Abstract
L-type lectins are involved in glycoproteins secretory pathways and are associated with many immune responses. There is growing evidence that L-type lectins are also involved in viral replication. In this study, a novel L-type lectin (named as PcL-lectin) was identified from red swamp crayfish (Procambarus clakii). Gene sequencing and phylogenetic tree analysis results showed that the PcL-lectin was a kind of endoplasmic reticulum Golgi intermediate compartment-53 (ERGIC-53). The expression level of PcL-lectin was significantly down regulated in crayfish after challenged with white spot syndrome virus (WSSV). Recombinant PcL-lectin protein facilitated the replication of WSSV in crayfish. In addition, WSSV replication was decreased when endogenous PcL-lectin was knocked down by RNA interference in crayfish. Furthermore, PcL-lectin may interact with VP24, an envelope protein of WSSV. Our results suggest that PcL-lectin may be required for the multiplication of WSSV, and will pave a new way for the developing of strategies against WSSV infection.
Keywords: Procambarus clarkii, White spot syndrome virus, Innate immunity, L-type lectin
Highlights
-
•
PcL-lectin was widely expressed in the several tissues.
-
•
PcL-lectin was downregulated expression after challenge with white spot syndrome virus (WSSV).
-
•
PcL-lectin knockdown inhibit the replication of WSSV, and it could be rescued by injected with recombinant PcL-lectin.
-
•
PcL-lectin may interact with VP24, an envelope protein of WSSV.
1. Introduction
Lectins are group of molecules which are highly specific for the binding of carbohydrate. It was earliest discovered in leguminous plants, therefore it was named legume lectin (or L-type lectin). Thereafter, many other members of lectins have been identified in most kinds of organisms, such as fungi, animals and plants. L-type lectins (LTLs) possess a luminal carbohydrate recognition domain which can bind to saccharides. In vertebrates, four kinds of LTLs have been reported, namely endoplasmic reticulum Golgi intermediate compartment-53 (ERGIC-53), ERGIC-53 like protein (ERGL), 36 kDa vesicular integral membrane protein (VIP36), and VIP36 like protein (VIPL) [1]. However, only ERGIC-53 and VIP36 were identified in invertebrates [2].
LTLs are associated with trafficking in the secretory pathway in endoplasmic reticulum Golgi by interaction with glycoproteins [3], [4], [5], [6], [7]. For example, VIP36 locates in the Golgi and transfers between Golgi and endoplasmic reticulum (ER). It serves as the export receptor in ER and is associated with export of glycoprotein [8]. VIP36 was involved in the regulation of the phagocytosis in macrophage [9]. Similarly, MjL-lectin promoted the hemocytes phagocytosis in kuruma shrimp (Marsupenaeus japonicus) [10]. ERGIC-53 is a mannose-specific membrane lectin operating as a cargo receptor for the transport of glycoproteins from the ER to the ERGIC.
White spot syndrome virus (WSSV) is the type species of the genus Whispovirus in the family Nimaviridae. WSSV has caused great economic loss in shrimp aquaculture industry worldwide. Up to date, there is lack of effective strategies against WSSV infection. The studies of WSSV replication and host-interactions would help to explore effective strategies for WSSV prevention and control. There is growing evidence that ERGIC-53 is closely related with virus infection. ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles [11]. In the present study, a LTL (named PcL-lectin) was identified from red swamp crayfish, Procambarus clarkii. Gene sequencing and phylogenetic tree analysis results showed that the PcL-lectin was a member of ERGIC-53. The expression level of PcL-lectin was significantly down regulated in crayfish after challenged with white spot syndrome virus (WSSV). The anti-viral properties of the PcL-lectin were further investigated by using recombinant PcL-lectin protein as well as RNA interference (RNAi). Viral proteins which might interact with the PcL-lectin were also studied. The results suggest that PcL-lectin may play an important role in response to WSSV infection in red swamp crayfish.
2. Materials and methods
2.1. Virus preparation
The WSSV was isolated and kept in our laboratory. The WSSV was maintained and multiplied by infection in red swamp crayfish using the method described previously [12]. One gram of the gill tissue from the infected crayfish was homogenized in 10 mL of phosphate-buffered saline (PBS, 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH = 7.4). Thereafter, it was centrifuged at 10,000 rpm for 30 min at 4 °C. The supernatant was stored at −80 °C until use after filtration with 0.45 μm filter.
2.2. Red swamp crayfish challenged with WSSV and tissue collection
Red swamp crayfish (8–12 g) were purchased from the Baishazhou market in Wuhan city, Hubei Province, China. The crayfish were kept in an aerated water tank at room temperature (23 ± 2 °C), and changed fresh water every day. Hemocyte, heart, hepatopancreas, gill, stomach and intestine were collected from healthy red swamp crayfish (3 crayfish) for the studying the tissue distribution of PcL-lectin. Total RNAs were extracted from these tissues by using RNAiso Plus (Takara, Dalian, China) according to the manufacturer’s protocol. Reverse transcription was carried out using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). The obtained cDNAs were stored at −20 °C. Meanwhile, total proteins were extracted from these samples, 100 μg tissues was homogenized in 1 mL RIPA buffer (Beyotime, Beijing, China), and then centrifuged at 10,000 rpm for 30 min at 4 °C. The supernatant was boiled with SDS loading buffer for 5 min, and stored at −20 °C until use.
