Abstract
Current technologies with next generation sequencing have revolutionized metagenomics analysis of clinical samples. To achieve the non-selective amplification and recovery of low abundance genetic sequences, a simplified Sequence-Independent, Single-Primer Amplification (SISPA) technique in combination with MiSeq platform was applied to target negative- and positive-sense single-stranded RNA viral sequences. This method allowed successful sequence assembly of full or near full length avian influenza virus (AIV), infectious bronchitis virus (IBV), and Newcastle disease virus (NDV) viral genome. Moreover, SISPA analysis applied to unknown clinical cases of mixed viral infections produced genome assemblies comprising 98% NDV and 99% of IBV genomes. Complete or near complete virus genome sequence was obtained with titers at or above 104.5 EID50/ml (50% embryo infectious dose), and virus identification could be detected with titers at or above 103 EID50/ml. Taken together, these studies demonstrate a simple template enrichment protocol for rapid detection and accurate characterization of avian RNA viruses.
Keywords: Random amplification, Sequence-independent single-primer amplification, Next generation sequencing, Avian influenza virus, Infectious bronchitis virus, Newcastle disease virus, RNA viruses, Poultry
Highlights
-
•
A simple, random priming technique was optimized to target viral RNA genomes.
-
•
This technique allows characterization of multiple viruses in single reaction.
-
•
Complete or near complete genome sequence with titers at or above 104.5 EID50/ml.
-
•
The detection limit with viral titers at or above 103 EID50/ml.
1. Introduction
RNA viruses are a large genetically-diverse group of infectious agents whose evolutionary diversity is driven by adaptability to its host (Sanjuán et al., 2010). Characterizing the genomes by sequence-based methods is the most widely used approach to determine RNA virus diversity and thereby determine the relationships between isolates within the population. Furthermore, to fully understand the mechanisms of viral adaptation and evolution, and to perform molecular diagnosis of emerging and re-emerging viral infections there is an increasing need to produce full length viral genomes sequences.
Utilization of next generation sequencing (NGS) in molecular epidemiological analysis of outbreak strains facilitates the rapid and accurate identification of etiologic agents and can be used to provide information on the origin of viruses (Gilchrist et al., 2015). In the absence of targeted sequencing protocols, as well as during outbreak situations, the random amplification of purified nucleic acids in combination with NGS can be a useful tool for monitoring viral evolution or diagnosis. Although most molecular detection methods are unable to detect mixed viral infections, NGS allows for both target dependent and target independent sequencing of viral genomes and provides an opportunity to detect the genome of multiple viruses simultaneously (Thorburn et al., 2015). However, the samples used in such studies are often available in limited quantities of virus.
Sequence-Independent, Single-Primer Amplification (SISPA) is one of the random priming methods develop by Reyes and Kim (1991) that allows enrichment of the viral genome in only a few steps (Djikeng et al., 2008). There have been several modifications of the SISPA method since its first implementation, including random-PCR (rPCR) (Froussard, 1993). This combines first-step reverse transcription followed by denaturation, annealing and amplification by using random hexamers tagged with a known sequence which is then used as a primer binding extension sequence. It was developed to yield double-stranded cDNA in sufficient abundance for cloning and then sequencing (Froussard, 1993, Zou et al., 2003). DePew et al. (2013) further modified the SISPA protocol to identify cultivable viruses from a single plaque using 454 and HiSeq Illumina platform without the need of a cloning step.
Previous studies have combined random priming approaches with NGS for identification of a novel mink astrovirus (Blomström et al., 2010), the partial sequencing of a novel paramyxovirus in penguins (Miller et al., 2010b), influenza viruses (Afonso, 2007), metagenomics analysis of Dengue virus infected mosquitoes (Bishop-Lilly et al., 2010), and viruses in human stool samples (Victoria et al., 2009). Moreover, the NGS approach has recently been applied for viral genome sequencing of archived serum samples from the oldest HIV-1 positive group genomes, resulting in identification of “Patient 0” (Worobey et al., 2016).
Single-stranded positive-sense RNA (ssRNA+) and negative sense RNA (ssRNA) viruses represent a large group of avian viral agents, including West Nile virus (WNV), avian metapneumovirus (aMPV), infectious bronchitis virus (IBV), Newcastle disease virus (NDV), and avian influenza virus (AIV) (Swayne, 2013). Among RNA viruses, AIV of Orthomyxoviridae family and NDV of Paramyxoviridae family are considered as the most devastating poultry diseases owing to their highly pathogenic forms as well as worldwide distribution and economic implications (Swayne, 2008; Miller et al., 2010a; Alexander et al., 2012). IBV of the Coronaviridae family also causes significant morbidity and economic losses to the poultry industry affecting primarily the respiratory tract, and can demonstrate a wide range of tissues tropism, including the renal and reproductive systems (Bande et al., 2016). Combining target independent genome amplification with the high sensitivity of NGS provides the opportunity not only for detection of these viral pathogens, but can also simultaneously provide complete genomic sequences to allow further genetic characterization. Nonetheless, the feasibility of applying these techniques to diagnosis and research of pathogens requires further evaluation and optimization study.
A simplified, sequence-independent technique of directional amplification in combination with NGS was used in this study. The protocol was tested against purified virus stocks and clinical samples. Viral genome sequences were determined from both negative- and positive-sense single stranded RNA viruses. The limit of detection necessary for virus identification using SISPA–NGS and metagenomics approaches was also determined.
