Summary
Numerous interventions are in clinical development for respiratory syncytial virus (RSV) infection, including small molecules that target viral transcription and replication. These processes are catalyzed by a complex comprising the RNA-dependent RNA polymerase (L) and the tetrameric phosphoprotein (P). RSV P recruits multiple proteins to the polymerase complex and, with the exception of its oligomerization domain, is thought to be intrinsically disordered. Despite their critical roles in RSV transcription and replication, structures of L and P have remained elusive. Here, we describe the 3.2-Å cryo-EM structure of RSV L bound to tetrameric P. The structure reveals a striking tentacular arrangement of P, with each of the four monomers adopting a distinct conformation. The structure also rationalizes inhibitor escape mutants and mutations observed in live-attenuated vaccine candidates. These results provide a framework for determining the molecular underpinnings of RSV replication and transcription and should facilitate the design of effective RSV inhibitors.
Keywords: RdRp, PRNTase, MTase, allostery, ALS-8176
Graphical Abstract

Highlights
-
•
Cryo-EM structure of RSV L bound by tetrameric RSV P solved to 3.2 Å
-
•
P tetramer adopts an asymmetric tentacular arrangement when bound to L
-
•
L priming loop adopts elongation-compatible state without PRNTase-RdRp separation
-
•
Structure rationalizes escape from small-molecule antivirals
Respiratory syncytial virus (RSV) remains a leading cause of bronchiolitis and hospitalization, especially of infants. Gilman et al. present a 3.2-Å cryo-EM structure of the RSV L polymerase in complex with the P phosphoprotein—components of the core viral replication machinery that represent an attractive target for the development of therapeutic agents.
Introduction
Respiratory syncytial virus (RSV) is a ubiquitous viral pathogen that infects nearly all children before 2 years of age (Glezen et al., 1986). It is estimated that RSV causes more than 60,000 in-hospital deaths per year in children less than 5 years old, with the majority of deaths occurring in developing regions (Shi et al., 2017). In addition, RSV infection can lead to serious complications in the elderly and high-risk adults and causes a disease burden similar to non-pandemic influenza A in these populations (Falsey et al., 2005). Currently, the only clinical intervention for RSV is passive prophylaxis with the monoclonal antibody Synagis, which reduces RSV-related hospitalization (The IMpact-RSV Study Group, 1998). However, the high cost and moderate efficacy of Synagis restrict its use to high-risk infants in developed regions of the world (Homaira et al., 2014, Shahabi et al., 2018, The IMpact-RSV Study Group, 1998). Thus, there is great interest in developing more effective therapeutic agents for RSV, including small-molecule antivirals.
RSV is an enveloped, negative-sense, single-stranded RNA virus that belongs to the Pneumoviridae family (Afonso et al., 2016). Only three viral proteins are absolutely required for replication of the RSV genome: the nucleoprotein (N), the large protein (L), and the phosphoprotein (P) (Grosfeld et al., 1995, Yu et al., 1995). N associates with viral RNA to form a tightly woven helical assembly that protects the RNA from cellular nucleases and recognition by the innate immune system (Bakker et al., 2013, Tawar et al., 2009). L harbors three conserved enzymatic domains: the RNA-dependent RNA polymerase (RdRp) domain, the polyribonucleotidyl transferase (PRNTase or capping) domain, and the methyltransferase (MTase) domain, which catalyzes cap methylation. These activities are all potential targets for inhibitor development. Nucleoside analogs that terminate RNA chain synthesis have been identified (Clarke et al., 2015, Deval et al., 2015, Wang et al., 2015), and one such compound, ALS-8176, has shown efficacy in RSV-infected adults (DeVincenzo et al., 2015). Several non-nucleoside small-molecule inhibitors have also been identified (Cockerill et al., 2019, Fearns and Deval, 2016), and although some (for example, BI-compound D) are known to disrupt RNA cap addition (Liuzzi et al., 2005), there are additional inhibitor classes for which the mechanism of action is not well understood (Duvall et al., 2016, McCutcheon et al., 2015).
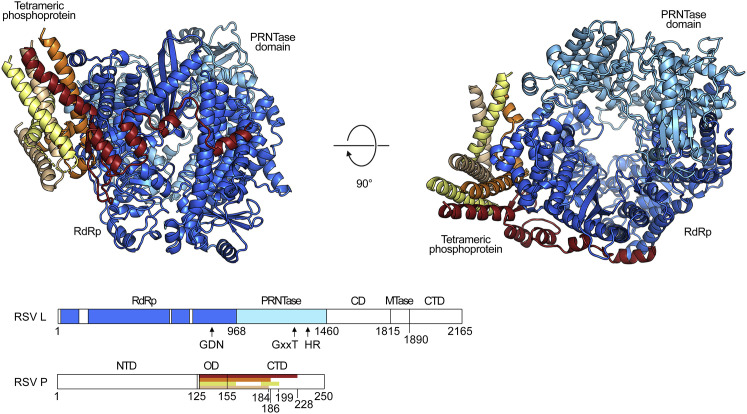
P serves as an essential polymerase cofactor that tethers L to the nucleoprotein-RNA complex (García et al., 1993, Grosfeld et al., 1995, Yu et al., 1995). P also acts as a chaperone that prevents the association of nascent N (N0) with host cell RNAs (Galloux et al., 2015, Pereira et al., 2017, Tran et al., 2007) and is responsible for recruiting the M2-1 protein, a processivity factor that is required for efficient transcription of viral RNA (Blondot et al., 2012, Collins et al., 1996, Mason et al., 2003). In addition, P recruits the cellular phosphatase PP1 to inclusion bodies to regulate viral transcription (Richard et al., 2018). Thus, P plays critical roles in regulating RNA replication and transcription through its interactions with multiple proteins. Structurally, RSV P contains a central oligomerization domain that is predicted to form a tetrameric coiled coil (Castagné et al., 2004, Llorente et al., 2008). Regions N- and C-terminal to the oligomerization domain are predicted to be intrinsically disordered and may only adopt defined conformations when bound to other proteins. The dynamic nature of RSV P has prevented determination of its structure (Pereira et al., 2017, Simabuco et al., 2011); thus, the molecular mechanisms by which P coordinates the activities of diverse viral components are not well understood.
To gain atomic-level information regarding RSV transcription and replication, we initiated structural studies of a purified polymerase complex comprising L and P. The resulting 3.2-Å cryoelectron microscopy (cryo-EM) structure reveals that P displays unique structural plasticity, with each monomer adopting a different conformation as it interacts with distinct regions on L. The variability in secondary structure of each P monomer indicates that this viral phosphoprotein exhibits characteristics of a “transformer” protein (Knauer et al., 2012). In addition, the surface on the RSV L RdRp domain that is recognized by RSV P is similar to the region bound by the ribosomal S1 protein cofactor on the RdRp of the distantly related Qβ bacteriophage polymerase, indicating that this interaction may be evolutionarily conserved. Our results also provide the first structural description of the RdRp and capping domains of RSV L and provide insights into the mechanism of viral escape from nucleoside analog and non-nucleoside inhibitors. Collectively, these results inform our understanding of viral polymerase complexes and should aid in the development of next-generation inhibitors of RSV infection.
Results
RSV L Co-expressed with RSV P Is Biochemically Active
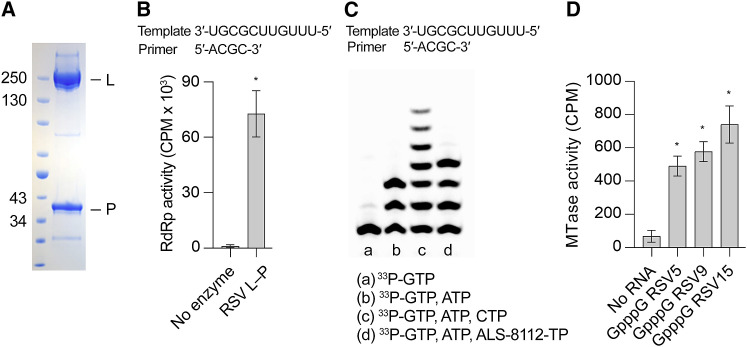
Human RSV L was co-expressed with RSV P in Sf9 cells. The purified protein was analyzed by size exclusion chromatography, and SDS-PAGE analysis confirmed the presence of both L (∼250 kDa) and P (∼40 kDa) in the final sample (Figure 1 A). Densitometric analysis and known oligomeric states of the proteins suggested a stoichiometry of 1:4 (L:P). To verify that the L-P complex was functional, RdRp activity was measured in an RNA template-dependent primer-extension assay. In the presence of GTP, ATP, and α33P-CTP, L-P incorporated 67-fold more α33P-CTP into the growing primer than what was detected in the control reaction (Figure 1B). L-P activity was also measured in a nucleotide-incorporation assay. When incubated with 33P-GTP, the polymerase incorporated one 33P-GMP (+1) into the primer (Figure 1C, lane a). With the addition of 33P-GTP and ATP, the enzyme extended the primer by three nucleotides (+3), GAA, complementary to the RNA template (Figure 1C, lane b). When incubated with 33P-GTP, ATP, and CTP, the enzyme extended the primer by seven nucleotides (+7), GAACAAA (Figure 1C, lane c). As a control, RSV L-P added only four nucleotides (+4) when incubated with 33P-GTP, ATP, and ALS-8112-TP, the active form of a known RSV polymerase nucleoside inhibitor that causes chain termination (Figure 1C, lane d) (Deval et al., 2015). The non-processive, abortive pattern of nucleotide addition and the chain termination property of ALS-8112-TP observed with RSV L-P was consistent with results reported previously (Deval et al., 2015), indicating that the recombinant RSV L-P has RNA-dependent RNA polymerase activity.
