Abstract
DNA recovery from ancient human remains has revolutionized our ability to reconstruct the genetic landscape of the past. Ancient DNA research has benefited from the identification of skeletal elements, such as the cochlear part of the osseous inner ear, that provides optimal contexts for DNA preservation; however, the rich genetic information obtained from the cochlea must be counterbalanced against the loss of morphological information caused by its sampling. Motivated by similarities in developmental processes and histological properties between the cochlea and auditory ossicles, we evaluate the ossicles as an alternative source of ancient DNA. We show that ossicles perform comparably to the cochlea in terms of DNA recovery, finding no substantial reduction in data quantity and minimal differences in data quality across preservation conditions. Ossicles can be sampled from intact skulls or disarticulated petrous bones without damage to surrounding bone, and we argue that they should be used when available to reduce damage to human remains. Our results identify another optimal skeletal element for ancient DNA analysis and add to a growing toolkit of sampling methods that help to better preserve skeletal remains for future research while maximizing the likelihood that ancient DNA analysis will produce useable results.
Ancient DNA has become an important tool for addressing key questions about human evolutionary and demographic history. Its rapid growth over the last decade has been driven largely by advances in isolating (Dabney et al. 2013; Rohland et al. 2018), preparing (Rohland et al. 2015; Gansauge et al. 2017), enriching (Fu et al. 2013, 2015; Haak et al. 2015; Mathieson et al. 2015), sequencing (Margulies et al. 2005), and analyzing (Briggs et al. 2007, 2010; Ginolhac et al. 2011; Skoglund et al. 2014) small quantities of degraded DNA. Although these methodological advances have contributed to an improvement in the quality and quantity of paleogenomic data obtained from ancient human remains, all ancient DNA research fundamentally depends upon access to biological material that has sufficient biomolecular preservation.
Skeletal tissue (i.e., bone or teeth) is the preferred biological material for human ancient DNA analysis owing to its ability to resist postmortem degradation better than other types of tissues, including skin and hair (Lindahl 1993; Smith et al. 2001, 2003; Collins et al. 2002). However, recent research has shown that not all bone elements are equally effective in preserving DNA and has identified the dense bone encapsulating the cochlea within the petrous pyramid of the temporal bone (referred to henceforth as the cochlea) (Gamba et al. 2014; Pinhasi et al. 2015), as well as the cementum layer in teeth roots (Damgaard et al. 2015; Hansen et al. 2017), as especially DNA-rich parts of the skeleton. The use of these skeletal elements that act as repositories for the long-term survival of DNA has proven to be particularly important for the analysis of biological samples recovered from regions where high temperatures and/or humidity increase the rate of molecular degradation and result in low concentrations of damaged DNA with reduced molecular complexity (e.g., Broushaki et al. 2016; Lazaridis et al. 2016; Schuenemann et al. 2017; Skoglund et al. 2017; Fregel et al. 2018; Harney et al. 2018; van de Loosdrecht et al. 2018).
Although use of the cochlea has contributed to the success of ancient DNA research across a growing range of geographic and temporal contexts, it is important to balance analytical goals with the irreparable damage to human skeletal remains that results from destructive analyses (Prendergast and Sawchuk 2018; Sirak and Sedig 2019). Ancient DNA is one of several such analyses that is now widely used in archaeology (others include radiocarbon dating and stable isotope analysis) (Hublin et al. 2008; Mays et al. 2013; Makarewicz et al. 2017; Pinhasi et al. 2019). To minimize damage to complete skulls from ancient DNA sampling while still accessing the rich genetic data in the cochlea, we developed a “cranial base drilling” method to limit damage to surrounding bone areas when a skull is intact (Sirak et al. 2017). However, even this method involves destructive sampling of the cochlea. Recent work has highlighted the fact that the morphological analysis of the inner ear part of the petrous pyramid (which includes the cochlea) can reveal population relationships and thus this part of the skeleton harbors some information about population history (e.g., Spoor et al. 2003; Ponce de León et al. 2018). Although genetic comparisons of samples involve analysis of tens of thousands of independent markers (single-nucleotide polymorphisms, or SNPs) that provide far higher statistical resolution than can be obtained by study of the smaller number of data points that can be extracted from morphological analysis, not all cochlear bone yields sufficient amounts of ancient DNA. The fact that there is morphological information in the petrous pyramid that will be destroyed through sampling of ancient DNA highlights the importance of being a careful steward of these elements.
As part of a search for alternative optimal sources for ancient DNA that can be used in place of the cochlea, we noted that auditory ossicles have similar developmental processes and histological properties as the osseous inner ear. We therefore tested whether the ossicles—the smallest bones in the human body—might serve as alternative optimal substrates for ancient DNA analysis.
Ossicle development and histology
The mechanism by which cochlear bone preserves endogenous DNA better than other skeletal elements or other regions of the same petrous pyramid is not well understood; however, it is likely related to the fact that human petrous bones are unique in being characterized by a near-absence of growth or remodeling following the completion of ossification by ∼24 wk in utero (Sølvsten Sørensen et al. 1992; Frisch et al. 1998; Hernandez et al. 2004). The inhibition of bone remodeling leads to the presence of a larger number of mineralized osteocytes that reside in lacunae within the bone tissue (Hernandez et al. 2004; Bell et al. 2008; Busse et al. 2010; Rask-Andersen et al. 2012). One hypothesis (Pinhasi et al. 2019) is that “microniches” created in the bone tissue by the maintenance of mineralized osteocytes, combined with the protected location of the cochlea, may act as repositories that encourage the long-term preservation of DNA (Bell et al. 2008; Kontopoulos et al. 2019). Ossicles are similar to the cochlea in this respect (see below), and we therefore hypothesized that they might also preserve high amounts of endogenous DNA.
