Highlights
-
•
A Chinese virulent genotype GIIb PEDV strain, CH/HNPJ/2017, was successfully separated and serially propagated in Vero cells.
-
•
The biological characteristics and pathogenicity of PEDV strain CH/HNPJ/2017 were determined.
-
•
The median pig diarrhea dose (PDD50) of Chinese PEDV strain was first determined.
-
•
The immune protective effect of PEDV strain CH/HNPJ/2017 as vaccine candidates was also be evaluated.
Keywords: Porcine epidemic diarrhea virus, Isolation, Pathogenicity, Immune protection effect
Abstract
Since October 2010, severe porcine epidemic diarrhea (PED) outbreaks caused by highly virulent PED virus (PEDV) strains have occurred continuously in the Chinese pig population and caused considerable economic losses. Although PEDV vaccines based on classical PEDV strains have been widely used in China in recent years, the morbidity and mortality in piglets remain high. Therefore, virulent genotype GII PEDV strains that are prevalent in the field should be isolated and used to develop next-generation vaccines. In the present study, a Chinese virulent genotype GIIb PEDV strain, CH/HNPJ/2017, was serially propagated in Vero cells for up to 90 passages. The S genes contained typical insertions and deletions that were also found in other recently isolated highly virulent PEDV strains from China and other countries and had two neighboring unique insertion mutations, which resulted in four amino acid changes in the S1 region of passages P10 and P60. Pig infection studies revealed that the CH/HNPJ/2017 strain was highly virulent in piglets, and the median pig diarrhea dose (PDD50) was 7.68 log10PDD50/3 mL. Furthermore, the cell-adapted CH/HNPJ/2017 strain elicited potent serum IgG and neutralizing antibody responses in immunized pigs when it was used as an inactivated vaccine candidate. In addition, the pigs that received the experimental inactivated vaccines were partially protected (3/5) against subsequent viral challenge. In brief, these data indicate that the CH/HNPJ/2017 strain is a promising candidate for developing a safe and effective PEDV vaccine in the future.
1. Introduction
Porcine epidemic diarrhea (PED), which is caused by porcine epidemic diarrhea virus (PEDV), is an acute and highly contagious enteric disease in swine characterized by watery diarrhea, vomiting, dehydration, anorexia, weight loss and high mortality in suckling pigs (Lee, 2015; Sun et al., 2012). Pigs of all ages can be infected with PEDV and show symptoms to varying degrees, but symptoms are especially severe in piglets, among which the mortality rate is up to 80–100% (Sun et al., 2012).
PEDV is an enveloped, single-stranded, positive-sense RNA virus that belongs to the order Nidovirales, the family Coronaviridae and the genus Alphacoronavirus (Lin et al., 2016a). The size of PEDV genomic RNA is approximately 28 kb long with a 5′ cap and a 3′ poly (A) tail and contains a 5′ untranslated region (UTR), seven open reading frames (ORFs; ORF1a, ORF1b, and ORF2-6) encoding viral proteins, and a 3′ UTR (Huang et al., 2013; Sun et al., 2016). Among these ORFs, ORF1a and ORF1b encode the viral polymerase; ORF3 encodes a nonstructural protein that is thought to be related to viral infectivity and pathogenicity (Wang et al., 2012). In addition, the other ORFs have specific names according to the proteins encoded in these regions, i.e., spike (S), envelope (E), matrix (M), and nucleocapsid (N) proteins (Jung and Saif, 2015; Song et al., 2015). Of the PEDV structural proteins, the S protein is considered the most antigenic; it is a glycosylated protein located on the envelope of the virus and consists of S1, which is a receptor-binding subunit, and S2, which is a membrane fusion subunit (Gillam et al., 2018; Hou et al., 2017). The S protein is also associated with growth adaptation in vitro and attenuation of PEDV virulence in vivo (Lin et al., 2017; Sato et al., 2011). The M protein is the most abundant component among viral proteins in the envelope and plays an important role in virus assembly by interacting with the S and N proteins (Jung and Saif, 2015; Vennema et al., 1996). The N protein of coronavirus binds RNA and packages viral genomic RNA into the nucleocapsid of viral particles (Spaan et al., 1983).
PED was first reported in pigs in the United Kingdom in 1971, but no PEDV was isolated from this outbreak (Chasey and Cartwright, 1978). In 1978, the PEDV CV777 strain was confirmed as the cause of a PED outbreak that occurred in 1977 in Belgium (Pensaert and de Bouck, 1978). During the 1980s and 1990s in Europe, outbreaks of PED occurred infrequently, but the virus continued to spread and persisted in an endemic form in the pig population (Van Reeth and Pensaert, 1994; Wang et al., 2016). PED was reported for the first time in China in 1973 (Huang et al., 1980), but the pathogen, PEDV, was not identified until 1984 using fluorescent-labeled antibody and serum neutralization (SN) tests (Xuan et al., 1984). Since then, PED outbreaks have occurred infrequently with only sporadic incidents in small regions of China (Li et al., 2012). In October 2010, a highly pathogenic variant PEDV strain was identified in China; this variant caused the worst recorded outbreak and soon swept throughout almost the entire country (Li et al., 2012). Subsequently, this PEDV variant caused a pandemic in the United States (US) starting in the spring of 2013 and then spread to Canada and Mexico (Niederwerder and Hesse, 2018). Severe PED outbreaks have recurred almost simultaneously in many Asian countries, such as the Philippines, Vietnam, Thailand, South Korea and Japan, and US prototype-like GII PEDV strains have been found to be accountable for these outbreaks (Cheun-Arom et al., 2016; Chung et al., 2016; Lee et al., 2017; Lin et al., 2014; Paraguison-Alili and Domingo, 2016; Van Diep et al., 2015). In addition, the US S INDEL-like strains have been described in many European countries, such as Belgium, France, Germany, Italy, Portugal, Slovenia and Netherlands (Choudhury et al., 2016; Pensaert and Martelli, 2016; Stadler et al., 2015). Therefore, PEDV has currently re-emerged as one of the most devastating viral diseases of swine in the world, resulting in tremendous economic losses to the global pig industry.
Currently, there are many commercially available PEDV vaccines in China derived from classical strains of the virus that are either inactivated/killed or attenuated. However, as demonstrated in clinical studies, even vaccinated pigs are not protected from PEDV infection, so vaccines based on classical strains are insufficiently protective (Chen et al., 2010; Li et al., 2012; Sun et al., 2016; Wang et al., 2016). The incomplete efficacies of the current PEDV vaccines might be ascribed to antigenic or genetic differences between the major S glycoprotein in the vaccine and those in the field epidemic strains. The results of epidemiological studies have shown that the prevalent strains responsible for the previous outbreak in China in late 2010 and those responsible for recent outbreaks Asia and North America belong to the GII genogroup (Wang et al., 2016). Therefore, new PEDV vaccines for swine based on the emerging variant strains are urgently needed. To achieve this objective, the epidemic variant strains should be isolated as soon as possible and their biological characteristics, pathogenicity and immune protective effects as vaccine candidates should also be elucidated. Therefore, in this study, a Chinese virulent genotype GIIb PEDV strain, CH/HNPJ/2017 (GenBank accession number: MF152604), was isolated and serially propagated in Vero cells for up to 90 passages. Furthermore, its biological characteristics, pathogenicity and immune protective effects as an inactivated vaccine candidate were clarified and evaluated. This strain could be used as an optimal inactivated or attenuated vaccine candidate for the prevention and control of PEDV in China.
