Abstract
Circular RNAs (circRNAs), a class of long noncoding RNAs, are known to be enriched in mammalian neural tissues. Although a wide range of dysregulation of gene expression in autism spectrum disorder (ASD) have been reported, the role of circRNAs in ASD remains largely unknown. Here, we performed genome-wide circRNA expression profiling in postmortem brains from individuals with ASD and controls and identified 60 circRNAs and three coregulated modules that were perturbed in ASD. By integrating circRNA, microRNA, and mRNA dysregulation data derived from the same cortex samples, we identified 8170 ASD-associated circRNA-microRNA-mRNA interactions. Putative targets of the axes were enriched for ASD risk genes and genes encoding inhibitory postsynaptic density (PSD) proteins, but not for genes implicated in monogenetic forms of other brain disorders or genes encoding excitatory PSD proteins. This reflects the previous observation that ASD-derived organoids show overproduction of inhibitory neurons. We further confirmed that some ASD risk genes (NLGN1, STAG1, HSD11B1, VIP, and UBA6) were regulated by an up-regulated circRNA (circARID1A) via sponging a down-regulated microRNA (miR-204-3p) in human neuronal cells. Particularly, alteration of NLGN1 expression is known to affect the dynamic processes of memory consolidation and strengthening. To the best of our knowledge, this is the first systems-level view of circRNA regulatory networks in ASD cortex samples. We provided a rich set of ASD-associated circRNA candidates and the corresponding circRNA-microRNA-mRNA axes, particularly those involving ASD risk genes. Our findings thus support a role for circRNA dysregulation and the corresponding circRNA-microRNA-mRNA axes in ASD pathophysiology.
Autism spectrum disorder (ASD) (Mendelian Inheritance in Man [MIM] 209850) is a heritable, complex, highly pervasive neurodevelopmental disorder that is characterized by limited social communication, restricted and ritualized interests, and repetitive behavior (Chen et al. 2015b; Ansel et al. 2017). Hundreds of genes affected by a variety of genomic variants have been reported to be associated with the etiology of ASD (Xu et al. 2012; Abrahams et al. 2013; Luo et al. 2018). These discoveries have provided valuable biological insights into the disorder. However, the contribution of these genetic factors to this complex disease is highly heterogeneous (Walsh et al. 2011), with each factor accounting for a very low percentage of the general ASD population. In addition to genetic factors, environment and gene–environment interactions have been widely shown to be associated with the development of ASD (Schendel et al. 2014; Kim and Leventhal 2015), but the underlying molecular mechanisms remain poorly understood (Geschwind 2011). Recently, using samples from postmortem ASD and control brains, several studies have reported a wide range of dysregulation of gene expression (Voineagu et al. 2011; Guan et al. 2016; Parikshak et al. 2016) in ASD. Moreover, epigenetic factors such as alternatively spliced transcripts (Voineagu et al. 2011; Irimia et al. 2014; Parikshak et al. 2016), long noncoding RNAs (Wang et al. 2015; Parikshak et al. 2016), microRNAs (miRNAs) (Ander et al. 2015; Wu et al. 2016), DNA methylation (Vogel Ciernia and LaSalle 2016), histone trimethylation (Shulha et al. 2012), and acetylation (Sun et al. 2016) were also implicated in ASD. Although these analyses based on transcriptome and genome sequencing data have provided important insights into the effects of regulatory genetic variants (Voineagu et al. 2011; Iossifov et al. 2014), the role of co-/post-transcriptional mechanisms, which are critical to the understanding of dysregulation of gene expression in ASD, remains largely unknown. Illuminating the underlying molecular mechanisms in the etiology of ASD awaits further investigation.
Circular RNAs (circRNAs) are a class of long noncoding RNAs produced by pre-mRNA back-splicing, which generates a structure of covalently closed loop (Ashwal-Fluss et al. 2014) of single-strand, non-polyadenylated circular molecules (Salzman et al. 2012; Jeck et al. 2013; Memczak et al. 2013). In general, circRNAs are more stable than their corresponding colinear mRNA isoforms (Jeck et al. 2013; Memczak et al. 2013; Zheng et al. 2016; Chuang et al. 2018). They are often expressed in temporal- and spatial-specific manners (Salzman et al. 2012, 2013; Jeck et al. 2013) and are especially abundant in neural tissues (Rybak-Wolf et al. 2015; You et al. 2015). The best understood function of circRNAs is the regulatory role of miRNA sponges (Hansen et al. 2013; Memczak et al. 2013), suggesting a previously underappreciated regulatory pathway in the form of circRNA-miRNA-mRNA axes. Several circRNAs are known to play important roles in development of the nervous system (Conn et al. 2015; Rybak-Wolf et al. 2015; Szabo et al. 2015). For example, CDR1-AS (also known as ciRS-7), the best-studied circRNA, is highly expressed and serves as a sponge for miRNA-7 in neuronal tissues (Hansen et al. 2013) and is implicated in the pathogenesis and progression of neurological diseases (Floris et al. 2017; Wang et al. 2018b). A recent study also reported that the interactions of circEFCAB2 with miR-485-5p and circDROSHA with miR-1252-5p were highly correlated with the expression of epilepsy-associated genes CLCN6 and ATP1A2, respectively (Li et al. 2018). These studies clearly indicate an important role of circRNA in epigenetic control over gene expression in neural tissues. Although the large-scale transcriptome analysis of miRNA-mRNA regulatory interactions have been reported (Hicks and Middleton 2016; Wu et al. 2016) in ASD, the role of circRNA-miRNA-mRNA regulatory axes in ASD has never been reported and remains largely unknown.
In this study, we investigated the potential regulatory role of circRNAs in ASD. Because circRNAs lack poly(A) tails, RNA-seq data derived from poly(A)-selected RNAs naturally are deprived of circRNA population. This greatly hampers the large-scale study of circRNAs in ASD. The recently released Synapse database (Parikshak et al. 2016), which provided a large brain sample size of both small RNA-seq data and RNA-seq data from total RNAs (rRNA-depleted RNAs without poly(A)-selection) of ASD cases and controls, brought an unprecedented opportunity for us to genome-wide investigate circRNA dysregulation in ASD and the corresponding ASD-associated circRNA-miRNA-mRNA regulatory axes. We performed genome-wide circRNA expression profiling in postmortem brains from ASD patients and non-ASD controls and thereby identified circRNAs and coregulated circRNA modules that were perturbed in ASD. By combining the miRNA dysregulation data previously detected in the same ASD and control cohort used in the current study (Wu et al. 2016), we constructed ASD-associated circRNA-miRNA-mRNA interactions according to the common target miRNAs of the circRNAs and mRNAs. Furthermore, we selected a circRNA-miRNA axis that was predicted to be an upstream regulator of several ASD risk genes and indeed confirmed the corresponding circRNA-miRNA-mRNA regulatory relationships in normal human astrocytes (NHAs) or primary human neural progenitor cells (hNPCs). Our findings thus help to unveil the molecular mechanism of aberrant circRNA expression and the corresponding circRNA-miRNA-mRNA regulations in idiopathic ASD.
Results
Identification of differentially expressed circRNAs in ASD brain
We retrieved the high-throughput RNA sequencing (RNA-seq) data of 236 postmortem samples of frontal cortex (FC) (Brodmann area 9), temporal cortex (TC) (Brodmann areas 22, 41, and 42), and cerebellar vermis (CV) from 48 individuals with ASD and 49 non-ASD controls (the Synapse database) (Table 1; Parikshak et al. 2016). After removing samples with low read depth, we then used NCLscan, which was reported to show the greatest precision among currently available circRNA-detection tools (Chuang et al. 2016; Zeng et al. 2017; Chen and Chuang 2019a,b), to identify circRNAs and identify differential expressed species in ASD versus control samples. We first found no significant difference in the number of identified circRNAs per million mapped RNA-seq reads between ASD and non-ASD control samples, regardless of the brain region (Fig. 1A; Supplemental Table S1). To increase the stringency of sample consistency, a sample was not considered in the following analysis if number of the identified circRNAs of this sample was one standard deviation below the mean of the sample set. After that, 202 samples (73 FC, 61 TC, and 68 CV) were retained. There were 53,427 NCLscan-identified circRNAs in the 202 samples (Supplemental Table S2). Several independent transcriptomics studies of ASD showed that the pathophysiology was present in cortex (Parikshak et al. 2013; Willsey et al. 2013), and the transcriptomic profiles of cortex and cerebellum were quite different. Changes in transcriptomic profiles were consistently stronger in the cortex than in the cerebellum, suggesting that the cortex was more selectively vulnerable to transcriptomic alterations as compared with the cerebellum (Parikshak et al. 2016). Like previous transcriptomics studies of miRNA (Wu et al. 2016) and mRNA (Parikshak et al. 2016) dysregulation in ASD, our following analysis focused on the frontal and temporal cortex samples (134 samples, including 73 samples from 45 ASD cases and 61 samples from 43 controls) (Fig. 1B). Of the 36,624 circRNAs identified in the 134 cortex samples, 40% (14,775 of 36,624) were observed in one sample only (Supplemental Table S2). To minimize potentially spurious events, we only considered the circRNAs observed in >50% of the 134 samples (Fig. 1B). A total of 1060 circRNAs were retained (Supplemental Table S3), of which 99.6% (1056 of 1060) have been previously collected in well-known circRNA databases—circBase (Glažar et al. 2014) or CIRCpedia (Dong et al. 2018)—and 99.1% (1050 of 1060) have been detected in human brain (Fig. 1C). Of note, 61.4% (651 events) and 47% (498 events) of the 1060 circRNAs were also detected in mouse and mouse brain, respectively (Fig. 1C; Supplemental Table S3), indicating the high conservation rate of circRNA expression between human and mouse. We performed principal component analysis (PCA) and showed that circRNA expression profiles of these 1060 circRNAs were very similar between two types of cortex regions but were quite distinct from that of cerebellum (Fig. 1D). This result is consistent with the patterns previously observed for mRNAs (Oldham et al. 2008; Voineagu et al. 2011) and miRNAs (Wu et al. 2016) between these brain regions.
Table 1.
Brain tissue samples used in this study

Figure 1.
