Abstract
Chromatin loops are a major component of 3D nuclear organization, visually apparent as intense point-to-point interactions in Hi-C maps. Identification of these loops is a critical part of most Hi-C analyses. However, current methods often miss visually evident CTCF loops in Hi-C data sets from mammals, and they completely fail to identify high intensity loops in other organisms. We present SIP, Significant Interaction Peak caller, and SIPMeta, which are platform independent programs to identify and characterize these loops in a time- and memory-efficient manner. We show that SIP is resistant to noise and sequencing depth, and can be used to detect loops that were previously missed in human cells as well as loops in other organisms. SIPMeta corrects for a common visualization artifact by accounting for Manhattan distance to create average plots of Hi-C and HiChIP data. We then demonstrate that the use of SIP and SIPMeta can lead to biological insights by characterizing the contribution of several transcription factors to CTCF loop stability in human cells. We also annotate loops associated with the SMC component of the dosage compensation complex (DCC) in Caenorhabditis elegans and demonstrate that loop anchors represent bidirectional blocks for symmetrical loop extrusion. This is in contrast to the asymmetrical extrusion until unidirectional blockage by CTCF that is presumed to occur in mammals. Using HiChIP and multiway ligation events, we then show that DCC loops form a network of strong interactions that may contribute to X Chromosome–wide condensation in C. elegans hermaphrodites.
High resolution Hi-C in human cells is able to find thousands of strong punctate signals that indicate the presence of loops formed by CTCF sites arranged in a convergent orientation (Rao et al. 2014). Based on this orientation preference, it has been proposed that CTCF loops are formed by a loop extrusion process mediated by cohesin (for review, see Rowley and Corces 2018). Indeed, depletion of cohesin in mammalian cells results in loss of CTCF loops (Rao et al. 2017). However, other transcription factors are also present at CTCF loop anchors, and it is unclear whether or not they play a role in loop extrusion or affect the frequency or stability of CTCF loops (Rao et al. 2014).
CTCF loops have been identified in mammals but have not been observed in other organisms. For example, Drosophila Hi-C maps do not display CTCF loops despite the existence of a conserved Drosophila homolog (Rowley et al. 2017). Instead, Drosophila contact maps in Kc167 cells contain a few hundred loops that lack CTCF, and their formation does not depend on cohesin (Rowley et al. 2019). Many nonvertebrate organisms, including Caenorhabditis elegans, lack a CTCF homolog (Heger et al. 2012). It is possible that proteins distinct from CTCF are able to form point-to-point interactions in these organisms, as is the case of Drosophila, or to stop the extrusion of SMC complexes to form loops. For example, C. elegans hermaphrodites regulate X Chromosome expression through the use of the DCC complex, which contains a condensin complex presumably able to extrude DNA (Lau and Csankovszki 2014). However, although published Hi-C contact maps reveal the presence of large self-interacting domains in the dosage-compensated X Chromosome and evidence of loop formation, current algorithms have not been successful at systematically annotating punctate signals corresponding to loops in C. elegans (Crane et al. 2015; Anderson et al. 2019). This has made it difficult to fully explore these features in nonmammalian organisms.
Here, we report a method of loop identification named Significant Interaction Peak caller (SIP) that relies on CPU-based image analysis of Hi-C contact maps to find loops. SIP detects additional functionally relevant loops in human cells and can be used to detect loops in a variety of other organisms. We also present a companion tool, SIPMeta, that creates average metaplots of loops. We show that current standard metaplots contain visual biases that SIPMeta corrects. Using SIP and SIPMeta, we test whether several transcription factors, including ZNF143, YY1, RNA Polymerase II, and CTCFL, affect the strength of CTCF loops. We then perform Hi-C and DPY-27 HiChIP in C. elegans hermaphrodites and show that the X Chromosome contains dozens of high-intensity loops configured into a complex network. These loops are associated with condensin I-DCC, suggesting the existence of extrusion-mediated non-CTCF loops. The results suggest the formation of a rosette-like structure that may be responsible for dosage compensation in this organism. Therefore, SIP and SIPMeta represent sensitive and versatile new methods for loop calling and analysis that can lead to the discovery of novel biological information from Hi-C data.
Results
SIP software
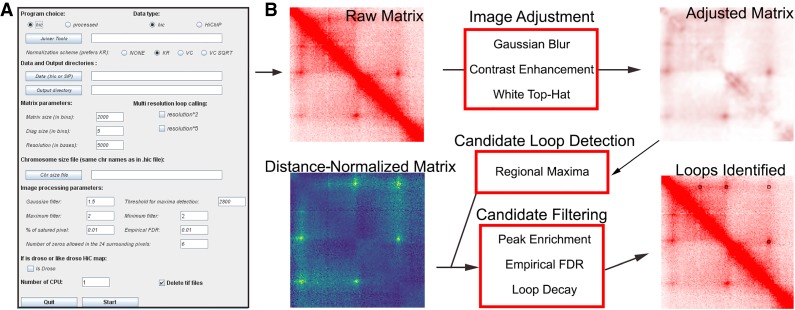
Loops present in Hi-C heat maps appear as intense saturated punctae (Rao et al. 2014). To identify these visibly evident interactions, we took advantage of image processing methods to create SIP. SIP includes options to use command line or graphical user interfaces (Fig. 1A). The SIP pipeline (Fig. 1B) reads Hi-C data in either the Juicer .hic format (Durand et al. 2016) or in a BEDPE-like format with distance-normalized signal. The genome is analyzed by sliding windows using image processing to identify potential loops, which are then filtered based on several aspects of the matrix. Images undergo a Gaussian blur, contrast enhancement, white top-hat, and a minimum-maximum filter. These steps provide a corrected image of the interactions (Fig. 1), which is used with a regional maxima detection algorithm to detect a preliminary list of candidate loops.
Figure 1.
Overview of SIP. (A) The graphical user interface provides options to specify a Juicer-derived .hic file or processed data, the output directory, and chromosome size file. It also allows adjustment of the various parameters shown. (B) SIP uses image-based detection to create a long list of candidate loops, which is further filtered based on properties of the distance-normalized matrix.
Candidate loops must then pass several filters that utilize the original distance-normalized signal in the Hi-C data. First, pixels near unmappable or repetitive regions are removed. Second, to remove isolated pixels representing noise, loops must display a decay such that the central pixel is the highest, followed by decreases at 1 and 2 pixels away from the center. The center must also be 1.2-fold higher than the average of nearby pixels and must pass a Poisson CDF filter such that the probability that the center is higher than other nearby pixels is greater than 0.9. Thus, SIP utilizes the local background to identify loops. While this is useful for identifying punctate signals, other programs that model enrichment over global background can be useful to create large lists of enhancer-promoter interactions (Ay et al. 2014). Finally, candidate loops are filtered based upon an empirical FDR calculated as the enrichment of loops versus random sites at equal distances.
Performance of SIP
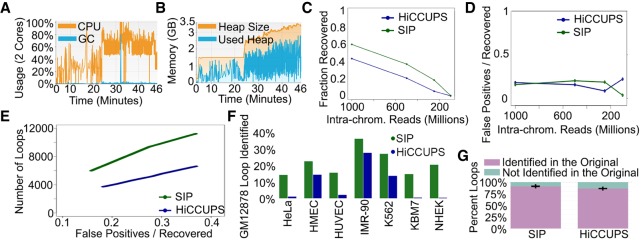
We tested the performance of SIP on Hi-C data from GM12878 cells containing approximately 2.4 billion intra-chromosomal reads (Rao et al. 2014). As a benchmark, we compared the time and memory usage of SIP to other interaction callers designed to identify CTCF loops (Durand et al. 2016; Heinz et al. 2018; Cao et al. 2019). SIP is intended to be used without the need of large computing power; therefore, we intentionally limited SIP to one thread and memory usage to 1 GB using java –Xmx1g for both SIP and HiCCUPS on Chromosome 1. In comparison, HOMER and cLoops used 62 and 103 GB, respectively, for Chromosome 1 (Supplemental Table S1). Even with these parameters, SIP identified loops 2×, 14×, or even 1057× faster than HiCCUPS, HOMER, and cLoops, respectively. We then tested SIP on a laptop with a 2-core processor and 4 GB of RAM, and we were able to call loops in the full data set, including all chromosomes, at 5-kb resolution in 46 min, including dumping data from the Juicer .hic file (Fig. 2A). To allow easier parameter optimization, users have the option of saving these dumped files and rerunning SIP, which took only 31 min. On a Linux machine using 23 cores, we were able to call loops in the full GM12878 data set in 12 min (Fig. 2B). A comparison of memory and time usage by SIP on different systems can be found in Supplemental Table S2.