Figure 1.
The Recombinant RSV L-P Complex Is Biochemically Active
(A) SDS-PAGE of the purified L-P complex. Molecular weights (in kilodaltons) of the ladder are shown on the left, and the L and P bands are labeled on the right.
(B) RNA synthesis activity of the RSV L-P complex was tested in a primer extension assay using an 11-mer RNA template and biotinylated 4-mer RNA primer. Primers were captured on streptavidin-coated plates, and extension was measured by incorporation of α33P-CTP. Bars indicate the mean, and error bars depict the SD of one independent experiment with 16 technical replicates (∗p < 0.001, Mann-Whitney U test).
(C) Polyacrylamide sequencing gel showing the results of the nucleotide incorporation assay for a single nucleotide (lane a) and multiple nucleotides (lanes b–d). ALS-8112-TP is a CTP analog that terminates elongation.
(D) Transfer of tritiated methyl groups from S-adenosyl methionine (SAM) molecules to a set of synthetic RNAs of varying length (5-, 9-, and 15-mers) that mimic the 5′ end of RSV mRNA was measured using a filter-binding assay. Bars indicate the mean, and error bars depict the SD of four independent experiments. Asterisks indicate means that were statistically different from that of the control sample as measured by Dunnett’s test (p < 0.001).
Because the L protein harbors multiple enzymatic domains, we sought to further validate the recombinant RSV L-P protein by measuring its MTase activity. The transfer of tritiated methyl groups from S-adenosyl methionine (SAM) to synthetic RNA molecules derived from the 5′ end of RSV mRNA was measured using a filter-binding assay. The L-P protein exhibited MTase activity for three synthetic GpppG RNAs of different lengths, with activity 7- to 10-fold higher than a control reaction lacking the synthetic RNAs (Figure 1D). Collectively, these results indicated that the recombinant L-P complex was functionally active and suitable for structural analysis by cryo-EM.
Determination of the 3.2-Å Cryo-EM Structure of RSV L in Complex with Tetrameric P
The L-P complex was plunge-frozen on UltrAuFoil 1.2/1.3 grids (Russo and Passmore, 2014), and a dataset was collected using a Titan Krios equipped with a K2 Summit detector (Figure S1 ; Table 1 ). After motion correction, contrast transfer function (CTF) estimation, 2D classification, and iterative rounds of heterogeneous 3D refinement, a total of 241,669 particles remained. Homogeneous 3D refinement of this particle stack resulted in a 3.4-Å map. After initial building of the L model and docking of the closely related human metapneumovirus (hMPV) P oligomerization domain structure into the map (Leyrat et al., 2013), it was possible to identify the portions of the density that corresponded to the P monomers. We then generated a map in which these regions were deleted. A low-pass filter was applied to this new map as well as to the original map, and both were used as input volumes for a final round of heterogenous 3D refinement to remove particles in which the tetrameric P had dissociated. The resulting stack of 196,720 RSV L-P particles was subjected to a homogeneous, non-uniform 3D refinement that yielded the final 3.2-Å map. The low sequence identity of vesicular stomatitis virus (VSV) L prevented use of the VSV L structure (Liang et al., 2015) as a starting model, but this structure was used to guide manual building of RSV L. Although the sharpened map was used for the majority of model building, the C-terminal portions of two P monomers were difficult to place in this map because of over-sharpening. However, these regions were visible in the unsharpened maps; therefore, both maps were used in combination to build these regions of the polymerase complex.
Figure S1.
Cryo-EM Data Processing, Related to Figure 2
Flow chart shows the data processing scheme used to obtain the 3.2 Å resolution map for the RSV L–P complex. A cutout of the cryo-EM map is shown as gray transparent density and the model built into the density is shown as blue sticks with oxygen and nitrogen atoms colored red and blue, respectively. The graph shows the FSC curves used to calculate the resolution of the final reconstruction with the FSC cutoff that was used (0.143) shown as a dotted line. Local resolution estimation was performed with MonoRes (Vilas et al., 2018). Scale bar = 500 Å.
Table 1.
Cryo-EM Data Collection and Validation Statistics
| Data Collection and Processing | |
|---|---|
| EMDB | EMD-20536 |
| Microscope | FEI Titan Krios |
| Magnification | 22,500 |
| Pixel size (Å) | 1.075 |
| Voltage (kV) | 300 |
| Detector | Gatan K2 Summit |
| Exposure (e−/Å2) | 48 |
| Defocus range (μm) | −1.5 to −2.5 |
| Final particles | 196,720 |
| Symmetry | C1 |
| Resolution (Å) | 3.2 |
| Model Refinement and Validation | |
| PDB ID | 6PZK |
| Model Composition | |
| Non-hydrogen atoms | 13,153 |
| RMSD bonds (Å) | 0.012 |
| RMSD angles (°) | 1.94 |
| Ramachandran | |
| Favored (%) | 96.2 |
| Allowed (%) | 3.8 |
| Outliers (%) | 0 |
| Rotamer outliers (%) | 1.95 |
| MolProbity score | 1.09 |
| EM Ringer score | 2.6 |
A total of 1,361 residues of L (residues Asn11–Leu133, Thr184–Gly618, Arg627–Ser658, and Asn690–Glu1460) were built into the 3.2-Å map (Figure 2 ). Based on homology with the VSV L structure, residues 1–968 belong to the RdRp domain, and residues 969–1460 belong to the PRNTase (capping) domain. Although the MTase domain, connector domain (CD), and C-terminal domain (CTD) of our construct were present when analyzed by SDS-PAGE after size exclusion chromatography, and the protein exhibited MTase activity in vitro, these domains were not visible in our map (Figure S2 ). This finding is not unexpected, however, given that the related VSV L is known to sample multiple states in which these three domains display various degrees of association with the RdRp and PRNTase domains (Rahmeh et al., 2010). In addition to the L core, we could build portions of four discrete monomers of P, ranging in length from 53–98 residues. The longest monomer of P spans residues 131–228, corresponding to the oligomerization domain and most of the C-terminal region of P.
Figure 2.
Structure of the RSV L Core Bound to Tetrameric P
RSV L is shown in ribbons colored in cool colors, and each monomer of tetrameric P is shown in ribbons colored in a unique warm color. The RNA-dependent RNA polymerase domain (RdRp) is colored dark blue, and the polyribonucleotidyltransferase (PRNTase) domain is shown in light blue. The linear maps show the portions of L (top) and P (bottom) that are resolved in the structure as colored bars. The connector domain (CD), methyltransferase domain (MTase) and C-terminal domain (CTD) of L are not observed in the structure. Arrows denote conserved residues required for function. NTD, N-terminal domain; OD, oligomerization domain. The linear schematics for L and P are not drawn to scale. See also Figure S1, Figure S2, Figure S4.
Figure S2.
The CD, MTase, and CTD Are Absent in the Cryo-EM Map, Related to Figure 2
(A) The model built for the L protein core of RSV and (B) the published structure of VSV L (Liang et al., 2015) are shown as ribbons colored in cool colors. The connector domain (CD), methyltransferase domain (MTase) and C-terminal domain (CTD) are colored white for VSV and are not observed in the RSV structure. The linear map shows the portions of L that are resolved in the RSV structure as colored bars.
The RSV P Tetramer Grips L Using an Unusual Tentacular Arrangement
As expected from previous experiments, residues 131–151 of RSV P formed a tetrameric coiled coil (Castagné et al., 2004, Llorente et al., 2006, Pereira et al., 2017). Historically, regions outside of this oligomerization domain have not been structurally well characterized because of their intrinsic flexibility (Longhi et al., 2017). However, the interaction of P with L stabilized ordered conformations of the C-terminal regions of P, allowing them to be resolved for the first time. Perhaps the most striking feature of the tetrameric P structure is the large degree of variation in the conformation of each of the four P monomers (Figure 3 ) (P1, P2, P3, and P4). These different conformations allow P to wrap around the RdRp of L in a tentacular fashion (Figure 4 ). Although P1 and P4 make extensive contacts with L, P3 makes minimal contacts with L, and P2 interacts almost exclusively with the other P monomers. The unique organization adopted in this complex allows a relatively small region of P (less than 100 residues) to contact more than 11 regions of the primary L sequence and bury more than 4,000 Å2 of surface area on L (Figures 4A and 4B).
Figure 3.
The RSV P Tetramer Wraps around L in a Tentacular Fashion
RSV L is shown as a white molecular surface, and P monomers are shown as ribbons. The P4 monomer is colored as a rainbow (blue to red, N to C terminus, respectively). All other monomers are colored to match P4.
See also Figures S6 and S7.
Figure 4.
Two Phosphoprotein Monomers Contact Three Discrete Regions on the RdRp Domain of RSV L
(A) RSV L and P are colored as in Figure 2 but are rotated by 90° about the vertical axis. The three windows represent the views shown in (C), (D), and (E).
(B) L and P are shown as molecular surfaces colored by electrostatic potential from red to blue, negative to positive, respectively. The surface of each is shown in an open-book representation. The approximate binding interface is outlined in yellow and black. A region of electrostatic complementarity is highlighted with a dashed oval.
(C) Magnified view of the P1–L interaction.
(D) Magnified view of the P4–L interaction.
(E) Magnified view of the C-terminal P4-L interaction.
Side chains involved in hydrogen bonds, salt bridges, or hydrophobic interactions are shown as sticks, with oxygen and nitrogen atoms colored red and blue, respectively. Hydrogen bonds and salt bridges are depicted as black dashed lines. See also Figure S3, Figure S5, Figure S6.