2. Material and methods
2.1. Viruses
Viruses used in these studies included two highly pathogenic AIV, A/duck/Vietnam/NCVD-672/2011(H5N1) (Dk/Vn), and A/turkey/Minnesota/15–12582-1/2015(H5N2) (Tk/Mn), two NDV isolates, A/duck/Vn/Long bien/78/02 (Dk/Vn/NDV) and Lasota B1 vaccine strain (LaS), and a single IBV strain, Ark99. The IBV (Ark99 strain) was kindly provided by Jackwood (University of Georgia, Athens, USA). All viruses were propagated in 9–11 day of embryonating specific pathogen free chicken eggs. Following three days of growth, the allantoic fluid was harvested for RNA extraction. Clinical samples, consisting of oral swabs, were collected from flocks of chickens in Jordan. The samples were kept under BSL3 facility in −80 °C until used. The SISPA-NGS method was applied to determine viral presence in clinical samples. Furthermore, clinical samples were used for comparison to purified virus stocks.
2.2. RNA isolation
Total RNA extraction from pure virus stocks and clinical samples was performed using RNeasy Mini Kit (QIAGEN, Valencia, USA) according to manufacturer's instruction. Optimal quality of the extracted RNA was verified by obtaining the OD260/OD280 values using NanoDrop (Thermo Scientific, Wilmington, USA).
2.3. Viral RNA quantification
For AIV, quantitative real time RT-PCR (qRRT-PCR) was performed as previously described (Kapczynski et al., 2013, Spackman et al., 2002). Briefly, qRRT-PCR reactions targeting the influenza virus M gene was conducted using AgPath-ID one-step RT-PCR Kit (Ambion, Austin, TX) and the ABI 7500 Fast Real-Time PCR system (Applied Biosystem, Calsbad, CA). The RT step conditions for reactions were 10 min at 45 °C and 95 °C for 10 min. The cycling conditions were 45 cycles of 15 s, 95 °C; 45 s, 60 °C. For virus quantification, a standard curve was established with RNA extracted from dilutions of the same titrated stock of the viruses. Ct (cycle threshold) values of each viral dilution were plotted against viral titers. The resulting standard curve had a high correlation coefficient (r2 > 0.99), and it was used to convert Ct values to EID50/ml.
For NDV, qRRT-PCR targeting the NDV M gene was conducted as described previously (Wise et al., 2004). The RT step was 30 min at 50 °C, followed by 15 min at 95 °C. The cycling conditions consisted of 40 cycles of 10 s of denaturation at 94 °C, 30 s of annealing at 52 °C, and extension at 72 °C for 10 s. For virus quantification, a standard curve was established with RNA extracted from dilutions of the titrated stock of the virus qRRT-PCR was conducted using AgPath-ID one-step RT-PCR Kit (Ambion) and the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Calsbad, CA).
For IBV, qRRT-PCR for IBV detection was performed as described previously (Callison et al., 2006). The reaction was conducted at 50 °C for 30 min; 95 °C for 15 min; 40 cycles of 94 °C for 1 s followed by 60 °C for 60 s. For virus quantification, a standard curve was established with RNA extracted from dilutions of the titrated stock of the virus. RRT-PCR was conducted using AgPath-ID one-step RT-PCR Kit (Ambion) and the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Calsbad, CA).
2.4. Random priming (RP)-mediated SISPA
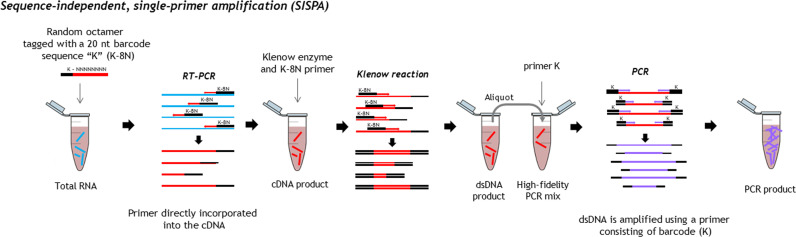
As shown in Fig. 1, first-strand cDNA was synthesized in a 20-μl reaction mixture with 5 μl of viral nucleic acids from each sample, 100 pmol of primer K-8N (GACCATCTAGCGACCTCCACNNNNNNNN) modified from (Rosseel et al., 2013), SuperScript IV Reverse Transcriptase (ThermoFisher scientific), and dNTPs (10 μM) following the manufacturer's instructions. To convert the first-strand cDNA into double-stranded cDNA, 20 μl of the first-strand cDNA was heated to 95 °C for 3 min and then cooled to 4 °C in the presence of 10 pmol of primer K-8N, and 10 μM dNTPs in 1× Klenow reaction buffer (NEB). Afterwards, 1 uL of Klenow fragment were added and incubated at 37 °C for 60 min (final volume, 25 μl). After conversion into dsDNA by Klenow polymerase (NEB), the products were purified using Agencourt AMPure XP beads (Beckman Coulter). The purified dsDNA of the Klenow reaction was subsequently used as a template for PCR amplification.
Fig. 1.
Overview of the SISPA strategy.
Sequence-independent PCR amplification was conducted with 5 μl of the double-stranded cDNA template in a final reaction volume of 50 μl, which contained 1× Phusion HF buffer, 200 μM deoxynucleoside triphosphate (dNTP), 10 μM primer K (GACCATCTAGCGACCTCCAC), and 0.5 U Phusion DNA polymerase (NEB). The PCR cycling was performed as follows: 98 °C for 30 s, followed by 35 cycles of 98 °C for 10 s, 55 °C for 30 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter). For quantification of the ds cDNA, the Qubit dsDNA HS assay (Invitrogen) was performed according to the manufacturer's instruction.
