Abstract
Global climate and land use change are altering plant and soil microbial communities worldwide, particularly in arctic and alpine biomes where warming is accelerated. The widespread expansion of woody shrubs into historically herbaceous alpine plant zones is likely to interact with climate to affect soil microbial community structure and function, however our understanding of alpine soil ecology remains limited. This study aimed to 1) determine whether the diversity and community composition of soil fungi vary across elevation gradients and to 2) assess the impact of woody shrub expansion on these patterns. In the White Mountains of California, sagebrush (Artemisia rothrockii) shrubs have been expanding upwards into alpine areas since 1960. In this study, we combined observational field data with a manipulative shrub removal experiment along an elevation transect of alpine shrub expansion. We utilized next generation sequencing of the ITS1 region for fungi and joint distribution modeling to tease apart effects of the environment and intra-community interactions on soil fungi. We found that soil fungal diversity declines and community composition changes with increasing elevation. Both abiotic factors (primarily soil moisture and soil organic C) and woody sagebrush range expansion had significant effects on these patterns. However, fungal diversity and relative abundance had high spatial variation, overwhelming the predictive power of vegetation type, elevation, and abiotic soil conditions at the landscape scale. Finally, we observed positive and negative associations among fungal taxa which may be important in structuring community responses to environmental change.
Keywords: fungi, soil, shrub expansion, alpine, joint distribution model, global change
Introduction
Changes in global climate and land use are having significant impacts on above and belowground organisms worldwide including plants and soil microbes (Wolters et al. 2000). This is particularly true in cold arctic and alpine biomes where warming is occurring at an accelerated pace (Rammig et al. 2010, Pepin et al. 2015). Alpine environments have been relatively poorly studied when considering the impacts of global change on belowground soil organisms (Lazzaro et al. 2015). This is especially true for soil fungi, as a large majority of soil microbial studies in alpine environments focus on bacteria and archaea (Siles and Margesin 2016). Therefore, it is necessary to improve our baseline knowledge of how soil fungal communities change across elevation and climate gradients in order to understand and predict how these patterns are being altered by global change.
Soil fungal communities generally change across elevation gradients, however these patterns are variable and mechanisms are still not well understood (Sundqvist et al. 2013). In particular, the relative importance of different mechanisms including soil abiotic conditions, plant associations, and interactions among taxa are difficult to disentangle. Soil fungal diversity often declines with increasing elevation in alpine environments (Schinner and Gstraunthaler 1981, Körner 2003). This is primarily due to a decline in plant species richness in high elevation ecosystems, as belowground fungal communities are known to be closely tied to plant species diversity and identity (Körner 2003, Bahram et al. 2012, Peay et al. 2013). In addition, fungal community composition in soils also changes along elevation gradients. Schinner and Gstraunthaler (1981) were among the first to describe this pattern in the central European Alps whereby soil fungal communities declined in diversity as elevation increased and fungal community composition and species dominance paralleled shifts in plant communities along this same gradient. More recent studies have confirmed these patterns, as soil fungal diversity declined strongly with elevation and paralleled declines in plant and bacterial diversity in tropical montane forests. (Nottingham et al. 2016). Other research however has suggested that fungal diversity and richness have no clear relationship with altitude (Coince et al. 2014, Siles and Margesin 2016) or that fungal community composition, but not alpha diversity and richness, vary across elevation gradients (Shen et al. 2014, Lanzén et al. 2016). In general however, it is known that elevation is an important predictor of fungal communities worldwide (Kivlin et al. 2011, Tedersoo et al. 2014). More studies of soil fungi across elevation gradients are required to understand how these patterns differ globally and the mechanisms that drive elevation-diversity relationships.
In addition to changing plant diversity and composition, the abiotic environment also changes significantly with elevation and may have important effects on fungal communities (Körner 2003, 2007, He et al. 2016). For example, mean annual temperature (MAT), soil moisture, soil organic carbon (SOC) and nitrogen (SON) and soil pH all influence the diversity and community structure of soil fungi along elevation gradients (Sundqvist et al. 2013), although not always in consistent ways. Fungal diversity may decline with mean annual temperature (MAT) at high elevation sites (Nottingham et al. 2016), or may increase due to greater soil moisture at high elevations irrespective of temperature (Pellissier et al. 2014). Soil pH was the most important predictor of fungal community structure in alpine soils in Northeast China (Shen 2014), and an equally important as MAT for root associated fungi in the French Alps and Pyrenees (Coince et al. 2014). Changes in abiotic soil parameters may also interact with vegetation in their effects on soil fungi (Sundqvist et al. 2013). For example, fungal diversity was inversely related to SOC and total soil N in alpine steppe of the Tibetan Plateau, however this trend was reversed in nearby alpine meadows, displaying a strong interaction between the dominant vegetation type and soil nutrients on fungal diversity (Zhang et al. 2017).
Individual fungal taxa may differ in their responses to elevation gradients due to their environmental tolerances and plant associations. For example, ectomycorrhizal (ECM) and arbuscular mycorrhizal fungi (AMF) decline in diversity at higher elevations because of declines in plant species hosts at high elevation sites (Wu et al. 2007, Bahram et al. 2012, Shen et al. 2014, Tedersoo et al. 2014). However, Dark Septate Endophytes (DSE) maintain high abundances in alpine environments where in general mycorrhizal abundance is low (Körner 2003, Newsham 2011, Schmidt et al. 2012). Free-living fungal taxa are also likely to vary across elevation gradients due to changes in abiotic conditions and plant resource quality and quantity, but these relationships are more poorly studied than for mycorrhizal groups. Two notable examples include evidence that Archaerhizomycetes have higher abundance at high elevation in tropical montane forests (Nottingham et al. 2016), and that Agaricomycete fungi increase in abundance with elevation at a global scale (Tedersoo et al. 2014).
Given the importance of plant communities for shaping fungal distributions, the expected shifts in alpine plant communities due to climate change could have large effects on fungal biogeography. One prevalent shift in alpine plant communities is that of woody plants, mainly shrubs and trees, expanding into historically herbaceous-dominated alpine grasslands and fellfields (Cannone et al. 2007). Woody plant encroachment can occur through a variety of global change drivers including warming temperatures, altered precipitation, and changes in grazing regimes. Because fungi are the primary decomposers of woody and other recalcitrant plant material, this is likely to have strong impacts on fungal diversity and community structure (Harmon et al. 1986, Bardgett et al. 2005, De Boer et al. 2005, Nielsen et al. 2015). Shifts from herbaceous to woody shrub cover may directly impact fungal communities by altering the quantity and quality of litter substrates, and indirectly by affecting the abiotic soil environment including carbon and N pools, pH, and water availability (Archer et al. 2001, Hollister et al. 2010). In arctic tundra, Ascomycota and Chytridiomycota were more abundant in grass tussock soils than shrub soils, while Zygo and Basidiomycete fungi were more abundant in shrub soils (Wallenstein et al 2007). This is likely due to higher levels of woody and lignin-rich litter in shrub soils, which promotes saprotrophic wood decomposer fungi common to the Basidiomycota (Boddy and Watkinson 1995). Because of the similarities in the “shrubification” of Arctic and alpine ecosystems with global climate and land use change (Myers-Smith et al. 2011), we may expect similar patterns in fungal communities under alpine shrub expansion scenarios.
Finally, interactions among members within microbial communities has become increasingly recognized as an important determinant of microbial community structure that is often missing from traditional analyses (Wardle 2006, Little et al. 2008, Cordero and Datta 2016). Both negative interactions such as resource competition and chemical antagonism and positive interactions including complementarity in enzyme production can be important drivers of community assembly and spatial aggregation of soil fungi (Gessner et al. 2010, Bell et al. 2013). Further, the Stress Gradient Hypothesis (SGH) is beginning to be applied to microbial interactions in soil communities and proposes that interactions between microbial taxa shift from competitive (negative) to facilitative (positive) as the abiotic stress of the soil environment increases (Callaway et al. 1997, Maestre et al. 2009, Li et al. 2013). Indeed, in biological soil crusts, interactions among microbial species were more neutral to positive in nutrient poor soils but shifted to strongly competitive as nutrient availability increased (Li et al. 2013). In alpine environments, facilitation among plant species in response to severe abiotic conditions is a well-established driver of plant community structure (Anthelme et al. 2014, Cavieres et al. 2016). Soil microbial communities in alpine soils may similarly tend towards positive interactions, however, interactions among microbial taxa are still poorly understood, particularly within natural communities (Bell et al. 2013) . How these interactions may change over abiotic stress gradients and with global change is an important next step in microbial ecology.
Overall, this study aims to 1) determine whether the diversity and community composition of soil fungi vary across elevation gradients and to 2) assess the impact of woody shrub expansion on these patterns. Alpine environments contain steep elevation gradients that offer a unique opportunity to understand how soil organisms respond to variability in both climate and vegetation (Sundqvist et al. 2013). We test three primary hypotheses: (i) Fungal diversity decreases and community composition changes with elevation in alpine soils; (ii) Vegetation more strongly influences fungal diversity and community structure than abiotic soil parameters as soil fungi are closely linked to plant identity; (iii) Interactions among fungal taxa will further shape community structure and positive interactions will dominate negative interactions due to high abiotic stress in alpine soils.
To test these hypotheses we combine observational field data with a manipulative shrub removal experiment along an elevation transect of alpine shrub expansion. We utilize next generation sequencing and joint distribution modeling in a novel way to tease apart effects of the environment and intra-community interactions on soil fungi.