The interaction of P with L is driven by a number of hydrophobic and electrostatic interactions (Figure 4B). The majority of electrostatic interactions are made between residues 167–179 of P1 and the RdRp domain of L. Two positively charged P1 residues, Arg167 and Arg174, form salt bridges with L residues Asp718 and Glu722 (Figure 4C). In addition, two negatively charged residues on P1, Glu176 and Glu179, form salt bridges with Arg523 and Lys529 on L. We predict that mutation of these residues would disrupt the L-P interaction and decrease the activity of L. Consistent with this hypothesis, two of these charged P residues (Arg174 and Glu176) have been shown previously to be critical for the activity of L in a minigenome assay (Lu et al., 2002). In addition, a temperature-sensitive virus with an E176G substitution in P reverted the glycine to a negatively charged residue when the virus was passaged at physiological temperatures (Lu et al., 2002). The RSV L-P structure thus provides a molecular explanation for the decreased activity of L because of these substitutions in P.
The P4 monomer, for which the largest number of residues could be built, contacts a completely different set of L residues than P1. Two residues on P4 (Ile181 and Leu198) form a hydrophobic cap above Phe452 on the β6 strand of L (Figure 4D). Additionally, a region of P4 forms a β-hairpin that hydrogen-bonds with β6 of L to form a small β sheet (Figure 4D). In P1 and P2, these same residues are within an α helix, and in P3, they are disordered, indicating that the conformation of this region of P is particularly malleable. Furthermore, at the base of the loop connecting the β strands of the P4 hairpin, Arg163 forms a salt bridge with Glu765 of L, and the main chain of Gly165 forms a hydrogen bond with Tyr772 of L (Figure 4D). The C-terminal region of P4 makes additional contacts with L, including hydrogen bonds between Asn217 of P and Asn362 and Thr365 of L, as well as hydrophobic interactions between Leu223 of P and surrounding hydrophobic residues on L (Figure 4E). Leu223, along with Leu216 and Leu227, are three hydrophobic residues on the C terminus of P shown previously to contribute to L binding. Mutation of these residues disrupts polymerase activity (Sourimant et al., 2015), demonstrating that these interactions between L and P, which are critical for function of the polymerase, can be explained by the structure presented here.
In addition to the contacts made between P and L, extensive interactions between P monomers are also present. Contacts within the oligomerization domain were expected (Castagné et al., 2004, Llorente et al., 2006, Pereira et al., 2017), but the structure reveals that interactions between P monomers also occur outside of the oligomerization domain. The C-terminal α helices of P2 and P3 rest on top of P1 to form a three-helix bundle (Figure 4A). In this arrangement, Ala169, Ile173, and Ile178 of P1 are capped by hydrophobic residues on P2 and P3. To validate the importance of these residues as well as of those described above, we generated a panel of 37 variants of P in which one or two residues were mutated to alanine and tested these variants in a minigenome replication assay. Variants with substitutions of residues between position 131 and 189 as well as position 205 showed less than 20% of the activity observed for wild-type P (Figure S3 ). For the P variants with reduced activity, 16 of the substitutions mapped to regions involved in formation of the coiled coil in the oligomerization domain, 8 were in regions involved in the interaction between P and L, and 8 mapped to residues involved in the interactions between P1, P2, and P3 that lie outside of the oligomerization domain. Collectively, these results demonstrate that the multifaceted interactions of P identified here are critical for its function in viral replication.
Figure S3.
Residues Involved in P-L and P-P Contacts Are Critical for Polymerase Function, Related to Figure 4
BSRT7/5 cells (Buchholz et al., 1999) were transfected with plasmids encoding the wild-type (WT) N, M2-1 and L proteins, the pMT/Luc minigenome, and WT or mutant P proteins, together with pCMV-βGal for transfection standardization. Viral RNA synthesis was quantified 24 h after transfection by measuring the luciferase activity after cell lysis. Each luciferase minigenome activity value was normalized based on β-galactosidase expression and is the mean of three independent experiments performed in triplicate. Error bars show standard deviations. The dotted line indicates 20% of wild-type activity. The color used to label each variant indicates the likely function of each residue based on the L–P structure.
Conserved Features of the RSV RdRp Domain
The RNA-dependent RNA polymerases of negative-sense RNA viruses contain four subdomains—the palm, fingers, thumb, and a subdomain thought to serve as a structural support (Figure 5 A)—and six conserved sequence motifs (motifs A–F) (Jácome et al., 2015, Poch et al., 1989). The majority of these motifs are found in the palm subdomain (Figure 5B), which is composed of two α helices (α29 and α30) and a β sheet comprising 5 strands (β9, β12, β13, β14, and β15). A conserved glycine residue (Gly877) just after the motif E β sheet connects the palm and thumb subdomains and may allow the palm domain to exhibit small degrees of motion relative to the thumb, as demonstrated for the enterovirus polymerase (Gong and Peersen, 2010; Figure 5B). Motif B forms one of two connections that are present between the palm and fingers. This motif is composed of a flexible loop that extends from the finger subdomain into the α29 helix of the palm. The presence of a glycine just before the α29 helix is known to be critical for catalysis in the RdRps of hepatitis C and encephalomyocarditis viruses (Lohmann et al., 1997, Sankar and Porter, 1992). RSV harbors two additional glycine residues that form a GGxxG motif also found in the RdRp domains of L proteins from hMPV, Ebola, VSV, and other non-segmented negative-sense RNA viruses (Figure S4 ). Thus, the palm subdomain is framed on either side by glycine residues that likely aid in flexibility of this region to accommodate the template RNA and incoming nucleoside triphosphates (NTPs) in the catalytic site (Garriga et al., 2013).
Figure 5.
Structure of the RSV L RdRp Domain
(A) The RdRp domain is shown in ribbons, with the fingers in blue, the thumb in green, the palm in red, and the remaining structural support of the RdRp domain in white. The dashed circle highlights the Gly-Asp-Asn (GDN) motif, with the Asp and Asn side chains shown as sticks.
(B) The same view as in (A), but with sequence motifs A–F highlighted in rainbow colors. The Cα atoms of conserved glycine residues are shown as spheres.
(C) Magnified view of the active site, colored as in (B). Select residues are shown as sticks, with oxygen atoms colored red, nitrogen atoms colored blue, and sulfur atoms colored yellow. The Cα atoms of conserved glycine residues are shown as spheres. The panel on the right shows the same view, but with a UTP molecule modeled based on alignment of the palm subdomain active site with the hepatitis C virus RNA polymerase structure (PDB: 1NB6).
(D) The same region as in (C), rotated by 180° to show three residues on motif B that are mutated in viruses that escape inhibition by ALS-8176.
See also Figure S5.
Figure S4.
Amino Acid Sequence Alignment of L Proteins, Related to Figure 2
The amino acid sequences of L from RSV strain A2 (P28887), human metapneumovirus (Q91L20), Zaire ebolavirus strain Mayinga-76 (Q05318) and vesicular stomatitis virus Indiana strain (P03523.2) were aligned using ESPript (Robert and Gouet, 2014). Secondary structure for RSV L is shown above the sequences.
The conserved catalytic motif (Gly810-Asp811-Asn812 [GDN]) of the RdRp is situated on the loop that connects β12 with β13 in motif C (Figure 5C). Another catalytic residue, Asp700, is located in motif A on the β9 strand. Together, Asp811, Asn812, and Asp700 are responsible for coordinating the two magnesium ions required for catalysis of phosphodiester bond formation (Noton et al., 2014). In our map, no density for magnesium ions is observed, but this may be due to the resolution or lack of magnesium during purification. A conserved aromatic residue, Phe704, is located in motif A on the α26 helix (Figure 5C). The side chain of Phe704 is oriented away from the active site and toward the α helix of Motif B, where it is neighbored by three hydrophobic residues (Leu701, Phe708, and Trp785). The presence of an aromatic residue at position 704 is well conserved, but it was previously unclear whether this residue is required for nucleotide binding in the active site or for stability of the L protein (Jácome et al., 2015). The RSV L-P structure suggests the latter.
The majority of the RSV RdRp finger subdomain is α-helical, but a small β sheet composed of β7, β8, and β10 contains the final motif required for polymerase activity (motif F) (Figure 5B). Although a portion of the density in this region is disordered in our map and prevented us from building residues 609–626, the base of the β7–β8 hairpin and the Arg627 side chain are visible and positioned to interact with negatively charged phosphate groups in the active site (Figure 5C). In addition to its role in coordinating the template strand and the incoming NTP for catalysis, the finger subdomain also interacts with the structural subdomain to form an electropositive pore that likely serves as the entry point for incoming NTPs (Figure S5 ), similar to what has been described for other viral polymerases (Choi et al., 2004, Lu et al., 2008, Tao et al., 2002).
Figure S5.
Positively Charged Channels on the RSV L Core, Related to Figure 5
L is shown as molecular surfaces colored by electrostatic potential (positive in blue, negative in red). Arrows highlight two positively charged channels. The right panel is the same as the left, but is rotated 30° about the vertical axis with the white regions of the molecular surface sliced away to clearly show the pores. Based on comparison to other RdRp structures, the two pores are proposed to accommodate an incoming NTP and the RNA template, with the opening at the top of the protein leading to the methyltransferase domain.
Viral Escape from a Nucleoside Analog Inhibitor Likely Occurs by an Allosteric Mechanism
Viral escape from the nucleoside inhibitor ALS-8176 has been mapped to the so-called “Quad” substitutions in RSV L (M628L/A789V/L795I/I796V) (Deval et al., 2015). The RSV L-P structure reveals that three of these residues are buried on the α29 helix of Motif B, oriented away from the active site of the polymerase, with one of the residues (A789V) in close proximity to Phe704 of motif A (Figure 5D). Previous reports have suggested that the Quad substitutions act by increasing the ability of the RdRp to discriminate the altered structure of the cytidine analog from that of cytidine, preventing its incorporation into the growing RNA (Deval et al., 2016). Our structure suggests that the A789V, L795I, and I796V substitutions act allosterically by altering the conformation of the polymerase active site without directly contacting either native NTPs or the cytidine analog. This is consistent with the finding that motif B is involved in sensing the correct NTP and allosterically regulating the activity of the RdRp (Garriga et al., 2013).