2.5. Sequence-dependent approach for AIV genome sequencing
One step RT-PCR was conducted with 5 μl and 20 μl of RNA template in a final reaction volume of 50 μl using OneTaq® One-Step RT-PCR Kit (NEB) with the primers Optil-F1, GTTACGCGCCAGCAAAAGCAGG, Optil-F2, GTTACGCGCCAGCGAAAG CAGG, and Optil-R1, GTTACGCGCCAGTAGAAACAAGG. The PCR cycling was performed as follows: 95 °C for 2 min, 42 °C for 60 min, 94 °C for 2 min, 5 cycles of 94 °C for 30 s, 44 °C for 30 s, and 68 °C for 3.5 min, followed by 26 cycles of 94 °C for 30 s, 57 °C for 30 s, 68 °C for 3.5 min with a final extension at 68 °C for 10 min (Zhou et al., 2009).
2.6. Genome sequencing
The Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and 0.2 ng/μl (1 ng total) of ds cDNA were used in this study to generate multiplexed paired-end sequencing libraries, according to the manufacturer's instructions. The dsDNA was fragmented and tagged with adapters by Nextera XT transposase. The Nextera XT transposome fragmented PCR amplicons with added adaptor sequences enabled a 12-cycle PCR amplification to append additional unique dual index (i7 and i5) sequences at the end of each fragmented DNA for cluster formation. PCR fragments were purified on Agencourt AMpure XP beads (Beckman Coulter). Fragments were analyzed on a High Sensitivity DNA Chip on the Bioanalyzer (Agilent Technologies) before loading on the sequencing chip. Briefly, the libraries were adjusted to 4 nM concentration and equal volumes of 5 μl of each library were pooled. The pool was denatured with NaOH (0.2 N final concentration) and further diluted to 10 pM. Control library (5% PhiX library, Illumina, USA) was added to the pool. The library pool was loaded in the flow cell of the 500 cycle MiSeq Reagent Kit v2 (Illumina, USA). The barcoded multiplexed library sequencing (2 × 250 bp) was performed on an Illumina MiSeq platform (Illumina, USA).
2.7. Sequence analysis
The de novo and directed assembly of genome sequences were performed using the Geneious 9.1.2 (http://www.geneious.com, Kearse et al., 2012) and GALAXY software (Afgan et al., 2016). The reads were directly mapped to the reference genomes (or all segments of reference genome): A/chicken/Vietnam/NCVD-A937/2011(H5N1) (8 segments, GenBank accession no. KP097848, KP097869, KP097890, KP097925, KP097946, KP097981, KP098002, KP098023); A/turkey/Minnesota/15-12582-1/2015(H5N2) (8 segments, GenBank accession no. KX351776- KX351783), A/chicken/Israel/554/2005(H9N2) (8 segments, GenBank accession no. EF492279, EF492227, EF492377, EF492336, EF492348, EF492308, EF492406, EF492250), Infectious bronchitis virus strain Arkansas Vaccine (GenBank accession no. GQ504721.2), Infectious bronchitis virus serotype H120, complete genome (GenBank accession no. JF950510.1, Newcastle disease virus strain JSG0210 (GenBank accession no. JF340367.1), and Newcastle disease virus strain go/CH/GD-QY/1997 (GenBank accession no. KJ782375.1). All sequence sets were analyzed by the Kraken Metagenomics classifier (Wood and Salzberg, 2014) installed at Illumina BaseSpace app using MiniKraken database version 20141208 (https://basespace.illumina.com/apps).
3. Results
3.1. Whole genome sequencing of pure virus stock
An optimized SISPA protocol was tested in this study to amplify the viral genome of AIV, NDV and IBV using viral preparation from allantoic fluid. The workflow for the SISPA–NGS technique is presented in Fig. 1. This modified SISPA protocol allowed successfully assembly of sequences into full or near full-length viral genomes representing negative- and positive-sense single stranded RNA ( Table 1). The genome coverage for the viral stock with the titer at or above 106 EID50/ml range from 99.8% for AIV to 100% for NDV Lasota B1 vaccine strain, and IBV Ark99 (Table 1). The percentage of reads assembled to the reference genomes was greater than 40% for all the viruses tested (Fig. S1). Although the average depth differed between the viruses tested, the number of reads were high and sufficient to perform polymorphism analysis (Table 1). The mean read depth metric performed with Geneious 9.1.2. for AIVs, IBV and NDV is shown in Fig. S2 and Fig. S3, respectively.
Table 1.
Quantitation of specific virus reads and sequence coverage in virus stocks.
| Virus | Virus titer (EID50/ml) | Gene segments | Coverage (%) | Mean depth of coverage | SD | Assembled reads/total paired readsa | Q30 |
|---|---|---|---|---|---|---|---|
| (%) | |||||||
| AIV H5N2 | 106 | PB2 | 100 | 12,180.5 | 12,784.2 | 112,856 | 96.5 |
| A/turkey/Minnesota/15–012582-1/2015 | PB1 | 98.9b | 4685.6 | 4207.6 | 46,495 | 96.5 | |
| PA | 100 | 7385.4 | 7727.8 | 79,192 | 87.4 | ||
| HA | 100 | 14,846.5 | 12,410.4 | 102,233 | 96.4 | ||
| NP | 100 | 1917.1 | 1125.9 | 20,574 | 75.9 | ||
| NA | 100 | 13,563.2 | 11,913.8 | 92,618 | 94 | ||
| M | 100 | 8656.1 | 6618.8 | 37,642 | 93.7 | ||
| NS | 100 | 3800.7 | 2176.8 | 14,017 | 97 | ||
| Genome | 505,627/ 718,922 | N/A | |||||
| AIV H5N1 | 105.9 | PB2 | 98.1c | 200.4 | 92.7 | 3007 | 95.8 |
| A/duck/Vn/NCVD-672/2011 | PB1 | 100 | 62.7 | 46.3 | 972 | 96.3 | |
| PA | 100 | 91.4 | 63.5 | 1296 | 94.9 | ||
| HA | 100 | 1330.9 | 817.2 | 15,448 | 67.4 | ||
| NP | 100 | 409.6 | 460.4 | 4233 | 86.7 | ||
| NA | 98.7d | 780.2 | 640.8 | 7302 | 95.7 | ||
| M | 100 | 1378.6 | 1061.9 | 9142 | 95.7 | ||
| NS | 100 | 149.9 | 79.6 | 758 | 95.8 | ||
| Genome | 163,661/ 421,418 | N/A | |||||
| NDV | 107 | NP | 100 | 625.6 | 203.9 | ||
| Lasota vaccine strain | P | 100 | 1898.2 | 1025.1 | |||
| M | 100 | 4280.4 | 2819.7 | ||||
| F | 100 | 3513.5 | 1078.1 | ||||
| HN | 100 | 23,703.1 | 20,025.1 | ||||
| L | 100 | 6408.3 | 3946.8 | ||||
| Genome | 100 | 7151 | 10,584.4 | 388,022/ 847,390 | 94.8 | ||
| 106.1 | N | 100 | 24,060 | 8087.3 | |||
| 5b | 100 | 9269.7 | 3608.6 | ||||
| IBV | 5a | 100 | 2272.4 | 990.8 | |||
| Ark99 | M | 100 | 490.6 | 114 | |||
| 3c | 100 | 597.9 | 49.9 | ||||
| 3b | 100 | 951.8 | 214.6 | ||||
| 3a | 100 | 1767.1 | 160.7 | ||||
| S | 100 | 1281.7 | 958.4 | ||||
| ORF1ab | 100 | 6382.4 | 15,046.8 | ||||
| Genome | 100 | 6503.4 | 13,883.2 | 576,259/ 772,020 | 96.6 | ||
SD, standard deviation; N/A, not applicable.