The infection experiment was performed as described previously [13]. Briefly, 25 μL of WSSV (about 1 × 107 copies) or PBS (as the control) was injected into the abdominal segment of the red swamp crayfish (60 crayfish each group), respectively. Hemocytes were collected from red swamp crayfish for the expression pattern analysis of PcL-lectin at 0, 6, 12, 24 and 48 h post of WSSV challenge and dissolved in 1/2 hemocyte volume of anticoagulant buffer [0.14 M NaCl, 0.1 M glucose, 30 mM trisodium citrate, 26 mM citric acid, and 10 mM ethylene diamine tetra acetic acid (EDTA), pH 4.6] [14]. The hemocytes were immediately centrifuged at 800 g for 8 min (4 °C). Thereafter, total RNAs and proteins were extracted as described above.
2.3. cDNA cloning and sequence analysis
Expressed sequence tag (EST) sequences similar to L-type lectin genes were obtained from transcriptomic sequencing of the heart of red swamp crayfish in our pilot experiments. The EST sequences were highly similar to the sequences of ERGIC-53 from Chinese mitten crab (Eriocheir sinensis) with around 86% amino acid sequence identity after BLAST analysis using the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). At the same time, the domain architecture of the PcL-lectin was analyzed by SMART (http://smart.embl-heidelberg.de/).
2.4. Recombinant PcL-lectin protein expression, purification and antiserum preparation
The lectin leg-domain of PcL-lectin gene was amplified from hemocytes using the ExPcL-lecF and ExPcL-lecR primers with EcoRI and XhoI restriction site (Table 1 ). The PCR procedure was 1 cycle at 95 °C for 5 min; 35 cycles at 94 °C for 30 s, 56 °C for 45 s, and 72 °C for 1 min; and 1 cycle at 72 °C for 10 min. PCR products were examined by 1.0% agarose gel electrophoresis. Then, the amplified DNA fragments were purified with gel extraction kit (Aidlab, Beijing, China). Both DNA fragments and the vector pGEX-KG were digested with the corresponding DNA restriction endonuclease at 37 °C for 1 h, and ligated in water bath overnight at 16 °C using T4 ligase (Fermentas, USA). The recombinant plasmids pGEX-KG-L-lectin was transformed into Eriocheir coli BL21 (DE3) cells, isopropyl-b-d-1-thiogalactopyranoside (IPTG) was used to induce the expression of the recombinant PcL-lectin (rPcL-lectin). Thereafter, it was purified from the inclusion body of the E. coli BL21. Specific polyclonal antibody of PcL-lectin was obtained by immunizing rabbit with the purified recombinant protein.
Table 1.
Sequences of the primers used in this study.
| Primers | Sequence (5′–3′) |
|---|---|
| ExPcL-lecF | TACTCAGAATTCCGTACCTGGCGCAGAAGG |
| ExPcL-lecR | TACTCACTCGAGTTACTTTCTGTACAGCATC |
| RT-PcL-lecF | CTTGGTGGCTGCCTTAGAG |
| RT-PcL-lecR | TCCTTCTCATTGTTTGTCATTC |
| 18S F | TCTTCTTAGAGGGATTAGCGG |
| 18S R | AAGGGGATTGAACGGGTTA |
| RNAi-PcL-lecF | GCGTAATACGACTCACTATAGGTTGAGATTGTTGCAAATGC |
| RNAi-PcL-lecR | GCGTAATACGACTCACTATAGGGCTAAAATATAAACCTACC |
| RNAi-GFP-F | GCGTAATACGACTCACTATAGGTGGTCCCAATTCTCGTGGAAC |
| RNAi-GFP-R | GCGTAATACGACTCACTATAGGCTTGAAGTTGACTTGATGCC |
2.5. Immune-fluorescent assay of PcL-lectin in hemocytes
Hemocytes were collected as above and Immune-fluorescent assay was performed to determine the location of PcL-lectin in the hemocytes as described previously [15].
2.6. Tissue distribution and expression profiles of PcL-lectin detected by quantitative real-time PCR (qRT-PCR) and Western blot
The tissue distributions and expression profiles of PcL-lectin were analyzed using qRT-PCR with the PcL-lectin specific primers RT-PcL-lecF and RT-PcL-lecR (Table 1). The 18S rRNA was used as the reference and was amplified with the primers 18S F and 18SR (Table 1). The qRT-PCR was programmed at 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 1 min. DNA melting analysis was performed to confirm the specificity of the amplified products. The obtained data were statistically analyzed and calculated using the threshold cycle method as previously described [16].
Meanwhile, the tissue distribution and expression profiles of the PcL-lectin protein were measured by Western blot using antiserum of PcL-lectin as described previously [13].
