Abstract
Plant endogenous small RNAs (sRNAs) are important regulators of gene expression. There are two broad categories of plant sRNAs: microRNAs (miRNAs) and endogenous short interfering RNAs (siRNAs). MicroRNA loci are relatively well-annotated but compose only a small minority of the total sRNA pool; siRNA locus annotations have lagged far behind. Here, we used a large data set of published and newly generated sRNA sequencing data (1333 sRNA-seq libraries containing more than 20 billion reads) and a uniform bioinformatic pipeline to produce comprehensive sRNA locus annotations of 47 diverse plants, yielding more than 2.7 million sRNA loci. The two most numerous classes of siRNA loci produced mainly 24- and 21-nucleotide (nt) siRNAs, respectively. Most often, 24-nt-dominated siRNA loci occurred in intergenic regions, especially at the 5′-flanking regions of protein-coding genes. In contrast, 21-nt-dominated siRNA loci were most often derived from double-stranded RNA precursors copied from spliced mRNAs. Genic 21-nt-dominated loci were especially common from disease resistance genes, including from a large number of monocots. Individual siRNA sequences of all types showed very little conservation across species, whereas mature miRNAs were more likely to be conserved. We developed a web server where our data and several search and analysis tools are freely accessible.
Plant regulatory small RNAs (sRNAs) play important roles in almost all biological processes. Endogenous sRNAs are 20–24 nucleotide (nt) in length and derive from longer RNA precursors that are processed by DICER-LIKE (DCL) ribonucleases. Once processed, they are loaded into Argonaute (AGO) proteins to form the RNA-induced silencing complex (RISC). Then, sRNAs guide the RISC complex to complementary sites on target RNAs, inducing either post-transcriptional or transcriptional gene silencing.
Endogenous sRNAs can be grouped into two broad classes based on their biogenesis and typical functions: microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Axtell 2013a). miRNAs are typically 21–22 nt long, processed from single-stranded RNA (ssRNA) stem-loop precursors by DCL1, and regulate gene expression post-transcriptionally, directing mRNA degradation and translational repression (Rogers and Chen 2013). siRNAs are processed from double-stranded RNA (dsRNA) precursors and are categorized in multiple subclasses. The most abundant subclass of siRNAs participates in the RNA-directed DNA methylation (RdDM) pathway, involving 24 or 21–22 nt siRNAs. Twenty-four-nucleotide siRNAs are derived from Polymerase IV (Pol IV) transcripts that are converted to dsRNAs by RNA-dependent RNA polymerase 2 (RDR2), which are then processed by DCL3. They act in canonical RdDM, primarily targeting transposable elements (TEs) and other repeats to induce DNA methylation and reinforce transcriptional silencing. In contrast, 21–22 nt siRNAs are derived from Pol II transcripts and are copied by RDR6 into dsRNAs and processed by DCL2/DCL4. They act in the noncanonical RdDM pathway to establish the silencing of young TEs, both transcriptionally and post-transcriptionally (Nuthikattu et al. 2013). Another major siRNA subclass is secondary siRNAs. Their biogenesis is triggered by a miRNA-directed cleavage of a coding or noncoding transcript. The transcript is then converted to dsRNA by RDR6 and processed by DCL proteins into secondary siRNAs in a phased pattern relative to the miRNA cut site. Phased secondary siRNAs (phasiRNAs) are typically 21 or 22 nt long, however, a specific population of 24-nt phasiRNAs has been detected in anthers of many angiosperms (Xia et al. 2019). TAS genes are an example of loci generating noncoding RNA precursors that produce secondary siRNAs, which act in trans (trans-acting siRNAs [tasiRNAs]) on other targets and direct their cleavage (Allen et al. 2005). Pentatricopeptide repeat (PPR) genes are the first reported protein-coding genes generating secondary siRNAs in Arabidopsis thaliana (Howell et al. 2007).
At the chromosomal level, sRNA distribution correlates with gene density, typically lower in the centromeric and pericentromeric regions and enriched in the distal euchromatic regions. This trend has been observed in maize (He et al. 2013), rice (Wei et al. 2014), tomato (The Tomato Genome Consortium 2012), hot pepper (Kim et al. 2014), upland cotton (Song et al. 2015), and sugar beet (Dohm et al. 2014). However, in a smaller number of species sRNAs mostly arise from centromeric and pericentromeric regions away from genes, as shown in A. thaliana (Kasschau et al. 2007; Ha et al. 2009), soybean (Schmitz et al. 2013), cucumber (Lai et al. 2017), and Brachypodium distachyon (The International Brachypodium Initiative 2010).
Despite differences in chromosomal distributions, the sRNA profiles near protein-coding genes are conserved among plant species, with 24-nt siRNAs preferentially found in gene-proximal regions but depleted in gene bodies themselves. This pattern has been described in maize (Gent et al. 2013), rice (Wei et al. 2014), rapeseed (Shen et al. 2017), Chinese cabbage (Woodhouse et al. 2014), soybean (Song et al. 2013), upland cotton (Song et al. 2015), and A. thaliana (Kasschau et al. 2007; Ha et al. 2009). Depending on the species, sRNAs are more frequently found in proximity of genes with different expression levels. In maize, 24-nt siRNAs are found with higher probability near expressed genes than nonexpressed genes (Gent et al. 2013; Lunardon et al. 2016). Here, siRNAs participate in RdDM to reinforce the silencing of TEs that are inserted upstream of genes, where the chromatin is accessible, therefore repressing the potentially deleterious Pol II transcription of TEs (Gent et al. 2014). In contrast, the siRNA-mediated silencing of TEs near genes is linked to lower expression of the genes in A. thaliana and Chinese cabbage (Hollister et al. 2011; Woodhouse et al. 2014). In addition to target TEs near genes, 24-nt siRNAs can also target TEs inserted inside genes, affecting their expression (Wei et al. 2014; Lunardon et al. 2016).
Genome-wide analyses in barley, soybean, Medicago truncatula, and Physcomitrella patens showed that 21-nt siRNAs are not enriched in gene-body regions (Lelandais-Brière et al. 2009; Schmitz et al. 2013; Coruh et al. 2015; Hackenberg et al. 2016). Nevertheless, there are many cases of well-characterized genes generating 21-nt phasiRNAs in dicots: nucleotide binding/leucine-rich repeat (NB-LRR) and receptor-like kinase (RLK) resistance genes, PPR genes, auxin-responsive factor (ARF) genes, MYB and NAC transcription factors, and F-BOX genes (Arikit et al. 2014; Hu et al. 2015a; Xia et al. 2015b). NB-LRR genes evolve rapidly by tandem duplication and transposition, and they are controlled by sRNA-mediated silencing to avoid their overexpression and prevent autoimmune responses (Freeling et al. 2008; Yang and Huang 2014). This mechanism is conserved in a large number of dicots: soybean, M. truncatula, common bean, chickpea, Populus trichocarpa, cassava, pima cotton, potato, and Norway spruce (Klevebring et al. 2009; Zhai et al. 2011; Xia et al. 2014; Formey et al. 2015; Hu et al. 2015b; Srivastava et al. 2015; Xia et al. 2015a). Among monocots, 21-nt phasiRNAs from NB-LRR genes have been only found in barley and wheat so far (Liu et al. 2014; Zhang et al. 2019). This is consistent with the fact that monocots, in contrast to dicots, produce phasiRNAs mainly from noncoding RNAs (Zheng et al. 2015; Komiya 2017).
The conservation of sRNAs across plants has been widely investigated for miRNAs. There are deeply conserved miRNA families together with their targets, suggesting common functional regulatory networks (Axtell and Bowman 2008). However, the majority of miRNA sequences are species-specific, indicating the presence of numerous young or still evolving miRNAs (Cuperus et al. 2011; Chávez Montes et al. 2014). Much less is known about conservation of siRNAs but a study comparing Arabidopsis thaliana and Arabidopsis lyrata suggested that individual siRNA sequences are not conserved even between closely related species (Ma et al. 2010). Moreover, although in most species analyzed so far, the 24-nt siRNAs are the most abundant expressed group of sRNAs, mosses, lycophytes, and conifers lack a strong peak of 24-nt siRNAs (Axtell and Bartel 2005; Dolgosheina et al. 2008; Banks et al. 2011).
There are several existing web-based resources that serve sRNA sequencing (sRNA-seq) data for multiple plants. The Cereal Small RNA Database contains maize and rice genome browsers with accessible sRNA-seq data (http://sundarlab.ucdavis.edu/smrnas/) (Johnson et al. 2007). The Pln24NT website stores annotations and sequences of 24-nt siRNA reads and loci for 10 species (http://bioinformatics.caf.ac.cn/Pln24NT/) (Liu et al. 2017). The Next-Gen Sequence Databases produced by the Meyers laboratory contain sRNA-seq and other high-throughput data with custom-built genome browsers and search functions for 27 species (https://mpss.danforthcenter.org) (Nakano et al. 2006). The miRBase database (http://www.mirbase.org) (Kozomara and Griffiths-Jones 2014) provides curated, comprehensive annotations of miRNA loci in a very large number of species. An equivalent database for the storage and distribution of reference annotations of siRNA-producing loci in a vast number of plant genomes does not exist (Coruh et al. 2014).
In this study, we used a large data set of published and newly generated sRNA-seq data that we processed with a consistent pipeline to create reference sRNA loci annotations for 47 plant species, including model plants and crops. We propose and use a systematic nomenclature and ontology for sRNA-producing loci that is consistent with their biology and easily traceable and updatable. We examined the genome-wide distribution of sRNA loci relative to protein-coding genes and compared it across species, providing insights into conserved sRNA functions. We organized the sRNA-seq alignment data and sRNA loci annotations in a freely available web-based database that represents an important public resource for future studies aimed to understand the biological function of sRNAs.
