Abstract
The deregulation of cellular long non-coding RNA (lncRNA) expression during a human adenovirus infection was studied by deep sequencing. Expression of lncRNAs increased substantially following the progression of the infection. Among 645 significantly expressed lncRNAs, the expression of 398 was changed more than 2-fold. More than 80% of them were up-regulated and 80% of them were detected during the late phase. Based on the genomic locations of the deregulated lncRNAs in relation to known mRNAs and miRNAs, they were predicted to be involved in growth, structure, apoptosis and wound healing in the early phase, cell proliferation in the intermediate phase and protein synthesis, modification and transport in the late phase. The most significant functions of cellular RNA-binding proteins, previously shown to interact with the deregulated lncRNAs identified here, are involved in RNA splicing, nuclear export and translation events. We hypothesize that adenoviruses exploit the lncRNA network to optimize their reproduction.
Keywords: Long non-coding RNA (lncRNA), Antisense RNAs, Cellular gene expression, Adenovirus infection, RNA sequencing, RNA-binding proteins (RBPs), lncRNA expression profile
Highlights
-
•
The expression of 398 lncRNAs showed a distinct temporal pattern during Ad2 infection.
-
•
80% of the deregulated lncRNAs were up-regulated during the late phase of infection.
-
•
The deregulated lncRNAs potentiallyinteract with 33 cellular RNA binding proteins.
-
•
These RBPs are involved in RNA splicing, nuclear export and translation.
-
•
Adenovirus exploits the cellular lncRNA network to optimize its replication.
1. Introduction
For decades adenoviruses have served as an outstanding model system to study eukaryotic gene structure and expression. They are non-enveloped, icosahedral particles containing a linear, double-stranded DNA molecule. Based on adenovirus gene expression, the replication cycle can be divided into two phases, an early and a late phase, defined by the start of viral DNA replication. Early viral proteins are involved in regulation of the cell cycle and suppression of the cellular antiviral response, whereas the viral structural proteins are synthesized during the late phase (see review by Shenk (1996)). Based on the cellular gene expression profiles in primary lung fibroblasts, the adenovirus infection can be further divided into four periods. Each period is characterized by deregulation of specific sets of genes. Genes involved in the inhibition of cell growth, control of the cell cycle, regulation of nucleic acid and protein synthesis, and cellular structure are deregulated during the first, second, third, and fourth period, respectively (Zhao et al., 2007). Most previous studies on the regulation of host cell gene expression during an adenovirus infection have been focused on the mRNA level. Recently, deregulation of cellular microRNA expression has also been addressed (Zhao et al., 2015).
In recent years, long non-coding RNAs (lncRNAs) have attracted a great deal of attention and have been shown to be important regulators of various biological processes. LncRNAs are transcripts of more than 200 nucleotides and most of them are 5′ capped, spliced, and poly-adenylated, but lack open reading frames (ORF) of significant lengths (Derrien et al., 2012). In general, lncRNAs lack strong conservation and only short regions of them are constrained by structure or sequence-specific interactions. Most of them are present at levels 10 to 100-fold lower than mRNAs although there are exceptions (Palazzo and Gregory, 2014, Palazzo and Lee, 2015). They are enriched in the nucleus, within the chromatin-associated fraction. They can interact with various chromatin regulatory proteins and recruit them to specific sites on DNA, thereby regulating gene expression (Wang and Chang, 2011). In addition, several lncRNAs have been shown to play important roles in organizing the nuclear structure, cell growth and survival, migration and differentiation, organismal development and various types of cancers (Gutschner and Diederichs, 2012, Imamachi et al., 2014, Nakagawa et al., 2013, Richards et al., 2015).
LncRNAs appear in diverse genomic contexts, such as intronic, antisense, intergenic, overlapping and pseudogenes. The most significant class of differentially expressed lncRNAs is antisense RNAs. Antisense RNAs are transcribed from the opposite strands of protein coding genes (sense RNAs). Thus, they have a complementary overlap with the sense RNAs. It has been shown that more than 70% of the protein-coding genes have antisense partners (Katayama et al., 2005, Rosok and Sioud, 2004). Because of their low evolutionary conservation and expression, they have been considered as junk until recently. Emerging evidence indicates that antisense RNA can regulate gene expression at different levels such as transcription, posttranscriptional processing, RNA stability, transport and translation (Lapidot and Pilpel, 2006, Pelechano and Steinmetz, 2013).
Few studies have so far examined lncRNA expression during a virus infection. By using deep sequencing, it was shown that the expression of approximately 500 annotated lncRNAs was changed in mice during a severe acute respiratory syndrome coronavirus (SARS-CoV) infection (Peng et al., 2010). About 40% of these changes were also detected in mice and mouse embryonic fibroblasts infected with influenza virus A and in response to interferon treatment. There were a higher proportion of down- vs up-regulated lncRNAs in SARS-CoV infected mice and in interferon stimulated cells. Deregulation of lncRNA expression has also been demonstrated in lung epithelial cells after influenza A virus infection and in human RD cells after an enterovirus 71 infection by using microarray analysis (Winterling et al., 2014, Yin et al., 2013). Deregulation of cellular lncRNAs expression was also revealed in HIV-1-infected T cells and eighteen out of 83 lncRNAs tested were differentially expressed (Zhang et al., 2013). Among them, up-regulation of NEAT1 is interesting. Knockdown of NEAT1 could enhance virus production by increasing nucleus-to-cytoplasm export of Rev-dependent instability element-containing HIV-1 transcripts. On the other hand NEAT1 is induced by influenza virus and herpes simplex virus infection (Imamura et al., 2014). Here, we extended these studies to human adenovirus type 2 (Ad2) and studied the deregulation of cellular lncRNA expression in human primary lung fibroblasts (IMR-90) during the course of an Ad2 infection by using deep sequencing.
2. Materials and methods
2.1. Cell culture and adenovirus infection
Human primary lung fibroblast cells (IMR-90), purchased from American Type Culture Collection (ATCC) were cultured in Eagle׳s minimum essential medium (ATCC) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. After reaching confluence, the cells were cultured for two more days in order to synchronize them. Over 95% of the cells were in the G0/G1 phase as indicated by FACS analysis (Zhao et al., 2007). Synchronized cells were mock-infected or infected with Ad2 at a multiplicity of infection (MOI) of 100 fluorescence-forming units (FFU) per cell in serum-free medium (Philipson, 1961). After 1 h adsorption at 37 °C, the medium was replaced with complete EMEM containing 10% FBS and incubated at 37 °C. Infected cells were collected at 6, 12, 24, and 36 hour post infection (hpi). Mock-infected cells were collected at 6 hpi.