Figure 2.
Performance of SIP. (A) CPU usage (orange) and GC (garbage collection, blue) over time using two cores during SIP loop calling. (B) Memory usage of SIP during loop calling. (C) Fraction of loops called using the full data set recovered by SIP (green) or by HiCCUPS (blue) in data down-sampled to different sequencing depths. (D) Ratio of false positives (loops not identified in the full data set) versus loops recovered by SIP (green) or HiCCUPS (blue) in down-sampled data. (E) Number of loops identified in down-sampled data (y-axis) for SIP (green) and HiCCUPS (blue) when parameters were adjusted to give the same false positive/recovery rate (x-axis). (F) Percentage of loops identified by SIP (green) or HiCCUPS (blue) in GM12878 cells that were identified in a different cell type. (G) Percentage of loops identified in each permutation down-sampling data (purple) versus new loops (i.e., false positives) (teal). Bars represent averages of 10 permutations with error bars representing standard deviation.
We compared loops called by SIP to those called by other loop identification programs (Supplemental Fig. S1A) and found that SIP identified more loops than existing tools (Supplemental Fig. S1B). An exception was Fit-Hi-C, which was designed to identify enhancer-promoter interactions rather than punctate spots present in Hi-C data and associated with CTCF loops (Supplemental Fig. S1A,B; Ay et al. 2014). We compared loops called by SIP versus HiCCUPS and observed good overlap. However, 33% of HiCCUPS loops were not identified by SIP, and 67% of SIP loops were not identified by HiCCUPS (Supplemental Fig. S1C). These show punctate signal in average metaplots and are therefore likely true loops that each program missed (Supplemental Fig. S1C). However, when we included cLoops, HOMER, and Fit-Hi-C in this analysis, we found that both SIP and HiCCUPS had more than 95% of loop calls identified in at least one other program (Supplemental Fig. S1D). In order to compare loops called by each program, we designated loops that were identified by at least two programs as pseudotrue positives, while loops that were unique to each program were designated as pseudofalse positives. SIP had a low pseudofalse positive rate and low pseudofalse negative rate in comparison to other programs (Supplemental Fig. S1E). These results indicate that, while current loop callers are unable to identify 100% of loops, SIP has an improved detection rate.
To further benchmark SIP, we evaluated three different aspects of the program—the ability to accurately capture loops with sparse data sets, the reproducibility of loop calls, and the resistance to noise. To test the ability to identify loops in data sets with fewer sequenced reads, we subsampled a data set with 2.4 billion intra-chromosomal paired reads and created contact maps with 1 billion, 500 million, 250 million, and 100 million reads. Regardless of the method, lower sequencing depth correlates with a decreased ability to identify loops at 5-kb resolution (Fig. 2C). However, SIP consistently identified a higher percentage of loops from the full data set than HiCCUPS (Fig. 2C). We also tested whether lower read counts resulted in identification of loops in the subsampled data set that were not identified in the full data set, i.e., likely false positives. We found that each method identified a low number of potential false positives with no correlation to sequencing depth (Fig. 2D). As a secondary test, we called loops with each method in the 1 billion-read data set but varied FDR parameters. For each FDR parameter tested, the false positive rate was calculated by the number of loops called in the subsampled data that were not called in the full data set. As expected, both methods displayed increased false positives with decreased FDR stringency. However, at similar false positive rates in the subsampled data, SIP was able to identify approximately twice the number of loops as HiCCUPS (Fig. 2E). Overall, we find that SIP is able to recover a high percentage of loops without increasing the false positive rate using Hi-C data sets with a low number of sequenced reads.
In order to determine the reproducibility of loop calls with different data sets, we called loops in Hi-C maps from eight distinct cell lines (Rao et al. 2014). Each of the eight data sets has different depths of sequencing, and, in general, the number of loops identified approximately matches the number obtained from down-sampled GM12878 data sets (Supplemental Fig. S1F). In comparison to HiCCUPS, SIP identified a larger number of loops in each data set that were also present in GM12878 cells (Fig. 2F; Supplemental Fig. S1G). Loops specific to each data set display Hi-C signal specific to that data set (Supplemental Fig. S1H). This suggests that SIP is able to reproducibly identify loops and that differences in loop calls between Hi-C maps are due to differences in looping. To further estimate the reproducibility of loop calls by SIP, we created distinct Hi-C data sets by random sampling the full data set down to 1 billion reads in independent iterations to create 10 different .hic maps. We then examined how many of the loops in each iteration were the same between data sets. Both SIP and HiCCUPS were able to reproducibly identify loops obtaining on average 91% (SIP) or 86% (HiCCUPS) of loops in each subsampled iteration that were consistent between data sets (Fig. 2G).
Next, we evaluated the ability of each method to identify loops in noisy data sets. We created Hi-C maps where noise was simulated by distributing random additional signal within the map (see Methods). We noticed that in maps with 50% additional noise signal, HiCCUPS called a large number of false positives at extreme distances crossing over the entire chromosome (Supplemental Fig. S2A, blue). These can be easily filtered using a distance cutoff. Thus, to more fairly benchmark SIP and HiCCUPS, we only examined loops <10 Mb in size. This noise model decreased the original loop signal versus background (Supplemental Fig. S2B), but both methods recovered a comparable fraction of the original loop calls despite the additional noise (Supplemental Fig. S2C,D). However, increased noise caused an increase in the false positive rate by HiCCUPS, while SIP remained consistently low (Supplemental Fig. S2C,E). While this noise model is purely artificial and may not recapitulate the true noise in a sample, these results indicate that SIP is at least partially resistant to these variations, while HiCCUPS is not.
Lastly, we examined the effects of bin size on loop calling by identifying loops at 5, 10, and 25 kb. We found that SIP and HiCCUPS had similar overlaps between these calls, but each program had loops uniquely identified at each resolution (Supplemental Fig. S2F). Therefore, as in the original HiCCUPS caller, it may be advantageous to call loops at multiple resolutions (Rao et al. 2014). Overall, these results suggest that SIP is memory- and time-efficient, identifies loops that are semiresistant to sequencing depth, has high reproducibility, and has high resistance to noise.
SIP and SIPMeta allow identification and visualization of loops in various organisms
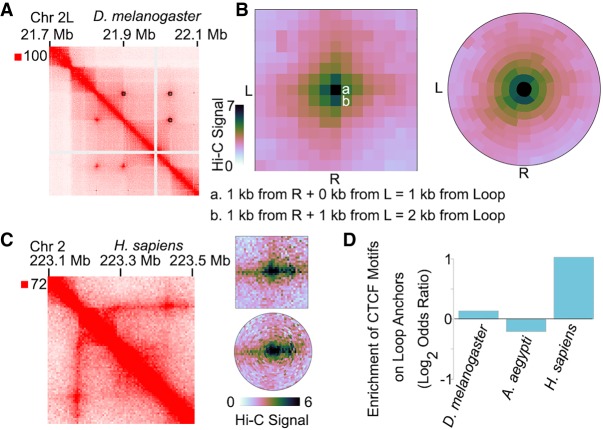
One problem with loop identification in Hi-C data sets has been that the training on one data set impacts loop calling on other data sets. This is the reason loop identification in Drosophila was done using separate custom scripts or by hand (Cubeñas-Potts et al. 2017; Eagen et al. 2017). We used SIP on Hi-C maps of Drosophila Kc167 cells and identified 143 high-intensity loops at 1-kb resolution (Fig. 3A). Visual inspection of these loops shows that they correspond to punctate signal (Fig. 3A). In comparison, other loop calling methods have a tendency to also call interactions near sparse signal that likely corresponds to repetitive regions (Supplemental Fig. S3A; see also Supplemental Fig. S2C). We then tested if anchors of loops identified by SIP were enriched in proteins previously found to be important for looping in Drosophila (Eagen et al. 2017; Ogiyama et al. 2018; Gutierrez-Perez et al. 2019). Indeed, Polycomb (Pc) and pipsqueak (Psq) are enriched on SIP loop anchors (Supplemental Fig. S3B).