2. Materials and methods
2.1. Clinical samples, cells, and antibodies
A total of 11 samples of porcine intestinal contents (9 from piglets and 2 from sows) were collected from a pig farm in Hunan Province, China, that was affected by PEDV in February 2017. The virus isolated from these samples was identified as PEDV by N gene-based reverse transcription PCR (RT-PCR). The samples of intestinal contents were homogenized with serum-free Dulbecco's Modified Eagle Medium (DMEM, Invitrogen, USA) containing 1% penicillin-streptomycin (10,000 units/mL of penicillin and 10,000 μg/mL of streptomycin) (Gibco™, USA) and 0.3% tryptose phosphate broth (TPB; Sigma, Germany) and then centrifuged at 3500 rmp at 4℃ for 30 min. The supernatant was filtered using 0.22-μm pore-size filter (Merck Millipore, Germany) to remove bacteria, and then frozen at −70 ℃ until use as an inoculum for virus isolation. Vero cells (ATCC CCL-81) were cultured in DMEM supplemented with 5% heat-inactivated fetal bovine serum (FBS; Invitrogen, Australia) and 1% antibiotics (10,000 units/mL of penicillin, 10,000 μg/mL of streptomycin, and 25 μg/mL of Fungizone®) (Gibco™, USA) and maintained at 37 °C in a humidified 5% CO2 incubator. Mouse anti-PEDV N protein monoclonal antibodies (McAbs) were prepared and stored in our laboratory.
2.2. Isolation and serial passage of the virus
Virus isolation of PEDV was performed using Vero cells as described previously, with some modifications (Oka et al., 2014). In detail, prior to inoculation, a 100% confluent Vero cell monolayer was washed with sterile phosphate-buffered saline (PBS; pH 7.2, Gibco™, USA) thrice to completely remove FBS. In the meantime, the supernatant of the samples of intestinal contents prepared as described above were diluted 10-fold in virus growth medium [DMEM supplemented with antibiotics (100 units/mL of penicillin and 100 mg/mL of streptomycin, Gibco™), 0.3% TPB (Sigma), and 20 μg/mL of trypsin (Gibco™, USA)] and vortexed briefly and then used as an inoculum for incubation. A total of 1 mL of the prepared inoculum was added to a T-25 flask. After incubation at 37 ℃ for one hour, 2 mL of virus growth medium was added without removing the inoculum. The inoculated cells were maintained at 37 °C in a humidified 5% CO2 incubator and monitored daily for cytopathic effects (CPE). When CPE were observed in >90% of Vero cells, the flask was subjected to three rounds of freezing and thawing. The cells and supernatants were mixed by pipetting and aliquoted and stored at −70 ℃. These cell culture harvests were used as seed stock for the next passage. For serial passaging, the culture scale was gradually increased, until finally T-75 flasks were used for propagation and serial passage of PEDV strains.
2.3. Virus titration
Viral titers were determined as the 50% tissue culture infectious dose (TCID50) on Vero cells in a 96-well plate, as described previously, with some modifications (Yang et al., 2015). In brief, Vero cells were seeded into 96-well plates, and after confluence, the monolayers were washed thrice with PBS (Gibco™, USA). In addition, 100 μl of 10-fold serially diluted virus suspensions containing 20 μg/mL of trypsin was inoculated in eight replicates per dilution. After absorption for 1 h, another 100 μl of virus growth medium was added to each well. Viral CPE was monitored for 3–5 days, and the viral titers were calculated according to the Reed and Muench method and expressed as TCID50/mL.
2.4. Electron microscopy assay
To image virion particles in Vero cell culture media, Vero cells infected with PEDV were harvested when CPE was observed in >90% of cells. The cell culture was frozen and thawed three times and then centrifuged at 10,000 rpm at 4 °C for 1 h. The supernatant was filtered through a 0.22-mm filter to remove the cell debris and then mixed with polyethylene glycol 8000 (PEG-8000; Solarbio, China) at 10% final concentration overnight. The mix was then ultracentrifuged at 12,000 rpm at 4 °C for 2 h to pellet the PEDV particles. The viral particle pellets were resuspended in tris buffered saline solution (TBS) and then negatively stained with 2% phosphotungstic acid and examined with a transmission electron microscope (JEOL, JEM-1200EX, Japan). To image virions in infected Vero cells, cells were fixed and imaged according to methods described in a previous study (Oka et al., 2014).
2.5. Immunofluorescence assay (IFA)
Vero cells in 6-well cell culture plates were mock infected or infected with PEDV at a multiplicity of infection (MOI) of 0.1. At 0, 12, 24 and 36 h postinfection, the cells were fixed using 4% paraformaldehyde at 4 °C for 30 min and then permeabilized with 0.25% Triton X-100 (Solarbio, China) for 10 min at room temperature (RT). After the plate was washed three times with PBS (Gibco™, USA), it was blocked with 5% bovine serum albumin (BSA; Solarbio, China) at RT for 1 h. Mouse anti-PEDV N protein McAbs #12 (prepared and stored in our laboratory) and Alexa Fluor® 488-conjugated goat anti-mouse IgG (Abcam, UK) were used as first and second antibodies, respectively. The cell nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI; Vectorlabs, USA) for 5 min at RT. After the cells were washed with PBS, and the stained cells were observed with a fluorescence microscope (Olympus, Japan).
2.6. Sequencing and phylogenetic analysis of the spike (S) gene
To monitor the sequence variation in the S gene during serial passaging, the parental CH/HNPJ/2017 strain (from the original intestinal content, P0) and its cell cultures at different passages (from P1 to P80 with 10 generation intervals) were determined by the Sanger method. Briefly, the total RNA was extracted from the original intestinal contents or cell culture samples using the RNeasy Mini Kit (QIAGEN, USA). All of the primers used to amplify genomic fragments of PEDV were designed and stored in our laboratory. Reverse transcription and PCR were performed with the avian myeloblastosis virus (AMV) reverse transcriptase (Promega, USA) and PrimeSTAR® GXL DNA polymerase, respectively. The PCR products were cloned using the TOPO® TA Cloning® Kit (Invitrogen, USA), and the resulting plasmids were sent to Sangon Biotech (Shanghai, China) for sequencing. The sequence fragments were assembled and analyzed with DNAStar 7.0 and BioEdit software, respectively. All of the complete sequences of the S gene and reference sequences obtained from GenBank were used in sequence alignments and phylogenetic analyses. Phylogenetic trees were constructed using the neighbor-joining method and MEGA version 6 (Tamura et al., 2013), with bootstrap values calculated for each node from 1000 replicates. All tree figures were produced using Mega 4.0 software.
2.7. Determination of the infectious titer and virulence in suckling piglets
To determine the infectious titer and virulence of the virus isolates, the median pig diarrhea dose (PDD50) was determined by using 4-day-old conventional suckling pigs as described in our previous study, with some modifications (Liu et al., 2015). In detail, three pregnant PEDV-naïve sows that tested seronegative for PEDV with a commercial enzyme-linked immunosorbent assay (ELISA) kit (Biovet, Canada) were obtained from a commercial pig farm with no previous herd history of PED outbreak or PEDV vaccination. After delivery, a total of healthy 30 suckling piglets were obtained and artificially fed with bovine milk from birth. All piglets were randomly assigned to 5 dose-groups (G1–G5) and one mock control group (Table 1 ). Each piglet was housed in an individual steel cage, and each group of pigs was housed in different room.
Table 1.
