Summary
How receptor tyrosine kinase (RTK) growth signaling is controlled physiologically is incompletely understood. We have previously provided evidence that the survival and mitotic activities of vascular endothelial cell growth factor receptor-2 (VEGFR2) signaling are dependent on C3a/C5a receptor (C3ar1/C5ar1) and IL-6 receptor (IL-6R)-gp130 joint signaling in a physically interactive platform. Herein, we document that the platelet derived and epidermal growth factor receptors (PDGFR and EGFR) are regulated by the same interconnection and clarify the mechanism underlying the dependence. We show that the joint signaling is required to overcome dominant restraint on RTK function by the combined repression of tonically activated PHLPP, SOCS1/SOCS3, and CK2/Fyn dependent PTEN. Signaling studies showed that augmented PI-3Kɣ activation is the process that overcomes the multilevel growth restraint. Live cell flow cytometry and single particle tracking indicated that blockade of C3ar1/C5ar1 or IL-6R signaling suppresses RTK growth factor binding and RTK complex formation. C3ar1/C5ar1 blockade abrogated growth signaling of four additional RTKs. Active relief of dominant growth repression via joint C3ar1/C5ar1 and IL-6R joint signaling thus enables RTK mitotic/survival signaling.
Keywords: VEGFR2 signaling, local complement, IL-6 receptor
Introduction
Cellular homeostasis and proliferation are regulated by growth factors which confer their activities via inducing receptor tyrosine kinase (RTK) signaling. In previous work, we studied signaling by vascular endothelial cell growth factor 2 (VEGFR2) and found that VEGFR2’s survival and growth functions in vascular endothelial cells (ECs) depend on joint signaling through two other receptor systems: C3a and C5a receptor (C3ar1/C5ar1) G protein coupled receptors (GPCRs), and IL-6 receptor (IL-6R) containing glycoprotein-130 (gp130) (1). Our experiments provided multiple lines of evidence that these receptor systems are physically interactive with VEGFR2 in a signaling platform. Studies in which each receptor system was disabled pharmacologically or genetically showed that concurrent signaling through all partners is needed for VEGFR2 auto-phosphorylation as well as for downstream VEGFR2 signaling via both of VEGFR2’s major canonical signaling pathways. Cell cycle assays showed that the joint signaling is needed for EC cell cycle progression. Studies of vascular tube formation and in three independent mouse models of angiogenesis including murine day 5 retinas documented that the results apply in vivo in growing ECs.
Epidermal growth factor receptor (EGFR) and platelet derived growth factor receptor (PDGFR) share many structural features of VEGFR2. Much literature has shown that they confer their viability and mitotic activities via the same canonical signaling pathways. Based on these structural and functional homologies, we hypothesized that the signaling interdependence of VEGFR2 with C3ar1/C5ar1 and IL-6R-gp130 signaling might also apply to EGFR and PDGFR.
The studies herein document that both major downstream growth signaling cascades of EGFR and PDGFR depend on coordinate C3ar1/C5ar1 and IL-6R-gp130 joint activation in signaling platforms in a fashion paralleling that of VEGFR2. Biophysical studies in which we performed live cell flow cytometry and single particle tracking analyses indicated that C3ar1/C5ar1 or IL-6R blockade impaired growth factor (GF) binding and increased RTK mobility (indicative of suppressed signaling complex recruitment) characteristic of inhibited growth signaling. The mechanism underlying the interdependent signaling emerged from findings that C3ar1/C5ar1 and IL-6R-gp130 transduction were required to coordinately repress the major inhibitors of growth signaling. Collectively, the data showed that while purified RTKs can auto-phosphorylate (2, 3) and bind signaling proteins in and of themselves in vitro, their efficient activation in intact cells requires the joint activation of the above signaling partners to repress growth inhibitory processes, which dominantly restrain RTK signaling and thereby prevent cellular overgrowth. Consequently, concurrent C3ar1/C5ar1 and IL-6R-gp130 signaling are obligate processes needed to lift tonic restraint on RTK viability and mitotic function in cells under physiological conditions.
Materials and Methods
Reagents and Antibodies
VEGF-A, EGF, PDGF-AA, and PDGF-BB were from ProSpec Bio (Ness Ziona, Israel) or Miltenyi Biotec (San Diego, CA). C3ar1 antagonist (C3ar1-A) and C5ar1 antagonist (C5ar1-A) are from Calbiochem (EMD Millipore, Billerica, MA) catalogue numbers C3ar1-A 559410–10mg and C5ar1-A-234415–5mg. Anti-C3a and anti-C5a mAbs were purchased from BD Biosciences (San Diego, CA). Anti-C3ar1 and anti-C5ar1 were purchased from Santa Cruz Biotech (Santa Cruz, CA). Endothelial cell growth supplement was purchased from BD Biosciences (San Diego, CA). Anti-phospho-VEGFR2 (p-Y1054/p-Y1059) was purchased from Invitrogen (Camarillo, CA). Anti-phospho-Stat3 and anti-phospho-Tyk2 were purchased from Cell Signaling Technologies (Danvers, MA). Mouse C3a was purchased from BD Biosciences (San Diego, CA). Mouse C5a was purchased from Hycult Biotech (Netherlands). Mouse C3a and C5a were biotinylated using EZ-Link NHS-Biotin kits from Thermo Scientific (Rockford, IL). AlexaFluor-488, −546, −555 and −647 labeling kits were purchased from Life Technologies (Camarillo, CA). For confocal microscopy, anti-C5ar1 and anti-C3ar1 were labeled with AlexaFluor-546, anti-VEGFR2 was labeled with AlexaFluor-488, and anti-IL-6R as well as anti-gp130 were labeled with AlexaFluor-647. SU5416 was purchased from Tocris (Bristol, UK). AG1478 and PDGFR tyrosine kinase inhibitor III were purchased from Calbiochem/Millipore (Billerica, MA). PI-3Kɣ inhibitor, AS252424, was purchased from Tocris (Bristol, UK).
Isolation of primary aortic ECs, SMCs and PMEFs
bEnd.3 cells and MS-1 murine EC lines were cultured in DMEM with 10% and 5% FBS, respectively. Primary mouse aorta ECs were isolated from WT Daf1−/−, C3ar1−/−C5ar1−/− C57BL/6 mice at ages from 1 to 4 m, by utilizing a non-mechanical and non-enzymatic method as described previously (4). Outgrowth of ECs from aortic rings occurred 72 h in the absence of antibiotics. After removing aortic rings, the ECs were maintained in DMEM/F12 medium containing 20% FBS, 2 mM L-glutamine, 1% nonessential amino acid, 0.09 mg/ml EC growth supplement, 1% antibiotic/antimycotic, 100 units/ml penicillin, 100 g/ml streptomycin, 5 ng/ml VEGF-A, and 0.09 mg/ml heparin until confluent.
Primary cultures of SMCs were isolated aortas in which ECs were removed using a scalpel. After this, the aorta was cut into small pieces and allowed to adhere to the bottom of the culture plate using 37°C incubation with the plate upside down. When the pieces were secure, complete smooth muscle cell media (Lonza) supplemented with 5% FBS was added. Cells were passaged, at maximum, 6–8 times.
PMEFs were isolated from 13 d embryos of pregnant female mice (8–10 days pregnant) euthanized by CO2 inhalation. The uterus was excised, rinsed in PBS, and embryos extracted into a dish of clean PBS after which the embryos were dissected, removing the heads, hearts, and livers, before being cut into small pieces and placed into a 0.25% Trypsin/EDTA (Invitrogen) solution. The solution was incubated at 37°C for 5 min, centrifuged, and the resulting pellet resuspended in DMEM supplemented with 10% FBS and plated. The media was changed the following day, as the first media became extremely acidic. Following that, cells were passaged several times.
Gene expression and flow cytometry
RNA was isolated by the TRIzol method (Invitrogen). Reverse transcription was achieved with Superscript-III reverse transcriptase (Invitrogen) using supplied oligo dT primers. qPCR was performed in a 24 μl volume with SYBR Green PCR mix (Applied Biosystems) using gene specific qPCR primers.
Flow cytometry was performed as described in Strainic 2013, Huang 2019. Breifly, after plating in respective medium supplemented with 10% FBS, cells were harvested with Versene (Invitrogen). The cells were stained with goat anti-C3ar1 or anti-C5ar1 mAbs or the antibody indicated in the Fig legend followed by biotin-labeled anti-goat and finally with streptavidin-APC. Stained cells were analyzed on a Becton Dickinson LSR-II.
Luminex (Multiplex) Assay:
Cells were stimulated for increasing times with agonists as described in the legends. After stimulation cells were assayed for p-Src, t-Src, p-AKT, t-AKT, p-Erk, and t-Erk using Millipore’s multiplex assay according to the manufacturer’s instructions. Values were normalized to β-tubulin. Briefly, cells were placed on ice immediately following incubation, centrifuged at 4°C, lysed in the buffer provided by the company, incubated with the capture beads followed by the detection beads, washed, then assayed on the Bioplex 2200 (Biorad, Hercules, CA).
Quantitation of Cell growth
For studies with bEnd.3, MS-1, NIH-3T3, and mIMCD3 cells, 2×104 cells were seeded in 24-well plates and allowed to adhere for 24 h. Following culturing in 0.5% FBS with DMEM for another 24 h, the cells were treated as described in the Figure legends. Growth was quantified manually at 24, 48 and 72 h with trypan blue exclusion. At least 95% of cells were viable in all experiments. VEGF-A was used at a concentration of 30 ng/ml, C3ar1-A and C5ar1-A at 10 ng/ml, and anti-C3a and anti-C5a mAbs at 1 μg/ml. The combinations used in different experiments are given in the Figure legends.
C3a/C5a/VEGF-A ELISAs
ELISAs were done in 96 well plates coated with 2 μg/ml of anti C3a, anti-C5a, anti-VEGF, or anti-IL-6 mAb. After blocking with 1% bovine serum albumin (BSA), C3a, C5a, or VEGF-A standards together with culture supernatants were added. Following incubation overnight, biotinylated secondary Ab and streptavidin HRP were added, the plates incubated at 37°C for 30 min, and color developed using enhanced TMB solution with H2O2. Stop solution consisted of 2 N H2SO4.
Propidium Iodide (PI) Staining
Propidium iodide staining (for dead cells) was performed per the manufacturer’s instructions using an Annexin V PI staining kit (BD Biosciences 556419). In brief, cells were serum starved for 24 h followed by addition of 1 μM colchicine (Sigma). Sixteen h later, cells were treated either with growth factor or with C5a (17 ng/ml each). After an additional 24 h, the cells were mobilized by Trypsin/EDTA and fixed in 0.25% formaldehyde for 10 min at 37°C. Following centrifugation, the fixed cells were resuspended in 90% methanol at 4°C until assayed. Prior to assays, methanol was aspirated, excess RNA removed via treatment with RNase (Sigma) and the cells stained with propidium iodide for 30 min at 4°C. The stained cells were analyzed on an Epics XL.
Three-D HUVEC tube formation
Fifty μL of the growth factor reduced Matrigel (BD Biosciences) was allowed to polymerize in 96-well plate at 37 °C for 30 min. Triplicates of 25,000 HUVEC were plated onto the prepared Matrigel in a volume of 150 μL of EBM-2 media (Lonza) in four different conditions: no growth factors, 30 ng/ml of VEGF-A, 30 ng/ml of VEGF-A ± 10 ng/ml each of C3aR-A and C5ar1-A and the complete set of factors. After 15 h, images were captured using a light microscope in high magnification.
Confocal Microscopy
bEnd.3 cells were grown overnight on ibiTreat 8 chamber μ-Slides (ibidi, Martinsried, Germany). Cells were treated with cytocholasin D for 3 hr after which they were stained with a cocktail containing 2 μg/ml of each of anti-C5ar1-AF647 (Abd Serotec), anti-VEGFR2-AF488 (BioLegend) + Streptavidin-Tx Red (Life Sciences), and anti-IL-6R-biotin (BioLegend) for 1 h. The slides were washed 3× with 1× PBS and fixed with 2% formaldehyde. Images were collected using a 63× (1.4 NA) oil immersion objective on an UltraVIEW Vox (Perkin Elmer, Waltham, MA) spinning disk confocal system attached to a Leica DMI 6000B inverted microscope. Resulting images were subjected to deconvolution using Autoquant software embedded in Metamorph imaging software (Downington, PA).
