Abstract
Background
The endolysosomal, non-selective cation channels, two-pore channels (TPCs) and mucolipins (TRPMLs), regulate intracellular membrane dynamics and autophagy. While partially compensatory for each other, isoform-specific intracellular distribution, cell-type expression patterns, and regulatory mechanisms suggest different channel isoforms confer distinct properties to the cell.
Scope of review
Briefly, established TPC/TRPML functions and interaction partners (‘interactomes’) are discussed. Novel TRPML3 interactors are shown, and a meta-analysis of experimentally obtained channel interactomes conducted. Accordingly, interactomes are compared and contrasted, and subsequently described in detail for TPC1, TPC2, TRPML1, and TRPML3.
Major conclusions
TPC interactomes are well-defined, encompassing intracellular membrane organisation proteins. TRPML interactomes are varied, encompassing cardiac contractility- and chaperone-mediated autophagy proteins, alongside regulators of intercellular signalling.
General significance
Comprising recently proposed targets to treat cancers, infections, metabolic disease and neurodegeneration, the advancement of TPC/TRPML understanding is of considerable importance. This review proposes novel directions elucidating TPC/TRPML relevance in health and disease. This article is part of a Special Issue entitled: ECS Meeting edited by Claus Heizmann, Joachim Krebs and Jacques Haiech.
Keywords: TRPML, TRPML1, TRPML3, TPC, TPC1, TPC2
Graphical abstract
Highlights
-
•
Endolysosomal ion channels regulate membrane trafficking and autophagy.
-
•
TPC interactomes converge upon the membrane fusion and viral trafficking machinery.
-
•
TPC1 interactome includes numerous regulators of luminal cation homeostasis.
-
•
TRPML1 interactome consists of cardiac and autophagy-related proteins.
-
•
TRPML3 interactome consists of cation pump complexes and signalling proteins.
1. Introduction
The Nobel Prize in Chemistry of 2012 was awarded to professors Lefkowitz and Kobilka for their pioneering discoveries of G protein-coupled receptor (GPCR) signalling. The pair disentangled the multifaceted signal transduction pathways, desensitisation processes, and regulatory mechanisms governing the world of adrenergic receptors [1]. Today, the GPCRs, kinase receptors, transporters, and voltage-/ligand-gated ion channels (VGIC/LGICs) are, to varying extents, understood. Whenever a novel protein family is characterised, it is invariably reaffirmed that molecular function is contextually defined. While adrenergic receptors were initially thought to signal from the plasma membrane, they are today recognised to reside in membrane microcompartments where their roles are dictated by neighbours; to be endocytosed where downstream signalling is drastically altered; and subjected to a plethora of regulatory mechanisms exerted by Arrestins, regulatory kinases, sorting- and adaptor proteins – All finely tuned to orchestrate appropriate response to stimuli. Unquestionably, one cannot claim to fully understand how a protein functions until its context-defining protein interactions have been described.
The endolysosomal non-selective cation channels, comprised by the mucolipins (TRPML1–3) and two-pore channels (TPC1/2), form a recently characterised protein family [[2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14]]. By virtue of their residency on intracellular membranes, the channels for long eluded efforts characterising membrane trafficking events and signal transmission pathways due to technical limitations. While recent developments in genetics such as cloning, in proteomics such as mass spectrometry (MS), or in electrophysiology such as endolysosomal patch clamping, have aided their characterisation, they remain enigmatic in function and importance. For example, both channel families are gated by the endolysosomal phosphoinositide PI(3,5)P2, conferring Na+ currents upon activation. TRPML channels are widely accepted to be Ca2+ permeable, yet the recent discovery of NAADP-activated TPC-mediated Ca2+ currents remains heavily debated, as at least two groups claim that TPCs are predominantly Na+ permeable channels activated by PI(3,5)P2, while several other groups show that TPCs are also Ca2+ permeable and can be activated by both PI(3,5)P2 and NAADP [[15], [16], [17], [18], [19], [20], [21]]. Resolution of these ongoing debates is of particular importance, as the channels are proposed therapeutic targets to treat disorders such as cancer [22,23], neurodegeneration [[24], [25], [26], [27]], metabolic/cardiovascular disorders [12,28], and infectious diseases [[29], [30], [31], [32]].
Since the best described proteins to date such as the GPCRs and LGICs are widely appreciated to serve context-driven functions, this paper will compare and contrast contexts of each endolysosomal cation channel to summarise what is known about their situations to date, and what future steps should be taken to delineate their biological importance. To achieve this, the established channel contexts will initially be discussed, followed by an unbiased proteomic meta-analysis summarising and discussing the to-date identified endolysosomal cation channel interaction partners.
1.1. TPCs and mTOR
Cang et al. [33] found, in an effort to locate endolysosomal ATP-sensitive channels, TPC-mediated Na+ currents to be mTORC1-regulated [33]. Through recording primary peritoneal macrophages, cardiomyocytes, fibroblasts, hepatocytes, and TPC1/TPC2-overexpressing HEK293 cells, the authors demonstrated endolysosomal ATP-sensitive Na+ currents to be attributable to activity of TPC1 and TPC2, but not TRPML1 (TRPML1 and mTOR are discussed in Section 1.3) [33]. Ogunbayo et al. [21] recently demonstrated mTORC1 to not only regulate lysosomal Na+ release, but also TPC2/NAADP-mediated Ca2+ release both in pulmonary arterial smooth muscle cells (PASMCs) and stably expressing HEK293 cells [21]. Indeed, rapamycin elicited similar Ca2+ signals as NAADP in wild-type PASMCs through lysosomal Ca2+ release, while neither NAADP nor rapamycin evoked similar signals in Tpc2 −/− PAMSCs [21]. While TPC2 sensitivity to ATP requires mTORC1 kinase activity, the mTOR target site on TPC2 remains uncharacterised. Recently however, a widely distributed human TPC2 gain-of-function polymorphism (G734E) was linked to decreased ATP sensitivity [34]. This could provide a stepping stone to further elucidate the exact mechanism of mTORC1-dependent TPC regulation.
1.2. TRPML1 and ALG-2
Li et al. [35] discovered a role of TRPML1 in lysosomal positioning and trafficking: Performing fluorescence recovery after photobleaching (FRAP) with the late endosomal (LE)/lysosomal (LY) marker LAMP1, the authors found an equivalence of Ca2+-dependent retrograde (periphery to perinucleus) and anterograde (perinucleus to periphery) LE/LY trafficking under resting conditions, shifting towards retrograde trafficking upon brief starvation [35]. Pharmacological TRPML activation enhanced retrograde LE/LY migration and perinuclear LE/LY accumulation, which was conversely suppressed by TRPML inhibition [35]. Overexpression of TRPML1, but not other TRPML isoforms, favoured peripheral LE/LY accumulation [35,36]. TRPML1 had previously been shown to interact Ca2+-dependently with ALG-2 (PDCD6) [36]. ALG-2 acts as a cytosolic penta-EF-hand Ca2+ sensor, permitting scaffolding upon binding Ca2+. Considering TRPML1 might regulate motility upon ALG-2 recruitment, ALG-2 overexpression was demonstrated to facilitate perinuclear LE/LY accumulation, while Ca2+-binding-deficient ALG-2 showed no effect on LE/LY distribution [35]. Consolidating the relevance of the TRPML1:ALG-2 interaction, both TRPML antagonism and TRPML1 knockout cells overexpressing ALG2-interaction-deficient TRPML1 showed abrogated ALG-2-induced perinuclear LE/LY accumulation [35]. Similarly, ALG-2 knockout cells were irresponsive to starvation- and TRPML activation-induced LE/LY retrograde trafficking [35]. LE/LY motility was demonstrated to depend on dynein motor proteins: Dynamitin – a constituent of the dynein cofactor complex – was co-immunoprecipitated with ALG-2 [35]. Dominant negative (DN) dynein expression and antagonism (ciliobrevin D) abrogated starvation- and TRPML1-induced LE/LY perinuclear redistribution [35]. The authors reasoned TRPML1 Ca2+ release in response to acute starvation raises perilysosomal [Ca2+], facilitating lysosomal recruitment of the ALG-2:dynactin:dynein motor complex, culminating in retrograde lysosomal movement [35].
1.3. TRPML1 and mTOR: debated interacTOR
Alike the TPCs, TRPML1 has been implicated in mTOR nutrient sensing. Acute pharmacological mTORC1 inhibition alleviates TRPML1 S51, S572, and S576 phosphorylation, phosphorylation of the latter two inhibiting TRPML1-mediated Ca2+ release [37]. Unlike TPCs however, TRPML1 currents on isolated endolysosomes are unaffected by ATP [33]. Lysosomal proteins, such as the v-type H+ ATPase, interact with mTOR under certain conditions, which could also be the case for TRPML1 [33]. As previously discussed (Section 1.2), TRPML1 interactions with the lysosomal motility complex appears driven by TRPML1-dependent Ca2+ release and ALG-2 scaffolding. Upon characterising TRPML1-dependent lysosomal ALG-2 recruitment, TRPML1 was shown to interact with Sec13 via ALG-2 [36]. Beyond facilitating Endoplasmic Reticulum (ER)-Golgi trafficking, Sec13 is a constituent of the GTPase-activating proteins towards Rags 2 (GATOR2) complex, a mTOR activator [38]. TRPML1 could thus potentially activate mTORC1 via ALG-2-dependent GATOR2 recruitment. In fact, TRPML1 has already been shown to induce mTOR activity, where TRPML1-mediated Ca2+ release activates mTORC1 via calmodulin (CaM) [39]. Furthermore, the possibility of mTOR-dependent TRPML1 regulation cannot be entirely discarded based on endolysosomal patch clamp data, as this regulation could require cytoplasmic factors eliminated upon endolysosomal isolation. Onyenwoke et al. [37] proposed mTOR regulation of TRPML1 could be mediated by a downstream kinase of mTOR such as S6K [37]. It is therefore of interest that the previously postulated mTOR phosphorylation motif on TRPML1, formed by S572 and S576 (CGRDPSEEHS), conforms better with the S6K target motif (RxR[T/R]xSx[S/T]xS) than with the mTOR-associated motif (xx[P/S][G/F]SPP[P/A][P/L]) [40,41].
