Abstract
The somatic macronucleus (MAC) and germline micronucleus (MIC) of Tetrahymena thermophila differ in chromosome numbers, sizes, functions, transcriptional activities, and cohesin complex location. However, the higher-order chromatin organization in T. thermophila is still largely unknown. Here, we explored the higher-order chromatin organization in the two distinct nuclei of T. thermophila using the Hi-C and HiChIP methods. We found that the meiotic crescent MIC has a specific chromosome interaction pattern, with all the telomeres or centromeres on the five MIC chromosomes clustering together, respectively, which is also helpful to identify the midpoints of centromeres in the MIC. We revealed that the MAC chromosomes lack A/B compartments, topologically associating domains (TADs), and chromatin loops. The MIC chromosomes have TAD-like structures but not A/B compartments and chromatin loops. The boundaries of the TAD-like structures in the MIC are highly consistent with the chromatin breakage sequence (CBS) sites, suggesting that each TAD-like structure of the MIC chromosomes develops into one MAC chromosome during MAC development, which provides a mechanism of the formation of MAC chromosomes during conjugation. Overall, we demonstrated the distinct higher-order chromatin organization in the two nuclei of the T. thermophila and suggest that the higher-order chromatin structures may play important roles during the development of the MAC chromosomes.
Chromatin conformation capture (3C) (Dekker et al. 2002)-related approaches, such as Hi-C and its further developed version, in situ Hi-C, have revealed that mammalian cells have hierarchical chromatin organizations, from larger scale A/B compartments to smaller scale topologically associating domains (TADs) and even smaller scale chromatin loops (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Rao et al. 2014). The binding of the insulator protein CTCF and the cohesin complex on chromatin are enriched at TAD boundaries in mammals, playing important roles in higher-order chromatin organization (Dixon et al. 2012; Rao et al. 2014, 2017). Removal of cohesin or cohesin-loading protein NIPBL eliminated chromatin loops and TADs while enhancing compartmentalization in mammalian cells (Rao et al. 2017; Schwarzer et al. 2017). However, recently a superresolution chromatin tracing technique revealed that the TAD-like structure is present in single cells after cohesin depletion, which indicates that cohesin is not required for maintaining the TAD-like structure in single cells (Bintu et al. 2018). Thus, the mechanism of TADs formation and maintenance is still largely unknown.
Tetrahymena thermophila, a ciliated protozoan, contains two nuclei in the same cell: the transcriptionally active polyploid macronucleus (MAC) and the transcriptionally inert diploid micronucleus (MIC) (Yao 1996; Chalker 2008; Mochizuki 2012). Although both the MAC and the MIC develop from the same zygotic nucleus during the sexual reproduction (conjugation) induced by starvation and share 70% of their genome sequences (Hamilton et al. 2016), their genomes differ in chromosome numbers, sizes, and functions. During conjugation, the new MAC develops from the zygotic nucleus and undergoes a programmed genome rearrangement which involves chromosome breakage at chromosome breakage sequence (CBS) sites and DNA elimination of internal eliminated sequences (IESs) (Noto and Mochizuki 2017, 2018). Ultimately, the MAC harbors about 181 stably maintained chromosomes derived from the five chromosomes in the zygote which remain unchanged in the MIC, and the MAC is ∼45–50 N (polyploid) while the MIC is 2 N (diploid) (Hamilton et al. 2016; Lin et al. 2016). However, the higher-order chromatin organization in T. thermophila is still unclear.
Recent work in mammals suggested that higher-order chromatin organization is associated with DNA double-strand breaks (DSBs) and chromosomal rearrangements (Canela et al. 2017). Since the development of the T. thermophila MAC during conjugation is associated with chromosome breakage at CBS sites and DNA rearrangement, we speculate that the higher-order chromatin organization in T. thermophila might be related to chromosome breakage and genome rearrangement in the MAC during conjugation. In addition, there is no CTCF in T. thermophila and the cohesin complex is only located in the MIC but not in the MAC (Howard-Till et al. 2013). Thus, T. thermophila provides a naturally controlled system in the same cell for both cohesin depletion (in the MAC) and presence (in the MIC). Therefore, we investigated the higher-order chromatin organization in the two different nuclei of T. thermophila.
Results
Determining Hi-C data for analyzing MAC or MIC higher-order chromatin structures
To examine the higher-order chromatin structures in the two distinct nuclei of T. thermophila, we planned to perform in situ Hi-C (Rao et al. 2014) with T. thermophila whole cells, isolated MICs, or isolated MACs, respectively, at different stages of conjugation, the sexual life cycle of T. thermophila. After starving the cells for 24 h, we mixed two different mating types of T. thermophila strains to induce conjugation and collected the T. thermophila cells at three different conjugation stages: 1.5 h post-mixing (hpm), when the cells were in premeiosis stage; 3 hpm, when the cells were in meiotic recombination stage, and the MIC underwent transcription and elongation, where the elongated meiotic MIC was also called crescent MIC; and 24 hpm, at the end of conjugation when the new MIC and the new MAC were formed and progenies would be produced upon feeding (Fig. 1A). We then performed in situ Hi-C with T. thermophila whole cells, isolated MACs, and isolated MICs at these three different conjugation stages (see Methods).
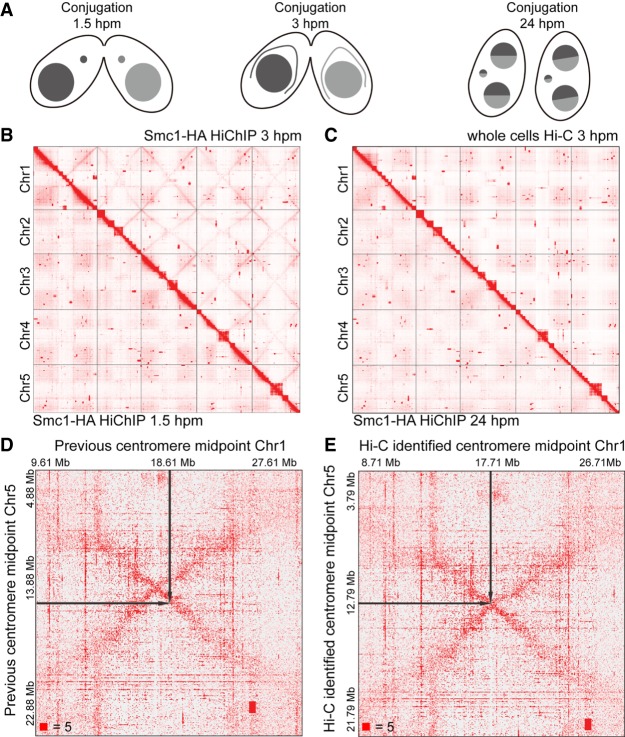
Figure 1.
