 3.8 Å (sequence identity 8%; Z-score: 15.0) was calculated (239 residues of 299 from 3D23, chain A, were aligned).
Dimerization and interfaces. Similar to what was shown previously for coronavirus 3CLpros, the CavV 3CLpro forms a tight dimer in the crystal structure. The arrangement of the two molecules in dimer A/B and C/D, respectively, is similar to the orientation reported for the two protomers that form the coronavirus 3CLpro dimer (Anand et al., 2002, 2003; Yang et al., 2003). Each monomer of the two dimers of the CavV 3CLpro structure reaches into the other, with its N-terminus (residues 1–16) interacting with residues of the dimer mate, forming 6 hydrogen bonds (A2-B147, A5-B136, A5-B136, A7-B147, A10-B134 (2x)) and 1 salt bridge (A1-B296) in the case of monomer A. For the entire A/B dimer, 20 hydrogen bonds and 5 salt bridges are formed. The large contact interfaces observed for dimers A/B (1764 Å2) and C/D (1640 Å2) suggest that CavV 3CLpro dimer formation is of functional relevance which remains to be confirmed in further studies, such as those performed for coronavirus 3CLpros, in which the role of dimerization for trans-cleavage activity was confirmed and characterized in significant detail (for a review, see (Xia and Kang, 2011)). In contrast to the A/B and C/D dimers described above, the formation of the tetramer A/B–C/D with an interface area of 494 Å2 (involving three salt bridges and eight hydrogen bonds between monomers A and C) likely represents a crystallographic artifact.
Substrate-binding pocket. The S1 pockets of CavV 3CLpro and coronavirus 3CLpros are similar and can accommodate a Gln residue as shown here for a peptide aldehyde corresponding to the P1-to-P4 residues of the C-terminal CavV 3CLpro autoprocessing site. The structure shows that, in addition to His168, a conserved Thr residue (Thr148) is located in the S1 subsite and suitably positioned to establish interactions with Gln (or Glu), both of which are common in the P1 position of mesonivirus (including CavV) 3CLpro substrates (Blanck and Ziebuhr, 2016). Interactions between the equivalent Thr/His residues conserved in other viral 3C/3CLpros, including arterivirus nsp4 (Barrette-Ng et al., 2002; Snijder et al., 1996), and the side chains of P1 Glu/Gln residues have been shown previously to be critically involved in the binding of Gln and Glu residues, respectively. Interestingly, mesonivirus 3CLpros do not appear to have a strong preference for Gln in the P1 position of substrates (which is a typical feature in coronavirus 3CLpros) nor do they have a preference for Glu over Gln as shown for arterivirus nsp4 enzymes (Ziebuhr et al., 2000). A possible explanation for this less pronounced specificity for Gln or Glu in mesonivirus 3CLpros may be that His and Thr do not establish the same type of interactions with nearby located residues that appear to be required for fine-tuning the P1 specificity for Gln and Glu, respectively (Mosimann et al., 1997; Anand et al., 2002; Bergmann et al., 1997). Thus, for example, the equivalent His residue in the S1 subsite of coronavirus 3CLpros was reported to establish interactions with two other residues (Phe and Tyr) (Anand et al., 2002). These latter interactions are thought to keep the His residue over a wide pH range in the neutral state required for interacting with the P1 Gln side chain. Compared to other viral 3C/3CLpros, mesonivirus 3CLpros have a less pronounced specificity toward the P1 residue and tolerate a range of different residues in this position. In addition to Gln and Glu, Lys and several other residues are found at the P1 position of predicted cleavage sites in the replicase polyprotein of CavV and several other mesonivirus 3CLpros (Blanck and Ziebuhr, 2016). Interestingly, a replacement of His168 with Ala in a bacterial fusion protein construct containing the CavV 3CLpro was previously shown to have differential effects on the cleavage of the N- and C-terminal 3CLpro autoprocessing sites (Blanck et al., 2014). While cleavage at the N-terminal autoprocessing site was retained, cleavage of the C-terminal processing site was abolished in a CavV 3CLpro H168A mutant. As the N-terminal autoprocessing site contains a Lys residue in the P1 position, while the C-terminal autoprocessing site contains Gln in this position, it is tempting to speculate that His168 is required for the cleavage of substrates containing Gln (and, likely, Glu) in the P1 position, while this His residue may be (largely) dispensable for the cleavage of substrates containing Lys in the P1 position. To test this hypothesis and, more generally, establish the structural details of substrate binding for 3CLpro substrates with other residues at the P1 (and other) position(s), additional studies of enzyme/inhibitor complexes with suitable peptidic inhibitors should be performed. Also, it may be worth testing if (and to what extent) specific P1 and other residues flanking the scissile bond affect the cleavage efficiency at specific polyprotein cleavage sites, thereby possibly contributing to the timely coordinated release of specific processing products from the viral replicase polyproteins. As indicated above, the P1 Gln of molecules A and C was found to (also) interact with the backbone carbonyl oxygen of Arg28 from a molecule of the other dimer. We however consider this a crystallographic artifact and, therefore, will not discuss this further.
Biochemical studies and comparative sequence analyses of mesonivirus replicase polyproteins (Blanck et al., 2014; Blanck and Ziebuhr, 2016) identified the P2 position as a key specificity determinant of mesonivirus 3CLpros. In CavV and most other members of the genus Alphamesonivirus, the P2 position of putative 3CLpro substrates is predominantly occupied by Asn. The crystal structure of the 3CLpro/inhibitor complex shows that the Asn side chain fits perfectly into the S2 subpocket, with its carboxamide functionality acting as hydrogen bond donor and acceptor in interactions with the main chain carbonyl oxygen of Asp216 and the Oγ atom of Ser52, respectively. The position of the strictly conserved Asp216 is stabilized by a salt bridge with the conserved Arg47 residue, suggesting that Asp216 has a dual functional role: besides its involvement in the coordination of the water molecule that interacts with the catalytic His48 residue, it is part of the S2 pocket, where it interacts with the carboxamide moiety of the Asn side chain. In addition to its specificity for Asn, the CavV 3CLpro S2 pocket would be suitable (and large enough) to accommodate other residues, such as Thr, which is found at the P2 position of one (out of 12) predicted CavV 3CLpro cleavage sites. In contrast, the P2 position of 3CLpro cleavage sites is either predominantly occupied by a Leu residue (in coronaviruses) or varies considerably (in arteriviruses).