In humans, the middle ear (the region of the ear located medial to the eardrum and lateral to the oval window of the inner ear) is enclosed within the temporal bone and contains the three auditory ossicles: the malleus, incus, and stapes (Fig. 1). The ossicles effectively allow humans to hear by transmitting sound-induced mechanical vibrations from the outer to the inner ear. Although the ossicles do not experience high-strain biomechanical loading, they are subject to unique vibrational patterns that impact their development and characteristics over the course of an individual's lifespan (Rolvien et al. 2018). In contrast to the majority of the human skeleton, but similar to the cochlea, the auditory ossicles present with their final size and morphology at birth following the onset of ossification of between 16 and 18 wk in utero and the completion of ossification around the 24-wk gestational age (Marotti et al. 1998; Yokoyama et al. 1999; Cunningham et al. 2000; Duboeuf et al. 2015; Richard et al. 2017). The ossicles and cochlea appear to follow the same developmental pattern of rapidly increasing bone volume through cortical thickening and densification, along with mineralization of the bony matrix (Richard et al. 2017).
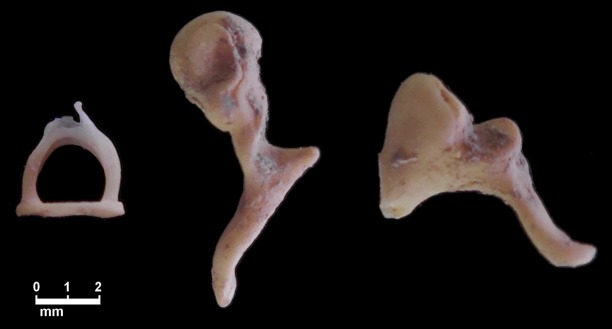
Figure 1.
The three auditory ossicles. From left to right, the stapes, malleus, and incus.
Like the cochlea, ossicular bone tissue is rapidly modeled around the time of birth; although it may undergo further postnatal maturation, there are no signs of bone remodeling observed above the age of 1 yr (Richard et al. 2017; Rolvien et al. 2018). The inhibition of bone remodeling of the auditory ossicles is evident from features such as the presence of a dense meshwork of collagenous fibers organized in an interlacing woven pattern, a smooth fibrous appearance, and limited vascular channels and viable osteocytes (Marotti et al. 1998; Chen et al. 2008). As in the case of the cochlea and in contrast to other skeletal elements, mineralized osteocytes appear to accumulate in the ossicles throughout an individual's life without resulting in increased bone absorption (Marotti et al. 1998; Kanzaki et al. 2006; Rolvien et al. 2018), likely conserving the overall architecture of the ossicles in order to maintain optimal sound transmission (Kanzaki et al. 2006; Rolvien et al. 2018). Although the consequences of inhibited bone remodeling and the accumulation of mineralized osteocytes have only been previously studied from a clinical perspective, we hypothesized that these features might contribute to an optimized DNA preservation similar to that in the cochlea by creating the “microniches” that enable long-term DNA survival (Bell et al. 2008).
Use of ossicles in ancient DNA research
Because of their small size and tendency to become dislodged from the skull, ossicles are only seldom recovered during excavation and are easily lost in collections excavated decades ago. Although ossicles are not recovered for every burial in every context, we have found that these bones may remain lodged within the middle ear of intact skulls or can be identified in the vicinity of a burial during excavation (Qvist 2000). Given the value of the ossicles as a substrate for ancient DNA analysis, as is shown in this study, we hope that more archaeologists, anthropologists, and museum curators will focus on preserving these elements.
It is important to recognize that ossicles, just like the cochlea, are morphologically informative. Indeed, there is a growing body of literature examining the comparative morphology and pathology of the ossicles (e.g., Rak and Clarke 1979; Arensburg et al. 1981, 2005; Siori et al. 1995; Spoor et al. 2003; Crevecoeur 2007; Quam and Rak 2008; Quam et al. 2013a,b; Stoessel et al. 2016). Although differences in metric and nonmetric features of the auditory ossicles may be taxonomically informative for comparisons across the genus Homo (e.g., Heim 1982; Spoor et al. 2003; Quam and Rak 2008; although see Arensburg et al. 1981), it is unclear whether phylogenetic and population relationship information can be retrieved from the auditory ossicles. In cases in which ossicle morphology may be a subject of future research, we encourage the anthropological study (including description and measurement) and surface or micro-CT scanning of the ossicles to collect metric and morphological information before ancient DNA analysis. Any ossicles that show visible pathologies should be avoided.
Although some anthropological attention has been given to the ossicles, we are not aware of previous genetic analyses of these bones. Only a single study has specifically attempted to analyze DNA from the ossicles, collecting the ossicles during medical autopsies of recently deceased individuals and determining them to be a reliable DNA source from bodies ranging from freshly deceased to highly putrefied (Schwark et al. 2015).