Crescent-specific interactions in the MIC of T. thermophila. (A) The schematic diagram of three different stages of T. thermophila during conjugation: 1.5 hpm, 3 hpm, and 24 hpm. (hpm) Hours post-mixing. (B) The heatmaps showing the genome-wide chromosome interactions on the MIC genome at 3 hpm (Smc1-HA HiChIP data, upper right) and 1.5 hpm (Smc1-HA HiChIP data, lower left). (C) The heatmaps showing the genome-wide chromosome interactions on the MIC genome at 3 hpm (whole cells Hi-C data, upper right) and 24 hpm (Smc1-HA HiChIP data, lower left). (D,E) The heatmaps showing the MIC inter-chromosome interactions between Chromosomes 1 and 5 at 3 hpm, either defined by a previous method (D) or by our Hi-C data (E). The black arrows on the heatmaps pointed to the midpoints of centromeres. Numbers at the lower left of heatmaps corresponded to the maximum signal in the matrix.
We first analyzed the reads covered on IESs among our Hi-C data to check the purity of the isolated MICs, as the IESs are only located in the MIC (Yao and Gorovsky 1974; Coyne et al. 1996; Hamilton et al. 2016; Noto and Mochizuki 2018). There are a total of ∼7500 IESs that are precisely mapped on the MIC genome with a total length of about 28.5 Mb (Hamilton et al. 2016; Noto and Mochizuki 2018). We divided the 7500 IESs into three classes with the same combined length (∼9.5 Mb) based on their sizes, and class 1 contained the largest IESs, while class 3 contained the shortest IESs. The results showed that the Hi-C data in T. thermophila whole cells had a similar low coverage on IESs compared to that in the isolated MACs (Supplemental Fig. S1A), which suggested that the Hi-C reads generated in the T. thermophila whole cells mainly came from the MAC DNA. We thus used the whole T. thermophila Hi-C data for the following MAC higher-order chromatin structures analysis with KR normalization (Knight and Ruiz 2012), as the nuclei isolation processes might influence the higher-order chromatin structures in the MAC.
For the Hi-C data generated in the isolated MICs, the coverage on IESs increased compared to that in the isolated MACs or the whole cells but was still lower than that of the other genomic regions (Supplemental Fig. S1), indicating that some of the Hi-C data from isolated MICs were contaminated with MAC genome. Since cohesin component protein Smc1 is only located in the MIC of T. thermophila (Howard-Till et al. 2013), to explore the higher-order chromatin organization in the MIC and rule out the effects from the MAC genome, we used HiChIP (Mumbach et al. 2016) with anti-HA antibodies in strains expressing tagged fusion protein Smc1-HA (Howard-Till et al. 2013). The reads coverage on IESs of Smc1-HA HiChIP was higher than that of the isolated MICs Hi-C (Supplemental Fig. S1). For the class 1 and class 2 IESs, the reads coverage of HiChIP data was comparable (or even higher for class 1) to that of the other genomic regions (Supplemental Fig. S1), which suggested that the cohesin HiChIP data mainly came from the MIC with little MAC contamination. We reasoned that IESs in class 3 were too small so that they were lost during the process of Hi-C or HiChIP experiments. We then used Smc1-HA HiChIP data for the following analysis of the MIC higher-order chromatin structures with KR normalization.
Crescent-specific interactions in the meiotic MIC of T. thermophila
Our Smc1-HA HiChIP data showed that there were genome-wide crescent-specific chromosome interactions in the MIC only at 3 hpm (Fig. 1B,C; Supplemental Fig. S2). The crescent-specific inter-arm chromosome interactions existed from telomeres to centromeres on all five chromosomes in the elongated crescent MIC (Fig. 1B). In addition, the crescent-specific interactions also existed between different chromosomes, which formed an X-shaped interaction pattern on the inter-chromosome heatmaps (Fig. 1B, upright; Supplemental Fig. S2B). These results suggested that the telomeres of all five MIC chromosomes interacted and clustered together, as did all of the five centromeres. Therefore, in the crescent MIC, the two telomeres of one chromosome gathered together, making the chromosome fold at the centromere in the middle, so that centromeres and telomeres were clustered at opposite ends of the nucleus. The X-shaped crescent-specific interactions revealed by our HiChIP data perfectly explained the existence of a meiotic chromosome bouquet in T. thermophila, which was organized by centromeres and promoted inter-homolog recombination (Loidl et al. 2012). We therefore demonstrated this phenomenon at the molecular level as a supplement to the imaging data previously supported by DNA FISH and IF (Mochizuki et al. 2008).
Optimizing identification of the centromere midpoint based on crescent-specific interactions
In addition to the above effects, the X-shaped crescent-specific chromosome interactions had other functions in the MIC. Currently, the midpoint of the centromere in T. thermophila is defined as follows: The two CBS sites that are closest to the centromere are designated as R-CBS (on the right chromosome arm) and L-CBS (on the left chromosome arm), respectively; the region between L-CBS and R-CBS is titled the putative centromere region, and the midpoint of this putative centromere region is considered as the midpoint of the centromere (Hamilton et al. 2016). Our cohesin HiChIP data showed that the centromeres were clustering together in the crescent MIC, and there were intra- and inter-chromosome interactions around the centromere region. Thus, the crossover points of the X-shaped crescent-specific chromosome interactions provided another, more accurate way to define the midpoints of centromeres (Fig. 1D,E; Supplemental Table S1). Our cohesin HiChIP data at the crescent stage therefore optimized the identification of the accurate midpoints of centromeres in the MIC.
