Abstract
Objective:
Schizophrenia is a multifactorial disorder. It is known that a combination of extensive multiple common alleles may be involved in its etiology, each contributing with a small to moderate effect, and, possibly, some rare alleles with a much larger effect size. We aimed to perform a systematic review of association studies between schizophrenia (and its subphenotypes) and polymorphisms in the CNR1 gene, which encodes cannabinoid receptors classically implicated in schizophrenia pathophysiology, as well as to present unpublished results of an association study in a Brazilian population.
Methods:
Two reviewers independently searched for eligible studies and extracted outcome data using a structured form. Papers were retrieved from PubMed and ISI Web of Knowledge using the search term schizophrenia in combination with CNR1 or CB1 or cannabinoid receptor. Twenty-four articles met our inclusion criteria. We additionally present data from a study of our own comparing 182 patients with schizophrenia and 244 healthy controls.
Results:
No consistent evidence is demonstrated.
Conclusion:
Some seemingly positive association studies stress the need for further investigations of the possible role of endocannabinoid genetics in schizophrenia.
Keywords: Cannabinoid receptors, cannabinoid receptors type-1, gene, association studies
Introduction
Schizophrenia is well known for its heterogeneous clinical symptom dimensions, interindividual variability in pharmacological treatment response, and the lack of an established understanding of its pathophysiology. It is a multifactorial disorder, as a complex interaction of genetic and environmental factors contribute to its etiology, and features high heritability (64-83%).1,2 Currently, it is known that a combination of extensive multiple common alleles - each one contributing with a small to moderate effect - and, possibly, some rare alleles with much larger effect size3 are implicated in schizophrenia. However, despite the progress achieved in recent years, particularly in genetics and neuroimaging, we still have more questions than answers regarding its causes.
Cannabis exposure, particularly during early adolescence, has long been considered a risk factor for psychosis and, more recently, for schizophrenia.4,5 However, the underlying neurobiological mechanisms responsible for this association are not well understood. Recent research has pointed toward a complex gene-environment interaction that involves several variables, such as time of exposure (usually in early life) and genetic predisposition to psychosis.6 This interaction would lead to disrupted brain maturation and abnormalities in molecular functioning.
Although dopaminergic and glutamatergic transmissions are the classical neurobiological pathways implicated in the pathophysiology of schizophrenia, the endocannabinoid system has emerged as a possible link between cannabis and psychosis.7 This interaction may consider the effects of exogenous and endogenous cannabinoid compounds alike on the brain. It is well established that the psychogenic properties of cannabis are mainly mediated by delta-9-tetrahydrocannabinol (THC) stimulation of type-1 cannabinoid receptors (CB1).8 CB1 receptor activation may also modulate other pathways, such as glutamate, dopamine, and gamma-aminobutyric acid (GABA).9 In addition, some studies have verified abnormalities in endocannabinoid ligands such as anandamide in patients with schizophrenia,10,11 while others observed a potential antipsychotic property of CB1 receptor antagonists.7,12 Neuroimaging findings have also associated cannabis use with progressive cortical thickness in areas rich in CB1 receptors during the initial years of schizophrenia.13 This body of evidence suggests possible influences of the endocannabinoid system in the pathophysiology of the disorder.
Therefore, considering the involvement of the endocannabinoid system in psychosis and the high heritability of schizophrenia, the CNR1 gene, which encodes CB1 receptors, has been proposed as one candidate gene that may contribute to the etiology of schizophrenia and related phenotypes. CNR1 is located on chromosome 6q14-1514 and is expressed widely in the central nervous system.15 Polymorphisms in this gene have been associated with schizophrenia16 and comorbid substance abuse,17 as well as with antipsychotic response18 and adverse effects19; however, other studies have reported negative findings,20,21 and these associations remain inconclusive.
Within this context, the aim of the present study was to conduct a systematic review of all association studies of the CNR1 gene with schizophrenia and its related subphenotypes, including clinical response and side effects of antipsychotic drugs. In addition, we report unpublished results of a comparison of CNR1 gene single nucleotide polymorphisms (SNPs) in a Brazilian sample of patients with schizophrenia and healthy controls.
Methods
Association study
This study was approved by the Ethics Committee of Universidade Federal de São Paulo (UNIFESP; protocol no. 1737/06). Written informed consent was obtained from all recruited participants or their caregivers, and all clinical and laboratory investigations were conducted in strict accordance with the principles expressed in the Declaration of Helsinki. All invited subjects agreed to enroll in this study. A total of 182 patients and 244 healthy controls were recruited through Programa de Esquizofrenia (PROESQ) and Laboratório Interdisciplinar de Neurociências Clínicas (LiNC), both at UNIFESP, São Paulo, Brazil. The diagnosis of schizophrenia was established according to the DSM-IV criteria, using the Structured Clinical Interview of the DSM-IV (SCID). Treatment-resistant (TR) status was defined in accordance with the International Psychopharmacological Criteria (IPAP; http://www.ipap.org) as failure to respond to 4- to 6-week trials of monotherapy with two different antipsychotics at adequate doses. Age- and gender-matched healthy controls with no family history of psychotic disease were recruited at UNIFESP. These subjects were also investigated using a modified version of SCID screening to exclude any with current or previous psychiatric diagnoses. The descriptive characteristics of the sample are presented in Table 1. Whole blood was collected from each subject and DNA was extracted using a Gentra Puregene kit (Qiagen, Germantown, United States). All four CNR1 SNPs (rs806380, rs806379, rs1049353, and rs806368) were genotyped by TaqMan probe-based real-time polymerase chain reaction (PCR) assays (Life Technologies, Foster City, United States) performed under standard conditions. For each reaction, at least one positive control for each genotype was included. Statistical analysis was based on logistic regression, including group (control and patient or TR-schizophrenia [TRS]. and non-TRS) as the dependent variable and genotypes, age, gender, and ethnicity as independent variables.
Table 1. Descriptive characteristics of the sample.
| Variable | Patients | Controls | Statistics |
|---|---|---|---|
| Gender | |||
| Male | 127 | 147 | |
| Female | 55 | 97 | χ2 = 4.129; df = 1; p = 0.042 |
| Age (years), mean (SD) | 35.71 (10.56) | 37.33 (12.40) | t = 1.43; df = 402.67; p = 0.153 |
| Ethnicity | |||
| European | 101 | 151 | |
| African | 42 | 61 | |
| Asian | 22 | 9 | |
| Not reported | 17 | 23 | χ2 = 10.984; df = 2; p = 0.004 |
| Treatment-resistant status | |||
| TR | 73 | ||
| Non-TR | 104 | ||
| Lifetime cannabis use | |||
| Yes | 56 | ||
| No | 116 | ||
| Lifetime frequency of cannabis use | |||
| < 10 times | 17 | ||
| 10-50 times | 9 | ||
| > 50 times | 30 | ||
| rs806380 | |||
| AA | 93 | 133 | |
| AG | 78 | 92 | |
| GG | 11 | 19 | p = 0.330 |
| rs806379 | |||
| AA | 50 | 75 | |
| AT | 89 | 122 | |
| TT | 43 | 47 | p = 0.157 |
| rs1049353 | |||
| CC | 119 | 163 | |
| CT | 55 | 74 | |
| TT | 8 | 7 | p = 0.638 |
| rs806368 | |||
| CC | 11 | 20 | |
| CT | 64 | 78 | |
| TT | 107 | 146 | p = 0.701 |
Data presented as n unless noted otherwise.