Summary information of the pig groups, inocula and pig diarrhea outcomes after inoculation.
| Pig group | Pig numbers | Inoculuma | Calculated inoculum infectious titers (log10 TCID50/mL)c |
Tested inoculum RNA titers (CT value)d |
Diarrhea (percent)e | Fecal virus RNA shedding (CT value) |
|---|---|---|---|---|---|---|
| G1 | 5 | 10−5 diluted P4b | −1.29 | 33.04 | 5/5 (100) | 19.95–25.96 |
| G2 | 5 | 10−6 diluted P4 | −2.29 | 34.99 | 5/5 (100) | 20.83–25.89 |
| G3 | 5 | 10−7 diluted P4 | −3.29 | 35.05 | 4/5 (80) | 18.62–21.58 |
| G4 | 5 | 10−8 diluted P4 | −4.29 | 35.66 | 2/5 (40) | 17.30–21.07 |
| G5 | 5 | 10−9 diluted P4 | −5.29 | 35.71 | 0/5 (0) | 35.67–35.95 |
| Mock Control | 5 | PBS | – | – | 0/5 (0) | – |
Each pig was inoculated orally with 3 mL of inoculum.
The fourth passage of cell culture-adapted CH/HNPJ/2017.
Titers were calculated based on the original titer of the P4 passage (104.29 TCID50/mL) and corresponding dilution times.
CT value: the mean cycle threshold value; a value greater than 35 was considered negative or below detection limit of real-time PCR.
Fecal scores of 3 as determined at 72 postinoculation hours (PIH) for pigs in G1–G5.
For the preparation of inocula for piglets, the fourth passage (P4) of cell culture-adapted CH/HNPJ/2017 (104·29 TCID50/mL) was 10-fold serially diluted in PBS (pH 7.2, Gibco™, USA) (from 10−1 to 10−9), mixed well, and vortexed. Each experimental group (G1–G5) of piglets was inoculated orally with 3 mL of the 10-fold serially diluted (from 10−5 to 10−9) virus at 4 days of age. The mock control group received PBS (Table 1). After inoculation, all of piglets were observed 3 times daily for clinical signs. Rectal swabs were collected daily from all piglets, tested by real-time PCR and scored for fecal consistency: 0 = normal; 1 = pasty; 2 = semiliquid; and 3 = liquid. Scores of 3 were considered watery diarrhea (Liu et al., 2015). To reduce the risk of cross-contamination among pigs, the piglets were euthanized at the onset of obvious clinical symptoms and PEDV RNA fecal shedding identified in the rectal swab samples. Furthermore, necropsy and histopathological examinations of small intestinal tissues were performed as described previously (Liu et al., 2015). The PDD50 was determined as the reciprocal of the virus dilution at which 50% of the pigs developed watery diarrhea at a given time point using the Reed and Muench method. The animal use protocols were reviewed and approved by the Institutional Animal Use and Care Committee of Lanzhou Veterinary Research Institute. All piglets used in the present study were humanely bred during the experiment and euthanized at the end of the experiment.
2.8. Histopathology and immunohistochemistry (IHC) of the small intestines
At necropsy, small intestinal tissue specimens were examined and collected from each piglet. After 48 h of fixation in 4% paraformaldehyde solution at RT, tissue specimens were processed and embedded in paraffin. The paraffin-embedded tissues were cut into thick sections on a microtome (Leica, Germany), deparaffinized with xylene, and washed in decreasing concentrations of ethanol. After that, the intestinal tissue specimens were routinely stained with hematoxylin and eosin (Baso, China) for histopathology or subjected to immunohistochemistry (IHC) using PEDV N-specific MAbs (prepared in our laboratory) as described previously (Lin et al., 2015a, 2015b).
2.9. Preparation of the experimental inactivated vaccines
The seventieth passage (P70) of cell culture-adapted CH/HNPJ/2017 was chemically inactivated using binary ethyleneimine (BEI). In brief, 107.29 TCID50/mL viruses were obtained from Vero cells cultured in T-75 flasks (Corning, USA). After three rounds of freezing and thawing, the liquid supernatant containing viruses was collected with centrifugal processes and then inactivated by addition of 0.2 M BEI to a final concentration of 2 mM at 30 °C for 24 h. After the reaction, the remaining BEI was neutralized by addition of 20% sodium thiosulfate. The effect of inactivation was assessed by the absence of viral growth in Vero cell cultures and inoculation of the piglets. The inactivated vaccines were prepared by mixing BEI-inactivated cell culture-adapted CH/HNPJ/2017 P70 with Freund's complete adjuvant (Sigma, Germany), according to the instruction manual, and stored at 4 °C until use.
2.10. Pig vaccination and challenge experiment
Fifteen PEDV-naïve pigs (5–6 weeks old) that tested seronegative for PEDV by a commercial ELISA kit (Biovet, Canada) were obtained from a commercial pig farm with no previous herd history of PED outbreak or PEDV vaccination. All of the pigs were raised in the laboratory animal facility at the Lanzhou Veterinary Research Institute. The animal use protocols were reviewed and approved by the Institutional Animal Use and Care Committee of Lanzhou Veterinary Research Institute. All pigs used in the present study were humanely bred during the experiment and euthanized at the end of the experiment.
All fifteen pigs were randomly divided into three groups: an experimental inactivated vaccine group (n = 5), a commercial inactivated vaccine group (n = 5) and a mock control group (n = 5). Each group was housed in separate rooms for the duration of the experiment. Pigs in the experimental inactivated vaccine group and commercial inactivated vaccine group were immunized once intramuscularly at 0 days post-vaccination (dpv) with 2 mL of the prepared experimental inactivated vaccines and commercial inactivated vaccine (based on a Chinese genotype GII PEDV strain), respectively. Pigs in the mock control group were immunized with PBS. Blood samples were collected on 0, 7, 14 and 21 dpv, and serum was isolated for antibody detection. At 21 dpv, all pigs were challenged orally with 4 mL of 100 TCID50 cell culture-adapted CH/HNPJ/2017 P4. After challenge, clinical signs of PEDV infection were observed daily for 10 days, and clinical scores of fecal consistency were determined according to the methods of previous study as follows: 0 = normal; 1 = pasty; 2 = semiliquid; and 3 = liquid (Liu et al., 2015). Fecal samples were collected daily for the duration of the challenge to monitor viral shedding in feces.
2.11. Enzyme-linked immunosorbent assay (ELISA)
PEDV-specific IgG antibody responses elicited by immunization with the BEI-inactivated cell culture-adapted CH/HNPJ/2017 P70 were assessed with a commercial indirect ELISA kit (Swinecheck PED indirect, BIOVET, GC Kerkrade, the Netherlands) according to the manufacturer’s instructions.
2.12. Serum neutralization (SN) test
The levels of PEDV-specific neutralizing antibodies in serum samples collected from pigs in all groups were determined using an SN test in 96-well microtiter plates (Corning, USA), according to a previously published method, with some modifications (Song et al., 2007). Briefly, serum samples were inactivated at 56 ℃ for 30 min and stored at −20 ℃ until use. Vero cells were grown in 96-well tissue culture plates for 1–2 days until a monolayer formed. Two-fold serial serum dilutions starting at 1:2 were coincubated at 37 ℃ for 1 h with equal volumes of viral stock containing 200 TCID50 of PEDV CH/HNPJ/2017 in 96-well plates (Corning, USA). Then, the mixture was inoculated into the Vero cell monolayers of a 96-well tissue culture plate, washed thrice with PBS and incubated at 37 °C for 1 h. After incubation, the mixture was discarded and washed thrice with PBS. Next, maintenance medium containing trypsin (20 μg/mL) was added to each well, and the plate was incubated for 5 days at 37 ℃. Neutralizing antibody titers were calculated as the reciprocal of the highest serum dilution that inhibited CPE.
