Abstract
An anti-influenza compound, DP2392-E10 based on inhibition of the nuclear export function of the viral nucleoprotein-nuclear export signal 3 (NP-NES3) domain was successfully identified by our previous high-throughput screening system. Here, we demonstrated that DP2392-E10 exerts its antiviral effect by inhibiting replication of a broad range of influenza A subtypes. In regard to the molecular mechanism, we revealed that DP2392-E10 inhibits nuclear export of both viral NP and nuclear export protein (NEP). More specifically, in vitro pull-down assays revealed that DP2392-E10 directly binds cellular CRM1, which mediates nuclear export of NP and NEP. In silico docking suggested that DP2392-E10 binds at a region close to the HEAT9 and HEAT10 domains of CRM1. Together, these results indicate that the CRM1-mediated nuclear export function of influenza virus represents a new potential target for antiviral drug development, and also provide a core structure for a novel class of inhibitors that target this function.
Abbreviations: NP, nucleoprotein; NEP, nuclear export protein; NES, nuclear export signal; vRNP, viral ribonucleoprotein; CRM1, chromosome region maintenance 1; AcGFP, Aequorea coerulescens green fluorescent protein; LMB, leptomycin B; IC50, half-maximal inhibitory concentration; CC50, half-maximal cytotoxic concentration; EC50, half-maximal effective concentration; MOE, Molecular Operating Environment
Keywords: Influenza A virus, Nucleoprotein, Nuclear export protein, Nuclear export signal, Chromosome region maintenance 1
Highlights
-
•
DP2392-E10 inhibits replication of a broad range of influenza A subtypes.
-
•
DP2392-E10 inhibits nuclear exports of NP and NEP via their NP-NES3 and NEP-NES2 domains, respectively.
-
•
DP2392-E10 is predicted to directly bind CRM1 in the region near the HEAT9 and HEAT10 repeats.
1. Introduction
Influenza virus, which belongs to family Orthomyxoviridae, replicates its single-stranded negative-sense genomic RNAs in the host nucleus. The viral genomic RNAs are then packed into viral ribonucleoproteins (vRNPs) with viral RNA-dependent RNA polymerase and nucleoproteins (NPs) and exported into cytoplasm for further viral particle assembly and budding at the plasma membrane (Eisfeld et al., 2015). vRNP nuclear export is mediated by the cellular chromosome region maintenance 1 (CRM1) pathway, with assistance from viral matrix protein 1 (M1) and nuclear export proteins (NEP or NS2) (Akarsu et al., 2003, Huang et al., 2013, O'Neill et al., 1998). The vRNP–M1–NEP–CRM1–RanGTP complex is then exported through the nuclear pore to the cytoplasm, where the viral cargo is spontaneously released upon hydrolysis of RanGTP to RanGDP (Eisfeld et al., 2015).
Several studies have described the role of NP in vRNP nuclear export. For example, leptomycin B (LMB), an inhibitor of the CRM1 nuclear export signal (NES) binding domain (Kudo et al., 1999), inhibits nuclear export of vRNP and NP, but not NEP or M1, in virally infected or transfected cells (Elton et al., 2001, Watanabe et al., 2001). In addition, CRM1 overexpression promotes the nuclear export of NP, but not NEP or M1 (Elton et al., 2001), and the interaction of NP with viral genomic RNA, and M1 is sufficient for nuclear export of vRNP (Huang et al., 2001). Moreover, NP directly binds to CRM1 (Chutiwitoonchai et al., 2014, Elton et al., 2001, Kakisaka et al., 2015), and the interaction between NP/CRM1 and a recent identified host factor, nuclear transport factor 2-like export protein 1 (NXT1), promotes nuclear export of NP and vRNA (Chutiwitoonchai and Aida, 2016). NP must also interact with cellular nucleolin, a major component of the nucleolar compartment, to facilitate the interaction between vRNP and the cellular nuclear export machinery (Terrier et al., 2016). In addition to its role in vRNP nuclear export, NP plays other important roles in the viral life cycle, including vRNP nuclear import and transcription/replication of the viral genome (Aida et al., 2012, Cianci et al., 2013, Portela and Digard, 2002, Sasaki et al., 2013). These functional studies support NP as a promising novel target for antiviral drug development.
Inhibition of vRNP nuclear export is an effective strategy for counteracting influenza replication. Treatment with LMB, an inhibitor that irreversibly covalently binds with the C258 of the CRM1 NES binding domain (Kudo et al., 1999), results in retention of vRNPs in the nuclei of infected cells (Elton et al., 2001, Ma et al., 2001, Watanabe et al., 2001). Similarly, Verdinixor, a representative of a new class of CRM1 inhibitor, forms a reversible covalent bond with C258 of CRM1 and thereby inhibits replications of influenza A subtypes H1N1, H5N1, and H7N9 (Perwitasari et al., 2014). Inhibition of vRNP nuclear export by an inhibitor with an unknown target, 14-deoxy-11,12-dehydroandrographolide (Cai et al., 2015), or NP-target inhibitors such as pyrimido-pyrrolo-quinoxalinedione analog (Lin et al., 2015) and 1,3,4,6-tetra-O-galloyl-β-D-glucopyranoside (Chang et al., 2016) can also potentially diminish replication of influenza viruses. Moreover, targeting of the third NES domain of NP (NP-NES3) by an inhibitor we discovered, RK424, results in antiviral effects in vitro and in vivo (Kakisaka et al., 2015). Recently, using our high-throughput CELAVIEW screening system, we identified another potential inhibitor, DP2392-E10, which targets the nuclear export function of the NP-NES3 domain (Kakisaka et al., 2016). Although the molecular mechanism of inhibition by DP2392-E10 was not validated at that time, our preliminary study demonstrated that the compound exerts an antiviral effect against influenza A/WSN/1933 (H1N1) replication (Kakisaka et al., 2016).
