Abstract
Niemann-Pick disease type C (NPC) is associated with mutations in NPC1 and NPC2, whose gene products are key players in the endosomal/lysosomal egress of low-density lipoprotein-derived cholesterol. NPC1 is also the intracellular receptor for Ebola virus (EBOV). Here, we present a 4.4 Å structure of full-length human NPC1 and a low-resolution reconstruction of NPC1 in complex with the cleaved glycoprotein (GPcl) of EBOV, both determined by single-particle electron cryomicroscopy. NPC1 contains 13 transmembrane segments (TMs) and three distinct lumenal domains A (also designated NTD), C, and I. TMs 2–13 exhibit a typical resistance-nodulation-cell division fold, among which TMs 3–7 constitute the sterol-sensing domain conserved in several proteins involved in cholesterol metabolism and signaling. A trimeric EBOV-GPcl binds to one NPC1 monomer through the domain C. Our structural and biochemical characterizations provide an important framework for mechanistic understanding of NPC1-mediated intracellular cholesterol trafficking and Ebola virus infection.
Graphical Abstract
Highlights
-
•
The cryo-EM structure of full-length human NPC1 was determined at 4.4 Å resolution
-
•
Structure-guided biochemical analysis of cholesterol transfer from NPC2 to NPC1
-
•
Low-resolution cryo-EM structure of NPC1 bound to GPcl of Ebola virus was obtained
-
•
A trimeric GPcl binds to one NPC1 through the crystal structure-revealed interface
Cryo-EM structures of full-length human Niemann-Pick type C1, a lipid and cholesterol transporter involved in lysosomal storage disease, reveals insights into cholesterol trafficking and Ebola infection.
Introduction
The Niemann-Pick type C (NPC) lysosomal storage disease is characterized by the accumulation of cholesterol, sphingomyelin, and other lipids in endosomes and lysosomes. NPC patients may develop progressive neurodegeneration, hepatosplenomegaly, and premature death (Vanier, 2010, Vanier and Millat, 2003). The disease is associated with NPC1 and NPC2 genes. NPC1 encodes a polytopic membrane protein that is located in the membranes of endosomes and lysosomes, and NPC2 is a small soluble protein in the lumen of endosomes and lysosomes or secreted from cell (Carstea et al., 1997, Loftus et al., 1997, Naureckiene et al., 2000). Genetic and biochemical characterizations revealed that NPC1 and NPC2 cooperate to export low density lipoprotein (LDL)-derived cholesterol from late endosomes and lysosomes to other cellular compartments (Infante et al., 2008, Sleat et al., 2004, Vanier, 2015).
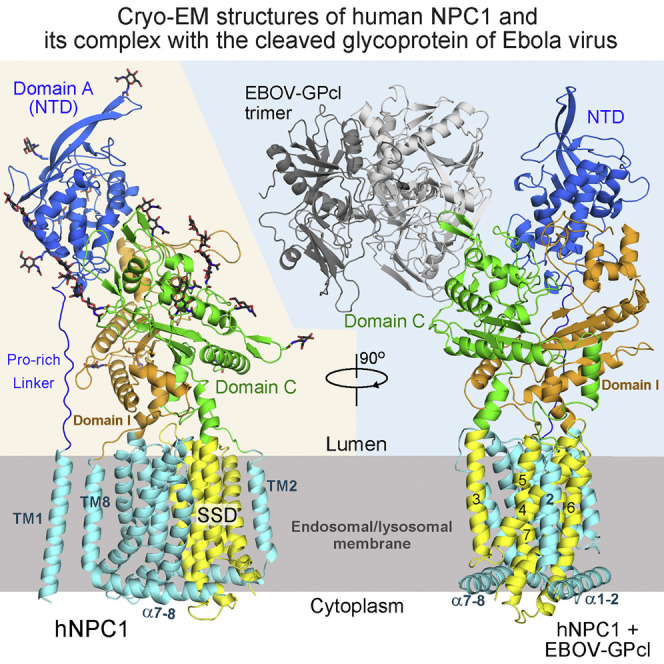
The human NPC1 (hNPC1) consists of 1,278 amino acids (aa) and is comprised of 13 transmembrane helices (TM) and 3 distinct lumenal domains A, C, and I (Davies and Ioannou, 2000). The amino terminal (N) domain A is also designated NPC1(NTD). The prevailing model suggests that cholesterol derived from endocytosed LDL is extracted by NPC2, which may be recruited to NPC1 by domain C and deliver the bound cholesterol to NPC1(NTD) through a “hydrophobic handoff” mechanism (Deffieu and Pfeffer, 2011, Kwon et al., 2009, Wang et al., 2010). The molecular basis for the ensuing intracellular trafficking of cholesterol remains largely elusive.
Sequence analysis suggested that NPC1 belongs to the resistance-nodulation-cell division (RND) superfamily (Davies et al., 2000, Scott and Ioannou, 2004, Tseng et al., 1999), whose members, exemplified by the bacterial multidrug resistance pump AcrB and MexB, generally catalyze substrate export in change of H+ influx (Delmar et al., 2014, Venter et al., 2015, Yamaguchi et al., 2015). NPC1 was reported to be a lipid exporter (Davies et al., 2000). TMs 3–7 of NPC1 were predicted to constitute the sterol-sensing domain (SSD), which is also identified in several other cholesterol metabolism or signaling-related membrane proteins, including the SREBP cleavage activating protein (SCAP) (Hua et al., 1996), the 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) (Luskey and Stevens, 1985), NPC1-Like 1 protein (NPC1L1) (Altmann et al., 2004), and the Hedgehog signaling pathway proteins Patched and Dispatched (Burke et al., 1999, Hooper and Scott, 1989, Nakano et al., 1989). The SSD acts as a regulatory domain in all these proteins (Doolman et al., 2004, Kuwabara and Labouesse, 2002, Malathi et al., 2004, Martín et al., 2001, Millard et al., 2005, Millat et al., 2001, Nohturfft et al., 1998, Strutt et al., 2001, Yabe et al., 2002). Mutations in NPC1-SSD eliminated binding with a photoactivatable cholesterol analog and failed to crosslink with a small compound inhibitor for cholesterol egress (Lu et al., 2015, Ohgami et al., 2004). The structure and functional mechanism of SSDs remain unclear.
In addition to cholesterol egress, NPC1 also serves as a critical component for cellular entry of Ebola virus (Carette et al., 2011, Côté et al., 2011). The surface glycoprotein GP of the endocytotic viruses undergoes proteolytic cleavage by cathepsin (Chandran et al., 2005, Schornberg et al., 2006). Binding of the cleaved GP (GPcl) to NPC1 through domain C is required for the subsequent release of the viral contents to the infected cells (Miller et al., 2012).
The crystal structures of individual NTD, domain C, NTD bound to sterols, and domain C in complex with GPcl of Ebola (EBOV-GPcl) were determined (Kwon et al., 2009, Wang et al., 2016, Zhao et al., 2016). However, the structure of the transmembrane domain and how the distinct domains are organized remain to be elucidated. Here, we report the 4.4 Å structure of full-length (FL) hNPC1 and a 6.6 Å reconstruction of hNPC1 in complex with EBOV-GPcl, both determined using single particle electron cryomicroscopy (cryo-EM).
