Abstract
Although several reports suggest that the entry of infectious bronchitis virus (IBV) depends on lipid rafts and low pH, the endocytic route and intracellular trafficking are unclear. In this study, we aimed to shed greater light on early steps in IBV infection. By using chemical inhibitors, RNA interference, and dominant negative mutants, we observed that lipid rafts and low pH was indeed required for virus entry; IBV mainly utilized the clathrin mediated endocytosis (CME) for entry; GTPase dynamin 1 was involved in virus containing vesicle scission; and the penetration of IBV into cells led to active cytoskeleton rearrangement. By using R18 labeled virus, we found that virus particles moved along with the classical endosome/lysosome track. Functional inactivation of Rab5 and Rab7 significantly inhibited IBV infection. Finally, by using dual R18/DiOC labeled IBV, we observed that membrane fusion was induced after 1 h.p.i. in late endosome/lysosome.
Keywords: IBV, Lipid rafts, Clathrin mediated endocytosis, Endosome, Membrane fusion
Highlights
-
•
Intact lipid rafts is involved in IBV entry.
-
•
Low pH in intracyplasmic vesicles is required for IBV entry.
-
•
IBV penetrates cells via clathrin mediated endocytosis.
-
•
IBV moves along with the classical endosome/lysosome track, finally fuses with late endosome/lysosome.
1. Introduction
Most viruses take advantage of cellular endocytic pathways to enter their host cells (Cossart and Helenius, 2014, Yamauchi and Helenius, 2013). Once internalized, virus particles move through a dynamic network of endocytic vesicles, which undergoes gradual sorting and complex maturation events. Endosome maturation, in turn, triggers conformational changes and dissociation events in the incoming viruses, which ultimately leads to the delivery of the viral genome and associated proteins into the cytoplasm. In general, while non-enveloped viruses are able to penetrate the plasma membrane or endosomal membrane by pore formation or lysis (Suomalainen and Greber, 2013), enveloped viruses usually fuse with the vesicle membranes to release the genome into the cytoplasm (Harrison, 2015). Numerous virus families utilize endocytosis to infect host cell, mediating virus internalization as well as trafficking to the site of replication. Different viruses employ various routes of endocytosis, and the route taken by a given virus largely depends on the receptor that the virus interacts with. The endocytic pathways utilized by viruses include clathrin-mediated endocytosis (CME), caveolin/raft-mediated endocytosis (CavME), macropinocytosis, and non-clathrin/non-caveolin mediated endocytosis. Among them, CME is the best understood endocytic pathway. In response to receptor mediated internalization signals, clathrin is recruited to plasma membrane, leading to the assembly of clathrin-coated pits (CCP) at the cytoplasmic side of the plasma membrane. The main scaffold component of clathrin coat is the clathrin heavy chain (CHC) and clathrin light chain (CLC), forming a three-legged trimers called triskelions (Keen, 1990). The adapter protein 2 (AP-2) provides a bridge between receptor cargo domain and the clathrin β subunit (Kirchhausen, 1999). Once assembled, CCPs pinch off from the plasma membrane and mature into clathrin-coated vesicles (Keen, 1990), with the helping of Eps15 and dynamin GTPase. Lots of viruses use CME to enter cells, including Semliki forest virus (Vonderheit and Helenius, 2005), vesicular stomatitis virus (VSV) (Cureton et al., 2010), hepatitis C virus (Benedicto et al., 2015), and adenovirus (Meier et al., 2002). The Caveolae is a relatively small vesicle with a diameter of 50–100 nm, which is a flask-shaped invasion formed by membrane retraction on the plasma membrane, and is a specialized lipid raft domain composed of caveolin and high levels of cholesterol (Hommelgaard et al., 2005, Insel et al., 2005). CavME is first observed for the cellular uptake of simian virus 40 (Pelkmans et al., 2001). After that, other viruses including some of the picornaviruses (Marjomaki et al., 2002), papillomaviruses (Bousarghin et al., 2003), filoviruses (Empig and Goldsmith, 2002), murine leukemia virus (Beer et al., 2005), human coronavirus 229E (HCoV-229E) (Nomura et al., 2004), and classical swine fever virus (Zhang et al., 2018) are found by utilizing CavME to enter into cells. Macropinocytosis is morphologically defined by the presence of membranous extensions of outwardly polymerizing actin termed membrane ruffles. It has been identified as an entry mechanism for several pathogens, including B2 adenovirus (Kalin et al., 2010), Coxsackie B virus (Inal and Jorfi, 2013), influenza A virus (de Vries et al., 2011), Ebola virus (Aleksandrowicz et al., 2011), Vaccinia virus (Rizopoulos et al., 2015), Nipah virus (Vogt et al., 2005), Kaposi’s Sarcoma-Associated Herpesvirus (Raghu et al., 2009), and Newcastle disease virus (Tan et al., 2016).
Lipid rafts are functional membrane microdomains which are also termed detergent-insoluble glycolipid-enriched complexes or detergent-resistant membranes, rich in cholesterol,sphingolipids, and proteins (Fivaz et al., 1999, Simons and Ikonen, 1997). They serve as domains to concentrate membrane-associated proteins that include receptors and signaling molecules (Simons and Ikonen, 1997). In addition to physiological roles in signal transduction of host cells (Simons and Toomre, 2000), lipid rafts often serve as a site for entry, assembly and budding of microbial pathogens (Kovbasnjuk et al., 2001, Rawat et al., 2003, Suomalainen, 2002). They are involved in the binding and entry of host cells for several enveloped and non-enveloped viruses, including human immunodeficiency virus type 1 (Viard et al., 2002), poliovirus (Danthi and Chow, 2004), human herpes virus 6 (Huang et al., 2006), West Nile virus (Medigeshi et al., 2008), foot-and-mouth disease virus (Martin-Acebes et al., 2007), and dengue virus (Diwaker et al., 2015). There is some evidence that lipid rafts are also involved in coronavirus entry. In the case of HCoV-229E, virus entry is inhibited by depletion of cholesterol, which results in the disruption of viral association with the cellular receptor CD13 (Nomura et al., 2004). Thorp and Gallagher show that lipid rafts are crucial for the entry of mouse hepatitis virus (MHV) (Thorp and Gallagher, 2004). Moreover, Guo et al., demonstrate that lipid rafts are involved in IBV entry (Guo et al., 2017).
Coronaviruses are enveloped, plus-strand RNA viruses belonging to the family Coronaviridae, which includes many human and animal pathogens of global concern, such as severe acute respiratory syndrome coronavirus (SARS-CoV), middle east respiratory syndrome virus (MERS-CoV), HCoV-229E, MHV, porcine epidemic diarrhea virus (PEDV), Feline infectious peritonitis virus (FIPV), and IBV. In most cases, they cause respiratory and/or intestinal tract disease. In general, coronaviruses enter cells by endocytosis, which undergoes pH-dependent membrane fusion. For example, CME as well as clathrin- and caveolae-independent entry pathways have been reported for SARS-CoV (Inoue et al., 2007, Wang et al., 2008); clathrin- and serine proteases-dependent uptake has been reported for PEDV (Park et al., 2014); FIPV is suggested to enter cells via a clathrin- and caveolae-independent endocytic route (Van Hamme et al., 2008); MHV2 enters cells via CME, but independent of Eps15 (Pu and Zhang, 2008). IBV belongs to the genus gamma coronavirus. Its genome is approximately 27.6 kilobases (kb) in length, encoding 2 polyproteins (1a, 1ab), four structural proteins, namely, spike protein (S), membrane protein (M), small envelope protein (E), nucleocapsid protein (N), and four accessory proteins named 3a, 3b, 5a, 5b. Polyprotein 1a and 1ab are cleaved by viral proteases into at least 15 non-structural proteins (nsp2-nsp16). Despite the importance of IBV as one of the dominant pathogens causing a highly contagious infectious bronchitis worldwide and leading to significant economic loss in the chicken industry, little information is available about its entry mechanisms. It has been suggested that the entry of IBV into cells depends on lipid rafts and low pH (Chu et al., 2006); however, the endocytic pathway that IBV hijacks is unclear.
The early step in the entry process of IBV into target cells is initiated by engagement of the S glycoprotein with the receptor. Here, using several cell lines permissive to IBV Beaudette strain, we report that IBV virion attaches to lipid rafts in the cell surface and the efficient entry of IBV into cells requires the presence of intact lipid rafts. We also address IBV entry pathway by systematically perturbing the function of key factors in the various endocytic routes by using chemical inhibitors, siRNA silence, and overexpression of dominant negative (DN) proteins. To achieve a successful tracking of virion attachment, entry, intracellular trafficking, and membrane fusion, octadecyl rhodamine (R18) and 3, 3'-Dihexyloxacarbocyanine Iodide (DiOC) were labeled on viral membrane as fluorescent tags. Immunofluorescence analysis was used to visualize the co-localization or co-trafficking of virus with cellular factors. Our results reveal that IBV is internalized into cells by CME, transports along early endosomes, late endosomes, and fuses with membrane in late endosome-lysosomes.
2. Materials and methods
2.1. Cells and viruses
Vero cells, Huh7 cells, BHK-21, and DF-1 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 4500 mg/l glucose, supplemented with 10% fetal bovine serum (FBS) (Hyclone, USA), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, USA). H1299 cells were maintained in Roswell Park Memorial Institute 1640 (RPMI 1640), supplemented with 10% FBS in the presence of 100 units/ml penicillin and 100 μg/ml streptomycin. Above cells were purchased from ATCC (USA) and cultured at 37 °C with 5% CO2.
The Beaudette strain of IBV (ATCC VR-22) adapted to Vero cells was used in this study. As a well characterized virus, Vesicular Stomatitis Virus (VSV) was used as control virus in this study. Virus stock was prepared by infecting monolayers of Vero cells with multiplicity of infection (MOI) of 0.1 in FBS free DMEM. After attaching at 37 °C for 1 h, the unbounded virus was removed and replaced with FBS free DMEM. The virus and cells were incubated at 37 °C until more than 90% cells were with cytopathic effect (CPE). After freezing and thawing the virus containing cells and medium for three times, cell debris was removed by centrifugation at 3000×rpm, and supernatant was aliquoted and stored at − 80 °C as virus stock. A control of Vero cell lysates from mock-infected cells was prepared in the same manner. The virus yield was assessed by tissue culture infective dose 50 (TCID50) titration.
2.2. Virus titration
Supernatant of IBV-infected cells collected were centrifuged at 5000×g for 15 min to remove cell debris. The viral titers were determined by TCID50 assay. Briefly, 10-fold serially diluted aliquots of virus were applied to confluent monolayers of Vero cells in 96-well plates. After 1 h of absorption at 37 °C, unbound viruses were removed, and the cells were washed twice with DMEM. The plates were incubated with DMEM at 37 °C and the cytopathic effect (CPE) was observed after 3 days. Each sample was titrated in triplicate. The TCID50 is calculated using Reed and Munch mathematical analysis (McHenry et al., 1938).
2.3. R18 and R18/DiOC labeling of virus
Vero cells were infected with viruses at MOI = 0.1 and incubated at 37 °C until more than 90% cells were with CPE. The culture medium with virus particles was clarified by centrifuged at 4000×g for 15 min. The supernatant was then centrifuged at 5000×g for 60 min and concentrated by 100-fold by using Amicon® Ultra-15 Centrifugal Filter Devices (10-kDa cutoff, Merk, Poland), which provides fast ultrafiltration. Mock infected Vero cells culture medium was concentrated in the same manner as control sample.
R18 labeling was prepared as described previously (Chu et al., 2006): 100 μl of concentrated virus or control sample was incubated with 1.7 mM R18 (Molecular Probes, USA) on a rotary shaker for 1 h at room temperature. R18/DiOC labeling was prepared as described (Krzyzaniak et al., 2013): 100 μl of concentrated virus or control sample was incubated with 3.3 mM DiOC (Molecular Probes, USA) and 6.7 mM R18 mixture (Molecular Probes, USA) with gentle shaking for 1 h at room temperature. After finishing the labeling, the virus and dye mixture was re-suspended in 8 ml phosphate-buffered saline (PBS), and the excess dye was removed with an Amicon® Ultra-15 Centrifugal Filter Devices (10-kDa cutoff, Merk, Poland) by centrifugating for 60 min. Finally, 100 μl of labeled virus or labeled mock sample were obtained.
2.4. Inhibitors and antibodies
The endocytotic pathway inhibitors Amiloride (S1811), Nystatin (S1934), and chlorpromazine (CPZ, S2456) were purchased from Selleck (USA). Actin monomer-sequestering drug cytochalasin D (CytoD, PHZ1063), actin polymer-stabilizing jasplakinolide (Jas, J7473) were purchased from Thermo Fisher Scientific (USA). Endosome acidification inhibitor NH4Cl (A9434), cholesterol depletion drug methyl-β-cyclodextrin (MβCD, C4555) and Cholesterol-Water Soluble (C4951) were purchased from Sigma-Aldrich (USA).
Anti-IBV S and N antibodies were obtained through immunization of rabbits with respective antigen, which are generous gifts from Prof Liu Dingxiang’s lab (South China Agricultural University, China). Anti-flotillin-1 (#18634), anti-GFP (#2956), anti-CHC (clathrin heavy chain) (#4769s), anti-Rab5 (#3547s), anti-Rab7 (#9367s), and anti-LAMP1 (#9091s) were purchased from Cell Signaling Technology (USA). Anti-VSV G (Ab50549) was obtained from Abcam (UK). Anti-β-actin (A1978) was purchased from Sigma Aldrich (USA). Cholera Toxin Subunit B (CTB, C34775) was purchased from Thermo Fisher Scientific (USA). Fluorescein isothiocyanate (FITC)-conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG), as well as horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit IgG were purchased from Cell Signaling Technology (USA). Alexa Fluor 488 Phalloidin (A-12379) was purchased from Thermo fisher (USA).
2.5. Virus infection and drug administration
To examine the effect of various inhibitors on IBV infection, Vero cells, H1299 cells, BHK-21 cells, Huh7 cells, or DF-1 cells were seeded into 6-well plates at 5 × 106 cells/well and cultured for 24 h until they reached 100% confluence. Cells were then pretreated with the indicated concentrations of NH4Cl, CPZ, Nystatin, Amiloride, Jas, or CytoD for 30 min at 37 °C, respectively. After treatment, the cells were inoculated with IBV or VSV at MOI = 1 and incubated for 1 h in the presence of corresponding compounds. The unbound virions were washed away with PBS and the cells were incubated with fresh medium without compounds for additional 2 h or 8 h at 37 °C. Virus internalization was determined by semi-quantitative real time RT-PCR at 2 h.p.i. by measuring the viral RNA genome, and virus replication was monitored by Western blot at 8 h.p.i. by checking the expression level of viral protein.
