Graphical abstract
Keywords: Hemagglutinin, Histidine, Influenza, Inhibitors, Protonation, Structure
Highlights
-
•
X-ray crystallography identifies a conserved histidine, HA1-H38, that changes side chain conformation at low pH.
-
•
We attribute the observed conformational change to cation-cation repulsion between protonated HA1-H18 and HA1-H38.
-
•
We suggest that the HA1-H18 and HA1-H38 pair plays a role in the pathway toward the postfusion conformation of HA.
Abstract
Hemagglutnin (HA) mediates entry of influenza virus through a series of conformational changes triggered by the low pH of the endosome. The residue or combination of residues acting as pH sensors has not yet been fully elucidated. In this work, we assay pH effects on the structure of H5 HA by soaking HA crystallized at pH 6.5 in a series of buffers with lower pH, mimicking the conditions of the endosome. We find that HA1-H38, which is conserved in Group 1 HA, undergoes a striking change in side chain conformation, which we attribute to its protonation and cation-cation repulsion with conserved HA1-H18. This work suggests that x-ray crystallography can be applied for studying small-scale pH-induced conformational changes providing valuable information on the location of pH sensors in HA. Importantly, the observed change in HA1-H38 conformation is further evidence that the pH-induced conformational changes of HA are the result of a series of protonation events to conserved and non-conserved pH sensors.
1. Introduction
Influenza hemagglutinin (HA), similarly to the analogous viral envelope proteins from Ebola, HIV, and MERS- and SARS-Coronavirus, mediates virus entry through binding to receptors on the human cell surface and then mediating fusion of the viral and target cell membranes, thereby enabling entry of the viral genetic material (Skehel and Wiley, 2000, Eckert and Kim, 2001, Harrison, 2008, Caffrey, 2011, Belouzard et al., 2012). Based on amino acid sequence and structure, HA glycoproteins fall into 2 phylogenetic groups (Nobusawa et al., 1991, Webster et al., 1992). Examples of Group 1 HA are those of circulating H1 and avian H5; examples of Group 2 HA are those of circulating H3 and avian H7. Influenza strains comprised of avian H5 or H7 HA are particularly concerning due to their relatively high mortality rates >40% in humans (Hui and Zumla, 2015). Notably, the critical role of HA function makes it an attractive target for the development of novel therapeutic agents (Lagoja and De Clercq, 2008).
HA is synthesized as the precursor protein HA0 and is subsequently cleaved by cellular proteases to form a disulfide-linked complex consisting of HA1, the receptor binding subunit, and HA2, the subunit that mediates membrane fusion (Skehel and Wiley, 2000, Eckert and Kim, 2001, Harrison, 2008). After binding to sialyllactose receptors, HA is internalized into the endosome where it undergoes a series of conformational changes, triggered by endosome acidification (Skehel and Wiley, 2000, Eckert and Kim, 2001, Harrison, 2008). Specifically, as the pH of the endosome decreases, HA1 partially dissociates from HA2 where HA2 becomes fully extended and inserts the hydrophobic N-terminal fusion peptide into the endosome membrane. Subsequently, HA2 folds back to bring the two membranes into close proximity whereby the membranes merge into one entity and a pore is formed to enable release of the viral RNA into the cytoplasm (Skehel and Wiley, 2000, Eckert and Kim, 2001, Harrison, 2008). The pH-based triggering of HA conformational changes is generally thought to be due to the protonation of amino acids that act as pH sensors. Since the membrane fusion event occurs in the pH range of 5–6, the most likely residues to function as pH sensors are histidines, aspartates and/or glutamates, which possess pKa in the appropriate pH range (Zhou et al., 2014).