While direct mTOR regulation of TRPML1 is debated, it is accepted that mTOR regulates TRPML1 transcription. Long-term mTOR inhibition induces endolysosomal TRPML1 currents in a manner dependent on de novo protein synthesis [42]. The transcription factor for lysosomal biogenesis, TFEB [43], appears responsible for starvation-induced TRPML1 transcription. Reciprocally, TRPML1 activates TFEB via Ca2+ release, activating the Calcineurin (PP2B) to dephosphosphorylate TFEB, permitting its nuclear translocation and transcriptional activation. Thereby, TRPML1 and TFEB form a positive feedback loop favouring lysosomal biogenesis and autophagy [44]. Additionally, a negative feedback loop appears evident, where sustained TRPML1 activity can suppress its own activity through Ca2+-dependent mTORC1 activation. The latter would prove crucial following starvation, where sustained TRPML1-induced Ca2+ release would reactivate mTOR, in turn suppressing excessive autophagy. The possible involvement of TRPML1-dependent lysosomal GATOR2 recruitment in this scenario remains to be validated.
The connection between TRPML1 and mTOR appears further relevant given the molecular functions of both converging at the cAMP-activated protein kinase A (PKA). The interaction between Ca2+ signalling, PKA and mTOR signalling has been postulated by several groups, where PKA phosphorylates Akt/PKB, which in turn activates mTOR [[45], [46], [47], [48], [49]]. In this context it is interesting to note that Vergarajauregui et al. [50] identified two PKA consensus motifs in the C-terminal tail of TRPML1, containing S557 and S559. The authors found PKA inhibition by H89 potently blocked phosphorylation of S557/S559, while adenylyl cyclase activation by forskolin causing cAMP generation increased wild-type TRPML1 phosphorylation, together suggesting PKA activity at TRPML1 S557 and S559. They also found that PKA-mediated phosphorylation inhibits TRPML1 activity [50]. The positive correlation between PKA and mTOR activity alongside negative correlations between PKA/mTOR activity and TRPML1 further consolidates the integration of TRPML1 within the PKA and mTOR signalling network.
Evidently, the endolysosomal non-selective cation channels appear central in regulating intracellular membrane trafficking and cellular adaptation to nutrient deprivation. This raises the question whether these novel, multifunctional channels are of importance in other areas of biology. To assess this possibility, this review will present novel TRPML3 interaction data and conduct an unbiased meta-analysis, gauging consistencies, variabilities and specificities of TRPML- and TPC interactions.
2. Methods
2.1. Proteomics-based TRPML3 interactome screen
The TRPML3 interaction screen was performed as previously described by Grimm et al. [12], using SILAC-labelled HEK293 cells overexpressing human TRPML3-YFP [25,51]. Proteomic datasets are available in the supplement (Supplementary Table 1, Supplementary Table 2). Where TRPML3 interaction data obtained in this publication is presented, it will be referred to by an asterisk ‘*’.
2.2. Protein interactome meta-analysis
Proteomic data was curated from published articles including experimentally obtained proteomic interaction data on either human or mouse TRPML1 (MCOLN1), TRPML2 (MCOLN2), TRPML3 (MCOLN3), TPC1 (TPCN1), and TPC2 (TPCN2). Publications containing interaction datasets were obtained by search engines Google Scholar, BioGRID 3.4 [52], and EMBL-EBI IntAct [53]. In silico interaction partner predictions were excluded from acquired data. Interaction partner identities were translated into Homo sapiens gene names, taking HGNC HUGO Homo sapiens standardised gene notation into consideration [54]. Sources of identified interactors are referenced where appropriate and in Supplementary Table 3. The analysis worksheet of mutual interaction partners is accessible in Supplementary Table 4. For visualisation of interaction networks, Cytoscape 3.6.1 [55] was employed alongside the following Cytoscape plug-in applications: For grouping by EBI GO biological processes gene ontology terms, ClueGO was employed, loading marker lists for Homo sapiens and applying GO term fusion (GO Biological Process EBI, release 20.11.2017) [[56], [57], [58]]. CluePedia was used to visualise interactions between channel interactors, loading marker lists for Homo sapiens, and visualising protein-level interactions (activation, binding, catalysis, inhibition, post-translational modifications, and reactions) [59]. Lone proteins were removed from obtained interaction networks. The TRPML2 interactome could not be visualised due to the limited number of TRPML2 partners identified, but the interactor list is available in Supplementary Table 3. CluePedia interaction networks were overlaid with gene ontology/protein family terms as identified by GeneCards [60], and disease relevance as identified by MalaCards [60,61].
3. Results
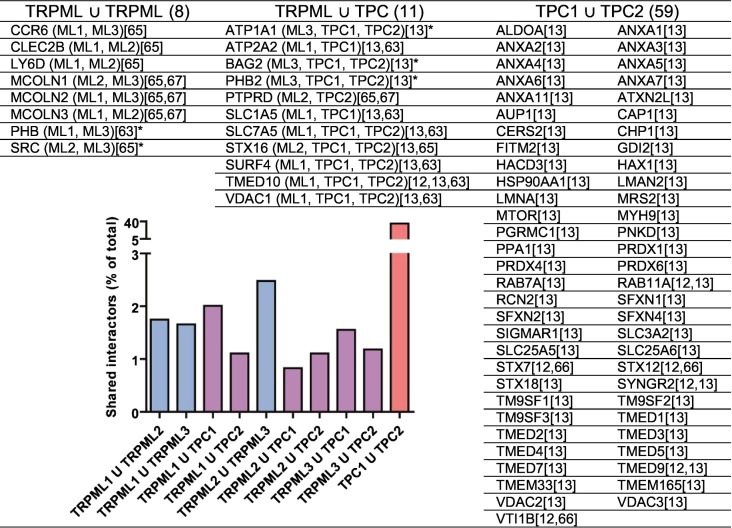
Aiming to assess the degree of interaction partners being shared between endolysosomal cation channels, we compared our databases of compiled experimentally obtained TRPML and TPC interactors (Table 1 ). Not surprisingly, most interaction partners were shared between TPC1 and TPC2 (35.8% of all identified interacting proteins) – a likely consequence of Lin Moshier et al. [13] having used the same methodology to rigorously compare and contrast interactions of the two. Shared TPC interactors are involved in membrane organisation (Annexins, GDI2, RABs, Syntaxins, SYNGR2, VTI1B), cytoskeletal organisation (CAP1, HAX1, LMNA, MYH9), transmembrane transport (MRS2, Sideroflexins, SLC3A2, SLC25A5/6, TM9SF1–3, TMEM165, VDAC2/3), and proteostasis (AUP1, HSP90AA1, MTOR, TMEDs, TMEM33). Similar ventures of rigorous, cross-isoform interaction landscape comparisons have not been performed for the TRPML channels, although this would provide valuable information both regarding general TRPML function, and isoform-specific roles. Still, it appears evident TRPMLs more frequently share interaction partners with other TRPMLs, than with the TPCs. The highest degree of interaction partners shared outside the TPC family occurs between TRPML2 and TRPML3 (2.48% of interactors), although this could be attributable to the few TRPML2 interactors identified to date. Intriguingly, the proto-oncogene SRC appears shared between the two, possibly reflecting SRC implications in endocytosis and, thereby, localisation proximal to early endosome (EE)/recycling endosome (RE) compartments of TRPML2 and TRPML3 residency. TRPMLs further share interactors implicated in the immune response, such as the chemokine receptor CCR6, the lymphoid adhesion molecule CLEC2B, and the B-cell specification marker LY6D. Of interest and possible clinical relevance, most within-family shared TRPML interaction partners are implicated in cancer.
Table 1.
Endolysosomal intra- and interfamily interactor promiscuity.
Characterised protein interactions of endolysosomal cation channels were compared between and within channel families (TRPMLs and TPCs). For TRPML U TRPML and TRPML U TPC, brackets denote interacting isoforms. Data obtained were obtained from referenced publications [9,12,13,[62], [63], [64], [65], [66], [67]]. BioGRID [52] was used to access unpublished interaction data from Huttlin et al. [65], EMBL-EBI IntAct [53] to access unpublished interaction. ‘*’ denotes data obtained in this publication. Inset visualises shared interactor frequency for specified isoform pairs. Blue columns indicate interactors shared within TRPMLs, red within TPCs, and purple across families.
3.1. Two-pore channels
3.1.1. TPC1
In silico analysis of the experimentally obtained TPC1 interaction network reveals that identified interactors [13,[65], [66], [67]] are associated with a narrow range of EBI GO biological processes [56,58], encompassing vesicle organisation (30.0%), organelle membrane fusion (26.7%), and cellular monovalent inorganic cation homeostasis (20.0%) (Supplementary Fig. 1).
The vesicle organisation- and organelle membrane fusion-associated TPC1 interactor terms principally regard vesicle organisation, vesicle fusion, and EE-LE/LE-LY vesicle transport. Vesicle organisation and fusion terms include the low-affinity, Ca2+-inducible membrane-binding Annexins (ANXA1 [13] and ANXA2 [13]), the LE trafficking-regulator RAB7A [13], and the vesicle-trafficking and -fusion SNARE complex constituents STX7 [66], STX8 [66], STX12 [66], and STX16 [13] as well as VTI1B [66]. Other vesicle organisation-proteins include the endosomal sorting complex required for transport-III (ESCRT-III) proteins CHMP2B [66] and CHMP3 [66]. EE-LE- and LE-LY-transporting proteins include CHMP3 [66], while RAB7A [13] and STX8 [66] are included specifically within the EE-LE transport term, and the protein-degradation/autophagy regulator VCP [13] and CHMP2B [66] within the LE-LY transport term.
In silico interaction screening demonstrates extensive interactions between TPC1 and endolysosomal fusion proteins as well as membrane-trafficking Annexins. From these interactors, RAB7A [13], STX7 [66], STX8 [66], and STX12 [66] have been validated by co-immunoprecipitation and western blotting (Supplementary Table 3). Considering the substantial overlap between identified TPC1- and TPC2-associated membrane trafficking partners, these will be discussed collectively (Section 3.1.2).
