Abstract
To subvert host antiviral immune responses, many viruses have evolved countermeasures to inhibit IFN signaling pathway. Porcine bocavirus (PBoV), a newly identified porcine parvovirus, has received attention because it shows clinically high co-infection prevalence with other pathogens in post-weaning multisystemic wasting syndrome (PWMS) and diarrheic piglets. In this study, we screened the structural and non-structural proteins encoded by PBoV and found that the non-structural protein NP1 significantly suppressed IFN-stimulated response element (ISRE) activity and subsequent IFN-stimulated gene (ISG) expression. However, NP1 affected neither the activation and translocation of STAT1/STAT2, nor the formation of the heterotrimeric transcription factor complex ISGF3 (STAT1/STAT2/IRF9). Detailed analysis demonstrated that PBoV NP1 blocked the ISGF3 DNA-binding activity by combining with the DNA-binding domain (DBD) of IRF9. In summary, these results indicate that PBoV NP1 interferes with type I IFN signaling pathway by blocking DNA binding of ISGF3 to attenuate innate immune responses.
Keywords: Porcine bocavirus (PBoV), Non-structural protein NP1, Type I IFN signaling
Highlights
-
•
Porcine bocavirus (PBoV) NP1 interferes with the IFN α/β signaling pathway.
-
•
PBoV NP1 does not prevent STAT1/STAT2 phosphorylation and nuclear translocation.
-
•
PBoV NP1 inhibits the DNA-binding activity of ISGF3.
-
•
PBoV NP1 interacts with the DNA-binding domain of IRF9.
Introduction
Innate immunity is the first line of host defense against invading pathogens. Type I interferons (IFNs), including IFN-α and IFN-β, represents the principal response of the innate immune system to virus invasion (Gonzalez-Navajas et al., 2012). During viral infection, host pattern recognition receptors (PRRs) recognize viral components or replication intermediates, known as pathogen-associated molecular patterns (PAMPs), and trigger signals that result in the expression of type I IFNs (Rathinam and Fitzgerald, 2011). Following binding of the IFN receptor complex, the synthesis and secretion of IFN-α/β initiates signaling through the Janus kinases/signal transducers and activators of transcription (JAK/STAT) pathway (O׳Sullivan et al., 2007), resulting in the phosphorylation of STATs (Leonard and O׳Shea, 1998, Pellegrini and Dusanter-Fourt, 1997). The phosphorylated STATs interact with the transcription factor IRF9 to form the heterotrimeric transcription factor complex, IFN-stimulated gene factor 3 (ISGF3). This complex translocates to the nucleus where it binds specific regulatory DNA sequences called IFN-stimulated response elements (ISREs) via IRF9 (Qureshi et al., 1995, Veals et al., 1993). This binding step drives the transcription of many IFN-stimulated genes (ISGs) involved in antiviral states, such as double-stranded RNA-dependent protein kinase R (PKR), 2′,5′-oligoadenylate synthetase (OAS), myxovirus resistance 1 (Mx1), ISG15, ISG56 and ISG56 (Borden et al., 2007, Fu et al., 1990).
To combat the antiviral effects of IFN response, viruses usually develop various strategies to disrupt the type I IFN system mainly by interfering with IFN induction and inhibiting IFN signaling. For example, at least eight proteins encoded by Severe acute respiratory syndrome coronavirus (SARS-CoV) and three IRF homolog proteins encoded by Kaposi׳s sarcoma-associated herpesvirus (KSHV) function as antagonists of IFN production (Totura and Baric, 2012); Several viral proteins, such as porcine reproductive and respiratory syndrome virus (PRRSV) NSP1β protein (Wang et al., 2013), foot-and-mouth disease virus (FMDV) 3C protein (Du et al., 2014) and Hepatitis C virus (HCV) NS5A (Bobardt et al., 2013), counteract host antiviral responses by interfering with IFN signaling.
Porcine Bocavirus (PBoV), a newly emergent parvovirus belonging to the genus bocavirus of the parvovirinae subfamily, was first identified in Swedish pigs with post-weaning multisystemic wasting syndrome (PMWS) (Allander et al., 2005, Blomstrom et al., 2009, Chen et al., 1986, Manteufel and Truyen, 2008). To date, a number of different strains of PBoV have been reported worldwide in PMWS, respiratory, diarrheic or asymptomatic swine (Blomstrom et al., 2009, Huang et al., 2014, Zhai et al., 2010, Zhang et al., 2014, Zhang et al., 2013). Although the pathogenesis of PBoV infection remains to be clarified, there is a significantly higher prevalence of co-infection with Porcine circovirus type 2 (PCV2), PRRSV, Porcine torque teno virus (PTTV), and classical Swine fever virus (CSFV) in PBoV-positive pigs, indicating that PBoV plays a role in the co-infection process (Huang et al., 2014, Zhai et al., 2010). Moreover, high prevalence of PBoV was detected in inguinal lymph node and spleen tissues in PBoV-positive pigs (Liu et al., 2014). Hence, it is speculated that PBoV is an immunosuppressive pathogen (Zhou et al., 2014). The full-length genome sequence of PBoV exceeds 5 kb and contains three potential open reading frames (ORFs) encoding two non-structural proteins (NS1 and NP1) and two structural proteins (VP1 and VP2). The genome organization is very similar to that of other bocavirus (Chen et al., 2010, Qiu et al., 2007). Despite extensive efforts, isolation of PBoV and the generation of an infectious clone have not yet been successful. To explore the possible mechanism(s) underlying the immunosuppressive effects of PBoV, we have focused on the effects of PBoV-encoded proteins on innate immunity. Recent study demonstrated that human bocavirus (HBoV) NP1 inhibits both Sendai virus- and poly(dA-dT)-induced IFN-β production (Zhang et al., 2012). We also found that PBoV NP1 could inhibit Sendai virus-induced IFN-β production (unpublished data). In this study, we demonstrated that, in addition to inhibiting IFN-β production, PBoV NP1 also antagonize IFN signaling by reducing the DNA-binding activity of ISGF3, exhibiting a broad-spectrum ability to inhibit innate immune responses.
