Abstract
New discoveries in Glioblastoma (GBM) biology have been made using genomics data. Genomic markers are routinely integrated into clinical neurosurgical practice. In this manuscript, we review the fundamentals of genomics such as the differences between first, second, and third generation sequencing technology. We also review the impact of single cell genomics in understanding the complex heterogenous GBM microenvironment. Finally, we will discuss advances in epigenetics that have lent insights into treatment resistance. The integration of genomics into neuro-oncology clinical practice is routine and will continue to expand with the expansion of precision of medicine. We provide a primer for clinicians.
Keywords: epigenetics, glioblastoma, genomics, microenvironment, Single-Cell Analysis, tumor microenvironment
Introduction
Glioblastoma Multiforme (GBM) remains the most common malignant primary brain tumor among adults and still portends a median survival of 15 months following standard therapy known as the Stupp protocol (i.e. surgery followed by radiation and temozolomide treatment). Despite over a decade of advances in cancer biology and corresponding clinical trials, little changes have been made in standard therapy for GBM since 20051,2. While glioblastoma remains one of the most malignant tumors of the body, likely due to a wide range of inter- and intratumor genetic heterogeneity contributing to treatment resistance, advances in technology have allowed for better understanding of the disease and classification.
In the early 2000’s, bioinformatics began developing as a field that uses computational methods to analyze data from genomic sequencing, proteomics, and systems biology3. Many researchers began to use bioinformatic techniques to understand the complexities of cancer biology including GBM. This use of integrated genotypic and phenotypic characteristics was one of the major updates to from the 2007 WHO CNS classifications to the 2016, updated 4th edition4. With this update, IDH status became key in the classification of GBM. IDH-wildtype represents the vast majority of cases clinically defined as primary/de novo glioblastoma, often occurring in patients later in life. IDH-mutant corresponds with “secondary glioblastoma”, patients with a history of prior low grade diffuse glioma, which arises earlier in life5.
Advances in genetic sequencing have also provided new insights into understanding GBM biology and the tumor microenvironment. These insights will hopefully lead to improvement in treatment, but there remains a chasm between clinicians and the practical utilization of the ever-growing field of genomics. In this review, we discuss the currently used genetic predictive markers in GBM and provide a primer in understanding genomics for clinicians. As the cost of genome sequencing continues to drop, the amount of data created is quickly becoming ‘big data’, on the same level as astronomy and other large-scale physics data 6. A basic understanding of genomics will be necessary to navigate the future of neuro-oncology practice.
2. Current Clinically Used Molecular Markers
2.1. IDH Mutation Status
Molecular markers have become routine to use clinically for the evaluation and treatment of GBM. Mutations in the Isocitrate Dehydrogenase 1 or 2 (IDH1/2) gene have become an important molecular biomarker used clinically. In 2016, the world health organization’s summary on tumors of the central nervous system incorporated IDH mutational status into the classification of GBM 7. Somatic mutations in these genes were frequently identified in patients with secondary glioblastoma (i.e. tumors that evolved from low grade gliomas)5. IDH1/2 mutations were found to be present in over 70% of low grade gliomas and secondary glioblastomas and do portend a better prognosis 8,9. Furthermore, IDH mutant gliomas undergo chromosomal scattering to a much larger extent and have distinct oncogenic pathways in comparison to IDH wildtype (wt) tumors 10.
Work by Lu et. al. was instrumental in elucidating the mechanisms of IDH mutations11. The IDH genes encode the enzymes needed for decarboxylation of isocitrate to α-ketoglutarate. These mutations lead to the production of a oncometabolite, 2-hydroxyglutarate (2-HG), that can modify cellular processes, such as, inhibition of histone demethylation that blocks the differentiation of non-transformed cells11,12. Levels of 2-HG correlate with increased mitotic activity, axonal disruption and vascular neoplasia 13.
Clinically, IDH mutational status confers a better prognosis and has become incorporated into predictive prognostic nomograms for patients with primary GBM 14. Studies of IDH-wt tumors demonstrate that IDH1 upregulation supports aggressive growth and treatment resistance in GBM 15. IDH-wt patients with a mutation in TElomerase Reverse Transcriptase gene promoter (TERTp) have a poor prognosis 16. Among IDH-wt patients, extent of surgical resection and MGMT promoter methylation status can still be used to predict overall and progression free survival 17.