2.2. RNA extraction, cDNA library preparation, and sequencing
Total RNA from adenovirus or mock-infected IMR-90 cells was extracted using TRIZOL Reagent (Invitrogen). The quality of the input RNA was controlled by the Agilent 2100 Bioanalyzer (Agilent Technologies). Then the total RNA was treated with RiboZero (Epicenter) to remove ribosomal RNA and cDNA libraries were constructed using ScriptSeq™ v2 RNA-Seq library preparation kit according to the manufacturer’s protocol (Epicenter). The cDNA libraries were sequenced using Illumina HiSeq 2000.
2.3. Bioinformatics analysis
After data cleaning, the reads were aligned to human genome sequences (GRCh38, Ensembl) with TopHat2 software (Imamachi et al., 2014). TopHat2 incorporates Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) algorithm to perform the alignment. We used default parameters which allowed a maximum of two mismatches when mapping the reads to the human genome. Cufflinks was then used to profile gene expression at each time point based on human gene annotation by Ensembl (Trapnell et al., 2012). Differentially expressed lncRNAs in adenovirus infected cells, compared to mock, were characterized by three statistical values. First, fold change was calculated by the FPKM (Fragments per kilobase per Million) values in different libraries; second, based on Poison distribution, p-values were used to present the significances of differentially expressed lncRNAs (Audic and Claverie, 1997). Using the NOIseq package the probability of a differentially expressed lncRNA was calculated (Tarazona et al., 2011). The hierarchical lncRNAs with different expression patterns were analyzed with uncentered correlation and centroid linkage method by Cluster and Tree View software.
2.4. Characterization of lncRNA interacting proteins
All the proteins that interacted with lncRNAs were downloaded from starBase v2.0 (http://starbase.sysu.edu.cn) (Li et al., 2014b). StarBase lists human lncRNA interacting proteins based on CHIP-Seq analysis.
2.5. Protein analysis using mass spectrometry (MS)
IMR-90 cells were grown in cell culture medium for stable isotope labeling by amino acids in cell culture (SILAC) for at least six population doublings. Cells labeled with heavy or light amino acids were then infected with Ad2 or mock infected, respectively. A biological replicate with swapped labeling was also performed. After cells were lysed, mock- and Ad2-infected lysates of different labeling were combined in a 1:1 protein ratio. Proteins were fractionated using SDS-PAGE and each lane was cut into ten pieces. Following in-gel tryptic digestion (Shevchenko et al., 2000), peptides were extracted and analyzed using QExactive Orbitrap Plus Mass spectrometer (ThermoFisher Scientific, Bremen, Germany) Acquired data (raw-files) were imported into MaxQuant software (version: 1.4) and searched against a FASTA-file containing both cellular and Ad2 proteins. The ratio of the chromatographic areas of heavy and light peptides matching to specific proteins was used for determining the protein expression levels. The reported protein expression is the average of the two biological replicates [manuscript in preparation].
2.6. Functional analysis
The biological significance of RNA-binding proteins (RBP) was analyzed using Bioinformatics Resources DAVID (The Database for Annotation, Visualization and Integrated Discovery) (Huang da et al., 2009). The symbols of the lncRNA interacting proteins were used as input. The functional annotation chart was selected to present the GO and KEGG pathway enrichment. The Benjamini method was used to globally correct enrichment p-values of individual term members.
3. Results
3.1. Expression of cellular lncRNAs in adenovirus infected IMR-90 cells
Using paired-end sequencing, the cellular lncRNA expression profiles were examined during the course of an adenovirus 2 (Ad2) infection in human primary lung fibroblasts (IMR-90). The infected cells were collected at 6, 12, 24, and 36 hours post infection (hpi). These time points represent different stages of the Ad2 infectious cycle, i.e. before any adenovirus gene expression occurs, when the immediate early gene (E1a) is activated, when the onset of adenoviral DNA replication starts, and after the adenovirus late genes begin to be expressed, respectively (Zhao et al., 2007). Furthermore, all our previous studies on cellular gene expression were performed under the same conditions (Zhao et al., 2015, Zhao et al., 2012, Zhao et al., 2007). Thus, we could correlate the expression profiles of lncRNAs with the profiles of cellular mRNA, miRNA and protein expression.
Total RNA extracts were subjected to deep sequencing, and all types of RNA except ribosomal RNAs and small RNAs were sequenced. About 30 million 255 bp long sequence reads per sample were generated. The most abundant RNAs in non-infected and in the early and middle phases of infection (6–24 hpi) was mRNA which accounted for 53–58% of all sequenced RNAs. LncRNAs accounted for around 37%, but they became the most abundant RNA type in the late phase (36 hpi) and accounted then for 57%. MicroRNAs and small nuclear RNAs accounted for less than 8%.
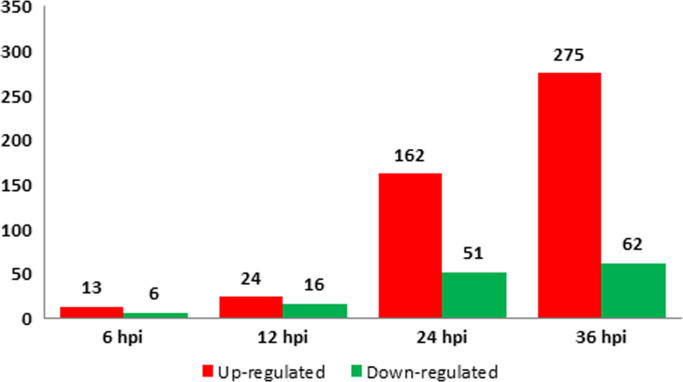
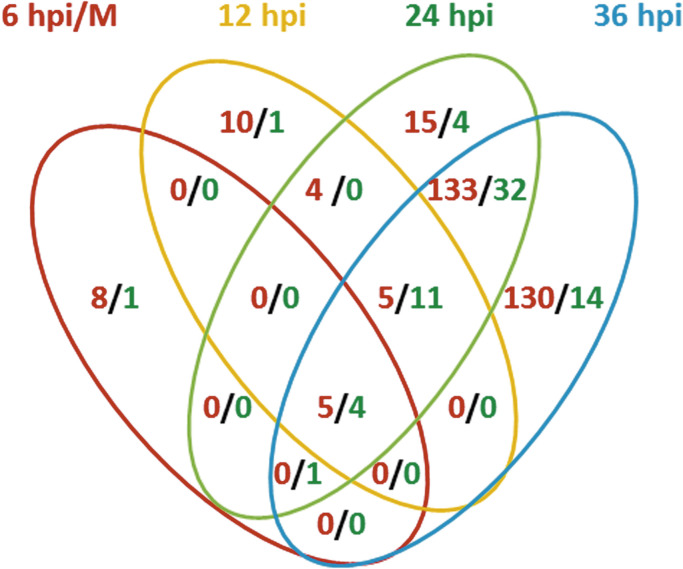
A comparison was made between the expressions of cellular lncRNAs at different stages of an Ad2 infection using lncRNAs in non-infected cells as reference. LncRNAs with a minimum of 10 FPKM were considered as significantly expressed and a 2-fold change was used as cut off for the scoring of differentially expressed lncRNAs. Thus, 645 cellular lncRNAs were detected, and among them 398 lncRNAs were identified as differentially expressed in infected cells at least at one of the time points. The majority (86%) of them had less than 100 FRKM. The complete list of the 398 differentially expressed lncRNAs is included in supplementary Table S1. Only 19 and 40 differentially expressed lncRNAs were identified at 6 and 12 hpi, respectively ( Fig. 1). The number increased to 213 and 337 at 24 and 36 hpi, respectively. The overlaps of differentially expressed lncRNAs at different time points are shown in Fig. 2.