Single Quantum Dot Tracking
HeLa cells were seeded at ~3×104 cells per well in an 8-well chamber coverglass slide (Lab-Tec, Thermo Scientific #155411). After allowing cells to adhere for ~8 h and serum starvation in serum-free medium overnight, cells were pre-treated for ~1 h in serum free medium alone or serum free medium plus 5 μg/ml each of anti-C3a and anti-C5a neutralizing mAbs. For single particle tracking, culture media was changed for Tyrodes buffer (135mM NaCl, 10mM KCl, 0.4mM MgCl2, 1mM CaCl2, 10mM HEPES, 20mM glucose, 0.1% BSA, pH 7.2). QD conjugates were prepared as previously described (5). Briefly, equal molar ratios of EGF-biotin (Life Technologies # E-3477) and QD 655 streptavidin (Molecular Probes #Q10121MP) were mixed in PBS with 1% BSA and incubated at 4 °C with agitation for ~4 h. Cells in pre-warmed Tyrodes buffer were incubated with 250 pM EGF-QD655 for 45–60 seconds, followed by extensive washing. Single particle tracking was carried out for up to 8 min after the addition of EGF to avoid tracking of internalized EGFR. Cells pre-treated with C3ar1-A/C5ar1-A were maintained, washed, and imaged in buffer that contained C3ar1-A/C5aR-A. To track unliganded EGFR, QD-655 was conjugated directly to a small, monovalent camelid antibody fragment against EGFR that is non-activating and does not compete for ligand binding (variable fragment of heavy chain antibody, or VhH). Alternatively, HeLa cells were transfected with HA-tagged EGFR (HA-EGFR) and tracked using QD conjugation to a biotinylated anti-HA Fab fragment. Briefly, ~106 HeLa cells were transfected with 2 μg HA-EGFR in pcDNA3.1(+) vector using Amaxa nucleofection. Twelve post-transfection, cells were seeded in 8-well chamber coverglass slides at ~3×104 cells per well, allowed to adhere for ~8 h, and serum starved overnight. Single particle tracking was carried out using biotinylated, monovalent anti-HA antibody-Fab fragments conjugated directly to QD 655 streptavidin. Cells in pre-warmed tyrodes buffer were incubated with 200pM VhH-QD655 for 4–5 min. Following extensive washing, the cells were treated without control or with EGF (100 nM) or human C5a (hC5a) (100 ng/ml) in Tyrodes buffer for 2 min to allow for ligand binding for 5 min, after which single particle tracking of EGFR was carried out up to ~8 min.
Wide field imaging for SPT was performed using an Olympus IX71 inverted microscope equipped with a 60× 1.2 N.A. water objective; and an objective heater (Bioptechs, Butler, PA) maintained at temperature (34–36 °C). Wide field excitation was induced by a mercury lamp with a 436/10 nm bandpass excitation filter and a 50/50 neutral density filter. Emission was collected by an electron multiplying CCD camera (Andor iXon 887). QD emission was collected using a 600 nm dichroic and 655/40 nm bandpass filter (Chroma, Rockingham, VT). An additional 1.6× magnification ratio with a single pixel equivalent to 166.7 nm was used and image processing was performed using Matlab (The MathWorks, Inc., Natick, MA) in conjunction with the image processing library DIPImage (Delft University of Technology). Details of the single particle tracking analysis are given in (5). Diffusion coefficients were calculated based on a weighted linear fit of the first twenty time lags, covering 1.0 second. R-squared values of the weighted linear fit were above 0.98.
AlexaFluor-Labeled Ligand binding assay
Human EGF was labeled with AlexaFluor647 (Life Technologies), human C5a was labeled with AlexaFluor 555, and human IL-6 was labeled with AlexaFluor 488 using Life Sciences microscale labeling kits per the manufacturer’s instructions. AlexaFluor 555-labeled transferrin (Life Technologies) was used as a control. 1×106 human aortic endothelial cells (HAEC) were preincubated with 10 μg/ml C3ar1-A/C5ar1-A for 30 min. Cells were then loaded onto the flow cytometer (BD LSR II) and baseline fluorescence was recorded for 90 sec. Tubes were removed and the labeled ligands were added and fluorescence was recorded for an additional 250 sec. The exported FCS files were processed in FlowJo 10.0.7 and data are presented as Median Fluorescence Intensity (MFI) with ‘moving average’ smoothing.
Co-immunoprecipitation Assays
Cells were lysed in 1× Cell Lysis Buffer (10X) (Cell Signal, cat#9803) supplemented with 1mM PMSF and 1 Complete Mini protease inhibitor tablet (Roche Cat# 11836153001) for 10 min on ice [1 mM Na Orthovanadate was used to inhibit phosphatase activity when lysates were to be assayed for kinase activity]. Lysates were then sonicated 3× for 2 min to break apart nucleic acids, after which the cells were centrifuged for 10 min at 12,000× G. Clean supernatants were transferred to new tubes and incubated with Protein-A/G beads to preclear the lysates and prevent nonspecific co-IP. Ab against VEGFR2, PDGFR, EGFR, C5ar1, C3ar1, PTEN, β-Arrestin, IL-6R, gp130, CK2, or CD31 were added and samples were incubated overnight at 4oC. Protein-A/G beads (Santa Cruz Biotechnology) were used to pull down Ab and IP’s were assayed by western blotting, ELISA, or protein activity via kits as described.
CK2 and PTEN activity assays
Following stimulation (as described, e.g. 5 min with VEGF or VEGF+C3ar1-A/C5ar1-A) cells were lysed and CK2 or PTEN were IP’d overnight at 4°C. CK2 activity was assayed by measuring transfer 32P to a CK2 substrate peptide via Millipore’s Casein Kinase 2 Assay Kit (Cat#17–132) per the manufacturer’s instructions. PTEN Activity in IPs was determined using Echelon’s (Salt Lake City, UT) PTEN Activity ELISA.
Statistics
Power calculations and animal numbers were determined using the information provided by ‘http://statpages.org’ and the therein linked Russ Lenth’s power and sample-size calculator obtainable through ‘http://www.stat.uiowa.edu/~rlenth/Power/index.html#Download_to_run_locally’. To achieve a true difference between means of 0.5 and a power of 0.2 testing the difference between 2 means via an unpaired, two-tailed Student’s T test, 10 mice are required in each group and there are 10 experimental groups. Statistical significance for all experimental data was determined by Student’s T test (unpaired, two-tailed) performed in Microsoft Excel, SigmaPlot or GraphPad Prizm 6 with a significance threshold values of p<0.05. Except where indicated, all experiments were repeated at least 3 times. Data are presented as mean values with SD. Statistical significance for all experimental data was determined by Student’s T test (unpaired, two-tailed) performed in Microsoft Excel, SigmaPlot or GraphPad Prizm 6 with a significance threshold values of p<0.05. All experiments were repeated at least 3 times. Data are presented as mean values plotted with SD unless otherwise noted in legend.
Results
Fibroblast and smooth muscle cell growth depends on autocrine C3ar1/C5ar1 signaling
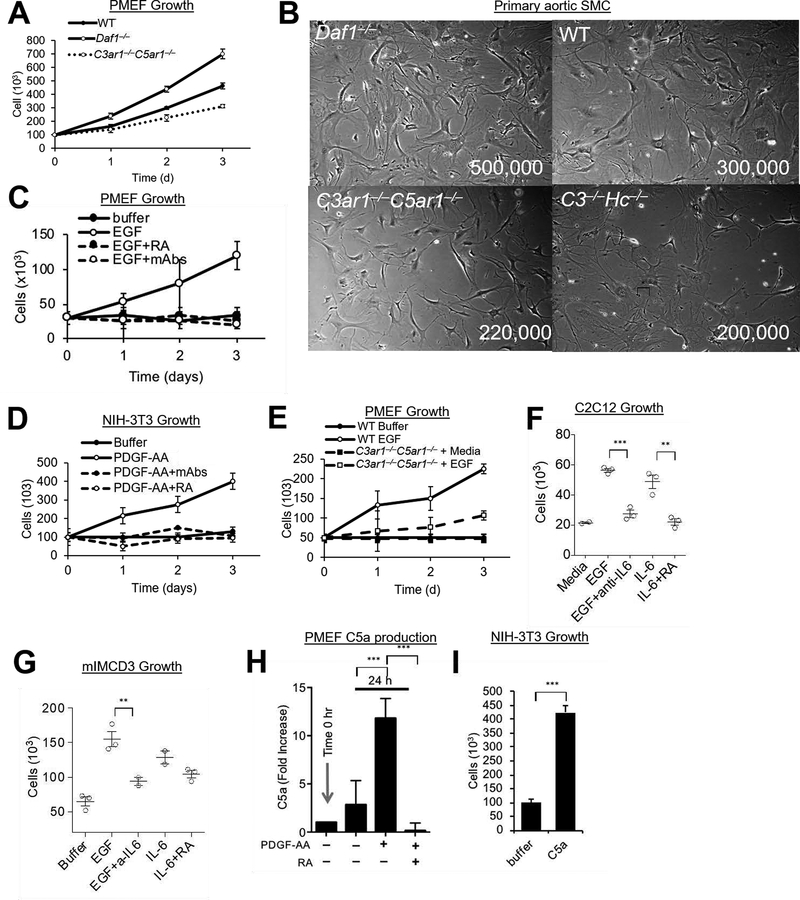
To study EGFR signaling, we established primary cultures of fibroblasts (PMEFs) from decay accelerating factor [(Daf1−/−) aka CD55−/−] mice [in which autocrine C3ar1/C5ar1 signaling is potentiated (6)], from WT mice (in which autocrine signaling is physiologically regulated), and from C3ar1−/−C5ar1−/− and C3−/−Hc−/− (Hc encodes C5) mice (in which C3ar1/C5ar1 signaling is disabled). For studies of PDGFR signaling, we established primary cultures of aortic smooth muscle cells (SMCs) from each genotype. Daf1−/− PMEFs and SMCs grew more rapidly than WT PMEFs and SMCs (Fig 1AB) indicative of autocrine C3ar1/C5ar1 signaling regulating PMEF and SMC growth. In accordance with this, deficiency of C3ar1/C5ar1 or of the (C3/C5) sources of the C3a/C5a ligands (Fig 1B) attenuated growth in each cell type. The C3ar1−/−C5ar1−/− and C3−/−Hc−/− PMEFs and SMCs retained some albeit slower growth than WT PMEFs and SMCs that was stable with passage (see Discussion). Trypan blue exclusion assays showed some apoptosis (see below). These results paralleled those for the constitutive growth of C3ar1−/−C5ar1−/− ECs (7). They are consistent with PMEFs and SMCs locally producing C3a/C5a from endogenously synthesized complement and autocrine C3ar1/C5ar1 signaling regulating growth as characterized in immune cells (8, 9). siRNA experiments in our prior study on VEGFR2 signaling (7) validated the integral role C3ar1/C5ar1 signaling in cell growth.
Figure 1. PDGFR and EGFR Signaling.
(A) Primary mouse embryonic fibroblasts (PMEF) (1 ×107) were cultured for 3 d on 10 cm plates in DMEM/F12 and cell numbers quantitated daily. (B) Primary smooth muscle cells (SMC) were isolated from the aortic rings of Daf1−/−, WT, C3−/−Hc−/−, and C3ar1−/−C5ar1−/− mice. Following growth for 2 wks in complete smooth muscle cell media (Lonza) supplemented with 5% FBS, 2 ×106 cells were grown in the same media for 3d after which cells were photographed. (C) Starved PMEFs were incubated for 3 d in DMEM/F12 medium containing 20% FBS, 2 mM L-glutamine, 1% nonessential amino acid, 0.09 mg/ml EC growth supplement, 1% antibiotic/antimycotic, 100 units/ml penicillin, 100 g/ml streptomycin, and 0.09 mg/ml heparin VEGF-A (30 ng/mL) ± anti-C3a/anti-C5a mAbs (5 μg/ml) or C3ar1-A/C5ar1-A (RA; 10 ng/ml ea) and cell numbers were quantified daily. (D) NIH-3T3 cells serum starved in 0.5% serum were incubated over 3 d with PDGF-AA (30 ng/mL) in the absence or presence of anti-C3a/C5a mAbs (5 μg/ml each) or RA (10 ng/ml each and their growth quantified daily. (E) Serum starved cultures of PMEFs from WT and C3ar1−/−C5ar1−/− mice were incubated with media alone or with EGF (30 ng/ml) and cell numbers counted daily. (F) C2C12 myocytes were alternatively cultured for 2 d with EGF (30 ng/ml), EGF+anti-IL-6 (5 μg/ml), IL-6 (10 ng/ml), or IL-6+ RA (10 ng/ml each), after which cell numbers were quantified. (G) mIMCD3 cells were cultured for 3 d with EGF (30ng/ml), EGF+anti-IL-6 (2 μg/ml), IL-6 (10 ng/ml), or IL-6+ RA (10 ng/ml each), after which cell numbers were quantified. (H) PMEFs were incubated for 24 h with media alone or with PDGF-AA (30 ng/ml) in the absence or presence of RA (10 ng/ml each) after which C5a in culture supernatants was assessed by ELISA. (I) NIH-3T3 cells were incubated with media in the absence or presence of added C5a (100 ng/ml) for 3 d after which cell numbers were quantified.
EGFR and PDGFR survival and mitotic activities depend on both C3ar1/C5ar1 and IL-6R-gp130 joint signaling.
We next tested whether autocrine C3ar1/C5ar1 signaling is essential for EGFR and PDGFR mitotic signaling in PMEFs and SMCs. For these studies, we used pharmaceutical C3ar1/C5ar1 antagonists in WT primary mouse and human cells and cell lines corresponding to the different cell lineages because this approach avoided genetic compensations that occur in the C3ar1−/−-C5ar1−/− knockouts. Disruption of C3ar1/C5ar1 signaling abolished EGF induced growth in WT PMEFs that express (prototypical) EGFR (Fig 1C). C3ar1/C5ar1 blockade had the same disruptive effect on EGF (not shown) or PDGF-AA induced growth of NIH-3T3 cells (which express both Her-2 and PDGFR-αα) (Fig 1D). It similarly blocked PDGF-BB induced growth of primary WT SMCs which express PDGFR-ββ (not shown). Genetic deficiency of C3ar1/C5ar1 signaling had a similar, albeit less complete abrogating effect (Fig 1E), as observed for VEGFR2 signaling (1) and linked to compensatory pathways.