The cellular monovalent inorganic cation homeostasis term includes the Na+/K+ ATPase subunit ATP1A1 [13], the H+-transporting v-type ATPase subunits ATP6V0C and ATP6V1C1 [66], the calcineurin-like regulator of the Na+/H+-exchanger CHP1 [13], the Golgi voltage-gated anion channel GPR89B [13], RAB7A [13], and the Golgi Ca2+/H+ antiporter TMEM165 [13]. In silico interaction screening reveals relatively weak interactions between the TPC1-associated cation transporters, suggesting TPC1 does not participate in a larger, defined protein complex with these (Fig. 1 ). Among the putative interactors involved in cation transport, only RAB7A has been validated (co-IP/WB) [13].
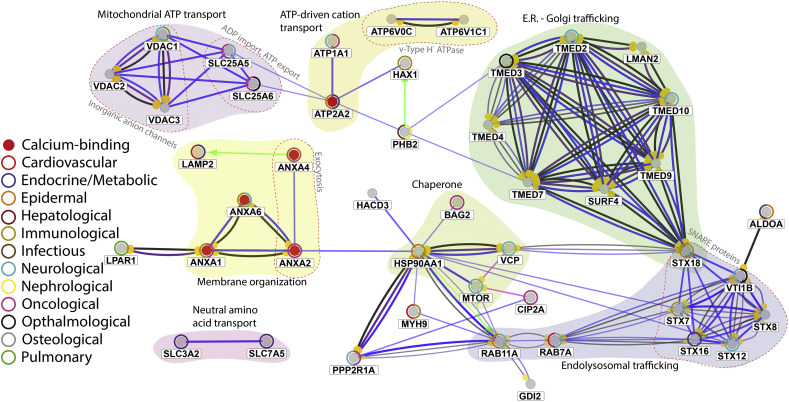
Fig. 1.
CluePedia visualisation of proteins found to interact with TPC1 through pull-down/MS [13,[65], [66], [67]]. Blue edges between nodes indicate binding, black edges a reaction, green edges activation, red edges inhibition, purple edges catalysis, and pink edges post-translational modifications. Direction is indicated by yellow spheres at the target end. The figure was manually clustered, based on protein functions and interactions (identified by GeneCards) [60]. Cluster functions were annotated by coloured, shaded overlays accompanied by text, denoting general GO terms, and by red dashed lines and lighter text denoting within-cluster distinctions. Additional annotations, based on GeneCards [60] and MalaCards [61] annotations, indicate whether displayed proteins bind Ca2+ (red nodes) or cause disease (coloured circles around nodes), respectively.
3.1.2. TPC2
In silico analysis of the experimentally obtained TPC2 interaction network reveals that identified interactors [12,13,65,67] are associated with well-defined EBI GO biological processes [56,58], encompassing membrane organisation (26.5%) and vesicle organisation (19.1%) (Supplementary Fig. 2). Therefore, TPC2 will be discussed in the following alongside TPC1 in the context of its numerous, interconnected, associated proteins mediating intracellular membrane organisation (Fig. 2 ).
Fig. 2.
CluePedia visualisation of proteins found to interact with TPC2 through pull-down/MS [12,13,65,67]. Blue edges between nodes indicate binding, black edges a reaction, green edges activation, red edges inhibition, purple edges catalysis, and pink edges post-translational modifications. Direction is indicated by yellow spheres at the target end. The figure was manually clustered, based on protein functions and interactions (identified by GeneCards) [60]. Cluster functions were annotated by coloured, shaded overlays accompanied by text, denoting general GO terms, and by red dashed lines and lighter text denoting within-cluster distinctions. Additional annotations, based on GeneCards [60] and MalaCards [61] annotations, indicate whether displayed proteins bind Ca2+ (red nodes) or cause disease (coloured circles around nodes), respectively.
To date, every TPC-centred proteomic publication has reported TPC1/2:SNAP/SNARE interactions (Table 2 ) [12,13,66]. The SNARE complex mediates membrane fusion through interactions between different SNAREs on the target membrane (t-SNARE) and on the fusing vesicle (v-SNARE). Originally classified as t- and v-SNAREs, SNARE proteins are now being classified as Q- (Qa, Qb, and Qc) and R-SNAREs [68,69]. SNAP/SNARE-mediated membrane fusion events have been thoroughly described in the context of synaptic vesicle fusion with the PM [70]. While synaptic SNAP/SNARE-mediated vesicle release is accepted to be Ca2+-induced through synaptotagmin/complexin interactions [70], the relevance of vesicular Ca2+ release in regulating ubiquitous SNAP/SNARE complexes is only starting to gain appreciation. The ubiquitous, Ca2+-regulated Synaptotagmin VII (SYT7), e-Synaptotagmins and Ferlins have all been proposed to link vesicular Ca2+ release to endosomal fusion events, although no members of these families have been found to interact with the TPCs [71]. In contrast, a wide range of Annexins, representing a family of Ca2+-activated membrane-binding proteins, have been identified as TPC-interactors (ANXA1–7), albeit lacking validation [13]. EE fusion depends on ANXA1 in a Ca2+-dependent manner, while ANXA2 has been demonstrated to Ca2+-dependently interact with SNAP23, another TPC2-associated SNARE (Table 2) [13,72]. ANXA5 and ANXA6 mediate fusion of autophagosomes/LY and LE/LY, respectively [73]. Other Annexins have been implicated in vesicular membrane trafficking, highlighting them as potential candidates to bridge TPC function and associated SNARE complex activity [[72], [73], [74]]. ANKRD27, a regulator of SNARE- and Rab-dependent melanosome enzyme trafficking is also a TPC2 interaction candidate [67]. In short, both TPC isoforms interact with an extensive repertoire of proteins orchestrating intracellular membrane dynamics, but the mechanistic relevance of such interactions remain to be elucidated. The observed, consistent, and validated interactions of TPCs with endosomal membrane trafficking proteins also appear functionally relevant: Castonguay et al. [66] demonstrated TPC1−/− cells, but not TPC2−/− cells, exhibit impaired bacterial toxin trafficking through EE/RE [66]. Similarly, Grimm et al. [12] found TPC2 to be necessary for endosomal trafficking of LDL and EGF/EGFR, TPC2−/− causing accumulation of EGF in Rab7+/LAMP1+ vesicles (LE/LY) [12]. The importance of TPCs in vesicular trafficking is further emphasised by the apparent dependence of particular viruses on channel activity for viral uptake and infectivity. TPC inhibition prevents infectivity of recombinant vesicular stomatitis virus strains (VSV) bearing filovirus glycoproteins (Ebola and Marburgvirus), but not VSV, Lassa virus, Venezuelan equine encephalitis virus, nor Rabies virus [29]. Recently, the middle east respiratory syndrome (MERS) coronavirus was also shown to depend on TPC1, TPC2, and NAADP signalling for endosomal trafficking and infectivity [75,76]. It remains unclear which other viruses may require TPC for uptake and transport, and which are TPC-independent (for instance, candidates from virus families Arenaviridae, Rhabdoviridae, and Togaviridae were shown not to) [29]. All in all, the potential to interfere with virus uptake through modulating TPC1/2-associated endosome functions appears evident, although the exact mechanism of viral entry and trafficking interference, alongside the particular pathogen targets of such interventions, require further characterisation.
Table 2.
TPC-interacting SNARE proteins.
The identified TPC-interacting SNARE-proteins are shown, alongside publications identifying interactions, validation status, implicated fusion events, and complexes involved in the respective fusion event.
3.2. TRPML1
In silico analysis of the experimentally obtained TRPML1 interaction network reveals that identified interactors [53,62,63,65,67] are associated with numerous of EBI GO biological processes [56,58], encompassing actin-mediated cell contraction and action potential regulation (54.6%) and regulation of transcription from RNA polymerase II promoter in response to stress (22.7%). TRPML1 will first be discussed in the context of cardiac cytoskeletal and action potential-associated interaction partners. Thereafter, due to the overarching topic of stress response among associated interactor terms, TRPML1 will be discussed in the context of associated chaperones and regulators of the TGFβ signalling pathway (Supplementary Fig. 3).
The actin-mediated cell contraction-associated TRPML1 interactor terms principally regard actin filament-based movement and regulation of action potential. Both terms include the cardiac Ca2+-transporting, sarco-/endoplasmic reticulum ATPase (SERCA) ATP2A2 [63], the desmosomal, Ca2+-regulated, cadherin-like DSG2 [63], and the Ca2+-inactivated Na+ V subunit SCN5A [63]. The former term also includes the gap junction protein GJA1, the myosin components MYL6 and MYL6B, and the sarcomere microfilament-connecting TTN, while the latter term includes the nAChR subunit CHRNB2 [63], and the myosin-binding, sarcoplasmic reticulum membrane-anchored protein SLMAP [63]. An in silico interaction screen further demonstrates the tight interaction between the TRPML1-interacting sarcomere constituents MYL6, MYL6B, and TTN, while a slightly weaker, yet functional, interaction appears between GJA1 and the voltage-gated cation channel subunits KCNG2, SCN5A, and SCN10A (Fig. 3). Neither of these interactions have been validated, nor further investigated in the context of TRPML1. It should also be emphasised the cardiac interaction network is constituted by data from a single publication, using mouse Trpml1 as bait in either murine RAW264.7 macrophages, or against a mouse heart cDNA library in a split ubiquitin Y2H assay (Supplementary Table 3) [63]. TRPML1 is highly expressed in the heart [77], yet little data exists on its importance in cardiac muscle. Mutations of TRPML1 causes the rare neurodegenerative disease mucolipidosis type IV (MLIV), marked by neurological, ophthalmological, and gastric manifestations, but no discernible cardiac phenotype [2]. The existence of sarcoplasmic reticulum (SR)/LY microdomains were however recently described in cardiomyocytes, placing TRPML1 in tight association with the cardiac contractile machinery [78]. However, until now TRPML1 has not been recognised for specifically vascular or cardiac functions [79].
Fig. 3.