Here, we extended our study of DP2392-E10 to characterize its broad range of anti-influenza effects, cytotoxicity, and efficiency of nuclear export inhibition. In addition, we investigated the molecular mechanism of inhibition by DP2392-E10 and found that it targets the CRM1 protein to block CRM1-mediated nuclear export of NP and NEP, thereby preventing vRNP nuclear export and viral replication.
2. Results
2.1. DP2392-E10 reduces replication of a broad range of influenza A subtypes
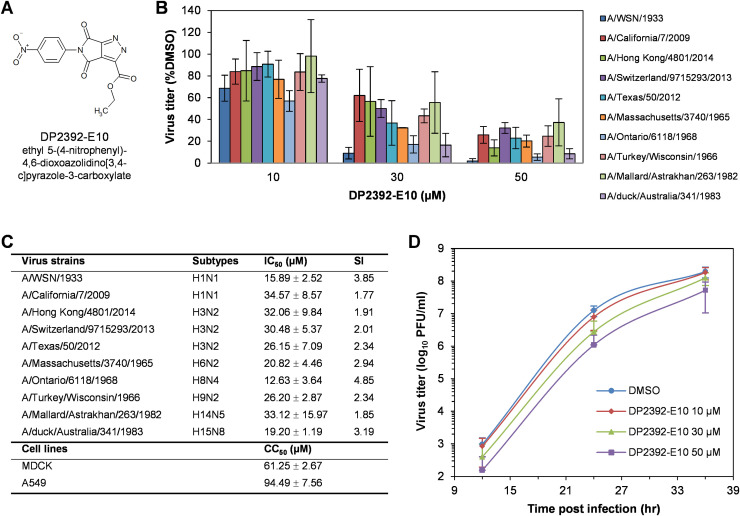
We previously succeeded in identifying a chemical compound, DP2392-E10 ( Fig. 1A), that inhibits the nuclear export function of NP-NES3 domain and decreased replication of influenza A/WSN/1933 (Kakisaka et al., 2016). To further characterize the broad-range inhibitory effect of DP2392-E10, we performed plaque assays to monitor the replications of seasonal influenza A H1N1 and H3N2 including other subtypes, H6N2, H8N4, H9N2, H14N5, and H15N8 in the presence ,,,,,,of the compound. The results revealed that DP2392-E10 decreased replication of all tested subtypes in a dose-dependent manner at the half-maximal inhibitory concentration (IC50) range between 12.63 and 34.57 µM, depending on the viral strains (Fig. 1B and C). Cytotoxicity tests were evaluated in Madin-Darby canine kidney (MDCK) and A549 (a human lung adenocarcinoma epithelial) cells indicating that the compound modestly affected cell viability at the half-maximal cytotoxic concentration (CC50) of 61.25±2.67 and 94.49±7.56 µM, respectively (Fig. 1C). Viral replication kinetic in the presence of DP2392-E10 showed that the compound decreased replication throughout the assay course (Fig. 1D). These results demonstrate that DP2392-E10 inhibits replication of a broad range of influenza A subtypes.
Fig. 1.
DP2392-E10 inhibits replication of a broad range of influenza A subtypes. (A) Chemical structure of DP2392-E10. (B) MDCK cells were infected with different strain of influenza A viruses and subjected to plaque assays in the presence of DMSO or DP2392-E10 at 10, 30, and 50 µM. After 48 h, the cells were stained with crystal violet. Plaques were counted, and the numbers were used to calculate the relative plaque forming units (PFU)/ml (%DMSO). Data represents means ±SD of virus titer from three independent experiments. (C) The half-maximal inhibitory concentration (IC50), half-maximal cytotoxic concentration (CC50), and selectivity index (SI = CC50/IC50) of DP2392-E10 calculated from the plaque assay in B and the Water Soluble Tetrazolium salt (WST-1) assay. (D) MDCK cells were infected with influenza A/WSN/1933 virus at a multiplicity of infection (MOI) of 0.0001 and treated with DMSO or DP2392-E10 at 10, 30, and 50 µM. Supernatant at 12, 24, and 36 h post infection (hpi) were collected for virus titration by plaque assay. Data represents means ±SD of virus titer from three independent experiments.
2.2. DP2392-E10 decreases virus replication by inhibiting nuclear export of viral NP and NEP
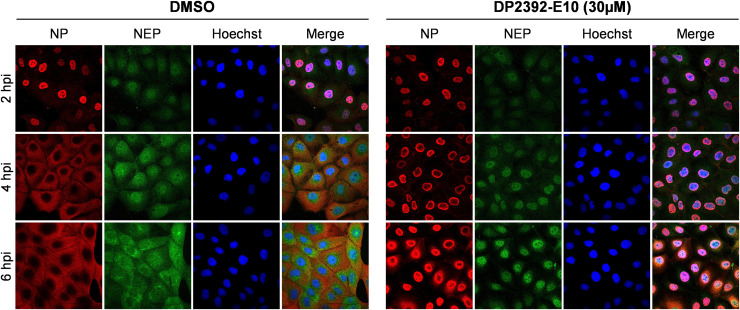
In a high-throughput screen using MDCK cells stably expressing AcGFP-NP-NES3, we identified DP2392-E10 as a compound that could inhibit the CRM1- dependent export of the fusion protein (Kakisaka et al., 2016). To validate the mechanism of inhibition by DP2392-E10, we monitored the intracellular localization of intact NP and another CRM1-dependent nuclear export protein, NEP (O'Neill et al., 1998), in infected cells treated with the compound for 2, 4, or 6 h. In the dimethyl sulfoxide (DMSO) control cells, NP and NEP were localized in the nucleus at 2 h post-infection (hpi), but most of the NP and some NEP had been observed in the cytoplasm at 4 and 6 hpi ( Fig. 2, left panel), reflecting nuclear export observation of NP and NEP in the infected cells. In the DP2392-E10–treated cells, NP and NEP were still localized in the nucleus at 4 and 6 hpi, indicating that the compound prevented nuclear export of both proteins (Fig. 2, right panel). Together with the results of the plaque assays described above, these data suggested that DP2392-E10 reduced virus replication by inhibition of NP and NEP nuclear export.