In addition, we found that there were some square-shaped chromosome interaction boxes that were located far away from the diagonal of the heatmaps (Supplemental Fig. S3), suggesting that these regions were very likely located near each other on the same chromosome but were mistakenly annotated to different chromosome regions. This result indicated that there might be some biases in the current MIC genome, although it did not affect our analysis in this work. Together, these results clearly showed that the higher-order chromatin organization information in the MIC of T. thermophila will be useful in assembling the MIC genome more accurately.
The MAC chromosomes lack A/B compartments, TADs, and chromatin loops
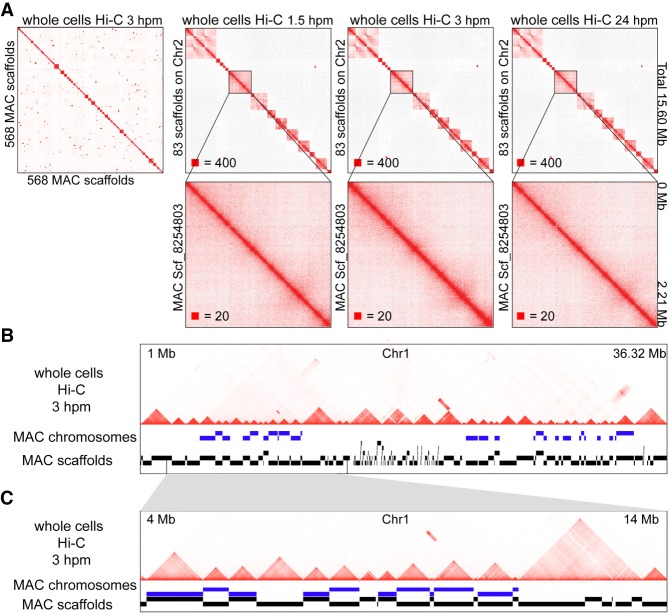
As there are two nuclei in the same T. thermophila cell, we analyzed our T. thermophila whole cells Hi-C data using both the MIC genome (Hamilton et al. 2016) and the MAC scaffolds (Coyne et al. 2008). Because the MAC genome assembly remains incomplete, we chose 568 relatively large (longer than 1 kb) and annotated MAC scaffolds, out of the total 1158, which occupy the majority of the genome (101.92 Mb out of 103.01 Mb). We reordered these 568 MAC scaffolds in the genome according to their relative location on the MIC chromosomes, based on the homology of MAC and MIC, for the following analysis. The results showed that there were square-shaped chromatin interaction patterns along the diagonal of the heatmaps, which were similar to the TADs in mammals, when we mapped the whole cells Hi-C data to the MAC scaffolds (Fig. 2A), but there were no square-shaped chromatin interaction patterns inside the MAC scaffolds, even in the largest ones (Fig. 2A). In addition, we found that every one of the MAC chromosomes, defined by the occupancy of double-end telomeres on the MAC scaffolds, was consistent with one of the TAD-like structures on the heatmaps when mapping the whole cells Hi-C data to the MIC genome (Fig. 2B,C). Thus, although these square-shaped interaction patterns along the diagonal of the heatmaps were similar to TAD structures in mammals, they were consistent with the MAC (whole) chromosomes but did not exist inside the MAC chromosomes, indicating that they were very similar to the chromosome territories in mammals. Additionally, there were no TADs identified by HiCExplorer (Ramírez et al. 2018) with the whole cells Hi-C data when we mapped them to the MAC scaffolds (Supplemental Fig. S4A), and the number of the TADs identified when mapping the whole cells Hi-C data to the MIC genome was close to the number of chromosomes in the MAC (Supplemental Fig. S4A). Thus, our data showed that the TAD-like structures might not exist in the MAC of T. thermophila, and the square-shaped interaction patterns along the diagonal of the heatmaps are the MAC chromosome territories.
Figure 2.
The square-shaped interaction patterns in the MAC of T. thermophila are chromosome territories. (A) Heatmaps showing the chromosome interactions on the 568 MAC scaffolds (ordered by their locations on the MIC genome) at 3 hpm (whole cells Hi-C data mapped to the MAC scaffolds), and the chromosome interactions of the 83 MAC scaffolds located on the MIC Chromosome 2 at 1.5 hpm, 3 hpm, and 24 hpm. Zoomed-in views of scf_8254803 are shown at the bottom. (hpm) Hours post-mixing. Numbers at the lower left of heatmaps correspond to the maximum signal in the matrix. (B,C) The MAC chromosomes (MAC scaffolds with telomere sequences at both ends) and the MAC scaffolds compared to the chromosome interactions on MIC Chromosome 1 at 3 hpm are shown (whole cells Hi-C data mapped to the MIC genome). Regions from 1 Mb to 36.32 Mb (B); zoomed in region from 4 Mb to 14 Mb (C).
In addition to TADs, in mammalian cells, Hi-C and related studies revealed that the chromatin is also organized into A/B compartments and chromatin loops (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Rao et al. 2014). PCA analysis of the T. thermophila whole cells Hi-C data did not show clear segmentation, whether mapping to the MIC genome or to the MAC scaffolds (Supplemental Fig. S4B), indicating that there were no A/B compartments in the MAC. Although we could identify chromatin loops with the T. thermophila whole cells Hi-C data by HiCCUPS (Durand et al. 2016), these chromatin loops could not be confirmed when visually compared to the chromatin loops identified in mammalian cells (Supplemental Fig. S4C,D), suggesting that chromatin loops might not exist in the MAC either. This result was consistent with the fact that there are no reports of distal enhancers or long-range gene regulation in T. theromphila. Together, our results showed that the gene-dense, fragmented MAC genome shows no obvious A/B compartments, TADs, and chromatin loops, but each MAC chromosome is a single isolated unit that is similar to the chromosome territory in mammals.