2.13. Statistical analysis
Statistical significance among the different experimental groups was determined using one-way ANOVA. Differences were considered significant when the P value was less than 0.05.
3. Results
3.1. Isolation and biological characteristics of the CH/HNPJ/2017 strain
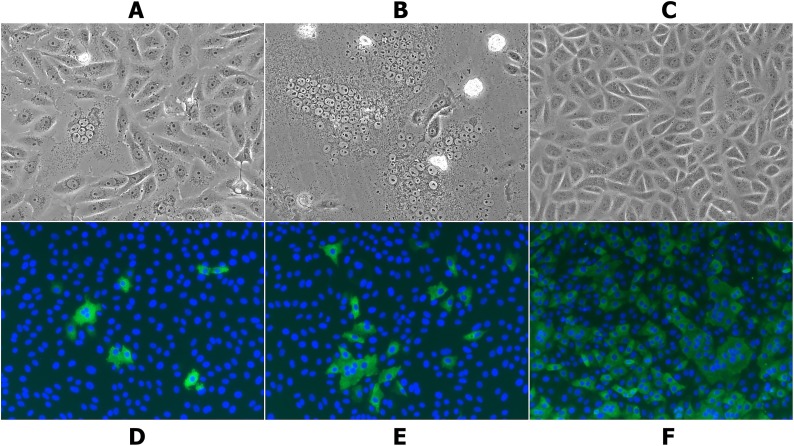
PEDV-positive samples were inoculated in Vero cells at a 10-fold dilution, and visible CPE were observed in the first passage 24 h after inoculation (Fig. 1 A). Complete CPE were observed at 72 h (Fig. 1B). Compared with uninoculated Vero cells, the PEDV-inoculated Vero cells were characterized by cell fusion with syncytial and vacuole formation in the initial stage, followed by shrinkage, detachment, and a lack of motility in the final stage (Fig. 1A–C). In the present study, the CH/HNPJ/2017 strain was isolated and serially propagated in Vero cells for up to the 90 passages. Furthermore, virus propagation of the 50th passage of the CH/HNPJ/2017 strain and cell nuclei was confirmed by detecting PEDV antigens with IFA using PEDV N-specific MAbs and DAPI. Distinct green signals were observed in the PEDV CH/HNPJ/2017 strain-infected Vero cells but not in the uninfected Vero cells beginning at 6 h; these signals tended to enhance significantly over time (12 h and 24 h) (Fig. 1D–F).
Fig. 1.
Isolation and detection of the PEDV CH/HNPJ/2017 strain in Vero cells. The upper and lower panels show light and immunofluorescence images, respectively, of Vero cells infected with the PEDV CH/HNPJ/2017 strain. (A) Cytopathic effects (CPE) caused by the 1st passage of the PEDV CH/HNPJ/2017 strain at 24 h after inoculation (400×). Vero cells were seeded into T-25 flasks and infected at a multiplicity of infection (MOI) of 0.1; (B) CPE at 72 h after inoculation; (C) Control (uninfected) Vero cells; (D) Immunofluorescence detection results for the 50th passage of the PEDV CH/HNPJ/2017 strain in infected Vero cells at 6 h after inoculation (400×). PEDV antigens and nuclei were detected with mouse anti-PEDV N protein monoclonal antibodies (McAbs) and 4′, 6-diamidino-2-phenylindole (DAPI), respectively; (E) Immunofluorescence detection results at 12 h after inoculation (400×); (F) Immunofluorescence detection results at 24 h after inoculation (400×).
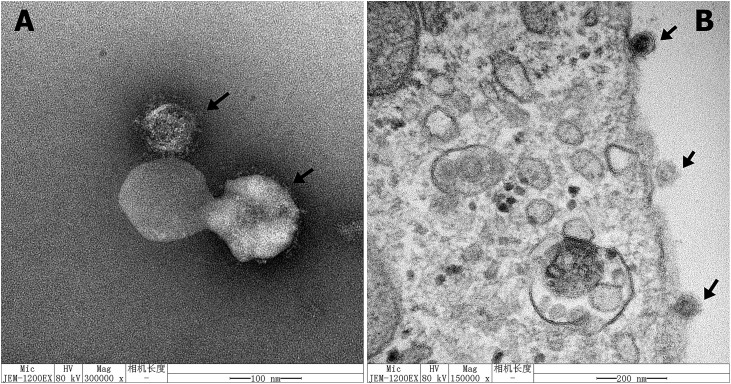
PEDV particles from the CH/HNPJ/2017 strain in Vero cell culture media and in infected Vero cells were imaged with transmission electron microscopy (TEM). As shown in Fig. 2 , the typical coronavirus particles were observed in the cell culture media (Fig. 2A) and in surface sections of PEDV-infected Vero cells (Fig. 2B). The virion was circular in shape and 80–120 nm in diameter, with surface projections characteristic of coronaviruses. The viral titers of the serially passaged PEDV CH/HNPJ/2017 strain were determined as the TCID50 at 10-passage intervals. The infectious titer of the cell-adapted virus ranged from 103·5 to 107.88 TCID50/mL (Fig. 3 ).
Fig. 2.
Electron micrograph of PEDV virions in cell culture media of infected Vero cells or on the cell surface of infected Vero cells. (A) Images of PEDV virions from cell culture media of Vero cells infected with the PEDV CH/HNPJ/2017 strain, as shown by the arrow. Scale bar = 100 nm; (B) Images of a PEDV-infected Vero cell. PEDV particles (arrow heads) on the cell surface of an infected Vero cell, as shown by the arrow. Scale bar = 200 nm.
Fig. 3.
Viral titers of the PEDV CH/HNPJ/2017 strain propagated in Vero cells after serial passage. All the results of a representative experiment performed with triplicate samples are shown.