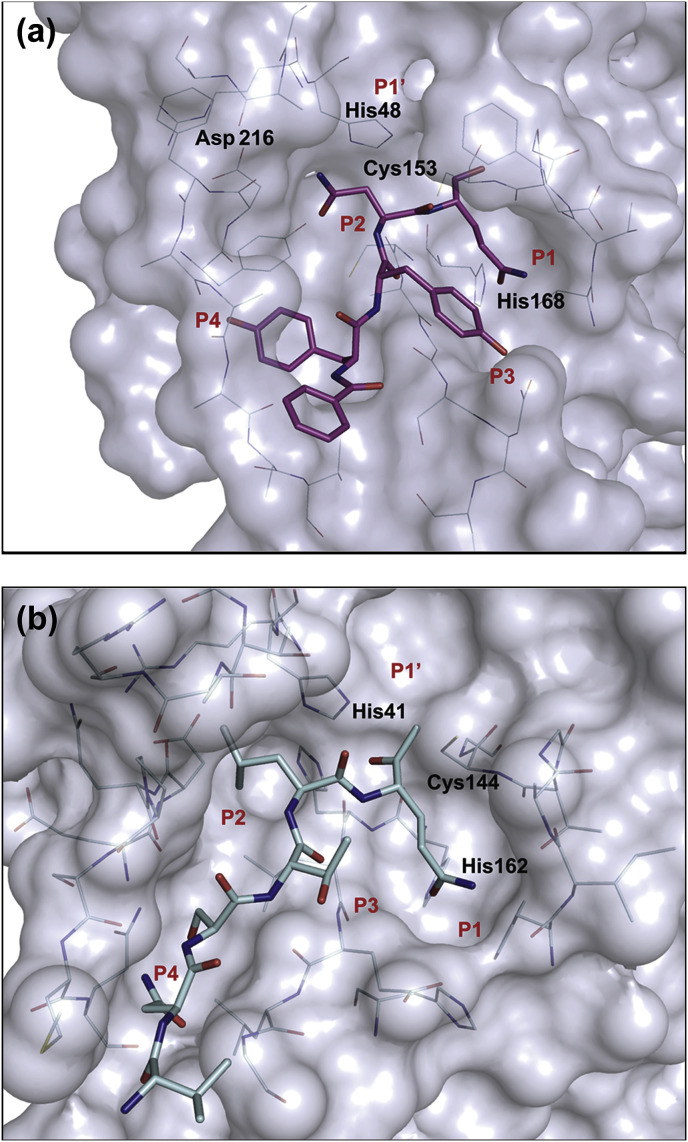
The direct interaction partner of the peptidic ligand in the S3 subpocket is Asp173, which forms a hydrogen bond to the phenolic hydroxyl of the tyrosine P3 residue. In this subpocket, also basic residues, such as Lys or Arg, as well as residues carrying a donor/acceptor function could establish a similar interaction pattern. This view is also supported by the occurrence of a range of other residues in the P3 position of CavV 3CLpro substrates (Blanck and Ziebuhr, 2016). Compared to the S3 and S1 subsites of coronavirus 3CLpros, the respective subsites are more spacious in the CavV 3CLpro (see Fig. 7 ).
Fig. 7.
Surface representation of mesonivirus and coronavirus 3CLpro substrate-binding sites using (a) the structure of the CavV 3CLpro/inhibitor complex (PDB ID: 5LAK; pink) and (b) the structure of TGEV 3CLpro in a complex with a peptidic inhibitor (Anand et al., 2003) (PDB ID: 1P9U; light blue). Ligands are represented as sticks and color-coded by atom types. Amino acids involved in substrate binding are shown as lines and color-coded by atom types. Catalytic residues and the conserved His residue in the S1 subsite are indicated (see text for details).
The rather hydrophobic S4 subpocket, consisting of Ile199, Ile207, Leu209, and Val222, is able to accommodate small hydrophobic residues including Leu, which is the most common residue at the P4 position of CavV 3CLpro substrates. However, as the loop formed by residues 216–221 covering the S4 pocket results in a semi-open pocket, larger residues with hydrogen bond donor functionalities, such as Tyr, can also be accommodated. In contrast to coronavirus 3CLpros, which offer only space for smaller residues, such as Ser, Val or Thr, in a fairly closed S4 pocket (Ziebuhr et al., 2000; Yang et al., 2003, 2005; Anand et al., 2002, 2003), the S4 subsite is larger in the CavV 3CLpro.
The main difference between the different chains present in the CavV 3CLpro/inhibitor crystal structure is that, in chain B (but not in chains A and C), a flipped Glu218 blocks the access to the S4 pocket, resulting in a deviating position of the benzoyl group which in turn enables an intramolecular hydrogen bond to the P2 substituent (Fig. 5). Also, no major differences are observed between the structures of the free enzyme and the enzyme/inhibitor complex. The superposition of 293 Cα atoms of 5LAC (chain B) and 5LAK (chain A) revealed an RMSD of 0.76 Å. The only major difference observed upon complex formation is a peptide flip of Phe150 enabling a hydrogen bond between the carbonyl oxygen of the inhibitor (Gln5) and the amide backbone of residue Gly151 (2.9 Å).