Fig. 1.
Numbers of up-regulated (red bars) and down-regulated (green bars) lncRNAs at different time points of an Ad2 infection. These lncRNAs were with a minimum of 10 FPKM and a 2-fold change between adenovirus infected and uninfected cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Overlaps of the up-regulated (red) and down-regulated (green) lncRNAs at 6, 12, 24 and 36 hpi. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
All data presented in the current paper were derived from a single experiment. However, a confirming biological duplicate was obtained by analyzing lncRNA expression in a previously reported sequencing data set obtained by single-end sequencing of 76 bp long reads (Zhao et al., 2012). This data set only contains data from two time points, 12 and 24 hpi. Nearly 90% of the deregulated lncRNAs at 24 hpi identified in the present study showed very similar expression profiles as in our previous data set (data not shown). Since pair-end sequencing generates more precise and quantitative data as compared to single-end sequencing, the results presented here are based on paired-end sequencing.
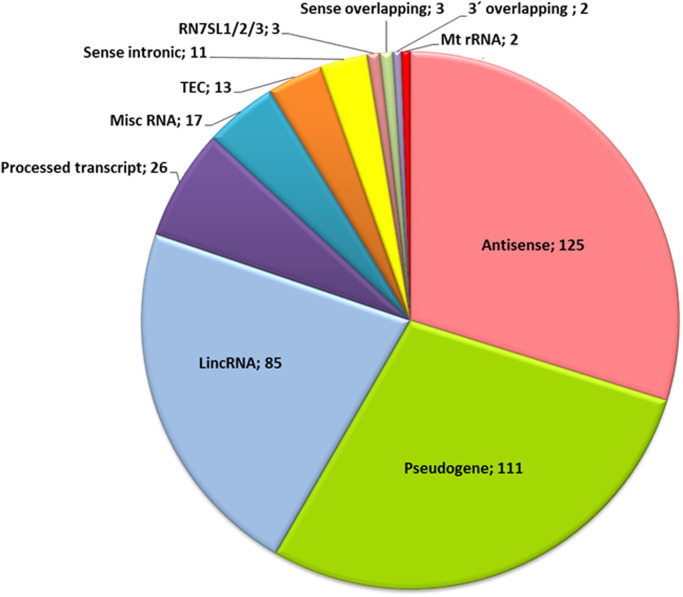
According to GENCODE (http://www.gencodegenes.org/gencode_biotypes.html), the deregulated lncRNAs belong to different classes ( Fig. 3). The largest class included 125 antisense transcripts that overlapped the genomic span of protein-coding genes on the opposite strand. A second class included 111 pseudogenes and the third class included 85 long intergenic non-coding RNAs (lincRNAs). In addition, there were 26 processed transcripts that lacked an ORF. Thirteen transcripts were classified as TEC (To be Experimentally Confirmed). These were expressed at a low level (less than 20 RPKM) except AC011558.5 and RP11-1148L6.8. The most strongly deregulated lncRNAs were 17 miscellaneous (misc) RNAs, including metazoan signal recognition particle RNAs. Eleven lncRNAs are located within introns of sense RNA and the remaining lncRNAs included a few lncRNAs which are located 3 prime of the sense RNA, mitochondrial RNAs and 3 highly abundant lncRNAs (RN7SL1, RN7SL2 and RN7SL3).
Fig. 3.
Numbers of differentially expressed lncRNAs that belong to different biotypes according to GENCODE.
3.2. Expression profiles of cellular lncRNAs
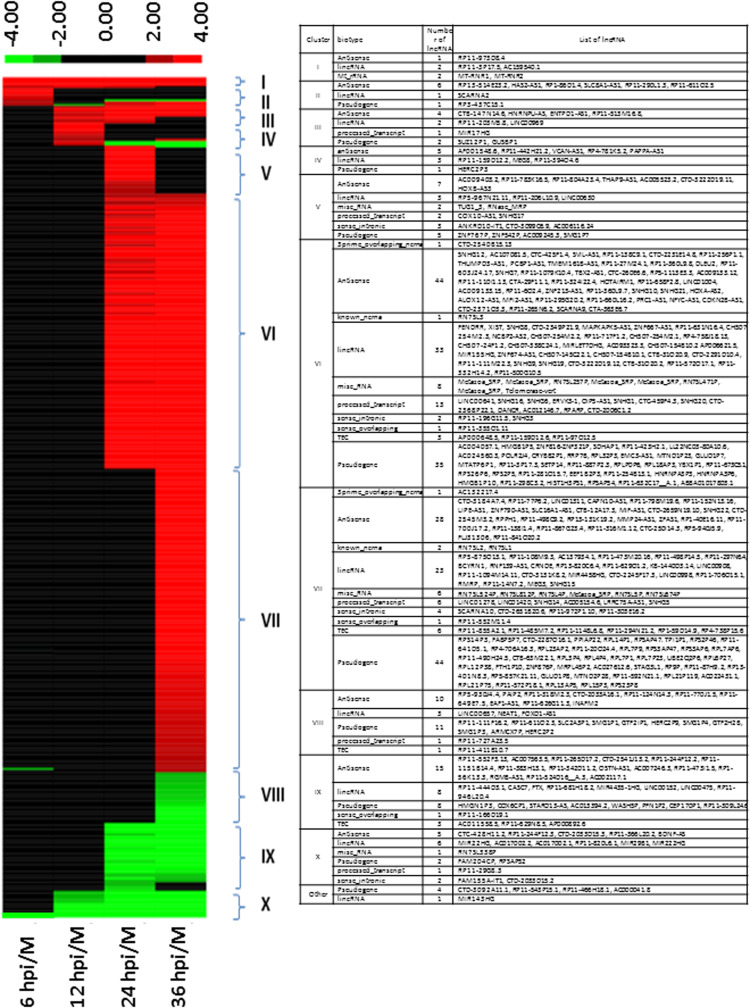
The differentially expressed lncRNAs fell into 10 major clusters as shown in Fig. 4. Clusters I and III contained lncRNAs that were up-regulated from 6 hpi or 12 hpi, respectively, and remained up-regulated until 36 hpi. LncRNAs in Cluster II, IV and V were transiently up-regulated at 6, 12 and 24 hpi, respectively. Clusters VI and VII were the two largest clusters and included lncRNAs that started to be up-regulated at 24 hpi and at 36 hpi, respectively. About 50% of these lncRNAs were not expressed at all or expressed at a much lower level in non-infected cells and in Ad2-infected cell during the early phase. More than 80% of the up-regulated lncRNAs at 24 hpi remained up-regulated until 36 hpi. Finally, clusters X, IX and VIII included lncRNAs that were down-regulated from 6, 12, 24 and 36 hpi, respectively. Together they represented less than 18% of all differentially expressed lncRNAs. Only 59 lncRNAs were differentially expressed during the early phase of infection (6 and 12 hpi) with three expression profiles; 1/3 remained up-regulated until the late phase, 1/3 remained down-regulated until the late phase and 1/3 was transiently up-regulated at 6 or 12 hpi.