Figure 3.
SIP and SIPMeta can be used to detect and analyze loops in different species. (A) Example locus for SIP loops detected in Hi-C for D. melanogaster. (B) Left: Metaplot of SIP loops illustrating that the Manhattan distance between a and b is different with respect to loops but is visually depicted as the same distance in square metaplots. (L) left anchor, (R) right anchor. Right: Metaplot of SIP loops illustrating the bullseye transformation performed by SIPMeta. (C) Left: Example locus in GM12878 cells for a stripe detected in Hi-C maps of mammals. Right: SIPMeta plots displaying how stripes appear in square versus bullseye plots. (D) Enrichment of CTCF motifs on loop anchors in D. melanogaster, A. aegypti, and H. sapiens.
A common approach to evaluating loops is through a metaplot analysis that averages the Hi-C signal at loops compared to the surrounding region (Rao et al. 2014; Rowley et al. 2019). Standard metaplots of Drosophila loops display central signal enrichment but with a crosshair-like pattern (Fig. 3B, left). This could be interpreted as evidence of extrusion, similar to enriched stripes in Hi-C maps of mammals that occur at some CTCF loops due to proximal loading of cohesin (Vian et al. 2018). However, it was previously found that depletion of cohesin or condensin II has no effect on Drosophila loop intensity (Rowley et al. 2019); thus, it is unlikely that these loops are formed via extrusion. When considering the crosshair pattern in square metaplots, we realized that the distance from the loop is different between pixels adjacent horizontally or vertically versus pixels adjacent diagonally. For example, one pixel to the right of the loop is 0 kb away from the left anchor and 1 kb away from the right anchor, equivalent to 1 kb Manhattan distance (Fig. 3B, left). However, one pixel diagonally away is 1 kb away from the left anchor and 1 kb away from the right anchor, equivalent to 2 kb Manhattan distance. Thus, the juxtaposition of pixels at different distances likely creates the observed crosshair pattern and could be potentially misinterpreted. To more accurately depict Hi-C signal versus distance from loops and thereby alleviate this common visualization issue, we created SIPMeta, which generates both the standard square metaplots, as well as “bullseye” plots where pixels in each ring represent the same Manhattan distance away from the loop (Fig. 3B, right). The bullseye visualization of Drosophila loops eliminates the crosshair pattern, demonstrating the potential usefulness and impact of SIPMeta on data interpretation. For comparison, we examined a “true” stripe found in human GM12878 cells (Fig. 3C, left) and found that the SIPMeta bullseye plot is able to display this stripe (Fig. 3C, right). Therefore, SIPMeta can distinguish extrusion-mediated stripes from crosshair patterns, which are due to Euclidean distance effects relative to the loop.
We then examined the ability of SIP to identify loops in an organism where they have not previously been characterized. We examined published Hi-C maps in the mosquito Aedes aegypti (Matthews et al. 2018) and detected visually apparent loops (Supplemental Fig. S3C). Using SIP, we identified 231 high-intensity loops that display central enrichment (Supplemental Fig. S3D). In this case, cLoops was also able to identify these peaks while other programs were not (Supplemental Fig. S3E). To test whether these loops correlate with the presence of CTCF at anchor sites, as is the case in mammals, or if they are similar to those found in Drosophila cells, we examined the enrichment of CTCF motifs at loop anchors. Unlike human cells, which display a large enrichment of CTCF motifs at loop anchors, we found that A. aegypti loop anchors are not enriched in CTCF motifs (Fig. 3D). Therefore, it is likely that A. aegypti loops are like those found in D. melanogaster and contain proteins other than CTCF at their anchors.
Characterization of SIP loops in human cells
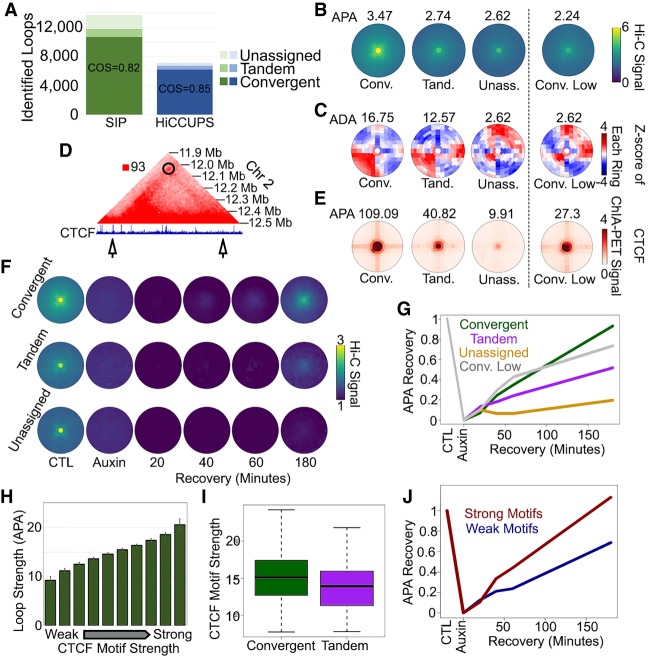
Having found that SIP identifies visually observable loops in D. melanogaster and A. aegypti, we next evaluated SIP loop calls in GM12878 human cells. SIP identified 13,692 loops in the 5 billion Hi-C contacts data set obtained in GM12878 cells at 5-kb resolution. This is nearly twice (1.93-fold) the 7101 loops identified by HiCCUPS using default parameters. We found a strong preference for loop anchors containing CTCF peaks assignable to a convergent orientation (10,663, 78%) (Fig. 4A). Compared to other programs, SIP and HiCCUPS detect the highest percentage of loops with convergent CTCF, which is indicative of their ability to identify these features (Supplemental Fig. S4A). Additionally, we examined CTCF ChIA-PET data (Tang et al. 2015) and found that SIP and HiCCUPS loops had the highest enrichment signal (Supplemental Fig. S4B). Of the loops identified by SIP, 1038 and 56 were assigned to tandem and divergent orientations, respectively, whereas 1935 (14%) did not coincide with detected CTCF peaks in any particular orientation (Fig. 4A). Analysis of metaplots of loops in all categories indicates that convergent loops are strongest, followed by tandem and then unassigned loops (Fig. 4B). We then tested the ability of each loop category to form domains by taking the Z-score values of each ring in the bullseye plot and calculating an aggregate domain analysis (ADA) score from the number of high Z-scores in the bottom right corner compared to the surrounding regions. This Z-score transformation and ADA calculation is included as an option in SIPMeta. Using this method, we found that loops between convergent CTCF sites form the strongest domains and tandem loops contain slightly weaker intra-domain interaction frequencies (Fig. 4C). Loops without identified CTCF peaks do not display an underlying domain (Fig. 4C). We tested whether the absence of an interaction domain is due to loop strength by examining convergent CTCF loops that display weak loop signal. Weak convergent loops do not display domain signal either, indicating that domain formation correlates with loop strength (Conv. Low, Fig. 4C).
Figure 4.
