Abstract
The common forms of metabolic diseases are highly complex, involving hundreds of genes, environmental and lifestyle factors, age-related changes, sex differences and gut–microbiome interactions. Systems genetics is a population-based approach to address this complexity. In contrast to commonly used ‘reductionist’ approaches, such as gain or loss of function, that examine one element at a time, systems genetics uses high-throughput ‘omics’ technologies to quantitatively assess the many molecular differences among individuals in a population and then to relate these to physiologic functions or disease states. Unlike genome-wide association studies, systems genetics seeks to go beyond the identification of disease-causing genes to understand higher-order interactions at the molecular level. The purpose of this review is to introduce the systems genetics applications in the areas of metabolic and cardiovascular disease. Here, we explain how large clinical and omics-level data and databases from both human and animal populations are available to help researchers place genes in the context of pathways and networks and formulate hypotheses that can then be experimentally examined. We provide lists of such databases and examples of the integration of reductionist and systems genetics data. Among the important applications emerging is the development of improved nutritional and pharmacological strategies to address the rise of metabolic diseases.
Most diseases and other traits exhibit complex forms of inheritance resulting from the combined effects of multiple genetic variants together with environmental factors. The sequencing of the human genome enabled genome-wide association studies (GWAS) that have now identified more than 6,000 genomic loci for common diseases (Box 1). These studies have revealed that most common disorders, such as obesity, diabetes and heart disease, have a genetic architecture that is highly heterogeneous, involving small contributions from hundreds or thousands of genetic variants, each explaining a tiny fraction of the total genetic susceptibility.
Box 1 |. Glossary of terms used in this review.
Biological networks
Representations of patterns of interaction between biological elements, typically shown as graphs consisting of nodes (elements) and edges (connections). For example, a protein interaction network consists of the proteins, with edges between each interacting protein pair.
BXD recombinant inbred (RI) strain set
A mouse reference population consisting of a set of more than 100 RI strains derived from the parental strains C57BL/6J (B) and DBA/2J (D).
Diversity Outbred (DO) population
A highly genetically diverse population of outbred mice derived from eight parental inbred strains.
Epistasis
A non-additive interaction between two or more genetic variations.
Expression QTL (eQTL)
Genetic loci associated with transcript levels. eQTL that reside near the gene whose expression is regulated are termed ‘local’ or ‘cis’ eQTL. Those that are distal are termed ‘trans’ eQTL.
Genome-wide association study (GWAS)
An approach used for mapping the genes underlying complex traits. Typically, large numbers of individuals (thousands or more) are examined for the trait, for hundreds of thousands of SNPs spanning the genome. Significant associations between SNP alleles and the trait are identified with various statistical tests.
Hybrid Mouse Diversity Panel (HMDP)
A reference population consisting of approximately 100 classical inbred strains of mice.
Inbred strain
A strain derived by brother–sister matings from a species for many generations (typically more than 20). Each member of an inbred strain is homozygous across the genome, and each member is identical to all others of that strain.
Reductionist approach
An approach to understanding complex traits by reducing them to the interactions of their parts, such as the use of mice engineered for specific mutations.
Linkage disequilibrium
The non-random association of alleles of variants (such as SNPs) that typically occur at genetic loci in populations. Such association complicates the identification of causal variation and genes in GWAS loci.
Quantitative trait loci (QTL)
Genetic loci contributing to a quantitative trait.
Single-nucleotide polymorphism (SNP)
A genetic variation affecting a single nucleotide. SNPs are the most common variety of genetic variants and are used for high-density genotyping in GWAS.
Mediation analysis
A statistical method to examine the causal relationships of traits associated with the same genetic variant.
Gene-by-environment interaction (GxE)
An interaction in which the effect of an environmental factor depends on the genetic background.
Systems genetics, also termed integrative genetics, was developed to address such complexity. Systems genetics uses high-throughput omics technologies, such as DNA sequencing, RNA sequencing or mass-spectrometry-based metabolomics, to quantify molecular phenotypes alongside the clinical phenotypes in populations of humans or experimental organisms. The data can then be integrated through correlation, co-mapping or various modelling approaches to generate hypotheses relating the molecular and clinical traits. The underlying concept is that genetic variation affects complex clinical traits by perturbing molecular traits, such as gene expression or metabolite levels, and thus through measuring these traits as a function of genetic variation (that is, in individuals in a population), their relationships can be understood1.
Systems genetics studies with populations of model organisms, such as rats, mice, flies or yeast, have been particularly useful for examining the overall architecture of complex traits, including issues such as gene-by-gene (GxG) and gene-by-environment (GxE) interactions. Studies in model organisms have an important advantage in that environmental factors and other sources of heterogeneity can be controlled. In addition, studies of model organisms allow access to relevant tissues, which is generally not feasible in human studies. The genetic loci contributing to clinical or physiological traits in animal models are generally termed quantitative trait loci (QTL; Box 1). The loci contributing to molecular traits are similarly designated expression QTL (eQTL) for transcript levels, protein QTL (pQTL) for protein levels and so forth.
Systems genetics approaches have recently been reviewed2–4, and our focus in this Review is discussing how systems genetics data can be of use to researchers in the metabolism field. We first provide an overview of systems genetics, including examples relevant to metabolism research. In particular, we have attempted to illustrate how systems genetics data can be useful to reductionist researchers. We then discuss certain specialized aspects such as network modelling, non-additive interactions and therapeutic applications. We conclude with a summary and some thoughts on future applications. We note that, owing to space limitations, we have not included a historical perspective of the field, and we have omitted discussion of systems genetics in organisms such as yeast and flies.
Systems genetics applications in metabolism research
Comparison of reductionist and systems genetics approaches
Metabolism research is now dominated by reductionist (traditional) approaches, such as gain- or loss-of-function studies in mice. These approaches are powerful in that they establish causality, but they have some important limitations that hinder full understanding of the architecture of complex traits, as discussed below. In contrast, systems genetics studies must generally be combined with experimental studies to conclusively establish causality. Therefore, a combination of the two approaches is most powerful.
One constraint with the purely reductionist approach is that it usually involves perturbation of a single gene in a single genetic background and thus is unlikely to detect genetic interactions, such as modifier genes5. In other words, a genetic variation acts in the context of the genetic background, and by examining the effect of a gain or loss of function in only a single genetic background, an incomplete view of the function of the gene will be obtained. For example, engineered mutations in mice often exhibit strikingly different phenotypes when examined in different strains, as discussed below6. Another consideration with some reductionist approaches is that the perturbations are often unrealistically extreme (for example, complete knockout) and thus do not correspond to the more subtle variations observed for complex traits in nature. A complete knockout can have very broad phenotypic effects that may perturb genes far from the core functional genes (as in the ‘omnigenic’ model discussed below).
An important feature of systems genetics approaches is that they are relatively unbiased. Reductionist scientists usually generate hypotheses based on results from previous studies, and thus some genes or pathways are explored in great depth, whereas others are ignored. A recent study7 has found that more than one-quarter of coding genes have never been the subject of a single paper, and most other genes have been largely neglected, whereas approximately 2,000 genes (less than 10% of the coding genome) have hogged most of the attention. Systems genetics hypotheses, in contrast, are driven by natural variation paired with global measures of omics data and are therefore relatively unbiased.
The power of natural variation derives from the multitude of genetic perturbations that occur in all combinations in a population. Generally, a large fraction of genes and pathways are perturbed. For example, in human populations, the expression of nearly every gene is influenced by genetic variation8. Thus, in terms of generating hypotheses, systems genetic approaches offer fairly complete coverage of biologic processes and incorporate both perturbations and interactions of realistic effect sizes. In a sense, systems genetics studies can be viewed as a global mutagenesis screen9.