Results
Identification and classification of sRNA loci in 47 plants
We obtained and analyzed 48 plant genome assemblies, representing 47 different species (two independent assemblies of Cuscuta campestris were analyzed) (Table 1). To facilitate succinct communication in figures and our database, a short code was designated for each assembly. The code begins with a three-letter prefix representing the genus and species, following the abbreviations established by miRBase (Kozomara et al. 2019). The second part of the code indicates the genome build (“-b”) version in use. These genome assemblies varied widely in size, contiguity, protein-coding gene number, and repeat content (Supplemental Figs. S1, S2). Most genome assemblies were from crops; others included the model plants Arabidopsis thaliana and Medicago truncatula, the parasitic plant Cuscuta campestris, and representatives of diverse lineages (Amborella trichopoda [basal angiosperm], Picea abies [gymnosperm], Physcomitrella patens [bryophyte], and Marchantia polymorpha [bryophyte]).
Table 1.
Genomes included in this study
We gathered sRNA-seq libraries from each genome (Fig. 1A). In most cases, these data were from public sequencing archives (Supplemental Table S1). In a few cases, we also generated novel sRNA-seq libraries (Zea mays, Spinacia oleracea, Daucus carota, Theobroma cacao) (Supplemental Table S1). We sought to annotate the full diversity of sRNA loci and thus selected libraries with the goal of including as many different tissues and conditions as possible. However, we excluded low-depth sRNA-seq data sets (less than two million reads aligned to the genome) and also excluded sRNA-seq data sets from mutants known to affect sRNA biogenesis or stability. For each given genome assembly, all cognate sRNA-seq libraries were aligned and then merged into a single master sRNA alignment which we call the “reference set” (Fig. 1A). Reference sets had considerable variation in both total number of sRNA reads (minimum: 2.1 × 106, median: 1.6 × 108, maximum: 4.1 × 109) and in number of contributing sRNA-seq libraries (minimum: 1, median: 11, maximum: 161) (Supplemental Fig. S3).
Figure 1.
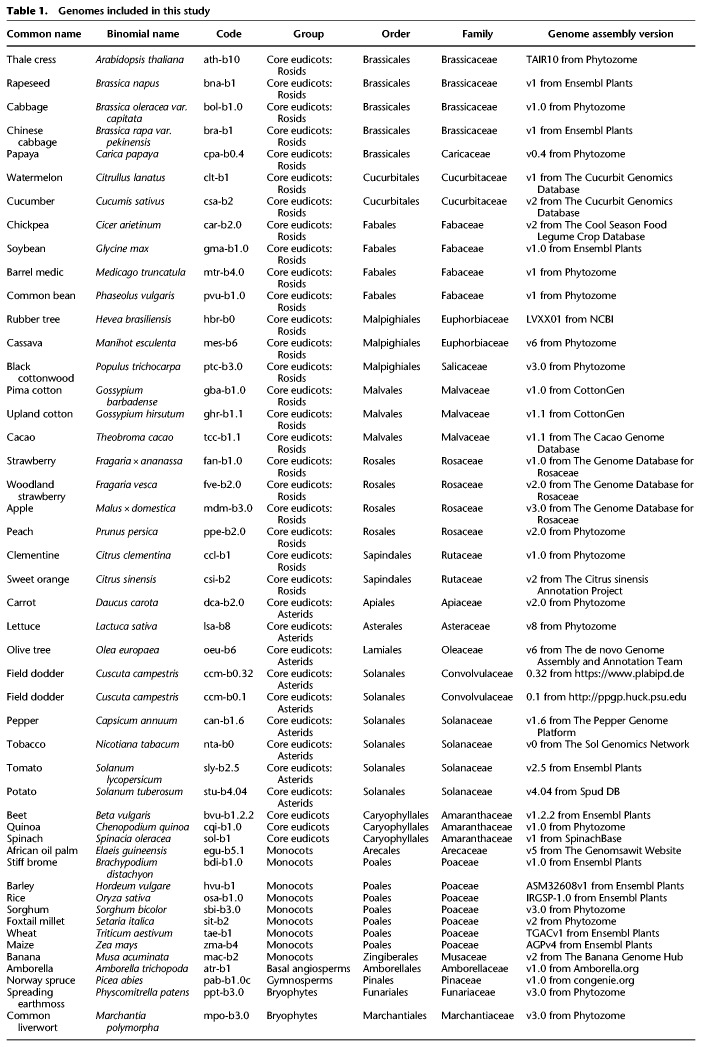
Overview of sRNA locus annotation pipeline and summary of annotated sRNA loci. (A) Schematic illustrating how multiple sRNA libraries from diverse plant tissues are merged to create a “reference set” of sRNAs for a given species. Accession numbers shown are fictional. (B) Distributions of the fractions of sRNAs between 20 and 24 nt in length (inclusive) within all loci in each genome. Gray line at 80% represents the cutoff used to discriminate silencing-related RNA loci from other types of sRNA-producing loci. (C) Flowchart illustrating the ontology used to classify sRNA-producing loci. Colors designating different locus types are used throughout this work. (D) Schematic illustrating the nomenclature used to annotate sRNA-producing loci. (E–H) Summary of annotated sRNA loci, by species and locus type, excluding the category “OtherRNA.” (E) Counts of annotated loci. (F) Proportions of annotated loci. (G) Total counts of aligned small RNAs in reference sets. (H) Proportions of small RNA total read counts in reference sets. See Table 1 for species codes. (Eu) eudicots; (BA) basal angiosperm; (G) gymnosperm; (Br) bryophyte.
For annotation, we first identified genomic regions producing sRNAs, independently in all sRNA-seq libraries with ShortStack (Axtell 2013b; Johnson et al. 2016). Then we compared the sRNA expression from different samples of the same species and identified the regions that were robustly expressing sRNAs in at least three separate samples. Millions of discrete sRNA clusters were annotated in this way and defined as sRNA-producing loci, which were then analyzed in the genome-aligned reference sets. Canonical plant miRNAs and siRNAs are between 20 and 24 nt in length, whereas other types of sRNA loci produce a broader range of RNA sizes. For each locus, we computed the fraction of aligned sRNA-seq reads that were 20–24 nt long. We found that these fractions had a consistent bimodal distribution in each individual genome (Fig. 1B). Based on these distributions, we used a cutoff of 80% to discriminate canonical siRNA/miRNA loci from “OtherRNA” loci (Fig. 1C). We then developed a simplified ontology to describe the siRNA and miRNA loci: “MIRNA” loci were those that met all miRNA annotation criteria, whereas “nearMIRNA” loci met most criteria except for that the exact predicted miRNA*, the complementary strand to the mature miRNA in the miRNA-miRNA* duplex, was not sequenced. The remaining loci were classified as siRNA loci based on the predominant length of aligned sRNAs within each locus (Fig. 1C). This ontology has the advantage of being applicable to any genome regardless of any other annotations or information. We also devised a simple nomenclature to systematically name the sRNA loci (Fig. 1D). In total, we annotated ∼2.7 × 106 sRNA-producing loci from the 48 genome assemblies (Supplemental Table S2; also see http://plantsmallrnagenes.science.psu.edu for easier access and more analysis options).
The “OtherRNA” category of loci, defined by having <80% of aligned reads with sizes between 20 and 24 nt in length, typically composed less than half of all loci in the flowering plants (Supplemental Fig. S4A,B). In contrast, the majority of loci identified in one gymnosperm and two bryophyte genomes were annotated as OtherRNA (Supplemental Fig. S4A,B). Across all taxa, OtherRNA loci typically contributed large fractions of total read abundance (Supplemental Fig. S4C,D). This is because many of the OtherRNA loci represented clusters of short fragments derived from highly abundant, longer RNAs, such as rRNAs, tRNAs, and plastid-derived mRNAs. There is evidence that some plant RNAs longer than 24 nt, or shorter than 20 nt, may function as gene-regulatory factors (Martinez et al. 2017); such loci will have been annotated in the OtherRNA category by our procedure. Nonetheless, we focused our subsequent analyses on the MIRNA, nearMIRNA, and siRNA loci dominated by 20–24 nt RNAs because these sizes are most clearly associated with production by DCL endonucleases and usage by AGO proteins. By default, ShortStack assigns a phasing score to the sRNA loci based on the algorithm described in Guo et al. (2015). However, an accurate annotation of the phasing would require a more complex study to avoid false positives that may be produced by the commonly used phasing-detecting algorithms (Polydore et al. 2018). Therefore, we did not further analyze the phasing of the sRNA loci in this analysis.
After excluding OtherRNA loci, the remaining loci were mostly designated siRNA24 in angiosperms (Fig. 1E,F). In contrast, and consistent with prior reports (Axtell and Bartel 2005; Dolgosheina et al. 2008), gymnosperm and bryophyte loci were less dominated by the siRNA24 type and instead had more siRNA21 loci. When tallied by sRNA abundance, MIRNA and siRNA21 loci made substantial contributions in all taxa (Fig. 1G,H). This indicates that a relatively small number of MIRNA and siRNA21 loci produce high levels of their respective sRNAs. In a number of species, the proportion of 22-nt siRNAs was also substantial and this trend was particularly consistent among the asterids (Fig. 1H). In most cases, angiosperms had more annotated sRNA loci compared to non-angiosperms (Fig. 1E; Supplemental Fig. S4A). However, that comparison is potentially complicated by the different amounts of input sRNA reads used for each species (Supplemental Fig. S3).