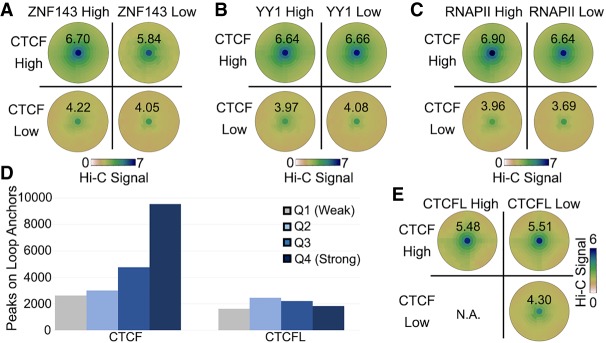
Characterization of CTCF loops with SIPMeta. (A) Number of loops in GM12878 cells at 5-kb resolution identified by SIP (green) or by HiCCUPS (blue) corresponding to CTCF sites in convergent, tandem, or in orientations that could not be assigned. The number of loops in divergent orientation was negligible and could not be depicted. (B,C) SIPMeta bullseye plots (B) or Z-score plots (C) of SIP loops in categories based on CTCF motif orientation. (APA) Aggregate peak analysis, (ADA) aggregate domain analysis. (D) Example of a SIP loop unassignable to any CTCF orientation. CTCF ChIP-seq signal is shown below. Arrows indicate loop anchors. (E) SIPMeta bullseye plots of CTCF HiChIP data for SIP loops in categories based on CTCF motif orientation. (F) Metaplots for SIP loops in each CTCF category in control, cohesin depletion, and recovery in Hi-C data obtained in HCT116 cells. (G) APA score changes after cohesin recovery relative to control and cohesin depletion Hi-C. (H) Average APA scores of convergent CTCF loops divided into 10 equal categories based on the strength of motifs found on loop anchors. Error bars indicate standard deviation. (I) CTCF motif scores on convergent versus tandem loops. (J) APA score changes after cohesin recovery relative to control and cohesin depletion Hi-C for convergent loops with the strongest (red) and weakest (blue) 10% motif scores.
SIP detects 1935 loops whose anchors seem to lack CTCF bound to its motif. This could be a result of the stringency of CTCF peak calling in ChIP-seq data. For example, an unassigned loop has a strong CTCF site on one anchor but has weak CTCF signal on the other (Fig. 4). Indeed, 1572 (81%) of these unassigned loops have an identifiable CTCF ChIP-seq peak on one anchor. Therefore, these loops are either interactions between CTCF and some other protein or loops where the second anchor shows weak CTCF ChIP-seq signal insufficient to call a peak but sufficient to form a loop. We examined CTCF ChIA-PET data (Tang et al. 2015) using SIPMeta and found enrichment of signal on convergent and tandem loops (Fig. 4E). Although unassigned loops display weaker CTCF ChIA-PET signal than even weak convergent loops, we still detect enrichment signal in the center compared to the surrounding region (Fig. 4E). Therefore, we believe that unassigned loops are CTCF loops where one anchor has low levels of CTCF. Thus, SIP and HiCCUPS identify similar types of features, although SIP is able to identify additional CTCF loops.
Next, we examined loops in published Hi-C data in HCT116 cells before and after cohesin depletion (Rao et al. 2017). Using SIPMeta, we examined changes in loops after RAD21 depletion and reintroduction of this protein (Fig. 4F). Loops in each context are lost after RAD21 depletion, confirming that loops in mammals generally depend on the presence of cohesin (Fig. 4F; Rao et al. 2017). We noticed that CTCF loops in tandem orientation or without an assignable CTCF peak are not able to recover as efficiently as convergent CTCF loops (Fig. 4F). Measuring APA scores after 180 min of cohesin recovery, convergent loops return to their original enrichment value almost fully (93% of APA score), whereas tandem and unassigned loops recover to only 52% and 20% of their original APA scores, respectively (Fig. 4G). This slow recovery is not due to weaker loop signal in the control, since weak convergent loops recover better than tandem and unassigned loops (Conv. Low, Fig. 4G).
CTCF is thought to block extrusion in an orientation-specific manner, so we reasoned that the strength of the motif may determine the strength of the loop. We examined CTCF motif strength versus loop strength and found that they are correlated (Fig. 4H). We also found that convergent loops display stronger CTCF motifs than tandem loops (Fig. 4I). To examine whether motif strength affects loop recovery after cohesin depletion and repletion, we examined convergent loops in the top and bottom 10% of motif strength. We found that convergent loops with weak motifs did not recover as quickly as convergent loops with strong motifs (Fig. 4J). Indeed, convergent loops with weak motifs recovered as slowly as tandem loops. These data fit with a model where strong convergent motifs efficiently dictate the orientation of the CTCF protein, resulting in more robust blockage of extrusion and quick recovery. Based on the results, weak convergent, tandem, or unassignable motifs are less efficient at dictating the orientation of the CTCF protein on chromatin, resulting in less blockage of extrusion and slower recovery. Therefore, we suggest that loops that appear to overlap tandem or no motifs could still be occupied by convergently oriented CTCF proteins.
Other transcription factors affect the strength of CTCF loops
Although CTCF has a major role in the establishment of loops in mammalian cells, other transcription factors present at loop anchors may affect the frequency of point-to-point interactions causing the formation of these loops. Using SIPMeta, we investigated several transcription factors whose binding sites have been previously shown to be present at CTCF loop anchors, including ZNF143, YY1, CTCFL, and RNA Polymerase II (RNAPII) (Rao et al. 2014; Tang et al. 2015). First, we examined loops with high levels of CTCF on both anchors and divided them into those with high or low levels of ZNF143. We find that when CTCF is high, loops with high ZNF143 are stronger than loops with low ZNF143 signal (Fig. 5A, top row). Loops with weak CTCF signal are also stronger when ZNF143 signal is high (Fig. 5A, bottom row), indicating that the presence of ZNF143 can enhance looping frequency. In contrast, we found no difference in loop signal between those containing high or low YY1 (Fig. 5B). However, we do detect small signal differences on CTCF loops with high or low RNAPII (Fig. 5C).
Figure 5.
Contribution of transcription factors to CTCF loops. (A–C) SIPMeta bullseye plots for loops with either high or low CTCF ChIP-seq signal on both anchors and either high or low ZNF143 (A), YY1 (B), or RNAPII (C) ChIP-seq signal on both anchors. Scores represent average distance-normalized Hi-C signal of the loop at the center of the bullseye plot. (D) Number ChIP-seq peaks for each strength quartile that overlaps loop anchors in K562 cells. (E) SIPMeta bullseye plots for loops with either high or low CTCF and CTCFL ChIP-seq signal on both anchors. (N.A.) An insufficient number of unambiguous loops in this category. Scores represent average distance-normalized Hi-C signal of the loop at the center of the bullseye plot.
CTCFL binds to the same motif as CTCF (Pugacheva et al. 2015). While GM12878 cells do not express CTCFL, many CTCF motifs in K562 cells display peaks of both CTCFL and CTCF. ChIP Re-ChIP experiments indicate that these sites are often bound by the two proteins at the same time (Pugacheva et al. 2015). We thus hypothesized that the presence of both proteins could affect looping. To test this, we examined published CTCFL ChIP-seq data in K562 cells (Pugacheva et al. 2015) and compared its presence at loop anchors to that of CTCF. While loop anchors preferentially contain strong CTCF peaks, there is equal presence of weak and strong CTCFL peaks at loop anchors (Fig. 5D). We could not identify enough loops with low CTCF and high CTCFL unambiguously (n = 2), but for loops with high CTCF we found no difference in signal when CTCFL was high or low (Fig. 5E). We should note that other programs were unable to categorize loops in this manner (Supplemental Fig. S5), thus highlighting the differences between loop callers. These results suggest that, despite a similar DNA binding domain, CTCFL is unable to form loops. Additionally, our results indicate that CTCFL does not interfere with looping when present at the same location as CTCF.
