Graphical abstract
Keywords: SARS CoV, SARS 3CL protease, Serine derivative, Docking simulation, Cathepsin B, Cytotoxicity
Abstract
Synthesis of serine derivatives having the essential functional groups for the inhibitor of SARS 3CL protease and evaluation of their inhibitory activities using SARS 3CL R188I mutant protease are described. The lead compounds, functionalized serine derivatives, were designed based on the tetrapeptide aldehyde and Bai’s cinnamoly inhibitor, and additionally performed with simulation on GOLD softwear. Structure activity relationship studies of the candidate compounds were given reasonable inhibitors ent-3 and ent-7k against SARS 3CL R188I mutant protease. These inhibitors showed protease selectivity and no cytotoxicity.
1. Introduction
Severe acute respiratory syndrome (SARS) is a contagious respiratory disease to humans that is caused by the SARS coronavirus (SARS-CoV). In 2003, an atypical pneumonia due to SARS-CoV lead to progressive respiratory failure in over 8,000 individuals and 800 deaths within a few months.1, 2, 3 Thereafter, the SARS epidemic was successfully controlled, but potential reemergence of pandemic SARS-CoV continues to be a risk, and new strains of SARS or a SARS-like virus could potentially be more infectious than the strains that led to the 2003 outbreak. Two human coronaviruses, NJ63 and HKU1, have been identified in patients around the world since 2003.4, 5, 6 A more recently identified human coronavirus, Middle East Respiratory Syndrome coronavirus (MERS-CoV), has infected 114 people since April 2012 with a nearly 50% mortality rate, and this number keeps rising daily.7, 8 Two Asian bat coronaviruses, BtCoV-HKU4 and BtCoV-HKU5, have been shown to be the closest relations to MERS-CoV.9 The possibility of a future SARS-like pandemic remains, and no vaccines or antiviral agents have yet been developed to prevent or treat SARS-like infections.10
SARS-CoV is a novel human coronavirus featuring the largest positive-stranded RNA genome known to date (27 kb for pp1a and 31 kb for pp1b). The key enzyme in the processing of polyproteins translated by the viral RNA genome of SARS-CoV is a 33 kDa protease called 3C-like protease (3CL protease). SARS 3CL protease is a cysteine protease containing a Cys-His catalytic dyad. It cleaves precursor polyproteins at as many as 11 conserved sites involving a conserved Gln at the P1 position and a small amino acid (Ser, Ala, or Gly) at the P′1 position. Due to its functional importance in the viral life cycle, SARS 3CL protease is considered to be an attractive target for drug design to treat SARS.
A variety of inhibitors against the SARS 3CL protease have been reported in the literature for the past decade, including substrate-based peptidemimetic,11 fravonoid derivatives,12 tannin derivatives,13 ML188,14 isatin derivatives,15 and decaisoquinolin,16 etc. However, no effective therapeutic drug or vaccine has been developed to date although many candidate anti-SARS CoV agents have been identified.
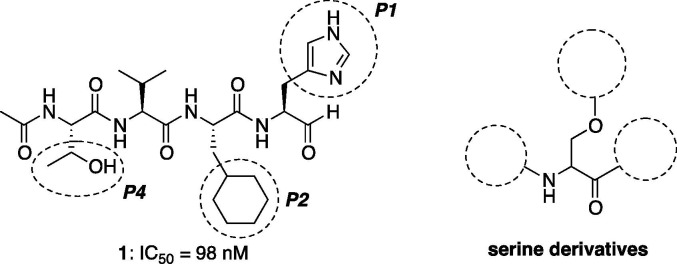
In the previous study, we reported that design and synthesis of the peptide aldehyde inhibitor Ac-Thr-Val-Cha-His-H (1) showed high inhibitory activity with an IC50 value of 98 nM toward 3CL R188I mutant protease as a substrate mimetic concept.17, 18 The inhibitor (1) was optimized by the screening of P1, P2 and P4 site residues of the previous reported peptide aldehyde inhibitor Ac-Ser-Ala-Val-Leu-Gln(Me)2-H, except at the P3 site where the side-chain directed outside and made no interactions with the protease guided by the X-ray crystal structure of the lead compound that bound the R188I mutant SARS 3CL protease. The interactions of inhibitor (1) at the P1 and P2 sites with the protease seemed very effective. Additionally, a new synthetic method using acetal to aldehyde conversion via thioacetal formation was described to afford the C-terminal peptide aldehyde.19, 20
Side chain structures at the P1, P2 and P4 sites and C-terminus aldehyde as the thiol capture of tetrapeptide inhibitor (1) were thought to have a critical role in its potent inhibitory activity. In contrast, there is generally enzymatic digestion of peptide chains and α-proton racemization and/or low specificity of aldehyde functionality. To develop non-peptidyl small molecular inhibitors of the SARS 3CL protease focusing on the P1, P2 and P4 site interactions, we aimed to design and synthesize serine derivatives having the essential functional groups and evaluated their inhibitory activity in the present study. Serine, a commercially available proteinogenic amino acid, has three variant reaction sites; alcohol, amine and carboxylic acid, which can be orthogonally connected to various functional groups (Fig. 1 ).
Figure 1.
Ac-Thr-Val-Cha-His-H (1) and concept for serine derivative.
2. Results and discussion
2.1. Design
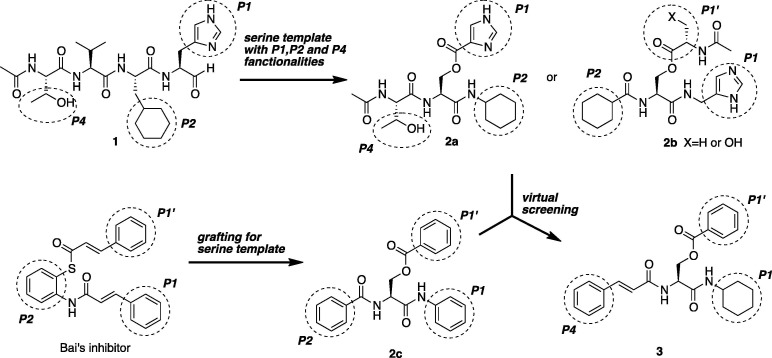
To determine whether a series of serine derivatives could adopt an energetically favorable conformation mimicking a tetrapeptide inhibitor (1), we performed a variety of molecular mechanics calculations with SPARTAN from Wavefunction and docking simulations of protein interactions by GOLD from CCDC. Firstly, imidazole, cyclohexyl and hydroxy groups, which were optimized functionalities with potent biological activities in the previous literature, were connected with l-serine to design the serine derivative (2a,b) as shown in Scheme 1 . However, it did not give good coverage of the substrate-recognition pocket of 3CL protease (PDB code 3AW1), that is, the interactions with optimized motifs and the corresponding pockets were different by binding mode, contrary to expectations. The cyclohexyl group of serine derivative (2) occupied the S1 pocket of the 3CL protease imperatively. Thus, the imidazole and hydroxy groups of serine derivative (2) expected for the interactions with the S1 and S4 pockets were not effective and consequently the numbers of interaction atom pairs between the inhibitor (2) and SARS-CoV 3CL protease were diminished. In contrast, Bai21 reported that the cinnamoyl derivatives inhibited SARS-CoV 3CL protease. In this case, the Bai’s inhibitor locates deep insides of the S′1, S1 and S2 pockets with appropriate cinnamoyl functionalities. This result was approximately identified with the Bai’s simulation employed by the AutoDock 3.0 estimated free energy of binding for the docking of the Bai’s inhibitor to the SARS 3CL protease (PDB code 1UJ1). Therefore, we examined molecular docking of the serine derivatives contained together with benzoyl and aniline moieties to give 2c, which was an attractive ligand for the 3CL protease. Furthermore, we investigated a reasonable structure for the inhibitor and mode hybrid compounds with Bai’s and our functionalities on the serine derivative (3). As a result, we concluded that reasonable structures are N-cinnamoyl derivatives with benzoate for the side chain on virtual screening by GOLD. Compound 3 has the following characteristics: (a) since the substrates by nature have involved small amino acids (Ser, Ala or Gly) at the P′1 position, it may be preferable to adopt the aromatic rings. (b) The proper placement of the cyclohexyl ring may be the result of development of a serine type inhibitor with functionality of the P1 position, as well as stability of interaction of other positions. (c) The S4 pocket is rather hydrophobic in nature and therefore the corresponding side chains, namely aromatic, hydrocarbon or neutral functionalities, make good contact with target regions. (d) Additionally, the region adjacent to the ester bond of 3 is catalytic thiol functionality (Cys145) of the enzyme (Scheme 1).
Scheme 1.
Virtual screening of the serine derivatives for SARS-CoV 3CL protease (PDB code 3AW1) on GOLD softwear.
2.2. Chemistry
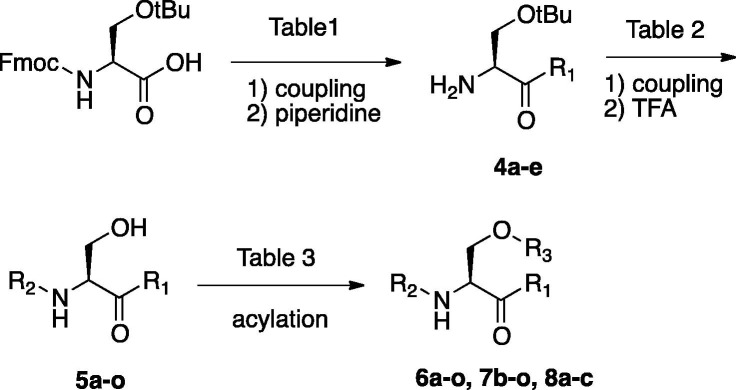
In view of the promising computational evaluation of the serine derivative (3) in terms of tetrapeptide mimetics, we plan to synthesize the target molecules. To evaluate inhibitory activity of the serine derivative (3) against SARS-CoV 3CL protease, a structure activity relationship study using the serine derivatives was attempted. As depicted in Scheme 2 , the serine derivatives attached with three functionalities were prepared. Each coupling partner to introduce the functionalities is shown in Table 1, Table 2, Table 3 . Coupling of Fmoc-l-Ser(tBu)-OH and amines as a P1 moiety with WSC/HOBt followed by the deprotection of Fmoc group by 20% piperidine/DMF gave the serine amide derivative (4) at acceptable chemical yields. The P4 moiety was introduced by coupling with carboxylic acid in the presence of coupling reagents or acylation using carboxylic anhydride or acyl chloride to afford diamide derivatives. Subsequently, treatment of TFA for the deprotection of the t-butyl group gave alcohol (5) at moderate yields. Finally, the esterification of a hydroxy group of 5 with a variety of acyl reagents was performed to give novel serine derivatives (6, 7 and 8) for evaluation of the small molecular inhibitors against SARS 3CL protease. Purification of all crude compounds was performed by silica gel column chromatography. The chemical structures for synthetic compounds were mainly determined by 1H and 13C NMRs, IR and mass spectra (Scheme 2).
Scheme 2.
Synthetic outline for the preparation of serine derivatives (6a–o and 7b–o).
Table 1.
Coupling of the P2 position and Fmoc-deprotection of Fmoc-Ser(tBu)-OH
| Entry | Aminea | Product |
|---|---|---|
| 1 | Cyclohexylamine | 4a (97%) |
| 2 | Piperidine | 4b (26%) |
| 3 | Morpholine | 4c (91%) |
| 4 | Benzylamine | 4d (19%) |
| 5 | Cyclohexylmethylamine | 4e (48%) |
Reagent and conditions: (1) amine, WSC, HOBt, CH2Cl2, (2) 20% piperidine/CH2Cl2.
Table 2.
Coupling of P4 position and deprotection of t-butyl group
| Entry | Subst | Conditionsa | Product | Entry | Subst | Conditionsa | Product |
|---|---|---|---|---|---|---|---|
| 1 | 4a |  |
5a (38%) | 9 | 4a |  |
5i (69%) |
| 2 | 4a | 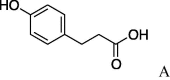 |
5b (79%) | 10 | 4a | 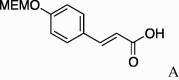 |
5j (55%) |
| 3 | 4a | 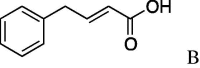 |
5c (37%) | 11 | 4a | 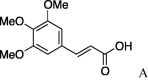 |
5k (29%) |
| 4 | 4a | 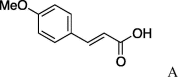 |
5d (43%) | 12 | 4b | 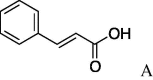 |
5l (26%) |
| 5 | 4a | 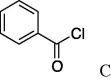 |
5e (80%) | 13 | 4c | 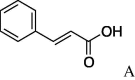 |
5m(91%) |
| 6 | 4a | 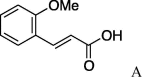 |
5f (7%) | 14 | 4d | 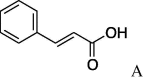 |
5n (19%) |
| 7 | 4a | 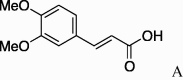 |
5g (23%) | 15 | 4e | 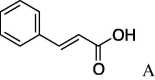 |
5o (48%) |
| 8 | 4a | 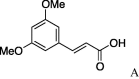 |
5h (29%) |
Reagents and conditions: (1) conditions A, B or C, DIPEA, CH2Cl2, rt, (2) 50% TFA/CH2Cl2, rt. (A) HATU, HOAt, (B) DMT-MM, (C) Et3N, DMAP.
Table 3.