χ2 = chi-square; df = degrees of freedom; SD = standard deviation; TR = treatment-resistant.
Systematic review
The PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and ISI Web of Knowledge (http://apps.webofknowledge.com) databases were searched from the first date available to June 2016, using the term schizophrenia in combination with CNR1 or CB1 or CB1. Two reviewers independently searched for eligible studies and extracted outcome data using a structured form designed for this purpose.
On PubMed, the final query resulted the following: ("schizophrenia" [MeSH Terms]. OR "schizophrenia" [All Fields].) AND (CNR1 [All Fields]. OR CB1 [All Fields]. OR ("receptors, cannabinoid" [MeSH Terms]. OR ("receptors" [All Fields]. AND "cannabinoid" [All Fields].) OR "cannabinoid receptors" [All Fields]. OR ("cannabinoid" [All Fields]. AND "receptor" [All Fields].) OR "cannabinoid receptor" [All Fields].)). On ISI Web of Knowledge the final query was: (schizophrenia AND (CNR1 or CB1 or cannabinoid receptor)). The results were downloaded and imported into EndNote X5 software (Thomson Reuters) to remove duplicates and select the papers that would be included in the systematic review.
Genetic analyses of CNR1 polymorphisms with schizophrenia or its subphenotypes as outcomes were eligible for inclusion. Studies that did not genotype CNR1 polymorphisms and did not have schizophrenia or its subphenotypes as outcomes were excluded.
Results
Association study (unpublished data)
The sample is described in Table 1. Our investigation did not show any significant association between schizophrenia and genotypes (Table 1), nor of TRS with rs806380 (p = 0.351), rs806379 (p = 0. 050), rs1049353 (p = 0.105), or rs806368 (p = 0.502). After considering cannabis use (yes/no) as a dependent variable in the logistic regression, we did not find any positive association with TRS: rs806380 (p = 0.375), rs806379 (p = 0.086), rs1049353 (p = 0.091), and rs806368 (p = 0.392). When considering frequency of cannabis use (< 10 times; 10-50 times; > 50 times) as a dependent variable, no association was found: rs806380 (p = 0.965), rs806379 (p = 0.266), rs1049353 (p = 0.985), and rs806368 (p = 0.684). We did not evaluate adverse effects of antipsychotic pharmacological treatment in our study population. As cannabis use was an exclusion criterion for the control group, we could not investigate the effect of genotypes and cannabis use on schizophrenia risk, although we could assess the influence of these factors on TRS.
Systematic review
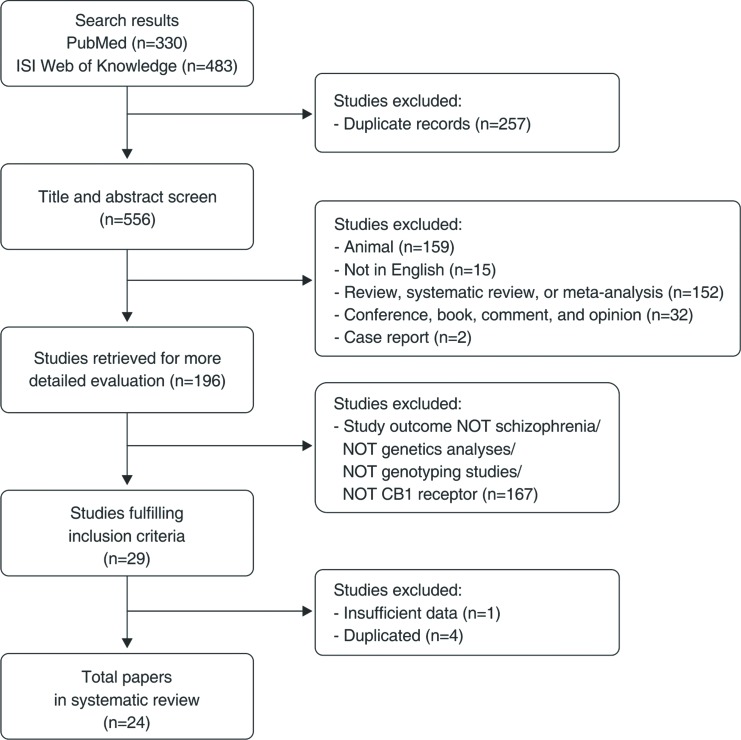
The final flow diagram is presented in Figure 1, and the selected studies listed in Table 2. A total of 24 studies involving CNR1 polymorphisms and schizophrenia were selected; we divided the results into three sections according to primary outcome: schizophrenia, substance abuse comorbidity, and pharmacological treatment.
Figure 1. Flow diagram of the systematic review. CB1 = type 1 cannabinoid receptors.
Table 2. Studies included in the systematic review.
| Author | Year | Main outcome | Study sample | Population | SNPs | Association (for main outcome) |
|---|---|---|---|---|---|---|
| Dawson22 | 1995 | SCZ | Multiplex SCZ pedigrees (23)SCZ patients (131)Controls (103) | Caucasian | (AAT)n | (-) |
| Tsai23 | 2000 | SCZ | SCZ patients (127) Controls (146) | Han Chinese | (AAT)n | (-) |
| Leroy24 | 2001 | SCZ; substance abuse; treatment response | SCZ patients (102) Controls (63) | French | rs1049353 | (+) for substance abuse in patients |
| Krebs25 | 2002 | SCZ; cannabis abuse | SCZ cannabis (102) Non-SCZ cannabis (85) Controls (89) | French | (AAT)n | (+) for cannabis-sensitive SCZ patients |
| Ujike16 | 2002 | SCZ; SCZ subcategories | SCZ patients (121) Controls (148) | Japanese | (AAT)n rs1049353 | (+) (AAT)n for hebephrenic SCZ |
| Martinez-Gras26 | 2005 | Substance abuse; PANSS scores; WCST (executive function) | SCZ patients (75 substance abusers/58 non-abusers) (133) | Spanish | (AAT)n | (+) for negative PANSS scale and WCST in substance abusers |
| Ballon17 | 2006 | SCZ; cocaine addiction | SCZ cocaine (45) Non-SCZ cocaine (97) Controls (88) | African-Caribbean | (AAT)n | (+) for cocaine addiction |
| Martinez-Gras5 | 2006 | SCZ; substance abuse | SCZ patients (113) Controls (111) | Spanish | (AAT)n | (+) for schizophrenia |
| Seifert27 | 2007 | SCZ | SCZ patients (104) Controls (140) | German | (AAT)n rs1049353 rs6454674 | (-) |
| Zammit28 | 2007 | SCZ; cannabis use; other phenotype characteristics | SCZ patients (797) Controls (688) | UK | rs1049353 | (-) |
| Chavarria-Siles29 | 2008 | SCZ; SCZ subcategories | SCZ patients (244) SCZ relatives (481) | Costa Rica | (AAT)n | (+) for hebephrenic SCZ |
| Hamdani18 | 2008 | SCZ; cannabis use; treatment response | SCZ patients (133) Controls (141) | French | rs806468 rs1049353 rs806379 rs806380 | (+) rs1049353 for treatment response |
| Monteleone30 | 2010 | SCZ; weight gain | SCZ patients (83) Controls (80) | Italian | rs1049353 | (-) |
| Tiwari19 | 2010 | Weight gain | SCZ patients (183) | German/U.S. | rs806368 rs1272071 rs1049353 rs806369 rs806370 rs806374 rs806375 rs806377 rs806378 rs2023239 rs806380 rs806381 rs7752758 rs12528858 rs12205430 rs6914429 rs2180619 rs754387 rs9450902 rs10485170 | (+) rs806378 in patients of European ancestry treated with clozapine or olanzapine |