The dosage-compensated X Chromosomes of C. elegans are organized in a network of loops
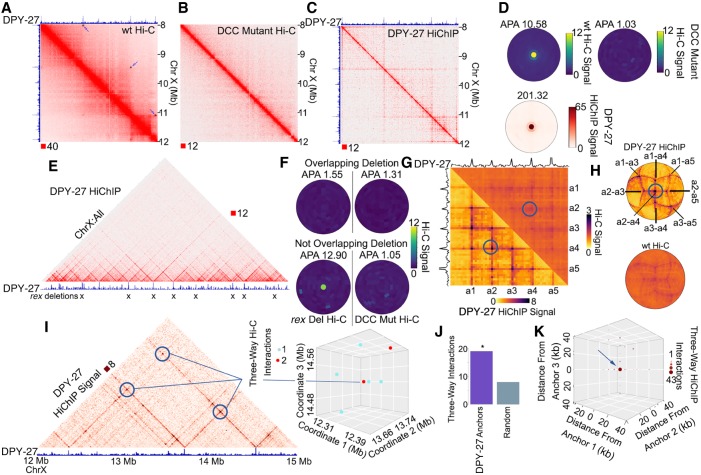
Results described above suggest that non-CTCF proteins can alter CTCF loop strength in mammals, and that non-CTCF loops can be observed in Hi-C data from organisms such as A. aegypti and D. melanogaster and C. elegans. These observations prompted us to investigate whether SIP is able to detect loops in Hi-C data from organisms where few loops have been previously detected. Previous Hi-C experiments in C. elegans embryos identified interaction domains in the X Chromosomes of hermaphrodites (Crane et al. 2015). These X Chromosomes are bound by a condensin I-containing dosage compensation complex (DCC) that remodels X Chromosome topology and down-regulates expression of genes chromosome-wide. This finding represents a significant advance in the understanding of the role of 3D chromatin architecture in the organization of dosage-compensated chromosomes. Borders separating these domains on the X Chromosome correspond to binding sites of the specialized condensin I-DCC (Crane et al. 2015; Anderson et al. 2019). However, it was difficult to determine whether these domains were formed by self-interactions, as is the case in Drosophila, or by point-to-point interactions between DCC sites to form loops by loop extrusion similar to those formed by CTCF and cohesin in mammals. To address this question, we performed Hi-C in C. elegans hermaphrodite embryos and used SIP to detect punctate signals (Supplemental Table S3). Recent experiments performed Hi-C in the same C. elegans hermaphrodite embryos (Anderson et al. 2019), and thus we combined Hi-C contacts reported by Anderson et al. with ours to obtain over 535 million usable Hi-C contacts. We then used SIP with this combined data set at 5-kb resolution and we were able to identify 41 loops (Fig. 6A). ChIP-seq for the DPY-27 subunit of condensin I-DCC shows the presence of this protein at loop anchors, suggesting its involvement in the establishment of loops in C. elegans (Fig. 6A, top track). SIP called zero loops in the Hi-C data for the DCC mutant (Fig. 6B; Anderson et al. 2019), indicating that the establishment of the 41 loops depends on the presence of DCC. To confirm that these loops are associated with condensin I-DCC, we performed HiChIP using a DPY-27 antibody (Fig. 6C). We detect enrichment of DPY-27 HiChIP signal on SIP loops identified by Hi-C, indicating that condensin I-DCC may play a role in the formation of loops in the X Chromosome of C. elegans hermaphrodites (Fig. 6D). In comparison to other loop callers, SIP loops have higher overlap with DPY27 HiChIP (Supplemental Fig. S6A,B). Anderson et al. found that deletion of eight rex sites at borders of domains results in the loss of these domains with no change to gene expression (Anderson et al. 2019). These deletions overlap with some of the loop anchors we detect (Fig. 6E). We examined average loop signal at sites where one anchor overlaps a deletion and found loss of these loops (Fig. 6F, top). However, our HiChIP data indicates that there are many DCC-mediated loop anchors that do not overlap these deletions (Fig. 6E). We examined loops where neither anchor overlaps a deleted site and found that these loops are still present and become stronger (cf. Fig. 6F, bottom and Fig. 6D). All of these loops are dependent on DCC (Fig. 6F, right). These additional DCC-dependent loops could explain the observation that disruption of DCC results in X Chromosome decondensation and increased gene expression, while deletion of eight rex sites does not (Anderson et al. 2019). A similar model has been proposed by Anderson et al. (2019).
Figure 6.
A network of condensin I-DCC loops in C. elegans. (A) Hi-C contact map showing domains and loops on the X Chromosome of C. elegans hermaphrodites. Black squares with arrows depict loops called by SIP. Top track displays the DPY-27 ChIP-seq signal across the region. (B) Hi-C in the DCC mutant sdc-2 (y93, RNAi) from Anderson et al. (2019) showing lack of loops on the X Chromosome. (C) DPY-27 HiChIP contact map depicting the enrichment of loops. Top track displays the DPY-27 ChIP-seq signal across the region. (D) SIPMeta bullseye plot of C. elegans SIP loops on the X Chromosome displaying the average wild-type Hi-C (top left), or DCC mutant Hi-C (top right), or DPY-27 HiChIP (bottom) signal. (E) X Chromosome–wide view of DPY-27 HiChIP signal. Bottom track displays the DPY-27 ChIP-seq signal across the region. Xs indicate sites deleted in Anderson et al. (2019). (F) SIPMeta bullseye plots of Hi-C data after eight rex site deletions (left) or after mutation of the DCC (right). SIP loops that overlap with deleted rex sites (top) or do not overlap with deleted rex sites (bottom). (G) Scaled metaplots of interactions between every DPY-27 loop anchor with its closest four others shown by Hi-C (top right) and by DPY-27 HiChIP (bottom left). The top and side tracks depict the median DPY-27 ChIP-seq signal. Blue circle indicates the point chosen as the center of bullseye plots shown later. (H) SIPMeta bullseye plots for DPY-27 HiChIP (top) and Hi-C (bottom) centered on the interaction between a2-a4. (I) DPY-27 HiChIP contact map depicting a network of two-way interactions between three anchors found to participate in three-way interactions. Bottom track shows DPY-27 ChIP-seq signal. 3D scatterplot of three-way interactions for the chromosome coordinates shown. (J) Number of three-way interactions discovered by Hi-C connecting DPY-27 loop anchors or the average of permutations using an equal number of random regions on the X Chromosome. (*) P < 0.05 Monte Carlo permutation test. (K) Profile of three-way interactions across all possible three-way DPY-27 loop anchor connections.
The presence of condensin I-DCC suggests that loops may be formed via extrusion in C. elegans. We examined motifs at loop anchors and found the MEX motif, which is enriched at DPY-27 peaks (Jans et al. 2009) and is at rex sites previously reported to be present at borders of domains (Crane et al. 2015; Anderson et al. 2019). We then tested whether loops occur between convergent MEX motifs, as is the case for CTCF loops in mammals, yet we found no bias in motif orientation (Supplemental Fig. S6C), consistent with what was reported for domain borders (Anderson et al. 2019). Thus, loop anchors in C. elegans likely represent bidirectional blocks to extrusion. In support of this notion, loop anchors generally form interactions both upstream of and downstream from the anchor (Fig. 6C,E). We detect no loops with visually apparent extrusion stripes in the Hi-C data (Fig. 6A). Stripes in mammalian Hi-C maps indicate unidirectional extrusion starting near one anchor (Vian et al. 2018). We ran molecular dynamics polymer simulations of unidirectional extrusion starting near loop anchors and detected strong stripes at loop anchors, which is consistent with an asymmetric extrusion model reported for CTCF loops in mammals (Supplemental Fig. S6D, bottom left; Vian et al. 2018). We then ran polymer simulations of bidirectional extrusion starting randomly and detected less striping and more filled-in domains (Supplemental Fig. S6D, top right). This pattern is more consistent with the absence of stripes at loops associated with the condensin I-DCC in C. elegans (Fig. 6A) and therefore suggests that loading is either random or takes place at many sites rather than just the high affinity rex sites.
