Abstract
Porcine deltacoronavirus (PDCoV), an emerging animal coronavirus causing enteric disease in pigs, belongs to the newly identified Deltacoronavirus genus in the Coronaviridae family. Although extensive studies have been carried out to investigate the regulation of interferon (IFN) responses by alphacoronaviruses, betacoronaviruses, and gammacoronaviruses, little is known about this process during deltacoronavirus infection. In this study, we found that PDCoV infection fails to induce, and even remarkably inhibits, Sendai virus- or poly(I: C)-induced IFN-β production by impeding the activation of transcription factors NF-κB and IRF3. We also found that PDCoV infection significantly suppresses the activation of IFN-β promoter stimulated by IRF3 or its upstream molecules (RIG-I, MDA5, IPS-1, TBK1, IKKε) in the RIG-I signaling pathway, but does not counteract its activation by the constitutively active mutant of IRF3 (IRF3–5D). Taken together, our results demonstrate that PDCoV infection suppresses RIG-I-mediated IFN signaling pathway, providing a better understanding of the PDCoV immune evasion strategy.
Keywords: Porcine deltacoronavirus (PDCoV), Interferon, RIG-I Signaling pathway
Highlights
-
•
PDCoV infection fails to induce IFN-β production in LLC-PK1 cells.
-
•
PDCoV infection suppresses Sendai virus-or poly(I: C)-induced IFN-β production.
-
•
PDCoV impedes Sendai virus- or poly(I: C)-induced activation of NF-κB and IRF3.
-
•
PDCoV interrupts RIG-I signaling pathway to inhibit IFN-β production.
1. Introduction
Coronaviruses (CoVs) are enveloped, single-stranded, positive-sense RNA viruses that can be divided into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and the newly identified Deltacoronavirus. Alphacoronavirus and Betacoronavirus mainly infect mammals and Gammacoronavirus generally infects birds (Chan et al., 2013), while Deltacoronavirus can be detected in both mammals and birds. Porcine deltacoronavirus (PDCoV) was first described in 2012 during a study to identify new coronaviruses in mammals and birds in Hong Kong, China (Woo et al., 2009, Woo et al., 2012). In early 2014, an outbreak of PDCoV was announced in some pig farms in the United States (Wang et al., 2014), and this novel porcine coronavirus has been demonstrated in at least 18 U. S. states (Marthaler et al., 2014a, Marthaler et al., 2014b, Thachil et al., 2015). Subsequently, the detection of PDCoV was also reported in fecal samples from piglets with diarrhea in Korea, Canada, and mainland China (Dong et al., 2015, Lee and Lee, 2014, Song et al., 2015). More recently, several groups have demonstrated that PDCoV can cause severe clinical diarrhea and intestinal pathological damage in roughly 10-day-old gnotobiotic and conventional piglets (Chen et al., 2015, Jung et al., 2015, Ma et al., 2015) adding to the increasing concern regarding the epidemiology, evolution, pathogenesis, and immunology of this emerging coronavirus.
Interferon (IFN) and the IFN-induced cellular antiviral response are the primary defense mechanisms against viral infection. In virus-infected cells, viral components or replication intermediates known as the pathogen-associated molecular patterns (PAMPs), can be recognized by host pattern-recognition receptors (PRRs), such as the cytoplasmic retinoic acid-inducible gene I (RIG-I) and melanoma differentiation gene 5 (MDA5). After recognition, RIG-I and/or MDA5 interact with the IFN-β promoter stimulator 1 (IPS-1, also known as MAVS/VISA/Cardif) via the caspase-recruiting domain (CARD)-like domain to activate the downstream κB kinase (IKK)-related kinases, such as TANK-binding kinase 1 and IKKε, leading to the activation of interferon regulation factor 3 (IRF3) and nuclear factor κB (NF-κB). Phosphorylated IRF3 and NF-κB translocate to nucleus and coordinately activate the type I IFN promoter (Hiscott et al., 2006, Ramos and Gale, 2011, Seth et al., 2005).