2.7. Recombinant PcL-lectin protein (rPcL-lectin) and the replication of WSSV
To analyze the relationship between PcL-lectin and the replication of WSSV, we performed following experiments in four groups of crayfish (5 crayfish each group). Group 1 (control): red swamp crayfish were injected with 50 μL PBS. Group 2 (PBS + WSSV): injected with 50 μL PBS containing WSSV (1 × 105 virions). Group 3 (GST + WSSV): injected with 50 μL mixture containing 200 μg GST protein and WSSV (1 × 105 virions). Group 4 (rPcL-lectin + WSSV): injected with 50 μL mixture containing 200 μg of rPcL-lectin and WSSV (1 × 105 virions). At 60 h post of challenge, the genomic DNA from gills of the treated red swamp crayfish were extracted using a genomic DNA extraction kit (CWbio, Beijing, China), qPCR was used to quantify the copy of WSSV genome in the crayfish with the vp28 primers RT-vp28F/R as described previously [17]. Meanwhile, proteins were extracted from the gills, and Western blot was performed using antiserum of VP28 as described previously [17].
2.8. RNA interference (RNAi) assay
Gene-specific primers for PcL-lectin (RNAi-PcL-lecF/R) and green fluorescent protein (GFP, RNAi-GFP-F/R) were incorporated with the T7 promoter showed in Table 1. T7 polymerase (Fermentas, USA) was used to synthesize dsRNA based on the method as described [18]. Crayfish were randomly divided into four groups (8 crayfish each group). The group 1 (control): without any treatment. The group 2 (PBS): each red swamp crayfish was injected with 25 μL PBS. The group 3 (dsGFP): injected with 25 μL dsGFP RNA. The group 4 (dsPcL-lectin): injected with 25 μL dsPcL-lectin RNA. At 48 h post of the treatment, total RNAs and proteins were extracted from the gills of the red swamp crayfish as mentioned above. qPCR and Western blotting were used to detect the efficiency of the RNAi. β-actin was used as control in western blot.
Infected experiment with WSSV after knock down of PcL-lectin was carried out. Crayfish were divided into four groups. The group 1 (normal): no treatment. The group 2 (dsPcL-lectin + WSSV): each crayfish was first injected with dsPcL-lectin RNA, 48 h later after the first injection, WSSV (1 × 105 virions) was injected. The group 3 (dsGFP + WSSV): first injected with dsGFP RNA, 48 h later, injected with WSSV (1 × 105 virions). The group 4 (PBS + WSSV): first injected with PBS, 48 h later, injected with WSSV (1 × 105 virions). At 60 h post of WSSV challenged, total proteins were extracted from the gills of the crayfish, and the expression of VP28 of WSSV was detected by Western blot.
2.9. Rescue assay with rPcL-lectin after RNAi treatment
The crayfish were divided into five groups and performed the following experiments (3 crayfish each group). The group 1 (PBS + WSSV): first injected nothing, after 48 h, PBS was injected, waited for 1 h, WSSV (1 × 105visions) was injected. The group 2 (dsPcL-lectin + rPcL-lectin + WSSV): first injected dsPcL-lectin, after 48 h, rPcL-lectin was injected, waited for 1 h, WSSV (1 × 105virions) was injected. The group 3 (dsPcL-lectin + GST + WSSV): first injected with dsPcL-lectin, after 48 h, GST was injected, waited for 1 h, then WSSV (1 × 105virions) was injected. The group 4 (dsGFP + GST + WSSV): first injected with dsGFP, after 48 h, GST was injected, waited for 1 h, then WSSV (1 × 105virions) was injected. The group 5 (dsGFP + rPcL-lectin + WSSV): first injected with dsGFP, after 48 h, rPcL-lectin was injected, waited for 1 h, WSSV (1 × 105virions) was injected. At 60 h after WSSV challenged, protein samples were extracted from the gills, and the amount of WSSV was revealed by Western blot using anti-VP28 antibody.
2.10. Pull-down and co-IP assays
To confirm the interaction of PcL-lectin with viral proteins, pull-down assays was performed using GST-tag fused PcL-lectin (GST-PcL-lectin) and His-tag fused viral proteins (His-VP15, His-VP19, His-VP24, His-VP26 and His-VP28). All these His-tag fused WSSV proteins were purified from E. coli which transformed pET-32 are combinant plasmids containing above viral genes. GST bind resin was used in this assay, and the pull-down was carried out as described previously [13]. Approximately 200 μg of purified GST-PcL-lectin was incubated with GST bind resin (GenScript, Nanjing, China) for 1 h, washed with PBS. About 400 μg of purified His-VP15 (His-VP19, His-VP24, His-VP26 or His-VP28) was added to the resin and incubated at 4 °C for 2 h. After being washed thoroughly with PBS, the proteins were eluted with elution buffer (GenScript, Nanjing, China) and then analyzed by 12% SDS-PAGE. GST bound resin was used as control, followed by washing with PBS and eluted with elution buffer.
To further analyze interaction of PcL-lectin with VP24, Co-IP assay was performed using antiserums of PcL-lectin and VP24 (VP24 antiserum was a gift of professor Jin-Xing Wang, Shandong University). A tube with antiserum-free protein A resin was used for control. The Co-IP assay was carried out as described previously [19]. Samples from crayfish gills challenged with WSSV were collected, and then were incubated with antiserum of PcL-lectin or VP24 overnight at 4 °C with rotation. The suspension was then incubated with 10 μL of Protein A beads for1 h at 4 °C under agitation. The beads were collected and washed 4 times, then were resuspended in 30 μL of SDS-PAGE sample buffer and boiled for10 min. The samples were analyzed by Western blot.