Identification of DE-circRNAs in the ASD cortex samples. (A) Comparison of normalized numbers of circRNAs in ASD and non-ASD control samples from different brain regions: (FC) frontal cortex, (TC) temporal cortex, (CV) cerebellar vermis. P-values were determined using two-tailed Wilcoxon rank-sum test. (B) Flowchart of the overall approach. To minimize potentially spurious events, only the circRNAs (1060 circRNAs) that were detected to be expressed in >50% of the samples examined are considered for the following analyses. (C) Comparisons of the 1060 circRNAs and the human/mouse circRNAs collected in well-known databases (i.e., circBase/CIRCpedia). (D) Principal component plots of circRNA expression profiles of the 1060 circRNAs in samples from FC, TC, and CV. (PC1/PC2) First and second principal components. (E) circRNA expression fold changes (>0 if higher in ASD; <0 if lower in ASD) between ASD and non-ASD control cortex samples, plotted against the percentile rank of mean expression levels of the 1060 circRNAs across 134 cortex samples used for differential expression analysis. The identified up-regulated (22) and down-regulated (38) circRNAs in the ASD cortex samples (LME model, P < 0.05 and |log2(fold change)| ≥ 0.5) are highlighted in red and green, respectively. (F) Comparison of circRNA expression fold changes in the FC and TC samples. The black line represents the regression line between fold changes in the FC and TC for the 60 DE-circRNAs. The Pearson correlation coefficient (R) and P-value are shown. (G) Comparison of circRNA expression fold changes in a small number of samples and all 134 samples combined: (left) samples with no Chromosome 15q11-13 duplication syndrome; (middle) samples with RIN ≥ 5; (right) samples with PMI ≤ 30 h). The black lines represent the regression lines between fold changes in the corresponding small number of samples and all samples combined for the 60 DE-circRNAs. (H) PCA based on the 60 DE-circRNAs. (I) Dendrogram representing hierarchical clustering of 134 cortex samples based on the 60 identified DE-circRNAs. Information on diagnosis, age, brain bank, PMI, brain region, sex, and RIN is indicated with color bars below the dendrogram according to the legend on the right. Heat map on the bottom represents scaled expression levels (color-coded according to the legend below the heat map) for the 60 DE-circRNAs.
We performed a linear mixed effects (LME) model to detect differentially expressed (DE)-circRNAs with controlling for potential confounding factors such as sex, age, brain region (FC or TC), RNA quality (RNA integrity number [RIN]), host gene expression, and so on (Methods). We calculated the effect size (Cohen's d) of each circRNA and estimated the needed sample size for detecting DE-circRNAs when considering multiple test correction. Our result suggested that it required 581 ASD samples versus 581 control samples to detect DE-circRNAs at the significance level of 80% power (Bonferroni-corrected P < 0.05) (Methods). Because there were only 73 ASD and 61 control samples in the examined data set, a larger sample size may be required to detect DE-circRNAs between ASD and control with considering multiple test correction. Therefore, we considered the 60 differentially expressed (P < 0.05 and |log2(fold change)| ≥ 0.5) for the following analyses. Of the 60 DE-circRNAs, 22 were up-regulated and 38 were down-regulated in ASD cortex (Fig. 1B,E; Supplemental Table S3). We found that the fold changes for the 60 DE-circRNAs were highly concordant between the FC and TC (Pearson correlation coefficient R = 0.75, P < 10−11) (Fig. 1F), and relatively marginally concordant between the cortex (FC/TC) and the cerebellum (R = ∼0.3, P < 0.05) (Supplemental Fig. S1).
To examine whether our result was biased by a small number of samples, we first performed resampling analysis with 100 rounds of random sampling of 70% of the samples examined and showed that the fold changes of DE-circRNAs for the resampled and the original sample sets were highly concordant between each other (Pearson's R = 0.95∼0.98, all P < 2.2 × 10−16) (Supplemental Fig. S2). In addition, we compared the fold changes for a small number of samples (from samples with no Chromosome 15q11-13 duplication syndrome, with RIN ≥ 5, or with postmortem interval [PMI] ≤ 30 h) with those for all samples combined and found a high concordance between each other (all Pearson's R = 0.99, P < 2.2 × 10−16) (Fig. 1G). Comparing the samples with RIN < 5 with those with RIN ≥ 5, the fold changes for the 60 DE-circRNAs were still highly concordant between each other (Pearson's R = 0.69, P < 10−9) (Supplemental Fig. S3A). Regarding the RNA quality of the transcript sequences where the DE-circRNAs located (measured by median transcript integrity number [TIN]) (Wang et al. 2016; Methods), we also found no significant difference between the samples with RIN < 5 and RIN ≥ 5 (P = 0.76 by two-tailed Wilcoxon rank-sum test) (Supplemental Fig. S3B). These observations revealed that our results were not biased by a small number of samples with removal of Chromosome 15q11-13 duplication syndrome, low RIN, or high PMI.
PCA based on the 60 DE-circRNAs revealed that ASD and non-ASD samples could be grouped into separate clusters (Fig. 1H). Hierarchical clustering further showed distinct clustering for the majority of ASD samples. We found two distinct groups for the clustering of the 134 cortex samples, in which 76% (44 of 58) and 38% (29 of 76) of cortex samples were ASD samples in the right and left groups, respectively (P-value < 0.0001 by two-tailed Fisher's exact test) (Fig. 1I). The confounding factors such as brain region, age, sex, RIN, brain bank, and PMI did not drive the similar clustering (Fig. 1I). Our result thus revealed a shared circRNA dysregulation signature among the majority of ASD samples.
Construction of signed coexpression networks of circRNA dysregulation in ASD
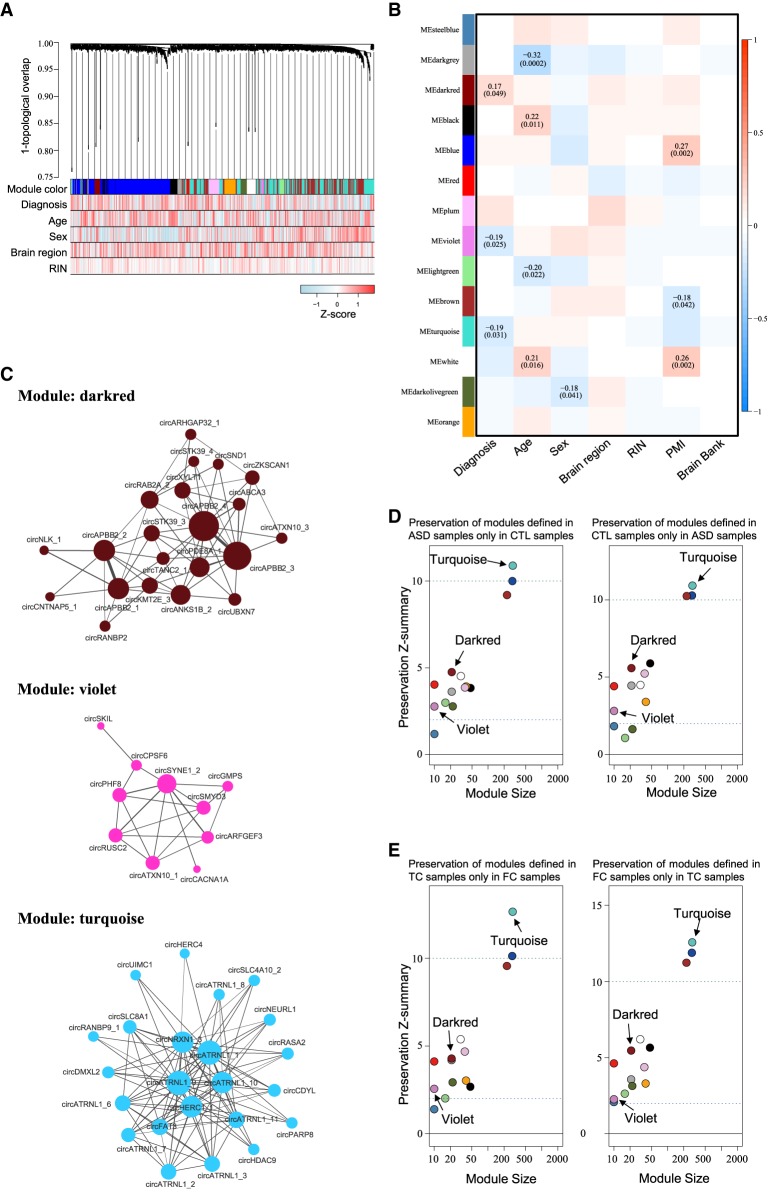
To probe the correlation between circRNA expression changes and disease status at the system level, we performed the weighted gene coexpression network analysis (WGCNA) to assign individual circRNAs to coexpression modules according to the 1060 circRNAs identified in the 134 cortex samples. As shown in Figure 2A, we identified 14 modules and estimated the module eigengene, that is, the first principal component (PC1) of the module's circRNA expression. By assessing the relationship between module eigengene and diagnosis status, three (darkred, violet, and turquoise) (Fig. 2B; Supplemental Table S3) modules were significantly correlated with ASD status (designated as “DE-modules”), of which the darkred module (including 21 circRNAs) was up-regulated and the violet (including 10 circRNAs) and turquoise (including 288 circRNAs) modules were down-regulated in ASD samples (Fig. 2C). Of note, the three modules were not correlated with important experimental covariates (age, sex, and brain region) and technique confounders (RIN, PMI, and brain bank) (Fig. 2B).
Figure 2.
Dysregulation of circRNA coexpression networks in ASD cortex. (A) Dendrogram of circRNA coexpression modules defined in 134 cortex samples. Consensus module color bar shows assignment based on 1000 rounds of bootstrapping. Diagnosis and potential confounders (age, sex, region, RIN) are treated as numeric variables to calculate their Pearson correlation coefficients with expression level for each circRNA. (B) Pearson's correlation between distinct covariates and module eigengenes in 134 cortex samples. (C) circRNAs are plotted according to their circRNA expression correlations; the circRNAs in violet and darkred modules are all plotted, but the circRNAs in the turquoise module are plotted with only kME ≥ 0.5 due to the sizable number of circRNAs. Node size is proportional to connectivity, and edge thickness is proportional to the absolute correlation between two circRNAs. (D,E) Module preservation defined in ASD samples only in non-ASD samples (CTL samples) (D, left), CTL samples only in ASD samples (D, right), TC samples only in FC samples (E, left), or FC samples only in TC samples (E, right).
To test the robustness of the three DE-modules, we then asked if the coexpression structure was similar (1) between ASD and non-ASD control samples and (2) between FC and TC. To this end, we performed module preservation tests (Langfelder et al. 2011) to examine preservation of modules defined in ASD samples only in control samples, in control samples only in ASD samples, in TC samples only in FC samples, and in FC samples only in TC samples, respectively. Indeed, we observed that the Zsummary scores, which were evaluated for each module to measure the preservation, of all three DE-modules were greater than or equal to two, indicating that the three modules were preserved across ASD and non-ASD control samples (Fig. 2D) and across FC and TC samples (Fig. 2E). Our results thus support the robustness of the three ASD-associated modules.