Results
Structural Determination of hNPC1
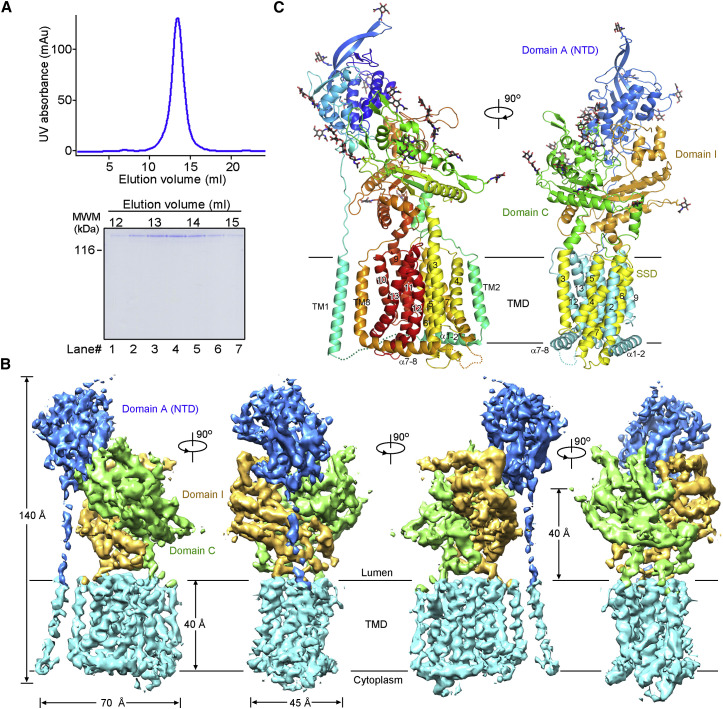
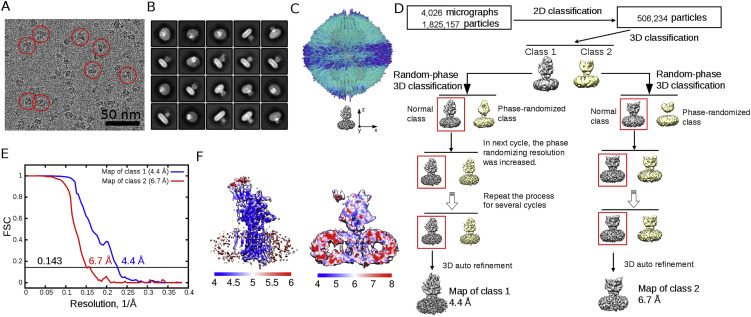
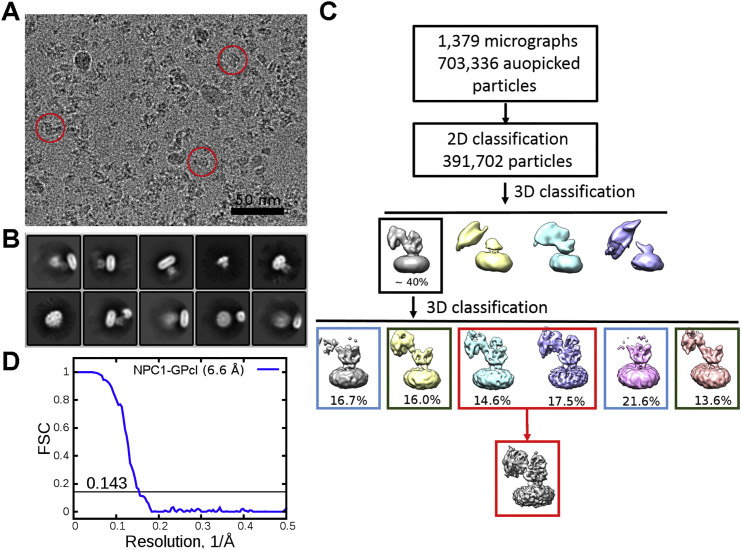
The details of recombinant expression, purification, and cryo-EM image acquisition of hNPC1 can be found in the Supplemental Experimental Procedures. The mature hNPC1 contains 1,254 aa after removal of the signal peptide. The protein appears to be a monomer when extracted and purified in the presence of digitonin (Figure 1 A). The small size of the protein represented a major challenge for structural analysis using cryo-EM. We developed a “random-phase 3D classification” method, which effectively eliminated “bad” particles (Table S1). Eventually, two maps were obtained, one at 4.4 Å out of 102,731 selected particles and the other at 6.6 Å out of 93,620 particles according to the gold-standard Fourier shell correlation (FSC) 0.143 criterion (Figure S1 and Table S2). We will mainly focus on the 4.4 Å reconstruction for structural analysis (Figure 1B).
Figure 1.
Overall Architecture of the Human NPC1
(A) The last step purification of full-length hNPC1 through size exclusion chromatography. The indicated fractions were applied to SDS-PAGE and visualized by Coomassie blue staining.
(B) The EM map of hNPC1 reconstructed at 4.4 Å resolution. The domain-colored maps were generated in Chimera (Pettersen et al., 2004). Unless otherwise indicated, the same domain color scheme is applied to all figures.
(C) The structural model of hNPC1. The structure on the left is rainbow colored with the amino and carboxyl termini colored blue and red, respectively. The invisible cytoplasmic segments connecting TM1 and α1-2 and between TM7 and α7-8 are indicated by dotted lines. The structure on the right is domain colored. The glycosyl groups are shown as black sticks. All structure figures were prepared with PyMol (DeLano, 2002).
Figure S1.
Cryo-EM Analysis of NPC1, Related to Figure 1 and Experimental Procedures
(A) Representative micrograph. Shown here is a cut from a cryo-EM micrograph with a number of particles marked with red circles.
(B) Representative 2D class averages. The box size is 26 nm.
(C) Euler angle distribution of the final 3D refinement of the Class 1 map.
(D) Flowchart of data processing. Please refer to Experimental Procedures and Table S1 for details.
(E) Gold standard Fourier shell correlation (FSC) curves of Class 1 (blue) and Class 2 (red) maps.
(F) Resolution maps of class 1 (left) and class 2 (right) reconstructions calculated with ResMap.
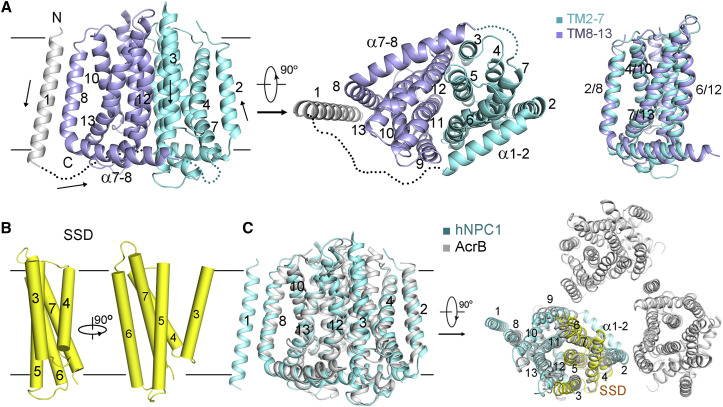
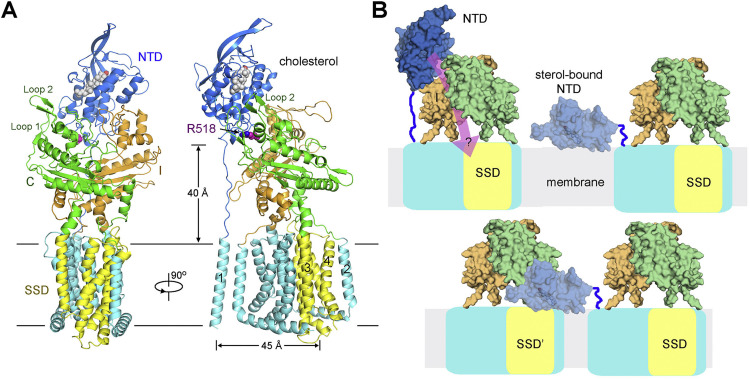
The overall structure, which is ∼140 Å high, contains a bulky lumenal region and a distinctive transmembrane domain (TMD) (Figure 1B). The reported crystal structures of NTD (Kwon et al., 2009), domain C (Wang et al., 2016, Zhao et al., 2016) and several bacterial RND members (Long et al., 2010, Murakami et al., 2002, Pak et al., 2013, Sennhauser et al., 2009) tremendously facilitated model building of hNPC1 at this modest resolution (Figures 1C and S2 ). The RND fold was readily discernible, leaving TM1 as a lone helix adjacent to TM8. A poly-Ala model was generated for the TMD, which is ∼40 Å high, 70 Å long, and 45 Å thick (Figures 1B and S3 A).
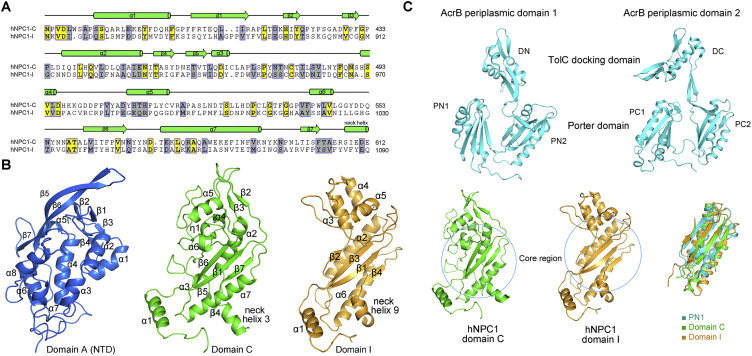
Figure S2.
Structures of the Individual Lumenal Domains, Related to Figures 1 and 3
(A) Domains C and I of hNPC1 share sequence similarity. The secondary structural elements of domain C is shown on top of the aligned sequences. Invariant and conserved residues are shaded yellow and gray, respectively.
(B) Crystal structures of the NTD and domain C (PDB: 3GKJ and PDB: 5HNS, respectively) were used for rigid body docking into the EM map with manual adjustment. A homologous model of domain I was generated based on domain C.
(C) Structural similarity of domains C and I with the periplasmic porter domains of AcrB. The core regions of domains C and I composed of the β sheet and two helices can be superimposed to the porter domains of AcrB.