Coupling of the P′1 position for the inhibitors
| Entry | Subst | Conditionsa | Product | Entry | Subst | Conditionsa | Product |
|---|---|---|---|---|---|---|---|
| 1 | 5a | 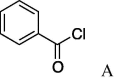 |
3 (71%) | 16 | 5a | 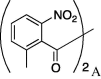 |
6o (36%) |
| 2 | 5a | 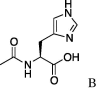 |
6a (3%) | 17 | 5b | 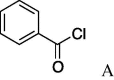 |
7b (7%) |
| 3 | 5a | 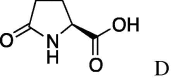 |
6b (25%) | 18 | 5c | 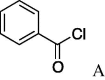 |
7c (17%) |
| 4 | 5a | 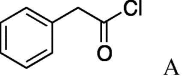 |
6c (78%) | 19 | 5d |  |
7d (22%) |
| 5 | 5a |  |
6d (43%) | 20 | 5e |  |
7e (43%) |
| 6 | 5a |  |
6e (47%) | 21 | 5f | 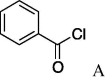 |
7f (57%) |
| 7 | 5a | 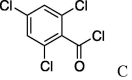 |
6f (27%) | 22 | 5g | 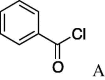 |
7g (37%) |
| 8 | 5a | 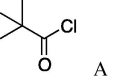 |
6g (70%) | 23 | 5h | 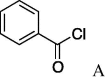 |
7h (19%) |
| 9 | 5a | Ac2O A | 6h (64%) | 24 | 5i | 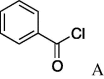 |
7i (57%) |
| 10 | 5a | 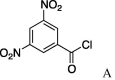 |
6i (26%) | 25 | 5j | 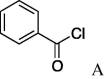 |
7j (52%) |
| 11 | 5a | 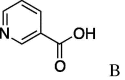 |
6j (47%) | 26 | 5k | 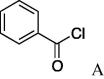 |
7k (81%) |
| 12 | 5a | 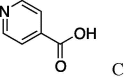 |
6k (95%) | 27 | 5l | 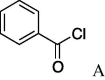 |
7l (44%) |
| 13 | 5a | 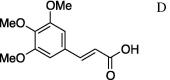 |
6l (4%) | 28 | 5m | 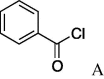 |
7m(75%) |
| 14 | 5a | 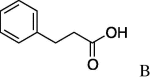 |
6m(13%) | 29 | 5n | 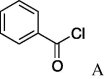 |
7n (44%) |
| 15 | 5a |  |
6n (26%) | 30 | 5o |  |
7o (57%) |
(A) carboxylic acid chloride or acetic anhydride, DMAP, Et3N, CH2Cl, (B) carboxylic acid, NMBA, DMAP, Et3N, CH2Cl2, (C) carboxylic acid, 2,4,6-trichlorobenzoic acid chloride, DMAP, DIPEA, CH2Cl2, (D) carboxylic acid, (COCl)2, DMF.
As depicted in Table 1, five cyclic functionalities were attached to the carboxylic acid of the serine template. As mentioned above, this was to investigate the importance of cyclohexane ring defined as the P1 position. Treatment of Fmoc-l-Ser(tBu)-OH and cyclohexylamine with WSC/HOBt and continuous Fmoc-deprotection afforded 4a in 97% yield (entry 1). Under the above-mentioned conditions, Fmoc-l-Ser(tBu)-OH was subjected to a coupling reaction with piperidine, morpholine, benzylamine or cyclohexylmethylamine to give 4b–e. Despite the poor yields in entries 2 and 4, it was available for the evaluation of inhibitory activities (Table 1).
To optimize P4 functionality, a variety of carboxylic acids, especially α,β-unsaturated carboxylic acids for 4a, were introduced. Coupling of 4a and cinnamic acid with HATU22/HOAt23/DIPEA in CH2Cl2 (condition A) followed by treatment with TFA for deprotection of the t-butyl group afforded 5a in 38% yield (entry 1). Treatment of 4a with 3-(p-hydroxyphenyl)propanoic acid in the presence of HATU/HOAt/DIPEA in CH2Cl2 also gave 5b in 79% yield after removal of t-butyl group (entry 2). Since 5c was not isolated by the condition of HATU/HOAt, we found that DMT-MM (condition B)24 as a coupling reagent was moderate to give 5c in 37% yield over 2 steps (entry 3). In an attempt to improve the chemical yields for the coupling reaction of cinnamic acid derivative and amines, we knew that HATU or DMT-MM as the coupling reagents were useful in preliminary studies. For this reason, a series of cinnamoyl derivatives (5d, 5f–o) were also prepared under identical conditions at moderate yields (entries 6–15). In addition, acylation of 4a with benzoyl chloride (condition C) was successfully performed to yield 5e without any problems (entry 5) (Table 2).
For the structure–activity relationship study of P′1 functionality of serine derivatives, we prepared the serine derivatives (3), (6a–o) and (7b–o) as depicted in Table 3. Acylation of the substrates (5a–o) was employed the Shiina25 and Yamaguchi26 conditions for the coupling of carboxylic acids or acyl chloride and acetic anhydride reagents. Using commercially available acyl chlorides and Ac2O as the acyl reagent gave the corresponding esters (3), (6c), (6d), (6g), (6h), (6i) and (6o) at moderate yields (entries 1, 4, 5, 8–10, 16). Esterification of 5a with N-acetyl-l-histidine by the Shiina reagent NMBA/DMAP afforded the His-compound (6a) at an extremely low yield (entry 2). In this case, 2-nitro-6-methylbenzote as a by-product was given at a moderate yield and it was difficult to improve the chemical yield even after attempting several conditions. Preparation of a pGlu-compound (6b), isonicotine (6e) and nicotine (6j) also resulted in similar situations (entries 3, 6 and 11). The Yamaguchi protocol was performed to give 6f and 6k (entries 7 and 12) and also acyl chlorides prepared by 3,4,5-trimethoxybenzolic acid, 3-phenylpropanoic acid or cinnamic acid with oxalyl chloride in the presence of DMF in situ reacted with 5a to afford 6l, 6m and 6n at low yields (entries 13–15). On the other hand, the treatment of a variety of alcohols (5b–o) with BzCl/Et3N/DMAP in CH2Cl2 converted to benzoyl esters (7b–o). Chemical yields of benzoyl esters (7b–o) were variable by used substrates (entries 17–30) (Table 3).
2.3. Biological assay
The inhibition of SARS 3CL R188I mutant protease was determined by the previous procedure using a synthetic decapeptide with the S01 cleavage sequence as a substrate. Synthetic l-serine derivatives from several structural subclasses were investigated; 5a, cinnamoyl-l-serine derivatives (6a–o) with modification at the P′1 position (Table 4 ), benzoyl-l-serine derivatives (7b–k) with modification at the P4 position (Table 5 ).
Table 4.
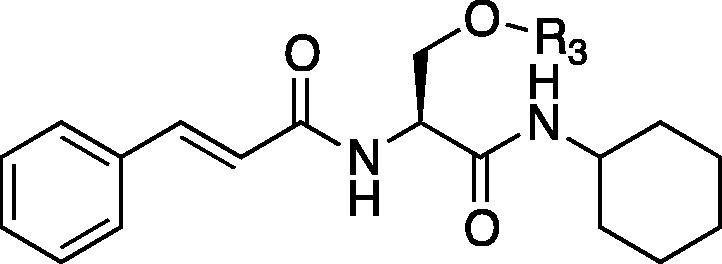
| Entry | Compd | R3 | IC50a | C log Pb | Entry | Compd | R3 | IC50a | C log Pb |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5a | H | 1500 | 2.233 | 10 | 6i | 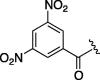 |
250 | 4.312 |
| 2 | 6a | 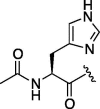 |
>3200 | 1.485 | 11 | 6j | 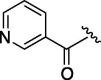 |
180 | 3.382 |
| 3 | 6b | 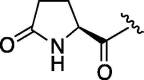 |
>1600 | 2.116 | 12 | 6k | 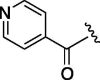 |
175 | 3.482 |
| 4 | 6c | 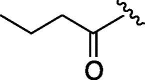 |
650 | 4.187 | 13 | 6l | 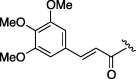 |
170 | 4.584 |
| 5 | 6d | 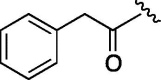 |
650 | 4.748 | 14 | 3 | 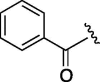 |
125 | 4.826 |
| 6 | 6e | 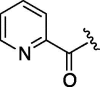 |
560 | 3.482 | 15 | 6m | 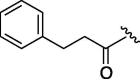 |
120 | 5.226 |
| 7 | 6f | 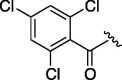 |
550 | 6.085 | 16 | 6n | 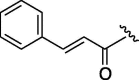 |
85 | 5.284 |
| 8 | 6g | 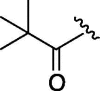 |
450 | 4.367 | 17 | 6o | 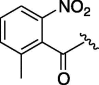 |
65 | 4.878 |
| 9 | 6h | 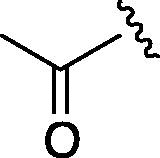 |
400 | 3.129 |
μM.
C log P was calculated by ChemBio3D Ultra 12.0 (PerkinElmer).
Table 5.
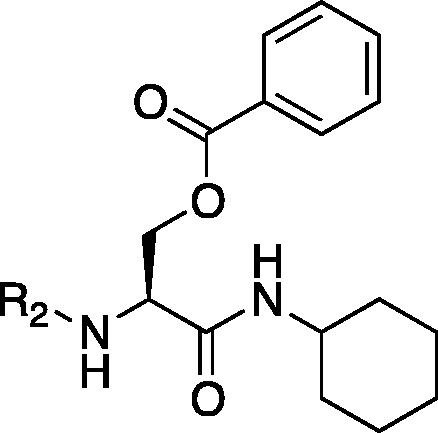
| Entry | Compd | R2 | IC50a | C log Pb | Entry | Compd | R2 | IC50a | C log Pb |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 7b | 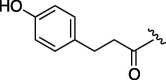 |
>3200 | 2.955 | 6 | 7g | 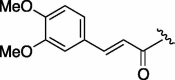 |
154 | 4.484 |
| 2 | 7c | 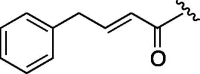 |
500 | 4.673 | 7 | 7h | 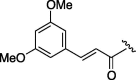 |
100 | 4.484 |
| 3 | 7d | 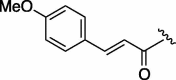 |
240 | 4.745 | 8 | 7i | 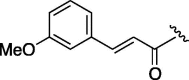 |
98 | 4.745 |
| 4 | 7e | 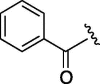 |
220 | 3.968 | 9 | 7j | 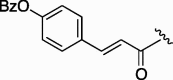 |
95 | 6.304 |
| 5 | 7f | 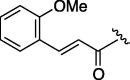 |
155 | 4.745 | 10 | 7k | 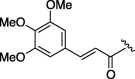 |
74 | 4.126 |
μM.
C log P was calculated by ChemBio3D Ultra 12.0 (PerkinElmer).
As depicted in Table 4, inhibitory potency of the synthesized cinnamoyl-l-serine derivatives (6a–o) with modification at the P′1 position was evaluated as IC50 values. The simulated compound (3) as a control exhibited an IC50 value of 125 μM (entry 14). Since the IC50 value of a previous reported peptide aldehyde (1) was reported to be 98 nM, imidazole of histidine or γ-lactam of pyroglutamic acid were important functionalities. However 6a and 6b that had been coupled with 5a and His or pGlu showed low inhibitory activities as expected from the docking simulations (entries 2 and 3). For optimization of the P′1 functional group of 6, different acyl units; alkyl, phenyl and pyridyl groups, were examined. Aliphatic groups-containing compounds (6c), (6g) and (6h) showed lower inhibitory activities than 3 (entries 4, 8 and 9). Pyridyl derivatives (6e), (6j) and (6k) were also similar (entries 6, 11 and 12). Aromatic rings with nitro, chloride and methoxy groups (6f), (6i) and (6l) gave moderate results (entries 7, 10 and 13). Phenylacetate (6d) also showed lower inhibitory activity than 3 (entry 5). In contrast, conversion of the phenylpropionate or cinnnamoyl groups at a P′1 site would be expected to have a dramatic effect on inhibitory activity. Especially, the cinnamoyl compound (6n) showed potent inhibitory activity at an IC50 value of 85 μM (entry 16). In addition, a 2-methyl-6-nitrophenyl derivative (6o) showed good potential as a 3CL protease inhibitor (entry 17). The IC50 value of saturated compound (6m) was decreased in comparison with those of the fixed unsaturated derivative (6n) (entry 15). These results suggest that serine type derivatives containing optimized functionalities would be useful as 3CL protease inhibitors. As the correlations between IC50 and C log P, we could assume that 3, 6m, 6n and 6o with potent inhibitory activities tend relatively to have high lipophilicity with the C log P values of 4.82–5.28 (Table 4).
To achieve further optimization using a serine template, a structure activity relationship study of the P4 functionality (R2) of a lead compound (3) was conducted. The aliphatic pivaloyl-containing compound (7b) substituted the cinnamoyl group of 3 showed no inhibitory activity and introduction of a phenyl-2-propenate (7c) and benzoyl group (7e) did not make a significant contribution (entries 1, 2 and 4). Next, modified cinnamoyl groups were designed because the moiety of the P4 site was replaced with the methoxy connecting cinnamoyl group to permit the moiety to fit into the S4 pocket more tightly than 3. Although each 2- or 3-methoxy functionalities were not effective (compounds 7d, 7f, entries 3 and 5), the inhibitors containing 3- and/or 4-methoxy groups on a phenyl ring at the P4 site exhibited markedly increased inhibitory activity (entries 6, 7 and 8). The IC50 value of 7k was 74 μM and so the results suggested that a planar aromatic ring and its hydrophobic functionality were essential factors to produce a reasonable 3CL protease inhibitor (entry 10). The correlations between IC50 and C log P are difficult to explain because these compounds are close in molecular formula (Table 5).