As reported previously, the primary structures of 3C-like cysteine proteases encoded by Coronavirinae, Roniviridae, and Mesoniviridae are poorly conserved except for two strictly conserved sequence signatures, RH and GxCG, that include the catalytic His and Cys residues (underlined) (Blanck et al., 2014; Nga et al., 2011; Zirkel et al., 2011, 2013; Ziebuhr et al., 2003). Our study suggests that this list of highly conserved residues may be extended by an Asp residue, Asp187 in the SARS-CoV-3CLpro and Asp216 in the CavV 3CLpro, that is shown to be conserved among the 3CLpros of all known coronavirus and mesonivirus 3CLpros in a similar position of the sequence (Fig. 2). Furthermore, previous comparative sequence analyses revealed that an equivalent Asp residue may also be conserved in ronivirus 3CLpros (Ziebuhr et al., 2003), suggesting a key function for this residue. In the first crystal structure reported for a coronavirus 3CLpro, this Asp (Asp186 in the TGEV 3CLpro sequence) was found to form a hydrogen bond to a water molecule located in a position that, in chymotrypsin and related serine proteases, is occupied by the side chain of the third member of the catalytic triad (typically Asp) (Anand et al., 2002). Asp186 was observed to form a salt bridge with Arg40. The strict conservation of the Asp and Arg residues in coronavirus 3CLpros was suggested to indicate an important role of Asp (and Arg) in maintaining the active-site geometry including the substrate-binding site, while a direct role for Asp186 in catalysis was considered unlikely. Our database searches revealed the presence of a water molecule that mediates a contact between the conserved Asp and the catalytic His residue in 70 out of 80 coronavirus 3CLpro structures available in the Protein Data Bank. In the remaining 10 structures, this water molecule is not visible, most likely because of insufficient resolution of the respective structure. The angle between Asp, the conserved water molecule and the catalytic His ranged between 119° and 145° (mean 130.3°) in the coronavirus 3CLpro structures and between 122.8° and 128.4° in the CavV 3CLpro/inhibitor complex structure (chains A-C) and 118.9° in the structure of the free enzyme (chain B), thus representing a nearly perfect angle for a hydrogen-bonded water bridge.
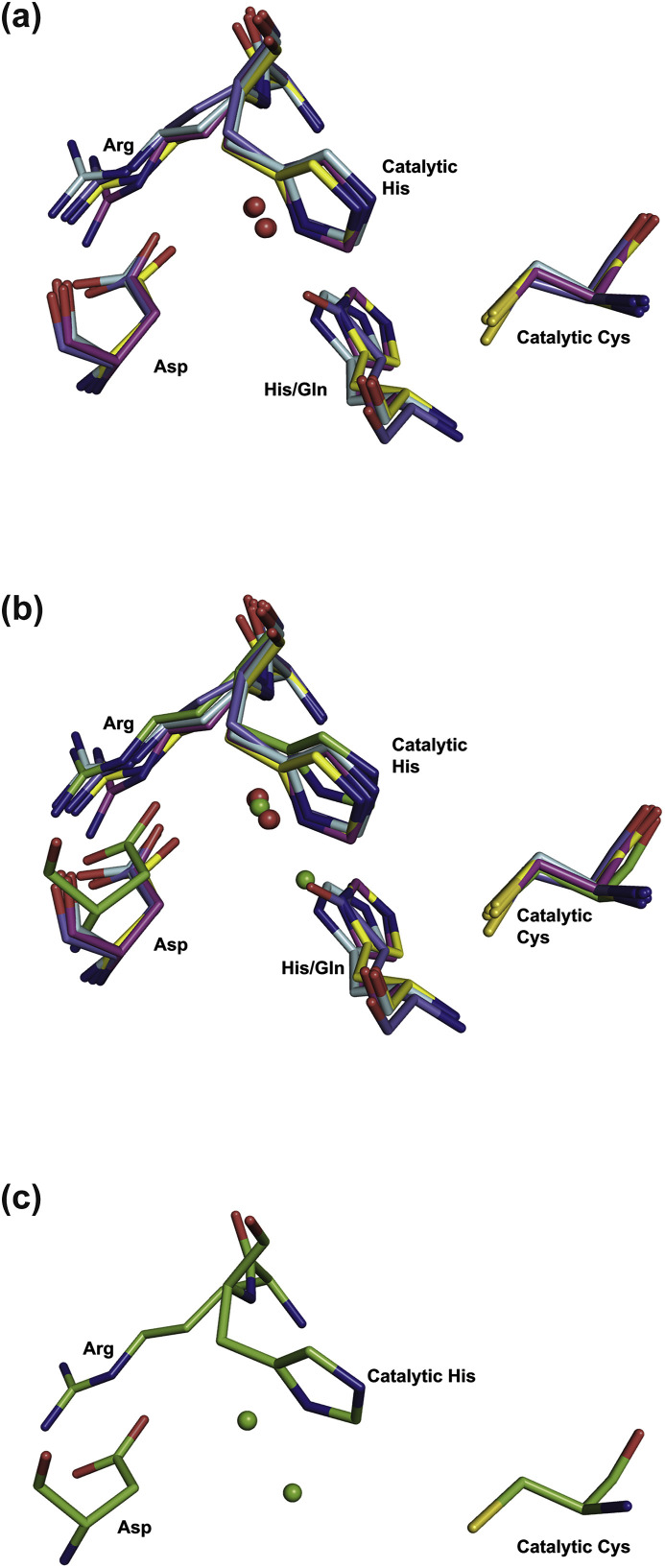
In all monomers of the two CavV 3CLpro structures presented in this study, Asp216 and Arg47 form a salt bridge, with Asp216 retaining its capability to act as a hydrogen bond acceptor for the buried water molecule. The conserved water forms a hydrogen bond network with several partners and serves as a kind of relay station. It forms hydrogen bonds to Asp216 (see above) and the backbone nitrogen of His48. Furthermore, the water molecule interacts with Nδ of His48, supporting a role in catalysis by stabilizing the protonated state of the catalytic His during the transition state (Fig. 4). In chains A and C of the structure of the CavV 3CLpro/inhibitor complex (PDB ID: 5LAK) and in chain B of the structure of the free enzyme (PDB ID: 5LAC), another conserved water molecule acts as a fourth partner. In coronavirus 3CLpro structures, the position of this second water molecule is occupied by His and Gln, respectively, each of them donating a hydrogen bond to the conserved water molecule (SARS-CoV 3CLpro His164; PDB ID: 3SNE (Zhu et al., 2011); MERS-CoV 3CLpro Gln167; PDB ID: 4WME (Needle et al., 2015)) (Fig. 8 ).