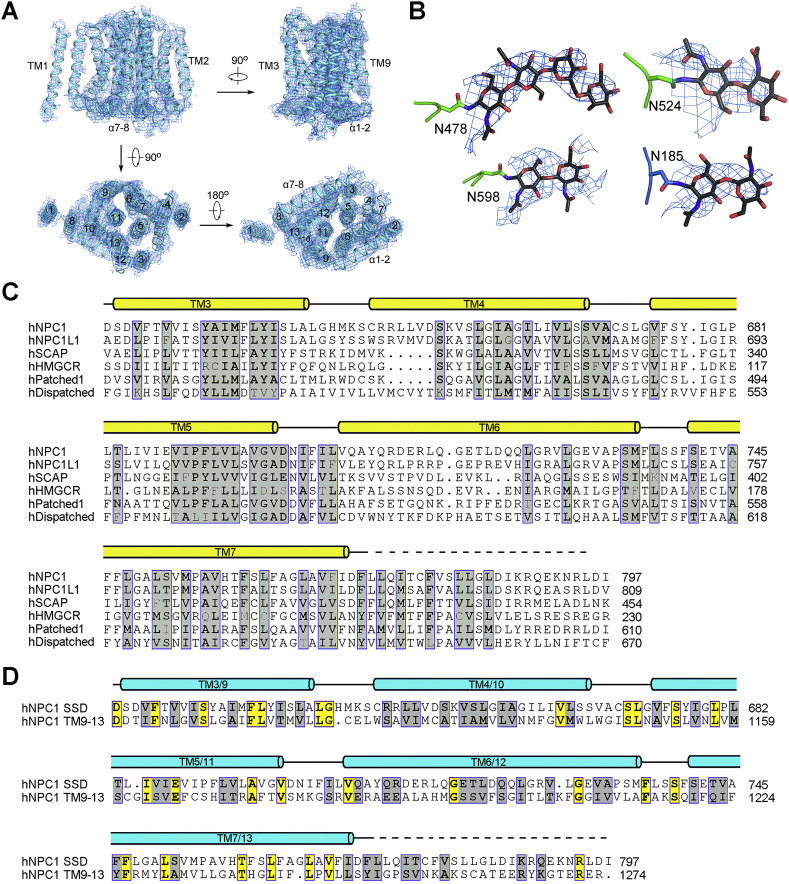
Figure S3.
TM3–7 Constitute the Sterol-Sending Domain, Related to Figure 2
(A) The EM map for the transmembrane domain of hNPC1.
(B) Representative densities of glycosylation sites in NTD and domain C. The maps were generated in PyMol and contoured at 5σ.
(C) Sequence comparison of the SSDs from human proteins involved in cholesterol metabolism or signaling. The UniProt ID numbers for the aligned human sequences are: hNPC1: “UniProt: O15118”; hNPC1L1: “UniProt: Q9UHC9”; hSCAP (SREBP cleavage activating protein): “UniProt: Q12770”; hHMGCR (3-hydroxy-3-methylglutaryl-CoA reductase): “UniProt: P04035”; hPatched1: “UniProt: Q13635”; hDispatched1: “UniProt: Q96F81”.
(D) TMs 9-13 of hNPC1 share sequence similarity with SSD. Shown here is the sequence comparison of SSD to the corresponding segments (TMs 9-13) in the second repeat of hNPC1. Invariant and conserved residues are shaded yellow and gray, respectively. Due to moderate resolution of the TMD, the boundaries of TM segments shown above the sequences may not be accurate.
The crystal structures of NTD and domain C (PDB: 3GKJ and PDB: 5HNS, respectively) were docked to the corresponding map with manual adjustment. The map of domain I resembles that of domain C, consistent with their 30% sequence similarity (Figures S2A and S2B). A homologous model of domain I was generated and docked into the map. 14 glycosylation sites were identified, 5 on NTD, 7 on domain C, and 2 on domain I. These glycosylation sites in turn validated sequence assignment (Figures 1C and S3B and Table S2).
The SSD-Containing TMD of hNPC1
TM2 and TM8 are each preceded by a long helix that attaches to the cytoplasmic membrane surface (Figure 2 A, left). The two transverse helices, designated α1-2 and α7-8, are almost parallel to each other and embrace TMs 2–13 on the cytoplasmic side (Figure 2A, middle). As observed in all the RND members of known structures (Long et al., 2010, Murakami et al., 2002, Pak et al., 2013, Sennhauser et al., 2009), the first six TMs (TMs 2–7 in hNPC1) are related to the C-terminal six TMs by 180-degree rotation around an axis that is perpendicular to the membrane plane (Figure 2A, right).
Figure 2.
Structure of the Transmembrane Domain TMD of hNPC1
(A) TMs 2–13 of hNPC1 exhibit a characteristic resistance-nodulation-cell division superfamily fold. The two repeats in the RND fold are colored cyan and light purple, respectively. The invisible cytoplasmic segments are shown as dotted lines. The arrows in the left panel indicate the N→C orientation of the corresponding TMs. Right panel: TMs 2–7 are related to TMs 8–13 by a 180-degree rotation around an axis that is perpendicular to the membrane plane.
(B) The sterol sensing domain (SSD). TMs 3–7 of hNPC1 constitute the SSD that is also found in several proteins involved in sterol metabolism or signaling. Two perpendicular side views are shown.
(C) Structural similarity between the TMDs of hNPC1 and the bacterial multidrug efflux transporter AcrB. Right panel: the SSD-corresponding TMs in AcrB are exposed to the lipid bilayer in the context of trimer (PDB: 1IWG).
The two repeats interact with each other through an extensive interface involving TMs 3/5/6/7 in the first repeat and the corresponding TMs 9/11/12/13 in the second one (Figure 2A). Notably, TMs 3–7 constitute the SSD (Davies and Ioannou, 2000) (Figure 2B). The lack of side chains precludes analysis of this regulatory domain in detail. Nevertheless, the structure shows that all the five SSD-forming TMs are exposed to the lipid bilayer to different extent, making SSD available for potential interactions with membrane-embedded sterols or other integral membrane proteins. The structural fold of SSD is likely preserved in all the other SSD-containing proteins (Figure S3C).
TMs 3–7 share 15% sequence identity and 43% similarity with TMs 9–13, suggesting that the latter is a SSD-like domain (Figures 2A and S3D). In essence, the interface between the two internal repeats is mediated through two SSDs (Figure 2A, middle). Notably, Patched and Dispatched were both predicted to be RND members comprising 12 TMs with internal 2-fold pseudosymmetry (Taipale et al., 2002, Tseng et al., 1999). These two proteins likely have the same TMD organization as NPC1. On the other hand, SCAP and HMGCR each contain 8 TMs, excluding the possibility of a second SSD or SSD-like domain. Nevertheless, the TMD of SCAP was shown to tetramerize (Radhakrishnan et al., 2004), and the catalytic domain of HMGCR also exists as a tetramer (Istvan et al., 2000). It remains to be investigated whether SSD in these two proteins are responsible for homotypic interactions.
The TMDs of hNPC1 and AcrB can be well superimposed (Figure 2C, left). AcrB and other bacterial RND members of known structures are all trimers, whereas hNPC1 is a monomer. When the structural comparison is carried out in the context of the trimer, none of the SSD-corresponding TMs in AcrB participate in trimerization. As in the monomeric hNPC1, the SSD-corresponding TMs in the trimeric AcrB are laterally exposed to the lipid bilayer (Figure 2C, right).
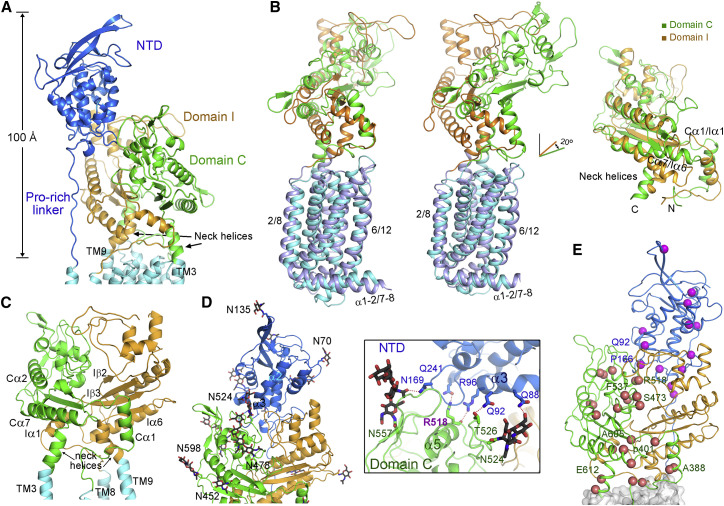
Organization of the Lumenal Domains
The three lumenal domains, which are separated in the primary sequence, interact with each other. The overall height of the lumenal region is ∼100 Å, allowing the NTD to potentially protrude above the glycocalyx (Neiss, 1984). As predicted, the NTD is connected to the lone TM1 through an elongated Pro-rich segment 249PKPQPPPPPAP259 (Figure 3 A) (Kwon et al., 2009). In the absence of NTD and TM1, the rest of the structure appears to have a 2-fold pseudosymmetry, a feature reminiscent of the bacterial RND proteins (Figure 3B, left). In fact, the core regions of domains C and I, including the central β sheet and two helices, are nearly identical to the periplasmic porter domains of AcrB, reflecting evolutionary conservation (Figure S2C). However, close examination shows that the two repeats of TMD are related to each other by 180° rotation, whereas the core regions of domains C and I are related by ∼160° rotation (Figure 3B).
Figure 3.
Organization of the Three Lumenal Domains of hNPC1
(A) The overall structure of the lumenal domains and their connections to TMD. The C-terminal helices in domains C and I whose extensions are TM3 and TM9, respectively, are designated the “Neck helices.”
(B) Structural similarity and difference between domains C and I. Left: in the absence of the NTD and TM1, the overall structure appears to have a 2-fold pseudosymmetry. Middle: the two halves are superimposed relative to TMD. Whereas the internal repeats of the TMD are related by 180° rotation, domains C and I are related by ∼160°. Right: when domains C and I are superimposed relative to the central β sheet, structural deviations occur to the peripheral helices. The secondary elements in domains C and I are labeled with prefix “C” or “I,” respectively.