The RSV L PRNTase Domain Adopts a Non-initiation Conformation
The PRNTase domain of RSV L is responsible for the addition of a 5′ cap to nascent viral mRNAs (Braun et al., 2017). In addition, the PRNTase domain contains the priming loop (1267–1282), which is thought to facilitate de novo initiation (Liang et al., 2015). In the structure determined here, the priming loop is flipped upward away from the RdRp domain (Figures 6A and 6B), removed from the RdRp active site. This is in contrast to what was observed previously in the VSV L structure, in which the priming loop reached out of the PRNTase domain toward the RdRp active site (Figure 6B; Liang et al., 2015). For the VSV L structure, the authors suggested that the conformation observed represents the initiation state and that the priming loop as well as the CD, MTase, and CTD domains would need to reorient for elongation to occur. The alternate positioning of the priming loop in the RSV L structure, combined with the lack of observable density for the CD, MTase, and CTD domains, suggests that the structure determined here may resemble an elongation conformation of RSV L.
Figure 6.
Structure of the RSV L PRNTase Domain
(A) The RSV L protein is shown as ribbons, with the RdRp domain in white and the PRNTase domain in light blue. The PRNTase domain of VSV L is aligned to that of RSV L and shown as purple ribbons.
(B) View defined by the box shown in (A). The priming loop and connecting regions are shown as ribbons colored light blue (RSV) and purple (VSV). Two threonine residues are shown as sticks. The Cα atoms of the conserved glycine residues of the GxxT motif are shown as spheres.
(C) The same view as in (A) but with conserved sequence motifs of the PRNTase domain highlighted in unique colors.
(D) Magnified view of the active site, colored as in (C). The Cα atoms of conserved glycine residues are shown as spheres. Transparent molecular surfaces are shown for two tyrosine residues, Tyr1331 and Tyr1336.
(E) View of two residues, Leu1421 and Ile1381, involved in viral escape from BI-D. A pocket near the PRNTase active site is shown as a transparent surface.
(F) View of the region surrounding Glu1269.
(G) View of the region surrounding the α helix that is mutated in temperature-sensitive viruses.
In all panels, select residues are shown as sticks, with oxygen and nitrogen atoms colored red and blue, respectively, and hydrogen bonds depicted as dashed lines.
Sequence analysis of the L proteins of non-segmented, negative-sense RNA viruses has identified several conserved residues in the PRNTase domain that have also been assigned motifs (motifs A–E) (Li et al., 2008, Neubauer et al., 2016). In RSV L, these residues are clustered around the center of the cavity formed between the RdRp and PRNTase domains (Figure 6C). Motif D (also referred to as the His-Arg or HR motif) consists of two catalytic residues that are critical for cap formation: His1338 and Arg1339 (Braun et al., 2017). The position of the priming loop in the RSV L structure results in a distance between motif D and motif B (also called the GxxT motif) of less than 5 Å (Figure 6D). Thus, flipping of the priming loop into a position likely associated with elongation is coupled with closing of the active site of the PRNTase domain into a compact conformation. Gly1264 marks the point around which the priming loop hinges (Figure 6B), and a variant with a substitution to a less flexible amino acid (alanine) at this position has been shown previously to have defects in initiation of replication, elongation of viral RNA, and cap addition (Braun et al., 2017). Collectively, these data suggest that motion of the priming loop is critical for all three processes, consistent with biochemical data that has demonstrated crosstalk between RNA synthesis and capping (Braun et al., 2017, Neubauer et al., 2016).
In addition to Gly1264, Trp1262 may also play a role in positioning the priming loop (Figure 6D). The presence of an aromatic residue at this position is conserved in L proteins from multiple non-segmented, negative-sense RNA viruses (Figure S4), likely because of hydrophobic interactions with residues N-terminal to motif E. However, the precise identity of this residue is not conserved, and the tolerance for different hydrophobic residues at this position depends on the viral background (Neubauer et al., 2016). The RSV L structure reveals that a hydrogen bond between the indole group of Trp1262 and the hydroxyl group of Ser1390 may help to anchor motif B and motif D together to form the PRNTase active site. In VSV L, the Trp-Ser interaction is replaced with a hydrogen bond that is formed between Tyr1152 and Gln1270. Thus, although Trp1262 is not conserved across viruses, the interaction between these two regions is structurally preserved.
Viral Escape from a Non-nucleoside Inhibitor
The non-nucleoside small-molecule inhibitor BI-compound D (BI-D) disrupts RNA cap addition, but its binding site on RSV L is currently unknown. Viruses that escape inhibition by BI-D harbor mutations encoding one of three substitutions in residues surrounding the PRNTase active site (I1381S, L1421F, or E1269D) (Liuzzi et al., 2005). Ile1381 and Leu1421 are in close proximity to one another on the surface of L. Ile1381 contributes to a small pocket on the surface of the PRNTase domain that includes motifs B, D, and E (Figure 6E). Although Leu1421 does not form a part of the cavity wall, it contacts Ile1381, and a substitution to the larger phenylalanine side chain may indirectly alter the shape of this pocket through its interaction with Ile1381. The location of these residues near a surface-exposed pocket within the active site of the PRNTase domain suggests that this pocket could be the binding site of BI-D.
The substitution present in the third escape variant occurs at Glu1269, which is approximately 20 Å away from Ile1381 and Leu1421. Glu1269 forms hydrogen bonds with three residues on L (Thr1341, Lys1294, and Trp1297), anchoring the priming loop to motif C (Figure 6F). In this conformation, Glu1269 is oriented away from the PRNTase active site. Substitution of Glu1269 with the shorter aspartate residue present in the escape mutant would disrupt this hydrogen bonding network and likely change the conformation of the priming loop. This change would be expected to affect the organization of the proposed BI-D binding site because Motif B contributes to the surface pocket and is located on the priming loop, just two residues from Glu1269. Thus, it seems unlikely that Glu1269 directly contacts BI-D, and the structure suggests that the E1269D substitution results in allosteric changes to the BI-D binding site.
A Temperature-Sensitive L Mutant Would Likely Have an Altered Motif D Conformation
A number of live-attenuated RSV vaccine candidates have mutations in L that render the virus temperature sensitive. One virus, called cpts530/1030, has been used as a background strain for engineering viruses with high levels of attenuation and genetic stability (Whitehead et al., 1999). The “1030” mutation in this virus refers to a substitution (Y1321N) that is located on α50 in the PRNTase domain, between motif C and the catalytic motif D. In the RSV L structure, Tyr1321 forms a hydrogen bond with Lys1305 on α49. This bond may help lock the position of these two α helices relative to one another but likely could not form with the shorter side chain of an asparagine residue (Figure 6G). The catalytic residues of motif D, as well as two aromatic residues (Tyr1331 and Tyr1336) that stack on either side of Trp1297 of motif C, are both located on the loop just C-terminal to α50. Motion of α50 away from α49 would be expected to disrupt the interaction of Tyr1331 and Tyr1336 with Trp1297 and alter the conformation of the loop that contains the catalytic residues. It is likely that the temperature sensitivity of the 1030 mutant virus is due to destabilization of this region, which can be compensated by growth at reduced temperatures. Mutational analysis has also demonstrated that, although the Y1321N substitution moderately attenuates viral replication in a mouse model of infection, insertion of a negatively charged residue into this position (Y1321E) results in more dramatic attenuation (Luongo et al., 2012). There are several negatively charged residues present on α48 and α49 that would repel a negatively charged residue if it were present on α50; thus, this substitution would be expected to have a more dramatic effect on the relative orientation of these three α helices.
Discussion
Replication and transcription of negative-sense RNA viruses rely on specialized RNA-dependent RNA polymerases that must be carried within the virus. These RdRps are essential for viral survival and are structurally dissimilar to host-cell polymerases, making them excellent targets for antivirals. To efficiently replicate and transcribe viral RNA, RdRps typically require one or more cofactors. Although understanding the interactions between viral polymerases and their cofactors can provide additional targets for inhibitor development, structural characterization of these interactions has often proven difficult. The structure presented here provides the first structural characterization of a viral phosphoprotein in complex with a viral L protein. The phosphoprotein of viruses in the Mononegavirales order are generally composed of an N-terminal domain, a central oligomerization domain, and a C-terminal domain and are known to tether multiple viral components to a centralized area where viral replication occurs (Cox and Plemper, 2015). Interactions with viral proteins have primarily been mapped to the N- and C-terminal domains, which are thought to be intrinsically disordered, but are capable of transiently adopting α-helical arrangements in solution or when bound to other proteins (Pereira et al., 2017, Selvaraj et al., 2018). Previous work has indicated that this structural plasticity is required for RSV P function (Noval et al., 2016). Intrinsically disordered proteins are thought to adopt “fuzzy” structures in which flexibility is retained even when these proteins interact with other binding partners (Arbesú et al., 2018). The regions of P that are not contacting L are not visible in our cryo-EM map, indicating that unliganded P is indeed dynamic. However, we show that regions of P can become ordered when bound to L and that the same residues of P monomers can adopt distinct conformations, indicating that RSV P shares properties with the “transformer” proteins (Knauer et al., 2012). This class of protein has also been identified in the Ebola virus, which uses different oligomeric states of the VP40 protein to assemble the viral matrix layer and to regulate viral transcription (Bornholdt et al., 2013). The ability to utilize one protein for multiple interactions via structural transformations helps maximize the efficiency of viruses with limited genome sizes, and our results suggest that this might be a common phenomenon among RNA viruses.