Identification of ASD-associated circRNA-miRNA-mRNA regulatory axes
We proceeded to identify potentially ASD-associated circRNA-miRNA-mRNA regulatory axes (Supplemental Fig. S4) based on the identified DE-circRNAs (and circRNAs from the identified DE-modules) and the DE-miRNAs previously identified by Wu et al. (2016). Of note, the DE-miRNAs (41 up-regulated and 17 down-regulated miRNAs) were derived from 95 human cortex samples (47 samples from 28 ASD cases and 48 samples from 28 non-ASD controls), of which 73 samples overlapped with the samples examined in this study. We first searched for the target sites of the DE-miRNAs in the circle sequences of the DE-circRNAs and circRNAs from the DE-modules and identified 808 circRNA-miRNA interactions (Supplemental Table S4). Next, we identified the potential targets of the 58 DE-miRNAs based on miRNA-target prediction tools or databases with collections of experimentally supported miRNA-target interactions (Methods). Thus, 36,512 miRNA-mRNA interactions were detected (Supplemental Table S4). On the basis of the integration of the identified circRNA-miRNA and miRNA-target interactions, we thereby determined 529,509 circRNA-miRNA-mRNA interactions according to the common miRNA target sites of the circRNAs and mRNAs (Supplemental Fig. S4). We also calculated the correlations between circRNA and miRNA expression, between miRNA and mRNA expression, and between circRNA and mRNA expression based on the same set of cortex samples (i.e., 73 samples). Since circRNAs often act as a miRNA sponge and the function can be enhanced through the long half-lives of circRNAs (Hansen et al. 2013; Memczak et al. 2013), this study focused on the circRNA-miRNA-mRNA interactions that simultaneously satisfied the following rules (Fig. 3A): (1) The circRNA-miRNA axes should be up-regulated circRNAs versus down-regulated miRNAs or down-regulated circRNAs versus up-regulated miRNAs (Supplemental Fig. S5); (2) both the circRNA-miRNA and miRNA-mRNA interactions should show a significantly negative correlation of expression profile between circRNAs and the corresponding predicted regulated miRNAs and between miRNAs and target mRNAs, respectively; (3) the circRNA expression should be positively correlated with the corresponding mRNA expression; and (4) the Fisher's combined P-values (Fisher 1932) of the aforementioned three independent Spearman's correlation tests should be less than 0.05. These criteria led to a total of 8170 (including 356 up-regulated and 7814 down-regulated circRNA-involved interactions) ASD-associated circRNA-miRNA-mRNA axes, which involved 2302 target genes (Table 2; Supplemental Table S5).
Figure 3.
Identification of ASD-associated circRNA-miRNA-mRNA regulatory interactions based on the 73 cortex samples overlapped with the samples examined in a previous miRNA study (Wu et al. 2016). (A) Schematic diagram representing the ASD-associated circRNA-miRNA-mRNA axes that satisfied our criteria (see the text). “−” represents the negative correlation between the expression of circRNAs and miRNAs and between the expression of miRNAs and mRNAs. “+” represents the positive correlation between the expression of circRNAs and mRNAs. The correlations are performed by Spearman's correlation tests. (B,C) Enrichment analyses of phenotype ontology (B) and 14 groups of gene list (C) among the target genes of the identified ASD-associated circRNA-miRNA-mRNA axes. The red dashed lines represent FDR Bonferroni-corrected P-values for enrichments with FDR < 0.05. For C, the P-values of gene enrichment analyses are determined using two-tailed Fisher's exact test (left) and empirical gene enrichment analysis (right), respectively. Top SFARI genes represent the genes with top ASD genetic risk factors from the SFARI database (score = 1–3 and syndromic). For the left panel, the enrichment odds ratios with FDR < 0.05 are provided in parentheses. For the right panel, the arrow represents FDR Bonferroni-corrected P-values for enrichments with FDR < 0.05. (D) The four categories of up-regulated (top) and down-regulated (bottom) circRNA-involved ASD-associated circRNA-miRNA-mRNA interactions. The Venn diagrams represent the overlap between the four categories of interactions, on which the numbers represent the numbers of target genes and the numbers of the corresponding circRNA-miRNA-mRNA interactions are shown in parentheses. A summary table representing the total number of interactions for each category is also provided (bottom right). (E) The 356 up-regulated circRNA-involved circRNA-miRNA-mRNA interactions plotted by the Cytoscape package.
Table 2.
ASD-associated circRNA-miRNA-mRNA regulatory axes identified in this study

To explore the relationship between ASD and the 2302 target genes of these ASD-associated circRNA-miRNA-mRNA interactions, we first assessed shared phenotypes among the target genes by enrichment for Human Phenotype Ontology terms (Chen et al. 2009). We found that the target genes were significantly enriched in the phenotype ontology terms of aplasia/hypoplasia involving the central nervous system (CNS) and abnormality of forebrain morphology, the cerebrum, the cerebral subcortex, and skull size (Fig. 3B), reflecting the brain morphometry differences between ASD patients and healthy individuals (van Rooij et al. 2018). Regarding the 2302 target genes, we further examined enrichment for ASD risk genes from Simons Foundation Autism Research Institutive (SFARI) AutDB (Abrahams et al. 2013) and AutismKB (Xu et al. 2012) databases (i.e., SFARI and AutismKB genes), which have been previously implicated in ASD through genetic syndromes, candidate gene studies, common variant association, and structural variation. We found that the target genes showed significant enrichment for both SFARI and AutismKB genes, particularly for top SFARI genes with score = 1–3 and syndromic (all P-values with Bonferroni correction [or FDR] < 0.05 by two-tailed Fisher's exact test and empirical gene enrichment analysis) (Methods), but not for genes implicated in monogenetic forms of schizophrenia, Alzheimer's disease, intellectual disability, and other brain disorders (Fig. 3C). These results suggest that targets of the identified circRNA-miRNA-mRNA axes are enriched for genes causally connected with autism, but less so for genes connected with other brain disorders. We also found that these target genes were significantly enriched (FDR < 0.05) for genes encoding inhibitory postsynaptic density (PSD) proteins, but not for those encoding excitatory PSD ones (Fig. 3C). This result is consistent with a previous observation that ASD-derived organoids show overproduction of inhibitory neurons (Mariani et al. 2015).
According to the target genes previously implicated in ASD or the experimental evidence of miRNA-mRNA binding, we further classified the identified circRNA-miRNA-mRNA interactions into four categories as follows (Fig. 3D):
Category 1: The target genes have been previously reported to be ASD risk genes (i.e., SFARI or AutismKB genes).
Category 2: The target genes were reported to be DE-genes in ASD (Parikshak et al. 2016) based on the samples overlapped with the samples examined in this study. The interactions should be either up-regulated circRNA-down-regulated miRNA-up-regulated mRNA or down-regulated circRNA-up-regulated miRNA-down-regulated mRNA interactions.
Category 3: The miRNA-mRNA interactions were supported by experimental data (see Methods).
Category 4: Other.
The ASD-affected circRNAs (DE-circRNAs and circRNAs in the DE-modules) of the interactions may provide an upstream regulation for the ASD-associated miRNA-mRNA axes and thereby potentially contribute to ASD susceptibility (Fig. 3E; Supplemental Fig. S6). Particularly, Category 1 interactions, which included 188 target ASD risk genes (Supplemental Table S5), may play an important regulatory role in ASD brain.
Experimental validation of the negative regulation of down-regulated miRNAs by up-regulated circRNAs in human neuronal cell lines
We then assessed the role of circRNA dysregulation in ASD-associated miRNA level alterations. Of the identified circRNA-miRNA-mRNA interactions (Fig. 3D), we selected a DE-circRNA (i.e., circARID1A, which was up-regulated in ASD) for the following experimental validations. circARID1A was predicted to harbor the largest number of target sites of one single down-regulated miRNA (seven miR-204-3p target sites) (Fig. 4A; Supplemental Fig. S5). The circRNA junction of circARID1A has not yet been experimentally confirmed previously. The role of circARID1A and its regulatory interaction with miR-204-3p is unknown and has not yet been studied experimentally. We first confirmed the non-co-linear (NCL) junction (or the back-spliced junction) of circARID1A. Because comparisons of different RTase products have been shown to effectively detect RT-based artificial NCL junctions (Houseley and Tollervey 2010; Wu et al. 2014; Yu et al. 2014; Chen et al. 2015a; Chuang et al. 2018), we used reverse transcription polymerase chain reaction (RT-PCR) using Avian Myeloblastosis Virus (AMV)- and Moloney Murine Leukemia Virus (MMLV)-derived RTase in parallel experiments in the examined cell lines/tissues, followed by Sanger sequencing of the RT-PCR amplicons to validate the NCL junction (Fig. 4A). Our result showed that the NCL junction of circARID1A was RTase-independent (Fig. 4A), supporting that the NCL junction was unlikely to be generated from an RT-based artifact. We further treated total RNA from the examined cell lines/tissues with RNase R and showed the RNase R-resistance of the NCL junction, supporting the existence of the circRNA event (Fig. 4B). Of note, we observed that circARID1A was commonly expressed across vertebrate (from primates to chicken) brains, indicating the evolutionary significance of circARID1A (Fig. 4C). We further examined the expression profiles of circARID1A and its corresponding colinear mRNA counterpart in various human tissues and found that these two isoforms showed different expression patterns (Fig. 4D). Although circARID1A was particularly enriched in the brain, its corresponding colinear mRNA counterpart was not (Fig. 4D). Regarding the relative expression of these two isoforms in the brain, circARID1A was significantly more abundant than its colinear form; in contrast, circARID1A was expressed at a relatively low level as compared to its colinear form in the non-brain tissues (Fig. 4E). We also found that circARID1A was widely expressed in 24 human brain regions (Supplemental Fig. S7). Together, these results suggested that circARID1A may play important biological roles in human brain.
Figure 4.