(C) Interactions between domains C and I. The extensive interface and the neck helices may confer structural stability to the two lumenal domains relative to their respectively connected transmembrane repeats.
(D) Interactions between the NTD with domains C and I. Inset: potential hydrogen-bonds between residues in NTD and domain C. Note that the sugar moieties attached to Asn524 and Asn557 may also participate in the inter-domain packing.
(E) Structural mapping of NPC disease-related mutations. The Cα atoms of disease-associated residues in NTD and domain C are shown as spheres. The residues that may be involved in interactions between lumenal domains are labeled. Due to the lack of reliable structural model, the mutations in domain I are not mapped.
Domain C and domain I each connect to the first TM in the internal repeat (TM2 and TM8, respectively) through an extended linker and end with a helix whose extension forms the second TM in the respective repeat (TM3 and TM9, respectively). For simplicity, we designate the last helices in domains C and I the “neck helices” (Figure 3C). The neck helices may represent the coupling elements between the respective lumenal domains and TMD repeats in case of concerted conformational changes (Eicher et al., 2014). The organization of domains C and I resembles folded arms, whose reciprocal interactions involve the first helix in one domain against the β sheet in the other, as well as contacts between the distal loop regions (Figure 3C). Detailed analysis of interactions involving domain I is precluded at this resolution.
The NTD is like the “head” on top of the shoulder and arms formed by domains C and I. A number of polar and charged residues appear to constitute the interface between NTD and domain C. In particular, helix α3 of NTD, which is enriched of polar residues, including Gln88/Gln92/Arg96, represents a major area contacting domain C. The sugar moieties attached to Asn524 and Asn557 of domain C may form hydrogen bonds with Gln88 and Asn169, respectively, in NTD (Figure 3D). It is of particular note that Arg518 is engaged in binding to NTD (Figure 3D). The single point mutations R518W or R518Q are disease related and led to reduced interaction between isolated domain C and NPC2 (Table S3) (Deffieu and Pfeffer, 2011).
Among the identified NPC disease-related mutations, 15 and 28 belong to NTD and domain C, respectively (Scott and Ioannou, 2004). Structural mapping suggested that nine residues are located at the interface between the lumenal domains (Figure 3E and Table S3). Whether some mutations lead to misfolding of NPC1 in the ER as a result of disruption of domain interactions, particularly between domains C and I, awaits further investigation. Asn92 and Arg518 will be analyzed in the next session.
Structure-Guided Analysis of Cholesterol Transfer from hNPC2 to hNPC1
The structural elucidation of FL hNPC1 offers an opportunity to investigate the mechanism of NPC1 and NPC2-mediated cholesterol trafficking and to provide potential molecular interpretations for reported biochemical observations. To establish the structure-function correlation, we employed two biochemical approaches: examination of cholesterol transfer from purified hNPC2 to hNPC1 variants in an in vitro assay system and measurement of the binding affinity between NPC1 variants and NPC2 using isothermal titration calorimetry (ITC).
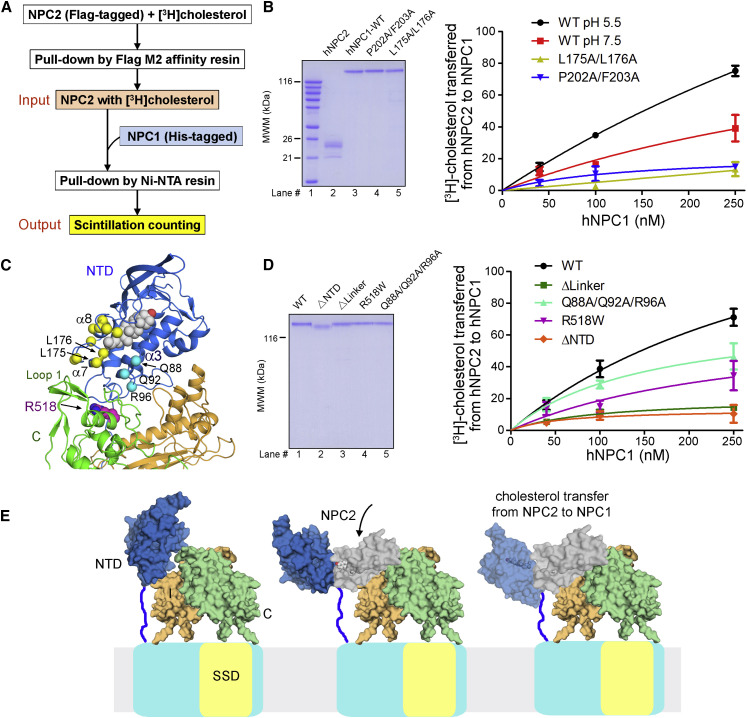
In vitro cholesterol transfer assays were conducted with NPC2 and isolated NPC1(NTD) (Infante et al., 2008, Kwon et al., 2009, Wang et al., 2010). Modifying the established protocol, we set up an in vitro assay to examine cholesterol transfer from NPC2 to NPC1 variants that were designed on the basis of structural analysis (Figure 4 A). Briefly, the FL NPC1 and NPC2 were respectively fused with His10 and FLAG tags and purified to homogeneity. The [3H]cholesterol-loaded NPC2 was incubated with NPC1 for cholesterol transfer to occur. The mixture was then applied to nickel-NTA (Ni) resin to remove non-specific binding, and the eluent was sent for scintillation counting, which indicated the [3H]cholesterol retained by the tested NPC1 variants. The ratio between the readings of the quantified Ni eluent and the input [3H]cholesterol-bound NPC2 was used to assess the cholesterol transfer activity of the tested NPC1 variants.
Figure 4.
Interface Residues between NTD and Domain C Are Involved in Cholesterol Transfer from NPC2 to NPC1
(A) The simplified protocol for an in vitro cholesterol transfer assay using purified full-length (FL) hNPC1 and hNPC2. Please refer to the Supplemental Experimental Procedures for details.
(B) Validation of the assay system with reported conditions and mutations. Left: the purified hNPC2 and hNPC1 variants used in the assay. Right: Ala substitution of the previously identified residues for sterol binding or transfer markedly reduced cholesterol transfer from NPC2 to FL NPC1 in our assay (Kwon et al., 2009). For each data point, the reading of the negative control, whereby NPC1 was omitted in the pull-down experiment, was subtracted from the output, and the resulted reading was normalized against the input and presented in percentage. Each data point is the average of three independent experiments. Error bars represent SD.
(C) Structural mapping of the NTD residues that are important for cholesterol transfer between NPC2 and NPC1(NTD). A cholesterol molecule, shown as silver spheres, was modeled based on the crystal structure of cholesterol-bound NTD (PDB: 3GKI). The Cα atoms of the previously characterized functional residues (Wang et al., 2010) and the ones identified in this study are shown as yellow and cyan spheres, respectively. Note that these residues are in the vicinity of the domain C residue Arg518, which is disease related and important for NPC2 recruitment (Deffieu and Pfeffer, 2011).
(D) The cholesterol transfer activities of NPC1 variants designed on the basis of the EM structure. Gln88/Gln92/Arg96 and Arg518 are on the interface between NTD and domain C (please refer to the inset of Figure 3D).
(E) The structure and biochemical characterizations suggest that sterol transfer between NPC2 and NPC1 necessitates proper accommodation of NPC2 by NPC1(NTD) and NPC1-C. Shown here are domain models, whose relative orientations and scales may be different from the real structure. Arrival of cholesterol-bound NPC2 may dislodge NTD from its interactions with domains C and I. NPC2 may be oriented by both the NTD and NPC1-C for the hydrophobic handoff of cholesterol to the NTD.
As a proof of principle, we first examined several reported parameters and mutations. The binding between NPC2 and domain C of NPC1 (NPC1-C) was shown to be pH dependent (Deffieu and Pfeffer, 2011), and the cholesterol transfer activity between NPC2 and NPC1(NTD) varied at different pH values (Infante et al., 2008). Consistent with these observations, the activity of cholesterol transfer from NPC2 to FL NPC1 at pH 7.5 was only half of that at pH 5.5 in our assay system (Figure 4B). Two well-characterized NTD mutants, P202A/F203A and L175A/L176A, which are deficient in cholesterol binding and transfer, respectively, were then examined (Kwon et al., 2009). Both mutants showed pronounced reduction of cholesterol transfer at pH 5.5 (Figure 4B).
The crystal structures of NPC1(NTD) bound to cholesterol or 25-hydroxycholesterol (Kwon et al., 2009) allow reliable modeling of a cholesterol molecule into the FL hNPC1, which would support structure-guided mutagenesis for identification of additional elements in cholesterol transfer (Figure 4C). The disease-related mutation R518W led to reduced binding between NPC2 and NPC1-C (Deffieu and Pfeffer, 2011). In our transfer assay, NPC1-R518W retained only half of the cholesterol transfer activity compared to the wild-type (WT) NPC1 at pH 5.5 (Figure 4D).