The numbers of assembled sequencing reads to a reference genome.
Gaps at nucleotide position 588–613.
Gaps at nucleotide position 1–44.
Gaps at nucleotide position 1–17.
3.2. The genome coverage of clinical samples
Next, the modified SISPA technique was tested against clinical field samples. The genome coverage of clinical samples with virus titers greater than 106 EID50/ml was similar to the genome coverage obtained after sequencing the viral stock and ranged from 98.1% for AIV, 94.9–98.1% for NDVs, to 98.8% for IBV ( Table 2). Moreover, near full length NDV chicken/Jordan/15/2004 and IBV Jordan/Mass/15/2004 viral genomes were obtained from a mixed infection clinical sample (Table 2). However, the mean depth of coverage decreased as compared to the virus stock sequencing, and was very low especially for the NP gene of NDV, and NS gene of AIV (Table 2). The percentage of reads assembled to the reference genomes was below 20% for clinical samples tested which was much lower as compared to the virus stock (Fig. S1), most likely due to overwhelming competition with non-specific or host genome sequences. The distribution of the reads classified by Kraken for mixed infection clinical sample is shown in Fig. 2. Out of 304,125 total reads, 40,329 reads were classified to the domain level, 35,744 to the family level, and 35,908 to the species level. An analysis of the classified reads reveals that 52.8% of reads were classified into virus domain. Newcastle disease virus and avian coronavirus represented 19% and 8% of total reads classified to the virus domain, and 11.26% and 4.99% of total reads classified to the species level, respectively.
Table 2.
Quantitation of specific virus reads and coverage in the clinical samples.
| Virus | Virus titer (EID50/ml) | Gene segment | Coverage (%) | Mean depth of coverage | SD | Assembled reads/total paired readsa | Q30 |
|---|---|---|---|---|---|---|---|
| (%) | |||||||
| AIV H9N2 | 107.7 | PB2 | 100 | 349 | 241 | 4879 | 93.7 |
| A/chicken/Jordan/13/2003 | PB1 | 100 | 100 | 263.7 | 139.8 | 94.4 | |
| PA | 100 | 71.6 | 27.6 | 909 | 94.2 | ||
| HA | 100 | 611.9 | 473.2 | 5959 | 94.1 | ||
| NP | 100 | 46.2 | 32.7 | 1024 | 94.4 | ||
| NA | 100 | 1092.4 | 981.7 | 9617 | 94.5 | ||
| M | 100 | 545.4 | 425.2 | 3214 | 93.4 | ||
| NS | 85.4b | 3.1 | 2 | 17 | 90.5 | ||
| Genome | 29,217/ 530,908 | N/A | |||||
| NDV | 107.1 | NP | 82.1 | 2.7 | 2.0 | ||
| chicken/Jordan/17/2004 | P | 100 | 21.3 | 13.4 | |||
| M | 99.3 | 16.7 | 8.0 | ||||
| F | 100 | 33.2 | 15.8 | ||||
| HN | 100 | 19.5 | 7.2 | ||||
| L | 99.7 | 21.3 | 19.6 | ||||
| Genome | 94.9c | 18.1 | 16.3 | 1378/ 534,158 | 92.1 | ||
| NDVf chicken/Jordan/15/2004 | 107.2 | NP | 90.1 | 2.1 | 2.6 | ||
| P | 100 | 100 | 34.1 | ||||
| M | 100 | 536.1 | 528 | ||||
| F | 100 | 48.8 | 17.3 | ||||
| HN | 100 | 281.6 | 280.6 | ||||
| L | 99.98 | 60 | 50.9 | ||||
| Genome | 98.1d | 874.3 | 3063 | 114,992/ 608,250 | 93.4 | ||
| IBVf Jordan/Mass/15/2004 | 106.2 | N | 100 | 176 | 43.9 | ||
| 5b | 100 | 100 | 69.1 | ||||
| 5a | 100 | 45.9 | 6.9 | ||||
| M | 100 | 24 | 5.1 | ||||
| 3c | 100 | 40.5 | 7.7 | ||||
| 3b | 100 | 73.9 | 14.9 | ||||
| 3a | 100 | 118 | 7.3 | ||||
| S | 100 | 68 | 61.7 | ||||
| ORF1ab | 98.5 | 23.7 | 31.7 | ||||
| Genome | 98.8e | 2468 | 6728 | 118,763/ 608,250 | 98 |
SD, standard deviation; N/A, not applicable.