To combat the antiviral effects of IFN, viruses have evolved various mechanisms to antagonize the host IFN responses. The molecular mechanisms of IFN antagonism have been extensively studied for alphacoronaviruses, betacoronaviruses, and gammacoronaviruses (Perlman and Netland, 2009). For example, porcine epidemic diarrhea virus (PEDV), a member of the Alphacoronavirus genus, inhibits dsRNA-induced IFN-β production by blockading the RIG-I-mediated pathway (Cao et al., 2015, Ding et al., 2014, Wang et al., 2016). Additionally, mouse hepatitis virus (MHV) and severe acute respiratory syndrome coronavirus (SARS-CoV), two representative members of the Betacoronavirus genus, interfere with the IFN response in various ways (Roth-Cross et al., 2007, Totura and Baric, 2012, Zhou and Perlman, 2007), and at least eight proteins encoded by SARS-CoV have been identified as IFN antagonists (Devaraj et al., 2007, Kopecky-Bromberg et al., 2007, Siu et al., 2009, Wathelet et al., 2007). The infectious bronchitis virus, a member of the Gammacoronavirus genus, induces a delayed activation of the IFN response (Kint et al., 2015). As a new member of coronavirus family, however, whether or not deltacoronaviruses antagonize IFN responses and, if they do, the details of this process are unclear.
Currently, PDCoV is the sole deltacoronavirus that has been successfully isolated in cell culture. In this study, we investigated the IFN responses after PDCoV infection of LLC-PK1 cells, a porcine kidney cell line. Our results show that PDCoV infection not only fails to activate IFN-β production, but it also inhibits Sendai virus (SeV)- or poly(I:C)-induced IFN-β production. We also demonstrate that PDCoV infection interrupts the RIG-I signaling pathway and impedes the activation of the critical transcription factors IRF3 and NF-κB.
2. Results and discussion
2.1. PDCoV proliferation characteristics in LLC-PK1 cells
To determine the kinetics of PDCoV propagation in LLC-PK1 cells, the cytopathic effects (CPEs) were examined and the virus titers were determined at different time points after PDCoV infection. To this end, LLC-PK1 cells were infected with PDCoV strain CHN-HN-2014 at a multiplicity of infection (MOI) of 0.01 and the CPEs were examined daily for up to three days. The infected cells were monitored by indirect immunofluorescence assays (IFAs) using a monoclonal antibody against PDCoV N protein at 6, 12, 18, 24, 30, and 36 h post-infection (hpi). As shown in Fig. 1A, only a small portion of cells were infected by 12 hpi, nearly all of the cells were PDCoV-positive at 18 hpi, and no cell detachment was observed at 24 hpi. The number of infected cells deceased rapidly up to 30 hpi, and obvious CPEs were observed along with serious cytopathy and a large number of detached cells at 36 hpi. A one-step growth curve for PDCoV strain CHN-HN-2014 in LLC-PK1 cells was also generated using TCID50 assays. As shown in Fig. 1B, virus titers presented a gradually upward tendency as the infection progressed, and at 24 hpi they reached a titer of 107.2 TCID50/mL. Together, these results show that PDCoV infection in LLC-PK1 cells achieves a high infection rate and titer without cell exfoliation at 24 hpi; thus, this time point was selected as the optimal time point for subsequent immunological studies.
Fig. 1.