Systems genetics study design
A typical systems genetics study involves the following steps: (1) identification of an important question, or set of questions, that could be addressed with a systems genetics study; (2) selection of an appropriate population, on the basis of the trait of interest and the required statistical power; (3) phenotyping of the population for physiological, pathological and molecular traits of interest; (4) integration of the resulting data through statistical methods and genetic mapping; (5) formulation of hypotheses, such as potential causal relationships; and (6) experimental perturbations, generally in a single genetic background, to test the hypotheses2.
An overriding consideration in developing such studies is the cost. Animal studies generally require hundreds of individuals, and human studies may involve thousands. Detailed phenotyping is critical to the success of a study, and the generation of omics-level data is usually very expensive. Therefore, the study must crucially be designed to enable important questions to be addressed, and it should have a clear purpose rather than simply aiming to collect a large amount of data. The study should also be designed so that the resulting data might be useful to other researchers in the field.
Several animal ‘reference’ populations have been developed specifically for systems genetics studies (Box 1 and Table 1). These include panels of diverse rodent inbred strains, such as the Hybrid Mouse Diversity Panel (HMDP)10, the C57BL/6J × DBA/2J (BXD) panel of recombinant inbred (RI) mouse strains4,11,12 and the Collaborative Cross (CC)13, a set of RI mouse strains derived from an intercross of eight highly diverse inbred strains. The eight strains used in the CC were also used to develop an outbred population known as the Diversity Outbred (DO) panel14, which is maintained at The Jackson Laboratory. The genetic structure of these reference populations is illustrated in Fig. 1. Each resource has certain advantages and disadvantages. The DO is much more diverse than the others and comprises approximately eight times as many total SNPs. However, because the HMDP and BXD RI strains are inbred, replication and time-course studies are feasible. There are many additional resources that are proving useful for systems genetics studies, such as various outbred mouse populations15,16, advanced intercross lines17, heterogeneous stock rats18 and congenic strains19.
Table 1 |.
Systems genetics resources
| Category | Name | Description |
|---|---|---|
| Population-based resources | BXD genetic reference panel | Diverse mouse panel derived from intercrosses of C57BL6J and DBA strains; http://www.genenetwork.org/mouseCross.html |
| HMDP | Collection of ~100 classical and recombinant inbred strains; https://systems.genetics.ucla.edu/ | |
| Attie Lab Diabetes Database | Several mouse crosses, molecular layers and phenotypes associated with diabetes development; http://diabetes.wisc.edu/ | |
| DO panel | Outbred population of mice derived from eight founder strains; http://churchill-lab.jax.org/website/ | |
| GTEx Portal | Collection of WGS, WES and RNA-sequencing data from multiple post-mortem tissues of ~1,000 people; https://www.gtexportal.org/home/index.html | |
| STARNET | Collection of genomic, multi-tissue transcriptomic and proteomic data from ~1,300 patients with, and 300 verified controls without, coronary artery disease; https://doi.org/10.1126/science.aad6970 | |
| METSIM | Longitudinal study in ~10,000 Finnish men, for whom genomic, transcriptomic (subcutaneous adipose tissue) and comprehensive metabolic phenotypes are available; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000743.v1.p1 | |
| TwinsUK | Collection of phenotypic, genomic, environmental and several intermediate molecular phenotype samples from ~1,200 sibling pairs in the United Kingdom; https://twinsuk.ac.uk/ | |
| UK Biobank | Large collection of genotypic, health and disease parameters from ~500,000 people; https://www.ukbiobank.ac.uk/ | |
| MUTHER (Multiple Tissue Human Expression Resource) | Collection of multi-tissue expression profiles and genomic data from ~850 UK twins; http://muther.ac.uk/ | |
| Statistical/modelling resources | Mergeomics | Integrative analysis to merge multiscale data at the pathway level and identify network key drivers; http://mergeomics.research.idre.ucla.edu/ |
| WGCNA | Network construction to generate coregulated modules and overlay with other data (for example, networks of genes to traits); https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/ | |
| MEGENA | Network construction to generate coregulated modules; https://doi.org/10.1371/journal.pcbi.1004574 | |
| MOFA | Approach to infer relationships between scales exclusively by using quantitative measures; https://github.com/bioFAM/MOFA | |
| ARACNE | Approach to generate co-expression networks, focused on eliminating indirect interactions; https://doi.org/10.1186/1471–2105-7-S1-S7 | |
| Pathway-analysis Resources | NURSA (Nuclear Receptor Signaling Atlas) | Collection of published data related to nuclear-receptor function and specific pathways; https://nursa.org/nursa/index.jsf |
| GO enrichment analysis | Deposited sets of functional annotations for gene and proteins; http://geneontology.org/ | |
| KEGG | Deposited sets of cellular annotations for genes, including localization and function; https://www.genome.jp/kegg/ | |
| IPA (Ingenuity Pathway Analysis) | Comprehensive pathway-annotation software; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ | |
| Reactome | Interaction database to query genes or proteins; https://reactome.org/ | |
| BRENDA | Database of enzymatic pathways, including structure and kinetics; https://brenda-enzymes.org/ | |
| Additional Resources | BioGPS | Expression profiles from many tissues and cell lines from mouse or human data; http://biogps.org/#goto=welcome |
| Systems-Genetics.org | Systems Genetics toolkits to assess gene function and trait relationships. https://systems-genetics.org/ | |
| Gene Networks | Database to investigate gene functions and/or trait relationships; http://gn2.genenetwork.org/ | |
| UniProt | Collection of published terms (for example, pathways and localization) for individual genes and proteins across many organisms; https://www.uniprot.org/ | |
| Human Protein Atlas | Combined proteomic, transcriptional and cellular localization data across human tissues and cell types; http://proteinatlas.org/ | |
| HMDB (The Human Metabolome Database) | Collection of deposited data from human metabolomics studies; http://hmdb.ca/ | |
| CMAP | Compound prediction using gene expression data; http://clue.io/cmap/ | |
| MGI (Mouse Genome Informatics) | Integrated database for laboratory mouse strains and lines; http://informatics.jax.org/ | |
| IMPC (International Mouse Phenotyping Consortium) | Curated database of pheonotyping outcomes from mouse knockout lines for 20,000 genes; http://mousephenotype.org/ | |
| PredictDB Repository | Database of imputation tools to investigate molecular layers contributing to GWAS results; http://predictdb.hakyimlab.org/ | |
| STRING (search tool for recurring instances of neighbouring genes) | Collection of protein-protein interactions from published studies; http://string-db.org/ | |
| BioGRID (Biological General Repository for Interaction Datasets) | Interaction database to query genes or proteins; http://thebiogrid.org/ | |
Fig. 1 |. Integration across biologic scales assayed in three different rodent reference populations.
a, Flow of information. Layers representing molecular or clinical phenotypes are shown as rectangles. b–d, Example systems genetics studies using mouse reference populations; different colours represent the haplotypes in three widely used rodent systems genetics cohorts: DO, HMDP and BXD RI strains. The two copies of a typical chromosome are shown for four DO (b) four HMDP (c) and four BXD (d) RI strains. b, The DO mice were derived by intercrossing eight diverse inbred strains of mice for many generations and are maintained as an outbred stock (top). In the study shown, Chick and colleges measured the liver transcriptome and proteome in DO mice30. Association mapping was applied to identify cis-eQTLS and pQTLs, and mediation analysis was used to model different causal interactions between these layers. For example, loci could be identified that map to a transcript only (left) or a protein (Prot) only (middle), or that drive expression of both a transcript and its corresponding protein (right). c, The HMDP consists of approximately 100 ‘classic’ inbred strains of mice and RI strains derived from some of these (top). In this example, Parker and colleagues examined natural variation in proteome and lipidome structure. They identified proteins showing strong correlation to multiple lipid species and validated the protein PSMD9 as a novel driver of hepatic lipid metabolism33. d, The BXD RI set of strains was derived by intercrossing the parental strains C57BL/6J and DBA/2J and then inbreeding pairs of mice from the F2 generation (top). In the example, Williams and colleagues integrated genomic, transcriptomic, proteomic, metabolomic and clinical-trait data obtained from the livers of BXD mice fed chow or high-fat diets12. Using a combination of mapping and correlation, the authors identified novel mechanisms of regulation of the hepatic mitochondrial proteome (bottom).