Experimental validation of the back-spliced junction of an up-regulated circRNA (circARID1A) and the corresponding circRNA-miRNA regulatory axis. (A) Validation of the NCL junction of circARID1A. (Top and middle) The schematic diagrams displaying seven predicted target sites of one single down-regulated miRNA (miR-204-3p) on the circle of circARID1A (top) and the designed divergent primers around the NCL junction of circARID1A (middle). (Bottom) Comparisons of two different RTase products (MMLV- and AMV-derived products) of circARID1A in TC/FC samples and four types of neuronal cell lines (hNPC [or ReN], NHA, SH-SY5Y, and U118), followed by Sanger sequencing the RT-PCR amplicons for the NCL event in the TC. (ReN) ReNcell VM. (B) Experimental validation of the circRNA (or back-spliced) junction of circARID1A. The figure shows the expression fold changes (as determined by qRT-PCR) for circARID1A, ARID1A mRNA, and GAPDH (negative control) in the indicated tissues/cell lines before and after RNase R treatment. (C) Experimental examination of the evolutionary conservation of circARID1A across the brains of vertebrate species from primates to chicken. Comparison of MMLV- and AMV-derived-RTase products (left) and the corresponding sequence chromatograms (right) for the circARID1A event in the brains of the indicated six species are shown. (D) Comparison of the expression profiles of circARID1A and its corresponding colinear mRNA counterpart in 10 normal human tissues. The expression levels of brain are used to normalize the relative expression values of the other tissues. (E) The relative expression of circARID1A and its corresponding colinear mRNA counterpart in 10 normal human tissues. (F) qRT-PCR analysis of the subcellular fractionation location for circARID1A and ARID1A mRNA. GAPDH and RNU6-1 snRNA are examined as a cytosol marker and a nucleus marker, respectively. The expression levels in nucleus are used to normalize the relative expression values in cytoplasm. (G,H) qRT-PCR analyses of the correlations between the expression of circARID1A and miR-204-3p after circARID1A knockdown (G) or overexpression (H) in various human neuronal cell lines. The top panels of G and H represent that circARID1A knockdown (G) or overexpression (H) did not significantly affect the ARID1A mRNA expression. (I) Luciferase reporter assay for the luciferase activity of GLuc-circARID1A in ReN, NHA, SH-SY5Y, and U118 cells transfected with miR-204-3p mimic and a scramble mimic (the negative control) to validate the binding between circARID1A and miR-204-3p. The entire circle sequence of circARID1A was cloned into the downstream region of the GLuc gene (i.e., GLuc-circARID1A; top). The luciferase activity of GLuc was normalized with secreted alkaline phosphatase (SEAP). (J) qRT-PCR analysis of the expression level of circARID1A in ReN and NHA cells after transfection with scramble and miR-204-3p mimics: (NC) negative control; (KD) knockdown; (OE) overexpression. All the qRT-PCR data are the means ±SD of three experiments. P-values are determined using two-tailed t-test. Significance: (*) P-value < 0.05; (***) P-value < 0.001; (NS) not significant.
To test if circARID1A regulates miR-204-3p activities, we first confirmed that circARID1A was indeed predominantly expressed in the cytoplasm in both NHAs and hNPCs (e.g., ReNcell VM [ReN]) (Fig. 4F). We then examined the correlation between expression of circARID1A and miR-204-3p in different types of neuronal cell lines. To this end, we disrupted circARID1A expression and overexpressed circARID1A, respectively. We found that circARID1A knockdown (Fig. 4G, top) and overexpression (Fig. 4H, top) did not significantly affect the expression of its corresponding colinear mRNA counterpart (ARID1A). However, miR-204-3p expression indeed significantly increased and decreased after circARID1A knockdown (Fig. 4G, bottom) and overexpression (Fig. 4H, bottom), respectively. To further examine if miR-204-3p can bind to circARID1A, we first performed luciferase reporter assay by cotransfecting miR-204-3p mimic or a scramble mimic with the luciferase reporters into NHAs or hNPCs. We showed that miR-204-3p significantly reduced the luciferase reporter activities as compared with the scrambled control (Fig. 4I). We also mutated miR-204-3p binding sites from the luciferase reporter and found that transfection of the miRNA had no significant effect on the luciferase activity of the luciferase-circARID1A mutated reporter as compared with the control vector (Supplemental Fig. S8). These experiments confirmed the binding between circARID1A and miR-204-3p. In addition, we showed that miR-204-3p overexpression did not significantly affect circARID1A expression (Fig. 4J). These results thus confirmed the regulatory role of circARID1A as a miR-204-3p sponge.
Regulation of ASD risk genes via the identified circRNA-miRNA axis
We have shown that circARID1A can function as a sponge to neutralize the activity of miR-204-3p (Fig. 4G–J). Of the identified circRNA-miRNA-mRNA regulatory interactions, we found 171 target genes in the circARID1A-miR-204-3p-involved interactions. The 171 genes included some genes previously implicated in ASD such as ASD risk genes (SFARI/AutismKB genes) or the genes regulated by RNA binding proteins (i.e., FMR1, ELAVL1 [or the so-called HuR], and RBFOX1) (Fig. 5A; Supplemental Table S6; Supplemental Fig. S9; Darnell et al. 2011; Mukherjee et al. 2011; Weyn-Vanhentenryck et al. 2014). We next experimentally tested the predicted up-regulation and down-regulation of target genes by circARID1A and miR-204-3p in ASD via in vitro perturbation of circARID1A or miR-204-3p in hNPCs. To this end, we performed circARID1A knockdown, circARID1A overexpressed, and miR-204-3p overexpression in hNPCs, respectively, and examined the fold changes of the corresponding target gene expression by microarray analysis (Supplemental Table S6). As expected, circARID1A overexpression significantly up-regulated the target genes as compared to those with circARID1A knockdown and those with miR-204-3p overexpression (both P < 0.05 by one-tailed paired t-test), whereas there was no significant difference between the mRNA fold changes after circARID1A knockdown and those after miR-204-3p overexpression (P = 0.19 by two-tailed paired t-test) (Fig. 5B). Regarding the mRNA fold changes of the targets, the Pearson correlation tests also showed (1) a negative correlation between the changes after circARID1A knockdown and those after miR-204-3p overexpression; (2) a negative correlation between the changes after circARID1A overexpression and those after miR-204-3p overexpression; and (3) a positive correlation between the changes after circARID1A knockdown and those after miR-204-3p overexpression (Fig. 5C). These results were consistent with our bioinformatics predictions.
Figure 5.
Experimental validation of the circARID1A regulatory role of miR-204-3p sponges and the corresponding circRNA-miRNA-mRNA regulatory interactions. (A) Schematic diagram representing the circARID1A-miR-204-3p regulatory axis and its corresponding targets (171 genes, including 12 ASD risk genes, two FMR1 targets, 102 ELAVL1 targets, and five RBFOX1 targets). (B–D) Microarray analysis of the target mRNA log2(fold change) in response to circARID1A knockdown, circARID1A overexpression, and miR-204-3p overexpression, respectively. (B) Distribution of the target mRNA log2(fold change) in response to circARID1A knockdown, circARID1A overexpression, and miR-204-3p overexpression. P-values are determined using one-tailed (circARID1A knockdown vs. circARID1A overexpression and circARID1A overexpression vs. miR-204-3p overexpression) or two-tailed (circARID1A knockdown vs. miR-204-3p overexpression) t-test. Significance: (*) P-value < 0.05; (**) P-value < 0.01; (NS) not significant. (C) The correlations between mRNA fold changes after circARID1A knockdown, circARID1A overexpression, and miR-204-3p overexpression. The Pearson correlation coefficient (R) and P-value are shown. (D) Heat map of the 12 ASD risk mRNA log2(fold change) in response to circARID1A knockdown, circARID1A overexpression, and miR-204-3p overexpression, respectively. The gene symbols highlighted in blue represent that the genes are also ELAVL1 targets. (E) qRT-PCR analyses of ASD risk gene expression in NHAs (top) and hNPCs (ReN cells) (bottom) after circARID1A knockdown, miR-204-3p mimics, and miR-204-3p mimics with circARID1A expression vector (p-circARID1A), respectively. (NC) Negative control. P-values are determined using two-tailed t-test. Significance: (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001. (F) Imaging of immunostained ReN cells after 2 wk of differentiation. Immunostaining shows undifferentiated (top) and differentiated (bottom) ReN cells with the cell proliferation marker MKI67 (left), the neuronal marker TUBB3 (middle), and the mature glial cell marker GFAP (right). Nuclei are stained with DAPI (blue). (Scale bar) 100 μm; (GFAP) glial fibrillary acidic protein. (G) Relative expression of circARID1A and two ASD risk genes (NLGN1 and STAG1) during hNPC differentiation. The Pearson correlation coefficients (R) between the expression of circARID1A and the two ASD risk genes and P-values are shown in parentheses. All the qRT-PCR data are the means ±SD of three experiments.
To further test whether the circARID1A-miR-204-3p axis can regulate genes implicated in ASD, we selected the 12 ASD risk genes and experimentally examined the regulatory interactions between the axis and the ASD risk genes in NHAs and hNPCs, respectively. Notably, of the 12 ASD risk genes, eight genes were also ELAVL1 targets (Fig. 5D; Supplemental Table S6). In NHAs, four SFARI genes (NLGN1, STAG1, HSD11B1, and VIP) were significantly down-regulated by the knockdown of circARID1A and the overexpression of miR-204-3p, respectively (Fig. 5E). The miR-204-3p-mediated repression of the four SFARI genes was rescued by ectopic expression of circARID1A. Similarly, in hNPCs, we found that three SFARI genes (NLGN1, STAG1, and UBA6) were significantly down-regulated by circARID1A knockdown and miR-204-3p mimics, and the miR-204-3p-mediated repression of the three genes was rescued by ectopic expression of circARID1A (Fig. 5E). Furthermore, we differentiated hNPCs into different neural cell types (Fig. 5F) and found that circARID1A expression was significantly positively correlated with the expression of NLGN1 and STAG1 during hNPC differentiation (Fig. 5G). Of note, both NLGN1 and STAG1 were also ELAVL1 targets (Fig. 5D). Because ELAVL1 targets were shown to be associated with neurodevelopmental defects (Kraushar et al. 2014), alteration of expression of ELAVL1 targets may contribute to the developing neocortex and then in autism pathogenesis (Popovitchenko et al. 2016). Taken together, these results revealed that circARID1A could regulate genes implicated in ASD through directly sponging miR-204-3p.