2.2. MGMT Promoter Methylation Status
Determination of the promoter methylation status of O(6)-Methylguanine-DNA Methyltransferase (MGMT) is another main prognostic indicator used by clinicians. Temozolomide, a DNA akylating agent, remains the primary chemotherapy in GBM and is used as part of standard treatment. Resistance to this chemotherapeutic can be mediated by the MGMT DNA repair enzyme and methylation of MGMT leads to silencing of this gene. MGMT methylated patients are therefore more responsive to the chemotherapy 18.
MGMT promoter methylation status has correlated with survival and treatment response in GBM patients. In the Nordic randomized clinical trial, which enrolled elderly patients with GBM, MGMT methylation status was a predictive marker to determine responsiveness to temozolomide treatment 19,20. Based on these results, some clinicians opt to forego temozolomide, however, there remains wide variability in practice paradigms globally 21. The evidence on this topic remains weak, therefore, use vs withholding of temozolomide in the elderly based on MGMT status alone is not recommended. Further consensus is needed to determine when temozolomide treatment is futile in the elderly popultion.
Further argument that MGMT methylation status should not be used as a sole guide for treatment and prognosis lies in the understanding of how the methylation status is assessed. Multiple methods exist for detection of MGMT methylation and cutoff values are also different among institutions. Immunohistochemistry is one method to assess methylation status, but there is concern for interobserver variability and how pathological processing can affect antigen specific binding 22. Methylation specific polymerase chain reaction (PCR) is the most common detection method, but optimal cutoff values remain controversial, and there is no standard value of methylation that correlates to a difference in clinical response. Furthermore, multiple trials have used different definitions for determining methylation status 23. Thus, it is important for clinicians to be aware of these limitations when deciding on the utility of temozolomide for certain patient populations. MGMT methylation status is not the only predictive biomarker and a comprehensive assessment is still needed.
2.3. Further Genetic Molecular Markers
Thus far, IDH mutant status and MGMT methylation are the main molecular markers that are used clinically to predict prognosis. However, clinicians should be aware of new genetic markers and subsets of GBM given their relevance to many ongoing clinical trials and their potential for further prediction of outcome. Epidermal Growth Factor Receptor (EGFR) is a tyrosine kinase receptor that is known to affect GBM development and progression. Amplification of the EGFR gene is present in up to 45% of GBMs. The variant III mutation of the epidermal growth factor receptor (EGFRvIII) is a neoantigen and can be used as a therapeutic target 24,25. EGFR expression has also been found to be a prognostic biomarker in some cohorts. A subgroup of patients with both unmethylated MGMT promoter status and high EGFR expression have the shortest survival in comparison to patients with other variations of these two markers 26
Mutations affecting the TERT promoter are also very common in GBM 27. Patients with a mutation in the TERT promoter had a shorter overall survival compared to TERT promoter-wt (13.8 months vs 18.4 months)28. In this cohort, when EGFR amplification or TERT promoter mutation was present, prognosis was poor (less than 1.5 years). For patients without these molecular markers, prognosis was much better especially for the IDH mutant patients (survival of more than 3 years) 28. These studies demonstrate that the best prognostication will be by combining the implications of multiple markers 29.
3. Implications of GBM Genomics
3.1. Next Generation Sequencing in GBM
All of these aforementioned genetic changes and markers in GBM have primarily been due to the advances in technology that have decreased cost and improved the speed of genetic sequencing techniques. With this new speed and relative affordability, analysis of large enough numbers of tissue samples to give statistically significant results is now possible.