Based on a number of studies, multiple pH sensors are involved. First, from biochemical, x-ray, EM and virological studies, HA is known to undergo multiple reversible conformational changes when exposed to low pH (Xu and Wilson, 2011, Fontana et al., 2012, Leikina et al., 2002). Second, despite a high degree of structural homology within HA subtypes, examination of HA sequences does not reveal absolutely conserved titratable residues (Zhou et al., 2014, Mair et al., 2014). Third, membrane fusion occurs at different pH values for different HA subtypes (Scholtissek, 1985, Puri et al., 1990, Korte et al., 2007). Fourth, mutagenesis studies have revealed conserved and non-conserved residues, located at diverse regions of the HA structure, that alter the pH of membrane fusion (Mair et al., 2014, Reed et al., 2009). Finally, Molecular Dynamics (MD) studies of HA suggest a large number of residues with titratable groups become protonated with diverse pKa values that are highly dependent on the local environment (Zhou et al., 2014). Taken together, these studies are consistent with the model of the successive protonation of multiple residues that result in destabilization of the prefusion conformations and stabilization of the postfusion conformation through cation-cation repulsion, cation-anion attraction, and anion-anion interaction (Harrison et al., 2013). However, despite the extensive study of HA, consensus on the role and involvement of individual residues in regulation of this highly coordinated process is still lacking. In the present manuscript we use x-ray crystallography to detect protonation events in different residues by visualizing small-scale conformational changes as a function of pH. Applying this approach to H5 HA we identified a conserved histidine pair that act as pH sensors.
2. Results and discussion
In the first step we prepared the H5 HA extracellular domain in insect cells as previously described with removal of the foldon-histidine tag at the C-terminus and full cleavage of HA0 to HA1-HA2 by furin (Antanasijevic et al., 2014). In the next step, HA was crystallized under conditions of an intermediate pH (100 mM cacodylate buffer/pH 6.5 + 200 mM NaCl + 2 M (NH4)2SO4 + 10% glycerol). To learn more about the nature of the prefusion pH states, crystals obtained at pH 6.5 were soaked in cryo solutions of identical chemical composition, differing only in the final pH (pH 7.0, 6.5, 6.0 and 5.5). Note that the buffering range of cacodylic acid (pH ~ 5–7) makes it ideally suitable for studying the pH range of interest. Following a 10 min incubation time in different cryo solutions, the crystals were frozen. Data acquisition and processing were performed as described in the Materials and Methods section and the final statistics are shown in Table 1 . For clarity, structures will be referred to according to the pH of the cryo solution applied. Accordingly, the structures for H5 HA at pH 5.5, 6.0, 6.5 and 7.0 were solved at 2.30, 2.11, 2.39 and 2.79 Å resolution, respectively.
Table 1.
Data collection and refinement statistics for H5 HA under different conditions.
| Structure | H5 HA (pH 5.5) | H5 HA (pH 6.0) | H5 HA (pH 6.5) | H5 HA (pH 7.0) |
|---|---|---|---|---|
| PDB Codes Data collection statistics |
6PD3 | 6PCX | 6PD5 | 6PD6 |
| X-ray source and detector | LS-CAT (ID-G) MAR CCD 300 |
LS-CAT (ID-G) MAR CCD 300 |
LS-CAT (ID-G) MAR CCD 300 |
LS-CAT (ID-G) MAR CCD 300 |
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.979 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å)a | 2.30 (2.43–2.30) | 2.11 (2.22–2.11) | 2.39 (2.52–2.39) | 2.79 (2.95–2.79) |
| Number of Reflections | ||||
| Observeda | 260,754 (37,761) | 356,742 (53,484) | 345,944 (51,268) | 161,037 (21,885) |
| Uniquea | 43,303 (6,554) | 55,355 (8,604) | 37,825 (5,929) | 24,399 (3,544) |