| Ho31 | 2011 | Cannabis abuse; MRI; neurocognitive assessment | SCZ patients (235) | U.S. | rs806365 rs7766029 rs806366 rs806368 rs12710071 rs1049353 rs806374 rs806375 rs806376 rs6454672 rs9450898 rs806380 | (+) rs7766029/rs12720071/rs9450898 for MRI findings |
| Park32 | 2011 | Weight gain (olanzapine) | SCZ patients (78) | Korean | rs806368 rs4707436 rs1049353 | (-) |
| van Winkel33 | 2011 | Psychosis; positive schizotypy; cannabis use | Patients with psychosis (801)Unaffected siblings (740) | Netherlands and Belgium | rs2023239 rs806379 rs1535255 rs6454674 rs806308 rs806377 rs1049353 rs6928499 | (-) |
| Schennach34 | 2012 | Treatment response | SCZ patients (274) Controls (427) | German | rs1049353 | (-) |
| Tiwari35 | 2012 | TD | SCZ patients (191) | U.S./Canadian | rs806369 rs1272071 rs1049353 rs806369 rs806370 rs806374 rs806375 rs806377 rs806378 rs2023239 rs806380 rs806381 rs7752758 rs12528858 rs12205430 rs6914429 rs2180619 rs754387 rs9450902 rs10485170 | (+) rs806374 |
| Costa36 | 2013 | SCZ | SCZ patients (170) Controls (350) | Italian | rs1049353 rs7766029 rs806366 rs12720071 rs806380 rs9450898 rs806375 rs806368 rs6454672 rs806374 rs806365 rs806376 | (-) |
| Yu37 | 2013 | MetS | SCZ patients (407) | Belgium | rs6454674 rs6928499 rs806379 rs1535355 rs806377 rs1049353 rs2023239 | (+) rs6928499 rs1535355 rs2023239 |
| Bae21 | 2014 | SCZ | SCZ patients (337) Controls (394) | Korean | rs806376 rs806368 rs806366 rs6689530 rs34570472 | (-) |
| Copoglu38 | 2015 | SCZ; symptoms | SCZ patients (66) Controls (65) | Turkish | rs6454674 rs806368 rs1049353 | (+) rs6454674 for SCZ symptoms |
| Suárez-Pinilla39 | 2015 | Cannabis use; MRI; BMI; clinical psychopathology | First-episode psychosis patients (65) | Spanish | rs1049353 rs1535255 rs2023239 | (+) all SNPs for MRI and clinical psychopathology |
BMI = body mass index; SCZ = schizophrenia; MetS = metabolic syndrome; MRI = magnetic resonance imaging; PANSS = Positive and Negative Syndrome Scale; SNP = single nucleotide polymorphism; TD = tardive dyskinesia; WCST = Wisconsin Card Sorting Test.
CNR1 gene polymorphisms and occurrence of schizophrenia and its symptoms
Studies of genetic variants affecting the CNR1 gene in different populations showed conflicting results regarding a possible association with schizophrenia. One of the first such studies investigated the CNR1 (AAT)n repeat polymorphism in Caucasians and did not find evidence for linkage or association to schizophrenia using 23 multiplex schizophrenia pedigrees, 131 individuals with the disease, and 103 controls.22
A few years later, Tsai et al.23 compared the genotype and allele frequencies of the (AAT)n repeat polymorphism in 127 patients with schizophrenia and 146 healthy controls from the Han Chinese population. The results indicated no significant association of this polymorphism with pathogenesis of schizophrenia in that population. Similarly, in a French Caucasian population, no significant association was found with the (AAT)n repeat in a comparison of 102 patients with schizophrenia and 89 controls.25
Ujike et al.16 conducted an association study with Japanese patients with schizophrenia (n=121) and age-matched healthy controls (n=148). Although the rs1049353 polymorphism was not associated with schizophrenia in this study, the allelic distributions of the (AAT)n repeat were significantly different between controls and individuals with schizophrenia (p = 0.046), especially for hebephrenia (p = 0.0028). Hebephrenic-type schizophrenia - characterized by predominance of negative symptoms such as blunted affect, disorganized thought, and deterioration of personality - was associated with significantly increased frequency of the 9 repeat allele (p = 0.032; odds ratio [OR]. = 2.30; 95% confidence interval [95%CI]. 1.91-2.69) and decreased frequency of the 17 repeat allele (p = 0.011; OR = 0.208; 95%CI 0.098-0.439). This was the first study to verify a possible influence of CNR1 gene variants on disorganized forms of schizophrenia.16
Later studies reported controversial findings regarding the association between this polymorphism and schizophrenia. Martinez-Gras et al.40 studied possible influences of the CNR1 gene in schizophrenia by analyzing the (AAT)n repeat in 113 Spanish patients with schizophrenia and 111 healthy controls. Comparison of allele frequencies showed significant differences between the two groups. Allele 4 was overrepresented in controls (chi-square [χ2]. = 7.858; degrees of freedom [df]. = 1; p = 0.005), suggesting that this allele could represent the protective variant of this gene for schizophrenia.40
In an African Caribbean population, Ballon et al.17 did not find differences in allele or genotype distribution of the (AAT)n polymorphism between cocaine dependents with schizophrenia (n=45) and those without schizophrenia (n=97), suggesting this polymorphism might not be associated with the disorder.
Chavarria-Siles et al.29 replicated the findings of Ujike et al.16 in a population of 244 subjects with a DSM-IV diagnosis of schizophrenia and 481 family members recruited from psychiatric hospitals and clinics in the Central Valley of Costa Rica. The study found a positive correlation of prominent lifetime scores for disorganization and negative symptoms (hebephrenia dimension) with the (AAT)n polymorphism (multiallelic p = 0.0368). These findings support the hypothesis that variation in the CNR1 gene confers risk for a hebephrenic type of schizophrenia.29 However, this study did not confirm the association between (AAT)n polymorphism and schizophrenia as a broad phenotype.