The plantsmallrnagenes.science.psu.edu server
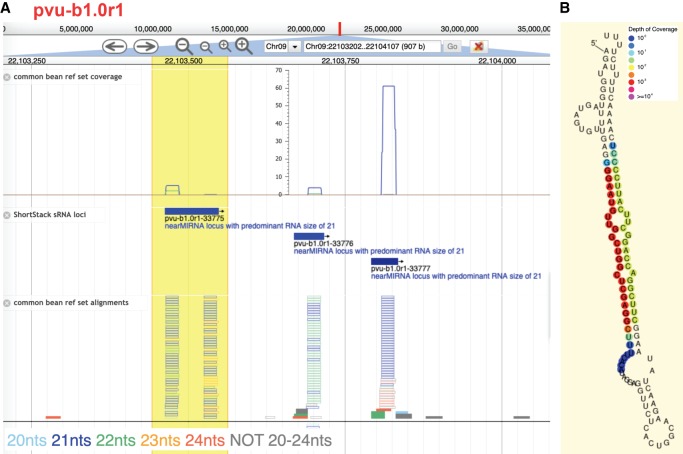
All data and analyses from this study have been systematically organized and are freely available at https://plantsmallrnagenes.science.psu.edu. Users can search for loci of interest by sRNA sequence, miRNA family name, locus name, or by BLAST-based homology searches. A JBrowse-based genome browser is available for each of the 48 genomes. Genome browsers are customized to display sRNA-seq data based on sRNA size, strand, and multimapping (Fig. 2A). Genome browsers also allow users to highlight a region of interest and perform on the fly analyses, including ShortStack (Axtell 2013b; Johnson et al. 2016) and visualization of possible miRNA hairpins using the tool strucVis (https://github.com/MikeAxtell/strucVis) (Fig. 2B; Supplemental Code 1). Bulk data are also available in standard, widely used formats: sRNA-seq alignments are in the BAM format, whereas annotations of sRNA loci are in the GFF3 format. It is our intention to maintain and expand this resource for the benefit of anyone interested in the analysis of plant sRNA-producing loci.
Figure 2.
Example screenshots from https://plantsmallrnagenes.science.psu.edu. (A) Screenshot of genome browser for a region of Phaseolus vulgaris Chromosome 9. Coverage track shows sRNA-seq alignment depths from the reference set, separated by sRNA lengths (indicated by colors). ShortStack sRNA loci track shows sRNA locus annotations. Alignments track shows individual sRNA reads from the reference set, with lengths indicated by colors. Hollow bars indicate multimapped reads; solid bars are uniquely mapped reads. A user-highlighted region is indicated in yellow. (B) Analysis of predicted RNA secondary structure with sRNA-alignment depths indicated by colors (powered by strucVis, which is embedded within the site and also available in standalone fashion at https://github.com/MikeAxtell/strucVis). This analysis is one of several that can be triggered by user selection of a region of interest (yellow region in A).
Chromosomal distribution of sRNA loci and association with protein-coding genes
Where feasible based on genome assembly quality, we compared the distribution of sRNA loci and genes across entire chromosomes and confirmed that the most common trend is a positive correlation between gene density and sRNA density (Supplemental Fig. S5), as has previously been shown in several prior species-specific studies (The Tomato Genome Consortium 2012; He et al. 2013; Dohm et al. 2014; Kim et al. 2014; Wei et al. 2014; Song et al. 2015). A. thaliana is unique in that it has a clear trend from telomeres to centromeres of decreasing gene density and increasing sRNA loci density (Kasschau et al. 2007; Ha et al. 2009). Rice showed a similar trend to A. thaliana (Supplemental Fig. S5). Chinese cabbage and sweet orange also showed a slight inverse correlation between the gene and the sRNA loci distributions. Finally, soybean had a general positive correlation between genes and sRNA loci but in the most distal segments of the chromosome arms it showed a local negative correlation (Supplemental Fig. S5).
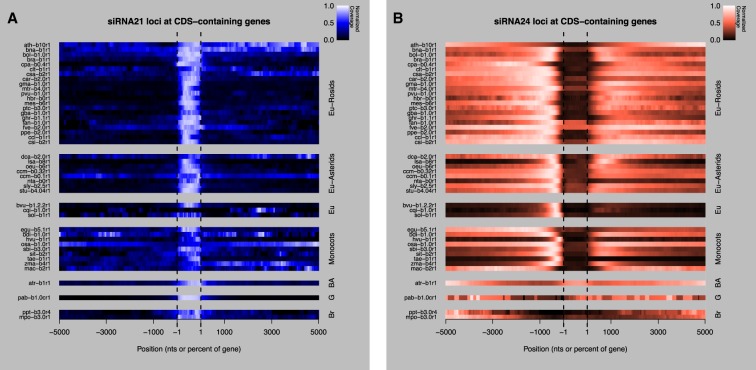
We examined siRNA21 loci and siRNA24 loci locations relative to protein-coding genes. Other types of sRNA loci were excluded because of their lower frequencies. Coverage of protein-coding genes and flanking 5-kb regions by siRNA21 or siRNA24 loci was calculated and normalized. siRNA21 loci had a strong tendency in nearly all taxa to overlap with protein-coding genes (Fig. 3A). In contrast, siRNA24 loci were strongly depleted in protein-coding genes in most angiosperms (Fig. 3B). siRNA24 loci were often strongly enriched in the 5′-proximal regions upstream of protein-coding genes. There were, however, some exceptions to this pattern. There was no upstream peak of siRNA24 loci in bryophytes and the gymnosperm (Fig. 3B), which is consistent with the generally low levels of siRNA24 loci in these taxa (Fig. 1). The basal angiosperm Amborella trichopoda was unusual in that siRNA24 loci were not depleted in gene bodies at all (Fig. 3B). Finally, the model plant A. thaliana also lacked a conspicuous upstream gene-proximal enrichment of siRNA24 loci. This observation, together with the unique chromosomal distribution of sRNA loci in A. thaliana, suggests that A. thaliana may not be representative of most angiosperms in its genome-wide patterns of sRNA loci. Protein-coding gene annotations used to perform this analysis were obtained from public resources and their different levels of accuracy could affect these results. For example, TEs can sometimes be erroneously annotated as protein-coding genes. For this reason, we highlight the possibility that the genome-wide distributions presented here could be refined in the future as more precise protein-coding gene annotations become available.
Figure 3.
Associations of siRNA21 loci and siRNA24 loci with protein-coding genes. (A) Heatmap showing normalized coverage of protein-coding genes ±5 kb by siRNA21 loci. Each row is a given species (for species codes, see Table 1), grouped taxonomically. (Eu) Eudicots; (BA) basal angiosperm; (G) gymnosperm; (Br) bryophyte. Negative and positive numbers are upstream and downstream regions, respectively (in nucleotides). The region from −1 to +1 represents the gene bodies, scaled to a uniform size of 1000 nominal units of 0.1% each. (B) As in A, except for siRNA24 loci.
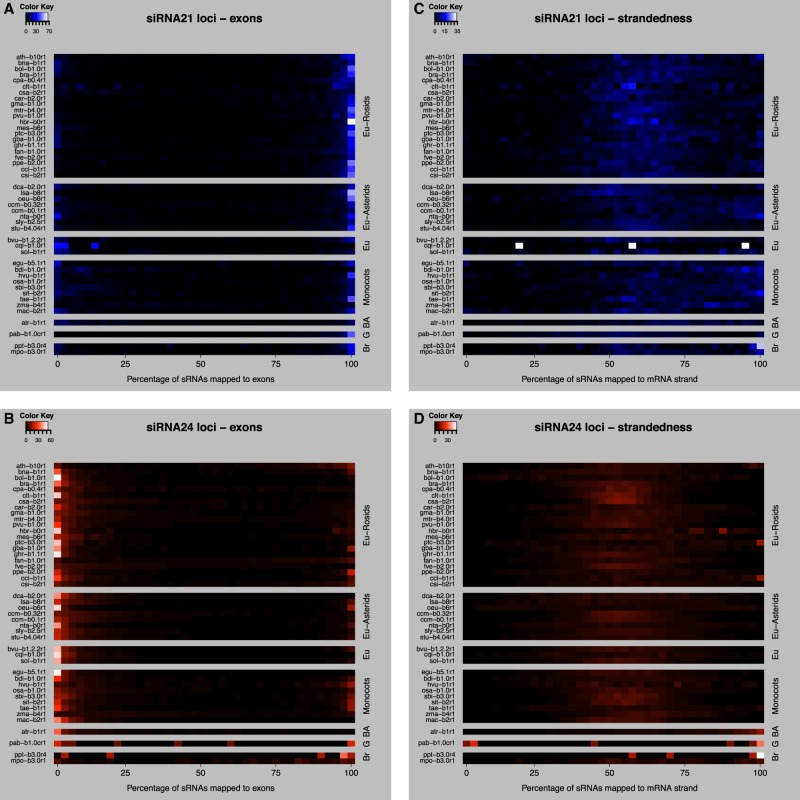
Distribution of sRNAs in exons and introns of protein-coding genes
We then analyzed the distribution of sRNAs mapped to protein-coding genes, relative to the mRNA exons/introns and relative to the coding/noncoding strand of the mRNA (Fig. 4). Although siRNA24 loci were generally depleted in mRNAs (Fig. 3), their very large numbers still resulted in many overlaps, and therefore they were included in this analysis (Supplemental Fig. S6). For each species, we calculated the proportion of mRNAs that have 0%–100% sRNAs mapped to the exons and the proportion of mRNAs that have 0%–100% sRNAs mapped to the same strand of the mRNA. The proportions were plotted separately for mRNAs containing siRNA21 and siRNA24 loci. mRNAs containing siRNA21 loci showed a strong association with sRNAs arising from exons in the vast majority of the species (Fig. 4A). These exonic 21-nt siRNAs are most likely secondary siRNAs derived from the processing of the mRNAs. In contrast, in the mRNAs containing siRNA24 loci, sRNAs were primarily generated from introns in nearly all species (Fig. 4B). Because 24-nt siRNAs are known to be enriched in TEs, these intronic 24-nt siRNAs could often be generated from intronic TE insertions. Some species showed a lesser association of siRNA24 loci with introns: This may be caused by differences in the annotation of TEs, which can sometimes be erroneously annotated as mRNAs. To verify these hypotheses, it would be necessary to analyze the genomic localization of high-confident TEs and their sRNA coverage. Accurate and specific TE annotations were only available for a very small number of species, and for this reason we did not analyze the general sRNA distribution on TEs and different TE families.
Figure 4.