Likewise, several human cohorts have been developed for the collection of broad clinical, metabolic and molecular phenotypes, such as the Metabolic Syndrome in Men cohort (METSIM)20, Twins UK21 and the Framingham Heart Study22–24. Some human studies, such as the Genotype-Tissue Expression (GTEx) project8 and the Stockholm–Tartu Atherosclerosis Reverse Networks Engineering Task (STARNET) study25, have generated global transcriptomic data from many human tissues. Primary, transformed and induced pluripotent cell lines derived from different individuals have also been used to integrate traits such as drug response and gene expression26–29.
Integration across biologic scales
Various biologic scales can be examined in a systems genetics study (illustration and examples in Fig. 1a). Among the scales, DNA variation is unique in that information only flows out, thus providing a causal anchor for modelling studies. Therefore, pairwise analysis of genetic variants such as single-nucleotide polymorphisms (SNPs) against intermediate molecular traits (such as those in the epigenome, spliceome, transcriptome, proteome and metabolome) can collectively reveal the information flow from DNA sequence to these additional layers (Fig. 1a). Information can also flow in reverse: just as metabolites, proteins and transcripts can affect the epigenome, so can proteins affect the transcriptome, and so on. Information also flows horizontally in a biologic scale; for example, proteins form complexes with each other. Biological processes occur as a cumulative result of interactions within and between layers, and molecular traits can be linked to physiologic and clinical traits.
There are three basic operations that can be used to integrate multi-omic data spanning multiple biologic scales, each of which is highlighted in Fig. 1b–d. The most straightforward is correlation. As information travels across scales, such as from transcript levels to protein levels, some degree of correlation would be expected in many cases. Correlation structure would also frequently be expected between a clinical trait and the genes that are either causal or reactive for that trait, although caution must be taken, because correlation can occur for many different reasons, including artefacts such as batch effects. Another important method, given sufficient power, is mapping traits from different scales (such as a clinical trait and a transcript-level trait) to the same genetic locus. Such co-mapping raises the possibility of one trait being causal for the second. A third approach for the integration of systems genetics data is statistical modelling, discussed below.
A nice illustration of how systems genetics approaches can dissect the flow of biological information is provided by some studies that have comprehensively examined the relationships between the levels of transcripts and the proteins that they encode as a function of genetic variation. These studies, using the HMDP and DO mouse populations (Fig. 1b and Box 1), have observed unexpectedly little correlation between transcript levels and protein levels, with an average correlation coefficient of approximately 0.3, and also correspondingly little overlap between eQTL and pQTL30,31. These discordances probably have several causes, as discussed in ref. 32. For example, protein turnover can be influenced by genetic variation, and thus some loci may affect protein levels (pQTL) but not transcript levels (eQTL) (Fig. 1b). Another important cause appears to be related to protein–protein interactions; for example, if several different proteins interact to form a complex, any excess subunits produced will probably be degraded. Recently, Parker and colleagues have extended such analyses and focused on correlation structures between protein levels and lipid levels, thus leading to the identification of some novel lipid-metabolism pathways33 (Fig. 1c).
Similarly, systems genetics can be used to connect molecular traits to physiologic functions. For example, in an elegant systems genetics study, Williams and colleagues12 have analysed the BXD mouse panel and revealed the regulation of mitochondria in the liver (Fig. 1d). The mice were subjected to normal chow or high-fat diets and then comprehensively characterized for global molecular phenotypes (transcriptome, proteome and metabolome) as well as clinical/physiologic traits (oral glucose tolerance and exercise regimen). The authors observed many co-regulated genes and proteins, whose integration with phenotypic traits highlighted a major role of mitochondrial function in mediating health status. Layering of molecular phenotypes on top of genetic associations allowed the authors to uncover genetic loci driving higher-order respiratory functions, such as the formation of electron-transport-chain supercomplexes in the liver. Recently, Jha and colleagues have used liver and plasma lipidomics from the same population to identify new lipid species and co-regulated modules34,35. By overlaying these data with phenotypic observations, the authors identified new lipid species and modules, thereby bridging previously studied molecular phenotypes (for example, proteomics) with mitochondrially mediated hepatic lipid metabolism. These observations were integrated with plasma lipidomics measures in the same mice to identify circulating biomarkers of lipid content in the liver, such as cardiolipin.
Systems genetics is also useful for the integration of genetic and environmental effects. Whereas GWAS results reflect only the heritable component of a trait, molecular and clinical phenotypes can capture both genetic and environmental factors36. The microbiome ‘scale’ is highly responsive to the environment and also strongly interconnected with the host metabolism37.
Integration through statistical modelling
According to the concept that information from DNA is unidirectional, causal pathways can be modelled, and whether certain ‘mediators’, such as transcript levels or chromatin marks, mediate the effect of DNA variation on a complex phenotype can be determined. For example, if both a clinical trait and the levels of a transcript are correlated and map to the same locus, researchers can condition on the transcript levels and ask whether a significant association between the locus and the clinical trait remains. If so, the results suggest that the effect on the clinical trait is not mediated by the transcript. Various causal inference tests have been developed and are typically referred to as mediation analysis38,39. Mendelian randomization is one form of mediation analysis that has especially strict criteria in that the mediator (such as the levels of a protein) is required to explain all of the association between a SNP and a complex trait. Whereas Mendelian randomization studies typically require quite large sample sizes (on the order of 100,000 individuals in humans), they have been particularly informative in dissecting causal influence between intermediate physiologic traits and pathophysiology. For example, mediation has been used to suggest that elevated plasma high-density-lipoprotein levels do not have a causal role in cardiovascular disease40.
Several statistical methods have been developed in recent years to facilitate multi-omics integration41,42. These methods can be broadly categorized into two types: those purely relying on data patterns across omics domains and those incorporating biological information. An example of the former category is Multi-omics Factor Analysis (MOFA)43, which uses dimension-reduction techniques to infer hidden factors reflecting biological and technical variability across multi-omics data types gathered from a study population. An example of the latter category is Mergeomics44,45, which builds on the explicit hypothesis that multi-omics modalities are functionally related and together can provide information on interconnected biological processes (discussed below).
An important advance in applying systems genetics to human populations has been the development of methods that integrate gene expression data with summary association statistics from GWAS to impute genes whose cis-regulated expression is associated with complex traits46. For example, Gusev and colleagues have used expression data from blood and adipose tissue to impute gene expression into large GWAS data; they have identified 69 genes significantly associated with obesity-related traits. Additional tools have been developed (http://predictdb.hakyimlab.org/) and compared47 to impute eQTL data onto larger GWAS datasets. Beyond eQTL data, imputation packages are being developed for protein and metabolite QTL measures. These packages also enable users to estimate variances explained by a given molecular layer to a trait of interest (https://github.com/hakyimlab/summary-gwas-imputation/).