Other studies in Caucasian populations did not show any association of CNR1 gene polymorphisms with occurrence of schizophrenia. Seifert et al.27 performed an association study in a sample of 104 German patients with schizophrenia and 140 healthy individuals, analyzing three known CNR1 polymorphisms (rs6454674, rs1049353, and the AAT-repeat). No statistically significant differences were found between the two groups.27
Zammit et al.28 conducted the association study of the CNR1 gene and schizophrenia with the largest sample to date. Individuals were recruited from outpatient and inpatient clinical settings and from volunteer support organizations in the United Kingdom. Overall, 797 participants with schizophrenia and 688 healthy controls were genotyped for the CNR1 rs1049353 polymorphism. Allele frequencies were compared between the two groups, and there was no evidence for any association of this polymorphism with schizophrenia (OR = 0.97; 95%CI 0.82-1.13). This study also failed to demonstrate any association between rs1049353 genotypes and various phenotypes within schizophrenia, such as age at onset.28
Two French studies also examined the allele frequencies of rs1049353 and other SNPs in patients with schizophrenia and controls. Both failed to demonstrate any association between the disorder and variations of CNR1 gene, but found possible influences on treatment response and comorbidity with drug abuse, as we will discuss further below.18,24
In an Italian population, two case-control association studies30,36 reported negative results. While Monteleone et al.30 investigated only one CNR1 SNP (rs1049353), Costa et al.36 tested 12 SNPs in the CNR1 gene. After running multiple comparisons analysis, none of the associations remained significant. Another study in 740 unaffected siblings of 801 patients with psychosis from the Netherlands and Belgium did not find a significant association between eight CNR1 SNPs and schizotypal symptoms and signs, though a marginal effect was observed for rs1049353 (p = 0.044).33
Bae et al.21 carried out a case-control association study in a Korean population and did not find any association between three SNPs in the CNR1 gene (rs806376, rs806368, and rs806366) and schizophrenia.21 A more recent study conducted in Turkey also found no differences in allele frequencies of three SNPs in CNR1 gene (rs6454674, rs806368, and rs1049353), though the authors reported a significant association between rs6454674 and schizophrenia symptoms assessed by the Positive and Negative Syndrome Scale (PANSS) and Clinical Global Impression Severity Scale (CGI-S).38 Investigating three CNR1 SNPs (rs1049353, rs1535255, and rs2023239) in a first-episode psychosis cohort of 65 patients, Suárez-Pinilla et al.39 reported a significant association of the rs2023239 polymorphism with symptoms of schizophrenia at 3-year follow-up. Specifically, T/T patients showed a significant improvement in positive (F1,61 = 8.76; p = 0.004) and negative symptoms (F1,61 = 4.51; p = 0.038) compared with T/C patients. Moreover, a significant association was observed between the interaction of these three SNPs and positive symptoms.
In total, of the 16 studies that explored the association between CNR1 polymorphisms and occurrence of schizophrenia, 15 failed to find a significant association with any SNP. However, some of the studies reported significant results for other categories or symptoms, such as an effect of the (AAT)n polymorphism on hebephrenia16,29 and of rs6454674 on PANSS and CGI-S scores.38 Moreover, a case-only study identified a significant association between CNR1 SNPs and positive and negative symptoms.39
CNR1 gene polymorphisms and comorbidity of schizophrenia with substance abuse
Substance abuse disorders, particularly cannabis abuse, are very common among patients with schizophrenia.41 Much research has been carried out in the search for possible neurobiological pathways and clinical implications underlying the high prevalence of this comorbidity.42,43 The endocannabinoid system appears as an interesting target due to its modulatory action and cross-talk with the dopaminergic system44; hence, this interaction could influence phenomena such as drug tolerance, craving, dependence, and relapse.45
A meta-analysis conducted by Benyamina et al.46 analyzed clinical studies regarding CNR1 polymorphisms and addictive disorders in the general population. They found that only the (AAT)n polymorphism was significantly associated with illicit substance dependence in Caucasian samples, using a risk allele of ≥ 16 repeats. The analysis also showed a small effect size (OR = 1.55; p = 0.045) with strong heterogeneity (Q = 19.87; p < 0.01 with I2 = 85%). This meta-analysis supports a minor implication of CNR1 AAT polymorphism in illicit substance abuse vulnerability, taking a multifactorial model into account. The same analysis also explored studies of other CNR1 polymorphisms, including rs1049353 and rs806379, and did not find a correlation with alcohol and drug dependence.46
As described above, Ballon et al.17 evaluated the (AAT)n polymorphism in 97 cocaine dependents without schizophrenia and 45 cocaine dependents with schizophrenia. They also compared both groups to 88 matched controls with no personal or family history of psychiatric disorders, including substance abuse. Although no significant difference was found between cocaine dependents with and those without schizophrenia, the frequency of the (AAT)12 repeat allele was increased in both groups of cocaine dependents vs. controls (cocaine dependents without schizophrenia, 25.3%; cocaine dependents with schizophrenia, 26.7%; controls, 5.7%; p < 0.001). The author concluded that the (AAT)n polymorphism near the CNR1 gene could be associated with predisposition to cocaine dependency, with the same risk for individuals with and those without schizophrenia.17
Other three studies, all conducted in France, investigated the association between schizophrenia/substance abuse and CNR1 polymorphisms. Leroy et al.24 found, as a tendency, a non-significantly lower frequency of the rs1049353 SNP G-allele in non-substance-abusing patients with schizophrenia compared to substance-abusing patients with schizophrenia (69% vs. 81%, p = 0.059). The GG-genotype was significantly less frequent in non-substance-abusing than in substance-abusing patients (48% vs. 69%; χ2 = 4.32; df = 1; p = 0.038), the frequency in the latter being similar to that of controls (65%).24 Later, the same group investigated another CNR1 polymorphism and found a significant lack of allele 8 of the (AAT)n repeat in “cannabis-sensitive” patients (i.e., patients with schizophrenia whose symptoms had been triggered or exacerbated in a context of cannabis consumption or misuse) when compared to the remaining patients (p = 0.0028) or to controls (p = 0.014).25 There were no differences in allele or genotype distributions between the whole group of patients with schizophrenia and controls. A third group of addicted patients without schizophrenia were not different from controls (p = 0.13).47 Finally, in an independent study, Hamdani et al.18 did not replicate this association between rs1049353 and substance abuse.
As mentioned, Martinez-Gras et al.40 observed an association between the (AAT)n polymorphism and schizophrenia; however, in the same study, they did not find significant differences with respect to substance abuse when comparing 68 patients with schizophrenia and substance abuse and 45 without substance abuse. Zammit et al.,28 in a larger sample, failed to find a significant effect of rs1049353 genotype on schizophrenia between those who did not use cannabis and those who did. However, van Winkel et al.,33 evaluating 740 unaffected siblings of patients with schizophrenia, observed an interaction between CNR1 SNPs rs806379 and rs806308 and earlier cannabis use, using positive schizotypy as the outcome, though the association was not significant after Bonferroni correction for multiple comparisons.
Evaluating other phenotypes, such as magnetic resonance imaging (MRI) findings and neurocognition, Martinez-Gras26 compared substance abusers and non-substance abusers with schizophrenia and reported that patients with CNR1 short alleles (from [AAT].n polymorphism) who abused substances exhibited significantly higher scores on the negative PANSS scale (p = 0.022) and completed less categories on the Wisconsin Card Sorting Test (WCST) (p = 0.039) than patients who did not abuse substances.