Results
NP1 interferes with the IFN α/β signaling pathway
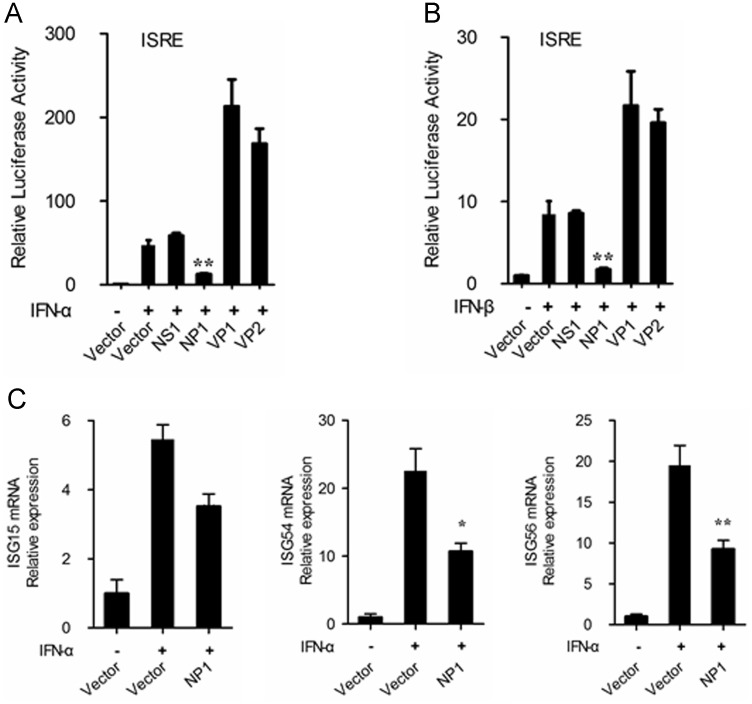
It is well-known that IFN-α/β initiates a series of signaling cascades through the JAK/STAT pathway, resulting in the expression of a set of ISGs, which collaboratively suppress the replication of viruses and contribute to the development of adaptive immune responses (Platanias, 2005). PBoV encodes two structural proteins (VP1, VP2) and two non-structural proteins (NS1, NP1). To identify whether these proteins influences type I IFN signaling, we examined the effects of PBoV encoding proteins expression on ISG promoter activity in response to IFN-α/β stimulation. To this end, HEK-293 cells were co-transfected with ISRE-Luc (containing a consensus sequence for the IFN-stimulated response element (ISRE) of the ISG promoter), pRL-TK (internal control plasmid) and the different PBoV protein expression vectors (VP1, VP2, NS1, NP1). Empty expression vectors were used as controls. At 24-h post-transfection, the cells were treated with IFN-α or IFN-β, ISRE-Luc was measured in whole-cell extracts using a dual luciferase reporter assay at 6 h after this treatment. As shown in Fig. 1A and B, the activation of ISRE promoter was markedly induced by IFN-α/β stimulation, while this induction was strongly impaired by NP1 (Fig. 1A and B). Conversely, overexpression of VP1 and VP2 enhanced the IFN-α/β-induced ISRE activation. Using real-time RT-PCR, we also analyzed the expression of several ISGs at the mRNA level in cells overexpressing NP1. As expected, ectopic expression of NP1 significantly reduced the expression of ISG15, ISG54 and ISG56 mRNA induced by IFN-α (Fig. 1C). These data indicated that PBoV NP1 functions as an antagonist of IFN signaling.
Fig. 1.
PBoV NP1 interferes with IFN α/β signaling pathway. (A and B) Overexpression of NP1 inhibited type I IFN-induced ISRE promoter activation. HEK-293T cells were co-transfected with ISRE-Luc, pRL-TK, and HA-tagged NS1, NP1, VP1 or VP2 expression plasmids. Twenty-four hours after the initial transfection, the cells were treated with IFN-α or IFN-β. Dual luciferase assays were performed after treatment. The resultant ratios were normalized to fold change values with reference to the untreated empty vector control. The results represent the means and standard deviations of data from three independent experiments. (C) Overexpression of NP1 inhibited IFN-α induced transcription of ISG15, ISG54 and ISG56. HEK-293T cells grown in 24-well plates were transfected with 1 μg of a plasmid encoding PBoV NP1 protein (pCAGGS-HA-NP1) or an empty vector. At 24-h post-transfection, the cells were treated with IFN-α and the cells were collected after a further 6 h. Total RNA was extracted from the cells, and the expression levels of the ISG15, ISG54 and ISG56, and GAPDH genes were evaluated by quantitative real-time RT-PCR. The results are expressed as increases in mRNA levels relative to those in transfected cells without IFN infection and were normalized to the expression level of the GAPDH housekeeping gene. One of three experiments is shown. *P<0.05, **P<0.01, Student׳s t-test.
NP1 does not prevent STAT1/STAT2 phosphorylation and nuclear translocation
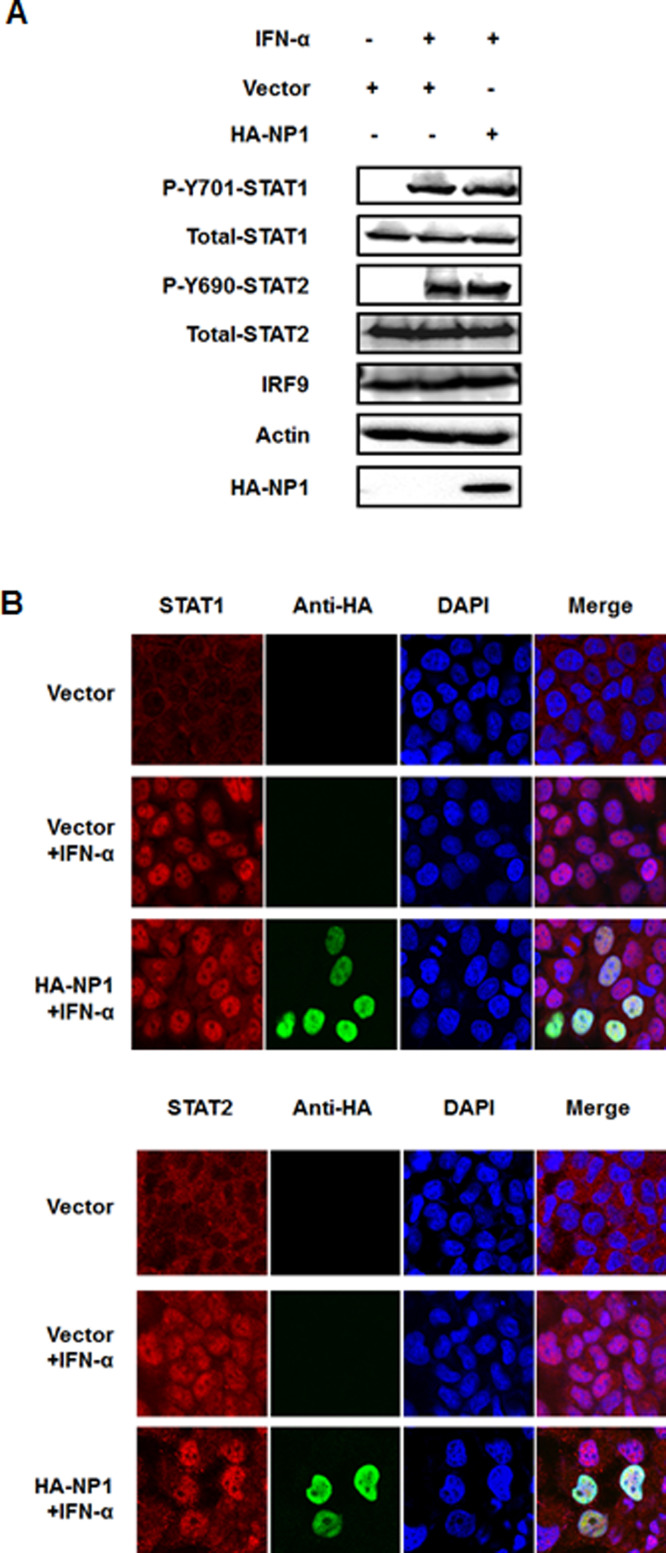
In response to type I IFN, the transcription factors STAT1 and STAT2 are phosphorylated at tyrosine residues by JAK1 and tyrosine kinase 2 (JAK1/Tyk2) and translocated to the nucleus (Darnell et al., 1994). Therefore, we investigated whether NP1 inhibits the phosphorylation of STAT1 or STAT2. To this end, HEK-293T cells were transfected with pCAGGS-HA-NP1 and then incubated with IFN-α. The results showed that phosphorylation of STAT1 (P-Y701-STAT1) and STAT2 (P-Y690-STAT2) were obviously induced by IFN-α in cells with or without NP1 expression (Fig. 2A). Similarly, the endogenous expression of STAT1, STAT2 and IRF9 was also maintained at a steady-state level in the presence or absence of NP1 (Fig. 2A). These observations indicated that NP1 did not prevent STATs phosphorylation or induce the degradation of STATs and IRF9.