Examples of the types of questions that can be asked by using available systems genetics data
Using systems genetics data is not just for geneticists or statisticians. Indeed, some of the most impactful science at present is performed by reductionist investigators using systems genetics data to generate hypotheses or support results. Crucially, systems genetics enables the formulation of unbiased hypotheses that can then be tested through experimental approaches. Below, we provide several examples of the types of questions that have been addressed with publicly available systems genetics data (Table 1).
Which gene at a GWAS locus is causal?
Identification of the causal gene underlying a GWAS locus can be challenging because of linkage disequilibrium (the correlation structure among genetic variants at the locus) and the ability of genetic variants to affect the expression of genes at distances up to hundreds of kilobases. In the absence of other evidence, the prime candidate is often assumed to be the one nearest to the peak SNP. One approach is to ask whether the lead SNP is associated with the expression of a gene at the locus, thus providing a potential mechanistic link. Variation in gene expression can be examined through technologies such as RNA sequencing if relevant tissue samples are available from some individuals in the population or can be imputed as discussed above48. In contrast to Mendelian disorders, in complex traits, genetic variation is most often regulatory, that is, involving enhancers or promoters rather than encoding protein49. In addition to testing for changes in gene expression, epigenetic- and chromosomal-interaction databases can be examined to identify regions that are likely to be enhancers or to bind specific transcription factors33,50. For example, Kessler and colleagues were interested in follow-up study of a GWAS locus for coronary artery disease that contained the candidate gene GUCY1A3, encoding a subunit of guanylyl cyclase51. They used previously published systems genetics data in both humans and mice to show that GUCY1A3 expression is regulated by the lead SNP (that is, that it constitutes a local eQTL). They also showed that the SNP is present in an enhancer region and affects the binding of the transcription factor ZED1. There are now many such examples of the use of systems genetics data to prioritize candidate genes at human, mouse and rat loci for metabolic and cardiovascular traits14,16,52,53.
What are the likely pathways contributing to a trait of interest?
Beyond suggesting candidate genes, systems genetics has also been used to uncover pathways underlying complex traits. For example, Kojima and colleagues have recently identified CD47 as a key regulator of efferocytosis in atherosclerotic lesions54. Given the complexity and heterogeneity of the disease, pinpointing the pathways underlying this efferocytotic signal would have been challenging through reductionist approaches. Therefore, the authors interrogated mouse (HMDP) and human (Biobank of Karolinska Endarterectomy (BiKE) study) atherosclerotic-plaque expression data, specifically looking for pathways enriched in genes correlated with CD47. This analysis suggested an inflammatory role of CD47, specifically in the expression of tumour necrosis factor, which was then validated experimentally.
Which tissue is likely to mediate the effects of genetic variation on disease susceptibility?
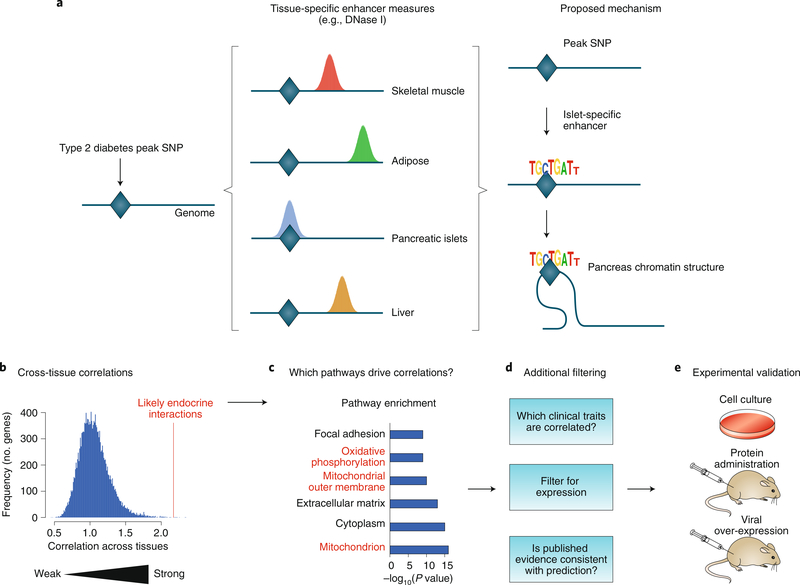
Because each cell type and tissue exhibits a specific set of regulatory elements and epigenetic modifications, thus resulting in differences in chromatin properties such as DNase I hypersensitivity, the locations of peak GWAS SNPs along the genome can provide information about the likely cell types and tissues in which the SNPs contribute to disease phenotypes. For example Mahajan and colleagues have generated a comprehensive dataset of GWAS loci contributing to type 2 diabetes and overlaid it with epigenomic data, thereby implicating pancreatic islets as a key regulatory tissue50. This study has identified several enhancers that are located within islets and may mechanistically link specific genetic variants to the progression of type 2 diabetes (Fig. 2a). A new nonparametric visualization tool has recently been reported in which users can upload GWAS SNPs and view cell-type-specific enrichment of chromatin marks available from ENCODE data55.
Fig. 2 |. Analysis of tissue-specific regulation and tissue–tissue cross-talk by using systems genetics.
a, Functional enrichment of human type 2 diabetes GWAS loci. GWAS loci associating with type 2 diabetes were identified (left) and overlaid with multiple open chromatin marks, such as DNase I hypersensitivity in four metabolically relevant tissues50. The authors observed notable overlap between the diabetes SNPs and chromatin marks specific for pancreatic islets (middle). Potential mechanisms driving type 2 diabetes were identified by focusing on known regulatory functions of islet-specific enhancer regions (right). b–e, Identification of novel endocrine circuits. b, Gene expression data from multiple tissues of the HMDP were used to identify correlations between the expression of secreted proteins in one tissue and overall gene expression in a second tissue. The transcripts exhibiting the strongest correlation (the right-hand skew) included many known endocrine factors as well as novel candidates. c, Pathway enrichment from the underlying strong correlations was used to identify processes likely to be perturbed by each candidate endocrine factor. d,e, Secreted proteins were filtered by using tissue-specific expression profiles, clinical traits and published literature, and then experimentally validated.
Are gain- or loss-of-function studies consistent with population data?
Studies on a single genetic background can often be difficult to translate to population-level variation. This translation can be accomplished in a relatively straightforward fashion by assessing mapping or correlation structure by using systems genetics data. For example, Rajbhandari and colleagues have recently identified a novel role of the cytokine IL-10 in suppressing obesity and insulin resistance through adipose-tissue beiging56. Because these studies were performed on a C57BL/6J background, the authors were interested in determining whether relationships between IL-10 and metabolic phenotypes persisted on a population scale. The authors confirmed a positive correlation among IL-10, insulin resistance (HOMA-IR score) and adiposity in both mouse (HMDP) and human (METSIM) systems genetics data.
Which molecular signatures account for cellular heterogeneity?
Cell types often exhibit substantial functional heterogeneity, which is often defined by a small set of markers. For example, various lymphocytes and monocyte or macrophage subtypes have historically been identified with various cell-surface markers. However, the overall functional heterogeneity of such subtypes has generally not been studied at the population level. Buscher and colleagues sought to understand the functional heterogeneity of macrophages and their responses to inflammatory mediators such as bacterial lipopolysaccharide57. The authors first surveyed published human and mouse macrophage expression data under normal or lipopolysaccharide-stimulated conditions. Whereas gene expression signatures showed striking heterogeneity between mouse strains or human subjects, the authors identified a set of core signature genes for the inflammatory insult. Expression of these core genes elucidated conserved regulators of NF-κB responsive elements and also predicted macrophage-associated tumour survival58.