To investigate the importance of the cyclohexyl group of 3 at the P1 site, we evaluated four derivatives combined with different amines; piperidine for 7l, morpholine for 7m, benzylamine for 7n and cyclohexylmethylamine for 7o. As a result, 7l and 7m showed no inhibitory activities. These compounds are shorter in terms of one carbon length of the P1 site than 3 and therefore the P1 position of the inhibitor showed decreased flexibility as expected and hardly fit the S1 pocket. On the other hand, aromatic benzyl amine (compound 7n) has a planer structure, which led to moderate inhibitory activity. Attempts to use cyclohexylmethylamine for 7o was acceptable to show IC50 = 180 μM. It is suggested that the cyclohexyl structure at a P1 site strictly interacts with the hydrophobic S1 pocket. As mentioned above, we optimized the functionalities at the P′1, P1 and P4 sites of an l-serine template based on reference compound 3, which was guided by virtual screening on GOLD. P′1 and P4 sites are preferred for their similar characteristics of aromatic rings, an sp2 planer structure and hydrophobicity.
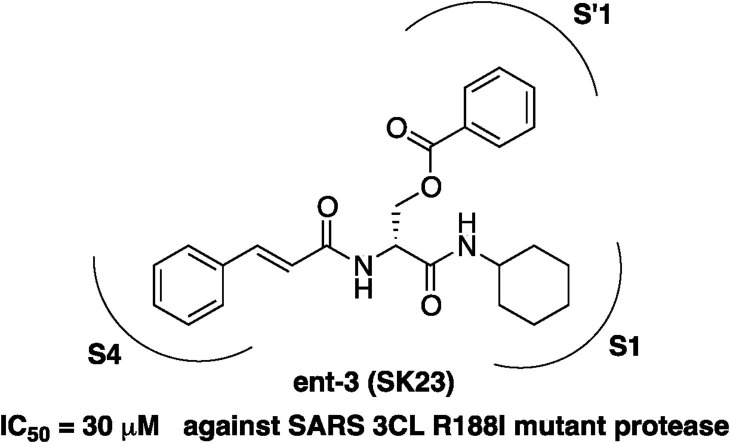
Next, the inhibitory activities of the selected serine derivatives with the d-form were evaluated based on the IC50 values. The IC50 values and tPSA of selected inhibitors are summarized in Table 6 . Preparation of d-serine derivatives was performed by synthetic protocols of the corresponding l-serine compounds without any problems. Fmoc-d-Ser(tBu)-OH as a starting material was selected and a 5 step sequence was performed as shown in Scheme 2. Interestingly, there were no extreme variants for IC50 values between the l- and d-forms except for 6n (entry 6). Both enantiomers of 5a, 6a and 7l showed no inhibitory activities (entries 1, 2 and 3). The inhibitory activity of the l-forms of 6h, 7o, 6j, 7n, 6k and 6o showed a tendency to be more potent than those of the d-forms (entries 4, 5, 7–10). In contrast, 7i, 7h, 7j, 7k and 3, which exhibited potent inhibitory activities against the 3CL protease, tended to have d-forms with more potent activities than l-forms (entries 11–15). We guessed the enantiomers of 7i, 7h, 7j, 7k and 3 fitted the active site of 3CL protease via different binding modes, that is, the P′1 site functionalities interact with the S4 pocket and consequently P4 site groups located in the S′1 pocket. These serine derivatives have high flexibility and therefore two hydrophobic spaces of the 3CL protease are recognized in a rigorous manner. The enantiomer of 3 (ent-3) exhibited the most potent inhibitory activity (IC50 = 30 μM) in the evaluated compounds. Though there are no correlation between the IC50 value and tPSA in Table 6, these tPSA values were acceptable toward the protease inhibitor for the target of catalytic domain (Table 6).
Table 6.
Inhibitory activities of d-serine derivatives
| Entry | Compd | IC50a | tPSAb | Entry | Compd | IC50a | tPSAb |
|---|---|---|---|---|---|---|---|
| 1 | ent-5a | >1600 | 78.43 | 9 | ent-6k | 210 | 96.86 |
| 2 | ent-7l | >1600 | 75.71 | 10 | ent-6o | 100 | 136.3 |
| 3 | ent-6a | 1600 | 138.0 | 11 | ent-7i | 85 | 93.73 |
| 4 | ent-6h | 550 | 84.50 | 12 | ent-7h | 80 | 103.0 |
| 5 | ent-7o | 430 | 84.50 | 13 | ent-7j | 68 | 110.8 |
| 6 | ent-6n | 340 | 84.50 | 14 | ent-7k | 65 | 112.2 |
| 7 | ent-6j | 240 | 96.86 | 15 | ent-3 | 30 | 84.50 |
| 8 | ent-7n | 225 | 84.50 |
μM.
tPSA was calculated by ChemBio3D Ultra 12.0 (PerkinElmer).
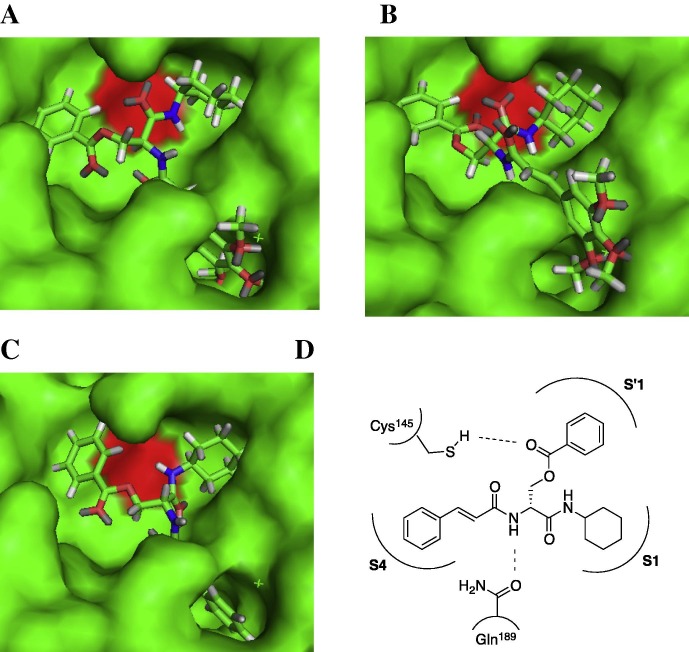
The binding mode of 7k, ent-7k and ent-3 to SARS 3CL protease was predicted by docking simulations using GOLD from CCDC. These results suggest that reasonable inhibitors assessed by docking simulation can be designed and lead effectively to acceptable structures. Interestingly, the interactions between the enzyme and both enantiomers of serine derivatives were similar. In other words, the functionalities at the P′1, P1 and P4 sites of the d-serine template were located on the S′1, S1 and S4 pockets, respectively. This suggests the flexibility of backbones and recognition of the functionalities for serine type inhibitors. Therefore, the enantiomers of the designed compounds were also significant candidates as SARS 3CL protease inhibitors. Moreover, we may consider that d-serine plays a critical role in digestion by proteases (Fig. 2 ).
Figure 2.
Docking simulation of selected inhibitors bound to SARS 3CL protease (PDB code 3AW1) using GOLD from CCDC. Molecular graphic image shown using PyMOL from Schrödinger; oxygen (red) and nitrogen (blue) of inhibitors; Cys145 (red) of SARS 3CL protease. (A) Surface mode with 7k, (B) with ent-7k, (C) with ent-3, (D) model of interaction with ent-3.
It is important to have enzyme selectivity for the development of protease inhibitors. We investigated inhibition of cathepsin B,27 which is a mammalian cysteine protease involved in numerous pathological processes. The cysteine protease inhibitor E-64 was hardly inhibited against SARS 3CL protease in our previous report. E-64, miraziridine A28 and tokaramide A29 were reasonable inhibitors to bind the active site of cathepsin B. For this reason, the inhibitors of SARS 3CL protease are thought to show specificity against endogenous protease. Therefore, we attempted to evaluate the inhibitory activity of the serine derivatives against cathepsin B. The inhibitory activity toward cathepsin B was determined with an assay using a Z-Arg-Arg-MCA substrate developed by Hiwasa et al.30, 31 The inhibition potency of serine derivatives was screened for cathepsin B inhibition at 1.0 mM concentration and subsequently, the IC50 values were determined only for the selected compounds. The reasonable inhibitors, 3, 7i, 7j, 7h and 7k and its enantiomers, for SARS 3CL R188I mutant protease and 6m, 7f and 7g, which had good potential as cathepsin B inhibitors, are shown in Table 7. As a result, the serine derivatives that exhibited potent inhibitory activity against SARS 3CL protease showed relatively less potent inhibitory effects against cathepsin B activity and had high protease selectivity (entries 1–10). On the other hand, three compounds were extracted by the screening for a cathepsin B inhibitor (entries 11–13). These compounds were useless for SARS 3CL protease inhibition (Table 7 ).
Table 7.
Inhibitory activities against cathpsin B
| Entry | Compd | Inhibitiona (%) | Entry | Compd | Inhibitiona (%) |
|---|---|---|---|---|---|
| 1 | 3 | 34 | 8 | ent-7h | 22 |
| 2 | ent-3 | NIb | 9 | 7k | 21 |
| 3 | 7i | 36 | 10 | ent-7k | 70 |
| 4 | ent-7i | 44 | 11 | 6m | 82 |
| 5 | 7j | 35 | 12 | 7f | 67 |
| 6 | ent-7j | 19 | 13 | 7g | 88 |
| 7 | 7h | NIb |
Inhibitory activity was measured by 1 mM.
NI: no inhibition.
The inhibitory activities of selected serine derivatives, 6m, 7f and 7g, were evaluated using the corresponding IC50 values. The IC50 value of synthetic 7f was 170 μM. Comparing IC50 values against SARS 3CL protease and cathepsin B, it was strongly suggested that the inhibitory activity is attributable mainly to the ester site of 7f.32 The results are consistent with the structural data on 7f in a complex with cathepsin B, which suggests that the P1 site of 7f can easily adopt a conformation that is similar to the binding mode of E-64d. Although the P2 site had a rather weak effect compared with the P1 site, the inhibitory activity of 7f was about 10 times that of 3. To estimate the inhibitory mechanism, an inhibitory kinetics experiment with 7f was performed by making a Lineweaver–Burk plot. The rate of cleavage for different amounts of substrate by cathepsin B in the absence or presence of 7f (100, 500 μM) was monitored using Fluorescence spectrometer and consequently competitive inhibition toward cathepsin B was assessed.
2.4. Cytotoxicity against Hela cells
The cytotoxic ability of serine derivatives was also important because any protease inhibitor needs to have an orthogonal relationship between anti-virus activity and cytotoxicity. A cytotoxicity assay was performed against HeLa cells using synthesized serine derivatives for cytotoxicity alongside rotenone, which served as the control. The cytotoxicities of these compounds were determined by measuring live-cell succinate-tetrazolium reductase activity (MTT assay). Although the serine derivatives, both enantiomers of 3, 7i, 7j, 7h and 7k at 200 μM concentrations were evaluated against Hela cells (5000 cells), the cell death ratio caused by these compounds was less than 20% and therefore showed practically no cytotoxicity.
3. Conclusion
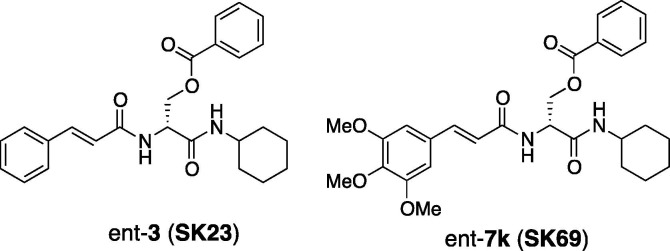
Inhibitory activities of ent-3 against SARS 3CL protease and cathepsin B showed an IC50 value of 30 μM and no inhibition at 1 mM concentration, respectively. Moreover, ent-7k has inhibitory effects with IC50 = 65 μM for SARS 3CL protease and IC50 = 500 μM for cathepsin B. These results indicate high specificity between the two proteases. In addition, the ent-3 and ent-7k inhibitors showed no cytotoxicity and therefore have good potential as drugs. Ent-3 and ent-7k were chosen for the next stages and designated as SK23 and SK69, respectively (Fig. 3 ).
Figure 3.
Optimized inhibitors ent-3 (SK23) and ent-7k (SK69).
In conclusion, we found effective serine derivatives toward the SARS 3CL protease inhibitor by the combination of docking simulation and a structure activity relationship study. Consequently, two attractive inhibitors, ent-3 (SK23) and ent-7k (SK69), which were derived from the tetrapeptide aldehyde (1) and Bai’s inhibitor were produced. These inhibitors showed protease selectivity and no cytotoxicity and therefore a useful methodology using a serine template will facilitate drug design. Especially, we testified that ent-3 (SK23) derived from the virtual screening was most reasonable inhibitor. To improve the inhibitory activity against the SARS 3CL protease, molecular design and evaluation are now underway.
4. Experimental
4.1. General information
All solvents were reagent grade. CH2Cl2 was distilled from CaH2. All commercial reagents were of the highest purity available. Amino acid derivatives were purchased from Watanabe Chemical Industries. Other chemicals were purchased from Aldrich, Wako chemical and Nacalai tesque. Optical rotations were determined with a JASCO DIP-371 polarimeter at the sodium D line. IR spectra of sample were obtained as films with a HORIBA FT-720 spectrometer. 1H (400 and 500 MHz) and 13C NMR (100 and 125 MHz) were determined on a JNM-ECX400 and JNM-ECX500. Chemical shifts are reported in ppm with reference to tetramethylsilane [1H NMR: TMS (0.00)], or solvent signals [1H NMR: CDCl3 (7.26), MeOH-d 4 (3.30); 13C NMR: CDCl3 (77.16), MeOH-d 4 (50.05)]. Mass spectra were recorded on a JEOL AccuTOF JMS-T100LC (ESI). Analytical TLC was performed on Merck Silica gel 60F254. Crude products were purified by column chromatography on Silica Gel 60 N [Kanto, particle size, (spherical, neutral) 63–210 μm or 100–200 μm]. HPLC system (monitored at 220 nm) equipped with Cosmosil 5C18 AR-II column (4.6 × 150 mm) using MeCN in 0.1% aqueous TFA. Absorbance for enzyme inhibition assay was measured with the HITACHI F-7000. Absorbance for cytotoxicity assay was measured with the CORONA MTP-310Lab.