As indicated (Introduction), our previous study on VEGFR2 signaling interlinked IL-6R-gp130 signaling as well as C3ar1/C5ar1 signaling as requisite processes in VEGFR2 signaling in ECs (1). Based on that, we next examined whether the same interdependence applies to EGFR and PDGFR signaling. Added IL-6, like added C5a, evoked the growth of C2C12 myoblasts and of mIMCD-3 kidney epithelial cells similarly to added EGF (Fig 1FG) and blockade of IL-6 signaling inhibited the EGF induced growth (lanes 3). IL-6 induction of growth in both cell types was reversed by blockade of C3ar1/C5ar1 signaling (lanes 5). PMEFs produced C5a constitutively (Fig 1H). Adding PDGF-AA or EGF (not shown) to PMEFs augmented C5a production (Fig 1H) in concert with inducing PMEF proliferation (not shown). NIH-3T3 cells similarly produced C5a constitutively and EGF increased C5a production (not shown) in concert with inducing cell growth (Fig 1I). Control studies showed that the effect of added C5a on cellular proliferation was abolished by blockade of C3ar1/C5ar1 signaling (Fig 1H). Consistent with autocrine C3ar1/C5ar1 signaling functioning jointly with PDGFR and EGFR signaling to enable cell growth, blockade of the C3a/C5a ligands had the same effect as blockade of C3ar1/C5ar1 (Supplementary Material Fig S1A). Together with the data in Figs 1FG, signaling studies that will be presented later (Figs 4EF, and 7 A–D and H and I documented the same requirement for IL-6R-gp130 signaling. Taken together, the data argued that the mitotic effects of EGFR and PDFR signaling depend on C3ar1/C5ar1 and IL-6R-gp130 joint signaling. This result parallels that found for VEGFR2 signaling (1).
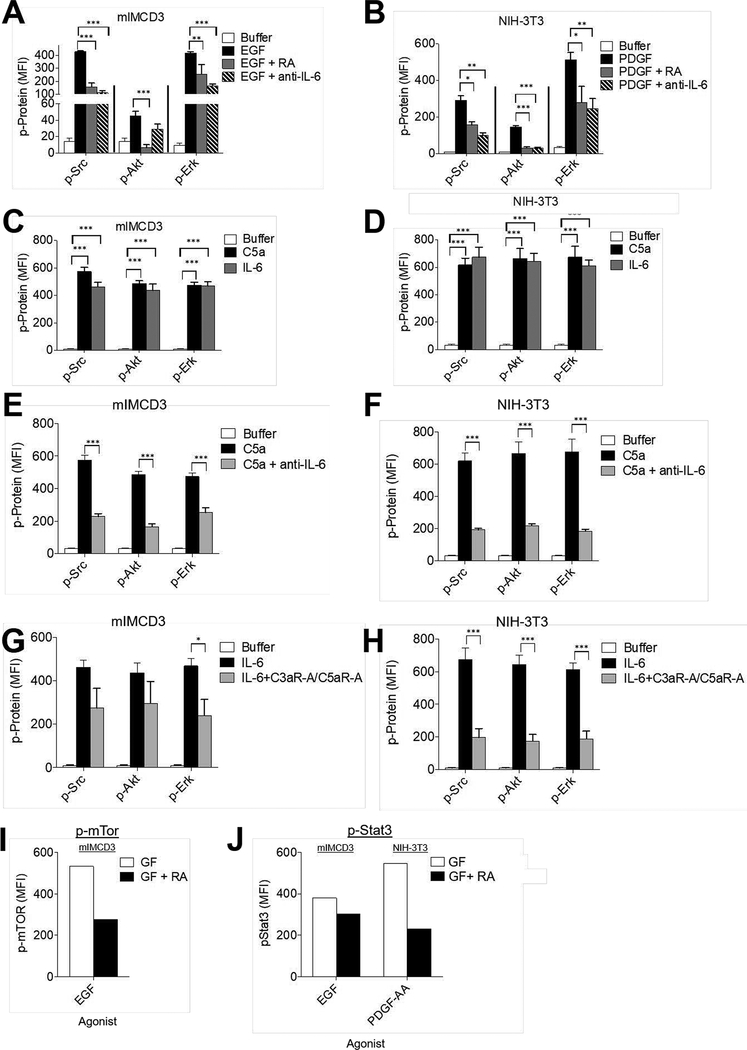
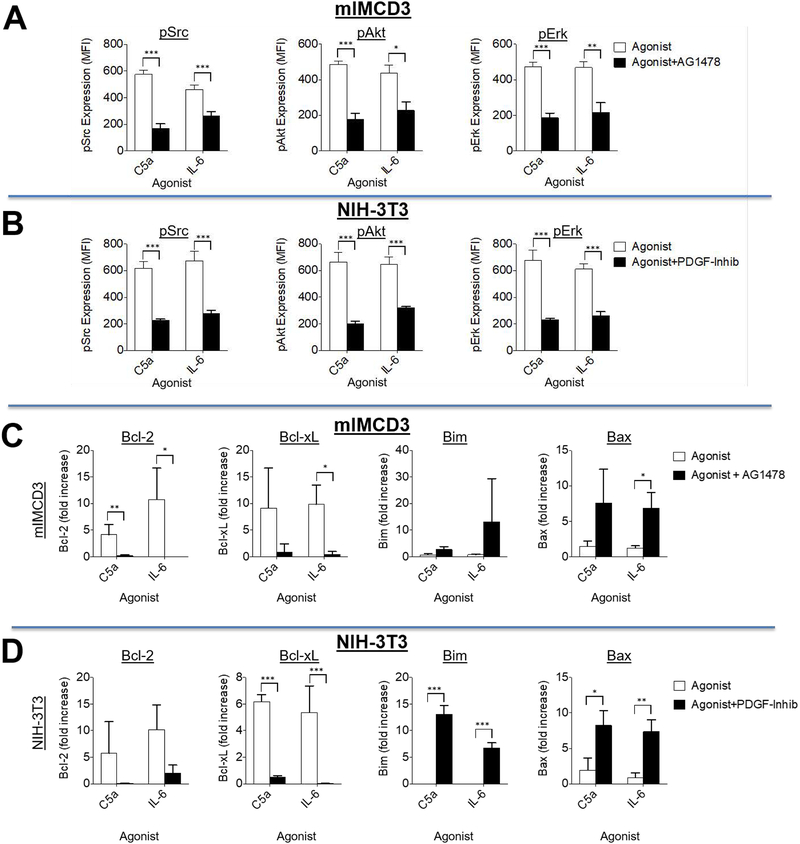
Figure 4. RTK signaling depends on C5ar1/C3ar1 and IL-6R-gp130 co-signaling.
(A) Serum-starved mIMCD3 and (B) serum-starved NIH-3T3 cells were incubated for 5 min with EGF (30 ng/ml) and PDGF-AA (30 ng/ml) respectively in the absence or presence of RA (10 ng/ml each) or anti-IL-6 mAb (10 ug/ml). Following extraction of the cells with the buffer provided by the manufacturer, the extracts were assayed for p-Src, p-Akt, and p-Erk by Luminex (multiplex) assay. (C) Serum-starved mIMCD3 and (D) serum-starved NIH-3T3 cells were incubated for 5 min with media alone, C5a (100 ng/ml) or IL-6 (100 ng/ml). Following extraction of the cells as in panels AB, the extracts were assayed for p-Src, p-Akt, and p-Erk by Luminex (multiplex) assay. (E) Serum-starved mIMCD3, and (F) serum-starved NIH-3T3 cells were incubated for 5 min with C5a (100 ng/ml) or C5a plus anti-IL-6 mAb (10 μg/ml). Following incubation, cells were assayed for p-Src, p-AKT and p-Erk by Luminex (multiplex) assay. (G)Serum-starved mIMCD3 and (H) serum-starved NIH-3T3 cells were incubated for 5 min with IL-6 (10 ng/ml) in the absence or presence of RA (10 ng/ml each. Following incubation for 5 min, cells were assayed for p-Src, p-AKT and p-Erk by Luminex (multiplex) assay. (I) Serum-starved mIMCD3 cells were incubated for 5 min with EGF (30 ng/ml) in the absence or presence of RA (10 ng/ml each). Following incubation for 5 min, cells were assayed for p-mTOR by intracellular FACS staining (system only assays phosphorylated mTOR). (J) Serum-starved mIMCD3 or serum-starved NIH-3T3 cells were incubated for 8 h with EGF (30 ng/ml) or PDGF-AA (30 ng/ml) respectively in the absence of presence of RA (10 ng/ml each), after which, phospho-STAT3 was assayed by flow cytometry (system only assays phosphorylated STAT3).
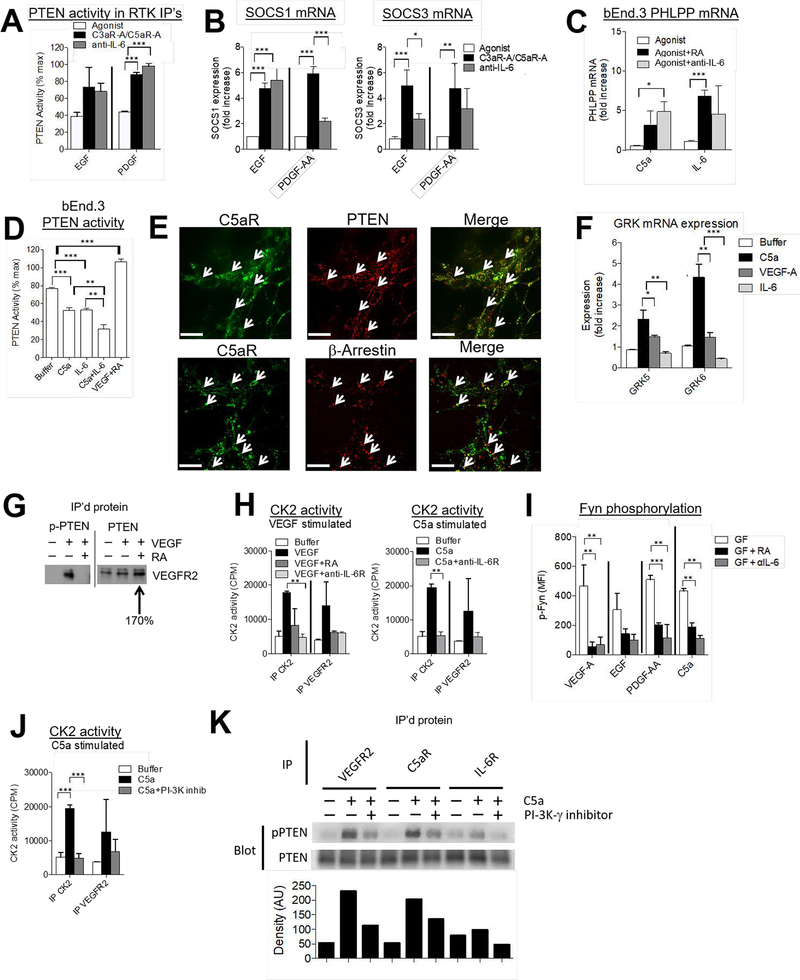
Figure 7. C3ar1/C5ar1 and IL-6R-gp130 co-signal transduction lifts the brake on RTK activation.
(A) Serum-starved mIMCD3 and serum-starved NIH-3T3 cells were incubated for 5 min with 30 ng/ml EGF or PDGF-AA, respectively in the absence of presence of RA (10 ng/ml each) or anti-IL-6 mAb (10 ug/ml). Each RTK was subsequently IP’d and PTEN enzymatic activity in the eluent of the washed IP was determined by ELISA as described in the Methods. (B) mIMCD3 and NIH-3T3 cells were incubated for 1 h with EGF or PDGF-AA in the absence or presence of RA (10 ng/ml each) or anti-IL-6 mAb (10 ug/ml), after which SOCS1/SOCS3 mRNA expression were assayed by qPCR. (C) bEnd.3 cells were incubated for 1 h with VEGF-A (30 ng/ml), C5a (100 ng/ml), or IL-6 (10 ng/ml) in the absence or presence of RA (10 ng/ml each) or anti-IL-6 mAb (10 ug/ml), after which PHLPP mRNA expression was assayed by qPCR. (D) bEnd.3 cells in complete media were incubated for 5 min with C5a (100 ng/ml), IL-6 (10 ng/ml), C5a (100 ng/ml) plus IL-6 (10 ng/ml), or VEGF-A (30 ng/ml) + RA (10 ng/ml each) after which PTEN was IP’d. Enzymatic activity in the eluent of the washed IP was assayed by ELISA as described in the Methods. (E) bEnd.3 cells serum starved on slides were preincubated for 3 h with cytochalasin D on ibiTreat 8 chamber μ-Slides (ibidi, Martinsried, Germany). They were then incubated for 20 min with VEGF-A as described in the Methods after which the cells were fixed and assayed for C5ar1, PTEN (upper) and β-arrestin (lower) expression by confocal microscopy (cells were visualized at 63×; Scale bar = 200 μm). (F) bEnd.3 cells in complete media were incubated for 60 min with C5a (100 ng/ml), IL-6 (10 ng/ml), or VEGF-A (30 ng/ml) after which mRNA levels for GRK5 and GRK 6 were assayed by RT-qPCR. (G) bEnd.3 cells were incubated for 5 min with VEGF-A (30 ng/ml) or VEGF-A (30 ng/ml) plus RA (10 ng/ml each). Following cell extraction, (left) anti- S380, T382, T383-phospho-PTEN (inactive) and (right) anti-PTEN (total) IPs were prepared. Immunoblots of the IP’d proteins were probed with anti-VEGFR2 Ab. (H) Left: bEnd.3 cells were incubated for 5 min with VEGF-A (30 ng/ml) in the absence of presence of RA (10 ng/ml each) or anti-IL-6R mAb (10 ug/ml). Following the incubation, anti-CK2 or anti-VEGFR2 IP’s were prepared and the IP’s assayed for CK2 kinase activity by radioassay as described in Methods. Right: In other wells on the plate, bEnd.3 cells were incubated for 5 min with C5a (100 ng/ml) in the absence or presence of anti-IL-6R mAb (10 ug/ml) and IPs similarly analyzed for enzyme activity. (I) bEnd.3, mIMCD3, or NIH-3T3 cells were incubated for 5 min with their corresponding GF, i.e., VEGF-A, EGF, or PDGF-AA (30 ng/ml each in the absence or presence of RA (10 ng/ml each) or anti-IL-6 mAb (10 ug/ml). The alternatively treated cells were assayed for p-Fyn by Luminex assay as described in the Methods. In other wells on the same plate C5a (10 ug/ml) was used in place of GF. (J) bEnd.3 cells were incubated for 5 min with C5a (100 ng/ml) in the absence or presence of the PI-3Kɣ inhibitor AS252424 (10 μM). Anti-CK2 or anti-VEGFR2 IPs were prepared and the IPs were assayed for CK2 kinase activity by radio assay as described in Methods. (K) bEnd.3 cells were incubated for 5 min with C5a (100 ng/ml) ± the PI-3Kɣ inhibitor AS252424 (10 μM), after which anti-VEGFR2, anti-C5ar1, and anti-IL-6R IPs were prepared and immunoblots of the IP’s proteins probed with anti-S380, T382, T383-phospho-PTEN (inactive) and anti-(total) PTEN mAb. Densitometry of phosphorylated PTEN is graphed below as arbitrary units (AU).