Fig. 2.
DP2392-E10 inhibits viral replication by preventing nuclear export of viral NP and NEP. MDCK cells were infected with influenza A/WSN/1933 at MOI of 2 and treated with DMSO or 30 µM of DP2392-E10. Intracellular localization of NP and NEP were observed at 2, 4, and 6 hpi by immunofluorescence staining with anti-NP/Alexa Fluor 594 and anti-NEP/Alexa Fluor 488 antibodies. Nuclei were counterstained with Hoechst 33342. Data are representative of two independent experiments.
2.3. DP2392-E10 inhibits CRM1-dependent nuclear exports of the NP-NES3 and NEP-NES2 domains
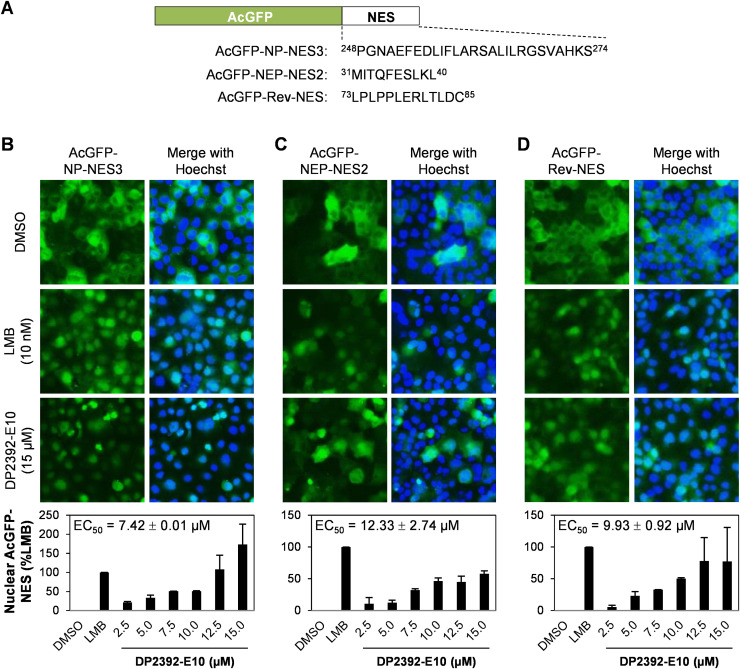
Influenza virus hijacks the cellular CRM1 nuclear export machinery to facilitate nuclear export of NP and NEP via the NP-NES3 and NEP-NES2 domains, respectively (Eisfeld et al., 2015, Huang et al., 2013, Yu et al., 2012). To further confirm that DP2392-E10 inhibits nuclear export of NP-NES3 and NEP-NES2 via the CRM1 pathway, well-known CRM1-dependent nuclear export marker, HIV-1 Rev (Dong et al., 2009, Fornerod et al., 1997), and a specific inhibitor of CRM1 nuclear export, LMB (Kudo et al., 1999), were used. MDCK cells stably expressing Aequorea coerulescens green fluorescent protein (AcGFP)-NP-NES3, AcGFP-NEP-NES2, or AcGFP-Rev-NES domain ( Fig. 3A) (Kakisaka et al., 2016) were treated for 8 h with DMSO, LMB, or DP2392-E10, and the nuclei were stained with Hoechst 33342. CELAVIEW quantification of the green fluorescence intensity from AcGFP-tagged NES domain protein in the nuclear region revealed that DP2392-E10 inhibited cytoplasmic localization of the NP-NES3 and NEP-NES2 domain proteins in a similar manner to LMB (Fig. 3B and C). As expected, DP2392-E10 also inhibited nuclear export of the positive control, the Rev-NES domain protein (Fig. 3D). The inhibition efficiency of DP2392-E10 against nuclear export of NP-NES3, NEP-NES2, and Rev-NES were at the half-maximal effective concentration (EC50) of 7.42±0.01, 12.33±2.74, and 9.93±0.92 µM, respectively. The comparable EC50 values suggest that DP2392-E10 targets the CRM1 protein to inhibit nuclear export of different viral NES domains. This result confirmed that DP2392-E10 inhibits CRM1-dependent nuclear export of NP and NEP via the NP-NES3 and NEP-NES2 domains, respectively, thereby decreasing virus replication.
Fig. 3.
