Summary
Enterovirus B (EV-B), a major proportion of the genus Enterovirus in the family Picornaviridae, is the causative agent of severe human infectious diseases. Although cellular receptors for coxsackievirus B in EV-B have been identified, receptors mediating virus entry, especially the uncoating process of echovirus and other EV-B remain obscure. Here, we found that human neonatal Fc receptor (FcRn) is the uncoating receptor for major EV-B. FcRn binds to the virus particles in the “canyon” through its FCGRT subunit. By obtaining multiple cryo-electron microscopy structures at different stages of virus entry at atomic or near-atomic resolution, we deciphered the underlying mechanisms of enterovirus attachment and uncoating. These structures revealed that different from the attachment receptor CD55, binding of FcRn to the virions induces efficient release of “pocket factor” under acidic conditions and initiates the conformational changes in viral particle, providing a structural basis for understanding the mechanisms of enterovirus entry.
Keywords: enterovirus, echovirus, human neonatal Fc receptor, FcRn, receptor, uncoating, attachment, cryo-EM
Graphical Abstract
Highlights
-
•
CRISPR screening identified FcRn as an essential and universal EV-B receptor
-
•
FcRn facilitates EV-B uncoating and CD55 for attachment
-
•
High-resolution Cryo-EM structures described the mechanism of virus entry
-
•
The molecular mechanism of dual (attachment versus uncoating) receptor-usage was illustrated
The human neonatal Fc receptor is the uncoating receptor that facilitates cellular viral entry for Enteroviruses, such as echovirus.
Introduction
The Enterovirus B (EV-B) group consists of echovirus, coxsackievirus B (CV-B), coxsackievirus A9 (CV-A9), and other newly identified enteroviruses (Marjomäki et al., 2015). EV-B infections cause viral encephalitis (VE), viral meningitis (VM), and viral meningo-encephalitis (VME), leading to substantial morbidity and mortality in children (Holmes et al., 2016, Khetsuriani et al., 2006). They are also the causative agents of acute flaccid paralysis (AFP), nonspecific rashes, pneumonitis, hepatitis, coagulopathy, and hand-foot-and-mouth disease (HFMD) (Abzug, 2014, Huang et al., 2015). In recent years, severe outbreaks of echovirus infections have been documented in America, Europe, and Asia (Croker et al., 2015, Kadambari et al., 2014, Kim et al., 2012, Mao et al., 2010). In particular, echoviruses and CV-A9 accounted for seven out of the top 15 most frequently occurring enterovirus serotypes from 2014–2016 in the United States (Abedi et al., 2018). In China, surveillance of children with VE and VM in several provinces revealed that EV-B, especially echovirus, is the dominant serotype in the cerebrospinal fluid of patients (Chen et al., 2017, Zhu et al., 2016). It remains obscure how EV-B viruses cross the blood-brain barrier to cause these neurological diseases. Thus, discovery of their potential receptor(s) and entry mechanism will provide insight into the underling mechanisms of EV-B crossing the blood-brain barrier.
According to our current understanding, there are two types of receptors, attachment and uncoating receptors, that are involved in non-enveloped virus entry and genome release (Rossmann et al., 2002). However, their differential roles remain undetermined. To date, several host receptors have been identified to facilitate the entry of enteroviruses, including CD155 (PVR) for poliovirus, intercellular adhesion molecule-1 (ICAM-1) for CV-A21 and CV-A24, scavenger receptor B2 (SCARB2) and P-selectin glycoprotein ligand-1 (PSGL-1) for EV-A71, and KREMEN1 for some of the group A enteroviruses (Abzug, 2014, Baggen et al., 2018, Mendelsohn et al., 1989, Nishimura et al., 2009, Staring et al., 2018, Tuthill et al., 2010, Xiao et al., 2001, Yamayoshi et al., 2009). For EV-B, CD55 has previously been reported as the receptor for CV-B1, -B3, -B5 and several other echovirus serotypes. Moreover, α2β1 integrin is the receptor for Echo 1, while αVβ6 and αVβ3 integrins facilitate CV-A9 entry (Bergelson et al., 1993, Bergelson et al., 1994, He et al., 2002, Ward et al., 1994, Williams et al., 2004). Among the above EV-B receptors, CD55, α2β1, αVβ6, and αVβ3 are responsible for virus attachment, but none of them induce conformational changes in the viral particle, a process required for virus uncoating and genome release (Shakeel et al., 2013, Tuthill et al., 2010). Although coxsackie and adenovirus receptor (CAR) serves as the uncoating receptor for CV-B (Bergelson et al., 1997), the uncoating receptors for other EV-B viruses are yet unknown.
Capsid uncoating and genome release are critical steps to fulfill the life cycle of non-enveloped virus after its attachment (Rossmann et al., 2002). Interestingly, most of the identified uncoating receptors for picornavirus (e.g., PVR, CAR, and ICAM-1) are Ig-like proteins (Bergelson et al., 1997, He et al., 2000, Rossmann et al., 2002), with the exception of SCARB2 (Wang et al., 2012). Most of the uncoating receptors bind to the “canyon” sites (deep depressions surrounding the five-fold axis) of the viral particle (Rossmann et al., 2002). This binding triggers the release of “pocket factor,” which is hypothesized to be a lipid accommodated within a pocket below the canyon to stabilize the virion structure, and subsequently induces conformational changes in the viral particle (Bergelson and Coyne, 2013, Rossmann et al., 2002). The general concepts for enterovirus entry have previously been established using cryo-electron microscopy (cryo-EM) structures of several enteroviruses in complexes with their receptors at moderate resolutions, including poliovirus, CV-B3, CV-A21, and CV-A24 (Baggen et al., 2018, Bubeck et al., 2005, He et al., 2001, Organtini et al., 2014, Strauss et al., 2015, Xiao et al., 2005, Zhang et al., 2008). However, structural information of enterovirus-receptor complexes at atomic resolution and the mechanism of “pocket factor” release are missing, which would provide a much more accurate and systematic insight into the process of enterovirus entry.
Here, we identified neonatal Fc receptor (FcRn) as a universal uncoating receptor for a large group of EV-B viruses by CRISPR-Cas9 library screening. As an immune factor, FcRn is hijacked by the virus as its uncoating receptor. FcRn-mediated uncoating was confirmed by FcRn-decorated liposome experiments. We also determined a series of high-resolution cryo-EM structures of Echo 6 virus in the apo form and in complex with either its attachment receptor CD55 or uncoating receptor FcRn, at both neutral and acidic pH conditions. By comparing these structures, we systematically dissected the differential roles of Echo 6 attachment and uncoating receptors. It is the uncoating receptor FcRn that triggers the “pocket factor” release at a low pH within the endosome.
Results
FcRn Is a Functional Receptor for Echo 6 Infection
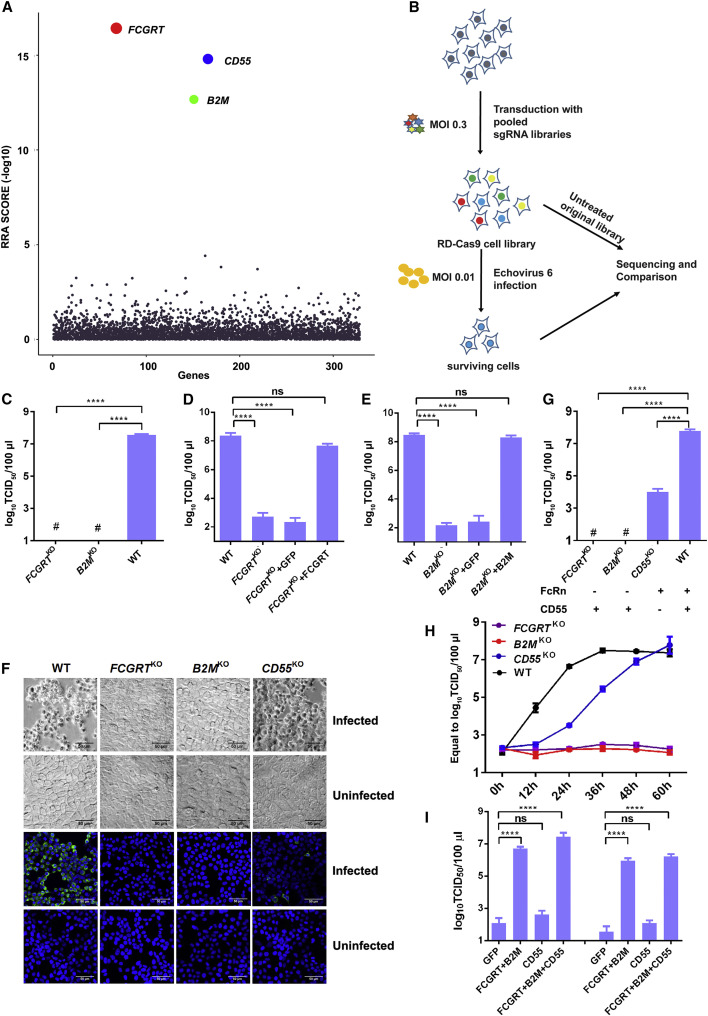
To identify the uncoating receptor for EV-B viruses, we performed a CRISPR knockout screen with Echo 6 strain SJZ-366 (Chen et al., 2017) in the RD-Cas9 cell line using a focused single guide RNA (sgRNA) library featuring human membrane protein-encoding genes. The sgRNA-coding regions were amplified from the extracted genomic DNA of library cells before and after Echo 6 infection, followed by next-generation sequencing (NGS) and data analysis using the MAGeCK method (Peng et al., 2015, Zhou et al., 2014, Zhu et al., 2017) (Tables S1 and S2). The top genes from the screen were: CD55, which has already been identified to encode the known Echo 6 receptor responsible for virus attachment (Bergelson et al., 1994, Ward et al., 1994); and FCGRT and B2M, encoding two subunits of FcRn, respectively (Figures 1A and 1B). FcRn is an MHC class I-like protein consisting of a heavy α chain encoded by the FCGRT gene, and a light β2-microglubulin (β2 m) chain encoded by the B2M gene. The α chain of FcRn contains three domains (α1, α2, and α3). The α3 and β2 m domains adopt immunoglobulin (Ig)-like folds (Oganesyan et al., 2014) (Figure S1 A).
Figure 1.
CRISPR Screening Uncovered FcRn, a Functional Receptor for Echo 6 Infection
(A) Dot plot illustrating the enriched genes from the CRISPR screen. Genes were rank-ordered by robust rank aggregation (RRA) scores based on MAGeCK analysis. The height and size of the top three genes, differently colored, was proportional to their RRA scores.
(B) Schematic diagram of sgRNA library construction and Echo 6 receptor screening.
(C and G) Echo 6 infection in FCGRTKO, B2MKO, CD55KO, and wild type (WT) HEK293T cells (C). Echo 6 infection in CD55KO (G), with the above three cell lines in (C) as controls. TCID50 was calculated at 48 h post infection (h.p.i). “+” and “-” indicate the presence or absence of FcRn or CD55 in the KO cell lines.
(D and E) Echo 6 infection in FCGRTKO cells complemented with lentivirus expressing FCGRT (D) and B2MKO cells complemented with lentivirus expressing β2 m (E). KO cell lines with lentivirus expressing GFP was used as a control. Cells were infected with Echo 6 (MOI 0.01), and supernatants were harvested for virus titration at 24 h.p.i.
(F) Light microscopy images showing CPE of FCGRTKO, B2MKO, CD55KO, and WT HEK293T cells at 24 h.p.i with Echo 6 (MOI 1). Immunofluorescence images of KO and WT cells infected by Echo 6 (MOI 10) for 8 h and then stained by anti-Echo 6 antibodies. The scale bar represents 50 μm.
(H) Growth curve of Echo 6 in FCGRTKO, B2MKO, CD55KO, and WT HEK293T cells.
(I) Echo 6 was inoculated onto non-susceptible CHO (left) and BHK (right) cells with ectopic expression of human FcRn (FCGRT+β2 m), CD55, or both.
The data depict means with SEM. TCID50 values were calculated by the Reed-Muench method. Experiments were repeated three times. # stands for undetected; ∗∗∗∗p < 0.0001; ns, not significant.
See also Figures S1 and S2 and Tables S1 and S2.
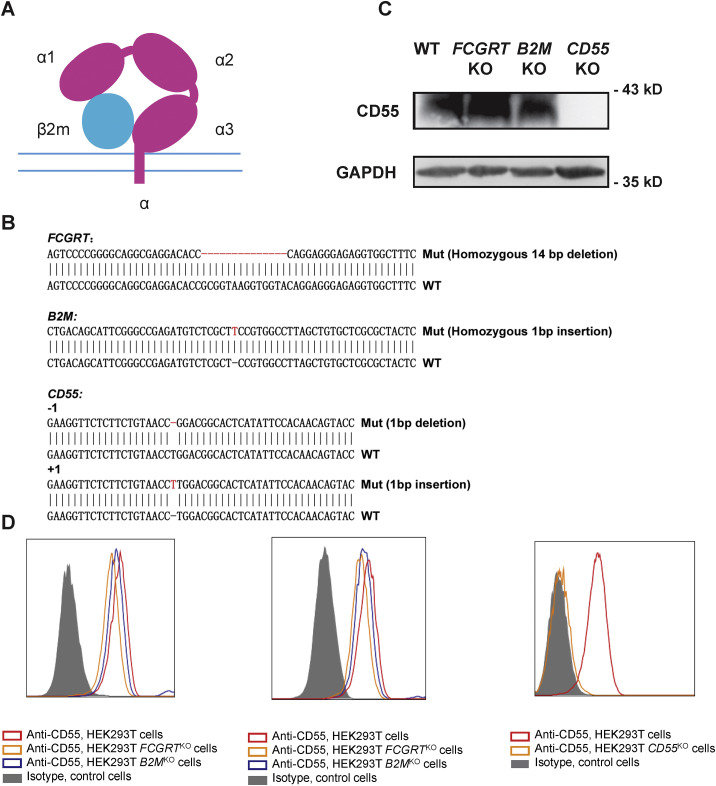
Figure S1.
FCGRT or B2M KO Has No Effect on CD55 Expression, Related to Figure 1
(A) Diagram of FcRn. The α chain expressed by FCGRT contains a transmembrane domain and three extracellular domains (α1, α2 and α3). The β2 m chain is expressed by B2M.
(B) Sanger sequencing of FCGRT, B2M and CD55 in KO cells. Sequencing data shows an alignment of WT sequence with the KO sequence.
(C) western blot of CD55 expression in WT, FCGRTKO, B2MKO and CD55KO cells.
(D) Flow cytometry analysis of CD55 expression in WT, FCGRTKO, B2MKO (left panel: fixed cell staining, middle panel: cell surface expression) and CD55KO cells (right panel).