Fig. 4.
Heatmap visualization of expression profiles of de-regulated lncRNAs based on their expression changes during the course of adenovirus infection. 398 differentially expressed lncRNAs were grouped into 10 major clusters I–X. The lncRNAs in each cluster are listed in the table on the right. The numbers of lncRNAs in each cluster that belonged to different biotypes are also indicated. Six lncRNAs with expression profiles that do not fit into any of the 10 clusters are included in “Other”.
The transient up-regulation of lncRNAs at 6 and 12 hpi must represent two waves of host cell response to the virus since few viral genes are expressed during this period. The lncRNAs that were transiently up-regulated at 6 hpi included 6 antisense RNAs (RP13-514E23.2, HAS2-AS1, RP1-86D1.4, SLC8A1-AS1, RP11-290L1.3 and RP11-611O2.5), one lincRNA (SCARNA2) and one pseudogene (RP3-437C15.1). These antisense RNAs overlap with protein coding genes for ARHGAP24, HAS2, PTPRK, SLC8A1, PHLDA1 and MDM2 (Table S1). The lncRNAs that were transiently up-regulated at 12 hpi comprised 5 antisense RNAs (AF001548.6, RP11-442H21.2, VCAN-AS1, RP4-781K5.2 and PAPPA-AS1), 3 lincRNAs (RP11-159D12.2, MEG8 and RP11-394O4.6), and one pseudogene (HERC2P3). The five antisense RNAs overlapped with protein coding genes for MYH11, DDIT4, VCAN, IRF2BP2 and PAPPA. Interestingly, these genes are involved in cell growth control (PTPRK), rearrangement of cellular structure (VCAN, ARHGAP24 and MYH11), transcription regulation (MSC and HEXIM1), apoptosis (PHLDA1) and wound healing (HAS2 and PAPPA). Only 14 lncRNAs remained up-regulated from early to late phase (Clusters I and III). Among them, RP11-973D8.4 is an antisense RNA and overlaps completely with the gene encoding CDK2, and MIR17HG is the precursor for microRNAs in the mir17-92 cluster. In addition two mitochondrial RNAs, MT-RNP1 and 2 were among the most highly expressed lncRNAs and their expression increased until the late phase.
In the late phase, the number of differentially expressed lncRNAs increased dramatically, and 162 lncRNAs were up-regulated at 24 hpi. Among them, the most significant biotype included 50 antisense RNAs. These antisense RNAs overlapped with genes involved in diverse cellular functions like transcriptional regulation (HOXA1, HOXA3, HOXA4, NFYC, MAZ, ZNF90, ZNF205, ZNF213, ZNF486 and TBX2), DNA, RNA, and protein metabolism (THUMPD3, PAGR1, PCBP1, PA2G2, CCT4, RAB8A, VPS11, GLRX5, MF12, DHCR7, GDE1, TRNAU1AP). Other genes are involved in cell cycle and growth control such as CDKN2A, CDKN2B, CCNY and PRC1. There were 38 lincRNAs but only FENDRR, XIST and FTX have been characterized. The interesting lncRNAs, MIRLET7DHG and MIR155HG are the parent RNAs for let-7d and miR-155 which have been shown to be highly and differentially expressed during the course of an Ad2 infection (Zhao et al., 2015). DANCR is crucial for tumor formation and progression (Yuan et al., 2015). Finally, the deregulation of a group of Small Nucleolar RNA Host Genes (SNHGs), SNHG1, 3, 6, 7, 8, 9, 10, 12, 16, 19, 20 and 22 was significant.
After 24 hpi, adenovirus takes over the control of the cellular metabolic machinery and uses it for its progeny production and at 36 hpi, replication of Ad2 DNA reaches its maximal rate. In addition to the lncRNAs that remained up-regulated from 24 hpi, 121 additional lncRNAs became up-regulated at 36 hpi. Among them 28 were antisense RNAs and the most significant functions of their overlapping protein-coding genes were involvement in protein metabolism, such as protein modification (ERO1L, P4HB, P4HTM), protein degradation (UBB, BPHL, PSMA3) and protein transport (HOOK2, RAB1B). Twenty-three were lincRNAs and most of them have not been characterized except CRNDE, RMRP and MEG3. Up-regulation of a group of ribosomal protein pseudogenes was also significant (RPL3P4, RPL4P4, RPL7P1, RPL7P23, RPL7AP6, etc.). Furthermore, the most highly expressed lncRNAs are known to be components of signal recognition particle, like RN7SL1/2/3, 4P, and 5P.
Only 18% of the lncRNAs were down-regulated during the infection. Cluster X included 17 lncRNAs that were down-regulated from the early phase to the late phase. Among them, five antisense RNAs, CTC-428H11.2, CTD-2033D15,3, RP11-366L20.2, BDNF-AS and RP11-244F12.3, overlap with protein-coding RNAs that are involved in signaling pathway (RASA1 and THBS1), cell cycle/growth regulation (HMGA2 and BDNF) and cytoskeleton organization (TPM1). Among other lncRNAs, MIR22HG and MIR29B1HG seem important because they are host RNAs for miR-22 and miR-29b which are down-regulated in the late phase (Zhao et al., 2015). Clusters IX and VIII included lncRNAs down-regulated after 24 and 36 hpi. The functions of these two clusters seem different. The most significant protein-coding genes that overlapped with antisense RNAs (AC007563,5, RP11-342D11,2, AC007246,3, RP1-56K13,3, RGMB-AS1) in cluster IX are involved in signaling pathways, such as IGFBP5, NRP1, MAP4K3, LASP1 and RGMB. The most significant protein-coding genes that overlapped with antisense RNAs (PAIP2 and RP11-770J1,5) in cluster VIII are involved in protein metabolism, like protein translation (PAIP2) and protein ubiquitination (UBE4A). Among other lncRNAs, NEAT1 (down-regulated from 36 hpi) and FTX (down-regulated from 24 hpi) have been studied previously (Saha et al., 2006, Zhao et al., 2014).