Discussion
On the basis of a large brain sample size of both small RNA-seq data and RNA-seq data from total RNAs (rRNA-depleted RNAs without poly(A)-selection) of ASD cases and controls, we conducted the first study, to the best of our knowledge, for systematically investigating circRNA dysregulation in ASD and the corresponding ASD-associated circRNA-miRNA-mRNA regulatory axes. Our genome-wide integrative analysis thus provided new insights into the role of circRNAs in ASD pathophysiology. We identified 60 DE-circRNAs and three DE-modules of circRNAs in ASD cortex, and showed a shared circRNA dysregulation signature among the majority of ASD samples. Regarding the identified ASD-associated circRNAs and the previously identified DE-miRNAs (Wu et al. 2016) derived from the same cortex samples used in this study, we identified the ASD-associated circRNA-miRNA regulatory axes and thus constructed 8170 ASD-associated circRNA-miRNA-mRNA interactions. For the target genes (i.e., 2302 genes) of the 8170 interactions, we observed significant enrichment for ASD risk genes (the top ASD risk genes particularly), but not for genes implicated in monogenetic forms of other brain disorders (such as epilepsy, which is often a comorbidity of ASD) (Fig. 3C). According to a most recently released data set of high-confidence ASD genetic risk genes (102 genes) (Satterstrom et al. 2020; Methods), the targets also showed significant enrichment for the high-confidence ASD genetic risk genes (P < 10−4 by two-tailed Fisher's exact test) (Supplemental Fig. S10). Moreover, we observed that target genes of the ASD-associated circRNA-miRNA-mRNA axes were significantly enriched in genes encoding inhibitory PSD proteins, but not in those encoding excitatory PSD ones (Fig. 3C). This result reflects the previous reports that there is an excitatory–inhibitory neuron imbalance in ASD (Casanova et al. 2003; Rubenstein 2010) and that inhibitory neurons are overproduced in ASD patient-derived organoids (Mariani et al. 2015). These results suggest that the identified ASD-associated circRNA-miRNA axes may serve as an alternative pathway for gene-disrupting mutations to perturb key transcript levels and thereby contribute to ASD susceptibility. Our experimental validation showed the negative correlation between the expression changes of ASD-affected circRNAs and miRNAs (Fig. 4G,H) and the regulation of the target ASD risk genes by the corresponding circRNA-miRNA axis (Fig. 5E). Our results thus suggest that circRNAs can function as efficient miRNA sponges for upstream regulation of the corresponding ASD risk genes.
Regarding the overall circRNA expression profiles, our PCA result represented that two types of cortex samples (FC and TC) were clustered together but cortex and CV samples were grouped into separate clusters (Fig. 1D). This result is similar to a new separate study for circRNA expression analysis (Gokool et al. 2020) and some previous observations for mRNA (Oldham et al. 2008; Voineagu et al. 2011) and miRNA (Wu et al. 2016) expression profiles. This also reflects the phenomenon that transcriptomic profiles are commonly clustered according to broad brain region (Oldham et al. 2008). Meanwhile, like the pattern observed by Gokool et al. (2020), our result (Supplemental Fig. S11) also revealed that ASD and non-ASD samples did not show distinct clustering based on the overall circRNA expression profiles of all circRNAs identified from the cortex samples. However, on the basis of the DE-circRNAs identified in this study, the two types of samples (ASD and non-ASD samples) can be grouped into two separate clusters by both PCA (Fig. 1H) and hierarchical clustering (Fig. 1I) analyses. We showed that the fold changes for the DE-circRNAs were concordant between the FC and TC (Fig. 1F) and were not biased by a small number of samples with removal of Chromosome 15q11-13 duplication syndrome, low RIN, or high PMI (Fig. 1G), supporting the robustness of the identified DE-circRNAs. We thus successfully provided a shared circRNA dysregulation signature among the majority of ASD samples.
Of the 8170 identified ASD-associated circRNA-miRNA-mRNA interactions, the circRNA-miRNA axes should be either up-regulated circRNA–down-regulated miRNA axes or down-regulated circRNA–up-regulated miRNA axes (Fig. 3A). To benefit the further studies for the identified circRNA-miRNA-mRNA regulatory interactions in ASD susceptibility, we divided these 8170 circRNA-miRNA-mRNA interactions into four categories according to the target genes previously implicated in ASD or the experimental evidence of miRNA-mRNA binding (Fig. 3D). Particularly, Category 1 interactions (780 interactions), in which the target genes have been previously implicated in ASD, provided a useful set of upstream regulations (circRNA-miRNA axes) for the ASD risk genes. Because ASD risk genes, which have been implicated through ASD rare or de novo variations, are predicted to disrupt protein function, down-regulated circRNAs may result in miRNA up-regulation in ASD and thereby contribute to ASD risk genes. Conversely, up-regulated circRNAs, which may act as a role of miRNA sponges, may result in miRNA down-regulation in ASD. A previous study (Wu et al. 2016) suggested that down-regulated miRNAs may play a compensatory or adaptive role for causing up-regulation of ASD risk genes. In addition to the ASD risk gene-involved interactions (i.e., Category 1), we emphasized that all the identified circRNA-miRNA-mRNA regulatory axes were derived from aberrant circRNA and miRNA expression in ASD and the correlations between circRNA, miRNA, and mRNA expression through the same cortex samples. The target genes of Categories 2–4 interactions, which were identified to be regulated by upstream ASD-affected circRNA-miRNA axes, may be valuable candidates for further studies in idiopathic ASD.
In this study, we not only provided a systems-level view of landscape of the circRNA expression in ASD cortex samples and the corresponding ASD-associated circRNA-miRNA-mRNA axes, but also experimentally characterized the targets of an up-regulated circRNA–down-regulated mRNA axis (i.e., the circARID1A-miR-204-3p axis) in NHAs and hNPCs. We confirmed that the expression of five SFARI genes (NLGN1, STAG1, HSD11B1, VIP, and UBA6) were regulated by circARID1A via sponging miR-204-3p in NHAs or hNPCs (Fig. 5E). The expression of both NLGN1 and STAG1 showed a significantly positive correlation with the circARID1A expression during hNPC differentiation (Fig. 5G). Of note, NLGN1 has been reported to play an important role in a variety of activity-dependent responses (Südhof 2017) and memory formation (Blundell et al. 2010; Dahlhaus et al. 2010; Katzman and Alberini 2018). Knockout of Nlgn1 in mice could cause increased repetitive behavior (Blundell et al. 2010). Loss or overexpression of Nlgn1 could impair spatial memory in transgenic mouse models; and alteration of NLGN1 expression in specific excitatory and inhibitory neuronal subpopulations can affect the dynamic processes of memory consolidation and strengthening (Katzman and Alberini 2018). Because excitatory–inhibitory neuron imbalances often accompany neuropsychiatric disorders, the dysregulation of NLGN1 may underlie the disorders. Therefore, the identified circARID1A-miR-204-3p axis, which regulates NLGN1 expression, may provide a useful circuitry/molecular mechanism of excitation and inhibition underlying long-term memory consolidation and strengthening for further developing potential therapeutic strategies to address these neuropsychiatric disorders, including ASD. In addition, Vip and Uba6 have been shown to play an essential regulatory role during rodent embryonic development (Hill et al. 2007; Lee et al. 2013). Vip is known as a regulator of embryogenesis of neural tube closure; interference with VIP can result in permanent effects on adult social behavior (Hill et al. 2007). It was shown that Uba6 brain-specific knockout mice showed social impairment and reduced vocalizations, representing a valid ASD mouse model (Lee et al. 2015). As a regulator of multiple ASD-associated genes, the circARID1A-miR-204-3p axis may be useful circuitry/molecular mechanism for further developing potential therapeutic strategies to address ASD.
It is noteworthy that circARID1A was validated to be widely expressed in the brain of multiple vertebrate species from human to chicken (Fig. 4C), suggesting the evolutionary importance of this circRNA across vertebrate species. This observation also implies a possibility that future studies may examine whether circARID1A serves as an early pathogenic feature during neurodevelopmental processes by altering circARID1A expression in appropriate transgenic animal models such as mice. In addition, we observed circARID1A was widely expressed in multiple brain regions (Supplemental Fig. S7). This raises an interesting question of whether circARID1A dysregulation could occur in multiple cell types or cell composition in ASD brains. Two recent studies have comprehensively investigated gene dysregulation in ASD (Velmeshev et al. 2019) and Alzheimer's disease (AD) (Mathys et al. 2019) in a cell-type-specific manner by single-nucleus RNA sequencing of cortical tissue from patients with ASD or AD, respectively. It is worthwhile investigating circRNAs in a cell-type-specific manner if single-cell sequencing data from non-poly(A)-selected RNA samples with a higher depth of single-cell sequencing reads are available. For example, only ∼70,000 reads per nucleus were gained in Velmeshev et al. (2019).
This study focused on the role of circRNAs in miRNA regulation because we can simultaneously obtain DE-circRNAs and DE-miRNAs in ASD and retrieve the corresponding expression profiles of circRNAs, miRNAs, and mRNAs from the same cortex samples. In addition to the regulatory role in miRNA activities, circRNAs may also function to interact with RNA-binding proteins, mediate alternative splicing, compete with their host genes for biogenesis during splicing, and translate into proteins (for review, see Chen et al. 2015a; Chen 2016; Salzman 2016; Wilusz 2018). It is worthwhile investigating the roles of circRNAs in ASD with respect to these issues in the future. Moreover, there is increasing evidence for the coexistence of positive and negative correlations between miRNA and mRNA expression, indicating that miRNAs can play not only an inhibitor of miRNA targets but also a complex bidirectional role in regulatory networks at the transcriptional level (Liu et al. 2018; O'Brien et al. 2018). On the basis of the DE-circRNAs and DE-miRNAs in ASD, the identified 529,509 circRNA-miRNA-mRNA interactions (Supplemental Fig. S4) derived from to the common miRNA target sites of the circRNAs and mRNAs, together with the correlations of expression profile between the corresponding circRNAs, miRNAs, and target mRNAs, may serve as a useful resource for further investigations of more complex circRNA-regulated miRNA-mRNA interactions in ASD.
Taken together, our study has provided a framework for assessing the functional involvement of circRNA in ASD and the corresponding ASD-associated circRNA-miRNA-mRNA regulatory axes. By integrating ASD candidate gene sets and circRNA, miRNA, and mRNA dysregulation data derived from the same ASD cortex samples, we have provided a rich set of ASD-associated circRNA candidates and the corresponding circRNA-miRNA-mRNA axes, particularly those involving ASD risk genes, for further investigation in ASD pathophysiology.