CluePedia visualisation of proteins found to interact with TRPML1 either through pull-down/MS [63,65,67] or by TRPML1 yeast two-hybrid screens against cDNA libraries [62,63]. Blue edges between nodes indicate binding, black edges a reaction, green edges activation, red edges inhibition, purple edges catalysis, and pink edges post-translational modifications. Direction is indicated by yellow spheres at the target end. The figure was manually clustered, based on protein functions and interactions (identified by GeneCards) [60]. Cluster functions were annotated by coloured, shaded overlays accompanied by text, denoting general GO terms, and by red dashed lines and lighter text denoting within-cluster distinctions. Additional annotations, based on GeneCards [60] and MalaCards [61] annotations, indicate whether displayed proteins bind Ca2+ (red nodes) or cause disease (coloured circles around nodes), respectively.
The transforming growth factor beta (TGF-β1)/stress response-associated TRPML1 interactor terms encompass negative regulation of TGF-β1 signalling and general stress response. Both terms include the TFEB-induced unfolded protein response (UPR) chaperone of lysosomal proteins HSPA5 (BiP, HSP70 family) [63] and the proteasome-directing ubiquitin, UBC. Furthermore, TRPML1 interacts with a component of the Ca2+-binding, TGF-β1-regulating microfibril subunit FBN1 [63]. Interactors from the stress response term include the autophagy-associated UPR chaperone DNAJB1 [63], the mitophagy-associated ubiquitin-conjugating UBE2D2 [63], and the mTORC1-regulated 26S proteasome subunits PSMB2 [63] and PSMD1 [63]. In silico simulations demonstrated extensive physical and functional interactions between TRPML1-interacting ubiquitin regulators and proteasomal proteins, chaperones, RNA processing-, DNA binding-, and phagocytosis-associated proteins (Fig. 3). Neither of the aforementioned interactors have been validated, and all arise from a single publication using mouse Trpml1 as bait in either murine RAW264.7 macrophages, or against a mouse heart cDNA library in a split ubiquitin Y2H assay (Supplementary Table 3) [63].
Interactions between TRPML1 and chaperones are extensively documented. Chaperone-mediated autophagy (CMA) appears impaired in MLIV (TRPML1−/−) fibroblasts, a possible consequence of downregulated LAMP2 (the lysosomal degradation-marked protein translocator) [80]. Overexpression of the HSP70-constituent hspA1L in TRPML knockout Drosophila neurons (trpml 1) rescues lethality and fly motility, suggesting functional relevance to TRPML1/chaperone interactions [81]. Several CMA proteins co-immunoprecipitate with TRPML1 (Hsp40, Hsc70, Hsc90 and Hop), two of these (HSP40 and HSC70) interacting with the luminal TRPML1 ECL1 [80]. The classical perception of chaperones situates them in the cytosol, shuttling degradation-targeted peptides towards the lysosome. However, HSC70 also functions intralumenally in degradative pathways, ‘pulling’ proteins across the lysosomal membrane, explaining its association with luminal TRPML1 domains [82]. Venugopal et al. [80] noted the TRPML1:HSC70 interaction to be Ca2+-dependent, where cytosolic [Ca2+] facilitates lysosomal HSC70 translocation [80]. Given reported roles of TRPML1 in nutrient sensing and adaptation to starvation [37,39,44] and an apparent reliance of chaperone function on TRPML1 [80,81], a functional relationship between the channel and CMA appears evident. As a lysosomal Ca2+ efflux channel, TRPML1 activity upon starvation could liberate Ca2+ necessary for lysosomal HSC70 recruitment [80], enhancing CMA and shifting metabolism from an anabolic towards a catabolic, nutrient-liberating state.
The stress response-implicated TRPML1 interactors are particularly interesting in the light of predicted interactions between TRPML1 and TGF-β1 regulators. As previously discussed, ER stress activates an UPR cascade which ultimately inhibits Golgi-ER retrograde transport while promoting PM translocation of HSPA5 (another putative interaction partner of TRPML1) [83]. PM-resident HSPA5 binds CD109, complexing TGF-β1 in calveolae to abrogate TGF signalling, and promoting proliferation [83]. Along similar lines, the observed interaction of FBN1 and TRPML1 is of interest, as FBN1 constitutes a versatile regulator of TGF-β1 signalling (full-length FBN1 sequesters TGF-β1 in microfibrils to prevent TGF signalling, while FBN1 fragments dissociate sequestered TGF-β1 to facilitate TGF signalling) [84]. While it was initially noted that trpml 1 Drosophila larvae show impaired synapse integrity reminiscent of a Drosophila mutant exhibiting impaired TGF-β regulation and synaptic recycling [81,85], the phenotype was later attributed to altered mTORC1/JNK signalling [85]. The possible interplay between TRPML1 activity and TGF-β1 signalling remains unexplored.
3.3. TRPML3
In silico analysis of the experimentally obtained TRPML3 interaction network reveals that identified interactors [64,65,67]* are associated with few EBI GO biological processes [56,58], encompassing cellular monovalent inorganic cation homeostasis (34.0%), ATP hydrolysis-coupled transmembrane transport (31.9%), rDNA chromatin silencing (14.9%) and the interleukin-7-mediated signalling pathway (6.38%) (Supplementary Fig. 4). First, the obtained interaction network will be discussed with respect to the more ubiquitous transmembrane transport term. Thereafter, since TRPML3 expression and function is established in immune cells, in particular macrophages [51,86] and the chromatin silencing term encompasses immunological differentiation, the TRPML3 interaction network will be discussed in the context of immunology.
The transmembrane transport-associated TRPML3 interactor terms (Fig. 4 ) predominantly regard ATP-coupled transmembrane transport and cellular monovalent inorganic cation homeostasis, the constituents of both terms largely overlapping. Reflecting this, both terms include Na+/K+ ATPase subunits (ATP1A1,* ATP1A2,* and ATP1A3*), the ATP synthase subunit ATP5B,* and v-type H+ ATPase subunits (ATP6V0A1,* ATP6V1A,* ATP6V1B1,* ATP6V1B2,* and ATP6V1E1*). Otherwise, the ATP-coupled transmembrane transport term also includes the SERCA subunit ATP2A3 [65,67], the ATP synthase subunit ATP5A1,* and the lysosomal P-type ATPase subunit ATP13A2 [65,67]. The monovalent inorganic cation homeostasis term also includes the Na+/K+ ATPase subunit ATP1B3.* An in silico interaction screen demonstrates the interaction of TRPML3 with interacting subunits of the Na+/K+ ATPase, the v-type H+ ATPase, and ATP synthase. Neither of these interactions have been validated. However, due to the multitude of TRPML3-associated Na+/K+ ATPase and v-type H+ ATPase subunits, these will be discussed in more detail.
Fig. 4.
CluePedia visualisation of proteins found to interact with TRPML3 through pull-down/MS [64,65,67].* Blue edges between nodes indicate binding, black edges a reaction, green edges activation, red edges inhibition, purple edges catalysis, and pink edges post-translational modifications. Direction is indicated by yellow spheres at the target end. The figure was manually clustered, based on protein functions and interactions (identified by GeneCards) [60]. Cluster functions were annotated by coloured, shaded overlays accompanied by text, denoting general GO terms, and by red dashed lines and lighter text denoting within-cluster distinctions. Additional annotations, based on GeneCards [60] and MalaCards [61] annotations, indicate whether displayed proteins bind Ca2+ (red nodes) or cause disease (coloured circles around nodes), respectively.
TRPML3 localises to EE and LE/LY, residing alongside Na+/K+ ATPase in PM/EE fractions, and alongside v-type H+ ATPase throughout the endosomal system [5,51]. TRPML3 activity is suppressed in acidic environments such as the LE/LY: In the presence of intraluminal [Na+], H+ exerts an irreversible inhibition, while in environments of lower [Na+], H+ inhibition becomes reversible [87]. Thus, v-type H+ ATPase activity is inversely correlated with TRPML3 activity. Functionally, this entails lysosomal TRPML3 being inactive in the acidified, Na+-rich lumen, whereas disruption of electrochemical membrane gradients decreasing intraluminal [H+] and [Na+] leads to TRPML3 activation and lysosomal exocytosis [86,88]. On the other hand, in an environment of low [H+] and high [Na+] such as the EE lumen, TRPML3 would be unaffected by Na+/K+ ATPase-mediated Na+ influx. Instead, the Na+/K+ ATPase would maintain an interior-positive membrane potential, opposing v-type H+ ATPase acidification [89] and sustaining TRPML3 activity. TRPML3 conversely appears to regulate endosomal pH: Wild-type [6], as well as dominant negative (DN) TRPML3 (DD458/459KK) overexpression and knockdown [90] are all found to increase the pH of endosomes. How this is mechanistically possible that both TRPML3 overexpression and knockdown disrupt endosomal pH remains unclear, but a tight control of TRPML3 expression may be critical. Evidently, a thorough validation of the above mentioned potential ATPase interaction partners is needed, alongside an investigation of the effects these may have on TRPML3 function or vice versa.
The immune-cell signalling-associated TRPML3 interactor terms (Fig. 4) largely encompass cellular response to platelet-derived growth factor (PDGF), the interleukin-7 (IL7)-mediated signalling pathway, and chromatin silencing at rDNA. Both PDGF- and IL7-signalling-associated proteins include the non-receptor tyrosine kinase (non-RTK) proto-oncogene FYN.* TRPML3 also associates with the non-RTK proto-oncogenes YES1,* SRC,* and LCK,* where the two former mediate PDGF response and the latter IL7 signalling. Again, neither of these interactions have been validated. However, while TRPML3 expression has not been directly linked to non-RTK activity, it has been implicated in receptor tyrosine kinase (RTK) signalling: The RTK EGFR is upon binding EGF internalised into EE where luminal acidification dissociates bound EGF. EGF shuttles to the lysosome for degradation, while the EGFR is either recycled or degraded [5]. TRPML3 overexpression has been shown to decrease intracellular EGF/EGFR accumulation and degradation, leading the authors to claim TRPML3 counteracts endocytosis [5]. Martina, Lelouvier and Puertollano [6] disputed this, arguing TRPML3 overexpression diminishes EGFR degradation through preventing its lysosomal delivery [6]. Nonetheless, the two groups consented TRPML3 regulates EGFR transit through the endosomal system [5,6]. The involvement of TRPML3 in RTK trafficking makes the association of TRPML3 with non-RTKs particularly interesting, as these often act downstream of RTK signalling. For example, the TRPML3-associated non-RTKs SRC and FYN promote EGF/EGFR endocytosis [91]. SRC furthermore enhances EGFR transit from EE to LE [92], contrasting the apparent TRPML3-induced EGFR retention from later endosomes, raising the question of whether non-RTKs mediate receptor trafficking through modulating TRPML3 activity.