What factors mediate tissue–tissue cross-talk?
Metabolic homeostasis involves tight interactions across multiple tissues, but elucidating the nature of such tissue–tissue cross-talk has been challenging. Seldin and colleagues59 have developed a statistical approach to identify novel tissue–tissue endocrine circuits by using expression data from multiple tissues of a mouse population (Fig. 2b–e). They postulated, on the basis of hundreds of secreted proteins having no known function, that many endocrine factors remain to be identified. The first step in the method was identifying secreted proteins in one tissue that exhibit high correlation with the total transcriptome of a second tissue (Fig. 2b). The next step was identifying which pathways underlie these strong correlations by determining whether gene-set enrichment was present for each potential endocrine factor (Fig. 2c). These candidate endocrine proteins were then assessed for tissue-specific enrichment and relationships with clinical-trait data (Fig. 2d). Finally, predicted tissue–tissue circuits were validated experimentally in cell-culture or mouse models (Fig. 2e). Using this approach, the authors have identified a novel adipose–skeletal muscle circuit mediated by Lcn5 that stimulates mitochondrial activity and insulin sensitivity in skeletal muscle. Other mechanisms of tissue–tissue communication, including novel factors mediating adipose-tissue thermogenesis and the cardiac starvation response, have also been uncovered59. Similar approaches could potentially be used to identify cross-talk mediated by metabolites, circulating microRNAs or exosomes; in addition, population-based single-cell-sequencing data could be used to examine cell–cell communication within a tissue60.
Specialized topics
Systems genetics and network modelling
Systems genetics emphasizes the interconnections among biological spaces, depicting how molecules are organized and function together in complex systems, thus making pathways and networks a natural and intuitive framework for systems genetics. Pathways depict cascades of reactions, interactions or signalling events among a group of biological molecules that perform a particular function. For instance, the cholesterol-biosynthesis pathway involves a series of enzymes, substrates and products that perform the function of synthesizing cholesterol, and the insulin signalling pathway depicts how insulin interacts with its receptors and triggers downstream signalling cascades and activation of various transcriptional programs that regulate glucose and lipid homeostasis. In recent decades, numerous databases have been established to curate various knowledgebased biological processes and pathways. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Biocarta, Reactome and MsigDB are among the most widely used (Table 1). Various network-modelling approaches have been compared and comprehensively reviewed61.
Weighted gene co-expression network analysis (WGCNA)
Among network-modelling approaches, WGCNA is the most commonly used. It uses the correlation patterns among molecular traits across a series of samples to search for higher-level co-regulation structures and to define cohesive ‘modules’, each containing a group of molecules that are not only directly correlated but also share similarities in their relationships with the other molecules62. Each module is biologically meaningful and contains genes that share regulatory mechanisms, perform similar functions or relate to similar diseases63,64. Numerous studies have applied WGCNA to studying the molecular mechanisms underlying metabolic disorders65. More recently, multiscale embedded gene co-expression network analysis (MEGENA) has emerged as a complementary approach to WGCNA for coexpression-based network construction. Although MEGENA is also based on correlation structure, it uses a different algorithm and addresses several limitations of WGCNA, including large and less coherent modules and mutually exclusive modules that are forced to include distinct molecules66.
Excellent examples of the use of WGCNA to derive mechanistic understanding include studies by Farber and colleagues on bone mineral density (BMD) in mice67–69. The authors initially quantified BMD and global transcript levels in a panel of inbred strains of mice (HMDP). They then generated co-expression networks by using WGCNA and layered them on top of trait-association and eQTL data. The results implicated the gene Asxl2 as a driver of a co-expression network of genes involved in the differentiation of osteoclasts, and this finding was experimentally confirmed67,69. Subsequent network biology studies led to insights into the cellspecific processes that regulate BMD68,70. In particular, Calabrese and colleagues have used the above network to predict causal genes in human BMD GWAS loci on the basis of the premise that genes underlying disease are often functionally related. In this way, the authors predicted and inferred the functions for 30 of 64 human GWAS loci and experimentally validated two of these68.
Mergeomics
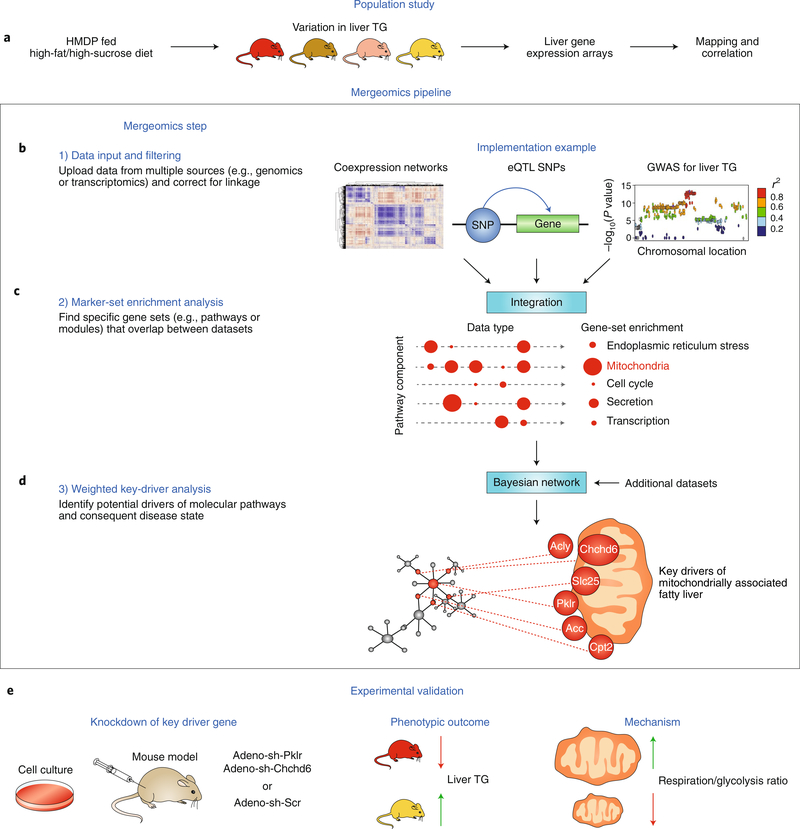
Mergeomics is another pathway- and network-based tool that has been successfully used to identify pathways and genes underlying complex metabolic disorders, as highlighted in Fig. 3. Unlike other tools that require all multi-omics data types to be derived from the same population, Mergeomics uses only summary-level multi-omics data, which can be derived from different studies or even species. Briefly, multi-layer disease-association signals are mapped to pathways or networks comprising interacting molecules to reveal pathogenic processes perturbed by individual omics variants as well as those affected by multiple omics layers. Recent applications of Mergeomics have yielded substantial insights into the tissue-specific biological processes and regulatory genes involved in individual diseases and those shared between diseases71.
Fig. 3 |. Application of Mergeomics to identify key regulators of liver and mitochondrial functions.
a, HMDP mice exhibited variations in hepatic triglyceride (TG) content after a high fat/high sucrose diet. Liver gene expression was measured across the panel to enable mapping of traits and analysis of correlation structure. b, Co-expression networks, eQTL and GWAS loci were generated with the data, which were then formatted and corrected for linkage disequilibrium. c, These data were integrated by using overlap of eQTL and clinical-trait loci to identify causal-gene sets (specific pathways or networks) involved in fatty liver. d, Weighted key-driver analysis was performed by incorporating the data into a Bayesian network to identify potential drivers of both mitochondrial networks and hepatic TG levels. e, Two selected key-driver genes were experimentally validated in cell culture and mouse models, thus leading to a proposed mechanism through which Pklr and Chchd6 drive fatty liver formation through effects on mitochondrial function.