For VSV L, an N-terminal fragment of the P was sufficient to favor the formation of a compact structure in which the CD, MTase, and CTD domains packed against the core of L (Liang et al., 2015, Rahmeh et al., 2012). However, VSV P was not clearly visible in the cryo-EM map obtained for the VSV L-P complex. In contrast, the RSV L-P structure described here shows that the oligomerization domain and C-terminal region of P, rather than the N-terminal region, interact with L. The P sequences of these two viruses vary substantially, and the region of VSV P used to stabilize the VSV L structure has low sequence homology to RSV P (Figure S6 ). These results suggest that the mechanisms by which P influences the structural organization of L may differ between RSV and VSV.
Figure S6.
Amino Acid Sequence Alignment of Phosphoproteins, Related to Figure 3
The amino acid sequences of P from RSV strain A2 (P03421), bovine RSV strain A51908 (P33454), ovine RSV strain WSU 83-1578 (Q83956), murine pneumonia virus strain 15 (Q5MKM7), human metapneumovirus (Q8B9Q8), avian metapneumovirus isolate Canada goose/Minnesota/15a/2001 (Q2Y2M5), and vesicular stomatitis virus Indiana strain (P03520) were aligned and displayed using ESPript (Robert and Gouet, 2014). The secondary structure for the P4 monomer is shown above the sequences. Both the oligomerization domain (residues 125–155) and regions in the C-terminal domain (156–190) are well conserved among pneumoviruses.
The results of our structural analysis allow us to update the interaction map that has been generated for RSV P (Pereira et al., 2017). Although structural studies have defined the interaction of M2-1 (Selvaraj et al., 2018) and N0 (Esneau et al., 2019, Galloux et al., 2015) with N-terminal peptides of RSV P, interactions in the oligomerization domain and C-terminal region are less well understood. Biochemical data have implicated P residues 216–239 in binding to L (Sourimant et al., 2015); however, the RSV L-P structure and minigenome assay results presented here demonstrate that, although these residues are important for binding to L, contacts between L and P are more extensive than previously thought and include the oligomerization domain and surrounding regions, particularly the charged residues from 167–179. It has also been suggested that the nucleocapsid and L binding sites on P may overlap (Lu et al., 2002, Pereira et al., 2017, Sourimant et al., 2015). Because the four different P monomers adopt different conformations and make unique contacts with L, it is possible that P could be bound simultaneously by L and nucleocapsid, with different monomers mediating the distinct interactions with each. Determining whether simultaneous binding of L and nucleocapsid occurs by such a mechanism will likely require structural information for the entire replication complex.
Although the sequence of the phosphoproteins within the Mononegavirales order show a large degree of diversity, their function is well conserved. The phosphoproteins, as well as polymerase cofactors from other viral families, serve multiple functions, all of which rely on their ability to interact with the viral polymerase to recruit it to the site of viral replication. Comparison of our structure with that of the recently described severe acute respiratory syndrome coronavirus (SARS-CoV) polymerase complex reveals that the region on the RdRp domain of RSV L that is bound by three of the four P monomers is similar to the region on the SARS-CoV polymerase that is occupied by residues in an N-terminal extension of the polymerase (Figure S7 ; Kirchdoerfer and Ward, 2019). Perhaps even more strikingly, comparison of the RSV L-P complex structure with the structure of the replicase holoenzyme complex from bacteriophage Qβ also reveals conserved features. During infection in bacteria, Qβ recruits the host-cell ribosomal S1 protein, which is composed of six oligonucleotide-oligosaccharide-binding (OB) domains, to initiate viral replication (Wahba et al., 1974). Two of these domains, OB1 and OB2, contact the Qβ replicase core but use different motifs to contact discrete regions and wrap around the replicase core (Gytz et al., 2015, Takeshita et al., 2014). Thus, although the structure of S1 and P share no homology, the modular domains of S1 and three of the monomers of P contact similar regions on the finger subdomain of their respective RdRp using a similar strategy (Figure S7). Contacts made with this region on the RdRp domains of these three viral polymerases may simply act to tether the replication complex and increase the interaction with viral RNA. However, the conserved location of this interface implicates these cofactors in regulating or maintaining the conformation of the replication machinery. This role can be filled by a domain of the polymerase itself (coronavirus), a secondary viral protein (RSV), or even a hijacked host cell protein (Qβ). Future studies will seek to determine how P influences the conformation of RSV L and further clarify the role of this evolutionarily conserved protein-binding interface in viral replication.
Figure S7.
Structural Similarity in Binding Modes of RSV P, SARS-CoV RdRp N-Terminal Extension, and Bacteriophage Qβ Accessory Protein, Related to Figure 3
(A) The RdRp domain of RSV L is shown as ribbons with the fingers colored blue, the thumb in green and the palm in red. All four monomers of P are shown as ribbons and colored orange. (B) The SARS coronavirus RdRp protein (PBD: 6NUR) was superimposed to the RdRp of RSV L using the active site of the palm domains. The RdRp domain is shown as ribbons with the fingers colored blue, the thumb in green, the palm in red, and the N-terminal extension in orange. (C) The RdRp domain of the bacteriophage Qβ replicase (PDB: 4R71) was superimposed to the RdRp of RSV using the active site of the palm domains. The RdRp domain is shown as ribbons with the fingers colored blue, the thumb in green and the palm in red. OB1 and OB2 are shown as ribbons and colored orange.
In addition to the interactions between P and L, our results provide the first structural information for the RdRp and capping domains of RSV L. The structure of the RdRp domain of RSV L is similar to that observed for VSV L (Liang et al., 2015), whereas the PRNTase domains of the two L proteins were observed in different states. In the previously determined VSV L structure, the priming loop was inserted into the RdRp domain, poised to serve a role in initiation. However, in the RSV L structure determined here, the priming loop is flipped away from the RdRp domain of RSV L, suggesting that the conformation identified here represents a non-initiation state and could more closely resemble the elongation conformation of the polymerase. The RSV L structure also rationalizes inhibitor escape mutants and mutations observed in live-attenuated vaccine candidates (Duvall et al., 2016, Liuzzi et al., 2005, Luongo et al., 2012, Whitehead et al., 1999). Several of the substitutions that enable escape from nucleoside and non-nucleoside inhibitors appear to allosterically alter the conformation of the inhibitor binding sites, although additional structural studies of inhibitor-bound complexes are needed to fully understand these mechanisms of escape. Atomic-level knowledge of these mechanisms should facilitate the design of antivirals with increased barriers to escape. The RSV L-P structure also provides detailed information for protein-protein interactions that could serve as novel targets for inhibitor development. Thus, our results lay the foundation for the design of novel antivirals and for the improvement of existing small-molecule inhibitors for RSV.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli DH10Bac | ThermoFisher Scientific | Cat#10361012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Human RSV L-P protein | This study | N/A |
| 33P CTP | ARC | Cat: ARP 0154-1 mCi |
| 3H-SAM | Perkin-Elmer | Adenosyl-L-methionine, S-[methyl-3H]-. NET155H001MC |
| Scintillation fluid | Perkin-Elmer | BETAPLATE SCINT. Cat#1205-440 |
| DEAE filtermats | Perkin-Elmer | Cat#1450-522 |
| Deposited Data | ||
| Cryo-EM map of RSV L–P complex | This study | EMDB: EMD-20536 |
| Coordinates for RSV L–P complex structure | This study | PDB ID: 6PZK |
| Experimental Models: Cell Lines | ||
| Spodoptera frugiperda (Sf9) | Thermofisher Scientific | CAT # 11496015 |
| BSRT7/5 | Dr KK Conzelmann, Department of Clinical Virology, Federal Research Center for Virus Diseases of Animals, D-72076 Tübingen, Germany. | Buchholz et al., 1999. |
| Oligonucleotides | ||
| 5′-pACGC | Deval et al., 2016 | https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0154097 |
| 5′-UUUGUUCGCGU | Deval et al., 2016 | https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0154097 |
| 5′-biotin-ACGC | Deval et al., 2016 | https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0154097 |
| 5′-Gppp GGG AC (RSV5), 5′-Gppp GGG ACA AAA (RSV9), 5′-Gppp GGG ACA AAA UGG AUC (RSV15) | Dr. Françoise DEBART and Dr. J-Jacques VASSEUR. IBMM, CNRS, Université Montpellier, ENSCM, Campus Triolet, Montpellier, France | Paesen et al., 2015 |
| Recombinant DNA | ||
| pFastbac Dual | Thermofisher Scientific | CAT#10712024 |
| pFastbac Dual L-P | This Study | N/A |
| pMT/Luc Minigenome | Dr J.-F. Eléouët, VIM, INRA, 78350 Jouy en Josas, France | Tran et al., 2009 |
| Software and Algorithms | ||
| cryoSPARC v2 | Punjani et al., 2017 | https://cryosparc.com/ |
| Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Adams et al., 2002, Afonine et al., 2018 | https://www.phenix-online.org/ |
| ISOLDE | Croll, 2018 | https://isolde.cimr.cam.ac.uk/what-isolde/ |
| ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| ImageQuant TL v8.1 | GE Healthcare Life Sciences | http://www.gelifesciences.com/en/us |
| Other | ||
| Streptavidin Flashplate | Perkin Elmer | Cat#SMP103A001PK |
| Strep-Tactin Superflow Plus Cartridge | QIAGEN | Cat#30060 |
| HiTrap Heparin HP | GE Healthcare | Cat#17-0406-01 |
| Superose 6 Increase | GE Healthcare | Cat#29091596 |
| Measuring luciferase | Tecan, Männedorf, Switzerland | Infinite 200 Pro microplate reader |
| Generation of P mutants | Stratagene | Quikchange site-directed mutagenesis kit |
Lead Contact and Materials Availability
Further information and requests for materials should be directed to and will be fulfilled by lead contact Jason S. McLellan (jmclellan@austin.utexas.edu).