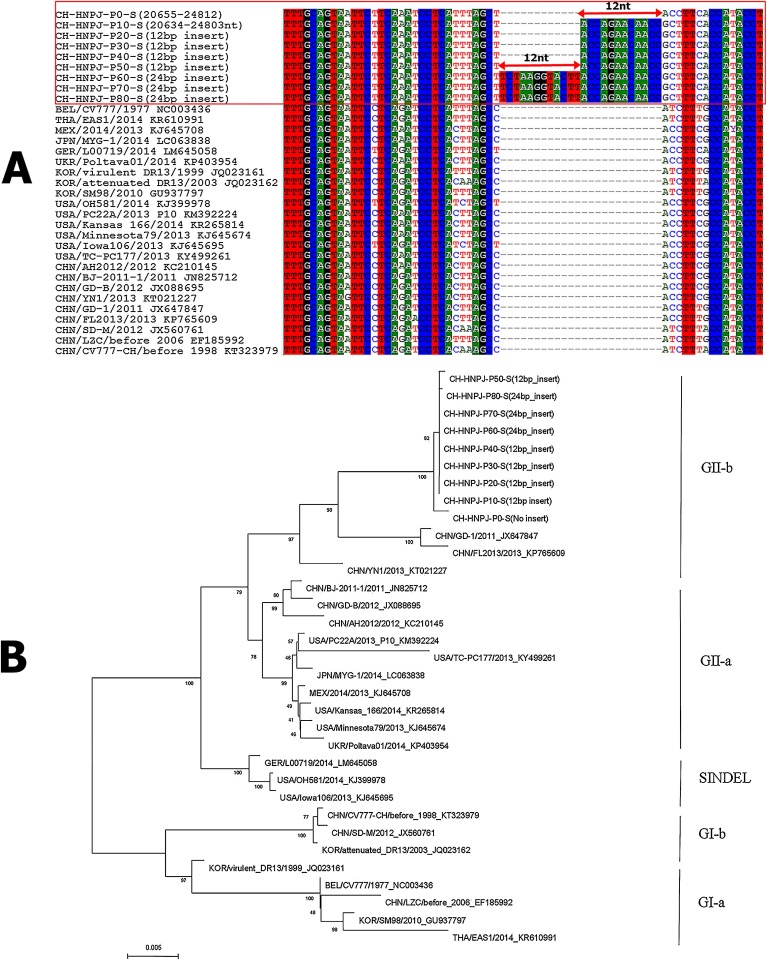
The results of the sequence analysis showed that compared with the S genes of classical or attenuated PEDVs, such as the CV777 strain, those of the parental (P0) and all cell culture-passaged CH/HNPJ/2017 strains (P1, P10, P20, P30, P40, P50, P60, P70 and P80) contained typical insertions and deletions that have also been found in other recently isolated highly virulent PEDV strains from China and other countries (data not shown). However, most importantly, two neighboring unique insertion mutations (12 bp in each, at positions nt 1074-1075 of the parental P0 virus) that resulted in four amino acid changes occurred in the S1 region of passages P10 and P60 (Fig. 4 A). In addition, these two unique insertion mutations existed only in cell culture-passaged CH/HNPJ/2017 strains but not in all the other strains isolated from China or other countries to date. This may be the result of the virus adapting to the host cell; however, its functions needs to be studied further using reverse genetics in the future. Additional phylogenetic analyses based on the full-length S gene indicated that all the cell culture-passaged CH/HNPJ/2017 strains could be divided into subgroup IIb along with the parental virus and global epizootic strains (Fig. 4B).
Fig. 4.
Nucleotide alignment and phylogenetic analysis of the S gene of the CH/HNPJ/2017 strain. (A) Alignments of the complete S gene of the parental (P0) and all cell culture-passaged CH/HNPJ/2017 strains with 10 generation intervals (P1, P10, P20, P30, P40, P50, P60, P70 and P80) and the reference sequence of the S gene of other PEDV strains are shown. The red solid box represents the CH/HNPJ/2017 sequence. The double-headed red arrows indicate inserted sequences. (B) Phylogenetic analysis based on the nucleotide sequences of the S gene of the parental (P0) and all cell culture-passaged CH/HNPJ/2017 strains with 10 generation intervals. The tree was constructed with the neighbor-joining method, and bootstrap values from 1000 resamplings are shown for each node. The country origin of the strains, years, GenBank accession numbers, genogroups and subgroups proposed in this study are shown (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.2. Pathogenicity of the CH/HNPJ/2017 strain
To determine the infectious titer and virulence of virus isolates, the PDD50 was determined by using 4-day-old conventional suckling pigs. Before inoculation, all piglets were lively, showed no clinical symptoms, had normal fecal consistency, and had no PEDV genetic material detected in fecal samples by PEDV-specific real-time PCR. During the whole experiment period, none of the negative control group piglets developed typical clinical signs of PEDV, and all their rectal swab samples were negative for PEDV RNA fecal shedding based on real-time PCR. AT 72 post-inoculation hours (PIH), viral RNA fecal shedding in samples from rectal swabs or intestinal contents became positive in G1–G4 piglets (10−5–10−8 diluted virus), with cycle threshold (CT) values ranging from 17.30 to 25.96 (Table 1). In detail, 100% (5/5) of pigs in G1 and G2, 80% (4/5) in G3, and 40% (2/5) in G4 had diarrhea, and no pigs (0/5) in G5 (10−9 diluted virus) or the control groups had diarrhea (Table 1). The cut-off timepoint was set as 72 PIH for the determination of the PDD50, which was 7.68 log10PDD50/3 mL. This value was much different than the TCID50 titer (104.29 TCID50/mL).
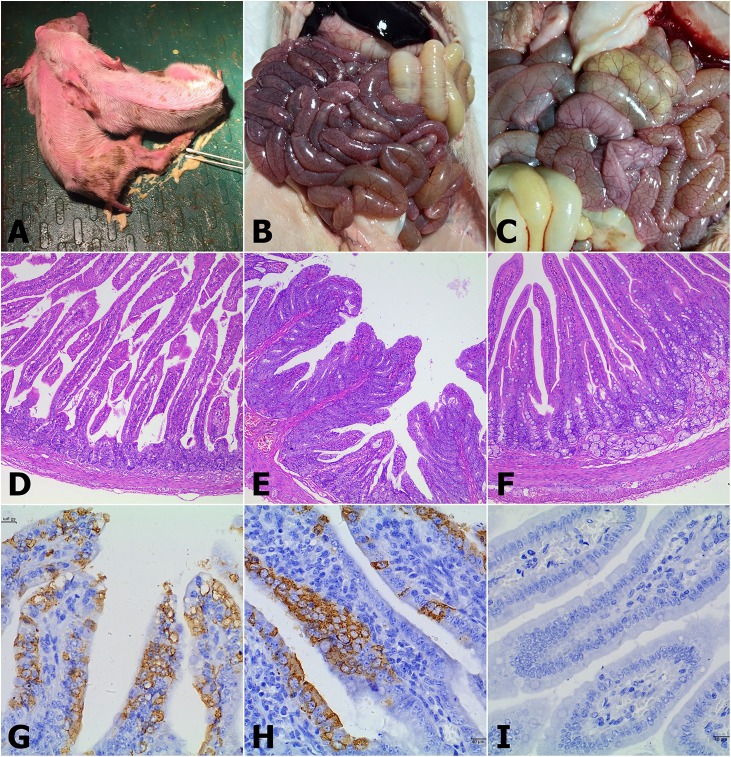
All of the diarrheal piglets in G1–G4 were anorexic and depressed, and their feces were watery (Fig. 5 A). At necropsy, typical macroscopic PED-like lesions were found in the intestinal tissues, especially in the small intestines; the intestinal tract was distended and transparent and filled with yellow fluid, and mesenteric congestion was present (Fig. 5B, C). The results of histopathological examination showed viral enteritis in all the diarrheal piglets, including vacuolation of the small intestinal enterocytes and shortening, fusion and sloughing of the small intestinal villi (Fig. 5D, E). The mock control group piglets exhibited normal intestinal histopathology (Fig. 5F). Furthermore, immunohistochemical examinations revealed that PEDV antigens were predominant in the cytoplasm of the epithelial cells of atrophied villi in some segments of the small intestines (Fig. 5G, H). No PEDV antigens were present in the small intestines in any piglets in the negative control group (Fig. 5I).
Fig. 5.