PDCoV proliferation characteristics in LLC-PK1 cells. (A) Indirect immunofluorescence assays were performed to examine the consequences of PDCoV infection in LLC-PK1 cells. LLC-PK1 cells were mock-infected or infected with PDCoV at a MOI of 0.01. At 6, 12, 18, 24, 30, or 36 h post-infection (hpi), the cells were fixed and incubated with a monoclonal antibody against PDCoV N protein (green). The nuclei of cells were stained with DAPI (blue). Fluorescent images were acquired with a confocal laser scanning microscope and representative images are shown here. (B) The growth curve of PDCoV strain CHN-HN-2014 in LLC-PK1 cells. LLC-PK1 cells were infected with PDCoV at a MOI of 0.01 and were collected at 6, 12, 18, 24, 30, or 36 hpi for the determination of virus titers via TCID50 assays. The mean titers and standard deviations were calculated from three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. PDCoV infection fails to activate IFN-β and interrupts SeV- or poly(I:C)-mediated IFN-β induction
Previous studies have demonstrated that Alphacoronavirus, Betacoronavirus, and Gammacoronavirus have evolved diverse mechanisms to evade or suppress the host's antiviral innate immunity, the most important of which are the IFN responses (Perlman and Netland, 2009). However, the evasion methods used by the deltacoronaviruses remain unclear. To explore if PDCoV antagonizes IFN-β production, the IFN-β promoter luciferase reporter system was used to analyze IFN-β expression after PDCoV infection. To this end, LLC-PK1 cells were co-transfected with the luciferase reporter plasmids IFN-β-Luc and the internal control plasmid pRL-TK, followed by mock-infection or infection with PDCoV at a MOI of 0.01. After 24 h of PDCoV infection, the cell lysates were harvested and the IFN-β promoter-driven luciferase activity was measured. The mock-infected cells were stimulated with 20 hemagglutinating activity units/well of SeV or 0.5 μg/well of poly(I:C), respectively, as positive controls to test whether or not LLC-PK1 cells are able to recognize SeV or poly(I:C) and activate IFN-β promoter activity in response. As shown in Fig. 2A and B, IFN-β promoter-driven luciferase activity was barely detectable in PDCoV-infected cells compared with the strong reporter signal in SeV-infected or poly(I:C)-transfected cells, indicating that PDCoV infection failed to activate IFN-β promoter activity.
Fig. 2.
The effect of PDCoV infection on IFN-β promoter activation and SeV- or poly(I:C)-induced IFN-β production. (A, B) LLC-PK1 cells were co-transfected with IFN-β-Luc and pRL-TK for 12 h, followed by PDCoV infection (MOI =0.01). At 24 hpi, cells were collected for a dual-luciferase assay as described in the Materials and Methods. The results are shown here as the fold induction of the IFN-β promoter activity. Cells infected with SeV (A) or transfected with poly(I:C) (B) were used as positive controls. (C, D) LLC-PK1 cells were co-transfected with IFN-β-Luc and pRL-TK for 12 h and then mock-infected or infected with PDCoV at different MOIs (1, 0.1, and 0.01). At 12 hpi, the cells were mock-infected or infected with SeV for an additional 12 h (C) or transfected with or without poly (I:C) for an additional 24 h (D), and then subjected to a dual-luciferase assay. The results are shown here as the fold induction of the IFN-β promoter activity. All data are presented as means±SD of three independent experiments (*p<0.05 and **p<0.01).
To further investigate if PDCoV inhibits SeV- or poly(I:C)-induced IFN-β promoter activity, LLC-PK1 cells were co-transfected with IFN-β-Luc and pRL-TK and then mock-infected or infected with PDCoV at different MOIs of 1, 0.1, or 0.01. At 12 h post- PDCoV infection, the infected cells were mock-infected or infected with SeV or transfected with or without poly(I:C), respectively. The cells were harvested and subjected to a dual-luciferase assay at 12 h after SeV inoculation or at 24 h after poly(I:C) transfection. As shown in Fig. 2(C), the IFN-β promoter was activated 80- to 100-fold when the PDCoV-mock-infected cells were stimulated with SeV, whereas this activation was significantly inhibited by PDCoV infection in a dose-dependent manner. Also, PDCoV infection significantly inhibited poly(I:C)-induced IFN-β promoter activity (Fig. 2D). These results suggest that PDCoV infection interrupts SeV- or poly(I:C)-mediated IFN-β production.