Method Details
Protein production and purification
Human RSV L protein (strain A2) with an N-terminal dual StrepTag and RSV P protein (strain A2) with a C-terminal 6x His-tag were co-expressed in Sf9 insect cells using pFASTbac Dual transfer vector (Life Technologies). The Sf9 cells expressing the L–P polymerase complex were lysed by two passes through a microfluidizer in Strep Buffer A (50 mM Tris-HCl, 300 mM NaCl, 10% glycerol, and 1 mM Tris (2-carboxyethyl) phosphine (TCEP), pH 8.0) supplemented with Complete EDTA-free Protease Inhibitor Cocktail (Roche) and 1 U/mL Benzonase (EMD Millipore). After clarification by high-speed centrifugation, the cell lysate was loaded onto a Strep-Tactin column (QIAGEN), and the bound protein was eluted using 10 mM desthiobiotin in Strep Buffer A. The eluted L–P complex was pooled and diluted with an equal volume of heparin buffer A (50 mM Tris-HCl, 10% glycerol, and 1 mM TCEP, pH 8.0), and was further purified using a heparin column (GE Healthcare). The L–P complex was eluted from the heparin column using a NaCl gradient to a final concentration of 500 mM. The protein was concentrated with an Amicon Ultra centrifugal filter, then loaded on to a size-exclusion column (Superpose 6 Increase 10/300, GE healthcare) equilibrated in 50 mM Tris-HCl, 500 mM NaCl, 10% glycerol, and 1 mM TCEP, pH 8.0. The fractions near the maximum height of the peak were combined, flash-frozen, and stored at −80°C.
Primer-extension assay
Primer-extension reactions contained 10 nM RSV L–P complex, incubated with 200 nM RNA (5′-UUUGUUCGCGU) and 4 μM 5′ biotin-labeled RNA primer (5′-biotin-pACGC) mixed in a buffer containing 20 mM Tris-HCl pH 7.5, 10 mM KCl, 6 mM MgCl2, 10% dimethyl sulfoxide (DMSO), 2 mM dithiothreitol (DTT), 10 μM GTP, 10 μM ATP, and 25 nM α33P-CTP. Each 10 μL reaction was incubated at 30°C for 2 hours and quenched with the addition of 0.1 M EDTA. Quenched reactions were transferred to a Flashplate (Perkin Elmer SMP103A001PK) and incubated for 1 hour at room temperature. The quenched reactions were then washed two times with 0.1% Tween-20, aspirated and read on a Microbeta Trilux (Perkin Elmer).
Nucleotide-incorporation assay
Each reaction contained 0.2 μM recombinant RSV L–P, 0.2 μM of an oligonucleotide template sequence derived from the RSV leader promoter (5′ UUUGUUCGCGU 3′) and 400 μM 5′-pACGC primer, mixed in a buffer containing 20 mM Tris-HCl pH 7.5, 10 mM KCl, 2 mM DTT, 0.01% Triton X-100, 10% DMSO, 0.2 U/μL RNasin (Ambion), and 6 mM MgCl2. Reactions were started by adding 100 nM α33P-GTP tracer with specific NTPs as described in the figure legend to a final volume of 10 μL, and incubated for 30 minutes at 30°C. Reactions were quenched by adding an equal volume of gel-loading buffer (Ambion). Samples were denatured at 95°C for 5 minutes and run on a 22.5% polyacrylamide urea sequencing gel for 1.5 hours at 80 W. The gel was dried, exposed to a phosphor screen, scanned on a Typhoon phosphorimager (GE Healthcare), and quantified using ImageQuant (GE Healthcare).
MTase activity assay
The methyltransferase activity was measured using a filter-binding assay, performed according to the method described previously (Paesen et al., 2015). A 50 nM solution of RSV L–P complex was incubated with 1.8 μM purified synthetic RNA, 0.17 μM S-adenosyl methionine (SAM) and 0.8 μM 3H-SAM (Perkin Elmer) in 50 mM Tris-HCl pH 8.0. After 3 h incubation at 30°C, reactions were quenched by a 20-fold dilution in cold water. Samples were transferred to DEAE filtermats (Perkin Elmer) using a Filtermat Harvester (Packard Instruments). The RNA-retaining mats were washed twice with 10 mM ammonium formate pH 8.0, twice with water and once with ethanol. They were then soaked with scintillation fluid (Perkin Elmer), and 3H-methyl transfer to the RNA substrates was determined using a Wallac MicroBeta TriLux Liquid Scintillation Counter (Perkin Elmer).
RNA synthesis
RNAs were chemically synthesized on a solid support using the method described previously (Paesen et al., 2015). After RNA elongation with 2′ O-pivaloyloxymethyl phosphoramidite monomers (Chemgenes, USA), the 5′-hydroxyl group was phosphorylated and the resulting H-phosphonate derivative was oxidized and activated into a phosphoroimidazolidate derivative to react with pyrophosphate (pppRNA) or guanosine diphosphate (GpppRNA). After deprotection and release from the solid support, RNA molecules were purified by IEX-HPLC (> 95% pure) and their identity was confirmed by MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization Time-of-Flight) spectrometry.
Cryo-EM data collection
A cryo-EM dataset of the RSV L–P complex was collected on a Titan Krios operating at 300kV and equipped with a K2 Summit detector. An initial dataset was collected on samples frozen in 1.2/1.3 holey carbon grids, but the resolution was limited to 3.7 Å. In an effort to improve the resolution, a second dataset was collected using UltrAuFoil 1.2/1.3 grids (Russo and Passmore, 2014). UltrAuFoil 1.2/1.3 grids (Electron Microscopy Sciences) were plasma cleaned for 30 s using a Gatan Solarus 950 with a 4:1 O2:H2 ratio. A 0.57 mg/mL solution of L–P complex in 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM TCEP and 10% glycerol was diluted with an equal volume of 20 mM Tris-HCl pH 8.0, 200 mM NaCl immediately before 3 μL was deposited onto grids and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Scientific) set to 100% humidity and 4°C, with a wait time of 0.5 s, a blot time of 4 s and a blot force of 1. Data were collected at 22,500x magnification, corresponding to a calibrated pixel size of 1.075 Å. A total of 30 frames were collected for each micrograph, with defocus values ranging from −1.5 μm to −2.5 μm, a total exposure time of 9 s, and a total electron dose of ∼48 e−/Å2.
Cryo-EM data processing
Motion correction, CTF estimation, template-based picking, 2D classification, heterogeneous 3D refinement, local motion correction, homogeneous 3D refinement, and non-uniform 3D refinement were performed in cryoSPARC v2 (Punjani et al., 2017, Rohou and Grigorieff, 2015, Rubinstein and Brubaker, 2015, Vilas et al., 2018). After motion correction and CTF estimation, a total of 3,144 micrographs were selected for subsequent processing. Template-based picking identified 1,455,759 particles, which was reduced to 622,521 particles after 2D classification and 241,669 particles after two iterative rounds of heterogeneous 3D refinement. Local motion correction was performed on the final particle stack (Rubinstein and Brubaker, 2015), followed by homogeneous 3D refinement, which resulted in a 3.4 Å map.
Upon initial building of the L protein model and docking of the closely related hMPV P oligomerization domain structure in our map, it was possible to identify the portions of the map that corresponded to the P protein monomers. We then generated a map in which these regions were deleted using Chimera (Pettersen et al., 2004). This map, as well as the unmodified map, were low-pass filtered and used as input volumes for a final round of heterogenous 3D refinement to remove particles in which the tetrameric P protein had dissociated. The resulting stack of 196,720 particles was subjected to homogeneous non-uniform 3D refinement that yielded the final 3.2 Å map.
The VSV L structure was used to guide manual building of RSV L (Liang et al., 2015). The map clearly revealed a coiled-coil tetramerization domain, which allowed placement of the hMPV P protein oligomerization domain into our map (Leyrat et al., 2013). The sharpened map was used for the majority of model building, with the exception of the C-terminal portions of two P monomers, which were built using both the sharpened and unsharpened maps. The region N-terminal to the RdRp Motif A that connects the palm and finger subdomains could not be accurately built in our model, but additional volume likely corresponding to residues ∼665–672 of RSV L was visible in the map. This region is predicted to contain a small α-helix that is visible in the related VSV structure. Manual model building was carried out using Coot (Emsley and Cowtan, 2004) and refinement of the coordinates was performed using Phenix (Adams et al., 2002, Afonine et al., 2018). Additional model optimization was performed using ISOLDE (Croll, 2018), accessed through ChimeraX (Goddard et al., 2018).
Minigenome assay
Plasmids for eukaryotic expression of the hRSV N, P, M2-1, and L proteins designated pN, pP, pM2-1 and pL, have been described previously (Fix et al., 2011, Tran et al., 2007). The pM/Luc subgenomic minigenome, which encodes the firefly luciferase (Luc) reporter gene under the control of the M/SH gene start sequence, was derived from the pM/SH subgenomic replicon (Hardy and Wertz, 1998) and has been described previously (Tran et al., 2009). Point mutations were introduced in pP by site-directed mutagenesis, using the Quikchange site-directed mutagenesis kit (Stratagene). Sequence analysis was carried out to check the integrity of all the constructs. Cells at 90% confluence in 48-well dishes were transfected with a plasmid mixture containing 125 ng of pM/Luc, 125 ng of pN, 125 ng of pP, 62.5 ng of pL, and 31 ng of pM2-1, as well as 31 ng of pRSV-β-Gal (Promega) to normalize transfection efficiencies (Tran et al., 2009). Transfections were done in triplicate, and each independent transfection was performed three times. Cells were harvested 24 h post-transfection, then lysed in luciferase lysis buffer (30 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT, 1% Triton X-100, and 15% glycerol). The luciferase activities were determined for each cell lysate with an Infinite 200 Pro (Tecan, Männedorf, Switzerland) and normalized based on β-galactosidase (β-Gal) expression.