Noting that Arg518 is mapped to the interface between the NTD and domain C, particularly in the vicinity of Leu175/Leu176 in NTD, we then examined the adjacent interface residues (Figures 3D and 4C). Deletion of the NTD (residues 25-257) abolished more than 90% of cholesterol transfer from NPC2 to NPC1-ΔNTD. The variant containing three point mutations on NTD (Q88A/Q92A/R96A) lost up to 40% of the transfer activity (Figure 4D). Interestingly, these three residues line up on helix α3 of NTD, which guards one side of the entrance to the cholesterol binding pocket, while the previously identified residues involved in cholesterol transfer, exemplified by Leu175 and Leu176, are mapped to the opposite side of the entrance.
Binding of NPC2 to NTD and Domain C Is Required for Cholesterol Transfer
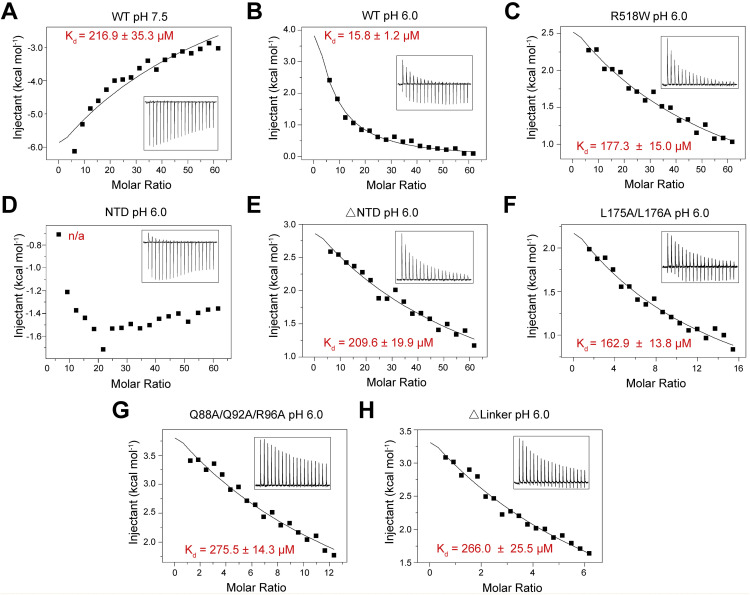
We next examined the binding affinities between NPC2 and several NPC1 variants using ITC (Table 1, Figure S4 ). Consistent with the reported pH-dependent association between NPC1-C and NPC2 (Deffieu and Pfeffer, 2011), the FL NPC1 binds to NPC2 with a Kd of ∼16 μM at pH 6.0 but ∼217 μM at pH 7.5. In addition, the single point mutation R518W reduced the affinity to ∼177 μM at pH 6.0 (Figures S4A–S4C). The binding affinities between NPC1 variants and NPC2 appear to be correlated with their cholesterol transfer activities (Figures 4B and 4D).
Table 1.
Binding Affinity between NPC2 and the NPC1 Variants Measured by ITC
| NPC1 Variants | Kd (μM) | NPC1 Variants | Kd (μM) |
|---|---|---|---|
| FL (pH 7.5) | 216.9 ± 35.3 | ΔNTD | 209.6 ± 19.9 |
| FL (pH 6.0) | 15.8 ± 1.2 | L175A/L176A | 162.9 ± 13.8 |
| R518W | 177.3 ± 15.0 | Δlinker | 266.0 ± 25.5 |
| NTD | n/a | Q88A/Q92A/R96A | 275.5 ± 14.3 |
Except for the first measurement, all the others were performed at pH 6.0. The indicated mutations were generated to full-length (FL) human NPC1. NTD: residues 1–252; ΔNTD: internal deletion of residues 25–257; ΔLinker: internal deletion of the Pro-rich segments 249PKPQPPPPP257.
Figure S4.
Measurement of the Binding Affinity between Human NPC2 (hNPC2) and hNPC1 Variants, Related to Table 1
Please refer to Supplemental Experimental Procedures for details of experiments and data fitting. A summary of the binding affinity is shown in Table 1.
(A and B) Measurement of the affinity between WT hNPC1 and hNPC2 at pH 7.5 (A) and pH 6.0 (B).
(C–H) Measurement of the binding affinity between hNPC2 and the indicated hNPC1 variants. All the measurements were conducted at pH 6.0. NTD: residues 1-252; ΔNTD: FL deletion of residues 25-257 from FL protein; ΔLinker: internal deletion of the Pro-rich segments 249PKPQPPPPP257.
Despite that the NPC1(NTD) is the acceptor for cholesterol handed over from NPC2, interaction between NPC2 and isolated NTD was not detected, suggesting that NTD alone may be insufficient for stable interaction with NPC2. Nevertheless, deletion of NTD marked reduced the binding between NPC1 and NPC2 (Figures S4D and S4E). Furthermore, consistent with the reduced cholesterol transfer activities, the hNPC1 variants L175A/L176A and Q88A/Q92A/R96A both showed decreased binding with NPC2 at pH 6.0 (Figures S4F and S4G), supporting the notion that NTD is required for the anchorage of NPC2 to NPC1.
The observation that the interface constituents between NTD and domain C, exemplified by Gln88/Gln92/Arg96 and Arg518, are important for both NPC2 binding and cholesterol transfer suggests that the structure presented here may represent an inactive state. It is thereby reasonable to postulate that structural rearrangements, presumably a displacement of the NTD, would occur to expose these interface residues for interaction with NPC2 (Figure 4E). Note that the NTD is invisible in the second class of EM reconstruction of NPC1 (Figures S1D and S1F), implying potential flexibility of this domain. On the other hand, the proximity between Leu175/Leu176 and the interface residues implicates that NTD and domain C may together constitute the docking site to orient NPC2 for cholesterol transfer to NTD (Figure 4E). Indeed, deletion of the Pro-rich linker (249PKPQPPPPP257), which would leave the NTD close to the membrane surface and away from domain C, led to reduced affinity with NPC2 (Table 1 and Figure S4H) and similar loss of cholesterol transfer activity as NPC1-ΔNTD (Figure 4D).
The structural analysis, cholesterol transfer assay, and ITC measurement support a mechanism whereby the NTD and domain C together constitute the scaffold to properly accommodate NPC2 for hydrophobic handoff of cholesterol to the pocket of NTD (Figure 4E). However, the molecular basis for the pH-dependent affinity between NPC1 and NPC2 or between domain C and NPC2 remains to be elucidated. Moreover, the mechanism of the subsequent cholesterol delivery from NTD to the membrane region is even more intriguing (Figure S5 ).
Figure S5.
Structure-Invoked Questions on the Sterol Transfer from NPC1(NTD) to Membrane, Related to Figure 4
(A) The NTD may not be able to reach SSD in the same molecule when it bends toward the membrane. A cholesterol molecule is modeled into the pocket of NTD based on the crystal structure of cholesterol-bound hNPC1(NTD) (PDB: 3GKI). Arg518 is shown as magenta spheres. The SSD is colored yellow. It is noteworthy that the NTD is on the opposite side of SSD in the structure. The distance between TM1 and the closest TM in SSD is > 45 Å while the linker between NTD and TM1 is about 40 Å. Furthermore, the relative stability of domains C and I with their respective anchoring TMD repeats may represent a spatial block to impede the placement of NTD to the proximity of SSD in the same protein molecule for direct delivery of the NTD-bound cholesterol to SSD.
(B) The molecular mechanism of cholesterol transfer from NPC1(NTD) to the membrane or SSD remains enigmatic. Shown here are speculative models. The pink arrow in the left panel of the upper row indicates the question of whether cholesterol can go from NTD through domains C and I to SSD if NPC1 functions like a RND transporter. Alternatively, NTD may directly deliver the bound cholesterol to membrane (upper right) or to the SSD of an adjacent NPC1 protein (lower row). However, the upper model cannot explain the loss of binding to cholesterol analog due to mutations in SSD unless SSD functions in later steps in cholesterol egress. The “transfer in trans” model in the bottom row is completely hypothetical especially when there is no evidence to support oligomerization of NPC1. The mechanism of cholesterol egress awaits further investigations.
Interaction between EBOV-GPcl and hNPC1
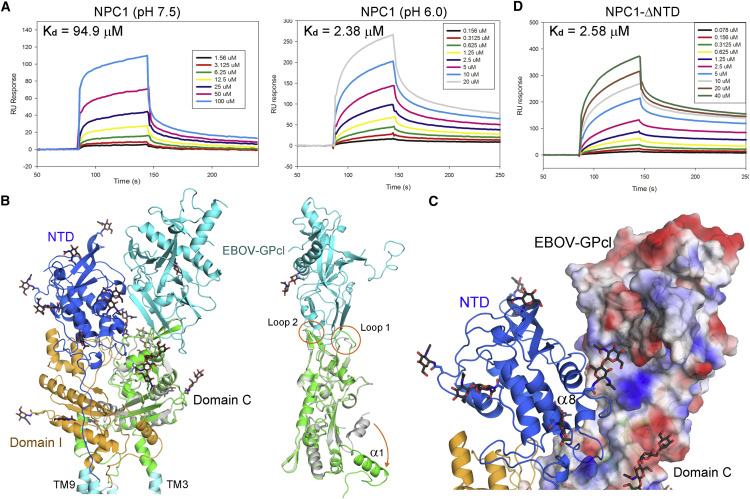
The structure of FL hNPC1 provides a template for examination of the interaction with EBOV-GPcl. The purified recombinant hNPC1 binds to GPcl, interestingly, also in a pH dependent manner as measured using surface plasmon resonance (SPR). At pH 7.5, the measured Kd value is ∼95 μM, whereas the affinity increases to 2.4 μM at pH 6.0, almost 40-fold higher (Figure 5 A).