Fig. 2.
PBoV NP1 does not degrade or prevent phosphorylation and nuclear translocation of STAT1/STAT2. (A) Effects of NP1 on STAT1/2 phosphorylation and expression. HEK-293T cells were mock-transfected or transfected with HA-tagged NP1 expression plasmid. At 24-h post-transfection, the cells were treated with IFN-α (1000 IU/ml) for 30 min. Cell lysates were collected for immunoblot analysis with antibodies directed against phosphorylated STAT1 (Y701), phosphorylated STAT2 (Y690), STAT1, STAT2, IRF9, HA, or β-actin. (B) NP1 did not prevent STAT1 or STAT2 translocation. HeLa cells were transfected with HA-tagged NP1 expression plasmid. At 24-h post-transfection, the cells were treated with IFN-α (1000 IU/ml) for 30 min. Cells were fixed and stained with mouse anti-HA specific for NP1 (green) and rabbit antibody for STAT1 or STAT2 (red). Cells were viewed under the confocal microscope (Olympus Fluoview ver. 3.1). One of three experiments is shown.
In addition to targeting STATs to reduce phosphorylation or stimulate degradation, some viruses subvert the IFN signaling pathway by modulating nuclear translocation of STATs (Palosaari et al., 2003, Reid et al., 2006). Consequently, we next examined the effects of NP1 on IFN-α-stimulated translocation of STAT1 or STAT2. To this end, HeLa cells were transfected with a HA-tagged NP1 construct or empty vector. At 24-h post-transfection, cells were treated with IFN-α for 20 min. As shown in Fig. 2B, treatment with IFN-α rapidly stimulated nuclear translocation of both STAT1 and STAT2, and this translocation was not affected in cells overexpressing NP1, demonstrating that NP1 has no effect on IFN-α-induced nuclear translocation of STAT1/2. Furthermore, similar to Bovine parvovirus (BPV), minute virus of canines (MVC), and HBoV (Chen et al., 2010, Lederman et al., 1984, Sukhu et al., 2013), PBoV NP1 was localized in the nucleus (Fig. 2B).
NP1 does not impair ISGF3 complex formation but inhibits its DNA-binding activity
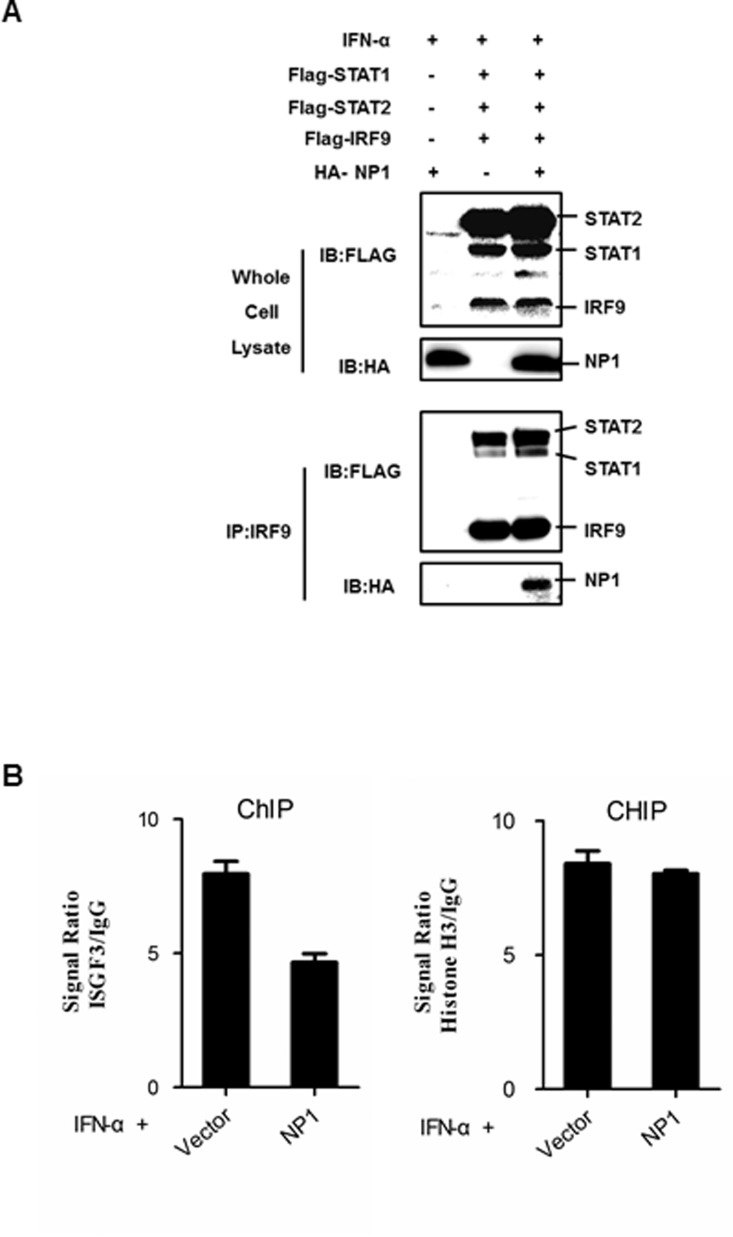
In the phosphorylated form, STATs bind to IRF9 to form a complex IFN-stimulated gene factor 3 (ISGF3) (Schindler et al., 1992), which acts as a transcription factor within ISRE sites in the promoter or enhancer regions of type I IFN responsive genes (Darnell et al., 1994). By using a reporter assay, we found that the IRF9 expression up-regulated ISRE activity and NP1 suppressed IRF9-induced ISRE activation. However, possibly due to a lack of the active form, non-phosphorylated STAT1 or STAT2 expression did not have any effect on ISRE activity (data not shown), indicating that NP1 targets at or downstream of the ISGF3 complex. It is known that essential contacts between IRF9 and STAT1 are required for ISGF3 stability and transcriptional activity (Horvath et al., 1996). As previously described, the human herpesvirus 6B (HHV-6B) IE1 protein disrupted ISGF3 complex formation and inhibited its binding to the ISRE by binding STAT2 within the 111–397 region (Jaworska et al., 2010). To address this issue, we used CoIP assays to determine whether NP1 alters ISGF3 complex formation. Constructs encoding Flag-tagged STAT1, STAT2, IRF9, or empty vector plasmid were co-transfected into HEK-293T cells with HA-tagged NP1 or empty vector. The transfected cells were stimulated with IFN-α for 6 h before lysates were prepared for immunoprecipitation with an anti-IRF9 antibody. As shown in Fig. 3A, Flag-IRF9 co-precipitated with Flag-STAT1 and Flag-STAT2, demonstrating that the Flag-tagged protein formed an ISGF3 complex after IFN treatment. This co-precipitation was not impaired by NP1 expression (Fig. 3A). Interestingly, the HA-tagged NP1 and ISGF3 complex were immunoprecipitated with the anti-IRF9 antibody (Fig. 3A), suggesting that NP1 interacts with the ISGF3 complex but does not impair its formation.