IL7 signalling is recognised for regulating CD4+ T-cell numbers (increasing numbers upon direct T-cell stimulation, decreasing numbers indirectly through dendritic stimulation) [93], and recently, for regulating tissue resident macrophage development [94]. Accordingly, TRPML3 appears highly expressed in the thymus [3,95], being transcribed in macrophages, CD4+ T-cells, and dendritic cells [95]. The TRPML3-interacting non-RTKs FYN and LCK are recruited and activated by IL7R upon IL7 signalling, yet their downstream pathways following IL7 stimulation remain uncharacterised [96]. FYN and LCK are better described in the context of T-cell receptor (TCR) signalling, and could in theory serve similar functions upon IL7 signalling: TCR-activated LCK recruits intracellular FYN and lipid rafts, culminating in outcomes such as thymocyte development, T-cell survival, or antigen recognition [97]. TRPML3 appears upregulated in squamous cell carcinoma and hepatocellular carcinoma, and downregulated in erythroleukemia and platelets of cancer patients [95]. The potential implications of TRPML3 in immunological and oncogenic signalling pathways may be of particular relevance here.
4. Summary
Recent technological advancements have permitted the biophysical characterisation of proteins residing on intracellular membranes and the proteins they interact with (their interactomes, or ‘contexts’). In an effort to further characterize mucolipins (TRPMLs) and two-pore channels (TPCs), this review has 1) summarized their established interactions and the relevance of these, 2) compared consistencies and contrasted differences between interaction signatures of each channel, and 3) outlined interactions of apparent functional relevance, which should be investigated further. The TPCs share several functionally comparable interaction partners mediating intracellular membrane trafficking, while the TRPMLs show more defined, isoform-specific interactions. Common for all endolysosomal cation channels investigated, is their numerous interactions with proteins implicated in diseases affecting all body systems (Fig. 1, Fig. 2, Fig. 3, Fig. 4). Several of these diseases currently lack treatment options, such as TRPML1 and TPCs interacting with the cystic fibrosis-associated VDAC1, or TRPML3 interacting with four proto-oncogenic non-RTKs. It should however be stressed that several of the described interactors lack validation following their initial proteomic identification. Similarly, further investigations of described interactors must be carried out before drawing final conclusions of their importance in a TPC/TRPML-associated context. Taken together, this review underscores the potential held by TPC/TRPML as future therapeutic targets to treat currently incurable diseases, while identifying approaches necessary to elucidate the multifaceted roles of these endolysosomal, non-selective cation channels [12,[22], [23], [24], [25],[29], [30], [31]].
Abbreviations
Organelles
Methodology
- cDNA
complementary DNA
- Co-IP
co-immunoprecipitation
- Cryo-EM
cryo-electron microscopy
- dKO
double knockout
- DN
dominant negative
- FRAP
fluorescence recovery after photobleaching
- MS
mass spectrometry
- rDNA
ribosomal DNA
- SILAC
stable isotype labelling by AA in cell culture
- WB
Western blotting
- Y2H
yeast two-hybrid
Miscellaneous
- BRV-UK
bovine rotavirus
- CMA
chaperone-mediated autophagy
- ECL1
extracellular loop 1
- HEK293
human embryonic kidney cell line
- JNCL
juvenile neuronal ceroid lipofuscinosis
- MERS
middle east respiratory coronavirus
- MLIV
mucolipidosis type IV
- PAMSC
pulmonary artery smooth muscle cell
- RAW264.7
murine macrophage cell line
- UPR
unfolded protein response
- VSV
vesicular stomatitis virus
Proteins
- ALDOA
aldolase, fructose-bisphosphate A
- ALG-2
asparagine-linked glycosylation 2 homolog
- ANKRD27
Ankyrin repeat domain-containing protein 27
- ANXA*
Annexin family
- ATP
adenosine triphosphate
- ATP1*
Na+/K+ transporting ATPase
- ATP13A2
PARK9 cation transporting ATPase 13A2
- ATP2*
Ca2+ transporting SR/ER ATPase
- ATP5*
ATP synthase family
- ATP6*
V-type H+ ATPase
- ATXN2L
Ataxin 2 like
- AUP1
ancient ubiquitous protein 1
- BAG2
BCL2 associated athanogene 2
- BK
big-conductance K+ channel
- BSG
basigin
- CaM
calmodulin
- CAP1
adenylyl cyclase-associated protein 1
- CCR6
C-C chemokine receptor type 6
- CD109
activated T-cell marker CD109
- CD4
T-cell surface glycoprotein CD4
- CERS2
ceramid synthase 2
- CHMP*
charged multivesicular body protein family
- CHP1
calcineurin like EF-hand protein 1
- CHRNB2
cholinergic receptor nicotinic beta 2
- CLEC2B
C-type lectin domain family 2 member B
- CLN3
ceroid lipofuscinosis, neuronal 3
- DNAJB1
DnaJ heat shock protein family (Hsp40) B1
- DSG2
desmoglein 2
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ESCRT*
endosomal sorting complex required for transport
- FBN1
fibrillin 1
- FITM2
fat storage-inducing transmembrane protein 2
- FLOT1
flotillin 1
- FYN
proto-oncogene c-Fyn
- GATOR2
GTPase-activating proteins towards Rags 2
- GDI2
GDP dissociation inhibitor 2
- GJA1
Gap junction alpha-1 protein
- GPCR
G-protein coupled receptor
- GPHR
Golgi pH regulator family
- GPR89B
putative Golgi PH regulator C
- H3F3A
H3 histone family member 3A
- HACD3
3-hydroxyacyl-CoA dehydratase 3
- HAX1
HCLS1 associated protein X-1
- HIST1H*
histone cluster 1 family
- HIST2H3A
histone cluster 2 family
- HSC70
heat shock cognate 71 kDa protein
- Hsc90
heat shock protein 90
- Hsp40
chaperone DnaJ
- HSP70
heat shock protein 70
- HSP90AA1
heat shock protein 90 alpha family class A1
- HSPA5
heat shock protein A5 or BiP
- IL7
interleukin 7
- IL7R
interleukin 7 receptor
- IP3R
inositol triphosphate receptor
- JNK
c-Jun N-terminal kinase
- KCNG2
potassium voltage-gated modifier subfamily G2
- LAMP1
lysosomal-associated membrane protein 1
- LAMP2
lysosomal-associated membrane protein 2
- LC3*
microtubule-associated protein 1 light chain 3
- LCK
proto-oncogene Lck
- LDL
low-density lipoprotein
- LGIC
ligand-gated ion channel
- LMAN2
lectin, mannose binding 2
- LMNA
lamin A/C
- LY6D
lymphocyte antigen 6 family member D
- MMP
matrix metalloproteinase
- MRS2
magnesium transporter
- mTOR*
mechanistic target of rapamycin
- MYH9
myosin heavy chain 9
- MYL6*
myosin light chain 6
- NAADP
nicotinic acid adenine dinucleotide phosphate
- NPC1
NPC intracellular cholesterol transporter 1
- p62
sequestosome 1
- PDGF
platelet derived growth factor
- PGRMC1
progesterone receptor membrane component 1
- PHB*
prohibitin family
- PI(3,5)P2
phosphatidylinositol 3,5-bisophosphate
- PKA
protein kinase A
- PNKD
paroxysmal nonkinesigenic dyskinesia
- PP2B
calcineurin
- PPA1
pyrophosphatase (inorganic) 1
- PRDX*
peroxiredoxin family
- PSM*
proteasome subunit family
- PTPRD
protein tyrosine phosphatase, receptor type D
- RAB*
Rab G-protein GTPase family
- RCN2
reticulocalbin 2
- RTK
receptor tyrosine kinase
- RyR
ryanodine receptor
- S6K
P70-S6 kinase 1
- SCN10A
sodium voltage-gated channel alpha subunit 10
- SCN5A
sodium voltage-gated channel alpha subunit 5
- Sec13
SEC13 homolog, nuclear pore and copii coat
- SERCA
sarco/endoplasmic reticulum atpase
- SFXN*
sideroflexin family
- SIGMAR1
sigma non-opioid intracellular receptor 1
- SLC1A5
sodium-dependent amino acid transporter 2
- SLC25A*
adenine nucleotide translocator family
- SLC3A2
amino acid transporter heavy chain, member 2
- SLC7A5
sodium-independent amino acid transporter
- SLMAP
sarcolemma associated protein
- SNAP*
soluble NSF attachment protein family
- SNARE
SNAP receptor
- SRC
proto-oncogene c-Src
- STX*
syntaxin family
- SURF4
surfeit 4
- SYNGR2
synaptogyrin 2
- SYT7
synaptotagmin-7
- TCR
T-cell receptor
- TFEB
transcription factor EB
- TGF*
transforming growth factor
- TM9SF*
transmembrane 9 superfamily
- TMED*
transmembrane p24 trafficking protein family
- TMEM165
transmembrane protein 165
- TMEM33
transmembrane protein 33
- TPC*
two-pore channel
- TRPML*
mucolipin
- TTN
titin
- UBC
polyubiquitin-C precursor
- UBE2D2
ubiquitin conjugating enzyme E2 D2
- VAMP*
vesicle associated membrane protein family
- VCP
vasolin-containing protein
- VDAC*
voltage dependent anion channel family
- VGIC
voltage-gated ion channel
- VTI1B
vesicle transport through T-SNAREs 1B
- YES1
proto-oncogene c-Yes
- YFP
yellow fluorescent protein
The following are the supplementary data related to this article.