Materials and Methods
Soil Sampling
Soils were sampled in August 2015 in the White Mountains of California, near Crooked Creek (3094 m; 37° 29’ 56” N, 118° 10’ 19” W) and Barcroft (3800 m; 37° 34’ 59” N, 118° 14’ 14” W) research stations. This mountain range runs up the far eastern side of California into Nevada and flanks the western edge of the Great Basin. It has a cold and dry climate receiving 150–450 mm of precipitation annually. Mean annual temperature and precipitation at the two ends of our sampling transect are 0.9 °C and 327mm at Crooked Creek Station and - 1.7 °C and 456 mm at Barcroft Station (Hall 1991). Sampling took place within a transition zone from sub-alpine sagebrush steppe into alpine fellfields dominated by prostrate cushion plants and perennial bunchgrasses. As described by Taylor (1974) and Travers (1993), plant communities here include Artemisia shrubland at low elevations and a mixture of Trifolium andersonii and Carex sp.-Eriogonum ovalifolium communities at high elevations. Artemisia shrubland (below 3657 m elevation) contains seventeen plant species with the three most common including Trifolium andersonii, Leptosiphon nuttallii, and Koeleria macrantha. Trifolium andersonii communities have a very similar species composition to Artemisia shrubland but with only 12 plant species present and no shrubs. Trifolium andersonii and Carex incurviformis are the two most common species. Finally, Carex sp.-Eriogonum ovalifolium communities have very low species diversity and are dominated by Carex incurviformis interspersed with Eriogonum ovalifolium.
We sampled under and outside of sagebrush canopies, and in 1-m2 sagebrush removal plots where shrubs were cut at the base of the stem and trimmed back yearly since 2011 at three elevation sites: 3100, 3500, and 3800 m (however sagebrush removal plots were only at 3100 and 3800 m elevations). This elevation gradient spans the observed sagebrush range expansion from subalpine (<3500 m) to alpine (>3500m) areas over the last 50 years (Kopp and Cleland 2014). In 1961, A. rothrockii was not present at the 3800 m site, and was found in moderate to low densities at the 3500 m site, while the subalpine (3100 m) site had historically high sagebrush cover (Mooney, Andre & Wright 1962; Kopp & Cleland 2014). Therefore, this elevation gradient can be considered a chronosequence, spanning a gradient from historically continuous cover of sagebrush at low elevations to recently established circular patches at high elevations. All sampling locations have granitic soils and east/southeast facing slopes to control for edaphic and aspect variation. Two replicate soil cores (1.3 cm diameter x 10 cm deep) were collected from directly under and outside 5 sagebrush individuals at each elevation site. In addition, two replicate soil cores were taken from five sagebrush removal plots at the low (3100 m) and high (3800 m) elevation sites. For soils characterizing non-shrub communities, cores were taken between 1 and 5 m from the edge of each sagebrush canopy, based on the sagebrush density at each site and distance to the next closest shrub canopy. We aimed to sample at distances outside of the direct influences of the sagebrush species. For shrub removal plots, only aboveground sagebrush biomass was removed in order to prevent significant disturbance to soil structure. Soil was placed in sterile specimen cups and stored at −80 ◦C prior to analysis.
Soil abiotic properties
Volumetric water content (VWC) and pH were measured at the same time and location of each soil core with a Campbell Scientific HS2 Hydrosense II probe (Campbell Scientific, Logan, UT, USA) and an Extech PH100 ExStik pH meter (Extech instruments, Nashua, NH, USA) at 10 cm depth. Total organic carbon and nitrogen (TOC, TON) for each soil sample were calculated using 5 g of field moist soil and 0.5 M K2SO4 extraction through a 1.2 μm glass fiber filter (Thomas C5500, Thomas Scientific, Swedesboro, NJ). Extracts were shipped overnight and analyzed on a Shimadzu TOC-L autoanalyzer (Shimadzu Scientific Instruments, Inc., Carlsbad, CA) at the EcoCore Analytical facility at Colorado State University, Fort Collins, CO. We also measured microbial biomass C and N from the same samples using chloroform fumigation-extraction (Brookes et al 1985). We subtracted unfumigated TOC/TON from paired fumigated samples and divided by the kEC and kEN coefficients of 0.45 and 0.69, respectively (Wu et al. 1990, Joergensen and Mueller 1996).
Molecular analyses
We extracted microbial DNA from 0.25 g of soil (±0.025 g) using a MO BIO Powerlyzer PowerSoil DNA Isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA, USA), and quantified the extracted DNA using a NanoDrop 2000 (Thermo Fisher Scientific Inc., Wilmington, DE, USA). We used modified versions of the universal fungal primers ITS1F and ITS2 described in Smith and Peay (2014) improved as part of the Earth Microbiome Project (Walters et al. 2015). While currently considered the most accurate for species identification of fungi, these primers do have certain limitations, particularly low resolution for arbuscular mycorrhizal fungi (Glomeromycota) (Schoch et al. 2012, Öpik et al. 2014) and poor mapping to phylogeny (Yarza et al. 2017).
PCR
We performed PCR amplification in 25 μl reactions including 1 μl of 10 μM for each primer (forward and reverse), 1 μl DNA, 12.5 μl of Taq 2X Master Mix (New England Biolabs), and 9.5 μl diH20. Thermocycler settings were 94⁰C for 3 minutes, followed by 35 cycles of 94⁰C for 45 seconds, 50⁰C for 60 seconds, and 72⁰C for 90 seconds, followed by 72⁰C for 10 minutes. Forward primers contained unique 12-base Golay barcodes as described in Walters et al. (2015); (see also Hamady et al. 2008). We then did PCR clean-up using a Nucleo Spin Gel-Extraction kit (Macherey-Nagel GmbH & Co. KG). Purified samples were pooled in equimolar concentrations and sequenced in a multiplexed 2- x 150-bp paired-end sequencing run on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) at the University of California Riverside (UCR) Genomics Core Facility.
Bioinformatics
Sequences were demultiplexed and processed with the split_libraries_fastq.py from QIIME 1.9.1 (Caporaso et al. 2010). Samples with less than 10,000 reads were removed. OTUs were generated from the forward ITS1 reads using open reference OTU assignment implemented in pick_open_reference_otus.py (QIIME 1.9.1) using UCLUST (Edgar 2010) and comparing to the v7 (ver7_dynamic_s_22.08.2016) database of UNITE (Kõljalg et al. 2005). OTUs were assigned taxonomy using QIIME BLAST at 97% similarity as defined by the UNITE v7 database. BIOM table files were generated from OTU tables and diversity analyses performed with QIIME core_diversity_analyses.py script where samples were rarefied to 10,000 sequences per sample. Samples were also normalized using cumulative sum scaling (CSS) in the Metagenomeseq (Bioconductor) package in R for analyses of differential abundance of taxa (HMSC analyses; see below).
Statistical analyses
Alpha and Beta Diversity were calculated using outputs from the core_diversity_analyses.py function in QIIME v1.9.1. For alpha diversity, we used both the number of observed OTUs (richness) and the Chao1 diversity metric for each sample. We used linear mixed effects models to test the relationship between alpha diversity (chao1 diversity and OTU richness) and elevation, vegetation type, and abiotic soil parameters. For abiotic models, Only TOC was used as for soil nutrients, as TOC and TON were correlated across samples (r=0.56, p<0.001). All above predictors were included as fixed effects while core replicate pair was included as a random effect of sampling location within sites. Models were fit using the ‘lmer’ function in the lme4 package in R (R core development team 2013). Models were assessed individually for significance of parameters and pairwise comparisons were run on significant predictors using a Tukey’s post hoc test in the ‘glht’ function of the multcomp package in R. In addition, because many abiotic variables co-vary with both elevation and vegetation type, we used a model selection approach for these parameters, assessing delta AIC of partial and full models via the ‘AICtab’ function in the bbmle package in R. Models are described in Table 1.
Table 1.
Model structure for alpha and beta diversity analyses. Linear mixed effects models were used for alpha diversity (chao1) and richness (observed_otus) via the function ‘lmer’. Elevation, vegetation type, their interaction, and abiotic soil variables were considered fixed effects and core replicate pair (PLOTID) was included as a random effect of sampling location within sites. Models are listed in order of best fit using delta AIC. Beta diversity (community composition) was assessed using non-metric multidimensional scaling (NMDS) of the Bray-Curtis dissimilarity metric (Bray) and Permutational multivariate analysis of variance (perMANOVA) via the function ‘adonis’. Elevation, vegetation type, their interaction were used as predictor variables. For abiotic models, elevation was used as a blocking variable (strata) to restrict permutations to within sites, and the relative influence of abiotic parameters vs. vegetation type was assessed using an interaction term. Models are listed in order of best fit (R2 values).
| Alpha diversity |
∆ AIC | R2 | df | |
|---|---|---|---|---|
| Full model | lmer(chao1~soil.pH + VWC + TOC + Vegetation*Elevation + (1∣PLOTID) | 0 | 0.20 | 13 |
| lmer(observed_otus ~ soil.pH + VWC + TOC + Vegetation*Elevation + (1∣PLOTID) | 0 | 0.273 | 13 | |
| Interaction | lmer(chao1 ~ Vegetation*Elevation + (1∣PLOTID); | 22.1 | 0.167 | 10 |
| lmer(observed_otus ~ Vegetation*Elevation + (1∣PLOTID) | 19.1 | 0.254 | 10 | |
| Abiotic variables | lmer(chao1~soil.pH + VWC + TOC + (1∣PLOTID) | 61.1 | 0.152 | 6 |
| lmer(observed_otus ~ soil.pH + VWC + TOC + (1∣PLOTID) | 57.9 | 0.202 | 6 | |
| Elevation | lmer(observed_otus ~ Elevation + (1∣PLOTID) | 67.2 | 0.088 | 5 |
| lmer(observed_otus ~ Elevation + (1∣PLOTID) | 62.5 | 0.154 | 5 | |
| Vegetation type | lmer(chao1 ~ Vegetation + (1∣PLOTID); | 70.4 | 0.026 | 5 |
| lmer(observed_otus ~ Vegetation + (1∣PLOTID) | 68.5 | 0.038 | 5 | |
| Beta diversity |
R2 | df | ||
| Elevation | adonis(Bray ~ Elevation) | 0.126 | 2 | |
| Vegetation type | adonis(Bray ~ Vegetation) | 0.087 | 2 | |
| Interaction | adonis(Bray ~ Elevation*Vegetation) | 0.081 | 3 | |
| Abiotic variables | adonis(Bray ~ VWC*Vegetation, strata= Elevation) | 0.07 (0.054-int) |
1 2 |
|
| adonis(Bray ~ TOC*Vegetation, strata= Elevation) | 0.047 (0.046-int) |
1 2 |
||
| adonis(Bray ~ soil.pH*Vegetation, strata= Elevation) | 0.022 (0.038-int) |
1 2 |
Beta diversity (community composition) was assessed using non-metric multidimensional scaling (NMDS) of the Bray-Curtis dissimilarity metric and Permutational multivariate analysis of variance (perMANOVA) in the vegan function ‘adonis’ in R (999 permutations; Oksanen et al. 2016). Vegetation type, elevation, and their interaction were included as predictor variables and checked for within group heterogeneity using the vegan functions ‘betadisper’ and ‘permutest.’ Additionally, abiotic parameters (TOC, pH, VWC) were tested in separate models and checked for heteroscedascity of predictors using a Breush-Pagan test. Elevation was used as a blocking variable (strata) to restrict permutations to within sites, and the relative influence of abiotic parameters vs. vegetation type was assessed using an interaction term (Table 1).