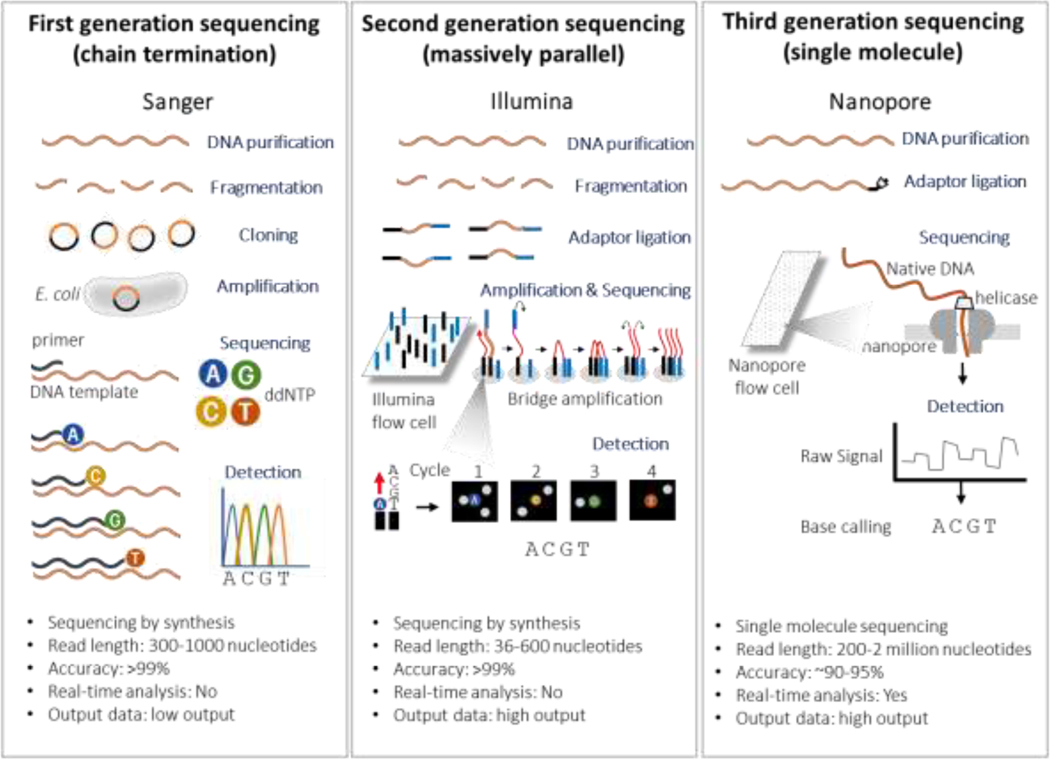
After the sequencing of the first proteins in the 1950s, critical refinement of cumbersome techniques led to the development of “first- generation” sequencing technology (figure 1) that allowed the first sequencing of the human genome in the 1970s30. This “first- generation” sequencing continued to be automated and improved. By 2005 (the same year as the development of the Stupp protocol for GBM treatment) a more high throughput system gave rise to “second-generation” also referred to as “next generation” sequencing (figure 1). This sequencing method is still the most commonly used technique in labs due to its availability, reliability, and high output. Developments in nanotechnology has ushered in third generation sequencing, which uses single molecule sequencing technology, without the need of DNA synthesis. While challenges still exist in implementing widespread use of third gen sequencing, it provides the hope of minimal pre-processing of samples and potential obtain direct detection of epigenetic markers 31,32 (Figure 1).
Figure 1.
Schematic Representation of first, second and third generation sequencing.
With this boom in available genetic data for analysis, GBM was the first cancer to be studied by the cancer genome atlas research network with the analysis of 206 samples in 2008. Genetic alterations identified in three main signaling pathways (RTK/RAS/PI-3K, p53, and RB) allowed for elucidation of gliomagenesis 33,34. Distinct pathways discovered are also involved in the development of supratentorial versus infratentorial GBM, implying that the pathogenesis of cerebellar GBM requires unique gliomagenesis pathways 35. Through collaborative efforts, many genomic sequences from GBM samples are now publicly available and can be analyzed using bioinformatic strategies. This collaborative analysis identified differential gene expression levels that were associated with GBM, in comparison to normal tissue samples, that were found to significantly affect prognosis 36. Further analyses of gene expression profiles from 1280 GBM patient samples found druggable genes (such as RRM2, MAPK9, and XIAP) that are related to carcinogenesis, giving new insight into the development of novel therapies37.
Advances in sequencing methodologies allowing sequencing on formalin fixed paraffin embedded (FFPE) tissue have made tumor genetics clinically important for all clinicians. This allows genetic sequencing to be part of the routine pathological analysis of tissue samples following surgical resection, as well as retrospective analysis on previously resected tumors. For clinicians, FFPE specimens can be sent to commercial vendors for genomic profiling, with information available in as little as five days 38,39. Typically, these vendors conduct an analysis on a panel of genes rather than the whole exome and summarize the findings for clinicians in order to match candidate patients to clinical trials. For some clinicians, sending genomic sequencing following surgical resection of de novo and recurrent tumors has become routine and is done prospectively 40,41. This allows for the identification of patients who may qualify for targeted therapy and/or clinical trials.