Fig. 8.
Superimposition of active-site residues of coronavirus and mesonivirus 3CLpros. Shown are the active sites of the 3CLpros of TGEV (Kim et al., 2012) (PDB ID: 4F49; light blue), IBV (Xue et al., 2008) (PDB ID: 2Q6D; pink), SARS-CoV (Zhu et al., 2011) (PDB ID: 3SNE; yellow), HCoV-HKU1 (Zhao et al., 2008) (PDB ID: 3D23; dark blue), and CavV (PDB ID: 5LAK; green). All structures are represented as sticks and color-coded by atom types. Water molecules of 4F49, 2Q6D, 3SNE and 3D23 are represented as red spheres and water molecules of 5LAK are represented as green spheres. (a) superimposition of 4F49, 2Q6D, 3SNE, and 3D23; (b) superimposition of the coronavirus 3CLpro active sites shown in panel (a) (PDB ID: 4F49, 2Q6D, 3SNE, and 3D23) with the CavV 3CLpro active site (PDB ID: 5LAK); (c) active site of the CavV 3CLpro alone (PDB ID: 5LAK).
These observations lead us to suggest that the 3CLpros of both Coronavirinae and Mesoniviridae share a conserved catalytic Cys-His dyad that is assisted by a water molecule interacting with an Asp side chain. In both coronavirus and mesonivirus proteases, the Asp residue forms a salt bridge with a conserved Arg that immediately precedes the catalytic His in the primary structure (Fig. 2). Furthermore, as revealed by the CavV 3CLpro/inhibitor structure (Fig. 6), the main chain carbonyl oxygen of this Asp residue contributes also to the S2 subsite of the substrate-binding pocket.
Taken together, the study provides interesting new insight into the molecular basis of substrate binding and catalysis of nidovirus 3CLpros. With less than 10% amino acid sequence identity, nidovirus 3CLpros are extremely diverse and include enzymes that either employ the classical catalytic triad of chymotrypsin-like serine proteases (Ser-His-Asp) as shown for members of the Arteriviridae and (former) Torovirinae (Barrette-Ng et al., 2002; Smits et al., 2006; Ulferts et al., 2011) or a noncanonical Cys-His catalytic dyad assisted by a water molecule that is oriented by a complex network of interactions involving a conserved Asp residue located in a region that connects domains II and III. Also, this work reveals the structural basis for the distinct specificities reported for coronavirus and mesonivirus 3CLpros, respectively.
4. Material and methods
Phylogeny reconstruction. Nidovirus phylogeny was reconstructed based on an MSA of the conserved core of the RdRp which was generated using the Viralis platform (Gorbalenya et al., 2010). Representatives of 67 species, 67 out of 88 delineated by DEmARC (Lauber and Gorbalenya, 2012) and recognized now by ICTV (Siddell et al., 2019; Adams et al., 2016; de Groot et al., 2013), were included in this analysis (Supplementary Table 1). IQ Tree 1.5.5 (Nguyen et al., 2015) with automatically selected LG + F + R7 evolutionary model was used. To estimate branch support, the Shimodaira-Hasegawa-like approximate likelihood ratio test (SH-aLRT) with 1000 replicates was conducted (Guindon et al., 2010).
Structure-based multiple sequence alignment. The structure-based MSA of coronavirus and CavV 3CLpros was built using the PyMOL Molecular Graphics System, version 2.2.3 (Schrödinger, LLC), command “extra_fit”, alignment method “cealign”, with only alpha carbon atoms considered. The TGEV 3CLpro structure (PDB entry 1LVO, chain A) served as a reference to which other structures (Supplementary Table 2) were aligned independently. For non-CavV mesoniviruses (Supplementary Table 1), the 3CLpro MSA was downloaded from the Viralis platform (Gorbalenya et al., 2010). The two MSAs were aligned by MUSCLE v3.8.31 in the profile mode (Edgar, 2004). The resulting MSA was manually adjusted to improve regional sequence and structure similarity, mostly in the C-terminal domain. This refinement took into account another structure-based alignment of the same enzymes which was generated using the protein structure comparison service PDBeFold at the European Bioinformatics Institute (Krissinel and Henrick, 2004) which superimposes multiple structures simultaneously (Krissinel et al., 2005). Secondary structures were retrieved from the DSSP database (Hekkelman and Vriend, 2005; Touw et al., 2015). The resulting alignment was visualized by ESPript 2.1 (Gouet et al., 2003; Robert and Gouet, 2014).
Production and purification of wild-type CavV 3CL pro . Wild type CavV 3CLpro was produced as described before (Blanck and Ziebuhr, 2016) using a slightly modified purification protocol. Briefly, a fusion protein, in which the maltose-binding protein (MBP) sequence was fused to the complete 3CLpro coding sequence (corresponding to the CavV pp1a/pp1ab residues 1387 to 1700, GenBank NC_015668), was produced in E. coli and purified by amylose affinity chromatography. Following cleavage with factor Xa, MBP and 3CLpro (the latter containing no extra residues) were separated by anion-exchange chromatography. Pooled peak fractions containing CavV 3CLpro were further purified by size-exclusion chromatography in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, 0.1 mM EDTA, and 1 mM DTT and using an ÄktaPURIFIER 10 chromatography system equipped with a HiLoad 16/60 Superdex 75 column (GE Healthcare). CavV 3CLpro eluted as a single peak after 60 mL. Following adjustment of the protein concentration to 7 mg/mL using Amicon® Ultra Filters (10 kDa NMWL), the protein was immediately used for crystallization experiments.