Another study genotyped 235 patients with schizophrenia and investigated 12 SNPs that account for most of the genetic variability of the CB1 coding region. Genotypes were correlated with high-resolution anatomic brain MRI and cognitive assessment. Comorbidity with DSM-IV diagnosis of cannabis abuse and dependence, taken together, was found in % of the sample. There were no differences in allele frequencies of any of the analyzed SNPs, including rs1049353, between patients with or without cannabis abuse/dependence. However, there were significant rs12720071 genotype-by-marijuana use interaction effects on white matter (WM) volumes and neurocognitive impairment. Notably, carriers of the rs12720071 G-allele with cannabis abuse/dependence had the smallest mean parietal WM volumes (p = 0.05) and the worst cognitive performance on a Problem Solving Test. The authors concluded that heavy cannabis use in the context of specific CNR1 genotypes might contribute to greater WM volume deficits and cognitive impairment, suggestive of gene-environment interactions conferring phenotypic abnormalities in schizophrenia.31
A more recent study investigated three CNR1 SNPs (rs1049353, rs1535255, and rs2023239) to verify their association with brain volumes, body mass index (BMI), or psychopathological scores in 65 first-episode psychosis patients.39 Data were obtained at the onset of first episode of psychosis and at 3-year follow-up. For those who were not cannabis users, the rs1535255 and rs2023239 polymorphisms had effects on lateral ventricle (LV) and LV and WM volume, respectively. Moreover, a significant interaction of the rs1049353 polymorphism with cannabis consumption on BMI was observed.
In total, 10 studies investigated CNR1 polymorphisms and substance abuse in individuals with schizophrenia. Six of these reported significant results. One associated the (AAT)n polymorphism with cocaine addiction17 and another with cannabis-sensitive schizophrenia.25 A third study reported a significant effect of the rs1049353 SNP and substance abuse in patients with schizophrenia.24 Moreover, three studies identified significant interaction effects on other phenotypes: Martinez-Gras26 observed an (AAT)n genotype-by-substance use interaction on psychopathology and neurocognition scores, Ho et al.31 reported a significant rs12720071 genotype-by-marijuana use interaction on MRI findings and neurocognition measures, and Suárez-Pinilla39 identified a rs1049353 genotype-by-marijuana use interaction on BMI and rs1535255 and rs2023239 genotypes-by-marijuana use interaction on MRI findings.
CNR1 gene polymorphisms and pharmacological treatment of schizophrenia
In this section, we will discuss studies that investigated possible influences of CNR1 gene polymorphisms on refractoriness to antipsychotic therapy and some adverse effects of these agents, including extrapyramidal symptoms and induced weight gain. This field clearly exemplifies the heterogeneity of schizophrenia in terms of treatment efficacy and tolerability. The future of psychopharmacology can be defined by the possibility of personalized treatment, in which clinicians could identify patients who might respond better to a certain drug or experience fewer adverse effects. This is crucial in a chronic disorder like schizophrenia, and could predict better prognosis and less functional impairment. In this sense, the field of pharmacogenomics seeks to identify biological markers for optimal treatment.48 The dopaminergic and serotoninergic systems have been the main targets of research into genetic determinants of schizophrenia refractoriness, but other systems (e.g., glutamatergic and endocannabinoid) are increasingly being studied. Authors have investigated possible influences of the CNR1 gene on treatment response and found controversial results. The first study to investigate this hypothesis was conducted by Leroy et al.24 in 102 patients with schizophrenia or schizoaffective disorder, 26 of whom were treatment-refractory. The authors analyzed the rs1049353 SNP and found that treatment-responding patients did not differ significantly from refractory patients with regard to allele frequency (χ2 = 0.03; df = 1; p = 0.86) or genotype distribution (χ2 = 2.69; df = 2; p = 0.26).24
Another study, also conducted in a French population sample by Hamdani et al.,18 recruited 133 patients with schizophrenia from two hospitals in the Paris area. All were treated with atypical antipsychotics (including clozapine) for at least 4 weeks at appropriate dosages. Four CNR1 SNPs were analyzed: rs806368, rs1049353, rs806379, and rs806380. The G-allele of rs1049353 was found in excess in patients with schizophrenia refractory to atypical antipsychotics compared with responders (85.6% vs. 71.6% ; χ2 = 7.420; p = 0.006). This observation was independent of the geographic region of patient origin. The G-allele appeared to exert a dose-related influence on therapeutic response, increasing the risk of refractoriness in up to 76% of GG homozygous subjects. The association appeared to be specific of the genetic block where the rs1049353 polymorphism is located, because the three other SNPs encompassing the CNR1 gene, but with no linkage disequilibrium (LD), showed no association with treatment refractoriness.18 In contrast to the previous study,24 these findings suggest that the G-allele of rs1049353 could be a pharmacogenetic factor for response to atypical antipsychotic drugs. In a recent letter to the editor, Schennach et al.34 reported having found no significant association between rs1049353 and early improvement or clinical response.
Other authors also looked at influences of the CNR1 gene on the development of adverse effects to antipsychotic treatment. Considering that CNR1 has been implicated in the regulation of feeding behavior and body weight,49 Tiwari et al.19 investigated the possible association of CNR1 polymorphisms with antipsychotic-induced weight gain. This study analyzed 20 SNPs of the CNR1 gene in a sample of 183 patients from three different sites in Germany and the U.S. Patients were treated with typical (haloperidol) and atypical (risperidone, olanzapine, and clozapine) agents and evaluated for antipsychotic-induced weight gain for up to 14 weeks. The results showed that the rs806378 polymorphism was associated with weight gain in patients of European ancestry treated with clozapine or olanzapine. T-allele carriers (CT + TT) had an additional weight gain of approximately 2.2 kg than patients homozygous for the CC genotype (4.33±3.89 kg vs. 2.21±4.51 kg; p = 0.022). The authors concluded that CNR1 may be associated with antipsychotic-induced weight gain in patients with chronic schizophrenia, but these results still need to be replicated in larger sample sets.19
Another study, conducted in Korea with a small sample (n=78), found no significant association between CNR1 polymorphisms (rs1049353, rs806368, and rs4707436) and olanzapine-induced weight gain in patients with chronic schizophrenia.32 Likewise, an Italian study found no association between CNR1 polymorphism rs1049353 and antipsychotic-induced weight gain.30
Besides weight gain, some studies have linked the CNR1 gene to obesity and various metabolic parameters.50,51 Yu et al.37 investigated whether genetic variation in CNR1 was associated with the metabolic syndrome (MetS). A total of 407 patients with schizophrenia, schizoaffective disorder, and schizophreniform disorder were compared regarding occurrence of MetS (n=143 with MetS and n=264 without MetS). Seven CNR1 SNPs were investigated (rs6454674, rs6928499, rs806379, rs1535355, rs806377, rs1049353, and rs3023239), and the minor alleles of rs6928499 (p = 0.006), rs1535255 (p = 0.006), and rs2023239 (p = 0.001) were significantly associated with lower risk of MetS. Further analysis revealed that this lower risk was attributable mainly to differences in HDL and fasting glucose.37
Regarding extrapyramidal side effects, Tiwari et al.35 evaluated the occurrence of tardive dyskinesia (TD) in a sample of 191 patients of European ancestry with schizophrenia (74 with TD). The authors analyzed 20 SNPs covering the CNR1 gene and found significant genotype and allele (p = 0.012) associations with the rs806374 (T > C) polymorphism. CC genotype carriers were more likely to be TD-positive (CC vs. TT + TC, OR = 3.4 [1.5-7.8].; p = 0.003). These results point toward a possible role of the CNR1 gene in the development of TD. However, the authors stress that these observations were marginal after correcting for multiple testing, and need to be replicated in a larger sample.35
A total of eight studies included in this review explored the association between CNR1 polymorphisms and pharmacological treatment of schizophrenia. Three of these investigated treatment response, and only one observed a significant effect.18 Three studies investigated antipsychotic-induced weight gain, and Tiwari et al.19 reported a positive association between rs806378 and weight gain in a subsample of patients of European ancestry treated with clozapine or olanzapine.19 Finally, one study investigated the association of CNR1 SNPs with MetS, and another investigated its association with TD; both reported significant results.