Figure 5.
Examination of the Interaction between Cleaved Glycoprotein of Ebola Virus (EBOV-GPcl) and FL NPC1
(A) The interaction between EBOV-GPcl and NPC1 is pH dependent. The binding affinity is measured using SPR.
(B) Docking of the domain C-GPcl complex (PDB: 5F1B) to the EM structure of hNPC1. Right: conformational changes of domain C. Note the pronounced rearrangement of helix α1 of domain C between the crystal structure of isolated domain (silver) and the EM structure of FL protein (green).
(C) The docking model suggested a potential interface between NPC1(NTD) and EBOV-GPcl. The domain C-GPcl1 complex is shown as semi-transparent surface electrostatic potential calculated in PyMol.
(D) The NTD-deleted hNPC1 binds to EBOV-GPcl with similar affinity as FL protein at pH 6.0.
When the crystal structure of the complex between NPC1-C and EBOV-GPcl is docked to the EM reconstruction of hNPC1, there is no clash between EBOV-GPcl and other domains. With the tip of EBOV-GPcl anchoring to the two loops of NPC1-C (Wang et al., 2016), EBOV-GPcl is far away from domain I (Figure 5B). Notably, a major rearrangement of helix α1 is observed between the crystal structures of isolated domain C and the EM architecture of the full-length protein. The helix, which connects domain C to TM2, folds back toward the central β sheet in the isolated domain (Figure 5B).
In the docking model, the side facet of GPcl appears to contact hNPC1(NTD) (Figure 5C). However, the NTD-deleted NPC1 binds to GPcl with a similar affinity as the FL protein (Figure 5D). In addition, the interaction between NTD and GPcl cannot be reliably detected by SPR (Figure S6 A), suggesting a negligible role of NTD in recruiting GPcl.
Figure S6.
Examination of the Interactions between hNPC1 and the Cleaved Glycoprotein of Ebola Virus (EBOV-GPcl), Related to Figure 5
(A) There is very weak, if any, binding between isolated hNPC1(NTD) and EBOV-GPcl. The binding affinity was measure by SPR. Please refer to Supplemental Experimental Procedures for details.
(B) Modeling of the EBOV-GPcl trimer bound to three hNPC1. When three FL hNPC1 molecules are docked to the structure of trimeric EBOV-GPcl in complex with three domain C as rigid body (PDB: 5F1B), the TMD of the three hNPC1 protomers are out of the same plane. TM1 in each protomer collides with TM3 in the adjacent one.
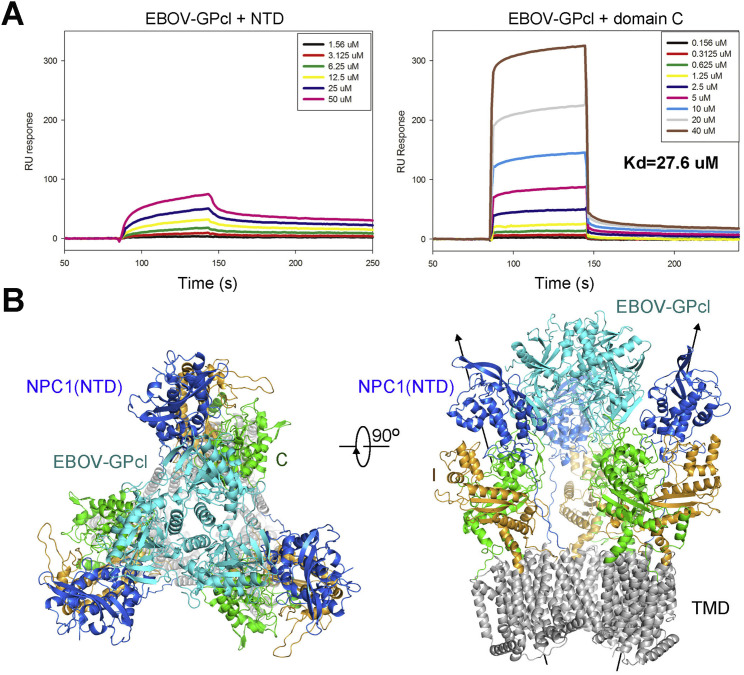
Another interesting question is the stoichiometric ratio between NPC1 and GPcl. Despite that purified hNPC1 exists as a monomer, the GPcl forms a trimer in isolation or in the complex with domain C (Lee et al., 2008, Wang et al., 2016). We attempted to dock three hNPC1 molecules to the trimeric complex of EBOV-GPcl and NPC1-C (Figure S6B). The lumenal domains of NPC1 are all compatible in the trimeric assembly without inter-protomer contact. However, the symmetry axis of the TMD in each protomer tilts away from the central axis of the trimer. Consequently, the TMDs in the three protomers no longer stay in the same membrane plane, and there is slight clash between TM1 in one protomer and TM3 in the adjacent one (Figure S6B). This structural model raised several possibilities. If multiple NPC1 molecules bind to a trimeric EBOV-GPcl simultaneously, the FL NPC1 would have to undergo conformational changes to avoid clash at TMDs. Alternatively, binding of EBOV would have to induce deformation of the membrane where NPC1 proteins are embedded. A third possibility is that the GPcl trimer may simply bind to one NPC1. Addressing these questions necessitates structural elucidation of the complex between the EBOV-GPcl and FL NPC1 (hereafter referred to as the NPC1-GPcl complex).
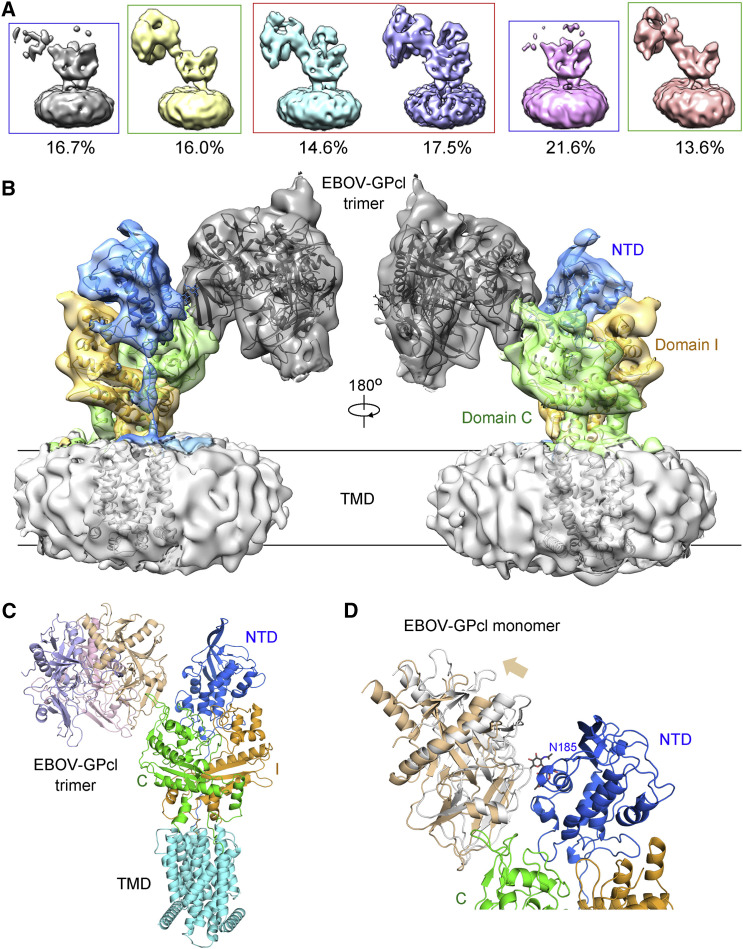
Cryo-EM Architecture of the NPC1-GPcl Complex
The hNPC1-GPcl complex was assembled at pH 5.5 and examined with cryo-EM (Figure S7 ). Three classes of reconstructions, in which the GPcl was clearly discernible, were obtained (Figure 6 A). At higher threshold, NTD becomes invisible in Class 2, while neither NTD nor GPcl can be seen in Class 3. Nevertheless, the presence of a GPcl trimer and a NPC1 monomer was unambiguous in all three classes with low-pass filter. Therefore, one detergent micelle-surrounded NPC1 monomer binds to one GPcl trimer (Figure 6A).
Figure S7.
Cryo-EM Analysis of the Complex between FL hNPC1 and EBOV-GPcl, Related to Figure 6
(A) Representative micrograph. Shown here is a cut from a cryo-EM micrograph with three particles marked with red circles.
(B) Representative 2D class averages. The box size is 33.45 nm.
(C) A brief flowchart of data processing.
(D) Gold standard Fourier shell correlation (FSC) curve of the reconstruction of the NPC1-GPcl complex.
Figure 6.
Cryo-EM Architecture of the Complex between EBOV-GPcl and FL hNPC1
(A) Summary of the 3D classifications of the NPC1-GPcl complex. Please refer to Figure S7 for cryo-EM data processing.