Production and purification of SeMet-CavV 3CL pro . The MBP-pp1a-1387-1700 coding sequence (Blanck et al., 2014) was subcloned into pET11d (Novagen) and selenomethionine-labeled MBP-3CLpro fusion protein was produced using the methionine auxotroph E. coli strain B834(DE3). Freshly transformed cells were grown in SM medium (2x M9 salts, 0.4% glucose, amino acids except methionine [each at 40 μg/mL], vitamins [1 μg/mL], adenine, thymine, uracil, guanine [each at 200 μg/mL] and trace elements) containing carbenicillin (75 μg/mL) and methionine (40 μg/mL). At an optical density at 600 nm of 0.6, the cells were harvested by low-speed centrifugation, resuspended in prewarmed SM medium without methionine and incubated for another 4 h at 37 °C. Then, seleno-l-methionine (40 μg/mL) was added to the medium and the culture was incubated for 30 min at 37 °C. Next, protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and the culture was incubated overnight at 16 °C under vigorous shaking (225 rpm). SeMet-CavV 3CLpro was purified as described above, concentrated to 7 mg/mL and used in crystallization trials.
Crystallization. Crystallization screens (1152 different conditions) were performed at the MarXtal laboratory (Philipps University Marburg) using CavV 3CLpro (7 mg/mL) or a mixture of CavV 3CLpro and N-benzoyl peptide (Bz-YYNQ-H; ThinkPeptides, Oxford, UK). The sequence of the peptide was derived from the CavV 3CLpro C-terminus and represents the P4–P3–P2–P1 residues of the C-terminal 3CLpro autoprocessing site. For crystallization of the enzyme/inhibitor complex, the protein solution (7 mg/mL) was pre-incubated with the peptide (1.5 mM) for 2 h to facilitate the formation of the thiohemiacetal with the sulfhydryl group of the catalytic Cys153. For the free enzyme, crystals suitable for diffraction experiments were obtained using 0.2 M lithium acetate, 24% polyethylene glycol (PEG3350). Crystals grew overnight using a mixture of 0.5 μL reservoir solution and 1 μL protein solution in a 15-well hanging-drop plate (Qiagen). Before flash-cooling in liquid nitrogen, crystals were incubated for 10–15 s in reservoir solution containing 20% glycerol. SeMet-CavV 3CLpro was crystallized using identical conditions. First crystals of the 3CLpro/inhibitor complex were observed after 6 days at 18 °C. The best crystals that were used for subsequent diffraction experiments were obtained by using 0.1 M bicine (pH 8.5) with 20% PEG6000 as the reservoir. Before flash-cooling, the crystals were incubated for 10–15 s in reservoir solution containing 20% PEG400.
X-ray diffraction. Preliminary diffraction experiments were carried out using a CuKɑ-X-ray source from Incoatec IμS with a Mar345dtb image plate detector, which rendered diffraction up to 2.8 Å for the CavV 3CLpro, the CavV 3CLpro/inhibitor complex, and the SeMet-CavV 3CLpro crystals. High resolution data sets for the CavV 3CLpro and SeMet-labeled CavV 3CLpro crystals, respectively, were collected at the MX beam line 14.2 at BESSY II (Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung, Helmholtz-Zentrum Berlin für Energie und Materialien, Berlin, Germany) (Mueller et al., 2015). Measurements were carried out at 100 K and a wavelength of 0.91841 Å. Data were processed using XDSAPP (Krug et al., 2012) resulting in a CavV 3CLpro SeMet dataset diffracting up to a resolution of 1.89 Å (see Table 1 ).
Table 1.
Collection statistics of MAD data obtained for a SeMet-labeled CavV 3CLpro (5LAC) protein crystal.
| Data collection | Peak | Inflection point | High energy remote (for phasing) |
|---|---|---|---|
| Wavelength (Å) | 0.97971 | 0.97990 | 0.91841 |
| Space group | P2(1)2(1)2 | ||
| Unit-cell parameters (Å) | a = 94.1, b = 111.3, c = 58.2 | ||
| Resolution range (Å)a | 50.0–2.00 (2.12-2-00) | 50.0–2.07 (2.19–2.07) | 50.0–1.89 (2.00–1.89) |
| No. of unique reflections | 78454 (12573) | 71073 (11493) | 93137 (15373) |
| Multiplicitya | 2.4 (2.3) | 2.4 (2.3) | 2.6 (2.5) |
| R sym (%)a,b | 5.3 (27.5) | 5.7 (30.0) | 5.9 (45.2) |
| CC (1/2) (%) | 99.7 (86.2) | 99.7 (83.1) | 99.8 (74.9) |
| Completeness (%)a | 98.1 (97.5) | 98.5 (98.8) | 98.0 (97.8) |
| {I/σ(I)} | 14.0 (3.7) | 13.6 (3.5) | 13.4 (2.5) |
Values in brackets are statistics for the highest resolution shell.
Rsym = where I is the observed intensity and Ī is the average intensity for multiple measurements.