2.6. Cell viability assay and pH assessment
Viability of drug-treated cells was measured using the WST-1 Cell proliferation and cytotoxicity assay kit according to the manufacturer’s instruction (Beyotime, Haimen, China). Cells were seeded in 96-well plate and treated with indicated drugs, 10 μl of WST-1 was added to each well and incubated for 1 h. The absorbance at 450 nm was monitored and the reference wavelength was set at 630 nm. The viability of cells was calculated by comparison to that of untreated cells.
To assess the effect of NH4Cl on the pH change of acidic intracellular vesicles, Vero cells were treated with increasing concentration of NH4Cl for 30 min at 37 °C, followed with 1 mg/ml acridine orange (A6014, Sigma) staining in serum free medium for 15 min at 37 °C. The cells were washed twice with PBS and stained with DAPI {2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride, Roche}. Images were taken with a Zeiss Axio Observer Z1 fluorescence microscope.
2.7. Cholesterol depletion and replenishment
Cells were incubated with 5 mM MβCD for 30 min at 37 °C at indicated infection time course. The cells exposed to DMSO for 30 min were set as control group. After incubation, the cells were washed twice with PBS and replaced with fresh culture medium. For cholesterol replenishment, cells were pretreated with 5 mM MβCD for 30 min at 37 °C, then supplemented with exogenous 1 mM cholesterol and incubated for 1 h at 37 °C. After extensive washing with PBS, the cells were infected with IBV.
2.8. Membrane flotation analysis
Cells (5 × 107) were incubated with IBV (MOI = 5) at 4 °C for 1 h. Mock-infected cells were set as control group. Cells were lysed on ice for 30 min in 1 ml TNE lysis buffer with 1% Triton X-100, supplemented with complete protease inhibitor cocktail. The cell lysates were centrifuged at 4000×g for 5 min at 4 °C, to remove cell debris and nuclei. The supernatant was collected and mixed with 1 ml of TNE buffer with 80% sucrose, placed at the bottom of the ultracentrifuge tube, overlaid with 6 ml of 30% and 3 ml of 5% sucrose in TNE buffer. The lysates were ultracentrifuged at 38,000 ×rpm for 18 h at 4 °C in an SW41 rotor (Beckman). After centrifugation, 12 fractions (1 ml each) were collected from the top to the bottom of the tube and subjected to 8% or 10% SDS−PAGE, followed by Western blotting using antibodies of anti-Flotillin-1, anti-N protein, or anti-S protein.
2.9. Plasmid transfection and siRNA transfection
Cells grown in 6-well plates were transfected with 3 µg of plasmid DNA, by using lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After 24 h post-transfection, cells were infected with virus at MOI = 1. To knock down Eps15, dynamin 1, Rab5, or Rab7, small interfering RNA (siRNA) duplexes at a concentration of 100 nM were transfected into the cells by using Lipofectamine 3000. At 36 h post-transfection, cells were infected with virus at MOI = 1. The level of IBV internalization, viral protein expression, and virus particle release were then measured by semi-quantitative real time RT-PCR, Western blot analysis, and TCID50 at indicated time points. The siRNA sequences were shown in Table 1.
Table 1.
siRNA sequence.
| Name | siRNA sequence (5′ to 3′) |
|---|---|
| CHC siRNA | 5′-GGAGGGAAGUUACAUAUUATT-3′ |
| sic | 5′-UAAUAUGUAACUUCCCUCCTT-3′ |
| Rab5 siRNA | 5′-GCCAGAGGAAGAGGAGTAGACCTTA-3′ |
| Rab7 siRNA | 5′-UACUGGUUCAUGAGCGAUGUCUUUC-3′ |
| Eps15 siRNA | 5′-AAACGGAGCUACAGAUUAU-3′ |
| Dynamin-1 siRNA | 5′-GCACUGCAAGGGAAAGAAATT-3′ |
| sic | 5′-AUGUUCUAAUGCACGCUGCTT-3′ |
CHC: Clathrin heavy chain
sic: Non-targeting control siRNA.
2.10. Measurement of virus gRNA by semi-quantitative real time RT-PCR
Viral internalization was detected as described previously (Zhu et al., 2012). Briefly, the cells in 6-well plates were incubated with IBV (MOI=1) at 4 °C for 1 h. The unbound virions were washed away with PBS, and the cells were incubated at 37 °C with fresh medium. At 2 h.p.i., cells were treated with 1 mg/ml proteinase K (Invitrogen) for 15 min to remove adsorbed but not internalized virus. Proteinase K was then inactivated with 2 mM phenylmethylsulfonyl fluoride (PMSF) in PBS with 3% bovine serum albumin. Cells were then washed three times with PBS and lysed in 1 ml of TRIZOL reagent (Invitrogen, USA) per well for RNA isolation. One fifth volume of chloroform was added into the cell lysates. The mixture was centrifuged at 13,000×rpm for 15 min at 4 °C, and the aqueous phase was then mixed with an equal volume of 100% isopropanol and incubated at -20 °C for 20 min. RNA was precipitated by centrifugation at 13,000×rpm for 10 min at 4 °C. RNA pellets were washed with 70% RNase-free ethanol and dissolved in RNase-free water.
The level of internalized viral +gRNA or -gRNA was determined by real time RT-PCR. Briefly, 3 µg of total RNA was used to perform reverse transcription using expand reverse transcriptase (Roche, USA) and oligo-dT or IBV specific primers. Equal volume of cDNAs was then PCR-amplified using SYBR green PCR master kit (Dongsheng Biotech, Guangdong, China). Genome copy numbers were normalized to β-actin level by using the comparative cycle threshold values determined in parallel experiment. Data were analyzed relative to control. All assays were performed in three replicates. The primers sequence used in RT-PCR were shown in Table 2.
Table 2.
Primer sequence for semi-quantitative real time RT-PCR.
| Name | Primer sequence (5′ to 3′) |
|---|---|
| IBV (+) gRNA | Forward 5′-TTTAGCAGAACATTTTGACGCAGAT-3′ |
| Reverse 5′-TTAGTAGAACCAACAAACACGACAG-3′ | |
| IBV (-) gRNA | Forward 5′-TTAGTAGAACCAACAAACACGACAG-3′ |
| Reverse 5′-TTTAGCAGAACATTTTGACGCAGAT-3′ | |
| Eps15 | Forward 5′-AAGCCAACACCCTTCCAGTA-3′ |
| Reverse 5′-GCCCAGCTCTCTCTGACACA-3′ | |
| Dynamin 1 | Forward 5′-CTCGGCATATTCTGTGGTTG-3′ |
| Reverse 5′-GCTCGAGAATTTCGTAGGCA-3′ | |
| β-actin | Forward 5′-GATCTGGCACCACACCTTCT-3′ |
| Reverse 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
2.11. Western blot analysis
Cells were lysed with 1x SDS loading buffer in the presence of 100 mM dithiothreitol and denatured at 100 °C for 5 min. Equivalent amounts of protein were separated by SDS-PAGE, followed by transferring onto polyvinylidenedifluoride (PVDF) membranes (Bio-Rad Laboratories, USA) by electroblotting. Immunoblot analysis was then performed by incubating membranes with blocking buffer for 1 h at room temperature, and with appropriate antibodies diluted in blocking buffer for another 1 h. After washing three times with PBST, membranes were incubated with HRP-conjugated secondary antibody for 1 h and washed with PBST thrice. Blots were developed with an enhanced chemiluminescence (ECL) detection system (GE Healthcare Life Sciences, USA) and exposed to Chemiluminescence gel imaging system (Tanon 5200, Shanghai, China). Membranes were stripped with stripping buffer (10 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris-Cl, pH 6.8) at 55 °C for 30 min before re-probing with other antibodies.
2.12. Immunostaining
Cells were seeded on 4-well chamber slides and infected with virus or R18/R18-DiOC labeled virus with MOI = 5. At indicated time points, cells were fixed with 4% paraformaldehyde for 10 min, washed thrice with PBS, permeabilized with 0.2% Triton X-100 for 10 min, and washed thrice with PBS. Cells were then blocked in 3% FBS for 1 h, and incubated with anti-Rab5, anti-Rab7, or anti-LAMP1 (1:200 diluted in PBS, 5% BSA), washed thrice with PBS, and then incubated with secondary antibody conjugating with FITC (DAKO) for 2 h (1:200 diluted in PBS, 5% BSA), followed by PBS washing. For the detection of GM1 by CTB-FITC, no cell permeabilization required, cells were incubated with CTB-FITC (5 µg/ml) at 4 °C for 2 h. For actin staining, after fixing and permeabilization, cells were incubated with Alexa Fluor 488 Phalloidin for 2 h. Cells were incubated with 0.1 µg/ml DAPI for 10 min and rinsed with PBS. Finally, the specimen was mounted with glass cover slips using fluorescent mounting medium (DAKO) containing 15 mMNaN3. Images were collected with a LSM880 confocal laser-scanning microscope (Zeiss).
2.13. Plasmid construction
Eps15, Rab5, and Rab7 were amplified by PCR from HeLa cellular cDNAs and cloned into vector pEGFPN1 between restrict enzyme Xho I and BamH I, under the control of a cytomegalovirus promoter, generating pEGFPN1-Eps15, pEGFPN1-Rab5, and pEGFPN1-Rab7 with EGFP-tag at N-terminus. EGFP-tagged pEGFPN1-Rab5-DN (S34N), pEGFPN1-Rab5-CA (Q79L), and pEGFPN1-Rab7-CA (Q67L) mutants were constructed by site-directed overlapped two round PCR mutagenesis. EGFP-tagged Rab7-DN (T22N) was constructed as described previously (Rizopoulos et al., 2015) using the TaKaRa Mutant BEST Kit (Takara Bio, Dalian, China, #R401). The pEGFPN1-Eps15-DN (EΔ95/295), pEGFPN1-Dynamin 1-WT and pEGFPN1-Dynamin 1-DN (K44A) constructs were generous gifts from Prof. Mao Xiang. All primers used for plasmid construction are shown in Table 3.
Table 3.
Primer sequence for plasmid construction.
| Name | Primer sequence (5′ to 3′) |
|---|---|
| Eps15 | Forward 5′-AAGCCAACACCCTTCCAGTA-3′ |
| Reverse 5′-GCCCAGCTCTCTCTGACACA-3′ | |
| Rab5 | Forward 5′-CCGCTCGAGGCCACCATGGCTAGTCGAGGCGCAACA-3′ |
| Reverse 5′-CGCGGATCCGTTACTACAACACTGATTCCT-3′ | |
| Rab7 | Forward 5′-CCGCTCGAGGCCACCATGACCTCTAGGAAGAAAGTGTTG-3′ |
| Reverse 5′-CGCGGATCCGCAACTGCAGCTTTCTGCCGA-3′ | |
| Rab5- DN(S34N) | Forward 5′-TCCGCTGTTGGCAAAAACAGCCTAGTGCTTCGT-3′ |
| Reverse 5′-AGGCGACAACCGTTTTTGTAGCATCACGAAGCA-3′ | |
| Rab5-CA ( Q79L) | Forward 5′-TGGGATACAGCTGGTCTAGAAGGATACCATAGC-3′ |
| Reverse 5′-ACCCTATGTCGACCAGATCTTCCTATGGTATCG-3′ | |
| Rab7-DN (T22N) | Forward 5′-TCTGGAGTCGGGAAGAACTCACTCATGAACCAG-3′ |
| Reverse 5′-AGACCTCAGCCCTTCTTGAGTGAGTACTTGCTC-3′ | |
| Rab7-CA (Q67L) | Forward 5′-TGGGACACAGCAGGACTAGAACGGTTCCAGTCT-3′ |
| Reverse 5′-ACCCTGTGTCGTCCTGATCTTGCCAAGGTCAGA-3′ |
2.14. Statistical analysis
All data are presented as means ± standard deviations (SD), as indicated. Student’s t-test was used to compare data from pairs of treated or untreated groups. Statistical significance is indicated in the figure legends. All statistical analyses and calculations were performed by using Graph Pad Prism 5 (Graph Pad Software Inc., La Jolla, CA).
2.15. Densitometry
The intensities of corresponding bands were quantified using Image J program (NIH) according to the manufacturer’s instruction.
3. Results
3.1. IBV entry is lipid raft-associated
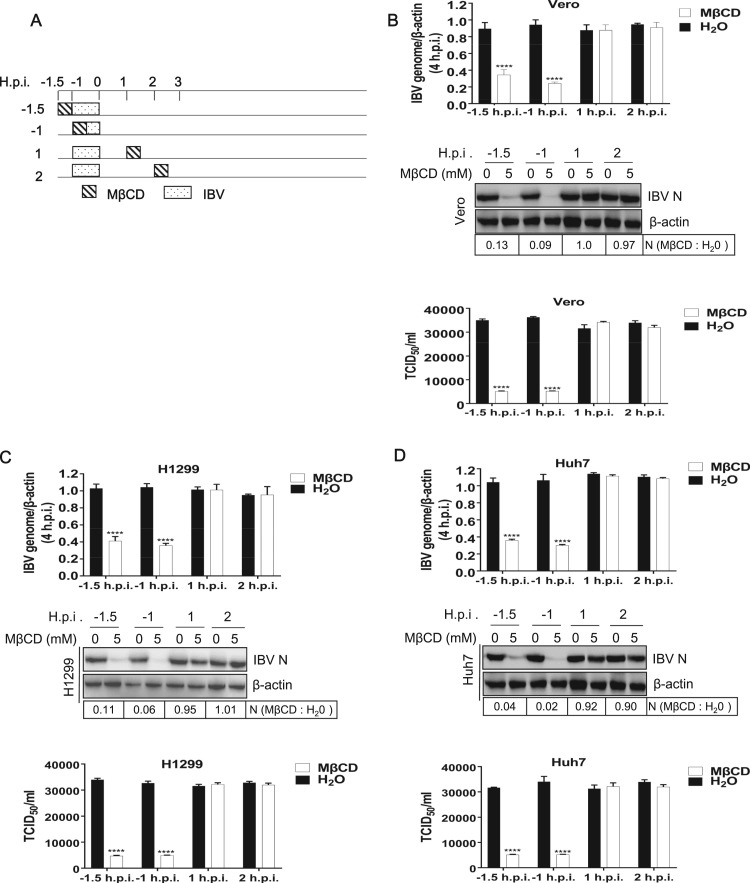
Lipid rafts are cholesterol-enriched microdomains, where many cellular proteins, including viral receptors, are preferentially localized at. A number of viruses enter host cells through lipid rafts (Fivaz et al., 1999). Previous report showed that IBV structural proteins migrate and integrate into lipid rafts during infection, while non-structural proteins do not integrate into rafts (Guo et al., 2017). This suggests that lipid rafts might participate in IBV entry or budding/release. To examine which stage of infection that lipid rafts are involved in, MβCD was used to disrupt lipid rafts at different stage of infection, in IBV Beaudette strain permissive cell lines. The subtoxic dose of the MβCD was determined by a cell proliferation assay (Supplementary Fig. 1). After determination of subtoxic dose, the IBV permissive Vero, H1299, and Huh7 cells were treated with 5 mM MβCD for 30 min at pre-, during-, or post- IBV infection at -1.5, -1, 1, 2 h.p.i. ( Fig. 1A). The effect of MβCD on virus infection was accessed by checking viral -gRNA, viral protein, and progeny virus release, respectively. As shown in Fig. 1B-D, treatment of various cell lines with MβCD at -1.5 and −1 h.p.i. acquired maximum blockage of virus genome production, as determined by checking viral -gRNA at 4 h.p.i.. Surprisingly, treatment with MβCD at 1 and 2 h.p.i. did not affect the virus genome production. Consistently, treatment with MβCD -1.5 and −1 h.p.i. obtained maximum inhibition effect on viral N protein expression, whereas treatment with MβCD at post-entry step did not affect the viral protein expression. Consequently, the progeny virus production was decreased by MβCD treatment at -1.5 and −1 h.p.i., but not by post-entry treatment. In all, above data demonstrate that MβCD inhibits virus entry step, but not post-entry process, suggesting that intact lipid rafts are required for virus entry.