(B) One EBOV-GPcl trimer binds to one NPC1 monomer. The 6.6 Å cryo-EM reconstruction of the hNPC1-GPcl complex obtained from the two classes indicated by the red box in (A).
(C) Structural model of the NPC1-GPcl complex. The trimeric crystal structure of EBOV-GPcl (PDB: 5F1B) and the EM structure of NPC1 were fitted to the cryo-EM map in Chimera.
(D) Deviation between the cryo-EM reconstruction and the docking model of GPcl in the complex. Despite the seeming contact between GPcl and NTD in the docking model (colored silver), the GPcl protomer (colored wheat) that contacts domain C slightly moves away from NTD in the cryo-EM reconstruction. Consequently, no direct contact between GPcl and NTD was seen in the EM map. It remains to be investigated whether glycosyl moieties may fill up the void between NTD and GPcl.
A 6.6 Å reconstruction of NPC1-GPcl complex was obtained out of 50,223 particles (Class 1), into which the crystal structures of GPcl and the 4.4 Å cryo-EM structure of NPC1 can be easily docked (Figure 6B and Table S4). One protomer of GPcl binds to Loop 1 and Loop 2 of NPC1-C (Figure 6C). Notably, there is no direct contact between GPcl and NPC1(NTD) in the EM map (Figure 6B). In fact, compared to the crystal structure of the complex between GPcl and NPC1-C, there is a slight tilt of GPcl that may be achieved through minor shift of the loops of NPC1-C (Figure 6D). Consequently, GPcl slightly moves away from NTD in the EM structure, unable to form the predicted contact as seen in the docking model (Figures 5C and 6D). Nevertheless, the NTD has a glycosylation site facing GPcl (Figures 5C and 6D). It is possible that glycosyl moieties may fill up the void between NTD and GPcl.
The EM architecture of NPC1-GPcl appears to support the biochemical characterizations (Figures 5D and S6A) and previous reports that NTD is not required for GPcl binding or Ebola infection (Miller et al., 2012). Nevertheless, GPcl binds to FL NPC1 and isolated NPC1-C with Kd values of 2.38 μM and 27.6 μM, respectively, at pH 6.0 (Figures 5A and S6A). The higher affinity with FL NPC1 may be achieved through a scaffolding role conferred by the other lumenal domains, which may orient NPC1-C to an optimal position for binding to GPcl. Supporting this notion, NPC1L1, which lacks filovirus receptor activity, became equally efficient as NPC1 in mediating Ebola virus infection when the NPC1L1-C was replaced by NPC1-C and more efficient than a minimal NPC1-C based receptor (Krishnan et al., 2012).
Ebola virus is attracted to host cell surface by diverse attachment factors such as T cell immunoglobulin and mucin domain-containing family molecules (Wang et al., 2015). After attachment, Ebola virus is internalized into endosome. NPC1 was suggested to serve as an essential high-affinity intracellular receptor for EBOV-GPcl (Hofmann-Winkler et al., 2012, Hunt et al., 2012, White and Schornberg, 2012). However, GPcl binds to isolated NPC1-C with a Kd of ∼140 μM at pH 7.5 (Wang et al., 2016), considerably lower than the nanomolar (nM) range of other highly pathogenic viruses such as SARS-CoV and MERS-CoV (Lu et al., 2013). The observation reported here that the binding affinity between NPC1 and GPcl was enhanced at lower pH may provide a plausible explanation to reconcile the discrepancy considering the intracellular environment for the endocytosed virus. Nevertheless, the structural basis for the pH dependence awaits future examination.
Despite the many remaining enigmas, the EM structures of hNPC1 and the hNPC1-GPcl complex, as well as the structure-guided biochemical characterizations reported here, represent a major step toward mechanistic understanding of NPC1-mediated intracellular cholesterol trafficking and Ebola virus infection. It is noteworthy that the molecular weight of hNPC1 protein is around 140 kDa. Structural resolution of the relatively small proteins by cryo-EM offers unprecedented opportunity for structural and mechanistic investigation of a broad range of biologically significant molecules. Furthermore, cryo-EM has the advantage of revealing structurally stable glycosylation sites. Hence, cryo-EM may also illuminate the avenue to the structural and functional characterizations concerning glycosylation (Li et al., 2015).
Experimental Procedures
Cryo-EM Data Collection and Processing of hNPC1
The details of recombinant expression, purification, and biochemical characterizations of hNPC1, hNPC2 and GPcl can be found in the Supplemental Experimental Procedures.
The cryo sample was prepared with FEI Vitrobot Mark IV. An aliquot of 4 μl purified hNPC1 at concentration of ∼15 mg/ml was applied to glow-discharged Quantifoil (1.2/1.3) 200 mesh Cu grid. After being blotted with filter paper for 3.0 s, the grid was plunged into liquid ethane cooled with liquid nitrogen. A total of 4,026 micrograph stacks were semi-automatically collected with UCSF Image4 on Titan Krios at 300 kV equipped with Gatan K2 Summit at a nominal magnification of 22,500× with a defocus range of 1.5–3.0 μm. Each stack was exposed for 8 s with an exposing time of 250 ms per frame, resulting in 32 frames per stack. The total dose rate was about 50 e−/Å2 for each stack. The drift of the stacks was corrected with dosef_driftcorr (Li et al., 2013). After motion correction, each frame in the stacks was filtered according to its exposing dose and added together with SumMovie (Grant and Grigorieff, 2015). The defocus was estimated with Gctf (Zhang, 2016).
A total of 1,825,157 particles were autopicked with RELION 1.4 (Scheres, 2012a, Scheres, 2012b, Scheres, 2015). After several rounds of 2D classification using RELION 1.4, a total of 506,234 particles were selected and subjected to 3D classifications. The 3D initial model was built from typical 2D class averages with SIMPLE (Elmlund and Elmlund, 2012). We attempted to 3D classify these particles with different number of groups, such as 4, 6, or 8 classes. The particles always fell into two major subsets, one with an additional lumenal domain (the NTD) and the other with a seemingly 2-fold pseudosymmetry without the protruding domain. We therefore classified the particles into two classes according to the visibility of this additional lumenal domain with global angular searching in RELION 1.4. Then each class was further 3D classified with local angular searching in RELION 1.4. During the local angular searching, a “random-phase 3D classification” method, which is described below, was applied to eliminate bad particles with home-modified RELION 1.4. The remaining particles of each class were subjected to 3D auto-refinement with RELION 1.4 separately. After 3D auto-refinement, the Class 1 particles of NPC1 were further polished with RELION 1.4 and then subjected to 3D auto-refinement. The final resolution was estimated with the gold-standard Fourier shell correlation criterion (Scheres and Chen, 2012) with high-resolution noise substitution method (Chen et al., 2013). Local resolutions were estimated with ResMap (Kucukelbir et al., 2014). The method of model building and refinement, as well as the cryo-EM analysis of NPC1-GPcl, can be found in the Supplemental Experimental Procedures.
Random-Phase 3D Classification
The source code and the testing details of the “random-phase 3D classification” method will be provided upon e-mail request addressed to Q. Z. Briefly, in each cycle of 3D classification, the particles were classified into two groups with sufficient number of iterations. In each iteration of the 3D classification, the second reference was the same as the first reference but phase-randomized above a specified resolution. After each cycle of 3D classification, the particles prone to be classified into the phase-randomized class were removed from the next cycle of 3D classification. The resolution above which the second reference was phase-randomized was gradually increased (Table S1). The remaining particles were then subjected to 3D auto-refinement using RELION 1.4.
Author Contributions
N.Y. and G.F.G. conceived the project. N.Y., X.G., and Q.Z. designed all experiments. G.F.G., T.W., Y.S., and P.W. designed experiments related to EBOV-GPcl. X.G., H.Q., X. Zhou, P.C., W.H. and X. Zhao generated NPC1 variants and carried out NPC1 and NPC2-related biochemical characterizations. T.W. and X.W. prepared GPcl and characterized the interactions between NPC1 and GPcl. Q.Z. developed the random-phase 3D classification method. Q.Z., H.Q., J.W. and X. Zhou conducted the cryo-EM analysis of NPC1 and NPC1-GPcl. All authors contributed to data analysis. X.G., J.W., X. Zhou, G.F.G, Y.S., T.W., and P.W. participated in the manuscript editing and discussion. N.Y. and Q.Z. wrote the manuscript.
Acknowledgments
We thank Jianlin Lei, Yanji Xu, and Xiaomei Li for technical support. We thank Xueming Li and Mingxu Hu for critical discussions. We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the facility support. The computation was completed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology. This work was supported by funds from the Ministry of Science and Technology of China (2015CB9101012014, ZX09507003006), National Natural Science Foundation of China (project 31321062 and 81590761), the special project of Ebola virus research from the President Foundation of Chinese Academy of Sciences, and Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08020100). The research of Nieng Yan was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and an endowed professorship from Bayer Healthcare.
Published: May 26, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.05.022.
Accession Numbers
The atomic coordinates of NPC1 and NPC1-GPcl complex have been deposited in the Protein Data Bank with the accession code PDB: 3JD8 and PDB: 5JNX, respectively. The 4.4 Å and 6.7 Å EM maps of NPC1 and the 6.6 Å EM map of NPC1-GPcl have been deposited in EMDB with accession codes EMD-6640, EMD-6641, and EMD-8169, respectively.