| Completeness (%) | 99.1 (94.7) | 99.4 (96.9) | 99.5 (98.6) | 98.2 (90.1) |
| Rmeas (%)a | 8.7 (78.5) | 6.3 (81.9) | 16.3 (176.4) | 16.6 (139.7) |
| CC1/2 (%)a | 99.8 (88.4) | 99.9 (81.9) | 99.7 (85.7) | 99.7 (54.2) |
| Average I/σ(I) a | 11.78 (2.09) | 14.59 (2.04) | 14.59 (2.04) | 14.79 (1.54) |
| Space group | H32 | H32 | H32 | H32 |
| Unit cell (Å): a, b, c | 109.38, 109.38, 421.03 | 108.54, 108.54, 419.79 | 108.06, 108.06, 419.82 | 109.55, 109.55, 421.56 |
| (°): α, β, γ | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Refinement statistics | ||||
| Refinement program | REFMAC5 | REFMAC5 | REFMAC5 | REFMAC5 |
| Rwork (%) | 19.86 | 20.69 | 21.32 | 21.22 |
| Rfree (%) | 22.89 | 25.55 | 26.07 | 28.63 |
| Resolution range (Å) | 30.00 – 2.30 | 30.00 – 2.10 | 30.00 – 2.39 | 30.00 – 2.79 |
| Protein molecules per a.u. | 1 | 1 | 1 | 1 |
| Number of atoms: | ||||
| Protein (Chain A, Chain B) | (2561, 1376) | (2561, 1382) | (2568, 1376) | (2561, 1371) |
| Water molecules | 122 | 129 | 105 | 66 |
| Ligands (NAG) | 56 | 56 | 56 | 56 |
| Sulfate + Glycerol | 78 | 78 | 73 | 73 |
| RMSD from ideal: | ||||
| Bond length (Å) | 0.0045 | 0.0043 | 0.009 | 0.005 |
| Bond angles (°) | 1.5443 | 1.5425 | 1.7159 | 1.3984 |
| Average B-factors (Å2) | ||||
| Protein (Chain A, Chain B) | (67.3, 69.5) | (65.8, 67.5) | (60.9, 64.9) | (84.5, 88.6) |
| Water molecules | 61.3 | 66.5 | 49.9 | 59.8 |
| Ligands (NAG) | 112.1 | 106.2 | 107.0 | 124.0 |
| Sulfate + Glycerol | 119.5 | 122.0 | 125.3 | 158.7 |
| Ramachandran plot (%): | ||||
| Most favored regions | 94 | 94 | 94 | 90 |
| Additionally allowed regions | 5 | 6 | 5 | 8 |
| Outlier regions | 0 | 0 | 0 | 1 |
Parenthesis denote the highest resolution shells.
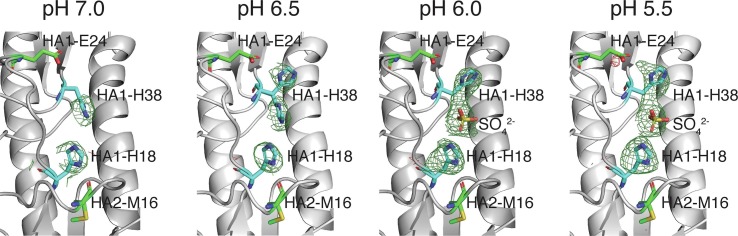
Overall, the 4 structures are found to be very similar. For example, the RMSD of the H5 HA backbone is ~ 0.23 Å between the pH 7.0 and 5.5 structures. However, as shown in Fig. 1 (and Supplementary Figure S1) an in depth comparison of the 4 structures reveals evidence of one important pH-sensitive effect, which involved a pair of conserved histidines found at the interface between the HA1 and HA2 subunits (and also in close proximity to the fusion peptide). At pH 7.0 HA1-H18 is forming a hydrogen bond with the main chain carbonyl of HA2–M16. This interaction positions the imidazole ring of HA1-H18 towards the HA1-H38 side chain. In the pH 6.0 and pH 5.5 structures, a ~120° rotation of the HA1-H38 side-chain around the Cα-Cβ bond (i.e. the Chi1 dihedral) is observed. Thus at the lower pH values the HA1-H38 side chain now faces away from HA1-H18 side chain. At lower pH the side chain of HA1-H38 is stabilized in the new conformation via an electrostatic interaction with the HA1-E24 carboxyl group (i.e. a stabilizing cation-anion interaction). Additionally, the increase in positive charge of this site at low pH attracts a negatively charged sulfate group (from the cryo solution). Importantly, the structure at pH 6.5 captures both states of the histidine in this transition. Specifically, the side chain of HA1-H38 exists in two conformers (facing towards and away from HA1-H18) with ~50% occupancy.
Fig. 1.
pH induced rotation of the side chain of HA1-H38. Structures (with electron densities) of the histidine cluster in 4 H5 HA structures were determined after freezing the crystals using cryo-solutions at different pH (corresponding pH shown in the top right corner of each structure). Fo - Fc difference maps are shown for each structure, with the contour level at 2.5σ (mesh) for select atoms. In these structures the closest approach of HA1-H18 to HA1-H38 is 4.7 Å at pH 7.0 and 7.8 Å at pH < 6.5. Information on the resolution and refinement parameters for each structure is shown in Table 1.