4.2. Coupling of P4 position and tBu-deprotection of amine
4.2.1. (S)-N-Cinnamoylserinecyclohexylamide (5a)
To a solution of amine (4a) (4.45 g, 11.3 mmol) in CH2Cl2 were added cinnamic acid (3.35 g, 22.6 mmol), HATU (8.59 g, 22.6 mmol), HOAt (3.08 g, 22.6 mmol) and DIPEA (7.80 ml, 45.2 mmol) and the mixture was stirred at room temperature for 8 h. After CH2Cl2 and H2O were added to the solution, the organic layer was washed with brine and dried over MgSO4. Organic layer was evaporated in vacuo and the residue was purified by silica gel column chromatography (hexane/AcOEt = 4:1) to give l-serine derivative (6.16 g, 16.5 mmol). To a solution of l-serine derivative was added TFA (3.0 ml) at room temperature. After being stirred for 2 h, the TFA was carried out azeotropic removal with MeOH and CH2Cl2 in vacuo. The residue was purified by silica gel column chromatography (CHCl3) to give alcohol (5a) (1.36 g, 4.30 mmol, 38%) as a white powder, mp 185–188 °C. [α]25 D −78°(c 0.13, CHCl3). IR (film) ν max cm−1: 3276, 2930, 1645, 1618, 1542, 1215, 1052, 975, 752, 666. 1H NMR (500 MHz, CDCl3) δ: 1.15–1.36 (6H, m), 1.56–1.72 (3H, m), 1.82–1.91 (2H, m), 3.69–3.77 (2H, m), 4.17 (1H, dd, J = 12.0, 3.0 Hz), 4.57 (1H, t, J = 3.5 Hz), 6.51 (1H, d, J = 15.0 Hz), 7.36–7.37 (3H, m), 7.49–7.51 (2H, m), 7.65 (1H, d, J = 16.0 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.7, 25.6, 32.8, 32.9, 48.5, 53.8, 63.1, 90.6, 119.7, 128.1, 129.1, 130.3, 134.6, 142.5, 166.9, 170.4. HR-MS (ESI) m/z. Calcd for C18H24N2O3Na [M+Na]+ 339.1658. Found 339.1700. ent-5a: mp 183–185 °C. [α]27 D +79° (c 0.27, CHCl3), HRMS (ESI) Found 339.1668.
4.2.2. (S)-N-(p-Hydroxyphenyl)propionylserinecyclohexylamide (5b)
(0.261 g, 0.780 mmol, 79%) as an oil. [α]28 D −2.7°(c 0.75, MeOH). IR (film) ν max cm−1: 3290, 2935, 2858, 1671, 1516, 1452, 1352, 1202, 1132, 1026, 892, 833, 758. 1H NMR (500 MHz, CD3OD) δ: 1.20–1.85 (10H, m), 2.61 (2H, t, J = 8.0 Hz), 2.79 (2H, t, J = 8.0 Hz), 3.61–3.88 (5H, m), 4.04–4.06 (1H, m), 4.30–4.33 (1H, m), 4.42–4.46 (1H, m), 6.65 (2H, d, J = 8.5 Hz), 6.97 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CD3OD) δ: 24.8, 25.2, 30.5, 32.3, 37.7, 48.3, 55.4, 61.8, 114.9, 129.0, 131.5, 155.5, 169.8, 174.1. HR-MS (ESI) m/z: calcd for C18H26N2O4Na [M+Na]+ 357.1790. Found 357.1800.
4.2.3. (S)-N-(E)-4-phenylbut-2-enoicserinecyclohexylamide (5c)
(0.278 g, 0.841 mmol, 37%) as a white powder, mp 150–152 °C. [α]26 D −3.0°(c 0.10, CHCl3). IR (film) ν max cm−1: 3282, 3059, 2933, 2854, 1749, 1655, 1583, 1545, 1475, 1456. 1H NMR (400 MHz, CD3OD) δ: 1.11–1.88 (11H, m), 4.58–4.67 (1H, m), 3.93 (2H, s), 4.36–4.42 (1H, m), 4.51–4.56 (1H, m), 6.33 (1H, d, J = 16.0 Hz), 6.53 (1H, d, J = 16.0 Hz), 7.17–7.21 (1H, m), 7.26–7.29 (2H, m), 7.37 (2H, d, J = 7.2 Hz). 13C NMR (100 MHz, CD3OD) δ: 24.8, 25.3, 26.3, 32.3, 37.8, 39.5, 54.3, 55.5, 57.4, 61.8, 122.5, 126.0, 127.2, 128.2, 133.5, 167.6, 170.0, 172.7. HR-MS (ESI) m/z: calcd for C19H26N2O3Na [M+Na]+ 353.1841. Found 353.1805.
4.2.4. (S)-N-(E)-4′-Methoxy-3-phenylprop-2-enoicserinecyclohexylamide (5d)
(0.228 g, 0.659 mmol, 43%) as a yellow powder, mp 226–228 °C. [α]23 D +1.0°(c 0.10, CHCl3). IR (film) ν max cm−1: 3282, 2933, 2856, 2777, 1724, 1657, 1603, 1552, 1485, 1412. 1H NMR (400 MHz, CDCl3) δ: 1.10–1.92 (11H, m), 4.02 (3H, s), 4.08–4.22 (2H, m), 4.51–4.54 (1H, m), 6.37 (1H, d, J = 15.6 Hz), 6.89 (2H, d, J = 9.2 Hz), 7.45 (2H, d, J = 8.7 Hz), 7.60 (1H, d, J = 15.6 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.3, 32.3, 48.7, 54.5, 55.6, 57.5, 61.8, 62.0, 114.0, 117.6, 129.2, 129.2, 140.8, 150.2, 161.4, 167.8, 169.8, 169.9, 170.2. HR-MS (ESI) m/z: calcd for C19H26N2O4Na [M+Na]+ 369.1790. Found 369.1783.
4.2.5. (S)-N-Benzoylserinecyclohexylamide (5e)
(0.247 g, 0.851 mmol, 80%) as a white powder, mp189–191 °C. [α]28 D +26°(c 0.10, CHCl3). IR (film) ν max cm−1: 3290, 3072, 2933, 2854, 2355, 2326, 1635, 1548, 1489, 1448. 1H NMR (400 MHz, CDCl3) δ: 1.10–1.95 (11H, m), 3.48–3.79 (2H, m), 4.23–4.28 (1H, m), 4.52–4.56 (1H, m), 7.43–7.56 (3H, m), 7,81 (2H, d, J = 11.6 Hz); 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.3, 32.3, 48.3, 56.0, 62.0, 127.2, 128.3, 131.6, 133.8, 168.7, 170.1. HR-MS (ESI) m/z: calcd for C16H22N2O3Na [M+Na]+ 313.1528. Found 313.1514.
4.2.6. (S)-N-(E)-2′-Methoxy-3-phenylprop-2-enoicserinecyclohexylamide (5f)
(34.0 mg, 98.2 μmol, 7.0%) as a white powder, mp 189–192 °C. [α]25 D +6.0°(c 0.10, MeOH). IR (film) ν max cm−1: 3282, 2933, 2846, 1649, 1539, 1489, 1452, 1248, 1201, 1180; 1H NMR (400 MHz, CDCl3) δ: 1.07–1.95 (11H, m), 3.62–3.78 (2H, m), 3.89 (3H, s), 4.19 (1H, dd, J = 12.0, 2.8 Hz), 4.48–4.54 (1H, m), 6.65 (1H, d, J = 16.0 Hz), 6.86–6.99 (2H, m), 7.30–7.37 (1H, m), 7.47 (1H, d, J = 8.0 Hz), 7.88 (1H, d, J = 16.0 Hz); 13C NMR (100 MHz, CDCl3) δ: 22.9, 24.8, 25.3, 29.3, 32.3, 54.7, 55.6, 62.0, 111.1, 120.4, 123.5, 128.1, 131.0, 136.4, 158.4, 167.8, 170.1. HR-MS (ESI) m/z: calcd for C19H26N2O4Na [M+Na]+ 369.1790. Found 369.1799.
4.2.7. (S)-N-(E)-3′,4′-Dimethoxy-3-phenylprop-2-enoicserinecyclohexylamide (5g)
(0.100 g, 0.266 mmol, 23%) as a yellow powder, mp 220 °C. [α]26 D +28°(c 0.10, MeOH). IR (film) ν max cm−1: 3276, 3099, 3001, 2931, 2846, 1643, 1610, 1543, 1514, 1452. 1H NMR (400 MHz, CDCl3) δ: 1.09–1.94 (11H, m), 3.66–3.81 (1H, m), 3.89 (6H, s), 6.43 (1H, d, J = 16.0 Hz), 6.80–6.89 (1H, m), 7.00–7.04 (1H, m), 7.08 (1H, d, J = 6.4 Hz), 7.58 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.7, 25.5, 32.6, 38.8, 48.4, 54.0, 56.1, 63.2, 109.6, 111.1, 117.3, 122.5, 127.4, 142.4, 149.3, 151.0, 167.3; HR-MS (ESI) m/z: calcd for C20H28N2O5Na [M+Na]+ 399.1896. Found 399.1907.
4.2.8. (S)-N-(E)-3′,5′-Dimethoxy-3-phenylprop-2-enoicserinecyclohexylamide (5h)
(0.236 g, 0.627 mmol, 29%) as a white powder, mp 175–177 °C. [α]26 D +22°(c 0.10, CHCl3). IR (film) ν max cm−1: 3269, 2924, 2846, 2355, 2326, 1647, 1606, 1558, 1541, 1522. 1H NMR (400 MHz, CDCl3) δ: 1.10–1.92 (11H, m), 3.62–3.76 (1H, m), 3.83 (6H, s), 4.12–4.21 (1H, m), 4.48–4.56 (1H, m), 6.41 (1H, d, J = 16.0 Hz), 6.47 (1H, s), 6.65 (2H, s), 7.56 (1H, d, J = 15.3 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.7, 25.5, 29.8, 32.8, 35.1, 48.3, 53.4, 55.5, 57.1, 63.0, 100.0, 102.0, 104.2, 105.9, 120.1, 126.3, 136.4, 142.3, 143.0, 161.1. HR-MS (ESI) m/z: calcd for C20H28N2O5Na [M+Na]+ 399.1896. Found 399.1872.
4.2.9. (S)-N-(E)-3′-Methoxy-3-phenylprop-2-enoicserinecyclohexylamide (5i)
(0.290 g, 0.838 mmol, 69% in 2 steps) as a white powder, mp 180–181 °C. [α]26 D +25°(c 0.10, MeOH). IR (film) ν max cm−1: 3269, 3086, 2931, 2854, 1647, 1620, 1577, 1547, 1491, 1448; 1H NMR (400 MHz, CDCl3) δ: 1.11–2.11 (11H, m), 3.63–3.68 (1H, m), 3.83 (3H, s), 4.19–4.25 (1H, m), 4.46–4.50 (1H, m), 6.45 (1H, d, J = 16.0 Hz), 6.76–6.83 (1H, m), 6.92 (1H, dd, J = 8.8, 2.8 Hz), 7.03 (1H, s), 7.11 (1H, d, J = 8.0 Hz), 7.62 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, MeOH-d 4) δ: 24.8, 25.3, 32.3, 54.4, 55.7, 62.0, 112.6, 115.3, 120.1, 120.5, 129.6, 136.3, 140.9, 160.2, 161.4, 167.1, 170.1. HR-MS (ESI) m/z: calcd for C19H26N2O4Na [M+Na]+ 369.1790. Found 369.1798.
4.2.10. (S)-N-(E)-4′-hydroxy-3-phenylprop-2-enoicserinecyclohexylamide (5j)
(0.170 g, 0.511 mmol, 55%) as a white powder, mp 220–222 °C. IR (film) ν max cm−1: 3338, 3269, 2918, 2854, 2495, 2432, 1655, 1641, 1606, 1570. 1H NMR (400 MHz, MeOH-d 4) δ: 1.10–1.90 (11H, m), 3.60–3.68 (1H, m), 3.76 (1H, d, J = 5.2 Hz), 4.49 (1H, t, J = 6.0 Hz), 6.52 (1H, d, J = 15.6 Hz), 6.77 (2H, d, J = 8.7 Hz), 7.41 (2H, d, J = 8.7 Hz), 7.46 (1H, d, J = 15.6 Hz). 13C NMR (100 MHz, MeOH-d 4) δ: 24.8, 25.3, 32.3, 48.6, 55.6, 62.0, 115.4, 116.6, 126.2, 129.4, 141.2, 159.4, 167.8, 170.1. HR-MS (ESI) m/z: calcd for C18H24N2O4Na [M+Na]+ 355.1634. Found 355.1639.
4.2.11. (S)-N-(E)-3′,4′,5′-Trimethoxy-3-phenylprop-2-enoicserinecyclohexylamide (5k)
(0.104 g, 0.256 mmol, 29%) as a white powder, mp 156–162 °C. [α]26 D +31°(c 0.10, MeOH). IR (film) ν max cm−1: 3298, 2933, 2846, 1649, 1616, 1583, 1542, 1506, 1452, 1419. 1H NMR (400 MHz, CDCl3) δ: 1.09–1.96 (11H, m), 3.60–3.78 (4H, m), 3.84 (9H, s), 4.10 (1H, dd, J = 8.4, 4.0 Hz), 4.57–4.64 (1H, m), 6.44 (1H, d, J = 16.0 Hz), 6.70 (2H, s), 7.04 (1H, d, J = 8.0 Hz), 7.12 (1H, d, J = 5.6 Hz), 7.52 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.7, 25.5, 32.8, 48.6, 54.4, 56.2, 61.0, 63.4, 105.2, 119.3, 130.0, 140.0, 142.1, 153.5, 166.8, 169.9. HR-MS (ESI) m/z: calcd for C21H30N2O6Na [M+Na]+ 429.2002. Found 429.2009.