Joint Distribution Models
CSS normalized read abundance of fungal OTUs at different taxonomic levels were analyzed using multivariate, joint distribution models (HMSC package in R; Ovaskainen et al. 2017). This approach uses a hierarchical Bayesian framework to fit a joint distribution model to occurrence and/or abundance data from multi-species communities. This approach is increasingly favored for analyzing plant and animal community data but is just beginning to be used for microbial community (sequencing) data (Aivelo and Norberg 2017). The primary motivation for these models is to simultaneously quantify the importance of environmental filtering (abiotic factors), biotic filtering (species interactions), and neutral processes (random effects) for shaping species distributions and structuring communities (Ovaskainen et al. 2017). Specifically, these models estimate fixed effects of environmental covariates, positive and negative species associations via a covariance matrix, and random effects based on study design.
We ran these community models using CSS-normalized read abundance data aggregated at the fungal class, order, and family levels. We included elevation, vegetation type, soil pH, VWC, TOC, TON, and Microbial biomass C and N as fixed effects, and specified soil core replicate and sampling location (“block”) as random effects. We used the default (flat) priors and Gibbs sampler as described in the supporting information of Ovaskainen et al. 2017 and ran models with a Gaussian distribution. MCMC chains were run for 10,000 iterations with the first 1,000 discarded and the remainder thinned for a total of 900 posterior samples. We checked for model convergence using visual assessment of trace plots and used the posterior distributions of each environmental covariate to calculate the probability that it was different from zero. We considered parameters to be “significant” when their posterior probabilities had a greater than 90% probability of being different from zero (p < 0.1). We calculated the relative proportion of the total model variance that could be attributed to each of our fixed and random effects using the ‘variPart’ function in the HMSC package. Finally, we estimated residual taxa associations using the “corRandomEff” function, which calculates pairwise correlation (r) matrices for all taxa. These associations represent the positive or negative associations among taxa after having accounted for the environmental effects, and may be influenced by both direct interactions among taxa and also common responses to unmeasured environmental variables.
Results
Molecular sequencing
Sequencing of soil fungal communities via the ITS1F and ITS2 yielded 1,590,851 total sequences and an average sequencing depth of 28,924 reads. Overall, these sequences made up 12 phyla, with Ascomycota making up the largest percentage (59.7%), followed by Basidiomycota (20.8%), unidentified fungi (8.5%), Zygomycota (2.9%), Chytridomycota (0.3%), Glomeromycota (0.3%), 0.5% Protists (Cercozoa) and Microsporidia-like organisms (Rozellomycota) (0.5%). Seven percent of the total sequences had no blast hit so taxonomy could not be assigned.
Alpha diversity
Fungal diversity and OTU richness decreased significantly with elevation, however elevation effects on richness were stronger than diversity (diversity: df=30.39, t-value= −2.063, p=0.047; richness: df=32.76, t-value= −2.555, p=0.015). The low elevation (3200 m) site had significantly higher richness than both the middle (3500m) and high (3800m) sites, while the latter two sites were not significantly different from each other (3500 vs. 3200: z value −2.531, p=0.031; 3800 vs. 3200: z value −2.555, p=0.028, Fig.1) and Chao1 diversity was slightly lower at the high vs. low elevation site (3800 vs. 3200: z-value= −2.063, p=0.097).
Fig. 1.
Alpha diversity results for the effect of vegetation type (OS=shrub interspace, S=shrub, SR= shrub removal) OTU richness for alpine soil fungal communities at three elevation sites (3200m, 3500m, and 3800m asl). Capital letters denote significant differences between elevation sites while lower case letters denote significant differences between vegetation types within a site and for the same vegetation type across sites.
Vegetation type (shrub, outside shrub, shrub removal) influenced fungal richness and diversity most at the high elevation site, where shrub soils had overall lower richness (df=22.97, t-value= −3.310, p=0.003, Fig. 1) and diversity (df=21.610, t-value= −2.688, p=0.013) and lower richness than shrub removal soils (z-value= −2.345, p=0.049). Within vegetation types, shrub soils at the high elevation site had lower richness than shrub soils at the low elevation site (z-value= −2.455, p=0.037, Fig. 1) and shrub interspace soils at the middle elevation site had lower richness than shrub interspace soils at the low elevation site (z-value= −2.380, p=0.045, Fig. 1).
For abiotic predictors, the full model incorporating VWC, TOC, and soil pH with elevation and vegetation type was the strongest model for both diversity and richness and was significantly better than the elevation x vegetation only model (∆ AIC: 22.1 diversity, 19.1 richness, Table 1). Additionally, models incorporating all abiotic parameters were significantly better than both elevation and vegetation type only models for diversity and richness (∆ AIC>>2, Table 1). No single abiotic parameter was a significant predictor of alpha diversity alone.
Beta diversity-community composition
Fungal community composition varied significantly across the elevation gradient. Beta diversity (bray Curtis dissimilarity) varied by vegetation type, elevation and their interaction (vegetation: df=2, F=2.92, p= 0.001, R2=0.087; elevation: df=2, F=4.19, p=0.001, R2=0.125, interaction: df=3, F=1.79, p=0.001, R2=0.81 respectively; Fig. 2). Across vegetation types shrub soil community composition was different from both interspace soil (p=0.001) and shrub removal soils (p=0.001), however shrub removal soil was not different than shrub interspace soils. Across elevations, low elevation soils differed from both middle (p=0.001) and high (p=0.001) elevation soils, and middle soils also differed from high soils (p=0.001)
Fig. 2.
Non-metric multidimensonal scaling (NMDS) of community dissimilarity (Bray-Curtis) of soil fungi (Stress value=0.145). Each point corresponds to a soil sample collected from one of three vegetation types (shape) or elevation sites (color). Points which are close together signify samples with similar fungal community compsoition. Colored ovals represent 95% confidence intervals of sample ordination grouped by elevation.
For abiotic drivers, VWC and TOC were significant predictors of fungal community composition (VWC: df=1 F=4.379,p= 0.001 R2=0.0703, TOC: df=1, F=2.755,p=0.001 R2=0.0467) while soil pH was not. In addition, both VWC and TOC had significant interactions with vegetation type on beta diversity, but the effect of vegetation type was stronger than either abiotic variable (VWC int: df=2 F=1.686, p= 0.004 R2=0.0541, TOC int: df=1, F=2.755,p=0.001 R2=0.0467)
Joint Distribution modeling
Joint distribution models through the HMSC framework provided information on the relative abundance of different fungal taxa in soils across our elevation and shrub expansion gradient. At the class level, relative abundance of different fungal taxa did not differ across elevation or vegetation types. At finer scales, the order Phyllachorales, a group commonly known to be foliar parasites (Silva-Hanlin and Hanlin 1998), were more abundant in shrub interspace soils (p=0.098). The order Rhizophlyctidales, a soil inhabiting, cellulose-degrading chytrid (Letcher 2008) was more abundant in shrub soils (p=0.0825). The corresponding families Phyllachoraceae and Rhizophlyctidaceae were also more abundant in interspace and shrub soils, respectively (p=0.11, 0.102) although these probability estimates were slightly higher than our proposed cutoff (Table 2, S1).
Table 2.
Description of significant fungal families from the HMSC analysis and relevant citations.
| Fungal family | Larger taxonomic group |
Increased relative abundance in |
Known function | Citation |
|---|---|---|---|---|
| Pucciniaceae | Basidiomycota (Pucciniomycetes) | Shrub interspace | Rust pathogen | (James et al. 2006) |
| Phyllachoraceae | Ascomycota (Sordariomycetes) | Shrub interspace | Foliar parasite | (Silva-Hanlin and Hanlin 1998) |
| Rhizophlyctidaceae | Chytridiomycota (Chytridiomycetes) | Shrub | Cellulose degradation | (Letcher et al. 2008) |
| Pluteaceae | Basidiomycota (Agaricomycetes) | Shrub | Saprotroph, litter decomposition | (Justo et al. 2011) |
| Lachnocladiaceae | Basidiomycota (Agaricomycetes) | Shrub removal | Wood decomposition | (Cannon and Kirk 2007) |
| Auriscalpiaceae | Basidiomycota (Agaricomycetes) | Shrub removal | Saprotrophic, wood decomposition | (Larsson and Larsson 2003) |
| Thelotremataceae | Ascomycota (Lecanoromycetes) | Shrub removal | Lichenized | (Mangold et al. 2008) |
| Botryobasidiaceae | Basidiomycota (Agaricomycetes) | High elevation | Wood, litter decomposition | (Larsson 2007) |
Family level models also revealed that the Puccinaceae, a Basidiomycete rust fungal pathogen, and the more abundant in shrub interspace soils (p=0.07), while the Pluteaceae, a family in the Agaricomycota closely related to Amanita, were more abundant in shrub soils (p=0.095). In shrub removal soils, both Lachnocladiaceae and Auriscalpiaceae, two families in the Russulales order of Agaricomycota, had higher relative abundance (p=0.1, 0.08), as well as the Thelotremataceae, a lichenized Pezizomycotina (p=0.07) (Table 2, S1).