Fig. 1.
Depletion of lipid rafts before or during virus absorption significantly reduces IBV entry in various cell lines. (A) Time frame of MβCD treatment. (B-D) Vero, H1299, and Huh7 cells were treated with 5 mM MβCD at indicated time points for 30 min, followed with IBV infection. H2O treated cells were included in a parallel experiment as control. At 4 h.p.i., the virus genome replication was determined by quantification of viral -gRNA with semi-quantitative real time RT-PCR; at 8 h.p.i., the virus N protein expression was determined with Western blot analysis; at 12 h.p.i. the release of virus particle was determined with TCID50 assay. The experiment was performed in triplicate, and average values with stand errors were presented in bar graph panel. The intensity of IBV N band was determined with Image J, normalized to β-actin, and shown as fold change of MβCD : H2O.
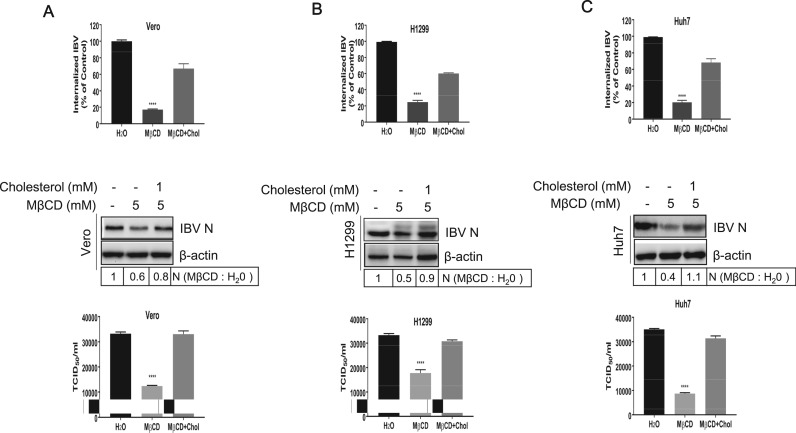
To check whether the effect of the MβCD on virus entry was solely due to cholesterol depletion, we examined the effect of exogenous cholesterol replenishment on virus entry after MβCD treatment. Vero, H1299, and Huh7 cells were pretreated with 5 mM MβCD, and supplemented with exogenous 1 mM cholesterol, followed with IBV infection. Results in Fig. 2A-C showed that MβCD treatment reduced virus gRNA internalization, however, cholesterol replenishment restored virus gRNA uptake. Accordingly, viral N protein expression was reduced by MβCD treatment and was restored by cholesterol replenishment. In accordance, progeny virus was rescued by cholesterol replenishment. Above results demonstrate that IBV entry depends on intact lipid rafts in various permissive cell lines.
Fig. 2.
Exogenous cholesterol replenishment restores virus infection. (A-C) Vero, H1299 cells, and Huh7 cells were pretreated with 5 mM MβCD for 30 min and supplemented with 1 mM cholesterol for 1 h before IBV infection. MβCD-treated cells without cholesterol replenishment were included in a parallel experiment as control. The entry of IBV was measured by quantification of viral gRNA at 2 h.p.i.; the expression of IBV N protein was measured at 8 h.p.i.; the release of virus particles in the medium was measured with TCID50 assay at 12 h.p.i. The experiment was performed in triplicate, and average values with stand errors were presented in bar graph panel. The intensity of IBV N band was determined with Image J, normalized to β-actin, and shown as fold change of MβCD : H2O.
3.2. Association of IBV with lipid rafts during the early stage of infection
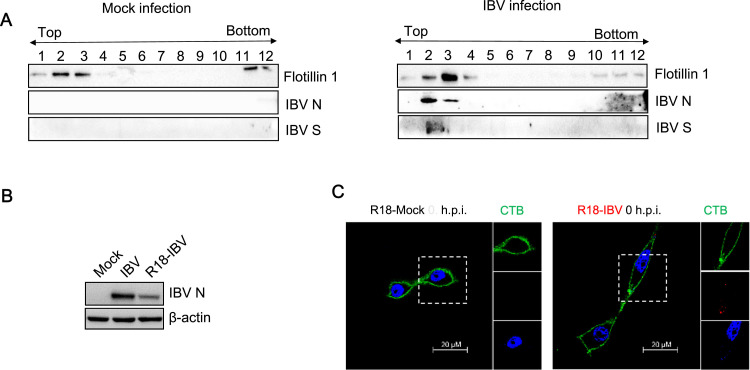
It should be mentioned that the receptor of IBV entry is controversial. Whereas some work states that heparan sulfate is a selective attachment factor for the IBV Beaudette strain (Madu et al., 2007), another report suggests that sialic acid is a receptor determinant of infection (Winter et al., 2006). Since membrane cholesterol plays an important role in early infection stages, the interaction of IBV with its receptor may occur in lipid rafts. To test this hypothesis, Vero cells were incubated with IBV at MOI = 5 for 1 h at 4 °C and lipid rafts were isolated by a sucrose flotation gradient. The individual fractions were analyzed for the presence of lipid rafts marker flotillin 1, IBV N protein, and IBV S protein by Western blotting. In IBV-infected cells, we found that S protein and N protein were co-localized with the rafts marker flotillin 1 in fraction 2 and 3 ( Fig. 3A, right panel). Thus, the biochemical fractionation analysis suggests the association of IBV with lipid rafts during attachment step.
Fig. 3.
IBV associates with lipid rafts during attachment. (A) Vero cells were incubated with IBV (MOI=5) at 4 °C for 1 h, followed by treatment with TNE buffer with 1% TX-100 at 4 °C for 30 min. Cells were subjected to membrane flotation analysis. Mock infection was included in a parallel experiment as control. The isolated membrane fractions were collected from top to bottom, and lipid rafts maker Flotillin-1, IBV N protein and S protein were detected by Western Blot analysis. (B) Vero cells were infected with IBV or R18-IBV at MOI = 5 and the expression of IBV N protein was analyzed with Western blot at 8 h.p.i.. (C) Vero cells were incubated with R18-IBV at MOI = 5 (red) or R18-mock for 1 h at 4 °C, followed with Alex Fluor 488 conjugated CTB staining (bind to lipid rafts marker GM1, green) for 1 h at 4 °C. After that, cells were fixed, permeablized, and stained with DAPI staining (blue) for 15 min at room temperature. Images were acquired by LSM880 confocal laser-scanning microscope (Zeiss). Representative images were shown. Scale bars = 20 µm.
To directly observe the binding of virus particles to cell surface lipid rafts, IBV were labeled with the lipophilic fluorescent dye R18, producing R18-IBV. The cell culture medium without virus infection was concentrated and mixed with R18 as control group (R18-mock), to eliminate the possibility of the mis-staining of R18 to cell components. Such R18-IBV retained moderate level of infectivity, compared to intact IBV, as determined by Western blotting (Fig. 3B). Immunofluoresence was then performed to check the interaction between R18-IBV and lipid rafts marker GM1. Vero cells were incubated with either R18-IBV or R18-mock at 4 °C for 1 h, followed with CTB-FITC (binds to lipid rafts marker GM1) staining. As shown in Fig. 3C, there was no R18 red signal in R18-mock treated cells, while the R18-IBV infected cells displayed red dot signals on the plasma membrane, suggesting that red dot signals indeed represent IBV virion. In addition, the R18-IBV red signals were co-localized with CTB-FITC, which marks the outline of lipid rafts. This result confirms the association of IBV with lipid rafts on cell surface during attachment step.
3.3. IBV entry is low pH dependent
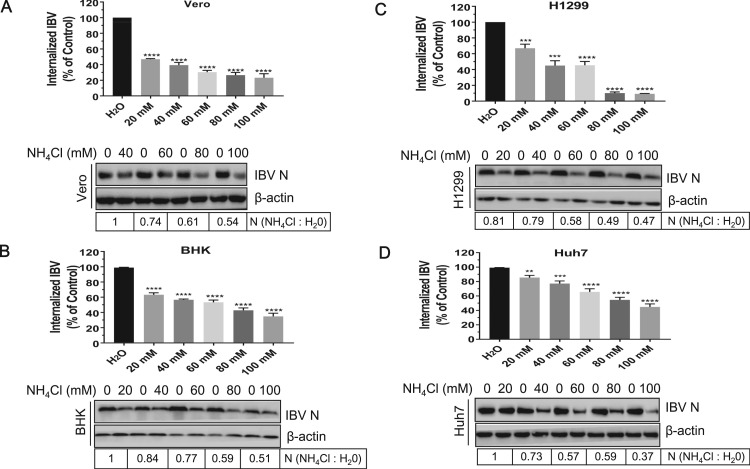
Although previous study demonstrates that the entry of IBV Beaudette strain is sensitive to pH increase in BHK cells (Chu et al., 2006), it also has been suggested that virus may employ different entry routes in different cell types (Frana et al., 1985). Thus, it is necessary to verify whether the low pH dependent entry of IBV is cell type specific or not. The subtoxic dose of endosome acidification inhibitor NH4Cl was determined by cell viability assay, which showed that Vero cells tolerated the treatment of NH4Cl up to 100 mM (Supplementary Fig. 2A). Increase of pH in intracellular vesicles was confirmed by acridine orange staining. In untreated cells, there was typical orange fluorescence, indicative of acidic compartments; as expected, the orange fluorescence was quenched along with increasing concentration of NH4Cl (Supplementary Fig. 2B). The basic mechanism of pH-dependent endocytosis for VSV has been well documented (Heine and Schnaitman, 1971, Matlin et al., 1982, Schlegel et al., 1981, Superti et al., 1987). Thus, VSV infection of Vero cells was used as a positive control to test the effectiveness of this chemical. Supplementary Fig. 2C showed that VSV G protein expression was decreased by NH4Cl treatment, suggesting that NH4Cl works well. After confirming the subtoxic dose and effectiveness of NH4Cl, we performed the experiment to check whether low pH dependence of IBV entry is universal in various cell lines. We increased the pH in Vero,BHK,H1299, and Huh7 cells by 0–100 mM NH4Cl pretreatment, then infected cells with IBV. BHK cells were used in this experiment as positive control. As shown in Fig. 4A-D, in all the cell lines we tested, virus internalization was significantly reduced by NH4Cl treatment in a dose dependent manner, compared to those in H2O treated cells. Accordingly, IBV N protein production was significantly decreased by NH4Cl treatment. Taken together, these results demonstrate that IBV entry requires low pH environment in various cell lines, consistent with the previous report (Chu et al., 2006).
Fig. 4.
IBV entry is hampered by increase of pH in the intracellular vesicles. (A-D) Vero, BHK, H1299, and Huh7 cells were treated with increasing concentrations (0–100 mM) of NH4Cl for 30 min, followed with IBV infection (MOI=1). The incoming virus level was determined by quantification of viral gRNA at 2 h.p.i. and virus N protein expression was determined by Western blot analysis at 8 h.p.i.. The experiment was performed in triplicate, and average values with stand errors were presented. The intensity of IBV N band was determined with Image J, normalized to β-actin, and shown as fold change of NH4Cl : H2O.