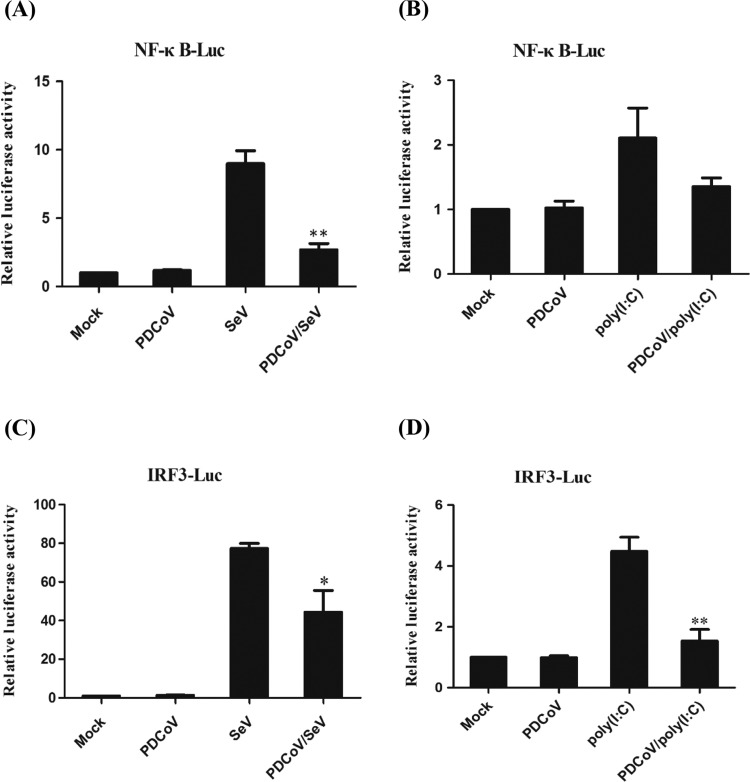
2.3. PDCoV impedes SeV- or poly(I:C)-mediated activation of NF-κB and IRF3
To investigate whether or not PDCoV impairs the activation of NF-κB and IRF3, LLC-PK1 cells were co-transfected with the luciferase reporter plasmids NF-κB-Luc or IRF3-Luc together with the internal control plasmid pRL-TK and, 12 h later, they were mock-infected or infected with PDCoV at a MOI of 0.01 for 12 h. The cells were then mock-infected or infected with SeV or transfected with or without poly(I:C), respectively. Cells were harvested 12 h after SeV infection or 24 h after poly(I:C) transfection and subjected to a dual-luciferase assay. As shown in Fig. 3, PDCoV infection failed to activate NF-κB promoter activity and significantly blocked the SeV-induced promoter activity of NF-κB (Fig. 3A) or partially blocked the poly(I:C)-induced promoter activity of NF-κB (Fig. 3B). PDCoV infection also failed to activate IRF3 promoter activity and partially blocked the SeV-induced promoter activity of IRF3 (Fig. 3B) or significantly blocked poly(I:C)-induced promoter activity of IRF3 (Fig. 3D). These results indicate that PDCoV impedes SeV- or poly(I:C)-mediated activation of the transcription factors NF-κB and IRF3, which are associated with the suppression of the IFN-β promoter after PDCoV infection.
Fig. 3.
The effect of PDCoV infection on the SeV- or poly(I:C)-mediated activation of NF-κB and IRF3. LLC-PK1 cells were first co-transfected NF-κB-Luc (A, B) or IRF3-Luc (C, D) together with pRL-TK for 12 h and then mock-infected or infected with PDCoV at a MOI of 0.01. At 12 hpi, the cells were mock-infected or infected with SeV for an additional 12 h or transfected with or without poly (I:C) for an additional 24 h. The cells were then harvested and subjected to a dual-luciferase assay, and the results are shown here as the fold induction of the NF-κB (A, B) or IRF3 (C, D) promoter activity. All data are presented as means±SD of three independent experiments (*p<0.05 and **p<0.01).