TAD-like structures are present in the MIC of T. thermophila
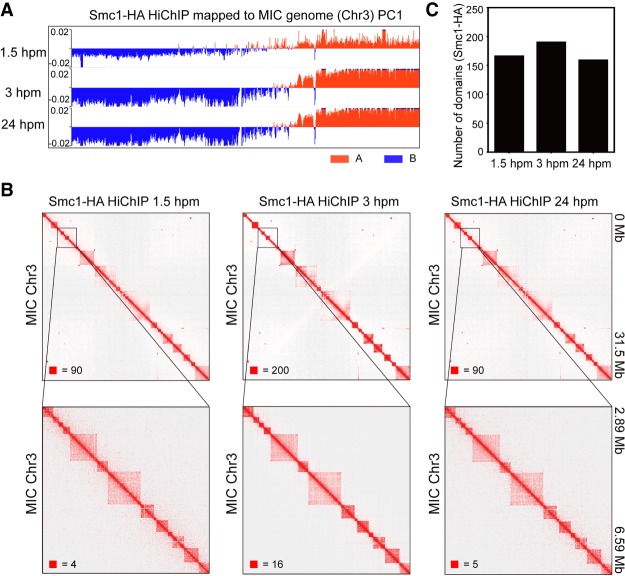
Next, we examined the A/B compartments, TADs, and chromatin loops in the MIC of T. thermophila using Smc1-HA HiChIP data. The results showed that there were no A/B compartments (Fig. 3A) or visible chromatin loops on the heatmaps either, similar to that in the MAC. The heatmaps of Smc1-HA HiChIP, however, clearly showed that there were square-shaped chromatin interaction patterns along the diagonal inside the chromosome (Fig. 3B), which we named TAD-like structures as they were similar to the TADs structure in mammals. There were 166, 190, and 159 TAD-like structures identified by HiCExplorer in the MIC at 1.5 hpm, 3 hpm, and 24 hpm, respectively (Fig. 3C). The number of TAD-like structures in the MIC was close to the number of chromosome territories in the MAC, indicating that TAD-like structures in the MIC might be related to the chromosomes in the MAC. However, although Smc1 is only localized in the MIC, there might still be some MAC contamination in the Smc1-HA HiChIP data since we could not guarantee 100% ChIP efficiency. To rule out the possibility that the highly similar feature between TAD-like structures in the MIC and the chromosome territories in the MAC might be due to the MAC contamination in our HiChIP data, the ideal way is to isolate 100% pure MICs to perform Hi-C, but it is not technically possible currently.
Figure 3.
TAD-like structures in the MIC of T. thermophila. (A) Compartment eigenvector of Smc1-HA HiChIP was plotted along Chromosome 3 of the MIC for three different conjugation stages and used as A (red) and B (blue) compartments segmentation. (B) Heatmaps showing the chromosome interactions on Chromosome 3 (50-kb bin) at three different times during conjugation (Smc1-HA HiChIP data, from left to right: 1.5 hpm, 3 hpm, and 24 hpm). Zoomed-in views (5-kb bin) are shown at the bottom. Numbers at the lower left of heatmaps correspond to the maximum signal in the matrix. (C) The number of chromatin domains found by HiCExplorer with HiChIP data of the three different conjugation stages.
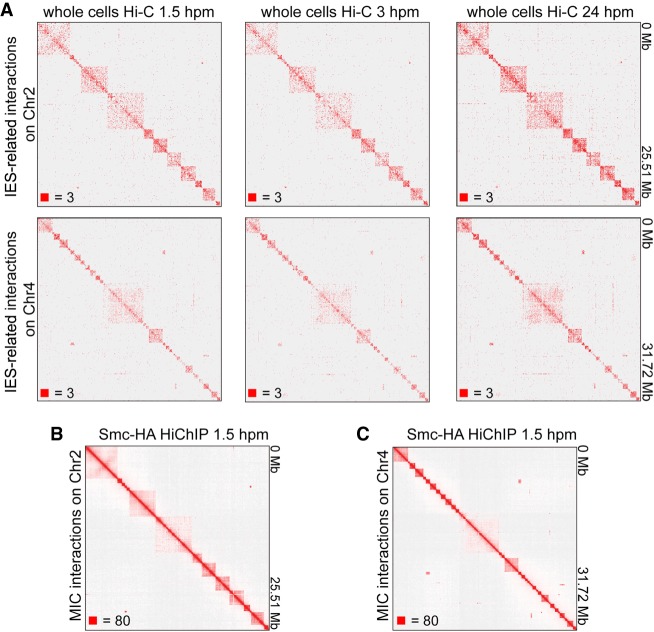
We then took another way to solve this potential MAC contamination problem. Since IESs are MIC-specific sequences (Hamilton et al. 2016; Noto and Mochizuki 2018), we selected the IES-associated interactions, which could only come from the MIC, from the whole cells Hi-C data. Although the IES-associated Hi-C interactions were only about 1.4% of the total paired reads, the IES-associated interaction heatmaps clearly showed that there were square-shaped chromosome interaction patterns along the diagonal of the heatmaps in the MIC (Fig. 4A), which were highly consistent with that on the heatmaps of HiChIP data (Fig. 4B,C). Thus, our data showed that there are no A/B compartments and chromatin loops in the MIC, while there are TAD-like structures in the MIC of T. thermophila.
Figure 4.
IES-associated interactions of T. thermophila show TAD-like structures in the MIC. (A) Heatmaps showing the IES-associated Hi-C interactions on Chromosome 2 (top) and Chromosome 4 (bottom) at 1.5 hpm, 3 hpm, and 24 hpm (left to right). (B,C) Heatmaps showing the chromosome interactions at 1.5 hpm on Chromosome 2 (B) and Chromosome 4 (C) (Smc1-HA HiChIP data). Numbers at the lower left of heatmaps correspond to the maximum signal in the matrix.