IDH mutational status is required for diagnosis and therefore is integrated into the pathology report of patients. At our institution, all glioblastoma patients also get tested for MGMT methylation and EGFR amplification. Since these biomarkers are integrated into the pathology workflow, they are often billed to and paid by insurance. Further genetic sequencing can be carried out by sending clinical samples to third-party companies. These sequencing data are typically filtered to provide a clinical report matching the patient to potential clinical trials. Medicare often covers these expenses for patients who qualify but private insurers may or may not pay for the extra sequencing costs. However many companies offer financial assistance.
3.2. Single Cell Sequencing and Tumor Immune Microenvironment
In genetic sequencing of spatially different regions of a GBM tumor, it was discovered that multiple subtypes were present within the same tumor 42. GBM is now known to be a highly heterogenous tumor, with multiple cell types, and a microenvironment with different regional niches of varying functions 43. Thus, a main hurdle in the understanding of GBM is this heterogeneity, as sequencing from a portion of the tumor bulk can lead to a biased understanding of the genetic cell lineages from different cell populations especially in GBM 44. In order to study genomics on a cellular level, single cell transcriptome analysis became possible in 2009 45,46. Now it is possible to understand the gene expression profiles from individual cells rather than a pool of cells. Single cell sequencing is therefore being used to understand subgroups within heterogenous tumors, making its application in GBM vital.
Single cell sequencing has already provided vital information to the pathophysiology of Glioblastoma. In one study, GBM samples demonstrated diverse ontogenic pathways and phenotype variability on the cellular level 47. Cells from the subventricular zone were also discovered that possess driver mutations, which guide the migration of these cells to other brain regions to develop into GBM 48 Infiltrating glial cancer cells at the migrating front of a GBM tumor also share gene signatures that give insight into mechanisms of glioma cell infiltration, a key pathological hallmark49. Transcriptionally distinct signatures were also present in myeloid populations residing in the tumor core and surrounding peritumoral space, highlighting the role of the tumor immune microenvironment49.
Thus, the glioma tumor microenvironment is diverse and composed of glioma cancer cells as well as stromal cells, multiple infiltrating inflammatory and immune cells, and stem cells 50,51. Of these, Tumor/Glioma Associated Macrophages/Microglia (GAMs) have been shown to account for up to 50% of the tumor cell mass52–54. The role of GAMs in the progression of GBM remains to be fully elucidated, but it is known that GAMs aid in glioma cancer cell proliferation, neovascularization, and invasion 55. GAMs and Myeloid Derived Suppressor Cells (MDSCs) also play a large role in directing immunosuppressive responses, which hinder efforts to employ immunotherapy as a treatment for GBM 56,57. Transcriptome analysis of human GAMs found expression signatures of upregulated genes necessary for cell migration, adhesion, and extracellular matrix organization 58. With this myeloid upregulation, studies have also demonstrated that GBM has a lymphocyte depleted immune landscape59.
Clinically, these insights into the GBM microenvironment provided by collaborative efforts between neurosurgeons and scientists are essential to integrate clinical trials and new therapies, especially immunotherapy, as the tumor immune microenvironment is one of the main hindrances to application of immunotherapy, such as CAR-T cell or NK therapy, in solid CNS tumors.
3.3. Glioblastoma Epigenetics
Epigenetics refers to the DNA modifications that affect gene activity without altering the DNA sequence itself. Two very popular and well-studied mechanisms for epigenetics are DNA methylation and post-translational modification of proteins (often histone modifications are studied, because these can have broad regulatory effects on gene expression levels). Epigenetic changes are integral for tumor progression and in mediating chemotherapy resistance 60. The most common example of this in GBM is related to the MGMT promoter methylation status which determines sensitivity or resistance to DNA alkylating agents as previously discussed in this review.