Data and Code Availability
Data Resources
Atomic coordinates for the RSV L–P complex structure have been deposited in the Protein Data Bank and assigned PDB: 6PZK. Cryo-EM maps have been deposited in the EMDB and assigned code EMD-20536.
Acknowledgments
We would like to thank Dr. Aguang Dai at the Sauer Structural Biology Laboratory at UT Austin for assistance with cryo-EM data collection. We acknowledge the University of Texas College of Natural Sciences and award RR160023 of the Cancer Prevention and Research Institute of Texas for support of the EM facility at the University of Texas at Austin. We acknowledge Dr. Jerome Deval for initial contributions to this project. We thank Françoise Debart and Jean-Jacques Vasseur for preparation of RNA molecules used in the methyltransferase assay. We also thank Johannes Langedijk, George Kukolj, and members of the McLellan Laboratory for providing helpful comments on the manuscript. Support for this work was provided by Janssen Pharmaceutical Companies of Johnson & Johnson.
Author Contributions
Conceptualization, D.R., P.R., J.-F.E., N.Y., P.J., C.L., and J.S.M.; Investigation and visualization, M.S.A.G., C.L., A.F., I.B., Z.J., P.S.-O., S.T., and J.S.; Writing – Original Draft, M.S.A.G.; Writing – Reviewing & Editing, M.S.A.G., E.D., J.-F.E., P.R., N.Y., C.L., Z.J., and J.S.M.; Supervision, P.R., D.R., Z.J., E.D., J.-F.E., and J.S.M.
Declaration of Interests
C.L., A.F., I.B., P.J., P.R., N.Y., D.R., and Z.J. are employees and possible shareholders of Janssen Pharmaceutical Companies of Johnson & Johnson.
Published: September 5, 2019
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso C.L., Amarasinghe G.K., Bányai K., Bào Y., Basler C.F., Bavari S., Bejerman N., Blasdell K.R., Briand F.X., Briese T. Taxonomy of the order Mononegavirales: update 2016. Arch. Virol. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbesú M., Iruela G., Fuentes H., Teixeira J.M.C., Pons M. Intramolecular Fuzzy Interactions Involving Intrinsically Disordered Domains. Front. Mol. Biosci. 2018;5:39. doi: 10.3389/fmolb.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker S.E., Duquerroy S., Galloux M., Loney C., Conner E., Eléouët J.F., Rey F.A., Bhella D. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J. Gen. Virol. 2013;94:1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondot M.L., Dubosclard V., Fix J., Lassoued S., Aumont-Nicaise M., Bontems F., Eléouët J.F., Sizun C. Structure and functional analysis of the RNA- and viral phosphoprotein-binding domain of respiratory syncytial virus M2-1 protein. PLoS Pathog. 2012;8:e1002734. doi: 10.1371/journal.ppat.1002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt Z.A., Noda T., Abelson D.M., Halfmann P., Wood M.R., Kawaoka Y., Saphire E.O. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M.R., Deflubé L.R., Noton S.L., Mawhorter M.E., Tremaglio C.Z., Fearns R. RNA elongation by respiratory syncytial virus polymerase is calibrated by conserved region V. PLoS Pathog. 2017;13:e1006803. doi: 10.1371/journal.ppat.1006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagné N., Barbier A., Bernard J., Rezaei H., Huet J.C., Henry C., Da Costa B., Eléouët J.F. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J. Gen. Virol. 2004;85:1643–1653. doi: 10.1099/vir.0.79830-0. [DOI] [PubMed] [Google Scholar]
- Choi K.H., Groarke J.M., Young D.C., Kuhn R.J., Smith J.L., Pevear D.C., Rossmann M.G. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. USA. 2004;101:4425–4430. doi: 10.1073/pnas.0400660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M.O., Mackman R., Byun D., Hui H., Barauskas O., Birkus G., Chun B.K., Doerffler E., Feng J., Karki K. Discovery of β-D-2′-deoxy-2′-α-fluoro-4′-α-cyano-5-aza-7,9-dideaza adenosine as a potent nucleoside inhibitor of respiratory syncytial virus with excellent selectivity over mitochondrial RNA and DNA polymerases. Bioorg. Med. Chem. Lett. 2015;25:2484–2487. doi: 10.1016/j.bmcl.2015.04.073. [DOI] [PubMed] [Google Scholar]
- Cockerill G.S., Good J.A.D., Mathews N. State of the Art in Respiratory Syncytial Virus Drug Discovery and Development. J. Med. Chem. 2019;62:3206–3227. doi: 10.1021/acs.jmedchem.8b01361. [DOI] [PubMed] [Google Scholar]
- Collins P.L., Hill M.G., Cristina J., Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R., Plemper R.K. The paramyxovirus polymerase complex as a target for next-generation anti-paramyxovirus therapeutics. Front. Microbiol. 2015;6:459. doi: 10.3389/fmicb.2015.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll T.I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J., Hong J., Wang G., Taylor J., Smith L.K., Fung A., Stevens S.K., Liu H., Jin Z., Dyatkina N. Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2′-Fluoro-4′-Chloromethyl-Cytidine Triphosphate. PLoS Pathog. 2015;11:e1004995. doi: 10.1371/journal.ppat.1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J., Fung A., Stevens S.K., Jordan P.C., Gromova T., Taylor J.S., Hong J., Meng J., Wang G., Dyatkina N. Biochemical Effect of Resistance Mutations against Synergistic Inhibitors of RSV RNA Polymerase. PLoS ONE. 2016;11:e0154097. doi: 10.1371/journal.pone.0154097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVincenzo J.P., McClure M.W., Symons J.A., Fathi H., Westland C., Chanda S., Lambkin-Williams R., Smith P., Zhang Q., Beigelman L. Activity of Oral ALS-008176 in a Respiratory Syncytial Virus Challenge Study. N. Engl. J. Med. 2015;373:2048–2058. doi: 10.1056/NEJMoa1413275. [DOI] [PubMed] [Google Scholar]
- Duvall J.R., VerPlank L., Ludeke B., McLeod S.M., Lee M.D., 4th, Vishwanathan K., Mulrooney C.A., Le Quement S., Yu Q., Palmer M.A. Novel diversity-oriented synthesis-derived respiratory syncytial virus inhibitors identified via a high throughput replicon-based screen. Antiviral Res. 2016;131:19–25. doi: 10.1016/j.antiviral.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Esneau C., Raynal B., Roblin P., Brûlé S., Richard C.A., Fix J., Eléouët J.F., Galloux M. Biochemical characterization of the respiratory syncytial virus N0-P complex in solution. J. Biol. Chem. 2019;294:3647–3660. doi: 10.1074/jbc.RA118.006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Hennessey P.A., Formica M.A., Cox C., Walsh E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- Fearns R., Deval J. New antiviral approaches for respiratory syncytial virus and other mononegaviruses: Inhibiting the RNA polymerase. Antiviral Res. 2016;134:63–76. doi: 10.1016/j.antiviral.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Fix J., Galloux M., Blondot M.L., Eléouët J.F. The insertion of fluorescent proteins in a variable region of respiratory syncytial virus L polymerase results in fluorescent and functional enzymes but with reduced activities. Open Virol. J. 2011;5:103–108. doi: 10.2174/1874357901105010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloux M., Gabiane G., Sourimant J., Richard C.A., England P., Moudjou M., Aumont-Nicaise M., Fix J., Rameix-Welti M.A., Eléouët J.F. Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J. Virol. 2015;89:3484–3496. doi: 10.1128/JVI.03666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García J., García-Barreno B., Vivo A., Melero J.A. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- Garriga D., Ferrer-Orta C., Querol-Audí J., Oliva B., Verdaguer N. Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 2013;425:2279–2287. doi: 10.1016/j.jmb.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Glezen W.P., Taber L.H., Frank A.L., Kasel J.A. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Peersen O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA. 2010;107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosfeld H., Hill M.G., Collins P.L. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gytz H., Mohr D., Seweryn P., Yoshimura Y., Kutlubaeva Z., Dolman F., Chelchessa B., Chetverin A.B., Mulder F.A., Brodersen D.E., Knudsen C.R. Structural basis for RNA-genome recognition during bacteriophage Qβ replication. Nucleic Acids Res. 2015;43:10893–10906. doi: 10.1093/nar/gkv1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.W., Wertz G.W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homaira N., Rawlinson W., Snelling T.L., Jaffe A. Effectiveness of Palivizumab in Preventing RSV Hospitalization in High Risk Children: A Real-World Perspective. Int. J. Pediatr. 2014;2014:571609. doi: 10.1155/2014/571609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jácome R., Becerra A., Ponce de León S., Lazcano A. Structural Analysis of Monomeric RNA-Dependent Polymerases: Evolutionary and Therapeutic Implications. PLoS ONE. 2015;10:e0139001. doi: 10.1371/journal.pone.0139001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S.H., Artsimovitch I., Rösch P. Transformer proteins. Cell Cycle. 2012;11:4289–4290. doi: 10.4161/cc.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C., Renner M., Harlos K., Grimes J.M. Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS ONE. 2013;8:e80371. doi: 10.1371/journal.pone.0080371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rahmeh A., Morelli M., Whelan S.P. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B., Li Z., Jenni S., Rahmeh A.A., Morin B.M., Grant T., Grigorieff N., Harrison S.C., Whelan S.P.J. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi M., Mason S.W., Cartier M., Lawetz C., McCollum R.S., Dansereau N., Bolger G., Lapeyre N., Gaudette Y., Lagacé L. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J. Virol. 2005;79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente M.T., García-Barreno B., Calero M., Camafeita E., López J.A., Longhi S., Ferrón F., Varela P.F., Melero J.A. Structural analysis of the human respiratory syncytial virus phosphoprotein: characterization of an α-helical domain involved in oligomerization. J. Gen. Virol. 2006;87:159–169. doi: 10.1099/vir.0.81430-0. [DOI] [PubMed] [Google Scholar]