In order to obtain phase information, a MAD experiment using crystals of SeMet-CavV 3CLpro, belonging to the space group P21212 (unit cell dimensions: a = 94.2 Å, b = 111.4 Å, c = 58.2 Å) with two molecules in the asymmetric unit, was carried out. Data were collected at the peak wavelength (0.97971 Å) with a significant anomalous signal up to 2.27 Å (d’’/sig(d’’) = 1.083–4.647), at the inflection point (λ = 0.97990 Å; 2.77 Å; d’’/sig(d’’) = 1.015–2.574), and at a high energy remote wavelength (λ = 0.91841 Å) with a resolution up to 1.89 Å. The anomalous signal allowed us to unambiguously determine 12 out of 14 possible SeMet sites using SHELXD (CCmax = 58.2) (Schneider and Sheldrick, 2002), followed by phasing performed with SHELXE (Sheldrick, 2002), all implemented in HKL2MAP (Pape and Schneider, 2004). An initial electron density map was calculated and a model (581 amino acids; 10 chains; 208 amino acids in the longest chain; score 0.960) was built by ARP/wARP (Langer et al., 2008). This model was further modified using COOT (Emsley et al., 2010) and refined using PHENIX (Adams et al., 2010).
A dataset for the co-crystallized CavV 3CLpro/inhibitor complex was collected at beam line BM30-A at the ESRF (European Synchrotron Radiation Facility, Grenoble, France). Measurements were carried out at 100 K at a wavelength of 0.979742 Å. The dataset was processed to a resolution of 2.3 Å using XDS (Kabsch, 2010). The crystals belong to space group P21 with cell dimensions a = 72.1 Å, b = 110.0 Å, c = 98.4 Å and β = 105.9°, featuring four molecules in the asymmetric unit.
Structure determination, model building, and refinement. Due to its higher resolution and superior data quality, the SeMet-CavV 3CLpro dataset was used for further model building and refinement. The refinement of the SeMet-CavV 3CLpro structure was carried out in PHENIX using xyz refinement, TLS refinement (from the TLSMD-Server; 12 groups), individual atomic displacement parameters (ADPs; isotropic), occupancy refinement, and finally NCS refinement (torsion angles). Water molecules were added in the automatic model building of ARP/wARP. For molecular replacement and first refinement cycles of the 3CLpro/inhibitor complex structure, an automated refinement script based on PHENIX was used (Schiebel et al., 2016). The structure of the SeMet-CavV 3CLpro was used as search model. For the refinement process in PHENIX, xyz refinement, TLS refinement (from the TLSMD-Server; 22 groups), grouped B-factors, occupancy refinement, and NCS refinement (torsion angles) were carried out. For R-free calculations, a 5% data fraction was used. COOT was used to add water molecules and for the fitting of amino acid side chains using σ-weighted 2Fo-Fc and Fo-Fc difference electron density maps. Restraints for ligands were obtained by using the Grade Web Server (http://grade.globalphasing.org/cgi-bin/grade/server.cgi). Average B-values were calculated using MOLEMAN (Kleywegt et al., 2006) and Ramachandran plot statistics were analyzed with PROCHECK (Laskowski et al., 1993) (see Table 2 ).
Table 2.
Data collection, refinement, and model statistics.
| Parameter | 5LAC | 5LAK |
|---|---|---|
| Wavelength (Å) | 0.91841 | 0.97974 |
| Space group | P2(1)2(1)2 | P2(1) |
| Unit-cell parameters (Å, °) | a = 94.2, b = 111.4, c = 58.2 | a = 72.1, b = 110.0, c = 98.4, ß = 105.9 |
| Resolution range (Å)a | 40.00–1.94 (1.99–1.94) | 50.00–2.30 (2.44–2.30) |
| Wilson B factor (Å2) | 24.1 | 35.1 |
| No. of unique reflections | 44979 (2800) | 65479 |
| Multiplicitya | 4.6 (4.7) | 3.2 (3.1) |
| R sym (%)a,b | 7.5 (46.8) | 7.0 (49.0) |
| Completeness (%)a | 97.0 (92.6) | 99.2 (98.3) |
| {I/σ(I)} | 21.1 (3.5) | 13.0 (2.6) |
| Resolution range in refinement (Å) | 36.99–1.94 | 47.61–2.30 |
| No. of reflections used for refinement | 44650 | 65456 |
| R cryst (%)c | 17.3 | 20.8 |
| R free (%)d | 20.7 | 23.0 |
| Refined residues | 599 | 1186 |
| Water molecules | 303 | 179 |
| Inhibitor atoms | – | 146 |
| Other ligands | 12 | 82 |
| R.m.s.d., angles (°) | 1.1 | 0.888 |
| R.m.s.d., distances (Å) | 0.008 | 0.004 |
| Ramachandran plot (%)e | ||
| Most favoured | 88.9 | 89.9 |
| Additionally allowed | 11.1 | 10.1 |
| Generously allowed | 0 | 0 |
| Outliers | 0 | 0 |
| Mean B (Å2) | ||
| All atoms | 28.1 | Chain A = 36.0 Chain B = 49.2 Chain C = 44.4 Chain D = 48.1 |
| Ligands | – | Ligand Chain A = 39.5 Ligand Chain B = 52.5 Ligand Chain C = 47.7 |
| Waters | 32.8 | 36.5 |
| Other solvents | 26.1 | 45.0 |
Values in brackets are statistics for the highest resolution shell.
Rsym = where I is the observed intensity and Ī is the average intensity for multiple measurements.
Rcryst = .
Rfree is calculated in the same manner as Rcryst but on 5% of the data that were not used for refinement.
from PROCHECK.
Coordinate accession numbers
Coordinates and structure factors for the CavV 3CLpro and the CavV 3CLpro/inhibitor complex have been deposited in the Protein Data Bank under the PDB IDs 5LAC and 5LAK, respectively.
Declarations of interest
None.
Acknowledgments
The work was supported by the Deutsche Forschungsgemeinschaft (SFB1021 A01, to JZ). The contribution of AAG and AEG was funded in part by the EU Horizon2020 EVAg 653316 project and the LUMC MoBiLe program (to AEG). We are very grateful to Prof. Dr. Wolfgang Buckel, Head of the Laboratory of Microbial Biochemistry and Max Planck Fellow of the MPI for Terrestrial Microbiology, Marburg, for helpful discussions regarding mechanistic aspects. We thank the beamline staff of BESSY II (Helmholtz-Zentrum Berlin) and ESRF for their outstanding help and support during data collection and acknowledge the generous support by travel grants from the Helmholtz-Zentrum für Materialien und Energie, Berlin. We also thank Ralf Pöschke (MarXtal, Philipps University Marburg) and Karin Schultheiß (Medical Virology, Justus Liebig University Giessen) for excellent technical assistance, and Igor Sidorov and Dmitry Samborskiy for assistance with the Viralis platform.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2019.05.001.