Recently, genome scale methylation maps of over one hundred GBM samples were constructed and linked to histopathological and radiographic data61. DNA methylation was linked to tumor immune cell infiltration and the microenvironment, and is largely preserved between primary and recurrent tumors 61. Further characterization of epigenetic changes in GBM have demonstrated that tumors with the CpG Island Methylator Phenotype (CIMP) have epigenomic aberrations that are related to pathogenicity62. How this phenotype is developed is not well understood, but IDH mutations are associated with widespread changes in the methylome which leads to activation of genetic processes62.
In cancer stem cells (also known as cancer progenitor cells), it has been demonstrated that epigenetic changes are critical in mediating differentiation and immortality of tumor cells 63. Key studies in GBM have shown cancer stem cells are resistant to radiation and chemotherapy and give rise to the cells that facilitate recurrence64,65. These epigenetic signatures found in cancer stem cells, which can even be unique among GBM subtypes, are related to important cellular pathways, such as DNA damage responses66,67.
Currently, epigenetic studies remain laborious and lack the feasibility to be carried out routinely, but one of the potential advantages to come of third generation sequencing, as previously discussed, is the possibility to obtain these methylation patterns routinely. Increased feasibility of epigenetic studies are important as epigenetic modulation may be a key to targeting treatment resistant glioma cell populations.
4. Conclusions
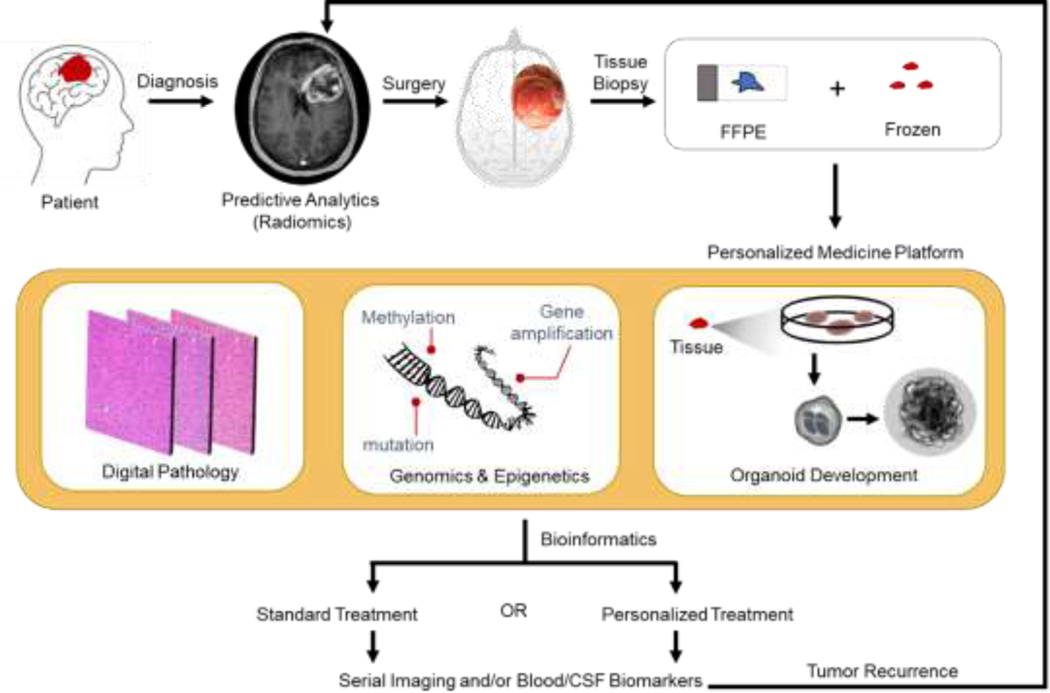
Genomics has been revolutionary in transforming the understanding of Glioblastoma biology. Bioinformatics and collaborative efforts between surgeons and scientists has been critical in leveraging the data and allowed for both the retrospective and prospective application of next generation sequencing. Molecular markers and mutational status has revolutionized the way we educate, plan, and treat patients with GBM. Advances, such as single cell sequencing, have furthered our understanding of the heterogeneity within GBM, and its complicated microenvironment. Studies in epigenetics and cancer stem cells have highlighted the importance these small molecular changes can make in determining tumor cell resilience and recurrence. These efforts are driving personalized treatment and prediction of survival and can be further be enhanced by the use of novel personalized treatment approaches. The use of patient derived organoids to assay for therapeutic sensitivity 68, and sequencing of circulating serum and cerebrospinal fluid biomarkers in GBM patients also provides the possibility of non-invasive disease course monitoring 69,70(Figure 2 outlines the likely neuro-oncology workflow of the future for patients with GBM).