4.2.12. (S)-N-Cinnamoylserinepiperidineamide (5l)
(95.4 mg, 26%) as an oil, [α]25 D +16° (c 0.40, MeOH). IR (film) ν max cm−1: 3286, 3060, 3027, 2939, 2860, 2245, 1660, 1608, 1549, 1450. 1H NMR (400 MHz, CDCl3) δ: 1.53–1.69 (6H, m), 3.50–3.56 (2H, m), 3.58–3.66 (1H, m), 3.82 (2H, s), 5.06–5.11 (1H, m), 6.50 (1H, d, J = 16.0 Hz), 7.32–7.38 (3H, m), 7.46–7.51 (2H, m), 7.62 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.4, 25.6, 26.5, 43.6, 47.0, 50.9, 52.0, 119.9, 128.0, 128.9, 130.0, 142.1, 166.5, 168.2. HR-MS (ESI) m/z: calcd for C17H22N2O3Na [M+Na]+ 325.1528. Found 325.1540. ent-5l: [α]25 D −16° (c 0.20, MeOH). HR-MS (ESI) m/z: found 325.1514.
4.2.13. (S)-N-Cinnamoylserinemorpholineamide (5m)
(196 mg, 0.644 mmol, 91%) as an oil, [α]25 D +8.5° (c 0.55, CHCl3). IR (film) ν max cm−1: 3303, 3059, 3016, 2966, 2931, 2862, 2368, 2305, 1736, 1620. 1H NMR (400 MHz, CDCl3) δ: 3.57–3.74 (8H, m), 3.79 (1H, dd, J = 11.2, 4.0 Hz), 3.88 (1H, dd, J = 11.6, 4.8 Hz), 4.38 (1H, s), 5.06–5.15 (1H, m), 6.52 (1H, d, J = 16.0 Hz), 7.30–7.40 (3H, m), 7.44–7.52 (2H, m), 7.62 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 42.8, 46.4, 51.1, 64.1, 66.7, 119.9, 128.7, 130.1, 134.5, 142.3, 166.4, 169.3. HR-MS (ESI) m/z: calcd for C16H20N2O4Na [M+Na]+ 327.1321. Found 327.1286. ent-5m: [α]27 D −7.1° (c 0.20, CHCl3). HR-MS (ESI) m/z: found 327.1302.
4.2.14. (S)-N-Cinnamoylserinebenzylamide (5n)
(111 mg, 0.342 mmol, 19%) as an oil. [α]26 D +46° (c 0.10, MeOH). IR (film) ν max cm−1: 3435, 3400, 3369, 3278, 1645, 1618, 1215, 1051. 1H NMR (400 MHz, CDCl3) δ: 3.64–3.72 (1H, m), 4.25–4.31 (1H, m), 4.41–4.49 (2H, m), 4.54–4.60 (1H, m), 4.46 (1H, d, J = 16.0 Hz), 6.78 (1H, s), 7.26–7.35 (4H, m), 7.38 (3H, t, J = 3.2 Hz), 7.50 (2H, d, J = 5.6 Hz), 7.65 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, MeOH-d 4) δ: 42.8, 55.9, 61.9, 120.4, 126.8, 127.6, 128.6, 129.6, 134.9, 138.4, 141.1, 167.4, 171.2. HR-MS (ESI) m/z: calcd for C19H20N2O3Na [M+Na]+ 347.1372. Found 347.1353. ent-5n: mp 171–176 °C. HR-MS (ESI) m/z: found 347.1361.
4.2.15. (S)-N-Cinnamoylserinecyclohexanemethylamide (5o)
(399 mg, 1.21 mmol, 48%) as a white powder, mp 183–184 °C. [α]26 D +21°(c 0.10, MeOH), IR (film) ν max cm−1: 3273, 3107, 3047, 2922, 2850, 2368, 2326, 1954, 1884, 1651. 1H NMR (400 MHz, CDCl3) δ: 0.83–0.97 (2H, m), 1.05–1.29 (3H, m), 1.40–1.53 (1H, m), 1.59–1.74 (6H, m), 3.05–3.14 (2H, m), 3.75–3.85 (2H, m), 3.75–3.84 (1H, m), 4.18 (1H, dd, J = 12.0, 4.0 Hz), 4.64–4.71 (1H, m), 6.57 (1H, d, J = 16.0 Hz), 7.32–7.40 (3H, m), 7.45–7.53 (2H, m), 7.65 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 25.8, 26.4, 30.8, 37.8, 38.8, 46.0, 54.2, 63.0, 119.3, 128.1, 129.0, 130.3, 134.3, 142.9, 166.1, 167.4, 171.2. HR-MS (ESI) m/z: calcd for C19H26N2O3Na [M+Na]+ 353.1841 Found 353.1818. ent-5o: mp 181–183 °C. [α]27 D −24°(c 0.10, MeOH), HR-MS (ESI) m/z: found 353.1818.
4.3. Coupling of P1′ position for the inhibitors
4.3.1. (S)-N-Cinnamoyl serine(benzoyl)cyclohexylamide (3)
To a solution of 5a (19.6 mg, 0.0619 mmol) in CH2Cl2 were added Benzoyl chloride (14.3 μl, 0.124 mmol), DMAP (15.1 mg, 0.124 mmol) and DIPEA (53.4 μl, 0.310 mmol) at 0 °C, and the mixture was stirred at room temperature for 3 h. After CH2Cl2 and H2O were added to the solution, the organic layer was washed with brine and dried over MgSO4. The solvent was evaporated in vacuo and the residue was purified by silica gel column chromatography (hexane/AcOEt = 4:1) to give 3 (18.6 mg, 0.0442 mmol, 71%) as a white powder, mp 200–203 °C. [α]24 D −1.5° (c 0.76, CHCl3). IR (film) ν max cm−1: 3282, 2930, 2849, 2364, 1727, 1646, 1617, 1541, 1450, 1270, 1112, 1025, 975, 710. 1H NMR (400 MHz, CDCl3) δ: 1.09–1.39 (6H, m), 1.66 (2H, m), 1.88 (2H, m), 3.77 (1H, m), 4.60 (1H, dd, J = 11.2, 5.6 Hz), 4.71 (1H, dd, J = 11.2, 6.4 Hz), 5.00 (1H, q, J = 6.0 Hz), 6.50 (1H, d, J = 7.2 Hz), 6.52 (1H, d, J = 15.6 Hz), 6.97 (1H, d, J = 7.6 Hz), 7.36–7.61 (8H, m), 7.64 (1H, d, J = 15.6 Hz), 8.02 (2H, d, J = 7.6 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 48.8, 52.8, 64.2, 120.1, 128.1, 128.6, 129.0, 129.9, 130.1, 130.3, 133.4, 133.6, 134.7, 136.8, 141.8, 142.2, 166.3, 166.5, 168.2, 170.4. HR-MS (ESI) m/z: calcd for C25H28N2O4Na [M+Na]+ 443.1947. Found 443.1913. ent-3: mp 205–206 °C. [α]27 D +1.4°(c 0.14, CHCl3). HR-MS (ESI) m/z: found 443.1906.
4.3.2. (S)-N-Cinnamoylserine{3-(R)-pyroglutamoyl}cyclohexylamide (6b)
(10.9 mg, 0.0255 mmol, 25%). [α]26 D +0.87° (c 0.23, CHCl3). IR (film) ν max cm−1: 3418, 2933, 2854, 2360, 2341, 1749, 1647, 1558, 1541, 1456, 1192. 1H NMR (400 MHz, CDCl3) δ: 1.15–1.88 (10H, m), 2.29–2.44 (4H, m), 3.74 (1H, br), 4.28–4.39 (2H, m), 4.53–4.57 (1H, m), 4.96 (1H, br), 6.51 (1H, d, J = 15.6 Hz), 6.79 (1H, d, J = 7.2 Hz), 7.12 (1H, d, J = 7.2 Hz), 7.31–7.50 (6H, m), 7.66 (1H, d, J = 15.6 Hz), 7.72 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.3, 24.4, 24.9, 25.5, 29.5, 32.9, 48.9, 52.1, 52.3, 65.3, 119.8, 128.0, 129.0, 130.2, 134.5, 142.5, 166.3, 167.5, 171.8, 178.4. HR-MS (ESI) m/z: calcd for C23H29N3O6Na [M+Na]+ 450.2005. Found 450.2041.
4.3.3. (S)-N-Cinnamoylserine(butanoyl)cyclohexylamide (6c)
(23.6 mg, 0.0611 mmol, 78%) as a white powder, mp 176–182 °C. [α]25 D −0.6° (c 0.17, CHCl3). IR (film) ν max cm−1: 3282, 3069, 2933, 2855, 1741, 1649, 1620, 1546, 1449, 1344, 1172, 1092, 977. 1H NMR (500 MHz, CDCl3) δ: 1.13–1.37 (11H, m), 1.60–1.72 (4H, m), 1.91 (2H, m), 3.73–3.79 (1H, m), 4.29 (1H, dd, J = 11.5, 5.0 Hz), 4.49 (1H, dd, J = 11.5, 6.0 Hz), 4.79 (1H, q, J = 6.3 Hz), 6.13 (1H, br), 6.45 (1H, d, J = 15.5 Hz), 6.59 (1H, br), 7.37–7.39 (3H, m), 7.51–7.52 (2H, m), 7.64 (1H, d, J = 15.5 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.9, 25.6, 27.3, 33.1, 39.0, 48.8, 53.0, 64.1, 119.9, 128.1, 129.0, 130.2, 134.6, 142.3, 166.1, 167.8, 178.8. HR-MS (ESI) m/z: calcd for C22H30N2O4Na [M+Na]+ 409.2103. Found 409.2080.
4.3.4. (S)-N-Cinnamoylserine(phenylacetyl)cyclohexylamide (6d)
(18.0 mg, 0.0414 mmol, 67%) as a white powder, mp 172–177 °C. [α]27 D 0° (c 0.10, MeOH). IR (film) ν max cm−1: 3649, 3583, 3282, 3064, 3030, 2931, 2854, 2362, 2312, 1749. 1H NMR (400 MHz, CDCl3) δ: 0.95–1.20 (5H, m), 1.21–1.40 (4H, m), 1.52–1.73 (4H, m), 1.76–1.89 (3H, m), 3.67–3.77 (1H, m), 4.27–4.35 (1H, m), 4.46–4.54 (1H, m), 4.73–4.82 (1H, m), 6.09–6.19 (1H, m), 6.34 (1H, d, J = 16.0 Hz), 6.49 (1H, d, J = 8.0 Hz), 7.26–7.33 (5H, m), 7.34–7.40 (3H, m), 7.46–7.52 (1H, m), 7.60 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 41.3, 48.7, 52.5, 64.3, 119.9, 127.3, 128.0, 128.8, 129.0, 129.4, 130.1, 133.7,134.6, 142.1, 166.0, 167.6, 171.5. HR-MS (ESI) m/z: calcd for C26H30N2O4Na [M+Na]+ 457.2103. Found 457.2079.
4.3.5. (S)-N-Cinnamoylserine(isonicotinoyl)cyclohexylamide (6e)
(13.2 mg, 0.0313 mmol, 47%) as a white powder, mp 210–212 °C. [α]24 D −6.1° (c 0.08, CHCl3). IR (film) ν max cm−1: 3282, 2929, 2854, 2360, 2341, 1741, 1645, 1616, 1560, 1539, 1211, 1144, 1066, 974. 1H NMR (400 MHz, CDCl3) δ: 1.09–1.39 (4H, m), 1.59–1.71 (4H, m), 1.89 (2H, m), 3.74–3.81 (1H, m), 4.64–4.72 (2H, m), 5.05 (1H, q, J = 6.4 Hz), 6.50 (1H, d, J = 15.6 Hz), 6.66 (1H, d, J = 7.6 Hz), 6.86 (1H, d, J = 7.6 Hz), 7.36–7.37 (3H, m), 7.48–7.50 (2H, m), 7.63 (1H, d, J = 15.6 Hz), 7.82 (2H, d, J = 6.0 Hz), 8.76 (2H, d, J = 5.6 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.8, 25.5, 33.0, 48.9, 52.6, 65.4, 119.8, 123.0, 128.1, 129.1, 130.3, 134.5, 136.9, 142.5, 150.8, 165.1, 166.2, 167.5. HR-MS (ESI) m/z: calcd for C24H27N3O4Na [M+Na]+ 444.1899. Found 444.1864.
4.3.6. (S)-N-Cinnamoylserine(2,4,6-trichlorobenzoyl)cyclohexylamide (6f)
(9.00 mg, 0.0172 mmol, 27%) as a white powder, mp 208–211 °C. [α]26 D −19° (c 0.67, CHCl3). IR (film) ν max cm−1: 3275, 2931, 2854, 2360, 2330, 1747, 1651, 1620, 1558, 1520, 1458, 1273, 1119, 1057, 972, 852, 752. 1H NMR (500 MHz, CDCl3) δ: 1.13–1.38 (4H, m), 1.58–1.69 (4H, m), 1.86–1.90 (2H, m), 3.70–3.79 (1H, m), 4.61 (1H, dd, J = 11.5, 5.0 Hz), 4.87 (1H, dd, J = 11.5, 6.5 Hz), 4.93 (1H, q, J = 6.3 Hz), 6.33 (1H, d, J = 8.5 Hz), 6.45 (1H, d, J = 16.0 Hz), 6.62 (1H, d, J = 8.0 Hz), 7.34–7.38 (5H, m), 7.49–7.51 (2H, m), 7.65 (1H, d, J = 15.5 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.8, 25.6, 32.9, 48.8, 52.6, 65.4, 119.7, 128.1, 128.3, 129.0, 130.3, 132.8, 134.5, 134.7, 142.6, 164.1, 166.3, 167.5. HR-MS (ESI) m/z: calcd for C25H25Cl3N2O4Na [M+Na]+ 545.0778. Found 545.0815.