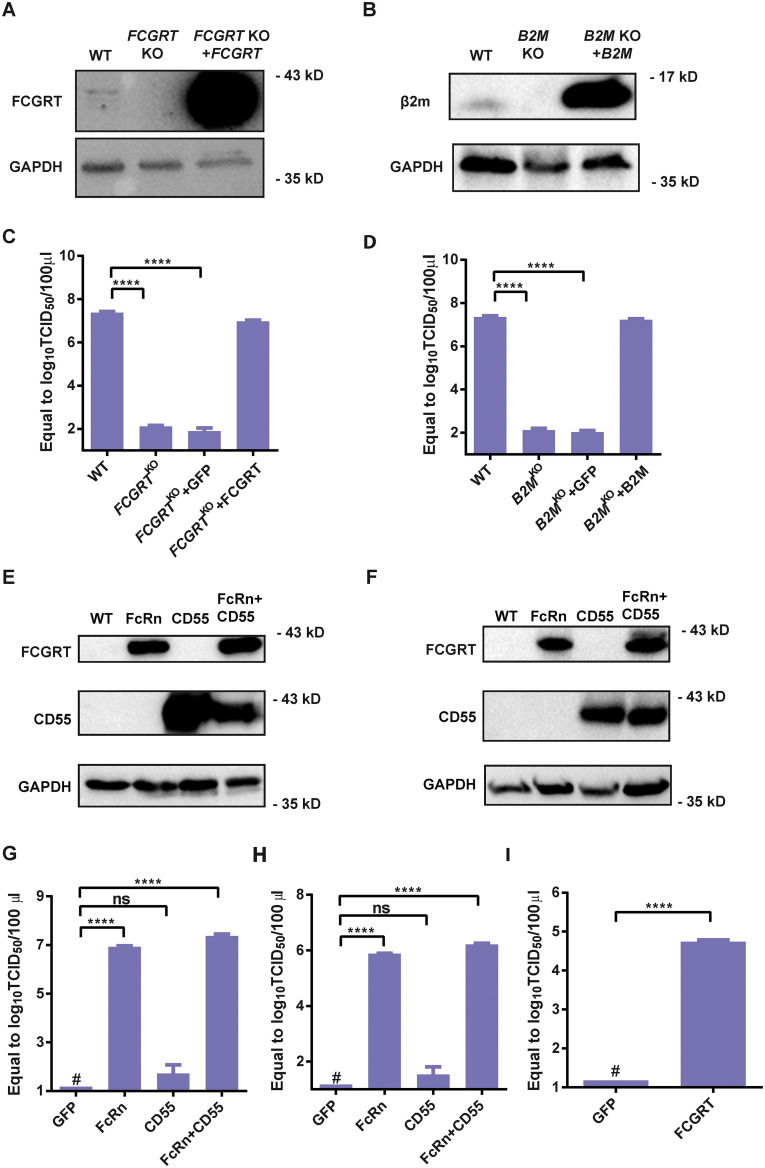
To verify FcRn’s role in echovirus infection, we infected FCGRT KO and B2M KO HEK293T cell lines with Echo 6 virus (Figure S1B). Cytopathic effect (CPE) was undetectable in either FCGRT KO or B2M KO HEK293T cells (Figures 1C and 1F, upper two panels). No Echo 6-positive signals were observed in either FCGRT KO or B2M KO HEK293T cells by immunofluorescence staining (Figure 1F, lower two panels), and no obvious viral replication was observed for both knockout (KO) cell lines spanning a 60-h growth (Figure 1H). Moreover, exogenous expression of the corresponding genes restored the susceptibility of both gene knockout lines to Echo 6 infection (Figures 1D, 1E, and S2 A–S2D). Furthermore, ectopic expression of human FcRn enabled efficient Echo 6 infection in two non-permissive cells, CHO and BHK (Renois et al., 2011) (Figures 1I and S2E–S2I).
Figure S2.
FcRn Complementation Can Restore Susceptibility; Ectopic Expression Makes Non-permissive Cells Susceptible to Echovirus, Related to Figure 1
(A and B) western blot of FCGRT (A) and β2 m (B) expression in WT, KO and supplementary cells.
(C and D) qPCR analysis of Echo 6 infection in FCGRTKO cells complemented with lentivirus expressing FCGRT (C). B2MKO cells complemented with lentivirus expressing β2 m (D). KO cell lines with lentivirus expressing GFP were used as a control. Cells were infected with 0.01 MOI Echo 6, and supernatants were taken for virus titration after 24 h.
(E and F) western blot of FCGRT and CD55 expression in WT, human FcRn, human CD55 and human FcRn+ CD55 ectopic expression CHO (E) or BHK (F) cells.
(G and H) qPCR analysis of Echo 6 in non-susceptible CHO (G) and BHK (H) cells with ectopic expression of human FcRn (FCGRT+B2M), CD55 or both.
(I) Echovirus infection in CHO cells with the ectopic expression of FCGRT only. (The data depict means with SEM. C, D, G and H: one-way ANOVA with multiple comparisons; I: unpaired two-tailed t test, with a Welch post-correction. # stands for undetected. ∗∗∗∗p < 0.0001; and ns, not significant.)
Importantly, knockout of neither FCGRT nor B2M downregulated CD55 expression (Figures S1C and S1D). As expected, CD55 KO reduced Echo 6 infection (Figure 1G), however, CD55 KO cells, different from FCGRT KO and B2M KO cells, were still permissive for virus infection (Figures 1F and 1G). Despite delayed viral growth kinetics, Echo 6 could eventually reach high titers in CD55 KO cells (Figure 1H).
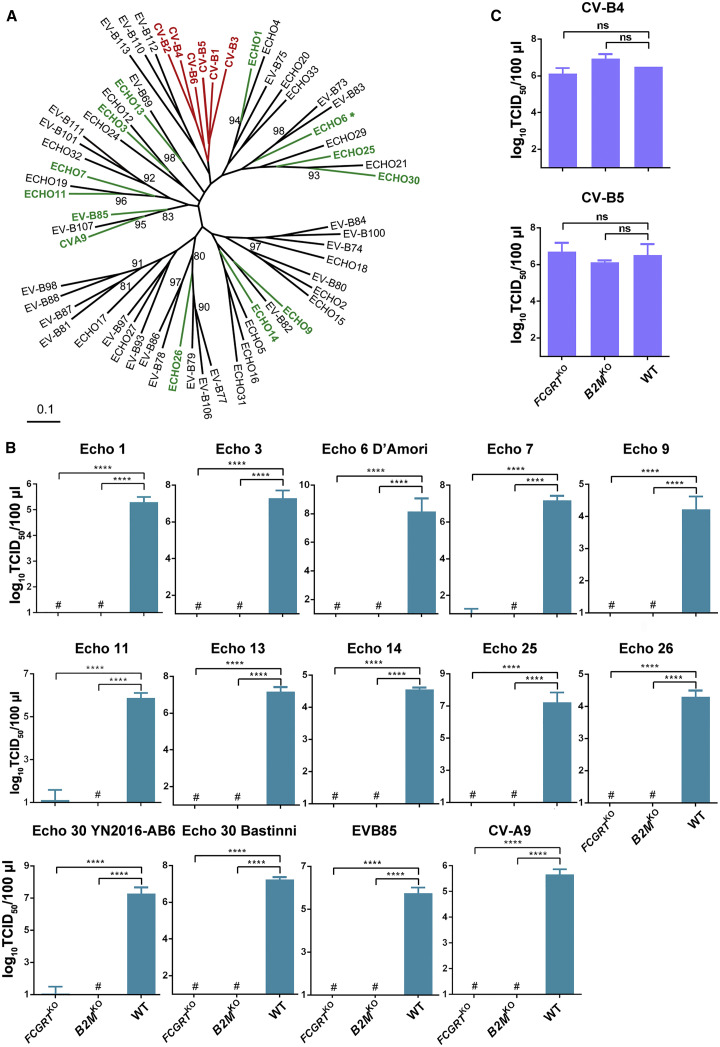
FcRn Is Essential for Major Serotypes of EV-Bs
Echo 6 belongs to the EV-B group, which includes echoviruses, CV-Bs, CVA-9, and several newly identified enterovirus serotypes (Figure 2 A) (Marjomäki et al., 2015). Given that CAR is the uncoating receptor for all six serotypes of CV-B viruses (Martino et al., 2000), we hypothesized that FcRn is also a universal uncoating receptor for other EV-B viruses. To test this hypothesis, we selected 11 echovirus serotypes in separate branches of the phylogenetic tree for further analysis: Echo 1, 3, 6, 7, 9, 11, 13, 14, 25, 26, and 30. EV-B85 and CVA-9 were also included because they are physiologically close to echoviruses (Figures 2A and 2B). For Echo 6 and 30, we tested both their epidemic and prototypic strains. FCGRT KO or B2M KO abolished the viral infection for EV-B85, CVA-9, and all of the above-mentioned echoviruses, suggesting that FcRn is a key host factor for these viruses (Figure 2B). In contrast, neither FCGRT KO nor B2M KO prevented CV-B4 or CV-B5 infections (Figure 2C). We thus propose that FcRn is the receptor candidate for the major group of EV-B viruses.
Figure 2.
FcRn Is Essential for Major Serotypes of EV-Bs
(A) Phylogenetic tree of all Enterovirus B serotypes. The representative serotypes proven to use FcRn as a receptor are shown in green, and the ones proven in previous works to use CAR as their uncoating receptor are shown in red. The remaining untested serotypes are shown in black.
(B) Infection of 12 echoviruses, EV-B85 and CVA-9 in FCGRTKO, B2MKO, and WT HEK293T cells. TCID50 was calculated at 48 h.p.i.
(C) Infection of CV-B4 and CV-B5 in FCGRTKO, B2MKO, and WT HEK293T cells. TCID50 was calculated at 48 h.p.i.
The data depict means with SEM. Experiments were repeated three times. # stands for undetected; ∗∗∗∗p < 0.0001; ns, not significant.
Direct Binding of FcRn to EV-B Virus
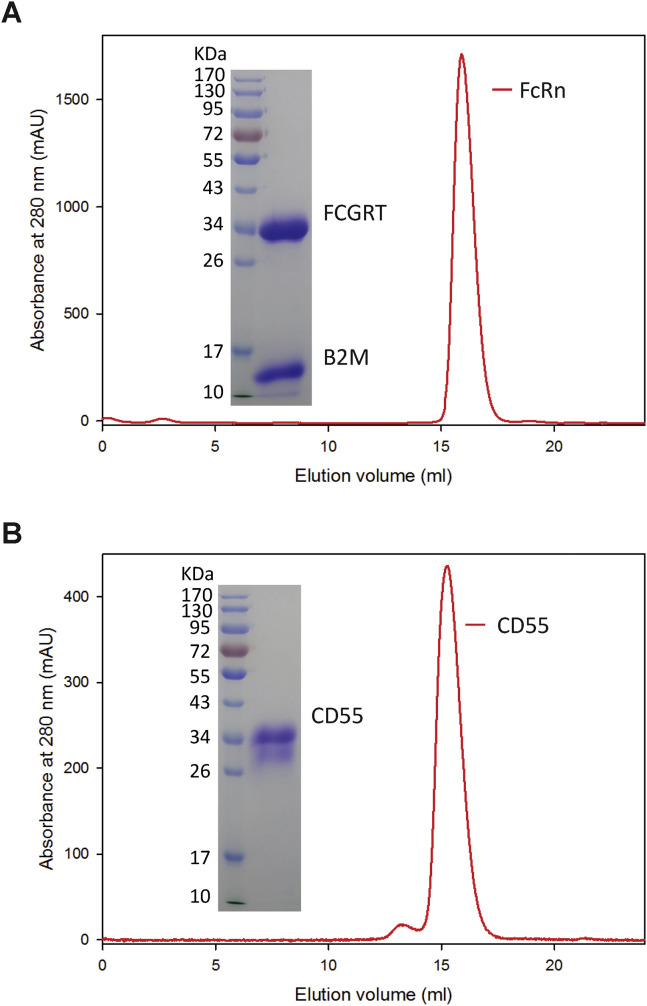
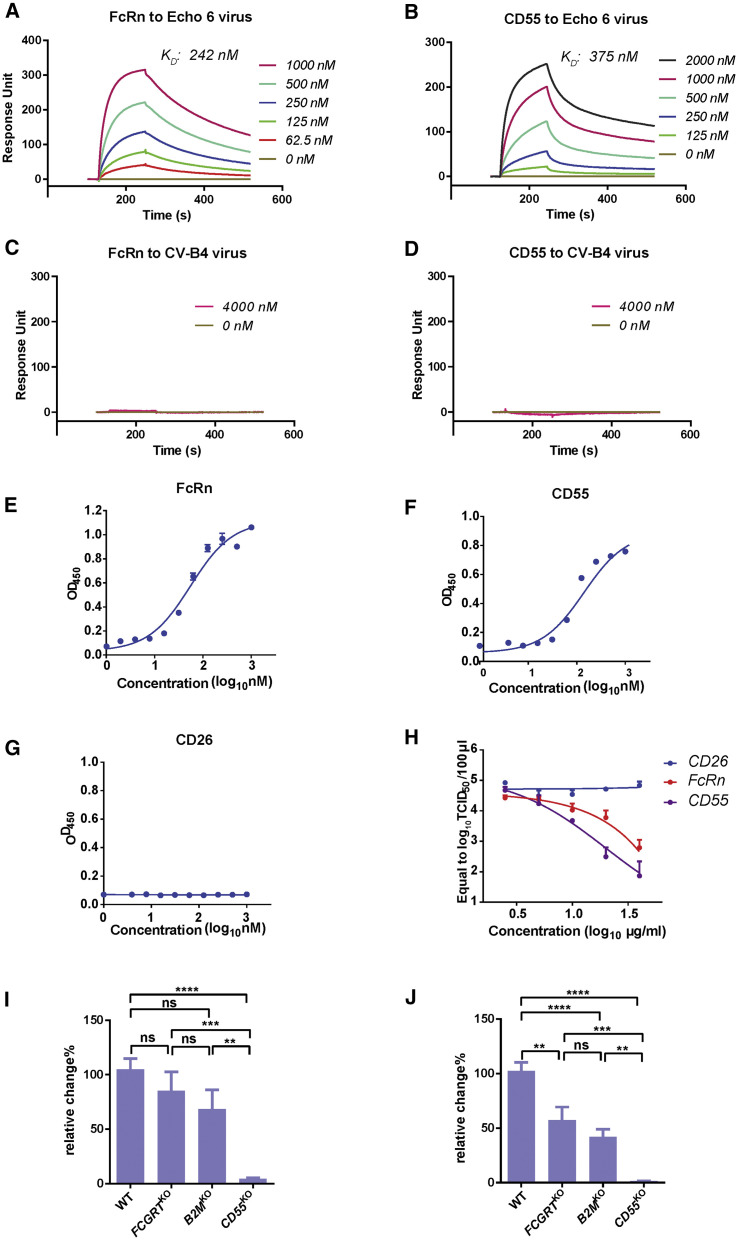
To verify if FcRn indeed serves as a receptor for and directly binds to EV-B viruses, we prepared the soluble extracellular domain of human FcRn protein and the extracellular domain of human CD55 protein (Figure S3 ). Surface plasmon resonance (SPR) experiments were performed to examine the interaction between Echo 6 viral particles and these recombinant proteins. We found that FcRn bound to Echo 6 virions with an affinity of approximately 242 nM, similar to the positive control CD55 (375 nM, Figures 3A and 3B). In comparison, no binding was detected for either FcRn or CD55 to CV-B4, which uses CAR as its receptor (Figures 3C and 3D) (Bergelson et al., 1997). ELISAs revealed dose-dependent binding of FcRn or CD55 to Echo 6 virions (Figures 3E and 3F). In comparison, CD26, the receptor for Middle East respiratory syndrome coronavirus (MERS-CoV), did not bind to Echo 6 (Figure 3G) (Lu et al., 2013). To examine whether the soluble FcRn or CD55 protein could block virus infection, we incubated Echo 6 virus with FcRn, CD55, or CD26 protein at various concentrations prior to the infection and found that incubation with either FcRn or CD55 (but not CD26) led to infection inhibition in a dose-dependent manner (Figure 3H). These results suggest that FcRn is the receptor for Echo 6 virus.