Discussion
The (AAT)n trinucleotide polymorphism was associated with schizophrenia, particularly the hebephrenic subtype, in several16,29,40 but not all studies included in this review. The same pattern was found for the rs1049353 SNP, which correlated positively with treatment refractoriness in a single study.18 Even in those studies, the authors did not establish how genetic variants could exert effects on disorder phenotype. The polymorphisms that were investigated are not capable of substituting amino acids and changing protein structure, and have not been found to alter mRNA expression or disrupt transcription at intronic promoter regions. Hence, the main possible explanation for these findings is the LD mechanism. Authors suggest that the tested variants could be in LD with another functional SNP along the CNR1 gene, and that haplotype analyses should be well measured in further studies.
In previous research, CNR1 variants have been associated with addictive disorders such as alcohol dependence and drug use.52,53 In a meta-analysis,46 only the (AAT)n polymorphism showed a significant association with illicit substance dependence in Caucasian population samples, but with a small effect size (OR = 1.55; p = 0.045). These data are in agreement with Ballon et al.,17 who concluded that the (AAT)n polymorphism near the CNR1 gene could be associated with predisposition to cocaine dependency, with the same risk for patients with and without schizophrenia. As pointed out by Martinez-Gras,26 the presence of CNR1 short alleles was associated with worse cognitive performance among substance abusers with schizophrenia. Based on these studies, we speculate that genetic variants involving the CNR1 gene could be reflected by neurobiological dysfunctions in the endocannabinoid system. Given the neuromodulatory nature of this system, acting on dopaminergic and GABAergic molecular pathways, those variants could influence the pathophysiology of psychosis, brain reward processes, and vulnerability to addictive disorders.54,55
Finally, gene-environment approaches integrate the evidence showing that CNR1 gene polymorphisms and cannabis misuse could lead to brain abnormalities (WM volume reductions) and cognitive deficits in schizophrenia.31 This modern reasoning directs the future of research in this field, but is still far from a final conclusion. Several gaps, such as differences in genetic ancestry among study populations, phenotypic variations of schizophrenia, and the heterogeneity of drug compounds (e.g., differential biological actions of THC and cannabidiol), should be better understood.55
Although our search for the systematic review was very restrictive, we may have failed to include some eligible studies, such as genome-wide association studies (GWAS). These studies probably would present negative results regarding the CNR1 gene, as they were not retrieved in our search. Searching for studies on schizophrenia in the GWAS catalog (https://www.ebi.ac.uk/gwas/) in June 2015, we found 45 studies with schizophrenia or response to treatment or substance dependence as outcome. Of these, none reported a significant association with CNR1 polymorphisms.
Studies on this gene are very heterogenous, reflecting the bias on investigating genotype associations using the candidate gene approach. The conflicting findings of this review, in part attributable to small sample sizes, do not allow us to reach a conclusion regarding the real influences of CNR1 SNPs on the pathophysiology of schizophrenia. Moreover, some unpublished data (including ours reported herein) might not have been accepted for publication, reflecting publication bias. However, we included the grey literature, such as meeting abstracts and letters.
In conclusion, our systematic review on potential associations of CNR1 polymorphisms with schizophrenia and its subphenotypes revealed conflicting data, with results tending towards a negative association. We also tested some CNR1 SNPs on a sample of Brazilian subjects and did not find a significant association. Further studies should also go beyond genes that encode brain receptors and investigate the genetic basis of endocannabinoid system enzymes (e.g., fatty acid amide hydrolase), endogenous ligands (anandamide), and transporters. GWAS with large samples are revealing more loci associated to schizophrenia, which may finally allow us to fully understand the role of endocannabinoid system malfunctioning in the pathophysiology of schizophrenia and other psychiatric disorders.
Disclosure
AG was on the speakers’ bureau and/or has acted as a consultant for Janssen-Cilag in the last 12 months. RAB has received research funding from Janssen, Eli Lilly, Lundbeck, Novartis, and Roche; has served as a speaker for AstraZeneca, Bristol, Janssen, and Lundbeck; and is a shareholder of Radiopharmacus Ltda. and Biomolecular Technology Ltda. The other authors report no conflicts of interest.
Acknowledgements
This study was funded by Fundação de Amparo è Pesquisa do Estado de São Paulo (FAPESP; grants 2010/08968-6, 2011/50740-5, and 2011/00030-1). AG has received research support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). RAB has received research funding from FAPESP, CNPq, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação Safra, and Fundação ABADS.
References
- 1.Cannon TD, Kaprio J, Lonnqvist J, Huttunen M, Koskenvuo M. The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch Gen Psychiatry. 1998;55:67–74. doi: 10.1001/archpsyc.55.1.67. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–9. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gejman PV, Sanders AR, Kendler KS. Genetics of schizophrenia: new findings and challenges. Annu Rev Genomics Hum Genet. 2011;12:121–44. doi: 10.1146/annurev-genom-082410-101459. [DOI] [PubMed] [Google Scholar]