4.3.7. (S)-N-Cinnamoylserine(pivaloyl)cyclohexylamide (6g)
(20.6 mg, 0.0514 mmol, 70%) as a white powder, mp 183–186 °C. [α]28 D +0.7° (c 0.28, CHCl3). IR (film) ν max cm−1: 3282, 2932, 2856, 2364, 1737, 1646, 1620, 1547, 1450, 1215, 1155, 979; 1H NMR (400 MHz, CDCl3) δ: 0.94 (3H, t, J = 7.4 Hz), 1.13–1.41 (5H, m), 1.60–1.69 (7H, m), 1.89 (2H, m), 2.32 (2H, t, J = 7.2 Hz), 3.78 (1H, m), 4.30 (1H, dd, J = 11.4, 5.5 Hz), 4.79 (1H, q, J = 6.3 Hz), 6.22 (1H, br), 6.46 (1H, d, J = 16.0 Hz), 6.64 (1H, br), 7.37–7.38 (3H, m), 7.51–7.52 (2H, m), 7.64 (1H, d, J = 15.6 Hz). 13C NMR (100 MHz, CDCl3) δ: 13.8, 18.5, 24.8, 25.6, 33.0, 36.1, 48.7, 52.9, 63.9, 119.9, 128.1, 129.0, 130.2, 134.6, 142.4, 167.8, 173.9. HR-MS (ESI) m/z: calcd for C23H32N2O4Na [M+Na]+ 423.2260. Found 423.2227.
4.3.8. (S)-N-Cinnamoylserine(acetyl)cyclohexylamide (6h)
(18.4 mg, 0.0513 mmol, 64%) as a white powder, mp 191–194 °C. [α]26 D −3.0° (c 0.81, CHCl3). IR (film) ν max cm−1: 3298, 3098, 3093, 2929, 2854, 2360, 2332, 1743, 1647, 1616, 1558, 1540, 1448, 1375, 1246, 1219, 1080, 1049, 974. 1H NMR (500 MHz, CDCl3) δ: 1.11–1.25 (3H, m), 1.29–1.40 (2H, m), 1.59–1.61 (1H, m), 1.68–1.73 (2H, m), 1.84–1.94 (2H, m), 2.07 (3H, s), 3.77 (1H, m), 4.34 (1H, dd, J = 11.5, 5.5 Hz), 4.48 (1H, dd, J = 11.5, 6.0 Hz), 4.90 (1H, q, J = 6.0 Hz), 6.53 (1H, d, J = 16.5 Hz), 6.66 (1H, d, J = 8.0 Hz), 6.95 (1H, d, J = 8.0 Hz), 7.34–7.36 (3H, m), 7.47–7.50 (2H, m), 7.47–7.50 (2H, m), 7.64 (1H, d, J = 15.5 Hz). 13C NMR (125 MHz, CDCl3) δ: 21.0, 24.9, 25.6, 33.0, 48.7, 52.6, 64.2, 120.1, 129.0, 130.1, 134.7, 142.1, 166.2, 167.9, 171.0. HR-MS (ESI) m/z: calcd for C20H26N2O4Na [M+Na]+ 381.1790. Found 381.1793. ent-6h: mp 185–187 °C. [α]27 D +2.4° (c 0.17, CHCl3). HRMS (ESI). Found 381.1749.
4.3.9. (S)-N-Cinnamoylserine(benzoyl)cyclohexylamide (6i)
(8.2 mg, 0.0161 mmol, 26%). [α]28 D −11°(c 0.23, CHCl3). IR (film) ν max cm−1: 3583, 3282, 2924, 2854, 2360, 2332, 1728, 1645, 1616, 1541, 1458, 1348, 1176. 1H NMR (500 MHz, CDCl3) δ: 1.25 (6H, m), 1.71–1.74 (2H, m), 1.94 (2H, m), 3.80 (1H, m), 4.72 (1H, dd, J = 11.5, 7.0 Hz), 4.76 (1H, dd, J = 11.5, 5.5 Hz), 5.01 (1H, q, J = 6.7 Hz), 6.16 (1H, d, J = 9.0 Hz), 6.47 (1H, d, J = 15.0 Hz), 6.59 (1H, d, J = 8.0 Hz), 7.37–7.39 (3H, m), 7.50–7.52 (2H, m), 7.65 (1H, d, J = 16.5 Hz), 9.13 (2H, d, J = 1.5 Hz), 9.23 (1H, m). HR-MS (ESI) m/z: calcd for C25H26N4O8Na [M+Na]+ 533.1648. Found 533.1600.
4.3.10. (S)-N-Cinnamoylserine(nicotinoyl)cyclohexylamide (6j)
(13.2 mg, 0.0313 mmol, 47%) as a white powder, mp 201–202 °C. [α]26 D −4.0° (c 0.45, CHCl3). IR (film) ν max cm−1: 3400, 2931, 2854, 2360, 2332, 1733, 1647, 1618, 1541, 1292, 1211, 1144, 974. 1H NMR (400 MHz, CDCl3) δ: 1.13–1.35 (6H, m), 1.68–1.71 (2H, m), 1.87–1.90 (2H, m), 3.73–3.82 (1H, m), 4.64 (1H, dd, J = 12.0, 5.6 Hz), 4.73 (1H, dd, J = 11.6, 6.0 Hz), 4.98 (1H, q, J = 6.4 Hz), 6.30 (1H, d, J = 8.4 Hz), 6.48 (1H, d, J = 16.0 Hz), 6.71 (1H, d, J = 7.6 Hz), 7.35–7.38 (4H, m), 7.48–7.51 (2H, m), 7.64 (1H, d, J = 15.6 Hz), 8.28 (1H, dt, J = 8.0, 2.0 Hz), 8.78 (1H, dd, J = 4.8, 1.6 Hz), 9.20 (1H, d, J = 1.2 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.9, 25.5, 33.0, 48.8, 52.7, 65.0, 119.9, 123.5, 125.7, 128.1, 129.0, 130.2, 134.6, 137.4, 142.5, 151.1, 153.9, 165.2, 166.2, 167.7. HR-MS (ESI) m/z: calcd for C24H27N3O4Na [M+Na]+ 444.1899. Found 444.1861. ent-6j: mp 197–201 °C. [α]27 D +3.0° (c 0.38, CHCl3). HRMS (ESI). Found 444.1866.
4.3.11. (S)-N-Cinnamoylserine(picolinoyl)cyclohexylamide (6k)
(25.8 mg, 0.0612 mmol, 95%) as a white powder, mp 202–204 °C. [α]28 D −6.0° (c 0.19, CHCl3). IR (film) ν max cm−1: 3260, 2933, 2854, 2360, 2341, 1734, 1653, 1622, 1558, 1541, 1290, 1217, 1130, 1090, 978. 1H NMR (500 MHz, CDCl3) δ: 1.10–1.36 (6H, m), 1.56–1.71 (2H, m), 1.83–1.90 (2H, m), 3.74–3.81 (1H, m), 4.51 (1H, q, J = 5.8 Hz), 4.86 (1H, q, J = 5.7 Hz), 4.97 (1H, q, J = 6.5 Hz), 6.50 (1H, d, J = 15.5 Hz), 6.66 (1H, d, J = 8.0 Hz), 7.02 (1H, d, J = 7.5 Hz), 7.36–7.38 (3H, m), 7.49–7.52 (3H, m), 7.64 (1H, d, J = 16.5 Hz), 7.86 (1H, dd, J = 7.5, 2.0 Hz), 8.14 (1H, d, J = 7.5 Hz), 8.72 (1H, dd, J = 4.0, 2.0 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.8, 25.6, 32.8, 32.9, 48.8, 52.4, 65.0, 120.1, 125.6, 127.5, 128.1, 129.0, 130.1, 134.7, 137.4, 142.1, 147.6, 149.8, 164.9, 166.2, 167.8. HR-MS (ESI) m/z: calcd for C24H27N3O4Na [M+Na]+ 444.1899. Found 444.1870. ent-6k: mp 205–208 °C. HRMS (ESI) Found 444.1866.
4.3.12. (S)-N-Cinnamoylserine{(E)-3′,4′,5′-trimethoxy-3-phenylprop-2-enly}cyclohexylamide (6l)
Oxalylchloride (27.3 μl, 0.316 mmol) and DMF (cat.) were added to (E)-3′,4′,5′-trimethoxy-3-phenylprop-2-enoic acid (37.6 mg, 0.158 mmol) at room temperature in CH2Cl2 under nitrogen atmosphere. The mixture was stirred at room temperature for 10 min, the solvent was evaporated in vacuo and the residue was added to the mixture of 5a (50.0 mg, 0.158 mmol), TEA (132 ml, 0.738 mmol) in CH2Cl2 and stirred at room temperature for 2 h. After CH2Cl2 and H2O were added to the solution, the organic layer was dried over MgSO4. The solvent was evaporated in vacuo and the residue was purified by silica-gel column chromatography (CHCl3) to give 6k (3.00 mg, 5.59 μmol, 3.9%) as a white powder, mp 227–229 °C. [α]27 D −9.8° (c 0.25, CHCl3). IR (film) ν max cm−1: 3283, 3073, 2931, 2841, 2370, 2337, 1720, 1640, 1621, 1582. 1H NMR (400 MHz, CDCl3) δ: 1.10–1.95 (11H, m), 3.87 (9H, s), 4.47–4.51 (1H, m), 4.60–4.67 (1H, m), 4.81–4.89 (1H, m), 6.21 (1H, d, J = 9.6 Hz), 6.34 (1H, d, J = 16.0 Hz), 6.47 (1H, d, J = 16.0 Hz), 6.68 (1H, d, J = 1.6 Hz), 6.73 (2H, s), 7.37 (3H, s), 7.50 (2H, s), 7.60–7.67 (2H, m). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 33.0, 48.6, 53.0, 56.2, 61.1, 64.2, 105.4, 116.4, 119.9, 128.0, 129.0, 129.7, 130.1, 134.5, 142.2, 146.0, 150.9, 153.5, 166.2, 167.0, 167.8. HR-MS (ESI) m/z: calcd for C30H37N2O7 [M+H]+ 537.2601 Found 537.2612.
4.3.13. (S)-N-Cinnamoylserine(phenylpropanoyl)cyclohexylamide (6m)
(4.2 mg, 9.36 μmol, 13%) as an oil. [α]27 D −19° (c 0.16, CHCl3). IR (film) ν max cm−1: 3286, 2931, 2854, 2360, 1736, 1651, 1624, 1543, 1454, 1362, 1215, 1173, 1080, 976. 1H NMR (500 MHz, CDCl3) δ: 1.10–1.39 (6H, m),1.68–1.71 (2H, m), 1.87–1.89 (2H, m), 2.67 (2H, t, J = 7.8 Hz), 2.95 (2H, t, J = 7.8 Hz), 3.75–3.76 (1H, m), 4.29 (1H, dd, J = 12.0, 5.5 Hz), 4.50 (1H, dd, J = 11.5, 6.5 Hz), 4.73 (1H, q, J = 6.3 Hz), 6.06 (1H, d, J = 8.0 Hz), 6.39 (1H, d, J = 15.5 Hz), 6.44 (1H, d, J = 7.0 Hz), 7.18–7.21 (2H, m), 7.27–7.30 (3H, m), 7.38–7.40 (3H, m), 7.50–7.52 (2H, m), 7.64 (1H, d, J = 15.5 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.8, 25.6, 30.9, 33.0, 35.7, 48.7, 52.8, 64.1, 119.8, 126.5, 128.1, 128.4, 128.7, 129.0, 134.6, 140.4, 142.4, 166.1, 167.7. HR-MS (ESI) m/z: calcd for C27H32N2O4Na [M+Na]+ 471.2260. Found 471.2264.
4.3.14. (S)-N-Cinnamoyl serine(cinnamoyl)cyclohexylamide (6n)
(4.00 mg, 0.00896 mmol, 26%) as a white powder, mp 192–198 °C. [α]27 D −6.9° (c 0.18, CHCl3). IR (film) ν max cm−1: 3279, 2929, 2856, 1716, 1643, 1617, 1541, 1447, 1311, 1172, 975, 769. 1H NMR (500 MHz, CDCl3) δ: 1.25–1.90 (10H, m), 3.80 (1H, m), 4.47 (1H, dd, J = 11.5, 5.0 Hz), 4.63 (1H, dd, J = 12.0, 6.5 Hz), 4.84 (1H, q, J = 6.2 Hz), 6.21 (1H, br), 6.44 (1H, d, J = 16.0 Hz), 6.47 (1H, d, J = 15.0 Hz), 6.69 (1H, br), 7.37–7.40 (6H, m), 7.51–7.52 (4H, m), 7.66 (1H, d, J = 16.0 Hz), 7.72 (1H, d, J = 16.0 Hz), 7.83 (2H, d, J = 8.0 Hz). 13C NMR (125 MHz, CDCl3) δ: 24.8, 25.6, 29.9, 33.0, 48.7, 53.1, 64.2, 117.2, 119.9, 128.1, 128.4, 129.0, 129.1, 130.2, 130.8, 134.3, 134.7, 142.4, 146.3, 166.2, 167.8. HR-MS (ESI) m/z: calcd for C27H30N2O4Na [M+Na]+ 469.2104. Found 469.2078. ent-6n: mp 207–211 °C. [α]27 D +9.0° (c 0.14, CHCl3). HRMS (ESI) Found 469.2064.