The numbers of assembled sequencing reads to a reference genome.
Gaps at nucleotide position 1–65; 603–763; 809–838.
Gaps at nucleotide position 1–196; 1148–1237;1419–1750; 3085–3316; 15,103– 15,190.
Gaps at nucleotide position 1–979; 14,939– 14,979.
Gaps at nucleotide position 1–302; 15,059– 15,063; 15,113– 15,213; 27,622– 27,641.
Mix viral infections obtained in a single reaction from clinical sample.
Fig. 2.
Taxonomic distribution of a mixed infection clinical sample of Newcastle disease virus and avian coronavirus classified by Kraken metagenomics tool.
The NDV and IBV viral sequences obtained from clinical samples were submitted to GenBank under accession numbers: NDV chicken/Jordan/17/2004, GenBank accession no. KY212126, NDV chicken/Jordan/15/2004, GenBank accession no. KY212127, IBV Jordan/Mass/15/2004, GenBank accession no. KY273667.
3.3. Limit of detection
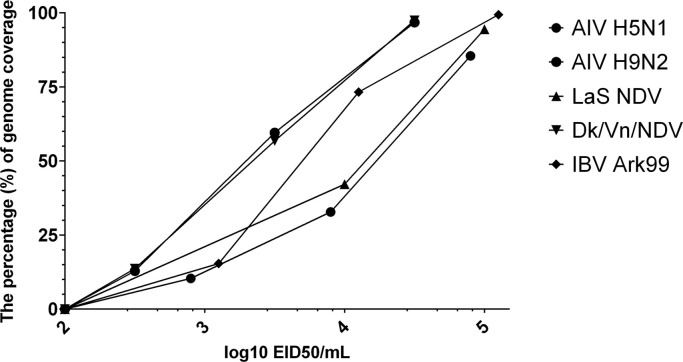
The second goal of the study was to assess the feasibility of virus identification using SISPA–NGS and to get an estimate of its limit of detection for diagnosis of viral infections. For this purpose, two AIV isolates (Dk/Vn H5N1 and AIV A/chicken/Jordan/13/2003 (H9N2) (Ck/JO)), two NDV isolates (Dk/Vn/NDV and LaS), and one IBV isolate (Ark99) were selected. Extracts of the dilution series of viral RNA from each strain was used (Table S1). The results demonstrate that initial viral titers greater than 104.5 EID50/ml allowed the generation from 85.5% to 99.4% of genome sequence ( Fig. 3). We were able to successfully assemble the sequences of AIV ( Table 3), NDV ( Table. 4) and IBV ( Table 5) into full or near full length of genomes. Detailed statistics on the NDV and IBV genomes assembly are provided in Supplementary Tables S2 and S3, respectively. Although the genome coverage of virus with titers below 103.0 EID50/ml was decreased, it was still possible to detect AIV and NDV viral genomes using Kraken metagenomics approach. For example, with the AIV clinical sample (102.5 EID50/ml), out of 105,544 total reads classified, 49.0% were classified to the domain level, 23.6% to the family level, and 24.3% to the species level. Influenza A virus represented 100% of ssRNA viruses, 23% of reads classified to the virus level and 0.6% of all reads classified. For NDV with the viral titer of 102.5 EID50/ml out of 117,062 total reads classified, 53.9% reads were categorized into domain level, 14.7% into family level, and 19.8% into species level. NDV represented 96% of reads classified to ssRNA viruses, 4.0% of reads classified to the virus level and 0.9% of all reads classified. IBV, was not detected when the virus titer was below 103 EID50/ml. The limit of detection for IBV by the Kraken metagenomics approach used lies at 103.1 EID50/ml. In this case, out of 34,254 reads classified, 25.6% were categorized into domain level, 22.3% into family level, and 18.8% into species level. Avian coronavirus represented 98% of positive ssRNA viruses and 2% of viruses.
Fig. 3.
Genome coverage comparison by EID50/ml for the viruses tested.
Table 3.
Sensitivity of SISPA – next generation sequencing for avian influenza viruses.
| Virus | Virus titer (EID50/ml) | Ct Value | Gene segment | Read mappeda | Genome coverage | Mean depth of coveragec | Contig (s) by de novo assemblyd | Detection by metagenomicse |
|---|---|---|---|---|---|---|---|---|
| (%)b | ||||||||
| AIV H5N1 | 104.9 | 26.4 | PB2 | 898 | 90.8 | 36.8 | ||
| A/duck/Vn/NCVD-672/2011 | PB1 | 755 | 66.3 | 83.6 | ||||
| PA | 60 | 78 | 3.7 | |||||
| HA | 202 | 89.3 | 16.6 | |||||
| NP | 104 | 99.1 | 10.7 | |||||
| NA | 101 | 84.8 | 12.8 | |||||
| M | 96 | 96.9 | 10.3 | |||||
| NS | 118 | 79.1 | 19.4 | |||||
| Genome | 85.5 | Y | Y | |||||
| 103.9 | 30.1 | PB2 | 39 | 42.1 | 2.1 | |||
| PB1 | 7 | 0 | 0 | |||||
| PA | 185 | 81.2 | 24.2 | |||||
| HA | 6 | 0 | 0 | |||||
| NP | 88 | 56.2 | 1.9 | |||||
| NA | 17 | 44.1 | 1.8 | |||||
| M | 0 | 0 | 0 | |||||
| NS | 18 | 39 | 1.4 | |||||
| Genome | 32.8 | N | Y | |||||
| 102.9 | 33.5 | Genome | 10.4 | N | N | |||
| AIV H9N2 | 104.5 | 28.7 | PB2 | 1894 | 100 | 179.5 | ||
| A/chicken/Jordan/13/2003 | PB1 | 737 | 97.7 | 74.6 | ||||
| PA | 269 | 100 | 29.4 | |||||
| HA | 1631 | 94.9 | 225.5 | |||||
| NP | 188 | 97.6 | 28.4 | |||||
| NA | 2154 | 92.9 | 358.7 | |||||
| M | 908 | 100 | 212.2 | |||||
| NS | 12 | 91.4 | 2.9 | |||||
| Genome | 96.8 | Y | Y | |||||
| 103.5 | 30.8 | PB2 | 532 | 82.6 | 47.4 | |||
| PB1 | 5940 | 97.2 | 244.9 | |||||
| PA | 254 | 66.7 | 18.1 | |||||
| HA | 28 | 36.3 | 2.7 | |||||
| NP | 35 | 55.2 | 3.8 | |||||
| NA | 355 | 81.1 | 44.6 | |||||
| M | 228 | 57.8 | 40.6 | |||||
| NS | 2 | 0 | 0 | |||||
| Genome | 59.6 | N | Y | |||||
| 102.5 | 34.3 | Genome | 12.9 | N | Y |
The numbers of assembled sequencing reads to a reference genome.