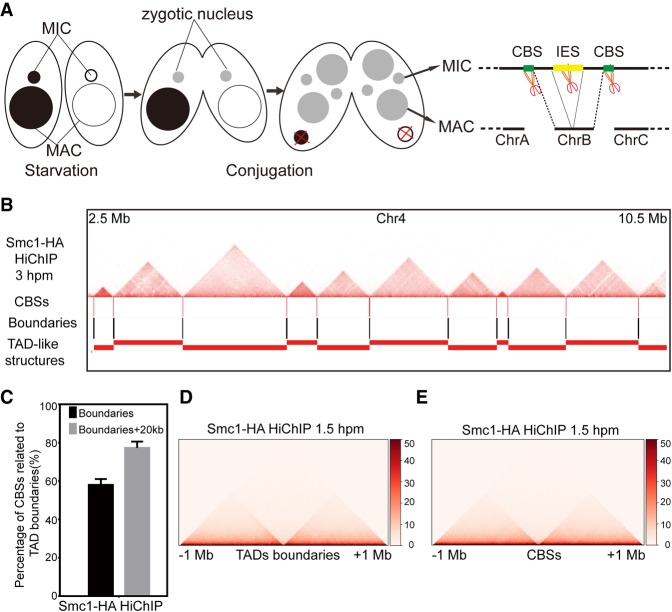
Boundaries of the TAD-like structures in the MIC coincide with CBS sites
During conjugation, both the MIC and the MAC are developed from the same zygotic nucleus, and the MIC possesses the five chromosomes as the zygotic nucleus, while the MAC undergoes programmed genome rearrangement with, finally, 181 stably maintained chromosomes (Hamilton et al. 2016; Lin et al. 2016). The MAC chromosomes are formed from the five zygotic chromosomes after chromosome breakage at CBS sites and elimination of IESs (Fig. 5A; Hamilton et al. 2016; Lin et al. 2016). As the number of TAD-like structures in the MIC was similar to the number of chromosomes in the MAC, we asked whether the TAD-like structures on the MIC chromosomes are associated with the formation of the MAC chromosomes. We found that the TAD-like structures in the MIC were all aligned to the chromatin regions between the CBS sites, and the boundaries of the TAD-like structures in the MIC were highly coincident with CBS sites (Fig. 5B; Supplemental Fig. S5). About 57% of CBS sites were exactly localized to the boundaries of TAD-like structures, which was increased to 77.3% when we included the 20-kb surrounding regions of the identified boundaries (Fig. 5C). The averaged chromatin contact frequency centered on boundaries of TAD-like structures or CBS sites were also highly consistent (Fig. 5D,E), suggesting that CBS sites on the MIC chromosomes are the boundaries of the TAD-like structures in the MIC.
Figure 5.
Boundaries of TAD-like structures in the MIC are consistent with CBS sites. (A) The schematic diagram shows the development of MAC and MIC from the same zygotic nuclei during conjugation. (B) The location of CBS sites, boundaries, and TAD-like structures compared to the heatmaps on the Chromosome 4 (2.5 Mb to 10.5 Mb) at 3 hpm. (C) The bar plot shows percentage of CBS sites localized to the boundaries of the TAD-like structures (10 kb) or 20 kb around the boundaries (total 50 kb). (D) Heatmaps showing averaged Hi-C interaction counts (Smc1-HA HiChIP, 1.5 hpm) around the boundaries of TAD-like structures. (E) Heatmaps showing averaged Hi-C interaction counts (Smc1-HA HiChIP, 1.5 hpm) around the CBS sites.
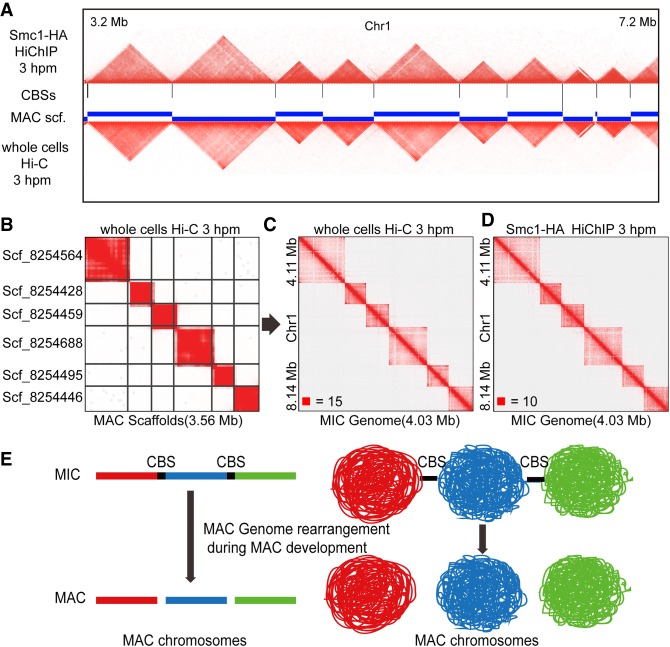
Chromosome territories in the MAC are consistent with TAD-like structures in the MIC
A comparison of the TAD-like structures in the MIC and the chromosome territories in the MAC revealed clear similarity (Fig. 6A). For some MAC scaffolds that have telomere sequences at both ends, which means that they are MAC chromosomes, the pattern of the whole cells Hi-C heatmaps mapping to the MAC scaffolds (Fig. 6B) or to the MIC genome (Fig. 6C) was highly consistent with the TAD-like pattern on the MIC genome (Fig. 6D). These results together suggested that the TAD-like structures in the MIC might play an important role in the formation of the MAC chromosomes during conjugation. Thus, we proposed a model for the genome organization in the two distinct nuclei of T. thermophila during conjugation: The higher-order genome structure (TAD-like structure) may form in the zygotic nucleus by cohesin and/or other factors before the new MAC and the new MIC differentiation; the MIC then retains cohesin and keeps its higher-order chromatin organization, while the cohesin complex disappears in the developing MAC; the higher-order genome structure of MAC chromosomes is directly formed from the higher-order genome structure of MIC chromosomes after chromosome breakage at CBS sites (and elimination at IESs) during the MAC development. Therefore, the chromosome territories in the MAC are similar to the TAD-like structures in the MIC, and these TAD-like structures may function in the development of the new MAC during conjugation (Fig. 6E).
Figure 6.