3. Results
3.1. Sequence characteristics of PcL-lectin
The obtained PcL-lectin sequence was 1680 bp with 1506 bp open reading frame (ORF) encoding a 501 amino acid protein with a predicted molecular weight of 56.8 kDa (Fig. S1). SMART analysis showed that the PcL-lectin contained a putative signal peptide of 19 amino acids, one lectin leg-like domain located at the N-terminal and one transmembrane domain at the C-terminal, one coiled coil region and one low complexity region in the middle (Fig. 4B). After BLAST in NCBI, the PcL-lectin was most related to ERGIC-53 members. Therefore, ERGIC-53 from other nine species were included to construct a phylogenetic tree. These species were common periwinkle (Littorina littorea), oyster (Crassostrea gigas), Chinese mitten crab (Eriocheir sinensis), fly (Drosophila melanogaster), large yellow croaker (Larimichthys crocea), frog (Xenopus tropicalis), green sea turtle (Chelonia mydas), woodpecker (Picoides pubescens), and human (Homo sapiens). The results showed that PcL-lectin was most similar with the Chinese mitten crab EsERGIC-53 with 86% amino acid identity (Fig. S2).
Fig. 4.
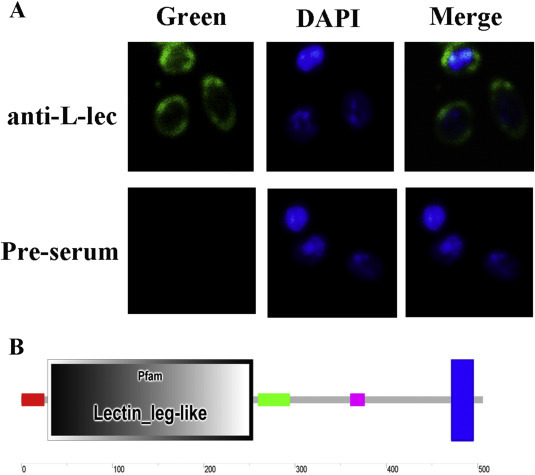
Immune-fluorescent assay of PcL-lectin in hemocytes. (A)Hemocytes were analyzed by immune-fluorescent assay using anti Pc-Lectin rabbit serum or serum (pre-serum) from rabbit without treated with any antigen. Nuclear was stained with DAPI. The green showed signal of PcL-lectin, the blue showed signal of nuclear. Merge showed the location of PcL-lectin and nuclear. (B) Domain structure of PcL-lectin, red showed signal peptide, grey showed lectin leg-like domain, green showed coiled-coil structure, purple showed low complexity structure, blue showed transmembrane domain. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. The location of PcL-lectin in the hemocytes of crayfish
To monitor the location of the PcL-lectin, the lectin leg-like domain (around 28 kDa) fused to GST to generate a recombinant PcL-lectin with molecular weight of about 54 kDa was used as antigen to generate rabbit anti-serum of PcL-lectin (Fig. 1 ). A specific band about 57 kDa which was identical the molecular weight of PcL-lectin was detected from the hemocytes of crayfish, indicating that the anti-serum could be used to detect the native PcL-lectin (Fig. 1). Immune-fluorescent assay was used to investigate the cellular localization of PcL-lectin in the hemocytes from non-infected crayfish. The results showed that PcL-lectin was mainly distributed on the membrane and the cytoplasm of hemocytes, which is identical with the existed of the predicted transmembrane domain of the PcL-lectin (Fig. 4).
Fig. 1.
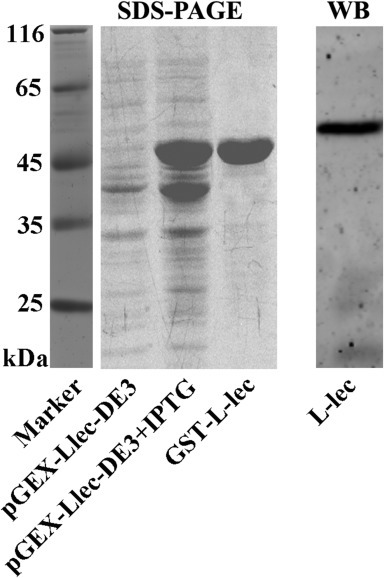
SDS–PAGE analysis of recombinant rPcL-lectin protein and the specificity of its anti-serum. pGEX-Llec-DE3 represents control group, rPcL-lectin protein was not expressed without IPTG induction. pGEX-Llec-DE3+IPTG represents experiment group, rPcL-lectin protein was expressed in E. coli with IPTG induction. GST-L-lec represents purified rPcL-lectin protein from E. coli, L-lec represents PcL-lectin, antiserum could recognize endogenous L-lectin from the crayfish.
3.3. Tissue distribution and expression profiles of PcL-lectin
The mRNA expressions of PcL-lectin from the tissues of hemocyte, heart, hepatopancreas, gill, stomach and intestine of crayfish were measured by qRT-PCR. The results showed that the mRNA of PcL-lectin was expressed in all tested tissues, with higher expression levels in the hemocytes, heart and stomach, and lower expression levels in hepatopancreas, gills and intestine (Fig. 2 A). Furthermore, the extracted proteins from the above six tissues were subjected to Western blot assay. The results showed that the expression patterns of PcL-lectin at both mRNA and protein levels were similar in the six tissues (Fig. 2).