Contributor Information
John Ziebuhr, Email: john.ziebuhr@viro.med.uni-giessen.de.
Wibke E. Diederich, Email: wibke.diederich@staff.uni-marburg.de.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M.J., Lefkowitz E.J., King A.M., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses (2016) Arch. Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaire M., Chernaia M.M., Malcolm B.A., James M.N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Banerjee K., Bhat R., Rao V.U.B., Nain A., Rallapalli K.L., Gangopadhyay S., Singh R.P., Banerjee M., Jayaram B. Toward development of generic inhibitors against the 3C proteases of picornaviruses. FEBS J. 2018 doi: 10.1111/febs.14707. ([Epub ahead of print]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrette-Ng I.H., Ng K.K., Mark B.L., Van Aken D., Cherney M.M., Garen C., Kolodenko Y., Gorbalenya A.E., Snijder E.J., James M.N. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 2002;277:39960–39966. doi: 10.1074/jbc.M206978200. [DOI] [PubMed] [Google Scholar]
- Bazan J.F., Fletterick R.J. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann E.M., Mosimann S.C., Chernaia M.M., Malcolm B.A., James M.N. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birktoft J.J., Blow D.M. Structure of crystalline alpha-chymotrypsin: V. The atomic structure of tosyl-alpha-chymotrypsin at 2 Å resolution. J. Mol. Biol. 1972;68:187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Blanck S., Ziebuhr J. Proteolytic processing of mesonivirus replicase polyproteins by the viral 3C-like protease. J. Gen. Virol. 2016;97:1439–1445. doi: 10.1099/jgv.0.000458. [DOI] [PubMed] [Google Scholar]
- Blanck S., Stinn A., Tsiklauri L., Zirkel F., Junglen S., Ziebuhr J. Characterization of an alphamesonivirus 3C-like protease defines a special group of nidovirus main proteases. J. Virol. 2014;88:13747–13758. doi: 10.1128/JVI.02040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damalanka V.C., Kim Y., Galasiti Kankanamalage A.C., Rathnayake A.D., Mehzabeen N., Battaile K.P., Lovell S., Nguyen H.N., Lushington G.H., Chang K.O., Groutas W.C. Structure-guided design, synthesis and evaluation of oxazolidinone-based inhibitors of norovirus 3CL protease. Eur. J. Med. Chem. 2018;143:881–890. doi: 10.1016/j.ejmech.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty W.G., Semler B.L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galasiti Kankanamalage A.C., Kim Y., Rathnayake A.D., Damalanka V.C., Weerawarna P.M., Doyle S.T., Alsoudi A.F., Dissanayake D.M.P., Lushington G.H., Mehzabeen N., Battaile K.P., Lovell S., Chang K.O., Groutas W.C. Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors. Eur. J. Med. Chem. 2017;126:502–516. doi: 10.1016/j.ejmech.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayathri P., Satheshkumar P.S., Prasad K., Nair S., Savithri H.S., Murthy M.R. Crystal structure of the serine protease domain of Sesbania mosaic virus polyprotein and mutational analysis of residues forming the S1-binding pocket. Virology. 2006;346:440–451. doi: 10.1016/j.virol.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J. Viral cysteine proteases. Perspect. Drug Discov. Des. 1996;6:64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Blinov V.M., Donchenko A.P. Poliovirus-encoded proteinase 3C: a possible evolutionary link between cellular serine and cysteine proteinase families. FEBS Lett. 1986;194:253–257. doi: 10.1016/0014-5793(86)80095-3. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Donchenko A.P., Blinov V.M., Koonin E.V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243:103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Lieutaud P., Harris M.R., Coutard B., Canard B., Kleywegt G.J., Kravchenko A.A., Samborskiy D.V., Sidorov I.A., Leontovich A.M., Jones T.A. Practical application of bioinformatics by the multidisciplinary VIZIER consortium. Antivir. Res. 2010;87:95–110. doi: 10.1016/j.antiviral.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Robert X., Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Hekkelman M.L., Vriend G. MRS: a fast and compact retrieval system for biological data. Nucleic Acids Res. 2005;33:W766–W769. doi: 10.1093/nar/gki422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G.J., Zou J.Y., Kjeldgaard M., Jones T.A. Around O. In: Rossmann M.G., Arnold E., editors. International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules. Springer; Dordrecht: 2006. pp. 353–356. [Google Scholar]
- Kräusslich H.G., Wimmer E. Viral proteinases. Annu. Rev. Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Multiple alignment of protein structures in three dimensions. In: Berthold M.R., Glen R., Diederichs K., Kohlbacher O., Fischer I., editors. Computational Life Sciences. CompLife 2005. Lecture Notes in Computer Science. Springer; Berlin, Heidelberg: 2005. pp. 67–78. [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krug M., Weiss M.S., Heinemann U., Mueller U. XDSAPP: a graphical user interphase for the convenient processing of diffraction data using XDS. J. Appl. Crystallogr. 2012;45:568–572. [Google Scholar]
- Kuo C.J., Liu H.G., Lo Y.K., Seong C.M., Lee K.I., Jung Y.S., Liang P.H. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett. 2009;583:549–555. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lauber C., Gorbalenya A.E. Partitioning the genetic diversity of a virus family: approach and evaluation through a case study of picornaviruses. J. Virol. 2012;86:3890–3904. doi: 10.1128/JVI.07173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Ziebuhr J., Junglen S., Drosten C., Zirkel F., Nga P.T., Morita K., Snijder E.J., Gorbalenya A.E. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157:1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., Parquet Mdel C., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9:e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm B.A. The picornaviral 3C proteinases: cysteine nucleophiles in serine proteinase folds. Protein Sci. 1995;4:1439–1445. doi: 10.1002/pro.5560040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.W., Sigler P.B., Henderson R., Blow D.M. Three-dimensional structure of tosyl-alpha-chymotrypsin. Nature. 1967;214:652–656. doi: 10.1038/214652a0. [DOI] [PubMed] [Google Scholar]
- Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- McGrath M.E., Wilke M.E., Higaki J.N., Craik C.S., Fletterick R.J. Crystal structures of two engineered thiol trypsins. Biochemistry. 1989;28:9264–9270. doi: 10.1021/bi00450a005. [DOI] [PubMed] [Google Scholar]
- Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Mueller U., Förster R., Hellmig M., Huschmann F.U., Kastner A., Malecki P., Pühringer S., Röwer M., Sparta K., Steffien M., Ühlein M., Wilk P., Weiss M.S. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: current status and perspectives. Eur Phys J Plus. 2015;130:141. [Google Scholar]
- Muhaxhiri Z., Deng L., Shanker S., Sankaran B., Estes M.K., Palzkill T., Song Y., Prasad B.V. Structural basis of substrate specificity and protease inhibition in Norwalk virus. J. Virol. 2013;87:4281–4292. doi: 10.1128/JVI.02869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needle D., Lountos G.T., Waugh D.S. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr D Biol Crystallogr. 2015;71:1102–1111. doi: 10.1107/S1399004715003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E.J., Morita K., Gorbalenya A.E. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn C.M., Jeeves M., Cliff M.J., Urquhart G.T., George R.R., Chao L.H., Tscuchia Y., Djordjevic S. Crystal structure of tobacco etch virus protease shows the protein C terminus bound within the active site. J. Mol. Biol. 2005;350:145–155. doi: 10.1016/j.jmb.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Otto H.H., Schirmeister T. Cysteine proteases and their inhibitors. Chem. Rev. 1997;97:133–172. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- Pape T., Schneider T.R. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. [Google Scholar]
- Phan J., Zdanov A., Evdokimov A.G., Tropea J.E., Peters H.K., 3rd, Kapust R.B., Li M., Wlodawer A., Waugh D.S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M.D., Flint M. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 1997;78(Pt 4):699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schiebel J., Krimmer S.G., Rower K., Knorlein A., Wang X., Park A.Y., Stieler M., Ehrmann F.R., Fu K., Radeva N., Krug M., Huschmann F.U., Glockner S., Weiss M.S., Mueller U., Klebe G., Heine A. High-Throughput crystallography: reliable and efficient identification of fragment hits. Structure. 2016;24:1398–1409. doi: 10.1016/j.str.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Schneider T.R., Sheldrick G.M. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Sheldrick G.M. Macromolecular phasing with SHELXE. Z. für Kristallogr. - Cryst. Mater. 2002;217:644–650. [Google Scholar]
- Siddell S.G., Walker P.J., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M., Rubino L., Sabanadzovic S., Sanfacon H., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Additional changes to taxonomy ratified in a special vote by the international committee on taxonomy of viruses (october 2018) Arch. Virol. 2019;164:943–946. doi: 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- Smits S.L., Snijder E.J., de Groot R.J. Characterization of a torovirus main proteinase. J. Virol. 2006;80:4157–4167. doi: 10.1128/JVI.80.8.4157-4167.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., van Dinten L.C., Spaan W.J., Gorbalenya A.E. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- Speroni S., Rohayem J., Nenci S., Bonivento D., Robel I., Barthel J., Luzhkov V.B., Coutard B., Canard B., Mattevi A. Structural and biochemical analysis of human pathogenic astrovirus serine protease at 2.0 A resolution. J. Mol. Biol. 2009;387:1137–1152. doi: 10.1016/j.jmb.2009.02.044. [DOI] [PubMed] [Google Scholar]
- Takahashi D., Hiromasa Y., Kim Y., Anbanandam A., Yao X., Chang K.O., Prakash O. Structural and dynamics characterization of norovirus protease. Protein Sci. 2013;22:347–357. doi: 10.1002/pro.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touw W.G., Baakman C., Black J., te Beek T.A., Krieger E., Joosten R.P., Vriend G. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 2015;43:D364–D368. doi: 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R., Mettenleiter T.C., Ziebuhr J. Characterization of Bafinivirus main protease autoprocessing activities. J. Virol. 2011;85:1348–1359. doi: 10.1128/JVI.01716-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerawarna P.M., Kim Y., Galasiti Kankanamalage A.C., Damalanka V.C., Lushington G.H., Alliston K.R., Mehzabeen N., Battaile K.P., Lovell S., Chang K.O., Groutas W.C. Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: structural, biochemical, spectroscopic, and antiviral studies. Eur. J. Med. Chem. 2016;119:300–318. doi: 10.1016/j.ejmech.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2:282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Li S., Xue F., Zou Y., Chen C., Bartlam M., Rao Z. Structure of the main protease from a global infectious human coronavirus. HCoV-HKU1. J Virol. 2008;82:8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antivir. Res. 2011;92:204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Bayer S., Cowley J.A., Gorbalenya A.E. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 2003;77:1415–1426. doi: 10.1128/JVI.77.2.1415-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Kurth A., Quan P.L., Briese T., Ellerbrok H., Pauli G., Leendertz F.H., Lipkin W.I., Ziebuhr J., Drosten C., Junglen S. An insect nidovirus emerging from a primary tropical rainforest. mBio. 2011;2 doi: 10.1128/mBio.00077-11. e00077-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Roth H., Kurth A., Drosten C., Ziebuhr J., Junglen S. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 2013;87:6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.