4.3.15. (S)-N-Cinnamoylserine(2-methyl-6-nitrobenzoyl)cyclohexylamide (6o)
(17.8 mg, 0.0371 mmol, 36%) as a white powder, mp 205–208 °C. [α]28 D −11° (c 0.23, CHCl3). IR (film) ν max cm−1: 3287, 2930, 2849, 2359, 1742, 1645, 1620, 1538, 1450, 1344, 1267, 1215, 1112, 1075, 975. 1H NMR (400 MHz, CDCl3) δ: 1.13–1.89 (10H, m), 2.42 (3H, s), 3.74 (1H, m), 4.73 (1H, dd, J = 11.6 Hz), 4.82 (1H, dd, J = 11.2, 4.8 Hz), 4.89–4.93 (1H, m), 6.32 (1H, m), 6.52 (1H, d, J = 15.2 Hz), 6.60 (1H, m), 7.37–7.39 (3H, m), 7.47–7.57 (4H, m), 7.68 (1H, d, J = 16.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 19.4, 24.8, 25.5, 32.9, 48.7, 52.7, 65.4, 119.7, 121.9, 128.2, 129.0, 130.0, 130.3, 136.5, 142.6, 166.4, 167.6. HR-MS (ESI) m/z: calcd for C26H29N3O6Na [M+Na]+ 502.1954 Found 502.1971. ent-6o: mp 205–208 °C. [α]27 D +12° (c 0.15, CHCl3). HRMS (ESI) Found 502.1926.
4.3.16. (S)-N-(E)-4-Phenylbut-2-enoicserine(benzoyl)cyclohexylamide (7c)
(11.1 mg, 25.5 μmol, 17%) as a white powder, mp 184–186 °C. [α]28 D +10° (c 0.10, CHCl3). IR (film) ν max cm−1: 3286, 3064, 3004, 2933, 2854, 2359, 1724, 1647, 1579, 1550. 1H NMR (400 MHz, CDCl3) δ: 1.02–1.89 (11H, m), 4.72–4.83 (1H, m), 3.92 (2H, s), 4.58–4.73 (2H, m), 4.88–4.96 (1H, m), 6.06 (1H, d, J = 7.6 Hz), 6.36 (1H, d, J = 7.6 Hz), 7.27–7.37 (1H, m), 7.38–7.43 (2H, m), 7.53–759 (1H, m), 7.93–8.01 (2H, m). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.4, 33.0, 48.7, 54.4, 55.0, 64.7, 128.6, 129.3, 129.8, 133.6, 166.4, 167.5, 167.9, 172.7. HR-MS (ESI) m/z: calcd for C26H30N2O4Na [M+Na]+ 457.2103. Found 457.2103.
4.3.17. (S)-N-(E)-4′-Methoxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamide (7d)
(5.8 mg, 12.9 μmol, 22%) as a white powder, mp 185–187 °C. [α]23 D +13° (c 0.10, CHCl3). IR (film) ν max cm−1: 3282, 2925, 2852, 2360, 2333, 1724, 1647, 1577, 1545, 1481. 1H NMR (400 MHz, CDCl3) δ: 1.02–1.90 (11H, m), 3.93 (3H, s), 4.60–4.73 (2H, m), 4.90–4.96 (1H, m), 6.03 (1H, d, J = 8.2 Hz), 6.28–6.35 (2H, m), 6.88 (1H, d, J = 8.7 Hz), 7.42–7.45 (3H, m), 7.56–7.61 (2H, m), 7.96–8.02 (2H, m). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.4, 33.0, 48.7, 54.4, 55.0, 64.7, 128.6, 129.3, 133.6, 166.4, 167.4, 167.9, 172.4, 172.6. HR-MS (ESI) m/z: calcd for C26H30N2O5Na [M+Na]+ 473.2052. Found 473.2099.
4.3.18. (S)-N-Benzoylserine(benzoyl)cyclohexylamide (7e)
(11.5 mg, 0.0292 mmol, 43%) as a white powder, mp 202–203 °C. [α]25 D +1.0° (c 0.10, MeOH). IR (film) ν max cm−1: 3292, 3072, 2924, 2852, 2359, 1734, 1649, 1635, 1558, 1541. 1H NMR (400 MHz, CDCl3) δ: 1.03–1.91 (11H, m), 3.70–4.81 (1H, m), 4.61–4.68 (1H, m), 4.73–4.81 (1H, m), 5.02 (1H, s), 6.50 (1H, s), 7.33–7.59 (6H, m), 7.81 (2H, d, J = 7.3 Hz), 8.01 (2H, d, J = 6.9 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 48.7, 53.2, 64.6, 127.3, 128.6, 128.8, 129.2, 129.9, 132.2, 133.5, 166.8, 167.6, 167.7. HR-MS (ESI) m/z: calcd for C23H25N2O4Na [M+Na]+ 417.1790. Found 417.1794.
4.3.19. (S)-N-(E)-2′-Methoxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamide (7f)
(16.6 mg, 36.9 μmol, 57%) (E/Z = 5:2) as a white powder, mp 201–204 °C. [α]25 D +11° (c 0.10, MeOH). IR (film) ν max cm−1: 3282, 3072, 2931, 2862, 1726, 1643, 1610, 1542, 1491, 1462. 1H NMR (400 MHz, CDCl3) δ: 1.04–1.94 (11H, m), 3.78 (1H, s), 3.87 (1H, s), 4.30 (0.33H, dd, J = 11.2, 5.2 Hz), 4.49 (0.33H, dd, J = 12.0, 5.6 Hz), 4.59 (0.67H, dd, J = 11.2, 5.2 Hz), 4.70 (0.67H, dd, J = 12.0, 6.4 Hz), 4.75–4.81 (0.17H, d, J = 16.0 Hz), 4.90–4.98 (0.83H, m), 6.03 (0.29H, d, J = 12.4 Hz), 6.60 (0.71H, d, J = 16.0 Hz), 6.76 (0.23H, d, J = 8.8 Hz), 6.81 (0.23H, t, J = 7.2 Hz), 6.81 (0.23H, t, J = 7.2 Hz), 6.88–6.98 (0.42H, m), 7.02 (0.15H, d, J = 12.4 Hz), 7.15–7.20 (0.21H, m), 7.32 (0.77H, t, J = 8.8 Hz), 7.39–7.48 (2.58 H, m), 7.56 (0.79H, t, J = 7.2 Hz), 7.86–7.92 (1.79H, m), 8.02 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 48.7, 52.8, 55.9, 56.0, 64.6, 109.7, 110.6, 111.1, 117.8, 121.0, 122.3, 123.2, 127.5, 128.5, 129.6, 129.7, 129.8, 133.4, 139.1, 142.0, 149.2, 150.9, 166.4, 166.6, 167.9. HR-MS (ESI) m/z: calcd for C26H30N2O5Na [M+Na]+ 473.2052. Found 473.2051.
4.3.20. (S)-N-(E)-3′,4′-Dimethoxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamide (7g)
(E:Z = 3:1) (9.3 mg, 19.4 μmol, 37%) as a white powder, mp 199–202 °C. [α]25 D +18° (c 0.10, MeOH). IR (film) ν max cm−1: 3282, 3080, 3022, 2931, 2846, 1728, 1643, 1614, 1516, 1452. 1H NMR (400 MHz, CDCl3) δ: 1.09–1.94 (11H, m), 3.66–3.81 (1H, m), 3.89 (6H, s), 4.46 (0.25H, dd, J = 12.0, 5.2 Hz), 4.56–4.63 (1H, m), 4.71 (0.75H, dd, J = 11.2, 6.0 Hz), 4.81–4.87 (0.25H, m), 4.93 (0.75H, dd, J = 12.8, 6.0 Hz), 5.87 (0.25H, d, J = 8.0 Hz), 6.07 (0.25H, d, J = 9.6 Hz), 6.26 (0.75H, d, J = 8.0 Hz), 6.34 (0.75H, d, J = 16.0 Hz), 6.53–6.59 (0.25H, m), 6.65 (0.75H, d, J = 8.8 Hz), 6.70–6.77 (0.5H, m), 6.84 (0.75H, d, J = 8.0 Hz), 7.02 (0.75H, s), 7.07 (1H, d, J = 8.0 Hz), 7.30 (0.25H, s), 7.43 (2H, t, J = 8.0 Hz), 7.53–7.61 (1.5H, m), 7.93 (0.5H, d, J = 8.0 Hz), 8.02 (1.5H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 48.7, 52.8, 55.9, 56.0, 64.6, 109.7, 111.1, 121.1, 122.3, 123.2, 127.6, 128.5, 129.6, 129.7, 129.8, 133.4, 142.0, 149.2, 150.9, 166.4, 166.6, 167.9. HR-MS (ESI) m/z: calcd for C27H32N2O6Na [M+Na]+ 503.2158. Found 503.2150.
4.3.21. (S)-N-(E)-3′,5′-Dimethoxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamide (7h)
(12.9 mg, 0.0194 mmol, 19%) as a white powder, mp 175–177 °C. [α]23 D +3.0° (c 0.10, MeOH). IR (film) ν max cm−1: 3396, 3276, 2924, 2846, 2360, 2333, 1726, 1645, 1614, 1558; 1H NMR (400 MHz, CDCl3) δ: 1.08–1.91 (11H, m), 3.96 (6H, s), 4.59 (1H, dd, J = 11.3, 6.4 Hz), 4.69 (1H, dd, J = 11.3, 6.0 Hz), 4.95–4.99 (1H, m), 6.43–6.47 (2H, m), 6.62 (2H, s), 6.80 (1H, d, J = 7.7 Hz), 7.42 (2H, t, J = 8.0 Hz), 7.52–7.57 (2H, m), 8.01 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 33.0, 48.7, 52.8, 55.5, 64.6, 102.3, 105.9, 120.5, 128.6, 129.5, 129.9, 133.5, 136.4, 142.2, 161.0, 166.0, 166.6, 167.7. HR-MS (ESI) m/z: calcd for C27H32N2O6Na [M+Na]+ 503.2158 Found 503.2170. ent-7h: mp 174–175 °C. [α]28 D −2.4° (c 0.25, MeOH). HR-MS (ESI) m/z: calcd for C27H33N2O6 [M+H]+ 481.2338. Found 481.2361.
4.3.22. (S)-N-(E)-3′-Methoxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamide (7i)
(16.6 mg, 0.0369 mmol, 57%) as a white powder, mp 192–194 °C. [α]27 D +1.8°(c 0.10, MeOH). IR (film) ν max cm−1: 3438, 3282, 2933, 2854, 2360, 2326, 1722, 1647, 1558, 1541. 1H NMR (400 MHz, CDCl3) δ: 1.06–1.96 (1H, m), 3.82 (3H, s), 4.56–4.62 (1H, m), 4.70 (1H, dd, J = 12.0, 6.0 Hz), 4.88–4.94 (1H, m), 6.17 (1H, s), 6.45 (1H, d, J = 16.0 Hz), 6.68 (1H, d, J = 7.2 Hz), 6.89–6.93 (1H, m), 7.00–7.02 (1H, m), 7.09 (1H, d, J = 8.0 Hz), 7.43 (2H, t, J = 8.0 Hz), 7.54–7.64 (2H, m), 8.02 (2H, d, J = 7.2 Hz). 13C NMR (100 MHz, CDCl3) δ: 15.4, 24.8, 25.5, 32.9, 48.7, 52.8, 55.3, 64.6, 65.9, 113.0, 115.9, 120.4, 120.6, 128.5, 129.6, 129.8, 129.9, 133.4, 136.0, 142.0, 160.0, 166.1, 166.5, 167.9; HR-MS (ESI) m/z: calcd for C26H30N2O5Na [M+Na]+ 473.2052. Found 473.2046. ent-7i: mp 194–196 °C. [α]28 D −2.4° (c 0.25, MeOH). HR-MS (ESI) m/z: found 473.2060.
4.3.23. (S)-N-(E)-4′-Benzoyloxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamine (7j)
(17.0 mg, 0.0314 mmol, 52%) as a white powder, mp 220–222 °C. [α]28 D −1.0° (c 0.40, CHCl3). IR (film) ν max cm−1: 3284, 2929, 2854, 2355, 2326, 1734, 1645, 1616, 1556, 1541. 1H NMR (400 MHz, CDCl3) δ: 1.08–1.93 (11H, m), 3.72–3.73 (1H, m), 3.93 (1H, s), 4.57–4.62 (1H, m), 4.69–4.73 (1H, m), 4.90–4.96 (1H, m), 6.21 (1H, d, J = 8.8 Hz), 6.45 (1H, d, J = 15.6 Hz), 6.73 (1H, d, J = 7.3 Hz), 7.43 (2H, t, J = 8.0 Hz), 7.49–5.59 (6H, m), 7.63–7.67 (2H, m), 8.02 (2H, d, J = 8.0 Hz), 8.19 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 48.7, 52.8, 64.8, 120.1, 122.4, 128.6, 128.7, 129.2, 129.9, 130.3, 132.5, 133.5, 133.9, 141.2, 152.2, 165.0, 166.0, 166.6, 167.8, 176.2. HR-MS (ESI) m/z: calcd for C32H32N2O6Na [M+Na]+ 563.2158. Found 563.2171. ent-7j: mp 238–240 °C. [α]28 D +3.0° (c 0.13, CHCl3). HR-MS (ESI) m/z: calcd for C32H33N2O6 [M+H]+ 541.2338. Found 541.2307.
4.3.24. (S)-N-(E)-3′,4′,5′-Trimethoxy-3-phenylprop-2-enoicserine(benzoyl)cyclohexylamide (7k)
(22.0 mg, 0.0431 mmol, 81%) as a white powder, mp 222–223 °C. [α]26 D +13° (c 0.10, MeOH). IR (film) ν max cm−1: 3583, 3290, 2931, 2854, 2355, 1722, 1651, 1583, 1506, 1271. 1H NMR (400 MHz, CDCl3) δ: 1.05–1.92 (11H, m), 3,84 (9H, s), 4.60 (1H, dd, J = 11.2, 5.2 Hz), 4.69 (1H, dd, J = 11.2, 6.0 Hz), 4.99–5.01 (1H, m), 6.41 (1H, d, J = 16.0 Hz), 6.65 (1H, d, J = 8.0 Hz), 6.69 (2H, s), 6.92 (1H, d, J = 8.0 Hz), 7.40 (2H, t, J = 8.0 Hz), 7.48–7.57 (2H, m), 8.01 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 24.8, 25.5, 32.9, 48.7, 52.7, 56.2, 61.0, 64.7, 105.1, 119.4, 128.5, 129.8, 130.1, 133.4, 142.1, 153.5, 166.0, 166.5, 167.8. HR-MS (ESI) m/z: calcd for C28H34N2O7Na [M+Na]+ 533.2264. Found 533.2269. ent-7k: mp 211–213 °C. [α]27 D −12° (c 0.15, MeOH). HR-MS (ESI) m/z: found 533.2274.