DP2392-E10 inhibits CRM1-mediated nuclear exports of NP-NES3 and NEP-NES2. (A) Schematic representation of Aequorea coerulescens green fluorescent protein (AcGFP)-NP-NES3, AcGFP-NEP-NES2, and AcGFP-Rev-NES constructs. (B-D) MDCK cells stably expressing AcGFP-NP-NES3, AcGFP-NEP-NES2, or AcGFP-Rev-NES were treated with DMSO, 10 nM LMB (positive control), or 2.5, 5.0, 7.5, 10.0, 12.5, or 15.0 µM DP2392-E10 for 8 h. The cells were then fixed with 4% paraformaldehyde, nuclei were stained with Hoechst 33342, and the mean fluorescent intensity of nuclear AcGFP was quantified on a CELAVIEW RS100. Data represents mean ±SD of nuclear AcGFP-NP-NES3 (B, bottom panel), AcGFP-NEP-NES2 (C, bottom panel), or AcGFP-Rev-NES (D, bottom panel) and the half-maximal effective concentration (EC50) values from two independent experiments. Each of triplicate samples in one experiment was visualized for 36 fields of observation.
2.4. DP2392-E10 prevents the NP/CRM1 interaction by directly targeting the CRM1 protein
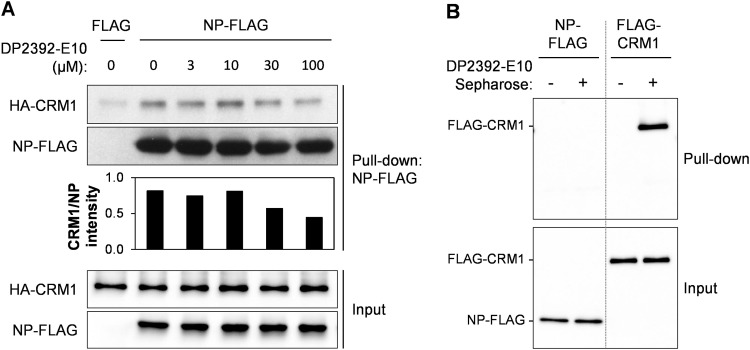
The nuclear export function of the NP-NES3 domain is CRM1-dependent (Yu et al., 2012), and NP directly binds CRM1 (Chutiwitoonchai et al., 2014, Elton et al., 2001, Kakisaka et al., 2016). To verify that DP2392-E10 inhibits nuclear export by blocking the NP/CRM1 (and NEP/CRM1) interaction, we performed in vitro pull-downs of NP and CRM1 proteins in the presence of the compound. For this purpose, NP-FLAG protein was immobilized on anti-FLAG monoclonal antibody (mAb)-conjugated agarose beads and incubated with purified CRM1-HA protein in the presence of DMSO or DP2392-E10 at 3, 10, 30, or 100 µM. Western blot analysis of the pull-down samples indicated that DP2392-E10 inhibited NP/CRM1 binding in a dose-dependent manner ( Fig. 4A).
Fig. 4.
DP2392-E10 inhibits NP/CRM1 binding by directly targeting CRM1. (A) FLAG– or NP-FLAG–immobilized agarose beads were incubated with purified CRM1-HA protein in the presence of DMSO or 3, 10, 30, or 100 µM of DP2392-E10 overnight, pulled down, washed, and subjected to western blot analysis with anti-HA and anti-FLAG antibodies. Intensity of the pulled-down CRM1 and NP were quantified using the ImageJ software. Data are representative of two independent experiments. (B) Photo-crosslinked Sepharose beads without DP2392-E10 (control) or with DP2392-E10 were co-incubated with purified NP-FLAG or FLAG-CRM1 proteins, pulled down, washed, and subjected to western blot analysis with anti-FLAG antibody. Data are representative of two independent experiments.
We next investigated whether CRM1 is the target of DP2392-E10 by generating DP2392-E10 photo-crosslinked Sepharose beads and subjecting them to the pull-down assay. Specifically, the beads were incubated with purified NP-FLAG or FLAG-CRM1 protein overnight, and then pulled down. Western blots revealed that DP2392-E10 directly bound CRM1, but not NP (Fig. 4B).
Together, these observations indicate that DP2392-E10 targets the CRM1 protein to inhibit the NP/CRM1 interaction, which is necessary for virus replication.
2.5. DP2392-E10 is predicted to interact with CRM1 in the region near the HEAT9 and HEAT10 repeats
The in vitro affinity binding assays described above indicated that DP2392-E10 directly binds to CRM1 protein. To obtain structural insights into the interaction between CRM1 and DP2392-E10, we performed an in silico study involving molecular modeling and docking simulations. Because our experimental assays used both human (293 T, for CRM1 protein purification) and canine (MDCK, for replication and immunofluorescence) cell lines, we constructed molecular models of unliganded CRM1 from both human and canine. The human CRM1 model retains the basic structural fold of twenty-one HEAT repeats, a common characteristic of CRM1 proteins ( Fig. 5A). We then used Molecular Operating Environment (MOE) to search for small-molecule binding pockets in these models. A total of 45 and 40 pockets were found in the human and canine CRM1, respectively, and the data suggested slight differences in the surface structures of the two proteins (Fig. 5B and C, left panels). Next, we evaluated the binding affinities and conformations of DP2392-E10 and each pocket, as reflected by U_dock docking scores. The highest U_dock score was identified at a pocket near the HEAT9–HEAT10 repeat regions in both human and canine CRM1s (human CRM1: −34.8 kcal/mol., canine CRM1: −31.0 kcal/mol) (Fig. 5B and C, right panel). These results suggest that the optimal binding sites for DP2392-E10 on human and canine CRM1 proteins are near the HEAT9 and HEAT10 repeats.
Fig. 5.