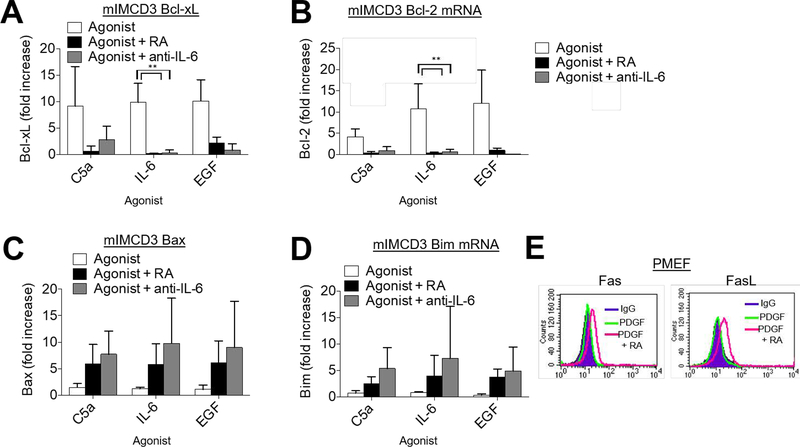
Assays of anti- and pro-apoptotic markers under serum starved conditions showed that the viability signaling of EGFR and PDGFR, like the mitotic signaling, is similarly dependent on coordinated C3ar1/C5ar1 and IL-6R signaling. Adding C5a-, IL-6-, or EGF to serum starved cultures of mIMCD3 cells upregulated anti-apoptotic Bcl-xl (Fig 2A) and Bcl-2 (Fig 2B) in a near identical fashion, whereas blockade of C3ar1/C5ar1 or IL-6R signaling abolished the upregulations. The C3ar1/C5ar1 or IL-6R blockades concurrently upregulated pro-apoptotic Bax (Fig 2C) and Bim (Fig 2D). Consistent with effects on both the extracellular and mitochondrial programmed cell death (PCD) pathways, blockade of C3ar1/C5ar1 in PDGF-treated PMEFs or SMCs (not shown) upregulated both Fas and FAS ligand (FAS-L) expression (Fig 2E). Comparable effects were observed with the mIMCD3 epithelial cells and NIH-3T3 cells (not shown). These findings that are consistent with our studies in immune cells (9) and ECs (1) that tonic C3ar1/C5ar1 signaling maintains viability. Together, the data pointed to combinatorial C3ar1/C5ar1 and IL-6R signaling participating in EGFR and PDGFR anti-apoptotic as well as mitotic signaling in PMEFs and SMCs. This signaling interconnection will be mechanistically examined in multiple ways in the experiments that follow.
Figure 2. Autocrine C3ar1/C5ar1 signaling provides survival signaling to non-immune cells.
(A-D) mIMCD3 cells were incubated for 1 h with EGF (30 ng/ml) in the absence or presence of RA (10 ng/ml each) or anti-IL-6 mAb after which (A) Bcl-xL, (B) Bcl-2, (C) Bax, and (D) Bim mRNA expression levels were quantified by qPCR. E) WT PMEFs were incubated in complete media with RA (10 ng/ml each) for 1 h after which cells were stained for Fas/FasL.
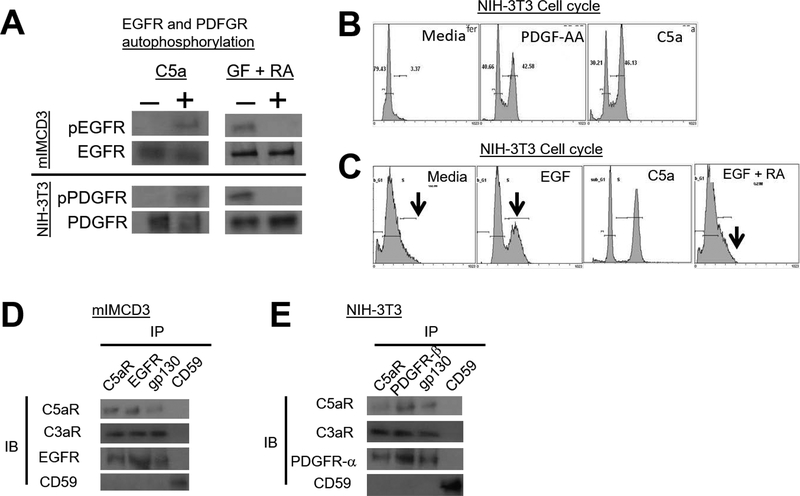
EGFR and PDGFR auto-phosphorylation and cell cycle progression depend on C3ar1/C5ar1 and IL-6R-gp130 joint signaling in a signaling platform
We first tested whether C3ar1/C5ar1 and IL-6R-gp130 are requisite participants in EGFR and PDGFR signaling and are jointly associated with the two RTKs in signaling platforms as found for VEGFR2. Similarly to C5a’s induction of VEGFR2 auto-phosphorylation in ECs (7), adding C5a to mIMCD3 cells induced auto-phosphorylation of EGFR at its auto-phosphorylation sites (Fig 3A left upper). Added C5a similarly induced auto-phosphorylation of PDGFR-αα at its auto-phosphorylation sites in NIH-3T3 cells (Fig 3A left lower). Consistent with these results, pharmaceutical blockade of C3ar1-A/C5ar1-A abolished EGFR (Fig 3A right upper) and PDGFR-αα (Fig 3A right lower) auto-phosphorylation induced by EGF or PDGF-αα, respectively. Adding C5a to serum starved NIH-3T3 cells (Fig 3B) or serum starved primary SMCs (not shown) triggered cell cycle transition from G0 into G2. Likewise, NIH-3T3 cell cycle entry induced by EGF (Fig 3C) or PDGF-AA (not shown) was abolished by C3ar1/C5ar1 blockade. The opposing effects of C3ar1/C5ar1 antagonism (RA) and of C5a on PDGFR and EGFR auto phosphorylation and cell cycle entry closely paralleled the opposing effects on cell growth in Figs 1FG (above). The opposing treatments likewise had opposite effects on downstream signaling to ERK and other signaling intermediates associated with PDGFR and EGFR activation (shown in Fig 4A–J below).
Figure 3. Autocrine C3ar1/C5ar1 signaling in non-immune cells is needed for PFGFR and EGFR autophosphorylation and cell cycle entry/progression.
(A) Upper: Serum-starved mIMCD3 (upper) or NIH-3T3 cells (lower) were incubated for 5 min with C5a (100 ng/ml) upper, or with GF (30 ng/ml) plus RA (10 ng/ml each) after which EGFR or PDGFR were IP’d and immunoblots of the IP’d EGFR and PDGFR probed for p-EGFR or p-PDGFR. (B) NIH-3T3 cells were incubated for 24 h with PDGF-AA (30 ng/ml) or C5a (100 ng/ml) and entry into cell cycles was assessed by propidium iodide incorporation. (C) Serum-starved NIH-3T3 cells were incubated for 24 h with EGF (30 ng/ml) in the absence or presence of RA (10 ng/ml each) and cell cycle entry was assessed by propidium iodide incorporation. (D) Serum-starved mIMCD3 and (E) serum-starved NIH-3T3 cells were incubated for 5 min with EGF (30 ng/ml) and PDGF-AA (30 ng/ml), respectively. Following detergent extraction of the cells with 1× Cell Lysis Buffer (10X) (Cell Signal, cat#9803) supplemented with 1mM PMSF and 1 Complete Mini protease inhibitor tablet (Roche Cat# 11836153001) and [1 mM Na Orthovanadate, anti-C5ar1, anti-EGFR, anti-PDGFR-α, anti-gp130, and anti-CD59 IPs were prepared. Immunoblots of the IP’d proteins were probed for C5ar1, C3ar1, EGFR, and CD59 and for C5ar1, C3ar1, PDGFR-α and CD59, respectively.
We showed that in ECs that anti-VEGFR2 immunoprecipitates (IPs) brought down C3ar1, C5ar1 and gp130 associated with IL-6R (7). Anti-EGFR IPs in mIMCD3 cells (Fig 3D) and anti-PDGF IPs in NIH-3T3 cells (Fig 3E) (both of which express IL-6R) similarly brought down C3ar1, C5ar1, and gp130, but did not bring down a non-relevant protein, i.e. CD59. The dependence of EGFR and PDGFR autophosphorylation on C3ar1/C5ar1 and IL-6R-gp130 signaling and the association (direct or indirect) of each RTK with C3ar1/C5ar1 and IL-6R-gp130 thus paralleled findings for VEGFR2.
Downstream EGFR and PDGFR signaling depends on joint activation of C3ar1/C5ar1 and IL-6R-gp130.
We used multiplex analyses to investigate how signaling via C3ar1/C5ar1 and IL-6R-gp130 impacts downstream signaling by EGFR and PDGFR. Blockade of C3ar1/C5ar1 or IL-6R signaling concomitantly with EGF stimulation of mIMCD3 cells (Fig 4A) or PDGF-AA stimulation of NIH-3T3 cells (Fig 4B) suppressed Src, AKT, and ERK activation by both RTKs by 50–90%. We used 5 min incubation because kinetic analyses in our studies of C3ar1/C5ar1 signaling in immune cells showed that it was the time of maximal kinase activity. Those studies (9) also showed that C3ar1/C5ar1 blockade prevented AKT phosphorylation for as long as 6 h. The suppressive effect of IL-6R blockade on AKT phosphorylation was more profound than that in mIMCD3 and NIH-3T3 cells in other cell types e.g., human HeLa cells, human umbilical endothelial cells (HUVEC) (Supplementary Material Fig S1BC) and murine bEnd.3 vascular endothelial cells (1). Complete blockade of AKT, Src, and ERK by the C3ar1/C5ar1 antagonists (RA) was observed with primary murine PMEFs (Fig 1C), primary human aortic endothelial cells (HAEC) and HUVEC in Fig S2. The incomplete suppressive effect of RA or IL-6R blockade in the transformed cell lines could reflect mutations in growth relevant genes. Conversely, the addition of C5a or IL-6 to mIMCD3 cells or NIH-3T3 cells induced the phosphorylation of all three signaling intermediates in both cells (Fig 4CD) as well as the other cell types (not shown). Consistent with C3ar1/C5ar1 and IL-6R signaling being interconnected with each other as well as with EGFR and PDGFR signaling respectively, the activations of Src, AKT, and ERK induced by added C5a were inhibited by blockade of IL-6R signaling (Fig 4EF) and the activations induced by added IL-6 were inhibited by C3ar1/C5ar1 blockade (Fig 4GH). Total levels of each signaling intermediate were not affected. Comparable results were obtained with human HeLa cells (Fig S2A) and with primary human aortic endothelial cells (HAEC) (Fig S2B). Activation of mTOR (Fig 4I) and phosphorylation of STAT3 (Fig 4J) in response to EGFR and PDGFR were similarly reduced by C3ar1/C5ar1 blockade.
C3ar1/C5ar1 and IL-6R-gp130 signaling depend on RTK signaling.
We next examined whether EGFR or PDGFR signaling are obligate participants in C3ar1/C5ar1 and IL-6R-gp130 signaling in the corresponding cell types. We cultured mIMCD3 cells and NIH-3T3 cells with C5a or IL-6 for 5 min in the absence or presence of pharmaceutical EGFR or PDGFR antagonists, AG1478 and PDGFR-inhib, and measured the above signaling intermediates. Each RTK antagonist abolished C5a or IL-6 activation of Src, AKT, and ERK in the corresponding cell type (Fig 5AB). Consistent with these data, culturing mIMCD3 cells with EGF in the presence of AG1478 blocked both endogenous C5a generation and endogenous IL-6 production (Supplementary Material Fig S1D). Corresponding abrogating effects on C5a and IL-6 production were observed for blockade of PDGFR-AA signaling in NIH-3T3 cells (not shown). These studies showed that EGFR or PDGFR signaling is needed for the reported abilities of added C3a/C5a or IL-6 to induce activation of Src, AKT and ERK in myeloid and NIH-3T3 cells, findings attributed to signaling through C3ar1/C5ar1 or IL-6R in and of themselves (10–12). Similarly, inhibition of EGFR or of PDGFR signaling reversed the reported (13, 14) anti-apoptotic effects of C5a and IL-6 (Fig 5CD).