Fig. 2.
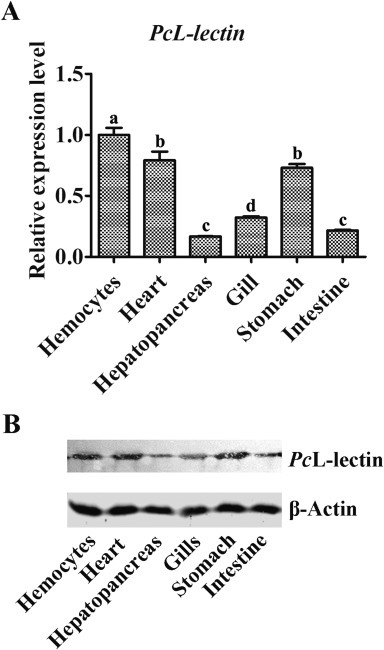
Tissues distribution of PcL-lectin. (A) The expressions of PcL-lectin mRNA in the six tissues of crayfish. The extracted total mRNAs from hemocytes, heart, hepatopancreas, gills, stomach and intestine were subjected to qRT-PCR assay. The 18S rRNA was used as reference. (B) The expression of PcL-lectin protein in the six tissues of crayfish. The extracted total proteins from the six tissues were subjected to Western blot assay. β-actin was used as internal control. The experiments were repeated three times. Different letters represented significant changes between groups, p < 0.05.
3.4. The effects of PcL-lectin on the multiplication of WSSV
Hemocyte was the major site for the multiplication of WSSV, therefore, we investigated the expression profiles of PcL-lectin in hemocytes from the swamp crayfish challenged with WSSV. The results showed that the overall mRNA of PcL-lectin was declined in hemocytes at 12 and 48 h post of challenge (Fig. 3 A). The overall expression profiles of PcL-lectin protein was similar with those of PcL-lectin mRNA, with a slight enhancement at 48 h post of challenge (Fig. 3B). These results showed that WSSV challenge was able to down-regulate the expression of PcL-lectin. To determine whether the PcL-lectin has an effect on the WSSV multiplication, the recombinant GST-fused PcL-lectin (rPcL-lectin) was injected into crayfish. Thereafter, the crayfish were challenged with WSSV. qRT-PCR and Western blot were performed to detect the multiplication of WSSV in gills. The results showed that the viral specific mRNA and protein in the rPcL-lectin injection group were markedly higher than those in the control groups (GST-WSSV, PBS-WSSV and WSSV) only (Fig. 5 A,B), indicating that PcL-lectin could facilitate the multiplication of WSSV.
Fig. 3.
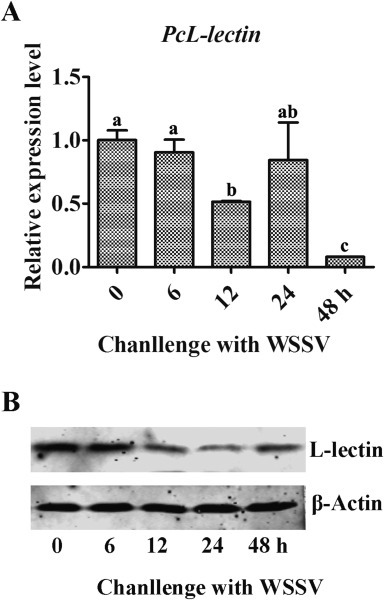
Expression pattern of PcL-lectin. The expression pattern of PcL-lectin was detected by qRT-PCR and Western blot assays after challenged with WSSV. (A) Expression pattern analysis of the PcL-lectin expression pattern in the hemocytes of the WSSV-challenged crayfish at 0, 6, 12, 24 and 48 h post of the challenge. (B) Western blot was used to detect the expression pattern of PcL-lectin. Each experiment was repeated three times. Different letters represented significant changes between groups, p < 0.05.
Fig. 5.
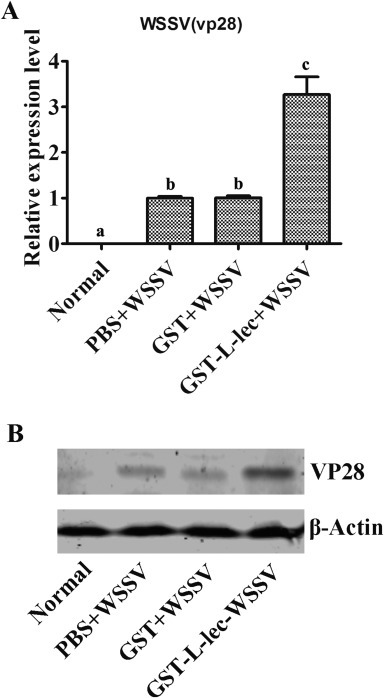
Recombinant PcL-lectin protein and WSSV replication. Crayfish was divided into four groups. Group 1 was control, Group 2 was injected with PBS and WSSV, Group 3 was injected with GST-tag and WSSV, Group 4 was injected with GST-PcL-lectin and WSSV. (A) Genome DNA was extracted from Gills, and qPCR was used to detect the amount of WSSV (vp28 mRNA as the marker for WSSV replication). (B)Total protein was extracted from gills of crayfish, and western blot was used to detect the amount of WSSV (VP28 as a marker for WSSV). Each experiment was repeated three times. Different letters represented significant changes between groups, p < 0.05.