- 4.Arseneault L, Cannon M, Poulton R, Murray R, Caspi A, Moffitt TE. Cannabis use in adolescence and risk for adult psychosis: longitudinal prospective study. BMJ. 2002;325:1212–3. doi: 10.1136/bmj.325.7374.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henquet C, Krabbendam L, Spauwen J, Kaplan C, Lieb R, Wittchen HU, et al. Prospective cohort study of cannabis use, predisposition for psychosis, and psychotic symptoms in young people. BMJ. 2005;330:11. doi: 10.1136/bmj.38267.664086.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspi A, Moffitt TE, Cannon M, McClay J, Murray R, Harrington H, et al. Moderation of the effect of adolescent-onset cannabis use on adult psychosis by a functional polymorphism in the catechol-O-methyltransferase gene: longitudinal evidence of a gene X environment interaction. Biol Psychiatry. 2005;57:1117–27. doi: 10.1016/j.biopsych.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Espejo E, Viveros MP, Nunez L, Ellenbroek BA, Rodriguez de Fonseca F. Role of cannabis and endocannabinoids in the genesis of schizophrenia. Psychopharmacology (Berl). 2009;206:531–49. doi: 10.1007/s00213-009-1612-6. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza DC, Abi-Saab WM, Madonick S, Forselius-Bielen K, Doersch A, Braley G, et al. Delta-9-tetrahydrocannabinol effects in schizophrenia: implications for cognition, psychosis, and addiction. Biol Psychiatry. 2005;57:594–608. doi: 10.1016/j.biopsych.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Wilson RI, Nicoll RA. Endocannabinoid signaling in the brain. Science. 2002;296:678–82. doi: 10.1126/science.1063545. [DOI] [PubMed] [Google Scholar]
- 10.Leweke FM, Giuffrida A, Wurster U, Emrich HM, Piomelli D. Elevated endogenous cannabinoids in schizophrenia. Neuroreport. 1999;10:1665–9. doi: 10.1097/00001756-199906030-00008. [DOI] [PubMed] [Google Scholar]
- 11.Giuffrida A, Leweke FM, Gerth CW, Schreiber D, Koethe D, Faulhaber J, et al. Cerebrospinal anandamide levels are elevated in acute schizophrenia and are inversely correlated with psychotic symptoms. Neuropsychopharmacology. 2004;29:2108–14. doi: 10.1038/sj.npp.1300558. [DOI] [PubMed] [Google Scholar]
- 12.Wong DF, Kuwabara H, Horti AG, Raymont V, Brasic J, Guevara M, et al. Quantification of cerebral cannabinoid receptors subtype 1 (CB1) in healthy subjects and schizophrenia by the novel PET radioligand [11C].OMAR. Neuroimage. 2010;52:1505–13. doi: 10.1016/j.neuroimage.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rais M, van Haren NE, Cahn W, Schnack HG, Lepage C, Collins L, et al. Cannabis use and progressive cortical thickness loss in areas rich in CB1 receptors during the first five years of schizophrenia. Eur Neuropsychopharmacol. 2010;20:855–65. doi: 10.1016/j.euroneuro.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Cao Q, Martinez M, Zhang J, Sanders AR, Badner JA, Cravchik A, et al. Suggestive evidence for a schizophrenia susceptibility locus on chromosome 6q and a confirmation in an independent series of pedigrees. Genomics. 1997;43:1–8. doi: 10.1006/geno.1997.4815. [DOI] [PubMed] [Google Scholar]
- 15.Glass M, Dragunow M, Faull RL. Cannabinoid receptors in the human brain: a detailed anatomical and quantitative autoradiographic study in the fetal, neonatal and adult human brain. Neuroscience. 1997;77:299–318. doi: 10.1016/s0306-4522(96)00428-9. [DOI] [PubMed] [Google Scholar]
- 16.Ujike H, Takaki M, Nakata K, Tanaka Y, Takeda T, Kodama M, et al. CNR1, central cannabinoid receptor gene, associated with susceptibility to hebephrenic schizophrenia. Mol Psychiatry. 2002;7:515–8. doi: 10.1038/sj.mp.4001029. [DOI] [PubMed] [Google Scholar]
- 17.Ballon N, Leroy S, Roy C, Bourdel MC, Charles-Nicolas A, Krebs MO, et al. (AAT)n repeat in the cannabinoid receptor gene (CNR1): association with cocaine addiction in an African-Caribbean population. Pharmacogenomics J. 2006;6:126–30. doi: 10.1038/sj.tpj.6500352. [DOI] [PubMed] [Google Scholar]
- 18.Hamdani N, Tabeze JP, Ramoz N, Ades J, Hamon M, Sarfati Y, et al. The CNR1 gene as a pharmacogenetic factor for antipsychotics rather than a susceptibility gene for schizophrenia. Eur Neuropsychopharmacol. 2008;18:34–40. doi: 10.1016/j.euroneuro.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari AK, Zai CC, Likhodi O, Lisker A, Singh D, Souza RP, et al. A common polymorphism in the cannabinoid receptor 1 (CNR1) gene is associated with antipsychotic-induced weight gain in Schizophrenia. Neuropsychopharmacology. 2010;35:1315–24. doi: 10.1038/npp.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman CA, Hopfer CJ, Haberstick B, Rhee SH, Crowley TJ, Corley RP, et al. The association between cannabinoid receptor 1 gene (CNR1) and cannabis dependence symptoms in adolescents and young adults. Drug Alcohol Depend. 2009;104:11–6. doi: 10.1016/j.drugalcdep.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae JS, Kim JY, Park BL, Kim JH, Kim B, Park CS, et al. Genetic association analysis of CNR1 and CNR2 polymorphisms with schizophrenia in a Korean population. Psychiatr Genet. 2014;24:225–9. doi: 10.1097/YPG.0000000000000047. [DOI] [PubMed] [Google Scholar]
- 22.Dawson E. Identification of a highly polymorphic triplet repeat marker for the brain cannabinoid receptor gene: use in linkage and association studies of schizophrenia. Schizophr Res. 1995;15:37. [Google Scholar]
- 23.Tsai SJ, Wang YC, Hong CJ. Association study of a cannabinoid receptor gene (CNR1) polymorphism and schizophrenia. Psychiatr Genet. 2000;10:149–51. doi: 10.1097/00041444-200010030-00008. [DOI] [PubMed] [Google Scholar]
- 24.Leroy S, Griffon N, Bourdel MC, Olie JP, Poirier MF, Krebs MO. Schizophrenia and the cannabinoid receptor type 1 (CB1): association study using a single-base polymorphism in coding exon 1. Am J Med Genet. 2001;105:749–52. doi: 10.1002/ajmg.10038. [DOI] [PubMed] [Google Scholar]
- 25.Krebs MO, Leroy S, Duaux E, Bourdel MC, Griffon N, Laqueille X, et al. Vulnerability to cannabis, schizophrenia and the (ATT)N polymorphism of the cannabinoid receptor type 1 (CNR1) gene. Schizophr Res. 2002;53:72. [Google Scholar]
- 26.Martinez-Gras I. Relationship between symptom dimension, neurocognitive functioning and CNR1 central cannabinoid receptor gene with susceptibility to schizophrenia and substance abuse. Eur Neuropsychopharmacol. 2005;15:S579–S80. [Google Scholar]
- 27.Seifert J, Ossege S, Emrich HM, Schneider U, Stuhrmann M. No association of CNR1 gene variations with susceptibility to schizophrenia. Neurosci Lett. 2007;426:29–33. doi: 10.1016/j.neulet.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Zammit S, Spurlock G, Williams H, Norton N, Williams N, O'Donovan MC, et al. Genotype effects of CHRNA7, CNR1 and COMT in schizophrenia: interactions with tobacco and cannabis use. Br J Psychiatry. 2007;191:402–7. doi: 10.1192/bjp.bp.107.036129. [DOI] [PubMed] [Google Scholar]
- 29.Chavarria-Siles I, Contreras-Rojas J, Hare E, Walss-Bass C, Quezada P, Dassori A, et al. Cannabinoid receptor 1 gene (CNR1) and susceptibility to a quantitative phenotype for hebephrenic schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2008;147:279–84. doi: 10.1002/ajmg.b.30592. [DOI] [PubMed] [Google Scholar]
- 30.Monteleone P, Milano W, Petrella C, Canestrelli B, Maj M. Endocannabinoid Pro129Thr FAAH functional polymorphism but not 1359G/A CNR1 polymorphism is associated with antipsychotic-induced weight gain. J Clin Psychopharmacol. 2010;30:441–5. doi: 10.1097/JCP.0b013e3181e742c5. [DOI] [PubMed] [Google Scholar]