Results
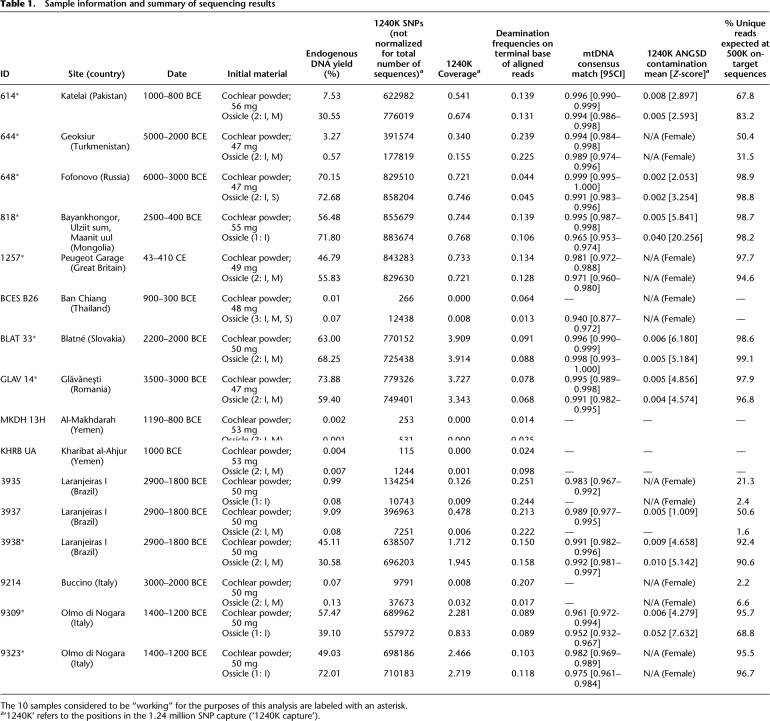
We performed pilot work (described in Supplemental Note S1) to assess if the quality and quantity of ancient DNA data recovered from the ossicles was approximately similar to that recovered from the cochlea. The results of this pilot work (Supplemental Table S1) suggested that ossicles perform comparably to the cochlea in metrics such as amount of endogenous human DNA recovered and frequency of damage at the terminal nucleotide of the DNA molecule (a commonly used measure of ancient DNA authenticity). Based on these results, we selected an additional 16 archaeological samples for further study from a wide range of geographic locations with varying climates and dated to between ∼6500 and 1720 yr before present (yBP) and with both petrous bone and ossicles present (Table 1; for detailed sample information, see Supplemental Table S2). Skeletal samples for both pilot work and the main study were excavated by professional archaeologists who held all appropriate permits and permissions from the institutions managing archaeological research in each region or country from which the samples originate; these samples were provided to us by the archaeologists for the purpose of paleogenomic analysis. No direct descendants or stakeholder communities were identified for any sample included in this work. Processing of all skeletal material was performed following suggested best practices for ancient DNA research, such as those outlined by Prendergast and Sawchuk (2018) and Sirak and Sedig (2019). To be included in this study, each specimen was required to have at least one ossicle as well as the cochlea of the petrous bone available for comparative analysis. Whenever possible, a petrous bone that had an antimere was chosen (Prendergast and Sawchuk 2018).
Table 1.
Sample information and summary of sequencing results
A summary of sequencing results for the 16 individuals reported in this paper is presented in Table 1 and Figure 2; for more detailed information, see Supplemental Table S2 and Supplemental Figure S1.
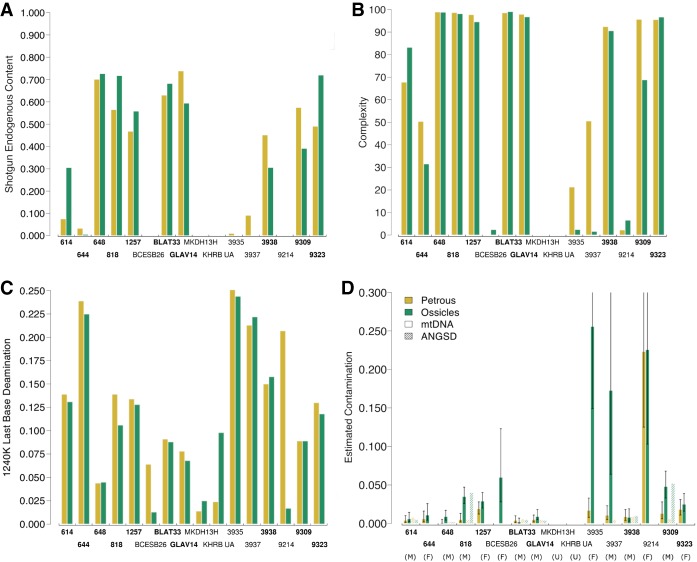
Figure 2.
Comparative results between cochlea (yellow) and ossicle (green) samples from the same individuals; bold font indicates the samples that were used in Wilcoxon signed-rank test. (A) Endogenous shotgun DNA ratios of the total reads. (B) Complexity as percentage of unique reads expected from 500,000 reads hitting targets. (C) Deamination frequencies on the terminal bases of sequences aligning to the human genome. (D) Contamination estimates calculated by subtracting the rate of mitochondrial matches to the consensus sequence from one (smooth bars) and based on the heterozygosity of the X Chromosome of male individuals (textured bars). Error bars, 95% confidence interval.
Out of 16 individuals included in this study, both the cochlea and ossicles produced enough data to call mitochondrial DNA (mtDNA) haplogroups, assess damage patterns at the terminal nucleotide of the molecule, and make estimations comparing contamination between the cochlea and the ossicles for 10 individuals. These individuals are henceforth referred to as the “working individuals.” Any individuals that did not meet these three criteria were considered to be “not working,” as we were unable to effectively carry out inter-element comparisons.
For four individuals, both the cochlea and ossicles were considered to have “failed” our analysis: One individual from Thailand produced marginal data for both skeletal elements with a large error interval for the mtDNA contamination estimate—calculated as one minus the rate of mitochondrial matches to the consensus sequence (Fu et al. 2013)—that was only assessible when the ossicles were used; two individuals, both from Yemen, did not produce enough data to allow for the determination of mtDNA haplogroup or contamination estimates; and a Copper Age individual from Italy produced a very large interval for the mtDNA contamination estimate and did not produce enough data to allow for the determination of mtDNA haplogroup. For two individuals from Late Holocene Brazil, 3935 and 3937, mtDNA haplogroups could not be determined, and contamination error intervals were large for the ossicles only.
We performed Wilcoxon signed-rank tests to compare the data generated using the ossicles and cochlear samples for the 10 working individuals (Supplemental Table S3). For completeness and confirmation, we present these results with and without including individuals 3935 and 3937 for the six absolute metrics analyzed (out of eight total metrics) (see Supplemental Table S3), because with the exception of contamination estimates, these individuals produced sufficient data to carry out statistical analysis.
With the Wilcoxon test, we obtained an average endogenous DNA yield of 47.83% for the 10 working cochlea samples and 50.01% for the corresponding ossicles, a nonsignificant difference (P = 0.557) (Table 1; Fig. 2A; Supplemental Table S3). The average complexity for cochlea and ossicles was 89.36% and 85.83%, respectively (Table 1; Fig. 2B); this difference is also nonsignificant (P = 0.275) (Supplemental Table S3). Complexity, defined here as the percentage of unique reads observed after down-sampling to 500,000 sequences that align to the approximately 1.24 million targeted SNPs, is a potentially more informative metric for comparing performance between the cochlea and ossicles because it is directly related to the maximum amount of sequencing data the library can possibly yield and is not biased by differences in sequencing depth across samples. Overall, these results suggest that the quantity of data generated using ossicles is comparable to that generated using the cochlea. Any minor differences are more likely owing to chance than to a systematic difference in DNA preservation between the cochlea and ossicles.