To further address the relationship between PcL-lectin and WSSV multiplication, we performed PcL-lectin knock-down and rescue experiments. Both mRNA and protein of PcL-lectin could be significantly knocked down by the injection of PcL-lectin specific dsRNA into the crayfish (Fig. 6 A,B). The multiplication of WSSV was inhibited in the crayfish which PcL-lectin has been knocked down (Fig. 6C). However, the inhibition could be rescued when recombinant PcL-lectin protein was injected into the crayfish which native PcL-lectin has been knocked down by RNAi. But the rescue was not observed in the crayfish which were treated with control RNAi (GFP RNAi) and injected with control protein (GST) (Fig. 6D), indicating that PcL-lectin may play a role in the multiplication of WSSV in crayfish.
Fig. 6.
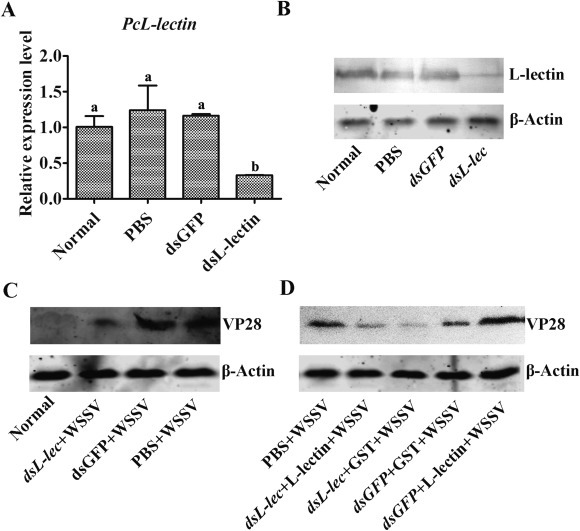
PcL-lectin knockdown inhibited WSSV in crayfish. The crayfish were subjected to RNA interference with dsGFP as the control. (A and B) Effect of PcL-lectin RNAi on gills determined using qPCR and Western blot analysis. (C)The crayfish were infected with WSSV after PcL-lectin knockdown, and the amount of WSSV was detected via western blot using VP28 as a marker. (D)The rescue experiment was also performed through the injection of rPcL-lectin into the PcL-lectin-silenced and GFP-silenced crayfish; GST-tag was injected into the PcL-lectin-silenced and GFP-silenced crayfish as controls. Different letters represented significant changes between groups, p < 0.05.
3.5. PcL-lectin bound to WSSV structural protein VP24
The above results suggested that PcL-lectin may affect the multiplication of WSSV, but its mechanism remains unclear. One possibility was that PcL-lectin might directly interact with the viral protein, so as to regulate the multiplication of WSSV. Therefore, the interaction of PcL-lectin with WSSV proteins was measured. Six His-tag fused proteins of WSSV (VP15, VP19, VP24, VP26 and VP28) were used in pull-down experiment with GST-tag fused PcL-lectin in vitro. The results showed that only the VP24 could bind to rPcL-lectin (Fig. 7 A). A Co-IP assay was also performed using the lysate of the gills from WSSV-challenged crayfish. The results showed that native PcL-lectin was able bind to VP24 (Fig. 7B). These results suggest that PcL-lectin might physically interact with VP24.
Fig. 7.
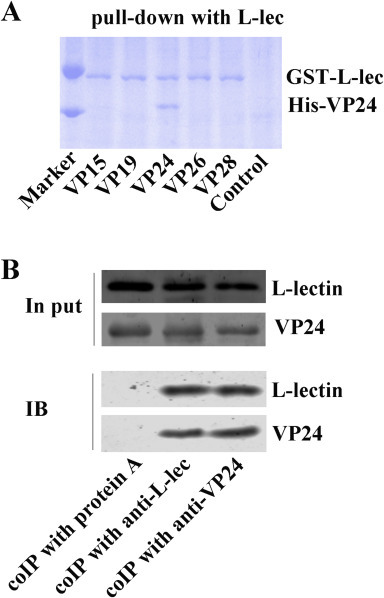
PcL-lectin interacts with VP24 of WSSV. (A) In vitro pull-down assays were performed to detect the interactions of His-tag fused VP15, VP19, VP24, VP26, and VP28 with GST-PcL-lectin. A tube with GST resin was used as the control. (B)The Co-IP assay was performed using lysates of gills from WSSV-challenged crayfish, PcL-lectin antiserum and VP24 antiserum. A tube with antiserum-free protein A resin was used as the control.
4. Discussion
ERGIC-53 is a kind of lectin which mainly locates in ER and Golgi of cells [20], [21], [22], [23], and serves as a regulator of export of glycoproteins [3], [8]. In the present study, we cloned and characterized a novel ERGIC-53 (named PcL-lectin) from red swamp crayfish. The PcL-lectin was ubiquitously expressed in all six detected tissues of the crayfish (Fig. 2). The PcL-lectin was ubiquitously expressed in all six detected tissues of the crayfish and located at the cytoplasm and membrane of hemocytes, which was identical to the existing of transmembrane domain of the PcL-lectin.