Raw data of SILAC immunoprecipitation experiments.
Processed data of SILAC immunoprecipitation experiments.
Meta-analysis of binary endolysosomal cation channel interactions.
Analysis of mutual endolysosomal cation channel interactions.
Supplementary figures 1–4.
Transparency document
Transparency document.
Acknowledgments
Acknowledgements
We thank Yu-Kai Chao and Dr. Cheng-Chang Chen for their valuable participation in discussions assisting the interpretation of previous publications and data published herein. We also thank Professor Dr. Reinhard Fässler, Dr. Ralph Böttcher, and Dr. Nagarjuna Nagaraj of the Max-Planck-Institute for Biochemistry, München, and Dr. Sami Hassan of Columbia University, New York, for performing TRPML3 mass spectrometric analysis. We are furthermore grateful for the helpful comments and suggestions made by Dr. Diego Medina of the Telethon Institute of Genetics and Medicine, Naples, as well as Professor Dr. Sandip Patel of the University College London, London.
Funding
This work was supported by the DFG (SFB/TRR152 P04, P06 and P12) awarded by the German Research Foundation to C.G., M.B., and C.W.-S., the NCL (Neuronal Ceroid Lipofuscinosis) Foundation Award 2016 (Hamburg, Germany) to C.G., the Mucolipidosis IV Foundation Grant [MDBR-17-120-ML4] to C.G., and the Care-for-Rare Foundation Award (Munich, Germany) 2017 to C.G. The funding sources were not involved in the writing of this review article.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Lefkowitz R.J. A brief history of G-protein coupled receptors (Nobel lecture) Angew. Chemie - Int. Ed. 2013;52:6366–6378. doi: 10.1002/anie.201301924. [DOI] [PubMed] [Google Scholar]
- 2.Sun M., Goldin E., Stahl S., Falardeau J.L., Kennedy J.C., Acierno J.S., Bove C., Kaneski C.R., Nagle J., Bromley M.C., Colman M., Schiffmann R., Slaugenhaupt S.A. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 3.Cuajungco M.P., Samie M.A. The varitint-waddler mouse phenotypes and the TRPML3 ion channel mutation: cause and consequence. Pflugers Arch. Eur. J. Physiol. 2008;457:463–473. doi: 10.1007/s00424-008-0523-4. [DOI] [PubMed] [Google Scholar]
- 4.Brailoiu E., Churamani D., Cai X., Schrlau M.G., Brailoiu G.C., Gao X., Hooper R., Boulware M.J., Dun N.J., Marchant J.S., Patel S. Essential requirement for two-pore channel 1 in NAADP-mediated calcium signaling. J. Cell Biol. 2009;186:201–209. doi: 10.1083/jcb.200904073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H.J., Soyombo A.A., Tjon-Kon-Sang S., So I., Muallem S. The Ca2+channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 2009;10:1157–1167. doi: 10.1111/j.1600-0854.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martina J.A., Lelouvier B., Puertollano R. The calcium channel mucolipin-3 is a novel regulator of trafficking along the endosomal pathway. Traffic. 2009;10:1143–1156. doi: 10.1111/j.1600-0854.2009.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brailoiu E., Rahman T., Churamani D., Prole D.L., Brailoiu G.C., Hooper R., Taylor C.W., Patel S. An NAADP-gated two-pore channel targeted to the plasma membrane uncouples triggering from amplifying Ca2+ signals. J. Biol. Chem. 2010;285:38511–38516. doi: 10.1074/jbc.M110.162073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitt S.J., Funnell T.M., Sitsapesan M., Venturi E., Rietdorf K., Ruas M., Ganesan A., Gosain R., Churchill G.C., Zhu M.X., Parrington J., Galione A., Sitsapesan R. TPC2 is a novel NAADP-sensitive Ca2+ release channel, operating as a dual sensor of luminal pH and Ca2+ J. Biol. Chem. 2010;285:35039–35046. doi: 10.1074/jbc.M110.156927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rietdorf K., Funnell T.M., Ruas M., Heinemann J., Parrington J., Galione A. Two-pore channels form homo- and heterodimers. J. Biol. Chem. 2011;286:37058–37062. doi: 10.1074/jbc.C111.289835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rybalchenko V., Ahuja M., Coblentz J., Churamani D., Patel S., Kiselyov K., Muallem S. Membrane potential regulates Nicotinic Acid Adenine Dinucleotide Phosphate (NAADP) dependence of the pH- and Ca2+-sensitive organellar two-pore channel TPC1. J. Biol. Chem. 2012;287:20407–20416. doi: 10.1074/jbc.M112.359612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cang C., Bekele B., Ren D. The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat. Chem. Biol. 2014;10:463–469. doi: 10.1038/nchembio.1522. [DOI] [PubMed] [Google Scholar]
- 12.Grimm C., Holdt L.M., Chen C.-C., Hassan S., Muller C., Jors S., Cuny H., Kissing S., Schroder B., Butz E., Northoff B., Castonguay J., Luber C.A., Moser M., Spahn S., Lullmann-Rauch R., Fendel C., Klugbauer N., Griesbeck O., Haas A., Mann M., Bracher F., Teupser D., Saftig P., Biel M., Wahl-Schott C. High susceptibility to fatty liver disease in two-pore channel 2-deficient mice. Nat. Commun. 2014;5:4699–4712. doi: 10.1038/ncomms5699. [DOI] [PubMed] [Google Scholar]
- 13.Lin-Moshier Y., Keebler M.V., Hooper R., Boulware M.J., Liu X., Churamani D., Abood M.E., Walseth T.F., Brailoiu E., Patel S., Marchant J.S. The two-pore channel (TPC) interactome unmasks isoform-specific roles for TPCs in endolysosomal morphology and cell pigmentation. Proc. Natl. Acad. Sci. 2014;111:13087–13092. doi: 10.1073/pnas.1407004111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C.S.K., Tong B.C.K., Cheng C.W.H., Hung H.C.H., Cheung K.H. Characterization of two-pore channel 2 by nuclear membrane electrophysiology. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep20282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calcraft P.J., Ruas M., Pan Z., Cheng X., Arredouani A., Hao X., Tang J., Rietdorf K., Teboul L., Chuang K.T., Lin P., Xiao R., Wang C., Zhu Y., Lin Y., Wyatt C.N., Parrington J., Ma J., Evans A.M., Galione A., Zhu M.X. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459:596–600. doi: 10.1038/nature08030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchant J.S., Patel S. Questioning regulation of two-pore channels by NAADP. Messenger. 2013;2:113–119. doi: 10.1166/msr.2013.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Zhang X., Dong X., Samie M., Li X., Clapham D.E., Ren D., Xu H., Wang Xu. TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. 2012;151(2013):372–383. doi: 10.1016/j.cell.2012.08.036.TPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jha A., Ahuja M., Patel S., Brailoiu E., Muallem S. Convergent regulation of the lysosomal two-pore channel-2 by Mg2+, NAADP, PI(3,5)P2and multiple protein kinases. EMBO J. 2014;33:501–511. doi: 10.1002/embj.201387035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan A.J., Galione A. Two-pore channels (TPCs): current controversies. BioEssays. 2014;36:173–183. doi: 10.1002/bies.201300118. [DOI] [PubMed] [Google Scholar]
- 20.Ruas M., Davis L.C., Chen C.-C., Morgan A.J., Chuang K.-T., Walseth T.F., Grimm C., Garnham C., Powell T., Platt N., Platt F.M., Biel M., Wahl-Schott C., Parrington J., Galione A. Expression of Ca2+-permeable two-pore channels rescues NAADP signalling in TPC-deficient cells. EMBO J. 2015;34:1743–1758. doi: 10.15252/embj.201490009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogunbayo O.A., Duan J., Xiong J., Wang Q., Feng X., Ma J., Zhu M.X., Evans A.M. MTORC1 controls lysosomal Ca2+release through the two-pore channel TPC2. Sci. Signal. 2018;11:1–6. doi: 10.1126/scisignal.aao5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen O.N.P., Grimm C., Schneider L.S., Chao Y.-K., Atzberger C., Bartel K., Watermann A., Ulrich M., Mayr D., Wahl-Schott C., Biel M., Vollmar A.M. Two-pore channel function is crucial for the migration of invasive cancer cells. Cancer Res. 2017;77:1427–1438. doi: 10.1158/0008-5472.CAN-16-0852. [DOI] [PubMed] [Google Scholar]
- 23.Grimm C., Bartel K., Vollmar A.M., Biel M. Endolysosomal cation channels and cancer—a link with great potential. Pharmaceuticals. 2018;11:1–8. doi: 10.3390/ph11010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen D., Wang X., Li X., Zhang X., Yao Z., Dong X., Yu T., Lieberman A.P., Showalter H.D., Xu H. Lipid storage disorders block lysosomal trafficking by inhibiting TRP channel and calcium release. Nat. Commun. 2012;3:731–751. doi: 10.1038/ncomms1735.Lipid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C.-C., Keller M., Hess M., Schiffmann R., Urban N., Wolfgardt A., Schaefer M., Bracher F., Biel M., Wahl-Schott C., Grimm C. A small molecule restores function to TRPML1 mutant isoforms responsible for mucolipidosis type IV. Nat. Commun. 2014;5:4681–4691. doi: 10.1038/ncomms5681. [DOI] [PubMed] [Google Scholar]
- 26.Hockey L.N., Kilpatrick B.S., Eden E.R., Lin-Moshier Y., Brailoiu G.C., Brailoiu E., Futter C.E., Schapira A.H., Marchant J.S., Patel S. Dysregulation of lysosomal morphology by pathogenic LRRK2 is corrected by TPC2 inhibition. J. Cell Sci. 2015;128:232–238. doi: 10.1242/jcs.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilpatrick B.S., Magalhaes J., Beavan M.S., McNeill A., Gegg M.E., Cleeter M.W.J., Bloor-Young D., Churchill G.C., Duchen M.R., Schapira A.H., Patel S. Endoplasmic reticulum and lysosomal Ca2+ stores are remodelled in GBA1-linked Parkinson disease patient fibroblasts. Cell Calcium. 2016;59:12–20. doi: 10.1016/j.ceca.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson S.M., Foote K., Kunuthur S., Gosain R., Tan N., Tyser R., Zhao Y.J., Graeff R., Ganesan A., Duchen M.R., Patel S., Yellon D.M. 2015. Inhibition of NAADP Signalling on Reperfusion Protects the Heart by Preventing Lethal Calcium Oscillations via Two-pore Channel 1 and Opening of the Mitochondrial Permeability Transition Pore; pp. 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakurai Y., Kolokoltsov A.A., Chen C.-C.C., Tidwell M.W., Bauta W.E., Klugbauer N., Grimm C., Wahl-Schott C., Biel M., Davey R.A. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science (80-) 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimm C. Endolysosomal cation channels as therapeutic targets - pharmacology of TRPML channels. Messenger. 2016;5:30–36. doi: 10.1166/msr.2016.1061. [DOI] [Google Scholar]