Distribution of sRNAs in the body region of protein-coding mRNAs. (A) Heatmap showing the proportion of mRNAs containing siRNA21 loci that have 0%–100% of their aligned sRNAs mapped to their exons: 0% means all sRNAs map to introns; 100% means all sRNAs map to exons. (B) As in A, except for siRNA24 loci. (C) Heatmap showing the proportion of mRNAs containing siRNA21 loci with 0%–100% of their aligned sRNAs mapped to the coding strand of the mRNA: 0% means all sRNAs map to the noncoding strand; 100% blue means all sRNAs map to the coding strand of the mRNA. (D) As in C, except for siRNA24 loci. Each row is a given species (for species codes, see Table 1), grouped taxonomically. (Eu) Eudicots; (BA) basal angiosperm; (G) gymnosperm; (Br) bryophyte.
The siRNAs at both siRNA21 and siRNA24 loci typically originated from both strands of their associated genes (Fig. 4C,D). This trend is consistent with processing from dsRNA precursors, as opposed to breakdown products from the mRNAs themselves.
Identity of genes associated with sRNA loci
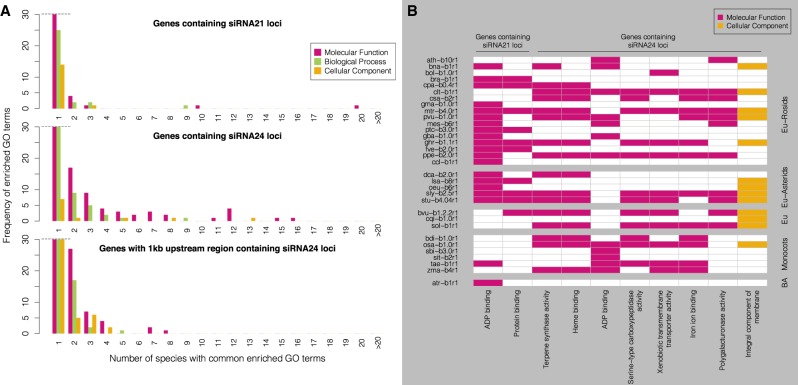
To begin to understand the function of genes associated with sRNA loci, we performed Gene Ontology (GO) enrichment analysis on the protein-coding genes that contained siRNA21 or siRNA24 loci within their gene body, or siRNA24 loci in their 1-kb upstream region (Fig. 5). For 38 of the 48 plant genomes, we were able to easily retrieve adequate GO annotations. These were used to perform Fisher's exact test in Blast2GO in each species (FDR < 0.05). We plotted the frequency at which the GO terms were found enriched among the species to find conserved terms (Fig. 5A). Enriched GO terms commonly found in at least 10 species were considered to be well conserved, because at this number the frequency distribution inverted after gradually decreasing to zero. Genes containing siRNA21 and siRNA24 loci had, respectively, two and eight well-conserved GO terms. In contrast, genes with siRNA24 loci within their 1-kb upstream region had no enriched GO term shared by 10 or more species. The species distribution of the well-conserved GO terms (Fig. 5B) revealed that the “ADP binding” term was enriched in genes containing siRNA21 loci in rosids, asterids, in A. trichopoda, and only in one monocot (wheat). Genes associated with the ADP binding function corresponded in all species with NB-LRR-type disease resistance genes, which are known to produce secondary siRNAs in many species and only in barley and wheat among the monocots (Liu et al. 2014; Zhang et al. 2019). The “protein binding” term was also enriched in genes containing siRNA21 loci, but the genes associated with this term had heterogeneous and variable annotations between species, therefore no single common pathway was identified. Nevertheless, a few gene families in the “protein binding” group were commonly found among species, for example F-box genes, PPR-containing genes, kinases, and SET domain-containing genes. Genes containing siRNA24 loci had well-conserved enriched GO terms mostly found in all clades and with different molecular functions (Fig. 5B): “Terpene synthase” and “heme binding” (mostly cytochromes P450 and other peroxidases) were the most conserved, followed by five others, including the “ADP binding” function. We hypothesize that the genes with these specific functions might be particularly frequent targets of intronic TE insertions silenced by 24-nt siRNAs.
Figure 5.
GO enrichment analysis of protein-coding genes associated with siRNA21 or siRNA24 loci. (A) Frequency of enriched GO terms in the 38 species analyzed (hbr-b0, tcc-b1.1, fan-b1.0, mdm-b3.0, csi-b2, ccm-b0.32, ccm-b0.1, can-b1.6, nta-b0, and hvu-b1 were excluded because no gene annotation or no GO annotation was available). (B) Species distribution of well-conserved enriched GO terms, common to 10 or more plant species (car-b2.0, egu-b5.1, mac-b2, pab-b1.0c, ppt-b3.0, and mpo-b3.0 were not displayed because they were not enriched in any of these terms). Each row is a given species (for species codes, see Table 1), grouped taxonomically. (Eu) Eudicots; (BA) basal angiosperm.
Disease resistance genes and other genes producing siRNAs in monocots
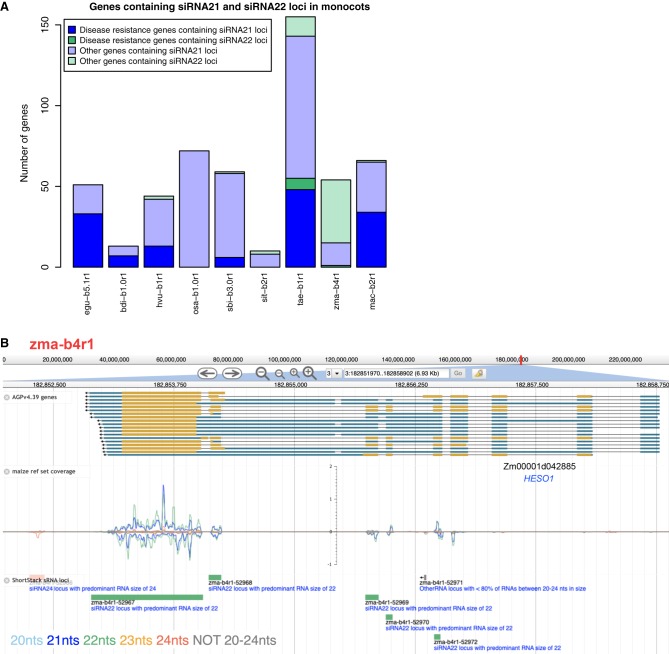
We further examined the nature of the genes containing exonic sRNA loci in monocots. This was of interest because the regulation of disease resistance genes by sRNAs in monocots has been described only in barley and wheat so far (Liu et al. 2014; Zhang et al. 2019). Genes containing exonic siRNA21 and siRNA22 loci were both studied, because within the monocots, maize produced high quantities of 22-nt siRNAs (Fig. 1), whose function is not well-understood. The genes were manually screened to discard those with stacks of sRNA reads mapped at only one or two unique positions, that could be alignment artifacts or miRNA-like sRNAs. Known miRNA matches, lowly expressed sRNA loci (<1 RPM [reads per million]), transposons, and inverted repeats were also discarded. In total, 524 genes in the nine monocots were selected as containing robust siRNA21 and siRNA22 loci (Fig. 6A; Supplemental Table S3). Maize was the only species in which the majority of genic siRNA loci were siRNA22 loci; wheat also had some genic siRNA22 loci. This suggests that in maize and maybe wheat, the 22-nt siRNAs could be a functionally active class of sRNAs in the regulation of genes, in addition to the 21-nt siRNAs.
Figure 6.
Genes containing siRNA21 and siRNA22 loci in nine monocot species. (A) Counts of genes containing siRNA21 and siRNA22 loci in monocots. (B) Screenshot of genome browser for maize HESO1 (Zm00001d042885). (Top row) mRNA structure: blue blocks for UTRs, yellow blocks for CDS, and black lines for introns. (Middle row) sRNA-seq coverage from the reference set across the gene. (Bottom row) ShortStack sRNA loci annotation.
Evidence of sRNA expression from genes annotated as or having sequence homology with disease resistance genes, was found in seven species (Supplemental Table S3). Confirming previous reports, 13 disease resistance genes in barley and 48 in wheat contained siRNA21 loci. In oil palm and banana, 33 and 34 disease resistance genes, produced 21-nt siRNAs, respectively. Disease resistance genes evolve rapidly by tandem duplications (Yang and Huang 2014), whose expression is controlled by sRNAs. In the banana genome we found an example of this where two clusters of disease resistance genes, both on Chromosome 3, contained 23 and 15 genes in tandem in a range of ∼137 and ∼130 kb, respectively, that were sources of 21-nt siRNAs. In B. distachyon and sorghum, we found seven and six resistance genes producing 21-nt siRNAs, respectively, and in maize only one resistance gene produced 22-nt siRNAs. In rice and foxtail millet there were no disease resistance genes associated with exonic 21- or 22-nt sRNAs. This result suggests that the sRNA-mediated regulation of resistance genes could be conserved in a larger number of monocots than just barley and wheat but be selectively absent in some other monocots like rice.
Genes with different functions than resistance genes also contained siRNA21 and siRNA22 loci in monocots, and a few were conserved in multiple species (Supplemental Table S3). Example of these genes include TAS3 genes, auxin-responsive genes, kinase genes, genes encoding transport inhibitor response 1-like (TIR1-like) proteins, predicted E3 ubiquitin ligase genes, genes encoding or similar to DNA-directed RNA polymerases, two-component response regulators, and methyl-CpG-binding domain-containing proteins. Genes participating in sRNA pathways were also found to be sources of siRNAs: HEN1 SUPPRESSOR1 (HESO1) (Fig. 6B) and AGO108 in maize, DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) in rice, a predicted AGO1B in sorghum, and three predicted copies of AGO2 in wheat. As it is visible by the sRNA alignment coverage in HESO1 (Fig. 6B), sRNAs were expressed from multiple adjacent exons. This pattern of sRNA expression that reflects the mature mRNA structure was observed in many genes and strongly suggests that these exonic 21- and 22-nt sRNAs are secondary siRNAs, originated from the processing of the mRNA by a DCL protein.