Figure 5. Suppression of RTK signaling inhibits C5a and IL-6 signal transduction.
(A) Serum-starved mIMCD3 cells and (B) serum-starved NIH-3T3 cells were incubated for 5 min with C5a or IL-6 in the absence or presence of the respective RTK antagonist AG1478 (for mIMCD3, 30 μM) or PDGFR tyrosine kinase Inhibitor III [PDGFR-inhib](for NIH-3T3; 10 μM) after which the cells were assayed for Src, Akt, and Erk phosphorylation by Luminex assay. (C) Serum-starved mIMCD3 cells and (D) serum-starved NIH-3T3 cells were incubated for 1 h with C5a (100 ng/ml) or IL-6 (10 ng/ml) in the absence or presence of the respective RTK antagonist AG1478 (for mIMCD3, 30 μM) or PDGFR tyrosine kinase Inhibitor III [PDGFR-inhib](for NIH-3T3; 10 μM), after which the cells were extracted with TRIZOL and Bcl-2, Bcl-xL, Bim, and Bax mRNA transcripts quantitated by qPCR.
These findings functionally implicate EGFR and PDGFR signaling as critical participants in C3ar1/C5ar1 and IL-6R signaling. This functional interdependence operating in the opposite direction further supports the proposition that these two RTKs confer their downstream signaling jointly with C3ar1/C5ar1 and IL-6R-gp130 signaling.
Signaling through C3ar1/C5ar1 and IL-6R affects the dynamic behavior of RTKs
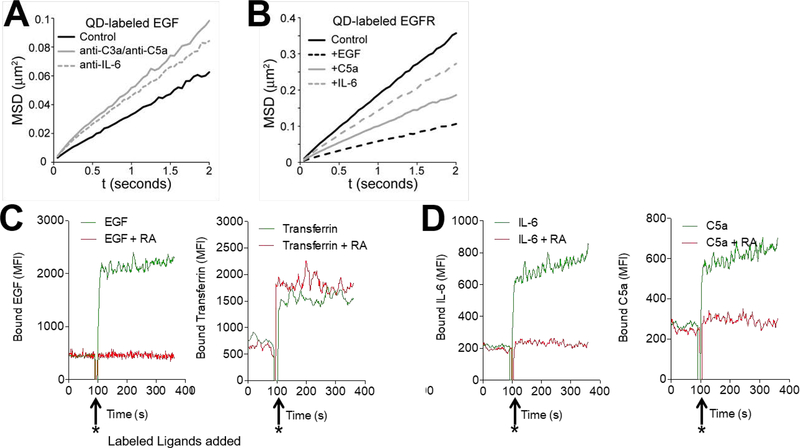
While the above experiments and our previous work on VEGFR2 signaling (1) provided in vitro and in vivo functional data as well as co-IP, confocal, ligand pull down, and BRET data indicating that the four receptors systems are functionally and physically interconnected, they utilized methods that in principle could have altered the cells. To assess whether the participation of C3ar1/C5ar1 or IL-6R signaling in RTK signaling and vice versa is accompanied by corresponding changes in the dynamic behavior of the RTK in the native cell membrane, we applied state-of-the-art single particle tracking methods to evaluate EGFR diffusion in the membrane of live cells. Our previous work has shown that changes in receptor diffusion are an indirect read-out of receptor phosphorylation state (Low-Nam 2011; Steinkamp 2014, Valley 2015), such that a reduced mobility reflects an increase in phosphorylation. Fluorescent quantum dots (QDs) were either directly conjugated to EGF ligand or alternatively conjugated to a non-activating anti-EGFR camelid (alpaca single chain antibody) fragment to quantify the diffusion of EGFR in a ligand-bound and unliganded state, respectively (Low-Nam et al, 2011). EGFR diffusion in the plasma membrane is then monitored by acquiring time series of single molecule motion, as described previously (Low-Nam 2011) (see Supplemental Material Movies). Mobility correlates with RTK phosphorylation status (5, 15) such that changes in diffusion reflect changes in the phosphorylation level. Pretreatment of HeLa cells with anti-C3a/anti-C5a mAbs or anti-IL-6 mAb prior to the addition of QD-labeled EGF (QD-EGF) to the Hela cells increased the mobility of ligand bound EGFR as compared to that in untreated HeLa cells (Fig 6A). The observed increase is consistent with reduced EGFR phosphorylation (as shown in Fig 3A) and decreased inner membrane recruitment of signaling intermediates. Conversely, the addition of EGF, C5a, or IL-6 to HeLa cells decreased the mobility of EGFR tracked by QD-labeled non-activating anti-EGFR camelid antibody fragments (Fig 6B), consistent with augmented EGFR phosphorylation (Fig 3A above) and increased recruitment of signaling intermediates. In an additional set of studies, C5a or IL-6 was added to HeLa cells expressing HA-tagged EGFR to track the unliganded receptor using a QD-labeled anti-HA Fab fragment. Added C5a or IL-6 caused decreased EGFR mobility (consistent with EGFR phosphorylation) relative to controls, whereas added C3ar1-A/C5ar1-A or anti-IL-6R mAb had the opposite effect (not shown). These changes in EGFR mobility are consistent with C3ar1/C5ar1 and IL-6R regulation of the EGFR phosphorylation levels
Figure 6. Effect of C3ar1/C5ar1 orIL-6R signaling on RTK Mobility and GF binding.
(A-B) Single QD tracking of EGFR on serum starved HeLa cells. EGFR mobility was quantified using mean squared displacement (MSD) analysis to calculate receptor diffusion coefficients. In the MSD plot, a steeper slope indicates higher mobility. (A): MSD plot performed using QD-labeled EGF in the absence or presence of anti-C3a/antiC5a mAbs (10 mg/ml each) or anti-IL-6 mAb (10 mg/ml) to track QD-EGF-EGFR. Interference of C3a/C5a (10 ng/ml each) or IL-6 (10 ng/ml) signaling caused an increase in the diffusion coefficient of QD-EGF-EGFR from 0.0087 ± 0.0004 μm2s−1 (control) to 0.0146 ± 0.0009 μm2s−1 with anti-C3a/anti-C5a or to 0.013 ± 0.0007 μm2s−1 with anti-IL-6. (B): MSD plot performed of QD-labeled anti-EGFR camelid Ab in the absence or presence of added EGF (30ng/ml), C5a (10 ng/ml), or IL-6 (10 ng/ml). Endogenous EGFR was tracked via QD-VhH. EGFR displayed the highest mobility in the absence of ligand (control, D = 0.0498 ± 0.0015 μm2s−1). Mobility slowed upon addition of EGF (D = 0.0159 ± 0.0007 μm2s−1). EGFR mobility was also reduced, though to a lesser extent, by added C5a (10 ng/ml) (D = 0.0277 ± 0.0012 μm2s−1) or added IL-6 (10 ng/ml) (D = 0.0388 ± 0.0013 μm2s−1). (C) HAEC were preincubated for 30 min in the absence or presence of RA (10 ng/ml each). Samples were placed in the pickup needle of the flow cytometer and basal fluorescence was recorded. After data acquisition for 90 s, AF647-labeled EGF (~10 ng/ml; left) or AF55-labeled transferrin (~10 ng/ml; right) was added (arrows) and fluorescence was recorded for an additional 270 s. D) HAEC were preincubated for 30 min in the absence or presence of RA (10 ng/ml each). Samples were placed in the pickup needle of the flow cytometer and basal fluorescence was recorded. After data acquisition for 90 s, AF488-labeled IL-6 (~10 ng/ml; left) or AF555-labeled C5a (~10 ng/ml; right) was added (arrows) and fluorescence was recorded for an additional 270 s.
In a second assay system, we investigated whether blockade of C3ar1/C5ar1 or IL-6R signaling affects GF occupation of its respective RTK using live cell flow cytometry. We added AlexaFluor647-labeled EGF or AlexaFluor555-labeled transferrin to primary HAECs pretreated with buffer control or with C3ar1/C5ar1 receptor antagonists and assayed the binding of the labeled EGF and that of labeled transferrin as control. The C3ar1/C5ar1 antagonists interfered with the binding of EGF (Fig 6C left), whereas the binding of transferrin was unaffected (Fig 6C right). In parallel studies, we tested 1) whether blockade of IL-6R would similarly interfere with EGF binding and 2) whether C3ar1/C5ar1 blockade would impact the binding of IL-6 to IL-6R. While anti-IL-6R mAb did not interfere with the binding of EGF (not shown), the C3ar1/C5ar1 antagonists inhibited the binding of IL-6 (Fig 6D left) with efficiency comparable to their inhibition of C5a binding to C5ar1 (Fig 6D right). These findings pointed to C3ar1/C5ar1 signaling affecting either EGFR and IL-6R conformation or their surface expression. Taken together with the biophysical tracking data, these assays argued that the joint activation process involves dynamic changes in the RTK consistent with physical modulation of C3ar1/C5ar1 and IL-6R on the native membrane regulating EGFR phosphorylation and consequent recruitment of signaling intermediates.
C3ar1/C5ar1 or IL-6R-gp130 joint activation operates to suppress inhibitory regulators
The above studies of EGFR and PDGFR in this paper and of VEGFR2 in our previous study (1) provided multiple lines of evidence documenting that the three RTKs are functionally interconnected with C3ar1/C5ar1 and IL-6R-gp130. They did not, however, clarify the signaling mechanism underlying the interconnection or explain the physiological relevance of the joint signaling. Because of the broad inhibition of RTK signaling intermediates evoked by C3ar1/C5ar1 or IL-6R blockade, we questioned whether these signaling partners might function coordinately to repress RTK inhibitory mechanisms. We further hypothesized that the repression of RTK signaling might be constitutively active and dominant. If so, interference with C3ar1/C5ar1 or IL-6R signaling would impair RTK induced growth by virtue of failing to lift restraint on RTK mitotic function.
Consistent with this idea, blockade of either C3ar1/C5ar1 or IL-6R signaling in EGF stimulated mIMCD3 cells, or in PDGF-AA stimulated NIH-3T3 cells up-regulated the lipid phosphatase activity (Fig 7A) and mRNA expression levels (Fig S4A) of PTEN, the major negative regulator of PtdIns 3,4,5-P3 assembly. Importantly, the PTEN upregulations were detectable in IPs of EGFR and PDGFR. Similar results were documented for VEGF-A stimulation of bEnd.3 ECs (not shown). In addition, blockade of either C3ar1/C5ar1 or IL-6R signaling concurrently upregulated the mRNA expression of SOCS1 and SOCS3, the negative regulators of Y705 STAT1/STAT3 nuclear translocation (Fig 7B left/right) in both cell types. Moreover, disabling either C3ar1/C5ar1 or IL-6R-gp130 signaling upregulated the mRNA expression of PHLPP, a key inactivator of p-AKT (Fig 7C).
In support of active repression of these growth inhibitors being required to function coordinately with growth factor engagement of the RTK to enable RTK growth signaling, added C5a or IL-6 conversely down-regulated PTEN enzymatic activity (Fig 7D), suppressed SOCS3 mRNA expression (Fig S4B) and suppressed PHLPP mRNA expression (Fig S4C). Consistent with the joint participation of both C3ar1/C5ar1 signaling and IL-6R signal transduction in repressing these growth inhibitors, the combination of C5a and IL-6 exerted additive down-regulatory effects on PTEN enzymatic activity (Fig 7D). In accordance with C5ar1 and IL-6R repression of the above negative regulators in each cell type functioning to enable RTK growth signaling, added C5a or IL-6 increased the activation of Src, AKT and ERK in both cell types (Fig 4CD).
PTEN and β-arrestin co-localize with C5ar1 following VEGF-A stimulation
Potentially relevant to the connection of C3ar1/C5ar1 signaling with PTEN activity, a prior study found that addition of C5a to polymorphonuclear cells (PMN) or macrophages induced recruitment of β-arrestin to C5ar1 (16). β-arrestin recruitment is well documented for many GPCRs and is induced by phosphorylation of the intracellular tail of the GPCR following its agonist occupation and G protein activation/release (3). Notably, a seemingly unrelated paper (17) reported that β-arrestin can both recruit and activate PTEN, but the significance of this and of the former observation as well as their potential connection were not clarified. Prompted by these findings, we performed confocal analyses of VEGF-A treated bEnd.3 cells that were stained for C5ar1, β-arrestin, and PTEN. These analyses showed that upon VEGF-A treatment of the bEnd.3 cells, PTEN as well as β-arrestin co-localized with C5ar1 (Fig 7E).
The intracellular GPCR phosphorylations induced following their agonist occupation are mediated by GPCR kinases (GRKs) that create “barcodes” for induction of different signaling pathways (18). The phosphorylations can alternatively bias signaling via the G protein versus the β-arrestin pathway (18). Six GRKs have been defined and three (GRK2, and GRK5/6) generate the barcode that recruits β-arrestin (18). Addition of C5a or VEGF-A but not IL-6 to cultures of bEnd.3 cells markedly upregulated GRK5/6 (Fig 7F). These findings, taken together with our prior findings in immune cells that C5a induces G-protein mediated activation of PI-3Kɣ, argued that C5ar1 signaling concurrently activates both the G-protein and β arrestin pathways.