In silico analysis of the interaction between DP2392-E10 and unliganded CRM1. Models of unliganded human and canine CRM1 proteins were constructed by homology modeling and subjected to docking simulation studies in the Molecular Operating Environment (MOE). (A) Structural model of human CRM1. The model consists of twenty-one HEAT repeats (H1–H21), each composed of two α-helices linked by a short loop (Fung and Chook, 2014, Monecke et al., 2013). HEAT repeats for a switch from the inactive to active conformation of CRM1 are highlighted by colors: H9 (magenta), H9 acid loop (orange), H11 (yellow), and H12 (cyan). (B and C) Left panels: 3D distribution of potential binding pockets on human (B) or canine (C) CRM1. The pockets were detected around all the HEAT repeats, except the H21 region. The pockets for the binding of small molecules are shown as grey spheres. Right panels: Distribution of docking scores of U_dock at individual pockets. In each pocket, several U_dock values representing distinct binding conformations were found. Only U_dock scores giving negative values are shown. A red bar at a given site indicates the average of all U_dock scores at the site.
3. Discussion
Nuclear export of vRNP was originally thought to be mediated by the NEP/CRM1 interaction (O'Neill et al., 1998), but several later studies indicated a role for NP in vRNP nuclear export: for example, LMB inhibits nuclear export of vRNP in a NEP-independent manner (Elton et al., 2001, Ma et al., 2001, Watanabe et al., 2001), and NES-3–defective NP decreases viral production and replication efficiency (Chutiwitoonchai et al., 2014, Yu et al., 2012). Therefore, we speculate that DP2392-E10 decreases viral replication by interfering with both NP/CRM1– and NEP/CRM1–mediated nuclear export of vRNP. The inhibitory effect of DP2392-E10 on both NP-NES3– and NEP-NES2–mediated nuclear export functions indicates that this molecule represents a promising lead compound for further drug development. Notably, however, DP2392-E10 may not affect the nuclear export function of the M1-NES domain, which is CRM1-independent (Cao et al., 2012). This study demonstrated a broad range inhibitory effect of DP2392-E10 against replication of seasonal influenza A H1N1 and H3N2 including some other subtypes. Because the NP-NES3 domain is conserved among human, avian, and swine influenza viruses (Chutiwitoonchai et al., 2014), we expect that DP2392-E10 will also be able to inhibit pandemic subtypes such as swine influenza H1N1 and avian influenza H5N1. The inhibitory effect of DP2392-E10 could be further improved by structure–activity relationship analysis, making it available for use in combination with, or as an alternative to, currently approved anti-influenza drugs.
Nuclear export mediated by CRM1 is initiated by an interaction between RanGTP and the HEAT9 loop of CRM1, triggering a conformational change in the NES binding site that permits NES–cargo protein binding (Dong et al., 2009, Fung and Chook, 2014). Our docking simulation results predicted that DP2392-E10 binds at the region around the HEAT9 and HEAT10 repeats of CRM1. This may directly cause a conformational change of the NES binding site of CRM1, or indirectly affect the folding of the HEAT9 loop that is critical for RanGTP binding. Thus, these unusual conformational changes of CRM1 decrease the affinity of binding with the NP-NES3 domain, interfere with the nuclear export function of NP, and inhibit viral replication. This proposed mechanism of inhibition by DP2392-E10 differs from that of LMB or Verdinixor, which directly bind the C258 of CRM1 NES binding site to prevent the CRM1/NES–cargo protein interaction (Kudo et al., 1999, Perwitasari et al., 2014).
Our CELAVIEW analysis indicated that DP2392-E10 also inhibits nuclear export of HIV-1 Rev-NES, which plays a well-defined role in nuclear export of HIV-1 unspliced and singly spliced RNAs for viral protein production and viral particle assembly (Fornerod et al., 1997). Antiviral activity against intact HIV-1 replication is further worthily verification that will provide a new class of anti-HIV-1 inhibitor based on the Rev nuclear export function for the highly active antiretroviral therapy. In addition, DP2392-E10 is expected to exert an antiviral effect against a broad range of viruses that exploit the cellular CRM1 nuclear export machinery, including severe acute respiratory syndrome coronavirus (Freundt et al., 2009), hepatitis viruses (Cerutti et al., 2011, Forgues et al., 2001), and herpes simplex virus (Williams et al., 2008).
In conclusion, we demonstrated that DP2392-E10 inhibits replication of a broad range of influenza A subtypes, including H1N1, H3N2, H6N2, H8N4, H9N2, H14N5, and H15N8. The compound reduces viral replication by directly targeting the CRM1 protein in the predicted region near the HEAT9 and HEAT10 domains, thereby preventing the CRM1/NP and CRM1/NEP interactions. These effects result in inhibition of the nuclear export functions of NP and NEP, which are important for vRNP nuclear export and viral assembly.
4. Materials and methods
4.1. Cells, viruses, plasmids, and DP2392-E10
MDCK, A549, and human embryonic kidney (HEK) 293 T cells were maintained in Dulbecco's Modified Eagle's Medium (Gibco, Beijing, China) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT, USA). MDCK stably expressing AcGFP-tagged influenza NP-NES3, AcGFP-NP-NES3 or HIV-1 Rev-NES, AcGFP-Rev-NES were established as previously described (Kakisaka et al., 2016). The influenza NEP-NES2 domain was derived from annealing of two oligonucleotides, 5'-CCGGTCAATGGAATAATAACACAGTTCGAGTCTCTGAAACTCTACAGATAAGC-3' and 5'-GGCCGCTTATCTGTAGAGTTTCAGAGACTCGAACTGTGTTATTATTCCATTGA-3', to establish stable AcGFP-NEP-NES2–expressing MDCK cells, as described (Kakisaka et al., 2016).