3.4. IBV enters cells via clathrin-medicated endocytosis (CME)
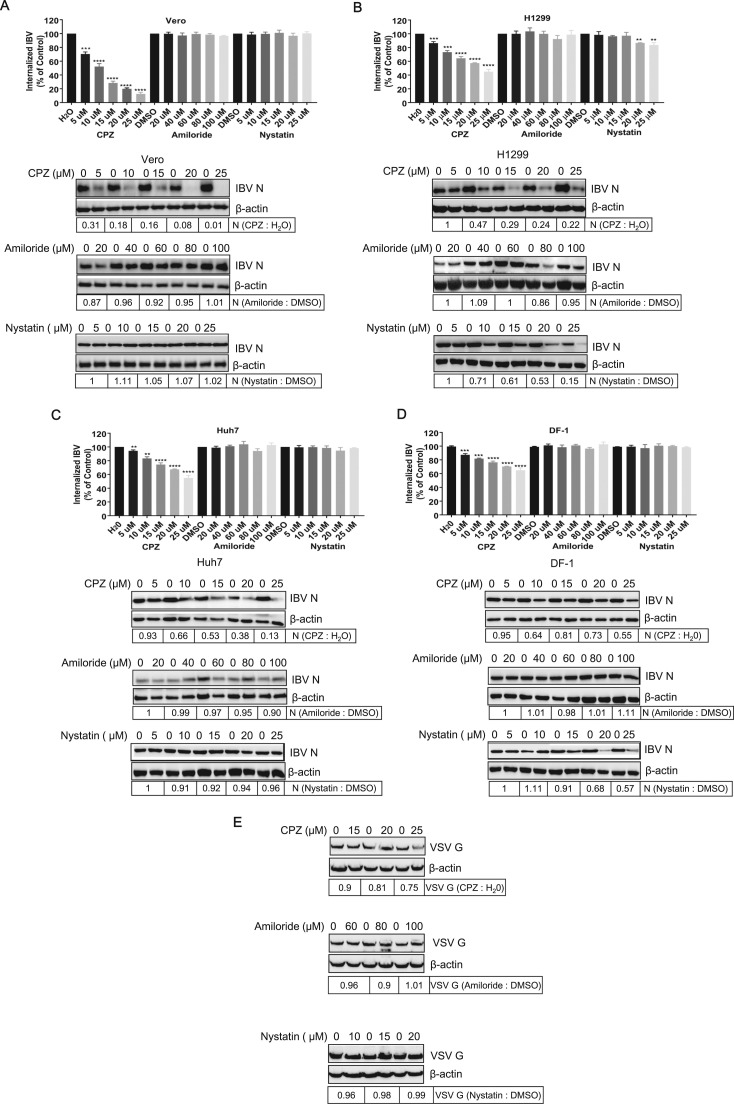
Several pH-dependent endocytic pathways are used by various viruses, including CME, CavME, macropinocytosis, and clathrin/caveolin-independent endocytosis (Marsh and Helenius, 2006). To identify the endocytic pathway employed by IBV, diverse pharmacological drugs pretreatments were carried out to block various endocytic pathways, and their effects on IBV entry were investigated. Prior to IBV infection, Vero, H1299, Huh7, and chicken cell DF-1 were exposed to increasing concentrations of chlorpromazine (CPZ), an inhibitor of clathrin coated pit formation (Martinez et al., 2007), Amiloride, a specific inhibitor of Na+/H+ exchanger activity important for macropinosome formation (Fretz et al., 2006, Kalin et al., 2010, Mercer and Helenius, 2008, Raghu et al., 2009), or Nystatin, a well known CavME inhibitor (Van Leeuwen et al., 2009). The toxicity of each inhibitor was carefully determined (data not shown) and the subtoxic dose was used to perform the experiment. It has been reported that VSV entry is mediated by CME (Superti et al., 1987). Thus, VSV infection was performed to validate the effectiveness of these inhibitors. Indeed, VSV G protein expression was inhibited by CPZ, but not Amiloride and Nystatin ( Fig. 5E). Similarly, CME inhibitor CPZ significantly inhibited the internalization of IBV gRNA in Vero, H1299, Huh7, and DF-1 cells, by a dose dependent manner, whereas macropinocytosis inhibitor Amiloride did not change the internalization of IBV in all these cell types (Fig. 5A-D). In accordance, the IBV N protein expression was significantly inhibited by CPZ, but not Amiloride. Thus, these results reveal that IBV enters cells via CME, but not via macropinocytosis, at least in the cell lines we tested. The involvement of CavME in IBV endocytosis is controversial. In Huh7 cell, which lacks functional caveolae (Beer et al., 2005, Vainio et al., 2002), efficient virus internalization and replication was observed (Fig. 5C), indicating that IBV entry is independent of CavME in this cell line. Indeed, Nystatin pretreatment did not alter the virus internalization and protein expression in both Vero and Huh7 cells (Fig. 5A and C). Thus, at least in these two cell lines, IBV does not employ CavME route to enter into cells. However, Nystatin slightly reduced the IBV internalization and protein expression in H1299 cells, especially in 20–25 µM concentration (Fig. 5B). It is probably that in H1299 cells, in addition to CME, CavME is also hijacked by IBV. In DF-1 cells, Nystatin did not inhibit virus internalization, but really reduced viral protein expression (20–25 µM) (Fig. 5D). The underlying mechanism of this phenomon is unclear. From above results, we conclude that IBV mainly employs CME to enter into cells; however, in some cell lines, IBV may also employ CavME to enter into cells, as a supplementary route.
Fig. 5.
IBV entry is mainly blocked by clathrin-mediated endocytosis (CME) inhibitor CPZ. (A-D) Vero, H1299, Huh7, and DF-1 cells were pretreated with increasing concentrations of CPZ (CME inhibitor), Amiloride (macropinocytosis inhibitor), or Nystatin (CavME inhibitor) for 30 min and infected with IBV at MOI= 1. H2O or DMSO pretreatment in parallel experiments was set as control. The incoming virus level was determined by quantification of viral gRNA at 2 h.p.i.; the expression level of IBV N protein was analyzed at 8 h.p.i.. The experiment was performed in triplicate, and average values with stand errors were presented in panel A, C, E, and G. (E) Vero cells pretreated with respective drug and infected with VSV at MOI = 1 were set as control. The expression of VSV G protein was analyzed by Western blot at 8 h.p.i.. The intensity of IBV N or VSV G band was determined with Image J, normalized to β-actin, and shown as fold change of CPZ : H2O, Amiloride : DMSO, or Nystatin : DMSO.
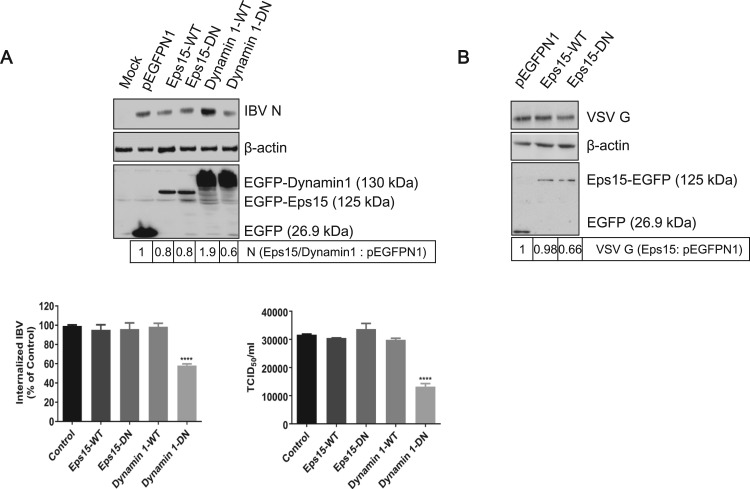
3.5. IBV entry is dependent on dynamin 1
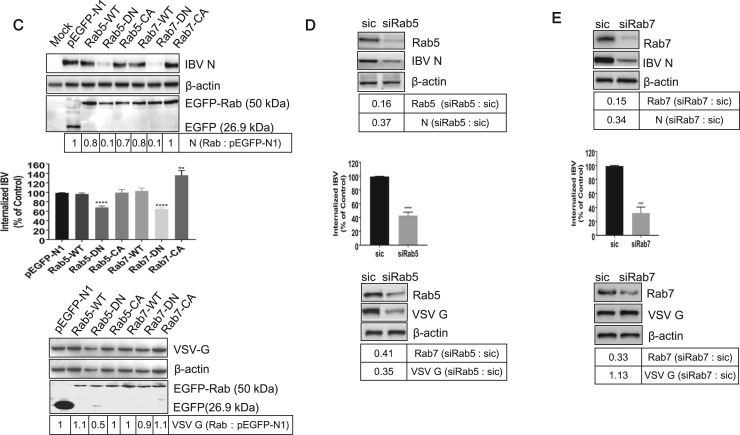
Eps15 is a regulatory protein which has been characterized by a ubiquitous and constitutive association with the AP-2 protein adapter. Microinjection of Eps15 antibody interfered with transferrin and EGFR internalization (Gallo et al., 1997), suggesting that Eps15 plays an important role in CME. Several reviews described previously that the expression of dominant negative Eps15 (Δ95/295) could efficiently block the uptake of Sindbis virus (Panda et al., 2013). Dynamin 1 is a GTPase facilitating membrane fission to generate endocytic vesicles in CME and CavME (Henley et al., 1998, Smith et al., 2007, Zhu et al., 2012). Many reviews describe that dynamin 1 is required for the internalization of many viruses, as the Dynamin 1-K44A, a dominant negative mutant, blocks virus entry (Cai et al., 2015, Holla et al., 2015). To examine the involvement of Eps15 and dynamin 1 in IBV entry, we constructed plasmids EGFP-Eps15-WT, EGFP-Eps15-DN (EΔ95/295), EGFP-Dynamin 1-WT, and EGFP-Dynamin 1-DN (K44A), with EGFP tag at the N-terminus. These plasmids were transfected into H1299 cells, respectively, followed with IBV infection. pEGFPN1 was expressed as control. H1299 cells were used in this experiment due to its high transfection efficiency. The successful expression of EGFP and EGFP-fused proteins was detected in Fig. 6A. It was noted that the expression level of EGFP-Eps15 is lower than that of EGFP-dynamin 1. Compared to that in EGFP expressing cells, the expression of IBV N protein was enhanced in EGFP-dynamin 1-WT expressing cell, whereas was reduced in EGFP-dynamin 1-DN expressing cells (Fig. 6A, upper panel). Consistently, the IBV internalization and progeny virus production was significantly reduced by the expression of EGFP-dynamin 1-DN (Fig. 6A, low panel). This result demonstrates the important role of dynamin 1 in IBV infection. Surprisingly, neither EGFP-Eps15-WT nor EGFP-Eps15-DN significantly changed the IBV internalization, protein expression, or progeny virus release, compared to those in EGFP expressing cells (Fig. 6A). It seems that Eps15 is not involved in IBV infection in H1299 cells. Whether the failure of blocking IBV infectivity by EGFP-Eps15-DN is due to its low expression level or other unknown mechanisms? It has been reported that VSV entry relies on functional Eps15 (Sun et al., 2005). Thus, the effect of Eps15 on VSV entry of Vero cells was examined as control experiment. As shown in Fig. 6B, although the expression level of EGFP-Eps15-DN was low, this mutant indeed inhibited VSV infection, as evidenced by the reduced level of VSV G protein (by 0.66-fold), compared to that in EGFP expressing cells. In all, above results suggest that in H1299 cells, IBV entry depends on functional dynamin 1, but not Eps15. It is probably due to IBV hijacks both CME and CavME to enter into H1299 cells, as evidenced in Fig. 5. When CME pathway is blocked by expression of EGFP-Eps15-DN, CavME becomes an alternative entry route for IBV. Another possibility is that IBV entry depends on CME, but does not require the help of Eps15.
Fig. 6.
IBV endocytosis is dependent on dynamin 1. (A) H1299 cells were transfected with constructs encoding EGFP tagged Eps15-WT, Eps15-DN, Dynamin 1-WT, Dynamin 1-DN, or vector pEGFP-N1, respectively. At 24 h post-transfection, cells were infected with IBV (MOI=1). Cells were harvested at 8 h.p.i.. The expression of EGFP, EGFP-Eps15-WT, EGFP-Eps15-DN, EGFP-Dynamin 1-WT, EGFP-Dynamin 1-DN, and IBV N protein was checked with Western blot analysis by using anti-GFP or anti-IBV N (upper panel); the internalized IBV gRNA was analyzed with semi-quantitative real time RT-PCR at 2 h.p.i. (low panel, left); the virus particle release was determined by TCID50 assay at 12 h.p.i. (low panel, right). The experiment was performed in triplicate, and average values with stand errors were presented in panel B and C. (B) Vero cells transfected with constructs encoding EGFP fused Eps15-WT, Eps15-DN, or vector pEGFP-N1, respectively. At 24 h post-transfection, cells were infected with VSV (MOI=1). Cells were harvested at 8 h.p.i.. The expression of EGFP, EGFP-Eps15-WT, EGFP-Eps15-DN, and VSV G protein was checked with Western blot analysis by using anti-GFP or anti-VSV G. The intensity of IBV N or VSV G band was determined with Image J, normalized to β-actin, and shown as fold change of Eps15/Dynamin 1 : pEGFP-N1 or Eps15 : pEGFP-N1.
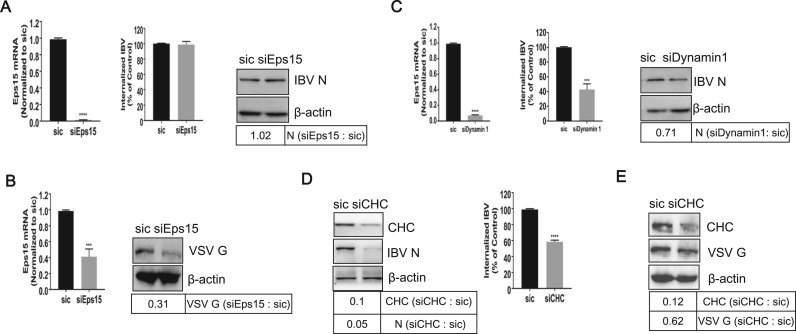
To further confirm above conclusion, we analyzed the effect of silencing Eps15 or dynamin 1 on IBV uptake. To this purpose, cells were transfected with non-targeting siRNA (sic) or siRNA targeting to either Eps15 or dynamin 1, and then infected with IBV (H1299 cells) or VSV (Vero cells, control group). The knock down efficiency of Eps15 or dynamin 1 and uptake of IBV gRNA were measured by semi-quantitative real time RT-PCR. As shown in Fig. 7A and B, successful silence of Eps15 mRNA was obtained by siEps15 in both H1299 and Vero cells. However, silence of Eps15 expression had no significant effect on the IBV gRNA internalization and protein expression in H1299 cells (Fig. 7A), although Eps15 depletion indeed inhibited VSV G expression (Fig. 7B). These data further suggest that when CME route is hampered by Eps15 depletion, CavME may be an alternative route for IBV entry in H1299 cells. As expected, knock down of dynamin 1 (by 92%) was accompanied by a significant reduction of the amount of incoming IBV gRNA (by 48%) and IBV protein expression (by 71%) (Fig. 7C). Thus, dynamin 1 is functionally required for IBV entry in H1299 cells, due to its crucial role in both CME and CavME.
Fig. 7.
Both dynamin 1 and CHC are involved in IBV endocytosis. (A-E) Cells were transfected with non-target siRNA (sic), siEps15, sidynamin 1, or siCHC for 36 h, followed with IBV infection (H1299 cells, A, C, D) or VSV infection (Vero cells, B, E). The knock down effect of Eps15, dynamin 1, and the internalization of IBV was analyzed by semi-quantitative real time RT-PCR at 2 h.p.i.. The knock down effect of CHC, the expression level of IBV N protein or VSV G protein, were determined with Western blot at 8 h.p.i.. The experiment was performed in triplicate, and average values with stand errors were presented in bar graph panel. The intensity of IBV N, VSV G, or CHC band was determined with Image J, normalized to β-actin, and shown as fold change of siEps15: sic, siDynamin 1 : sic, or siCHC : sic.