PTEN phosphatase activity and stability are regulated by phosphorylations at multiple sites within the PTEN molecule (see Discussion). Studies by others (19) found that PTEN phosphorylations at S380, T382, and T383 are associated with disabled PTEN enzymatic activity as well as reduced PTEN stability. In view of these findings, we incubated bEnd.3 ECs with VEGF-A alone or with VEGF-A plus C5ar1-A/C5ar1-A and prepared anti-p-S380, p-T382, p-T383 and anti-total PTEN IPs of the cell extracts. Examination of the anti-p-S380, T382, T383 PTEN IP’d proteins (Fig 7G left) showed that in VEGF-A treated cells, p-S380, p-T382, p-T383 PTEN (hereafter termed inactive PTEN) was brought down and that it blotted for VEGFR2. A control blot documented co-association of inactive PTEN protein in the anti-VEGFR2 blotted band (not shown). This finding was indicative of VEGF-A induced generation of inactive PTEN protein and its association with VEGFR2 (a condition that enables growth signaling). In contrast, in VEGF-A treated cells in which C3ar1/C5ar1 signaling was blocked, no inactive PTEN molecules blottable for VEGFR2 brought down in the anti-p-S380, T382, T383 IPs. This finding was indicative of no inactive PTEN associated with VEGFR2 under conditions of C3ar1/C5ar1 blockade. In contrast, the anti-total PTEN IPs showed 170% increased amounts of active (non-phosphorylated S380, T382, T383) PTEN associated with VEGFR2 (Fig 7G right) (conditions which disable growth signaling) compared with cells in which C3ar1/C5ar1 signaling was not blocked (conditions which enable growth signaling). These data thus showed that the VEGF-A-VEGFR2 complex was associated with inactive PTEN, and that C3ar1/C5ar1 blockade de-repressed the VEGFR2-induced inactivating phosphorylations of PTEN.
PTEN inactivation occurs as a result of C5ar1 dependent PI-3Kɣ activation of CK2 and Fyn
Phosphorylation of the S380, T382, and T383 residues of PTEN that inhibit PTEN phosphatase activity have been linked to activation of casein kinase 2 (CK2) (19, 20) and/or activation of Fyn [(19) see Discussion]. To determine whether the activation of CK2 is mechanistically linked with C3ar1/C5ar1 and/or IL-6R signaling, we incubated bEnd.3 cells with VEGF-A in the absence or presence of C3ar1/C5ar1 or IL-6R blockade. We then prepared anti-CK2 and anti-VEGFR2 IPs and assayed the IP’d proteins for CK2 kinase activity consistent with the ability to inactivate PTEN. The anti-CK2 IPs from the bEnd.3 cells treated with VEGF-A alone showed kinase activity, whereas those from the VEGF-A treated bEnd.3 cells that were disabled in C3ar1/C5ar1 or in IL-6R signaling showed no kinase activity (Fig 7H left). Consistent with C3ar1/C5ar1 and IL-6R signaling functioning in concert with the RTK signaling to regulate CK2 activation, added C5a induced CK2 kinase activity comparably to added VEGF-A, and the induction of kinase activity was abolished by the presence of anti-IL-6R blocking mAb (Fig 7H right).
We next investigated whether activation of Fyn is similarly dependent on C3aR/C5ar1 and IL-6R signaling. Multiplex analyses of bEnd.3 cells, mIMCD3 cells, or NIH-3T3 cells incubated with VEGF-A, EGF, or PDGF-AA, respectively, showed activated Fyn consistent with the ability to inactivate PTEN, whereas cells incubated with the respective GF plus C3ar1-A/C5ar1-A or anti-IL-6R mAb showed inactive Fyn (Fig 7I left and middle). As found for CK2, the same results were obtained when C5a was used in place of GF (Fig 7I right).
To clarify the molecular mechanism underlying the linkage of C3ar1/C5ar1 signaling with CK2 activation and its PTEN inactivation, we incubated bEnd.3 cells with C5a in the absence or presence of the PI-3Kɣ inhibitor AS252424. We then assayed CK2 enzymatic activities in anti-CK2 IPs (of the C5a treated bEnd.3 cells possessing un-manipulated or inhibited PI-3Kɣ). We also assayed CK2 enzymatic activities in anti-VEGFR2 IPs of the same cells. The PI-3Kɣ blockade abolished the CK2 activation as assessed in the anti-CK2 IP’s (Fig 7J left) as well as in anti-VEGFR2 IP’s (Fig 7J right). We next treated bEnd.3 cells with added C5a in the absence or presence of the PI-3-Kɣ inhibitor AS252424. We prepared an anti-VEGFR2 IP, and probed an immunoblot of the IP with anti-p-S380, p-T382, p-T383 PTEN (inactive PTEN) mAb. The inhibitory anti-p-S380, p-T382, p-T383 PTEN phosphorylations were induced by C5a, but they were markedly attenuated if PI-3Kɣ enzymatic activity was antagonized (Fig 7K). Densitometry is shown below the bands.
The above data indicate that inhibition of PTEN depends on C3ar1/C5ar1 signaling, and the mechanism underlying the dependence is activation of PI-3Kɣ. Inactive PTEN bearing the PI-3Kɣ dependent inhibitory phosphorylations was uniformly detectable in eluates of anti-VEGFR2, anti-C5ar1, and anti-IL-6R IPs (Fig 7K), in accordance with the PTEN inhibition occurring within the VEGFR2-C3ar1/C5ar1-IL-6R signaling complex. These findings thus directly link C5ar1 activation of PI-3Kɣ with activation of the CK2 and Fyn kinases that inactivate VEGFR2 associated PTEN and thereby enable RTK induced growth.
Joint activation may be relevant for other RTKs
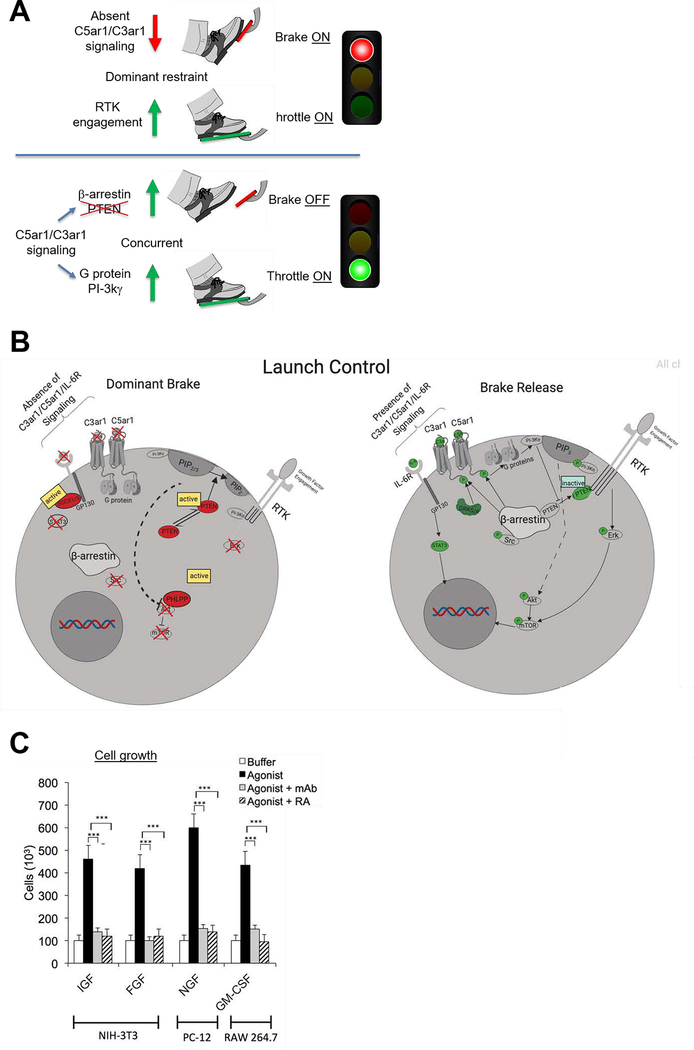
A cartoon diagramming the obligate role of C3ar1/C5ar1 and IL-6R signaling in lifting dominate restraint of PTEN and other phosphatases on RTK signaling and thereby enabling RTK mitotic function is shown in Fig 8A. A more detailed diagram of the signaling mechanisms that underlie depression of dominant restrain on RTK mitotic function by joint C3ar1/C5ar1 and IL-6R-gp130 signaling is given Fig 8B.
Figure 8. Diagram of the role of C3ar1/C5ar1 and IL-6R-gp130 co-signaling in RTK function and potentially broad relevance.
(A). Diagram explaining the requirement of coordinate C3ar1/C5ar1 and IL-6R-gp130 signaling to lift dominant restraint on RTK signaling. C3ar1/C5ar1 and IL-6R co-signaling is needed to lift dominate restraint (the brake) on RTK signaling (throttle). (B) Cartoon of signaling interconnections. When C3ar1/C5ar1 or IL-6R-gp130 signaling is absent, RTK mitotic function is blocked despite occupation of GF. (C) Data implicating the requirement of this signaling interconnection in the function of other RTKs: IGF= insulin dependent growth factor, FGF=fibroblast growth factor, NGF= nerve growth factor, GM-CSF=granulocyte-monocyte colony stimulating factor.
Based on the common findings in this paper on EGFR and PDGFR signaling, along with our previous work on VEGFR2 signaling, we hypothesized that the same functional interdependence might apply to other RTKs. Data implicating the above characterized joint activation mechanism in the functions of insulin like growth factor (IGF), nerve growth factor (NGF), fibroblast growth factor (FGF), and granulocyte monocyte colony stimulating factor (GM-CSF) are given in (Fig 8C).
Discussion
The data in this paper show that EGFR and PDGFR signaling (and perhaps a broader range of RTKs), like VEGFR2 signaling, are dependent on joint activation of C3ar1/C5ar1 and IL-6R-gp130 in signaling scaffolds. We explain this dependence mechanistically by showing that C3ar1/C5ar1 and IL-6R-gp130 signaling are essential to suppress dominant PTEN, SOCS1/3, and PHPLP function, all of which actively restrain growth signaling homeostatically. We show that the critical process that lifts this dominant restraint and enables downstream RTK transduction is activation of PI-3Kɣ which is C3ar1/C5ar1 and IL-6R-gp130-dependent. A useful analogy would be the throttle of a locomotive in which constant pressure by the engineer is needed to simultaneously release the brake (C3ar1/C5ar1 and IL-6R-gp130) and engage the engine (via GF). Equivalent to the effects of disabled C3ar1/C5ar1 and IL-6R-gp130 on RTK signaling, loss of pressure on the throttle results in automatic reengagement of the brake.
While most other studies on RTK function have focused on PI-3Kα which is activated downstream of RTK signaling (3), our data indicate that PI-3Kɣ activation also is required for RTK function. As highlighted in our work on VEGFR2 signaling, the joint activation could be required for optimal assembly of PtdIns 3,4,5-P3 (1). Alternatively, PI-3Kα could be associated with RTK mitotic function, whereas PI-3Kɣ could be associated with lifted restraint by PTEN and the other repressors, or both isoforms could participate in both functions.
Our data provide physiological understanding for the finding by others (21) that PI-3Kɣ directly inactivates PP2A, a phosphatase that negatively regulates β-adrenergic receptor signaling and is implicated in hypertrophy of cardiac myocytes. Our findings that regulation of PTEN activity occurs in a platform containing the RTK and β-arrestin associated C5ar1 also provide mechanistic and functional clarification of previous unlinked observations (17, 22) that in GF stimulated cells, PTEN is recruited by β-arrestin and this recruitment is connected with PTEN inactivation.
PTEN is expressed in all cell types and its growth suppressive activity is essential for homeostasis. While it functions in multiple cellular locations, its main activity is inhibition of RTK signaling. It does this via its recruitment from the cytoplasm to plextrin homology (PH) domain binding sites on PtdIns 3,4,5-P3 in combination with inner-leaflet phosphatidyl-serine (PS) and/or to phosphorylated sites on RTKs themselves. Phosphorylation of PTEN converts PTEN from a closed to open conformation exposing its N terminal phosphatase domain. Although multiple kinases are involved in its activation as well as its stability, phosphorylation at p-S380, T382, and T383 have been identified as functionally important to maintain PTEN in its closed inactive form (19) at these three sites which is induced by CK2 (19, 20, 23) and by LKB1 (24), the latter of which is activated by p-Fyn (19, 25). Our data show that both CK2 and Fyn activation are dependent on C3ar1/C5ar1 and IL-6R-gp130 signaling. Our data further show that PTEN molecules are physically associated with β-arrestin and C5ar1 as well as the RTK and that autocrine C5ar1 and IL-6R-gp130 signaling represses p-S380, T382, T383 de-phosphorylation connected with PTEN activation that would inhibit RTK growth signaling. Two other inactivating phosphorylations, i.e. Y240 and Y315, of PTEN are mediated by activated Src, an activation which our data show also is dependent on C3ar1/C5ar1 and IL-6R-gp130 signaling. Relevant to our linkage of absent C3ar1/C5ar1 signaling with PTEN activation in this manuscript, and our prior linkage of C3ar1/C5ar1 signaling with phosphorylation of AKT in immune cells (Strainic 2008), past studies have shown that PHLPP which dephosphorylates AKT can be co-recruited with PTEN (26). The data in this manuscript show that PHLPP inactivation is also dependent on C3ar1/C5ar1 and IL-6R-gp130 signaling. An important insight is that the growth repression of these multiple inhibitory regulators is a homeostatically active process which must be suppressed by associated C3ar1/C5ar1 and IL-6R partners of the RTK in concert with RTK signaling to enable growth induction.