Fig. 3.
PBoV NP1 does not impair ISGF3 complex formation but inhibits its DNA-binding activity. (A) NP1 did not affect formation of the ISGF3 complex. 293T cells were transfected with FLAG-IRF9, FLAG-STAT1, FLAG-STAT2 or empty vector plasmid and HA-tagged NP1 or empty vector plasmid. At 24-h post-transfection, the cells were incubated with IFN-α for 6 h. Cells were lysed and subjected to immunoprecipitation (IP) using rabbit anti-IRF9 antibody. The whole-cell lysates and immunoprecipitation complexes were analyzed by immunoblotting using mouse anti-Flag or mouse anti-HA antibodies. (B) ChIP analysis of ISGF3 binding to the IFN-β promoter. HEK-293T cells were transfected with NP1 or empty expression vector (10 μg each). At 24 h post-transfection, the cells were incubated with IFN-α for 6 h. Cells were lysed and subjected to immunoprecipitation (IP) using rabbit anti-IRF9 antibody. ChIP assays were then performed as described in the Materials and methods section. Real-time PCR analysis of the relative binding of ISGF3 to the ISG56 promoter was performed. The results were expressed as a signal ratio, which represents the signal to the background (IgG) ratio. One of three experiments is shown.
Active ISGF3 was identified as an ISRE-binding factor present in extracts from IFN-α-treatment (Stark and Kerr, 1992). Due to the same cellular localization, we speculated that NP1 inhibits the transcription factor function of ISGF3. Therefore, we examined whether NP1 inhibits the DNA-binding activity of ISGF3. HEK-293T cells were transfected with HA-tagged NP1 or empty vector. After transfection, the cells were treated with IFN-α for 6 h and ChIP assays were performed. Our results showed that NP1 inhibited the IFN-induced binding of ISGF3 to the ISG56 promoter ( Fig. 3B). In contrast, the DNA-binding activity of a positive control, trimethyl-histone H3 was not affected by NP1 (Fig. 3B), indicating that NP1 blocks the binding activity of ISGF3 to the ISG56 promoter. Taken together, these observations indicated that NP1 inhibits IFN signaling by impairing ISGF3 DNA-binding activity.
NP1 interacts with the IRF9 DNA-binding domain
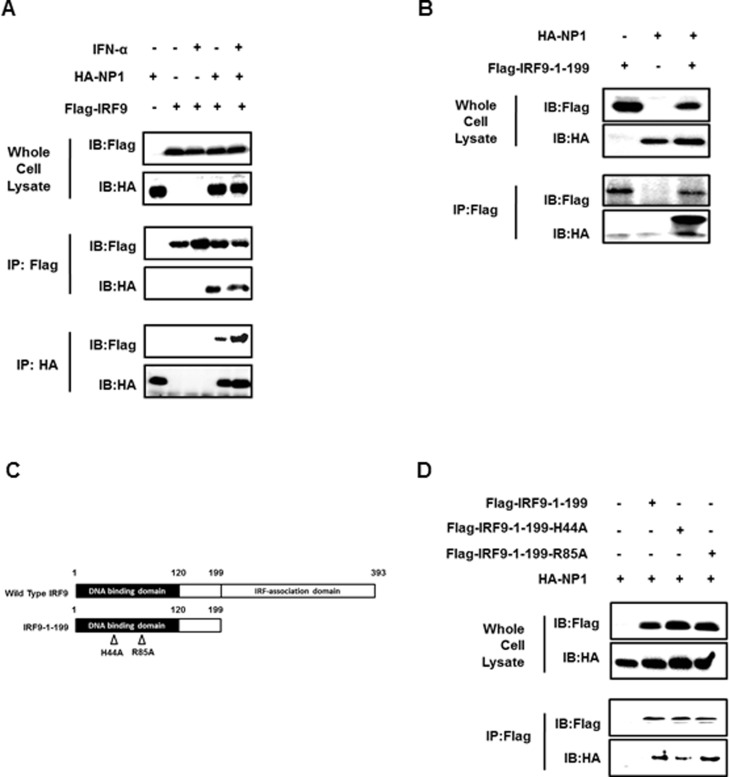
The DNA-binding components of ISGF3 provide the specificity for binding to the promoter in an association that is mediated by IRF9 and STAT1. However, IRF9 is required to provide the main contact with the core of the ISRE consensus sequence, while STAT1 has only limited DNA-binding affinity (Darnell et al., 1994, Veals et al., 1993). Because there is an obvious interaction between NP1 and ISGF3, it is possible that NP1 inhibits the DNA-binding step of ISGF3 by interacting with IRF9. To test this hypothesis, HEK-293T cells were transfected with HA-tagged NP1 and Flag-tagged IRF9 and then treated or mock treated with IFN-α for 6 h. Co-IP assays were performed to determine the interaction between NP1 and IRF9. As shown in Fig. 4A, co-immunoprecipitation of IRF9 and NP1 was observed in assays using both anti-Flag tag and anti-HA antibodies (middle and lower panels), thus confirming the interaction between NP1 and IRF9.
Fig. 4.
PBoV NP1 interacts with the IRF9 DNA-binding domain. (A) PBoV NP1 protein interacts with IRF9. HEK-293T cells were transfected with expression constructs encoding HA-tagged NP1 protein and Flag-tagged IRF9. At 24 h post-transfection, the cells were treated or mock-treated with IFN-α for 6 h. Cells were then lysed and subjected to immunoprecipitation (IP) using mouse anti-FLAG tag or mouse anti-HA tag antibodies. The whole-cell lysates and immunoprecipitation (IP) complexes were analyzed by immunoblotting (IB) using rabbit anti-Flag or mouse anti-HA antibodies. (B) PBoV NP1 protein interacts with IRF9 DNA binding domain. HEK-293T cells were transfected with HA-tagged NP1 expression constructs and Flag-tagged IRF9-1-199 deletion mutants for 24 h. Cell lysates were immunoprecipitated using mouse anti-Flag tag. The whole-cell lysates and immunoprecipitation (IP) complexes were analyzed by immunoblotting (IB) using mouse anti-Flag or mouse anti-HA. (C) Schematic representation of the N-terminal domain of IRF9 and their derivatives (IRF9-1-199-H44A and IRF9-1-199-R85A). (D) Co-IP analysis of the associations of the N-terminal mutants of IRF9 with NP1. HA-tagged NP1 expression constructs were co-transfected with Flag-tagged IRF9-1-199-H44A or Flag-tagged IRF9-1-199-R85A mutants in HEK-293T cells for 24 h. Cell lysates were immunoprecipitated using mouse anti-FLAG tag antibody. The whole-cell lysates and immunoprecipitation (IP) complexes were analyzed by immunoblotting (IB) using mouse anti-Flag or mouse anti-HA antibodies. One of three experiments is shown.