The application of Mergeomics to identify pathways and genes underlying steatosis in a mouse model of nonalcoholic fatty liver disease is illustrated in Fig. 3. The tool has been used to identify pathway and ‘key-driver’ genes, most of which converged on mitochondrial functions. Experimental perturbation of several of the novel key-driver genes including Pklr and Chchd6, confirmed their effects on liver fat and mitochondrial oxidation72. In another study, von Scheidt and colleagues have used Mergeomics to integrate data from mouse and human GWAS studies along with expression profiling to identify pathways contributing to atherosclerosis, revealing ~70% sharing of disease pathways between the two species73.
GWAS applications
Boyle and Pritchard have recently proposed an omnigenic model positing that gene regulatory networks are sufficiently dense to cause some genetic variations to ‘percolate’ throughout the network in a relevant tissue74. The resulting GWAS loci may thus represent genetic variation in genes whose functions are only distantly unrelated to traits. In this model, ‘core’ genes that are more central in the networks are more likely to have a major effect on diseases and serve as more effective targets to modulate susceptibility or outcome. If the omnigenic model indeed proves correct, network modelling will be useful for the identification of these core genes.
Genetic interactions
One important application of systems genetics is to help understand genetic interactions. The term epistasis refers to the phenomenon in which variations in different genes combine and result in a phenotype different from the expectation based on the individual variation (that is, the effects are not additive). Thus, epistasis involves GxG interactions. Genetic variations can also interact in a non-additive manner with environmental factors (GxE) or sex (GxSex). Such genetic interactions are commonly observed in studies of experimental organisms, such as mice, for which the genetic background and the environment can be rigorously controlled (Fig. 4). However, these interactions have been difficult to study in humans, in which complications include the small effect sizes of most common genetic factors as well as the inability to assess the environment75. Thus, with some exceptions, most human GWAS studies have revealed little evidence of non-additive risk effects76–78.
Fig. 4 |. Examples of genetic interactions involved in metabolic traits.
a, GxG interactions. Gene-targeted mutations exhibit strikingly different effects on traits (methamphetamine sensitivity, blood glucose or acoustic startle response) depending on the genetic background. b, GxE interactions. Striking differences in fat-mass gain in response to a high-fat/high-sucrose diet were observed among HMDP strains of mice. c, GxSex interactions. Isolated mitochondria from adipose tissue of males and females of three HMDP mouse strains were monitored for oxygen consumption with a Seahorse bioanalyzer. Image reprinted from ref. 84, with permission from Elsevier. Whereas differences between sexes in C57BL/6 were modest, A/J and C3H/HeJ showed large sex effects. OCR, oxygen-consumption ratio.
A clear example of the importance of GxG interactions has come from experiments on targeted mutations studied on two or more genetic backgrounds in mice. For example, Sitting and colleagues6 have examined the effects of three different engineered mutations on behavioural traits in multiple genetic backgrounds and observed striking differences, ranging from strong to negligible, in each case (Fig. 4a). Such dependence on the genetic background appears to be the rule rather than the exception. These findings suggest that the effects of an engineered mutation often cannot be generalized to even different individuals of the same species, and thus, unsurprisingly, attempts to generalize from rodents to humans frequently fail.
The occurrence of GxE interactions in studies with mice or rats is also pervasive. Examples of experimental perturbations that have been studied include responses to diet, drugs, noise, temperature and forced exercise, as well as many other perturbations. For example, when the HMDP population of 100 inbred strains was exposed to a high-fat, high-sucrose diet for 8 weeks, the changes in body fat ranged from no increase whatsoever to an approximately sixfold increase79 (Fig. 4b). The integration of these phenotypes with microbiome and gene expression data has led to the identification of genes and microbes contributing to dietary responsiveness80.
Sex differences can have profound effects on complex traits and susceptibility to diseases81,82. Unfortunately, they have been greatly understudied in metabolism. In fact, studies using model organisms such as mice have frequently examined only males, with exceptions such as ageing studies, which have tended to focus only on females83. A recent study has systematically examined GxSex interactions for approximately 50 metabolic traits, including body fat, insulin resistance, plasma lipids and organ weights in the HMDP resource84. All traits with the exception of blood-cell parameters exhibited sex differences, and the effects of sex were often dependent on the genetic background. Whereas male mice of certain strains gained more fat than female mice in response to a high-fat diet, the reverse was true for certain other strains. Integration of the clinical-trait data with adipose gene expression data across the strains indicated an important role of adipose mitochondrial function in these sex differences. Indeed, studies of isolated mitochondria from several of the strains validated striking GxSex differences that were also associated with traits such as diet-induced obesity and insulin resistance84 (Fig. 4c). Notably, in contrast to many other strains, C57BL/6J mice exhibited no significant differences in adipose mitochondrial activity and abundance between sexes, thus illustrating the limitations of exclusively studying a single genotype.
Why might studies in experimental organisms demonstrate pervasive non-additive interactions, whereas studies in humans reveal only modest evidence for such interactions? Basic biologic differences may contribute but would seem unlikely to entirely account for the discrepancy. In a recent essay, Sackton and Hartl75 have distinguished ‘statistical epistasis’ and ‘physiologic epistasis’ and argue that the latter can be pervasive and still result in negligible levels of the former. Because additive models are fit by least squares, some of the effects of epistasis are tallied with additive or dominant inheritance. In addition, the ability to detect statistical epistasis depends on the frequencies of the multi-locus genotypes. Thus, one possible explanation for why non-additive interactions are missed in human studies compared with animal studies is the relatively smaller effect sizes of the loci contributing to complex traits and the greater heterogeneity, which is difficult to control. Indeed, studies in human populations of variations with large effect sizes, such as Mendelian traits or eQTLs, have provided strong evidence of non-additive interactions5,76
Therapeutic and diagnostic applications
An approach to drug targeting termed The Connectivity Map (CMAP)85 is ideally suited to systems genetics data. The approach uses global expression data obtained after treatment of cell lines by many different drugs. A more recent version of CMAP called LINC1000 has been developed, incorporating more drugs and more cell lines86. The concept is that if a disorder exhibiting a pattern of expression opposite from that of one of the surveyed drugs, that drug or a related drug might mitigate or reverse the disorder. Conversely, if a disease-gene pattern mimics that of a drug, the drug may contribute to toxicity or side effects. For example, in one study, endoplasmic reticulum stress pathways were induced by either injection of 4-phenyl butyrate or overexpression of X-box-binding protein 1 in genetically obese ob/ob mice. Liver gene expression patterns in the mice were then analysed with CMAP, which prioritized Celastrol as a potential regulator of endoplasmic reticulum stress pathways. The drug was then experimentally validated to have potent anti-obesity effects in mice85,87. Systems genetics approaches would appear to offer many advantages for not only the identification of novel therapeutics but also understanding of off-target effects and variations in responses among individuals.
Systems genetics can also be useful in the identification of novel biomarkers for diagnostic applications. For example, on the basis of a strong correlation found between heart-failure traits and the expression of Gpnmb in the heart in a mouse cohort, Lin and colleagues have postulated that the protein may be a useful biomarker for the disease88. Indeed, the authors observed a striking correlation between heart-failure traits and plasma protein levels in both mice and human subjects.