The MAC chromosomes are related to the MIC TAD-like structures during conjugation. (A) The heatmap regions (Chromosome 1: 3.2 Mb to 7.2 Mb) of the MIC (Smc1-HA HiChIP at 3 hpm, upper) and the MAC (whole cells Hi-C at 3 hpm, below) compared to the CBS sites on the MIC and the MAC scaffolds are plotted. (B) The heatmap showing the Hi-C interactions on six of the MAC scaffolds at 3 hpm (whole cells Hi-C data mapped to the MAC scaffolds). (C) The heatmap showing the Hi-C interactions on the MIC Chromosome 4 (4.11 Mb to 8.14 Mb) at 3 hpm (whole cells Hi-C data mapped to the MIC genome). (D) The heatmap showing the Hi-C interactions on the MIC Chromosome 4 (4.11 Mb to 8.14 Mb) at 3 hpm (Smc1-HA HiChIP data mapped to the MIC genome). (E) A model for the MAC chromosomes development during the conjugation of T. thermophila. The TAD-like structures established in the zygotic nuclei were followed by chromosome breakage at CBS sites.
Discussion
T. thermophila has two distinct nuclei in the same cell, with a different genome and function, while these two nuclei both develop from the same zygotic nucleus during conjugation. However, little is known about the higher-order chromatin organization and its function in this process. Here, we explored the higher-order chromatin organization in T. thermophila and found that there are no A/B compartments and chromatin loops in both of the MAC and the MIC. There are also no TADs in the MAC, while the MIC has TAD-like structures. The boundaries of the TAD-like structures in the MIC are consistent with the CBS sites where zygotic chromosomes break to develop the MAC chromosomes, suggesting that the TAD-like structures may function in the MAC development during conjugation.
During conjugation, the MIC keeps the same genome as the zygotic nucleus, while the MAC undergoes a programmed genome rearrangement which includes chromosome breakage at CBS sites and DNA elimination of IESs (Mochizuki 2012). Chromosome breakage at CBS sites is followed by de novo telomere formation, which is important for the formation of the MAC chromosomes, but the mechanism of chromosome breakage is still largely unknown (Mochizuki 2012; Hamilton et al. 2016; Lin et al. 2016). We showed in this study that in the MAC, one chromosome is one chromosome territory and the interaction pattern of each MAC chromosome territory is consistent with each TAD-like structure in the MIC, indicating that one TAD-like structure in the MIC may give rise to one chromosome in the MAC. Thus, the MAC chromosomes may be derived from zygotic TAD-like structures after chromosome breakage at CBS-related regions. Future studies pursuing the role and mechanism of zygotic TAD-like structures in the MAC chromosome reorganization will help to optimize the assembly of the MAC genome, which will also promote the understanding of the function of the 3D genome in other chromosomal processes such as DNA DSBs.
In addition, we noticed that the TAD-like structures in the MIC of T. thermophila have clearer boundaries compared to the TADs identified in mammals. Considering recent advances about TADs in mammals, these clearer boundaries might be the nature of TAD-like structures in the MIC of T. thermophila. Recently, single-cell 3D genome studies showed that there are TAD-like structures in the single cell and the TADs detected by Hi-C with a pool of cells are the average of TAD-like structures in individual mammalian cells (Flyamer et al. 2017; Bintu et al. 2018). In a single mammalian cell, any genomic position may be the boundary of a TAD-like structure, while the boundaries are likely to be binding sites of CTCF and cohesin (Bintu et al. 2018). The binding of CTCF and cohesin is highly dynamic in individual mammalian cells with active transcription. Therefore, the TADs obtained in the Hi-C data of a population of cells (the average result of the superposition of TAD-like structures in each single cell) have the enrichment of CTCF and cohesion at the TAD boundaries, which will be, in turn, a region rather than a point. In contrast, the MIC of T. thermophila is transcriptionally inert (except for the transient transcription activity in crescent stage), and its genome may be much more stable compared to the transcriptionally active genome in mammals. Thus, the TAD-like structures in the MIC have clearer boundaries (boundaries are limited to individual points of the CBS sites, not to relative wide region as in mammals).
In mammals, TADs are reorganized during spermatogenesis and are almost lost during meiosis (Vara et al. 2019; Wang et al. 2019). However, we observed that TAD-like structures were still clear in the MIC at 3 hpm during conjugation when T. thermophila cells enter meiosis. There were, however, crescent-specific X-shaped chromosome interactions in the MIC at this stage which was similar to the X-shaped chromosome interactions in meiotic prophase chromosomes during mouse spermatogenesis (Patel et al. 2019). The TADs are lost from mouse meiotic prophase chromosomes when the X-shaped chromosome interactions formed (Patel et al. 2019), and we speculated that the TAD-like structures might also be lost or attenuated from the MIC chromosomes at 3 hpm when the crescent-specific X-shaped chromosome interactions formed. The reason for the apparent TAD-like structures at 3 hpm is very likely the fact that the percentage of the crescent MICs in the conjugation is <40% (Loidl et al. 2012). Thus, we could not obtain pure crescent MICs, and the crescent-specific interactions were generated from the crescent MICs while the TAD-like structures might come from the MICs at other stages during conjugation. Hi-C or HiChIP with 100% pure crescent MICs or single-cell Hi-C methods (Nagano et al. 2013, 2017; Ramani et al. 2017; Bintu et al. 2018) may help us to answer these questions in the future.
CTCF is found to play important roles in TADs organization in mammals (Dixon et al. 2012; Phillips-Cremins et al. 2013; Rao et al. 2014), and TADs are almost lost after depletion of CTCF in mammalian cells (Nora et al. 2017). However, there are also some species that have TAD-like structures but do not have a CTCF ortholog (Le et al. 2013; Hsieh et al. 2015; Marbouty et al. 2015), similar to T. thermophila. However, in budding yeast or bacteria, which have no CTCF ortholog, the boundaries of the TAD-like structures are enriched in genes with high transcription activities (Le et al. 2013; Hsieh et al. 2015). Thus, there is a CTCF-dependent pathway for TADs formation in mammalian cells and also a CTCF-independent but transcription-dependent pathway for TAD-like structures formation in some species without a CTCF ortholog. The MIC of T. thermophila does not even have transcription activity at most times, suggesting that the formation of the TAD-like structures in the MIC may be independent of CTCF or transcription. We showed that the boundaries of TAD-like structures in the MIC were consistent with CBS sites, indicating that the CBS sites-related proteins or other factors, such as chromatin modifiers, may function in the formation of TAD-like structures in the MIC of T. thermophila. Future works to identify binding proteins and chromatin features at CBS sites and their nearby regions will disclose the underlying mechanism of TAD-like structures formation in the MIC of T. thermophila, which will also help to understand the mechanism of the formation of MAC chromosomes during conjugation. Taken together, our work provided the first genome-wide view of the higher-order chromatin organization in T. thermophila, which carries two distinct nuclei in the same cell, and revealed the relationship between the TAD-like structures in the MIC and the chromosomes in the MAC. These findings have the potential to reveal unique insights into how higher-order chromatin organization is established and maintained.