Figure 2.
Schematic of the Future of Neuro-Oncology Practice following diagnosis of GBM. Patients will likely have preoperative screening with radiomics and from tumor tissue have an array of assays designed for personalized medicine. This process will be iterative as recurrence remains inevitable.
GBM remains a deadly tumor with minimal treatment options available. Higher order bioinformatic genetic processing of patient samples and integration with clinical parameters have allowed for novel insights into gliomagenesis. Beyond characterization and survival prediction, the next steps are to provide personalized therapies for patients.
Acknowledgments
Funding: Support was provided to A.R. by NIH grant P20 GM 109005O
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl_4):iv1-iv86. doi: 10.1093/neuonc/noy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Can T Introduction to Bioinformatics. In: Methods in Molecular Biology (Clifton, N.J.). Vol 1107 ; 2014:51–71. doi: 10.1007/978-1-62703-748-8_4 [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Perry A, Reifenberger Guido, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 5.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res.2013;19(4):764–772. doi: 10.1158/1078-0432.CCR-12-3002 [DOI] [PubMed] [Google Scholar]
- 6.Stephens ZD, Lee SY, Faghri F, et al. Big Data: Astronomical or Genomical? PLoS Biol. 2015;13(7):e1002195. doi: 10.1371/journal.pbio.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network, Brat DJ, Verhaak RGW, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen A, Sato M, Aldape K, et al. DNA copy number analysis of Grade II–III and Grade IV gliomas reveals differences in molecular ontogeny including chromothripsis associated with IDH mutation status. Acta Neuropathol Commun. 2015;3(1):34. doi: 10.1186/s40478-015-0213-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward PS, Patel J, Wise DR, et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting α-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jalbert LE, Elkhaled A, Phillips JJ, et al. Metabolic Profiling of IDH Mutation and Malignant Progression in Infiltrating Glioma. Sci Rep. 2017;7(1):44792. doi: 10.1038/srep44792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng W, Zhang C, Ren X, et al. Treatment strategy and IDH status improve nomogram validity in newly diagnosed GBM patients. Neuro Oncol. 2017;19(5):736–738. doi: 10.1093/neuonc/nox012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvert AE, Chalastanis A, Wu Y, et al. Cancer-Associated IDH1 Promotes Growth and Resistance to Targeted Therapies in the Absence of Mutation. Cell Rep. 2017;19(9):1858–1873. doi: 10.1016/j.celrep.2017.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Koh J, Kim S-I, et al. The frequency and prognostic effect of TERT promoter mutation in diffuse gliomas. Acta Neuropathol Commun. 2017;5(1):62. doi: 10.1186/s40478017-0465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gessler F, Bernstock JD, Braczynski A, et al. Surgery for Glioblastoma in Light of Molecular Markers: Impact of Resection and MGMT Promoter Methylation in Newly Diagnosed IDH-1 Wild-Type Glioblastomas. Neurosurgery. 2018;84(1):190–197. doi: 10.1093/neuros/nyy049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wick W, Weller M, van den Bent M, et al. MGMT testing--the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. doi: 10.1038/nrneurol.2014.100 [DOI] [PubMed] [Google Scholar]
- 19.Malmström A, Grønberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13(9):916–926. doi: 10.1016/S1470-2045(12)70265-6 [DOI] [PubMed] [Google Scholar]
- 20.Wick W, Platten M, Meisner C, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13(7):707–715. doi: 10.1016/S1470-2045(12)70164-X [DOI] [PubMed] [Google Scholar]
- 21.Palmer JD, Bhamidipati D, Mehta M, et al. Treatment recommendations for elderly patients with newly diagnosed glioblastoma lack worldwide consensus. J Neurooncol. 2018;140(2):421–426. doi: 10.1007/s11060-018-2969-3 [DOI] [PubMed] [Google Scholar]