Across elevation, there was an increase in relative abundance for the family Botryobasidiaceae, an Agaricomycete of the Cantharellales order (p=0.054). In addition, this family and closely related Cantharellales (family incertae sedis) increased in relative abundance with higher pH (p=0.05, 0.077) (Table 2, S1).
Variance partitioning
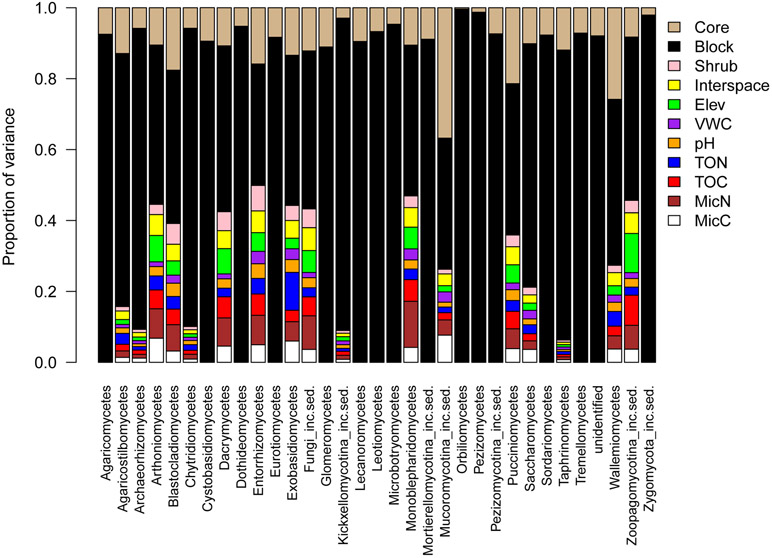
Variance partitioning of the relative abundance of different fungal classes revealed that random effects including sampling location (block) and core replicate (core) explained the majority of variation in the data. Block explained between 34–99% of the variation dependent on fungal class with an average of 74%, while core explained between 0.4%−37% of the variance with an average of 10% (Fig. 3, Table S2). Overall, fixed effects including elevation, vegetation type, and biotic and abiotic soil parameters explained between 1–50% of total variation (Fig. 3, Table S2). Vegetation type explained ~3.6% of the variation (shrub and shrub interspace only), while elevation explained ~2%. Other biotic and abiotic soil parameters explained on average 2% of the variation in the data, with Microbial biomass N explaining the most (~2.7%) and Volumetric water content (VWC) explaining the least (~1%).
Fig. 3.
Results of variance partitioning for the variation in fungal relative abundance (at the class level) in response to vegetation type (shrub and interspace), elevation, soil pH, VWC, TOC, TON, and Microbial biomass C and N. Core replicate as well as sampling location (“block”) were included as random effects.
Taxa associations
After accounting for fixed effects, there were significant positive and negative associations among individual fungal taxa. At a correlation (r) level of +/− 0.3 or greater, 25 fungal orders showed varying positive and negative relationships (Fig. 4). Out of these 25, 10 fungal orders had correlations (r) of +/− 0.4 or greater. Specifically, the Wallemiales order was negatively correlated with four other taxa including Pezizomycotina (inc. sedis), Mytilindiales, Hymenochetales, and Arthoniales, as well as positively correlated with Myriangiales and Amylocorticales. The Pezizomycotina (inc. sedis) were positively correlated with four other taxa including Mytilinidiales, Diversisporales, Coniochaetales, and Agaricostilbales (Fig. 4, Table S3).
Fig. 4.
Estimates of associations among fungal taxa (at the order level) based on residual correlations after accounting for fixed predictors. Taxa have a positive (red), negative (blue), or neutral (white) association with other taxa and the strength of this association (Pearson correlation coefficient, r) is depicted by the shade of the color. Shown here are the subset of fungal orders (25 total) that had at least one association that was greater than or less than +/− 0.3. 85 fungal orders did not have any associations meeting this criterion.
Discussion
In this study, we assessed how elevational patterns in soil fungal diversity and community composition are altered by global change driven shrub expansion in an alpine environment. We found at least partial support for our three hypotheses. First, we observed that fungal diversity declined and community composition shifted with elevation as has been demonstrated in other alpine research. Next, both vegetation type and abiotic soil parameters were important predictors of fungal alpha diversity and community composition. Vegetation type however was a stronger predictor of beta diversity, explaining more variation than any abiotic parameter. Finally, we found both positive and negative associations among fungal taxa after controlling for environmental covariates, and positive interactions were more common, implying that facilitation, and to a lesser degree competition, may mediate how fungal communities are structured and adapt to abiotic stress in alpine soils. Understaing how soil fungal communities respond to global change, both directly through abiotic controls, and indirectly through plant species range shifts, will be critical as alpine ecosystems continue to undergo rapid warming and land use changes.
Alpha and Beta diversity
Fungal diversity, including the Chao1 index and OTU richness, declined with increasing elevation, exhibiting the strongest decline from the subalpine (3200m) to alpine sites (3500, 3800m). Beta diversity (community composition) was also distinct among elevations, and high elevation soils had less variation in community composition compared to low elevation sites (reflected in the width of respective circles in the NMDS plots, Fig. 2). These results are in accordance with previous alpine research highlighting that fungal diversity declines with increasing elevation (Körner 2003, Sundqvist et al. 2013). Elevation, in and of itself, is not a mechanistic driver (Körner 2007), but is nonetheless useful in determining large scale patterns in fungal communities in response to biotic and abiotic factors. Instead, a decline in plant species diversity and biomass at high elevations is a proposed mechanism for changing fungal diversity across elevation gradients (Tedersoo et al. 2014). Indeed, the plant community at our study sites changes from more speciose subalpine Artemisia shrubland at low elevations to lower diversity alpine grasslands at high elevations, including the Carex sp.-Eriogonum ovalifolium and Trifolium andersonii vegetation communities (Taylor 1976, Travers 1993). In a previous study at this location, we observed that Shannon diversity was unaffected and OTU richness slightly increased with elevation for bacteria and archaea (Collins et al. 2016), signifying very different controls over bacterial and fungal alpha diversity across elevation gradients.
Vegetation type and abiotic soil properties also influenced fungal diversity and community composition, and their relative importance differed for alpha and beta diversity. Shrub soils at high elevation sites had lower richness than shrub soils at the low elevation site; at the high elevation site, shrub soils had the lowest OTU richness of all three vegetation types (although not significantly lower than shrub interspace soils). This supports the idea that fungal diversity at high elevation sites declines even more as shrubs move into alpine areas. Additionally, shrub removal soils had higher alpha diversity than shrub soils, and equally high diversity as shrub interspace soils at the high elevation site, demonstrating that alpine fungal communities can shift rapidly in response to changes in plant communities.
Among abiotic variables, VWC, TOC, and soil pH all influenced alpha diversity in combination, although no abiotic parameters were significant predictors alone. This was confirmed by our model selection approach in which the best model incorporated abiotic parameters, elevation and vegetation type. This model was a significant improvement to the elevation by vegetation interaction model, implying that the combined abiotic conditions of the soil environment directly influence fungal community diversity in addition to their influence via elevation and plant community shifts.
Fungal beta diversity was also regulated by VWC in addition to SOC, but VWC was a stronger predictor, explaining a higher proportion of model variation (7% vs. 4.7%). VWC significantly increases with elevation along our transect, primarily due to increased precipitation as snow at high elevations, therefore soil moisture likely plays a key role in the observed diversity-elevation trend in soil fungi. Hawkes et al. (2011) found decreases in fungal diversity with increased precipitation in a rainfall manipulation experiment in California grasslands. We see similar results for both alpha and beta diversity in that low elevation sites were more taxonomically diverse within and across sampling locations. Because this is a dry alpine ecosystem, drought is common, particularly in subalpine sites with low annual precipitation. This abiotic stress may ameliorate competition and promote stable coexistence among different fungal groups, thereby increasing overall taxonomic diversity and diversity across the landscape (Hawkes et al. 2011). Another potential mechanism is that dry soil increases heterogeneity of the soil matrix via decreased connectivity among soil pores. This may increase resource hotspots and diversity of niches, promoting more distinct fungal communities across sites (Frey 2007, Classen et al. 2015).
There is reasonable evidence that fungi are more closely associated with plant species identity than other microbial groups, particularly prokaryotes, which are predominantly regulated by abiotic soil properties (Nielsen et al. 2010, Cassman et al. 2016). This is likely due to the major role that soil fungi play in plant litter decomposition, especially because fungi produce lignin-degrading enzymes absent in most prokaryotes (Hammel 1997, De Boer et al. 2005, Thorn and Lynch 2007). Our data partially supported this hypothesis, with vegetation type being a stronger predictor (higher R2) of fungal community composition than any abiotic factor alone. Abiotic soil parameters including soil moisture and organic nutrients (but not pH) also interacted with vegetation type in their influence on fungal community structure, suggesting indirect effects of plants on soil fungi via shifts in the soil environment. In a previous study, we found that both soil moisture and TOC/TON are enhanced in soils below sagebrush canopies and that this indirectly affects soil bacterial diversity and richness (Collins et al. 2016). Trends in fungal diversity and community composition in suggest that shrub expansion may affect community structure through shifts in soil organic nutrient pools, likely resulting from the accumulation of low quality woody litter. In addition, enhanced soil moisture below shrub canopies may further promote decomposition of soil organic matter and thereby impacting fungal community composition in this arid environment. In the same way, alpine cushion plants influence soil fungal communities via enhanced soil moisture and nutrients, as well as buffering changes in soil pH, which then has a stronger influence on fungal communities in adjacent open soils (Roy et al. 2013). We similarly found that soil pH played a smaller role than water and nutrients in determining fungal community composition, which may reflect a strong influence of plant communities on the abiotic conditions for soil fungi.