There is growing evidence that the members of lectin family have been involved in virus infection. LdlrLec1 and LdlrLec1 interacted with VP28, and inhibited the infection and replication of WSSV in M. japonicus [24]. MjsvCL, DC-SIGN and L-SIGN were served as receptor candidates in the infection of WSSV or influenza A virus [25], [26]. Lavvy, a kind of CTLs, was response to WSSV infection in Litopenaeus vannamei. MjsvCL together with calreticulin, was identified as a receptor in the infection of WSSV in M. japonicus [25]. However, all these lectins identified from shrimps are all C-type lectins. It has been reported that ERGIC-53 was related with the replication of some RNA viruses, such as arenavirus, coronavirus, and filovirus [11]. To better understand the function of PcL-lectin in crayfish during WSSV infection, recombinant PcL-lectin was further used for the studies. The results showed that recombinant PcL-lectin was able to enhance the multiplication of WSSV in crayfish (Fig. 5) and it could rescues the multiplication of WSSV in the crayfish which their endogenous PcL-lectin was knocked down by RNAi (Fig. 6), suggesting that PcL-lectin may play a role in the multiplication of WSSV. The PcL-lectin was down regulated in crayfish infected with WSSV at the early stage (Fig. 3), and recombinant PcL-lectin protein could facilitate WSSV replication (Fig. 5). The mortality of the crayfish was enhanced 2 d post of the challenge with WSSV [13]. It suggested that WSSV may utilize the PcL-lectin which stored in cell at the early stage of infection. On the other hand, crayfish may also resist WSSV infection by down regulated the expression of PcL-lectin. The reduction of PcL-lectin could not completely suppress the replication of WSSV.
Virus structural proteins play an essential role in virus infection [27]. It has been reported that WSSV envelope protein VP28 interacted with host protein and helped it entry into the host cell [28], VP26 and VP24 were first identified as nuclear proteins and then they were found at envelope of WSSV [29], [30], VP24 together with VP28 could participate in the infection of WSSV [31]. In this study, we showed that PcL-Lectin interacted with VP24, but not VP28, VP26 (Fig. 7A and B). The exact mechanism underlying PcL-lectin-VP24 interaction remains enigmatic and remains to be deciphered in the future.
Acknowledgement
This work was supported by grants from National Natural Science Foundation of China (No. 31502199, 31572657, 31372563) and the Fundamental Research Funds for the Central Universities (2015QC019, 2014PY035). Key Laboratory of Fishery Drug Development, Ministry of Agriculture (201401). Antiserum of VP24 was a gift of professor Jin-Xing Wang (Shandong University).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.fsi.2016.05.020.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Nufer O., Mitrovic S., Hauri H.P. Profile-based data base scanning for animal L-type lectins and characterization of VIPL, a novel VIP36-like endoplasmic reticulum protein. J. Biol. Chem. 2003;278:15886–15896. doi: 10.1074/jbc.M211199200. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y., Huang X., Wang Z., Tan J.M., Hui K.M., Wang W., Ren Q. Function of two novel single-CRD containing C-type lectins in innate immunity from Eriocheir sinensis. Fish. Shellfish Immunol. 2014;37:313–321. doi: 10.1016/j.fsi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Hauri H.P., Kappeler F., Andersson H., Appenzeller C. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 2000;113:587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- 4.Appenzeller-Herzog C., Roche A.C., Nufer O., Hauri H.P. pH-induced conversion of the transport lectin ERGIC-53 triggers glycoprotein release. J. Biol. Chem. 2004;279:12943–12950. doi: 10.1074/jbc.M313245200. [DOI] [PubMed] [Google Scholar]
- 5.Moussalli M., Pipe S.W., Hauri H.P., Nichols W.C., Ginsburg D., Kaufman R.J. Mannose-dependent endoplasmic reticulum (ER)-Golgi intermediate compartment-53-mediated ER to Golgi trafficking of coagulation factors V and VIII. J. Biol. Chem. 1999;274:32539–32542. doi: 10.1074/jbc.274.46.32539. [DOI] [PubMed] [Google Scholar]
- 6.Nyfeler B., Reiterer V., Wendeler M.W., Stefan E., Zhang B., Michnick S.W., Hauri H.P. Identification of ERGIC-53 as an intracellular transport receptor of alpha1-antitrypsin. J. Cell Biol. 2008;180:705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vollenweider F., Kappeler F., Itin C., Hauri H.P. Mistargeting of the lectin ERGIC-53 to the endoplasmic reticulum of HeLa cells impairs the secretion of a lysosomal enzyme. J. Cell Biol. 1998;142:377–389. doi: 10.1083/jcb.142.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neve E.P., Svensson K., Fuxe J., Pettersson R.F. VIPL, a VIP36-like membrane protein with a putative function in the export of glycoproteins from the endoplasmic reticulum. Exp. Cell Res. 2003;288:70–83. doi: 10.1016/s0014-4827(03)00161-7. [DOI] [PubMed] [Google Scholar]