The average mtDNA coverage was approximately 423× (range, approximately 61×–1244×) for the 10 working cochlea samples and 388× for the corresponding ossicles (range, approximately 31×–1286×) (Supplemental Table S2), this is also a nonsignificant difference (P = 0.275) (Supplemental Table S3). The average coverage of the approximately 1.24 million targeted SNPs from across the genome was 1.72× for the working cochlea samples, and 1.58× for the ossicles (Table 1); on average, 711,916 SNPs were called when the cochlea was used and 696,454 were called when the ossicles were used (without normalizing for total number of sequences) (Table 1). Consistent with other results, both of these differences were nonsignificant when assessed with the Wilcoxon signed-rank test (P = 1.000 and 0.770, respectively) (Supplemental Table S3).
For the 10 working samples, the average deamination frequency was slightly reduced from 12.31% to 11.5% when the ossicles were used (Table 1; Fig. 2C; see Supplemental Fig. S1), a decrease that, although small, was marginally statistically significant (P = 0.049) (Supplemental Table S3). We note that if we include individuals 3935 and 3937, considered to be “not working” by our previously described parameters but for which we were able to generate data using the cochlea, this statistic loses significance (P = 0.0923) (Supplemental Table S3). This is the only metric for which there is a change in significance when including individuals 3935 and 3937.
Mitochondrial contamination estimates (inferred by identifying mismatches to the mtDNA consensus sequence) (Fu et al. 2013) increased from an average of 0.80% when the cochlea was used to 1.80% when the ossicles were used (Table 1; Fig. 2D); this difference has a significant P-value of 0.014 (Supplemental Table S3). Contamination estimates based on the heterozygosity rate of the X Chromosome (a test only applicable to males) (Korneliussen et al. 2014) averaged 0.58% for the cochlea and 1.70% for the ossicles, a nonsignificant change (P = 0.688) (Table 1; Fig. 2; Supplemental Table S3). The overall low levels of contamination are also supported by consistency in the estimation of mtDNA haplogroups and molecular sex for all cochlea–ossicle pairs (Table 1; Fig. 2; Supplemental Table S2).
Discussion
DNA recovery from the auditory ossicles
This study presents a direct comparison of DNA recovery from the ossicles and corresponding cochlear bone using archaeological specimens that originate from varying geographic and temporal contexts, and offers several new insights. First, we show that the ossicles perform comparably to the cochlea in terms of ancient DNA recovery regardless of sample preservation. Focusing on 10 “working” individuals from whom we were able to generate enough ancient DNA data to call mtDNA haplogroups, assess damage pattern, and make contamination estimates for both the cochlea and ossicles, we find that the use of the cochlea or ossicles from each individual produces similar amounts of endogenous DNA, mtDNA coverage, nuclear SNP coverage, and number of SNPs called. We show that there is no substantial reduction in data quantity or complexity associated with the analysis of the ossicles instead of the cochlea, despite a smaller absolute quantity of bone material used. Second, although we find that the ossicles show a marginal reduction in the frequency of deamination (a signal of ancient DNA authenticity) compared with the corresponding cochlea, the magnitude of difference is not large enough to affect whether we consider the DNA to be authentically ancient, and this change becomes nonsignificant when we use a larger comparison pool of 12 individuals for which this metric was available. This finding, which may be an artifact of sample size or possibly related to an aspect of preservation that is currently unknown, should continue to be investigated using additional samples in future research.
We observe a modest but significant increase in the amount of estimated contamination between the cochlea and ossicles using mtDNA (from 0.80% to 1.80%), and a nonsignificant increase using heterozygosity on the X Chromosome (from 0.58% to 1.70%). These results suggest the possibility that ossicle extractions may be more susceptible to contamination than petrous bones extractions, as might be expected by the greater difficulty of cleaning the samples before ancient DNA analysis. However, we emphasize that the average mtDNA contamination in the ossicles in our analysis remains well under the accepted levels typically found in the ancient DNA literature (e.g., requiring a >95% match to the consensus sequence) (Fu et al. 2013). Thus, our results support the conclusion that ossicles are a useful and effective alternative to petrous samples in ancient DNA research. Future cleaning methods for ossicles may be able to reduce contamination rates even further.
Although four of the six individuals classified as “not working” did not produce sufficient amounts of data for either the cochlea or ossicles, only the cochlea produced sufficient amounts of authentic data for two first-degree relatives from the same site in Brazil (3935 and 3937). Although it is possible that the nine- and 31-fold decrease in complexity between the cochlea and ossicles of each individual (respectively) results from use of the ossicles, other variables such as burial- or sample-specific diagenesis need to be considered as well. In fact, the ossicles and cochlea from another individual (3938) from the same site produced comparable data for the two skeletal elements. The opposite scenario, in which the ossicles outperform the cochlea, has also been noted in the case of a previously published Pastoral Neolithic individual from Tanzania (Prendergast et al. 2019), for which high-quality data were recovered only from the ossicles (e.g., coverage of targeted SNPs increased from 0.233× using the cochlea to 1.58× using the ossicles).
Our data support no consistent differences between the ossicles and cochlea in terms of ancient DNA data recovered, suggesting at most only minimal differences between data quality associated with the analysis of the ossicles instead of the cochlea. We therefore conclude that the auditory ossicles, when present, are an alternative optimal skeletal element that can be used in ancient DNA research in place of the cochlea.