A plasmid set for reverse genetics of infectious influenza A/WSN/1933 (H1N1) virus was gift from Dr. Yoshihiro Kawaoka (Institute of Medical Science, University of Tokyo, Japan) (Neumann et al., 1999). Seasonal influenza A/California/7/2009, A/Hong Kong/4801/2014, A/Switzerland/9715293/2013, and A/Texas/50/2012 viruses were provided by Dr. Shinji Watanabe (Center for Influenza Virus Research, National Institute of Infectious Diseases, Japan). Influenza A/Massachusetts/3740/1965 (H6N2), A/Ontario/6118/1968 (H8N4), A/Turkey/Wisconsin/1966 (H9N2), A/Mallard/Astrakhan/263/1982 (H14N5), and A/duck/Australia/341/1983 (H15N8) viruses were gifts from Dr. Hiroshi Kida and Dr. Yoshihiro Sakoda (Graduate School of Veterinary Medicine, Hokkaido University, Japan).
The pCAGGS/NP-FLAG, pCAGGS/FLAG-CRM1, and pCAGGS/CRM1-HA plasmids were constructed as described previously (Chutiwitoonchai and Aida, 2016, Chutiwitoonchai et al., 2014).
DP2392-E10 was provided by the Drug Discovery Initiative at the University of Tokyo (http://www.ddi.u-tokyo.ac.jp/en/) or synthesized by Vitas-M Laboratory, Ltd. (Mongkok, Kowloon, Hong Kong) and GreenPharma (Allée du Titane, Orléans, France). DMSO was purchased from Nacalai Tesque, Inc. (Kyoto, Japan) and used to dissolve DP2392-E10 including normalization of the compound volume at each tested concentration.
4.2. Antiviral activity assay
MDCK cells (5×105/well) were seeded overnight onto 6-well plates before infection with various influenza A subtypes, and then subjected to plaque assays as previously described (Chutiwitoonchai et al., 2014) in the presence of DMSO (0.5% total volume) or 10, 30, or 50 µM DP2392-E10. The IC50 was calculated from linear regression of virus titer vs. compound concentration. DMSO was defined as 100% infection.
Replication kinetic assay was performed as described previously (Chutiwitoonchai et al., 2014) in MDCK cells infected with influenza A/WSN/1933 virus at a multiplicity of infection (MOI) of 0.0001 and treated with DMSO or DP2392-E10 at 10, 30, or 50 µM. Viruses in supernatant at 12, 24, and 36 hpi were titrated by plaque assay.
4.3. Cytotoxicity assay
MDCK or A549 cells (3×104/well) were seeded overnight in 96-well plates and treated with DMSO (1% total volume) or 1, 10, 30, 50, 70, or 100 µM DP2392-E10 for 48 h. Water Soluble Tetrazolium salt (WST-1) assays were performed as described previously (Chutiwitoonchai and Aida, 2016). The CC50 was calculated from linear regression of OD450 vs. compound concentration. DMSO was defined as 100% cell viability.
4.4. Immunofluorescence staining
MDCK cells (4×104 cells) were seeded overnight on cover glass in 12-well plates before infection with influenza A/WSN/33 virus at MOI of 2, and then treated with DMSO (0.3% total volume) or 30 µM DP2392-E10 for 2, 4, or 6 h. The infected cells were subjected to immunofluorescence staining with anti-NP mouse mAb (Santa Cruz Biotechnology, Dallas, TX, USA), anti-NEP rabbit polyclonal antibody (GeneTex, Irvine, CA, USA), Alexa Fluor 594 goat anti-mouse IgG (Invitrogen, Waltham, MA, USA), and Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen), as described previously (Chutiwitoonchai and Aida, 2016).
4.5. CELAVIEW analysis
MDCK cells (2×104/well) stably expressing AcGFP-NP-NES3, AcGFP-NEP-NES2, or AcGFP-Rev-NES (Kakisaka et al., 2016) were seeded overnight in 96-well plates before treatment for 8 h with DMSO, 10 nM LMB (Sigma-Aldrich, Saint Louis, MO, USA; as positive control), or DP2392-E10 at 2.5, 5.0, 7.5, 10.0, 12.5, or 15.0 µM. Nuclear localization of the AcGFP-NES proteins was quantitated on a CELAVIEW RS100 (Olympus) as previously described (Kakisaka et al., 2016). The mean fluorescence intensity of nuclear NES was subtracted with DMSO control and normalized against the value from the LMB control. The EC50 was calculated from linear regression of mean fluorescence intensity vs. compound concentration. LMB was defined as 100% inhibition.
4.6. Pull-down assays
NP-FLAG–immobilized agarose beads and purified CRM1-HA protein from transfected HEK293T cells were prepared as previously described (Chutiwitoonchai et al., 2014). Pull-down assays of NP/CRM1 interaction with DP2392-E10 were performed by incubation of 15 µl of NP-FLAG–immobilized agarose beads (50% slurry), 2 µg of purified CRM1-HA protein, and either DMSO or 3, 10, 30, or 100 µM DP2392-E10 in bind/wash buffer (10 mM Tris-Cl [pH 7.8], 150 mM NaCl, 0.05% NP-40) at 4 °C overnight. The beads were pulled down, washed five times with bind/wash buffer, and subjected to western blot analysis with anti-HA mouse mAb (Medical and Biological Laboratories, Nagoya, Japan) and anti-FLAG rabbit mAb (Sigma-Aldrich). The intensity of CRM1 and NP bands were quantitated by ImageJ 1.50 software (National Institute of Health, Bethesda, MD, USA).