Methods
T. thermophila culture, starvation, and conjugation
T. thermophila strains B2086, Cu428, B2086 with Smc1-HA (gift from Dr. Josef Loidl), and Cu428 with Rec8-GFP (gift from Dr. Josef Loidl) were used for Hi-C and HiChIP experiments. Cells were cultured at 30°C in 1× SPP medium (10 g protease peptone, 2 g dextrose, 1 g yeast extract, 0.03 g Sequestrene (Fe-EDTA), adding water to final 1 L and autoclave prior to use). For conjugation, cells were first starved in 10 mM Tris-HCl pH 7.4 to a final density of ∼2.5 × 105 cells/mL for 24 h at 30°C without shaking. Equal numbers of cells representing two different mating types were mixed and incubated at 30°C without disturbing, until cells were collected at 1.5 h, 3 h, or 24 h for the subsequent experiments.
Collection of mating T. thermophila cells for Hi-C
We added 37% formaldehyde (Sigma-Aldrich) to mating cells in 10 mM Tris-HCl pH 7.4 to make final a 1% concentration, mixed well, and incubated for 10 min at room temperature, then quenched the formaldehyde with final 125 mM glycine (Sigma-Aldrich), mixed well, and incubated for 5 min at room temperature, followed by 15 min on ice. We centrifuged the cells at 800g at 4°C and discarded the solution, then washed the cell pellet with ice-cold 10 mM Tris-HCl pH 7.4. We then centrifuged again and discarded the solution, transferred cells to 1.5-mL tubes and removed as much of the residual solution as possible. The cell samples were stored at −80°C for future experiments or immediately used.
T. thermophila MAC and MIC isolation
We resuspended 2 × 107 fixed cells in 1 mL of Hi-C lysis buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 20 mM KCl, 1.5 mM MgCl2, 1% NP-40, 1× PIC), and incubated at 4°C for 30 min with slow rotation. Then, we centrifuged at 800g 4°C for 2 min and washed once with ice-cold lysis buffer. Next, we resuspended cells in 400 μL of 0.5% SDS (Sigma-Aldrich) and incubated at 62°C for 6–12 min with shaking (checked by DAPI stain) until cell membranes were broken (not waiting for all cell membranes to be broken; we stopped when 30%–50% were broken). We put the tubes on ice and quenched the SDS by adding 400 μL of 5% Triton X-100 (Sigma-Aldrich). Nuclei were checked by DAPI stain, using dounce (Sigma-Aldrich) if necessary. Then, we centrifuged at 780g for 10 min at 4°C, transferred the solution which contains MIC to a new tube (the pellet which contains MAC was also collected and washed twice with 1× NEBuffer 2.1), centrifuged the tubes with MIC again at 780g for 10 min at 4°C, repeated this step 3–4 times and checked by DAPI stain to see if the MIC was pure. Then, we spun at 8000g at 4°C for 10 min to pellet the MIC for the following Hi-C experiments.
Hi-C and HiChIP
The detailed Hi-C method was described previously (Rao et al. 2014). In brief, we resuspended T. thermophila whole cells (after Hi-C lysis), isolated MIC, or isolated MAC in 1× NEBuffer 2.1, and digested overnight with HindIII or MboI (NEB) at 37°C. After inactivating HindIII or MboI by incubating at 65°C for 10 min, we filled in chromosome ends with dATP (Thermo Fisher Scientific), dTTP (Thermo Fisher Scientific), dGTP (Thermo Fisher Scientific), and biotin-14-dCTP (Thermo Fisher Scientific) at 37°C for 45–60 min with shaking. The DNA fragments were ligated with T4 DNA ligase (NEB) at room temperature for 4 h with slow rotation. After ligation, we incubated at 65°C overnight in the presence of Proteinase K (Thermo Fisher Scientific) to de-crosslink and then purified the ligated DNA. The Hi-C DNA was then sheared to 200–500 bp and selected by Dynabeads MyOne Streptavidin C1 beads (Thermo Fisher Scientific). Then, we prepared the sequencing libraries on beads, with end-repair, dA-tailing, and adapter ligation. After PCR, we purified the DNA between 300 and 500 bp with AMPure XP beads (Beckman Coulter). The Hi-C libraries were sequenced using the Illumina HiSeq platform. For HiChIP, we generated Smc1-HA HiChIP using standard methods with HindIII or MboI digestion (Mumbach et al. 2016). The antibody used in HiChIP is: anti-GFP antibody-ChIP grade (Abcam ab290).
Hi-C and HiChIP data processing
The reads coverage on IES regions was calculated by deepTools (Ramírez et al. 2014) and normalized by reads per kilobase per million mapped reads (RPKM). The paired-end Hi-C and HiChIP sequencing data were mapped and processed through HiC-Pro 2.10.0 (Servant et al. 2015) to build the contact matrix. The paired reads were mapped to the MIC genome (Hamilton et al. 2016) or the MAC scaffolds (Coyne et al. 2008) reordered by their localizations on the MIC chromosome by Bowtie 2 (Langmead and Salzberg 2012). The Hi-C matrices were then transferred to Juicebox .hic format for visualization or further analysis with KR normalization. The IES-associated interactions were selected from the whole cells Hi-C data by HiC-Pro 2.10.0 while setting the IESs as the capture targets; then, the IES-associated Hi-C matrices were transferred to Juicebox .hic format for visualization with KR normalization.