- Llorente M.T., Taylor I.A., López-Viñas E., Gomez-Puertas P., Calder L.J., García-Barreno B., Melero J.A. Structural properties of the human respiratory syncytial virus P protein: evidence for an elongated homotetrameric molecule that is the smallest orthologue within the family of paramyxovirus polymerase cofactors. Proteins. 2008;72:946–958. doi: 10.1002/prot.21988. [DOI] [PubMed] [Google Scholar]
- Lohmann V., Körner F., Herian U., Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi S., Bloyet L.M., Gianni S., Gerlier D. How order and disorder within paramyxoviral nucleoproteins and phosphoproteins orchestrate the molecular interplay of transcription and replication. Cell. Mol. Life Sci. 2017;74:3091–3118. doi: 10.1007/s00018-017-2556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Brazas R., Ma C.H., Kristoff T., Cheng X., Jin H. Identification of temperature-sensitive mutations in the phosphoprotein of respiratory syncytial virus that are likely involved in its interaction with the nucleoprotein. J. Virol. 2002;76:2871–2880. doi: 10.1128/JVI.76.6.2871-2880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., McDonald S.M., Tortorici M.A., Tao Y.J., Vasquez-Del Carpio R., Nibert M.L., Patton J.T., Harrison S.C. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo C., Winter C.C., Collins P.L., Buchholz U.J. Increased genetic and phenotypic stability of a promising live-attenuated respiratory syncytial virus vaccine candidate by reverse genetics. J. Virol. 2012;86:10792–10804. doi: 10.1128/JVI.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S.W., Aberg E., Lawetz C., DeLong R., Whitehead P., Liuzzi M. Interaction between human respiratory syncytial virus (RSV) M2-1 and P proteins is required for reconstitution of M2-1-dependent RSV minigenome activity. J. Virol. 2003;77:10670–10676. doi: 10.1128/JVI.77.19.10670-10676.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon K.M., Jordan R., Mawhorter M.E., Noton S.L., Powers J.G., Fearns R., Cihlar T., Perron M. The Interferon Type I/III Response to Respiratory Syncytial Virus Infection in Airway Epithelial Cells Can Be Attenuated or Amplified by Antiviral Treatment. J. Virol. 2015;90:1705–1717. doi: 10.1128/JVI.02417-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer J., Ogino M., Green T.J., Ogino T. Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme, domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic Acids Res. 2016;44:330–341. doi: 10.1093/nar/gkv1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton S.L., Aljabr W., Hiscox J.A., Matthews D.A., Fearns R. Factors affecting de novo RNA synthesis and back-priming by the respiratory syncytial virus polymerase. Virology. 2014;462-463:318–327. doi: 10.1016/j.virol.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noval M.G., Esperante S.A., Molina I.G., Chemes L.B., Prat-Gay Gd. Intrinsic Disorder to Order Transitions in the Scaffold Phosphoprotein P from the Respiratory Syncytial Virus RNA Polymerase Complex. Biochemistry. 2016;55:1441–1454. doi: 10.1021/acs.biochem.5b01332. [DOI] [PubMed] [Google Scholar]
- Paesen G.C., Collet A., Sallamand C., Debart F., Vasseur J.J., Canard B., Decroly E., Grimes J.M. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat. Commun. 2015;6:8749. doi: 10.1038/ncomms9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira N., Cardone C., Lassoued S., Galloux M., Fix J., Assrir N., Lescop E., Bontems F., Eléouët J.F., Sizun C. New Insights into Structural Disorder in Human Respiratory Syncytial Virus Phosphoprotein and Implications for Binding of Protein Partners. J. Biol. Chem. 2017;292:2120–2131. doi: 10.1074/jbc.M116.765958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rahmeh A.A., Schenk A.D., Danek E.I., Kranzusch P.J., Liang B., Walz T., Whelan S.P. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. USA. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh A.A., Morin B., Schenk A.D., Liang B., Heinrich B.S., Brusic V., Walz T., Whelan S.P. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2012;109:14628–14633. doi: 10.1073/pnas.1209147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard C.A., Rincheval V., Lassoued S., Fix J., Cardone C., Esneau C., Nekhai S., Galloux M., Rameix-Welti M.A., Sizun C., Eléouët J.F. RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription. PLoS Pathog. 2018;14:e1006920. doi: 10.1371/journal.ppat.1006920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320-4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein J.L., Brubaker M.A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Russo C.J., Passmore L.A. Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar S., Porter A.G. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J. Biol. Chem. 1992;267:10168–10176. [PubMed] [Google Scholar]
- Selvaraj M., Yegambaram K., Todd E.J.A.A., Richard C.A., Dods R.L., Pangratiou G.M., Trinh C.H., Moul S.L., Murphy J.C., Mankouri J. The Structure of the Human Respiratory Syncytial Virus M2-1 Protein Bound to the Interaction Domain of the Phosphoprotein P Defines the Orientation of the Complex. MBio. 2018;9 doi: 10.1128/mBio.01554-18. e01554-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahabi A., Peneva D., Incerti D., McLaurin K., Stevens W. Assessing Variation in the Cost of Palivizumab for Respiratory Syncytial Virus Prevention in Preterm Infants. Pharmacoeconom. Open. 2018;2:53–61. doi: 10.1007/s41669-017-0042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T., McAllister D.A., O’Brien K.L., Simoes E.A.F., Madhi S.A., Gessner B.D., Polack F.P., Balsells E., Acacio S., Aguayo C., RSV Global Epidemiology Network Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simabuco F.M., Asara J.M., Guerrero M.C., Libermann T.A., Zerbini L.F., Ventura A.M. Structural analysis of human respiratory syncytial virus p protein: identification of intrinsically disordered domains. Braz. J. Microbiol. 2011;42:340–345. doi: 10.1590/S1517-83822011000100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourimant J., Rameix-Welti M.A., Gaillard A.L., Chevret D., Galloux M., Gault E., Eléouët J.F. Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein. J. Virol. 2015;89:4421–4433. doi: 10.1128/JVI.03619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita D., Yamashita S., Tomita K. Molecular insights into replication initiation by Qβ replicase using ribosomal protein S1. Nucleic Acids Res. 2014;42:10809–10822. doi: 10.1093/nar/gku745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Farsetta D.L., Nibert M.L., Harrison S.C. RNA synthesis in a cage--structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- Tawar R.G., Duquerroy S., Vonrhein C., Varela P.F., Damier-Piolle L., Castagné N., MacLellan K., Bedouelle H., Bricogne G., Bhella D. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- The IMpact-RSV Study Group Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- Tran T.L., Castagné N., Bhella D., Varela P.F., Bernard J., Chilmonczyk S., Berkenkamp S., Benhamo V., Grznarova K., Grosclaude J. The nine C-terminal amino acids of the respiratory syncytial virus protein P are necessary and sufficient for binding to ribonucleoprotein complexes in which six ribonucleotides are contacted per N protein protomer. J. Gen. Virol. 2007;88:196–206. doi: 10.1099/vir.0.82282-0. [DOI] [PubMed] [Google Scholar]
- Tran T.L., Castagné N., Dubosclard V., Noinville S., Koch E., Moudjou M., Henry C., Bernard J., Yeo R.P., Eléouët J.F. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner. J. Virol. 2009;83:6363–6374. doi: 10.1128/JVI.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas J.L., Gómez-Blanco J., Conesa P., Melero R., Miguel de la Rosa-Trevín J., Otón J., Cuenca J., Marabini R., Carazo J.M., Vargas J., Sorzano C.O.S. MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure. 2018;26:337–344.e4. doi: 10.1016/j.str.2017.12.018. [DOI] [PubMed] [Google Scholar]
- Wahba A.J., Miller M.J., Niveleau A., Landers T.A., Carmichael G.G., Weber K., Hawley D.A., Slobin L.I. Subunit I of G beta replicase and 30 S ribosomal protein S1 of Escherichia coli. Evidence for the identity of the two proteins. J. Biol. Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- Wang G., Deval J., Hong J., Dyatkina N., Prhavc M., Taylor J., Fung A., Jin Z., Stevens S.K., Serebryany V. Discovery of 4′-chloromethyl-2′-deoxy-3′,5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J. Med. Chem. 2015;58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- Whitehead S.S., Firestone C.Y., Karron R.A., Crowe J.E., Jr., Elkins W.R., Collins P.L., Murphy B.R. Addition of a missense mutation present in the L gene of respiratory syncytial virus (RSV) cpts530/1030 to RSV vaccine candidate cpts248/404 increases its attenuation and temperature sensitivity. J. Virol. 1999;73:871–877. doi: 10.1128/jvi.73.2.871-877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Hardy R.W., Wertz G.W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data Resources
Atomic coordinates for the RSV L–P complex structure have been deposited in the Protein Data Bank and assigned PDB: 6PZK. Cryo-EM maps have been deposited in the EMDB and assigned code EMD-20536.