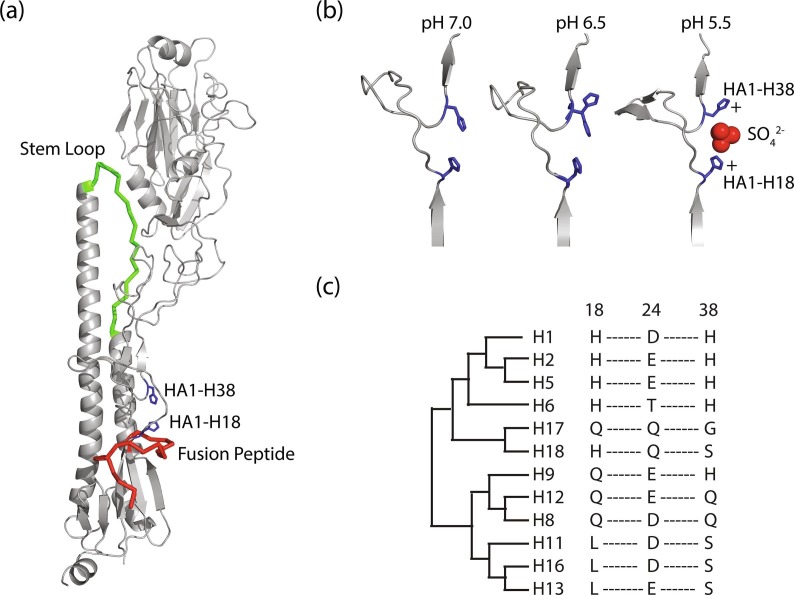
The protonation of HA histidine residues is thought to be one of the triggers for the large conformational changes necessary for fusion of the viral and target membranes (Zhou et al., 2014, Xu and Wilson, 2011, Mair et al., 2014). Notably histidines can participate in stabilizing cation-anion and destabilizing cation-cation interactions in proteins (Harrison et al., 2013). Importantly, the kinetic barrier for the transformation of HA prefusion conformation to intermediate and postfusion states would be lowered by destabilizing cation-cation interactions (Harrison et al., 2013). Although this work used crystals crystallized in the same condition and only exposed to differing pH conditions after formation, we were still able to find one striking change in the conformation of H5 HA between pH 5.5–7.0: the side chain interactions between HA1-18 and HA1-H38. As shown by Fig. 2 a, the HA1-H18 and HA1-H38 pair brings together 2 discreet regions that are in close proximity to the stem loop, which becomes helical at low pH, and the fusion peptide, which inserts into the endosome membrane (Skehel and Wiley, 2000, Eckert and Kim, 2001). Notably, at neutral pH the closest approach of the histidine side chains is 4.7 Å. This is consistent with other conserved histidine pH sensor pairs found in other proteins, where the side chain separations are < 7 Å (Harrison et al., 2013). We attribute the rotation of HA1-H38 at lower pH to be due to protonation of both histidines and the resulting cation-cation repulsion as modeled in Fig. 2b. However, an alternative explanation is that the rotation of HA1-H38 is driven by an attractive interaction with HA1-E24 (closest approach is 3.9 Å at pH 6.0) or some combination of cation-cation repulsion and cation-anion attraction. Our observation of the HA1-H38 in two conformations at pH 6.5 (Fig. 1) implies that the pKa of HA1-H38 and/or HA1-H18 is ~6.5. Previous work has shown that the HA fusion peptide becomes more highly exposed and dynamic at lower pH (Garcia et al., 2015). Thus we suggest that charge repulsion between the two histidines plays a role in the increased dynamics and solvent exposure of the region proximal to the fusion peptide. As shown by Fig. 2c, H18 and H38 (as well as an acidic amino acid at position 24) of the HA1 subunit are conserved in many Group 1 HA but not all, suggesting that alternative pH sensors are present in other HA. Nonetheless, we note that previous mutagenesis studies of HA1-H18 have suggested that this residue plays an important role in regulating the pH of virus fusion (Reed et al., 2009). Moreover, MD studies have suggested that the neutralizing antibody CR6261 inhibits entry by lowering the HA1-H18 pKa by ~1 unit (Zhou et al., 2014). In addition, studies on species dependent sensitivity of HA subtypes to the neutralizing antibody C179 have suggested that HA1-H38 plays a critical role in sensitivity to this antibody (Doud et al., 2018). Finally, we note that the irreversible transition of H5 HA to the postfusion form occurs at pH ~5.4 (Galloway et al., 2013). Consequently, the observations presented herein for pH > 5.4 clearly apply to the prefusion forms on the pathway toward the fusion conformation.