Although they are small, often isolated, and can be accessed without significant impact to larger and more morphologically informative parts of the skeleton, the use of ossicles for ancient DNA analysis still requires the destruction of human skeletal material that may be anthropologically valuable. Ossicles have previously been used in studies of comparative morphology; most notably, they have provided insight into morphological differences and functional similarities in the middle ear of Neandertals and anatomically modern humans, which has implications for understanding the auditory capacity of extinct hominins (e.g., Stoessel et al. 2016). For this reason, we encourage all researchers contemplating ancient DNA analysis to balance their analytical goals with the impact that sampling for ancient DNA analysis will have on future availability of material.
In light of these findings, we suggest that archaeologists and curators attempt to identify and preserve auditory ossicles whenever possible. Ideally, ossicles would be identified and collected during archaeological recovery of human skeletal remains in a way that minimizes the introduction of contamination, particularly given some evidence of this contamination to have downstream effects during the analysis of the ossicles. Methods for proactively reducing contamination include wearing disposable medical gloves that are changed frequently when handling samples, avoiding washing skeletal material with water, and storing samples in a cold, dry place as soon as possible (Llamas et al. 2017).
The use of ossicles for ancient DNA analysis will contribute to the successful analysis of skeletal material that does not have a petrous bone present or of sets of remains that have a petrous bone that cannot be processed in a destructive manner for ancient DNA research (e.g., those that may be morphologically intact and displayed in museum collections). On a broader level, the identification of the ossicles as an alternative optimal skeletal element for ancient DNA analysis contributes to the reduction in the amount of damage inflicted to human skeletal samples for the purposes of ancient DNA analysis. It is another step toward the preservation of DNA-rich and anthropologically valuable skeletal material for future studies that may benefit from future methodological improvements.
Methods
Sample selection and preparation
The number of ossicles collected for each of the archaeological samples varied (see Table 1), but the incus and malleus were identified and collected most frequently (n = 10 and n = 8, respectively), whereas the stapes was identified and collected least frequently (n = 2), likely owing to its diminutive size and fragility. In most cases, we recovered the ossicles while following the standard cochlea sampling procedure (Pinhasi et al. 2019). In other cases, we intentionally dislodged the ossicles from the skull for the purpose of this study; in most of these instances, the ossicles were partially visible within the external auditory meatus. To dislodge the ossicles, we cleaned a small engraving burr (as described by Sirak et al. 2017) by wiping it with a diluted bleach solution (∼10% concentration). We placed the cleaned burr inside the external auditory meatus and gently manipulated it within the inner ear canal. This caused no apparent damage to the ossicles or to the cranium from which they were retrieved. All ossicles were immediately placed into a sterile 2.0-mL tube upon their removal from the ear canal.
The preparation of all skeletal material for ancient DNA analysis was performed in dedicated cleanrooms at University College Dublin (UCD) or at the University of Vienna following standard anticontamination protocols (e.g., Hofreiter et al. 2001; Poinar 2003; Llamas et al. 2017). All petrous bones were processed following a standard protocol (Pinhasi et al. 2019). This protocol uses a dental sandblaster to systematically locate, isolate, and clean the cochlea, which is then milled to homogeneous bone powder. Approximately 50 mg of bone powder from the cochlea (range, 47–56 mg) was aliquoted for DNA extraction. Complete auditory ossicles were decontaminated through exposure to UV irradiation for 10 min on each side. A DNA clean room was used for DNA extraction and preparation of sequencing libraries.
DNA extraction
DNA was extracted from the cochlear bone powder and the whole auditory ossicles in ancient-DNA facilities at the University of Vienna following a standard ancient DNA extraction protocol (Dabney et al. 2013) with a modification (Korlević et al. 2015) that uses the tube assemblies from a high pure viral nucleic acid large-volume kit (Roche 05114403001). The intact ossicles were placed in the extraction buffer and completely dissolved during the incubation period in most cases. Lysates were mixed with binding buffer and applied to the column assembly. DNA was bound to the silica matrix of the columns by centrifugation and was washed twice with 650 µL of PE buffer (Qiagen) and spun through the columns at 6000 rpm for 1 min each time. After the column was put in a fresh 1.5-mL collection tube, 25 µL of TET buffer was pipetted on the dry spun columns’ silica membrane. After a 10-min incubation at room temperature, the columns were spun at maximum speed for 1 min. The elution step was repeated to give a final volume of 50 µL of DNA extract. A negative control that contained no bone material was included with each extraction batch.
Library preparation
High-throughput sequencing libraries were prepared in ancient DNA facilities at Harvard Medical School from all extracts and controls included in the main study using a library preparation method optimized for ancient DNA (Rohland et al. 2015). This protocol uses a partial-UDG treatment that causes characteristic C-to-T ancient DNA damage to be restricted to the terminal molecules while nearly eliminating it in the interior of the DNA molecules so that the library can be used to test for ancient DNA authenticity. Ten microliters of DNA extract was used as input during library preparation. Libraries were enriched for approximately 1.2 million nuclear sites across the genome (“1240K capture”) in addition to sites on the human mitochondrial genome (Fu et al. 2013, 2015; Haak et al. 2015; Mathieson et al. 2015). Enriched libraries were sequenced on an Illumina NextSeq 500 instrument, with 2 × 76 cycles and an additional 2 × 7 cycles used for identification of indices. In addition, a small proportion of reads was generated from unenriched versions of each library. These unenriched (“shotgun”) data were used to estimate the proportion of endogenous molecules in each library.
Data processing
Following sequencing, we trimmed molecular adapters and barcodes from sequenced reads before merging forward and reverse reads using publicly available software and pipelines (Prendergast et al. 2019; https://github.com/DReichLab/ADNA-Tools). We allowed up to three mismatches at positions of low base quality (lower than 20) and up to one mismatch at higher base quality (20 or greater); in cases of base matching, we kept the highest base quality in the overlap region, whereas in cases in which bases differed, we retained the higher quality base with reduced base quality of the difference of the two base qualities. We aligned reads to the mitochondrial RSRS genome (Behar et al. 2012) and to the hg19 human reference sequence with the samse command in BWA (v0.7.15) (Li and Durbin 2009). We use the hg19 build of the human genome to avoid potential reference bias possible when performing population genetics analysis with other samples previously aligned to hg19, which is the historical norm for ancient DNA studies. Other versions of the human reference are expected to have nearly identical numbers of SNPs in aligned reads, but those SNPs may be biased relative to hg19.