Supplemental Information
References
- Altmann S.W., Davis H.R., Jr., Zhu L.J., Yao X., Hoos L.M., Tetzloff G., Iyer S.P.N., Maguire M., Golovko A., Zeng M. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- Burke R., Nellen D., Bellotto M., Hafen E., Senti K.A., Dickson B.J., Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H., Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.P., Ioannou Y.A. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- Davies J.P., Chen F.W., Ioannou Y.A. Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science. 2000;290:2295–2298. doi: 10.1126/science.290.5500.2295. [DOI] [PubMed] [Google Scholar]
- Deffieu M.S., Pfeffer S.R. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc. Natl. Acad. Sci. USA. 2011;108:18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. (2002). The PyMOL Molecular Graphics System on World Wide Web http://www.pymol.org.
- Delmar J.A., Su C.C., Yu E.W. Bacterial multidrug efflux transporters. Annu. Rev. Biophys. 2014;43:93–117. doi: 10.1146/annurev-biophys-051013-022855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolman R., Leichner G.S., Avner R., Roitelman J. Ubiquitin is conjugated by membrane ubiquitin ligase to three sites, including the N terminus, in transmembrane region of mammalian 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for sterol-regulated enzyme degradation. J. Biol. Chem. 2004;279:38184–38193. doi: 10.1074/jbc.M405935200. [DOI] [PubMed] [Google Scholar]
- Eicher T., Seeger M.A., Anselmi C., Zhou W., Brandstätter L., Verrey F., Diederichs K., Faraldo-Gómez J.D., Pos K.M. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife. 2014;3:e03145. doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund D., Elmlund H. SIMPLE: Software for ab initio reconstruction of heterogeneous single-particles. J. Struct. Biol. 2012;180:420–427. doi: 10.1016/j.jsb.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Grant T., Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Winkler H., Kaup F., Pöhlmann S. Host cell factors in filovirus entry: novel players, new insights. Viruses. 2012;4:3336–3362. doi: 10.3390/v4123336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J.E., Scott M.P. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Hua X., Nohturfft A., Goldstein J.L., Brown M.S. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Hunt C.L., Lennemann N.J., Maury W. Filovirus entry: a novelty in the viral fusion world. Viruses. 2012;4:258–275. doi: 10.3390/v4020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante R.E., Wang M.L., Radhakrishnan A., Kwon H.J., Brown M.S., Goldstein J.L. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istvan E.S., Palnitkar M., Buchanan S.K., Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A., Miller E.H., Herbert A.S., Ng M., Ndungo E., Whelan S.P., Dye J.M., Chandran K. Niemann-Pick C1 (NPC1)/NPC1-like1 chimeras define sequences critical for NPC1’s function as a flovirus entry receptor. Viruses. 2012;4:2471–2484. doi: 10.3390/v4112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara P.E., Labouesse M. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- Kwon H.J., Abi-Mosleh L., Wang M.L., Deisenhofer J., Goldstein J.L., Brown M.S., Infante R.E. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Deffieu M.S., Lee P.L., Saha P., Pfeffer S.R. Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc. Natl. Acad. Sci. USA. 2015;112:14876–14881. doi: 10.1073/pnas.1520490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus S.K., Morris J.A., Carstea E.D., Gu J.Z., Cummings C., Brown A., Ellison J., Ohno K., Rosenfeld M.A., Tagle D.A. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- Long F., Su C.C., Zimmermann M.T., Boyken S.E., Rajashankar K.R., Jernigan R.L., Yu E.W. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Liang Q., Abi-Mosleh L., Das A., De Brabander J.K., Goldstein J.L., Brown M.S. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife. 2015;4:e12177. doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskey K.L., Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J. Biol. Chem. 1985;260:10271–10277. [PubMed] [Google Scholar]
- Malathi K., Higaki K., Tinkelenberg A.H., Balderes D.A., Almanzar-Paramio D., Wilcox L.J., Erdeniz N., Redican F., Padamsee M., Liu Y. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. J. Cell Biol. 2004;164:547–556. doi: 10.1083/jcb.200310046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Carrillo G., Torroja C., Guerrero I. The sterol-sensing domain of Patched protein seems to control Smoothened activity through Patched vesicular trafficking. Curr. Biol. 2001;11:601–607. doi: 10.1016/s0960-9822(01)00178-6. [DOI] [PubMed] [Google Scholar]
- Millard E.E., Gale S.E., Dudley N., Zhang J., Schaffer J.E., Ory D.S. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 2005;280:28581–28590. doi: 10.1074/jbc.M414024200. [DOI] [PubMed] [Google Scholar]
- Millat G., Marçais C., Tomasetto C., Chikh K., Fensom A.H., Harzer K., Wenger D.A., Ohno K., Vanier M.T. Niemann-Pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am. J. Hum. Genet. 2001;68:1373–1385. doi: 10.1086/320606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.H., Obernosterer G., Raaben M., Herbert A.S., Deffieu M.S., Krishnan A., Ndungo E., Sandesara R.G., Carette J.E., Kuehne A.I. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Nakashima R., Yamashita E., Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Guerrero I., Hidalgo A., Taylor A., Whittle J.R.S., Ingham P.W. A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature. 1989;341:508–513. doi: 10.1038/341508a0. [DOI] [PubMed] [Google Scholar]
- Naureckiene S., Sleat D.E., Lackland H., Fensom A., Vanier M.T., Wattiaux R., Jadot M., Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- Neiss W.F. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 1984;80:603–608. [PubMed] [Google Scholar]
- Nohturfft A., Brown M.S., Goldstein J.L. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc. Natl. Acad. Sci. USA. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgami N., Ko D.C., Thomas M., Scott M.P., Chang C.C.Y., Chang T.Y. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J.E., Ekendé E.N., Kifle E.G., O’Connell J.D., 3rd, De Angelis F., Tessema M.B., Derfoufi K.M., Robles-Colmenares Y., Robbins R.A., Goormaghtigh E. Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter. Proc. Natl. Acad. Sci. USA. 2013;110:18484–18489. doi: 10.1073/pnas.1318705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A., Sun L.P., Kwon H.J., Brown M.S., Goldstein J.L. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol. Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C., Ioannou Y.A. The NPC1 protein: structure implies function. Biochim Biophys Acta. 2004;1685:8–13. doi: 10.1016/j.bbalip.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Sennhauser G., Bukowska M.A., Briand C., Grütter M.G. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Sleat D.E., Wiseman J.A., El-Banna M., Price S.M., Verot L., Shen M.M., Tint G.S., Vanier M.T., Walkley S.U., Lobel P. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc. Natl. Acad. Sci. USA. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Thomas C., Nakano Y., Stark D., Neave B., Taylor A.M., Ingham P.W. Mutations in the sterol-sensing domain of Patched suggest a role for vesicular trafficking in Smoothened regulation. Curr. Biol. 2001;11:608–613. doi: 10.1016/s0960-9822(01)00179-8. [DOI] [PubMed] [Google Scholar]
- Taipale J., Cooper M.K., Maiti T., Beachy P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- Tseng T.T., Gratwick K.S., Kollman J., Park D., Nies D.H., Goffeau A., Saier M.H., Jr. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- Vanier M.T. Niemann-Pick disease type C. Orphanet J. Rare Dis. 2010;5:16. doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier M.T. Complex lipid trafficking in Niemann-Pick disease type C. J. Inherit. Metab. Dis. 2015;38:187–199. doi: 10.1007/s10545-014-9794-4. [DOI] [PubMed] [Google Scholar]
- Vanier M.T., Millat G. Niemann-Pick disease type C. Clin. Genet. 2003;64:269–281. doi: 10.1034/j.1399-0004.2003.00147.x. [DOI] [PubMed] [Google Scholar]
- Venter H., Mowla R., Ohene-Agyei T., Ma S. RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front Microbiol. 2015;6:377. doi: 10.3389/fmicb.2015.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.L., Motamed M., Infante R.E., Abi-Mosleh L., Kwon H.J., Brown M.S., Goldstein J.L. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010;12:166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qi J., Liu N., Li Y., Gao J., Zhang T., Chai Y., Gao F., Zhang H., Li X. Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors. Chin. Sci. Bull. 2015;60:3438–3453. [Google Scholar]
- Wang H., Shi Y., Song J., Qi J., Lu G., Yan J., Gao G.F. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Schornberg K.L. A new player in the puzzle of filovirus entry. Nat. Rev. Microbiol. 2012;10:317–322. doi: 10.1038/nrmicro2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D., Xia Z.P., Adams C.M., Rawson R.B. Three mutations in sterol-sensing domain of SCAP block interaction with insig and render SREBP cleavage insensitive to sterols. Proc. Natl. Acad. Sci. USA. 2002;99:16672–16677. doi: 10.1073/pnas.262669399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Nakashima R., Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:327. doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ren J., Harlos K., Stuart D.I. Structure of glycosylated NPC1 luminal domain C reveals insights into NPC2 and Ebola virus interactions. FEBS Lett. 2016;590:605–612. doi: 10.1002/1873-3468.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.