IRF9, as a member of the IRF family, contains two major domains, an N-terminal DNA-binding domain (DBD) and a C-terminal domain, which are largely responsible for its interactions. The DNA-binding region for the ISRE have been identified in the well-conserved DBD (Yanai et al., 2012). As mentioned above, the PBoV NP1 protein binds IRF9 and blocks the binding of IRF9 to responsive DNA sequences within the promoter (Fig. 3A and Fig. 4A). These observations indicated that this inhibition may involve in the IRF9 DBD. To confirm this speculation, we investigated the capacity of NP1 to interact with the IRF9 DBD. The N-terminal mutant containing the DBD region (IRF9-1-199) was constructed and the results of Co-IP assay showed that NP1 bound to the IRF9 DBD ( Fig. 4B).
The DBD contains a signature repeat of tryptophan residues within a unique helix-turn-helix structure which is responsible for DNA-binding activity (Escalante et al., 2007, Fujii et al., 1999). Knowing the structure loop 1 (L1) and α-helix 3 (α3) of the IRFs DBD play important role in DNA binding, it is possible that PBoV NP1 blocks this binding surface to inhibit IRF9 binding activity. Although the crystal structure of the IRF9/DNA complex has not yet been resolved, a Histidine (H) residue on L1 and a Arginine (R) residue on α3 are completely conserved in the IRF family, and which were identified as an important DNA recognition sites in IRF2, IRF3 and IRF7 (Escalante et al., 2007, Fujii et al., 1999). Thus, we speculated that H44 on L1 and R85 on α3 of IRF9 perform binding function in IRF9/DNA-association. To investigate whether the binding surface of IRF9 is required for interaction with NP1, we used two constructs by incorporating mutations in Loop1 or α-helix 3 of the IRF9 DBD in Co-IP assays (Fig. 4C). As shown in Fig. 4D, compared with the wild-type controls, less NP1 was co-immunoprecipitated with IRF9-1-199-H44A, suggesting that the conserved Histidine residue contributes to NP1 binding. However, similar amounts of NP1 were immunoprecipitated with IRF9-1-199-R85A and the wild-type IRF9-1-199 control (Fig. 4D), possibly because Arginine 85 of IRF9 is not required for DNA binding. Taken together, these data further demonstrate a mechanism by which PBoV NP1 disrupts the IFN signaling pathway by interaction with the DNA-binding domain of IRF9.
Discussion
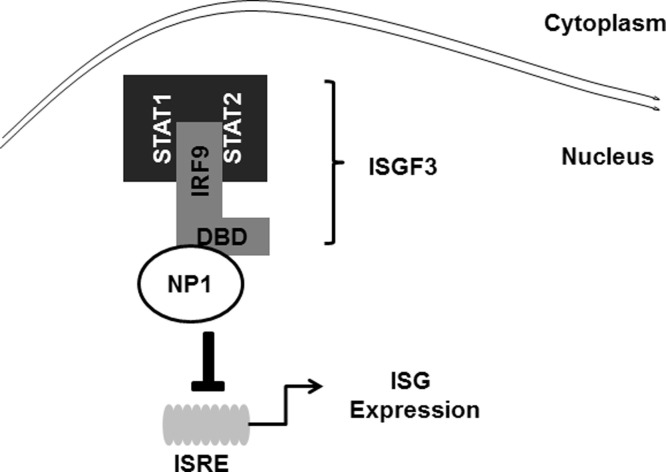
In this study, we demonstrated that PBoV NP1 antagonizes the antiviral innate immune by interfering with type I IFN signaling. Our data also revealed that PBoV NP1 interacts with the DBD of IRF9, and blocks the DNA-binding activity of ISGF3, thus impeding ISG expression ( Fig. 5).
Fig. 5.
The proposed model for association between PBoV NP1 and the IRF9 DNA-binding domain. PBoV NP1 interacts with the DNA-binding domain of IRF9 to block the DNA-binding step of ISGF3, resulting in reduction of ISG expression.
Members in the Parvoviridae family infect a wide range of hosts, and their evasion of the innate immune system has rarely been reported. Infection with the minute virus of mice (MVM) inhibited poly(I:C)-induced IL-6 expression in mouse embryonic fibroblasts (Mattei et al., 2013) and porcine parvovirus (PPV) infection blocked IFN-β production by NS2 inhibition of poly(I:C)-induced IFN-β promoter activation (Lin et al., 2013). Previous studies have demonstrated the HBoV NS1 and NP1 proteins block SeV- and poly(dA-dT)-induced IFN-β production (Zhang et al., 2012), and the structural protein VP2 up-regulated IFN-β production (Luo et al., 2013). To determine whether bocavirus could antagonize the IFN signaling pathway, we screened the effects of PBoV encoding proteins on IFN signaling in present study. We found PBoV structural proteins, VP1 and VP2, further enhanced IFN-induced ISRE activation, while non-structural protein NP1 inhibited transcript levels of ISGs and ISRE promoter activity. In addition, our experiments demonstrated PBoV NS1 and NP1 both inhibits type I IFN production (data not shown). These results indicate that non-structural proteins provide type I IFN resistance against bocavirus infection, while structural proteins contribute to host immune protection.
The type I IFN system provides a powerful antiviral response that protects the host from virus infection. To subvert the antiviral effects of IFN-induced ISGs, viruses were shown to express certain proteins that negatively regulate type I IFN signaling by targeting the JAK-STAT pathway at different levels, including impeding IFN receptor activation (Bisson et al., 2009), degrading STATs (Tay et al., 2013, Ulane and Horvath, 2002), inhibiting STATs phosphorylation (Caignard et al., 2007, Devaux et al., 2007), blocking STAT1/STAT2 nuclear translocation (Chen et al., 2013, Xu et al., 2014), and disturbing ISGF3 formation (Jaworska et al., 2010, Lin et al., 2006). In this study, we screened the components of JAK-STAT pathway and found that PBoV NP1 does not affect the amounts of STATs and IRF9 in IFN-α-treated cells, nor does it alter the IFN-induced phosphorylation of STAT1/STAT2. Furthermore, NP1 hardly affected the nuclear translocation of STATs. Interestingly, we found NP1 interacted with ISGF3 without any effect on its formation but inhibited ISGF3 DNA-binding activity. Thus, we identified PBoV NP1 as an antagonist of the type I IFN system to mediate negative regulation of IFN signaling by targeting the DNA-binding step of ISGF3. Because we did not investigate the potential interaction of NP1 with CREB-binding protein (CBP) in the present study, we could not exclude the possibility that NP1 may impair CBP function. Given that binding of transcription factors to promoters and combining with CBP are two independent events, we mainly focused on the mechanism by which the DNA-binding of ISGF3 is inhibited.