- 31.Grimm C., Butz E., Chen C., Wahl-Schott C., Biel M. From mucolipidosis type IV to Ebola: TRPML and two-pore channels at the crossroads of endo-lysosomal trafficking and disease. Cell Calcium. 2017;67:148–155. doi: 10.1016/j.ceca.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Patel S., Kilpatrick B.S. Two-pore channels and disease. BBA Mol. Cell Res. 2018;1865:1678–1686. doi: 10.1016/j.bbamcr.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cang C., Zhou Y., Navarro B., Seo Y.J., Aranda K., Shi L., Battaglia-Hsu S., Nissim I., Clapham D.E., Ren D. MTOR regulates lysosomal ATP-sensitive two-pore Na+channels to adapt to metabolic state. Cell. 2013;152:778–790. doi: 10.1016/j.cell.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao Y.-K., Schludi V., Chen C.-C., Butz E., Nguyen O.N.P., Müller M., Krüger J., Kammerbauer C., Ben-Johny M., Vollmar A.M., Berking C., Biel M., Wahl-Schott C.A., Grimm C. TPC2 polymorphisms associated with a hair pigmentation phenotype in humans result in gain of channel function by independent mechanisms. Proc. Natl. Acad. Sci. 2017;114:E8595–E8602. doi: 10.1073/pnas.1705739114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X., Rydzewski N., Hider A., Zhang X., Yang J., Wang W., Gao Q., Cheng X., Xu H. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 2016;18:404–417. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vergarajauregui S., Martina J.A., Puertollano R. Identification of the penta-EF-hand protein ALG-2 as a Ca2+-dependent interactor of Mucolipin-1. J. Biol. Chem. 2009;284:36357–36366. doi: 10.1074/jbc.M109.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onyenwoke R.U., Sexton J.Z., Yan F., Diaz M.C.H., Forsberg L.J., Major M.B., Brenman J.E. The mucolipidosis IV Ca2+ channel TRPML1 (MCOLN1) is regulated by the TOR kinase. Biochem. J. 2015;470:331–342. doi: 10.1042/BJ20150219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maki M., Takahara T., Shibata H. Multifaceted roles of ALG-2 in Ca2+-regulated membrane trafficking. Int. J. Mol. Sci. 2016;17:1–23. doi: 10.3390/ijms17091401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R.J., Xu J., Fu C., Zhang J., Zheng Y.G., Jia H., Liu J.O. Regulation of mTORC1 by lysosomal calcium and calmodulin. elife. 2016;5:1–16. doi: 10.7554/eLife.19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Safaei J., Maňuch J., Gupta A., Stacho L., Pelech S. Prediction of 492 human protein kinase substrate specificities. Proteome Sci. 2011;9:1–13. doi: 10.1186/1477-5956-9-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Gao Q., Yang M., Zhang X., Yu L., Lawas M., Li X., Bryant-Genevier M., Southall N.T., Marugan J., Ferrer M., Xu H. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc. Natl. Acad. Sci. 2015;112:E1373–E1381. doi: 10.1073/pnas.1419669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennario V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., Banfi S., Parenti G., Cattaneo E., Ballabio A. A gene network regulating lysosomal biogenesis and function. Science (80-) 2009;325:473–478. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 44.Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., Settembre C., Wang W., Gao Q., Xu H., Sandri M., Rizzuto R., De Matteis M.A., Ballabio A. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia Y., Wen H.Y., Young M.E., Guthrie P.H., Taegtmeyer H., Kellems R.E. Mammalian target of rapamycin and protein kinase a signaling mediate the cardiac transcriptional response to glutamine. J. Biol. Chem. 2003;278:13143–13150. doi: 10.1074/jbc.M208500200. [DOI] [PubMed] [Google Scholar]
- 46.Mavrakis M., Lippincott-Schwartz J., Stratakis C.A., Bossis I. mTOR kinase and the regulatory subunit of protein kinase a (PRKAR1A) spatially and functionally interact during autophagosome maturation. Autophagy. 2007;3:151–153. doi: 10.4161/auto.3632. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q., Heimberg H., Pipeleers D., Ling Z. Glibenclamide activates translation in rat pancreatic beta cells through calcium-dependent mTOR, PKA and MEK signalling pathways. Diabetologia. 2008;51:1202–1212. doi: 10.1007/s00125-008-1026-8. [DOI] [PubMed] [Google Scholar]
- 48.De Joussineau C., Sahut-Barnola I., Dumontet T., Drelon C., Batisse-Lignier M., Tauveron I., Pointud J., Lefranc A., Stratakis C.A. mTOR pathway is activated by PKA in adrenocortical cells and participates in vivo to apoptosis resistance in primary pigmented nodular adrenocortical disease (PPNAD) Hum. Mol. Genet. 2014;23:5418–5428. doi: 10.1093/hmg/ddu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B., Guo F., Ma Y., Song Y., Lin R., Shen F., Jin G. 2017. Activation of D1R/PKA/mTOR Signaling Cascade in Medial Prefrontal Cortex Underlying the Antidepressant Effects of l-SPD; pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vergarajauregui S., Oberdick R., Kiselyov K., Puertollano R. Mucolipin 1 channel activity is regulated by protein kinase A-mediated phosphorylation. Biochem. J. 2008;410:417–425. doi: 10.1042/BJ20070713. [DOI] [PubMed] [Google Scholar]
- 51.Chen C.-C., Butz E.S., Chao Y.-K., Biel M., Wahl-Schott C., Grimm C. Small molecules for early endosome-specific patch clamping, cell. Chem. Biol. 2017;24:1–10. doi: 10.1016/j.chembiol.2017.05.025. [DOI] [PubMed] [Google Scholar]
- 52.Stark C. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., Del-Toro N., Duesbury M., Dumousseau M., Galeota E., Hinz U., Iannuccelli M., Jagannathan S., Jimenez R., Khadake J., Lagreid A., Licata L., Lovering R.C., Meldal B., Melidoni A.N., Milagros M., Peluso D., Perfetto L., Porras P., Raghunath A., Ricard-Blum S., Roechert B., Stutz A., Tognolli M., Van Roey K., Cesareni G., Hermjakob H. The MIntAct project - IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:358–363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S., Bruford E.A. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017;45:D619–D625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon P., Markiel A., Ozier Owen, Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003:2498–2504. doi: 10.1101/gr.1239303.metabolite. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carbon S., Dietze H., Lewis S.E., Mungall C.J., Munoz-Torres M.C., Basu S., Chisholm R.L., Dodson R.J., Fey P., Thomas P.D., Mi H., Muruganujan A., Huang X., Poudel S., Hu J.C., Aleksander S.A., McIntosh B.K., Renfro D.P., Siegele D.A., Antonazzo G., Attrill H., Brown N.H., Marygold S.J., Mc-Quilton P., Ponting L., Millburn G.H., Rey A.J., Stefancsik R., Tweedie S., Falls K., Schroeder A.J., Courtot M., Osumi-Sutherland D., Parkinson H., Roncaglia P., Lovering R.C., Foulger R.E., Huntley R.P., Denny P., Campbell N.H., Kramarz B., Patel S., Buxton J.L., Umrao Z., Deng A.T., Alrohaif H., Mitchell K., Ratnaraj F., Omer W., Rodríguez-López M., Chibucos M.C., Giglio M., Nadendla S., Duesbury M.J., Koch M., Meldal B.H.M., Melidoni A., Porras P., Orchard S., Shrivastava A., Chang H.Y., Finn R.D., Fraser M., Mitchell A.L., Nuka G., Potter S., Rawlings N.D., Richardson L., Sangrador-Vegas A., Young S.Y., Blake J.A., Christie K.R., Dolan M.E., Drabkin H.J., Hill D.P., Ni L., Sitnikov D., Harris M.A., Hayles J., Oliver S.G., Rutherford K., Wood V., Bahler J., Lock A., De Pons J., Dwinell M., Shimoyama M., Laulederkind S., Hayman G.T., Tutaj M., Wang S.J., D'Eustachio P., Matthews L., Balhoff J.P., Balakrishnan R., Binkley G., Cherry J.M., Costanzo M.C., Engel S.R., Miyasato S.R., Nash R.S., Simison M., Skrzypek M.S., Weng S., Wong E.D., Feuermann M., Gaudet P., Berardini T.Z., Li D., Muller B., Reiser L., Huala E., Argasinska J., Arighi C., Auchincloss A., Axelsen K., Argoud-Puy G., Bateman A., Bely B., Blatter M.C., Bonilla C., Bougueleret L., Boutet E., Breuza L., Bridge A., Britto R., Hye-A-Bye H., Casals C., Cibrian-Uhalte E., Coudert E., Cusin I., Duek-Roggli P., Estreicher A., Famiglietti L., Gane P., Garmiri P., Georghiou G., Gos A., Gruaz-Gumowski N., Hatton-Ellis E., Hinz U., Holmes A., Hulo C., Jungo F., Keller G., Laiho K., Lemercier P., Lieberherr D., Mac-Dougall A., Magrane M., Martin M.J., Masson P., Natale D.A., O'Donovan C., Pedruzzi I., Pichler K., Poggioli D., Poux S., Rivoire C., Roechert B., Sawford T., Schneider M., Speretta E., Shypitsyna A., Stutz A., Sundaram S., Tognolli M., Wu C., Xenarios I., Yeh L.S., Chan J., Gao S., Howe K., Kishore R., Lee R., Li Y., Lomax J., Muller H.M., Raciti D., Van Auken K., Berriman M., Stein L., Kersey Paul, Sternberg P.W., Howe D., Westerfield M. Expansion of the gene ontology knowledgebase and resources: the gene ontology consortium. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Iny Stein T., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., Rappaport N., Safran M., Lancet D. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 2016;2016:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 61.Rappaport N., Nativ N., Stelzer G., Twik M., Guan-Golan Y., Stein T.I., Bahir I., Belinky F., Morrey C.P., Safran M., Lancet D. MalaCards: an integrated compendium for diseases and their annotation. Database. 2013;2013:1–14. doi: 10.1093/database/bat018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E.E. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 63.Spooner E., McLaughlin B.M., Lepow T., Durns T.A., Randall J., Upchurch C., Miller K., Campbell E.M., Fares H. Systematic screens for proteins that interact with the mucolipidosis type IV protein TRPML1. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Varjosalo M., Keskitalo S., Vandrogen A., Nurkkala H., Vichalkovski A., Aebersold R., Gstaiger M. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 2013;3:1306–1320. doi: 10.1016/j.celrep.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 65.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L.P., Ordureau A., Rad R., Erickson B.K., Wühr M., Chick J., Zhai B., Kolippakkam D., Mintseris J., Obar R.A., Harris T., Artavanis-Tsakonas S., Sowa M.E., De Camilli P., Paulo J.A., Harper J.W., Gygi S.P. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castonguay J., Orth J.H.C., Müller T., Sleman F., Grimm C., Wahl-Schott C., Biel M., Mallmann R.T., Bildl W., Schulte U., Klugbauer N. The two-pore channel TPC1 is required for efficient protein processing through early and recycling endosomes. Sci. Rep. 2017;7:1–15. doi: 10.1038/s41598-017-10607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H., Szpyt J., Tam S., Zarraga G., Pontano-Vaites L., Swarup S., White A.E., Schweppe D.K., Rad R., Erickson B.K., Obar R.A., Guruharsha K.G., Li K., Artavanis-Tsakonas S., Gygi S.P., Harper J.W. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inoue H., Tani K., Tagaya M. SNARE-associated proteins and receptor trafficking. Recept. Clin. Investig. 2016;3 doi: 10.14800/rci.1377. [DOI] [Google Scholar]
- 69.Teng F.Y., Wang Y., Tang B.L. The syntaxins. Genome Biol. 2001;2:3012.1–3012.7. doi: 10.1186/gb-2001-2-11-reviews3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scheller R.H. In search of the molecular mechanism of intracellular membrane fusion and neurotransmitter release. Nat. Med. 2013;19:1232–1235. doi: 10.1038/nm.3339. [DOI] [PubMed] [Google Scholar]
- 71.Johnson C.P. Emerging functional differences between the synaptotagmin and ferlin calcium sensor families. Biochemistry. 2017;56:6413–6417. doi: 10.1021/acs.biochem.7b00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang P., Chintagari N.R., Gou D., Su L., Liu L. Physical and functional interactions of SNAP-23 with annexin A2. Am. J. Respir. Cell Mol. Biol. 2007;37:467–476. doi: 10.1165/rcmb.2006-0447OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Enrich C., Rentero C., Grewal T. Annexin A6 in the liver: from the endocytic compartment to cellular physiology. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:933–946. doi: 10.1016/j.bbamcr.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 74.Enrich C., Rentero C., Meneses-Salas E., Tebar F., Grewal T. Annexins: Ca2+ effectors determining membrane trafficking in the late endocytic compartment. In: Krebs J., editor. Membr. Dyn. Calcium Signal. Springer International Publishing; Cham: 2017. pp. 351–385. [DOI] [PubMed] [Google Scholar]
- 75.Gunaratne G.S., Johns M.E., Hintz H.M., Walseth T.F., Marchant J.S. A screening campaign in sea urchin egg homogenate as a platform for discovering modulators of NAADP-dependent Ca2+ signaling in human cells. Cell Calcium. 2018;75:42–52. doi: 10.1016/j.ceca.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gunaratne G.S., Yang Y., Li F., Walseth T.F., Marchant J.S. NAADP-dependent Ca2+ signaling regulates Middle East respiratory syndrome-coronavirus pseudovirus translocation through the endolysosomal system. Cell Calcium. 2018;75:30–41. doi: 10.1016/j.ceca.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Falardeau J., Kennedy J., Acierno J., Sun M., Stahl S., Goldin E., Slaugenhaupt S. Cloning and characterization of the mouse Mcoln1 gene reveals an alternatively spliced transcript not seen in humans. BMC Genomics. 2002;3:3. doi: 10.1186/1471-2164-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aston D., Capel R.A., Ford K.L., Christian H.C., Mirams G.R., Rog-Zielinska E.A., Kohl P., Galione A., Burton R.A.B., Terrar D.A. High resolution structural evidence suggests the sarcoplasmic reticulum forms microdomains with acidic stores (lysosomes) in the heart. Sci. Rep. 2017;7:1–15. doi: 10.1038/srep40620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inoue R., Jensen L.J., Shi J., Morita H., Nishida M., Honda A., Ito Y. Transient receptor potential channels in cardiovascular function and disease. Circ. Res. 2006;99:119–131. doi: 10.1161/01.RES.0000233356.10630.8a. [DOI] [PubMed] [Google Scholar]
- 80.Venugopal B., Mesires N.T., Kennedy J.C., Curcio-Morelli C., Laplante J.M., Dice J.F., Slaugenhaupt S.A. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J. Cell. Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 81.Venkatachalam K., Long A.A., Elsaesser R., Nikolaeva D., Broadie K., Montell C. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 2008;135:838–851. doi: 10.1016/j.cell.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arias E., Cuervo A.M. Chaperone-mediated autophagy in protein quality control. Curr. Opin. Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsai Y.-L., Ha D.P., Zhao H., Carlos A.J., Wei S., Pun T.K., Wu K., Zandi E., Kelly K., Lee A.S. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF-β signaling. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4245–E4254. doi: 10.1073/pnas.1714866115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaudhry S.S., Cain S.A., Morgan A., Dallas S.L., Shuttleworth C.A., Kielty C.M. Fibrillin-1 regulates the bioavailability of TGFβ1. J. Cell Biol. 2007;176:355–367. doi: 10.1083/jcb.200608167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong C.O., Palmieri M., Li J., Akhmedov D., Chao Y., Broadhead G.T., Zhu M.X., Berdeaux R., Collins C.A., Sardiello M., Venkatachalam K. Diminished MTORC1-dependent JNK activation underlies the neurodevelopmental defects associated with lysosomal dysfunction. Cell Rep. 2015;12:2009–2020. doi: 10.1016/j.celrep.2015.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miao Y., Li G., Xu H., Abraham S.N., Miao Y., Li G., Zhang X., Xu H., Abraham S.N. TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell. 2015;161:1306–1319. doi: 10.1016/j.cell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou X., Li M., Su D., Jia Q., Li H., Li X., Yang J. Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nat. Struct. Mol. Biol. 2017;24:1146–1154. doi: 10.1038/nsmb.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hirschi M., Herzik M.A., Wie J., Suo Y., Borschel W.F., Ren D., Lander G.C., Lee S.Y. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature. 2017;550:411–414. doi: 10.1038/nature24055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cain C.C., Sipe D.M., Murphy R.F. Regulation of endocytic pH by the Na+, K+-ATPase in living cells. Proc. Natl. Acad. Sci. 1989;86:544–548. doi: 10.1073/pnas.86.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lelouvier B., Puertollano R. Mucolipin-3 regulates luminal calcium, acidification, and membrane fusion in the endosomal pathway. J. Biol. Chem. 2011;286:9826–9832. doi: 10.1074/jbc.M110.169185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Riento K., Frick M., Schafer I., Nichols B.J. Endocytosis of flotillin-1 and flotillin-2 is regulated by Fyn kinase. J. Cell Sci. 2009;122:912–918. doi: 10.1242/jcs.039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oda K., Matsuoka Y., Funahashi A., Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005;1:E1–E17. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guimond M., Veenstra R.G., Grindler D.J., Zhang H., Cui Y., Murphy R.D., Kim S.Y., Na R., Hennighausen L., Kurtulus S., Erman B., Matzinger P., Merchant M.S., Mackall C.L. Interleukin 7 signaling in dendritic cells regulates the homeostatic proliferation and niche size of CD4+T cells. Nat. Immunol. 2009;10:149–157. doi: 10.1038/ni.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beaudin A.E., McCann T., Leung G., Forsberg E.C. IL7r-alpha fate-mapping labels distinct adult tissue resident macrophages. Exp. Hematol. 2017;53:S126. doi: 10.1016/j.exphem.2017.06.318. [DOI] [Google Scholar]
- 95.Papatheodorou I., Fonseca N.A., Keays M., Tang Y.A., Barrera E., Bazant W., Burke M., Füllgrabe A., Fuentes A.M.P., George N., Huerta L., Koskinen S., Mohammed S., Geniza M., Preece J., Jaiswal P., Jarnuczak A.F., Huber W., Stegle O., Vizcaino J.A., Brazma A., Petryszak R. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46:D246–D251. doi: 10.1093/nar/gkx1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Page T.H., Lali F.V., Foxwell B.M.J. Interleukin-7 receptor in primary human T cells. Eur. J. Immunol. 1995;25:2956–2960. doi: 10.1002/eji.1830251036. [DOI] [PubMed] [Google Scholar]
- 97.Palacios E.H., Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data of SILAC immunoprecipitation experiments.
Processed data of SILAC immunoprecipitation experiments.
Meta-analysis of binary endolysosomal cation channel interactions.
Analysis of mutual endolysosomal cation channel interactions.
Supplementary figures 1–4.
Transparency document.