We further analyzed the effect of silencing CHC expression on IBV uptake. Cells were transfected with siRNA targeting CHC and then infected with IBV (H1299 cells) or VSV (Vero cells, control group). As shown in Fig. 7D-E, depletion of CHC was accompanied with a significant reduction of IBV gRNA internalization and IBV/VSV protein expression. This result suggests that a functional CME pathway is required for efficient IBV infection. Thus, CME is the main entry pathway of IBV.
3.6. IBV entry leads to active actin rearrangement
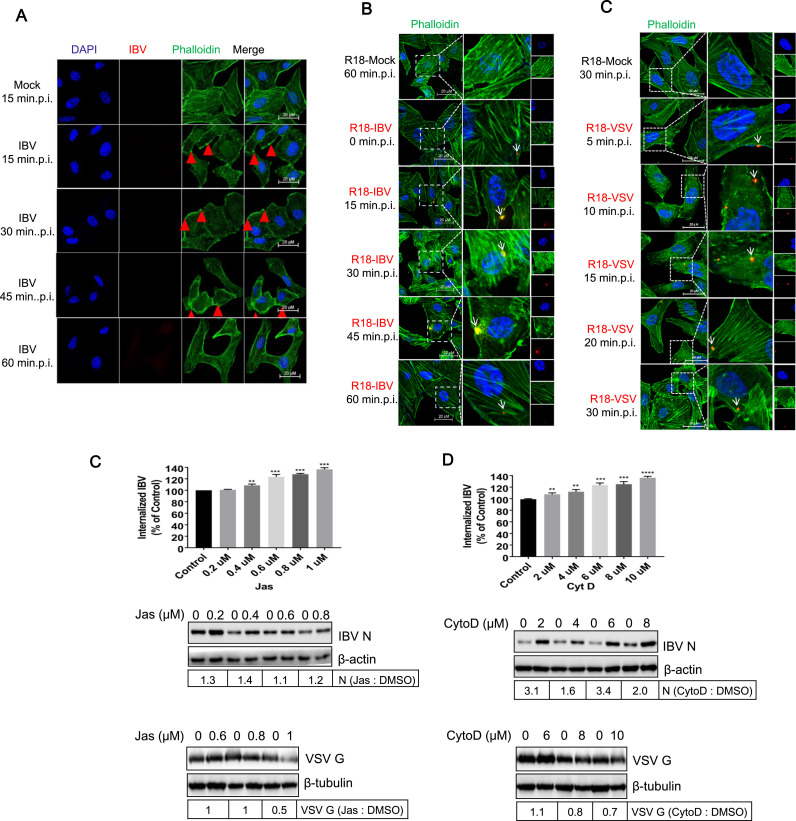
In Saccharomyces cerevisae, clathrin-coated patches on the cell surface mature into ∼ 200nm tubular invaginations with coat proteins at their tip. The actin cytoskeleton machinery is recruited after coat formation and provides an essential force for membrane deformation and internalization (Kaksonen et al., 2005). Recent studies have revealed that the rhabdoviruses (Regan and Whittaker, 2013), VSV (Cureton et al., 2010), RABV (Piccinotti et al., 2013), and IHNV (Liu et al., 2011) are internalized into cells via CME pathway and actin filaments are involved in. Although we have demonstrated that IBV enter cells by hijacking clathrin dependent route, it is unclear whether actin filaments are involved in IBV uptake. To examine the role of actin filaments in IBV entry, we first examined the actin polymerization during IBV internalization. Vero cells were incubated with IBV at 4°C for 1h, allowing attachment. R18-mock treated cells were included as control. After removing the unbound virus by replacing with fresh DMEM, the temperature was shifted to 37°C to allow the synchronous internalization. Actin filaments were stained with Alexa Fluor 488-phalloidin (green) and IBV virus particles were stained with N antibody (red) with 15 min time interval. As shown in Fig. 8A, in mock-infected cells, the actin stress fibers were visible clearly. From 15–45min.p.i., the number of actin stress filaments dramatically decreased, the cells rounded up, and cells surface displayed significant blebbing (red arrows). The cell morphology and actin distribution returned to normal at 60min.p.i.. Above observation indicates that IBV infection results in actin de-polymerization and re-arrangement. Unfortunately, IBV N protein antibody failed to detect the incoming virus. To make the incoming virus particles visible, R18-IBV was applied to Vero cell. Again, the decrease of actin filaments was observed at 15, 30, and 45min.p.i.. R18-IBV was detected at cell surface at 0min.p.i., displayed as red signal (white arrow). After 0min.p.i., R18-IBV entered cells and associated with filaments at 15, 30, and 45min, shown as yellow signals (white arrows). At 60min.p.i., R18-IBV particles disassociated with actin filaments, displayed as red signal again (whie arrow). This observation suggests that IBV enters cells via actin cytoskeleton. Here, we also incubated the well characterized R18-VSV with Vero cells as control group, ensuring above experiment was performed properly. As R18-VSV entry is quite fast, we chose short time interval. Image in Fig. 8C showed that R18-VSV infection also caused actin filaments de-polymerization, and the cells were rounded up at 5, 10, and 15min.p.i.. The cell morphology and actin distribution returned to normal at 20 and 30min.p.i.. Meanwhile, R18-VSV associated with actin fibers at 5, 10, 15, 20, and 30min.p.i. (white arrows), in consistence with previous report (Mire et al., 2010). Above experiment demonstrates that IBV induces actin rearrangement and move along with actin filaments after internalization, similar to VSV.
Fig. 8.
IBV entry causes actin rearrangement. (A-C) Vero cells were incubated with IBV, R18-IBV, or R18-VSV (MOI=5) at 4 °C for 1 h to allow virus attachment, and the unbound virus particles were washed away with DMEM. Mock infection or R18-mock infection was set as control group. Temperature was shifted to 37 °C to allow virus synchronous internalization. Cells were fixed at indicated time points, and incubated with Alexa Fluor 488-phalloidin (green) for 1 h to stain actin filaments, followed with IBV N protein staining (for IBV infection group, red) and nuclei staining by DAPI (blue). The signals were observed under LSM880 confocal laser-scanning microscope (Zeiss). Representative images were shown. Scale bars = 20 µm. Red signal: R18-IBV; green signal: actin; blue signal: nuclei. (C-D) Vero cells were pretreated with increasing concentrations of CytoD or Jas for 30 min and infected with IBV or VSV (MOI=5). The internalized IBV gRNA was assayed at 2 h.p.i. (upper panel), and the expression of IBV N protein (middle panel) or VSV (low panel) was determined at 8 h.p.i.. β-actin, was detected as loading control. The experiment was performed in triplicate, and average values with stand errors were presented in bar graph panel. The intensity of IBV N or VSV G was determined with Image J, normalized to β-actin, and shown as fold change of Jas : DMSO or CytoD : DMSO.
We further investigated the role of the actin cytoskeleton on IBV entry by using chemical inhibitors to interfere with actin polymerization or de-polymerization. Before being infected with virus, Vero cells were pretreated with Jas, an actin polymer-stabilizing drug (Bubb et al., 1994, Ou et al., 2002, Spector et al., 1999), or CytoD, which binds to the growing ends of actin filaments and prevents the polymerization (Sampath and Pollard, 1991). The VSV infection was included in a parallel experiment as control, ensuring these inhibitors work. Fig. 8D showed that when actin polymers were stabilized by Jas, the IBV internalization were increased in a dose-dependent manner (upper panel). Consistently, the expression of IBV N protein was also slight enhanced (middle panel). Fig. 8E showed that when CytoD prevented the formation of actin polymers, IBV internalization and N protein expression were also significantly increased (upper and middle panels). This observation is self-contradictory and out of expectation. In the VSV infection group, both Jas and CytoD reduced VSV G protein expression (Fig. 8D-E, low panels), consistent with previous report (Mire et al., 2010). These results imply that the inhibitors indeed interfere with actin polymerization or de-polymerization. It has been reported that SFV infection was also enhanced by CytoD pretreatment (Schelhaas et al., 2012). Thus, the enhancement of IBV infection by Jas or CytoD is not the only case. How do these inhibitors increase IBV entry? The underlying mechanisms remain open question.
3.7. IBV moves across the entire endo-lysosomal system
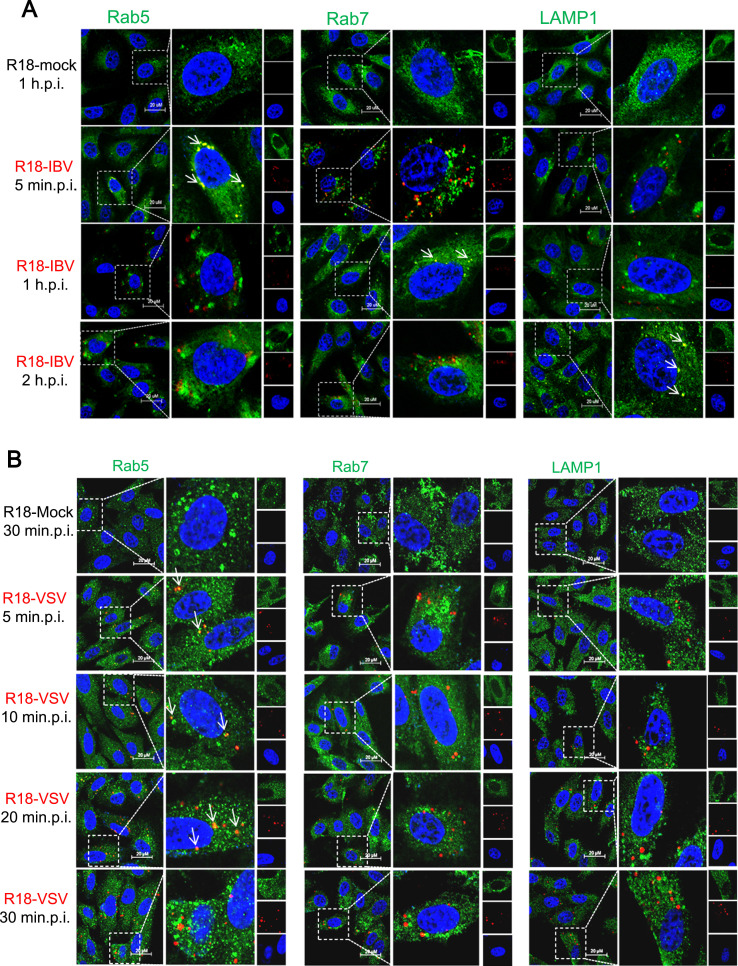
After identification of the endocytic pathway hijacked by IBV, we next tracked the transport vesicles utilized by incoming IBV. R18-IBV infected cells were subjected to immunostaining by using antibodies against early endosome marker Rab5, late endosome marker Rab7, or lysosome marker LAMP1. Rab5 and Rab7 are GTPases that are required for the movement of endocytosed cargo to early or late endosomes (Feng et al., 1995a, Gruenberg, 2001). The co-localization of R18-IBV with transport vesicles markers was observed from 5 min.p.i. to 2 h.p.i.. As shown in Fig. 9A, there was no red signal detected in R18-mock treated cells (first panel); the overlapped signals of R18-IBV and early endosome marker Rab5 were mainly observed at 5 min.p.i. (yellow dots with white arrows in second panel); however, there was no co-image of R18-IBV with Rab5 at 1 h.p.i. and 2 h.p.i. (third and fourth panels). This indicates that IBV enters into early endosomes in a few minutes when the temperature is shifted to 37 °C, and dissociates with early endosomes at 1 and 2 h.p.i.. Moreover, the co-localization of virus particles and Rab7 was mainly observed at 1 h.p.i. (yellow dots with white arrow in third panel), suggesting that R18-IBV enters late endosomes at 1 h.p.i.. Furthermore, R18-IBV was found in LAMP1 containing lysosomes at 2 h.p.i. (yellow dots with white arrows in fourth panel). Previous study showed that VSV was internalized into early endosomes in a few minutes via CME (Johannsdottir et al., 2009). To ensure the feasibility and reliability of this tracking assay, we used R18-VSV to do the infection and immunostaining. As shown in Fig. 9B, R18-VSV co-localized well with early endosome marker Rab5 at 5, 10, and 20 min.p.i. (yellow dots with white arrows in second, third, and fourth panels). There was no yellow dot of R18-VSV with Rab5 at 30 min.p.i. (fifth panel), indicating that R18-VSV left early endosomes at this time point. Moreover, there was almost no co-image of R18-VSV with Rab7/LAMP1 from 5 to 30 min.p.i.. These observations verify that VSV enters early endosomes, but is not delivered into late endosomes and lysosomes (Mire et al., 2010), demonstrating this tracking assay is reliable. Altogether, above results reveal that IBV enters early endosomes within a few minutes after endocytosis, passes through late endosomes at around 1 h.p.i., and reaches lysosomes at 2 h.p.i..
Fig. 9.
The incoming R18-IBV transports along the entire endocytic pathway. (A-B) Vero cells were infected with R18-IBV or R18-VSV (MOI=5) at 4 °C for 1 h, the unbound virus particles were washed away by DMEM, and temperature was shifted to 37 °C to allow synchronous internalization. Cells were immunostained with Rab5, Rab7, or LAMP1 at indicated time points, followed with DAPI staining. R18-mock infection was included in a parallel experiment as control. The signals were observed under LSM880 confocal laser-scanning microscope. Representative images were shown. Scale bars = 20 µm. Red signal: R18-IBV or R18-VSV; green signal: early endosome (Rab5), later endosome (Rab7), or lysosome (LAMP1); blue signal: nuclei. (C) Cells were transiently transfected with constructs encoding EGFP, EGFP-Rab5-WT, EGFP-Rab5-DN (S34N), EGFP-Rab5-CA (Q79L), EGFP-Rab7-WT, EGFP-Rab7-DN (T22N), and EGFP-Rab7-CA (Q67L) for 24 h, followed with IBV infection (H1299 cells) or VSV infection (Vero cells). The expression of IBV N protein, VSV G protein, EGFP, EGFP-Rab5, and EGFP-Rab7 were detected with corresponding antibodies at 8 h.p.i. (upper panel and low panel); the incoming virus level was determined by quantification of viral gRNA at 2 h.p.i. (middle panel). (D-E) Cells were transfected with siRNAs targeting to either Rab5 or Rab7, respectively. Non-target siRNA (sic) was transfected as control. At 36 h post-transfection, cells were infected with IBV (H1299 cells) or VSV (Vero cells). The knock down effect of Rab5 and Rab7, the expression of IBV N and VSV G was detected by Western Blot analysis at 8 h.p.i. (middle panel). The incoming IBV level was determined by measurement of viral gRNA at 2 h.p.i. (middle panel). Above experiments were performed in triplicate, and average values with stand errors were presented in graph bar panel. The intensity of IBV N or VSV G was determined with Image J, normalized to β-actin, and shown as fold change of Rab : pEGFP-N1, siRab5 : sic, or siRab7 : sic.