Acting as key transcription factors in the IFN signaling pathway, the activated ISGF3 recognize and bind directly to the conserved DNA sequences in the nucleus. Multiple levels of regulation occur during ISGF3 translocation to the nucleus. However, to the best of our knowledge, only the rabies virus P protein has been shown to inhibit ISGF3 binding to the ISRE directly, whereas it also blocks STAT1 nuclear translocation (Vidy et al., 2007). It is unclear at present how the PBoV NP1 protein inhibits the binding of ISGF3 to the DNA. We speculate that there are two possibilities: 1) NP1 interacts directly with the IRF9 DBD to block the binding step; 2) NP1 interacts directly with the promoter to block the binding site on ISRE. In this study, we confirmed that NP1 interacts with IRF9 DBD. This result is consistent with a previous report (Zhang et al., 2012), which described the interaction of the HBoV NP1 protein with the IRF3 DBD. Furthermore, we noted that a potential DNA binding site (H44) on Loop1 of IRF9 contributes to IRF9 and NP1 association, indicating that NP1 acts as an obstruction on the DNA-binding interface of IRF9, and in turn interferes with the IRF9 DNA-binding activity. Considering the structural similarity among the different IRF DBDs and the histidine residue on Loop1 is also highly conserved in IRFs family, it is possible that NP1 interacts with similar binding sites on the DBDs of other IRFs to mediate broad-spectrum impairment of the IFN system. However, we cannot exclude the possibility that NP1 interacts directly with the responsive DNA-binding site of the promoter. Indeed, as previously shown, the KSHV K-bZIP protein, which is an IFN antagonist, interacts directly with the PRDIII-I region of IFN-β and the ISRE region of RANTES (Lefort et al., 2007). Considering that bocavirus NP1 has effects on RNA processing and DNA replication (Sukhu et al., 2013, Sun et al., 2009) and the existence of a potential nucleic acid recognition site at the N-terminal region of PBoV NP1 (http:// http://smart.embl-heidelberg.de/; http://pfam.xfam.org), it is possible that PBoV NP1 recognizes and binds to the DNA sequence on the promoter.
Bocavirus NP1 contains a highly phosphorylated N-terminal and a conserved C-terminal. Previous study demonstrated HBoV and BPV NP1 are both able to recover MVC replicative form DNA as same as MVC NP1, suggesting that functions of NP1 could be cross-complemented among Bocavirus (Sun et al., 2009). In agreement, HBoV NP1 interacts with IRF3 DBD (Zhang et al., 2012), while PBoV NP1 interacted with the IRF9 DBD in current study. Therefore, it can be speculated that a functionally conserved domain in NP1 is responsible for its IRF DBD binding activity. Moreover, the same localization of NP1 and activated ISGF3 in the nucleus lays the foundation for their interaction, suggesting that the subcellular location of NP1 may affect its function as an IFN antagonist. Further studies are required to fully elucidate the function of NP1 as an IFN antagonist.
Materials and methods
Cells and virus
HEK-293T cells and HeLa cells were maintained in RPMI 1640 (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37 °C in a humidified 5% CO2 incubator.
Plasmids
The luciferase reporter plasmid pISRE-Luc has been described previously (Zhong et al., 2013). The internal control plasmid pRL-TK was purchased from Promega. All primers used for the construction of plasmids are listed in Table 1. To construct the expression plasmids encoding individual PBoV proteins, DNA fragments encoding the full-length NS1, NP1, VP1, VP2 proteins were amplified by PCR from the almost full-length cDNA clone of bocavirus pig/SX/China/2010 (GenBank accession number HQ223038). The fragments were then subcloned into the pCAGGS-HA vector. The full-length cDNA of IRF9 (GenBank accession number NM_006084) was amplified from total RNA extracted from HEK-293T cells and cloned into the pCMV-Tag2B expression vector to generate pCMV-IRF9. Truncation of IRF9 (IRF9-1-199; residues 1–199) was constructed by PCR and cloned into the pCMV-Tag2B expression vector. Constructs encoding mutants of IRF9-1-199 (IRF9-1-199-H44A and IRF9-1-199-R85A) were generated by overlapping extension PCR using specific mutagenic primers (available upon request). The expression plasmids Vim-flag-STAT1 and Vim-flag-STAT2 were purchased from Vigene (China).
Table 1.
Primers used for plasmid constructs.
| symbol | Forward primer | Reverse primer |
|---|---|---|
| NS1 | ccgctcgagatggctctacttcacttcaag | ccggaattcttacttacgtccgtcgtccc |
| NP1 | ccgctcgagatgagtgggcatcacagccac | ccggaattcttattttccagcttcagcttc |
| VP1 | ccgctcgagatgaatcaattgtttcctgtg | ccggaattcttacaacacttggttgattc |
| VP2 | ccgctcgagatgtccgcacaggggggcg | ccggaattcttacaacacttggttgattc |
| IRF-3-1-197 | ccggaattcatgggaaccccaaagccacggat | ccgctcgagttacaccaacagccgcttcag |
| IRF-9 | ccggaattcatggcatcaggcagggcac | ccctcgagctacaccagggacagaatgg |
| IRF-9-1-199 | ccggaattcatggcatcaggcagggcac | ggctcgagttaggcctcagttgtgtctgtaactt |
Transfection and luciferase reporter assay
HEK-293T cells and HeLa cells were transfected using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer׳s instructions. For luciferase reporter assays, HEK-293T cells in 24-well plates were co-transfected with 0.1-μg/well reporter plasmid ISRE-Luc with 0.05-μg/well internal control plasmid pRL-TK. At 24 h after the initial co-transfection, the cells were further stimulated with IFN-α ( Catalog no. 11101-2, PBL assay science) or IFN-β (Catalog no. 300-02BC, Peprotech) for 6 h. Luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega), according to the manufacturer׳s protocol. Data represent relative firefly luciferase activity normalized to Renilla luciferase activity and are representative of three independently conducted experiments.
RNA extraction and quantitative real-time RT-PCR
Total RNA was extracted from the cells with TRIzol reagent (Invitrogen), and an aliquot (1 μg) was reverse transcribed to cDNA by using avian myeloblastosis virus (AMV) reverse transcriptase (Roche). To determine the effects of NP1 protein on the expression of ISG15, ISG54 and ISG56, HEK-293T cells in 24-well plates were transfected with 1 μg of the empty vector or a plasmid encoding the NP1 protein. After 24 h, the cells were mock stimulated or stimulated with IFN-α. The cDNA (1 μl of the 20-μl RT reaction mixture) was then used as the template in a SYBR green PCR assay (Applied Biosystems). The abundance of the individual mRNA transcripts in each sample was assayed three times and normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (the internal control).
Co-immunoprecipitation and immunoblot assays
Briefly, HEK-293T cells cultured in 60-mm dishes were transfected with the appropriate plasmids. After 28 h, the cells were harvested by the addition of 150-μl lysis buffer (4% SDS, 3% dithiothreitol [DTT], 0.065 mM Tris–HCl [pH 6.8], 30% glycerin), and the protein concentrations in the whole-cell extracts were measured. Equal amounts of samples were then subjected to SDS-PAGE and analyzed for the expression of STAT1, p-STAT1, STAT2, p-STAT2 and IRF9 proteins by immunoblotting using the following antibodies (raised in rabbit): anti-STAT1, anti-p-STAT1 (both Santa Cruz), anti-STAT2, anti-p-STAT2 (both ABclone), and anti-IRF9 antibody (Santa Cruz), respectively. An anti-hemagglutinin (HA) antibody (MBL) was used for immunoblotting to confirm the expression levels of HA-tagged PBoV NP1 protein. The expression of β-actin was detected with a mouse anti-β-actin monoclonal antibody (MBL) to demonstrate equal protein sample loading.