Analysis of sRNA conservation across plant species
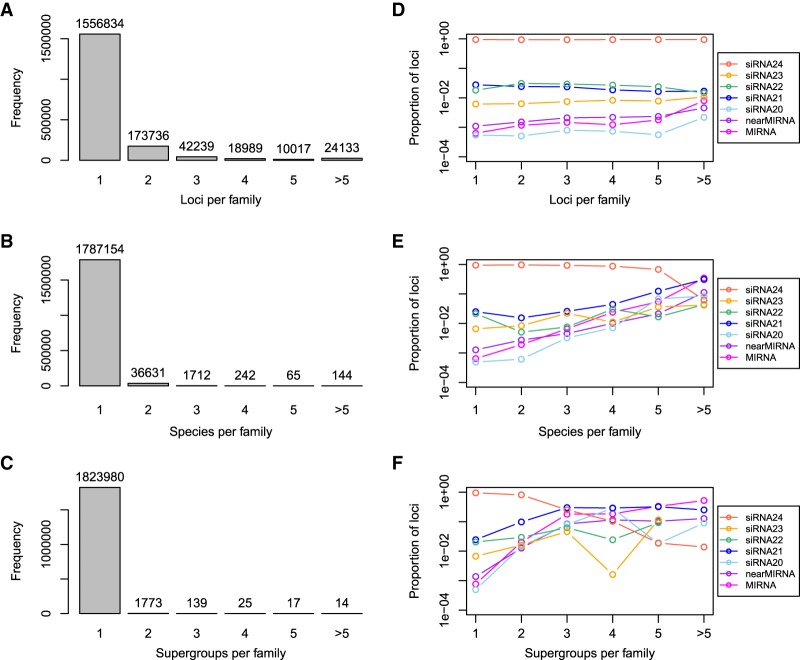
Annotated sRNA loci were grouped into putative families based on the sequences of the most abundant single sRNA (the “major RNA”) produced by each locus (Supplemental Table S4). Loci were considered to be members of the same family if the sequences of their major RNAs had up to two mismatches with each other; these criteria are similar to those commonly used to group miRNA loci into families. Most of the resulting families (1,556,834; 85.3%) had only a single locus (Fig. 7A), and relatively few families (38,794; 2.1%) were present in more than a single species (Fig. 7B). Even fewer families (1968; 0.1%) were present in more than one major taxonomic group (Fig. 7C). In general, the proportions of MIRNA, nearMIRNA, and siRNA21 loci were higher for more extensively conserved families (Fig. 7D–F); at the most extreme levels of conservation, MIRNA loci and siRNA21 loci predominated.
Figure 7.
Conservation of sRNA loci in plants. (A) Frequency distribution of number of sRNA loci per putative sRNA family. (B) Frequency distribution of number of distinct plant species per putative sRNA family. (C) Frequency distribution of number of plant “supergroups” per putative sRNA family. The supergroups defined in this study are rosids, asterids, other eudicots, monocots, basal angiosperms, gymnosperms, and bryophytes. (D) Proportions of types by number of loci per putative sRNA family. (E) Proportions of types by number of distinct plant species per putative sRNA family. (F) Proportions of types by number of plant “supergroups” per putative sRNA family.
Discussion
A public resource on sRNAs for the scientific community
We created an extensive resource for a large number of plant genomes that allows users to freely and easily retrieve, visualize and analyze sRNA loci, including not only miRNA annotations but also siRNA annotations. Our research extended into non-model systems, including many species of horticultural importance. For three economically important plants, spinach, carrot, and cacao, we annotated for the first time miRNA and siRNA loci. Recently, a study published the first miRNA annotation in carrot using high-throughput sequencing, but the siRNAs were not examined (Bhan et al. 2019). In many published works, sRNA-seq is used to annotate and profile miRNAs but not individual siRNA loci. We used the vast amount of available sRNA-seq data sets to exploit all this unrevealed information and annotate the entire population of sRNAs in 48 plant genomes.
Our database and analyses are limited by the quality and quantity of the available genomic annotations and sRNA-seq data (Supplemental Figs. S1–S3). For example, sRNAs expressed in specific tissues/cell types or growth conditions that were not represented in our sRNA-seq data set are by consequence absent from our reference annotations, but this does not necessarily imply that they are missing from the plant. This might be the case of the reproductive phasiRNAs (Zhai et al. 2015; Fei et al. 2016; Xia et al. 2019): For most of the analyzed species we did not have any or enough sRNA-seq libraries from reproductive tissues to allow the specific annotation of this sRNA population. For this reason, we did not investigate the reproductive sRNAs in our work. Another example is the limited number of manually curated TE annotations that are only available for a small number of species. TEs are the primary source/target of sRNAs. Therefore, precisely describing the pattern of sRNAs mapped to TEs is of importance to understand their regulative role. Because of the limited available data we did not analyze the sRNA distribution on TE families. Our database has the potential to be expanded in the future to include new plant genomes, new annotations, and new sRNA-seq data that are of interest for the plant biology community.
Overall, we created a resource that will be useful for future sRNA studies. Owing to the standard annotation and classification methods followed for all genomes, our sRNA annotations and alignments can be directly visualized or downloaded from our web server and compared between species. Our web server is a practical way to quickly interrogate existing plant sRNA data in a usable format and will enable scientists to rapidly search for evidence of sRNA expression in specific regions in a species or investigate the conservation of single sRNA sequences across species.
Multiple protein-coding gene families are sources of 21-nt siRNAs in dicots and monocots
The best characterized case of protein-coding genes generating secondary siRNAs are the disease resistance genes, whose expression is kept under control by secondary siRNA production to avoid fitness loss (Yang and Huang 2014). We confirmed expression of 21-nt siRNAs from exons of resistance genes in the rosid and asterid clades and expanded the number of monocot species that also showed this evidence, suggesting that this pathway might be more broadly conserved than what is known. In none of the three studied Caryophyllales species were the protein-coding genes containing siRNA21 loci enriched in the GO:ADP binding term, characteristic of resistance genes. This could result from incomplete gene/GO annotations in spinach, sugar beet, and quinoa, missing real resistance genes. Alternatively, these secondary siRNAs might be reduced in Caryophyllales because the number of disease resistance genes in this clade is lower compared to the typical expansion of this gene family in rosids and asterids or because in Caryophyllales, specific subfamilies of resistance genes have expanded that might be differentially regulated (Dohm et al. 2014; Xu et al. 2017; Funk et al. 2018).
In the literature, the number of known protein-coding genes producing secondary siRNAs in monocots is smaller than in dicots. Accordingly, from our analyses, the enrichment of siRNA21 loci in protein-coding genes was less evident in monocots compared to dicots and also the tendency of 21-nt siRNAs to map to exons was smaller in monocots. For these reasons, we decided to manually screen the monocot species for evidence of 21-nt siRNA production from protein-coding genes. We described a number of gene families, more or less conserved in the nine monocots, that produced 21-nt siRNAs, and also 22-nt siRNAs in maize and wheat. In many cases, the siRNAs were expressed specifically from multiple adjacent exons, supporting the hypothesis that they are secondary siRNAs processed from mature mRNAs. Some of the genes found were previously described as sources of secondary siRNAs in other species, for example kinase genes (Zheng et al. 2015; Reyes-Chin-Wo et al. 2017), TIR1-like genes (Si-Ammour et al. 2011; Xia et al. 2015a; Seo et al. 2018), and AGO2 (Arikit et al. 2014). In addition to AGO2, there are more genes participating in siRNA biogenesis and function that are themselves known targets of siRNA regulation: DCL1 (Xie et al. 2003; Xia et al. 2014; Hu et al. 2015b), DCL2 (Zhai et al. 2011; Arikit et al. 2014), AGO1 (Vaucheret et al. 2006), and SUPPRESSOR OF GENE SILENCING 3 (Arikit et al. 2014). We found evidence of siRNA expression from four additional genes involved in siRNA pathways: in maize, from AGO108 (also named AGO5d), highly expressed in ears but not well functionally characterized (Zhai et al. 2014), and HESO1, a nucleotidyl transferase that uridylates unmethylated sRNAs to trigger their degradation (Zhao et al. 2012); in sorghum, from a predicted AGO1B; and in rice from DRM2. DRM2 is a known target of miR820 in rice (Nosaka et al. 2012), which could be the trigger miRNA for the production of the observed 21-nt siRNAs. We reported many more genes in monocots that spawned 21- or 22-nt-long siRNAs, belonging to different families. These genes represent an interesting set to research in the future to better characterize the nature of genic siRNAs. The next obvious step will be searching for possible miRNA triggers and examining the phasing pattern of siRNA expression in each specific gene, to confirm that these siRNAs are secondary siRNAs.
Different hypotheses on 22-nt siRNA functions
We found that asterids consistently had considerable proportions of siRNA22 loci, but in the other clades, only certain species (soybean, cassava and maize) had this same trend. There are several hypotheses that could explain the presence of 22-nt siRNAs in a genome: They could originate from miRNA or miRNA-like loci that were missed by our annotation method, from endogenous direct or inverted repeats (Kasschau et al. 2007), or from protein-coding genes, as we observed in maize. Alternatively, these siRNA22 loci could express siRNAs involved in the noncanonical RdDM pathway to silence active TEs (Matzke and Mosher 2014), as it was proposed for maize (Nobuta et al. 2008). Active retrotransposons have been described in asterids, for example the Tto1 element or the Tnt1 element, which have many copies that are still transcriptionally active in tobacco (Casacuberta et al. 1997) and lettuce (Mazier et al. 2007). In this hypothesis, what still remains unclear is why we observed expression of 22-nt siRNAs most often in the asterids and not in the grasses, where retrotransposon transcription is most prevalent (Vicient et al. 2001). If the 22-nt siRNAs come from active retrotransposons, then the ability to detect their expression could depend on the specific samples analyzed, because retrotransposons are only active during certain stages of plant development or stress conditions (Flavell et al. 1992). Last, 22-nt siRNAs could target Endogenous Viral Elements, virus segments that are integrated in the host genome, that form inverted repeats (Pooggin 2018). To understand the role of the siRNA22 loci, the next step in future research will be the genome-wide profiling of the genomic regions where these loci map, discriminating between genes, intergenic regions, and different classes of TEs.