2.4. PDCoV interrupts the RIG-I signaling pathway
Both SeV and dsRNA are critical inducer of the RIG-I-like receptor (RLR)-mediated IFN-β signaling pathway (Meylan et al., 2005, Pythoud et al., 2012). It is possible that PDCoV interrupts SeV- or ploy(I:C)-mediated IFN-β production by blocking some of the individual members of the RIG-I signaling pathway. To test this possibility and to identify potential target molecules, we investigated the effect of PDCoV infection on the activity of a series of molecules in the RIG-I signaling pathway: RIG-I, MDA5, IPS-1, TBK1, IKKε, and IRF3. To this end, LLC-PK1 cells were mock-infected or infected with PDCoV at a MOI of 0.01 for 6 h, followed by co-transfection with a series of expression constructs encoding RIG-I, RIG-IN (a constitutively active mutant of RIG-I), MDA5, IPS-1, TBK1, IKKε, IRF3, and IRF3(5D) (a constitutively active mutant of IRF3) together with the luciferase reporter plasmids IFN-β-Luc and the internal control plasmid pRL-TK. At 28 h post-transfection, the cell lysates were harvested and IFN-β promoter-driven luciferase activities were measured. As shown in Fig. 4(A)–C, overexpression of any molecule of the RIG-I signaling pathway induced a significant activation of the IFN-β promoter in mock-infected cells. Interestingly, the activation of the IFN-β promoter induced by IRF3 and its upstream molecules (RIG-I/RIG-IN, MDA-5, IPS-1, TBK1 and IKKε) was blocked by PDCoV infection (Fig. 4A–C). In contrast, the activation of the IFN-β promoter induced by IRF3(5D) was not affected by PDCoV infection (Fig. 4C). These results provided evidence supporting the hypothesis that PDCoV interrupts SeV- or poly(I:C)-mediated IFN-β induction by blocking the activity of molecules in the RIG-I signaling pathway. Signaling components downstream of IRF3 remained intact in the PDCoV-infected cells. Based on these results, IRF3 appears to be the target protein of PDCoV suppression.
Fig. 4.
PDCoV interrupts the activation of IRF3 in RIG-I signaling pathway. LLC-PK1 cells were mock-infected or infected with PDCoV at a MOI of 0.01 for 6 h, and then the cells were co-transfected with an IFN-β promoter luciferase reporter and the indicated plasmid expressing RIG-I, RIG-IN, MDA5, IPS-1 (A), TBK1, IKKε (B), IRF3, IRF3(5D) (C), or an empty vector for 28 h. The cell lysates were harvested and subjected to a dual-luciferase assay, and the results are shown here as the fold induction of the IFN-β promoter activity. All data are presented as means ± SD of three independent experiments (*p<0.05 and **p<0.01).
2.5. PDCoV blocks SeV-induced phosphorylation and nuclear translocation of IRF3 and p65
Because our initial results showed that PDCoV blocked SeV-induced IRF3-dependent promoter activity, we further investigated the possible mechanism(s) for this inhibition. IRF3 and NF-κB are considered to be essential transcription factors for IFN-β production, and phosphorylation is a key step during their activation that in turn leads to nuclear translocation. Together, phosphorylation and nuclear translocation are the hallmarks of IRF3 and NF-κB activation (Ramos and Gale, 2011). Therefore, we explored the effect of PDCoV infection on the phosphorylation and nuclear translocation of IRF3 and NF-κB.
LLC-PK1 cells were mock-infected or infected with PDCoV at a MOI of 0.01 for 12 h followed by mock-infection or infection with SeV. At 12 h post-SeV infection, cells were harvested, and their phosphorylation levels of IRF3 and the NF-κB p65 subunit were examined. As shown in Fig. 5A, the total protein levels of IRF3 and p65 were almost equal between the mock-infected and PDCoV-infected cells. SeV infection markedly enhanced the IRF3 phosphorylation (p-IRF3) and p65 phosphorylation (p-p65) levels in comparison with the amounts in unstimulated cells. As expected, the SeV-mediated increase was significantly reduced in the PDCoV-infected cells. We also used IFA and confocal microscopy to analyze the translocation of IRF3 and p65 after PDCoV infection. As shown in Fig. 5B and C, consistent with our observations from western blot analyses, IRF3 and p65 were located exclusively in the cytoplasm in unstimulated LLC-PK1 cells, but they rapidly translocated to the nucleus after the cells were infected with SeV. In contrast, nuclear IRF3 and p65 translocation did not occur in PDCoV-infected cells. Moreover, PDCoV infection blocked the nuclear translocation of IRF3 and p65 otherwise induced by SeV infection. Collectively, our data clearly support the hypothesis that PDCoV inhibits SeV-induced IRF3-dependent promoter activity by blocking the phosphorylation and nuclear translocation of IRF3 and p65.