Figure S3.
FcRn and CD55 Soluble Protein Expression and Purification, Related to Figure 3
(A) FcRn soluble protein was expressed by co-transfection of constructs encoding the FCGRT extracellular domain and B2M in HEK293T cells. The protein was eluted as a single peak from gel filtration on Superdex 200 column (GE Healthcare), and two bands showing FCGRT and B2M were evident by SDS-PAGE.
(B) Gel filtration and SDS-PAGE of CD55 soluble protein purification.
Figure 3.
Direct Binding of FcRn to Echo 6 Virus
(A and B) BIAcore diagram of soluble FcRn protein (A) or CD55 protein (B) bound to Echo 6 virus. The KD values were calculated by the BIAcore 3000 analysis software (BIAevaluation version 4.1).
(C and D) The binding affinity of CV-B4 virus with FcRn (C) or CD55 protein (D) was measured by SPR, which served as a control.
(E–G) Binding of FcRn (E), CD55 (F), or CD26 (G, control) in 2-fold serial dilutions with Echo 6 virus was assayed using ELISAs.
(H) Dose dependence of soluble FcRn or CD55 protein to block Echo 6 virus infection. CD26 was used as a control protein.
(I and J) Echo 6 was incubated with FCGRTKO, B2MKO, CD55KO, or WT HEK293T cells at 4°C for 1 h (I) or plus 37°C for 30 min (J). Cells were collected, and RNA was extracted for qPCR analysis.
The data depict means with SEM. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
See also Figures S3 and S4.
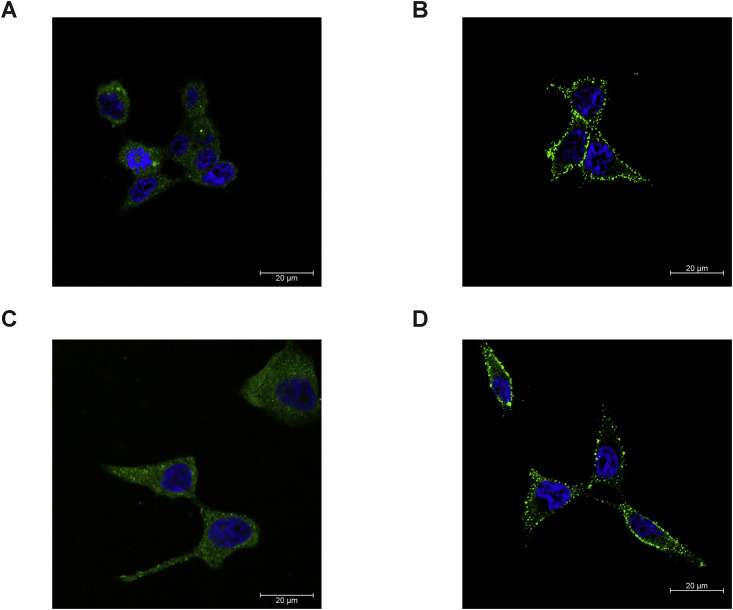
Next, we investigated the potential function of FcRn in EV-B entry. In contrast to CD55’s role in facilitating echovirus attachment to the cell surface, FcRn did not seem to be involved in viral attachment because FCGRT KO or B2M KO had little effect on Echo 6 binding to the cell surface (Figure 3I). Viral internalization was reduced in both FCGRT KO and B2M KO cells (Figure 3J). However, substantial amounts of virus still entered the host cells in the absence of FcRn (Figure 3J). These results suggest that FcRn plays its essential role after viral attachment. Consistently, CD55 was mainly localized on the cell surface, whereas FcRn displayed substantial expression within the cytoplasm, presumably on the endosome membrane (Figure S4 ).
Figure S4.
Confocal Analysis of FcRn and CD55 Expressed in HEK293T and RD Cells, Related to Figure 3
(A and C) Immunofluorescence staining of FcRn in HEK293T cells (A) and RD cells (C). FcRn is expressed both on the cell surface and in the cytoplasm.
(B and D) Immunofluorescence staining of CD55 in HEK293T cells (B) and RD cells (D). CD55 was mostly expressed on the cell surface.
Structural Dissection of Echo 6 Virus with Its Receptors
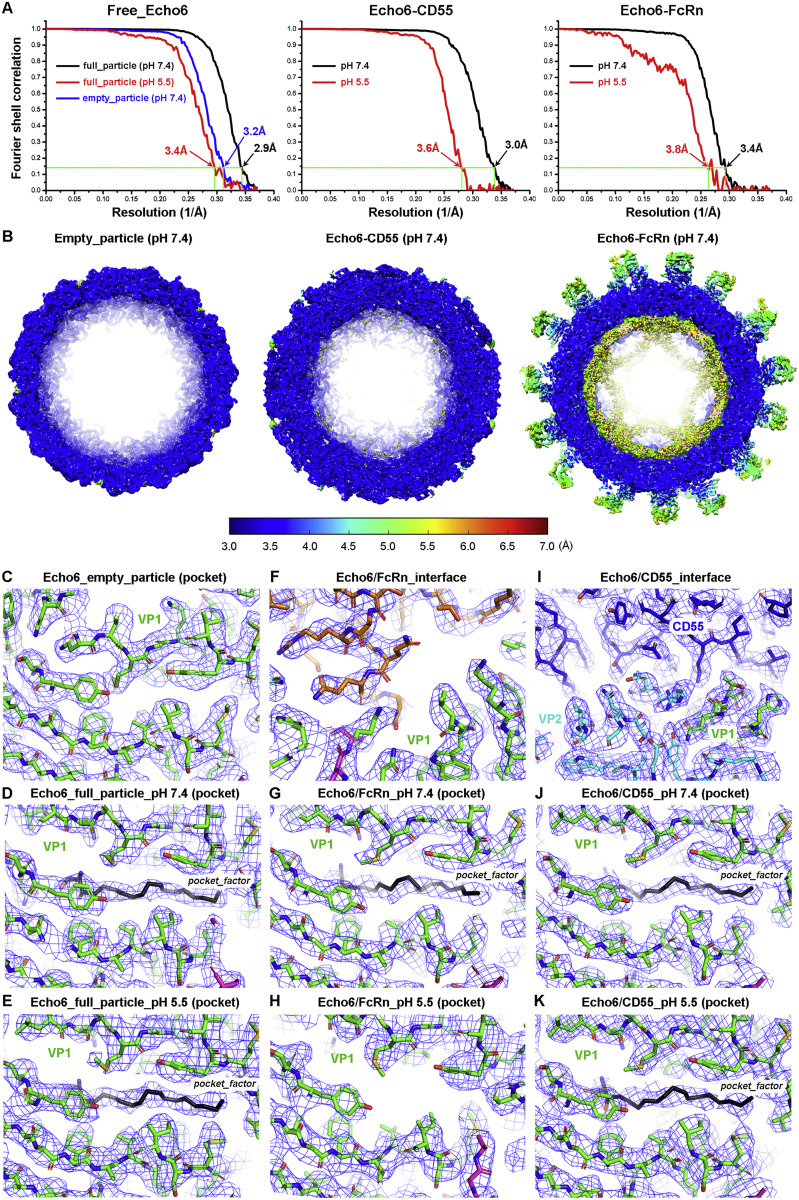
To elucidate the mechanism of FcRn in EV-B entry, we performed cryo-EM analysis to determine the structures of Echo 6 virus alone and in complex with CD55 or FcRn (Figure 4 ). As enterovirus entry is mediated by endocytosis and the uncoating process occurs in the endosome, the structures were thus determined under both neutral (pH 7.4) and acidic (pH 5.5) conditions. With near-atomic or atomic resolutions, we obtained detailed characterization of the molecular events during Echo 6 entry (Figure S5 ; Table S3).
Figure 4.
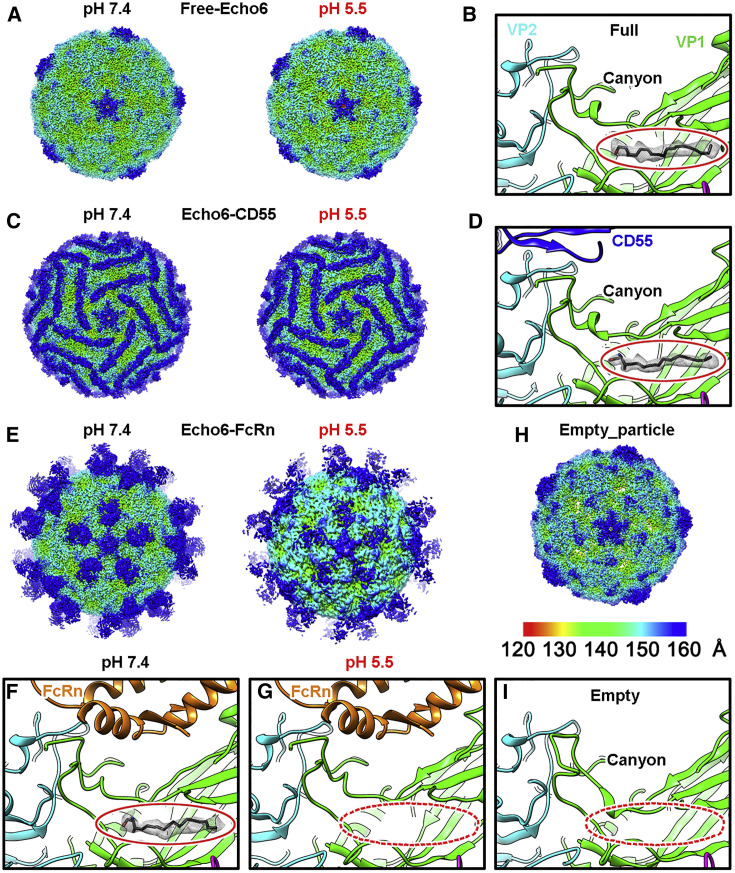
Cryo-EM Structures of the Echo 6 Viral Particle and Its Complex with Attachment (CD55) or Uncoating (FcRn) Receptor
(A) The density maps of free Echo 6 full particles at pH 7.4 (2.9 Å) and pH 5.5 (3.4 Å).
(B) The close-up view of the hydrophobic pocket and the “pocket factor” inside.
(C and D) Cryo-EM maps (C) of Echo 6 virus in complex with CD55 at pH 7.4 (3.0 Å) and pH 5.5 (3.6 Å), as well as the close-up view (D) of the “pocket factor” with the receptor binding site shown in the same style as (B).
(E–G) Cryo-EM maps (E) of Echo 6 virus in complex with FcRn at pH 7.4 (3.4 Å) and pH 5.5 (3.8 Å), as well as the close-up view showing the hydrophobic pocket of Echo 6 virus in complex with FcRn at pH 7.4 (F) and pH 5.5 (G). The receptor binding sites are shown in the same style as in (B) and (D).
(H and I) Cryo-EM map of Echo 6 virus empty particle (H) at pH 7.4 (3.2 Å) and its collapsed pocket without “pocket factor” (I).
All density maps are colored by radius as shown in the legend in (H). Atomic models of proteins are shown in ribbons and colored by chains with VP1, VP2, VP3, CD55, and FcRn heavy chain in green, cyan, magenta, blue, and orange, respectively. The “pocket factor” is shown in sticks, and the corresponding density map is shown at the 18σ contour level, which is highlighted by red ovals in close-up views. In the structures of the Echo 6 virus-FcRn complex at pH 5.5 and free Echo 6 virus empty particle, the pocket factor is absent, and its supposed positions are indicated by red dashed ovals.
See also Figures S5, S6, and Table S3.
Figure S5.
Cryo-EM Analysis of Echo 6 Virus and Its Complex with Receptors, Related to Figure 4
(A) Gold-standard Fourier shell correlation (FSC) curves of the structures of Echo 6 virus alone or in complex with its receptors. The 0.143 cut-off value is shown to indicate the resolution of each reconstruction.
(B) Local resolution maps of representative density maps of Echo 6 virus or its complex with receptors. In all of these structures, most regions reach 3.0 Å and allowed the atomic details to be resolved.
(C-K) Representative density maps and atomic models of the pocket region or receptor binding interface. Most side chains of key residues within the pocket are clearly resolved. The “pocket factor” is preserved in structures of free Echo 6 full-particle or in complex with CD55 at both pH 7.4 and pH 5.5. In the structures with FcRn binding, it is well accommodated at pH 7.4, but is released at pH 5.5.
Echo 6 virus is an icosahedral particle of ∼30 nm in diameter, in which the viral protein (VP)1, VP2, and VP3 form the outer shell, and VP4 is shielded underneath (Figures 4A and 4B). Each viral particle contains 60 copies of VP1, VP2, VP3, and VP4, which highly resembles the architecture of other enteroviruses (Rossmann et al., 2002). Similar to other enteroviruses, the Echo 6 virus contains both full and empty particles (lacking the “pocket factor,” VP4 subunit and genome inside) (Figures 4H and 4I). We found that only the full particles could bind the receptors, indicating that the empty particles are the relics of virus uncoating without infectivity. This is consistent with previous hypotheses based on EV71 studies (Wang et al., 2012). Our biochemical studies and cell-based assays suggested that the role of FcRn in mediating Echo 6 entry is different from that of CD55, which enables virus attachment at the early stage of infection (Powell et al., 1997). Based on these cryo-EM structures, binding of CD55 or FcRn at neutral pH did not induce evident conformational changes in the Echo 6 virions, in which the “pocket factor” is well accommodated (Figures 4C–4F). These observations indicated that both receptors can mediate virus attachment, and FcRn plays a role in inducing virus uncoating.