Roles of 24-nt siRNAs in regulating protein-coding gene expression
We assumed that the distribution of the total sRNA loci across the chromosome length reflected the distribution of the siRNA24 loci, because these accounted for the vast majority of loci in angiosperms. A. thaliana and Chinese cabbage are two of the few species in which siRNA24 loci and gene densities were negatively correlated. In both species, siRNA regulation of TEs near genes was previously linked to lower expression of the genes (Hollister et al. 2011; Woodhouse et al. 2014). It would be informative to test if the same link occurs in the other species with inverse correlation between siRNA24 locus and gene densities, like sweet orange. Differences in siRNA24 locus distribution and influence on gene expression might be directly explained by differences in TE composition between genomes. Accordingly, it was previously suggested that the transcription of gene networks can be balanced by the genome distribution of TEs (Freeling et al. 2015), which can be highly variable among species (Vicient and Casacuberta 2017). In many cases, a few TE families have increased their copy number in one lineage (Baidouri and Panaud 2013). For example, a single type of LTR retrotransposon is responsible for most of the hot pepper genome expansion (Park et al. 2012).
The angiosperms analyzed were strongly enriched in siRNA24 loci in the 5′-proximal regions upstream of protein-coding genes. In A. thaliana, this distribution was much less strong but the enrichment of siRNA24 loci in the 5′ upstream region compared to the gene-body region was still evident. The function of siRNA24 loci at these sites has been widely studied in maize: Near genes, 24-nt siRNAs engage RdDM, blocking the spread of open, active chromatin into adjacent transposons (Li et al. 2015). In addition to silencing TEs, the RdDM activity near genes in A. thaliana can also affect the expression levels of some genes (Zhong et al. 2012; Zheng et al. 2013), likely by changing the chromatin landscape at gene promoters and influencing the ability of transcription factors to bind to the promoters and stimulate transcription. In maize on the contrary, no obvious direct effects on gene expression were detected as a consequence of the loss of gene proximal 24-nt siRNAs (Lunardon et al. 2016). Finally, in A. thaliana, it has been speculated that the RdDM activity near genes can influence their expression by inhibiting interactions between the promoters and their potential distant regulatory elements (Rowley et al. 2017). Similarly, most angiosperms were also enriched in siRNA24 loci at the 3′-proximal regions downstream from genes. For some genes, the RdDM activity at their 3′-proximal downstream region was suggested to reduce the readthrough transcription by Pol II into neighboring genes or TEs (Erhard et al. 2015).
When siRNA24 loci were found inside protein-coding genes, they were mostly in introns. A few gene families were most commonly targeted by 24-nt siRNAs in both dicots and monocots. Two possible reasons might explain why these specific genes were a common target of 24-nt siRNAs. On one side, families like disease resistance genes evolve rapidly, creating high numbers of partial genes and pseudogenes (Luo et al. 2012) that might be suppressed by the activity of 24-nt siRNAs (Kasschau et al. 2007). This could also be the case of polygalacturonases that are encoded by a large gene family. An accurate study of the protein-coding gene annotations, precisely separating genes from pseudogenes, would be necessary to verify this hypothesis. On the other side, gene families like disease resistance genes control adaptive responses to the environment, making them frequent targets of TE transposition events (Quadrana et al. 2016). Although the majority of TE insertions in genes are deleterious, they can be advantageous and therefore be retained as a source of variability, which is essential in environmental response genes to adapt to changing conditions. As a consequence, new TE insertions are overrepresented in genes that respond to environmental stresses (Grover et al. 2003; Miyao et al. 2003). Also in cytochrome P450s, a family known to participate in stress responses, frequent TE insertions were described as a strategy for variability (Chen and Li 2007) and this could explain why these genes were frequent targets of 24-nt siRNAs. Likewise, serine-type carboxypeptidases, which participate in protein degradation, and xenobiotic transmembrane transporters, which work in xenobiotic detoxification pathways together with cytochromes P450, both play pivotal roles in plant defense responses and therefore could be frequent targets of TE insertions controlled by 24-nt siRNAs. To verify if the intronic 24-nt siRNAs influence the regulation of the genes that they target, it will be informative in the future to examine mutants lacking the production of 24-nt siRNAs and observe if these gene families tend to be altered in their expression.
Conservation of siRNAs
The sequence comparison of the most abundant sRNA expressed from each locus revealed a very low level of conservation of siRNAs across species, not just between distant species but also between close relatives. Studying the conservation of siRNAs is complicated by the fact that the siRNA population can vary substantially between different organs of the same plant species (Ha et al. 2009). Nonetheless, our result is in line with previous observations (Ma et al. 2010). If we consider plants that all have a strong peak of 24-nt siRNAs and have a functional RdDM pathway, the genomic TE composition and organization can significantly differ between different species and even between different varieties of the same species (Brunner et al. 2005; Quadrana et al. 2016). This might explain why the siRNA sequences that target the TEs are also poorly conserved. Much of our knowledge regarding sRNAs comes from model plants like A. thaliana, which has a low amount of TEs that are not active in wild-type plants. Crop genomes, instead, have high TE loads and some TEs are active in wild-type genetic backgrounds in maize and rice (Jiang et al. 2003; Nakazaki et al. 2003; Lisch 2012). Because of these differences it is important to study sRNAs in non-model systems, because lineage- or species-specific sRNAs might be associated to traits that other plants lack or have not evolved (Chen et al. 2018).
Methods
Plant material and sRNA sequencing
Leaves of Theobroma cacao (line Scavina 6) were kindly provided by Dr. M. Guiltinan of The Pennsylvania State University, from plants grown in greenhouse conditions. The tips of leaves at the immature green leaf stage were collected. Daucus carota (cultivar “Burpee”) was grown in a growth room at 22°C, 16 h light 8 h dark regime, and leaves and roots from 5- and 6-wk-old plants, respectively, were sampled. Spinacia oleracea Sp75 inbred line seeds were kindly provided by Dr. Z. Fei of the Boyce Thompson Institute, Cornell University, and grown in a growth room at 22°C, 16 h light 8 h dark regime. Leaves from 3- and 5-wk-old plants were collected. Zea mays B73 inbred line seeds were germinated on ProMix B, then transferred to soil in pots and grown in greenhouse conditions with occasional Osmocote fertilization. The fifth and the sixth leaves from V5 plants, mature pollen, and 21–27 days after pollination (DAP) embryo tissue were collected from a pool of plants. All samples were flash frozen in liquid nitrogen, stored at −80°C, and then ground with liquid nitrogen–cooled mortar and pestle. For carrot, spinach, and maize, the RNA was extracted with TRI Reagent (Sigma-Aldrich) per manufacturer instructions, adding a second sodium-acetate–ethanol precipitation and ethanol wash step. For cacao, the RNA was extracted with PureLink Plant RNA Reagent (Thermo Fisher Scientific) following the manufacturer's suggestions. Sequencing libraries were prepared using the NEB Next sRNA-seq library preparation kit for Illumina (NEB, E7300S) following the manufacturer's suggestions. Reactions were purified and size selected for sRNAs 15–40 nt in length by PAGE. Extracted bands were quantified by qPCR and quality-controlled by high-sensitivity DNA chip (Agilent). Sequencing was performed on a HiSeq 2500 (Illumina) in rapid run mode (50 nt, single-end, single barcode) by the Penn State genomics core.
sRNA-seq data processing
sRNA-seq raw FASTQ files were downloaded from the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) and the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) databases (Supplemental Table S1). The libraries were processed to remove the 3′ adapter with cutadapt (Martin 2011) (cutadapt -a 3′_adapter_sequence --discard-untrimmed -m 15 -o output_file.fastq input_file.fastq). Reads containing the 5′ adapter were removed with cutadapt (cutadapt -g 5′_adapter_sequence --discard-trimmed -m 15 -o output_file.fastq input_file.fastq). Low quality reads were discarded with FASTX-Toolkit (fastq_quality_ filter -q 20 -p 85 -Q 33 -v -i input_file.fastq -o output_file.fastq) (http://hannonlab.cshl.edu/fastx_toolkit/). Finally, read quality was checked with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc): If additional sequencing adapters were overrepresented among reads, they were eliminated from the FASTQ files with a custom Perl script (Supplemental Code 3).
Pipeline to create reference sRNA loci annotations
For each species, the reference annotation of sRNA loci was created with the following steps. Each individual library was aligned to the genome (see https://plantsmallrnagenes.science.psu.edu for list of genome assemblies used) using ShortStack v3.8.1 (Supplemental Code 2; Axtell 2013b; Johnson et al. 2016) with default parameters. By default, ShortStack handles multimapping sRNAs in this way: (1) Extreme multimapping reads with more than 50 possible best-match alignments are marked as unmapped (--bowtie_m 50) (Langmead et al. 2009); (2) the other multimapping reads are aligned by the unique-weighting mode (ShortStack --mmap u), that uses the frequencies of uniquely mapping reads within the vicinity of the multimapper to determine its proper placement (Johnson et al. 2016). Libraries with fewer than 2 million mapped reads were discarded. Clusters of sRNAs were de novo identified in each library independently with ShortStack (ShortStack --bamfile alignment_file.bam --mincov 2rpm --genomefile genome_file.fa). The sRNA cluster files from all libraries of the same species were intersected with the BEDTools function “multiIntersectBed” (Quinlan and Hall 2010) with default parameters. Only genomic intervals with annotated sRNA clusters common to at least three libraries were kept and merged with BEDTools, with 25 nt as maximum distance allowed between the intervals to be merged into sRNA loci (mergeBed -d 25 -i input_intervals_file.bed > output_merged_intervals_file.bed). sRNA loci with length <15 nt were removed with a custom Perl script (Supplemental Code 4). Finally, sRNA loci whose expression was <0.5 RPM in all libraries were also removed. The sRNA loci that were selected after applying these filters represented the reference annotation for each species.