- 22.Lipp ES, Healy P, Austin A, et al. MGMT: Immunohistochemical Detection in High-Grade Astrocytomas. J Neuropathol Exp Neurol. 2019;78(1):57–64. doi: 10.1093/jnen/nly110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansouri A, Hachem LD, Mansouri S, et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol. September 2018. doi: 10.1093/neuonc/noy132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hicks MJ, Chiuchiolo MJ, Ballon D, et al. Anti-Epidermal Growth Factor Receptor Gene Therapy for Glioblastoma. PLoS One. 2016;11(10):e0162978. doi: 10.1371/journal.pone.0162978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegi ME, Rajakannu P, Weller M. Epidermal growth factor receptor. Curr Opin Neurol. 2012;25(6):774–779. doi: 10.1097/WCO.0b013e328359b0bc [DOI] [PubMed] [Google Scholar]
- 26.Tini P, Pastina P, Nardone V, et al. The combined EGFR protein expression analysis refines the prognostic value of the MGMT promoter methylation status in glioblastoma. Clin Neurol Neurosurg. 2016;149:15–21. doi: 10.1016/j.clineuro.2016.07.023 [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Wu G, Shan Y, Hartmann C, von Deimling A, Xing M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle. 2013;12(10):1637–1638. doi: 10.4161/cc.24662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labussiere M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83(13):1200–1206. doi: 10.1212/WNL.0000000000000814 [DOI] [PubMed] [Google Scholar]
- 29.Stichel D, Ebrahimi A, Reuss D, et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018;136(5):793–803. doi: 10.1007/s00401-018-1905-0 [DOI] [PubMed] [Google Scholar]
- 30.Shendure J, Balasubramanian S, Church GM, et al. DNA sequencing at 40: past, present and future. Nature. 2017;550(7676):345–353. doi: 10.1038/nature24286 [DOI] [PubMed] [Google Scholar]
- 31.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38. doi: 10.1016/j.cell.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 33.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng K, Kim R, Kesari S, Carter B, Chen CC. Genomic profiling of glioblastoma: convergence of fundamental biologic tenets and novel insights. J Neurooncol. 2012;107(1):1–12. doi: 10.1007/s11060-011-0714-2 [DOI] [PubMed] [Google Scholar]
- 35.Cho HJ, Zhao J, Jung SW, et al. Distinct genomic profile and specific targeted drug responses in adult cerebellar glioblastoma. Neuro Oncol. 2019;21(1):47–58. doi: 10.1093/neuonc/noy123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang J, He D, Yang P, He J, Zhang Y. Genome-wide expression profiling of glioblastoma using a large combined cohort. Sci Rep. 2018;8(1):15104. doi: 10.1038/s41598-018-33323-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brahm CG, Walenkamp AME, Van Linde ME, Verheul HMW, Fehrmann RSN. Identification of novel therapeutic targets in glioblastoma with functional genomic mRNA profiling. J Clin Oncol. 2017;35(15_suppl):2018–2018. doi: 10.1200/JCO.2017.35.15_suppl.201828453411 [DOI] [Google Scholar]
- 38.Sahm F, Schrimpf D, Jones DTW, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131(6):903–910. doi: 10.1007/s00401-015-1519-8 [DOI] [PubMed] [Google Scholar]
- 39.Movassaghi M, Shabihkhani M, Hojat SA, et al. Early experience with formalin-fixed paraffin-embedded (FFPE) based commercial clinical genomic profiling of gliomas-robust and informative with caveats. Exp Mol Pathol. 2017;103(1):87–93. doi: 10.1016/j.yexmp.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 40.Neilsen BK, Sleightholm R, McComb R, et al. Comprehensive genetic alteration profiling in primary and recurrent glioblastoma. J Neurooncol. December 2018:1–8. doi: 10.1007/s11060-018-03070-2 [DOI] [PubMed] [Google Scholar]
- 41.Nørøxe DS, Skjøth-Rasmussen J, Brennum J, et al. GENE-50. GENOMIC PROFILING AND PRECISION MEDICINE IN GLIOBLASTOMA - A PROSPECTIVE STUDY. Neuro Oncol. 2017;19(suppl_6):vi103-vi103. doi: 10.1093/neuonc/nox168.422 [DOI] [Google Scholar]
- 42.Sottoriva A, Spiteri I, Piccirillo SGM, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hambardzumyan D, Bergers G. Glioblastoma: Defining Tumor Niches. Trends in cancer. 2015;1(4):252–265. doi: 10.1016/j.trecan.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alves JM, Prieto T, Posada D. Multiregional Tumor Trees Are Not Phylogenies. Trends in cancer. 2017;3(8):546–550. doi: 10.1016/j.trecan.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- 46.Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):96. doi: 10.1038/s12276-018-0071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH, Lee JE, Kahng JY, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560(7717):243–247. doi: 10.1038/s41586-018-0389-3 [DOI] [PubMed] [Google Scholar]