Histopathology and immunohistochemistry (IHC) of the small intestines of piglets inoculated with P4 of the cell culture-adapted Chinese PEDV CH/HNPJ/2017 strain (104.29 TCID50/mL). (A) The infected pigs were anorexic and depressed and had severe diarrhea. (B–C) Necropsy examinations of CH/HNPJ/2017-infected piglets. Severe hyperemia, swelling and transparency were present in the intestinal tissues, especially in the small intestines. (D–F) Hematoxylin and eosin-stained tissue sections of jejunum from CH/HNPJ/2017-infected and mock control piglets (100 × magnification). (D) Severe villous abruption was observed in the infected piglets. (E) Severe villous atrophy and fusion and degeneration and necrosis of mucosal epithelial cells were observed in the infected piglets. (F) The normal villous epithelium of the jejunum from mock control piglets. (G–I) Detection of PEDV antigen by IHC analysis of jejunal tissue sections from CH/HNPJ/2017-infected and mock control piglets (400 × magnification). (G–H) PEDV antigen signals appear brown and were detected in jejunal epithelial cells from CH/HNPJ/2017-infected piglets. (I) No PEDV antigen was detected in jejunum from mock control piglets.
3.3. Immune protective effect of the CH/HNPJ/2017 strain as an inactivated vaccine candidate
Virus inactivation was verified by the absence of viral growth in Vero cell cultures and viral shedding in inoculated piglets. Thus, no typical PEDV-like CPE appeared in Vero cell cultures within 96 h after inoculation, and none of the piglets had diarrhea within 7 days after inoculation (data not shown). Further test results using real-time PCR showed no PEDV RNA in the harvested cell cultures and rectal swab fecal samples from pigs (data not shown). These results indicated that the virus was completely inactivated, and no live virus was present in the prepared vaccine.
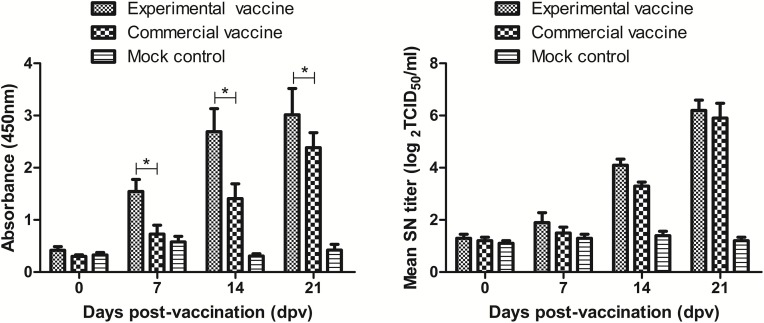
The levels of PEDV-specific IgG and neutralizing antibodies in serum induced by the experimental vaccines were assessed with indirect ELISA (I-ELISA) and SN, respectively. The I-ELISA results showed that the experimental inactivated vaccine and commercial inactivated vaccine induced significant PEDV-specific serum IgG responses to PEDV compared with PBS (P < 0.05, Fig. 6 A). The mean IgG levels in the two vaccine groups increased continuously from 7 dpv and were significantly higher than those in the mock control group (Fig. 6A). In addition, importantly, the mean IgG levels were significantly higher in the experimental inactivated vaccine group than in the commercial inactivated vaccine group at 7, 14 and 21 dpv (P < 0.05, Fig. 6A), indicating that the experimental inactivated vaccine induced a stronger PEDV-specific IgG response than the commercial inactivated vaccine in pigs. The SN titers also increased continuously in the two vaccine groups from 7 dpv and were significantly higher than those in the mock control group (P < 0.05, Fig. 6B). However, there was no significant difference between the experimental inactivated vaccine and commercial inactivated vaccine at 7 and 21 dpv (P > 0.05, Fig. 6B), except for at 14 dpv (P < 0.05, Fig. 6B). These findings indicated that the experimental inactivated vaccine induced a stronger humoral immune response in pigs.
Fig. 6.
Antibody responses against PEDV from serum samples. Pigs were vaccinated at 0 days post vaccination (dpv) and then challenged orally with 4 mL of 100 TCID50 of cell culture-adapted CH/HNPJ/2017 P4 at 21 dpv. Serum samples were collected at 0, 7, 14 and 21 dpv. (A) Antibody responses against PEDV from serum samples from pigs using IgG-specific ELISA before challenge exposure. At each serum collection time point, the number of IgG-specific antibody positive pigs for experimental vaccine group experimental vaccine, commercial vaccine and mock control group as following: at 0 dpv, all three of group are 0/5; at 7 dpv, three of group are respectively 3/5, 0/5 and 0/5; at 14 dpv, three of group are respectively 4/5, 2/5 and 0/5; at 21 dpv, three of group are respectively 5/5, 5/5 and 0/5; The asterisk means significant difference (P < 0.05). (B) Antibody responses against PEDV from serum samples from pigs using the SN test before challenge exposure.
All three groups of pigs were apparently healthy and had no clinical symptoms before oral challenge. At 21 dpv, all pigs were challenged orally with 4 mL of 100 TCID50 of cell culture-adapted CH/HNPJ/2017 P4. During 10 day after challenge, the viral RNA fecal shedding in rectal swabs samples from ten diarrheal pigs (the experimental inactivated vaccine, the commercial inactivated vaccine and mock control group were 2, 3 and 5 pigs, respectively) became positive, with cycle threshold (CT) values ranging from 15.25 to 22.58. Therefore, in the experimental inactivated vaccine group and the commercial inactivated vaccine group, 60% (3/5) and 40% (2/5) of pigs, respectively, were protected during observation period. The protective efficacy of the experimental inactivated vaccine was better than that of the commercial inactivated vaccine. All pigs in the mock control group had diarrhea within 72 h.
4. Discussion
Although PEDV infection did occur in Chinese pig farms from the time of first report in 1973 until October 2010, outbreaks were sporadic and regional, and there were no reports of large-scale outbreaks (Sun et al., 2016). However, a large-scale outbreak clinically characterized by acute diarrhea and a high mortality rate of 80–100% among piglets occurred in the southern provinces in October 2010 and soon swept throughout the country, causing enormous economic losses (Li et al., 2012; Sun et al., 2012; Tian et al., 2014; Wang et al., 2013). Currently, PED outbreaks in China are caused by field epidemic G2 variant strains that differ genetically from the classical PEDV strain, CV777 (Lee, 2015). Although PEDV vaccines based on the classical PEDV strain CV777, such as the inactivated, bivalent transmissible gastroenteritis virus (TGEV) and PEDV vaccine (1999 to present) and the attenuated, bivalent TGEV and PEDV vaccine (2003–2006), have been widely used in the Chinese pig population, PED outbreaks have still occurred in vaccinated herds with a high mortality rate in neonatal piglets, indicating that vaccines derived from classical strains cannot provide adequate immune protection against the currently prevalent strains (GII genogroup) (Ayudhya et al., 2012). This failure of protection may be caused by dramatic mutation of the virus, which poses a major challenge for the prevention and control of PED in China (Wang et al., 2016). In addition to molecular evidence, antigenic variations between classical and emerging highly virulent (non-S INDEL) PEDV strains were demonstrated in several serological cross reactivity assays (Kim et al., 2015; Lin et al., 2015a, 2015b; Wang et al., 2015). There are also some studies showed that the highly virulent (non-S INDEL) PEDV strains could provide piglets against homologous challenge (Baek et al., 2016; Lee et al., 2018; Lin et al., 2016b ; Park et al., 2018). Therefore, there is an urgent need for more research aimed at understanding the biological characteristics, pathogenicity, and immune protective effects of the field prevalent strain in the GII genogroup. For this purpose, the epidemic variant strain CH/HNPJ/2017 (GenBank accession number: MF152604) belonging to the GIIb genogroup was isolated in Vero cells and its biological characteristics were ascertained in the present study. The CH/HNPJ/2017 strain was serially propagated in Vero cells for up to the 90 passages. We are currently continuing serial passages of the CH/HNPJ/2017 strain in an effort to develop a live attenuated PEDV vaccine that exerts better protective effects than the currently available vaccines against GII epidemic PEDV in China.