4.7. Photo-crosslinked small-molecule affinity binding assay
DP2392-E10 was photo-crosslinked onto Sepharose beads as described previously (Kanoh et al., 2005). The photo-crosslinked Sepharose beads without compound were used as control. NP-FLAG and FLAG-CRM1 proteins were purified as described (Chutiwitoonchai and Aida, 2016, Chutiwitoonchai et al., 2014) from transfected HEK293T cells. Pull-down assays to assess the DP2392-E10/NP or DP2392-E10/CRM1 interaction were performed by incubation of 20 µl of DP2392-E10–photo-crosslinked Sepharose beads (50% slurry) and 2 µg of purified NP-FLAG or FLAG-CRM1 protein in bind/wash buffer at 4 °C overnight. The beads were pulled down, washed five times with bind/wash buffer, and subjected to western blot analysis with anti-FLAG rabbit mAb (Sigma-Aldrich).
4.8. In silico docking
Molecular models of human and canine CRM1 in the unliganded state were constructed using homology modeling in the MOE (Chemical Computing Group Inc., Montreal, Canada). The X-ray crystal structure of the unliganded CRM1 from Chaetomium thermophilum at a resolution of 2.94 Å (Protein Data Bank accession number 4FGV) was used as the template for homology modeling. Homo sapiens CRM1 (NP_003391) and Canis lupus familiaris (XP_531839) amino acid sequences were retrieved from the NCBI database. After the CRM1 models were constructed, they were refined based on reference to the Ramachandran plot. Potential binding sites on each CRM1 were identified using the site finder application of MOE. Binding affinities and conformation between DP2392-E10 and each pocket of the CRM1s were evaluated using the ASEDock application of MOE (Goto et al., 2008) as described (Yokoyama et al., 2010).
Acknowledgments
We thank the Platform for Drug Discovery, Informatics, and Structural Life Science of the Ministry of Education, Culture, Sports, Science and Technology, Japan, for providing DP2392-E10. We are also grateful to Dr. Shinji Watanabe (Center for Influenza Virus Research, National Institute of Infectious Diseases, Japan), Dr. Hiroshi Kida, and Dr. Yoshihiro Sakoda (Graduate School of Veterinary Medicine, Hokkaido University, Japan) for giving influenza A subtype viruses, Dr. Yoshihiro Kawaoka (Institute of Medical Science, University of Tokyo, Japan) for providing the plasmids used for reverse genetics, and Ms. Kaori Honda (Bio-Active Compounds Discovery Research Unit, RIKEN CSRS, Japan) for technical support with DP2392-E10 photo-crosslinked Sepharose beads. This work was supported in part by a research project of the Ministry of Agriculture, Forestry and Fisheries of Japan aimed at improving food safety and animal health (grant number 13406421), and by the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship (grant number 15F15416).
References
- Aida Y., Sasaki Y., Hagiwara K. Discovery of novel antiviral agents directed against the influenza A Virus Nucleoprotein. In: Arbuthnot P., editor. Antiviral Drugs - Aspects of Clinical Use and Recent Advances. InTech; Rijeka, Croatia: 2012. pp. 99–120. [Google Scholar]
- Akarsu H., Burmeister W.P., Petosa C., Petit I., Muller C.W., Ruigrok R.W., Baudin F. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2) EMBO J. 2003;22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Li Y., Chen S., Wang M., Zhang A., Zhou H., Chen H., Jin M. 14-Deoxy-11,12-dehydroandrographolide exerts anti-influenza A virus activity and inhibits replication of H5N1 virus by restraining nuclear export of viral ribonucleoprotein complexes. Antivir. Res. 2015;118:82–92. doi: 10.1016/j.antiviral.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Cao S., Liu X., Yu M., Li J., Jia X., Bi Y., Sun L., Gao G.F., Liu W. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. J. Virol. 2012;86:4883–4891. doi: 10.1128/JVI.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A., Maillard P., Minisini R., Vidalain P.O., Roohvand F., Pecheur E.I., Pirisi M., Budkowska A. Identification of a functional, CRM-1-dependent nuclear export signal in hepatitis C virus core protein. PloS One. 2011;6:e25854. doi: 10.1371/journal.pone.0025854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.Y., Park J.H., Kim Y.H., Kang J.S., Min J.Y. A natural component from Euphorbia humifusa Willd displays novel, broad-spectrum anti-influenza activity by blocking nuclear export of viral ribonucleoprotein. Biochem. Biophys. Res. Commun. 2016;471:282–289. doi: 10.1016/j.bbrc.2016.01.123. [DOI] [PubMed] [Google Scholar]
- Chutiwitoonchai N., Aida Y. NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway. Viruses. 2016:8. doi: 10.3390/v8080209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutiwitoonchai N., Kakisaka M., Yamada K., Aida Y. Comparative analysis of seven viral nuclear export signals (NESs) reveals the crucial role of nuclear export mediated by the third NES consensus sequence of nucleoprotein (NP) in influenza A virus replication. PLoS One. 2014;9:e105081. doi: 10.1371/journal.pone.0105081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci C., Gerritz S.W., Deminie C., Krystal M. Influenza nucleoprotein: promising target for antiviral chemotherapy. Antivir. Chem. Chemother. 2013;23:77–91. doi: 10.3851/IMP2235. [DOI] [PubMed] [Google Scholar]