Fig. 2.
(a) Location of the histidine pair within the H5 HA structure. The histidine pair is colored blue, the fusion peptide is colored red, and the stem loop region is colored green. For clarity only one monomer of the HA trimer is shown. (b) Model for pH induced rotation of HA1-H38 side chain with histidine side chains colored blue and a stabilizing sulfate ion found in the low pH structure colored red. (c) Cladogram showing the conservation of HA1-18 and HA1-38 within Group 1 HA subtypes. UniProtKB entries: Q9WFX3 (H1), P03451 (H2), Q14RX4 (H5), Q80RX6 (H6), H6QM73 (H17), U5N1D3 (H18), Q9J4A2 (H9), P03446 (H12), P03456 (H8), P04661 (H11), Q5DL24 (H16), and P13103 (H13).
3. Materials and Methods
3.1. Protein expression and purification
H5 HA was prepared as previously described (Antanasijevic et al., 2014). Briefly H5 HA extracellular domain was expressed in SF9 insect cells grown in SF-900 II serum free media (Life Technologies). The cells were co-transfected with a pAcGP67 plasmid containing a H5 HA (A/Vietnam/1203/ 04 (H5N1)) expression construct and BD BaculoGold linearized baculovirus DNA (BD Biosciences). The H5 HA protein construct has altered N and C-termini. The mammalian cell secretion signal (residues Met-1 to Ser-16) was replaced with the GP67 secretion signal (pAcGP67 vector). Transmembrane and cytosolic regions of the protein (residues V521 to R564 of HA0 were removed and replaced with an artificial trimerization domain (the foldon from T4 fibritin), and a His-tag as previously described (Stevens et al., 2004). Cell handling, transfection and protein expression were performed as recommended by the BD BaculoGold starter package kit (BD Biosciences). Viral titers were monitored using the BacPAKTM qPCR Titration Kit (Clontech Laboratories). For expression, fresh SF9 cells at 3–4 × 106 cells/mL confluency were infected with H5 HA-containing baculovirus solution at MOI between 2 and 4. 4 days later the suspension was collected and the cells were removed by centrifugation. H5 HA is secreted into the insect cell media and purified by Ni-affinity chromatography. Given the relatively low processing percentage of HA expressed in this insect cell line, furin protease (NEB) was used to cleave the HA0 into HA1 and HA2. After 48 h at 4 °C, furin was inactivated using Furin Inhibitor I from EMD Millipore. The protein concentrate was then subjected to Sephacryl S300 gel filtration column with phosphate buffer (50 mM sodium phosphate/pH 8.1 and 50 mM NaCl) as a running buffer. Protein fractions were pooled and concentrated and the final yield was ~ 3 mg of protein per liter of SF9 cells.
3.2. X-ray crystallography
For the crystallography experiments additional thrombin processing was performed on HA construct after the Ni-NTA purification to remove the foldon domain and His-tag present at the C-terminus of HA2. Specifically, thrombin at 10–50 ng/mL was applied to remove the foldon domain. HA was then purified on Sephacryl S300 gel filtration column using 50 mM phosphate (pH 8.2) + 50 mM NaCl as the running buffer. Protein was then concentrated to 8 mg/mL and dialyzed to 50 mM Tris-HCl (pH 8.2) + 50 mM NaCl. Initial crystallization trials were performed with H5 HA using JSSG+, Protein Complex and PEGs suites. Multiple hits were observed and optimized for hanging drop vapor diffusion method. For the pH-based experiments we used crystals obtained under the following reservoir conditions: 100 mM Cacodylate buffer, pH 6.5 + 200 mM NaCl + 2 M (NH4)2SO4 + 10% Glycerol. The drops were initially set by mixing 1 μL of 8 mg/mL H5 HA solution and 1 μL of reservoir solution but different combinations were also attempted. For crystal freezing we used the cryo-solution containing 100 mM Cacodylate buffer + 200 mM NaCl + 2 M (NH4)2SO4 + 20% Glycerol. 4 versions of this solution were made with the only difference in the final pH value (pH 7.0, pH 6.5, pH 6.0, pH 5.5). The crystals were soaked in appropriate cryo-solutions for 10 min before freezing and data acquisition. Diffraction datasets were collected at the stations of Life Sciences Collaborative Access Team at the Advanced Photon Source, Argonne, Illinois. Initial data processing was performed using XDS. For molecular replacement PDB entry 2FK0 was used. Molecular replacement and structure refinement were done in CCP4 (Winn et al., 2011) and PHENIX (Adams et al., 2010), and all manual refinement was performed with Coot (Emsley et al., 2010). The PyMOL program package was used for structure comparison and generation of figures.