3.3. Deregulation of antisense RNAs and their overlapping sense RNAs
The largest group of differentially expressed lncRNAs was 125 antisense RNAs. Among them, 115 overlapped with nearby sense RNAs and 66 of them overlapped completely with their sense partners. Ten were located head to head or tail to tail relative to their nearby sense RNAs. The potential effects of antisense RNAs on the expression of nearby sense genes were analyzed by comparing their expression profiles. Eighty-six mRNAs showed similar expression profiles as their antisense RNAs (Table S1) and only 18 mRNAs showed different expression profiles than their antisense RNAs. Twenty-one mRNAs were not expressed or expressed at a level below 6 FPKM (not included in Table S1). Among 18 antisense RNAs that showed different expression profile with their overlapping mRNAs, 9 were up- (CTB-12A17,3, CTD-3222D19,11, HOXB-AS3 and PCBP1-AS1) or down-regulated (CTD-2033A16,1, PAIP2, RP11-124N14,3, RP11-770J1,5 and RP5-930J4,4), while the expression of their overlapping mRNAs was unchanged. Nine antisense RNAs showed an opposite expression profile compared to their sense RNA partners (ATP5L, MVP, ZNF143, TRIM13, CCNY, SVIL, TBX2, THUMPD3 and ZNFX1).
3.4. LncRNA interacting proteins and their expression
Most, if not all, lncRNAs carry out their functions by interacting with RNA-binding proteins (RBPs), thereby directing them to specific targets (Guttman and Rinn, 2012, Wang and Chang, 2011). According to StarBase, 146 (36.7%) out of 398 differentially expressed lncRNAs have been experimentally proven to be associate with RBPs (supplementary Table S2). The most abundant lncRNAs with RBP binding sites comprised a group of SNHGs (such as SNHG1, 3, 5, 6, 7, 8, 12, 15 and 16). Furthermore, 118 lncRNAs bind multiple RBPs. For example, LINC00657, SNHG1 and HNRNPU-AS1 bind 31 different RBPs, whereas XIST and OIP5-AS1 bind 29 different RBPs. However, for most lncRNAs, their interacting RBP have not yet been identified. In total, only 33 RBPs have previously been shown to interact with the differentially expressed lncRNAs identified here (supplementary Table S3). Among them, EIF4AIII, UPF1 and FUS were the most widely interacting RBPs and could bind 107, 98 and 92 differentially expressed lncRNAs, respectively (Table S2). Other RBPs like U2AF65, DGCR8, hnRNPC, TAIL1, PTB and FMRP bind more than 40 differentially expressed lncRNAs.
Twenty-one out of the 33 RBPs could be detected at the RNA level (Table S2). Among them, hnRNPC, EWSR1, CAPRIN1, TIA1, IGF2BP3, FXR2 and eIF4AIII were the most abundant. Twelve RBPs (U2AF65, PTB, FMRP, LIN28, LIN28A, LIN28B, TDP43, HuR, SFRS1, C22ORF28, FUS mutant and TNRC6) were neither expressed in IMR-90 cells nor induced by the virus. Expression of these RBPs at the protein level was studied by SILAC (stable isotope labeling by amino acids in cell culture)-based quantitative mass spectrometry (MS) (manuscript in preparation). Among 21 RBPs that were expressed at the RNA level, 15 were also detected at the protein level. The lower number is most likely due to the limited dynamic range of MS or difficulties in extracting certain proteins. Similar RNA and protein expression profiles were observed for eIF4AIII, UPF1, FUS, IGF2BP1, MOV10, FXR1 and EWSR1. The expression of hnRNPC, QKI, TIAL1, IGF2BP2 and IGF2BP3 was stable at the protein level although their expression at the RNA level was up- (hnRNPC) or down-regulated (TIAL1, IGF2BP2, IGF2BP3, FXR2, TAF15 and QKI). In contrast, expression of CAPRIN1 was slightly up-regulated at the protein level, but its RNA remained constant during infection. Gene ontology analysis showed that the most significant functions of these RBPs were posttranscriptional gene regulation, including translation regulation and posttranscriptional RNA processing (Table S3).
4. Discussion
Previously, we have studied changes in host cell mRNA and miRNA expression during the course of a human adenovirus infection by deep sequencing. IMR-90 cells have been used in our studies because of the slow progression of the adenovirus infectious cycle which offers possibilities to examine the cellular gene expression changes during different phases of an infection. Our results have shown that deregulation of mRNA and miRNA occurs in a stepwise manner, switching from genes involved in the cell cycle, growth control, and antiviral response in the early phase to those required for DNA, RNA and protein synthesis in the late phase. Using paired-end sequencing, 645 cellular lncRNAs were identified to be expressed in IMR-90 cell before or after adenovirus infection. Among them, the expression of 398 cellular lncRNAs changed more than 2-fold during infection. Two features characterized the change in lncRNA expression; first, the most striking deregulation occurred during the late phase, and second, the majority of the altered lncRNAs were up-regulated (76% and 82% lncRNAs were up-regulated at 24 and 36 hpi, respectively). The fact that 80% of the differentially expressed lncRNAs showed an increasing expression in the late phase suggests that adenovirus genes are responsible for the regulation. Since the biological functions of the bulk of the lncRNAs are unknown, the functional predictions of deregulated lncRNAs were focused on those lncRNAs which were antisense to known genes and those which have been characterized previously. Antisense RNAs are the most noteworthy class of differentially expressed lncRNAs. They have been shown to exert their effects on their sense partners in cis or in trans (Berretta et al., 2008, Camblong et al., 2009, Guttman and Rinn, 2012). Here, we only studied effects of antisense RNAs on the expression of sense genes in cis.