The breadth of coverage of a target genome (%).
The average number of times that each base in the reference is covered by aligned reads.
The formation of contigs in de novo genome assembly.
Detection of AIV viral genomes using Kraken metagenomics approach.
Table 4.
Sensitivity of SISPA – next generation sequencing for Newcastle disease viruses.
| Virus | Virus titer (EID50/ml) | Ct Value | Total paired reads | Read mappeda | Genome coverageb | Mean depth of coveragec | Contig (s) by de novo assemblyd | Detection by metagenomicse |
|---|---|---|---|---|---|---|---|---|
| NDV A/duck/Vn/Long bien/78/02 | 104.5 | 25.02 | 494,904 | 8431 | 97.5 | 82.3 | Y | Y |
| 103.5 | 28.34 | 348,868 | 3636 | 56.8 | 41.1 | N | Y | |
| 102.5 | 31.46 | 344,538 | 1307 | 13.7 | 15.8 | N | Y | |
| 101.5 | 34.45 | 565,646 | N/A | N/A | N/A | N | N | |
| NDV Lasota | 107 | 18.5 | 847,390 | 388,022 | 100 | 7151 | Y | Y |
| 106 | 21.4 | 766,972 | 369,177 | 99.6 | 6273.2 | Y | Y | |
| 105 | 24.7 | 854,782 | 4623 | 94.5 | 72.1 | Y | Y | |
| 104 | 27.5 | 1,061,980 | 320 | 42.2 | 4.4 | N | Y | |
| 103 | 30.5 | 1,036,364 | 32 | N/A | N/A | N | N | |
| 102 | 33.7 | 1,028,688 | N/A | N/A | N/A | N | N |
N/A, not applicable.
The numbers of assembled sequencing reads to a reference genome.
The breadth of coverage of a target genome (%).
The average number of times that each base in the reference is covered by aligned read.
The formation of contigs in de novo genome assembly.
Detection of IBV viral genomes using Kraken metagenomics approach.
Table 5.
Sensitivity of SISPA – next generation sequencing for infectious bronchitis virus.
| Virus | Virus titer (EID50/ml) | Ct Value | Total paired reads | Read mappeda | Genome coverageb | Mean depth of coveragec | Contig (s) by de novo assemblyd | Detection by metagenomicse |
|---|---|---|---|---|---|---|---|---|
| IBV Ark99 | 106.1 | 19.5 | 772,020 | 576,259 | 100 | 6503.4 | Y | Y |
| 105.1 | 22.5 | 898,634 | 192,805 | 97.3 | 1813.1 | Y | Y | |
| 104.1 | 25.8 | 336,150 | 4339 | 73.3 | 29.6 | Y | Y | |
| 103.1 | 28.8 | 259,626 | 222 | 15.4 | 1.3 | N | Y | |
| 102.1 | 32.4 | 275,462 | N/A | N/A | N/A | N | N |
N/A, not applicable.
The numbers of assembled sequencing reads to a reference genome.
The breadth of coverage of a target genome.
The average number of times that each base in the reference is covered by aligned read.
The formation of contigs in de novo genome assembly.
Detection of IBV viral genomes using Kraken metagenomics approach.
3.4. Comparison of sequence-dependent and sequence independent approaches for AIVs
Finally, the sequence-dependent sequencing and SISPA technique were compared using AIV. For this purpose, Dk/Vn H5N1 and Ck/JO H9N2 viruses were used in the experiment (Table S4). Viral particle titers greater than 106 EID50/ml allowed generation of the complete genome of all 8 segments with high depth of coverage, with a range from 52,928 reads/bp for M gene to 544.9 reads for NA gene. Since there was no overwhelming competition with other non-specific sequences as observed in sequence-independent approach, the depth of coverage in sequence-dependent sequencing was higher than that of SISPA–NGS method. However, below 106 viral particles per RT-PCR reaction, assembled sequences of AIVs were missing nearly 50% of one of the genes (the coverage of PB1 segment of Dk/Vn H5N1 was 54.2% and NA gene of A/chicken/Jordan/13/2003 was 52.9%). The genome coverage of each segment was substantially decreased in samples with the titer below 105 viral particles per RT-PCR reaction, especially for gene longer than 1.7 kb, such as PB2, PB1, PA, and HA (Table S4). We were not able to amplify and sequence the samples with titers below 104 EID50/ml (data not shown), suggesting that it has lower limit of detection than SISPA-NGS method.