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 6PD3, 6PCX, 6PD5 and 6PD6
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This research was supported by the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsb.2019.107412.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M. HIV envelope: challenges and opportunities for the discovery of entry inhibitors. Trends Microbiol. 2011;19:191. doi: 10.1016/j.tim.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa E., Aoyama T., Kato H., Suzuki Y., Tateno Y., Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., Zumla A. Emerging respiratory tract viral infections. Curr. Opin. Pulm. Med. 2015;21:284. doi: 10.1097/MCP.0000000000000153. [DOI] [PubMed] [Google Scholar]
- Lagoja I.M., De Clercq E. Anti-influenza virus agents: synthesis and mode of action. Med. Res. Rev. 2008;28:1. doi: 10.1002/med.20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wu C., Zhao L., Huang N. Exploring the early stages of the pH-induced conformational change of influenza hemagglutinin. Proteins. 2014;82:2412. doi: 10.1002/prot.24606. [DOI] [PubMed] [Google Scholar]
- Xu R., Wilson I.A. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 2011;85:5172. doi: 10.1128/JVI.02430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J., Cardone G., Heymann J.B., Winkler D.C., Steven A.C. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J. Virol. 2012;86:2919. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E., Ramos C., Markovic I., Zimmerberg J., Chernomordik L.V. Reversible stages of the low-pH-triggered conformational change in influenza virus hemagglutinin. EMBO J. 2002;21:5701. doi: 10.1093/emboj/cdf559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair C.M., Meyer T., Schneider K., Huang Q., Veit M., Herrmann A. A histidine residue of the influenza virus hemagglutinin controls the pH dependence of the conformational change mediating membrane fusion. J. Virol. 2014;88:13189. doi: 10.1128/JVI.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C. Stability of infectious influenza A viruses at low pH and at elevated temperature. Vaccine. 1985;3(3 Suppl):215. doi: 10.1016/0264-410x(85)90109-4. [DOI] [PubMed] [Google Scholar]
- Puri A., Booy F.P., Doms R.W., White J.M., Blumenthal R. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J. Virol. 1990;64:3824. doi: 10.1128/jvi.64.8.3824-3832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte T., Ludwig K., Huang Q., Rachakonda P.S., Herrmann A. Conformational change of influenza virus hemagglutinin is sensitive to ionic concentration. Eur. Biophys. J. 2007;36:327. doi: 10.1007/s00249-006-0116-0. [DOI] [PubMed] [Google Scholar]
- Reed M.L., Yen H.L., DuBois R.M., Bridges O.A., Salomon R., Webster R.G., Russell C.J. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J. Virol. 2009;83:3568. doi: 10.1128/JVI.02238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.S., Higgins C.D., O'Meara M.J., Koellhoffer J.F., Kuhlman B.A., Lai J.R. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. 2013;21:1085. doi: 10.1016/j.str.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antanasijevic A., Ramirez B., Caffrey M. Comparison of the sensitivities of WATERLOGSY and STD NMR experiments. J. Biomolec. NMR. 2014;60:37. doi: 10.1007/s10858-014-9848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia N.K., Guttman M., Ebner J.L., Lee K.K. Dynamic changes during acid-induced activation of influenza hemagglutinin. Structure. 2015;23:65. doi: 10.1016/j.str.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud M.B., Lee J.M., Bloom J.D. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 2018;9:1386. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E., Reed M.L., Russell C.J., Steinhauer D.A. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J., Corper A.L., Basler C.F., Taubenberger J.K., Palese P., Wilson I.A. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol Crystallogr. 2011;67:235. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.