Methods
Identification of circRNAs in the cortex samples
The RNA-seq data of human brain were obtained by request from Synapse (https://www.synapse.org) under the accession number syn4587609, which included 236 postmortem samples of FC, TC, and CV from 48 ASD-affected individuals and 49 non-ASD-affected controls (Table 1). On the basis of human reference genome (GRCh38) and the Ensembl annotation (version 90), we detected circRNAs in these ASD and non-ASD control samples using NCLscan (version 1.6; https://github.com/TreesLab/NCLscan) (Chuang et al. 2016) with default parameters. Here, we only considered intragenic non-co-linear RNA events. Numbers of the detected circRNAs per million mapped RNA-seq reads in the ASD and non-ASD control samples from the three brain regions (FC, TC, and CV) are provided in Supplemental Table S1. The expression levels of circRNAs were calculated using the number of supporting circRNA junction reads per million uniquely mapped reads (RPM) (Venø et al. 2015). For accuracy, a sample was not considered in the following analysis if the number of the identified circRNAs of this sample was one standard deviation below the mean of the sample set. Therefore, 202 samples (73 FC, 61 TC, and 68 CV samples) were retained, including 53,427 NCLscan-identified circRNAs (Supplemental Table S2). Our analysis focused on the 134 cortex samples (73 FC and 61 TC samples) and the 36,624 identified circRNAs (Fig. 1B). To minimize potentially spurious events, we only considered the circRNAs that were detected in >50% of the 134 samples examined. Thus, a total of 1060 circRNAs (Supplemental Table S3) were used in the following analyses. The previously identified human and mouse circRNAs were downloaded from two publicly accessible data sets: circBase (www.circbase.org) (Glažar et al. 2014) and CIRCpedia v2 (https://www.picb.ac.cn/rnomics/circpedia/) (Dong et al. 2018). Because the genomic coordinates of human circRNAs in circBase were defined on the hg19 assembly, we used the liftOver tool (Hinrichs et al. 2006) to obtain the genomic coordinates of circRNAs on the GRCh38 assembly. Regarding the mouse circRNAs collected in circBase (mm9) and CIRCpedia v2 (mm10), we also performed the liftOver tool to obtain the corresponding orthologous mouse coordinates of circRNAs on the GRCh38 assembly.
Identification of differentially expressed circRNAs in ASD
We then used the nlme program in the R package (version 3.4.2) (R Core Team 2017) to perform a LME model and detect DE-circRNAs between ASD and control samples (diagnosis) with controlling for potential confounding factors including sex, age, brain region (FC or TC), RIN, host gene expression, sequencing batch, and brain bank batch as follows:
Here, each individual sample was set as a random effect; the other factors were set as fixed effects. Significant results of the diagnosis factor were reported at P < 0.05. The expression levels of the host genes were measured by log2 normalized FPKM (which accounts for gene read counts, GC content, gene length, and library size) using the cqn program in the R package (Hansen et al. 2012). The read counts of the host genes were calculated by the STAR aligner (Dobin et al. 2013), followed by the RSEM tool (Li and Dewey 2011).
The TIN score across transcripts has been shown to be an accurate measurement of RNA integrity at the transcript level for samples from clinical tissues (Wang et al. 2016). We used the RSeQC software (v2.6.4) (Wang et al. 2012) to calculate the median TIN scores of the corresponding host genes for the 60 DE-circRNAs for each examined sample. According to the median TIN scores, we compared the RNA quality of the transcript sequences where the DE-circRNAs located for the cortex samples with RIN < 5 and RIN ≥ 5.
To estimate the needed sample size for detecting DE-circRNAs when considering multiple test correction, we first calculated the effect size (Cohen's d) of each circRNA using whereas beta was calculated by the LME model stated above, StdErr was the standard error of beta, and n was the number of all samples examined. After that, the sample size given an effect size of circRNAs was estimated using pwr.t.test function in R package pwr with the following parameters: d = maximal effect size of tested circRNAs (i.e., 0.289); sig.level = 0.05/number of circRNAs tested; power = 0.8, type = c(“two.sample”); and alternative = c(“two.sided”).
Weighted gene coexpression network analysis (WGCNA)
To build the signed compression network, we used the R package WGCNA (Zhang and Horvath 2005; Langfelder and Horvath 2008), a dynamic tree-cutting algorithm, to identify the modules of correlated genes. We followed the similar approach of Wu et al. (2016). Illustration of networks was plotted by the Cytoscape package (https://cytoscape.org/). The detailed procedures are described in Supplemental Methods.
Identification of ASD-associated circRNA-miRNA-mRNA regulatory axes
We identified potentially ASD-associated circRNA-miRNA-mRNA regulatory axes based on the identified DE-circRNAs (and circRNAs from the identified DE-modules) and the 58 DE-miRNAs previously identified by Wu et al. (2016). The DE-miRNAs (41 up-regulated and 17 down-regulated miRNAs) were derived from 95 human cortex samples (47 samples from 28 ASD cases and 48 samples from 28 non-ASD controls), of which 73 samples overlapped with the samples examined in this study. Of the 58 DE-miRNAs, 37 were well-annotated and 21 were newly identified by Wu et al. (2016). We identified ASD-associated circRNA-miRNA-mRNA regulatory axes using the following processes (Supplemental Fig. S4). Because RNA22 (Miranda et al. 2006) can predict highly accurate miRNA-mRNA binding sites in both protein-coding sequences (CDSs) and untranslated regions (UTRs) whether the targeting miRNAs are well-annotated or unknown (Loher and Rigoutsos 2012; Akhtar et al. 2016), we first searched for the target sites of the 58 miRNAs in the circle sequences of the identified DE-circRNA and the circRNAs defined in the DE-modules (the sequences may be derived from the CDSs or UTRs of their host genes) using RNA22 with default parameters (version 2.0; https://cm.jefferson.edu/rna22/). Thus, a total of 808 potential circRNA-miRNA interactions were identified (Supplemental Table S4). Next, we identified the target genes of the 58 DE-miRNAs (Supplemental Methods; Supplemental Fig. S4) and determined 36,512 miRNA-mRNA interactions (Supplemental Table S4). By integrating the 808 circRNA-miRNA interactions with the 36,512 miRNA-mRNA interactions, we therefore determined 529,509 circRNA-miRNA-mRNA interactions according to the common miRNA target sites of the circRNAs and mRNAs. We then calculated the correlations between circRNA and miRNA expression, between miRNA and mRNA expression, and between circRNA and mRNA expression based on the same set of cortex samples (i.e., 73 samples). Here, the expression level of the target gene was measured by log2 normalized FPKM, which accounted for gene read counts, GC content, gene length, and library size, using the cqn program in the R package (Hansen et al. 2012). The expression levels of the examined miRNAs were kindly provided by Prof. Daniel H. Geschwind and Ye E. Wu (Wu et al. 2016), which were measured by log2 normalized read counts for library size, GC content, batch effect, and other technical covariates (RIN, PMI, and batch bank). Only the circRNA-miRNA-mRNA interactions were considered if they simultaneously satisfied the following rules: (1) Both the circRNA-miRNA and miRNA-mRNA interactions should show a significantly negative correlation (one-tailed Spearman's P < 0.05) of expression profile between circRNAs and the corresponding predicted regulated miRNAs and between miRNAs and target mRNAs, respectively; (2) the circRNA expression should be positively correlated with the corresponding mRNA expression; and (3) the Fisher's combined P-values (Fisher 1932) of the aforementioned three independent Spearman's correlation tests should be less than 0.05. Finally, we identified 8170 ASD-associated circRNA-miRNA-mRNA regulatory axes (Fig. 3D; Supplemental Fig. S4) with 2302 target genes (Supplemental Table S5). Of note, the experimentally supported miRNA-mRNA binding events were extracted from Ingenuity Expert Findings in IPA (Krämer et al. 2014) or DIANA-TarBase (version 8) (Karagkouni et al. 2018).
Gene set enrichment analysis
The SFARI (Abrahams et al. 2013) gene list was downloaded from https://gene.sfari.org/ (December 2018). The high-confidence ASD genetic risk genes (102 genes) were downloaded from the study of Satterstrom et al. (2020), which were derived from an enhanced Bayesian analytic framework based on a large data set of whole-exome sequencing (35,584 ASD subjects). The epilepsy-related gene list was downloaded from the EpilepsyGene database (Ran et al. 2015) at http://www.wzgenomics.cn/EpilepsyGene/index.php (all epilepsy-related genes). The schizophrenia-related gene list was downloaded from the SZgene database (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). The genes associated with human height were downloaded from the study of Lango Allen et al. (2010). The gene lists of Autism_KB genes, iPSD, ePSD, and other brain disorder risk genes were downloaded from the study of Wang et al. (2018a). Phenotype ontology analysis (Fig. 3B) was performed using the ToppFun module of ToppGene Suite software at https://toppgene.cchmc.org/enrichment.jsp (version 2019-Oct-08 21:31) (Chen et al. 2009). False discovery rate (FDR) adjusted P-values were calculated using Bonferroni correction. For gene set enrichment analyses in the left panel of Figure 3C, we used the 20,070 protein-coding genes, which were expressed in the cortex (based on the RNA-seq data examined in this study), as the background set. For the 2302 targets of the identified circRNA-miRNA-mRNA interactions, 2299 genes were considered for the following analyses because three of the 2302 targets were not protein-coding genes. We took the analysis of SFARI gene enrichment as an example. We created a two-way contingency table with rows containing numbers of SFARI and non-SFARI genes and columns containing numbers of targets and nontargets of the identified circRNA-miRNA-mRNA interactions. The statistical significance and odds ratio were determined using a two-tailed Fisher's exact test with the fisher.test R function. Finally, P-values were FDR adjusted across 14 target groups for each gene list using Bonferroni correction. For empirical gene enrichment analysis in the right panel of Figure 3C, we asked whether the targets of the identified circRNA-miRNA-mRNA interaction (2299 genes) had a higher proportion (proportionobs) of the disease-related gene lists (e.g., SFARI genes) compared to a null distribution of the proportion observed in the 100,000 times of random sampling. For each time, the equivalent number of genes (2299 genes) were randomly selected from the background set (20,070 genes), and the proportions (proportioni) in response to 14 target groups for each gene list were calculated. The empirical P-value for each gene list was calculated as
Empirical P-values were then FDR adjusted across 14 target groups for each gene list using Bonferroni correction.
Microarray analysis
The microarray hybridization and the data collection were performed by the Affymetrix GeneChip System Service center in Genomics Research Center, Academia Sinica. Purity and RIN of total RNA were evaluated by a 2100 Bioanalyzer (Agilent Technologies). Total RNA was hybridized to Affymetrix Human Genome Plus 2.0 Array (Affymetrix). The microarray raw data were analyzed using Transcriptome Analysis Console (TAC 4.0) software (Affymetrix). Microarray results of the fold changes for the target genes of the circARID1A-miR-204-3p-involved interactions with the treatments of circARID1A knockdown/overexpression and miR-204-3p overexpression are provided in Supplemental Table S6. The FMR1, ELAVL1, and RBFOX1 target gene lists were downloaded from the study of Lee et al. (2019).