4. Discussion
Whole-genome sequencing provides an unsurpassed level of genetic information for virus discovery, diagnosis, characterization and genotypic classification that would not be available by using classical methodologies. Moreover, NGS technologies provide opportunities to screen for pathogens in outbreak situations from known and novel or unexpected pathogens in a single reaction. Thus, developing the simple, universal and fast amplification protocol that yields double-stranded cDNA in sufficient abundance can facilitate and expedite the whole–genome sequencing and might be a useful procedure for use not only in research but also in routine clinical diagnosis. So far, several research groups have optimized and adapted modifications of SISPA method of viral genomes and coupled it to the construction of a plasmid library generating full or near full length viral genomes (Allander et al., 2001, Djikeng et al., 2008). Palmenberg et al. (2009) have successfully applied the SISPA method to obtain the complete genome sequence of human RNA viruses, including the full length genome sequences of a total of 78 rhinovirus isolates available at the American Type Culture Collection (ATCC). In turn, Rosseel et al. (2012) applied the DNase SISPA method to detect Schmallenberg virus in infected field samples. Other groups have also used the DNase- SISPA technique for the detection of bovine and human viruses (Allander et al., 2001, Djikeng and Spiro, 2009), for the identification of a novel human coronavirus (van der Hoek et al., 2004) and for viral discovery in the plasma of HIV infected patients (Jones et al., 2005).
In this study, we present a simplified sequence-independent technique of directional amplification, in combination with illumina MiSeq NGS Platform that permitted us to generate the viral genome sequences of AIV, NDV and IBV. The main advantage of this modified SISPA protocol is its simplicity and possibility to detect multiple RNA viruses in single reaction without the need to propagate the viruses, thereby greatly reducing time and costs necessary to characterize viral genomes. It was possible to extract and amplify RNA in 5 h using modified SISPA protocol and identify the viral genomes in 3 days (including 49 h Illumina Miseq run). The protocol presented here, do not require a pre-processing step, duplication of amplification, or pooling of samples (Depew et al., 2013). We applied a denaturation step before conversion of cDNA into dsDNA by Klenow polymerase and size selection after the Klenow reaction and PCR amplification, which improved the yield of double-stranded cDNA and increased the average size of fragments. The PCR product obtained after amplification can be directly processed to generate multiplexed paired-end sequencing libraries. We have demonstrated that the method is efficient, easy to perform, and needs modest virus titers to detect and obtain complete or nearly complete genome sequence. It is predominantly useful for obtaining genome sequences from RNA viruses, uncharacterized RNA viruses, or investigating complex clinical samples (such as mixed infections in single reaction) as no prior sequence information is needed. As shown in Table 2 and Fig. 2, the SISPA method in combination with NGS can be a useful random priming method for a molecular detection of mixed viral infections for disease diagnosis in a single reaction.
Not unexpectedly, the detection limit was dependent on viral load in the samples. Based on these findings, a minimum initial virus titer of 104.5 EID50/ml was needed to obtain the full or nearly full genome sequence as the titer of the virus correlated positively with the genome coverage. Secondly, for diagnostic detection, identification of already known agents required a minimum titer of 103 EID50/ml. We also demonstrate that non-specific bacterial or host genes in clinical samples can greatly reduce the efficiency of viral genome amplification and sequencing. Metagenomics analysis of samples that contain less than 103.0 EID50/ml of the viruses was possible, but could vary greatly and result in low reproducibility if the sample contains a high amount of non-viral or host genes. In this case a pre-processing step in the sample preparation, such as DNase treatment and filtration, could improve the efficacy of viral amplification and sequencing.
The SISPA technique was further compared with sequence-dependent amplification for AIV (Zhou et al., 2009). The sequence-dependent AIV sequencing allowed us to generate complete genome sequence when the sample contained at least 106 EID50/ml of virus, and the depth of coverage was higher than that obtained from SISPA amplification. The limitation of this method is its relatively higher limit of detection and the need of primer sequences at the 5′ and 3′ ends of the genes. Comparison of sequence-dependent and sequence-independent sequencing approaches for AIV showed that both of the methods can result in complete genome sequences when the virus titer is higher than 105.0 EID50/ml. In this case, the method of choice depends on the purpose of the study (screening, virus discovery, diagnosis, SNP analysis, etc.). Below 105.0 EID50/ml, sequence-independent amplification can result in lower depth of coverage, but better overall coverage for large segments. In addition, both methods showed uneven distribution of sequence reads between segments. Optimization of PCR condition could be helpful to improve this limitation by extending the random oligomer sequence (Rosseel et al., 2013), using different 5′ defined tag sequences (Zhou et al., 2009), and using augmented PCR cycling for long segments.
In conclusion, NGS approaches are now leading to a more in-depth analysis of virus genetic diversity and thereby increase our overall knowledge of viral epidemiology and evolutionary dynamics what is crucial for understanding viral ecology. In this study, the simplified SISPA protocol in combination with NGS efficiently generated full length RNA viral genomes of AIV, NDV and IBV. It will be useful to detect the emergence of novel virus strains and provide information on their genetic features, enabling a better diagnosis and disease control.
Acknowledgments
This work was supported by USDA, United States Department of Agriculture, ARS CRIS funds 6040-32000-062-00D and USDA-NIFA grant number 60-6040-5-005.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2017.06.019.