As IBV co-localized extensively with endo-lysosomal compartments, we asked whether all these vehicles are really involved in IBV infectious route. To achieve this, we took advantage of the fact that, in addition to providing organelle identity, Rab5 and Rab7 serve critical role in regulation of cargo transport within the endosomal system. Accordingly, dominant negative mutants (GTP-binding defective mutants) of these proteins have been shown to block cargo transport (Feng et al., 1995b, Stenmark et al., 1994). Expression of Rab5-DN prevents fusion of endocytic vesicles with early endosomes, while Rab7-DN prevents cargo movement from early to late endosomes. H1299 cells were transfected with pEGFPN1, EGFP fused Rabs-WT, Rabs-DN, or Rabs-CA plasmid, respectively, followed with IBV infection. Rabs-CA are constitutively active mutants with constitutive GTP-binding activity and were expressed to enhance the cargo transport ability of endosome. The cells were harvested for Western blot to check the expression of Rabs and IBV N protein. As shown in Fig. 9C upper panel, all these EGFP-Rabs protein were successfully expressed in comparable level. Compared with EGFP expressing group, expression of EGFP-Rab5-WT, EGFP-Rab5-CA, EGFP-Rab7-WT, and EGFP-Rab7-CA has only minimal effect on the expression of IBV N, whereas expression of EGFP-Rab5-DN and EGFP-Rab7-DN caused significantly reduction (0.1-fold) in IBV protein expression (Fig. 9C, upper panel). Accordingly, Rab5-DN and Rab7-DN decreased internalization of IBV gRNA by 0.6-fold, respectively (Fig. 9C, middle panel). It was noted that expression of Rab7-CA slightly increased virus internalization (Fig. 9C, middle panel). These data show that IBV could not efficiently infect cells which express Rab5-DN or Rab7-DN, indicating that transport through the early and late endosome is necessary for successful infection. To ensure our Rabs construct was functional, we examined the effect of Rabs on VSV infection. In consistence with previous report (Mire et al., 2010), Rab5-DN, but not Rab7-DN, inhibited VSV infection (Fig. 9C, low panel). Collectively, these data demonstrate that both early and late endosome are involved in IBV intracellular transport.
Next, knock down of Rab5 or Rab7 was carried out to confirm above conclusion. H1299 cells were transfected with siRab5 or siRab7, followed with IBV infection. The knock down effect of Rabs and the expression of IBV N protein were examined by Western blot. Fig. 9D-E upper panels showed that both Rab5 and Rab7 expression were successfully knocked down (0.16-fold and 0.15-fold, respectively), accompany with lower expression of IBV N protein (0.37-fold and 0.34-fold, respectively), comparing to those in sic tranfected cells. Quantification of incoming virus genome showed that knock down of Rab5 or Rab7 significantly blocked virus internalization (Fig. 9D-E, middle panels). Above results demonstrate that the virus infection is inefficiency when the transport pathway is disabled by removing Rab5 or Rab7. To ensure siRabs was indeed functional, we examined the effect of Rabs on VSV infection in Vero cells. Western blot result showed that knock down of Rab5 reduced VSV G protein expression by 0.35-fold (Fig. 9D, low panel), while knock down of Rab7 had no effect on VSV infection (Fig. 9E, low panel). Altogether, these data further confirm that early and late endosomes are required for intracellular transport of IBV.
3.8. IBV fuses with late endosomes or lysosomes
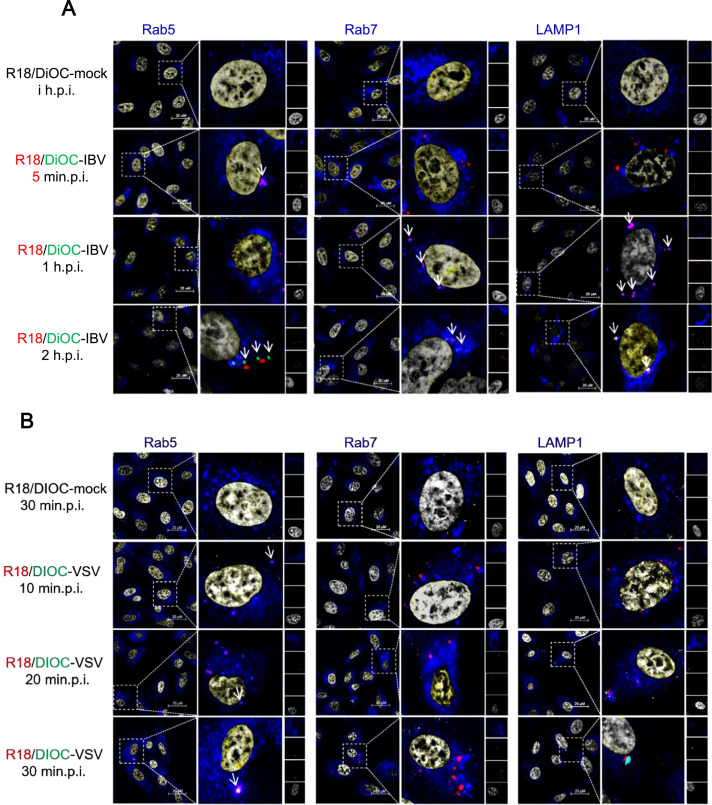
To dissect the fusion of IBV envelope with intracellular vesicle membranes, we labeled IBV virion with two fluorescent lipids, R18 (red) and DiOC (green), a labeling method developed by Sakai and coworkers (Sakai et al., 2006). The intact virus membrane displays the red color as the high concentration of R18 quenches the fluorescence signal emitted by the DiOC. When the virus membranes fuses with cellular membranes, the two lipids were diluted and the green fluorescence of DiOC was no longer quenched by R18, red and green color will be displayed respectively. To rule out the possibility of mislabeling cell debris membranes, cell culture medium was labeled with R18 and DiOC as control (R18/DiOC-mock). Vero cells were infected with R18/DiOC-mock for 1 h, or infected with R18/DiOC-IBV for 5 min, 1 h, and 2 h, followed with Rab5, Rab7, or LAMP1 staining. As shown in Fig. 10A, in mock infection group, there was neither red signal nor green signal observed (first panel). In R18/DiOC-IBV infected cells, at 5 minp.i. and 1 h.p.i., there was no green signals detected, only red color spots observed (second and third panels). Moreover, most of the red spots were co-localized with early endosome marker Rab5 at 5min.p.i (pink spots with white arrow in second panel), and transferred to Rab7 containing late endosomes or LAMP1 containing lysosomes at 1 h.p.i. (pink spots with white arrows in third panel). Gradually, green spots (membrane fusion signals) were observed and increased in numbers at 2 h.p.i., overlapped with either Rab7 or LAMP1, without overlapping with Rab5 (green and red spots with white arrows in fourth panel). These results imply that IBV membranes undergo fusion with late endosome/lysosome membranes at 2 h.p.i.. It is possible that the fusion process may happen at earlier time of 1 h.p.i. to 2 h.p.i. We also performed above fusion assay by using R18/DiOC-VSV, which has been reported to fuse with early endosomes. As shown in Fig. 10B second panel, pink spots overlapped with Rab5 were observed at 10 min.p.i. (pink spots with white arrow); there was no virus particles overlapped with Rab7 or LAMP1 at this time point (virus particles were displayed as red spots); and no green spots observed. At 20 min.p.i. and 30 min.p.i. (Fig. 10B, third and fourth panels), green spots were detected, without overlapping with Rab7 or LAMP1, but co-localized well with Rab5 (white spot with white arrow), consistent with previous report (Mire et al., 2010). In all, we demonstrate that IBV fuses with late endosome/lysosome at 1–2 h.p.i., and its intracellular trafficking is slower than VSV.
Fig. 10.
IBV fuses with late endosomes or lysosomes membranes. (A-B) Vero cells were infected with R18-DiOC-mock, R18-DiOC-IBV, or R18-DiOC-VSV (MOI=5) at 4 °C for 1 h, the unbound virus particles were washed away by DMEM, and the temperature was shifted to 37 °C to allow synchronous internalization. At indicated time points, cells were subjected to immunostaining by using Rab5, Rab7, or LAMP1 antibody. The separation of R18 (red) and DiOC (green) signals, and co-localization with vehicle markers (blue) were observed under LSM880 confocal laser-scanning microscope. Representative images were shown. Scale bars = 20 µm. Red signal: R18-DiOC-virus or R18-virus; green signal: DiOC-virus; blue signal: early endosome (Rab5), later endosome (Rab7), or lysosome (LAMP1); grey signal: nuclei.
4. Discussion
Viruses must enter the host cell to start their life cycle. Thus, characterization of viral entry pathway is important for understanding of virus pathogenesis and design of anti-viral drugs. Envelope viruses penetrate cells by two different ways: fusion with cell plasma membrane or receptor-mediated endocytosis. Direct membrane fusion on the cell surface is independent of low pH, and access through the endocytosis pathway usually relies on the low pH of the endocytic vesicles. Different endocytosis pathways have been characterized over the past decade, in which the CME is one of the most commonly used by many envelope viruses (Sieczkarski and Whittaker, 2002). Coronavirus entry is mediated by S protein, which is responsible for receptor binding and membrane fusion. Previously, it has been demonstrated that SARS-CoV is internalized by CME in human hepatoma HepG2 cell line (Inoue et al., 2007), however, in HEK293E-ACE2-GFP cells, the entry of SARS-CoV is clathrin- and caveolae-independent (Wang et al., 2008); MHV entry is mediated by CME through the endo-/lysosomal pathway in proteolysis-dependent manner (Burkard et al., 2014; Eifart et al., 2007; Nash and Buchmeier, 1997; Pu and Zhang, 2008; Qiu et al., 2006); PHEV enters into cells via CME in Rab5, cholesterol, and low pH dependent manner (Li et al., 2017); HCoV-NL63 requires CME for successful entry into the LLC-MK2 cells (Milewska et al., 2017); for the HCoV-229E, a CavME uptake has been suggested (Nomura et al., 2004), however, the clinical isolates of HCoV-229E bypass the endosome for cell entry (Shirato et al., 2017). Increasing evidence has demonstrated that coronavirus entry mechanisms are quite complex: they may enter cells via multiple pathways, or employ a specific route to enter into a specific cell line, or different virus strains or serotypes utilize different entry pathways. Thus, study on group gamma coronavirus IBV entry mechanisms is notable for understanding infection process. As mentioned previously, IBV infection on BHK cells is inhibited by NH4Cl treatment (Chu et al., 2006), demonstrating that IBV entry requires low pH triggered membrane fusion. In this study, we made an effort to systematically examine every step of this process. First, we find that IBV particles initially attach to lipid rafts on plasma membrane, and enter cells via CME, which requires the help of CHC and Dynamin 1. Additionally, we demonstrate that IBV particles are transported via early and late endosome, and fuse with late endosome-lysosome membranes to release the nucleocapsid.
Lipid rafts are involved in regulation of different biological events, serve as organizing centers of signaling molecules assembly, and influence membrane fluidity and membrane protein trafficking (Pike, 2009). The involvement of lipid rafts in virus entry, assembly and budding has been demonstrated by either the co-localization study of the viral structural proteins and lipid rafts or the effects of lipid rafts disrupting agents on the replication processes of several viruses (Suzuki and Suzuki, 2006). The best characterized lipid rafts-related non-enveloped viral entry is simian virus 40 (SV40) and echovirus type 1 (EV1) (Chazal and Gerlier, 2003, Pietiainen et al., 2005). Binding of SV40 with MHC class I receptors triggers receptor clustering and redistribution, more caveolae is recruited from the cytoplasm to the site of entry, resulting in caveolae-mediated endocytosis in about 20 min (Pietiainen et al., 2005). In some cell types, SV40 can enter the caveosomes directly from lipid rafts in non-coated vesicles (Rajendran and Simons, 2005). Similar to SV40, EV1 attachment and binding with cells triggers clustering and relocation of α2β1-integrin receptors from lipid rafts to the caveolae-like structures (Pietiainen et al., 2005). Depletion of cholesterol in lipid rafts inhibits EV1 infection (Chazal and Gerlier, 2003). For envelope virus, Semliki Forest virus (SFV) and Sindbis virus (SIN) require cholesterol and sphingolipids in target membrane lipid rafts for envelope glycoprotein-mediated membrane fusion and entry (Rawat et al., 2003). Many envelope virus receptors are located in lipid rafts or would be relocated into lipid rafts after infection, such as Human T-lymphotropic virus Type I (HTLV-1) receptor glucose transporter 1 (GLUT-1), Ebola virus and Marburg virus folate receptor-α (FRα), Hepatitis B virus complement receptor type 2 (CR2), Human herpesvirus 6 (HHV-6) receptor CD46 (Santoro et al., 1999). An alternative receptor for Human Immunodeficiency virus (HIV-1) envelope glycoprotein on epithelial cells is glycosphingolipid galactosyl-ceramide (GalCer), which also enriches at lipid rafts (Alving et al., 2006, Campbell et al., 2001). Membrane cholesterol is also involved in the first stages of flavivirus infection (Carro and Damonte, 2013, Zhu et al., 2012). Guo et al. (2017) demonstrate that IBV structural proteins, but not non-structural proteins, migrate and integrate into lipid rafts after viral protein translation. The cholesterol enriched microdomains were only involved in IBV attachment, not virus replication or virus particle release. However, the involvement mechanisms of lipid rafts on IBV infection are still unclear. In our study, we checked the involvement of lipid rafts on virus life cycle via disruption of lipid rafts by using chemicals at pre-, during-, or post-infection steps, confirming that lipid rafts were involved in virus entry step. Cholesterol replenishment experiment further confirmed that lipid rafts were necessary for IBV entry. Membrane flotation assay showed the co-fractionation of IBV particles and lipid rafts during absorption step. This was further validated by immunofluoresence experiment, displaying as association of R18-IBV with lipid rafts on the cells surface. Above evidence substantiates that IBV initiates the entry step by attaching to lipid rafts.