Clustering analyses showed that deregulation of lncRNA expression correlated with the progression of the infection. Our previous transcriptome studies have shown that the host cell response to the incoming virus is very rapid (Granberg et al., 2006). Thus, the transiently up-regulated lncRNAs during the early phase (Clusters II and IV) must represent a host cell immediate response to the incoming virus since it occurs before viral gene expression has commenced. The lncRNAs in clusters II and IV overlapped with genes that are involved in cell growth control, rearrangement of cellular structure, transcriptional regulation, apoptosis, as well as wound healing. The up-regulation of lncRNAs in the period between the early and the late phase of infection (Clusters I and III) should be caused by early viral gene activity. The up-regulation of these lncRNAs lasted until the late phase, thus their functions would benefit virus replication. A characteristic of adenovirus is its ability to force infected cells to enter the S-phase, thereby providing optimal condition for viral DNA replication. Among 13 lncRNAs in clusters I and III, RP11-973D8.4 and MIR17HG are most noteworthy. RP11-973D8.4 is an antisense RNA which overlaps with the CDK2 gene. In a complex with cyclin E, which also is up-regulated (Zhao et al., 2012), CDK2 plays a critical role in the transition from G1 to S phase of the cell cycle. Consistently, similar expression profiles were observed for CDK2 mRNA and RP11-973D88.4 although there was a slight delay. MIR17HG is a precursor for the miRNA in mir17-92 cluster, a well characterized oncogenic miRNA known as oncomir-1. It comprises miR-17, miR-18a, miR-19a, miR-20a and miR-92a and has been shown to be the most significantly deregulated miRNA cluster during an adenovirus infection (Zhao et al., 2015). Their main targets include E2F, PTEN and BIM which are critical for cell proliferation and apoptosis (Mogilyansky and Rigoutsos, 2013).
The down-regulation of lncRNAs from the early to the late phase of infection appears to be more complicated and should result from both host cell and adenovirus activities. Among the most significant lncRNAs in this category were the antisense RNAs CTC-428H11.2, CTD-2033D15.3, RP11-366L20.2 and BDNF-AS which overlap with RASA1, THBS1, HMGA2 and BDNF, respectively. These genes showed very similar expression profiles at the RNA level as their antisense RNAs. Both RASA1 and THBS1 are negative regulators of signaling pathways and have been implicated in the regulation of cell growth, proliferation and survival. RASA1 acts as a suppressor of RAS function, thus inhibiting cellular proliferation and differentiation. The role of THBS1 in control of cell growth remains controversial (Sid et al., 2004). HMGA2 functions as a transcriptional regulator by binding to the CCNB2 promoter, activating its transcription (De Martino et al., 2009). HMGA2 activates E2F1 by interacting with pRB and by displacing HDAC1 from the pRB/E2F1 complex. Other significantly down regulated lncRNAs were MIR22HG and MIR29B1, host genes for miR-22 and miR-29b. Expression of miR-22 followed the MIR22HG expression changes, whereas miR-29b was slightly up-regulated at 12 hpi before becoming down-regulated at 24 hpi (Zhao et al., 2015). miR-22 is known as a tumor suppressor. It inhibits the growth of several different cancer cell lines by targeting the c-Myc binding partner Max and by silencing the c-Myc binding protein MYCBP, a positive regulator of c-Myc (Ting et al., 2010, Xiong et al., 2010). miR-29 plays diverse roles in different types of cancers and its overexpression has been shown to interfere with bovine viral diarrhea virus replication (Fu et al., 2015).
In the late phase, expression of lncRNAs increased dramatically. The most significant functions of the protein-coding genes that overlap with the antisense lncRNAs were regulators of transcription, DNA, RNA, and protein metabolism as well as cell cycle and growth control. Genes involved in protein synthesis, modification, and transport became more prominent at 36 hpi. These results were consistent with our earlier transcriptome studies (Zhao et al., 2012, Zhao et al., 2003). More than 75% of the antisense RNAs displayed similar expression changes as the mRNAs from the overlapping protein-coding genes. Several differentially expressed lincRNAs identified here have been characterized previously, including MEG3, XIST, FENDRR, NEAT1, RMRP and CRNDE. It has been shown that the expression of MEG3, a putative tumor suppressor, is lost in many human tumors and tumor cell lines (Zhou et al., 2012). Re-expression of MEG3 results in accumulation of p53 protein, as well as stimulation of transcription from p53-dependent promoters, inhibiting tumor cell proliferation in culture. Thus we speculated that up-regulation of MEG3 in the late phase is likely to represent a host antiviral response. Increased MEG3 expression could stimulate p53-dependent transcription and prevent adenovirus-induced cell growth. XIST and FENDRR showed similar expression levels and profiles, both being up-regulated from 24 hpi and remaining increased until 36 hpi. It has been shown that they bind the chromatin-modifying complex PRC2 (polycomb-repressive complex 2) and silence gene expression (Wutz, 2011). XIST plays a role in transcriptional silencing of the X chromosome by recruiting transcriptional silencers and maintaining the inactive state by recruiting PRC2 (Riising et al., 2014). FENDRR plays an essential role in mammalian embryogenesis (Grote and Herrmann, 2013). Expression of FENDRR has been detected in the lung, and is associated with a lethal lung developmental disorder (Stankiewicz et al., 2009).
Deregulation of NEAT1 (Nuclear Enriched Abundant Transcript 1), also known as virus inducible lncRNAs (VINC), is noteworthy (Saha et al., 2006). It was first identified in the mouse central nervous system after infection by Japanese encephalitis virus or Rabies virus. Subsequent studies revealed that its expression also is induced after HIV, herpes simplex virus 1 (HSV-1), and measles virus (MV) infection (Imamura et al., 2014, Zhang et al., 2013). Further studies showed that NEAT1 plays an important role in the innate immune response by facilitating the expression of antiviral genes including cytokines like IL-8. We show here that expression of NEAT1 is slightly up-regulated (1.5-fold) at 12 hpi, and then became down-regulated at 36 hpi. Up-regulation of NEAT1 at 12 hpi is likely to be a host antiviral response which subsequently was suppressed by the virus during the late phase. Consistently, the expression of IL-8 increased more than 9-fold at 6 hpi, before decreasing 2-, 7- and 15-fold at 12, 24 and 36 hpi (data not shown here).
The expression of RMRP (the RNA component of mitochondrial RNA processing endoribonuclease) was high in IMR-90 cells. After adenovirus infection, it was slightly down-regulated at 6 hpi before becoming up-regulated at 36 hpi. RMRP influences multiple cellular RNA processing events. In yeast, it is involved in ribosome synthesis, the generation of RNA primers for mitochondrial DNA replication and the degradation of cell cycle–regulated mRNA (Gill et al., 2004, Lygerou et al., 1996, Welting et al., 2004). In contrast, up-regulation of CRNDE in IMR-90 cells took place after 24 hpi. Up-regulation of CRNDE has been shown in several cancers (Ellis et al., 2012, Graham et al., 2011). The elevated levels of CRNDE expression results in increased cellular metabolism, e.g. glucose synthesis, lactates secretion and lipid production. It is likely that up-regulation of CRNDE is mediated by adenovirus.