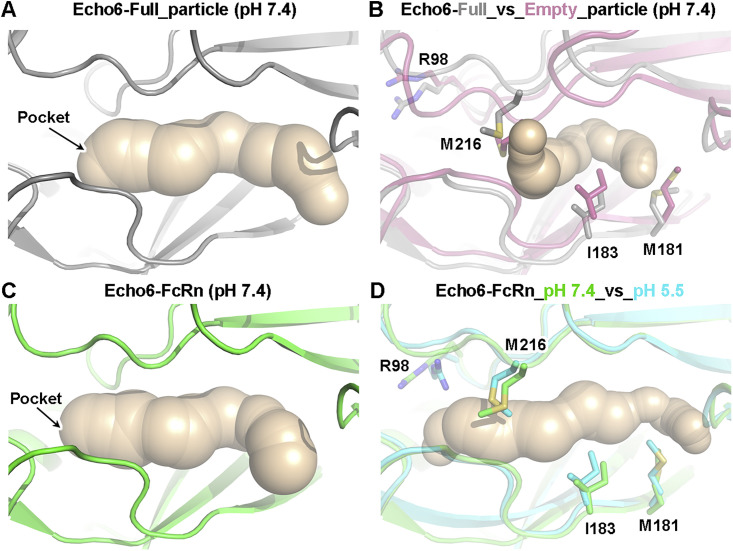
To further verify this, we examined the structures of Echo 6 virus in complex with either CD55 or FcRn at pH 5.5, which mimics the endosomal environment. The structure of CD55-bound Echo 6 virus at pH 5.5 was highly similar to that at pH 7.4. The “pocket factor” was still preserved, and the viral proteins did not exhibit significant structural rearrangements (Figures 4C and 4D). In contrast, the acidic condition caused dramatic changes to FcRn-bound Echo 6 viral particles. In this structure, the bound FcRn molecules began to dissociate from the virions, resulting in significantly reduced receptor densities due to low occupancies (Figure 4E). In comparison to the apo form and the CD55-bound Echo 6 virions, the “pocket factor” in FcRn-bound viral particles at pH 5.5 was released, and the pocket began to collapse with slight conformational changes on some residues within the “pocket” region (Figures 4G and S6 ). Furthermore, the structure of the FcRn-Echo 6 complex under acidic conditions was also different from that of the empty viral particles. The pocket was not fully collapsed, and the VP4 subunits and viral genome inside were still preserved, suggesting that the FcRn-Echo 6 structure is an intermediate in the process of virus uncoating (Figure S6). This structural evidence demonstrates that FcRn mediates viral binding, “pocket factor” release, and capsid uncoating, whereas CD55 is solely responsible for the virus attachment.
Figure S6.
Conformational Change Surrounding the “Pocket” of Echo 6 Virus and Echo 6 Virus with Its Receptor FcRn, Related to Figure 4
(A) Backbone traces of atomic models showing the conformation of the “pocket” in Echo 6 full particles in pH 7.4. Backbone of Echo 6 full particles is shown in gray. The inner pocket is shown in surface model.
(B) Conformational difference of the protein surrounding the “pocket” region of Echo 6 full-particle with empty-particle at pH 7.4. The backbone of Echo 6 full particle is shown in gray, and empty particle is shown in pink. The remarkable residues involved in conformational changes are shown as sticks and colored by elements. The pocket in the empty-particle is obviously collapsed as compared to that in the full viral particle.
(C) Backbone traces of atomic models showing the conformation of the “pocket.” Backbone of Echo 6 particle in the Echo 6-FcRn complex at pH 7.4 is shown in green. The pocket is highly similar to that in the full viral particle and allows the accommodation of the “pocket factor.”
(D) The conformational difference of the Echo 6-FcRn complex at pH 7.4 and pH 5.5. Backbones of the Echo 6 particle in the Echo 6-FcRn complex at pH 7.4 and pH 5.5 are shown in green and cyan, respectively. The “pocket” in the Echo 6-FcRn complex at pH 5.5 began to shrink compared to that at pH 7.4, but is not fully collapsed as observed in the empty particle. The remarkable residues involved in the conformational change are shown as sticks and colored by elements.
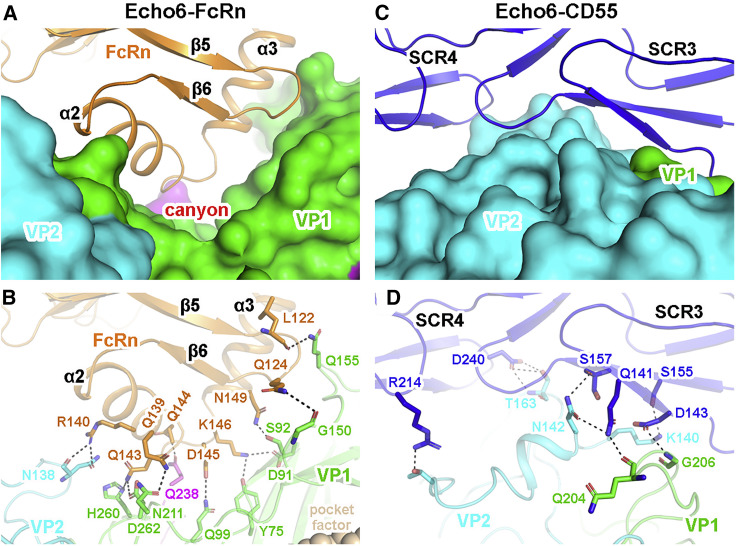
Molecular Interactions between Echo 6 Virus and Receptors
FcRn binds to Echo 6 virus in its “canyon” site, with mainly the α2 and α3 helices in the α2 domain interacting with VP1, VP2, and VP3. VP1 dominates the interactions (Figures 5A and 5B). In contrast, CD55 binds across VP1 and VP2 outside of the “canyon,” in which VP2 accounts for most of the interface (Figures 5C and 5D).
Figure 5.
Molecular Interactions between Echo 6 Virus and FcRn/CD55 Receptors
(A) The atomic model of Echo 6-FcRn in the main contacting interface. The viral proteins are shown in surface models, and the receptor is represented as ribbon model. VP1, VP2, and VP3 of Echo 6 are shown in green, cyan, and magenta, respectively. The interacting FCGRT subunit of FcRn is colored in orange. The “canyon” is indicated by a red asterisk.
(B) The interaction details between Echo 6 virus and FcRn receptor. The proteins are colored by chains as in (A), and the contacting residues are shown as sticks and colored by elements. Hydrogen bonds are represented by black dashed lines.
(C) The atomic model of the Echo 6-CD55 in the main contacting interface. The viral proteins (colored by chains) are shown in surface models, and CD55 (blue) is represented as ribbons. The interaction is mainly mediated by the SCR3 and SCR4 domains.
(D) The interaction details between Echo 6 virus and CD55 in the same style of FcRn in (B).
See also Figure S7.
The binding of FcRn to Echo 6 virus is mainly mediated by polar interactions with the α2 and α3 helices in the α2 domain, forming two interfaces on both sides of the canyon (Figure 5A). In the junction region of the α2 and α3 helices of FcRn, residues Q139, R140, Q143, and Q144 form consecutive hydrogen bond networks with VP1, VP2, and VP3 subunits, comprising the core of the FcRn-Echo 6 interface (Figure 5B). Two residues, D145 and K146, insert into the canyon site and form hydrogen bonds with Y75, D91, and Q99 of the VP1 subunit, which might be the key switch governing the transportation of the “pocket factor” (Figure 5B).
The interactions between CD55 and Echo 6 virus are mainly mediated by CD55’s SCR3 and SCR4 domains (Figure 5C). In the terminal SCR4 domain, residue R214 forms a hydrogen bond with T154 of the VP2 subunit. Meanwhile, other residues within the 230-strand contact the VP2 subunit by extensive van der Waals interactions. Residue D240 in the SCR4 domain forms a center of interaction and contacts both the main chain and side chain of T163 in the VP2 subunit by three hydrogen bonds (Figure 5D). The SCR3 domain of CD55 interacts with both VP1 and VP2 subunits of Echo 6 virus. Residues S155 and S157 form two hydrogen bonds with K140 and N142 of the VP2 subunit, and Q141 connects Q204 of VP1 and N142 of VP2 by two consecutive hydrogen bonds. In addition, D143 interacts with G206 of VP1 in the periphery to further stabilize the interface (Figure 5D).
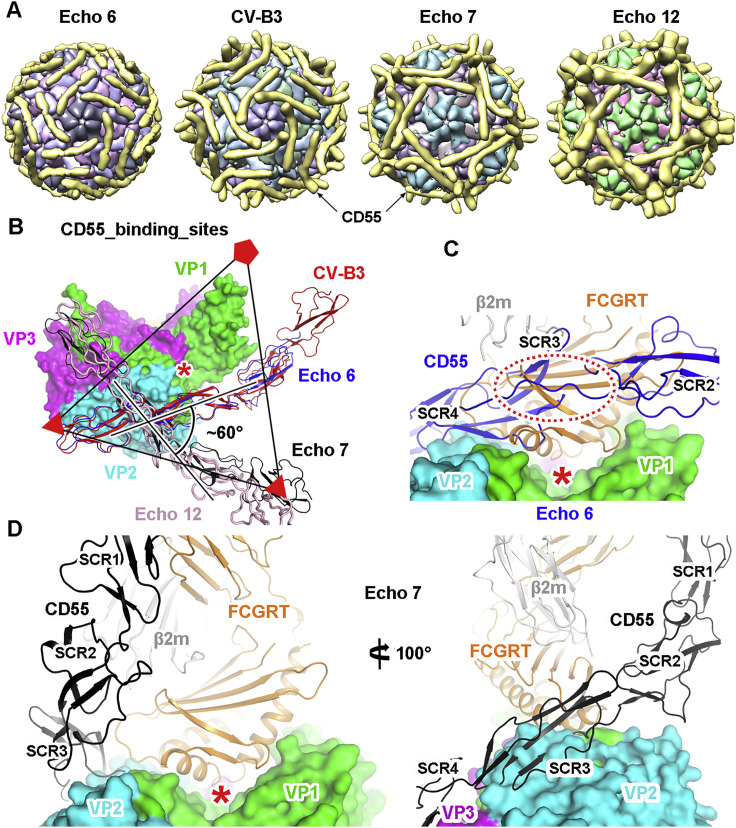
Because CD55 is a universal attachment receptor for several enteroviruses, we compared its binding sites on different viruses with the available virus-receptor complex structures (Bhella et al., 2004, Plevka et al., 2010, Yoder et al., 2012). Among them, the binding site of CD55 on Echo 6 virus is highly similar to that on CV-B3 but different from those on Echo 7 and Echo 12 viruses (Figures S7 A and S7B). CD55 is rotated by ∼60° around the SCR3 domain on the Echo 7 and Echo 12 viruses and mainly binds to VP2 and VP3, leaving the “canyon” fully exposed (Figure S7B). As CD55 is not able to mediate uncoating of Echo 6 virus, it may dissociate from the virion before FcRn binding or bind to another icosahedral asymmetric unit as the canyon is shielded by CD55 to render steric hindrance for the access of FcRn (Figure S7C). For Echo 7 and Echo 12 viruses, however, the scenario is probably different as the two receptors could bind simultaneously within the same icosahedral asymmetric unit (Figure S7D).
Figure S7.
Comprehensive Comparison of CD55 and FcRn Binding Sites on Different Enteroviruses, Related to Figure 5
(A) Cartoon diagrams of CD55 bound to different enteroviruses determined by cryo-EM. Each component is represented with a unique color, and CD55 is colored in gold.
(B) Superimposition of the atomic models of CD55 bound to different enteroviruses within an asymmetric unit. The icosahedral axes are shown as triangles and pentangles. The binding site of CD55 on Echo 6 virus is similar to that on CV-B3, but different from those on the Echo 7 and Echo 12 viruses. The “canyon” is indicated by a red asterisk.
(C) Comparison of the binding sites of CD55 and FcRn on Echo 6 virus. The viral proteins are shown in surface models, and the receptors are shown as ribbons. The position of the “canyon” is indicated by a red asterisk. The steric clash between CD55 and FcRn is highlighted by a dashed oval.
(D) Comparison of the binding sites of CD55 and FcRn on Echo 7 virus. The two receptors could bind simultaneously on the virus without clashing. Two different views of the superimposition are shown to reveal the compatibility of the two receptors in space.
VP1, VP2 and VP3 of the virus are shown in green, cyan and magenta, respectively. CD55 molecules bound to Echo 6, Echo 7, Echo 12 and CV-B3 are shown in blue, black, pink and red, respectively. The FCGRT subunit of FcRn is shown in orange, and β2 m is in gray.
FcRn-Decorated Liposomes Induce Echo 6 Uncoating
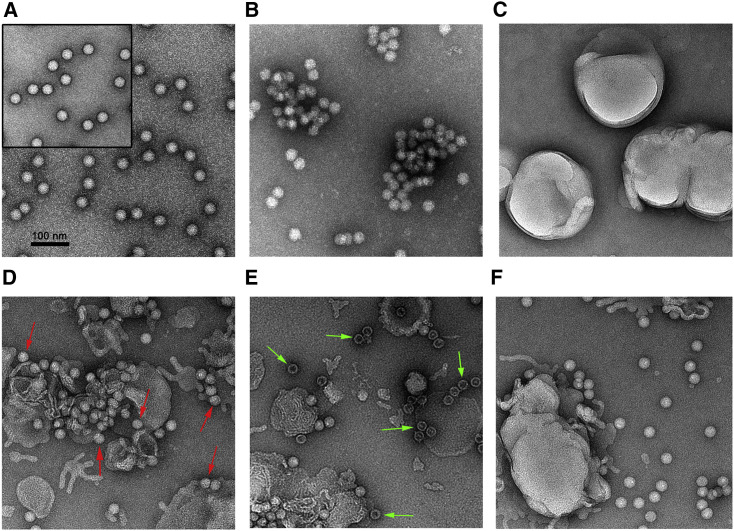
To further verify that FcRn-binding facilitates EV-B uncoating, we tested the FcRn-induced morphology changes of Echo 6 virions under 37°C, pH 5.5 conditions, which mimic the physiological conditions within the endosome (Figure 6 ). Negative-stain EM analysis showed that binding of FcRn soluble protein triggered the aggregation of Echo 6 viral particles (Figure 6B). The same phenomenon was not observed in either free Echo 6 particles or the CD55-Echo 6 complex (Figure 6A). To further confirm that FcRn mediates virus capsid uncoating, a receptor-decorated liposome model was used as designed for a PVR-induced poliovirus uncoating study (Groppelli et al., 2017, Tuthill et al., 2006). His-tagged FcRn or CD55 was incubated with nickel-charged liposomes to form receptor-decorated liposomes. Echo 6 particles attached to the surface of both FcRn- and CD55-decorated liposomes (Figures 6D and 6E) but not undecorated liposomes (Figure 6F). In addition, only the FcRn-decorated liposomes were able to induce structural transitions of Echo 6 particles (Figure 6E). A considerable proportion of empty particles were clearly observed after full Echo 6 particles were incubated with FcRn-decorated liposomes (Figure 6E). These data further demonstrate that FcRn is able to mediate EV-B uncoating under physiological conditions.