Further experiment showed that treatment of chemical inhibitor (NH4Cl) of endosome acidification in various susceptible cells reduced the IBV internalization and protein expression, suggesting the low pH in endosomes is required for IBV infection. To further confirm our observations, we used inhibitor CPZ to interfere with CME in various susceptible cells, or knocked down CHC siRNA to specifically perturb CME in H1299 cells. We found that IBV internalization and protein expression were significantly decreased when CME was interfered. This evidence determines the importance of CME in IBV entry, which is widely hijacked by various envelope viruses. Moreover, the importance of dynamin 1, a GTPase that helps to snap the vesicle from plasma membrane, was determined on IBV entry. Lipid rafts is often considered to be involved in CavME. To explain these contradictory observations, Huh7, a caveolin deficient cell line (Damm et al., 2005, Vela et al., 2007), was used in the cholesterol depletion experiment. When cholesterol was depleted by chemicals in Huh7 cells, similar inhibition effect on IBV entry was observed as those in Vero and H1299 cells. These results confirm that although IBV attachment and entry depend on the intact lipid rafts, CME is the main endocytotic pathway for internalization. In H1299 cells and DF-1 cells, Nystatin also inhibited IBV entry at a certain level, suggesting IBV may hijack CavME as an alternative entry route in these two cell lines.
Our final research question is the virus trafficking route. By using R18-labeled IBV to infect cells, we found that IBV attachment lead to actin rearrangement and the R18-IBV moved along with actin filaments after internalization. This indicates that virus internalization requires the actin de-polymerization or polymerization, to facilitate the virus containing vesicles to move along actin filaments. However, stabilization of the actin filaments by using Jas or inhibition of actin polymerization by using CytoD resulted in increase of IBV entry, which remain as open questions. By using R18-labeled virus and staining the endosomal/lysosomal vesicles with their specific protein markers, we found that after internalization, the incoming IBV particles entered into early endosomes within a few minutes; after that, virus particles were delivered into late endosomes (1 h.p.i.); finally, virus particles reached lysosome (2 h.p.i.), across the entire endocytic network. Rab5 and Rab7 GTPases are the key regulators for cargo transport to early and late endosomes. Thus, ablation of these two Rabs has been widely used to study virus entry. The importance of Rab5 and Rab7 protein on IBV entry were examined by overexpression or RNA interference technology. The results showed that perturbation of early/late endosomes transport by overexpression of Rab5/7-DN or knock down of Rab5/7, significantly reduced IBV internalization and viral protein expression. This confirms that IBV trafficking really depends on both early and late endosomes. Also, by using the R18 and DiOC double labeled virus, we revealed that the membrane fusion happens at 1–2 h.p.i., the time point of virus reaches late endosomes or lysosomes. We speculate that IBV particles were first transported to early endosomes in a Rab5-demepdent manner, and were guided to late endosomes in a Rab7 dependent manner. Also, the Rab7 dependence indicates that membrane fusion requires endosomal maturation, suggesting that low pH environment in late endosome or lysosome may provide an important signal for membrane fusion and IBV gRNA release.
A recent report showed that MHV infection also depends on CME, endosomal maturation, and late endosome-to-lysosome trafficking. The membrane fusion requires the cleavage of MHV S protein by lysosomal proteases. Introduction of furin cleavage site just upstream of the fusion peptide in MHV S protein renders MHV independent of lysosomal proteases and fuses in early endosomes. Analysis of the upstream sequence of fusion peptide of several alpha, beta, and gamma coronaviruses showed that MERS-CoV and IBV-Beaudette contain a minimal furin cleavage site Arg-X-X-Arg just upstream of the fusion peptide. Indeed, MERS-CoV infection only requires furin mediated cleavage but not lysosomal proteases, and does not require trafficking to lysosomes for efficient entry. The sequence immediately upstream of the fusion peptide in S protein might be a key determinant of the intracellular site of fusion, and the cleavage of the fusion proximal position is likely to be a general requirement for coronavirus entry (Burkard et al., 2014). Based on this hypothesis, it predicts IBV-Beaudette is cleaved by furin and fuses in early endosomes. However, in our experiment, we show that IBV-Beaudette particles require trafficking to the low pH environment of late endosomes or lysosomes to achieve membrane fusion. Thus, in addition to furin cleavage (Yamada and Liu, 2009), proteases in late endosomes or lysosomes may be involved in IBV S cleavage. The timing and location study of pesudovirus bearing SARS-CoV S shows that this virus enter into late endosomes and lysosomes after 30 min.p.i., and the membrane fusion peaks at 2 h.p.i.(Mingo et al., 2015). Study on PHEV entry also shows this procine coronavirus moves together with Rab5- or Rab7-positive endosomes before 45 min.p.i., and enter into endolysosomes widely by 1.5 h.p.i. (Li et al., 2017). Therefore, the timing of intracellular trafficking and fusion of IBV-Beaudette is similar to those coronaviruses which passes through late endosome and lysosome.
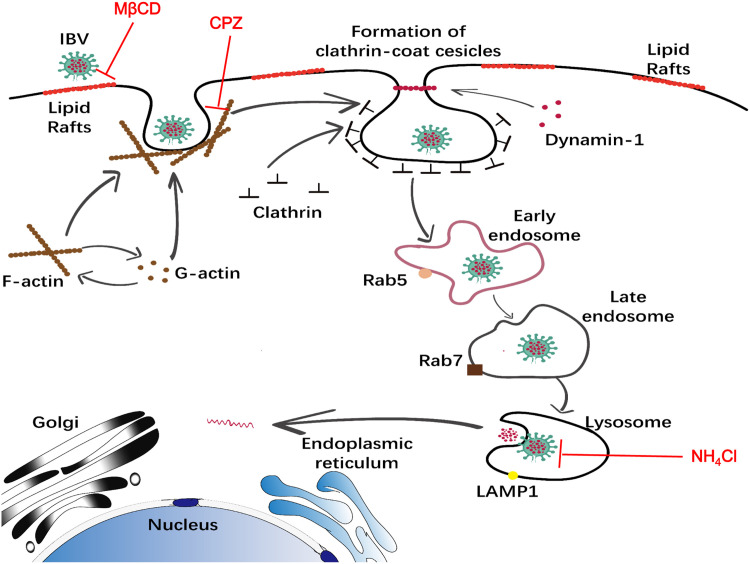
In conclusion, we conduct a systematic study to dissect IBV entry process in susceptible cell lines. The evidence presented here indicates that IBV attaches to lipid rafts, induces actin de-polymerization, internalizes into clathrin coated vesicles via dynamin 1 snapping, transports along early/late endosome, and lysosome, finally fuses with late endosome-lysosome membrane, releases genome in to cytoplasm ( Fig. 11). An understanding of the cellular components involved in IBV invasion into the cell, is likely to open new opportunities to develop novel therapeutic approaches. It is worth noting that the cell adaptive IBV-Beaudette strain was used for study here. Bickerton et al. reports that three amino acids surrounding the Beaudette S2 cleavage site are determinant of Vero cells and human cells tropism. Introduction of Beaudette-associated amino acids into M41 is sufficient to confer tropism for growth in Vero cells, either by facilitating the binding of virus to additional host attachment factors or by inducing membrane fusion as an additional protease cleavage site (Bickerton et al., 2018). It is widely accepted that the protease cleavage site on S protein is a determinant of coronavirus entry and fusion, and the sequence difference of S protein in various IBV strains may determine not only the cell tropsim but also the entry route/fusion site. Thus, it is worth to note that our study may not reflect the situation with field strains of IBV in primary cells and tissues in birds.
Fig. 11.
Model of IBV internalization, trafficking, and fusion. IBV attaches to lipid rafts on the cell surface and is internalized from the plasma membrane via CME, with the help of dynamin 1 and actin de-polymerization. The virus containing clathrin coated vesicles were taken up into Rab5-containing early endosomes, transferred into late endosomes and lysosomes. The low pH environment in late endosomes-lysosomes induces virus-cell membrane fusion. MβCD, CPZ, NH4Cl target to indicated entry steps to block virus entry.
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant No. 31772724), Chinese Special Fund for Agricultural Sciences Research in the Public Interest (Grant No. 2014JB16; Grant No. 2017JB05), China Ministry of Science and Technology (Grant No. 2017YFD0500802), Elite Youth Program of Chinese Academy of Agricultural Sciences (Grant No. 20170260401), and National Natural Science Foundation of China (Grant No. 31530074).
We acknowledge the gifts of IBV Beaudette strain and viral protein antibodies from Prof. Liu Dingxiang’s lab in South China Agricultural University, China.
Acknowledgments
Conflict of interests statement
The authors have no conflict of interests with the content of this article.
Footnotes
Rab proteins as membrane organizers.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2018.12.012
Contributor Information
Chan Ding, Email: shoveldeen@shvri.ac.cn.
Ying Liao, Email: liaoying@shvri.ac.cn.
Appendix A. Supplementary material
Supplementary material
References2
- Aleksandrowicz P., Marzi A., Biedenkopf N., Beimforde N., Becker S., Hoenen T., Feldmann H., Schnittler H.J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 2011;204(Suppl. 3):S957–S967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alving C.R., Beck Z., Karasavva N., Matyas G.R., Rao M. HIV-1, lipid rafts, and antibodies to liposomes: implications for anti-viral-neutralizing antibodies. Mol. Membr. Biol. 2006;23:453–465. doi: 10.1080/09687860600935348. [DOI] [PubMed] [Google Scholar]
- Beer C., Andersen D.S., Rojek A., Pedersen L. Caveola-dependent endocytic entry of amphotropic murine leukemia virus. J. Virol. 2005;79:10776–10787. doi: 10.1128/JVI.79.16.10776-10787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedicto I., Gondar V., Molina-Jimenez F., Garcia-Buey L., Lopez-Cabrera M., Gastaminza P., Majano P.L. Clathrin mediates infectious hepatitis C virus particle egress. J. Virol. 2015;89:4180–4190. doi: 10.1128/JVI.03620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton E., Maier H.J., Stevenson-Leggett P., Armesto M., Britton P. The S2 subunit of infectious bronchitis virus beaudette Is a determinant of cellular tropism. J. Virol. 2018:92. doi: 10.1128/JVI.01044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousarghin L., Touze A., Sizaret P.Y., Coursaget P. Human papillomavirus types 16, 31, and 58 use different endocytosis pathways to enter cells. J. Virol. 2003;77:3846–3850. doi: 10.1128/JVI.77.6.3846-3850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb M.R., Senderowicz A.M., Sausville E.A., Duncan K.L., Korn E.D. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J., Bosch B.J., de Haan C.A. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Postnikova E.N., Bernbaum J.G., Yu S.Q., Mazur S., Deiuliis N.M., Radoshitzky S.R., Lackemeyer M.G., McCluskey A., Robinson P.J., Haucke V., Wahl-Jensen V., Bailey A.L., Lauck M., Friedrich T.C., O'Connor D.H., Goldberg T.L., Jahrling P.B., Kuhn J.H. Simian hemorrhagic fever virus cell entry is dependent on CD163 and uses a clathrin-mediated endocytosis-like pathway. J. Virol. 2015;89:844–856. doi: 10.1128/JVI.02697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S.M., Crowe S.M., Mak J. Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J. Clin. Virol. 2001;22:217–227. doi: 10.1016/s1386-6532(01)00193-7. [DOI] [PubMed] [Google Scholar]
- Carro A.C., Damonte E.B. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res. 2013;174:78–87. doi: 10.1016/j.virusres.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Chazal N., Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.C., McElroy L.J., Chu V., Bauman B.E., Whittaker G.R. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J. Virol. 2006;80:3180–3188. doi: 10.1128/JVI.80.7.3180-3188.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P., Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 2014:6. doi: 10.1101/cshperspect.a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cureton D.K., Massol R.H., Whelan S.P., Kirchhausen T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010;6:e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm E.M., Pelkmans L., Kartenbeck J., Mezzacasa A., Kurzchalia T., Helenius A. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 2005;168:477–488. doi: 10.1083/jcb.200407113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danthi P., Chow M. Cholesterol removal by methyl-beta-cyclodextrin inhibits poliovirus entry. J. Virol. 2004;78:33–41. doi: 10.1128/JVI.78.1.33-41.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries E., Tscherne D.M., Wienholts M.J., Cobos-Jimenez V., Scholte F., Garcia-Sastre A., Rottier P.J., de Haan C.A. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7:e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwaker D., Mishra K.P., Ganju L., Singh S.B. Protein disulfide isomerase mediates dengue virus entry in association with lipid rafts. Viral Immunol. 2015;28:153–160. doi: 10.1089/vim.2014.0095. [DOI] [PubMed] [Google Scholar]
- Eifart P., Ludwig K., Bottcher C., de Haan C.A., Rottier P.J., Korte T., Herrmann A. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 2007;81:10758–10768. doi: 10.1128/JVI.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empig C.J., Goldsmith M.A. Association of the caveola vesicular system with cellular entry by filoviruses. J. Virol. 2002;76:5266–5270. doi: 10.1128/JVI.76.10.5266-5270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Press B., Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.X., Fu W., Winter A.J., Levin J.G., Rein A. Multiple regions of Harvey sarcoma virus RNA can dimerize in vitro. J. Virol. 1995;69:2486–2490. doi: 10.1128/jvi.69.4.2486-2490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivaz M., Abrami L., van der Goot F.G. Landing on lipid rafts. Trends Cell Biol. 1999;9:212–213. doi: 10.1016/s0962-8924(99)01567-6. [DOI] [PubMed] [Google Scholar]
- Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretz M., Jin J., Conibere R., Penning N.A., Al-Taei S., Storm G., Futaki S., Takeuchi T., Nakase I., Jones A.T. Effects of Na+/H+ exchanger inhibitors on subcellular localisation of endocytic organelles and intracellular dynamics of protein transduction domains HIV-TAT peptide and octaarginine. J. Control Release. 2006;116:247–254. doi: 10.1016/j.jconrel.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Gallo R., Provenzano C., Carbone R., Di Fiore P.P., Castellani L., Falcone G., Alema S. Regulation of the tyrosine kinase substrate Eps8 expression by growth factors, v-Src and terminal differentiation. Oncogene. 1997;15:1929–1936. doi: 10.1038/sj.onc.1201344. [DOI] [PubMed] [Google Scholar]
- Gruenberg J. The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell Biol. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- Guo H., Huang M., Yuan Q., Wei Y., Gao Y., Mao L., Gu L., Tan Y.W., Zhong Y., Liu D., Sun S. The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS One. 2017;12:e0170123. doi: 10.1371/journal.pone.0170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine J.W., Schnaitman C.A. A method for the isolation of plasma membrane of animal cells. J. Cell Biol. 1971;48:703–707. doi: 10.1083/jcb.48.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley J.R., Krueger E.W., Oswald B.J., McNiven M.A. Dynamin-mediated internalization of caveolae. J. Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holla P., Ahmad I., Ahmed Z., Jameel S. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic. 2015;16:398–416. doi: 10.1111/tra.12260. [DOI] [PubMed] [Google Scholar]