STAT3 is activated by JAK (Tyk)-complexed with gp130. SOCS1 and SOCS3 inhibit STAT3 phosphorylation by blocking the ATP site in the JAK (Tyk)-gp130 complex thereby preventing STAT3 binding. Both are expressed in low levels tonically. Unlike PTEN, their activities are regulated by their own expression levels by virtue as serving as transcription factors (TFs) for their inactivators. Following their induction, STAT expression levels decline within 20–140 min additionally via ubiquitination (27). SOCS3 is the predominant regulator (28). Thus, activated STAT3, like PtdIns 3,4,5-P3, serves as a sensor which regulates its own activation. Although activation of STAT3 previously has been separately connected with IL-6R and with RTK signaling (29, 30), the data herein show that STAT3 activation is dependent on C3ar1/C5ar1 joint signaling. Additionally, our data document that C3ar1/C5ar1 signal transduction maintains STAT3 activation through repression of its SOCS1 and SOCS3 regulators.
Our experiments showed that C5a induction of C5ar1 signaling caused phosphorylation of VEGFR2 (1) as well as EGFR, and PDGFR (this paper). Although this process initially might be thought to be analogous to the previously described phenomenon of “transactivation” in which GPCRs transmit signals to EGFR or other RTKs (31, 32), fundamental differences exist. First, the findings in this paper show that RTKs signal to the C3ar1/C5ar1 GPCRs and IL-6R receptors, a process which involves the flow of information in two opposite directions via a previously undefined signaling mechanism. This signaling interconnection is required for RTK viability and growth induction rather than RTKs exclusively functioning as a GPCR signaling circuit. We thus propose the term “joint activation” for this process. Second, the mechanism differs from the previously defined process in that our co-IP, biotin-C5a pull down, confocal data, BRET analyses and single particle tracking show that it is not a linear process (termed by some as “triple membrane passing signal”) (33) involving sequential activation of separated subunits. The analyses herein rather support the concept that it involves components which have a requisite interaction with each other within a signaling scaffold. The joint activation process described herein also differs from a second previously proposed “transactivation” mechanism (34) involving direct phosphorylation of the RTK following activation of the GPCR. This second hypothetical mechanism did not envision signaling operating in the opposite direction from RTKs to GPCRs or implicate the common involvement of C3ar1/C5ar1 and IL-6R-gp130 in a signaling complex (22, 35). While some other studies have provided evidence for signaling from the RTK to the GPCR (36–38) and/or for physical association (33), none have implicated this process as being an obligatory circuit for concurrent repression of growth inhibitors in concert with GF activation of RTKs, nor have they provided a molecular mechanism for lifting this active restraint. Importantly, our data differ from the concept of biased GPCR signaling via the G protein or via β-arrestin in that we show that joint signaling through the G protein is needed to activate the RTK and β-arrestin is needed to inactivate recruited PTEN. Our findings thus provide an important insight into a newly uncovered biological purpose for the “transactivation” process.
Our live cell flow cytometric studies of EGF binding to HAEC primary cells revealed that C3ar1/C5ar1 antagonism prevented EGF binding to EGFR. This finding provides one mechanism for our biochemical findings that interfering with C3ar1/C5ar1 signaling reduces EGFR signaling. Our biophysical measurements with single QD tracking support this model by showing that suppression or augmentation of C3ar1/C5ar1 signaling alters EGFR mobility in a manner consistent with decreased or increased EGFR auto-phosphorylation respectively, a process which subsequently alters p-EGFR dependent recruitment of signaling intermediates (5, 15). Our documentation of the tonic production of GF, C3a/C5a and IL-6 by serum starved cells together with our confocal analyses and co-IP studies at time zero in the absence of added agonist (1) suggests a model in which the three receptor systems may shuttle in and out of association. The increase in any one of the agonists could increase the balance in favor of association. While our co-IP, confocal analyses, BRET analyses, and live cell EGF binding studies could be consistent with direct interactions of the signaling partners, further studies will be needed to determine whether C3ar1/C5ar1 and IL-6R-gp130 signaling modulate RTK activation via steric changes or through intermediate pathways, such as cross-talk via Src activation or by an as yet unidentified adaptor protein. Our live cell flow cytometry showing that the C3ar1/C5ar1 antagonists interfered with EGF binding argue that they may exert their inhibitory effect not only be repressing PI-3Kɣ activation but also physically by interfering with GF binding. More studies will be needed to more fully clarify this and precisely determine the physical points of interactions between the interactive receptors or adaptor protein.
Past studies of β-adrenergic (β2aR) signaling (39) together with the study herein that C5ar1, PTEN, β arrestin, and the RTK co-localize raise the possibility that β arrestin could be the adaptor protein. Our findings that C5ar1 signaling upregulates GRK5/6 which enables β-arrestin uptake also would be consistent. Our findings of active vs inactive PTEN regulating RTK function would provide a newly uncovered mechanism for PI-3Kɣ regulating desensitization vs resensitization (40).
Our results may relate to findings that Gαq/11 signaling (which activates PLC) functions independently of VEGF-A to induce VEGFR2 phosphorylation (41). We showed previously that C3ar1/C5ar1 signaling can activate PLC (Hwang et al). Our prior studies with the THP-1 macrophage line showed that both C5ar1 and C3ar1 signaling induce a Ca++ signal [(9) supplement]. C5ar1 is traditionally regarded as being a Gαi coupled (pertussis toxin sensitive) G protein. While several studies (42–45) have reported that C5ar1 can also couple to Gαq, controversy has existed about whether PLC activation is activated via the Gβɣ subunits of C5ar1 or by the putatively coupled Gαq. Possibly relevant to our results are findings (46, 47) that C5a can synergize with Gαq-11 coupled UDP receptor to activate PLCβ3 or couple with fMLP-R to augment Gαq-16 signaling (48).
Since deficiency of PDGFR and EGFR is embryonically lethal, why then do C3ar1−/−C5ar1−/− and IL-6R−/− mice survive. This question was addressed at length in our previous work on VEGFR2 signaling (Hwang et al). Briefly, one answer lies in genetic compensations involving heightened RTK signaling and upregulation of other GF signaling pathways including IL-6R signaling, which is directly implicated herein in lifting restraint on RTK signaling. Regarding IL-6R deficiency, there is evidence (49, 50) that while global IL-6R knockout (which leads to compensations) has a small effect, cell selective conditional IL-6R knockout leads to apoptosis. Moreover, IL-6R deficiency has been connected with impaired proliferation and increased gp130 expression. Another answer may lie in redundant function. For example, IL-6R deficiency has been connected with upregulation of leukemia inhibitory factor (LIF), another gp-130 ligand which can compensate for IL-6R deficiency (51, 52). Our linkage of autocrine C3ar1/C5ar1 signaling with RTK function sheds light on previous observations that mice deficient in C5ar1, C5, C3ar1 or C3 have defects in cell growth, e.g. liver regeneration following partial hepatectomy (53, 54). It is noteworthy that our results explain puzzling reported linkages of C5a and/or IL-6 with PDGF or EGF that have been ascribed to serum C5a and IL-6, as examples (55, 56). In no cases in these associations, have molecular mechanisms been delineated or the involvement of C3ar1/C5ar1 and IL-6R-gp130 in a signaling complex been suggested.
More studies will be needed to examine how joint C3ar1/C5art, IL-6R and RTK signaling relates to reported processes connected with RTK signaling. In addition to classical transactivation, e.g. angiotensin, bradykinin, and thrombin to EGFR (57, 58) and the finding that Src in a complex with β- arrestin activates ERK (39), studies will be needed to identify the precise receptor subunits, i.e. Gαq or Gβɣ that interact directly or indirectly with the RTK and IL-6R or alternatively interact with β-arrestin or another scaffold protein.
It is tempting to speculate that RTK joint signaling might have relevance to cell-cell interactions such as angiogenesis which is not only dependent on ECs but also on neighboring cells i.e., SMCs (59). The evidence that C3ar1/C5ar1 and IL-6R-gp130 signaling participate in both PDGFR-ββ induced SMC proliferation and VEGFR2 induced EC proliferation and both express VEGFR2 and PDGFR-ββ (60, 61) as well as C3ar1/C5ar1 and IL-6R-gp130 raise the possibility that VEGF-A, PDGF, C5a, or IL-6 generated by the adjacent ECs and SMCs augment the growth of each other via agonist exchange or effects on adhesion or other molecules. This could be relevant to many processes, as examples atherosclerosis and cancer.
Supplementary Material
Acknowledgments
The authors thank Scott Howell for imaging assistance in the confocal studies, Fengqi An for generation of the knockouts, Jacob Paiano for growth studies with C2C12 cells, and Sathyamangla v Naga Prasad for critical analysis proof reading. Biotinylated VhH was kindly provided by Paul van Bergen en Henegouwen (Utrecht University). DSL and CCV were supported by the New Mexico Spatiotemporal Modeling Center (NIH P50GM085273) and NIH R35GM126934. We thank William Kanagy and Elton Jhamba for assistance in generating the SPT movies. We gratefully acknowledge use of the University of New Mexico Comprehensive Cancer Center fluorescence microscopy facility supported by NIH P30CA118100.