We used the tool ContamMix (Fu et al. 2014) to determine the rate of matching between the consensus RSRS sequence and reads that aligned to the mitochondrial genome. We determined the rate of C-to-T substitution at the terminal ends of each molecule using PMDtools (Skoglund et al. 2014; https://github.com/pontussk/PMDtools). We used the tool ANGSD (Korneliussen et al. 2014) to determine the amount of contamination in the X Chromosome of individuals identified as genetically male. The complexity of the sample was assessed by quantifying the number of unique reads expected from a predetermined number of reads hitting target.
Data access
All sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB32751.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was partially supported by a European Research Council starting grant ADNABIOARC 263441 to R.P., a National Science Foundation (NSF) doctoral dissertation research improvement grant BCS-1613577 to K. Sirak, an Irish Research Council grant GOIPG/2013/36 to D.F., a graduate student fellowship from the Max Planck-Harvard Research Center for the Archaeoscience of the Ancient Mediterranean (MHAAM) to E.H., and Russian Foundation for Basic Research grants 18-00-00360, 18-09-00349 to V.M. T.H. and T.S. were supported by grants from the Hungarian Research, Development and Innovation Office, project numbers FK128013 and TÉT_16-1-2016-0020. D.R. is an investigator of the Howard Hughes Medical Institute. We thank the Canterbury Archaeological Trust for permission to analyze sample 1257 (I4491); full details about this sample can be found in the CAT PXA report (Helm et al. 2017). We also thank Museu do Homem do Sambaqui “Pe. João Alfredo Rohr” for permission to analyze the samples from Brazil included in this analysis.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.260141.119.
References
- Arensburg B, Harell M, Nathan H. 1981. The human middle ear ossicles: morphometry, and taxonomic implications. J Hum Evol 10: 199–205. 10.1016/S0047-2484(81)80018-8 [DOI] [Google Scholar]
- Arensburg B, Belkin V, Wolf M. 2005. Middle ear pathology in ancient and modern populations: incudal osteoma. Acta Otolaryngol 125: 1164–1167. 10.1080/00016480510043431 [DOI] [PubMed] [Google Scholar]
- Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli E-L, Silva NM, Kivisild T, Torroni A, Villems R. 2012. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet 90: 675–684. 10.1016/j.ajhg.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LS, Kayser M, Jones C. 2008. The mineralized osteocyte: a living fossil. Am J Phys Anthropol 137: 449–456. 10.1002/ajpa.20886 [DOI] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci 104: 14616–14621. 10.1073/pnas.0704665104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Meyer M, Krause J, Kircher K, Pääbo S. 2010. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res 38: e87. 10.1093/nar/gkp1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broushaki F, Thomas MG, Link V, López S, van Dorp L, Kirsanow K, Hofmanová Z, Diekmann Y, Cassidy LM, Díez-del-Molino D, et al. 2016. Early neolithic genomes from the eastern fertile crescent. Science 353: 499–503. 10.1126/science.aaf7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse B, Djonic D, Milovanovic P, Hahn M, Püschel K, Ritchie RO, Djuric M, Amling M. 2010. Decrease in the osteocyte lacunar density accompanied by hypermineralized lacunar occlusion reveals failure and delay of remodeling in aged human bone. Aging Cell 9: 1065–1075. 10.1111/j.1474-9726.2010.00633.x [DOI] [PubMed] [Google Scholar]
- Chen H, Okumura T, Emura S, Shoumura S. 2008. Scanning electron microscopic study of the human auditory ossicles. Annals of Anatomy - Anatomischer Anzeiger 190: 53–58. 10.1016/j.aanat.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Collins MJ, Nielsen–Marsh CM, Hiller J, Smith CI, Roberts JP, Prigodich RV, Wess TJ, Csapò J, Millard AR, Turner–Walker G. 2002. The survival of organic matter in bone: a review. Archaeometry 44: 383–394. 10.1111/1475-4754.t01-1-00071 [DOI] [Google Scholar]
- Crevecoeur I. 2007. New discovery of an Upper Paleolithic auditory ossicle: the right malleus of Nazlet Khater 2. J Hum Evol 52: 341–345. 10.1016/j.jhevol.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Cunningham C, Scheuer L, Black S. 2000. Developmental juvenile osteology. Academic Press, San Diego. [Google Scholar]
- Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci 110: 15758–15763. 10.1073/pnas.1314445110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard PB, Margaryan A, Schroeder H, Orlando L, Willerslev E, Allentoft ME. 2015. Improving access to endogenous DNA in ancient bones and teeth. Sci Rep 5: 11184 10.1038/srep11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboeuf F, Burt-Pichat B, Farlay D, Suy P, Truy E, Boivin G. 2015. Bone quality and biomechanical function: a lesson from human ossicles. Bone 73: 105–110. 10.1016/j.bone.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Fregel R, Méndez FL, Bokbot Y, Martín-Socas D, Camalich-Massieu MD, Santana J, Morales J, Ávila-Arcos MC, Underhill PA, Shapiro B, et al. 2018. Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe. Proc Natl Acad Sci 115: 6774–6779. 10.1073/pnas.1800851115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch T, Sølvsten Sørensen M, Overgaard S, Lind M, Bretlau P. 1998. Volume-referent bone turnover estimated from the interlabel area fraction after sequential labeling. Bone 22: 677–682. 10.1016/S8756-3282(98)00050-7 [DOI] [PubMed] [Google Scholar]
- Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Pääbo S. 2013. DNA analysis of an early modern human from Tianyuan Cave, China. Proc Natl Acad Sci 110: 2223–2227. 10.1073/pnas.1221359110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prüfer K, de Filippo C, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514: 445–449. 10.1038/nature13810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, et al. 2015. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524: 216–219. 10.1038/nature14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővari I, Pap I, Anders A, et al. 2014. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun 5: 5257 10.1038/ncomms6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansauge M-T, Gerber T, Glocke I, Korlević P, Lippik L, Nagel S, Riehl L, Schmidt A, Meyer M. 2017. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res 45: e79 10.1093/nar/gkx033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginolhac A, Rasmussen M, Gilbert MTP, Willerslev E, Orlando L. 2011. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics 27: 2153–2155. 10.1093/bioinformatics/btr347 [DOI] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522: 207–211. 10.1038/nature14317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HB, Damgaard PB, Margaryan A, Stenderup J, Lynnerup N, Willerslev E, Allentoft ME. 2017. Comparing ancient DNA preservation in petrous bone and tooth cementum. PLoS One 12: e0170940 10.1371/journal.pone.0170940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney E, May H, Shalem D, Rohland N, Mallick S, Lazaridis I, Sarig E, Stewardson K, Nordenfelt S, Patterson N, et al. 2018. Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nat Commun 9: 3336 10.1038/s41467-018-05649-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim JL. 1982. Les Enfants Néandertaliens de La Ferrassie. Publié sous les auspices de la Fondation Singer-Polignac. Bulletins et Mémoires de la Société d'Anthropologie de Paris 9: 347. [Google Scholar]
- Helm R, Allison E, Anderson I, Barber L, Broadley R, Carruthers W, Locker A, Lyne M, McIntyre L, McNee B, Richardson A, Smith I, Wilson T. 2017. Former Peugeot Garage, Rhodaus Town (A28), Canterbury, Kent CT1 2RH. Post-excavation assessment, client report no. 2017/107. Canterbury Archaeological Trust, Canterbury, UK. [Google Scholar]
- Hernandez CJ, Majeska RJ, Schaffler MB. 2004. Osteocyte density in woven bone. Bone 35: 1095–1099. 10.1016/j.bone.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. 2001. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res 29: 4793–4799. 10.1093/nar/29.23.4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hublin JJ, Pääbo S, Derevianko AP, Doronichev VB, Golovanova LV, Freiss M, Froment A, Hoffmann A, Jillani Kachache NE, Kullmer O, et al. 2008. Suggested guidelines for invasive sampling of hominid remains. J Hum Evol 55: 756–757. 10.1016/j.jhevol.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Kanzaki S, Ito M, Takada Y, Ogawa K, Matsuo K. 2006. Resorption of auditory ossicles and hearing loss in mice lacking osteoprotegerin. Bone 39: 414–419. 10.1016/j.bone.2006.01.155 [DOI] [PubMed] [Google Scholar]
- Kontopoulos I, Penkman K, McAllister GD, Lynnerup N, Damgaard PB, Hansen HB, Allentoft ME, Collins MJ. 2019. Petrous bone diagenesis: a multi-analytical approach. Palaeogeogr Palaeoclimatol Palaeoecol 518: 143–154. 10.1016/j.palaeo.2019.01.005 [DOI] [Google Scholar]
- Korlević P, Gerber T, Gansauge M-T, Hajdinjak M, Nagel S, Aximu-Petri A, Meyer M. 2015. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. BioTechniques 59: 87–93. 10.2144/000114320 [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15: 356 10.1186/s12859-014-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536: 419–424. 10.1038/nature19310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715. 10.1038/362709a0 [DOI] [PubMed] [Google Scholar]
- Llamas B, Valverde G, Fehren-Schmitz L, Weyrich LS, Cooper A, Haak W. 2017. From the field to the laboratory: controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. STAR: Science & Technology of Archaeological Research 3: 1–14. 10.1080/20548923.2016.1258824 [DOI] [Google Scholar]
- Makarewicz C, Marom N, Bar-Oz G. 2017. Palaeobiology: Ensure equal access to ancient DNA. Nature 548: 158 10.1038/548158a [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotti G, Farneti D, Remaggi F, Tartari F. 1998. Morphometric investigation on osteocytes in human auditory ossicles. Ann Anat 180: 449–453. 10.1016/S0940-9602(98)80106-4 [DOI] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Alpaslan-Roodenberg S, Harney E, Stewardson K, Fernandes D, Novak M, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528: 499–503. 10.1038/nature16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays S, Elders J, Humphrey L, White W, Marshall P. 2013. Science and the dead: a guideline for the destructive sampling of archaeological human remains for scientific analysis. Advisory Panel on the Archaeology of Burials in England, English Heritage Publishing, Swindon, UK. [Google Scholar]
- Pinhasi R, Fernandes DM, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromov A, Raczky P, et al. 2015. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10: e0129102 10.1371/journal.pone.0129102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fernandes DM, Sirak K, Cheronet O. 2019. Isolating the human cochlea to generate bone powder for ancient DNA analysis. Nat Protoc 14: 1194–1205. 10.1038/s41596-019-0137-7 [DOI] [PubMed] [Google Scholar]
- Poinar HN. 2003. The top 10 list: criteria of authenticity for DNA from ancient and forensic samples. In International Congress Series 1239: 575–579. 10.1016/S0531-5131(02)00624-6 [DOI] [Google Scholar]
- Ponce de León MS, Koesbardiati T, Weissmann JD, Milella M, Reyna-Blanco CS, Suwa G, Kondo O, Malaspinas A-S, White TD, Zollikofer CPE. 2018. Human bony labyrinth is an indicator of population history and dispersal from Africa. Proc Natl Acad Sci 115: 4128–4133. 10.1073/pnas.1717873115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast ME, Sawchuk E. 2018. Boots on the ground in Africa's ancient DNA ‘revolution’: archaeological perspectives on ethics and best practices. Antiquity 92: 803–815. 10.15184/aqy.2018.70 [DOI] [Google Scholar]