- 49.Darmanis S, Sloan SA, Croote D, et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017;21(5):1399–1410. doi: 10.1016/j.celrep.2017.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59(8):1169–1180. doi: 10.1002/glia.21136 [DOI] [PubMed] [Google Scholar]
- 51.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simmons GW, Pong WW, Emnett RJ, et al. Neurofibromatosis-1 heterozygosity increases microglia in a spatially and temporally restricted pattern relevant to mouse optic glioma formation and growth. J Neuropathol Exp Neurol. 2011;70(1):51–62. doi: 10.1097/NEN.0b013e3182032d37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutmann DH, McLellan MD, Hussain I, et al. Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res. 2013;23(3):431–439. doi: 10.1101/gr.142604.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi ML, Hughes JT, Esiri MM, Coakham HB, Brownell DB. Immunohistological study of mononuclear cell infiltrate in malignant gliomas. Acta Neuropathol. 1987;74(3):269–277. doi: 10.1007/bf00688191 [DOI] [PubMed] [Google Scholar]
- 55.Zhou W, Bao S. Reciprocal Supportive Interplay between Glioblastoma and Tumor-Associated Macrophages. Cancers (Basel). 2014;6(2):723–740. doi: 10.3390/cancers6020723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussain SF, Yang D, Suki D, Aldape K, Grimm E, Heimberger AB. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006;8(3):261–279. doi: 10.1215/15228517-2006-008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kamran N, Kadiyala P, Saxena M, et al. Immunosuppressive Myeloid Cells’ Blockade in the Glioma Microenvironment Enhances the Efficacy of Immune-Stimulatory Gene Therapy. Mol Ther. 2017;25(1):232–248. doi: 10.1016/j.ymthe.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szulzewsky F, Pelz A, Feng X, et al. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2):e0116644. doi: 10.1371/journal.pone.0116644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strauss J, Figg WD. Using epigenetic therapy to overcome chemotherapy resistance. Anticancer Res. 2016;36(1):1–4. [PMC free article] [PubMed] [Google Scholar]
- 61.Klughammer J, Kiesel B, Roetzer T, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med. 2018;24(10):1611–1624. doi: 10.1038/s41591-018-0156-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lapinska K, Faria G, McGonagle S, Macumber KM, Heerboth S, Sarkar S. Cancer Progenitor Cells: The Result of an Epigenetic Event? Anticancer Res. 2018;38(1):1–6. doi: 10.21873/anticanres.12184 [DOI] [PubMed] [Google Scholar]
- 64.Chen J, Li Y, Yu T-S, et al. A restricted cell population propagates glioblastoma growth following chemotherapy. Nature. 2012;488(7412):522. doi: 10.1038/NATURE11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 66.Zhou D, Alver BM, Li S, et al. Distinctive epigenomes characterize glioma stem cells and their response to differentiation cues. Genome Biol. 2018;19(1):43. doi: 10.1186/s13059018-1420-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pangeni RP, Zhang Z, Alvarez AA, et al. Genome-wide methylomic and transcriptomic analyses identify subtype-specific epigenetic signatures commonly dysregulated in glioma stem cells and glioblastoma. Epigenetics. 2018;13(4):432–448. doi: 10.1080/15592294.2018.1469892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aboulkheyr Es H, Montazeri L, Aref AR, Vosough M, Baharvand H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018;36(4):358–371. doi: 10.1016/j.tibtech.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 69.Ebrahimkhani S, Vafaee F, Hallal S, et al. Deep sequencing of circulating exosomal microRNA allows noninvasive glioblastoma diagnosis. NPJ Precis Oncol. 2018;2:28. doi: 10.1038/s41698-018-0071-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. doi: 10.1038/s41586-019-0882-3 [DOI] [PMC free article] [PubMed] [Google Scholar]