The viral titers of the parental and serially passaged PEDV strain CH/HNPJ/2017 revealed that the cell-adapted CH/HNPJ/2017 strain grew more efficiently and had higher titers than the parental virus in Vero cells. The viral titers were higher than 107.88 TCID50/mL at 30 passages, indicating that the cell-adapted CH/HNPJ/2017 strain had superior viral fitness in Vero cells. In addition, sequencing results of the S gene showed two neighboring unique insertion mutations (12 bp in each, at positions nt 1074-1075 of the parental P0 virus) in the S1 region of passages P10 and P60. These mutations can be explained as a survival mechanism by which the viruses adapted to the host cells. The mechanism of these mutations will be clarified in our future study by reverse genetics.
Based on epidemiological and clinical observations in the field since 2010, the emerging non-S insertion-deletion (INDEL) PEDV strains are highly pathogenic (Lin et al., 2016a). Experimental infection with highly virulent PEDV strains in different types of pigs resulted in consistent outcomes and conclusions (Chen et al., 2016; Lin et al., 2015a, 2015b, 2016a; Liu et al., 2015; Madson et al., 2016, 2014; Thomas et al., 2015). In this study, the infectious titer and virulence of the fourth passage of the cell culture-adapted CH/HNPJ/2017 strain (104.29 TCID50/mL) was determined by using 4-day-old conventional suckling pigs. At 72 PIH, viral RNA fecal shedding identified in rectal swab samples or samples of intestinal content became positive in G1–G4 piglets (10−5–10−8 diluted virus), with CT values ranging from 17.30 to 25.96 (Table 1). The PDD50 was determined to be 7.68 log10PDD50/3 mL, and it similar to that of the US highly virulent PEDV PC22A strain (7.83 log10PDD50/3 mL, 24 PIH), indicating that the Chinese PEDV CH/HNPJ/2017 strain is highly virulent as well. In addition, highly virulent PEDV strains can induce severe villous atrophy (Lin et al., 2016a). Similar pathological findings were also presented in the intestinal tissues of piglets infected with the CH/HNPJ/2017 strain. On histopathological examination, viral enteritis was present in all diarrheal piglets, as evidenced by vacuolation of the small intestinal enterocytes and shortening, fusion and sloughing of the small intestinal villi (Fig. 5D, E).
Pregnant sows are suitable models for evaluating the effectiveness of inactivated PEDV vaccines in target populations, such as neonatal piglets, because transfer of maternal antibodies to piglets via colostrum may play an important role in the development of protective immunity against PEDV in suckling piglets (Goede et al., 2015). However, pregnant sows usually cannot be used in vaccination experiments because of breeding conditions and because experiment numbers and experimental expenditure are limited. Therefore, in this study, we used piglets to evaluate the protective efficacy of our experimental inactivated vaccine because pigs of all ages are susceptible to PEDV infection (Lee et al., 2018). The detection of serum antibodies showed that the experimental inactivated vaccine induced significant PEDV-specific serum IgG and neutralizing antibody responses to PEDV compared with mock control at 7, 14 and 21 dpv (P < 0.05, Fig. 6), indicating that the experimental inactivated vaccine had better immunogenicity and could induce a stronger PEDV-specific antibody response than the commercial inactivated vaccine in pigs. Furthermore, the challenge study showed that the experimental inactivated vaccine could protect 60% (3/5) of pigs against 100 TCID50 of cell culture-adapted CH/HNPJ/2017 P4 challenge. The protective efficacy of the experimental inactivated vaccine was better than that of the commercial inactivated vaccine, indicating that the experimental inactivated vaccine could be used as a good vaccine candidate in future clinical practice. Maternally derived lactogenic immunity (IgG and IgA) plays an important role in protecting neonatal piglets against PEDV infection (Song et al., 2015). Therefore, to achieve this goal, sow vaccination and challenge experiments will be carried out to evaluate the protective efficacy of the vaccine candidate against virulent PEDV and to assess any safety concerns for use in neonatal piglets in our future studies.
5. Conclusions
In conclusion, we successfully isolated and identified a novel Chinese virulent genotype GIIb PEDV CH/HNPJ/2017 strain that had unique variations in the S gene in cell-adapted passages. The CH/HNPJ/2017 strain has good cellular adaptation to Vero cells and is highly virulent in piglets. To our knowledge, this is the first report to describe the PDD50 of a Chinese PEDV strain, and the results will be very helpful for future pig infection experiments. In addition, the CH/HNPJ/2017 strain had better immune protective effects as an inactivated vaccine candidate than the commercially available vaccine. The results of this study are important for understanding the characteristics of Chinese PEDV and for the development of novel effective vaccines against PED in China.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 31602095), the National Key Research and Development Program (2016YFD0501505), the China Agriculture Research System (CARS-35) and the Central Public Interest Scientific Institution Basal Research Fund (Y2016CG23).
Contributor Information
Xinsheng Liu, Email: liuxinsheng@caas.cn.
Qiaoling Zhang, Email: 876835192@qq.com.
Liping Zhang, Email: zhanglp2319@163.com.
Peng Zhou, Email: zhoupeng02@caas.cn.
Jun Yang, Email: yangjunpro@163.com.
Yuzhen Fang, Email: fangyuzhen@caas.cn.
Zhaoliang Dong, Email: dzlgsdx@163.com.
Donghong Zhao, Email: zhaodonghong123@163.com.
Weiyan Li, Email: liweiyan160129@163.com.
Jiaxin Feng, Email: fengjiaxin0524@163.com.
Baofeng Cui, Email: 33644264@163.com.
Yongguang Zhang, Email: zhangyongguang@caas.cn.
Yonglu Wang, Email: wangyonglumd@hotmail.com.