- Prendergast ME, Lipson M, Sawchuk EA, Olalde I, Ogola CA, Rohland N, Sirak KA, Adamski N, Bernardos R, Broomandkhoshbacht N, et al. 2019. Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science 365: eaaw6275 10.1126/science.aaw6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quam R, Rak Y. 2008. Auditory ossicles from southwest Asian Mousterian sites. J Hum Evol 54: 414–433. 10.1016/j.jhevol.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Quam R, Martínez I, Arsuaga JL. 2013a. Reassessment of the La Ferrassie 3 Neandertal ossicular chain. J Hum Evol 64: 250–262. 10.1016/j.jhevol.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Quam R, de Ruiter DJ, Masali M, Arsuaga J-L, Martínez I, Moggi-Cecchi J. 2013b. Early hominin auditory ossicles from South Africa. Proc Natl Acad Sci 110: 8847–8851. 10.1073/pnas.1303375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvist M. 2000. Auditory ossicles in archaeological skeletal material from medieval Denmark. Acta Otolaryngol 120: 82–85. 10.1080/000164800454053 [DOI] [PubMed] [Google Scholar]
- Rak Y, Clarke RJ. 1979. Ear ossicle of Australopithecus robustus. Nature 279: 62–63. 10.1038/279062a0 [DOI] [PubMed] [Google Scholar]
- Rask-Andersen H, Liu W, Erixon E, Kinnefors A, Pfaller K, Schrott-Fischer A, Gluckert R. 2012. Human cochlea: anatomical characteristics and their relevance for cochlear implantation. Anat Rec 295: 1791–1811. 10.1002/ar.22599 [DOI] [PubMed] [Google Scholar]
- Richard C, Courbon G, Laroche N, Prades JM, Vico L, Malaval L. 2017. Inner ear ossification and mineralization kinetics in human embryonic development - microtomographic and histomorphological study. Sci Rep 7: 4825 10.1038/s41598-017-05151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. 2015. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos Trans R Soc Lond B Biol Sci 370: 20130624 10.1098/rstb.2013.0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Glocke I, Aximu-Petri A, Meyer M. 2018. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat Protoc 13: 2447–2461. 10.1038/s41596-018-0050-5 [DOI] [PubMed] [Google Scholar]
- Rolvien T, Schmidt FN, Milovanovic P, Jähn K, Riedel C, Butscheidt S, Püschel K, Jeschke A, Amling M, Busse B. 2018. Early bone tissue aging in human auditory ossicles is accompanied by excessive hypermineralization, osteocyte death and micropetrosis. Sci Rep 8: 1920 10.1038/s41598-018-19803-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann VJ, Peltzer A, Welte B, van Pelt WP, Molak M, Wang C-C, Furtwängler A, Urban C, Reiter E, Nieselt K, et al. 2017. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat Commun 8: 15694 10.1038/ncomms15694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwark T, Modrow JH, Steinmeier E, Poetsch M, Hasse J, Fischer H, von Wurmb-Schwark N. 2015. The auditory ossicles as a DNA source for genetic identification of highly putrefied cadavers. Int J Legal Med 129: 457–462. 10.1007/s00414-015-1177-3 [DOI] [PubMed] [Google Scholar]
- Siori MS, Monchietto MJ, Masali M. 1995. Morphometrics of human auditory ossicles from Antinoe Necropolis (Egypt). Int J Anthropol 10: 29–36. 10.1007/BF02447595 [DOI] [Google Scholar]
- Sirak KA, Sedig JW. 2019. Balancing analytical goals and anthropological stewardship in the midst of the paleogenomics revolution. World Archaeology 10.1080/00438243.2019.1617190 [DOI] [Google Scholar]
- Sirak K, Fernandes DM, Cheronet O, Novak M, Gamarra B, Balassa T, Bernert Z, Cséki A, Dani J, Gallina JZ, et al. 2017. A minimally-invasive method for sampling human petrous bones from the cranial base for ancient DNA analysis. BioTechniques 62: 283–289. 10.2144/000114558 [DOI] [PubMed] [Google Scholar]
- Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Pääbo S, Krause J, Jakobsson M. 2014. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. ProcNatl Acad Sci 111: 2229–2234. 10.1073/pnas.1318934111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Thompson JC, Prendergast ME, Mittnik A, Sirak K, Hajdinjak M, Salie T, Rohland N, Mallick S, Peltzer A, et al. 2017. Reconstructing prehistoric African population structure. Cell 171: 59–71.e21. 10.1016/j.cell.2017.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CI, Chamberlain AT, Riley MS, Cooper A, Stringer CB, Collins MJ. 2001. Neanderthal DNA: not just old but old and cold? Nature 410: 771–772. 10.1038/35071177 [DOI] [PubMed] [Google Scholar]
- Smith CI, Chamberlain AT, Riley MS, Stringer CB, Collins MJ. 2003. The thermal history of human fossils and the likelihood of successful DNA amplification. J Hum Evol 45: 203–217. 10.1016/S0047-2484(03)00106-4 [DOI] [PubMed] [Google Scholar]
- Sølvsten Sørensen M, Bretlau P, Balslev Jørgensen M. 1992. Quantum type bone remodeling in the human otic capsule: morphometric findings. Acta Otolaryngol 112: 4–10. 10.3109/00016489209136839 [DOI] [Google Scholar]
- Spoor F, Hublin JJ, Braun M, Zonneveld F. 2003. The bony labyrinth of Neanderthals. J Hum Evol 44: 141–165. 10.1016/S0047-2484(02)00166-5 [DOI] [PubMed] [Google Scholar]
- Stoessel A, Gunz P, David R, Spoor F. 2016. Comparative anatomy of the middle ear ossicles of extant hominids—introducing a geometric morphometric protocol. J Hum Evol 91: 1–25. 10.1016/j.jhevol.2015.10.013 [DOI] [PubMed] [Google Scholar]
- van de Loosdrecht M, Bouzouggar A, Humphrey L, Posth C, Barton N, Aximu-Petri A, Nickel B, Nagel S, Talbi EH, El Hajraoui MA, et al. 2018. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360: 548–552. 10.1126/science.aar8380 [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Iino Y, Kakizaki K, Murakami Y. 1999. Human temporal bone study on the postnatal ossification process of auditory ossicles. Laryngoscope 109: 927–930. 10.1097/00005537-199906000-00016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.