Joint modeling of fungal communities
The amount of taxa-specific distribution data generated from sequencing, combined with the joint distribution modeling approach, offer remarkable new potential to understand what controls the distributions of soil organisms. In particular, we can begin assembling a unique understanding of the relative importance of environmental variation, species interactions, and random spatial assembly processes for determining belowground communities.
This approach showed that the relative abundance of particular fungal taxa differed among vegetation types and elevations, and that there are significant residual associations (positive and negative) among many taxa. The trends varied by classification level (i.e. class vs. order vs. family). At the class level, no significant trends were detected, however taxa at the order level, and most strongly, taxa at the family level had increased relative abundance in soils across elevation and vegetation types. Our ability to detect stronger trends at finer taxonomic scales posits that these broader groups (class, order) contain taxa with distinct environmental responses (Lu et al. 2016) and therefore are not ecologically equivalent. This is likely most relevant for very large classes with many fungal families, such as the Agaricomycetes or Sordariomycetes, which was corroborated in our variance partitioning analysis (below).
Relative abundance of fungal taxa across elevation and vegetation types presented several trends. First, Agaricomycetes and close relatives had higher relative abundance in shrub, shrub removal and high elevation soils. Agaricomycetes are commonly saprotrophic, wood or litter decaying fungi (Lyncht and Thorn 2006, Zak et al. 2011) and also include mycorrhizal species. They are important decomposers in cold, dry environments, as has been shown in arctic studies (Ludley and Robinson 2008) and are dominant in forest floor communities (Edwards and Zak 2010). The increased relative abundance of these fungal groups may result from increased woody litter accumulation from shrubs, both above and belowground, may provide important substrate for decomposer fungi, in particular at high elevation sites. This is especially relevant in shrub removal soils as root systems decompose gradually after aboveground sagebrush removal. Shrub removal therefore is likely to promote an initial proliferation in wood decay fungi which will decline over time. By sampling 4 years after shrub removal, we were able to characterize how fungal communities may recover over time after disturbance.
Next, shrub interspace soils had increased relative abundance of two pathogenic fungal families, the Pucciniaceae, a known plant pathogen of rust fungi and the Phyllachoraceae, an Ascomycete family of mostly foliar parasites. Higher relative abundance of pathotrophs in shrub interspace soils was consistent across elevations. Members of the family Pucciniaceae are particularly strong plant pathogens which are commonly used as bio-control for agricultural weeds (Stubbs and Kennedy 2012). Because shrub interspace plant communities have been historically present in alpine environments, species-specific soil pathogens may have developed over time in the rhizosphere of these plants but have not yet accumulated underneath the newly-arrived shrubs (Colautti et al. 2004, Diez et al. 2010).
In addition, we observed increased relative abundance of a cellulose degrading chytrid (Rhizophlyctidaceae) in shrub soils and lichenized Pezizomycotina (Thelotremataceae) in shrub removal soils. The high relative abundance of Rhizophlyctidaceae suggests that shrub soils provide substrates that promote saprotrophic decomposer taxa such as these cellulose-degrading, soil-inhabiting chytrids (Letcher et al. 2008). Additionally, the Thelotremataceae is a large family in the Lecanoromycetes, known to form soil crusts on bare soil surfaces. Sagebrush removal led to high levels of newly exposed soil, which is ideal for lichen establishment. Zumsteg et al. (2012) found this group to be an important colonizer of barren substrate after glacial retreat, revealing its opportunistic life strategy and tolerance of cold, dry environments.
Interpretation of joint distribution model results need to be made cautiously however, as read abundances of fungal OTUs are normalized relative to the sequence count within a given sample. While CSS-normalized read abundances account for several common issues including amplification biases and under-sampling (Paulson et al. 2013) any attempt to estimate true biological abundance from sequence read abundance is imperfect (Weiss et al. 2017). Nonetheless, we use and interpret these differential abundance data in a manner consistent with other recent analyses of the environmental effects on microbial composition across samples (Ghanbari et al. 2017, Timonen et al. 2017) .
Variance partitioning
Despite the significant effects of vegetation and elevation on fungal communities, variance partitioning revealed that sampling location “block” was a substantially better predictor of the relative abundance of fungal groups than any measured environmental covariate. In addition, replicate core pair was the second best predictor of relative group abundance, suggesting that the particular spatial location within the landscape is more influential than abiotic soil properties, plant community, or elevation. These results propose that there is remarkable heterogeneity in the relative abundance of fungal taxa at the landscape scale, which may be related to both small microsite variation in environmental variables and processes such as dispersal limitation and priority effects. Feinstein and Blackwood (2012) found high spatial variation in forest floor fungal communities and little explanatory power of plant traits or plant species identity. Rather, neutral models (zero-sum) had the highest predictive power for species abundance and distribution, indicating the critical role of neutral processes in community assembly of saprotrophic fungi. This parallels observed patterns at a global scale, where community composition of soil fungi is highly variable, with often very few shared OTUs across geographic regions (Meiser et al. 2014). Nonetheless, fixed effects explained up to half of the total variation in relative abundance for some taxa, so it appears that the relative importance of environmental and stochastic effects may vary among taxa. We found stronger environmental effects for the more narrow or smaller taxonomic groups, such as the Entorrhizomycetes, a fungal class with a single order and family (Fig. 3), in which individual members are likely to have more conserved environmental responses (Lu et al. 2016).
Taxa associations
After controlling for direct responses to environmental variation, many significant positive and negative associations remained among taxa, reflecting either important interactions among fungal groups or common responses to unmeasured environmental variables (Clark et al. 2014, Ovaskainen et al. 2017). As hypothesized, interactions tended to be positive rather than negative (Fig. 4), proposing the importance of facilitative interactions among taxa in this stressful alpine environment. Facilitative interactions among fungi are common during decomposition, including the process by which some taxa break down complex plant tissues (lignin, cellulose) into simpler forms which in turn are decomposed by other taxa (Gessner et al. 2010). For example, in our study the Pezizomycotina (inc. sedis) tended to have positive associations with other fungal orders. Pezizomycotina are among the most abundant and diverse group of ascomycete fungi in forest floor communities (Edwards and Zak 2011), and proliferate during and directly after peak plant biomass in alpine soils (Zinger et al. 2009) offering a potentially important facilitative role for this group of saprotrophic soil fungi (Damon et al. 2010). Although less common, we observed negative interactions among several fungal orders, in particular, the Wallemiales had primarily negative associations with other orders. This small group is comprised of highly xerophillic Basidiomycete fungi (Zalar et al. 2005). Surviving in very dry environments is a rare trait for Basidiomycota, and establishment of these hyphal forming fungi may prevent colonization by other more common xerophillic taxa, in particular ascomycetes. Colonization of a substrate (e.g. leaf, piece of wood) by saprophytic fungal taxa may prevent other fungi from utilitzing that same substrate, highlighting a negative (competitive) interaction between taxa within a trophic guild (Wardle 2006). Certainly priority effects of colonizing fungal taxa are influential in structuring subsequent community composition in saprotrophic wood rot communities either via direct spatial exclusion or alteration of resource pools (Hiscox et al. 2015, Maynard et al. 2017). Release of microbial antibiotics or allelochemicals into the surrounding soil matrix is another example of such interaction, common for lichens in particular (Stark et al. 2007). Thus, these results suggest that both positive and negative interactions among taxa may help regulate community structure, and positive interactions may help to buffer abiotic stress for soil fungi in this ecosystem. The underlying causes of these associations will remain uncertain using observational data, but regardless can prove useful in developing further hypotheses about interactions among specific taxa that may then be experimentally tested.
Comparison across microbial groups
Because soil bacterial and fungal communities are intricately linked and play synergistic roles in decomposition (De Boer et al. 2005) and well as interactions with plants in the rhizosphere (Artursson et al. 2006) , it important to know how our results compare to other microbial groups, particularly bacteria. Our previous work in this alpine ecosystem has shown that bacterial diversity and community composition are weakly linked to elevation, and that shrub expansion increases bacterial alpha diversity. Shrub expansion also altered bacterial community composition indirectly by causing shifts in abiotic soil parameters including soil moisture and organic nutrients. In addition, pH was a strong driver of bacterial community structure, as has been shown in other research (Lauber et al. 2009, Siles and Margesin 2016) although was not linked to elevation or vegetation type in our system (Collins et al. 2016). This contrasts to the patterns observed in fungal communities in that elevation was a much stronger predictor of alpha diversity and that shrub expansion promoted a decrease in fungal diversity, particularly at high elevation sites. Additionally, unlike for bacteria, pH was a relatively unimportant abiotic driver for soil fungi, however interactions between vegetation type and soil water and nutrients did similarly influence fungal community structure. Finally, both bacterial and fungal communities in this ecosystem showed remarkable community resilience and were able to revert back to similar levels of diversity and community composition after 4 years of shrub removal (Collins et al. 2016).