Deregulation of a group of SNHGs is noteworthy. Seventeen SNHGs were differentially expressed with very similar expression patterns, e.g. up-regulated during the late phase except SNHG17. Among them, up-regulation of SNHG19 was very dramatic and reached 23- and 63-fold high at 24 hpi and 36 hpi, respectively. Furthermore, 10 out of 17 SNHGs have been demonstrated to interact with more than 10 RBPs. However, the biological functions of these SNHGs are unknown except for SNHG1, also known as U22HG, which is essential for the maturation of 18S rRNA (Tycowski et al., 1994).
Several deregulated lncRNAs are host genes for miRNAs or involved in regulation of miRNA expression. Besides MIR17HG, MIR22HG, MIRLET7DHG, MIR155HG and MIR29B1 discussed above, FTX harbors 4 miRNAs in its introns, miR-374a, miR-374b, miR-421 and miR-545. In addition, it has been shown that FTX regulates XIST expression, thus playing a major role in X chromosome inactivation (Chureau et al., 2011). Here, we showed that expression of FTX was down-regulated during the late phase of infection. However, 4 miRNAs generated from FTX intron showed different expression profiles (Zhao et al., 2015). miR-374a and miR-545 were up-regulated from 12 to 24 hpi and then their expression decreased at 36 hpi. Expression of miR-421 was up-regulated at 36 hpi, whereas miR-374b remained stable during infection. Another miRNA related lncRNA is DANCR, which was up-regulated after 24 hpi. Although DANCR does not encode any miRNA, it blocks miR-320a, miR-199a and miR-214 mediated repression of catenin beta-1 (CTNNB1) expression (Yuan et al., 2015). CTNNB1 is a subunit of the cadherin protein complex and acts as an intracellular signal transducer in the Wnt signaling pathway. A battle between adenovirus and its host in the control the Wnt signaling pathway has been shown in a previous study (Zhao et al., 2007). Many cellular genes involved in Wnt signaling were differentially expressed at different stages of infection. An inhibitory effect of the host cell on the Wnt pathway in the early phase is suppressed by the virus during the late phase. Here, we suggest that one of adenovirus mediates activation of Wnt signaling by up-regulation of DANCR.
Using high-throughput sequencing of immunoprecipitated RNA after cross-linking (CLIP-Seq), many lncRNA-protein interactions have been identified and included in the Starbase database (Li et al., 2014a). Based on information in this database, the differentially expressed lncRNAs identified here should have the potential to interact with 33 RBPs of which 21 were expressed at the RNA level in IMR-90 cells and 15 of them were detected at the protein level. The most important RBPs were eIF4AIII, UPF1 and FUS. They are predicted to interact with more than 50 differentially expressed lncRNAs identified here. Their expression increased during the late phase of the infection. EIF4AIII is a nuclear matrix protein, a member of the DEAD box protein family of putative RNA helicases that are characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD). They are implicated in a number of cellular processes involving alteration of RNA secondary structure, translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. UPF1 is part of a post-splicing multiprotein complex involved in both mRNA nuclear export and mRNA surveillance. FUS belongs to the FET family of RNA-binding proteins which have been implicated in cellular processes, such as regulation of gene expression, maintenance of genomic integrity, as well as mRNA and microRNA processing. The facts that more than 80% of the lncRNAs were up-regulated in the late phase and that many of them are involved in posttranscriptional regulation suggests that cellular lncRNAs play important roles in the late stages of adenovirus replication. The most significantly deregulated lncRNAs and their potential targets, as well as functions are summarized in Fig. 5.
Fig. 5.
Summary of the most noteworthy differentially expressed lncRNAs during adenovirus infection and their potential functions. The left panel includes selected lncRNAs from each cluster. Most of them are antisense RNAs. LincRNAs are in bold text. Red and green bars/arrows represent up- and down-regulated lncRNAs. The right panel shows the mRNAs that overlap with antisense RNAs or miRNAs that are imbedded in the lincRNAs and their known/predicted biological functions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Acknowledgments
Sequencing was performed at the SNP&SEQ Technology Platform at Uppsala University and University Hospital. We thank Ulrika Liljedahl and Johanna Lagensjö for excellent sequencing. Martin Dahlö at UPPMAX is acknowledged for helping to make the code run on the UPPMAX resources. This work was supported by the Kjell and Märta Beijer Foundation (UP), Åke Wiberg Foundation (SBL) and Magnus Bergvall Foundation (SBL).
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.virol.2016.02.017.
Appendix A. Supplementary material
Supplementary material
References
- Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Berretta J., Pinskaya M., Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L.M., Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C., Chantalat S., Romito A., Galvani A., Duret L., Avner P., Rougeulle C. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 2011;20:705–718. doi: 10.1093/hmg/ddq516. [DOI] [PubMed] [Google Scholar]
- De Martino I., Visone R., Wierinckx A., Palmieri D., Ferraro A., Cappabianca P., Chiappetta G., Forzati F., Lombardi G., Colao A., Trouillas J., Fedele M., Fusco A. HMGA proteins up-regulate CCNB2 gene in mouse and human pituitary adenomas. Cancer Res. 2009;69:1844–1850. doi: 10.1158/0008-5472.CAN-08-4133. [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., Lagarde J., Veeravalli L., Ruan X., Ruan Y., Lassmann T., Carninci P., Brown J.B., Lipovich L., Gonzalez J.M., Thomas M., Davis C.A., Shiekhattar R., Gingeras T.R., Hubbard T.J., Notredame C., Harrow J., Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis B.C., Molloy P.L., Graham L.D. CRNDE: a long non-coding RNA involved in CanceR, Neurobiology, and DEvelopment. Front. Genet. 2012;3:270. doi: 10.3389/fgene.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Shi H., Ni W., Shi M., Meng L., Zhang H., Ren Y., Guo F., Wang P., Qiao J., Jia B., Chen C. Lentivirus-mediated Bos taurus bta-miR-29b overexpression interferes with bovine viral diarrhoea virus replication and viral infection-related autophagy by directly targeting ATG14 and ATG9A in Madin–Darby bovine kidney cells. J. Gen. Virol. 2015;96:85–94. doi: 10.1099/vir.0.067140-0. [DOI] [PubMed] [Google Scholar]