For co-immunoprecipitation experiments, HEK-293T cells in 100-mm dishes were transiently transfected with the indicated plasmids using Lipofectamine 2000 according to the manufacturer׳s instructions (Invitrogen). Transfected HEK-293T cells from each dish were washed with phosphate-buffered saline (PBS) and lysed for 20 min at 4 °C in 1-ml lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 20 nM phenylmethylsulfonyl fluoride [PMSF]), and the protein concentration was measured and adjusted. For each immunoprecipitation, 500 μg of cell lysate protein was incubated with 2-μg mouse anti-flag (MBL), mouse anti-HA (MBL) or 2-μg rabbit anti-IRF9 (Santa Cruz) and 25-μl protein A/G-agarose (Beyotime, China) overnight at 4 °C. The Sepharose beads were then washed three times with 1-ml lysis buffer. The precipitates were subjected to 10% SDS-PAGE and subsequent immunoblot analysis using the indicated antibodies.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using a ChIP assay kit (Millipore, Billerica, MA, USA) according to the manufacturer׳s instructions. Approximately 5×107 HEK-293T cells were transfected with the PBoV NP1 protein expression plasmid or the empty vector plasmid for use in each assay. For determination of the ISRE element in the ISG56 promoter, cells were treated with IFN-α for 6 h. Cell extracts were then prepared, and chromatin was sonicated 10 times (30 s at 30% of the maximum setting each time). After sonication, protein–DNA complexes were then immunoprecipitated from nuclear extracts using a rabbit polyclonal IRF9 antibody (Santa Cruz), normal rabbit IgG or anti-polymerase II or mouse IgG, followed by capture on protein A/G Sepharose beads. After IP and elution, the DNA was amplified by real-time PCR. The results are expressed as a signal ratio, which represents the signal to background (IgG) ratio. The sequences of the primers used for amplification of ISG56 promoter are as follows: pISG56-F: 5′-CTGGCCAGTCATTGGGTTTC-3′; pISG56-R: 5′-GAGCTAAACAGCAGCCAATGG-3′.
Indirect immunofluorescence
HeLa cells were seeded onto microscope coverslips (NEST Co. 801007) and placed into 24-well dishes. At approximately 70% to 80% confluence, the cells were transfected with the PBoV NP1 protein expression plasmid. After transfection for 28 h, cells were mock-treated or treated with recombinant human IFN-α (1000 U/ml) for 30 min for detection of STAT1 or STAT2. The cells were then fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 15 min at room temperature. After three washes with PBS, the cells were blocked with PBS containing 5% goat serum (Sigma Co. G6767) for 1 h. The cells were incubated separately with rabbit polyclonal antibodies directed against STAT1 (1:200 Santa Cruz) or STAT2 (1:200 Santa Cruz) and a mouse monoclonal antibody directed against the HA tag (1:200 MBL) for 1 h at room temperature. The cells were then treated with FITC-labeled goat anti-mouse (1:500 Beyotime) or Alexa 594-conjugated goat anti-rabbit IgG (1:1000 Santa Cruz) for 1 h and then stained with DAPI for 15 min at room temperature. After three washes, images were obtained using an Olympus Fluoview ver. 3.1 confocal laser scanning microscope.
Statistical analysis
Data are presented as mean±standard deviations (SD). P-values <0.05 were considered significant, and P-values <0.01 were considered highly significant.
Conclusion
Here, we identified PBoV NP1 protein as an antagonist of interferon signaling and demonstrated that PBoV NP1 antagonizes type I IFN signaling by targeting the IRF9 DNA-binding domain to inhibit the DNA-binding activity of ISGF3. This is an efficient mechanism for viral protein to antagonize IFN signaling, providing a possible explanation of innate immune suppression for the pathogenesis of PBoV co-infection.
Acknowledgments
This work was supported by the Natural Science Foundation of Hubei Province (2014CFA009), the National Natural Sciences Foundation of China (31470268) and the Fundamental Research Funds for the Central Universities (2013PY043).
References
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson S.A., Page A.L., Ganem D. A Kaposi׳s sarcoma-associated herpesvirus protein that forms inhibitory complexes with type I interferon receptor subunits, Jak and STAT proteins, and blocks interferon-mediated signal transduction. J. Virol. 2009;83:5056–5066. doi: 10.1128/JVI.02516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomstrom A.L., Belak S., Fossum C., McKillen J., Allan G., Wallgren P., Berg M. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009;146:125–129. doi: 10.1016/j.virusres.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Bobardt M., Hopkins S., Baugh J., Chatterji U., Hernandez F., Hiscott J., Sluder A., Lin K., Gallay P.A. HCV NS5A and IRF9 compete for CypA binding. J. Hepatol. 2013;58:16–23. doi: 10.1016/j.jhep.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden E.C., Sen G.C., Uze G., Silverman R.H., Ransohoff R.M., Foster G.R., Stark G.R. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caignard G., Guerbois M., Labernardiere J.L., Jacob Y., Jones L.M., Infectious Mapping Project I.M., Wild F., Tangy F., Vidalain P.O. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology. 2007;368:351–362. doi: 10.1016/j.virol.2007.06.037. [DOI] [PubMed] [Google Scholar]
- Chen A.Y., Cheng F., Lou S., Luo Y., Liu Z., Delwart E., Pintel D., Qiu J. Characterization of the gene expression profile of human bocavirus. Virology. 2010;403:145–154. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu M., Zhang X., Zhang W., Zhang Z., Chen L., He J., Zheng Y., Chen C., Wang F., Hu Y., Zhou X., Wang C., Xu Y., Lu M., Yuan Z. Hepatitis B virus polymerase impairs interferon-alpha-induced STA T activation through inhibition of importin-alpha5 and protein kinase C-delta. Hepatology. 2013;57:470–482. doi: 10.1002/hep.26064. [DOI] [PubMed] [Google Scholar]
- Chen K.C., Shull B.C., Moses E.A., Lederman M., Stout E.R., Bates R.C. Complete nucleotide sequence and genome organization of bovine parvovirus. J. Virol. 1986;60:1085–1097. doi: 10.1128/jvi.60.3.1085-1097.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E., Jr., Kerr I.M., Stark G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Devaux P., von Messling V., Songsungthong W., Springfeld C., Cattaneo R. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology. 2007;360:72–83. doi: 10.1016/j.virol.2006.09.049. [DOI] [PubMed] [Google Scholar]
- Du Y., Bi J., Liu J., Liu X., Wu X., Jiang P., Yoo D., Zhang Y., Wu J., Wan R., Zhao X., Guo L., Sun W., Cong X., Chen L., Wang J. 3Cpro of foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2014;88:4908–4920. doi: 10.1128/JVI.03668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante C.R., Nistal-Villan E., Shen L., Garcia-Sastre A., Aggarwal A.K. Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer. Mol. Cell. 2007;26:703–716. doi: 10.1016/j.molcel.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Fu X.Y., Kessler D.S., Veals S.A., Levy D.E., Darnell J.E., Jr. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y., Shimizu T., Kusumoto M., Kyogoku Y., Taniguchi T., Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Navajas J.M., Lee J., David M., Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.M., Stark G.R., Kerr I.M., Darnell J.E., Jr. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Mor S.K., Erber J., Voss E., Goyal S.M. Detection and characterization of porcine bocavirus in the United States. Arch. Virol. 2014;159:1797–1801. doi: 10.1007/s00705-013-1972-4. [DOI] [PubMed] [Google Scholar]