Fig. 5.
The effect of PDCoV infection on the SeV-induced phosphorylation and nuclear translocation of IRF3 and p65. (A) LLC-PK1 cells were mock-infected or infected with PDCoV at a MOI of 0.01 for 12 h and then mock-infected or infected with SeV. After 12 h of SeV infection, the cells were collected for western blot analyses with specific antibodies against IRF3, p-IRF3, p65, p-p65, or PDCoV N protein. An anti-β-actin antibody was used as a control for sample loading. (B, C) LLC-PK1 cells were mock-infected or infected with PDCoV at a MOI of 0.01 for 12 h and then mock-infected or infected with SeV. After 12 h of SeV infection, the cells were fixed for indirect immunofluorescence assays with the following antibodies: mouse anti-PDCoV N (green) and rabbit anti-IRF3 (red) (B) or rabbit anti-p65 (red) (C). Cellular nuclei (blue) were counterstained with 1 μg/mL of DAPI. Fluorescence was observed under a Fluoview ver. 3.1 confocal fluorescence microscope (Olympus) and representative images are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As a newly identified coronavirus, the immune evasion strategy utilized by PDCoV remains largerly unclear. Previous studies have shown that other coronaviruses, including alphacoronaviruses, betacoronaviruses, and gammacoronaviruses, can suppresses IFN-β production by blocking the activation of transcription factor IRF3 (Perlman and Netland, 2009). In this study, we demonstarted that PDCoV infection also blocks IRF3 activation. It is possible that blockade of IRF3 activation is a common strategey utilized by coronaviruses to antagonize IFN-β production. In addition, multiple proteins encoded by coronaviruses have been identified as IFN antagonists (Roth-Cross et al., 2007, Totura and Baric, 2012). Interestingly, different mechanisms are used by some homologous proteins of different coronaviruses. For example, the nucleocapsid (N) protein of PEDV antagonizes IFN production by sequestering the interaction between IRF3 and TBK1 (Ding et al., 2014); MHV A59 N protein antagonizes IFN activity by interfering with the RNase L activity associated with the induction of 2′−5′-oligoadenylate synthetase (Ye et al., 2007); SARS-CoV N protein blocks a very early step in IFN production, probably at the RNA-sensor recognition step (Lu et al., 2011). PDCoV also encodes the nucleocapsid protein, and whether PDCoV N protein antagonize IFN activity and what mechanisms are used by PDCoV N protein are very interesting and these issues are currently under investigation in our laboratory.
3. Conclusions
In summary, we showed that PDCoV infection fails to induce IFN-β production in LLC-PK1 cells. Furthermore, PDCoV can interfere with the RIG-I-mediated signaling pathway. Mechanistically, PDCoV infection suppresses IFN-β production by blocking the activation of transcription factors IRF3 and NK-κB. To our knowledge, PDCoV is currently the only isolated deltacoronavirus able to be propagated in a cell culture system; thus, it is an important model for studying the interaction between deltacoronaviruses and the innate immune system. Our data provide a novel insight into the immune evasion strategy of PDCoV. Future studies to further identify the PDCoV-encoded IFN antagonists and to understand the mechanism of action of each antagonist could yield novel therapeutic targets and more effective vaccines.