A chromosome-wide view of DPY-27 HiChIP shows a network of loops spanning the X Chromosome (Fig. 6E). This loop network can also be seen in scaled metaplots of distance-normalized Hi-C data corresponding to DPY-27 peaks (Fig. 6G). The formation of a rosette structure by the compensated X Chromosome is supported by viewing this network as a bullseye plot (Fig. 6H). Since Hi-C and HiChIP are performed on a population of cells, the apparent network of DPY-27 loops could either represent multiway interactions occurring in the same cell or individual two-way interactions occurring in different cells. If all the loops are present simultaneously in all cells, the results would suggest that these nested loops can occur between five anchors or more (Fig. 6E,G). To distinguish between these two possibilities, we examined Hi-C reads containing multiple interacting fragments. Because the Hi-C protocol involves digestion with DpnII followed by ligation and sonication of fragments for library preparation, sequenced reads can contain several ligation events. Therefore, we examined paired-end reads in which we could determine ligations between three different genomic regions (see Methods). For example, DPY-27 loop anchors at chromosomal coordinates 12.35 Mb, 13.70 Mb, and 14.52 Mb were ligated together, indicating that these loops occur within the same cell (Fig. 6I). Analysis of multiway Hi-C interactions shows enrichment of contacts among multiple DPY-27 loop anchors (Fig. 6J). Three-way interactions between DPY-27 loop anchors are 2.4-fold higher (P < 0.05) than permutations on sets of random loci at similar distances on the X Chromosome. To improve the ability to detect condensin I-DCC-mediated three-way ligations, we examined DPY-27 HiChIP data obtained from 250-bp paired-end sequencing (Supplemental Table S4). We then examined three-way ligations in DPY-27 HiChIP data and found enrichment of multiway DPY-27 anchor interactions compared to Hi-C and compared to random regions (Supplemental Fig. S6E). Additionally, in a metaplot of all possible three-way interactions between DPY-27 loop anchors and the surrounding regions, we found enrichment at DPY-27 loop anchors (Fig. 6K). Altogether, our observations suggest that loops identified by SIP in C. elegans may represent nested interconnected DCC interactions mediated by condensin I-DCC, implying that the dosage-compensated X Chromosome of hermaphrodites is organized in a rosette-like structure.
Discussion
Hi-C data sets containing billions of contacts have allowed the identification of thousands of loops representing point-to-point interactions between CTCF sites in mammals (Rao et al. 2014). However, there are very few methods capable of identifying these loops, and sometimes it has been more feasible to annotate loops by eye (Eagen et al. 2017). SIP utilizes image processing and the local background to identify loops. Here, we demonstrate the utility of SIP as a loop caller in identifying additional CTCF loops in mammals, non-CTCF loops in D. melanogaster and A. aegypti, and condensin I-DCC loops in C. elegans. The high accuracy of SIP in loop identification allows the detection of nearly double the CTCF loops from the same data set as well as detection of loops in nonmammalian species. With the companion tool SIPMeta, SIP can facilitate discovery of novel aspects of 3D chromatin architecture. We intend SIP to be easily usable by anyone performing analysis of Hi-C data on a variety of platforms and have given users the ability to alter most parameters to facilitate custom loop calling.
While CTCF has been the major focus of studies of loop formation, other chromatin-bound factors may also affect this process. For example, non-CTCF loops are evident in D. melanogaster, and we are also able to detect loops in A. aegypti in this study using SIP. Although we cannot determine the nature of the proteins forming loops in A. aegypti, these loops appear similar to Pc/Psq loops in D. melanogaster, and similar proteins are likely involved in their establishment. In mammals, CTCF loop strength may also be affected by other proteins, since cohesin sliding has been shown to be delayed by other DNA-bound complexes in vitro, including quantum dot labeled catalytically inactive EcoRI and dCas9 (Davidson et al. 2016; Stigler et al. 2016). Thus, DNA-bound proteins at specific sites in the genome may affect the loop extrusion process and thereby affect loop strength. The involvement of ZNF143 in the establishment of CTCF loops (Wen et al. 2018; Jung et al. 2019) is supported by the increased strength of loops detected by SIP when CTCF and ZNF143 are both present at interacting anchors. The inability of CTCFL to form loops has been confirmed by a recent study that indicates that CTCFL cannot stop cohesin extrusion (Pugacheva et al. 2020).
Using SIP, we find that condensin I-DCC in C. elegans forms dozens of loops along the dosage-compensated X Chromosome of hermaphrodites. Previous studies of the structure of metaphase chromosomes in chicken cells found that condensin II creates the axial scaffold, while condensin I creates clusters of nested loops (Gibcus et al. 2018). We speculate that, during dosage compensation in C. elegans, condensin I-DCC may perform a similar function along the X Chromosome, creating a rosette-like structure and thereby compacting the chromosome sufficiently to decrease transcription by twofold. This hypothesis agrees with microscopy data showing that the X Chromosome of hermaphrodites occupies a smaller nuclear volume than autosomes and that mutations in DCC result in decreased compaction (Lau et al. 2014). However, deletion of eight of the rex sites that are important for the establishment of contact domains observed in Hi-C data reportedly has no effect on chromosome compaction or gene expression (Anderson et al. 2019). While we find that there are more than eight loop anchors, and thus not all loops were lost when removing eight of them, it is curious that removal of a portion of loops did not affect gene expression. It has been suggested that loss of each anchor results in continued extrusion until the next anchor (Anderson et al. 2019). This would keep the overall network or rosette-like structure intact but containing larger loops.
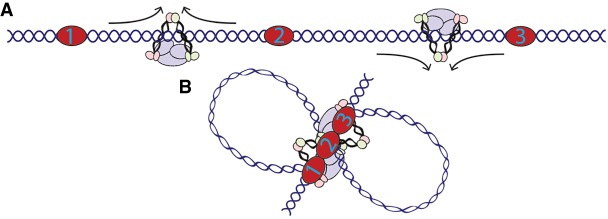
CTCF loop anchors in mammals often coincide with a stripe of intense interaction signal in Hi-C maps (Vian et al. 2018). This observation prompted the suggestion of a “loop gun” model of extrusion, where cohesin is loaded near loop anchors and proceeds asymmetrically until reaching the other anchor. Our study provides in vivo evidence of condensin I-mediated extrusion in animals. Unlike the formation of CTCF loops in mammals, our analysis suggests an alternate method of extrusion-mediated looping in C. elegans. Our results indicate that SMC proteins are loaded randomly on the X Chromosome of C. elegans hermaphrodites and extrude until reaching stopping points from either direction, in a fashion similar to what was suggested by Anderson et al. (2019). This may occur via a single condensin complex undergoing bidirectional extrusion or by unidirectional extrusion of multiple condensin complexes, recreating a bidirectional effect. While condensin complexes from yeast display unidirectional extrusion and compaction in vitro (Ganji et al. 2018; Kong et al. 2019), recent work indicates mammalian condensins perform bidirectional extrusion (Kong et al. 2019). Studies performed in vitro suggest that condensin complexes can pass over each other during extrusion and thereby form a z-loop structure (Kim et al. 2020). Therefore, if condensin I-DCC moves unidirectionally, the interaction-enriched interior of the domain may form by randomly placed z-loops in the population of cells. In either case, looping both upstream of and downstream from each anchor indicates that extrusion is blocked from both sides. In a simplified three-anchor example where each anchor is a bidirectional block, extrusion on both sides of the middle anchor would naturally cause all three anchors to come into close proximity (Fig. 7A). This indicates that these bidirectional blocks could create the network of interactions observed by Hi-C and HiChIP to form the axis of a rosette-like structure (Fig. 7B).
Figure 7.
Model of condensin extrusion in C. elegans. (A) Extrusion in the X Chromosome of C. elegans likely begins at random locations and proceeds until blocked. Depicted here is bidirectional extrusion through one complex, but unidirectional extrusion through multiple complexes is also possible. (B) Loop anchors for DCC in C. elegans represent bidirectional blocks resulting in proximity of each anchor with every other.