- 9.Shirakabe K., Hattori S., Seiki M., Koyasu S., Okada Y. VIP36 protein is a target of ectodomain shedding and regulates phagocytosis in macrophage Raw 264.7 cells. J. Biol. Chem. 2011;286:43154–43163. doi: 10.1074/jbc.M111.275586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu S., Wang L., Wang X.W., Zhao Y.R., Bi W.J., Zhao X.F., Wang J.X. L-Type lectin from the kuruma shrimp Marsupenaeus japonicus promotes hemocyte phagocytosis. Dev. Comp. Immunol. 2014;44:97–405. doi: 10.1016/j.dci.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Klaus J.P., Eisenhauer P., Russo J., Mason A.B., Do D., King B., Taatjes D., Cornillez-Ty C., Boyson J.E., Thali M., Zheng C., Liao L., Yates J.R., Zhang B., Ballif B.A., Botten J.W. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe. 2013;14:522–534. doi: 10.1016/j.chom.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie X., Li H., Xu L., Yang F. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 2005;108:63–67. doi: 10.1016/j.virusres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Lan J.F., Li X.C., Sun J.J., Gong J., Wang X.W., Shi X.Z., Shi L.J., Weng Y.D., Zhao X.F., Wang J.X. Prohibitin Interacts with envelope proteins of white spot syndrome virus and prevents infection in the red swamp crayfish, Procambarus clarkii. J. Virol. 2013;87:12756–12765. doi: 10.1128/JVI.02198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Söderhäll K., Smith V.J. Separation of the haemocyte populations of Carcinus maenas and other marine decapods, and prophenoloxidase distribution. Dev. Comp. Immunol. 1983;7:229–239. doi: 10.1016/0145-305x(83)90004-6. [DOI] [PubMed] [Google Scholar]
- 15.Chen A.J., Gao L., Wang X.W., Zhao X.F., Wang J.X. SUMO-conjugating enzyme E2 UBC9 mediates viral immediate-early protein SUMOylation in crayfish to facilitate reproduction of white spot syndrome virus. J. Virol. 2013;87:636–647. doi: 10.1128/JVI.01671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Lan J.F., Dai Y.J., Lin L. Quantitative analysis of white spot syndrome virus in the tissues of cultured red swamp crayfish (Procambarus clarkii ) J. Fish. China. 2016;40:56–63. [Google Scholar]
- 18.Wang S., Liu N., Chen A.J., Zhao X.F., Wang J.X. TRBP homolog interacts with eukaryotic initiation factor 6 (eIF6) in Fenneropenaeus chinensis. J. Immunol. 2009;182:5250–5258. doi: 10.4049/jimmunol.0802970. [DOI] [PubMed] [Google Scholar]
- 19.Wang X.W., Zhao X.F., Wang J.X. C-type lectin binds to β-integrin to promote hemocytic phagocytosis in an invertebrate. J. Biol. Chem. 2014;289:405–414. doi: 10.1074/jbc.M113.528885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shima D.T., Scales S.J., Kreis T.E., Pepperkok R. Segregation of COPI-rich and anterograde-cargo-rich domains in endoplasmic-reticulum-to-Golgi transport complexes. Curr. Biol. 1999;9:821–824. doi: 10.1016/s0960-9822(99)80365-0. [DOI] [PubMed] [Google Scholar]
- 21.Scales S.J., Pepperkok R., Kreis T.E. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- 22.Klumperman J., Schweizer A., Clausen H., Tang B.L., Hong W., Oorschot V., Hauri H.P. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J. Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- 23.Aridor M., Bannykh S., Rowe T., Balch W.E. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y.H., Bi W.J., Wang X.W., Zhao Y.R., Zhao X.F., Wang J.X. Two novel C-type lectins with a low-density lipoprotein receptor class A domain have antiviral function in the shrimp Marsupenaeus japonicus. Dev. Comp. Immunol. 2014;42:323–332. doi: 10.1016/j.dci.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Wang X.W., Xu Y.H., Xu J.D., Zhao X.F., Wang J.X. Collaboration between a soluble C-type lectin and calreticulin facilitates white spot syndrome virus infection in shrimp. J. Immunol. 2014;193:2106–2117. doi: 10.4049/jimmunol.1400552. [DOI] [PubMed] [Google Scholar]
- 26.Gillespie L., Roosendahl P., Ng W.C., Brooks A.G., Reading P.C., Londrigan S.L. Sci. Rep. 2016;6:19428. doi: 10.1038/srep19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chazal N., Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Y.S., Liu W.J., Lee C.C., Chou T.L., Lee Y.T., Wu T.S., Huang J.Y., Huang W.T., Lee T.L., Kou G.H., Wang A.H., Lo C.F. A 3D model of the membrane protein complex formed by the white spot syndrome virus structural proteins. PLoS One. 2010;5:e10718. doi: 10.1371/journal.pone.0010718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X., Wu J., Sivaraman J., Hew C.L. Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship. J. Virol. 2007;81:6709–6717. doi: 10.1128/JVI.02505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X., Xu L., Yang F. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 2006;80:10615–10623. doi: 10.1128/JVI.01452-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X., Yang F. White spot syndrome virus VP24 interacts with VP28 and is involved in virus infection. J. Gen. Virol. 2006;87:1903–1908. doi: 10.1099/vir.0.81570-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