Cell culture
Normal human astrocytes (NHA) cell line was purchased from Gibco (N7805100) and cultured in human astrocytes growth medium (Cell Applications). Human neuroblastoma (SH-SY5Y) cell line was obtained from ATCC (CRL-2266) and cultured in DMEM/F12 medium (Gibco). Human glioblastoma (U118) cell line was purchased from ATCC (HTB-15) maintained in Dulbecco's Modified Eagle's Medium (Gibco). In addition, human neural progenitor cells (hNPCs; ReNcell VM) (Takahashi and Yamanaka 2006; Donato et al. 2007) were commercially purchased from Sigma-Aldrich (SCC008). The cells were grown on 20 μg/mL laminin (Merck)-coated culture plates containing ReNCell NSC maintenance medium (Merck) supplemented with 20 ng/mL of bFGF and EGF (Merck). All culture media were supplemented with 10% fetal bovine serum (FBS) and 1× antibiotic-antimycotic (Gibco) at 37°C with 5% CO2. Cells were passaged when the confluence reached 80% of the culture plate every 3 d. Briefly, cells were rinsed with PBS and then incubated in Accutase (Millipore) for 3–5 min until the cell detached. We used the culture medium to inhibit enzymatic reaction and centrifuged the suspension at 500g for 5 min. We then resuspended the cell pellet in fresh medium and counted the cell number.
Vector construction and design of oligonucleotide
For overexpression of circARID1A, we constructed the putative exon sequence of circARID1A (E2-E3-E4) into Circle3 vector (provided by Drs. S. K. Gupta and L. Yen) (Hsiao et al. 2017), which can circularize the transcript to produce circular RNA. The putative circARID1A sequence was amplified by PCR, and the PCR product was inserted into the Circle3 vector between MfeI and AgeI sites. Sanger sequencing verified recombinant plasmid construction. For ReNcell transfection, cells were infected with lentiviruses that were constructed by the National RNAi Core Facility in Academia Sinica (Taipei, Taiwan). The lentiviruses were carrying three different vectors: circARID1A-shRNA (pLKO_TRC005), circARID1A-cDNA (pLAS2w.Ppuro), and dual-luciferase with circARID1A cDNA (pLAS2.1w.PeGFP-I2-Puro). All constructs were verified by Sanger sequencing (Supplemental Table S7).
Cell transfection and lentiviral transduction
Cells were transfected with siRNA (MDBio) or miRNA mimics (TOOLS) at final concentration of 10 nM using Lipofectamine RNAiMAX reagent (Invitrogen). The sequences are listed in Supplemental Table S7. When transferring plasmids into the cells, the transfection was carried out using TransIT-LT1 Reagent (Mirus) according to the transfection manufacturer's instructions. For lentiviral transduction, the cells were infected for 24 h (MOI = 2) and then washed three times to remove the remaining virus. After transduction, cells were selected by culture medium containing 0.25 μg/mL puromycin for 4 d. The effects of knockdown or overexpression were examined by RT-qPCR. To efficiently knockdown circARID1A expression in different cell types, the siRNA and shRNA targeting the circARID1A back-spliced junction were designed. NHA, SH-SY5Y, and U118 were transfected with si-circARID1A; and hNPCs were infected with lentivirus carrying sh-circARID1A vector.
Total RNA isolation
Total RNA from cells was extracted using TRIzol reagent with PureLink RNA Mini Kit (Ambion) and PureLink DNase Set (Ambion). Total RNAs of normal human tissues were purchased from Invitrogen. Human brain cDNA array was purchased from Amsbio (HBRT501). Brain total RNA of monkey, mouse, and rat were purchased from BioChain. Brain total RNA of rabbit and chicken were purchased from Zyagen. The information of purchased RNA was provided in Supplemental Table S7.
RNase R treatment and subcellular fractionation
For RNase R treatment, total RNA was incubated with or without RNase R (Epicentre) for 45 min at 37°C to deplete linear and enrich circular RNAs. To validate the subcellular localization preference, we used the NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific) to separate nuclear and cytoplasmic RNAs. Total RNA was then extracted using TRIzol reagent according to the manufacturer's instructions. qRT-PCR analyses were performed to examine the relative expression of cytoplasmic and nuclear localization for each gene. GAPDH was used as a cytosol marker and RNU6-1 was used as a nucleus marker.
cDNA synthesis and quantitative real-time PCR analysis
For mRNA and circRNA quantitation, RNAs were reverse transcribed into cDNA using SuperScript III with random hexamer and oligo(dT)18 primers (Thermo Fisher Scientific). qRT-PCR was performed using Luminaris Color HiGreen High ROX qPCR Master Mix (Thermo Fisher Scientific). Quantity of gene expression was normalized to the expression level of GAPDH. For miRNA quantitation, cDNA synthesis is carried out by a miRCURY LNA RT Kit (Qiagen). qRT-PCR was performed by miRCURY LNA SYBR Green PCR Kit (Qiagen) with miRCURY LNA miRNA PCR assays (Qiagen) for each candidate miRNAs, and RNU6-1 small nuclear RNA (snRNA) served as an internal control. All reactions were performed three times per experiment and detected by StepOnePlus Real-Time PCR Systems (Thermo Fisher Scientific). All primers used in this study were listed in Supplemental Table S7.
Luciferase reporter assays
The luciferase reporter was constructed by subcloning the circARID1A sequence into the Secrete-Pair Dual Luminescence vector (GeneCopoeia). Cells were seeded in a 24-well plate and incubated for 24 h before transfection. Dual Luminescence vectors that contained circARID1A sequence were cotransfected into human neuronal cells with miR-204-3p mimic or a scramble mimic (the negative control) by TransIT-X2 transfection reagent (Mirus). After 72 h of transfection, cell culture medium was collected. The luciferase activity was measured by Secrete-Pair Dual Luminescence Assay kit (GeneCopoeia) according to the manufacturer's protocol. The luciferase activity of Gaussia luciferase (GLuc) was normalized with secreted alkaline phosphatase (SEAP). The fold change was measured by miR-204-3p mimic versus the negative control. To further examine the specificity of the binding between circARID1A and miR-204-3p, we selected four of the seven RNA22-identified miR-204-3p binding sites in the circle sequence of circARID1A (Fig. 4A) and then mutated these binding sites in the GLuc-circARID1A reporter vector using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies 210518). The four binding sites were selected because they were also identified by another miRNA-target prediction tool (i.e., MiRanda, version 3.3a) (Enright et al. 2003). The mutated GLuc-circARID1A reporter construct was verified by Sanger sequencing. The primers used for mutations of these miR-204-3p binding regions were listed in Supplemental Table S7.
Neuronal differentiation and immunostaining
ReNcell VM (Sigma-Aldrich SCC008) was incubated with ReNCell NSC maintenance medium (Millipore SCM005) without containing FGF2 and EGF growth factors for differentiation. Maintenance basal media was changed every 3 d for 2 wk. After 2 wk, we categorized the neuronal subtypes by immunofluorescence. Cells were plated on glass slides coated with laminin overnight. Cells were fixed with 4% formaldehyde for 25 min at 37°C, permeabilized with 0.05% Triton X-100 for 15 min, and incubated with 3% FBS for blocking for 1 h. Cells were then incubated overnight at 4°C with primary antibody against MKI67 (the cell proliferation marker; Abcam ab15580), βIII-tubulin (neuronal marker; Abcam ab18207), and GFAP (glial marker; Invitrogen 13-0300), respectively. Next day, cells were incubated with fluorescently labeled secondary antibodies (Supplemental Table S7) for 1.5 h at 37°C. Nuclei were stained with SlowFade Diamond Antifade Mountant with DAPI (Invitrogen S36968).
Data access
The microarray raw data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE145417. The codes used in this study were deposited in Supplemental Code and GitHub at https://github.com/TreesLab/circRNA_ASD. The 529,509 identified circRNA-miRNA-mRNA interactions and the related data are also publicly accessible at GitHub (https://github.com/TreesLab/circRNA_ASD). Of note, the coordinates used in this study are 1-based.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This work was supported by Genomics Research Center (GRC), Academia Sinica, Taiwan; and the Ministry of Science and Technology (MOST), Taiwan, under the contract MOST 107-2311-B-001-046 and MOST 108-2311-B-001-020-MY3 (both to T.-J.C.). S.K.G. was supported by Cancer Prevention Research Institute of Texas (CPRIT) training grant RP160283. L.Y. was supported by CPRIT High-Impact/High-Risk Research Awards (HIHRRA) RP160795. We thank the National RNAi Core Facility (Academia Sinica) for providing shRNA reagents and related services and Affymetrix Facility for microarray analysis (GRC, Academia Sinica), respectively. We especially thank Profs. Michael Hsiao, Ying-Chih Chang, and Jean Lu for providing experimental assistance, and Prof. Daniel H. Geschwind and Ye E. Wu for providing the related data of miRNAs examined.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.255463.119.