SIP and SIPMeta greatly facilitate the analysis of Hi-C data and the detection of point-to-point interactions. Loops formed by these interactions represent an important aspect of the three-dimensional organization of the mammalian genome. Our evaluation indicates that sequencing depth is an important factor in loop calling; thus, methods that increase signal by data imputation may become valuable tools (Zhang et al. 2018). As sequencing costs decrease and high resolution Hi-C data sets become standard, memory- and time-inefficient methods will perform worse as the matrix processing becomes more complex. However, using images instead of matrices along with image processing should limit the increased memory costs associated with deeper sequencing. The ability to call loops and quantitatively measure their strength using SIP will facilitate the discovery of the biological significance of 3D nuclear architecture.
Methods
The SIP program and the loop calling process
SIP retrieves raw Hi-C signal stored in Juicer .hic files using Juicer Tools (Durand et al. 2016) at the resolution and with the normalization scheme chosen by the user. The genome is analyzed by sliding windows, the size of which depends on the resolution and matrix size specified by the user. For example, we used 5-kb resolution data with KR normalization and a matrix size of 2000 for all experiments involving GM12878 or HCT116 cells. This creates 10-Mb snapshots sliding over 5 Mb each step. Observed-expected (o-e) values are used to create images. Later, in postfiltering, retrieved data is distance-normalized by the formula valuenormalized = 1 + ((value − expected)/expected + 1), which is used to compute the central loop value. SIP then uses image processing methods to create a list of candidate loops that will be filtered later. Because even with o-e normalized values the diagonal represents extremes in the data, outliers within 2 bins along the diagonal are removed if the value is higher than the average +1 std dev of the image signal. The first image processing step utilizes Gaussian blurring to smooth the Hi-C signal in order to avoid detection of outlier pixel signals. Afterward, contrast enhancement is used to increase the contrast between the background and the signal of interest (Schneider et al. 2012). White top-hat, a mathematical morphology method from the MorpholibJ plugin, is used to homogenize the background and make bright structures easier to detect (Legland et al. 2016). Because loops appear as bright punctate signal in images, we use this top-hat method to transform the grayscale intensity values of each Hi-C image, causing the bright structures to have increased contrast from the background. The last step uses a minimum and maximum filter (Schneider et al. 2012) combination to remove isolated pixels and further homogenize the background. These steps provide a corrected image of the interactions (Fig. 1). This corrected image is then used with the regional maxima detection algorithm available from ImageJ (Schneider et al. 2012) to detect a long list of candidate loops.
Candidate loops must then pass several filters that utilize the distance-normalized signal from the original matrix before image processing (Fig. 1). The first step is to exclude pixels near columns and rows with insufficient data; the default is to filter any with ≥6 pixels with zero values in the surrounding 24-pixel neighborhood. The second filter is to remove pixels without increased interactions compared to the surrounding 8-pixel neighborhood and the 24-pixel neighborhood. To remove isolated enriched pixels, loops must display decay between the central pixel and the surrounding neighborhood pixels. Candidate loops are then filtered so that the center pixel's KR value ≥0.3 and >1.2-fold higher than nearby pixels (PA score). Loops are then filtered such that the probability that the Poisson CDF function of the center pixel being higher than the nearby pixels is greater than 0.9. Finally, candidate loops are filtered if their PA score is lower than the PA scores of a top percentage of random sites. This percentage is specified by the user.
Parameters for SIP loop calling in D. melanogaster used a threshold of 6000, with -nbZero 10, matrix size 500, resolution 1 kb, -d 20, –fdr 0.05, and –isDroso true. Analysis of A. aegypti Hi-C data was performed using parameters–g 1.5, -mat 2000, -d 5 -res 5 kb, -t 5000, –nbZero 4, -fdr 0.05, and –isDroso true. Human Hi-C maps were originally published with genome build hg19, and we ensured that all data used was mapped to the same genome build. Remapping everything to GRCh38 will not alter these results. CTCF motif enrichment was calculated in 5-kb windows using the formula
Expected values were derived using random loci.
Choosing parameters
SIP was designed for quick and memory-efficient loop calling so that loops can be visually inspected for parameter optimization. While we have listed the specific parameters that we used for each map, we recommend users to optimize loop calls using their own criteria. We recommend calling loops at 5 kb, but if sequencing depth is limited, it may be advantageous to call loops at 5 kb, 10 kb, and 25 kb. We suggest using KR normalization (Rao et al. 2014), but depending on sequencing depth, this normalization scheme may not be available for all chromosomes. In this case, other normalization schemes included in the Juicer tool set, such as VC_SQRT (Durand et al. 2016), are acceptable alternatives. Users may also wish to alter the –d option, which removes signal near the diagonal, depending on the diagonal signal specific to the Hi-C map or especially if using resolutions other than 5 kb. For example, the default –d 6 will remove interactions at <30 kb at 5-kb resolution, but 60 kb at 10-kb resolution.
The parameters we recommend altering when optimizing loop calls are the –g and –fdr options. Raising –g will increase the blur for the initial loop calls, thereby filtering out more isolated speckles that are potentially not true loops. However, this can also blur actual looping signal. Because loops appear more punctate at 10 kb and 25 kb, we suggest decreasing –g to reduce the blur and thereby identify more speckled signal. Altering –fdr will change how many loops pass the second filter. As SIP is processing, it outputs the number of loops identified before fdr filtering so that users can determine how many spots identified by the first pass are filtered by the second pass. This can serve as a gauge for altering the fdr parameter. The final parameter we recommend changing is –nbZero which filters pixels near areas with low coverage. If loops are erroneously identified near repetitive regions, we suggest increasing –nbZero. We recommend optimizing the SIP parameters by visual inspection of a single chromosome first, and then using those parameters for all the chromosomes.
Performance testing
Comparison of loops between data sets containing various numbers of Hi-C contacts was performed by random picking intra-chromosomal reads. Bootstrapping was performed by down-sampling the full data set to 1 billion intra-chromosomal reads 10 different times. Noise was simulated to follow the same distance decay as the Hi-C data. Additional details can be found in the Supplemental Methods. Recovery rates were calculated by the number of loops obtained in the down-sampled or noise-added data sets that were within two pixels of the loops identified in the full data set. False positive rates were calculated under the assumption that loops called in the down-sampled or noise-added data sets that do not overlap with loops in the full data set are false.
SIPMeta
SIPMeta is implemented in Java and includes a choice between command line options or a graphical user interface (GUI). SIPMeta first reads a loop file from which the bin size (resolution) is inferred. If the images are not present in the input directory, SIPMeta makes images from the SIP-derived BEDPE file corresponding to distance-normalized signals. Alternatively, users can specify a .hic file from which values are retrieved. Then, SIPMeta examines all signal within a specified distance surrounding the loop, computes an APA score as previously described (Rao et al. 2014), and outputs a matrix of averaged values. This matrix can be run through bullseye.py to obtain both square and bullseye plots.
The bullseye transformation of a heat map is a visualization technique intended to more accurately represent the secondary interactions around a strong loop in the genome. The plot is a simple transformation of the rectangular heat map such that each bin's Euclidean distance to the center now directly corresponds to its Manhattan distance in the original map. Each ring in the bullseye plot has segments corresponding to the 4 × N bins with a Manhattan distance of N from the central bin. Each bin in a ring takes up exactly the same angular area and they are evenly distributed around the circle. Although this represents some distortion from their actual angles in the original plot, this creates the same visual area for each bin. Z-score transformation is done for each ring separately and the ADA score is obtained by percentage of Z-scores > 1 in the bottom right quarter versus the total plot.
Contribution of transcription factors to loop strength
Overlaps between transcription factors and loop anchor sites were assigned if ChIP-seq peaks were within 2 pixels of the loop anchors. Loops categorized as overlapping high ChIP-seq peaks were those where both anchors overlap peaks in the top quartile of ChIP-seq signal. Loops where anchors do not overlap peaks in either of the top two quartiles were categorized as low. Loops were also separated into 10 equal categories based on motif scores overlapping the two anchors. Because the purpose of the test is to approximate the role of the motif in loop strength, the lower of the two motif values corresponding to the two anchors was assigned as the motif score.