- Hommelgaard A.M., Roepstorff K., Vilhardt F., Torgersen M.L., Sandvig K., van Deurs B. Caveolae: stable membrane domains with a potential for internalization. Traffic. 2005;6:720–724. doi: 10.1111/j.1600-0854.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- Huang H., Li Y., Sadaoka T., Tang H., Yamamoto T., Yamanishi K., Mori Y. Human herpesvirus 6 envelope cholesterol is required for virus entry. J. Gen. Virol. 2006;87:277–285. doi: 10.1099/vir.0.81551-0. [DOI] [PubMed] [Google Scholar]
- Inal J.M., Jorfi S. Coxsackievirus B transmission and possible new roles for extracellular vesicles. Biochem. Soc. Trans. 2013;41:299–302. doi: 10.1042/BST20120272. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel P.A., Head B.P., Ostrom R.S., Patel H.H., Swaney J.S., Tang C.M., Roth D.M. Caveolae and lipid rafts: G protein-coupled receptor signaling microdomains in cardiac myocytes. Ann. N.Y. Acad. Sci. 2005;1047:166–172. doi: 10.1196/annals.1341.015. [DOI] [PubMed] [Google Scholar]
- Johannsdottir H.K., Mancini R., Kartenbeck J., Amato L., Helenius A. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 2009;83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Toret C.P., Drubin D.G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kalin S., Amstutz B., Gastaldelli M., Wolfrum N., Boucke K., Havenga M., DiGennaro F., Liska N., Hemmi S., Greber U.F. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J. Virol. 2010;84:5336–5350. doi: 10.1128/JVI.02494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen J.H. Clathrin and associated assembly and disassembly proteins. Annu. Rev. Biochem. 1990;59:415–438. doi: 10.1146/annurev.bi.59.070190.002215. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 1999;15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- Kovbasnjuk O., Edidin M., Donowitz M. Role of lipid rafts in Shiga toxin 1 interaction with the apical surface of Caco-2 cells. J. Cell Sci. 2001;114:4025–4031. doi: 10.1242/jcs.114.22.4025. [DOI] [PubMed] [Google Scholar]
- Krzyzaniak M.A., Zumstein M.T., Gerez J.A., Picotti P., Helenius A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 2013;9:e1003309. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao K., Lan Y., Lv X., Hu S., Guan J., Lu H., Zhang J., Shi J., Yang Y., Song D., Gao F., He W. Porcine hemagglutinating encephalomyelitis virus enters neuro-2a cells via clathrin-mediated endocytosis in a Rab5-, cholesterol-, and pH-dependent manner. J. Virol. 2017:91. doi: 10.1128/JVI.01083-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu Y., Liu S., Pang D.W., Xiao G. Clathrin-mediated endocytosis in living host cells visualized through quantum dot labeling of infectious hematopoietic necrosis virus. J. Virol. 2011;85:6252–6262. doi: 10.1128/JVI.00109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Chu V.C., Lee H., Regan A.D., Bauman B.E., Whittaker G.R. Heparan sulfate is a selective attachment factor for the avian coronavirus infectious bronchitis virus beaudette. Avian Dis. 2007;51:45–51. doi: 10.1637/0005-2086(2007)051[0045:HSIASA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Marjomaki V., Pietiainen V., Matilainen H., Upla P., Ivaska J., Nissinen L., Reunanen H., Huttunen P., Hyypia T., Heino J. Internalization of echovirus 1 in caveolae. J. Virol. 2002;76:1856–1865. doi: 10.1128/JVI.76.4.1856-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M., Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Acebes M.A., Gonzalez-Magaldi M., Sandvig K., Sobrino F., Armas-Portela R. Productive entry of type C foot-and-mouth disease virus into susceptible cultured cells requires clathrin and is dependent on the presence of plasma membrane cholesterol. Virology. 2007;369:105–118. doi: 10.1016/j.virol.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Martinez M.G., Cordo S.M., Candurra N.A. Characterization of Junin arenavirus cell entry. J. Gen. Virol. 2007;88:1776–1784. doi: 10.1099/vir.0.82808-0. [DOI] [PubMed] [Google Scholar]
- Matlin K.S., Reggio H., Helenius A., Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- McHenry E.W., Reedman E.J., Sheppard M. The physiological properties of ascorbic acid: an effect upon the weights of guinea-pigs. Biochem. J. 1938;32:1302–1304. doi: 10.1042/bj0321302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigeshi G.R., Hirsch A.J., Streblow D.N., Nikolich-Zugich J., Nelson J.A. West Nile virus entry requires cholesterol-rich membrane microdomains and is independent of alphavbeta3 integrin. J. Virol. 2008;82:5212–5219. doi: 10.1128/JVI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O., Boucke K., Hammer S.V., Keller S., Stidwill R.P., Hemmi S., Greber U.F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J., Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang-Bujnowicz A., Berniak K., Wojarski J., Zeglen S., Baster Z., Rajfur Z., Pyrc K. Entry of human coronavirus NL63 to the cell. J. Virol. 2017 doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingo R.M., Simmons J.A., Shoemaker C.J., Nelson E.A., Schornberg K.L., D'Souza R.S., Casanova J.E., White J.M. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J. Virol. 2015;89:2931–2943. doi: 10.1128/JVI.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire C.E., White J.M., Whitt M.A. A spatio-temporal analysis of matrix protein and nucleocapsid trafficking during vesicular stomatitis virus uncoating. PLoS Pathog. 2010;6:e1000994. doi: 10.1371/journal.ppat.1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash T.C., Buchmeier M.J. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology. 1997;233:1–8. doi: 10.1006/viro.1997.8609. [DOI] [PubMed] [Google Scholar]
- Nomura R., Kiyota A., Suzaki E., Kataoka K., Ohe Y., Miyamoto K., Senda T., Fujimoto T. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 2004;78:8701–8708. doi: 10.1128/JVI.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G.S., Chen Z.L., Yuan M. Jasplakinolide reversibly disrupts actin filaments in suspension-cultured tobacco BY-2 cells. Protoplasma. 2002;219:168–175. doi: 10.1007/s007090200018. [DOI] [PubMed] [Google Scholar]
- Panda D., Rose P.P., Hanna S.L., Gold B., Hopkins K.C., Lyde R.B., Marks M.S., Cherry S. Genome-wide RNAi screen identifies SEC. 61A and VCP as conserved regulators of Sindbis virus entry. Cell Rep. 2013;5:1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res. 2014;191:21–29. doi: 10.1016/j.virusres.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L., Kartenbeck J., Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 2001;3:473–483. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- Piccinotti S., Kirchhausen T., Whelan S.P. Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J. Virol. 2013;87:11637–11647. doi: 10.1128/JVI.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietiainen V.M., Marjomaki V., Heino J., Hyypia T. Viral entry, lipid rafts and caveosomes. Ann. Med. 2005;37:394–403. doi: 10.1080/07853890510011976. [DOI] [PubMed] [Google Scholar]
- Pike L.J. The challenge of lipid rafts. J. Lipid Res. 2009;50(Suppl.) doi: 10.1194/jlr.R800040-JLR200. (S323-328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y., Zhang X. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J. Virol. 2008;82:8112–8123. doi: 10.1128/JVI.00837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80:5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu H., Sharma-Walia N., Veettil M.V., Sadagopan S., Chandran B. Kaposi's sarcoma-associated herpesvirus utilizes an actin polymerization-dependent macropinocytic pathway to enter human dermal microvascular endothelial and human umbilical vein endothelial cells. J. Virol. 2009;83:4895–4911. doi: 10.1128/JVI.02498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L., Simons K. Lipid rafts and membrane dynamics. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- Rawat S.S., Viard M., Gallo S.A., Rein A., Blumenthal R., Puri A. Modulation of entry of enveloped viruses by cholesterol and sphingolipids (Review) Mol. Membr. Biol. 2003;20:243–254. doi: 10.1080/0968768031000104944. [DOI] [PubMed] [Google Scholar]
- Regan A.D., Whittaker G.R. Entry of rhabdoviruses into animal cells. Adv. Exp. Med. Biol. 2013;790:167–177. doi: 10.1007/978-1-4614-7651-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizopoulos Z., Balistreri G., Kilcher S., Martin C.K., Syedbasha M., Helenius A., Mercer J. Vaccinia virus infection requires maturation of macropinosomes. Traffic. 2015;16:814–831. doi: 10.1111/tra.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Ohuchi M., Imai M., Mizuno T., Kawasaki K., Kuroda K., Yamashina S. Dual wavelength imaging allows analysis of membrane fusion of influenza virus inside cells. J. Virol. 2006;80:2013–2018. doi: 10.1128/JVI.80.4.2013-2018.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P., Pollard T.D. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry. 1991;30:1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- Santoro F., Kennedy P.E., Locatelli G., Malnati M.S., Berger E.A., Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- Schelhaas M., Shah B., Holzer M., Blattmann P., Kuhling L., Day P.M., Schiller J.T., Helenius A. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 2012;8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R., Willingham M., Pastan I. Monensin blocks endocytosis of vesicular stomatitis virus. Biochem. Biophys. Res. Commun. 1981;102:992–998. doi: 10.1016/0006-291x(81)91636-3. [DOI] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the edosome for cell entry. J. Virol. 2017:91. doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieczkarski S.B., Whittaker G.R. Dissecting virus entry via endocytosis. J. Gen. Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Smith J.L., Campos S.K., Ozbun M.A. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J. Virol. 2007;81:9922–9931. doi: 10.1128/JVI.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I., Braet F., Shochet N.R., Bubb M.R. New anti-actin drugs in the study of the organization and function of the actin cytoskeleton. Microsc. Res. Tech. 1999;47:18–37. doi: 10.1002/(SICI)1097-0029(19991001)47:1<18::AID-JEMT3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Parton R.G., Steele-Mortimer O., Lutcke A., Gruenberg J., Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13(1287–1296) doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Yau V.K., Briggs B.J., Whittaker G.R. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Suomalainen M. Lipid rafts and assembly of enveloped viruses. Traffic. 2002;3:705–709. doi: 10.1034/j.1600-0854.2002.31002.x. [DOI] [PubMed] [Google Scholar]
- Suomalainen M., Greber U.F. Uncoating of non-enveloped viruses. Curr. Opin. Virol. 2013;3:27–33. doi: 10.1016/j.coviro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Superti F., Seganti L., Ruggeri F.M., Tinari A., Donelli G., Orsi N. Entry pathway of vesicular stomatitis virus into different host cells. J. Gen. Virol. 1987;68(Pt 2):387–399. doi: 10.1099/0022-1317-68-2-387. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Suzuki Y. Virus infection and lipid rafts. Biol. Pharm. Bull. 2006;29:1538–1541. doi: 10.1248/bpb.29.1538. [DOI] [PubMed] [Google Scholar]
- Tan L., Zhang Y., Zhan Y., Yuan Y., Sun Y., Qiu X., Meng C., Song C., Liao Y., Ding C. Newcastle disease virus employs macropinocytosis and Rab5a-dependent intracellular trafficking to infect DF-1 cells. Oncotarget. 2016;7:86117–86133. doi: 10.18632/oncotarget.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp E.B., Gallagher T.M. Requirements for CEACAMs and cholesterol during murine coronavirus cell entry. J. Virol. 2004;78:2682–2692. doi: 10.1128/JVI.78.6.2682-2692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio S., Heino S., Mansson J.E., Fredman P., Kuismanen E., Vaarala O., Ikonen E. Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep. 2002;3:95–100. doi: 10.1093/embo-reports/kvf010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hamme E., Dewerchin H.L., Cornelissen E., Verhasselt B., Nauwynck H.J. Clathrin- and caveolae-independent entry of feline infectious peritonitis virus in monocytes depends on dynamin. J. Gen. Virol. 2008;89:2147–2156. doi: 10.1099/vir.0.2008/001602-0. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen M.R., Golovina E.A., Dijksterhuis J. The polyene antimycotics nystatin and filipin disrupt the plasma membrane, whereas natamycin inhibits endocytosis in germinating conidia of Penicillium discolor. J. Appl. Microbiol. 2009;106:1908–1918. doi: 10.1111/j.1365-2672.2009.04165.x. [DOI] [PubMed] [Google Scholar]
- Vela E.M., Zhang L., Colpitts T.M., Davey R.A., Aronson J.F. Arenavirus entry occurs through a cholesterol-dependent, non-caveolar, clathrin-mediated endocytic mechanism. Virology. 2007;369:1–11. doi: 10.1016/j.virol.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viard M., Parolini I., Sargiacomo M., Fecchi K., Ramoni C., Ablan S., Ruscetti F.W., Wang J.M., Blumenthal R. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J. Virol. 2002;76:11584–11595. doi: 10.1128/JVI.76.22.11584-11595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt C., Eickmann M., Diederich S., Moll M., Maisner A. Endocytosis of the Nipah virus glycoproteins. J. Virol. 2005;79:3865–3872. doi: 10.1128/JVI.79.6.3865-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheit A., Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C., Schwegmann-Wessels C., Cavanagh D., Neumann U., Herrler G. Sialic acid is a receptor determinant for infection of cells by avian Infectious bronchitis virus. J. Gen. Virol. 2006;87:1209–1216. doi: 10.1099/vir.0.81651-0. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Helenius A. Virus entry at a glance. J. Cell Sci. 2013;126:1289–1295. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- Zhang Y.N., Liu Y.Y., Xiao F.C., Liu C.C., Liang X.D., Chen J., Zhou J., Baloch A.S., Kan L., Zhou B., Qiu H.J. Rab5, Rab7, and Rab11 are required for caveola-dependent endocytosis of classical swine fever virus in porcine alveolar macrophages. J. Virol. 2018:92. doi: 10.1128/JVI.00797-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.Z., Xu Q.Q., Wu D.G., Ren H., Zhao P., Lao W.G., Wang Y., Tao Q.Y., Qian X.J., Wei Y.H., Cao M.M., Qi Z.T. Japanese encephalitis virus enters rat neuroblastoma cells via a pH-dependent, dynamin and caveola-mediated endocytosis pathway. J. Virol. 2012;86:13407–13422. doi: 10.1128/JVI.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material