- Gill T., Cai T., Aulds J., Wierzbicki S., Schmitt M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L.D., Pedersen S.K., Brown G.S., Ho T., Kassir Z., Moynihan A.T., Vizgoft E.K., Dunne R., Pimlott L., Young G.P., Lapointe L.C., Molloy P.L. Colorectal Neoplasia Differentially Expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer. 2011;2:829–840. doi: 10.1177/1947601911431081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granberg F., Svensson C., Pettersson U., Zhao H. Adenovirus-induced alterations in host cell gene expression prior to the onset of viral gene expression. Virology. 2006;353:1–5. doi: 10.1016/j.virol.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Grote P., Herrmann B.G. The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imamachi N., Tani H., Mizutani R., Imamura K., Irie T., Suzuki Y., Akimitsu N. BRIC-seq: a genome-wide approach for determining RNA stability in mammalian cells. Methods. 2014;67:55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., Kai C., Yada T., Suzuki Y., Yamada T., Ozawa T., Kaneki K., Inoue T., Kobayashi M., Kodama T., Wada Y., Sekimizu K., Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Suzuki H., Carninci P., Hayashizaki Y., Wells C., Frith M., Ravasi T., Pang K.C., Hallinan J., Mattick J., Hume D.A., Lipovich L., Batalov S., Engstrom P.G., Mizuno Y., Faghihi M.A., Sandelin A., Chalk A.M., Mottagui-Tabar S., Liang Z., Lenhard B., Wahlestedt C., Group R.G.E.R., Genome Science G., Consortium F. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Lapidot M., Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.H., Liu S., Zheng L.L., Wu J., Sun W.J., Wang Z.L., Zhou H., Qu L.H., Yang J.H. Discovery of protein-lncRNA interactions by integrating large-scale CLIP-Seq and RNA-Seq datasets. Front. Bioeng. Biotechnol. 2014;2:88. doi: 10.3389/fbioe.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Pluk H., van Venrooij W.J., Seraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky E., Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Endo H., Yokoyama M., Abe J., Tamai K., Tanaka N., Sato I., Takahashi S., Kondo T., Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- Palazzo A.F., Gregory T.R. The case for junk DNA. PLoS Genet. 2014;10:e1004351. doi: 10.1371/journal.pgen.1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A.F., Lee E.S. Non-coding RNA: what is functional and what is junk? Front. Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., Bradel-Tretheway B.G., Korth M.J., Castle J.C., Biery M.C., Bouzek H.K., Haynor D.R., Frieman M.B., Heise M., Raymond C.K., Baric R.S., Katze M.G. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson L. Adenovirus assay by the fluorescent cellcounting procedure. Virology. 1961;15:263–268. doi: 10.1016/0042-6822(61)90357-9. [DOI] [PubMed] [Google Scholar]
- Richards E.J., Zhang G., Li Z.P., Permuth-Wey J., Challa S., Li Y., Kong W., Dan S., Bui M.M., Coppola D., Mao W.M., Sellers T.A., Cheng J.Q. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J. Biol. Chem. 2015;290:6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising E.M., Comet I., Leblanc B., Wu X., Johansen J.V., Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Rosok O., Sioud M. Systematic identification of sense-antisense transcripts in mammalian cells. Nat. Biotechnol. 2004;22:104–108. doi: 10.1038/nbt925. [DOI] [PubMed] [Google Scholar]
- Saha S., Murthy S., Rangarajan P.N. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- Shenk T. 3rd edition. Lippincott-Raven, Publishers; Philadelphia: 1996. Adenoviridae: The Viruses and their Replication. [Google Scholar]
- Shevchenko A., Chernushevich I., Wilm M., Mann M. De Novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. Methods Mol. Biol. 2000;146:1–16. doi: 10.1385/1-59259-045-4:1. [DOI] [PubMed] [Google Scholar]
- Sid B., Sartelet H., Bellon G., El Btaouri H., Rath G., Delorme N., Haye B., Martiny L. Thrombospondin 1: a multifunctional protein implicated in the regulation of tumor growth. Crit. Rev. Oncol./Hematol. 2004;49:245–258. doi: 10.1016/j.critrevonc.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P., Sen P., Bhatt S.S., Storer M., Xia Z., Bejjani B.A., Ou Z., Wiszniewska J., Driscoll D.J., Maisenbacher M.K., Bolivar J., Bauer M., Zackai E.H., McDonald-McGinn D., Nowaczyk M.M., Murray M., Hustead V., Mascotti K., Schultz R., Hallam L., McRae D., Nicholson A.G., Newbury R., Durham-O׳Donnell J., Knight G., Kini U., Shaikh T.H., Martin V., Tyreman M., Simonic I., Willatt L., Paterson J., Mehta S., Rajan D., Fitzgerald T., Gribble S., Prigmore E., Patel A., Shaffer L.G., Carter N.P., Cheung S.W., Langston C., Shaw-Smith C. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am. J. Hum. Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S., Garcia-Alcalde F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y., Medina D.J., Strair R.K., Schaar D.G. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem. Biophys. Res. Commun. 2010;394:606–611. doi: 10.1016/j.bbrc.2010.03.030. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Shu M.D., Steitz J.A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welting T.J., van Venrooij W.J., Pruijn G.J. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterling C., Koch M., Koeppel M., Garcia-Alcalde F., Karlas A., Meyer T.F. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014;11:66–75. doi: 10.4161/rna.27504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Xiong J., Yu D., Wei N., Fu H., Cai T., Huang Y., Wu C., Zheng X., Du Q., Lin D., Liang Z. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- Yin Z., Guan D., Fan Q., Su J., Zheng W., Ma W., Ke C. lncRNA expression signatures in response to enterovirus 71 infection. Biochem. Biophys. Res. Commun. 2013;430:629–633. doi: 10.1016/j.bbrc.2012.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S.X., Wang J., Yang F., Tao Q.F., Zhang J., Wang L.L., Yang Y., Liu H., Wang Z.G., Xu Q.G., Fan J., Liu L., Sun S.H., Zhou W.P. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2015 doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen C.Y., Yedavalli V.S., Jeang K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4:e00596–00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Chen M., Tellgren-Roth C., Pettersson U. Fluctuating expression of microRNAs in adenovirus infected cells. Virology. 2015;478:99–111. doi: 10.1016/j.virol.2015.01.033. [DOI] [PubMed] [Google Scholar]
- Zhao H., Dahlo M., Isaksson A., Syvanen A.C., Pettersson U. The transcriptome of the adenovirus infected cell. Virology. 2012;424:115–128. doi: 10.1016/j.virol.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Zhao H., Granberg F., Elfineh L., Pettersson U., Svensson C. Strategic attack on host cell gene expression during adenovirus infection. J. Virol. 2003;77:11006–11015. doi: 10.1128/JVI.77.20.11006-11015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Granberg F., Pettersson U. How adenovirus strives to control cellular gene expression. Virology. 2007;363:357–375. doi: 10.1016/j.virol.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Li T., Qi J., Liu J., Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS One. 2014;9:e109782. doi: 10.1371/journal.pone.0109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material