Hi-C and HiChIP in C. elegans
Hi-C and HiChIP libraries were prepared as previously described (Crane et al. 2015; Rowley et al. 2019); details can be found in the Supplemental Methods. Two biological replicates were obtained for Hi-C and HiChIP experiments and processed using Juicer (Durand et al. 2016) with the ce10 genome. Mapping statistics can be found in Supplemental Tables S1 and S2.
Loops in C. elegans Hi-C were identified using SIP with parameters -g 1.5, -d 5, -fdr 0.05, -res 5000 -mat 500. Because DPY-27 HiChIP data showed enrichment across the X Chromosome, we identified potential anchors by peaks in the coverage normalization vector. Network metaplots were done by taking every anchor with the closest five others and scaling the region in between each anchor as well as the same distance upstream of and downstream from the first and last anchors. Median Hi-C or HiChIP signal was profiled within these regions. Bullseye plots were generated from this scaled matrix recentered on the a2-a4 matrix coordinate. Polymer simulations were performed as described (Vian et al. 2018).
Three-way interactions were obtained by scanning Hi-C or HiChIP FASTQ files for the ligation sequence GATCGATC and mapping each side to the ce10 genome using Bowtie 2 (Langmead and Salzberg 2012). Paired-end reads with at least three different sections mapping to different genomic locations at least 50 kb apart were used in downstream analysis. Overlaps were done with all possible three-way combinations of DPY-27 loop anchors in 10-kb bins or with the same number of random 10-kb bins following the same distance distribution. P-values were derived from Monte Carlo permutations.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE132640. The latest release of SIP can be obtained from https://github.com/PouletAxel/SIP/releases and SIPMeta from https://github.com/PouletAxel/SIPMeta/releases, including usage documentation and a separate script for the bullseye transformation of matrices. Source code for SIP and SIPMeta, including bullseye.py, is also provided as Supplemental Code. Any issues with the program can be reported on GitHub or directly via email.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the HudsonAlpha Institute for Biotechnology Genomic Services Lab for their help in Illumina Sequencing. We also thank Drs. William Noble and Doug Phanstiel for constructive discussions during the optimization of SIP. This work was supported by National Institutes of Health (NIH) Pathway to Independence Award K99/R00 GM127671 (M.J.R.) and U.S. Public Health Service Award R01 GM035463 (V.G.C.) from the National Institutes of Health. B.J.B. was supported by NIH T32 GM008490. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.257832.119.
References
- Anderson EC, Frankino PA, Higuchi-Sanabria R, Yang Q, Bian Q, Podshivalova K, Shin A, Kenyon C, Dillin A, Meyer BJ. 2019. X Chromosome domain architecture regulates Caenorhabditis elegans lifespan but not dosage compensation. Dev Cell 51: 192–207.e6. 10.1016/j.devcel.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay F, Bailey TL, Noble WS. 2014. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res 24: 999–1011. 10.1101/gr.160374.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chen Z, Chen X, Ai D, Chen G, McDermott J, Huang Y, Guo X, Han JJ. 2019. Accurate loop calling for 3D genomic data with cLoops. Bioinformatics 36: 666–675. 10.1093/bioinformatics/btz651 [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeñas-Potts C, Rowley MJ, Lyu X, Li G, Lei EP, Corces VG. 2017. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res 45: 1714–1730. 10.1093/nar/gkw1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Goetz D, Zaczek MP, Molodtsov MI, Huis in ‘t Veld PJ, Weissmann F, Litos G, Cisneros DA, Ocampo-Hafalla M, Ladurner R, et al. 2016. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J 35: 2671–2685. 10.15252/embj.201695402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagen KP, Aiden EL, Kornberg RD. 2017. Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc Natl Acad Sci 114: 8764–8769. 10.1073/pnas.1701291114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. 2018. Real-time imaging of DNA loop extrusion by condensin. Science 360: 102–105. 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. 2018. A pathway for mitotic chromosome formation. Science 359: eaao6135 10.1126/science.aao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Perez I, Rowley MJ, Lyu X, Valadez-Graham V, Vallejo DM, Ballesta-Illan E, Lopez-Atalaya JP, Kremsky I, Caparros E, Corces VG, et al. 2019. Ecdysone-induced 3D chromatin reorganization involves active enhancers bound by Pipsqueak and Polycomb. Cell Rep 28: 2715–2727.e5. 10.1016/j.celrep.2019.07.096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. 2012. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci 109: 17507–17512. 10.1073/pnas.1111941109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. 2018. Transcription elongation can affect genome 3D structure. Cell 174: 1522–1536.e22. 10.1016/j.cell.2018.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23: 602–618. 10.1101/gad.1751109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Kremsky I, Gold HB, Rowley MJ, Punyawai K, Buonanotte A, Lyu X, Bixler BJ, Chan AWS, Corces VG. 2019. Maintenance of CTCF- and transcription factor-mediated interactions from the gametes to the early mouse embryo. Mol Cell 75: 154–171.e5. 10.1016/j.molcel.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kerssemakers J, Shaltiel IA, Haering CH, Dekker C. 2020. DNA-loop extruding condensin complexes can traverse one another. Nature 10.1038/s41586-020-2067-5 [DOI] [PubMed]
- Kong M, Cutts E, Pan D, Beuron F, Thangavelu K, Xue C, Morris E, Musacchio A, Vannini A, Greene EC. 2019. Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. bioRxiv 10.1101/683540 [DOI] [PMC free article] [PubMed]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AC, Csankovszki G. 2014. Condensin-mediated chromosome organization and gene regulation. Front Genet 5: 473 10.3389/fgene.2014.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AC, Nabeshima K, Csankovszki G. 2014. The C. elegans dosage compensation complex mediates interphase X chromosome compaction. Epigenetics Chromatin 7: 31 10.1186/1756-8935-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legland D, Arganda-Carreras I, Andrey P. 2016. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32: 3532–3534. 10.1093/bioinformatics/btw413 [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. 2018. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563: 501–507. 10.1038/s41586-018-0692-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G. 2018. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell 71: 73–88.e5. 10.1016/j.molcel.2018.05.032 [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Rivero-Hinojosa S, Espinoza CA, Méndez-Catalá CF, Kang S, Suzuki T, Kosaka-Suzuki N, Robinson S, Nagarajan V, Ye Z, et al. 2015. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol 16: 161 10.1186/s13059-015-0736-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, Strunnikov AV, Zentner GE, Ren B, Lobanenkov VV. 2020. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc Natl Acad Sci 117: 2020–2031. 10.1073/pnas.1911708117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Huang S-C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. 2018. Organizational principles of 3D genome architecture. Nat Rev Genet 19: 789–800. 10.1038/s41576-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell 67: 837–852.e7. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Lyu X, Rana V, Ando-Kuri M, Karns R, Bosco G, Corces VG. 2019. Condensin II counteracts cohesin and RNA polymerase II in the establishment of 3D chromatin organization. Cell Rep 26: 2890–2903.e3. 10.1016/j.celrep.2019.01.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigler J, Çamdere GÖ, Koshland DE, Greene EC. 2016. Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep 15: 988–998. 10.1016/j.celrep.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. 10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vian L, Pękowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. 2018. The energetics and physiological impact of cohesin extrusion. Cell 175: 292–294. 10.1016/j.cell.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Huang ZT, Zhang R, Peng C. 2018. ZNF143 is a regulator of chromatin loop. Cell Biol Toxicol 34: 471–478. 10.1007/s10565-018-9443-z [DOI] [PubMed] [Google Scholar]
- Zhang Y, An L, Xu J, Zhang B, Zheng WJ, Hu M, Tang J, Yue F. 2018. Enhancing Hi-C data resolution with deep convolutional neural network HiCPlus. Nat Commun 9: 750 10.1038/s41467-018-03113-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.