Pirie and colleagues89 have recently used systems genetics to examine the pharmacokinetics and pharmacodynamics of antisense oligonucleotides (ASOs), which can be used to modify the expression of genes in vivo and have become widely used therapeutic agents. ASOs exhibit variation in efficacy in patient populations, and the authors have used transcriptomic analysis and genome-wide association in the HMDP mouse population to identify several genes associated with the uptake and potency of ASOs.
Conclusions and future directions
Systems genetics approaches are now being applied to many areas of metabolism research, and several powerful reference cohorts, both rodent and human, have been developed. In addition, such studies have generated large datasets, many of which are publicly available. To date, only a small fraction of researchers are taking advantage of the available datasets, but this utilization will ideally change as more investigators become aware of the complementary nature of systems genetics and reductionist approaches.
Given the great technological and analytical advances in human genetics over the past 20 years90, studies in animal models have been suggested to be potentially unnecessary or even misleading, in terms of understanding common diseases. However, we feel strongly that studies in animal models will continue to be critical, given their advantages such as the ability to control the environment, access tissues, direct experimental follow-up and engineer mutations91,92. With the identification of GWAS loci for complex human traits, examination of the overlap between mouse and human genes and pathways has become possible. This overlap appears to be extensive for traits such as diabetes, obesity and atherosclerosis26,79,93, thus supporting the conclusion that mouse models indeed capture much of the pathophysiology of humans.
The development of new technologies is a key driving force in systems genetics. The various omics technologies have greatly improved over the past decade, and new technologies applicable to systems genetics have been developed. One particularly powerful technology is single-cell RNA sequencing94, in which a fraction of the expressed transcripts from a single cell can be measured quantitatively. Single-cell RNA sequencing has been successfully applied to various tissues to uncover rare cell populations, such as niche stem cells in the liver95 or intestine96, and to infer spatial cell population diversity information in a complex and heterogeneous tissues, such as brain tissue97. Beyond RNA, technologies for other single-cell omics domains, such as single-cell assay for transposase-accessible chromatin using sequencing (ATAC-seq) for epigenome profiling and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) for protein measurements, have also been developed98–101. Analysing these single-cell profiles across diverse individuals could provide substantial information regarding the genomic regulation of cell identity and composition, when they are overlaid with additional data. One recent study has highlighted the presence of single-cell QTLs by mapping these data onto the genome in blood cells of ~40 individuals102. Similar application of this technology to other tissues and populations, as well as overlaying with other ‘layers’ of biology, offers the potential to reveal which specific cells and pathways are relevant for disease and function, to provide mechanistic insights at single-cell resolution.
Several statistical advances have been made, as discussed above. One tool likely to become increasingly useful in systems genetics is machine learning, which allows for identification of interconnections within datasets that might be missed through traditional linear or nonlinear approaches, such as correlation. Zeevi and colleagues103 have measured gut microbiota composition together with blood glucose levels, dietary habits and physical activity to predict variable responses in glucose levels after meals. Machine learning was used to integrate the data and develop an algorithm that accurately predicted glycaemic responses from microbial composition.
The most important future challenges in metabolism are likely to include the areas of nutrition, exercise and ageing. Inter-individual differences in response to a dietary challenge are clearly mediated not only by host genetics but also by the gut microbiome37,104,105. Large-scale systems genetics studies in humans and rodents are likely to be key in dissecting such complex host–microbiome–environment interactions. Like diet, exercise clearly has a large effect on health, but the mechanisms linking persistent exercise to protective effects against disease, apart from weight gain, are largely unknown106. The past decade has seen dramatic advances in the identification of mechanisms contributing to ageing, and systems genetics studies have indicated the key roles of caloric restriction107 and mitochondrial ribosomal abundance108. At the genomic level, population-based approaches suggest that the ability to maintain a healthy metabolic state and prolonged lifespan can be attributed to a specific ‘resilience’ network of interactions to buffer detrimental mutations109.
Acknowledgements
We are grateful to our colleagues, particularly M. Mehrabian, B. Pasaniuc, H. Allayee, C. Pan and K. Chella Krishnan for useful discussions, and to R. Chen for help in manuscript preparation. This work was supported by NIH grants HL28481, GM115318, HL144651, DK117850, HL147883 (A.J.L.), HL138193 (M.S.) and DK104363 (X.Y.).
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information Primary Handling Editor: Pooja Jha.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. © Springer Nature Limited 2019
References
- 1.Civelek M & Lusis AJ Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 15, 34–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasin Y, Seldin M & Lusis A Multi-omics approaches to disease. Genome Biol. 18, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karczewski KJ & Snyder MP Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H et al. An integrated systems genetics and omics toolkit to probe gene function. Cell Syst. 6, 90–102.e104 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Riordan JD & Nadeau JH From peas to disease: modifier genes, network resilience, and the genetics of health. Am. J. Hum. Genet. 101, 177–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sittig LJ et al. Genetic background limits generalizability of genotype-phenotype relationships. Neuron 91, 1253–1259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoeger T, Gerlach M, Morimoto RI & Nunes Amaral LA Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 16, e2006643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battle A, Brown CD, Engelhardt BE & Montgomery SB Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).The GTEx project’s characterization of variations in gene expression levels across individuals and 44 tissues of the human body.
- 9.Heinz S et al. Effect of natural genetic variation on enhancer selection and function. Nature 503, 487–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lusis AJ et al. The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J. Lipid Res. 57, 925–942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreux PA et al. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell 150, 1287–1299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams EG et al. Systems proteomics of liver mitochondria function. Science 352, aad0189 (2016).Detailed phenotypic, molecular and genetic analyses of BXD animals fed normal or high-fat diets, uncovering new regulatory pathways of hepatic mitochondrial function and clinical outcomes.
- 13.Threadgill DW, Miller DR, Churchill GA & de Villena FP The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J. 52, 24–31 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Bogue MA, Churchill GA & Chesler EJ Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database. Mamm. Genome 26, 511–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicod J et al. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat. Genet. 48, 912–918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker CC et al. Genome-wide association study of behavioral, physiological and gene expression traits in outbred CFW mice. Nat. Genet. 48, 919–926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzales NM & Palmer AA Fine-mapping QTLs in advanced intercross lines and other outbred populations. Mamm. Genome 25, 271–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holl K et al. Heterogeneous stock rats: a model to study the genetics of despair-like behavior in adolescence. Genes Brain Behav. 17, 139–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchner DA & Nadeau JH Contrasting genetic architectures in different mouse reference populations used for studying complex traits. Genome Res. 25, 775–791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laakso M et al. The Metabolic Syndrome in Men study: a resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 58, 481–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moayyeri A, Hammond CJ, Hart DJ & Spector TD The UK Adult Twin Registry (TwinsUK Resource). Twin Res. Hum. Genet. 16, 144–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedman AK et al. Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circ. Cardiovasc Genet 10, e001487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huan T et al. Integrative network analysis reveals molecular mechanisms of blood pressure regulation. Mol. Syst. Biol. 11, 799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huan T et al. Dissecting the roles of microRNAs in coronary heart disease via integrative genomic analyses. Arterioscler. Thromb. Vasc. Biol. 35, 1011–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talukdar HA et al. Cross-tissue regulatory gene networks in coronary artery disease. Cell Syst. 2, 196–208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller MP et al. Genetic drivers of pancreatic islet function. Genetics 209, 335–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romanoski CE et al. Network for activation of human endothelial cells by oxidized phospholipids: a critical role of heme oxygenase 1. Circ. Res. 109, e27–e41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanoski CE et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am. J. Hum. Genet. 86, 399–410 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X et al. Interrogation of the atherosclerosis-associated SORT1 (Sortilin 1) locus with primary human hepatocytes, induced pluripotent stem cell-hepatocytes, and locus-humanized mice. Arterioscler. Thromb. Vasc. Biol. 38, 76–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chick JM et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534, 500–505 (2016).A detailed integration of transcriptomic and proteomic data in DO mice.