4. Materials and methods
4.1. Viruses, cells, and reagents
PDCoV strain CHN-HN-2014 (GenBank accession number KT336560), which was isolated from a suckling piglet with acute diarrhea in China in 2014, was used in this study. SeV was acquired from the Centre of Virus Resource and Information at the Wuhan Institute of Virology. LLC-PK1 cells, purchased from ATCC, were cultured at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, and these cells were used to amplify PDCoV. Poly(I:C) was purchased from Sigma-Aldrich as a sodium salt and dissolved in water to obtain a stock solution of 1 mg/mL. Rabbit polyclonal antibodies against NF-κB p65, phosphorylated NF-κB p65 (p-p65), IRF3, and phosphorylated IRF3 (p-IRF3) were purchased from ABclone (China). Mouse monoclonal antibodies (mAbs) against β-actin were purchased from Medical and Biological Laboratories (Japan). The monoclonal antibody used for the detection of PDCoV N protein was produced from hybridoma cells derived from Sp2/0 myeloma cells and the spleen cells of BALB/c mice immunized with the recombinant N protein from PDCoV strain CHN-HN-2014.
4.2. Plasmids
The generation of the luciferase reporter plasmids IFN-β-Luc, 4×PRDII-Luc (referred to as NF-κB-Luc), and 4×PRDIII/I-Luc (referred to as IRF3-Luc) have been described previously (Wang et al., 2008, Wang et al., 2010). The luciferase reporter plasmids 4×PRDII-Luc and 4×PRDIII/I-Luc contain four copies of the NF-κB- or IRF-binding motif, respectively, of the porcine IFN-β promoter that are upstream of the firefly luciferase reporter gene. The generation of the cDNA expression constructs encoding porcine RIG-I and its constitutively active mutant (RIG-IN), MDA5, IPS-1, TBK1, IKKε, IRF3, and IRF3–5D have also been described previously (Wang et al., 2008, Wang et al., 2011, Wang et al., 2010).
4.3. Virus titrations by TCID50 assay
To determine viral one-step growth curves, LLC-PK1 cells in 24-well plates were inoculated with PDCoV (MOI =0.01). The whole cell samples were collected at 6, 12, 18, 24, 30, or 36 post-infection (hpi) by freezing and thawing three times, followed by centrifugation at 2500 r/min for 10 min to collect the supernatant, and the samples were stored at −80 °C until virus titrations were performed. The virus titers for each time point were determined by performing TCID50 assays in LLC-PK1 cells as described previously (Hu et al., 2015). Virus titers were calculated using the Reed-Muench method from the results of three independent experiments.
4.4. Luciferase reporter gene assay
LLC-PK1 cells grown in 24-well plates were transfected with a reporter plasmid (IFN-β-Luc, NK-κB-Luc, or IRF3-Luc) and pRL-TK (an internal control for normalization of the transfection efficiency) using Lipofectamine 2000, and the cells were also mock-infected or infected with PDCoV and/or SeV (20 hemagglutinating activity units/well) or transfected with or without poly(I:C) (0.5 μg/well). In selected experiments, cells were also transfected with an expression plasmid (RIG-I, RIG-IN, MDA5, IPS-1, TBK1, IKKε, IRF3, or IRF3–5D) or an empty control plasmid. The cells were then lysed, and the firefly luciferase and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega). Data are shown as the relative firefly luciferase activities normalized to the Renilla luciferase activities from three independently conducted experiments.
4.5. Western blotting
LLC-PK1 cells were cultured in 60-mm dishes and mock-infected or infected with PDCoV and/or SeV. At 24 hpi, the cells were harvested by adding lysis buffer (4% SDS, 3% DTT, 0.065 mM Tris–HCl [pH 6.8], and 30% glycerin) supplemented with a protease inhibitor cocktail, phenylmethylsulfonyl fluoride (PMSF), and a phosphatase inhibitor cocktail. The lysates were subjected to SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad). The membranes were then analyzed for the expression of p65, p-p65, IRF3, and p-IRF3 proteins by immunoblotting using rabbit anti-p65, anti-p-p65, anti-IRF3, and anti-p-IRF3 antibodies, respectively. An anti-PDCoV N protein monoclonal antibody was used for immunoblotting to confirm the expression levels of the PDCoV N protein. An anti-β-actin monoclonal antibody was used to detect the expression of β-actin to confirm equal protein sample loading.