Figure 6.
FcRn-Decorated Liposomes Induce Echo 6 Uncoating
Negative staining EM detection of:
(A) Purified Echo 6 full particles (upper left) and Echo 6-CD55 complexes.
(B) Echo 6-FcRn complexes.
(C) Nickel-charged liposomes.
(D) Echo 6 incubated with CD55-decorated liposomes. Red arrows indicate the particles that attached to the surface of the liposomes.
(E) Echo 6 incubated with FcRn-decorated liposomes. Green arrows indicate empty particles.
(F) Echo 6 incubated with nickel-charged liposomes.
All samples were treated for 10 min at 37°C, pH 5.5. The scale bar represents 100 nm.
Discussion
Potential Mechanisms of Enterovirus Entry
In this work, we identified FcRn as an uncoating receptor for echoviruses, CV-A9, and other physiologically closely related novel EV-B viruses. FcRn bound echovirus particles and induced conformational changes. FcRn-mediated Echo 6 capsid uncoating was confirmed with an FcRn-decorated liposome model, which mimics the physiological condition of FcRn-expressing endosomes. Our findings are consistent with previous reports that the binding of CD55 to echovirus does not induce conformational changes of the viral particles (He et al., 2002, Plevka et al., 2010, Powell et al., 1997). As FcRn is also present on the cell surface in addition to its expression on endosome membrane (Roopenian and Akilesh, 2007), it is possible that CD55, as an initial attachment receptor, “hands off” the virus to FcRn on the cell membrane, which then mediates virus uncoating, just like CAR for CV-B entry (Whitton et al., 2005). It has been speculated by several groups that echoviruses should have an Ig-like receptor to mediate uncoating (Plevka et al., 2010, Powell et al., 1997, Rossmann et al., 2002). Moreover, β2 m has been found to play a crucial role for the infections of several echoviruses and CV-A9, but it is unclear how it works (Heikkilä et al., 2010, Marjomäki et al., 2002, Ward et al., 1998). Our findings verify that β2 m plays an essential role for echovirus uncoating as a subunit of the FcRn receptor.
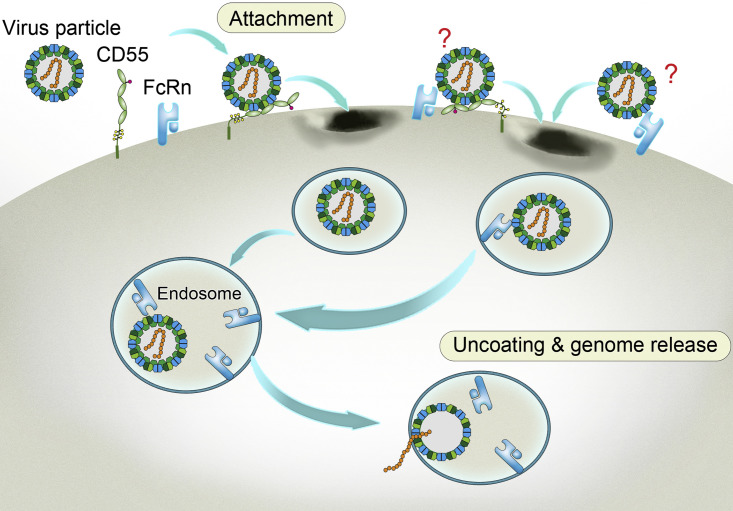
Most enteroviruses and some other picornaviruses use two different types of receptors at different stages of virus entry: an attachment receptor and an uncoating receptor (Rossmann et al., 2002, Tuthill et al., 2010, Whitton et al., 2005). Previous work has proposed functional differences between the attachment and uncoating receptors (Powell et al., 1997, Rossmann et al., 2002). However, high-resolution structural evidence was missing to comprehensively understand the different functions of these receptors at the stage of virus entry. In addition, it has been suggested that uncoating receptor-mediated release of “pocket factor” allows picornavirus genome release into the cytosol, though more detailed evidence is required to better understand this process (Rossmann et al., 2002). In this study, we unambiguously observed “pocket factor” release in the enterovirus at acidic pH after binding to the viral uncoating receptor, in high-resolution cryo-EM structures. Together with the cryo-EM structures at other stages of virus entry, we describe a working model for enterovirus entry (Figure 7 ).
Figure 7.
Model of Echovirus Entry into the Cell
The four structural proteins (VP1, VP2, VP3, and VP4) comprising of viral capsid are indicated in blue, dark olive green, forest green, and pigment green, respectively. The viral RNA genome is shown in orange. CD55 and FcRn are displayed in light green and sky blue, respectively. The two undefined receptor binding modes on the viral surface are labeled with question marks.
Taking Echo 6 as an example, the virus binds to the attachment receptor CD55 on the cell surface, which triggers viral endocytosis and internalization. Viral particles are subsequently captured by the uncoating receptor FcRn within endosomes. The low pH environment of the endosomes prompts the extrusion of “pocket factor” after FcRn binding, resulting in virus uncoating and genome release. Alternatively, viral particles could also be captured by FcRn at the cell membrane, as FcRn bound at both neutral and acidic pH, and further induces endocytosis and uncoating in the endosome (Figure 7).
The Expression Distribution of FcRn Coincides with EV-B Tropism
It is known that FcRn facilitates the transport of maternal IgG across the placenta to the fetus and across the intestinal epithelium to the bloodstream of the newborn, as well as mediating the recycling of IgG for adults (Roopenian and Akilesh, 2007). Our findings indicate that FcRn can be exploited by a large group of EV-B viruses for entry into the cell. There is substantial expression of FcRn in the gut, kidneys, lungs, placenta, hematopoietic cells, vascular endothelium, and at the blood-brain barrier (Roopenian and Akilesh, 2007, Schlachetzki et al., 2002). This is in accordance with the tissue tropism of EV-B, as the gut is the major target for EV-B, where the viruses transmit to many other organs such as the CNS, liver, spleen, lungs, bone marrow, and heart to cause several types of diseases (Choudhary, 2017). As functional receptor, FcRn possibly helps EV-B viruses cross both the blood-brain barrier and the blood-placenta barrier.
Implications for Antiviral Drug Design and Enterovirus-Based Oncolytic Therapeutics
Thus far, there is no approved drug or vaccine specifically against EV-B infection. Identification of FcRn as the functional receptor for a large group of EV-B viruses paves the way for effective vaccine and drug development. Most echoviruses and other physiologically closely related viruses in the EV-B group have no animal model, except for Echo 1 whose mouse model expresses human VLA-2 (Dalldorf et al., 1955, Hughes et al., 2003). Lack of animal models greatly hinders studying the pathogenesis of EV-B as it relates to VE and VM, as well as the development of antiviral drugs and vaccines. Ectopic expression of CD55 is insufficient to make rodent cells susceptible to echovirus infection (Renois et al., 2011). We now know why and could restore the echovirus susceptibility in rodent cells by expressing human FcRn, shedding light on the development of VE and VM animal models.
Further, echoviruses such as Echo 1, Echo 7, and Echo12 possess oncolytic activity (Haley et al., 2009, Israelsson et al., 2011, Shafren et al., 2005, Ylä-Pelto et al., 2016). RIGVIR, the Echo 7-based oncolytic drug, has been approved for clinical use in Latvia for melanoma treatment (Chumakov et al., 2012, Doniņa et al., 2015). However, the lack of receptor information for Echo 7 hampered our understanding and evaluation of this type of oncolytic therapy (Chumakov et al., 2012, Tilgase et al., 2018). The discovery of FcRn as its receptor will improve the development of echovirus-based oncolytic therapeutics.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti-Mouse IgG H&L (Alexa Fluor® 488) | Abcam | Cat#ab150113; RRID: AB_2576208 |
| Anti-FCGRT antibody - Extracellular domain | Abcam | Cat#ab193148; RRID: AB_2801386 |
| Anti-CD55 antibody | Abcam | Cat#ab33111; RRID: AB_726605 |
| Anti-CD55 antibody | Abcam | Cat#ab133684; RRID: AB_2801387 |
| Fluorescein-Conjugated Goat anti-Rabbit IgG (H+L) | ZSGB-BIO | Cat#ZF-0311; RRID: AB_2571576 |
| FcRn-Specific Antibody | Proteintech | Cat#16190-1-AP; RRID: AB_2231751 |
| Primary mouse monoclonal antibody against His tag | ZSGB-BIO | Cat#TA-02; RRID: AB_2801388 |
| Goat anti-rabbit IgG/HRP | ZSGB-BIO | Cat#ZB-2301; RRID: AB_2747412 |
| Goat anti-mouse IgG/HRP | ZSGB-BIO | Cat#ZB-2305; RRID: AB_2747415 |
| Anti- β2 m antibody | biorbyt | Cat#orb153541; RRID: AB_2801389 |
| Anti-GAPDH antibody | Santa Cruz | Cat#sc-47724; RRID: AB_627678 |
| Anti-GAPDH antibody | cwbio | Cat#CW0100M; RRID: AB_2801390 |
| Bacterial and Virus Strains | ||
| Echo 6-SJZ-366, GENBANK: MH830353 | Isolated from patient; Chen et al., 2017 | N/A |
| Echo 1-XZ2005-T48 | China CDC | N/A |
| Echo 3-GS08-072 | China CDC | N/A |
| Echo 4-XZ10-KA66 | China CDC | N/A |
| Echo 6-D’Amori | China CDC | N/A |
| Echo 7-HN09-524 | China CDC | N/A |
| Echo 9-XZ99-120 | China CDC | N/A |
| Echo 11-SD03-478 | China CDC | N/A |
| Echo 13-2011-XJ-HT-YTH-63 | China CDC | N/A |
| Echo 14-GS09-522F | China CDC | N/A |
| Echo 16-XJ11-653128012 | China CDC | N/A |
| Echo 25-GS10-142T | China CDC | N/A |
| Echo 26-2006-X122 | China CDC | N/A |
| Echo 30-YN2016-AB6 | China CDC | N/A |
| Echo 30-Bastinni | China CDC | N/A |
| EV-B85-XJ2011-3-29 | China CDC | N/A |
| CV-A9-HB10-221 | China CDC | N/A |
| CV-B4-NM07-714 | China CDC | N/A |
| CV-B5-SJZ-097 | China CDC | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM basic | Thermo Fisher Scientific | Cat#C11995500BT |
| FBS | Invitrogen | Cat#16000044 |
| Lipofectamine 2000 | Invitrogen | Cat#11668019 |
| XtremeGENE HP DNA transfection reagent | Roche | Cat#06366546001 |
| Sucrose | Sigma | Cat#V900116-500 g |
| Fixation/Permeabilization Solution Kit | BD | Cat#554722 |
| Human protein CD26 | Lu et al., 2013 | N/A |
| Human FcRn soluble protein, GENBANK: NM_001136019.2 and NM_004048 | This paper | N/A |
| Human CD55 soluble protein GENBANK: NM_000574.3 | This paper | N/A |
| Critical Commercial Assays | ||
| HisTrap HP 5 ml column | GE Healthcare | Cat#17524802 |
| Hiload 16/600 Superdex 200 PG column | GE Healthcare | Cat#28989335 |
| Membrane concentrator | Millipore | Cat#UFC9010961 |
| Series S Sensor Chip CM5 | GE Healthcare | Cat#29149603 |
| Deposited Data | ||
| Cryo-EM Echovirus 6 bound to FcRn (pH 7.4) | This paper | EMDB-9687, PDB: 6ILM |
| Cryo-EM Echovirus 6 bound to FcRn (pH 5.5) | This paper | EMDB-9686, PDB: 6ILL |
| Cryo-EM Echovirus 6 full particle (pH7.4) | This paper | EMDB-9690, PDB: 6ILP |
| Cryo-EM Echovirus 6 empty particle (pH7.4) | This paper | EMDB-9689, PDB: 6ILO |
| Cryo-EM Echovirus 6 full particle (pH5.5) | This paper | EMDB-9688, PDB: 6ILN |
| Cryo-EM Echovirus 6 bound to CD55 (pH 7.4) | This paper | EMDB-9685, PDB: 6ILK |
| Cryo-EM Echovirus 6 bound to CD55 (pH 5.5) | This paper | EMDB-9684, PDB: 6ILJ |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | ATCC CRL-3216 |
| Human: rhabdomyosarcoma (RD) cells | ATCC | ATCC CCL-136 |
| Hamster: Chinese hamster ovary (CHO-K1) cells | ATCC | ATCC CCL-61 |
| Hamster: baby hamster kidney (BHK-21) cells | ATCC | ATCC CCL-10 |
| Human: HEK293T FCGRTKO | Edigene | Cat#CL0029124702A |
| Human: HEK293T B2MKO | Edigene | Cat#CL0049664423A |
| Human: HEK293T CD55KO | This paper | N/A |
| Oligonucleotides | ||
| Primers: CaslibF: TATCTTGTGGAAAGGACGAAACACC; CaslibR: AATACGGTTATCCACGCGGC |
Zhou et al., 2014 | N/A |
| sgRNA sequence: for CD55 KO: GTTCTCTTCTGT AACCTGGA |
This paper | N/A |
| Primers: for CD55 KO detection: CD55_1F: TTTAGGTAGC TGCGAGGTGC, CD55_1R: ATTTCCCCCAAACACCAGA TG; CD55_2F: TGCATCCCTCAAACAGCCTT, CD55_2R: CCCAAACACCAGATGGTTGAAA; CD55_3F: GCAGGA CATTAGTCCAGGGG, CD55_3R: CCCCAAACACCAGA TGGTTGAA |
This paper | N/A |
| Oligonucleotides used for the membrane protein sgRNA library construction | This paper | Table S1 |
| Recombinant DNA | ||
| pMD2.G | Laboratory of Didier Trono | Addgene #12259 |
| pCMVR8.74 | Laboratory of Didier Trono | Addgene #22036 |
| lentiCas9-Blast | Sanjana et al., 2014 | Addgene #52962 |
| pLVX-IRES-ZsGreen1 | Clontech | Cat#632187 |
| pLenti-MCS-BSD | Zhou et al., 2014 | N/A |
| pLVX-DsRed-Monomer-N1 | Clonetech | Cat#632152 |
| pLVX-IRES-ZsGreen1-FCGRT | This paper | N/A |
| pLenti-MCS-BSD-B2M | This paper | N/A |
| pLVX- Monomer-N1-CD55 | This paper | N/A |
| Beta-2 microglobulin cDNA ORF Clone, Human, N-His tag | Sino biological | Cat# HG11976-NH |
| pCMV3-FCGRT | This paper | N/A |
| pCMV3-CD55 | This paper | N/A |
| Software and Algorithms | ||
| Model-based Analysis of Genomewide CRISPR/Cas9 Knockout (MAGeCK) | Li et al., 2014 | https://sourceforge.net/p/mageck/wiki/Home |
| TIDE | Brinkman et al., 2014 | https://tide.nki.nl/ |
| MEGA program (version 5.0) | Tamura et al., 2011 | N/A |
| BIAcore 3000 Evaluation software | GE Healthcare | N/A |
| Graphpad Prism 6 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/ |
| Flowjo 7.6.1 | FLOWJO | https://www.flowjo.com/solutions/flowjo/downloads |
| MotionCor2 | Zheng et al., 2017 | N/A |
| CTFFIND4 | Rohou and Grigorieff, 2015 | N/A |
| EMAN2 | Tang et al., 2007 | N/A |
| Relion-2.0 | Scheres, 2012 | N/A |
| PyMOL software | DeLano WL PyMOL molecular graphics system | https://pymol.org/2/ |
| Chimera | Pettersen et al., 2004 | N/A |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| MolProbity | Chen et al., 2010 | N/A |
| e2boxer.py | Tang et al., 2007 | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, George F. Gao (gaof@im.ac.cn).