- Dong X., Biswas A., Chook Y.M. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat. Struct. Mol. Biol. 2009;16:558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld A.J., Neumann G., Kawaoka Y. At the centre: influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015;13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton D., Simpson-Holley M., Archer K., Medcalf L., Hallam R., McCauley J., Digard P. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 2001;75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgues M., Marrogi A.J., Spillare E.A., Wu C.G., Yang Q., Yoshida M., Wang X.W. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 2001;276:22797–22803. doi: 10.1074/jbc.M101259200. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Park E., Lenardo M.J., Xu X.N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung H.Y., Chook Y.M. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin. Cancer Biol. 2014;27:52–61. doi: 10.1016/j.semcancer.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto J., Kataoka R., Muta H., Hirayama N. ASEDock-docking based on alpha spheres and excluded volumes. J. Chem. Inf. Model. 2008;48:583–590. doi: 10.1021/ci700352q. [DOI] [PubMed] [Google Scholar]
- Huang S., Chen J., Chen Q., Wang H., Yao Y., Chen J., Chen Z. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J. Virol. 2013;87:767–778. doi: 10.1128/JVI.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Liu T., Muller J., Levandowski R.A., Ye Z. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology. 2001;287:405–416. doi: 10.1006/viro.2001.1067. [DOI] [PubMed] [Google Scholar]
- Kakisaka M., Mano T., Aida Y. A high-throughput screening system targeting the nuclear export pathway via the third nuclear export signal of influenza A virus nucleoprotein. Virus Res. 2016;217:23–31. doi: 10.1016/j.virusres.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Kakisaka M., Sasaki Y., Yamada K., Kondoh Y., Hikono H., Osada H., Tomii K., Saito T., Aida Y. A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein. PLoS Pathog. 2015;11:e1005062. doi: 10.1371/journal.ppat.1005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh N., Honda K., Simizu S., Muroi M., Osada H. Photo-cross-linked small-molecule affinity matrix for facilitating forward and reverse chemical genetics. Angew. Chem. 2005;44:3559–3562. doi: 10.1002/anie.200462370. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E.P., Wolff, B., Yoshida, M., Horinouchi, S., 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. In: Proceedings of the National Academy of Sciences of the United States of America 96, pp. 9112–9117. [DOI] [PMC free article] [PubMed]
- Lin M.I., Su B.H., Lee C.H., Wang S.T., Wu W.C., Dangate P., Wang S.Y., Huang W.I., Cheng T.J., Lin O.A., Cheng Y.S., Tseng Y.J., Sun C.M. Synthesis and inhibitory effects of novel pyrimido-pyrrolo-quinoxalinedione analogues targeting nucleoproteins of influenza A virus H1N1. Eur. J. Med. Chem. 2015;102:477–486. doi: 10.1016/j.ejmech.2015.08.016. [DOI] [PubMed] [Google Scholar]
- Ma K., Roy A.M., Whittaker G.R. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology. 2001;282:215–220. doi: 10.1006/viro.2001.0833. [DOI] [PubMed] [Google Scholar]
- Monecke, T., Haselbach, D., Voss, B., Russek, A., Neumann, P., Thomson, E., Hurt, E., Zachariae, U., Stark, H., Grubmuller, H., Dickmanns, A., Ficner, R., 2013. Structural basis for cooperativity of CRM1 export complex formation. In: Proceedings of the National Academy of Sciences of the United States of America 110, pp. 960–965. [DOI] [PMC free article] [PubMed]
- Neumann, G., Watanabe, T., Ito, H., Watanabe, S., Goto, H., Gao, P., Hughes, M., Perez, D.R., Donis, R., Hoffmann, E., Hobom, G., Kawaoka, Y., 1999. Generation of influenza A viruses entirely from cloned cDNAs. In: Proceedings of the National Academy of Sciences of the United States of America 96, pp. 9345–9350. [DOI] [PMC free article] [PubMed]
- O'Neill R.E., Talon J., Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwitasari O., Johnson S., Yan X., Howerth E., Shacham S., Landesman Y., Baloglu E., McCauley D., Tamir S., Tompkins S.M., Tripp R.A. Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo. J. Virol. 2014;88:10228–10243. doi: 10.1128/JVI.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A., Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. General. Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Hagiwara K., Kakisaka M., Yamada K., Murakami T., Aida Y. Importin alpha3/Qip1 is involved in multiplication of mutant influenza virus with alanine mutation at amino acid 9 independently of nuclear transport function. PloS One. 2013;8:e55765. doi: 10.1371/journal.pone.0055765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier O., Carron C., De Chassey B., Dubois J., Traversier A., Julien T., Cartet G., Proust A., Hacot S., Ressnikoff D., Lotteau V., Lina B., Diaz J.J., Moules V., Rosa-Calatrava M. Nucleolin interacts with influenza A nucleoprotein and contributes to viral ribonucleoprotein complexes nuclear trafficking and efficient influenza viral replication. Sci. Rep. 2016;6:29006. doi: 10.1038/srep29006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Takizawa N., Katoh M., Hoshida K., Kobayashi N., Nagata K. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 2001;77:31–42. doi: 10.1016/s0168-1702(01)00263-5. [DOI] [PubMed] [Google Scholar]
- Williams P., Verhagen J., Elliott G. Characterization of a CRM1-dependent nuclear export signal in the C terminus of herpes simplex virus type 1 tegument protein UL47. J. Virol. 2008;82:10946–10952. doi: 10.1128/JVI.01403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama M., Mori H., Sato H. Allosteric regulation of HIV-1 reverse transcriptase by ATP for nucleotide selection. PLoS One. 2010;5:e8867. doi: 10.1371/journal.pone.0008867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Liu X., Cao S., Zhao Z., Zhang K., Xie Q., Chen C., Gao S., Bi Y., Sun L., Ye X., Gao G.F., Liu W. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. J. Virol. 2012;86:4970–4980. doi: 10.1128/JVI.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]