- Jaworska J., Gravel A., Flamand L. Divergent susceptibilities of human herpesvirus 6 variants to type I interferons. Proc. Natl. Acad. Sci. USA. 2010;107:8369–8374. doi: 10.1073/pnas.0909951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman M., Patton J.T., Stout E.R., Bates R.C. Virally coded noncapsid protein associated with bovine parvovirus infection. J. Virol. 1984;49:315–318. doi: 10.1128/jvi.49.2.315-318.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort S., Soucy-Faulkner A., Grandvaux N., Flamand L. Binding of Kaposi׳s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 2007;81:10950–10960. doi: 10.1128/JVI.00183-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W.J., O׳Shea J.J. Jaks and STATs: biological implications. Annu. Rev. Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Lin W., Kim S.S., Yeung E., Kamegaya Y., Blackard J.T., Kim K.A., Holtzman M.J., Chung R.T. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J. Virol. 2006;80:9226–9235. doi: 10.1128/JVI.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Qiu Z., Liu Q., Cui S. Interferon induction and suppression in swine testicle cells by porcine parvovirus and its proteins. Vet. Microbiol. 2013;163:157–161. doi: 10.1016/j.vetmic.2012.11.032. [DOI] [PubMed] [Google Scholar]
- Liu M., Li Y., Sun D., Xia Y., Huang J., Guo L. Detection and genetic analysis of porcine bocavirus in different swine herds in North Central China. Sci. World J. 2014;2014:947084. doi: 10.1155/2014/947084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Zhang Z., Zheng Z., Ke X., Zhang X., Li Q., Liu Y., Bai B., Mao P., Hu Q., Wang H. Human bocavirus VP2 upregulates IFN-beta pathway by inhibiting ring finger protein 125-mediated ubiquitination of retinoic acid-inducible gene-I. J. Immunol. 2013;191:660–669. doi: 10.4049/jimmunol.1202933. [DOI] [PubMed] [Google Scholar]
- Manteufel J., Truyen U. Animal bocaviruses: a brief review. Intervirology. 2008;51:328–334. doi: 10.1159/000173734. [DOI] [PubMed] [Google Scholar]
- Mattei L.M., Cotmore S.F., Tattersall P., Iwasaki A. Parvovirus evades interferon-dependent viral control in primary mouse embryonic fibroblasts. Virology. 2013;442:20–27. doi: 10.1016/j.virol.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O׳Sullivan L.A., Liongue C., Lewis R.S., Stephenson S.E., Ward A.C. Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol. Immunol. 2007;44:2497–2506. doi: 10.1016/j.molimm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Palosaari H., Parisien J.P., Rodriguez J.J., Ulane C.M., Horvath C.M. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini S., Dusanter-Fourt I. The structure, regulation and function of the Janus kinases (JAKs) and the signal transducers and activators of transcription (STATs) Eur. J. Biochem. 1997;248:615–633. doi: 10.1111/j.1432-1033.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Qiu J., Cheng F., Johnson F.B., Pintel D. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J. Virol. 2007;81:12080–12085. doi: 10.1128/JVI.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S.A., Salditt-Georgieff M., Darnell J.E., Jr. Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. USA. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Fitzgerald K.A. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S.P., Leung L.W., Hartman A.L., Martinez O., Shaw M.L., Carbonnelle C., Volchkov V.E., Nichol S.T., Basler C.F. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Fu X.Y., Improta T., Aebersold R., Darnell J.E., Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Kerr I.M. Interferon-dependent signaling pathways: DNA elements, transcription factors, mutations, and effects of viral proteins. J. Interferon Res. 1992;12:147–151. doi: 10.1089/jir.1992.12.147. [DOI] [PubMed] [Google Scholar]
- Sukhu L., Fasina O., Burger L., Rai A., Qiu J., Pintel D.J. Characterization of the nonstructural proteins of the bocavirus minute virus of canines. J. Virol. 2013;87:1098–1104. doi: 10.1128/JVI.02627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Chen A.Y., Cheng F., Guan W., Johnson F.B., Qiu J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 2009;83:3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Y., Fraser J.E., Chan W.K., Moreland N.J., Rathore A.P., Wang C., Vasudevan S.G., Jans D.A. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir. Res. 2013;99:301–306. doi: 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulane C.M., Horvath C.M. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- Veals S.A., Santa Maria T., Levy D.E. Two domains of ISGF3 gamma that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol. Cell. Biol. 1993;13:196–206. doi: 10.1128/mcb.13.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidy A., El Bougrini J., Chelbi-Alix M.K., Blondel D. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 2007;81:4255–4263. doi: 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Nan Y., Yu Y., Zhang Y.J. Porcine reproductive and respiratory syndrome virus Nsp1beta inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-alpha1 degradation. J. Virol. 2013;87:5219–5228. doi: 10.1128/JVI.02643-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Edwards M.R., Borek D.M., Feagins A.R., Mittal A., Alinger J.B., Berry K.N., Yen B., Hamilton J., Brett T.J., Pappu R.V., Leung D.W., Basler C.F., Amarasinghe G.K. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014;16:187–200. doi: 10.1016/j.chom.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H., Negishi H., Taniguchi T. The IRF family of transcription factors: inception, impact and implications in oncogenesis. Oncoimmunology. 2012;1:1376–1386. doi: 10.4161/onci.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai S., Yue C., Wei Z., Long J., Ran D., Lin T., Deng Y., Huang L., Sun L., Zheng H., Gao F., Zheng H., Chen S., Yuan S. High prevalence of a novel porcine bocavirus in weanling piglets with respiratory tract symptoms in China. Arch. Virol. 2010;155:1313–1317. doi: 10.1007/s00705-010-0698-9. [DOI] [PubMed] [Google Scholar]
- Zhang B., Tang C., Yue H., Ren Y., Song Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J. Gen. Virol. 2014;95:1603–1611. doi: 10.1099/vir.0.063743-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Hu R., Tang X., Wu C., He Q., Zhao Z., Chen H., Wu B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013;158:1631–1636. doi: 10.1007/s00705-013-1659-x. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zheng Z., Luo H., Meng J., Li H., Li Q., Zhang X., Ke X., Bai B., Mao P., Hu Q., Wang H. Human bocavirus NP1 inhibits IFN-beta production by blocking association of IFN regulatory factor 3 with IFNB promoter. J. Immunol. 2012;189:1144–1153. doi: 10.4049/jimmunol.1200096. [DOI] [PubMed] [Google Scholar]
- Zhong H., Wang D., Fang L., Zhang H., Luo R., Shang M., Ouyang C., Ouyang H., Chen H., Xiao S. Ubiquitin-specific proteases 25 negatively regulates virus-induced type I interferon signaling. PLoS One. 2013;8:e80976. doi: 10.1371/journal.pone.0080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Sun H., Wang Y. Porcine bocavirus: achievements in the past five years. Viruses. 2014;6:4946–4960. doi: 10.3390/v6124946. [DOI] [PMC free article] [PubMed] [Google Scholar]