- 31.Ghazalpour A et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7, e1001393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Beyer A & Aebersold R On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Parker BL et al. An integrative systems genetic analysis of mammalian lipid metabolism. Nature 567, 187–193 (2019).Combined proteomic and lipidomic analyses of HMDP livers paired with experimental validation, identifying novel mechanisms of hepatic lipid regulation.
- 34.Jha P et al. Systems analyses reveal physiological roles and genetic regulators of liver lipid species. Cell Syst. 6, 722–733.e726 (2018).An analysis of hepatic and plasma lipidomes in BXD RI strains, identifying new regulatory mechanisms and providing insight into human disease.
- 35.Jha P et al. Genetic regulation of plasma lipid species and their association with metabolic phenotypes. Cell Syst. 6, 709–721.e706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romanov N et al. Disentangling genetic and environmental effects on the proteotypes of individuals. Cell 177, 1308–1318.e1310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeevi D et al. Structural variation in the gut microbiome associates with host health. Nature 568, 43–48 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Schadt EE et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37, 710–717 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Voight BF et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet 380, 572–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie MD, Holzinger ER, Li R, Pendergrass SA & Kim D Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 16, 85–97 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Sun YV & Hu YJ Integrative analysis of multi-omics data for discovery and functional studies of complex human diseases. Adv. Genet. 93, 147–190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Argelaguet R et al. Multi-Omics Factor Analysis: a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arneson D, Bhattacharya A, Shu L, Mäkinen VP & Yang X Mergeomics: a web server for identifying pathological pathways, networks, and key regulators via multidimensional data integration. BMC Genomics 17, 722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu L et al. Mergeomics: multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genomics 17, 874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gusev A et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fryett JJ, Inshaw J, Morris AP & Cordell HJ Comparison of methods for transcriptome imputation through application to two common complex diseases. Eur. J. Hum. Genet. 26, 1658–1667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albert FW & Kruglyak L The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16, 197–212 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Pickrell JK Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94, 559–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahajan A et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).Survey of human type 2 diabetes GWAS SNPs and integration with open chromatic marks, highlighting pancreatic islet mechanisms as potential key drivers of disease.
- 51.Kessler T et al. Functional characterization of the GUCY1A3 coronary artery disease risk locus. Circulation 136, 476–489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett BJ et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20, 281–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hui ST et al. The genetic architecture of NAFLD among inbred strains of mice. eLife 4, e05607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kojima Y et al. CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 536, 86–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iotchkova V et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet. 48, 1303–1312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajbhandari P et al. IL-10 signaling remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell 172, 218–233.e217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buscher K et al. Natural variation of macrophage activation as diseaserelevant phenotype predictive of inflammation and cancer survival. Nat. Commun. 8, 16041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gentles AJ et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seldin MM et al. A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metab. 27, 1138–1155.e1136 (2018).A systems genetics application for the discovery of novel endocrine factors on the basis of correlation structure of expression data across tissues.
- 60.Thomou T et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang JK et al. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. 6, 484–495.e485 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emilsson V et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Keller MP et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 18, 706–716 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song WM & Zhang B Multiscale embedded gene co-expression network analysis. PLoS Comput. Biol. 11, e1004574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calabrese G et al. Systems genetic analysis of osteoblast-lineage cells. PLoS Genet. 8, e1003150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calabrese GM et al. Integrating GWAS and co-expression network data identifies bone mineral density genes SPTBN1 and MARK3 and an osteoblast functional module. Cell Syst. 4, 46–59.e44 (2017).A beautiful example of the application of network modelling of systems genetics data to identify novel genes and pathways underlying the complex trait of BMD.
- 69.Farber CR et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 7, e1002038 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mesner LD et al. Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density. J. Clin. Invest. 124, 2736–2749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shu L et al. Shared genetic regulatory networks for cardiovascular disease and type 2 diabetes in multiple populations of diverse ethnicities in the United States. PLoS Genet. 13, e1007040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chella Krishnan K et al. Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst. 6, 103–115.e107 (2018).Application of Mergeomics to pinpoint mitochondrial function as a key contributor to hepatic triglyceride accumulation.
- 73.von Scheidt M et al. Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab. 25, 248–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boyle EA, Li YI & Pritchard JK An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sackton TB & Hartl DL Genotypic context and epistasis in individuals and populations. Cell 166, 279–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemani G et al. Detection and replication of epistasis influencing transcription in humans. Nature 508, 249–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Lenz TL et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet. 47, 1085–1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Visscher PM et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parks BW et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17, 141–152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Org E et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25, 1558–1569 (2015).Analysis of the genetics of gut microbiota composition in HMDP mice, demonstrating high heritability and GxE interactions.
- 81.Karp NA et al. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat. Commun 8, 15475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ober C, Loisel DA & Gilad Y Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 9, 911–922 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arnold AP, van Nas A & Lusis AJ Systems biology asks new questions about sex differences. Trends Endocrinol. Metab. 20, 471–476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Norheim F et al. Gene-by-sex interactions in mitochondrial functions and cardio-metabolic traits. Cell Metab. 29, 932–949.e4 (2019).Demonstration of the importance of adipose-tissue respiration in the mediation of GxSex interactions in cardio-metabolic traits.
- 85.Lamb J et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Subramanian A et al. A next generation connectivity map: l1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e1417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J, Lee J, Salazar Hernandez MA, Mazitschek R & Ozcan U Treatment of obesity with celastrol. Cell 161, 999–1011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin LY et al. Systems genetics approach to biomarker discovery: Gpnmb and heart failure in mice and humans. G3 (Bethesda) 8, 3499–3506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pirie E et al. Mouse genome-wide association studies and systems genetics uncover the genetic architecture associated with hepatic pharmacokinetic and pharmacodynamic properties of a constrained ethyl antisense oligonucleotide targeting Malat1. PLoS Genet. 14, e1007732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.FitzGerald G et al. The future of humans as model organisms. Science 361, 552–553 (2018). [DOI] [PubMed] [Google Scholar]
- 91.Attie AD, Churchill GA & Nadeau JH How mice are indispensable for understanding obesity and diabetes genetics. Curr. Opin. Endocrinol. Diabetes Obes. 24, 83–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nadeau JH & Auwerx J The virtuous cycle of human genetics and mouse models in drug discovery. Nat. Rev. Drug Discov. 18, 255–272 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Parks BW et al. Genetic architecture of insulin resistance in the mouse. Cell Metab. 21, 334–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Svensson V, Vento-Tormo R & Teichmann SA Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018). [DOI] [PubMed] [Google Scholar]
- 95.Halpern KB et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haber AL et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chappell L, Russell AJC & Voet T Single-cell (multi)omics technologies. Annu. Rev. Genomics Hum. Genet. 19, 15–41 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Macaulay IC, Ponting CP & Voet T Single-cell multiomics: multiple measurements from single cells. Trends Genet. 33, 155–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mezger A et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun. 9, 3647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van der Wijst MGP et al. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat. Genet. 50, 493–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeevi D et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Kasahara K et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat. Microbiol. 3, 1461–1471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Z et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoffman NJ et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 22, 922–935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liao CY, Rikke BA, Johnson TE, Diaz V & Nelson JF Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9, 92–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Houtkooper RH et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen R et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 34, 531–538 (2016). [DOI] [PubMed] [Google Scholar]