Experimental Model and Subject Details
Cells and viruses
Human rhabdomyosarcoma (RD) cells, HEK293T, Chinese hamster ovary (CHO) cells and baby hamster kidney (BHK) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 100 U/mL of penicillin-streptomycin with 5% CO2 at 37°C.
Viruses used in this work include: Echo 1 (XZ2005-T48), Echo 3 (GS08-072), Echo 6 (strains SJZ-366 and D’Amori), Echo 7 (HN09-524), Echo 9 (XZ99-120), Echo 11 (SD03-478), Echo 13 (2011-XJ-HT-YTH-63), Echo 14 (GS09-522F), Echo 25 (GS10-142T), Echo 26 (2006-X122), Echo 30 (strains YN2016-AB6 and Bastinni), EV-B85 (XJ2011-3-29), CV-A9 (HB10-221), CV-B4 (NM07-714) and CV-B5 (SJZ-097). All viruses were propagated on RD cells. The anti-Echo 6 sera production in mice was performed according to the procedures approved by the Chinese Academy of Sciences and comply with all relevant ethical regulations regarding animal research.
Method Details
Construction of the Cas9/sgRNA library
We constructed a focused Cas9/sgRNA library targeting 3,283 genes encoding human membrane proteins. This library consisted of 30,848 sgRNAs, among which 400 were non-targeting sgRNAs as negative controls. The sgRNA plasmid library was packaged in HEK293T cells after co-transfection with pMD2.G, a plasmid expressing vesicular stomatitis virus glycoprotein (VSV-G), and pCMVR8.74, a lentiviral packaging plasmid (Addgene), with the X-tremeGENE HP DNA transfection reagent (Roche). Supernatants were collected and filtered with a syringe filter with a 0.22 μm pore size.
A single clone of RD cells stably expressing S. pyogenes cas9 (SpCas9) was constructed by transduction of a lentiviral vector expressing spCas9 and selection by blasticidin (Addgene #52962) (Sanjana et al., 2014). The cleavage activity of Cas9 nuclease in this cell line was confirmed by T7E1 assays. RD-Cas9 cells were transduced with the packaged lentiviral sgRNA library at a multiplicity of infection (MOI) of 0.3, followed by a puromycin selection for ∼2 weeks (Peng et al., 2015, Zhu et al., 2017).
CRISPR-Cas9 screening and data analysis
Echo 6 strain SJZ-366 (MOI of 0.01) was introduced onto approximately 108 RD-Cas9 cells harboring the sgRNA library in DMEM with 2% FBS. After 24 h, > 99% cells were detached, with cytopathic effect (CPE) observed. The medium was exchanged for fresh DMEM with 2% FBS at 24 h and 48 h.p.i (hours post inoculation). The medium was then exchanged with 10% FBS-supplemented DMEM for the resistant cells to grow at day 4 post infection. The medium was exchanged with fresh 10% DMEM every 2 days, and surviving cells were passed into a 6-well plate. Cell populations were further expanded to reach ∼107, and genomic DNA was extracted. The sgRNA-coding regions within the genomic DNA from both experimental groups (two biological replicates) and untreated cell libraries (two biological replicates) were amplified by PCR (CaslibF primer: TATCTTGTGGAAAGGACGAAACACC; CaslibR primer: AATACGGTTA TCCACGCGGC) and subsequently identified by deep-sequencing analysis. The screening data were analyzed with the published Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) method (Li et al., 2014). sgRNAs with low read-counts (≤10) in both control libraries were excluded from data analysis.
KO cell line establishment and validation
Both FCGRT KO and B2M KO cell lines were ordered from Edigene, Beijing. The HEK293T CD55KO cell line was constructed by transient co-transfection of a plasmid expressing sgRNA targeting the CD55 gene and a plasmid expressing spCas9. The sgRNA sequence was GTTCTCTTCTGTAACCTGGA. After selection with puromycin for ∼2 weeks, single cells were seeded in 96-well plates. After cell expansion, the cell clones in each well were transferred to 12-well plates for further culture. The cell clones were screened for CD55 expression by western blot analysis, and the clones lacking CD55 expression were selected. The CD55 DNA fragments of target loci were independently amplified by PCR with three primer pairs (CD55_1F: TTTAGGTAGCTGCGAGGTGC and CD55_1R: ATTTCCCCCAAACACCAGA TG; CD55_2F: TGCATCCCTCAAACAGCCTT and CD55_2R: CCCAAACACCA GATGGTTGAAA; and CD55_3F: GCAGGACATTAGTCCAGGGG and CD55_3R: CCCCAAACACCAGATGGTTGAA). The purified PCR products were sequenced (Ruibiotech, Beijing), and the insertions and deletions (indels) within the CD55 gene caused by sgRNA/Cas9 were analyzed with an online tool (https://tide.nki.nl/) (Brinkman et al., 2014).
Viral TCID50 analysis in KO cell lines
Enteroviruses were propagated in RD cells and collected when > 80% CPE was observed. Serially diluted viruses (from 10−1- 10−10) were inoculated onto WT and KO HEK293T cells. CPE was observed at 48 h.p.i., and TCID50 was calculated using the Reed-Muench method.
Plasmid construction and lentivirus generation
The C-terminally FLAG-tagged human FCGRT coding sequence with a human IL2 signal sequence was cloned into the EcoRI and BamHI sites of the pLVX-IRES-ZsGreen1 vector (Clontech, 632187). The N-terminally His-tagged human β2 m coding sequence with a human IL2 signal sequence was cloned into the XbaI and MluI sites of pLenti-MCS-BSD (Zhou et al., 2014). C-terminally HA-tagged human CD55 with a human IL2 signal sequence was cloned into the EcoRI and XbaI sites of pLVX-DsRed-Monomer-N1 (Clontech, 632152) with a puromycin-resistant element. Lentiviruses were packaged in HEK293T cells by co-transfection of each of the above-mentioned plasmid with pLP1, pLP2 and VSV-g (Invitrogen) at a ratio of 20:20:13:5 using Lipofectamine 2000 (Invitrogen, 11668019).
Trans-complementation
The packaged lentiviruses were introduced into corresponding KO cell lines. HEK293T FCGRT KO +Lenti-FCGRT cells were sorted by flow cytometry. B2M KO +Lenti-B2M cells were selected with 5 μg/mL blasticidin for 7 days. FCGRT KO +Lenti-GFP and B2M KO +Lenti-GFP were used as controls. WT, FCGRT KO/ B2M KO cells, FCGRT KO +Lenti-FCGRT/ B2M KO +Lenti-B2M and FCGRT KO +Lenti-GFP/ B2M KO +Lenti-GFP were seeded in 12-well plates. The Echo 6 strain SJZ-366 (MOI 0.01) was inoculated onto these cells, and supernatants were collected for TCID50 and qPCR analyses after 24 h. TCID50 assays were performed on RD cells. Viral RNA from supernatants was extracted for qPCR analysis (qPCR primers: PEVF, CCTGAATGCGGCTAATCC and PEVR; TTGTCACCATWAGCAGYCA PEV Probe, FAM-CCGACTACTTTGGGWGTCCGTGT-TAMRA).
Ectopic expression
After seeding in 12-well plates, CHO and BHK cells were transduced with Lenti-FCGRT + Lenti-B2M, Lenti-CD55, Lenti-FCGRT + Lenti-B2M+ Lenti-CD55, or Lenti-GFP. Positive cells were selected with antibiotics and/or flow cytometry sorting (Lenti-FCGRT positive: FITC positive in flow cytometry; Lenti-B2M positive: 5 μg/mL blasticidin selection for 7 days; and Lenti-CD55 positive: 5 μg/mL puromycin selection for 7 days). Echo 6 strain SJZ-366 (MOI of 0.1) was inoculated into the cells for 1 h, then removed and washed twice with PBS. Supernatants were collected for TCID50 and qPCR analyses 24 h.p.i.
Growth curve analysis
FCGRT KO, B2M KO, CD55 KO and WT HEK293T cells were seeded in 12-well plates. Echo 6 strain SJZ-366 (MOI 0.01) was added to these cells, and supernatants were collected at 0, 12, 24, 48 and 60 h.p.i. Viral RNA was extracted for qPCR analysis.
Phylogenic analysis
Pairwise alignment of the nucleotide sequences of EV-B was performed using the MEGA program (version 5.0) (Tamura et al., 2011). Phylogenetic trees were constructed by the maximum likelihood algorithms implemented in the MEGA program using the Tamura-Nei model. Regions containing alignment gaps were omitted from the analysis. The branch lengths of the dendrogram were determined from the topologies of the trees and were obtained by majority rule consensus among 1000 bootstrap replicates. Bootstrap values > 80% were considered statistically significant for grouping. The output tree files were visualized using the FigureTree program.
Bright light image of CPE
For light images, FCGRT KO, B2M KO, CD55KO and WT HEK293T cells were infected with Echo 6 at a MOI of 1. Twenty-four hours later, CPE was observed, and pictures were taken.
Echo 6 antibody generation
Echo 6 was inactivated by 0.05% β-propiolactone at 4°C for 48 h, and β-propiolactone was then inactivated at 37°C for 2 h. Eight-week-old BALB/c mice were intramuscularly immunized with 200 μL inactivated Echo 6. After boosting twice, sera were collected, and IgG was purified with a protein A column (GE Healthcare).
Immunofluorescence
For immunofluorescence analysis of Echo 6 infection, FCGRT KO, B2M KO, CD55KO and WT HEK293T cells were seeded onto coverslips in 24-well plates at a density of 10,000 cells per well overnight and infected by Echo 6 (MOI 10). Eight hours later, cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 in PBS for 20 min. After blocking with 3% BSA, cells were incubated for 1 h with anti-Echo 6 primary antibody at a dilution of 1:200, followed by incubation with an Alex 488-labeled secondary antibody (Abcam, ab150113, 1:100 dilution). After DAPI staining for 1 h, samples were mounted on microscope slides. Images were taken with a Leica TCS-SP confocal microscope and processed using ZEN software.
For FcRn and CD55 staining, cells were stained by anti-FCGRT (Abcam, ab193148) or anti-CD55 (Abcam, ab33111) primary antibody at a dilution of 1:100. FITC-labeled anti-rabbit secondary antibody (ZSGB-BIO, ZF-0311, 1:100 dilution) and Alex 488-labeled anti-mouse secondary antibody (Abcam, ab150113, 1:100 dilution) were subsequently used, respectively.
Gene cloning, expression, and protein purification
The human FCGRT coding fragment (A16–L282) or the human CD55 coding fragment (D35-G285) was inserted into pCMV3 (Sino biological, China) with a human IL2 signal sequence at the N terminus and six histidines at the C terminus using the HindIII and BamHI sites, respectively. Plasmids expressing human FCGRT and human β2 m were transiently co-transfected into HEK293T cells for human FcRn protein expression. Plasmid expressing human CD55 was transfected into HEK293T cells for CD55 expression. In each case, the supernatant was collected, and the protein was subsequently purified by Ni-nitrilotriacetic acid (NTA) chromatography (GE Healthcare) and gel filtration using a Hiload 16/600 Superdex 200 PG column (GE Healthcare) with a buffer containing 20 mM Tris·HCl and 150 mM NaCl (pH 8.0).
Virus purification
Echo 6 strain SJZ-366 was propagated in RD cells for 36 h at 37°C after inoculation with a MOI of ∼0.1. The supernatant was collected and centrifuged at 12,000 x g for 60 min to remove cell debris. Initial purification was performed by ultracentrifugation through a 30% (w/v) sucrose cushion using a type 45Ti rotor at 140,000 x g for 2 h. The virus was resuspended in PBS (phosphate buffer saline, pH 7.4) and loaded onto a sucrose gradient (7.5%–45%, w/v) for further purification by centrifugation using a SW41 rotor at 4°C for 5 h. Fractions containing viral particles were pooled and buffer-exchanged into PBS buffer and concentrated using an Amicon Ultra-6 100 kDa cut-off centrifugal concentrator (Millipore).
SPR
SPR binding experiments were performed using an BIAcore 3000 device (GE Healthcare) at room temperature (25°C) to measure the kinetics and affinity of FcRn or CD55 binding to Echo 6 virus. The buffers for all proteins used for kinetic analyses were exchanged to PBST consisting of PBS and 0.005% (v/v) Tween-20 (pH 7.4) via gel filtration. Purified Echo 6 or CV-B4 viruses were immobilized on a CM5 chip with the Standard EDC/NHS coupling method to about 6,000 response units (RU). Serial dilutions of FcRn or CD55 were prepared and sequentially injected at a rate of 30 μL/min, and the data were collected over time. After each cycle, a short injection of 4 M NaCl was employed to regenerate the sensor surface. The binding kinetics was analyzed, and affinity constants were calculated using BIAcore 3000 analysis software (BIAevaluation Version 4.1).
ELISA-based binding assay
The binding of FcRn or CD55 to Echo 6 virions was determined by ELISAs. The purified virions were immobilized at 1.5 × 1010 TCID50/well in carbonate-bicarbonate buffer (pH 9.6) on 96-well plates at 4°C overnight. Then, wells were washed with PBS-T (0.01 M PBS with 0.1% (v/v) Tween 20) for 5 × 5 min. After blocking with 5% non-fat milk at room temperature (RT) for 1 h, plates were washed once with PBS-T (0.01 M PBS with 0.1% (v/v) Tween 20). FcRn, CD55, or the irrelevant human protein CD26 (Lu et al., 2013) was serially diluted in PBS and then added and incubated at RT for 1 h. The wells were washed with PBST for 5 × 5 min. After that, anti-FcRn (Proteintech, 16190-1-AP) or anti-His (ZSGB-BIO, TA-02) antibodies were diluted with PBS and added to the wells for 1 h at RT. The plates were washed with PBST for 5 × 5 min. Subsequently, goat anti-rabbit IgG/HRP (ZSGB-BIO, ZB-2301) or goat anti-mouse IgG/HRP (ZSGB-BIO, ZB-2305) diluted with PBS to 1:2000 was added and incubated at RT for 1 h, followed by washing with PBST for 5 × 5 min. TMB (3, 3′, 5′, 113 5′- tetramethylbenzidine) substrate (CWBio) was added, and 2 M HCl was applied to stop the reaction. Absorbance at 450 nm was measured and recorded.