Abbreviations
- C3ar1.C5ar1 aka CD80/86
C3a and C5a receptor
- C3ar1-A/C5ar1-A aka RA
C3ar1.C5ar1 pharmaceutical antizionists
- C5
Hc
- IL-6R-gp130
IL-6 receptor
- RTK
receptor tyrosine kinase
- VEGFR2
vascular endothelial cell growth factor receptor
- EGFR
epidermal growth factor receptor
- PDGFR
platelet derived growth receptor
- ECs
vascular endothelial cells
- SMCs
smooth muscle cells
- PMEFs
primary fibroblasts
- HUVEC
human umbilbical endothelial cells
- HAECs
human aortic endothelial cells
- bEnd.3 cells
murine EC cell line
- mIMCD3 cells
murine epithelial cell line
- DAF aka CD55
decay accelerating factor
- CD59
plasma membrane inhibitor of C5b-9 mediated lysis
- PI-3Kγ
phosphoinositide-3 kinase γ
- PI-3
phosphoinositide-3 kinase γ
- PtdIns (3,4,5)P3
inner leaflet phosphatidylinositol 3,4,5 trisphosphate
- mTOR
mammalian target of rapamycin
- PHLPP
phosphatase that dephosphorylates AKT
- SOCS1/SOCS3
inhibitors of STAT3 activation
- casein kinase 2 (CK20) and Fyn
kinases that inactivate PTEN
- STAT3
signal transducer and activation of transcription 3
- Tyk2
JAK kinase that activates STAT3
- AG1418
specific antagonists of EGFR signaling
- AS253434
PI-3kg inhibitor
- β-arrestin
adaptor protein
- QD
fluorescent quantum dot conjugate
- camelid antibody
single chain camel antibody
- IGF
insulin like growth factor
- NGF
nerve growth factor
- FGF
fibroblast growth factor
- GM-CSF
granulocyte monocyte colony stimulating factor
- PH
plextrin homology domain
References
- 1.Hwang MS, Strainic MG, Pohlmann E, Kim H, Pluskota E, Ramirez-Bergeron DL, Plow EF, and Medof ME (2019) VEGFR2 survival and mitotic signaling depends on joint activation of associated C3ar1/C5ar1 and IL-6R-gp130. J Cell Sci 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlessinger J (2014) Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb Perspect Biol 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Morgan D, Raff MC, Roberts K, Walter P, Wilson J, and Hunt T (2014) Molecular biology of the cell, Garland Science [Google Scholar]
- 4.Mahabeleshwar GH, Somanath PR, and Byzova TV (2006) Methods for isolation of endothelial and smooth muscle cells and in vitro proliferation assays. Methods Mol Med 129, 197–208 [DOI] [PubMed] [Google Scholar]
- 5.Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PM, Wilson BS, and Lidke DS (2011) ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nature structural & molecular biology 18, 1244–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Lin F, Strainic MG, An F, Miller RH, Altuntas CZ, Heeger PS, Tuohy VK, and Medof ME (2008) IFN-gamma and IL-17 production in experimental autoimmune encephalomyelitis depends on local APC-T cell complement production. J Immunol 180, 5882–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang MS, Strainic MG, Pohlmann E, Pluskota E, Ramirez-Bergeron DL, Plow EF, and Medof ME (2019) VEGFR2 survival and mitotic signaling depends on joint-activation of associated C3ar1/C5ar1 and IL-6R-gp130. J Cell Sci [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strainic MG, Shevach EM, An F, Lin F, and Medof ME (2013) Absence of signaling into CD4(+) cells via C3aR and C5aR enables autoinductive TGF-beta1 signaling and induction of Foxp3(+) regulatory T cells. Nature immunology 14, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strainic MG, Liu J, Huang D, An F, Lalli PN, Muqim N, Shapiro VS, Dubyak GR, Heeger PS, and Medof ME (2008) Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity 28, 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wegiel B, Bjartell A, Culig Z, and Persson JL (2008) Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int J Cancer 122, 1521–1529 [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Berriguete G, Prieto A, Fraile B, Bouraoui Y, de Bethencourt FR, Martinez-Onsurbe P, Olmedilla G, Paniagua R, and Royuela M (2010) Relationship between IL-6/ERK and NF-kappaB: a study in normal and pathological human prostate gland. Eur Cytokine Netw 21, 241–250 [DOI] [PubMed] [Google Scholar]
- 12.Huang YH, Yang HY, Hsu YF, Chiu PT, Ou G, and Hsu MJ (2014) Src contributes to IL6-induced vascular endothelial growth factor-C expression in lymphatic endothelial cells. Angiogenesis 17, 407–418 [DOI] [PubMed] [Google Scholar]
- 13.Leu CM, Wong FH, Chang C, Huang SF, and Hu CP (2003) Interleukin-6 acts as an antiapoptotic factor in human esophageal carcinoma cells through the activation of both STAT3 and mitogen-activated protein kinase pathways. Oncogene 22, 7809–7818 [DOI] [PubMed] [Google Scholar]
- 14.Spets H, Stromberg T, Georgii-Hemming P, Siljason J, Nilsson K, and Jernberg-Wiklund H (2002) Expression of the bcl-2 family of pro- and anti-apoptotic genes in multiple myeloma and normal plasma cells: regulation during interleukin-6(IL-6)-induced growth and survival. Eur J Haematol 69, 76–89 [DOI] [PubMed] [Google Scholar]
- 15.Steinkamp MP, Low-Nam ST, Yang S, Lidke KA, Lidke DS, and Wilson BS (2014) erbB3 is an active tyrosine kinase capable of homo- and heterointeractions. Molecular and cellular biology 34, 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bamberg CE, Mackay CR, Lee H, Zahra D, Jackson J, Lim YS, Whitfeld PL, Craig S, Corsini E, Lu B, Gerard C, and Gerard NP (2010) The C5a receptor (C5aR) C5L2 is a modulator of C5aR-mediated signal transduction. The Journal of biological chemistry 285, 7633–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima-Fernandes E, Enslen H, Camand E, Kotelevets L, Boularan C, Achour L, Benmerah A, Gibson LC, Baillie GS, Pitcher JA, Chastre E, Etienne-Manneville S, Marullo S, and Scott MG (2011) Distinct functional outputs of PTEN signalling are controlled by dynamic association with beta-arrestins. Embo j 30, 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Qiao Y, and Li Z (2018) New Insights into Modes of GPCR Activation. Trends Pharmacol Sci 39, 367–386 [DOI] [PubMed] [Google Scholar]
- 19.Fragoso R, and Barata JT (2014) Kinases, tails and more: Regulation of PTEN function by phosphorylation. Methods [DOI] [PubMed] [Google Scholar]
- 20.Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, and Sellers WR (2001) Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. The Journal of biological chemistry 276, 48627–48630 [DOI] [PubMed] [Google Scholar]
- 21.Vasudevan NT, Mohan ML, Gupta MK, Hussain AK, and Naga Prasad SV (2011) Inhibition of protein phosphatase 2A activity by PI3Kgamma regulates beta-adrenergic receptor function. Molecular cell 41, 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilley DG, Kim IM, Patel PA, Violin JD, and Rockman HA (2009) beta-Arrestin mediates beta1-adrenergic receptor-epidermal growth factor receptor interaction and downstream signaling. The Journal of biological chemistry 284, 20375–20386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres J, and Pulido R (2001) The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. The Journal of biological chemistry 276, 993–998 [DOI] [PubMed] [Google Scholar]
- 24.Mehenni H, Lin-Marq N, Buchet-Poyau K, Reymond A, Collart MA, Picard D, and Antonarakis SE (2005) LKB1 interacts with and phosphorylates PTEN: a functional link between two proteins involved in cancer predisposing syndromes. Hum Mol Genet 14, 2209–2219 [DOI] [PubMed] [Google Scholar]
- 25.Yamada E, and Bastie CC (2014) Disruption of Fyn SH3 domain interaction with a proline-rich motif in liver kinase B1 results in activation of AMP-activated protein kinase. PloS one 9, e89604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, and Georgescu MM (2012) PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 31, 1264–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams JJ, Munro KM, and Palmer TM (2014) Role of Ubiquitylation in Controlling Suppressor of Cytokine Signalling 3 (SOCS3) Function and Expression. Cells 3, 546–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babon JJ, and Nicola NA (2012) The biology and mechanism of action of suppressor of cytokine signaling 3. Growth factors 30, 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegenka UM, Lutticken C, Buschmann J, Yuan J, Lottspeich F, Muller-Esterl W, Schindler C, Roeb E, Heinrich PC, and Horn F (1994) The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family. Molecular and cellular biology 14, 3186–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartoli M, Platt D, Lemtalsi T, Gu X, Brooks SE, Marrero MB, and Caldwell RB (2003) VEGF differentially activates STAT3 in microvascular endothelial cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 17, 1562–1564 [DOI] [PubMed] [Google Scholar]
- 31.Daub H, Weiss FU, Wallasch C, and Ullrich A (1996) Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature 379, 557–560 [DOI] [PubMed] [Google Scholar]
- 32.Gschwind A, Zwick E, Prenzel N, Leserer M, and Ullrich A (2001) Cell communication networks: epidermal growth factor receptor transactivation as the paradigm for interreceptor signal transmission. Oncogene 20, 1594–1600 [DOI] [PubMed] [Google Scholar]
- 33.Wetzker R, and Bohmer FD (2003) Transactivation joins multiple tracks to the ERK/MAPK cascade. Nature reviews. Molecular cell biology 4, 651–657 [DOI] [PubMed] [Google Scholar]
- 34.Roudabush FL, Pierce KL, Maudsley S, Khan KD, and Luttrell LM (2000) Transactivation of the EGF receptor mediates IGF-1-stimulated shc phosphorylation and ERK1/2 activation in COS-7 cells. The Journal of biological chemistry 275, 22583–22589 [DOI] [PubMed] [Google Scholar]
- 35.Luttrell LM, Della Rocca GJ, van Biesen T, Luttrell DK, and Lefkowitz RJ (1997) Gbetagamma subunits mediate Src-dependent phosphorylation of the epidermal growth factor receptor. A scaffold for G protein-coupled receptor-mediated Ras activation. The Journal of biological chemistry 272, 4637–4644 [DOI] [PubMed] [Google Scholar]
- 36.Hobson JP, Rosenfeldt HM, Barak LS, Olivera A, Poulton S, Caron MG, Milstien S, and Spiegel S (2001) Role of the sphingosine-1-phosphate receptor EDG-1 in PDGF-induced cell motility. Science 291, 1800–1803 [DOI] [PubMed] [Google Scholar]
- 37.El-Shewy HM, Johnson KR, Lee MH, Jaffa AA, Obeid LM, and Luttrell LM (2006) Insulin-like growth factors mediate heterotrimeric G protein-dependent ERK1/2 activation by transactivating sphingosine 1-phosphate receptors. The Journal of biological chemistry 281, 31399–31407 [DOI] [PubMed] [Google Scholar]
- 38.Takabe K, Paugh SW, Milstien S, and Spiegel S (2008) “Inside-out” signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev 60, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, Caron MG, and Lefkowitz RJ (1999) Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science 283, 655–661 [DOI] [PubMed] [Google Scholar]
- 40.Gupta MK, Mohan ML, and Naga Prasad SV (2018) G Protein-Coupled Receptor Resensitization Paradigms. Int Rev Cell Mol Biol 339, 63–91 [DOI] [PubMed] [Google Scholar]
- 41.Zeng H, Zhao D, Yang S, Datta K, and Mukhopadhyay D (2003) Heterotrimeric G alpha q/G alpha 11 proteins function upstream of vascular endothelial growth factor (VEGF) receptor-2 (KDR) phosphorylation in vascular permeability factor/VEGF signaling. The Journal of biological chemistry 278, 20738–20745 [DOI] [PubMed] [Google Scholar]
- 42.Jiang H, Kuang Y, Wu Y, Smrcka A, Simon MI, and Wu D (1996) Pertussis toxin-sensitive activation of phospholipase C by the C5a and fMet-Leu-Phe receptors. The Journal of biological chemistry 271, 13430–13434 [DOI] [PubMed] [Google Scholar]
- 43.Lee CH, Katz A, and Simon MI (1995) Multiple regions of G alpha 16 contribute to the specificity of activation by the C5a receptor. Molecular pharmacology 47, 218–223 [PubMed] [Google Scholar]
- 44.Buhl AM, Eisfelder BJ, Worthen GS, Johnson GL, and Russell M (1993) Selective coupling of the human anaphylatoxin C5a receptor and alpha 16 in human kidney 293 cells. FEBS letters 323, 132–134 [DOI] [PubMed] [Google Scholar]
- 45.Amatruda TT 3rd, Gerard NP, Gerard C, and Simon MI (1993) Specific interactions of chemoattractant factor receptors with G-proteins. The Journal of biological chemistry 268, 10139–10144 [PubMed] [Google Scholar]
- 46.Flaherty P, Radhakrishnan ML, Dinh T, Rebres RA, Roach TI, Jordan MI, and Arkin AP (2008) A dual receptor crosstalk model of G-protein-coupled signal transduction. PLoS computational biology 4, e1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roach TI, Rebres RA, Fraser ID, Decamp DL, Lin KM, Sternweis PC, Simon MI, and Seaman WE (2008) Signaling and cross-talk by C5a and UDP in macrophages selectively use PLCbeta3 to regulate intracellular free calcium. The Journal of biological chemistry 283, 17351–17361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burg M, Raffetseder U, Grove M, Klos A, Kohl J, and Bautsch W (1995) G alpha-16 complements the signal transduction cascade of chemotactic receptors for complement factor C5a (C5a-R) and N-formylated peptides (fMLF-R) in Xenopus laevis oocytes: G alpha-16 couples to chemotactic receptors in Xenopus oocytes. FEBS letters 377, 426–428 [DOI] [PubMed] [Google Scholar]
- 49.Durlacher-Betzer K, Hassan A, Levi R, Axelrod J, Silver J, and Naveh-Many T (2018) Interleukin-6 contributes to the increase in fibroblast growth factor 23 expression in acute and chronic kidney disease. Kidney international [DOI] [PubMed] [Google Scholar]
- 50.Kuhn KA, Manieri NA, Liu TC, and Stappenbeck TS (2014) IL-6 stimulates intestinal epithelial proliferation and repair after injury. PloS one 9, e114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z, Sakamoto T, Yokomuro S, Ezure T, Subbotin V, Murase N, Contrucci S, and Demetris AJ (2000) Acute obstructive cholangiopathy in interleukin-6 deficient mice: compensation by leukemia inhibitory factor (LIF) suggests importance of gp-130 signaling in the ductular reaction. Liver 20, 114–124 [DOI] [PubMed] [Google Scholar]
- 52.Ezure T, Sakamoto T, Tsuji H, Lunz JG 3rd, Murase N, Fung JJ, and Demetris AJ (2000) The development and compensation of biliary cirrhosis in interleukin-6-deficient mice. Am J Pathol 156, 1627–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phieler J, Garcia-Martin R, Lambris JD, and Chavakis T (2013) The role of the complement system in metabolic organs and metabolic diseases. Semin Immunol 25, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markiewski MM, Mastellos D, Tudoran R, DeAngelis RA, Strey CW, Franchini S, Wetsel RA, Erdei A, and Lambris JD (2004) C3a and C3b activation products of the third component of complement (C3) are critical for normal liver recovery after toxic injury. J Immunol 173, 747–754 [DOI] [PubMed] [Google Scholar]
- 55.Kato K, Otsuka T, Kondo A, Matsushima-Nishiwaki R, Natsume H, Kozawa O, and Tokuda H (2012) AMP-activated protein kinase regulates PDGF-BB-stimulated interleukin-6 synthesis in osteoblasts: involvement of mitogen-activated protein kinases. Life sciences 90, 71–76 [DOI] [PubMed] [Google Scholar]
- 56.Schraufstatter IU, Trieu K, Sikora L, Sriramarao P, and DiScipio R (2002) Complement c3a and c5a induce different signal transduction cascades in endothelial cells. J Immunol 169, 2102–2110 [DOI] [PubMed] [Google Scholar]
- 57.Zwick E, Hackel PO, Prenzel N, and Ullrich A (1999) The EGF receptor as central transducer of heterologous signalling systems. Trends Pharmacol Sci 20, 408–412 [DOI] [PubMed] [Google Scholar]
- 58.Zwick E, Wallasch C, Daub H, and Ullrich A (1999) Distinct calcium-dependent pathways of epidermal growth factor receptor transactivation and PYK2 tyrosine phosphorylation in PC12 cells. The Journal of biological chemistry 274, 20989–20996 [DOI] [PubMed] [Google Scholar]
- 59.Chung AS, and Ferrara N (2011) Developmental and pathological angiogenesis. Annu Rev Cell Dev Biol 27, 563–584 [DOI] [PubMed] [Google Scholar]
- 60.Hellstrom M, Kalen M, Lindahl P, Abramsson A, and Betsholtz C (1999) Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055 [DOI] [PubMed] [Google Scholar]
- 61.Ishida A, Murray J, Saito Y, Kanthou C, Benzakour O, Shibuya M, and Wijelath ES (2001) Expression of vascular endothelial growth factor receptors in smooth muscle cells. Journal of cellular physiology 188, 359–368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.