In other ecosystems, comparisons of soil bacterial and fungal communities across elevation gradients are likewise complex. Siles and Margesin (2016) observed that bacterial diversity decreased from submontane to alpine sites while fungal diversity did not change but the relative abundance of soil fungi increased. Across two sufbalpine mountain transects in China, bacterial diversity peaked at mid elevations rather than at either end of the climatic gradient and that differences in relative abundance of taxa across the transect were much stronger for bacteria than fungi (Meng et al. 2013, Ren et al. 2018). Due to inconsistencies across studies, it has been argued that bacteria simply do not exhibit the elevation-diversity patterns present in other Eukaryotic organisms (Fierer 2011), however high spatial heterogeneity within soil sampling locations as well as low sampling intensity within transects may obscure the ability to detect trends across larger elevation gradients (Rowe and Lidgard 2009, Nottingham et al. 2016). We found similarly in this study, that sampling location and spatial heterogeneity across the landscape were dominant drivers of soil fungal community structure and composition, and it is likely that increased sampling intensity within could help explain a larger proportion of the variation in these communities.
Although not examined in this study, seasonal fluctuations are another important driver of soil microbial community structure and relative abundance of taxa in alpine environments (Lazzaro et al. 2015) . While elevation and vegetation type have significant influence on abiotic soil properties, seasonal fluctuations in resources can be equally important or stronger predictors of microbial community composition (Shahnavaz et al. 2012, Lazzaro et al. 2015). Further, bacteria and fungi respond very differently to seasonal events including snowpack, snowmelt and peak growing season (Zinger et al. 2009, Lazzaro et al. 2015). In general, annual cycles of biomass, diversity, and turnover of particular taxa are more pronounced for bacteria than fungi, as fungi tend to be more cold tolerant, and can utilize more recalcitrant plant compounds to maintain their biomass under winter snowpack (Zinger et al. 2009, Lazzaro et al. 2015). As inter-annual seasonal variability increases with climate change (Nicholls and Alexander 2007) these annual cycles may become much less predictable, increasing our need to understand the mechanisms driving diversity and biogeographic patterns of alpine soil microbial communities.
Conclusion
Overall, we found support for our hypothesis that soil fungal diversity declines and community composition changes with increasing elevation. In addition, both abiotic factors (particularly soil moisture and soil organic C and N) and woody sagebrush range expansion had significant effects on these patterns. In the context of global change, it is particularly striking that the negative effect of shrubs on alpha diversity was strongest in high elevation sites where shrubs have only recently colonized. However fungal communities displayed a relatively rapid ability to recover this diversity after 4 years of shrub removal. Moreover, the increased relative abundance of saprotrophic Agaricomycete fungi at high elevations portends ongoing changes to soil community function as shrubs continue moving uphill. Nevertheless, while fungal diversity and distribution were significantly affected by vegetation type, elevation, and abiotic conditions, the residual spatial variation overwhelmed these fixed effects, highlighting the extreme heterogeneity in fungal communities at the landscape scale. Finally, positive and negative associations between fungal taxa may be important in structuring community responses to environmental change, particularly facilitative interactions in alpine environments. These within-community interactions are difficult to quantify and typically absent in studies of microbial biogeography (Martiny et al. 2006, Kivlin et al. 2011). As more studies integrate sequencing data, manipulative experiments, and joint distribution models, we may test more general hypotheses about the nature and importance of these associations in a global change context.
Supplementary Material
Acknowledgements
We thank Derreck Carter-House for assistance with laboratory analyses and sequencing prep, Christopher Kopp for the use of Sagebrush removal plots for soil sampling and Teresa Bohner for statistical advice and feedback on the manuscript. Funding was provided by the University of California Riverside, Department of Botany and Plant Sciences, and Department of Plant Pathology and Microbiology as well as from a White Mountain Research Center mini-grant and a graduate fellowship from the University of California’s Institute for the Study of Ecological and Evolutionary Climate Impacts (ISEECI), funded by a Presidential Research Catalyst Award. Fungal ITS primers were made available through the Sloan Built Environment Program and sequencing supported by funds through United States Department of Agriculture - National Institute of Food and Agriculture Hatch project CA-R-PPA-5062-H to Jason Stajich. This work was also supported by a National Science Foundation Doctoral Dissertation Improvement Grant (DDIG) Award Number 1701979 to Courtney Collins, and Nuttapon Pombubpa was supported by a Royal Thai government fellowship.
Footnotes
Data Accessibility
Raw data for all nonmolecular analyses including sampling location, soil nutrient, moisture, pH and microbial biomass may be found in the Dryad repository https://doi.org/10.5061/dryad.rh61t5g. Raw sequences may be found in the NCBI Short Read Archive (SRA) Accession no. SRP133697 and BioProject PRJNA434743.
References
- Aivelo T, and Norberg A. 2017. Parasite-microbiota interactions potentially affect intestinal communities in wild mammals. Journal of Animal Ecology:1–10. [DOI] [PubMed] [Google Scholar]
- Anthelme F, Cavieres LA, and Dangles O. 2014. Facilitation among plants in alpine environments in the face of climate change. Frontiers in Plant Science 5:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S, Boutton TW, and Hibbard KA. 2001. Trees in grasslands: Biogeochemical consequences of woody plant expansion. Pages 115–137 Global Biogeochemical Cycles in the Climate System. [Google Scholar]
- Artursson V, Finlay RD, and Jansson JK. 2006. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environmental Microbiology 8:1–10. [DOI] [PubMed] [Google Scholar]
- Bahram M, Põlme S, Kõljalg U, Zarre S, and Tedersoo L. 2012. Regional and local patterns of ectomycorrhizal fungal diversity and community structure along an altitudinal gradient in the Hyrcanian forests of northern Iran. New Phytologist 193:465–473. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, Hopkins DW, Usher MB, and British Ecological Society. 2005. Biological diversity and function in soils. Cambridge University Press. [Google Scholar]
- Bell T, Callender K, Whyte L, and Greer C. 2013. Microbial Competition in Polar Soils: A Review of an Understudied but Potentially Important Control on Productivity. Biology 2:533–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy L, and Watkinson SC. 1995. Wood decomposition, higher fungi, and their role in nutrient redistribution. Canadian Journal of Botany 73:1377–1383. [Google Scholar]
- De Boer W, Folman LB, Summerbell RC, and Boddy L. 2005. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiology Reviews 29:795–811. [DOI] [PubMed] [Google Scholar]
- Callaway RM, Walker LR, Callaway ‘and RM, and Walker LR2. 1997. Competition and Facilitation: A Synthetic Approach to Interactions in Plant Communities. Source: Ecology Ecology 7812514473:1958–1965. [Google Scholar]
- Cannon PF, and Kirk PM. 2007. Fungal Families of the World. CABI. [Google Scholar]
- Cannone N, Sergio S, and Guglielmin M. 2007. Unexpected impacts of climate change on alpine vegetation. Frontiers in Ecology and the Environment 1953:360–364. [Google Scholar]
- Cassman NA, Leite MFA, Pan Y, De Hollander M, Van Veen JA, and Kuramae EE. 2016. Plant and soil fungal but not soil bacterial communities are linked in long-term fertilized grassland. Scientific Reports 6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavieres LA, Hernández-Fuentes C, Sierra-Almeida A, and Kikvidze Z. 2016. Facilitation among plants as an insurance policy for diversity in Alpine communities. Functional Ecology 30:52–59. [Google Scholar]
- Clark JS, Gelfand AE, Woodall CW, and Zhu K. 2014. More than the sum of the parts: Forest climate response from joint species distribution models. Ecological Applications 24:990–999. [DOI] [PubMed] [Google Scholar]
- Classen A, Sundqvist MK, Henning JA, Newman GS, M Moore JA, Cregger MA, Moorhead LC, and Patterson CM. 2015. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 6:art130. [Google Scholar]
- Coince A, Cordier T, Lengellé J, Defossez E, Vacher C, Robin C, Buée M, and Marçais B. 2014. Leaf and root-associated fungal assemblages do not follow similar elevational diversity patterns. PLoS ONE 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colautti RI, Ricciardi A, Grigorovich IA, and MacIsaac HJ. 2004. Is invasion success explained by the enemy release hypothesis?