Blocking assays with FcRn or CD55 soluble protein
RD cells were seeded into 48-well plates. Serial dilutions of purified FcRn or CD55 soluble protein were incubated with Echo 6 (0.1 MOI) for 2 h at 4°C in a volume of 100 μL. The mixture was then added to RD cells for 1 h incubation at 37°C, removed, and washed twice with PBS. DMEM with 2% FBS was added to the cells and incubated for another 12 h. Supernatants were collected for viral RNA extraction and qPCR analysis.
Virus binding and internalization assays
The assays were performed in suspension as described previously (Zhang et al., 2018) with modifications. Briefly, FCGRT KO, B2M KO, CD55KO, and WT HEK293T cells were collected in suspension. Echo 6 strain SJZ-366 (MOI 50) was incubated with the cells (1 × 106) on ice for 1 h. After three cycles of washing using ice-cold PBS, cells were collected, and RNAs were extracted for qPCR analysis. For the internalization assays, after on-ice incubation, the cell-virus mixture was directly placed at 37°C for 30 min. Cells were washed once with PBS and then treated with 500 ng/mL proteinase K on ice for 1 h. After three washes, cells were collected for RNA extraction and qPCR analysis. Virus titers were detected using PEV primers and probes as described above. GAPDH was included as an internal control (qPCR Primers: HGapdhF, CCACTCCTCCACCTTTGAC and HGapdhR, ACCCTGTTGCT GTAGCCA; and HGapdhProbe, FAM- TTGCCCTCAACGACCACTTTGTC-TAMRA)
Cryo-EM sample preparation and data collection
To prepare the Echo 6-CD55/FcRn complexes, purified Echo 6 particles (1.8 mg/mL) were incubated with excess CD55/FcRn protein for 8 h and 10 min at 4°C, respectively. A 3 μL aliquot of free-Echo6 virus, Echo6-CD55, or Echo6-FcRn complex at pH 7.4 was placed on freshly glow-discharged copper grid coated with a layer of ultra-thin carbon film. After a 30 s incubation for particle absorption, the excess buffer solution was manually blotted, and the grid was plunge-frozen using a Vitrobot Mark IV operated at 100% humidity. For the low pH samples, a drop (4 μL) of acidic buffer (20 mM MES and 150 mM NaCl, pH 5.5) was immediately added to the grid after the first blotting, the samples were incubated for an additional 10 min at 4°C before blotting and plunge-freezing.
Cryo-EM data was manually collected using a 300 kV Titan Krios (FEI) microscope equipped with a K2 summit camera. Images were recorded in super-resolution counting mode with a calibrated pixel size of 1.35 Å. Each exposure was performed with an accumulative dose of 40 e-/Å2, which was fractionated into 32 frames for each image stack. The final defocus range of the micrographs was −1.0 to −3.0 μm.
Image processing
The movie-frames were aligned and summed using the program MotionCor2 (Zheng et al., 2017). The initial contrast transfer function (CTF) parameters were estimated with CTFFIND4 (Rohou and Grigorieff, 2015), and particles were semi-automatically boxed using the e2boxer.py tool in the EMAN2 (Tang et al., 2007) program package. Seven structures were determined using Relion-2.0 (Scheres, 2012) with icosahedral symmetry applied. Under physiological conditions (pH 7.4), the structures of free-Echo 6 full and empty particles, Echo 6-CD55 complex, and Echo 6-FcRn complex were determined at 2.9, 3.2, 3.0, and 3.4 Å resolution, respectively. Under acidic conditions (pH 5.5), the structures of free-Echo6 full particle, Echo 6-CD55, and Echo 6-FcRn complexes were determined at 3.4, 3.6, and 3.8 Å resolution, respectively. The resolutions were estimated by the gold-standard Fourier shell correction 0.143 cut-off values (Scheres, 2012).
Model building and refinement
The crystal structure of Echovirus 1 (PDB: 1EV1) was used as the initial model to fit the free-Echo 6 density map (pH 7.4, 2.9 Å) using Chimera (Pettersen et al., 2004). After initial fitting, the model was manually adjusted in Coot (Emsley et al., 2010) to improve map fitting and update the sequence registers. The density map of an asymmetric unit (ASU) was then segmented using Chimera (Pettersen et al., 2004) and placed into a pseudo crystallographic unit cell (P1 space group) for model refinement. The atomic model was refined against the ASU map in real space using Phenix with secondary structure restraints applied. Model statistics (including bond lengths, bond angles, and all-atom clash), rotamer statistics, and Ramachandran plot statistics were monitored during the refinement procedure. After several rounds of iterative model building and refinement, the model coordinates converged and fit well in the density map by visual inspection. At this point, the individual B factors and occupancies of each atom were refined using the standard reciprocal space refinement procedure in Phenix (Adams et al., 2010) The stereochemical quality of the final model was assessed using MolProbity (Chen et al., 2010). The Echo 6-CD55/FcRn complexes were modeled with the free-Echo6 structure as the initial model following a similar procedure as described above. All data collection, processing, model building and refinement statistics are summarized in Table S3.
Structure analysis and visualization
The reconstructed maps and atomic models were visualized using Chimera (Pettersen et al., 2004) and analyzed using the wrapped applications. All EM density figures were rendered by Chimera, and cartoon representations of atomic models were generated with PyMOL software (DeLano WL PyMOL molecular graphics system. Available from: URL: http://www.pymol.org).
Preparation of receptor-decorated liposomes
Liposomes consisting of phosphatidylethanolamine, phosphatidylcholine, sphingomyelin, cholesterol, and phosphatidic acid were supplied in chloroform and mixed in molar ratios of 1:1:1:1.5:0.3, respectively (Strauss et al., 2013). Nickel-chelating liposomes, for binding of the His-tagged receptor, were generated by the inclusion of the nickel salt of 1,2-dioleoyl-sn-glycero-3-{[N(5-amino-1-carboxypentyl) iminodiacetic acid] succinyl}, at a final concentration of 20% (w/w). The chloroform was evaporated under argon gas to produce a lipid film, which was dried under vacuum overnight. Dried lipid was rehydrated at a concentration of 3 mg/mL in 50 mM HEPES (pH 7.3) and 50 mM NaCl. The nickel-chelating liposomes were made by extruding lipids through a membrane filter with 0.4-μm diameter pores and then incubated with His-tagged CD55 or FcRn at a final concentration of 100 μg/mL at room temperature for 30 min.
Incubation assay of receptor-decorated liposomes with Echo 6 particles
High-purity full-Echo 6 particles were obtained by 15%–45% (w/v) sucrose density gradient centrifugation. Receptor-decorated liposomes and full-Echo 6 particles were separately and rapidly buffer-exchanged and concentrated using an Amicon Ultra-6 100 kDa cutoff centrifugal concentrator into acidic buffer (20 mM MES and 150 mM NaCl, pH 5.5) at 4°C. Samples of receptor-decorated liposomes containing approximately 2 mg/mL lipid and 0.3 mg/mL bovine serum albumin (BSA) were mixed at 37°C with equal volume (100 μL) of full-Echo 6 particles (0.8 mg/mL). After a 10-min incubation, the mixed samples were used for negative staining. The specimens were loaded onto an FEI Tecnai G20 transmission electron microscope operating at 120 kV for imaging.
Western blotting
Cells were collected, lysed, and electrophoresed using 12% SDS-PAGE. Proteins were transferred onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk for 2 h at 4°C, then stained at 4°C overnight using the following primary antibodies: anti-FcRn (Proteintech, 16190-1-AP), anti-CD55 (Abcam, ab133684), anti- β2 m (biorbyt, orb153541) and anti-GAPDH (Santa Cruz, sc-47724; cwbio, CW0100M). After three washes with PBST (PBS with 0.1% Tween-20), the membranes were incubated with secondary antibody (goat anti-mouse IgG/HRP (ZSGB-BIO, ZB-2305) or goat anti-rabbit IgG/HRP (ZSGB-BIO, ZB-2301)) for 1 h at RT. After another set of washes, the blots were analyzed using SuperSignal West Pico Chemiluminescent Substrate (Life Technologies).
Flow cytometry
Cells were washed with PBS, trypsinized, and pelleted. For fixed cell staining, cells were fixed using a Fixation/Permeabilization Solution Kit (BD, 554722) for 20 min at 4°C and washed twice with PBS. For cell surface staining, cells were washed and directly sent for primary antibody staining. The cells were stained with CD55 (Abcam, ab33111) or isotype antibody at a dilution of 1:100, followed by three washes with PBS, and incubated with an Alex 488 labeled secondary antibody (Abcam, ab150113, 1:100 dilution) for 1 h. Flow cytometry acquisition was performed with a BD FACSDiva-driven BD FACSCANTO I. Data were analyzed with FlowJo software.
Quantification and Statistical Analysis
Statistical significance was determined as p < 0.05 using GraphPad Prism 6. Experiments were analyzed by unpaired two-tailed t tests with the Welch post-correction or ANOVA with multiple comparisons.
Data and Software Availability
Data Resources
The cryo-EM density maps and corresponding atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB), respectively. The accession numbers for the cryo-EM structures reported in this paper are: Echovirus 6 full particle at pH7.4 (EMDB-9690, PDB: 6ILP); Echovirus 6 empty particle at pH7.4 (EMDB-9689, PDB: 6ILO); Echovirus 6 full particle at pH5.5 (EMDB-9688, PDB: 6ILN); Echovirus 6 bound to CD55 at pH 7.4 (EMDB-9685, PDB: 6ILK); Echovirus 6 bound to CD55 at pH 5.5 (EMDB-9684, PDB: 6ILJ); Echovirus 6 bound to FcRn at pH 7.4 (EMDB-9687, PDB: 6ILM); Echovirus 6 bound to FcRn at pH 5.5 (EMDB-9686, PDB: 6ILL).
Acknowledgments
We thank the Center of Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences (CAS), Beijing, for assistance with cryo-EM data collection. We thank Dr. Junjie Hu (National Laboratory of Biomacromolecules, Institute of Biophysics, CAS) for the help on nickel-charged liposome preparation. We are grateful to Dr. Guopeng Wang (the core facility at School of Life Sciences, Peking University), Dr. Tie Yang, and the staff in the EM department of the State Key Laboratory of Membrane Biology, Institute of Zoology, CAS, Beijing, for technical support in electron microscope operation. We thank Gary Wong for the proofreading. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010000 to G.F.G.), the China National Grand S&T Special Project (2015ZX09102024 to G.F.G.), the National Key R&D Program of China (2016YFC1200304 to X.Z.), NSFC (81601766 to X.Z. and 31430025 to W.W.), funds from Beijing Municipal Science & Technology Commission (Z181100001318009 to W.W.), Beijing Advanced Innovation Center for Genomics at Peking University (to W.W.), the Peking-Tsinghua Center for Life Sciences (to W.W.), Capital Clinical Feature Project of Beijing Technology Program (Z151100004015046 to Z.X.), postdoctoral fellowship from the Peking-Tsinghua Center for Life Sciences (to G.Z.), Beijing Natural Science Foundation (7184208 to X.C.), and Youth Innovation Promotion Association CAS (2018113 to L.D.). G.F.G. is supported partly as a leading principal investigator of the NSFC Innovative Research Group (81621091).
Author Contributions
G.F.G., W.W., and X.Z. designed and supervised the study. X.Z., G.Z., X.C., P.Y., and Z.L. did the library construction, screening, and sequencing data analysis. G.Z. established and certified the KO cell lines. X.Z., S. Li, Z.G., and M.Z. performed the infection analysis. X.Q. and Y.L. did the imaging, fluorescence-activated cell sorting (FACS), and confocal analysis. X.Z. and N.D. did the construction design and stable cell line establishment. H.S., C.L, Z.S., and H.W. designed and expressed the soluble protein and performed the SPR detection. S. Liu, J.Q., X.Z., and X.C. performed the virus propagation and purification. S. Liu and R.P. did the cryo-EM analysis. Y.Z. provided key reagents and did phylogenic analysis. X.Z., Y.W., J.Q., Y.B., J.Y., Z.X., R.P., and W.W. designed and directed the experiments. X.Z, G.Z., H.S., R.P., W.W., and G.F.G. wrote the manuscript. L.D., Y.S., S.G., X.C., Y.Z., and Z.X. participated in the manuscript editing and discussion.
Declaration of Interests
W.W. serves as a scientific advisor for EdiGene.
Published: May 16, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.04.035.
Contributor Information
Zhengde Xie, Email: xiezhengde@bch.com.cn.
Wensheng Wei, Email: wswei@pku.edu.cn.
George F. Gao, Email: gaof@im.ac.cn.