- 31.Ho BC, Wassink TH, Ziebell S, Andreasen NC. Cannabinoid receptor 1 gene polymorphisms and marijuana misuse interactions on white matter and cognitive deficits in schizophrenia. Schizophr Res. 2011;128:66–75. doi: 10.1016/j.schres.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park YM, Choi JE, Kang SG, Koo SH, Kim L, Geum D, et al. Cannabinoid type 1 receptor gene polymorphisms are not associated with olanzapine-induced weight gain. Hum Psychopharmacol. 2011;26:332–7. doi: 10.1002/hup.1210. [DOI] [PubMed] [Google Scholar]
- 33.van Winkel R; Genetic Risk and Outcome of Psychosis (GROUP) Investigators Family-based analysis of genetic variation underlying psychosis-inducing effects of cannabis: sibling analysis and proband follow-up. Arch Gen Psychiatry. 2011;68:148–57. doi: 10.1001/archgenpsychiatry.2010.152. [DOI] [PubMed] [Google Scholar]
- 34.Schennach R, Zill P, Obermeier M, Hauer D, Dehning S, Cerovecki A, et al. The CNR1 gene in depression and schizophrenia - is there an association with early improvement and response? Psychiatry Res. 2012;196:160. doi: 10.1016/j.psychres.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 35.Tiwari AK, Zai CC, Likhodi O, Voineskos AN, Meltzer HY, Lieberman JA, et al. Association study of cannabinoid receptor 1 (CNR1) gene in tardive dyskinesia. Pharmacogenomics J. 2012;12:260–6. doi: 10.1038/tpj.2010.93. [DOI] [PubMed] [Google Scholar]
- 36.Costa M, Squassina A, Congiu D, Chillotti C, Niola P, Galderisi S, et al. Investigation of endocannabinoid system genes suggests association between peroxisome proliferator activator receptor-alpha gene (PPARA) and schizophrenia. Eur Neuropsychopharmacol. 2013;23:749–59. doi: 10.1016/j.euroneuro.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Yu W, De Hert M, Moons T, Claes SJ, Correll CU, van Winkel R. CNR1 gene and risk of the metabolic syndrome in patients with schizophrenia. J Clin Psychopharmacol. 2013;33:186–92. doi: 10.1097/JCP.0b013e318283925e. [DOI] [PubMed] [Google Scholar]
- 38.Copoglu US, Igci M, Bozgeyik E, Kokacya MH, Igci YZ, Ozden A, et al. Cannabinoid receptor 1 (cnr1) gene polymorphisms in schizophrenia patients: Rs6454674 polymorphism is associated with disease severity. Klinik Psikofarmakol Bülteni. 2015;25:341–7. [Google Scholar]
- 39.Suárez-Pinilla P, Roiz-Santianez R, Ortiz-García de la Foz V, Guest PC, Ayesa-Arriola R, Cordova-Palomera A, et al. Brain structural and clinical changes after first episode psychosis: focus on cannabinoid receptor 1 polymorphisms. Psychiatry Res. 2015;233:112–9. doi: 10.1016/j.pscychresns.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Gras I, Hoenicka J, Ponce G, Rodriguez-Jimenez R, Jimenez-Arriero MA, Perez-Hernandez E, et al. (AAT)n repeat in the cannabinoid receptor gene, CNR1: association with schizophrenia in a Spanish population. Eur Arch Psychiatry Clin Neurosci. 2006;25:437–41. doi: 10.1007/s00406-006-0665-3. [DOI] [PubMed] [Google Scholar]
- 41.Regier DA, Farmer ME, Rae DS, Locke BZ, Keith SJ, Judd LL, et al. Comorbidity of mental disorders with alcohol and other drug abuse. Results from the Epidemiologic Catchment Area (ECA) Study. JAMA. 1990;264:2511–8. [PubMed] [Google Scholar]
- 42.Chambers RA, Krystal JH, Self DW. A neurobiological basis for substance abuse comorbidity in schizophrenia. Biol Psychiatry. 2001;50:71–83. doi: 10.1016/s0006-3223(01)01134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volkow ND. Substance use disorders in schizophrenia--clinical implications of comorbidity. Schizophr Bull. 2009;35:469–72. doi: 10.1093/schbul/sbp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voruganti LN, Slomka P, Zabel P, Mattar A, Awad AG. Cannabis induced dopamine release: an in-vivo SPECT study. Psychiatry Res. 2001;107:173–7. doi: 10.1016/s0925-4927(01)00104-4. [DOI] [PubMed] [Google Scholar]
- 45.Robledo P, Berrendero F, Ozaita A, Maldonado R. Advances in the field of cannabinoid--opioid cross-talk. Addict Biol. 2008;13:213–24. doi: 10.1111/j.1369-1600.2008.00107.x. [DOI] [PubMed] [Google Scholar]
- 46.Benyamina A, Kebir O, Blecha L, Reynaud M, Krebs MO. CNR1 gene polymorphisms in addictive disorders: a systematic review and a meta-analysis. Addict Biol. 2011;16:1–6. doi: 10.1111/j.1369-1600.2009.00198.x. [DOI] [PubMed] [Google Scholar]
- 47.Krebs MO, Morvan Y, Jay T, Gaillard R, Kebir O. Psychotomimetic effects at initiation of cannabis use are associated with cannabinoid receptor 1 (CNR1) variants in healthy students. Mol Psychiatry. 2014;19:402–3. doi: 10.1038/mp.2013.188. [DOI] [PubMed] [Google Scholar]
- 48.Frank J, Lang M, Witt SH, Strohmaier J, Rujescu D, Cichon S, et al. Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol Psychiatry. 2015;20:913. doi: 10.1038/mp.2015.52. [DOI] [PubMed] [Google Scholar]
- 49.Russo P, Strazzullo P, Cappuccio FP, Tregouet DA, Lauria F, Loguercio M, et al. Genetic variations at the endocannabinoid type 1 receptor gene (CNR1) are associated with obesity phenotypes in men. J Clin Endocrinol Metab. 2007;92:2382–6. doi: 10.1210/jc.2006-2523. [DOI] [PubMed] [Google Scholar]
- 50.Benzinou M, Chevre JC, Ward KJ, Lecoeur C, Dina C, Lobbens S, et al. Endocannabinoid receptor 1 gene variations increase risk for obesity and modulate body mass index in European populations. Hum Mol Genet. 2008;17:1916–21. doi: 10.1093/hmg/ddn089. [DOI] [PubMed] [Google Scholar]
- 51.de Luis DA, Gonzalez Sagrado M, Aller R, Izaola O, Conde R. Relation of G1359A polymorphism of the cannabinoid receptor (CB1) gene with metabolic syndrome by ATP III classification. Diabetes Metab Res Rev. 2011;27:506–11. doi: 10.1002/dmrr.1200. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt LG, Samochowiec J, Finckh U, Fiszer-Piosik E, Horodnicki J, Wendel B, et al. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend. 2002;65:221–4. doi: 10.1016/s0376-8716(01)00164-8. [DOI] [PubMed] [Google Scholar]
- 53.Comings DE, Muhleman D, Gade R, Johnson P, Verde R, Saucier G, et al. Cannabinoid receptor gene (CNR1): association with i.v. drug use. Mol Psychiatry. 1997;2:161–8. doi: 10.1038/sj.mp.4000247. [DOI] [PubMed] [Google Scholar]
- 54.Solinas M, Yasar S, Goldberg SR. Endocannabinoid system involvement in brain reward processes related to drug abuse. Pharmacol Res. 2007;56:393–405. doi: 10.1016/j.phrs.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fusar-Poli P, Crippa JA, Bhattacharyya S, Borgwardt SJ, Allen P, Martin-Santos R, et al. Distinct effects of {delta}9-tetrahydrocannabinol and cannabidiol on neural activation during emotional processing. Arch Gen Psychiatry. 2009;66:95–105. doi: 10.1001/archgenpsychiatry.2008.519. [DOI] [PubMed] [Google Scholar]