4.6. Indirect immunofluorescence assay (IFA)
IFAs were performed to examine the subcellular localization of IRF3 and p65 in LLC-PK1 cells. LLC-PK1 cells were seeded onto microscope coverslips, placed into 24-well dishes, and allowed to reach approximately 80% confluence. The cells were then mock-infected or infected with PDCoV and/or SeV. At 24 hpi, the cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with methyl alcohol for 10 min at room temperature. After three washes with TBST, the cells were blocked with TBST containing 5% bovine serum albumin (BSA) for 1 h and then incubated separately with a rabbit polyclonal antibody against p65 (1:100) or against IRF3 (1:100) or a mouse monoclonal antibody against the PDCoV N protein (1:100) for 1 h. The cells were then treated with Alexa Fluor 488-labeled anti-mouse secondary antibody or Alexa Fluor 594-labeled anti-rabbit secondary antibody for 1 h at room temperature and subsequently treated with 4ʹ,6-diamidino-2-phenylindole (DAPI) for 15 min at room temperature. Fluorescent images were visualized and examined by using a confocal laser scanning microscope (Fluoview ver. 3.1; Olympus, Japan).
4.7. Statistical analysis
Data are expressed as the mean±SD of three independent experiments. Student's t-tests were performed, and p-values of <0.05 were considered statistically significant.
Acknowledgements
This work was supported by the Key Technology R&D Programme of China (2015BAD12B02), the Natural Science Foundation of Hubei Province (2014CFA009), and the Fundamental Research Funds for the Central Universities (2013PY043).
References
- Cao L., Ge X., Gao Y., Herrler G., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015;12:127. doi: 10.1186/s12985-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Tse K.K., Jin H., Yuen, K.Y D.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine Deltacoronavirus in Mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J., Lin R., Nakhaei P., Paz S. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 2006;12:53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint J., Fernandez-Gutierrez M., Maier H.J., Britton P., Langereis M.A., Koumans J., Wiegertjes G.F., Forlenza M. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J. Virol. 2015;89:1156–1167. doi: 10.1128/JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Complete genome characterization of Korean Porcine Deltacoronavirus Strain KOR/KNU14-04/2014. Genome Announc. 2014;2 doi: 10.1128/genomeA.01191-14. e01191–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Pan J., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio. 2015;6:e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Jiang Y., Collins J., Rossow K. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc. 2014;2 doi: 10.1128/genomeA.00218-14. e00218–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pythoud C., Rodrigo W.W., Pasqual G., Rothenberger S., Martinez-Sobrido L., de la Torre J.C., Kunz S. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKepsilon. J. Virol. 2012;86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos H.J., Gale M., Jr. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Martinez-Sobrido L., Scott E.P., Garcia-Sastre A., Weiss S.R. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 2007;81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and Characterization of MAVS, a mitochondrial Antiviral signaling protein That Activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Siu K.L., Kok K.H., Ng M.H., Poon V.K., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thachil A., Gerber P.F., Xiao C.T., Huang Y.W., Opriessnig T. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies. Plos One. 2015;10:e0124363. doi: 10.1371/journal.pone.0124363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Li T., Luo R., Xie L., Jiang Y., Chen H., Xiao S. Molecular cloning and functional characterization of porcine IFN-beta promoter stimulator 1 (IPS-1) Vet. Immunol. Immunopathol. 2008;125:344–353. doi: 10.1016/j.vetimm.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Wang D., Fang L., Liu L., Zhong H., Chen Q., Luo R., Liu X., Zhang Z., Chen H., Xiao S. Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-lambda1 pathway. Mol. Immunol. 2011;49:407–412. doi: 10.1016/j.molimm.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Wang D., Fang L., Luo R., Ye R., Fang Y., Xie L., Chen H., Xiao S. Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem. Biophys. Res. Commun. 2010;399:72–78. doi: 10.1016/j.bbrc.2010.07.044. [DOI] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine coronavirus HKU15 detected in 9 US states. Emerg. Infect. Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hauns K., Langland J.O., Jacobs B.L., Hogue B.G. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Perlman S. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 2007;81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]