A/B compartments, TADs, and chromatin loops analysis
We also used the HiCExplorer 2.1.4 (Ramírez et al. 2018) to process the Hi-C and HiChIP raw data. The A/B compartments and TADs analysis were performed with KR balanced matrix by HiCExplorer. Briefly, the raw sequencing data were mapped to the MIC genome (Hamilton et al. 2016) or the MAC scaffolds (Coyne et al. 2008) reordered by their localizations on the MIC chromosome by BWA (Li and Durbin 2009). The Hi-C matrix was then generated by hicBuildMatrix tool in HiCExplorer with 10-kb bin size and then KR balanced by hicCorrectMatrix tool. The A/B compartments analysis was performed with the hicPCA tool, while the TADs and boundaries were found by the hicFindTADs tool. For chromatin loops, we identified chromatin loops at 5-kb, 10-kb, and 25-kb resolution and merged in our Hi-C and HiChIP data of T. thermophila together with mES Hi-C data (Bonev et al. 2017) using CPU version of HiCCUPS (Durand et al. 2016).
Data access
The raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE140429.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Dr. Josef Loidl (University of Vienna) for providing the T. thermophila strains B2086 with Smc1-HA and Cu428 with Rec8-GFP. We also thank Dr. Jie Xiong (Institute of Hydrobiology, Chinese Academy of Sciences), Dr. Wei Miao (Institute of Hydrobiology, Chinese Academy of Sciences), and Dr. Yong Zhang (Institute of Zoology, Chinese Academy of Sciences) for their constructive suggestions, and Dr. Leonard Lipovich (Wayne State University) and Dr. Zachary Smith (University of Science and Technology of China) for their help in editing the manuscript. This work was supported by grants from the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0406-2 to X.S.), and the National Natural Science Foundation of China (31671490 and 31472059 to X.S.).
Author contributions: X.S. conceived and designed the project. Z.L., T.H., and Q.X. performed the experiments. Z.L., T.H., H.J., R.W., and S.Z. analyzed the data. All authors discussed and interpreted results. X.S., Z.L., and T.H. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.241687.118.
References
- Bintu B, Mateo LJ, Su J-H, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362: eaau1783 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J-P, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171: 557–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. 2017. Genome organization drives chromosome fragility. Cell 170: 507–521.e18. 10.1016/j.cell.2017.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL. 2008. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim Biophys Acta 1783: 2130–2136. 10.1016/j.bbamcr.2008.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao M-C. 1996. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet 30: 557–578. 10.1146/annurev.genet.30.1.557 [DOI] [PubMed] [Google Scholar]
- Coyne RS, Thiagarajan M, Jones KM, Wortman JR, Tallon LJ, Haas BJ, Cassidy-Hanley DM, Wiley EA, Smith JJ, Collins K, et al. 2008. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9: 562 10.1186/1471-2164-9-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, et al. 2016. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. eLife 5: e19090 10.7554/eLife.19090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. 2013. A single cohesin complex performs mitotic and meiotic functions in the protist Tetrahymena. PLoS Genet 9: e1003418 10.1371/journal.pgen.1003418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T-HS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. 2015. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162: 108–119. 10.1016/j.cell.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PA, Ruiz D. 2012. A fast algorithm for matrix balancing. IMA J Numer Anal 33: 1029–1047. 10.1093/imanum/drs019 [DOI] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. 2013. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342: 731–734. 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CG, Lin IT, Yao MC. 2016. Programmed minichromosome elimination as a mechanism for somatic genome reduction in Tetrahymena thermophila. PLoS Genet 12: e1006403 10.1371/journal.pgen.1006403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Lukaszewicz A, Howard-Till RA, Koestler T. 2012. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J Cell Sci 125(Pt 23): 5873–5880. 10.1242/jcs.112664 [DOI] [PubMed] [Google Scholar]
- Marbouty M, Le Gall A, Cattoni DI, Cournac A, Koh A, Fiche J-B, Mozziconacci J, Murray H, Koszul R, Nollmann M. 2015. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol Cell 59: 588–602. 10.1016/j.molcel.2015.07.020 [DOI] [PubMed] [Google Scholar]
- Mochizuki K. 2012. Developmentally programmed, RNA-directed genome rearrangement in Tetrahymena. Dev Growth Differ 54: 108–119. 10.1111/j.1440-169X.2011.01305.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Novatchkova M, Loidl J. 2008. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J Cell Sci 121: 2148–2158. 10.1242/jcs.031799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. 2016. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 13: 919–922. 10.1038/nmeth.3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Várnai C, Dudley C, Leung W, Baran Y, Cohen NM, Wingett S, Fraser P, Tanay A. 2017. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547: 61–67. 10.1038/nature23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Mochizuki K. 2017. Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open Biol 7: 170172 10.1098/rsob.170172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Mochizuki K. 2018. Small RNA-mediated trans-nuclear and trans-element communications in Tetrahymena DNA elimination. Curr Biol 28: 1938–1949.e5. 10.1016/j.cub.2018.04.071 [DOI] [PubMed] [Google Scholar]
- Patel L, Kang R, Rosenberg SC, Qiu Y, Raviram R, Chee S, Hu R, Ren B, Cole F, Corbett KD. 2019. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat Struct Mol Biol 26: 164–174. 10.1038/s41594-019-0187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong C-T, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. 10.1016/j.cell.2013.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J. 2017. Massively multiplex single-cell Hi-C. Nat Methods 14: 263–266. 10.1038/nmeth.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42(Web Server issue): W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E. 2015. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16: 259 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vara C, Paytuví-Gallart A, Cuartero Y, Le Dily F, Garcia F, Salvà-Castro J, Gómez HL, Julià E, Moutinho C, Aiese Cigliano R, et al. 2019. Three-dimensional genomic structure and cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep 28: 352–367.e9. 10.1016/j.celrep.2019.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang H, Zhang Y, Du Z, Si W, Fan S, Qin D, Wang M, Duan Y, Li L, et al. 2019. Reprogramming of meiotic chromatin architecture during spermatogenesis. Mol Cell 73: 547–561.e6. 10.1016/j.molcel.2018.11.019 [DOI] [PubMed] [Google Scholar]
- Yao M-C. 1996. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet 12: 26–30. 10.1016/0168-9525(96)81385-0 [DOI] [PubMed] [Google Scholar]
- Yao M-C, Gorovsky MA. 1974. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma 48: 1–18. 10.1007/bf00284863 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.