Analysis of sRNA loci occupancy relative to protein-coding genes
Locations of protein-coding genes were determined from public GFF3 files from each genome. Intergenic regions were calculated using BEDTools “complement,” computationally cut in half, and associated with their nearest protein-coding genes using BEDTools “closest.” The regions were marked as upstream or downstream based on the orientation of their nearest flanking gene. Per-nucleotide overlap between upstream, downstream, and gene-body regions versus small RNA loci were calculated using BEDTools “overlap.” The lengths of gene bodies were scaled to 1000 arbitrary units (each such unit is 0.1% of the gene length). Coverage was summarized in 25 nt/unit bins, and normalized to a scale of 0 to 1, where 1 represented the maximum fraction occupancy observed in that genome.
Analysis of sRNA distribution in exons and introns of protein-coding mRNAs
Only protein-coding mRNAs having at least one intron and overlapping with siRNA21 and siRNA24 loci were studied. Each mRNA was either classified as containing siRNA21 or siRNA24 loci: In case of overlap with both siRNA21 and siRNA24 loci, the longest sRNA locus was considered. The number of sRNAs mapped to exons and to the same strand of protein-coding mRNAs containing one or more introns were calculated with the BEDTools function “coverageBed -counts” (parameters added for exons: “-F 1”; for the same strand: “-F 1 -s”). The number of sRNAs mapped to introns and to the opposite strand of the mRNAs were also calculated for the final ratios (parameters added for the opposite strand: “-F 1 -S”). The percentage of sRNAs mapped to exons was calculated based on the ratio “number of reads mapped to exons/(number of reads mapped to exons + number of reads mapped to introns).” The percentage of sRNAs mapped to the same strand of the mRNA was calculated based on the ratio “number of reads mapped to the same strand/(number of reads mapped to the same strand + number of reads mapped to the opposite strand).” Here and in the other analyses of siRNAs, sRNA loci classified as MIRNA and nearMIRNA or whose most abundant sequence had a perfect match with a high-confidence plant miRNA hairpin annotated in miRBase v22 (Kozomara and Griffiths-Jones 2014) were not included.
GO enrichment analysis
Protein-coding genes, both containing single and multiple exons, were classified as containing siRNA21 or siRNA24 loci and as flanked in their 1-kb upstream region by siRNA21 or siRNA24 loci: When the same gene/upstream region overlapped with both siRNA21 and siRNA24 loci, the longest sRNA locus determined the classification. The GO enrichment analysis was performed with Blast2GO (Götz et al. 2008), using the Fisher's exact test with default parameters (FDR < 0.05). Only the species for which we were able to retrieve a GO annotation were analyzed, this excluded hbr-b0, tcc-b1.1, fan-b1.0, mdm-b3.0, csi-b2, ccm-b0.32, ccm-b0.1, can-b1.6, nta-b0, and hvu-b1.
Analysis of genes containing siRNA21 and siRNA22 loci in monocots
To find all genes, including single and multiple exon genes, containing siRNA21 and siRNA22 loci in exons we used BEDTools (intersectBed -wao -F 0.75 -a exons_file.gff3 -b sRNA_loci_file.gff3 > output_intersection_file.txt). When the same gene contained both siRNA21 and siRNA22 loci, if it contained a greater number of siRNA21 loci than siRNA22 loci it was classified as containing siRNA21 loci. In case there were the same number of siRNA21 and siRNA22 loci, the gene was classified based on the longest locus. The description of the genes (Supplemental Table S3) was copied from the gene annotation files retrieved from the same online resources used for the genome sequences (see https://plantsmallrnagenes.science.psu.edu for sources of genomes and gene annotations files). For species without available gene annotations, the function of the genes was predicted using BLAST (Camacho et al. 2009) on the gene sequence and considering the best result.
Data access
All sRNA-seq libraries used, published, and newly generated are available in the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and/or Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra); see Supplemental Table S1 for a complete list of accession numbers. The newly generated data have been submitted under the following accession numbers: GSM2805293–GSM2805297, GSM2055763–GSM2055772. All data and analyses are hosted at https:// plantsmallrnagenes.science.psu.edu.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the Penn State Genomics Core Facility for small RNA sequencing services, and the Eberly College of Science Information Technology office for providing server hosting services for this project. We thank Matthew Jones-Rhoades for supervision and mentoring of undergraduate researchers and for comments on the manuscript. This work was supported by an award from the U.S. National Science Foundation (Award 1339207) to M.J.A.
Author contributions: A.L. and M.J.A. generated most primary annotations and most analyses, with some primary annotations also contributed by S.P. The project website was developed by M.J.A. N.R.J., E.H., T.P., and C.C. contributed small RNA sequencing results. The manuscript was written by A.L. and M.J.A. with input from N.R.J. and C.C.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.256750.119.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdés-López O, Prince S, Musket TA, Nguyen HT, et al. 2014. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26: 4584–4601. 10.1105/tpc.114.131847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013a. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159. 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. 2013b. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19: 740–751. 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. 2005. Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673. 10.1105/tpc.105.032185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. 2008. Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349. 10.1016/j.tplants.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Baidouri M El, Panaud O. 2013. Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol Evol 5: 954–965. 10.1093/gbe/evt025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, DePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. 10.1126/science.1203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan B, Koul A, Sharma D, Manzoor MM, Kaul S, Gupta S, Dhar MK. 2019. Identification and expression profiling of miRNAs in two color variants of carrot (Daucus carota L.) using deep sequencing. PLoS One 14: e0212746 10.1371/journal.pone.0212746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A. 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. 10.1105/tpc.104.025627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta JM, Vernhettes S, Audeon C, Grandbastien M-A. 1997. Quasispecies in retrotransposons: a role for sequence variability in Tnt1 evolution. Genetica 100: 109–117. 10.1023/A:1018309007841 [DOI] [PubMed] [Google Scholar]
- Chávez Montes RA, De Fátima Rosas-Cárdenas F, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martínez N, Meyers BC, Green PJ, De Folter S. 2014. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 5: 3722 10.1038/ncomms4722 [DOI] [PubMed] [Google Scholar]
- Chen S, Li X. 2007. Transposable elements are enriched within or in close proximity to xenobiotic-metabolizing cytochrome P450 genes. BMC Evol Biol 7: 46 10.1186/1471-2148-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zeng Z, Liu Z, Xia R. 2018. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic Res 5: 63 10.1038/s41438-018-0072-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruh C, Shahid S, Axtell MJ. 2014. Seeing the forest for the trees: annotating small RNA producing genes in plants. Curr Opin Plant Biol 18: 87–95. 10.1016/j.pbi.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruh C, Cho SH, Shahid S, Liu Q, Wierzbicki A, Axtell MJ. 2015. Comprehensive annotation of Physcomitrella patens small RNA loci reveals that the heterochromatic short interfering RNA pathway is largely conserved in land plants. Plant Cell 27: 2148–2162. 10.1105/tpc.15.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. 2011. Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442. 10.1105/tpc.110.082784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, et al. 2014. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549. 10.1038/nature12817 [DOI] [PubMed] [Google Scholar]
- Dolgosheina EV, Morin RD, Aksay G, Sahinalp SC, Magrini V, Mardis ER, Mattsson J, Unrau PJ. 2008. Conifers have a unique small RNA silencing signature. RNA 14: 1508–1515. 10.1261/rna.1052008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard KF Jr, Talbot JE, Deans NC, McClish AE, Hollick JB. 2015. Nascent transcription affected by RNA polymerase IV in Zea mays. Genetics 199: 1107–1125. 10.1534/genetics.115.174714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Yang L, Liang W, Zhang D, Meyers BC. 2016. Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. J Exp Bot 67: 6037–6049. 10.1093/jxb/erw361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A. 1992. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res 20: 3639–3644. 10.1093/nar/20.14.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formey D, Iñiguez LP, Peláez P, Li YF, Sunkar R, Sánchez F, Reyes JL, Hernández G. 2015. Genome-wide identification of the Phaseolus vulgaris sRNAome using small RNA and degradome sequencing. BMC Genomics 16: 423 10.1186/s12864-015-1639-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D. 2008. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res 18: 1924–1937. 10.1101/gr.081026.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Xu J, Woodhouse M, Lisch D. 2015. A solution to the C-value paradox and the function of junk DNA: the genome balance hypothesis. Mol Plant 8: 899–910. 10.1016/j.molp.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Funk A, Galewski P, McGrath JM. 2018. Nucleotide-binding resistance gene signatures in sugar beet, insights from a new reference genome. Plant J 95: 659–671. 10.1111/tpj.13977 [DOI] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK. 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637. 10.1101/gr.146985.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, Kent MR, Zhang X, Stam M, McGinnis KM, Dawe RK. 2014. Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917. 10.1105/tpc.114.130427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435. 10.1093/nar/gkn176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover D, Majumder PP, Rao CB, Brahmachari SK, Mukerji M. 2003. Nonrandom distribution of Alu elements in genes of various functional categories: insight from analysis of human chromosomes 21 and 22. Mol Biol Evol 20: 1420–1424. 10.1093/molbev/msg153 [DOI] [PubMed] [Google Scholar]
- Guo Q, Qu X, Jin W. 2015. PhaseTank: genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics 31: 284–286. 10.1093/bioinformatics/btu628 [DOI] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ. 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci 106: 17835–17840. 10.1073/pnas.0907003106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Rueda A, Gustafson P, Langridge P, Shi BJ. 2016. Generation of different sizes and classes of small RNAs in barley is locus, chromosome and/or cultivar-dependent. BMC Genomics 17: 735 10.1186/s12864-016-3023-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Chen B, Wang X, Li X, Li J, He H, Yang M, Lu L, Qi Y, Wang X, et al. 2013. Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol 14: R57 10.1186/gb-2013-14-6-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS. 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci 108: 2322–2327. 10.1073/pnas.1018222108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. 2007. Genome-Wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. 10.1105/tpc.107.050062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Rashotte AM, Singh NK, Weaver DB, Goertzen LR, Singh SR, Locy RD. 2015a. The complexity of posttranscriptional small RNA regulatory networks revealed by in silico analysis of Gossypium arboreum L. leaf, flower and boll small regulatory RNAs. PLoS One 10: e0127468 10.1371/journal.pone.0127468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Yu D, Liu H. 2015b. Bioinformatics analysis of small RNAs in pima (Gossypium barbadense L.). PLoS One 10: e0116826 10.1371/journal.pone.0116826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Brachypodium Initiative. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. 10.1038/nature08747 [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR. 2003. An active DNA transposon family in rice. Nature 421: 163–167. 10.1038/nature01214 [DOI] [PubMed] [Google Scholar]
- Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. 2007. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res 35: D829–D833. 10.1093/nar/gkl991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NR, Yeoh JM, Coruh C, Axtell MJ. 2016. Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6: 2103–2111. 10.1534/g3.116.030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57 10.1371/journal.pbio.0050057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, et al. 2014. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46: 270–278. 10.1038/ng.2877 [DOI] [PubMed] [Google Scholar]
- Klevebring D, Street NR, Fahlgren N, Kasschau KD, Carrington JC, Lundeberg J, Jansson S. 2009. Genome-wide profiling of Populus small RNAs. BMC Genomics 10: 620 10.1186/1471-2164-10-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R. 2017. Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J Plant Res 130: 17–23. 10.1007/s10265-016-0878-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res 47: D155–D162. 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YS, Zhang X, Zhang W, Shen D, Wang H, Xia Y, Qiu Y, Song J, Wang C, Li X. 2017. The association of changes in DNA methylation with temperature-dependent sex determination in cucumber. J Exp Bot 68: 2899–2912. 10.1093/jxb/erx144 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, Gouzy J, Crespi M. 2009. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796. 10.1105/tpc.109.068130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, et al. 2015. RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci 112: 14728–14733. 10.1073/pnas.1514680112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. 2012. Regulation of transposable elements in maize. Curr Opin Plant Biol 15: 511–516. 10.1016/j.pbi.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu D, Xu W, Wise R, Shen QH. 2014. The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet 10: e1004755 10.1371/journal.pgen.1004755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, DIng C, Chu Y, Zhang W, Guo G, Chen J, Su X. 2017. Pln24NT: a web resource for plant 24-nt siRNA producing loci. Bioinformatics 33: 2065–2067. 10.1093/bioinformatics/btx096 [DOI] [PubMed] [Google Scholar]
- Lunardon A, Forestan C, Farinati S, Axtell MJ, Varotto S. 2016. Genome-wide characterization of maize small RNA loci and their regulation in the required to maintain repression6-1 (rmr6-1) mutant and long-term abiotic stresses. Plant Physiol 170: 1535–1548. 10.1104/pp.15.01205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhang Y, Hu Q, Chen J, Li K, Lu C, Liu H, Wang W, Kuang H. 2012. Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol 159: 197–210. 10.1104/pp.111.192062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Coruh C, Axtell MJ. 2010. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103. 10.1105/tpc.110.073882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martinez G, Choudury SG, Slotkin RK. 2017. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45: 5142–5152. 10.1093/nar/gkx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Mazier M, Botton E, Flamain F, Bouchet J-P, Courtial B, Chupeau M-C, Chupeau Y, Maisonneuve B, Lucas H. 2007. Successful gene tagging in lettuce using the Tnt1 retrotransposon from tobacco. Plant Physiol 144: 18–31. 10.1104/pp.106.090365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. 2003. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780. 10.1105/tpc.012559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. 2006. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res 34: D731–D735. 10.1093/nar/gkj077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T. 2003. Mobilization of a transposon in the rice genome. Nature 421: 170–172. 10.1038/nature01219 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. 2008. Distinct size distribution of endogenous siRNAs in maize: evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci 105: 14958–14963. 10.1073/pnas.0808066105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka M, Itoh JI, Nagato Y, Ono A, Ishiwata A, Sato Y. 2012. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet 8: e1002953 10.1371/journal.pgen.1002953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. 2013. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol 162: 116–131. 10.1104/pp.113.216481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Park J, Kim S, Kwon JK, Park HM, Bae IH, Yang TJ, Lee YH, Kang BC, Choi D. 2012. Evolution of the large genome in Capsicum annuum occurred through accumulation of single-type long terminal repeat retrotransposons and their derivatives. Plant J 69: 1018–1029. 10.1111/j.1365-313X.2011.04851.x [DOI] [PubMed] [Google Scholar]
- Polydore S, Lunardon A, Axtell MJ. 2018. Several phased siRNA annotation methods can frequently misidentify 24 nucleotide siRNA-dominated PHAS loci. Plant Direct 2: e00101 10.1002/pld3.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin MM. 2018. Small RNA-omics for plant virus identification, virome reconstruction, and antiviral defense characterization. Front Microbiol 9: 2779 10.3389/fmicb.2018.02779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Silveira AB, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V. 2016. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 5: e15716 10.7554/eLife.15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C, Xia L, Froenicke L, Lavelle DO, Truco MJ, et al. 2017. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun 8: 14953 10.1038/ncomms14953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Rothi MH, Böhmdorfer G, Kuciński J, Wierzbicki AT. 2017. Long-range control of gene expression via RNA-directed DNA methylation. PLoS Genet 13: e1006749 10.1371/journal.pgen.1006749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, He Y, Valdés-López O, Khan SM, Joshi T, Urich MA, Nery JR, Diers B, Xu D, Stacey G, et al. 2013. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res 23: 1663–1674. 10.1101/gr.152538.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Kim T, Park JH, Yeom SI, Kim S, Seo MK, Shin C, Choi D, Tabata S. 2018. Genome-wide comparative analysis in Solanaceous species reveals evolution of microRNAs targeting defense genes in Capsicum spp. DNA Res 25: 561–575. 10.1093/dnares/dsy025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Sun S, Hua S, Shen E, Ye CY, Cai D, Timko MP, Zhu QH, Fan L. 2017. Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J 91: 874–893. 10.1111/tpj.13605 [DOI] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F, Vazquez F. 2011. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol 157: 683–691. 10.1104/pp.111.180083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Lu X, Li QT, Chen H, Hu XY, Ma B, Zhang WK, Chen SY, Zhang JS. 2013. Genome-wide analysis of DNA methylation in soybean. Mol Plant 6: 1961–1974. 10.1093/mp/sst123 [DOI] [PubMed] [Google Scholar]
- Song Q, Guan X, Chen ZJ. 2015. Dynamic roles for small RNAs and DNA methylation during ovule and fiber development in allotetraploid cotton. PLoS Genet 11: e1005724 10.1371/journal.pgen.1005724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Zheng Y, Kudapa H, Jagadeeswaran G, Hivrale V, Varshneyc RK, Sunkara R. 2015. High throughput sequencing of small RNA component of leaves and inflorescence revealed conserved and novel miRNAs as well as phasiRNA loci in chickpea. Plant Sci 235: 46–57. 10.1016/j.plantsci.2015.03.002 [DOI] [PubMed] [Google Scholar]
- The Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP. 2006. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129–136. 10.1016/j.molcel.2006.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Casacuberta JM. 2017. Impact of transposable elements on polyploid plant genomes. Ann Bot 120: 195–207. 10.1093/aob/mcx078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Jääskeläinen MJ, Kalendar R, Schulman AH. 2001. Active retrotransposons are a common feature of grass genomes. Plant Physiol 125: 1283–1292. 10.1104/pp.125.3.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Gu L, Song X, Cui X, Lu Z, Zhou M, Wang L, Hu F, Zhai J, Meyers BC, et al. 2014. Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci 111: 3877–3882. 10.1073/pnas.1318131111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci 111: 5283–5288. 10.1073/pnas.1402475111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zeng C, Chen Z, Zhang K, Chen X, Zhou Y, Song S, Lu C, Yang R, Yang Z, et al. 2014. Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in Cassava. BMC Genomics 15: 634 10.1186/1471-2164-15-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Xu J, Arikit S, Meyers BC. 2015a. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol Biol Evol 32: 2905–2918. 10.1093/molbev/msv164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Ye S, Liu Z, Meyers BC, Liu Z. 2015b. Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiol 169: 594–610. 10.1104/pp.15.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Chen C, Pokhrel S, Ma W, Huang K, Patel P, Wang F, Xu J, Liu Z, Li J, et al. 2019. 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat Commun 10: 627 10.1038/s41467-019-08543-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. 2003. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13: 784–789. 10.1016/S0960-9822(03)00281-1 [DOI] [PubMed] [Google Scholar]
- Xu C, Jiao C, Sun H, Cai X, Wang X, Ge C, Zheng Y, Liu W, Sun X, Xu Y, et al. 2017. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun 8: 15275 10.1038/ncomms15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang H. 2014. Roles of small RNAs in plant disease resistance. J Integr Plant Biol 56: 962–970. 10.1111/jipb.12200 [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, de Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553. 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Sun W, Zhang K, Jia H, Liu L, Liu Z, Teng F, Zhang Z. 2014. Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J Integr Plant Biol 56: 1042–1052. 10.1111/jipb.12205 [DOI] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan GL, Walbot V, Meyers BC. 2015. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci 112: 3146–3151. 10.1073/pnas.1418918112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang S, Hao W, Song G, Li Y, Li W, Gao J, Zheng Y, Li G. 2019. Lineage-specific evolved microRNAs regulating NB-LRR defense genes in Triticeae. Int J Mol Sci 20: 3128 10.3390/ijms20133128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. 2012. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 22: 689–694. 10.1016/j.cub.2012.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Rowley MJ, Böhmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT. 2013. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J 73: 179–189. 10.1111/tpj.12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang Y, Wu J, Ding B, Fei Z. 2015. A dynamic evolutionary and functional landscape of plant phased small interfering RNAs. BMC Biol 13: 32 10.1186/s12915-015-0142-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. 2012. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19: 870–875. 10.1038/nsmb.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.