Appendix A. Supplementary material
Supplementary material
References
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvierm D., Čech M., Chilton J., Clements D., Coraor N., Eberhard C., Grüning B., Guerler A., Hillman-Jackson J., Von Kuster G., Rasche E., Soranzo N., Turaga N., Taylor J., Nekrutenko A., Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso C.L. Sequencing of avian influenza virus genomes following random amplification. Biotechniques. 2007;43:188–192. doi: 10.2144/000112529. [DOI] [PubMed] [Google Scholar]
- Alexander D.J., Aldous E.W., Fuller C.M. The long view: a selective review of 40 years of Newcastle disease research. Avian Pathol. 2012;41:329–335. doi: 10.1080/03079457.2012.697991. [DOI] [PubMed] [Google Scholar]
- Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA. 2001;98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bande F., Arshad S.S., Omar A.R., Bejo M.H., Abubakar M.S., Abba Y. Pathogenesis and diagnostic approaches of avian infectious bronchitis. Adv. Virol. 2016;2016:4621659. doi: 10.1155/2016/4621659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop-Lilly K.A., Turell M.J., Willner K.M., Butani A., Nolan N.M., Lentz S.M., Akmal A., Mateczun A., Brahmbhatt T.N., Sozhamannan S., Whitehouse C.A., Read T.D. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl. Trop. Dis. 2010;4(11):e878. doi: 10.1371/journal.pntd.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström A.L., Widén F., Hammer A.S., Belák S., Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010;48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callison S.A., Hilt D.A., Boynton T.O., Sample B.F., Robison R., Swayne D.E., Jackwood M.W. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods. 2006;138:60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePew J., Zhou B., McCorrison J.M., Wentworth D.E., Purushe J., Koroleva G., Fouts D.E. Sequencing viral genomes from a single isolated plaque. Virol. J. 2013;10:181. doi: 10.1186/1743-422X-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Spiro D. Advancing full length genome sequencing for human RNA viral pathogens. Future Virol. 2009;4:47–53. doi: 10.2217/17460794.4.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Halpin R., Kuzmickas R., Depasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., Spiro D.J. Viral genome sequencing by random priming methods. BMC Genom. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froussard P. rPCR: a powerful tool for random amplification of whole RNA sequences. PCR Methods Appl. 1993;2:185–190. doi: 10.1101/gr.2.3.185. [DOI] [PubMed] [Google Scholar]
- Gilchrist C.A., Turner S.D., Riley M.F., Petri W.A., Hewlett E.L. Whole-genome sequencing in outbreak analysis. Clin. Microbiol. Rev. 2015;28:541–563. doi: 10.1128/CMR.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.S., Kapoor A., Lukashov V.V., Simmonds P., Hecht F., Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapczynski D.R., Pantin-Jackwood M., Guzman S.G., Ricardez Y., Spackman E., Bertran K., Suarez D.L., Swayne D.E. Characterization of the 2012 highly pathogenic avian influenza H7N3 virus isolated from poultry in an outbreak in Mexico: pathobiology and vaccine protection. J. Virol. 2013;87:9086–9096. doi: 10.1128/JVI.00666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Mentjies P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.J., Decanini E.L., Afonso C.L. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 2010;10:26–35. doi: 10.1016/j.meegid.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Miller P.J., Afonso C.L., Spackman E., Scott M.A., Pedersen J.C., Senne D.A., Brown J.D., Fuller C.M., Uhart M.M., Karesh W.B., Brown I.H., Alexander D.J., Swayne D.E. Evidence for a new avian paramyxovirus serotype 10 detected in rockhopper penguins from the Falkland Islands. J. Virol. 2010;84:11496–11504. doi: 10.1128/JVI.00822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A.C., Spiro D., Kuzmickas R., Wang S., Djikeng A., Rathe J.A., Fraser-Liggett C.M., Liggett S.B. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes G.R., Kim J.P. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol. Cell. Probes. 1991;5:473–481. doi: 10.1016/s0890-8508(05)80020-9. [DOI] [PubMed] [Google Scholar]
- Rosseel T., Scheuch M., Höper D., De Regge N., Caij A.B., Vandenbussche F., Van Borm S. DNase SISPA-next generation sequencing confirms Schmallenberg virus in Belgian field samples and identifies genetic variation in Europe. PLoS One. 2012;7(7):e41967. doi: 10.1371/journal.pone.0041967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosseel T., Van Borm S., Vandenbussche F., Hoffmann B., van den Berg T., Beer M., Höper D. The origin of biased sequence depth in sequence-independent nucleic acid amplification and optimization for efficient massive parallel sequencing. PLoS One. 2013;8(9):e76144. doi: 10.1371/journal.pone.0076144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E., Senne D.A., Myers T.J., Bulaga L.L., Garber L.P., Perdue M.L., Lohman K., Daum L.T., Suarez D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E. Blackwell Publishing Ltd; Oxford, UK: 2008. Avian Influenza. [Google Scholar]
- Swayne D.E. 13th ed. John Wiley & Sons; Ames, IA: 2013. Diseases of Poultry. [Google Scholar]
- Thorburn F., Bennett S., Modha S., Murdoch D., Gunson R., Murcia P.R. The use of next generation sequencing in the diagnosis and typing of respiratory infections. J. Clin. Virol. 2015;69:96–100. doi: 10.1016/j.jcv.2015.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J.G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M.G., Suarez D.L., Seal B.S., Pedersen J.C., Senne D.A., King D.J., Kapczynski D.R., Spackman E. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004;42:329–338. doi: 10.1128/JCM.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M., Watts T.D., McKay R.A., Suchard M.A., Granade T., Teuwen D.E., Koblin B.A., Heneine W., Lemey P., Jaffe H.W. 1970s and 'Patient 0' HIV-1 genomes illuminate early HIV/AIDS history in North America. Nature. 2016;539:98–101. doi: 10.1038/nature19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Donnelly M.E., Scholes D.T., St George K., Hatta M., Kawaoka Y., Wentworth D.E. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J. Virol. 2009;83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou N., Ditty S., Li B., Lo S.C. Random priming PCR strategy to amplify and clone trace amounts of DNA. BioTechniques. 2003;35:758–765. doi: 10.2144/03354st06. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material