4.3.25. (S)-N-Cinnamoylserine(benzoyl)piperidineamide (7l)
(19.5 mg, 49.9 μmol, 75%) as an oil. [α]26 D −2.5° (c 0.40, MeOH). IR (film) ν max cm−1: 3288, 3060, 3025, 3006, 2941, 2857, 2360, 2331, 1722, 1616. 1H NMR (400 MHz, CDCl3) δ; 1.52–1.74 (6H, m), 3.47–3.71 (4H, m), 4.46 (1H, dd, J = 12.0, 5.6 Hz), 4.58 (1H, dd, J = 12.0, 4.0 Hz), 5.46–5.52 (1H, m), 6.48 (1H, d, J = 16.0 Hz), 6.92 (1H, d, J = 8.0 Hz), 7.32–7.57 (7H, m), 7.60 (1H, d, J = 16.0 Hz), 8.00 (2H, d, J = 6.8 Hz). 13C NMR (100 MHz, CDCl3) δ: 14.3, 24.5, 25.6, 26.5, 43.7, 46.9, 48.7, 60.5, 64.7, 120.4, 128.0, 128.5, 128.9, 129.7, 129.9, 133.3, 141.7, 165.4, 166.8. HR-MS (ESI) m/z: calcd for C24H26N2O4Na [M+Na]+ 429.1793. Found 429.1782. ent-7l: [α]26 D +1.6° (c 0.15, MeOH). HR-MS (ESI) m/z: found 429.1770.
4.3.26. (S)-N-Cinnamoylserine(benzoyl)morpholineamide (7m)
(13.0 mg, 0.0318 mmol, 31%) as an oil. [α]25 D +43° (c 0.11, CHCl3). IR (film) ν max cm−1: 3649, 3583, 2362, 1718, 1558, 1541, 1508, 1473, 1419, 1313. 1H NMR (400 MHz, CDCl3) δ: 3.65–3.78 (8H, m), 4.49 (1H, dd, J = 12.0, 6.4 Hz), 4.58 (1H, dd, J = 11.6, 4.4 Hz), 5.44–5.52 (1H, m), 6.48 (1H, d, J = 16.0 Hz), 6.81 (1H, d, J = 8.0 Hz), 7.32–7.58 (5H, m), 7.61 (1H, d, J = 16.0 Hz), 8.00 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 42.8, 46.3, 48.5, 64.5, 66.7, 120.1, 128.0, 128.6, 128.9, 129.9, 133.4, 134.7, 142.0, 165.5, 167.5. HR-MS (ESI) m/z: calcd for C23H24N2O5Na [M+Na]+ 431.1583. Found 431.1557. ent-7m: [α]28 D −31° (c 0.20, CHCl3). HR-MS (ESI) m/z: found 431.1569.
4.3.27. (S)-N-Cinnamoylserine(benzoyl)benzylamide (7n)
(11.4 mg, 0.0292 mmol, 44%) as a white powder, mp 207–208 °C. [α]27 D +26° (c 0.10, MeOH). IR (film) ν max cm−1: 3585, 3286, 2360, 1728, 1647, 1624, 1540, 1450, 1394, 1338. 1H NMR (400 MHz, CDCl3) δ: 4.47 (2H, d, J = 6.0 Hz), 4.64 (1H, dd, J = 11.6, 6.0 Hz), 6.45 (1H, d, J = 16.0 Hz), 6.74 (2H, d, J = 6.8 Hz), 7.22 (4H, s), 7.34–7.44 (5H, m), 7.46–7.50 (2H, m), 7.62 (1H, d, J = 16.0 Hz), 7.97 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 43.8, 53.0, 64.5, 119.7, 127.7, 128.1, 128.6, 128.8, 128.9, 129.9, 130.1, 133.5, 142.5, 166.3, 168.8. HR-MS (ESI) m/z: calcd for C26H24N2O4Na [M+Na]+ 451.1634. Found 451.1603. ent-7n: mp 207–209 °C. [α]23 D −25° (c 0.11, MeOH). HR-MS (ESI) m/z: found 451.1594.
4.3.28. (S)-N-Cinnamoylserine(benzoyl)cyclohexanemethylamide (7o)
(15.0 mg, 0.0345 mmol, 57%) as a white powder, mp 208 °C. [α]27 D +27° (c 0.10, MeOH). IR (film) ν max cm−1: 3583, 3290, 2918, 2852, 2360, 2341, 1730, 1647, 1622, 1545. 1H NMR (400 MHz, CDCl3) δ: 0.81–0.94 (2H, m), 1.03–1.18 (3H, m), 1.36–1.48 (1H, m), 1.55–1.69 (6H, m), 3.05–3.21 (2H, m), 4.62 (1H, dd, J = 12.0, 4.8 Hz), 4.73 (1H, dd, J = 11.2, 6.0 Hz), 4.97–5.04 (1H, m), 6.48 (1H, d, J = 16.0 Hz), 7.33–7.38 (3H, m), 7.42 (2H, t, J = 8.0 Hz), 7.46–7.52 (2H, m), 7.55 (1H, t, J = 8.0 Hz), 7.63 (1H, d, J = 16.0 Hz), 8.01 (2H, d, J = 8.0 Hz). 13C NMR (100 MHz, CDCl3) δ: 25.8, 26.3, 30.8, 37.9, 46.0, 53.0, 64.6, 119.8, 128.1, 128.6, 129.0, 130.1, 133.5, 134.5, 142.4, 166.2, 166.8, 168.8. HR-MS (ESI) m/z: calcd for C26H30N2O4Na [M+Na]+ 457.2103. Found 457.2153. ent-7o: mp 202–203 °C. [α]27 D −27° (c 0.15, MeOH). HR-MS (ESI) m/z: found 457.2052.
4.4. Measurement of inhibitory activity against SARS 3CL R188I mutant protease
Enzyme assays were carried out using recombinant SARS 3CL R188I mutant protease at an enzyme concentration of 120 nM. The reaction mixture (40 mM AcONa buffer, pH 4.0, containing 10% grecerol, 10 mM DTT and 4 M NaCl) was analyzed on a Cosmosil 5C18-AR-II column (4.6 × 150 mm), employing a linear gradient of MeCN (10–40%, 30 min) in aq 0.1% TFA. Each IC50 value was obtained from a sigmoidal dose–response curve obtained from the decrease of the substrate in the reaction mixture. Each experiment was repeated three times.
4.5. Measurement of inhibitory activity against cathesin B
Bovine spleen cathepsin B was purchased from Sigma Chemical company (St. Louis, MO). 10 mU of the cathepsin B was preincubated at 40 °C for 10 min in 90 μl of 50 mM MES (pH 6.0), 2 mM DTT, and 0.1% Brij-35. The solution was then mixed with 20 mM Z-Arg-Arg-MCA and each inhibitor (1 nM to 1 mM) in DMSO (10 μl), and the mixture was incubated at 40 °C for 10 min. The reaction was stopped by adding 100 μl of 100 mM sodium monochloroacetate, 30 mM sodium acetate, and 70 mM acetic acid (pH 4.3). Each IC50 value was obtained from a sigmoidal dose–response curve obtained from the decrease of the substrate in the reaction mixture. Each experiment was repeated three times.
4.6. Evaluation of the cytotoxicity by MTT assay
HeLa cells were maintained in a suspension culture of DMEM supplemented with 5% FBS (Fetal Bovine Serum) containing 1% of a penicillin–streptomycin mixture. A 100 μl aliquot of HeLa cells (5000 cells/ml) was added to a 96 well plate and incubated for 24 h at 37 °C in a humidified incubator containing 5% CO2 in air. After 24 h, a 10 μl aliquot of compound serine derivatives (concentrations varying in the range of 5, 4, 3, 2, 1, 0.1 μM) and rotenone as a control was added to each of the 96 wells and incubated for 24 h. A 10 μl WST-8 solution (mixture of WST-8 and 1-Methoxy PMS) was added to each well and the incubation continued for 2 h. The visible absorbance at 450 nm and 630 nm as the reference wavelength of each well was quantified using a microplate reader.
Acknowledgement
This work was supported in part by Adaptable and Seamless Technology transfer Program (A-STEP) through target driven R&D, Japan Science and Technology Agency.
Footnotes
Supplementary data (additional experimental procedures, analytical data for library compounds) associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2016.01.052.
Supplementary data
Additional experimental procedures, analytical data for library compounds.
References and notes
- 1.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F. N. Engl. J. Med. 2003;348:1986. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., Ven der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolensnikova L., Fouchier R.A.M., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D.E.E., Schemitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin R.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh G.J., Guarner J.Y., Paddock C.D., Rota P., Fields B., DeRisi L.J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellinim W.J., Anderson J.W. N. Engl. J. Med. 2003;348:1953. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Pyrc K., Berkhout B., Van der Hoek L. J. Virol. 2007;81:3051. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacBride R., Fielding B.C. Future Microbiol. 2011;6:153. doi: 10.2217/fmb.10.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui L.-J., Zhang C., Zhang T., Lu R.-J., Xie Z.-D., Zhang L.-L., Liu C.-Y., Zhou W.-M., Ma X.-J., Tan W.-J. Adv. Virol. 2011 doi: 10.1155/2011/129134. No. 129134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. N. Engl. J. Med. 1814;2012:367. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Bermingham A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopai R., Newsholme W., Drosten C., Fouchier R.A., Zambon M. Euro. Surveill. 2012;17:20290. [PubMed] [Google Scholar]
- 9.Van Boheemen S., Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D.M., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A.M. MBio. 2012;3 doi: 10.1128/mBio.00473-12. e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weiβbrich B., Snijider E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J.J. J. Gen. Virol. 2003;84:2305. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 11.Thanigaimaalai P., Konno S., Yamamoto T., Koiwai Y., Taguchi A., Takayama K., Yakushiji F., Akaji K., Kiso Y., Kawasaki Y., Chen S.-E., Naser-Tavakolian A., Scheön A., Freire E., Hayashi Y. Eur. J. Med. Chem. 2013;65:436. doi: 10.1016/j.ejmech.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen T.T.H., Woo H.-J., Kang H.-K., Nguyen V.D., Kim Y.-M., Kim D.-W., Ahn S.-A., Xia Y., Kim D. Biotechnol. Lett. 2012;34:831. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J.-Y., Kim J.H., Kwon J.M., Kwon H.-J., Jeong H.J., Kim Y.M., Kim D., Lee W.S., Ryu Y.B. Bioorg. Med. Chem. 2013;21:3730. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.C., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. J. Med. Chem. 2013;56:534. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W., Zhu H.-M., Niu G.-J., Shi E.-Z., Chen J., Sun B., Chen W.-Q., Zhou H.-G., Yang C. Bioorg. Med. Chem. 2014;22:292. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Bioorg. Med. Chem. 2015;23:876. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akaji K., Konno H., Onozuka M., Makino A., Nosaka K. Bioorg. Med. Chem. 2008;16:9400. doi: 10.1016/j.bmc.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akaji K., Konno H., Mitsui H., Teruya K., Shimamoto Y., Hattori Y., Ozaki T., Kusunoki M., Sanjoh A. J. Med. Chem. 2011;54:7962. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 19.Konno H., Sema Y., Ishii M., Hattori Y., Nosak K., Akaji K. Tetrahedron Lett. 2013;54:4848. doi: 10.1016/j.tetlet.2013.06.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konno H., Sema Y., Tokairin Y. Tetrahedron. 2015;71:3433. [Google Scholar]
- 21.Bai D., Yang Q., Chem L., He X., Gao Z., Shen X. Chem. Pharm. Bull. 2008;56:1400. doi: 10.1248/cpb.56.1400. [DOI] [PubMed] [Google Scholar]
- 22.Carpino L.A. J. Am. Chem. Soc. 1993;115:4397. [Google Scholar]
- 23.Carpino L.A., Imazumi H., El-Faham A., Ferrer F.J., Zhang C., Lee Y., Foxman B.M., Henklein P., Hanay C., Mugge C., Wenschuh H., Klose J., Beyerman M., Bienert M. Angew. Chem., Int. Ed. 2002;41:442. doi: 10.1002/1521-3773(20020201)41:3<441::aid-anie441>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 24.Kunishima M., Wamachi C., Hioki K., Terao K., Tani S. Tetrahedron. 2001;57:1551. [Google Scholar]
- 25.Shiina I. Bull. Chem. Soc. Jpn. 2014;87:196. [Google Scholar]
- 26.Inanaga J., Hirata K., Saeki H., Katsuki Y., Yamaguchi M. Bull. Chem. Soc. Jpn. 1979;52:1989. [Google Scholar]
- 27.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. J. Med. Chem. 2013;56:534. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konno H., Nosaka K., Akaji K. Tetrahedron. 2011;67:9067. [Google Scholar]
- 29.Konno K., Kubo K., Makabe H., Toshiro E., Nosaka K., Akaji K. Tetrahedron. 2007;63:9502. [Google Scholar]
- 30.Hiwasa T., Fujita-Yoshigaki J., Shirouzu M., Koide H., Sawada T., Sakiyama S., Yokoyama S. Cancer Lett. 1993;69:161. doi: 10.1016/0304-3835(93)90169-a. [DOI] [PubMed] [Google Scholar]
- 31.Barrett A.J., Kirschke H. Method Enzymol. 1981;380:535. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- 32.Lim I.T., Meroueh S.O., Lee M., Heeg M.J., Mobashery S. J. Am. Chem. Soc. 2004;126:10271. doi: 10.1021/ja0489240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional experimental procedures, analytical data for library compounds.