Freely available online through the Genome Research Open Access option.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. 2013. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4: 36 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MM, Micolucci L, Islam MS, Olivieri F, Procopio AD. 2016. Bioinformatic tools for microRNA dissection. Nucleic Acids Res 44: 24–44. 10.1093/nar/gkv1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ander BP, Barger N, Stamova B, Sharp FR, Schumann CM. 2015. Atypical miRNA expression in temporal cortex associated with dysregulation of immune, cell cycle, and other pathways in autism spectrum disorders. Mol Autism 6: 37 10.1186/s13229-015-0029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel A, Rosenzweig JP, Zisman PD, Melamed M, Gesundheit B. 2017. Variation in gene expression in autism spectrum disorders: an extensive review of transcriptomic studies. Front Neurosci 10: 601 10.3389/fnins.2016.00601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56: 55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Sudhof TC, Powell CM. 2010. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci 30: 2115–2129. 10.1523/JNEUROSCI.4517-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden D, Gomez J. 2003. Disruption in the inhibitory architecture of the cell minicolumn: implications for autism. Neuroscientist 9: 496–507. 10.1177/1073858403253552 [DOI] [PubMed] [Google Scholar]
- Chen LL. 2016. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17: 205–211. 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- Chen CY, Chuang TJ. 2019a. Comment on “A comprehensive overview and evaluation of circular RNA detection tools”. PLoS Comput Biol 15: e1006158 10.1371/journal.pcbi.1006158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chuang TJ. 2019b. NCLcomparator: systematically post-screening non-co-linear transcripts (circular, trans-spliced, or fusion RNAs) identified from various detectors. BMC Bioinformatics 20: 3 10.1186/s12859-018-2589-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311. 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Chen CY, Chuang TJ. 2015a. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 6: 563–579. 10.1002/wrna.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JA, Peñagarikano O, Belgard TG, Swarup V, Geschwind DH. 2015b. The emerging picture of autism spectrum disorder: genetics and pathology. Annu Rev Pathol 10: 111–144. 10.1146/annurev-pathol-012414-040405 [DOI] [PubMed] [Google Scholar]
- Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY. 2016. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res 44: e29 10.1093/nar/gkv1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TJ, Chen YJ, Chen CY, Mai TL, Wang YD, Yeh CS, Yang MY, Hsiao YT, Chang TH, Kuo TC, et al. 2018. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res 46: 3671–3691. 10.1093/nar/gky032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Dahlhaus R, Hines RM, Eadie BD, Kannangara TS, Hines DJ, Brown CE, Christie BR, El-Husseini A. 2010. Overexpression of the cell adhesion protein neuroligin-1 induces learning deficits and impairs synaptic plasticity by altering the ratio of excitation to inhibition in the hippocampus. Hippocampus 20: 305–322. 10.1002/hipo.20630 [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R, Miljan EA, Hines SJ, Aouabdi S, Pollock K, Patel S, Edwards FA, Sinden JD. 2007. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci 8: 36 10.1186/1471-2202-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma XK, Li GW, Yang L. 2018. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics 16: 226–233. 10.1016/j.gpb.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. 2003. MicroRNA targets in Drosophila. Genome Biol 5: R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1932. Statistical methods for research workers, 4th ed Oliver and Boyd, London. [Google Scholar]
- Floris G, Zhang L, Follesa P, Sun T. 2017. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol 54: 5156–5165. 10.1007/s12035-016-0055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH. 2011. Genetics of autism spectrum disorders. Trends Cogn Sci 15: 409–416. 10.1016/j.tics.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glažar P, Papavasileiou P, Rajewsky N. 2014. circBase: a database for circular RNAs. RNA 20: 1666–1670. 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokool A, Anwar F, Voineagu I. 2020. The landscape of circular RNA expression in the human brain. Biol Psychiatry 87: 294–304. 10.1016/j.biopsych.2019.07.029 [DOI] [PubMed] [Google Scholar]
- Guan J, Yang E, Yang J, Zeng Y, Ji G, Cai JJ. 2016. Exploiting aberrant mRNA expression in autism for gene discovery and diagnosis. Hum Genet 135: 797–811. 10.1007/s00439-016-1673-7 [DOI] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z. 2012. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13: 204–216. 10.1093/biostatistics/kxr054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hicks SD, Middleton FA. 2016. A comparative review of microRNA expression patterns in autism spectrum disorder. Front Psychiatry 7: 176 10.3389/fpsyt.2016.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JM, Cuasay K, Abebe DT. 2007. Vasoactive intestinal peptide antagonist treatment during mouse embryogenesis impairs social behavior and cognitive function of adult male offspring. Exp Neurol 206: 101–113. 10.1016/j.expneurol.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2010. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One 5: e12271 10.1371/journal.pone.0012271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. 2017. Non-coding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res 77: 2339–2350. 10.1158/0008-5472.CAN-16-1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515: 216–221. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O'Hanlon D, et al. 2014. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159: 1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, et al. 2018. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Res 46: D239–D245. 10.1093/nar/gkx1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman A, Alberini CM. 2018. NLGN1 and NLGN2 in the prefrontal cortex: their role in memory consolidation and strengthening. Curr Opin Neurobiol 48: 122–130. 10.1016/j.conb.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Leventhal BL. 2015. Genetic epidemiology and insights into interactive genetic and environmental effects in autism spectrum disorders. Biol Psychiatry 77: 66–74. 10.1016/j.biopsych.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J Jr, Tugendreich S. 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30: 523–530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushar ML, Thompson K, Wijeratne HR, Viljetic B, Sakers K, Marson JW, Kontoyiannis DL, Buyske S, Hart RP, Rasin MR. 2014. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc Natl Acad Sci 111: E3815–E3824. 10.1073/pnas.1408305111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. 2011. Is my network module preserved and reproducible? PLoS Comput Biol 7: e1001057 10.1371/journal.pcbi.1001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. 10.1038/nature09410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Dodart JC, Aron L, Finley LW, Bronson RT, Haigis MC, Yankner BA, Harper JW. 2013. Altered social behavior and neuronal development in mice lacking the Uba6-Use1 ubiquitin transfer system. Mol Cell 50: 172–184. 10.1016/j.molcel.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kwak M, Lee PC. 2015. Impairment of social behavior and communication in mice lacking the Uba6-dependent ubiquitin activation system. Behav Brain Res 281: 78–85. 10.1016/j.bbr.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Lee C, Kang EY, Gandal MJ, Eskin E, Geschwind DH. 2019. Profiling allele-specific gene expression in brains from individuals with autism spectrum disorder reveals preferential minor allele usage. Nat Neurosci 22: 1521–1532. 10.1038/s41593-019-0461-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin H, Sun Z, Kong G, Yan X, Wang Y, Wang X, Wen Y, Liu X, Zheng H, et al. 2018. High-throughput data of circular RNA profiles in human temporal cortex tissue reveals novel insights into temporal lobe epilepsy. Cell Physiol Biochem 45: 677–691. 10.1159/000487161 [DOI] [PubMed] [Google Scholar]
- Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. 2018. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer 17: 64 10.1186/s12943-018-0765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loher P, Rigoutsos I. 2012. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics 28: 3322–3323. 10.1093/bioinformatics/bts615 [DOI] [PubMed] [Google Scholar]
- Luo W, Zhang C, Jiang YH, Brouwer CR. 2018. Systematic reconstruction of autism biology from massive genetic mutation profiles. Sci Adv 4: e1701799 10.1126/sciadv.1701799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al. 2015. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162: 375–390. 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. 2019. Single-cell transcriptomic analysis of Alzheimer's disease. Nature 570: 332–337. 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. 2006. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217. 10.1016/j.cell.2006.07.031 [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43: 327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J, Hayder H, Zayed Y, Peng C. 2018. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9: 402 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. 2008. Functional organization of the transcriptome in human brain. Nat Neurosci 11: 1271–1282. 10.1038/nn.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155: 1008–1021. 10.1016/j.cell.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LT, Huang J, et al. 2016. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540: 423–427. 10.1038/nature20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovitchenko T, Thompson K, Viljetic B, Jiao X, Kontonyiannis DL, Kiledjian M, Hart RP, Rasin MR. 2016. The RNA binding protein HuR determines the differential translation of autism-associated FoxP subfamily members in the developing neocortex. Sci Rep 6: 28998 10.1038/srep28998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran X, Li J, Shao Q, Chen H, Lin Z, Sun ZS, Wu J. 2015. EpilepsyGene: a genetic resource for genes and mutations related to epilepsy. Nucleic Acids Res 43: D893–D899. 10.1093/nar/gku943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org/. [Google Scholar]
- Rubenstein JL. 2010. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol 23: 118–123. 10.1097/WCO.0b013e328336eb13 [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58: 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Salzman J. 2016. Circular RNA expression: its potential regulation and function. Trends Genet 32: 309–316. 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9: e1003777 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, Peng M, Collins R, Grove J, Klei L, et al. 2020. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180: 568–584.e23. 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel DE, Grønborg TK, Parner ET. 2014. The genetic and environmental contributions to autism: looking beyond twins. JAMA 311: 1738–1739. 10.1001/jama.2014.3554 [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Cheung I, Whittle C, Wang J, Virgil D, Lin CL, Guo Y, Lessard A, Akbarian S, Weng Z. 2012. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry 69: 314–324. 10.1001/archgenpsychiatry.2011.151 [DOI] [PubMed] [Google Scholar]
- Südhof TC. 2017. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171: 745–769. 10.1016/j.cell.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Poschmann J, Cruz-Herrera Del Rosario R, Parikshak NN, Hajan HS, Kumar V, Ramasamy R, Belgard TG, Elanggovan B, Wong CC, et al. 2016. Histone acetylome-wide association study of autism spectrum disorder. Cell 167: 1385–1397.e11. 10.1016/j.cell.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. 2015. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16: 126 10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- van Rooij D, Anagnostou E, Arango C, Auzias G, Behrmann M, Busatto GF, Calderoni S, Daly E, Deruelle C, Di Martino A, et al. 2018. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD Working Group. Am J Psychiatry 175: 359–369. 10.1176/appi.ajp.2017.17010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, Kriegstein AR. 2019. Single-cell genomics identifies cell type–specific molecular changes in autism. Science 364: 685–689. 10.1126/science.aav8130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. 2015. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16: 245 10.1186/s13059-015-0801-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel Ciernia A, LaSalle J. 2016. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat Rev Neurosci 17: 411–423. 10.1038/nrn.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474: 380–384. 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Elsabbagh M, Bolton P, Singh I. 2011. In search of biomarkers for autism: scientific, social and ethical challenges. Nat Rev Neurosci 12: 603–612. 10.1038/nrn3113 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28: 2184–2185. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao X, Ju W, Flory M, Zhong J, Jiang S, Wang P, Dong X, Tao X, Chen Q, et al. 2015. Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl Psychiatry 5: e660 10.1038/tp.2015.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Nie J, Sicotte H, Li Y, Eckel-Passow JE, Dasari S, Vedell PT, Barman P, Wang L, Weinshiboum R, et al. 2016. Measure transcript integrity using RNA-seq data. BMC Bioinformatics 17: 58 10.1186/s12859-016-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao D, Lachman HM, Zheng D. 2018a. Enriched expression of genes associated with autism spectrum disorders in human inhibitory neurons. Transl Psychiatry 8: 13 10.1038/s41398-017-0058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Qu L, Chen X, Zhao YH, Luo Q. 2018b. Progress in understanding the relationship between circular RNAs and neurological disorders. J Mol Neurosci 65: 546–556. 10.1007/s12031-018-1125-z [DOI] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6: 1139–1152. 10.1016/j.celrep.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. 2013. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155: 997–1007. 10.1016/j.cell.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. 2018. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA 9: e1478 10.1002/wrna.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Yu CY, Chuang CY, Hsiao M, Kao CF, Kuo HC, Chuang TJ. 2014. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res 24: 25–36. 10.1101/gr.159483.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Parikshak NN, Belgard TG, Geschwind DH. 2016. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat Neurosci 19: 1463–1476. 10.1038/nn.4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LM, Li JR, Huang Y, Zhao M, Tang X, Wei L. 2012. AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic Acids Res 40: D1016–D1022. 10.1093/nar/gkr1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18: 603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Liu HJ, Hung LY, Kuo HC, Chuang TJ. 2014. Is an observed non-co-linear RNA product spliced in trans, in cis or just in vitro? Nucleic Acids Res 42: 9410–9423. 10.1093/nar/gku643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Lin W, Guo M, Zou Q. 2017. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 13: e1005420 10.1371/journal.pcbi.1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article17 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. 2016. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7: 11215 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.