References
- Ayudhya S.N., Assavacheep P., Thanawongnuwech R. One world--one health: the threat of emerging swine diseases. An Asian perspective. Transbound. Emerg. Dis. 2012;59(Suppl. 1):9–17. doi: 10.1111/j.1865-1682.2011.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek P.S., Choi H.W., Lee S., Yoon I.J., Lee Y.J., Lee du S., Lee S., Lee C. Efficacy of an inactivated genotype 2b porcine epidemic diarrhea virus vaccine in neonatal piglets. Vet. Immunol. Immunopathol. 2016;174:45–49. doi: 10.1016/j.vetimm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasey D., Cartwright S.F. Virus-like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978;25(2):255–256. doi: 10.1016/S0034-5288(18)32994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155(9):1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H.Y., Zheng Y., Li G.W., Zhang J.Q. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J Gen. Virol. 2016;97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Cheun-Arom T., Temeeyasen G., Tripipat T., Kaewprommal P., Piriyapongsa J., Sukrong S., Chongcharoen W., Tantituvanont A., Nilubol D. Full-length genome analysis of two genetically distinct variants of porcine epidemic diarrhea virus in Thailand. Infect. Genet. Evol. 2016;44:114–121. doi: 10.1016/j.meegid.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury B., Dastjerdi A., Doyle N., Frossard J.P., Steinbach F. From the field to the lab - an European view on the global spread of PEDV. Virus Res. 2016;226:40–49. doi: 10.1016/j.virusres.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.C., Lee J.H., Nguyen V.G., Huynh T.M.L., Lee G.E., Moon H.J., Park S.J., Kim H.K., Park B.K. New emergence pattern with variant porcine epidemic diarrhea viruses, South Korea, 2012–2015. Virus Res. 2016;226:14–19. doi: 10.1016/j.virusres.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam F., Zhang J., Zhang C. Hepatitis B core antigen based novel vaccine against porcine epidemic diarrhea virus. J. Virol. Methods. 2018;253:61–69. doi: 10.1016/j.jviromet.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a "mild" strain of porcine epidemic diarrhea virus confers protection against infection with a "severe" strain. Vet. Microbiol. 2015;176(1-2):161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Hou Y., Lin C.M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J., Wang Q. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017;91(14) doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.J., Liu H.Y., Qian Y.Q., Wang Z.J., Tang Q.J., Cao W.M. Study on the transmissible gastroenteritis coronavirus. Agric. Sci. Technol. Shanghai. 1980;2:42–45. (in Chinese) [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4(5):e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204(2):134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lee J.M., Jung J., Kim I.J., Hyun B.H., Kim H.I., Park C.K., Oem J.K., Kim Y.H., Lee M.H., Lee K.K. Genetic characterization of porcine epidemic diarrhea virus in Korea from 1998 to 2013. Arch. Virol. 2015;160(4):1055–1064. doi: 10.1007/s00705-015-2353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Son K.Y., Noh Y.H., Lee S.C., Choi H.W., Yoon I.J., Lee C. Genetic characteristics, pathogenicity, and immunogenicity associated with cell adaptation of a virulent genotype 2b porcine epidemic diarrhea virus. Vet. Microbiol. 2017;207:248–258. doi: 10.1016/j.vetmic.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Yang D.K., Kim H.H., Cho I.S. Efficacy of inactivated variant porcine epidemic diarrhea virus vaccines in growing pigs. Clin. Exp. Vaccine Res. 2018;7(1):61–69. doi: 10.7774/cevr.2018.7.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18(8):1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.N., Chung W.B., Chang S.W., Wen C.C., Liu H., Chien C.H., Chiou M.T. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013–2014. J. Vet. Med. Sci. 2014;76(9):1297–1299. doi: 10.1292/jvms.14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Annamalai T., Liu X., Gao X., Lu Z., El-Tholoth M., Hu H., Saif L.J., Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 2015;46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q., Saif L.J. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J. Virol. 2015;89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Chen L., Gao L., Yuan X., Ma Z., Fan H. Epidemic strain YC2014 of porcine epidemic diarrhea virus could provide piglets against homologous challenge. Virol. J. 2016;13:68. doi: 10.1186/s12985-016-0529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Hou Y., Marthaler D.G., Gao X., Liu X., Zheng L., Saif L.J., Wang Q. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet. Microbiol. 2017;201:62–71. doi: 10.1016/j.vetmic.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lin C.M., Annamalai T., Gao X., Lu Z., Esseili M.A., Jung K., El-Tholoth M., Saif L.J., Wang Q. Determination of the infectious titer and virulence of an original US porcine epidemic diarrhea virus PC22A strain. Vet. Res. 2015;46:109. doi: 10.1186/s13567-015-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson D.M., Magstadt D.R., Arruda P.H., Hoang H., Sun D., Bower L.P., Bhandari M., Burrough E.R., Gauger P.C., Pillatzki A.E., Stevenson G.W., Wilberts B.L., Brodie J., Harmon K.M., Wang C., Main R.G., Zhang J., Yoon K.J. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet. Microbiol. 2014;174(1–2):60–68. doi: 10.1016/j.vetmic.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Madson D.M., Arruda P.H., Magstadt D.R., Burrough E.R., Hoang H., Sun D., Bower L.P., Bhandari M., Gauger P.C., Stevenson G.W., Wilberts B.L., Wang C., Zhang J., Yoon K.J. Characterization of porcine epidemic diarrhea virus isolate US/Iowa/18984/2013 infection in 1-day-old cesarean-derived colostrum-deprived piglets. Vet. Pathol. 2016;53(1):44–52. doi: 10.1177/0300985815591080. [DOI] [PubMed] [Google Scholar]
- Niederwerder M.C., Hesse R.A. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018;65(3):660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173(3-4):258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraguison-Alili R., Domingo C.Y. Phylogenetic tracking of current porcine epidemic diarrhea virus (PEDV) strains in the Philippines. Arch. Virol. 2016;161(9):2601–2604. doi: 10.1007/s00705-016-2938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Kang K.J., Ryu J.H., Park J.Y., Jang H., Sung D.J., Kang J.G., Shin H.J. Porcine epidemic diarrhea vaccine evaluation using a newly isolated strain from Korea. Vet. Microbiol. 2018;221:19–26. doi: 10.1016/j.vetmic.2018.05.012. [DOI] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., Martelli P. Porcine epidemic diarrhea: a retrospect from Europe and matters of debate. Virus Res. 2016;226:1–6. doi: 10.1016/j.virusres.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takeyama N., Katsumata A., Tuchiya K., Kodama T., Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011;43(1):72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Oh J.S., Kang B.K., Yang J.S., Moon H.J., Yoo H.S., Jang Y.S., Park B.K. Oral efficacy of vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82(1):134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin. Exp. Vaccine Res. 2015;4(2):166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B.A., Siddell S.G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler J., Zoels S., Fux R., Hanke D., Pohlmann A., Blome S., Weissenbock H., Weissenbacher-Lang C., Ritzmann M., Ladinig A. Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Vet. Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wang X., Wei S., Chen J., Feng L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet. Med. Sci. 2016;78(3):355–363. doi: 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.T., Chen Q., Gauger P.C., Gimenez-Lirola L.G., Sinha A., Harmon K.M., Madson D.M., Burrough E.R., Magstadt D.R., Salzbrenner H.M., Welch M.W., Yoon K.J., Zimmerman J.J., Zhang J.Q. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P.F., Jin Y.L., Xing G., Qv L.L., Huang Y.W., Zhou J.Y. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg. Infect. Dis. 2014;20(10):1735–1738. doi: 10.3201/eid2010.140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Diep N., Norimine J., Sueyoshi M., Lan N.T., Hirai T., Yamaguchi R. US-like isolates of porcine epidemic diarrhea virus from Japanese outbreaks between 2013 and 2014. SpringerPlus. 2015;4:756. doi: 10.1186/s40064-015-1552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K., Pensaert M. Prevalence of infections with enzootic respiratory and enteric viruses in feeder pigs entering fattening herds. Vet. Rec. 1994;135(25):594–597. [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586(4):384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.M., Niu B.B., Yan H., Gao D.S., Yang X., Chen L., Chang H.T., Zhao J., Wang C.Q. Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch. Virol. 2013;158(12):2487–2494. doi: 10.1007/s00705-013-1767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J., Shi D., Shi H., Zhang X., Yuan J., Jiang S., Feng L. Immunogenicity and antigenic relationships among spike proteins of porcine epidemic diarrhea virus subtypes G1 and G2. Arch. Virol. 2015;161:537–547. doi: 10.1007/s00705-015-2694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan H., Xing D.K., Wang D.Y., Zhu W.Z., Zhao F.Y., Gong H.J., Fei S.G. Study on the culture of porcine epidemic diarrhea virus adapted to fetal porcine intestine primary cell monolayer. Chin. J. Vet. Sci. 1984;4(3):202–208. (in Chinese) [Google Scholar]
- Yang F., Liu X., Zhou Y., Lyu W., Xu S., Xu Z., Zhu L. Histopathology of porcine kobuvirus in Chinese piglets. Virol. Sin. 2015;30(5):396–399. doi: 10.1007/s12250-015-3608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]