- Collins CG, Carey CJ, Aronson EL, Kopp CW, and Diez JM. 2016. Direct and indirect effects of native range expansion on soil microbial community structure and function. Journal of Ecology 104:1271–1283. [Google Scholar]
- Cordero OX, and Datta MS. 2016. Microbial interactions and community assembly at microscales. Current Opinion in Microbiology 31:227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon C, Barroso G, Férandon C, Ranger J, Fraissinet-Tachet L, and Marmeisse R. 2010. Performance of the COX1 gene as a marker for the study of metabolically active Pezizomycotina and Agaricomycetes fungal communities from the analysis of soil RNA. FEMS Microbiology Ecology 74:693–705. [DOI] [PubMed] [Google Scholar]
- Diez JM, Dickie I, Edwards G, Hulme PE, Sullivan JJ, and Duncan RP. 2010. Negative soil feedbacks accumulate over time for non-native plant species. Ecology Letters 13:803–809. [DOI] [PubMed] [Google Scholar]
- Edgar RC 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Edwards IP, and Zak DR. 2010. Phylogenetic similarity and structure of Agaricomycotina communities across a forested landscape. Molecular Ecology 19:1469–1482. [DOI] [PubMed] [Google Scholar]
- Edwards IP, and Zak DR. 2011. Fungal community composition and function after long-term exposure of northern forests to elevated atmospheric CO2 and tropospheric O3. Global Change Biology 17:2184–2195. [Google Scholar]
- Feinstein LM, and Blackwood CB. 2012. Taxa-area relationship and neutral dynamics influence the diversity of fungal communities on senesced tree leaves. Environmental Microbiology 14:1488–1499. [DOI] [PubMed] [Google Scholar]
- Frey SD 2007. Spatial Distribution of Soil Organisms Pages 283–300 Soil Microbiology, Ecology and Biochemistry (Third Edition). Academic Press, San Diego. [Google Scholar]
- Gessner MO, Swan CM, Dang CK, McKie BG, Bardgett RD, Wall DH, and Hättenschwiler S. 2010. Diversity meets decomposition. Trends in Ecology and Evolution 25:372–380. [DOI] [PubMed] [Google Scholar]
- Ghanbari M, Shahraki H, Kneifel W, and Domig KJ. 2017. A first insight into the intestinal microbiota of snow trout (Schizothorax zarudnyi). Symbiosis 72:183–193. [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, and Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods 5:235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel KE 1997. Fungal degradation of lignin Pages 33–45 in Cadisch G and Giller KE, editors. Driven by nature: plant litter quality and decomposition. CAB International, Wallingford. [Google Scholar]
- Harmon ME, Franklin JF, Swanson FJ, Sollins P, V Gregory S, Lattin JD, Anderson NH, Cline SP, Aumen NG, and Sedell JR. 1986. Ecology of Coarse Woody Debris in Temperate Ecosystems. Advances in Ecological Research 15. [Google Scholar]
- Hawkes CV, Kivlin SN, Rocca JD, Huguet V, Thomsen M. a., and Suttle KB. 2011. Fungal community responses to precipitation. Global Change Biology 17:1637–1645. [Google Scholar]
- He X, Hou E, Liu Y, and Wen D. 2016. Altitudinal patterns and controls of plant and soil nutrient concentrations and stoichiometry in subtropical China. Scientific Reports 6:24261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J, Savoury M, Müller CT, Lindahl BD, Rogers HJ, and Boddy L. 2015. Priority effects during fungal community establishment in beech wood. The ISME Journal 9:2246–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister EB, Schadt CW, Palumbo AV, James Ansley R, and Boutton TW. 2010. Structural and functional diversity of soil bacterial and fungal communities following woody plant encroachment in the southern Great Plains. Soil Biology and Biochemistry 42:1816–1824. [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung G-H, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüßler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, and Vilgalys R. 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443:818–822. [DOI] [PubMed] [Google Scholar]
- Joergensen RG, and Mueller T. 1996. The fumigation-extraction method to estimate soil microbial biomass: Calibration of the kEN value. Soil Biology and Biochemistry 28:33–37. [Google Scholar]
- Justo A, Vizzini A, Minnis AM, Menolli N, Capelari M, Rodríguez O, Malysheva E, Contu M, Ghignone S, and Hibbett DS. 2011. Phylogeny of the Pluteaceae (Agaricales, Basidiomycota): Taxonomy and character evolution. Fungal Biology 115:1–20. [DOI] [PubMed] [Google Scholar]
- Kivlin SN, Hawkes CV, and Treseder KK. 2011. Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biology and Biochemistry 43:2294–2303. [Google Scholar]
- Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, H??iland K, Kj??ller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vr??lstad T, and Ursing BM. 2005. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytologist 166:1063–1068. [DOI] [PubMed] [Google Scholar]
- Kopp CW, and Cleland EE. 2014. Shifts in plant species elevational range limits and abundances observed over nearly five decades in a western North America mountain range. Journal of Vegetation Science 25:135–146. [Google Scholar]
- Körner C 2003. Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems. Springer; Berlin Heidelberg. [Google Scholar]
- Körner C 2007. The use of ‘ altitude ‘ in ecological research. Trends in Ecology & Evolution 22. [DOI] [PubMed] [Google Scholar]
- Lanzén A, Epelde L, Blanco F, Martín I, Artetxe U, and Garbisu C. 2016. Multi-targeted metagenetic analysis of the influence of climate and environmental parameters on soil microbial communities along an elevational gradient. Scientific Reports 6:28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, and Larsson K-H. 2003. Phylogenetic relationships of russuloid basidiomycetes with emphasis on aphyllophoralean taxa. Mycologia 95:1037–1065. [DOI] [PubMed] [Google Scholar]
- Larsson KH 2007. Re-thinking the classification of corticioid fungi. Mycological Research 111:1040–1063. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, and Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology 75:5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro A, Hilfiker D, and Zeyer J. 2015. Structures of microbial communities in alpine soils: Seasonal and elevational effects. Frontiers in Microbiology 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher PM, Powell MJ, Barr DJS, Churchill PF, Wakefield WS, and Picard KT. 2008. Rhizophlyctidales-a new order in Chytridiomycota. Mycological Research 112:1031–1048. [DOI] [PubMed] [Google Scholar]
- Li H, Colica G, pei Wu P, Li D, Rossi F, De Philippis R, and Liu Y. 2013. Shifting Species Interaction in Soil Microbial Community and Its Influence on Ecosystem Functions Modulating. Microbial Ecology 65:700–708. [DOI] [PubMed] [Google Scholar]
- Little AEF, Robinson CJ, Peterson SB, Raffa KF, and Handelsman J. 2008. Rules of Engagement: Interspecies Interactions that Regulate Microbial Communities. Annual Review of Microbiology 62:375–401. [DOI] [PubMed] [Google Scholar]
- Lu H-P, Yeh Y-C, Sastri AR, Shiah F-K, Gong G-C, and Hsieh C. 2016. Evaluating community–environment relationships along fine to broad taxonomic resolutions reveals evolutionary forces underlying community assembly. The ISME Journal 10:2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludley KE, and Robinson CH. 2008. ‘ Decomposer ‘ Basidiomycota in Arctic and Antarctic ecosystems. Soil biology & biochemistry 40:11–29. [Google Scholar]
- Lyncht MDJ, and Thorn RG. 2006. Diversity of basidiomycetes in Michigan agricultural soils. Applied and Environmental Microbiology 72:7050–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre FT, Callaway RM, Valladares F, and Lortie CJ. 2009. Refining the stress-gradient hypothesis for competition and facilitation in plant communities. Journal of Ecology 97:199–205. [Google Scholar]
- Mangold A, Martín MP, Lücking R, and Lumbsch HT. 2008. Molecular phylogeny suggests synonymy of Thelotremataceae within Graphidaceae (Ascomycota: Ostropales). Taxon 57:476–486. [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, a Fuhrman J, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreås L, Reysenbach A-L, Smith VH, and Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nature reviews. Microbiology 4:102–112. [DOI] [PubMed] [Google Scholar]
- Maynard DS, Bradford MA, Lindner DL, Van Diepen LTA, Frey SD, Glaeser JA, and Crowther TW. 2017. Diversity begets diversity in competition for space. Nature Ecology and Evolution 1. [DOI] [PubMed] [Google Scholar]
- Meiser A, Bálint M, and Schmitt I. 2014. Meta-analysis of deep-sequenced fungal communities indicates limited taxon sharing between studies and the presence of biogeographic patterns. New Phytologist 201:623–635. [DOI] [PubMed] [Google Scholar]
- Meng H, Li K, Nie M, Wan J-R, Quan Z-X, Fang C-M, Chen J-K, Gu J-D, and Li B. 2013. Responses of bacterial and fungal communities to an elevation gradient in a subtropical montane forest of China. Applied Microbiology and Biotechnology 97:2219–2230. [DOI] [PubMed] [Google Scholar]
- Myers-Smith IH, Forbes BC, Wilmking M, Hallinger M, Lantz T, Blok D, Tape KD, Macias-Fauria M, Sass-Klaassen U, Lévesque E, Boudreau S, Ropars P, Hermanutz L, Trant A, Collier LS, Weijers S, Rozema J, a Rayback S, Schmidt NM, Schaepman-Strub G, Wipf S, Rixen C, Ménard CB, Venn S, Goetz S, Andreu-Hayles L, Elmendorf S, Ravolainen V, Welker J, Grogan P, Epstein HE, and Hik DS. 2011. Shrub expansion in tundra ecosystems: dynamics, impacts and research priorities. Environmental Research Letters 6:045509. [Google Scholar]
- Newsham KK 2011. A meta-analysis of plant responses to dark septate root endophytes. New Phytologist:783–793. [DOI] [PubMed] [Google Scholar]
- Nicholls N, and Alexander L. 2007. Has the climate become more variable or extreme? Progress 1992–2006. Progress in Physical Geography 31:77–87. [Google Scholar]
- Nielsen UN, Osler GHR, Campbell CD, Burslem DFRP, and van der Wal R. 2010. The influence of vegetation type, soil properties and precipitation on the composition of soil mite and microbial communities at the landscape scale. Journal of Biogeography 37:1317–1328. [Google Scholar]
- Nielsen UN, Wall DH, and Six J. 2015. Soil Biodiversity and the Environment. Annual Review of Environment & Resources 40:63–90. [Google Scholar]
- Nottingham A, Fierer N, Turner B, Whitaker J, Ostle N, McNamara N, Bardgett R, Leff J, Salinas N, Ccahuana A, Silman M, and Meir P. 2016. Temperature drives plant and soil microbial diversity patterns across an elevation gradient from the Andes to the Amazon. bioRxiv 44:79996. [Google Scholar]
- Oksanen J, Blanchet F, Kindt R, Legendre P, and O’Hara R. 2016. Vegan: community ecology package. R package 2.3–3:Available at: https://cran.r-project.org/web/packa.