Supplemental Information
References
- Abedi G.R., Watson J.T., Nix W.A., Oberste M.S., Gerber S.I. Enterovirus and Parechovirus Surveillance - United States, 2014-2016. MMWR Morb. Mortal. Wkly. Rep. 2018;67:515–518. doi: 10.15585/mmwr.mm6718a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abzug M.J. The enteroviruses: problems in need of treatments. J. Infect. 2014;68(Suppl 1):S108–S114. doi: 10.1016/j.jinf.2013.09.020. [DOI] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen J., Hurdiss D.L., Zocher G., Mistry N., Roberts R.W., Slager J.J., Guo H., van Vliet A.L.W., Wahedi M., Benschop K. Role of enhanced receptor engagement in the evolution of a pandemic acute hemorrhagic conjunctivitis virus. Proc. Natl. Acad. Sci. USA. 2018;115:397–402. doi: 10.1073/pnas.1713284115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M., Coyne C.B. Picornavirus entry. Adv. Exp. Med. Biol. 2013;790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- Bergelson J.M., St John N., Kawaguchi S., Chan M., Stubdal H., Modlin J., Finberg R.W. Infection by echoviruses 1 and 8 depends on the alpha 2 subunit of human VLA-2. J. Virol. 1993;67:6847–6852. doi: 10.1128/jvi.67.11.6847-6852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M., Chan M., Solomon K.R., St John N.F., Lin H., Finberg R.W. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. USA. 1994;91:6245–6248. doi: 10.1073/pnas.91.13.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J.M., Cunningham J.A., Droguett G., Kurt-Jones E.A., Krithivas A., Hong J.S., Horwitz M.S., Crowell R.L., Finberg R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Bhella D., Goodfellow I.G., Roversi P., Pettigrew D., Chaudhry Y., Evans D.J., Lea S.M. The structure of echovirus type 12 bound to a two-domain fragment of its cellular attachment protein decay-accelerating factor (CD 55) J. Biol. Chem. 2004;279:8325–8332. doi: 10.1074/jbc.M311334200. [DOI] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck D., Filman D.J., Hogle J.M. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol. 2005;12:615–618. doi: 10.1038/nsmb955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li J., Guo J., Xu W., Sun S., Xie Z. An outbreak of echovirus 18 encephalitis/meningitis in children in Hebei Province, China, 2015. Emerg. Microbes Infect. 2017;6:e54. doi: 10.1038/emi.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary M.C. Echovirus Infection. eMedicine. 2017 https://emedicine.medscape.com/article/216564-overview [Google Scholar]
- Chumakov P.M., Morozova V.V., Babkin I.V., Baĭkov I.K., Netesov S.V., Tikunova N.V. [Oncolytic enteroviruses] Mol. Biol. (Mosk.) 2012;46:712–725. [PubMed] [Google Scholar]
- Croker C., Civen R., Keough K., Ngo V., Marutani A., Schwartz B., Centers for Disease Control and Prevention (CDC) Aseptic meningitis outbreak associated with echovirus 30 among high school football players--Los Angeles County, California, 2014. MMWR Morb. Mortal. Wkly. Rep. 2015;63:1228. [PubMed] [Google Scholar]
- Dalldorf G., Enders J.F., Hammon W.M., Sabin A.B., Syverton J.T., Melnick J.L. ENTERIC cytopathogenic human orphan (ECHO) viruses. Science. 1955;122:1187–1188. [PubMed] [Google Scholar]
- Doniņa S., Strēle I., Proboka G., Auziņš J., Alberts P., Jonsson B., Venskus D., Muceniece A. Adapted ECHO-7 virus Rigvir immunotherapy (oncolytic virotherapy) prolongs survival in melanoma patients after surgical excision of the tumour in a retrospective study. Melanoma Res. 2015;25:421–426. doi: 10.1097/CMR.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppelli E., Levy H.C., Sun E., Strauss M., Nicol C., Gold S., Zhuang X., Tuthill T.J., Hogle J.M., Rowlands D.J. Picornavirus RNA is protected from cleavage by ribonuclease during virion uncoating and transfer across cellular and model membranes. PLoS Pathog. 2017;13:e1006197. doi: 10.1371/journal.ppat.1006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley E.S., Au G.G., Carlton B.R., Barry R.D., Shafren D.R. Regional administration of oncolytic Echovirus 1 as a novel therapy for the peritoneal dissemination of gastric cancer. J. Mol. Med. (Berl.) 2009;87:385–399. doi: 10.1007/s00109-008-0433-0. [DOI] [PubMed] [Google Scholar]
- He Y., Bowman V.D., Mueller S., Bator C.M., Bella J., Peng X., Baker T.S., Wimmer E., Kuhn R.J., Rossmann M.G. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA. 2000;97:79–84. doi: 10.1073/pnas.97.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Chipman P.R., Howitt J., Bator C.M., Whitt M.A., Baker T.S., Kuhn R.J., Anderson C.W., Freimuth P., Rossmann M.G. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 2001;8:874–878. doi: 10.1038/nsb1001-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Lin F., Chipman P.R., Bator C.M., Baker T.S., Shoham M., Kuhn R.J., Medof M.E., Rossmann M.G. Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex. Proc. Natl. Acad. Sci. USA. 2002;99:10325–10329. doi: 10.1073/pnas.152161599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkilä O., Susi P., Tevaluoto T., Härmä H., Marjomäki V., Hyypiä T., Kiljunen S. Internalization of coxsackievirus A9 is mediated by beta2-microglobulin, dynamin, and Arf6 but not by caveolin-1 or clathrin. J. Virol. 2010;84:3666–3681. doi: 10.1128/JVI.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C.W., Koo S.S.F., Osman H., Wilson S., Xerry J., Gallimore C.I., Allen D.J., Tang J.W. Predominance of enterovirus B and echovirus 30 as cause of viral meningitis in a UK population. J. Clin. Virol. 2016;81:90–93. doi: 10.1016/j.jcv.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhou Y., Lu H., Yang H., Feng Q., Dai Y., Chen L., Yu S., Yao X., Zhang H. Characterization of severe hand, foot, and mouth disease in Shenzhen, China, 2009-2013. J. Med. Virol. 2015;87:1471–1479. doi: 10.1002/jmv.24200. [DOI] [PubMed] [Google Scholar]
- Hughes S.A., Thaker H.M., Racaniello V.R. Transgenic mouse model for echovirus myocarditis and paralysis. Proc. Natl. Acad. Sci. USA. 2003;100:15906–15911. doi: 10.1073/pnas.2535934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson S., Jonsson N., Gullberg M., Lindberg A.M. Cytolytic replication of echoviruses in colon cancer cell lines. Virol. J. 2011;8:473. doi: 10.1186/1743-422X-8-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadambari S., Okike I., Ribeiro S., Ramsay M.E., Heath P.T., Sharland M., Ladhani S.N. Seven-fold increase in viral meningo-encephalitis reports in England and Wales during 2004-2013. J. Infect. 2014;69:326–332. doi: 10.1016/j.jinf.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Khetsuriani N., Lamonte-Fowlkes A., Oberst S., Pallansch M.A., Centers for Disease Control and Prevention Enterovirus surveillance--United States, 1970-2005. MMWR Surveill. Summ. 2006;55:1–20. [PubMed] [Google Scholar]
- Kim H.J., Kang B., Hwang S., Hong J., Kim K., Cheon D.S. Epidemics of viral meningitis caused by echovirus 6 and 30 in Korea in 2008. Virol. J. 2012;9:38. doi: 10.1186/1743-422X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao N., Zhao L., Zhu Z., Chen X., Zhou S., Zhang Y., Cui A., Ji Y., Xu S., Xu W. An aseptic meningitis outbreak caused by echovirus 6 in Anhui province, China. J. Med. Virol. 2010;82:441–445. doi: 10.1002/jmv.21707. [DOI] [PubMed] [Google Scholar]
- Marjomäki V., Pietiäinen V., Matilainen H., Upla P., Ivaska J., Nissinen L., Reunanen H., Huttunen P., Hyypiä T., Heino J. Internalization of echovirus 1 in caveolae. J. Virol. 2002;76:1856–1865. doi: 10.1128/JVI.76.4.1856-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjomäki V., Turkki P., Huttunen M. Infectious Entry Pathway of Enterovirus B Species. Viruses. 2015;7:6387–6399. doi: 10.3390/v7122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino T.A., Petric M., Weingartl H., Bergelson J.M., Opavsky M.A., Richardson C.D., Modlin J.F., Finberg R.W., Kain K.C., Willis N. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology. 2000;271:99–108. doi: 10.1006/viro.2000.0324. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C.L., Wimmer E., Racaniello V.R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Shimojima M., Tano Y., Miyamura T., Wakita T., Shimizu H. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 2009;15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- Oganesyan V., Damschroder M.M., Cook K.E., Li Q., Gao C., Wu H., Dall’Acqua W.F. Structural insights into neonatal Fc receptor-based recycling mechanisms. J. Biol. Chem. 2014;289:7812–7824. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organtini L.J., Makhov A.M., Conway J.F., Hafenstein S., Carson S.D. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J. Virol. 2014;88:5755–5765. doi: 10.1128/JVI.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Zhou Y., Zhu S., Wei W. High-throughput screens in mammalian cells using the CRISPR-Cas9 system. FEBS J. 2015;282:2089–2096. doi: 10.1111/febs.13251. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plevka P., Hafenstein S., Harris K.G., Cifuente J.O., Zhang Y., Bowman V.D., Chipman P.R., Bator C.M., Lin F., Medof M.E., Rossmann M.G. Interaction of decay-accelerating factor with echovirus 7. J. Virol. 2010;84:12665–12674. doi: 10.1128/JVI.00837-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R.M., Ward T., Evans D.J., Almond J.W. Interaction between echovirus 7 and its receptor, decay-accelerating factor (CD55): evidence for a secondary cellular factor in A-particle formation. J. Virol. 1997;71:9306–9312. doi: 10.1128/jvi.71.12.9306-9312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renois F., Hong S.S., Le Naour R., Gafa V., Talmud D., Andréoletti L., Lévêque N. Development of a recombinant CHO cell model for the investigation of CAR and DAF role during early steps of echovirus 6 infection. Virus Res. 2011;158:46–54. doi: 10.1016/j.virusres.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopenian D.C., Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat. Rev. Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., He Y., Kuhn R.J. Picornavirus-receptor interactions. Trends Microbiol. 2002;10:324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlachetzki F., Zhu C., Pardridge W.M. Expression of the neonatal Fc receptor (FcRn) at the blood-brain barrier. J. Neurochem. 2002;81:203–206. doi: 10.1046/j.1471-4159.2002.00840.x. [DOI] [PubMed] [Google Scholar]
- Shafren D.R., Sylvester D., Johansson E.S., Campbell I.G., Barry R.D. Oncolysis of human ovarian cancers by echovirus type 1. Int. J. Cancer. 2005;115:320–328. doi: 10.1002/ijc.20866. [DOI] [PubMed] [Google Scholar]
- Shakeel S., Seitsonen J.J., Kajander T., Laurinmäki P., Hyypiä T., Susi P., Butcher S.J. Structural and functional analysis of coxsackievirus A9 integrin αvβ6 binding and uncoating. J. Virol. 2013;87:3943–3951. doi: 10.1128/JVI.02989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staring J., van den Hengel L.G., Raaben M., Blomen V.A., Carette J.E., Brummelkamp T.R. KREMEN1 Is a Host Entry Receptor for a Major Group of Enteroviruses. Cell Host Microbe. 2018;23:636–643. doi: 10.1016/j.chom.2018.03.019. [DOI] [PubMed] [Google Scholar]
- Strauss M., Levy H.C., Bostina M., Filman D.J., Hogle J.M. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M., Filman D.J., Belnap D.M., Cheng N., Noel R.T., Hogle J.M. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J. Virol. 2015;89:4143–4157. doi: 10.1128/JVI.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tilgase A., Patetko L., Blāķe I., Ramata-Stunda A., Borodušķis M., Alberts P. Effect of the oncolytic ECHO-7 virus Rigvir® on the viability of cell lines of human origin in vitro. J. Cancer. 2018;9:1033–1049. doi: 10.7150/jca.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill T.J., Bubeck D., Rowlands D.J., Hogle J.M. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 2006;80:172–180. doi: 10.1128/JVI.80.1.172-180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill T.J., Groppelli E., Hogle J.M., Rowlands D.J. Picornaviruses. Curr. Top. Microbiol. Immunol. 2010;343:43–89. doi: 10.1007/82_2010_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Peng W., Ren J., Hu Z., Xu J., Lou Z., Li X., Yin W., Shen X., Porta C. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 2012;19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T., Pipkin P.A., Clarkson N.A., Stone D.M., Minor P.D., Almond J.W. Decay-accelerating factor CD55 is identified as the receptor for echovirus 7 using CELICS, a rapid immuno-focal cloning method. EMBO J. 1994;13:5070–5074. doi: 10.1002/j.1460-2075.1994.tb06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T., Powell R.M., Pipkin P.A., Evans D.J., Minor P.D., Almond J.W. Role for beta2-microglobulin in echovirus infection of rhabdomyosarcoma cells. J. Virol. 1998;72:5360–5365. doi: 10.1128/jvi.72.7.5360-5365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J.L., Cornell C.T., Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 2005;3:765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- Williams C.H., Kajander T., Hyypiä T., Jackson T., Sheppard D., Stanway G. Integrin alpha v beta 6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 2004;78:6967–6973. doi: 10.1128/JVI.78.13.6967-6973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Bator C.M., Bowman V.D., Rieder E., He Y., Hébert B., Bella J., Baker T.S., Wimmer E., Kuhn R.J., Rossmann M.G. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 2001;75:2444–2451. doi: 10.1128/JVI.75.5.2444-2451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Bator-Kelly C.M., Rieder E., Chipman P.R., Craig A., Kuhn R.J., Wimmer E., Rossmann M.G. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure. 2005;13:1019–1033. doi: 10.1016/j.str.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Yamayoshi S., Yamashita Y., Li J., Hanagata N., Minowa T., Takemura T., Koike S. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 2009;15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- Ylä-Pelto J., Tripathi L., Susi P. Therapeutic Use of Native and Recombinant Enteroviruses. Viruses. 2016;8:57. doi: 10.3390/v8030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.D., Cifuente J.O., Pan J., Bergelson J.M., Hafenstein S. The crystal structure of a coxsackievirus B3-RD variant and a refined 9-angstrom cryo-electron microscopy reconstruction of the virus complexed with decay-accelerating factor (DAF) provide a new footprint of DAF on the virus surface. J. Virol. 2012;86:12571–12581. doi: 10.1128/JVI.01592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Mueller S., Morais M.C., Bator C.M., Bowman V.D., Hafenstein S., Wimmer E., Rossmann M.G. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc. Natl. Acad. Sci. USA. 2008;105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Kim A.S., Fox J.M., Nair S., Basore K., Klimstra W.B., Rimkunas R., Fong R.H., Lin H., Poddar S. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature. 2018;557:570–574. doi: 10.1038/s41586-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Zhou X., Liu J., Xia L., Pan Y., Chen J., Luo N., Yin J., Ma S. Molecular identification of human enteroviruses associated with aseptic meningitis in Yunnan province, Southwest China. Springerplus. 2016;5:1515. doi: 10.1186/s40064-016-3194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Zhou Y., Wei W. Genome-Wide CRISPR/Cas9 Screening for High-Throughput Functional Genomics in Human Cells. Methods Mol. Biol. 2017;1656:175–181. doi: 10.1007/978-1-4939-7237-1_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Resources
The cryo-EM density maps and corresponding atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB), respectively. The accession numbers for the cryo-EM structures reported in this paper are: Echovirus 6 full particle at pH7.4 (EMDB-9690, PDB: 6ILP); Echovirus 6 empty particle at pH7.4 (EMDB-9689, PDB: 6ILO); Echovirus 6 full particle at pH5.5 (EMDB-9688, PDB: 6ILN); Echovirus 6 bound to CD55 at pH 7.4 (EMDB-9685, PDB: 6ILK); Echovirus 6 bound to CD55 at pH 5.5 (EMDB-9684, PDB: 6ILJ); Echovirus 6 bound to FcRn at pH 7.4 (EMDB-9687, PDB: 6ILM); Echovirus 6 bound to FcRn at pH 5.5 (EMDB-9686, PDB: 6ILL).