- Öpik M, Davison J, Moora M, and Zobel M. 2014. DNA-based detection and identification of Glomeromycota: the virtual taxonomy of environmental sequences 1. Botany 92:135–147. [Google Scholar]
- Ovaskainen O, Gleb Tikhonov, Norberg A, Blanchet FG, Duan L, Dunson D, Roslin T, and Abrego N. 2017. How to make more out of community data? A conceptual framework and its implementation as models and software. Ecology letters:1–16. [DOI] [PubMed] [Google Scholar]
- Paulson JN, Stine OC, Bravo HC, and Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nature methods 10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peay KG, Baraloto C, and a Fine PV. 2013. Strong coupling of plant and fungal community structure across western Amazonian rainforests. The ISME journal 7:1852–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellissier L, Niculita-Hirzel H, Dubuis A, Pagni M, Guex N, Ndiribe C, Salamin N, Xenarios I, Goudet J, Sanders IR, and Guisan A. 2014. Soil fungal communities of grasslands are environmentally structured at a regional scale in the Alps. Molecular Ecology 23:4274–4290. [DOI] [PubMed] [Google Scholar]
- Pepin N, Bradley RS, Diaz HF, Baraer M, Caceres EB, Forsythe N, Fowler H, Greenwood G, Hashmi MZ, Liu XD, Miller JR, Ning L, Ohmura A, Palazzi E, Rangwala I, Schöner W, Severskiy I, Shahgedanova M, Wang MB, Williamson SN, and Yang DQ. 2015. Elevation-dependent warming in mountain regions of the world. Nature Climate Change 5:424–430. [Google Scholar]
- Rammig A, Jonas T, Zimmermann NE, and Rixen C. 2010. Changes in alpine plant growth under future climate conditions. Biogeosciences 7:2013–2024. [Google Scholar]
- Ren C, Zhang W, Zhong Z, Han X, Yang G, Feng Y, and Ren G. 2018. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Science of The Total Environment 610–611:750–758. [DOI] [PubMed] [Google Scholar]
- Rowe RJ, and Lidgard S. 2009. Elevational gradients and species richness: Do methods change pattern perception? Global Ecology and Biogeography 18:163–177. [Google Scholar]
- Roy J, Albert CH, Ibanez S, Saccone P, Zinger L, Choler P, Clément J-C, Lavergne S, and Geremia RA. 2013. Microbes on the cliff: alpine cushion plants structure bacterial and fungal communities. Frontiers in microbiology 4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinner F, and Gstraunthaler G. 1981. Adaptation of microbial activities to the environmental conditions in alpine soils. Oecologia 50:113–116. [DOI] [PubMed] [Google Scholar]
- Schmidt SK, Naff CS, Lynch RC, and Newsham KK. 2012. Fungal communities at the edge : Ecological lessons from high alpine fungi. Fungal Ecology 5:443–452. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, Miller AN, Wingfield MJ, Aime MC, An K-D, Bai F-Y, Barreto RW, Begerow D, Bergeron M-J, Blackwell M, Boekhout T, Bogale M, Boonyuen N, Burgaz AR, Buyck B, Cai L, Cai Q, Cardinali G, Chaverri P, Coppins BJ, Crespo A, Cubas P, Cummings C, Damm U, de Beer ZW, de Hoog GS, Del-Prado R, Dentinger B, Dieguez-Uribeondo J, Divakar PK, Douglas B, Duenas M, Duong TA, Eberhardt U, Edwards JE, Elshahed MS, Fliegerova K, Furtado M, Garcia MA, Ge Z-W, Griffith GW, Griffiths K, Groenewald JZ, Groenewald M, Grube M, Gryzenhout M, Guo L-D, Hagen F, Hambleton S, Hamelin RC, Hansen K, Harrold P, Heller G, Herrera C, Hirayama K, Hirooka Y, Ho H-M, Hoffmann K, Hofstetter V, Hognabba F, Hollingsworth PM, Hong S-B, Hosaka K, Houbraken J, Hughes K, Huhtinen S, Hyde KD, James T, Johnson EM, Johnson JE, Johnston PR, Jones EBG, Kelly LJ, Kirk PM, Knapp DG, Koljalg U, Kovacs GM, Kurtzman CP, Landvik S, Leavitt SD, Liggenstoffer AS, Liimatainen K, Lombard L, Luangsa-ard JJ, Lumbsch HT, Maganti H, Maharachchikumbura SSN, Martin MP, May TW, McTaggart AR, Methven AS, Meyer W, Moncalvo J-M, Mongkolsamrit S, Nagy LG, Nilsson RH, Niskanen T, Nyilasi I, Okada G, Okane I, Olariaga I, Otte J, Papp T, Park D, Petkovits T, Pino-Bodas R, Quaedvlieg W, Raja HA, Redecker D, Rintoul TL, Ruibal C, Sarmiento-Ramirez JM, Schmitt I, Schussler A, Shearer C, Sotome K, Stefani FOP, Stenroos S, Stielow B, Stockinger H, Suetrong S, Suh S-O, Sung G-H, Suzuki M, Tanaka K, Tedersoo L, Telleria MT, Tretter E, Untereiner WA, Urbina H, Vagvolgyi C, Vialle A, Vu TD, Walther G, Wang Q-M, Wang Y, Weir BS, Weiss M, White MM, Xu J, Yahr R, Yang ZL, Yurkov A, Zamora J-C, Zhang N, Zhuang W-Y, and Schindel D. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences 109:6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnavaz B, Zinger L, Lavergne S, Choler P, and Geremia RA. 2012. Phylogenetic Clustering Reveals Selective Events Driving the Turnover of Bacterial Community in Alpine Tundra Soils. Arctic, Antarctic, and Alpine Research 44:232–238. [Google Scholar]
- Shen C, Liang W, Shi Y, Lin X, Zhang H, Wu X, Xie G, Chain P, Grogan P, and Chu H. 2014. Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecology 95:3190–3202. [Google Scholar]
- Siles JA, and Margesin R. 2016. Abundance and Diversity of Bacterial, Archaeal, and Fungal Communities Along an Altitudinal Gradient in Alpine Forest Soils: What Are the Driving Factors? Microbial Ecology 72:207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Hanlin DMW, and Hanlin RT. 1998. The order phyllachorales: Taxonomic review. Mycoscience 39:97–104. [Google Scholar]
- Smith DP, and Peay KG. 2014. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark S, Kytöviita MM, and Neumann AB. 2007. The phenolic compounds in Cladonia lichens are not antimicrobial in soils. Oecologia 152:299–306. [DOI] [PubMed] [Google Scholar]
- Stubbs TL, and Kennedy AC. 2012. Microbial Weed Control and Microbial Herbicides. Herbicides - Environmental Impact Studies and Management Approaches. [Google Scholar]
- Sundqvist MK, Sanders NJ, and Wardle D. a.. 2013. Community and Ecosystem Responses to Elevational Gradients: Processes, Mechanisms, and Insights for Global Change. Annual Review of Ecology, Evolution, and Systematics 44:261–280. [Google Scholar]
- Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou S, Wardle DA, and Lindahl BD. 2014. Disentangling global soil fungal diversity. Science 346:1052–1053. [DOI] [PubMed] [Google Scholar]
- Thorn RG, and Lynch MDJ. 2007. 6 - FUNGI AND EUKARYOTIC ALGAE A2 - PAUL, ELDOR A. BT - (Third Edition). Pages 145–162 Soil Microbiology, Ecology and Biochemistry. Academic Press, San Diego. [Google Scholar]
- Timonen S, Sinkko H, Sun H, Sieti OM??, Rinta-Kanto JM, Kiheri H, and Heinonsalo J. 2017. Ericoid Roots and Mycospheres Govern Plant-Specific Bacterial Communities in Boreal Forest Humus. Microbial Ecology 73:939–953. [DOI] [PubMed] [Google Scholar]
- Wallenstein MD, McMahon S, and Schimel J. 2007. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiology Ecology 59:428–435. [DOI] [PubMed] [Google Scholar]
- Walters W, Hyde ER, Berg-lyons D, Ackermann G, Humphrey G, Parada A, a Gilbert J, and Jansson JK. 2015. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal InternalTranscribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 1:e0009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle DA 2006. The influence of biotic interactions on soil biodiversity. Ecology Letters 9:870–886. [DOI] [PubMed] [Google Scholar]
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, and Knight R. 2017. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters V, Silver WL, Bignell DE, Coleman DC, and Lavelle P. 2000. Effects of Global Changes on Above- and Belowground Biodiversity in Terrestrial Ecosystems : Implications for Ecosystem Functioning. BioScience 50. [Google Scholar]
- Wu B, Hogetsu T, Isobe K, and Ishii R. 2007. Community structure of arbuscular mycorrhizal fungi in a primary successional volcanic desert on the southeast slope of Mount Fuji. Mycorrhiza 17:495–506. [DOI] [PubMed] [Google Scholar]
- Wu J, Joergensen RG, Pommerening B, Chaussod R, and Brooks PC. 1990. Short Communication Measurement of Soil Microbial Biomass C by Fumigation- Extraction – An Automated Procedure. Soil biology & biochemistry 22:1167–1169. [Google Scholar]
- Yarza P, Yilmaz P, Panzer K, Glöckner FO, and Reich M. 2017. A phylogenetic framework for the kingdom Fungi based on 18S rRNA gene sequences. Marine Genomics. [DOI] [PubMed]
- Zak DR, Pregitzer KS, Burton AJ, Edwards IP, and Kellner H. 2011. Microbial responses to a changing environment: Implications for the future functioning of terrestrial ecosystems. Fungal Ecology 4:386–395. [Google Scholar]
- Zalar P, Sybren de Hoog G, Schroers HJ, Frank JM, and Gunde-Cimerman N. 2005. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie van Leeuwenhoek, International Journal of General and Molecular Microbiology 87:311–328. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dong S, Gao Q, Liu S, Ganjurjav H, Wang X, Su X, and Wu X. 2017. Soil bacterial and fungal diversity differently correlated with soil biochemistry in alpine grassland ecosystems in response to environmental changes. Scientific Reports 7:43077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinger L, Shahnavaz B, Baptist F, Geremia RA, and Choler P. 2009. Microbial diversity in alpine tundra soils correlates with snow cover dynamics. ISME Journal 3:850–859. [DOI] [PubMed] [Google Scholar]
- Zumsteg A, Luster J, Göransson H, Smittenberg RH, Brunner I, Bernasconi SM, Zeyer J, and Frey B. 2012. Bacterial, Archaeal and Fungal Succession in the Forefield of a Receding Glacier. Microbial Ecology 63:552–564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.